Abstract
Parkinson’s disease is a complex neurodegenerative disorder that is about 1.5 times more prevalent in males than females. Extensive work has been done to identify the genetic risk factors behind Parkinson’s disease on autosomes and more recently on Chromosome X, but work remains to be done on the male-specific Y chromosome. In an effort to explore the role of the Y chromosome in Parkinson’s disease, we analysed whole-genome sequencing data from the Accelerating Medicines Partnership—Parkinson’s disease initiative (1466 cases and 1664 controls), genotype data from NeuroX (3491 cases and 3232 controls) and genotype data from UKBiobank (182 517 controls, 1892 cases and 3783 proxy cases), all consisting of male European ancestry samples. We classified sample Y chromosomes by haplogroup using three different tools for comparison (Snappy, Yhaplo and Y-LineageTracker) and meta-analysed this data to identify haplogroups associated with Parkinson’s disease. This was followed up with a Y-chromosome association study to identify specific variants associated with disease. We also analysed blood-based RNASeq data obtained from the Accelerating Medicines Partnership—Parkinson’s disease initiative (1020 samples) and RNASeq data obtained from the North American Brain Expression Consortium (171 samples) to identify Y-chromosome genes differentially expressed in cases, controls, specific haplogroups and specific tissues. RNASeq analyses suggest Y-chromosome gene expression differs between brain and blood tissues but does not differ significantly in cases, controls or specific haplogroups. Overall, we did not find any strong associations between Y-chromosome genetics and Parkinson’s disease, suggesting the explanation for the increased prevalence in males may lie elsewhere.
Keywords: chromosome Y, Parkinson’s disease, genetics
Grenn et al. analysed Y-chromosome haplogroups, variants and genes to determine the role of the Y chromosome in Parkinson’s disease, a disease more prevalent in males than females. Grenn et al. report no associations between Y-chromosome genetics and Parkinson’s disease.
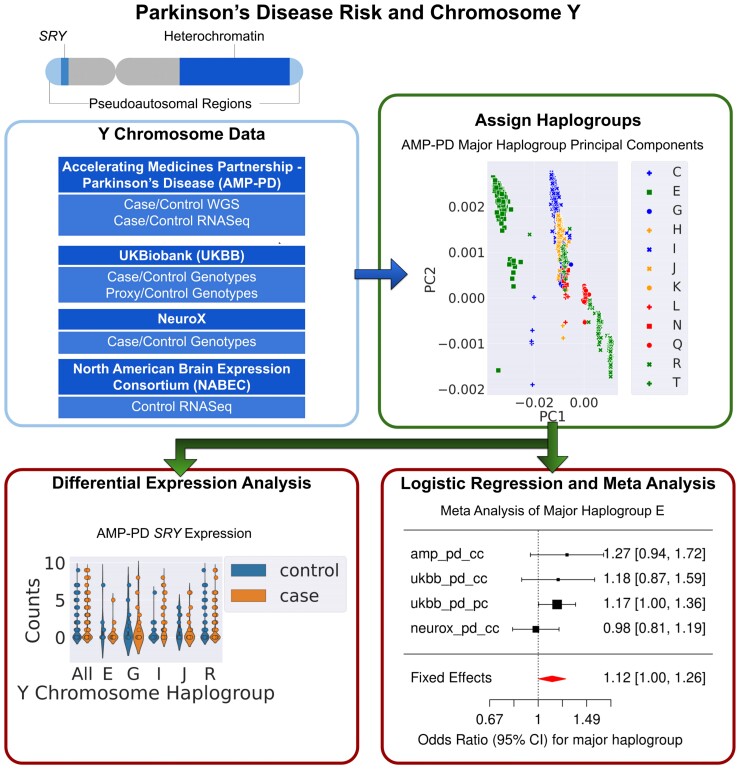
Graphical Abstract
Graphical abstract.
Introduction
Parkinson’s disease is a progressive movement disorder that includes symptoms such as tremors, slowed speech, bradykinesia, loss of balance and muscle stiffness. While age is the primary risk factor for Parkinson’s disease, genetic and likely environmental factors contribute to its onset and progression.1 Recent genetic studies have identified 92 Parkinson’s disease risk variants in European and Asian ancestry populations, but common variation findings from genome-wide association studies (GWASs) only account for 16–36% of heritable risk.2,3 Environmental factors such as pesticide exposure, smoking and caffeine intake have been linked to Parkinson’s disease aetiology, but the underlying mechanisms are not fully understood.4–7
Parkinson’s disease is ∼1.5 times more prevalent in males than in females with European ancestry.8 However, it is unclear whether this difference is due to environmental or genetic factors or a combination of the two. Studies have shown that there are sex-specific differences in Parkinson’s disease clinical presentation and progression.9 To date, no significant autosomal genetic differences have been found between male and female Parkinson’s disease cases.10 A recent Chromosome X GWAS did identify potential GWAS hits but no significant differences in Parkinson’s disease risk between sexes.11 This leaves the male-specific Chromosome Y as a potential candidate for sex-specific Parkinson’s disease risk.
Chromosome Y is frequently excluded from large-scale GWAS due to its size and unique characteristics.12 The Y chromosome makes up ∼2% of the total DNA in a human male cell. Additionally, quality control filters typically used in GWAS, such as Hardy–Weinberg equilibrium, do not apply to the hemizygous alleles of Chromosome Y. Lastly, Y-chromosome reference panels for imputation are not widely available for GWAS.13 Therefore, autosomes and sex chromosomes are typically analysed separately.
Chromosome Y is unique in that it is passed exclusively from father to son and only recombines in the pseudoautosomal regions (PARs) that make up ∼5% of its DNA. As a result, ∼95% of Chromosome Y is identical between father and son, with the exception of random mutations, making it easy to identify ancestry and assign entire Y chromosomes to haplogroups. These haplogroups, sometimes referred to as clades, are defined by unique genetic markers and have been used to identify associations with disease. For example, specific Y-chromosome haplogroups have been associated with AIDS progression, coronary artery disease and infertility in males.14–17 Therefore, the use of Y-chromosome haplogroups to identify associations with Parkinson’s disease is a valid strategy when using large sample sizes. Here, we take advantage of the structure of Chromosome Y to identify haplogroups, variants and gene expression patterns potentially associated with Parkinson’s disease risk in males using multiple large cohorts.
Methods
Accelerating Medicines Partnership for Parkinson’s Disease data
We used Y-chromosome whole-genome sequencing (WGS) data from the Accelerating Medicines Partnership—Parkinson’s Disease initiative (AMP-PD, https://amp-pd.org/), which includes data from multiple cohorts.18 This data contained a total of 184 317 Y-chromosome variants and 9887 samples, including 4146 controls, 2844 Parkinson’s disease cases, 2628 Lewy body dementia cases and 269 samples with a neurological disorder other than Parkinson’s disease or Lewy body dementia. Plink (v1.9) was used to filter this data.19 Female samples were removed, leaving 5470 male samples totalling 1895 controls, 1751 Parkinson’s disease cases, 1668 Lewy body dementia cases and 156 samples with another neurological disorder. Of these 5470 male samples, 5352 had European ancestry, which were included for further analyses. Heterozygous Y-chromosome variants were set to missing and completely removed from all samples if they were heterozygous in over 10% of the male samples, bringing the variant count down to 183 109. To match the hg19 reference genome used in the haplogroup calling tools, we used the UCSC liftover web tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver) to lift the hg38 AMP-PD data to the hg19 human genome reference build. We removed variants that failed to convert (41 081 variants) and variants that converted to a different chromosome (345 variants), leaving us with a total of 141 683 variants. Non-European samples, samples with Lewy body dementia, Parkinson’s disease genetic carriers of known disease-causing variants identified from biased recruitment and samples with a neurological disorder other than Parkinson’s disease were removed from the AMP-PD Parkinson’s disease case/control data set, leaving a total of 3130 samples, including 1466 cases and 1664 controls.
