Abstract
The discovery of synthetic lethal interactions with genetic deficiencies in cancers has highlighted several candidate targets for drug development, with variable clinical success. Recent work has unveiled a promising synthetic lethal interaction between inactivation/inhibition of the WRN DNA helicase and tumours with microsatellite instability, a phenotype that arises from DNA mismatch repair deficiency. While these and further studies have highlighted the therapeutic potential of WRN inhibitors, compounds with properties suitable for clinical exploitation remain to be described. Furthermore, the complexities of MSI development and its relationship to cancer evolution pose challenges for clinical prospects. Here, we discuss possible paths of MSI tumour development, the viability of WRN inhibition as a strategy in different scenarios, and the necessary conditions to create a roadmap towards successful implementation of WRN inhibitors in the clinic.
Subject terms: Targeted therapies, Drug development, Cancer therapy, Biomarkers, Cancer therapeutic resistance
Synthetic lethality – accelerating precision oncology
Precision oncology aims to tailor cancer treatments to the specific biology of patients and their underlying tumours through genomic profiling, biomarker-mediated stratification, and choice of selected therapies1. Therefore, the potential for precision oncology hinges on improvement of profiling strategies, identification of therapeutically relevant biomarkers, and development of novel drugs to selectively target different cancer types2. Advances in next-generation sequencing technologies, such as deep-sequencing approaches and epigenomic profiling, are constantly improving our ability to genetically map tumour heterogeneity and enable better patient stratification3–5. However, the discovery of actionable candidates for targeted therapy and downstream drug development processes is time consuming, expensive, and often unsuccessful6,7. Consequently, identification of molecular signatures that can be precisely targeted by potent and specific drugs with a high likelihood of clinical success is paramount to make drug development a worthwhile investment8,9.
The ultimate goal of anti-cancer drug development is the discovery of chemicals that can eliminate cancer cells without harming the patient’s normal cells10. To this end, pharmaceutical companies and research laboratories alike are investing major resources in identifying actionable synthetic lethal interactions11. Synthetic lethality occurs when simultaneous mutations in two genes causes cell death, but a single mutation in either gene is viable12,13. Due to the nature of these interactions, inhibiting the products of genes that have a synthetic lethal relationship with prevalent genetic mutations in cancer cells should specifically kill cancer cells, while sparing normal cells14. Exploiting the selective vulnerabilities of cancer cells bearing specific mutations and/or pathway dysfunctions, through inhibition of their synthetic lethal partners has produced various levels of success in the clinic15–17. The most established and successful of these endeavours, thus far, is the development of Poly (ADP-ribose) polymerase (PARP) inhibitors to selectively target BRCA1/2 mutated cancers and other cancers with underlying defects in DNA repair by homologous recombination18–21. Such developments have fuelled focused studies and large-scale projects to systematically map cancer-specific dependencies22.
Mismatch repair deficiency leads to microsatellite instability
DNA mismatch repair (MMR) is a conserved mechanism that contributes to maintenance of genome stability by removing errors generated during DNA replication, long-tract DNA repair synthesis, and recombination23. Germline or somatic mutations and epigenetic alterations in the genes of MMR components lead to a hypermutator phenotype characterized by cancer predisposition and high genomic instability, particularly at repetitive regions of the genome known as microsatellites24. Microsatellites, or short-tandem repeats, are short (1–6 base pair) repetitive DNA sequences distributed along coding and non-coding regions that constitute approximately 3% of the human genome25. Due to their repetitive nature, microsatellites are prone to DNA polymerase slippage events, producing INDELs (insertions and deletions) that are mainly recognized and repaired by MMR26. Consequently, MMR deficiencies lead to a phenotype termed microsatellite instability (MSI), characterized by the accumulation of repeat-length alterations at microsatellite regions27. Detection of MSI is clinically relevant, since patients with MSI cancers have a better overall prognosis and reduced metastatic potential compared to patients with microsatellite stable (MSS) cancers28. This seems, in part, to reflect the high mutational burden of MSI tumours causing production of neoantigens that increase immunogenicity and sensitivity to immune checkpoint inhibitors29–32. Nevertheless, a significant proportion of MSI tumours do not respond, or evolve resistance, to immunotherapy and chemotherapy, highlighting the need for more and improved targeted and combinatorial treatments33–35. Key aspects in the relationship between MMR deficiency and cancer are outlined in Box 1 and current MSI detection methods are shown in Table 1.
