Abstract
Background
Most patients with SARS-CoV-2 are non-infectious within 2 weeks, though viral RNA may remain detectable for weeks. However there are reports of persistent SARS-CoV-2 infection, with viable virus and ongoing infectivity months after initial detection. Beyond individuals, viral evolution during persistent infections may be accelerated, driving emergence of mutations associated with viral variants of concern. These patients often do not meet inclusion criteria for clinical trials, meaning clinical and virologic characteristics, and optimal management strategies are poorly evidence-based.
Methods
We analysed cases of SARS-CoV-2 infection from a regional testing laboratory in South-West England between March 2020 and December 2021, with at least two SARS-CoV-2 positive samples separated by ≥ 56 days were identified. Excluding those with confirmed or likely re-infection, we identified patients with persistent infection, characterised by an ongoing clinical syndrome consistent with COVID-19 alongside monophyletic viral lineage of SARS-CoV-2. We examined clinical and virologic characteristics, treatment, and outcome. We further performed a literature review investigating cases of persistent SARS-CoV-2 infection, reviewing patient characteristics and treatment.
Results
We identified six patients with persistent SARS-CoV-2 infection. All were hypogammaglobulinaemic and had underlying haematological malignancy, with four having received B-cell depleting therapy. Evidence of viral evolution, including accrual of mutations associated with variants of concern, was demonstrated in five cases. Four patients ultimately cleared SARS-CoV-2. In two patients, clearance followed treatment with casirivimab/imdevimab. Both survived beyond thirty days following viral clearance, having experienced infections of 305- and 269-days duration respectively, after failed attempts at clearance with alternative therapies. We found 60 cases of confirmed persistent infection in the literature, with a further 31 probable cases. Of those, 80% of patients treated with monoclonal antibodies cleared SARS-CoV-2, and none died.
Conclusion
Haematological malignancy and patients receiving B-cell depleting therapies represent key groups at risk of persistent SARS-CoV-2 infection. Throughout persistent infection, SARS-CoV-2 can evolve rapidly, giving rise to significant mutations, including those implicated in variants of concern. Monoclonal antibodies appear to be a promising therapeutic option, potentially in combination with antivirals, crucial for individuals, and for public health.
Keywords: Persistent infection, SARS-CoV-2, Persistent SARS-CoV-2 infection, Viral evolution, Variant of concern, Monoclonal antibody, Casirivimab/Imdevimab, Haematological malignancy, Immunocompromised, Hypogammaglobulinaemia
Introduction
Though most patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are non-infectious after 10–14 days, viral RNA may remain detectable for several weeks (Cevik et al., 2021). The vast majority do not have active infection with SARS-CoV-2 (Sethuraman et al., 2020), however ongoing RNA detection could represent persistent coronavirus disease 2019 (COVID-19) (Brown et al., 2022). With B-cells and production of neutralising antibodies crucial components in the immune response to SARS-CoV-2, persistent infection has been described in patients with antibody deficiency or secondary B-cell depletion, often due to therapies such as anti-CD20 monoclonal antibodies (mAbs) (Brown et al., 2022, Roeker et al., 2020, Haidar and Mellors, 2021). Cases display evidence of ongoing viral replication, viable virus on culture far into illness, and ongoing symptoms or relapses for months (Brown et al., 2022, Choi et al., 2020, Kemp et al., 2021, Leung et al., 2022). Patients may experience delayed symptom onset, more severe disease, and higher mortality (Arcani et al., 2021).
Whilst such cases are challenging for individual patient care, they also generate epidemiological concern due to the potential development of SARS-CoV-2 genome mutations (Choi et al., 2020, Kemp et al., 2021, Leung et al., 2022). This accelerated viral evolution within a single host potentially contributes to the development of viral variants of concern (VOCs) (Moran et al., 2021), with important implications for infection control and public health.
Various treatments, including convalescent plasma, antivirals and mAbs have been investigated (Taha et al., 2021, RECOVERY Collaborative Group, 2022) with variable findings of their utility. Therefore, there is no consensus on optimal treatment strategy.
We sought to investigate cases of persistent SARS-CoV-2 infection in three hospitals in South-West England, describing the clinical and virologic characteristics and the management of cases, correlated with existing literature.
Methods
We included cases of persistent SARS-CoV-2 infection in this retrospective study, initially identified by searching cases of SARS-CoV-2 RNA positivity with ≥ 2 positive results > 56 days apart, within a regional testing laboratory in south-west England from March 2020 to December 2021.
SARS-CoV-2 RNA positivity was detected through sampling nasopharyngeal swabs, tested initially on the closed platform system, Panther Hologic using the Aptima SARS-CoV-2 transcription mediated assay (TMA), followed by semi-quantitative analysis using SARS-CoV-2 reverse transcription polymerase chain reaction (RT-PCR) (NeuMoDx SARS CoV-2 assay [Qiagen] or RealStar SARS-CoV-2 RT-PCR Kit 1.0 [Altona Diagnostics]) where possible, allowing cycle threshold (Ct) values to be recorded for viral load assessment. Thereafter, VOC genotyping (ThermoFisher) and whole genome sequencing (WGS) of positive samples was performed via the COG-UK consortium (COG-UK, 2020).
Clinical and biochemical data were collected from hospital records, regarding symptomatology, underlying immunosuppression and respiratory conditions, serum immunoglobulin levels, presence of serum SARS-CoV-2 antibodies (Liaison XL anti-SARS-CoV-2 S1/S2 IgG assay, DiaSorin, Italy), course of COVID-19 clinical illness, treatment and outcome.
Using clinical and testing data, cases were reviewed to confirm the presence of persistent virus alongside a clinical syndrome in keeping with persistent SARS-CoV-2 infection. Cases due to confirmed or probable re-infection were excluded, as evidenced by VOC genotyping or WGS, or cases where there was no evidence of clinical disease (e.g. hospital admission) or samples were > 100 days apart with no intermediate sampling.
For the literature review, we searched the MEDLINE database for case reports or series describing persistent SARS-CoV-2 infection. The search was performed on 8 June 2022, using a search strategy of ‘(prolong* OR persist* OR chronic) AND (SARS-CoV-2 OR covid* OR coronavirus*) AND (immunocomp* OR immunodef* OR “immune comp*” OR “immune def*” OR lymphom* OR myelom* OR leukaem* OR leukem* OR malignan*)’. Peer-reviewed and pre-print reports were included.
Results
Overview of cases
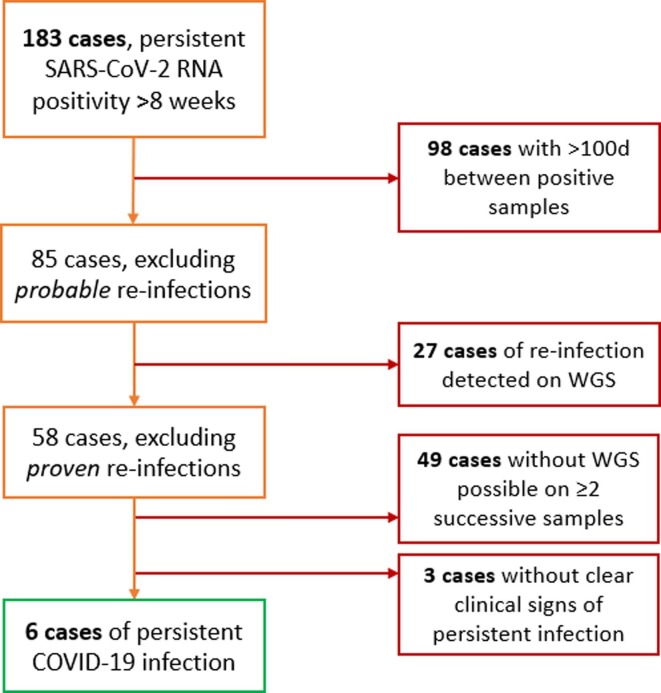
We identified 183 patients with at least two SARS-CoV-2 RNA positive samples > 56 days apart. Having excluded those due to probable and proven re-infection, and those with no ongoing clinical signs of persistent infection (fever, raised inflammatory markers and ongoing respiratory illness), we identified six cases of persistent SARS-CoV-2 infection (Fig. 1 ).
Fig. 1.
Identification of persistent SARS-CoV-2 infection cases.