UK biobank data
Y-chromosome un-imputed genotype data were obtained from the UK Biobank (UKBB).20 This contained 488 377 samples and 691 Y-chromosome variants. Parkinson’s disease phenotypes were based on data field 131023 (Codes 40, 41, 50, 51, 20, 30 or 31) and proxy Parkinson’s disease phenotypes (individuals without Parkinson’s disease but with an affected father with Parkinson’s disease) were based on data field 20107 (Code 11). Female (based on Genetic Sex Code 22001) and non-European samples were removed using Plink (v1.9) leaving 188 192 samples. Of the remaining male European samples, there were 182 517 controls, 1892 Parkinson’s disease cases and 3783 proxy cases. There were about twice as many proxy samples as cases in the UKBB data, so one-third of the total of 182 517 controls were randomly selected for the UKBB Parkinson’s disease case/control data set, leaving 1892 Parkinson’s disease cases and 60 839 controls. The remaining two-thirds of the controls were included in the UKBB proxy/control data set, including 3783 Parkinson’s disease proxies and 121 678 controls.
NeuroX data
NeuroX data were downloaded from dbGAP (phs000918.v1.p1).21 This data set included 7791 cases and 9036 controls. The NeuroX data contained 139 Y-chromosome genotyped variants. After standard sample level, quality control was performed, including ancestry and relatedness, as reported elsewhere,2 3491 cases and 3232 controls were left, all of which had European ancestry.
Haplogroup calling tools
After sample and variant level quality control, AMP-PD, UKBB and NeuroX data were used to assign haplogroups to each sample. Three haplogroup calling tools were applied for comparison. Plink binary files were used as input for the Single-Nucleotide Assignment of Phylogenetic Parameters on the Y-chromosome (Snappy) tool.22 Plink binary files were converted to vcf format for the Yhaplo and Y-LineageTracker tools.23,24 Using output from these tools, each sample was assigned a major haplogroup by extracting the first character from each full haplogroup name (e.g. R1a1a1b to R, J2a1b1 to J, etc.) to follow the grouping used by the International Society of Genetic Genealogy Y-DNA Haplogroup Tree.25
Each of the three tools included its own set of reference data used to identify Y-chromosome haplogroup based on Y-chromosome variation. These three sets of reference data were compared with determine if the use of original reference data would lead to unbiased comparisons between tools. Y-LineageTracker reference data were reformatted for use in the Snappy and Yhaplo tools and used to reassign full and major haplogroups to all samples. These results were used in all downstream analysis to reduce bias between the tools.
To verify the accuracy of the haplogroups assigned to each AMP-PD sample, we compared the haplogroups of all related samples. Plink2 was used to filter for a minimum allele count of two and a Hardy–Weinberg equilibrium of 0.0001 before calculating relatedness coefficients using the Kinship-based Inference for GWAS tool.19,26 We identified 129 pairs of related male samples after filtering for a minimum relatedness coefficient of 0.088. Of these, 124 pairs were assigned the same major haplogroup and five pairs had different major haplogroups (Supplementary Table 1). The relatedness coefficients for these five samples were between 0.0884 and 0.177, suggesting they were second-degree relatives who inherited their Y chromosome from different fathers and are related through female relatives.
Statistical analyses
Plink was used to calculate Y-specific principal components for all AMP-PD samples, including genetic Parkinson’s disease carriers, using only Y-chromosome variants to determine if samples cluster by major haplogroup. AMP-PD and NeuroX autosomal principal components were calculated with Plink and calculated with flashpca/2.0 for UKBB data27 using autosomal variants from all four data sets, including AMP-PD case/control, UKBB case/control, UKBB proxy/control and NeuroX case/control data sets. The Python statsmodels/0.12.1 package was used to perform logistic regression on four data sets to determine if haplogroup could predict disease status in males. The first five autosomal principal components for each data set and age (age at baseline for AMP-PD, age at recruitment for UKBB and age at onset and age at recruitment for NeuroX cases and controls, respectively) were included as covariates in this analysis.
The results of the logistic regression for each major haplogroup were meta-analysed using inverse variance weighting under a fixed effect model with R/3.6 and the metafor/3.0–2 package.28 The AMP-PD case/control, UKBB case/control, UKBB proxy/control and NeuroX data sets were included in this meta-analysis. Data for a major haplogroup were excluded if there were <50 samples with the major haplogroup in the data set.
Logistic regression was also performed for each full haplogroup that was in >50 samples in any of the four data sets. Results from the AMP-PD case/control, UKBB case/control, UKBB proxy/control and NeuroX data sets were meta-analysed using inverse variance weighting under a fixed effect model with R/3.6 and the metafor/3.0-2 package.28 Full haplogroups were only included in this meta-analysis if they were present in >50 samples in at least two of the data sets, where the UKBB case/control and UKBB proxy/control data sets counted as one. This meta-analysis included 12 unique haplogroups that were present in >50 samples.
Single variant testing was performed for each data set independently using logistic regression. A minor allele frequency filter of 0.05 was applied to each data set before performing logistic regression with covariates of age, one-hot encoded major haplogroup and the first five autosomal principal components. One-hot encoded major haplogroups were only included as covariates if the major haplogroup was present in 50 or more samples in the data set to reduce instances of collinearity. Few genetic markers were available for both UKBB (691 variants) and NeuroX (139 variants), but AMP-PD WGS data had full Chromosome Y sequencing data available (141 683 variants). Association results across the AMP-PD case/control, NeuroX case/control, UKBB case/control and UKBB proxy/control data sets were combined and filtered to include only variants that were present in at least two of these data sets, bringing the variant count from 3387 down to 29. These 29 variants were meta-analysed using inverse variance weighting under a fixed effect model with METAL29 and annotated with ANNOVAR.30 The AMP-PD case/control data set was analysed on its own since it was the only data set with full coverage of the genome. This separate analysis utilized data prior to lifting over to hg19 to include more variants (183 109 variants). Before performing association analyses, insertions and deletions were removed, leaving 91 115 variants, followed by the removal of multiallelic variants, leaving 74 775 variants, due to the relatively high-sequencing false-positive rate of these variant types. Age, the first five autosomal principal components and one-hot encoded major haplogroups for major haplogroups G, I and J were included as covariates in this analysis. A minor allele frequency filter of 0.05 was applied to the results, leaving 1289 variants.