Table 1.
MSI detection methods.
| Method | Accuracy | Property tested |
|---|---|---|
| Fluorescent multiplexed PCR and capillary electrophoresis66 | ~100% (standard) | Instability in 5 distinct microsatellites |
| Next-generation sequencing67 | ~92–94% | Instability in ~100 distinct microsatellites |
| Single-molecule molecular inversion probes68 | ~90–95% | Instability in 111 distinct microsatellites |
| Immunohistochemistry69 | ~95% (standard) | Presence of MMR proteins |
Box 1 DNA mismatch repair deficiency and cancer.
Lynch syndrome
Heterozygous germline alterations in certain DNA mismatch repair genes (MLH1, MSH2, MSH6, and PMS2) give rise to Lynch syndrome, also known as hereditary nonpolyposis colorectal cancer (HNPCC)70. Lynch syndrome is an autosomal dominant disorder that accounts for 3–5% of colorectal cancer cases and approximately 2.5% of endometrial cancer cases71,72. Diagnosing Lynch syndrome patients is clinically relevant since these patients have an 80% lifetime risk of developing colorectal cancer, and an increased risk of developing endometrial, ovarian, urinary tract, and gastric cancers among others73. It is estimated that Lynch syndrome affects 1/300 to 1/500 individuals in the general population, making it one of the most common genetic predispositions to cancer74.
Constitutional mismatch repair deficiency syndrome
Constitutional mismatch repair deficiency (CMMRD) arises from homozygous alterations in DNA mismatch repair genes and is characterized by a drastic predisposition to cancer75. In contrast to the relatively low prevalence of tumours in Lynch syndrome patients in the early stages of life, almost all individuals with CMMRD develop cancer in childhood and early adolescence, with a generally poor prognosis76,77. The most common cancers developed by CMMRD patients are colorectal, brain, and blood cancers, and CMMRD patients have a high likelihood of developing multiple cancers throughout their lives78.
Microsatellite instability in cancer
Microsatellite instability (MSI) is present in at least 27 different tumour types, with the prevalence of MSI ranging from ~31% in endometrial carcinoma to 0.25% in glioblastoma multiforme79. Moreover, MSI is detected in ~15% of colorectal cancers; approximately 3% of these are associated with Lynch syndrome and the other 12% are caused by somatic alterations in MMR, most often promoter hypermethylation of the MLH1 gene80,81. Importantly, MSI is present in most tumours associated with Lynch syndrome55,56. Cancers with MSI show aggressive histological features, but paradoxically favourable prognosis82.
WRN inactivation is synthetic lethal with MSI
In 2019, a set of high-impact publications independently unveiled a therapeutically promising synthetic lethal relationship between the RecQ-like family helicase protein, WRN, and MSI tumours36–39. WRN is a multifunctional enzyme with helicase and exonuclease activities and plays roles in various cellular processes crucial for the maintenance of genome stability, including DNA replication, transcription, DNA repair, and telomere maintenance40–43. Through CRISPR-Cas9 and RNAi screens, WRN was identified as the top hit for preferential dependency in MSI but not MSS cancer cell lines36,37,39. Further analysis revealed that WRN depletion causes cell cycle arrest, DNA damage, mitotic defects, chromosome shattering, and apoptosis specifically in MSI cells36–39. Moreover, WRN depletion reduced xenograft growth and tumour formation in mice transplanted with MSI cells39. Strikingly, acute depletion of various MMR components did not induce WRN dependency in MSS cells; conversely, genetic rescue experiments in MSI cells re-introducing the missing MMR component failed to rescue the synthetic lethal relationship36. These experiments suggested that the WRN synthetic lethal relationship develops via ensuing mutational consequences of MMR dysfunction rather than through MMR deficiency per se. Additionally, dissection of the various enzymatic activities of WRN using loss-of-function mutations within the helicase domain, exonuclease domain, or both, demonstrated that WRN dependency in MSI cells is linked only to its helicase function37.