Of these, the median time from first to final positive samples was 133.5 days (range 58–305 days, 2 patients > 250 days), with four patients ultimately clearing SARS-CoV-2. Two patients died remaining positive. The median patient age was 63.5 years (range 42–80 years), with all six patients having an underlying haematological malignancy, being hypogammaglobulinaemic, and having received immunosuppressive medication, such as anti-CD20 mAbs. Three of six patients were tested for anti-SARS-CoV-2 antibodies, none of whom had an antibody response. No patients had received anti-SARS-CoV-2 vaccination before presentation. Baseline patient characteristics are described in Table 1 , with Table 2 describing clinical aspects of infections and treatment received.
Table 1.
Characteristics of persistent SARS-CoV-2 infected patients.
| Patient | Age Sex (M/F) | Primary disease | Immunosuppression | Co-morbidities | SARS-CoV-2 positivity duration (days) | Time to SARS-CoV-2 negativity (days) | Serum immuno-globulins | SARS-CoV-2 antibodies | Mortality (from last positive SARS-CoV-2 sample) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 71 M |
CLL | FCR chemotherapy (6 cycles, completed 6 m pre-COVID) |
Resp: NSIP/HP Other: hypo-gammaglobulinaemia post-chemotherapy |
305 | 310 | IgA + IgM low | Initially negative | 30-day: N 90-day: N 1-year: N |
| 2 | 53 F |
AML | Ongoing ciclosporin + Sorafenib Bone marrow transplant (6 months pre-COVID) |
Resp: ex-smoker Other: GvHD |
154 | Died while still positive | IgA + IgM low | Not tested | 30-day: Y 90-day: Y 1-year: Y |
| 3 | 58 F |
NHL | Obinutuzumab |
Resp: asthma, ex-smoker Other: nil |
269 | 296 | IgA + IgM low | Negative | 30-day: N 90-day: N 1-year: awaited |
| 4 | 80 F |
CLL | Previous Ibrutinib (stopped 1 m pre-COVID) |
Resp: ex-smoker Other: malignant melanoma, neutropaenia |
94 | Died while still positive | IgA low IgM normal |
Negative | 30-day: Y 90-day: Y 1-year: Y |
| 5 | 42 M |
ALL | Bone marrow transplant - haploidentical allograft (14 months pre-COVID) |
Resp: progressive pulmonary fibrosis Other: T2DM |
58 | 61 | IgA + IgM low | Not tested | 30-day: N 90-day: Y 1-year: Y |
| 6 | 69 F |
DLBCL | R-CHOP chemotherapy |
Resp: asthma Other: nil |
113 | 122 | IgA + IgM low | Not tested | 30-day: Y 90-day: Y 1-year: Y |
ALL - acute lymphoblastic leukaemia; AML - acute myeloid leukaemia; CLL - chronic lymphocytic leukaemia; DLBCL - diffuse large B-cell lymphoma; FCR - Fludarabine/cyclophosphamide/rituximab; GvHD - graft vs host disease; NHL - Non-Hodgkin lymphoma; NSIP/HP – non-specific interstitial pneumonitis/ hypersensitivity pneumonitis; T2DM - Type 2 diabetes mellitus.
Table 2.
Summary of 6 patients’ COVID-19 presentation, testing, and treatment.
| Patient | Initial date of SARS-CoV-2 positivity | Severity of illness | Concurrent diagnoses | Attempted SARS-CoV-2 elimination therapies (Day 0 = D0 = date of first positive sample) |
|---|---|---|---|---|
| 1 | 09/05/2020 | Hospitalised | Pulmonary bacterial super-infection | Remdesivir (5 days, D213-217) IV immunoglobulin (D216) Casirivimab/imdevimab (D266) |
| 2 | 21/05/2020 | Hospitalised | Nil | Remdesivir |
| 3 | 11/01/2021 | Hospitalised | Nil | Remdesivir (21 days, D57-77) Remdesivir (5 days, D273-277) + Casirivimab/imdevimab (D277) |
| 4 | 11/01/2021 | Hospitalised | Neutropaenic sepsis Pulmonary bacterial super-infection |
|
| 5 | 26/01/2021 | Intensive Care Unit care required | NTM respiratory infection | Remdesivir (5 days, D52-56) Tocilizumab (D52) |
| 6 | 11/01/2021 | Hospitalised | NTM skin infection Pulmonary bacterial super-infection |
NTM – Non-tuberculous mycobacterium.
Virologic analysis
WGS was attempted on SARS-CoV-2 positive samples, with success and sequencing quality dramatically reduced in samples with a Ct value ≥ 30. If the sample did not meet 50 % CLIMB sequencing threshold, these were not uploaded to CLIMB and did not meet the 50% sequencing coverage threshold, as per the COG-UK protocol (COG-UK, 2020).
Viral evolution of SARS-CoV-2 was evident in patients 1–5 (P1-5), with less certainty regarding viral evolution in P6. Phylogenetic analysis demonstrated viral lineages of B.52 (P1), B1.1.7 (alpha VOC, P2,4,5 + 6) and B1.177 (P3), which were maintained throughout the course of infections. Details are outlined below, with P1′s course also visually presented in Fig. 2 , and P2-5 in Fig. 3 .
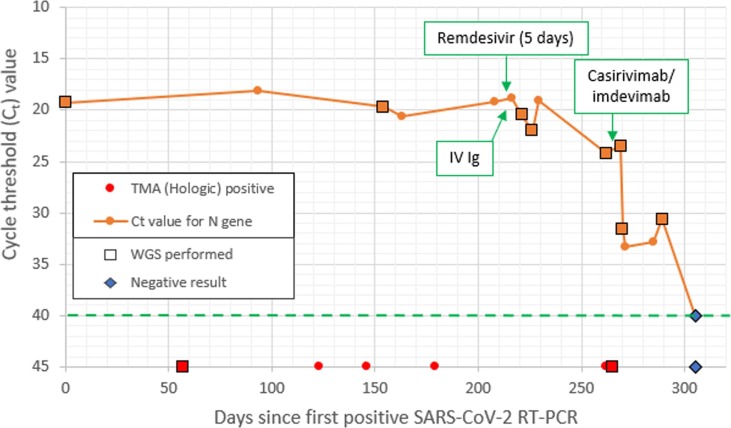
Fig. 2.
Clinical and virological course of patient 1. Ct values plotted from results of NeuMoDx RT-PCR, N gene target, where semi-quantitation was possible. For ease of viewing, though TMA (Hologic) was also positive on each occasion semi-quantitation was possible, TMA positives are only plotted when these were positive, but semi-quantitation was not performed. The green dotted line represents the line above which Ct values would be detectable as a ‘positive’ result. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
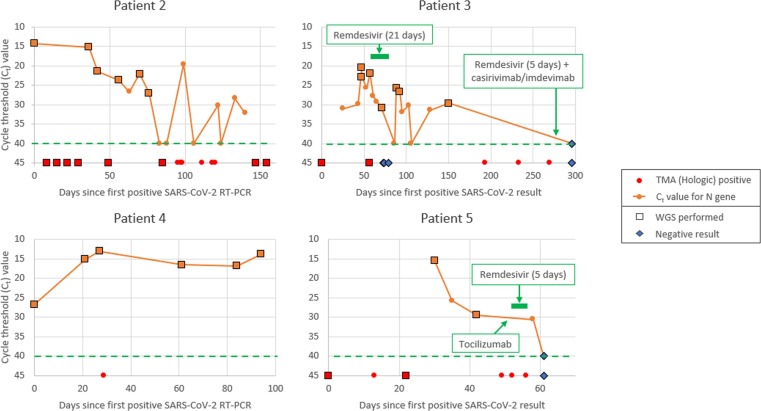
Fig. 3.