RNA sequencing data
RNA sequencing (RNASeq) data from the North American Brain Expression Consortium (NABEC) were used to quantify gene expression in the frontal cortex in 171 male samples, consisting entirely of population controls.31 Gene expression was quantified twice to determine if the exclusion of Y-chromosome PARs, the only regions of the Y chromosome that recombine with the X chromosome, alters expression levels. To do so, we used two different reference genomes, a version of the GENCODE release 38 hg38 reference genomes with Y PARs masked and the default GENCODE release 38 hg38 reference genome, which includes Y PARs by default.32
XYalign was used to mask Y PARs in one of the reference genome files.33 The genomeGenerate runMode from STAR was then used to generate a genome file containing masked Y PARs using the output from the XYalign tool and an annotated transcript file from GENCODE.34 We followed the same parameters used by AMP-PD to generate reference genomes in anticipation of comparing expression between NABEC and AMP-PD samples. Similar steps were taken to generate a reference genome file including Y PARs, the only difference being that the default GENCODE files include Y PARs as input. NABEC male samples were mapped to both the Y PAR masked reference genome and the default reference genome using the alignReads runMode in STAR. The featureCounts tool from the Subread package was used to count mapped reads for genes in all samples,35 following the same parameters used in the AMP-PD RNASeq pipelines. The featureCount data were combined into two expression matrices, one containing counts for samples mapped to the reference genome with Y PARs masked and one containing counts for the samples mapped to the default reference genome. Each count matrix included a total of 171 samples and 60 708 genes, 566 of which were located on Chromosome Y. The edgeR R package was used to filter out genes with low expression, leaving a total of 22 049 genes and identify genes differentially expressed between the Y PAR masked and unmasked data.36 This analysis was followed up by thresholded testing using the glmTreat function, which repeated the test for differential expression but relative to a minimum log-fold change of two, instead of the default log-fold change of zero. This thresholded testing was applied to all other differential expression analyses as well.
Blood-based RNASeq data were obtained from AMP-PD to compare Y-chromosome gene expression in blood with the previously quantified brain expression data. Expression data quantified with featureCounts were available at baseline for 563 Y-chromosome genes and 1020 AMP-PD samples after removing Parkinson’s disease genetic carriers and non-European samples. This was combined with our re-quantified NABEC frontal cortex expression data generated using a reference genome including Y PARs. A total of 553 genes were common between the two data sets, including 1020 blood samples and 171 brain samples. EdgeR was used to filter out genes with low expression, leaving a total of 275 genes and to identify genes differentially expressed in brain and blood tissues in all samples while adjusting for major haplogroup.
EdgeR was used to identify Y-chromosome genes differentially expressed between Parkinson’s disease cases and controls in AMP-PD samples while adjusting for both sample age and major haplogroup. Genes with low expression were removed prior to analysis, leaving 278 genes for the 1020 AMP-PD samples, including 722 cases and 298 controls.
A similar analysis was done to identify genes differentially expressed between major haplogroups in AMP-PD samples while adjusting for case/control status. Genes with low expression were filtered out prior to performing this analysis, leaving a total of 240 genes for the 1020 AMP-PD samples. This was only performed for major haplogroups present in at least 40 samples, leaving major haplogroups E, G, I, J and R. Gene expression for samples with each of these major haplogroups was compared with expression for all samples with a different major haplogroup. Gene counts were plotted for Y-chromosome genes of interest along with case/control status and sample major haplogroup obtained from Y-LineageTracker to visualize differences in gene expression. This was repeated with the NABEC frontal cortex expression, excluding case/control status because all samples were controls.
Data and code availability
All AMP-PD (https://amp-pd.org/) and UKBB (https://www.ukbiobank.ac.uk/) data are available via application on their websites and the NeuroX data are available via dbGap at phs000918.v1.p1. NABEC data are available via dbGap at phs001354.v1.p1. All codes are available on the GitHub page: https://github.com/neurogenetics/chrY_haplogroups_PD.
Results
Identification of Chromosome Y haplogroups
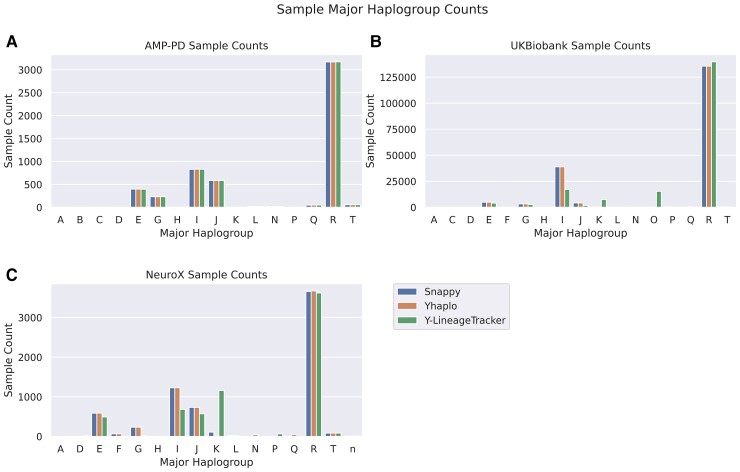
In total, we included 6849 Parkinson’s disease cases, 3783 proxy cases and 187 413 controls derived from AMP-PD, UKBB and NeuroX cohorts. We called haplogroups for all included samples using three different tools: Snappy, Yhaplo and Y-LineageTracker (Fig. 1). Full haplogroup frequencies, major haplogroup frequencies and the number of unique haplogroups identified by each tool using each cohort were recorded to compare across tools (Supplementary Tables 2–8, Fig. 2). Overall, concordance was high between tools within the AMP-PD cohort but comparably lower within the UKBB and NeuroX cohorts.
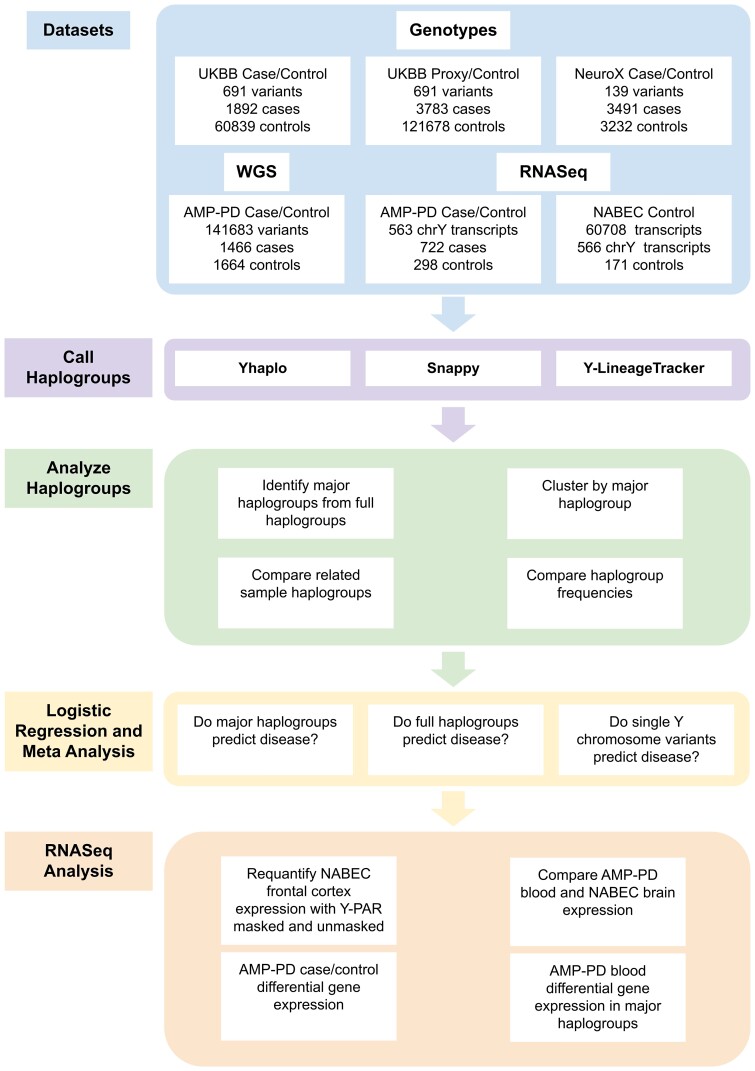
Figure 1.