Mechanistic insights into the WRN dependency of MSI tumours
Microsatellites can adopt non-B form DNA secondary structures in a sequence- and length-dependent manner44. Interestingly, one of the initial publications unveiling the synthetic lethality between WRN and MSI hypothesized that the potential mechanism driving this dependency was an increase in noncanonical secondary DNA structures that require WRN for their resolution38. Indeed, a seminal publication by van Wietmarschen et al. in Nature later demonstrated that TA-dinucleotide repeats are highly unstable in MMR deficient cells and undergo large-scale expansions in this setting, ultimately forming non-B form DNA secondary structures45. In Escherichia coli and yeast, expanded (TA)n repeats can form cruciform structures when their length exceeds roughly 20 repeat units46,47. Furthermore, long (TA)n tracts cause replication fork stalling and chromosome fragility at common fragile sites (CFSs)48,49. Importantly, WRN can resolve various non-B DNA substrates, including forks, flaps, bubbles, Holliday junctions, displacement loops (D-loops), and G-quadruplexes50,51. Accordingly, WRN depletion was found to induce replication fork collapse and DNA double-strand break formation precisely at expanded (TA)n repeats in MSI cells45. Mechanistically, expanded (TA)n microsatellites likely form cruciform structures that cause replication fork stalling, activating the apical ATR kinase, and causing the recruitment of WRN to resolve these structures via its helicase activity45. However, in the absence of WRN, expanded (TA)n repeats are unresolved and cleaved by the structure-specific endonucleases MUS81-EME1 and SLX4, leading to extensive chromosome shattering and ensuing cell death45. These findings suggest that WRN is uniquely able to resolve non-B form DNA secondary cruciform structures that form from (TA)n expansions in MSI cells and provide a mechanistic explanation for the synthetic lethal relationship between WRN and MSI.
Clinical potential of WRN inhibitors – lost in translation?
The synthetic lethal relationship between WRN and MSI has nurtured interest by academic groups and companies to develop WRN inhibitors to selectively target MSI cancers, with some drug development programmes already underway11,52. Importantly, WRN dependence appears to be conserved within heterogenous MSI tumour models, albeit only when MSI is derived from MLH1 or MSH2 deficiencies53. Of note, the small number of MSI models that can tolerate WRN loss appear to lack the MSI genomic (TA)n repeat expansion characteristics that invoke WRN dependence, supporting the likely clinical relevance of this relationship53. These findings suggest WRN dependency is influenced by the underlying MMR gene altered and the degree of MMR deficiency conferred. Collectively, mounting evidence supports the potential of future WRN inhibitors to selectively treat a subset of MSI tumours. Nevertheless, the data gathered so far might not be painting the full picture.
All reported work studying WRN dependence in MSI tumours, has been done in cellular models that fail to capture the full extent of the intratumour heterogeneity observed in patients. Understanding clonal diversity within MMR deficient cancers throughout their evolution is crucial to be able to accurately stratify patients, develop robust chemotherapeutic strategies, and attempt to avoid resistance to therapy54. A key issue to note is that, while MMR deficiency nurtures MSI, (TA)n repeat expansions, and tumourigenic mutations in tumour suppressors and oncogenes, these outcomes are largely stochastic and independent of one another. Indeed, MMR deficiency does not invariably, at the cellular level, lead to MSI or cancer development, as evidenced by experimental studies showing that acute MMR dysfunction does not cause MSI or WRN dependence, the subset of Lynch syndrome patients that do not develop cancer in their lifetimes, and the presence of MMR deficient cancers that are not MSI55–58. Consequently, the spatiotemporal development of MSI with (TA)n repeat expansions and cancer in MMR deficient clones in a tissue context is still poorly understood and is an elusive, but fundamental, issue to explore.