Clinical and virological courses of patients 2–5. Ct values taken from results of NeuMoDx RT-PCR, N gene target, wherever semi-quantitation was possible. For ease of viewing, though TMA (Hologic) was also positive on each occasion semi-quantitation was possible, TMA positives are only plotted when these were positive, but semi-quantitation was not performed. The green dotted line on each graph represents the line above which Ct values are detectable as a ‘positive’ result. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
P1 had RNA positivity for 305 days, the longest duration of our cohort. Multiple therapies were trialled attempting elimination of SARS-CoV-2, including Remdesivir, intravenous immunoglobulin (IVIg) and casirivimab/imdevimab, with eventual viral clearance after 310 days, Day-45 (D45) following casirivimab/imdevimab therapy, coinciding with a resolution in symptoms. High levels of viral RNA were detected throughout, with Ct values ranging from 18 to 24 from D1-D270, before a gradual increase in Ct value until clearance (Fig. 2). This case has previously been described in a case report (Kavanagh Williamson et al., 2021). The viral lineage was B.52 variant, remaining monophyletic other than the final successful WGS (D290), which revealed viral lineage B1.1.7 (alpha VOC), raising the possibility that this final detection was a brief re-infection. However given subsequent viral clearance, further WGS was not possible. By D155, substantial viral evolution had occurred, with accrual of mutations ΔH69/ΔV70 alongside spike protein substitution H655Y. Evolution continued, with a peak of 14 single nucleotide polymorphisms (SNPs) when compared with the originally sequenced sample. One month following treatment with Remdesivir and IVIg, 6 non-synonymous and 3 synonymous alterations were found however following treatment with casirivimab/imdevimab no further viral evolution was detected, indeed SARS-CoV-2 cleared thereafter.
P2 remained positive for 154 days, first presenting within six months of a haematopoietic stem cell transplant. While SARS-CoV-2 viral load gradually fell, especially after D99, they remained RNA positive at the time of their death. Sequence analysis again revealed accrual of mutations, with 47 SNP differences from first to final sample, including amino acid replacements (T205I) associated with the beta VOC, and persistent accrual of an M86V mutation. As it was unclear when Remdesivir was administered, its relationship to mutation accrual is unclear.
P3 was positive for 269 days, becoming negative by D296. An initial prolonged (21-day) course of Remdesivir did not clear the virus, although there was a transient period of negativity and suppression of symptoms towards the end of the course (D73-79), before retesting RNA positive within a week (Fig. 3). The virus later cleared 17 days after combination therapy with Remdesivir and casirivimab/imdevimab. There were 11 SNPs throughout the course of illness, alongside 7 amino acid replacements. Following the initial course of Remdesivir, persistent amino acid substitutions arose, such as E420D in the nsp13 protein before viral clearance soon after combination therapy.
P4 was positive for 94 days, remaining so until their death, while still displaying symptoms and radiological changes consistent with COVID-19. This was in keeping with gradually falling Ct values (increasing viral load). It is not clear from clinical notes why P4 did not receive any COVID-19 specific therapeutics. P5 remained positive for 58 days, clearing SARS-CoV-2 by D61, within 10 days of receiving Remdesivir and Tocilizumab. Due to the short duration of positivity, and failure to successfully sequence beyond D42, we assumed persistent infection while RT-PCR remained positive. Both P4 + 5 gained an E484K mutation through the course of disease (D84 - P4, D22 - P5), prior to any COVID-19 therapeutics, which persisted throughout subsequent sequencing.
WGS data for P6 was available on three samples (D0, D14 and D87), however D87 data had significant areas of poor coverage throughout WGS and therefore it could not be confirmed that the D87 virus, which had 10 mutations was derived from the same virus as was detected on D0 and D14. As the patient thereafter cleared the virus, further analysis was not possible.
Literature review
From our database search, 1502 articles were identified, with 69 case reports/series ultimately included for further analysis. Using our definition of persistent infection, displaying evidence of clinical and virologic persistence, we identified 60 cases (46 articles) of persistent SARS-CoV-2 infection. The details of these are outlined in Table 3 . Further, we identified 31 further cases (23 case reports/series) with evidence of clinical persistence, however without clear documentation of virologic persistence. These cases are felt likely to represent persistent SARS-CoV-2 infection but given the lack of virologic evidence we have termed these “probable persistent SARS-CoV-2 infection”. These are outlined in Supplementary Table S1.
Table 3.
Cases of persistent SARS-CoV-2 infection identified by literature review.
| Case # | Authors | Age,Sex | Immunosuppression |
Persistence |
Elimination Rx attempted | Anti-SARS CoV-2 Abs detected? | Virologic outcome | Mortality | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Primary disease | Secondary causes | Virologic duration | Clinical symptoms | |||||||
| 1 | Baang et al. (2021) | 60 M | MCL | CD20 bispecific Ab + 2nd B-cell directed Ab, as part of chemotherapy regimen | 131 days | Cough, Dyspnoea, Fever | RDV + CP (x2) | Minimally post-CP, otherwise negative | Likely cleared (unstated) | No |
| 2 | Montejano et al. (2022) | 30 M |
HIV + DLBCL |
Anti-CD20-containing chemotherapy (RIX) CAR-T-cell therapy |
97 days | Fever, “pulmonary infiltrates“ | RDV + CP, mAb (Sotrovimab) | N/A | Cleared | No |
| 3 | Morel et al. (2022) | 74 M |
SOT (renal) ITP |
Tacrolimus, MMF, steroidsRIX (2 m pre-COVID) |
3 months | Dyspnoea, Fever, hypoxia | – | N/A | Cleared | No |
| 4 | Ko et al. (2022) | 53 M |
AML GvHD |
Allogenic SCT Ruxolitinib + steroids |
94 days | Cough, fever, dyspnoea, | RDV (x3), Cas/Imd | N/A | Persisted | No |
| 5 | Ko et al. (2022) | 67 F |
MZL Evans syndrome |
Splenectomy RIX + steroids |
97 days | Dyspnoea | RDV (x2), IVIg, mAb (Cas/Imd), Tocilizumab | N/A | Persisted | No |
| 6 | Schenker et al. (2021) | 61 M |
CLL | Anti-CD20-containing chemotherapy (RIX) |
148 days | Cough, Diarrhoea, Fever, Myalgia |
RDV (x2), CP | Negative | Persisted | No |
| 7 | Arai et al. (2022) | 71 M |
FL | Anti-CD20-containing chemotherapy (RIX) | >8 weeks | Fever | RDV | Negative | Cleared | No |
| 8 | Leung et al. (2022) | 21 F |
ALL | Allogenic SCT, BlinatumomabAnti-CD22 mAb (inotuzumab) |
98 days | Dyspnoea, fever, hypoxia | RDV | Negative | Persisted | Yes (in-hospital) |
| 9 | Martinot et al. (2021) | 76 F |
CLL | Anti-CD20-containing chemotherapy (RIX) | 66 days | Fever, hypoxia | RDV, CP, HCQ, IVIg | Negative | Cleared | No |
| 10 | Morishita et al. (2022) | 51 M |
NHL | RIX | 134 days | Anosmia, fever | RDV | Negative | Cleared | No |
| 11 | Bailly et al. (2022) | 23 M |
ALL | Allogenic SCTRIX (EBV-reactivation) |
385 days | Dyspnoea, fever | CP (x4), RDV, mAb (Cas/Imd) (x2) | N/A | Cleared | No |
| 12 | Cabañero-Navalon et al. (2020) | 22 M |