Analysis flow chart of included data and performed analyses.
Figure 2.
Comparison of major haplogroup counts in Y-chromosome haplogroup calling tools. Major haplogroup counts using AMP-PD whole-genome sequencing data (A), major haplogroup counts using UKBiobank genotype data (B) and major haplogroup counts using NeuroX genotype data (C).
Y-chromosome major haplogroup frequencies were obtained from six different studies to compare our frequencies with those of European populations in specific countries (Supplementary Table 9). These included Belgium, The Netherlands,37 Poland,38 Spain, Portugal,39 Belarus,40 the UK,41 Algeria, Egypt and Italy.42 Major haplogroup frequencies identified in European ancestry samples using Y-LineageTracker for the AMP-PD, UKBiobank and NeuroX cohorts were similar to those of other European countries.
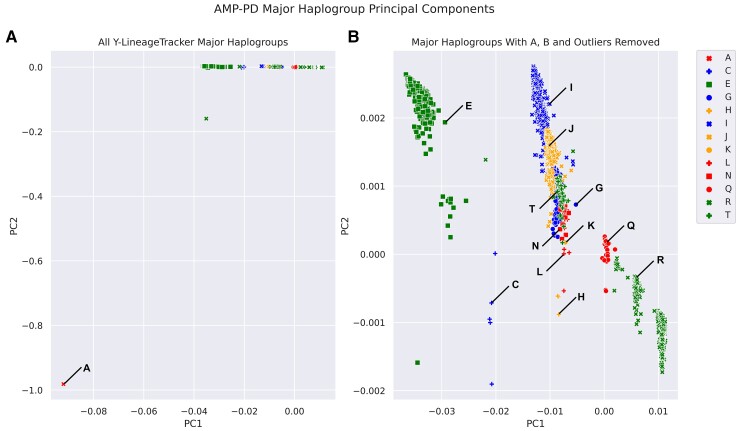
To visualize the differences between haplogroups, Y-specific principal components were calculated for AMP-PD samples using Y-chromosome data. The first two principal components were plotted with the major haplogroups obtained from all three tools. Initial clustering focused on major haplogroups A and B since they were the earliest haplogroups to split off from the most recent common ancestor, making them two of the oldest major haplogroups43,44 (Fig. 3A, Supplementary Fig. 1). Removal of these haplogroups and outlier samples shows clear clustering based on identified major haplogroups (Fig. 3B, Supplementary Fig. 1).
Figure 3.
Y-chromosome genetic principal components with Y-chromosome major haplogroups. The first two principal components are plotted with all major haplogroups (A) and with major haplogroups A, B and one outlier sample removed (B).
Case/control Chromosome Y associations
Logistic regression was performed on each data set for each major haplogroup obtained from Y-LineageTracker to determine if a major haplogroup can predict disease status (Supplementary Table 10). These results were combined to perform a meta-analysis for each major haplogroup (Supplementary Table 11). The meta-analyses only included data sets with at least 50 samples of the major haplogroup. Major haplogroup E was nominally significant (P < 0.05) but did not pass multiple test correction (P < 0.05/10).
Logistic regression was conducted on each data set for each full haplogroup obtained from all three haplogroup calling tools to identify variants making up haplogroups that predict disease status. Logistic regression was only performed on haplogroups that had 50 or more samples in the data set (Supplementary Table 12). These results were combined to perform a meta-analysis for each full haplogroup present in at least two of the cohorts. No full haplogroups were significant after multiple test correction (P < 0.05/12 = 0.00416; Supplementary Table 13).
Single variant testing
Plink logistic regression was performed to identify Y-chromosome variants associated with Parkinson’s disease while adjusting for covariates of age, one-hot encoded major haplogroup and the first five autosomal principal components. METAL was used to meta-analyse the AMP-PD case/control, UKBB case/control, UKBB proxy/control and NeuroX case/control data sets together. However, given the lack of Chromosome Y coverage in the UKBB and NeuroX cohorts, very few variants could be meta-analysed (Supplementary Table 14). In total, we meta-analysed 29 variants and none passed correction for multiple testing. A separate analysis of the AMP-PD data identified a total of 241 Y-chromosome variants with a P-value of <0.05, but none passed multiple test correction (Supplementary Table 15).
Gene expression assessments of different Y-chromosome haplogroups
In addition to assessing genetic associations between chromosome Y and Parkinson’s disease, we investigated (i) if the PARs affect gene quantifications of Y-chromosome genes, (ii) whether blood and brain gene expression of the Y chromosome is comparable and (iii) whether gene expression differences of the Y chromosome are associated with Parkinson’s disease or Y-chromosome haplogroup.
Previous studies have suggested that the removal of the Y chromosome, including Y PARs, from the reference genome improves mapping quality and alters downstream variant calling results across the X chromosomes of female samples.33 To determine if similar behaviour exists on the Y chromosome of male samples, we measured differences in Y-chromosome gene expression when masking Y PARs in male samples. To do so, we re-quantified frontal cortex expression in NABEC samples using a Y PAR masked reference genome and a reference genome including Y PARs. EdgeR was used to compare the results of these two methods. Thresholded testing was applied using the glmTreat function to test for differential expression relative to a minimum log-fold change of two, instead of the default log-fold change of zero. This resulted in 17 differentially expressed genes (Supplementary Table 16). All of these genes were upregulated in samples mapped to the Y PAR masked reference genome and were located in PARs. Therefore, we concluded that the presence of Y PARs would not impact any additional analysis of Y-chromosome gene expression.
Next, we conducted a linear regression with edgeR to identify Y-chromosome genes differentially expressed between brain and blood tissues to assess whether blood expression could be used as a proxy for brain gene expression. Using blood expression from the 1020 AMP-PD samples and brain expression from the 171 NABEC samples, we identified a total of 208 genes upregulated in blood tissues, 53 genes upregulated in brain tissues and 14 genes that were not differentially expressed in either of the tissues. Thresholded testing relative to a log-fold change of two was applied, resulting in a total of 163 genes upregulated in blood tissues, 27 genes upregulated in brain tissues and 85 genes that were not differentially expressed, suggesting blood is a poor proxy for brain tissue when quantifying gene expression (Supplementary Table 17).
Finally, we assessed whether gene expression differences on the Y chromosome are associated with Parkinson’s disease or Y-chromosome haplogroup. Genes differentially expressed in Parkinson’s disease cases and controls were identified with edgeR using the blood-derived AMP-PD data. One pseudogene, KDM5DP1, was significantly upregulated in some cases but did not remain significant after thresholded testing relative to a log-fold change of two was applied (Supplementary Table 18). Next, differences in Y-chromosome gene expression between major haplogroups were identified in AMP-PD samples using edgeR. Only major haplogroups present in at least 40 samples were compared, leaving major haplogroups E, G, I, J and R (Supplementary Fig. 2). After filtering out genes with low expression, we identified a total of 64 genes upregulated in individuals with major haplogroup G and eight genes upregulated in samples with major haplogroup J. Four of these 72 genes had a log-fold change greater than two (Supplementary Fig. 3). None of the total 240 genes were differentially expressed in major haplogroup E, I and R samples when compared with samples with different haplogroups. However, when we applied thresholded testing relative to a log-fold change of two, no genes were differentially expressed in any major haplogroup (Supplementary Tables 19–23).
Discussion
Here we assessed whether Y-chromosome variation contributes to Parkinson’s disease using the largest publicly available case/control Parkinson’s disease data sets containing Y-chromosome genotypes. We used three different tools, each with their own algorithm and reference data, to call Y haplogroups. Reference data from each tool were compared to determine if the use of each tool’s reference data would allow for an unbiased comparison between haplogroup calling tools (Supplementary Fig. 4). Y-LineageTracker included more variants compared with the other tools (73 223 for Y-LineageTracker, 16 551 for Yhaplo and 29 586 for Snappy; Supplementary Fig. 4B). Additionally, the AMP-PD data set, the data set with the most variants, had more variants in Y-LineageTracker’s reference data compared with the other two tools (8863 for Y-LineageTracker, 2712 for Yhaplo and 3961 for Snappy; Supplementary Fig. 4C). However, Yhaplo had the largest percentage of variants in its reference file included in all three cohorts (∼16% for AMP-PD, ∼1.3% for UKBB, ∼0.3% for NeuroX) (Supplementary Fig. 4D). In spite of this, reference data from Y-LineageTracker were used for downstream analysis because it was the most up-to-date and the largest of the three reference data sets. Haplogroup frequency comparisons between data sets with default and updated Y-LineageTracker reference data showed little change between major haplogroups (∼87 to ∼99% samples with the same major haplogroup when using original versus updated reference data), but full haplogroups showed a much larger change (∼0.2 to ∼43% samples with the same full haplogroups when using original versus updated reference data) and an increase in the number of unique full haplogroups, confirming our hypothesis that use of each tool’s original reference data would lead to biased results when comparing tools (Supplementary Table 8).
The three haplogroup calling tools had the highest concordance rates for major haplogroups in the AMP-PD cohort. Differences were more pronounced in the UKBB and NeuroX cohorts; however, the major haplogroup was typically consistent (Fig. 2). This is likely due to the smaller number of variants included in these genotyped cohorts compared with the WGS AMP-PD cohort.
Major haplogroup R was the most common in all three of our cohorts (AMP-PD, UKBiobank and NeuroX), being present in at least half of all samples. Our comparison with other studies shows that the same is applicable for European countries such as Belgium, The Netherlands, the UK, Poland, Spain, Portugal and Belarus (Supplementary Table 9). All included cohorts and these countries had the same major haplogroup with the second highest frequency (named I), with the exception of Spain and Portugal. It should be noted that major haplogroup frequencies in our included cohorts were distinct from those found in the Northern African countries of Algeria and Egypt and in areas of southern Europe, including Italy. Samples included in our study were obtained from European ancestry populations, so these comparisons support the accuracy and reliability of the Snappy, Yhaplo and Y-LineageTracker haplogroup calling tools for determining major haplogroups.
However, the same cannot necessarily be stated for the comparability of these tools when assigning full Y-chromosome haplogroups. Of the three tools, Y-LineageTracker identified the most unique full haplogroups in the AMP-PD and NABEC cohorts and Yhaplo identified the most unique full haplogroups in the UKBB and NeuroX cohorts (Supplementary Table 8). This contrast is likely due to differences in algorithms used by the tools and in Chromosome Y variant coverage across included cohorts. However, only a small percentage (<20%) of the variants explored through each tool were present in each of the three cohorts. Therefore, it is possible that precise Y-chromosome haplogroups were incorrectly assigned for some samples. However, the similarity of major haplogroup frequencies with past studies suggests these tools are very reliable when identifying major haplogroups.
Y-LineageTracker was used to determine major haplogroups in our analyses because it identified the most unique full haplogroups in the WGS AMP-PD data set and included the most comprehensive and up-to-date reference data. Interestingly, we observed a large difference between cohorts when comparing the percentage of variants included in the Y-LineageTracker reference data (∼12.1% for AMP-PD, ∼0.42% for UKBB and ∼0.07% for NeuroX).
The large difference in percentages suggests that WGS data, such as the AMP-PD data, are a much better fit for our analyses than genotype data, such as the UKBB and NeuroX data. The performance of these tools will likely increase as methods to identify Y-chromosome variants improve, such as the development of Y-chromosome imputation panels, allowing for a better understanding of the relationship between Y-chromosome haplogroups and Parkinson’s disease.
The meta-analysis of the logistic regression per haplogroup identified no major haplogroups associated with Parkinson’s disease after multiple test correction (Supplementary Table 11). Likewise, a meta-analysis focusing on the more specific full haplogroups resulted in no full haplogroups associated with Parkinson’s disease after multiple test correction (Supplementary Table 13). No Y-chromosome variants were found to be associated with disease after multiple test correction in the analysis of AMP-PD data or in a meta-analysis of all data sets (Supplementary Tables 14 and 15). Therefore, our analyses identified neither Y-chromosome haplogroups nor Y-chromosome variants significantly associated with Parkinson’s disease.
While Y-chromosome gene expression was overall low in both brain and blood tissues, we identified clear differences in Y-chromosome gene expression between these tissues both before and after applying thresholded testing with a log-fold change filter to the edgeR results (Supplementary Table 17). This is in line with previous studies that have shown Y-chromosome genes to be most highly expressed in sex-specific tissues.45
A large proportion of genes were removed from the data sets used in each differential expression analysis when genes with zero expression in all samples were removed and when the filterByExpr edgeR function was used. After applying these filters, a total of 22 049 of the 60 708 (including autosomal genes) NABEC genes remained for the Y PAR analysis, 275 of the 553 genes remained for the NABEC brain versus AMP-PD blood analysis, 278 of the 563 AMP-PD genes remained for the Parkinson’s disease case/control analysis and 240 of the 563 AMP-PD genes remained for the major haplogroup analysis.
Comparison of gene expression between samples mapped to a reference genome without Y PARs and samples mapped to a reference genome with Y PARs identified a total of 17 genes significantly upregulated in the samples without Y PARs. All of these genes were located in PAR regions on the X chromosome. This was expected, because Y PARs were not present in the reference genome used for these samples. Gene expression was not significantly different for genes in any other areas of the genome. This suggests that masking PARs when quantifying gene expression likely does not significantly alter expression levels of genes outside of PARs when using brain derived data sets.