CVID | RIX (3y ago) Note: regular IVIg |
77 days | Dyspnoea, fever | IVIg, RDV, CP | N/A | Cleared | No |
| 13 | Moutinho-Pereira et al. (2021) | 39 F |
SLE | RIX HCQ, Leflunomide |
94 days | Dyspnoea, fever | CP | Negative | Likely cleared (unstated) | No |
| 14 | Taha et al. (2021) | 55 M |
FL | Anti-CD20-containing chemotherapy (RIX) 2nd anti-CD20/anti-CD3) bispecific Ab (Glofitamab) |
210 days | Fever | RDV, CP, mAb (Cas/Imd) | Negative | Cleared | No |
| 15 | Nussenblatt et al. (2022) | 48 F |
DLBCL | CAR-T cell therapy | 335 days | “worsening respiratory symptoms” | CP, RDV | Negative | Cleared | No |
| 16 | Sonnleitner et al. (2022) | ‘60 s’ F |
SCLL EBV re-activation |
Anti-CD20-containing chemotherapy (RIX) |
207 days | Cough, fatigue, fever | IVIg (x3), COVID-19 mRNA vaccine (2 doses) | Only post-IVIg | Cleared | No |
| 17 | Drouin et al. (2021) | 59 M |
FL | Anti-CD20-containing chemotherapy (OBZ) |
312 days | Hypoxia, cough, dyspnoea | HCQ, Lop/Rit, CP (x2), RDV, IVIg (x2), mAb (Cas/Imd) (x2) | Negative | Cleared | No |
| 18 | Keitel et al. (2021) | 25 F |
SCID Graft failure |
HSCT Note: Regular IVIg |
61 days | “persistent respiratory symptoms” | RDV, CP (x2) | Only post-CP | Cleared | No |
| 19 | Ueda et al. (2022) | 63 F |
FL | Anti-CD20-containing chemotherapy (OBZ) | 56 days | Fever, hypoxia | RDV | Negative | Cleared | No |
| 20 | Shoji et al. (2022) | 61 M |
FL | Anti-CD20-containing chemotherapy (RIX) |
100 days | Hypoxia, pneumonia (on imaging) | Favipiravir, RDV, IVIg | Negative | Cleared | No |
| 21 | Bronstein et al. (2021) | 33 M |
HL | Nil | 3 months | Fever | mAb (Bamlanivimab) | N/A | Likely cleared (unstated) | No |
| 22 | Gandhi et al. (2022) | 70 F |
NHL | Anti-CD20-containing chemotherapy (RIX) | 217 days | Fever, anosmia | IVIg, RDV, mAb (Cas/Imd) | Negative | Cleared | No |
| 23 | Hanssen et al. (2021) | 77 M |
CLL | Anti-CD20-containing chemotherapy (RIX), ibrutinib Note: regular IVIg |
64 days | Cough, dyspnoea, fever | RDV, CP (x3) | Negative | Cleared | No |
| 24 | Zimmerli et al. (2021) | 74 M |
CLL | Anti-CD20-containing chemotherapy (RIX) |
78 days | Cough, diarrhoea, fever | CP (x4) | Only post-CP | Cleared | No |
| 25 | Choi et al. (2020) | 45 M |
Anti-phos-pholipid syndrome | Cyclophosphamide, RIX, eculizumab (anti-complement 5 mAb) | 151 days | Abdominal pain, fever, hypoxia | RDV (x3), IVIg, ruxolitinib, cyclophosphamide. | N/A | Persisted | Yes (in-hospital) |
| 26 | Zhabokritsky et al. (2022) | 76 M |
HIV | Nil | 142 days | Cough, fever, dysphagia (oral thrush) | Nil specific | N/A | Persisted | Yes |
| 27 | Kemp et al. (2021) | 70 s M |
MZL | Anti-CD20-containing chemotherapy (RIX) | 102 days | Fever, hypoxia | RDV (x3), CP (x3), Tocilizumab | Only post-CP | Persisted | Yes (in-hospital) |
| 28 | Pérez-Lago et al. (2021) | 52 M |
FL | Anti-CD20-containing chemotherapy (RIX) |
188 days | Fever | CP (x2), HCQ (x2), anakinra, RDV (x3), Lop/Rit, Tocilizumab | Negative | Persisted | Yes |
| 29 | Pérez-Lago et al. (2021) | 47 M |
FL | Anti-CD20-containing chemotherapy (RIX) |
141 days | Fever, “respiratory failure” | HCQ (x2), RDV (x2), Lop/Rit (x3), CP, Tocilizumab, IVIg | Transiently post-CP | Cleared | No |
| 30 | Pérez-Lago et al. (2021) | 63 F |
FL | Anti-CD20-containing chemotherapy (RIX) | 69 days | Not clear | HCQ, Lop/Rit (x3), IVIg, RDV (x2), CP | Negative | Persisted | Yes |
| 31 | Khatamzas et al. (2021) | 70 s F |
FL | Anti-CD20-containing chemotherapy (OBZ) | 156 days | “Persistent infection” | CP (x5) | Transiently post-CP | Persisted | Yes |
| 32 | Yasuda et al. (2021) | 61 F |
FL | Anti-CD20-containing chemotherapy (prev OBZ, current RIX) | “10 months” | Cough, fever | IVIg (multiple) | Negative | Cleared | No |
| 33 | Ciuffreda et al. (2021) | 23 M |
XLA | Note: Regular IVIg | 149 days | Dyspnoea, fever | RDV (x2), Lop/Rit, CP | Negative | Persisted | Yes |
| 34 | Scherer et al. (2022) | 40 s F |
DLCBL | Anti-CD20-containing chemotherapy (RIX) | 75 days | Dyspnoea, fever, hypoxia | mAb (Bamlanivimab), RDV (x2) | Only post-mAb | Persisted | Yes |
| 35 | Scherer et al. (2022) | 30 s F |
MZL | Anti-CD20-containing chemotherapy (RIX) | 109 days | Cough, fever, dyspnoea | IVIg (x2), RDV (x2), CP |
Negative | Cleared | No |
| 36 | Scherer et al. (2022) | 30 s F |
Myelo-dysplastic syndrome GvHD |
SCT RIX, MMF, steroids Note: Regular IVIg |
302 days | Dyspnoea | mAb (Bamlanivimab), IVIg, RDV | Negative | Cleared | No |
| 37 | Scherer et al. (2022) | 40 s M |
Thymoma | Thymectomy | 200 days | Cough, fever, dyspnoea | RDV (x2), CP, IVIg | Negative | Cleared | No |
| 38 | Berktas and Koyunc (2022) | 71 M |
MCL | Anti-CD20-containing chemotherapy (RIX) Autologous bone marrow transplant |
164 days | Fever, hypoxia | Favipiravir, RDV, Anakinra, CP | Negative | Persisted | No |
| 39 | Thornton et al. (2022) | 58 M |
FL | Anti-CD20-containing chemotherapy (RIX) |
184 days | Fever, hypoxia | RDV (x3), IVIg, mAb (Bamlanivimab) | N/A | Cleared | No |
| 40 | Purpura et al. (2022) | 69 F |
SOT (heart) | Recent IVIg + eculizumab Tacrolimus, MMF, steroids |
134 days | Hypoxia | HCQ (x2), Tocilizumab, RDV | Negative initially, +ve at D72 |
Persisted | Yes |
| 41 | Sepulcri et al. (2021) | 70 M |
MCL | Anti-CD20-containing chemotherapy (RIX) | 268 days | Fever | Darunavir/Rit, CP, HCQ, Tocilizumab (x2), IVIg, RDV (x4) | Negative | Persisted | Yes |
| 42 | Lynch et al. (2021) | 52 F |
FL | Anti-CD20-containing chemotherapy (OBZ) Note: Monthly IVIg |
95 days | Fever, hypoxia | HCQ | N/A | Persisted | No |
| 43 | Gibson et al. (2021) | 46 F |
Multiple sclerosis | Ocrelizumab (anti-CD20 mAb) | 70 days | Dyspnoea, fever | CP, RDV | Negative | Cleared | No |
| 44 | Lee et al. (2022) | 30 s M |
DLBCL | Anti-CD20-containing chemotherapy (RIX) | 328 days (at least) | Cough, fever, dyspnoea | Not recorded | N/A | Not listed | Yes |
| 45 | Lee et al. (2022) | 70 s F |
CLL | Anti-CD20-containing chemotherapy (OBZ) | 61 days (at least) | Dyspnoea, fever | Not recorded | N/A | Not listed | No |
| 46 | Lee et al. (2022) | 40 s F |
Myelo-fibrosis | Allogenic SCT | 63 days (at least) |
Diarrhoea, fever | Not recorded | N/A | Not listed | No |
| 47 | Lee et al. (2022) | 60 s M |
DLBCL | CAR-T-cell therapy | 64 days (at least) | Dyspnoea, fever | Not recorded | N/A | Not listed | No |
| 48 | Lee et al. (2022) | 50 s M |
FL | Anti-CD20-containing chemotherapy (OBZ) | 112 days (at least) | Cough, fever, dyspnoea | Not recorded | N/A | Not listed | No |
| 49 | Lee et al. (2022) | 50 s M |
MCL | CAR-T-cell therapy | 95 days (at least) | Dyspnoea, fever | Not recorded | N/A | Not listed | No |
| 50 | Hensley et al. (2021) | 73 M |
Multiple myeloma | CAR-T cell therapy | 72 days | Dyspnoea, hypoxia | CP, RDV | Negative | Persisted | Yes |
| 51 | Riddell et al. (2022) | 40 s ? |
HIV |
Nil | 61 days | Dyspnoea, fever | Not recorded | Negative | Not listed | No |
| 52 | Riddell et al. (2022) | 40 s ? |
HIV DLBCL |
Anti-CD20-containing chemotherapy (RIX) | 111 days | Hypoxia | Not recorded | Negative | Not listed | No |
| 53 | Riddell et al. (2022) | 30 s ? |
HIV | Nil | 255 days | Fever | RDV | Yes, from initial infection | Not listed | No |
| 54 | Truong et al. (2021) | 21 M |
ALL | CAR-T-cell therapy | 250 days | Cough, fever | RDV (x2), CP (x9) | Yes, from D136 | Persists | No |
| 55 | Truong et al. (2021) | 2 M |