Differences in gene expression between major haplogroups were identified in major haplogroups G and J (Supplementary Tables 20 and 22). Genes with high log-fold change values displayed clear differences in expression levels between samples (Supplementary Fig. 3). However, none of these genes remained significant after thresholded testing was applied, suggesting gene expression patterns are not specific to Y-chromosome haplogroups. There was a similar lack of differentially expressed genes in the Parkinson’s disease case/control analysis, suggesting Y-chromosome gene expression patterns are not specific to disease status (Supplementary Table 18).
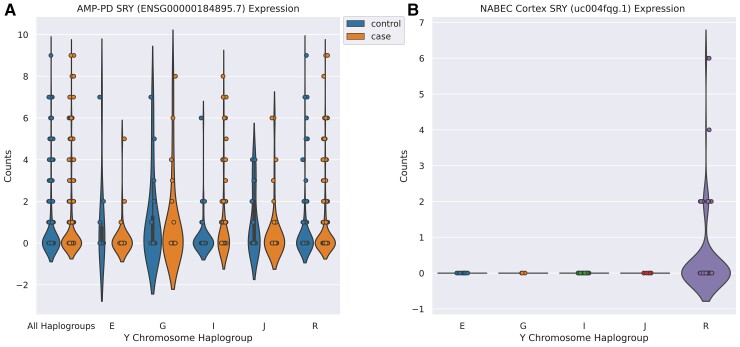
To-date, chromosome Y Parkinson’s disease studies have focused on several genes, including SRY. In vitro studies suggest upregulation of SRY protects against 6-hydroxydopamine induced Parkinson’s disease in male dopaminergic neurons.46 An in vivo study has demonstrated that inhibition of SRY diminishes dopaminergic cell damage in 6-hydroxydopamine and rotenone induced Parkinson’s disease rat models.47 A study of Parkinson’s disease in Asian populations has found no significant association between SRY variants and disease risk.48 In concordance with this study, we found no significant SRY variants to be associated with Parkinson’s disease and expression patterns of SRY in brain and blood tissue was overall low, suggesting this gene does not play a major role in Parkinson’s disease (Fig. 4). Mouse studies have shown that SRY expression is precisely regulated during embryonic development, going from high to low expression in a few days, to properly trigger male development.49 This suggests an assessment of SRY expression in the brain of older individuals, like in our study, may be of limited use. Additionally, SRY indirectly leads to the production of testosterone, a steroid found to be significantly lowered in human Parkinson’s disease cases.50 Findings such as these suggest SRY, or other genes involved in sex determination or hormone regulation, may indirectly influence male neurobiology, potentially explaining the increased prevalence of Parkinson’s disease in males.51
Figure 4.
SRY expression in blood from AMP-PD samples and frontal cortex from NABEC samples. SRY counts were obtained from featureCounts grouped by case/control status and major haplogroup in blood tissues using AMP-PD data (A) and in brain tissues using NABEC frontal cortex data (B).
While our analyses included a large number of samples and Y-chromosome variants, further inspection of the included variants reveals that a large portion of Chromosome Y remains to be explored (Supplementary Fig. 5). This empty region, spanning ∼30 megabases, is a known heterochromatic region of Chromosome Y, often referred to as Yq12 and consists mostly of DYZ1 and DYZ2 repeats.52 This region, covering ∼50% of Chromosome Y, and the two PARs were not covered in our analyses due to lack of data. Therefore, our assigned haplogroups only account for the euchromatic and non-recombining areas of Chromosome Y. Interestingly, relatively few variants overlapped between each cohort and the Y-LineageTracker reference data used to assign haplogroups, suggesting single variant analyses are a logical next step after haplogroup association to investigate variants not included in haplogroup assignment (Supplementary Fig. 4C).
The heterochromatic region on the long arm of Chromosome Y has typically been left out of sequencing studies due to the difficulties involved in sequencing highly repetitive regions, leading some to assume this region is genetically inert and unimportant.53 However, numerous studies have shown that this may not be the case. For example, noncoding RNA transcribed from heterochromatin on Yq12 has been shown to regulate isoforms of CDC2L2, a chromosome one gene.54 Variation in Yq12 heterochromatin has been reported in humans, but its significance is unknown.55 Additionally, partial Y chromosomal heterochromatin deletions have been shown to induce increased vulnerability to stress in mice and reduced neurogenesis in the hippocampus.56 These findings support previous predictions that heterochromatin may be involved in the regulation of euchromatic gene expression.57 Methods such as long read sequencing are likely required to properly characterize the heterochromatic and repetitive Yq12 region of Chromosome Y.58 Future studies will need to take this into account when reassessing the role of Chromosome Y in Parkinson’s disease.
Overall, our data suggest that genetic variation on Chromosome Y does not have a major effect on risk for Parkinson’s disease. However, disease association may exist in Y-chromosome variants not included in this study. Additionally, regulatory Y-chromosome variants may indirectly affect Parkinson’s disease risk. Future research will need to reassess these scientific questions once additional WGS data and more genetically diverse data are available.