ALL | Non-anti-CD20 chemotherapy | 162 days | Fever | RDV | Yes, only on D84, D176 | Cleared | No |
| 56 | Helleberg et al. (2020) | 50 s M |
CLL | Anti-CD20-containing chemotherapy (RIX) | 56 days | Fever | CP, RDV (x2) | Negative | Cleared | No |
| 57 | Reuken et al. (2021) | 56 F |
FL | Anti-CD20-containing chemotherapy (RIX) | 4 months | Cough, fever | RDV, CP (x2), Infliximab | Only post-CP | Cleared | No |
| 58 | Nakajima et al. (2021) | 47 M |
FL | Anti-CD20-containing chemotherapy (OBZ) | 59 days | Fever | Favipiravir, Lop/Rit | Negative | Cleared | No |
| 59 | Borges et al. (2021) | 61 F |
NHL | “immunosuppressive therapy” | 197 days | Cough, fever, dyspnoea | RDV | Negative | Likely cleared (unstated) | No |
| 60 | Monrad et al. (2021) | 75 M |
CLL | Anti-CD20-containing chemotherapy (RIX), Ibrutinib | 333 days | Dyspnoea, fever | RDV (x2), CP (x2) | Negative | Persisted | No |
Key: ALL = acute lymphoblastic leukaemia CAR = chimeric antigen receptor (T-cell therapy); Cas/Imd = casirivimab/imdevimab (REGN-COV2) dual monoclonal antibody; CLL = chronic lymphocytic leukaemia; CP = convalescent plasma; CVID = common variable immunodeficiency; DLBCL = diffuse large B-cell lymphoma; FL = follicular lymphoma; GvHD = graft versus host disease; HCQ = hydroxychloroquine; HIV = human immunodeficiency virus; HL = Hodgkin lymphoma; ITP = idiopathic thrombocytopaenic purpura; IVIg = intravenous immunoglobulin; Lop/Rit = lopinavir/ritonavir; (m)Ab = (monoclonal) antibody; MCL = mantle cell lymphoma; MMF = mycophenolate mofetil; MZL = marginal zone lymphoma; NHL = non-Hodgkin lymphoma; OBZ = obinutuzumab; RDV = remdesivir; RIX = rituximab; SCID = severe combined immunodeficiency; SCLL = small cell lymphocytic lymphoma; SCT = stem cell transplant; SLE = systemic lupus erythematosus; SOT = solid organ transplant; XLA = X-linked agammaglobulinaemia.
Discussion
This retrospective cohort of 6 cases of persistent SARS-CoV-2 infection describes the clinical and virologic characteristics associated with such infections. In addition, our literature review comprehensively summarises similar cases previously described. In a cohort for which prospective research is challenging, this can help provide insight into risk factors associated with persistent SARS-CoV-2 infection as well as potential treatment modalities.
There is wide variability in the classification of persistent infection, due to challenges in differentiating persistency from re-infection (Choudhary et al., 2021). In this study, viral culture was not widely available, though was performed for P1, therefore we used evidence of viral RNA persistence with successive accumulation of mutations, coupled with an ongoing COVID-19 clinical syndrome, to indicate persistent infection.
Our cohort all had haematological malignancies, echoing the findings of our literature review in which 48 patients (80%) had haematological malignancy, higher than in a UK-wide case series (Brown et al., 2022) in which 42% of cases had this underlying diagnosis, with other persistent cases in patients with primary immunodeficiencies or early HIV infection. However several authors assert that any immunodeficiency may pre-dispose to persistent SARS-CoV-2 infection (Brown et al., 2022, Haidar and Mellors, 2021). Other immuno-depleting factors are therapeutics, such as B-cell therapies, used in four of our patients, more specifically anti-CD20 mAbs (3 of our cases, 42 cases (70%) of those from our literature review). While patients with haematological malignancies produce antibodies, those with lymphoma or receiving B-cell depleting therapies may produce fewer neutralising antibodies (Cattaneo et al., 2021). Alongside this, Rituximab or other B-cell therapies lead to hypogammaglobulinaemia (Roberts et al., 2015) and may impact the immune system’s ability to produce anti-SARS-CoV-2 total antibodies (Mrak et al., 2021), as was the case in our cohort. Due to each patient’s status as “clinically vulnerable”, all would have been eligible for COVID-19 vaccination, however these six persistent infections pre-dated the widespread roll-out of the UK’s COVID-19 vaccination program (Department of Health Social Care, 2020). While significant morbidity is associated with persistent infection (Brown et al., 2022), mortality has not been widely reported. In our cohort 50% of cases died during the hospital admission, with 66% mortality at 30 days following the final SARS-CoV-2 positive result. Mortality for patients with haematological malignancy and persistent SARS-CoV-2 infection may be even worse than the already poor prognosis for acute COVID-19 in similar patients (Arcani et al., 2021). Having said that, in our literature review mortality was 22% for this persistently infected cohort (16% if probable infections are included). However, it may be that some patients died before the 56-day cut-off used in our definition, or the mortality in the acute phase of COVID-19 is higher in these multiple co-morbid patients.
Our cohort also demonstrated extensive viral evolution, including several acquisitions of spike protein mutations indicative of subsequent VOCs, such as the E484K mutation in P4 + 5 (beta and gamma), as well as H655Y (gamma) and ΔH69/ΔV70 (alpha) in P1. Several of the mutations occurring in our patient group are commonly observed in other viral sequences, often causing predicted changes to known T and B cell viral epitopes, highlighting the risk that immune escape variants may arise in this patient group. We did not observe mutations associated with resistance to COVID-19 antivirals utilising the COG-UK/Mutation explorer (Wright et al., 2022) (manuscript in preparation). The E484K mutation, the commonest mutation found in a genomic analysis of persistent infections (Wilkinson et al., 2022), affects the receptor-binding domain of the spike protein, a region critical in neutralisation of SARS-CoV-2 (Cele et al., 2022). In contrast to Scherer et al. (2022) who felt that persistent infection alone was not enough to drive emergence of spike mutations, we found the emergence of significant mutations irrespective of elimination strategies, particularly in P4 who never received any elimination therapies. Similarly, P5 developed the E484K mutation prior to Remdesivir administration. However P1 experienced rapid viral evolution before, and following Remdesivir and IVIg, though mutations of therapeutic significance largely arose pre-Remdesivir, and no further mutations were detected following later dual mAb therapy. Thus, while therapeutics may play a role in driving accelerated viral evolution, from our cohort it appears that persistent infection alone was sufficient opportunity for mutations to occur. Given the rate of mutations and SNPs in our cohort particularly affecting the spike protein, alongside variable virologic response to elimination therapies, it is likely that SARS-CoV-2 has various approaches to avoiding neutralisation in persistent infection, increasing virulence, or enhancing immune escape. Given concerns of where the Omicron variant has arisen (Mallapaty, 2022), as well as the potential for further, highly mutated variants of SARS-CoV-2 in future, if chronically infected individuals are implicated in the development of VOCs also suggests that control of such infections is crucial in controlling the SARS-CoV-2 pandemic.
Two patients (P1 + P3) have survived beyond 6 months since viral clearance, have recovered and remain negative. Both cleared SARS-CoV-2 following dual mAb therapy with casirivimab/imdevimab, with P3 having received combination therapy with Remdesivir. Given the prolonged, persistent infection prior to the administration of mAb therapy, alongside subsequent rapid resolution of clinical symptoms and SARS-CoV-2 from the upper respiratory tract, we suspect mAbs played a crucial role. This is in keeping with other reports (Brown et al., 2022, Taha et al., 2021) and our literature revealed that in cases of confirmed persistent infection, 80% (12/15) of those receiving mAb therapy cleared SARS-CoV-2 by the time of publication. No patients who had received mAbs for persistent SARS-CoV-2 infection died. Both P1 + 3 had previously failed to clear SARS-CoV-2 with Remdesivir monotherapy. Multiple therapeutics have been trialled, with Remdesivir monotherapy, convalescent plasma, and IVIg in persistently infected patients having delivered variable results (Brown et al., 2022, Kemp et al., 2021, Rüfenacht et al., 2022), occasionally leading to a very brief period of improvement virologically or clinically before rapid relapse (Schenker et al., 2021, Morishita et al., 2022), exactly as occurred for P3. Further, it has been suggested that a relapse of infection following the administration of convalescent plasma can drive accelerated accrual of escape mutations (Kemp et al., 2021), however this has not always been replicated in subsequent cases (Taha et al., 2021). Clinical data on the use of convalescent plasma is inconsistent (RECOVERY Collaborative Group, 2021, Sullivan et al., 2022), though successful viral clearance has been described in certain populations and may be dependent on further factors such as dose responses. However, combination therapy, such as the role of Remdesivir alongside mAbs may be valuable (Taha et al., 2021, Dioverti et al., 2022), and was the treatment approach used for P3, with rapid viral clearance. Further trials would need to address this question as currently there remains a lack of strong evidence. Given the role which anti-CD20 therapies seem to play in persistent infection, it is also critical to consider whether any alternative therapies for haematological malignancy are possible, to reduce the B-cell depletion which seems such a crucial pre-disposing factor (Von-Lilienfeld-Toal et al., 2020). The long-term sustainability of monoclonal therapy for the eradication of persistent infection is in question following the replacement of Delta by the Omicron coronavirus variant. Casirivimab/imdevimab was quickly shown to have negligible neutralising activity against Omicron (Bruel et al., 2022) and its use quickly advised against in the UK and USA among other jurisdictions (Department of Health Social Care, 2021). Nonetheless, newer synthetic antibody therapies such as tixagevimab-cilgavimab and sotrovimab, may provide therapeutic options against newer variants of SARS-CoV-2 and their utility continues to be investigated (ACTIV-3-Therapeutics for Inpatients with COVID-19 (TICO) Study Group, Pogue and McCreary, 2022). We are yet to see whether the widespread adoption of early antiviral medications to prevent severe disease in high-risk groups, such as those with haematological malignancy, will also prevent the development of persistent infection.
Limitations to this study include its retrospective nature and the small numbers included, reflecting the prevalence of persistent infection on anything below a national level. There are few statistics possible beyond descriptive analysis of this cohort, potentially limiting generalisability. However, reports such as this are crucial to drive recognition of small, yet significant cohorts, not only for individuals but for the wider approach to the pandemic. Further, our literature review seeks to collate and describe the most comprehensive body of cases of persistent SARS-CoV-2 infection yet described. Finally, while viral culture would be optimal in confirming the presence of live virus, our approach of correlating clinical and virologic parameters represents a realistic approach to recognising and monitoring persistent infection in clinical practice.
Conclusions
We describe clinical and virologic characteristics of persistent SARS-CoV-2 infection, a small but crucial cohort of patients given the potential impact not only for individuals but also to public health. Haematological malignancy and B-cell depletion, often secondary to anti-CD20 mAb therapy are important risk factors to recognise, to ensure appropriate investigation, such as regular semi-quantitative SARS-CoV-2 analysis. Previous concerns regarding accelerated viral evolution, particularly relating to potential intra-host evolution driving VOCs, were evidenced in this study, irrespective of SARS-CoV-2 therapeutics. Given this, rapid viral elimination in persistently positive patients is critical. Our cohort mimics many of the findings of our literature review, which describes 60 cases of confirmed infection, though given our further finding of 31 cases of probable infection, a clearer definition and approach to diagnosing persistent SARS-CoV-2 infection is also important going forwards. Combination therapy, with antivirals and mAbs appears a promising therapeutic approach, though larger-scale trials are required.
Ethical statement
As the study was retrospective in nature, with no impact on the clinical care of patients, specific ethical approval was not required for this study.
CRediT authorship contribution statement
David Hettle: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing. Stephanie Hutchings: Methodology, Investigation, Data curation, Writing – review & editing. Peter Muir: Conceptualization, Methodology, Writing – review & editing, Supervision. Ed Moran: Conceptualization, Methodology, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clinpr.2022.100210.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- ACTIV-3-Therapeutics for Inpatients with COVID-19 (TICO) Study Group, 2022. Tixagevimab-cilgavimab for treatment of patients hospitalised with COVID-19: a randomised, double-blind, phase 3 trial. Lancet Respir Med 2022;10(10):972-84. [DOI] [PMC free article] [PubMed]
- Arai T., Mukai S., Kazama R., Gawa Y., Nichida K., Hatanaka H., et al. Persistent viral shedding of severe acute respiratory syndrome coronavirus 2 after treatment with bendamustine and rituximab: a case report. J Infect Chemother. 2022;28(6):810–813. doi: 10.1016/j.jiac.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcani R., Colle J., Cauchois R., Koubi M., Jarrot P.A., Jean R., et al. Clinical characteristics and outcomes of patients with haematologic malignancies and COVID-19 suggest that prolonged SARS-CoV-2 carriage is an important issue. Ann Hematol. 2021;100(11):2799–2803. doi: 10.1007/s00277-021-04656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baang J.H., Smith C., Mirabelli C., Valesano A.L., Manthei D.M., Bachman M.A., et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis. 2021;223(1):23–27. doi: 10.1093/infdis/jiaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly B., Péré H., Veyer D., Berceanu A., Daguindau E., Roux P., et al. Persistent coronavirus disease 2019 (COVID-19) in an immunocompromised host treated by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific monoclonal antibodies. Clin Infect Dis. 2022;74(9):1706–1707. doi: 10.1093/cid/ciab868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berktas B.M., Koyuncu A. Case report: Unremitting COVID-19 pneumonia, viral shedding and failure to develop anti-SARS-CoV-2 antibodies for more than 6 months in patient with mantle cell lymphoma treated with rituximab. Am J Trop Med Hyg. 2022;106(4):1104–1107. doi: 10.4269/ajtmh.21-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V., Isidro J., Cunha M., Cochicho D., Martins L., Banha L., et al. Long-term evolution of SARS-CoV-2 in an immunocompromised patient with non-Hodgkin lymphoma. mSphere. 2021;6(4) doi: 10.1128/mSphere.00244-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein Y., Adler A., Katash H., Halutz O., Herishanu Y., Levytskyi K. Evolution of spike mutations following antibody treatment in two immunocompromised patients with persistent COVID-19 infection [published online ahead of print 13 Nov 2021] J Med Virol. 2021 doi: 10.1002/jmv.27445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L.K., Moran E., Goodman A., Baxendale H., Bermingham W., Buckland M., et al. Treatment of chronic or relapsing COVID-19 in immunodeficiency. Ann Clin Microbiol Antimicrob. 2022;149(2):557–561. doi: 10.1016/j.jaci.2021.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I., et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med. 2022;29:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- Cabañero-Navalon M.D., Garcia-Bustos V., Ruiz-Rodriguez P., Comas I., Coscollá M., Martinez-Priego L., et al. Persistent SARS-CoV-2 infection with repeated clinical recurrence in a patient with common variable immunodeficiency. Clin Microbiol Infect. 2020;28(2):308–310. doi: 10.1016/j.cmi.2021.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo C., Cancelli V., Imberti L., Dobbs K., Sottini A., Pagani C., et al. Production and persistence of specific antibodies in COVID-19 patients with hematologic malignancies: role of rituximab. Blood Cancer J. 2021;11(9):151. doi: 10.1038/s41408-021-00546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Karim F., Lustig G., San J.E., Hermanus T., Tegally H., et al. SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe. 2022;30:154–162. doi: 10.1016/j.chom.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. NEJM. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary M.C., Crain C.R., Qiu X., Hanage W., Li J.Z. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequence characteristics of coronavirus disease 19 (COVID-19) persistence and reinfection. Clin Inf Dis. 2021;74(2):237–245. doi: 10.1093/cid/ciab380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffreda L., Lorenzo-Salazar J.M., Alcoba-Florez J., Rodriguez-Pérez H., Gil-Campesino H., Iñigo-Campos A., et al. Longitudinal study of a SARS-CoV-2 infection in an immunocompromised patient with X-linked agammaglobulinemia. J Infect. 2021;83(5):607–635. doi: 10.1016/j.jinf.2021.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19 Genomics UK (COG-UK) consortiumcontact@cogconsortium.uk. An integrated national scale SARS-CoV-2 genomic surveillance network. Lancet Microbe 2020;1(3):e99-e100. [DOI] [PMC free article] [PubMed]
- Department of Health & Social Care. Joint Committee on Vaccination and Immunisation: advice on priority groups for COVID-19 vaccination (2020, December 30). Available at: https://www.gov.uk/government/publications/priority-groups-for-coronavirus-covid-19-vaccination-advice-from-the-jcvi-30-december-2020/joint-committee-on-vaccination-and-immunisation-advice-on-priority-groups-for-covid-19-vaccination-30-december-2020 [Accessed 3 October 2022].