Supplementary Material
Acknowledgements
The authors thank all of the subjects who donated their time and biological samples to be part of this study. The authors are grateful to members of the North American Brain Expression Consortium for contributing DNA samples. This research has been conducted using the UK Biobank Resource under Application Number 33601. Data used in the preparation of this article were obtained from the Accelerating Medicine Partnership® (AMP®) Parkinson’s Disease (AMP-PD) Knowledge Platform. For up-to-date information on the study, visit https://www.amp-pd.org. The AMP®-PD programme is a public–private partnership managed by the Foundation for the National Institutes of Health and funded by the National Institute of Neurological Disorders and Stroke (NINDS) in partnership with the Aligning Science Across Parkinson’s (ASAP) initiative; Celgene Corporation, a subsidiary of Bristol–Myers Squibb Company; GlaxoSmithKline plc (GSK); The Michael J. Fox Foundation for Parkinson’s Research; Pfizer Inc.; Sanofi US Services Inc.; and Verily Life Sciences. Accellerating Medicines Partnership and AMP are registered service marks of the US Department of Health and Human Services. Clinical data and biosamples used in preparation of this article were obtained from the (i) Michael J. Fox Foundation for Parkinson’s Research (MJFF) and NINDS BioFIND study, (ii) Harvard Biomarkers Study (HBS), (iii) National Institute on Aging (NIA) International Lewy Body Dementia Genetics Consortium Genome Sequencing in Lewy Body Dementia Case–control Cohort (LBD), (iv) MJFF LRRK2 Cohort Consortium (LCC), (v) NINDS Parkinson’s Disease Biomarkers Programme (PDBP), (vi) MJFF Parkinson’s Progression Markers Initiative (PPMI) and (vii) NINDS Study of Isradipine as a Disease-modifying Agent in Subjects With Early Parkinson Disease, Phase 3 (STEADY-PD3) and (viii) the NINDS Study of Urate Elevation in Parkinson’s Disease, Phase 3 (SURE-PD3). BioFIND is sponsored by MJFF with support from the NINDS. The BioFIND Investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit michaeljfox.org/news/biofind. Genome sequence data for the LBD were generated at the Intramural Research Programme of the US National Institutes of Health. The study was supported in part by the NIA (programme #: 1ZIAAG000935) and the National Institute of Neurological Disorders and Stroke (programme #: 1ZIA-NS003154). The Harvard Biomarker Study (HBS) is a collaboration of HBS investigators (full list of HBS investigators found at https://www.bwhparkinsoncenter.org/biobank/) and funded through philanthropy and NIH and non-NIH funding sources. The HBS Investigators have not participated in reviewing the data analysis or content of the manuscript. Data used in preparation of this article were obtained from The Michael J. Fox Foundation sponsored LCC. The LCC Investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit https://www.michaeljfox.org/biospecimens. PPMI is sponsored by MJFF and supported by a consortium of scientific partners: 4D Pharma, AbbVie Inc., AcureX Therapeutics, Allergan, Amathus Therapeutics, ASAP, Avid Radiopharmaceuticals, Bial Biotech, Biogen, BioLegend, Bristol–Myers Squibb, Calico Life Sciences LLC, Celgene Corporation, DaCapo Brainscience, Denali Therapeutics, The Edmond J. Safra Foundation, Eli Lilly and Company, GE Healthcare, GSK, Golub Capital, Handl Therapeutics, Insitro, Janssen Pharmaceuticals, Lundbeck, Merck & Co., Inc., Meso Scale Diagnostics, LLC, Neurocrine Biosciences, Pfizer Inc., Piramal Imaging, Prevail Therapeutics, F. Hoffmann-La Roche Ltd and its affiliated company Genentech Inc., Sanofi Genzyme, Servier, Takeda Pharmaceutical Company, Teva Neuroscience, Inc., UCB, Vanqua Bio, Verily Life Sciences, Voyager Therapeutics, Inc. and Yumanity Therapeutics, Inc. The PPMI investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit www.ppmi-info.org. The PDBP consortium is supported by the NINDS at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. The PDBP investigators have not participated in reviewing the data analysis or content of the manuscript. The Study of Isradipine as a Disease-modifying Agent in Subjects with Early Parkinson Disease, Phase 3 (STEADY-PD3) is funded by the NINDS at the National Institutes of Health with support from The Michael J. Fox Foundation and the Parkinson Study Group. For additional study information, visit https://clinicaltrials.gov/ct2/show/study/NCT02168842. The STEADY-PD3 investigators have not participated in reviewing the data analysis or content of the manuscript. The Study of Urate Elevation in Parkinson’s Disease, Phase 3 (SURE-PD3) is funded by the NINDS at the National Institutes of Health with support from The Michael J. Fox Foundation and the Parkinson Study Group. For additional study information, visit https://clinicaltrials.gov/ct2/show/NCT02642393. The SURE-PD3 investigators have not participated in reviewing the data analysis or content of the manuscript.
Abbreviations
- AMP-PD =
Accelerating Medicines Partnership for Parkinson’s Disease
- GWAS =
genome-wide association study
- NABEC =
North American Brain Expression Consortium
- PAR =
pseudoautosomal region
- Snappy =
Single-Nucleotide Assignment of Phylogenetic Parameters on the Y Chromosome
- UKBB =
United Kingdom Biobank
- WGS =
whole-genome sequencing
Contributor Information
Francis P Grenn, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA.
Mary B Makarious, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA; Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, London, UK; UCL Movement Disorders Centre, University College London, London, UK.
Sara Bandres-Ciga, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA.
Hirotaka Iwaki, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA; Center for Alzheimer’s and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, USA; Data Tecnica International, Washington, DC, USA.
Andrew B Singleton, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA; Center for Alzheimer’s and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, USA.
Mike A Nalls, Molecular Genetics Section, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA; Center for Alzheimer’s and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, USA; Data Tecnica International, Washington, DC, USA.
Cornelis Blauwendraat, Center for Alzheimer’s and Related Dementias (CARD), National Institute on Aging and National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, USA; Integrative Neurogenomics Unit, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD, USA.
Funding
This work was supported in part by the Intramural Research Programmes of the National Institute on Aging (NIA) part of the National Institutes of Health, Department of Health and Human Services; project numbers 1ZIA-NS003154, Z01-AG000949-02, ZO1 AG000535 and Z01-ES101986.
Competing interests
M.A.N.’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research, he also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc.
Supplementary material
Supplementary material is available at Brain Communications online.
References
- 1. Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of Parkinson’s disease. Lancet Neurol. 2020;19(2):170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nalls MA, Blauwendraat C, Vallerga CL, et al. . Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18(12):1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Foo JN, Chew EGY, Chung SJ, et al. . Identification of risk loci for Parkinson disease in Asians and comparison of risk between Asians and Europeans: A genome-wide association study. JAMA Neurol. 2020;77(6):746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nandipati S, Litvan I. Environmental exposures and Parkinson’s disease. Int J Environ Res Public Health. 2016;13(9):E881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gallo V, Vineis P, Cancellieri M, et al. . Exploring causality of the association between smoking and Parkinson’s disease. Int J Epidemiol. 2019;48(3):912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ascherio A, Weisskopf MG, O’Reilly EJ, et al. . Coffee consumption, gender, and Parkinson’s disease mortality in the cancer prevention study II cohort: The modifying effects of estrogen. Am J Epidemiol. 2004;160(10):977–984. [DOI] [PubMed] [Google Scholar]
- 7. Noyce AJ, Bestwick JP, Silveira-Moriyama L, et al. . Meta-analysis of early nonmotor features and risk factors for Parkinson disease. Ann Neurol. 2012;72(6):893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moisan F, Kab S, Mohamed F, et al. . Parkinson disease male-to-female ratios increase with age: French nationwide study and meta-analysis. J Neurol Neurosurg Psychiatry. 2016;87(9):952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwaki H, Blauwendraat C, Leonard HL, et al. . Differences in the presentation and progression of Parkinson’s disease by sex. Mov Disord. 2021;36(1):106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blauwendraat C, Iwaki H, Makarious MB, et al. . Investigation of autosomal genetic sex differences in Parkinson’s disease. Ann Neurol. 2021;90(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Guen Y, Napolioni V, Belloy ME, et al. . Common X-chromosome variants are associated with Parkinson disease risk. Ann Neurol. 2021;90(1):22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parker K, Erzurumluoglu AM, Rodriguez S. The Y chromosome: A complex locus for genetic analyses of complex human traits. Genes (Basel). 2020;11(11):E1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anderson K, Cañadas-Garre M, Chambers R, Maxwell AP, McKnight AJ. The challenges of chromosome Y analysis and the implications for chronic kidney disease. Front Genet. 2019;10:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sezgin E, Lind JM, Shrestha S, et al. . Association of Y chromosome haplogroup I with HIV progression, and HAART outcome. Hum Genet. 2009;125(3):281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Charchar FJ, Bloomer LD, Barnes TA, et al. . Inheritance of coronary artery disease in men: An analysis of the role of the Y chromosome. Lancet Lond Engl. 2012;379(9819):915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ran J, Han TT, Ding XP, et al. . Association study between Y-chromosome haplogroups and susceptibility to spermatogenic impairment in Han People from southwest China. Genet Mol Res. 2013;12(1):59–66. [DOI] [PubMed] [Google Scholar]
- 17. Eales JM, Maan AA, Xu X, et al. . Human Y chromosome exerts pleiotropic effects on susceptibility to atherosclerosis. Arterioscler Thromb Vasc Biol. 2019;39(11):2386–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iwaki H, Leonard HL, Makarious MB, et al. . Accelerating medicines partnership: Parkinson’s disease. Genetic resource. Mov Disord. 2021;36(8):1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bycroft C, Freeman C, Petkova D, et al. . The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nalls MA, Bras J, Hernandez DG, et al. . NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol Aging. 2015;36(3):1605.e7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Severson AL, Shortt JA, Mendez FL, Wojcik GL, Bustamante CD, Gignoux CR. Snappy: Single nucleotide assignment of phylogenetic parameters on the Y chromosome. Published online October 29, bioRxiv 2018:454736. [Google Scholar]
- 23. Poznik GD. Identifying Y-chromosome haplogroups in arbitrarily large samples of sequenced or genotyped men. Published online November 19, bioRxiv 2016:088716. [Google Scholar]
- 24. Chen H, Lu Y, Lu D, Xu S. Y-LineageTracker: A high-throughput analysis framework for Y-chromosomal next-generation sequencing data. BMC Bioinf. 2021;22(1):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. International Society of Genetic Genealogy . Y-DNA Haplogroup Tree 2019, Version: 15.73. Published online July 11, 2020. http://www.isogg.org/tree/. Accessed August 20, 2021.