- Department of Health & Social Care. COVID-19 therapeutic alert: Neutralising monoclonal antibodies in the treatment of COVID-19 in hospitalised patients (2021, 16 December). Available at: https://www.cas.mhra.gov.uk/ViewandAcknowledgment/ViewAlert.aspx?AlertID=103187 [Accessed 1 July 2022].
- Dioverti, M.V., Gaston, D.C., Morris, C.P., Huff, C.A., Jain, T., Jones, R., et al., 2022. Combination therapy with casirivimab/imdevimab and remdesivir for protracted SARS-CoV-2 infection in B-cell-depleted patients. Open Forum Infect Dis 2022;9(6):ofac064. [DOI] [PMC free article] [PubMed]
- Drouin A.C., Theberge M.W., Liu S.Y., Smither A.R., Flaherty S.M., Zeller M., et al. Successful clearance of 300 day SARS-CoV-2 infection in a subject with B-cell depletion associated prolonged (B-DEAP) COVID by REGEN-COV anti-spike monoclonal antibody cocktail. Viruses. 2021;13(7):1202. doi: 10.3390/v13071202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S., Klein J., Robertson A.J., Peña-Hernandez M.A., Lin M.J., Roychoudhury P., et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat Commun. 2022;13(1):1547. doi: 10.1038/s41467-022-29104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, E.G., Pender, M., Angerbauer, M., Cook, C., Jones, B., Spivak, A.M., et al., 2021. Prolonged SARS-CoV-2 illness in a patient receiving ocrelizumab for multiple sclerosis. Open Forum Infect Dis 2021;8(7):ofab176. [DOI] [PMC free article] [PubMed]
- Haidar G., Mellors J.W. Improving the outcomes of immunocompromised patients with COVID-19. Clin Inf Dis. 2021;73(6):e1397–e1401. doi: 10.1093/cid/ciab397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen J.L.J., Stienstra J., Boers S.A., Pothast C.R., Zaaijer H.L., Tjon J.M., et al. Convalescent plasma in a patient with protracted COVID-19 and secondary hypogammaglobulinaemia due to chronic lymphocytic leukaemia: buying time to develop immunity? Infect Dis Rep. 2021;13(4):855–864. doi: 10.3390/idr13040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleberg M., Niemann C.U., Moestrup K.S., Kirk O., Lebech A.M., Lane C., et al. Persistent COVID-19 in an immunocompromised patient temporarily responsive to two courses of remdesivir therapy. J Infect Dis. 2020;222(7):1103–1107. doi: 10.1093/infdis/jiaa446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley M.K., Bain W.G., Jacobs J., Nambulli S., Parikh U., Cillo A., et al. Intractable coronavirus disease 2019 (COVID-19) and prolonged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication in a chimeric antigen receptor-modified T-cell therapy recipient: a case study. Clin Infect Dis. 2021;73(3):e815–e821. doi: 10.1093/cid/ciab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh Williamson M., Hamilton F., Hutchings S., Pymont H.M., Hackett M., Arnold D., et al. Chronic SARS-CoV-2 infection and viral evolution in a hypogammaglobulinaemic individual. MedRxiv. 2021 doi: 10.1101/2021.05.31.21257591. [DOI] [Google Scholar]
- Keitel V., Bode J.G., Feldt T., Walker A., Muller L., Kunstein A., et al. Case report: Convalescent plasma achieves SARS-CoV-2 viral clearance in a patient with persistently high viral replication over 8 weeks due to severe combined immunodeficiency (SCID) and graft failure. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.645989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatamzas E., Rehn A., Muenchhoff M., Hellmuth J., Gaitzsch E., Weiglein T., et al. Emergence of multiple SARS-CoV-2 mutations in an immunocompromised host. MedRxiv. 2021 10.1101.2021.01.10.20248871. [Google Scholar]
- Ko, K.K.K., Yingtaweesittikul, H., Tan, T.T., Wijaya, L., Cao, D.Y., Goh, S.S., et al., 2022. Emergence of SARS-CoV-2 spike mutations during prolonged infection in immunocompromised hosts [published online ahead of print, 11 May 2022]. Microbiol Spectr. DOI: 10.1128/spectrum.00791-22. [DOI] [PMC free article] [PubMed]
- Lee C.Y., Shah M.K., Hoyos D., Solovyov A., Douglas M., Taur Y., et al. Prolonged SARS-CoV-2 infection in patients with lymphoid malignancies. Cancer Discov. 2022;12(1):62–73. doi: 10.1158/2159-8290.CD-21-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.F., Chorlton S., Tyson J., Al-Rawahi G.N., Jassem A.N., Prystajecky N., et al. COVID-19 in an immunocompromised host: persistent shedding of viable SARS-CoV-2 and emergence of multiple mutations: a case report. Int. J. Infect. Dis. 2022;2022(114):178–182. doi: 10.1016/j.ijid.2021.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Macori G., Fanning S., O’Regan E., Hunt E., O’Callaghan D., et al. Genomic evolution of SARS-CoV-2 virus in immunocompromised patient, Ireland. Emerg Infect Dis. 2021;27(9):2499–2501. doi: 10.3201/eid2709.211159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. The hunt for the origins of Omicron. Nature. 2022;602:26–28. doi: 10.1038/d41586-022-00215-2. [DOI] [PubMed] [Google Scholar]
- Martinot M., Jary A., Fefi-Kremer S., Leducq V., Delagreverie H., Garnier M., et al. Emerging RNA-dependent RNA-polymerase mutation in a remdesivir-treated B-cell immunodeficient patient with protracted coronavirus disease 2019. Clin Infect Dis. 2021;73(7):e1762–e1765. doi: 10.1093/cid/ciaa1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monrad I., Sahlertz S.R., Nielsen S.S.F., Pedersen L.Ø., Petersen M.S., Kobel C.M., et al. Persistent severe acute respiratory syndrome coronavirus 2 infection in immunocompromised host displaying treatment induced viral evolution. Open Forum Infect Dis. 2021;8(7):ofab295. doi: 10.1093/ofid/ofab295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montejano R., Marcelo C., Falces-Romero I., del Valle L.G., De Soto T., Garcia-Rodríguez J., et al. Efficacy of sotrovimab for persistent coronavirus disease-2019 in a severely immunocompromised person living with HIV. AIDS. 2022;36(5):751–753. doi: 10.1097/QAD.0000000000003179. [DOI] [PubMed] [Google Scholar]
- Moran E., Cook T., Goodman A.L., Gupta R.K., Jolles S., Menon D.K., et al. Persistent SARS-CoV-2 infection: the urgent need for access to treatment and trials. Lancet Infect Dis. 2021;21(10):1345–1347. doi: 10.1016/S1473-3099(21)00464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A., Imbeaud S., Scemla A., Péré H., Fourgeaud J., Amrouche L., et al. Severe relapse of SARS-CoV-2 infection in a kidney transplant recipient with negative nasopharyngeal SARS-CoV-2 RT-PCR after rituximab [published online ahead of print, 1 Mar 2022] Am J Transplant. 2022 doi: 10.1111/ajt.17000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita M., Suzuki M., Matsunaga A., Ishizhima K., Yamamoto T., Kuroda Y., et al. Prolonged SARS-CoV-2 infection associated with long-term corticosteroid use in a patient with impaired B-cell immunity. J Infect Chemother. 2022;28(7):971–974. doi: 10.1016/j.jiac.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Pereira S., Calisto R., Sabio F., Guerreiro L. High-titre convalescent plasma therapy for an immunocompromised patient with systemic lupus erythematosus with protracted SARS-CoV-2 infection. BMJ Case Rep. 2021;14(8):e244853. doi: 10.1136/bcr-2021-244853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrak D., Tobudic S., Koblischke M., Graninger M., Radner H., Sieghart D., et al. SARS-CoV-2 vaccination in rituximab-treated patients: B cells promote humoral immune responses in the presence of T-cell-mediated immunity. Ann Rheum Dis. 2021;80:1345–1350. doi: 10.1136/annrheumdis-2021-220781. [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Ogai A., Furukawa K., Arai R., Anan R., Nakano Y., et al. Prolonged viral shedding of SARS-CoV-2 in an immunocompromised patient. J Infect Chemother. 2021;27(2):387–389. doi: 10.1016/j.jiac.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenblatt V., Order A.E., Das S., de Wit E., Youn J.H., Banakis S., et al. Yearlong COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B-cell depletion. J Infect Dis. 2022;225(7):1118–1123. doi: 10.1093/infdis/jiab622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Lago L., Aldámiz-Echevarria T., García-Martinez R., Pérez-Latore L., Herranz M., Sola-Campoy P.J. Different within-host viral evolution dynamics in severely immunocompromised cases with persistent SARS-CoV-2. Biomedicines. 2021;9(7):808. doi: 10.3390/biomedicines9070808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue J.M., McCreary E.K. Monoclonals for patients hospitalised with COVID-19. Lancet Respir Med. 2022;10(10):928–930. doi: 10.1016/S2213-2600(22)00222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purpura L.J., Chang M., Annavajhala M.K., Mohri H., Liu L., Shah J., et al. Prolonged severe acute respiratory syndrome coronavirus persistence, attenuated immunologic response, and viral evolution in a solid organ transplant patient. Am J Transplant. 2022;22(2):649–653. doi: 10.1111/ajt.16837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group Convalescent plasma in patients admitted to hospital with COVID-19 (RECOVERY): a randomised controlled, open-label, platform trial. Lancet. 2021;397(10289):2049–2059. doi: 10.1016/S0140-6736(21)00897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RECOVERY Collaborative Group (2022, 15 March). Randomised evaluation of COVID-19 therapy: Study Protocol. Available at: https://www.recoverytrial.net/files/recovery-protocol-v23-1-2022-03-15.pdf [Accessed: 30 May 2022].
- Reuken P.A., Stallmach A., Pletz M.W., Brandt C., Andreas N., Hahnfeld S., et al. Severe clinical relapse in an immunocompromised host with persistent SARS-CoV-2 infection. Leukemia. 2021;35(3):920–923. doi: 10.1038/s41375-021-01175-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddell A.C., Kele B., Harris K., Bible J., Murphy M., Dakshina S., et al. Generation of novel SARS-CoV-2 variants on B1.1.7 lineage in three patients with advanced HIV disease [published online ahead of print 25 May 2022] Clin Infect Dis. 2022 doi: 10.1093/cid/ciac409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.M., Jones R.B., Smith R.M., Alberici F., Kumaratne D.S., Burns S., et al. Rituximab-associated hypogammaglobulinaemia: incidence, predictors and outcomes in patients with multi-system autoimmune disease. J Autoimmunity. 2015;57:60–65. doi: 10.1016/j.jaut.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Roeker L.E., Knorr D.A., Pessin M.S., Ramanathan L.V., Thompson M.C., Leslie L.A., et al. Anti-SARS-CoV-2 antibody response in patients with chronic lymphocytic leukemia. Leukemia. 2020;34:3047–3049. doi: 10.1038/s41375-020-01030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüfenacht S., Gantenbein P., Boggian K., Flury D., Kern L., Dollenmaier G., et al. Remdesivir in coronavirus disease 2019 patients treated with anti-CD20 monoclonal antibodies: a case series. Infection. 2022;50(3):783–790. doi: 10.1007/s15010-022-01821-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenker C., Hirzel C., Walti L.N., Zeerleder S.S., Andres M., Ramette A., et al. Convalescent plasma and remdesivir for protracted COVID-19 in a patient with chronic lymphocytic leukaemia: a case report of late relapse after initial rapid response [published online ahead of print, Aug 30 2021] Br J Haematol. 2021 doi: 10.1111/bjh.17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer E.M., Babiker A., Adelman M.W., Allman B., Key A., Kleinhenz J.M., et al. SARS-CoV-2 evolution and immune escape with immunocompromised patients treated with exogenous antibodies. medRxiv. 2022 doi: 10.1101/2022.04.12.22273675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcri C., Dentone C., Mikulska M., Bruzzone B., Lai A., Fenoglio D., et al. The longest persistence of viable SARS-CoV-2 with recurrence of viraemia and relapsing symptomatic COVID-19 in an immunocompromised patient – a case study. Open Forum Infect Dis. 2021;8(11):ofab2017. doi: 10.1093/ofid/ofab217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Shoji K., Suzuki A., Okamoto M., Tsinda E.K., Sugawara N., Sasaki M., et al. Prolonged shedding of infectious viruses with haplotype switches of SARS-CoV-2 in an immunocompromised patient. J. Infect. Chemother. 2022;28(7):1001–1004. doi: 10.1016/j.jiac.2022.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner S.T., Prelog M., Sonnleitner S., Hinterbichler E., Halbfurter H., Kopecky D.B.C., et al. Cumulative SARS-CoV-2 mutations and corresponding changes in immunity in an immunocompromised patient indicate viral evolution with the host. Nat. Commun. 2022;13(1):2560. doi: 10.1038/s41467-022-30163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D.J., Gebo K.A., Shoham S., Bloch E.M., Lau B., Shenoy A.G., et al. Early outpatient treatment for COVID-19 with convalescent plasma. N Engl. J. Med. 2022;386(18):1700–1711. doi: 10.1056/NEJMoa2119657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha Y., Wardle H., Evans A.B., Hunter E.R., Marr H., Osborne W., et al. Persistent SARS-CoV-2 infection in patients with secondary antibody deficiency: successful clearance following combination casirivimab and imdevimab (REGN-COV2) monoclonal antibody therapy. Ann. Clin. Microbiol. Antimicrob. 2021;20:85. doi: 10.1186/s12941-021-00491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C.S., Huntley K., Berenger B.M., Bristow M., Evans D.H., Fonseca K., et al. Prolonged SARS-CoV-2 infection following rituximab treatment: clinical course and response to therapeutic interventions correlated with quantitative viral cultures and cycle threshold values. Antimicrob. Resist. Infect Control. 2022;11:28. doi: 10.1186/s13756-022-01067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong T.T., Ryutov A., Pandey U., Yee R., Goldberg L., Bhojwani D., et al. Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, Y., Asakura, S., Wada, S., Saito, T., Yano, T., 2022. Prolonged COVID-19 in an immunocompromised patient treated with Obinutuzumab and Bendamustine for follicular lymphoma [published online ahead of print, 31 May 2022]. Intern Med. DOI: 10.2169/internalmedicine.9136-21. [DOI] [PMC free article] [PubMed]
- Von-Lilienfeld-Toal M., Vehreschild J.J., Cornely O., et al. Frequently asked questions regarding SARS-CoV-2 in cancer patients-recommendations for clinicians caring for patients with malignant diseases. Leukemia. 2020;34:1487–1494. doi: 10.1038/s41375-020-0832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, S.A.J., Richter, A., Casey, A., Osman, H., Mirza, J.D., Stockton, J., et al., Recurrent SARS-CoV-2 mutations in immunodeficient patients. medRxiv 2022.03.02.22271697; DOI: https://doi.org/10.1101/2022.03.02.22271697. [DOI] [PMC free article] [PubMed]
- Wright, D.W., Harvey, W.T., Hughes, J., et al. Tracking SARS-CoV-2 mutations and variants through the COG-UK-Mutation Explorer. Virus Evol. 2022;8(1):veac023. Available at: https://sars2.cvr.gla.ac.uk/cog-uk/ [Accessed 3 October 2022]. [DOI] [PMC free article] [PubMed]
- Yasuda H., Mori Y., Chiba A., Bai J., Murayama G., Matsushita Y., et al. Resolution of one-year persisting COVID-19 pneumonia and development of immune thrombocytopenia in a follicular lymphoma patient with preceding rituximab maintenance therapy: a follow-up report and literature review of case with prolonged infections. Clin Lymphoma Myeloma Leuk. 2021;21(10):e810–e816. doi: 10.1016/j.clml.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhabokritsky, A., Mubareka, S., Kozak, R.A., Maguire, F., Yip, L., Yip, P., et al., 2022. Persistent infection with severe acute respiratory coronavirus virus 2 (SARS-CoV-2) in a patient with untreated human immunodeficiency virus (HIV) [published online ahead of print, 20 May 2022]. Infect. Control Hosp. Epidemiol. DOI: 10.1017/ice.2022.140. [DOI] [PMC free article] [PubMed]
- Zimmerli A., Monti M., Fenwick C., Eckerle I., Beigelman-Aubry C., Pellaton C., et al. Case report: stepwise anti-inflammatory and anti-SARS-CoV-2 effects following convalescent plasma therapy with full clinical recovery. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.613502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.