- 26. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinforma Oxf Engl. 2010;26(22):2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abraham G, Qiu Y, Inouye M. Flashpca2: Principal component analysis of Biobank-scale genotype datasets. Bioinforma Oxf Engl. 2017;33(17):2776–2778. [DOI] [PubMed] [Google Scholar]
- 28. Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. [Google Scholar]
- 29. Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinforma Oxf Engl. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang K, Li M, Hakonarson H. Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gibbs JR, van der Brug MP, Hernandez DG, et al. . Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6(5):e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frankish A, Diekhans M, Jungreis I, et al. . Gencode 2021. Nucleic Acids Res. 2021;49(D1):D916–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Webster TH, Couse M, Grande BM, et al. . Identifying, understanding, and correcting technical artifacts on the sex chromosomes in next-generation sequencing data. GigaScience. 2019;8(7):giz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dobin A, Davis CA, Schlesinger F, et al. . Star: Ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47(8):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson MD, McCarthy DJ, Smyth GK. Edger: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Larmuseau MHD, Vanderheyden N, Jacobs M, Coomans M, Larno L, Decorte R. Micro-geographic distribution of Y-chromosomal variation in the central-western European region Brabant. Forensic Sci Int Genet. 2011;5(2):95–99. [DOI] [PubMed] [Google Scholar]
- 38. Grochowalski Ł, Jarczak J, Urbanowicz M, et al. . Y-chromosome genetic analysis of modern polish population. Front Genet. 2020;11:567309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adams SM, Bosch E, Balaresque PL, et al. . The genetic legacy of religious diversity and intolerance: Paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am J Hum Genet. 2008;83(6):725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kushniarevich A, Sivitskaya L, Danilenko N, et al. . Uniparental genetic heritage of Belarusians: Encounter of rare middle eastern matrilineages with a central European mitochondrial DNA pool. PLoS One. 2013;8(6):e66499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bowden GR, Balaresque P, King TE, et al. . Excavating past population structures by surname-based sampling: The genetic legacy of the Vikings in northwest England. Mol Biol Evol. 2008;25(2):301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bekada A, Fregel R, Cabrera VM, et al. . Introducing the Algerian mitochondrial DNA and Y-chromosome profiles into the North African landscape. PLoS One. 2013;8(2):e56775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karmin M, Saag L, Vicente M, et al. . A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res. 2015;25(4):459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Y Chromosome Consortium . A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12(2):339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Godfrey AK, Naqvi S, Chmátal L, et al. . Quantitative analysis of Y-chromosome gene expression across 36 human tissues. Genome Res. 2020;30(6):860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Czech DP, Lee J, Correia J, Loke H, Möller EK, Harley VR. Transient neuroprotection by SRY upregulation in dopamine cells following injury in males. Endocrinology. 2014;155(7):2602–2612. [DOI] [PubMed] [Google Scholar]
- 47. Lee J, Pinares-Garcia P, Loke H, Ham S, Vilain E, Harley VR. Sex-specific neuroprotection by inhibition of the Y-chromosome gene, SRY, in experimental Parkinson’s disease. Proc Natl Acad Sci U S A. 2019;116(33):16577–16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pan H, Wang Y, Zhao Y, et al. . No relationship between SRY variants and risk of Parkinson’s disease in Chinese population. Neurobiol Aging. 2021;100:119.e3–119.e6. [DOI] [PubMed] [Google Scholar]
- 49. Larney C, Bailey TL, Koopman P. Switching on sex: Transcriptional regulation of the testis-determining gene SRY. Dev Camb Engl. 2014;141(11):2195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toczylowska B, Zieminska E, Michałowska M, Chalimoniuk M, Fiszer U. Changes in the metabolic profiles of the serum and putamen in Parkinson’s disease patients—In vitro and in vivo NMR spectroscopy studies. Brain Res. 2020;1748:147118. [DOI] [PubMed] [Google Scholar]
- 51. Kopsida E, Stergiakouli E, Lynn PM, Wilkinson LS, Davies W. The role of the Y chromosome in brain function. Open Neuroendocrinol J. 2009;2:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manz E, Alkan M, Bühler E, Schmidtke J. Arrangement of DYZ1 and DYZ2 repeats on the human Y-chromosome: A case with presence of DYZ1 and absence of DYZ2. Mol Cell Probes. 1992;6(3):257–259. [DOI] [PubMed] [Google Scholar]
- 53. Bachtrog D, Charlesworth B. Towards a complete sequence of the human Y chromosome. Genome Biol. 2001;2(5):REVIEWS1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jehan Z, Vallinayagam S, Tiwari S, et al. . Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2. Genome Res. 2007;17(4):433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cotter PD, Norton ME. Y chromosome heterochromatin variation detected at prenatal diagnosis. Prenat Diagn. 2005;25(11):1062–1063. [DOI] [PubMed] [Google Scholar]
- 56. Dey SK, Kamle A, Dereddi RR, et al. . Mice with partial deletion of Y-heterochromatin exhibits stress vulnerability. Front Behav Neurosci. 2018;12:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. [DOI] [PubMed] [Google Scholar]
- 58. De Bustos A, Cuadrado A, Jouve N. Sequencing of long stretches of repetitive DNA. Sci Rep. 2016;6:36665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All AMP-PD (https://amp-pd.org/) and UKBB (https://www.ukbiobank.ac.uk/) data are available via application on their websites and the NeuroX data are available via dbGap at phs000918.v1.p1. NABEC data are available via dbGap at phs001354.v1.p1. All codes are available on the GitHub page: https://github.com/neurogenetics/chrY_haplogroups_PD.