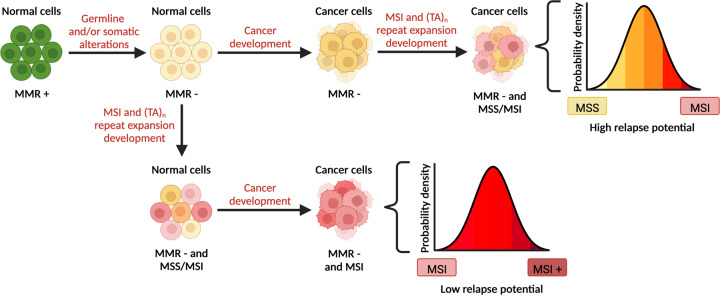
The likely impact of future WRN inhibitors will ultimately be in MSI cells that carry the molecular signature of (TA)n repeat expansions. Consequently, the prevalence of this molecular signature within MSI tumours and normal tissues is important to investigate and define. In this regard, it is critical to discuss the potential scenarios of MSI and cancer development in cells with somatically acquired or germline MMR deficiency (Fig. 1). Cells that have acquired MMR deficiency can gather tumourigenic mutations and become cancerous before developing MSI (Fig. 1, top branch), or develop MSI before becoming cancerous (Fig. 1, bottom branch). On one hand, if cancerous cells arise first followed by MSI development, the overall population of cells in the ensuing tumour would have various degrees of microsatellite instability ranging from MSS to MSI (Fig. 1, top branch). Since WRN inhibition would be effective only on the MSI cells that have developed (TA)n repeat expansions, a considerable proportion of the cells in such a tumour could be unaffected, likely causing failure to respond or relapse. Nevertheless, combinatorial therapies with immune checkpoint inhibitors or other chemotherapies might still enable favourable outcomes. On the other hand, if MSI development occurs first followed by cancer evolution from an MSI clone, the entire cancer cell population would be MSI and, thereby, WRN dependent (Fig. 1, bottom branch). Although WRN inhibition in this context might effectively eliminate the cancer cells, full cancer eradication would only ensue if the MSI cells had accumulated sufficient (TA)n repeat expansions to render them WRN dependent before becoming transformed. The extent to which such a scenario exists clinically, however, remains to be established. Crucially, even a small number of MSI cancer cells with low levels of (TA)n repeat expansions could lead to tumour relapse. In this regard, it would be interesting to investigate whether MSI cancer cells with (TA)n repeat expansions have the potential to evolve resistance to WRN inhibition, by altering the length and/or sequences of the (TA)n repeat expansions, by inactivating MUS81-EME1 and/or SLX4 which may alleviate the DSB formation and chromosomal shattering, or by up-regulating genome stability processes, perhaps involving promiscuous DNA helicases that might compensate for WRN inhibition in this context. Importantly, reversion mutations of the underlying MMR defect would not confer resistance to WRN inhibition since the MSI phenotype would already be established59.
Fig. 1. Temporal differences in MSI and cancer development might lead to contrasting clinical outcomes upon WRN inhibition.
Schematic outlining the possible paths towards developing MSI tumours with (TA)n repeat expansions that can be targeted with WRN inhibitors. Acquiring MMR deficiency, through germline and/or somatic alterations in MMR genes, is a necessary first step towards MSI and tumour development. MMR deficient cells can become cancerous before developing MSI and (TA)n repeat expansions (top branch) or develop MSI and (TA)n repeat expansions before becoming cancerous (bottom branch) potentially leading to different clinical outcomes. The therapeutic window of WRN inhibitors ultimately depends on MSI status and the degree of (TA)n repeat expansions. Microsatellite stable (MSS) and MSI cells with (TA)n repeat expansions are highlighted in yellow and red respectively. Figure was created with BioRender.com.
Paving the way – towards a roadmap for clinical success
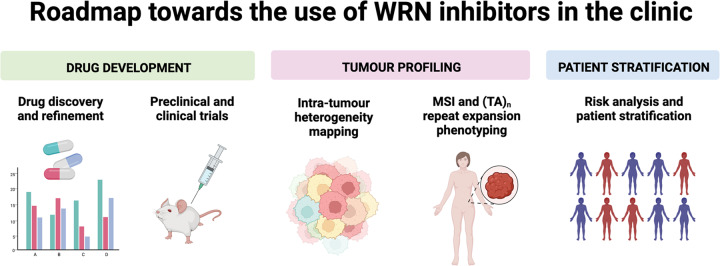
The scenarios of MSI tumour development outlined above pose real challenges in the clinic that could seriously limit the prospects for effective use of WRN inhibitors in treating a subset of MSI cancers. Furthermore, the initial hurdle of developing a suitable compound for clinical exploitation which is specific, potent, and bioavailable must not be understated60. Moreover, the lack of clarity and specificity when diagnosing a complex phenotype like MSI with (TA)n repeat expansions, gives rise to various issues that need to be addressed before fully grasping WRN dependence in this context. Mapping of intratumour heterogeneity and normal tissue heterogeneity regarding MSI and (TA)n repeat status, will invariably provide clarity and enable better patient stratification for the future use of WRN inhibitors. Recent advances in tumour mapping and identification using liquid biopsies, deep sequencing, and bioinformatic modelling of cancer evolution will be useful tools towards predicting patient outcomes61–65. In sum, the roadmap towards clinical applications of WRN inhibitors is paved by roadblocks in drug development, tumour profiling, and patient stratification. Surmounting these will be a highly sought and worthy goal for the researchers, drug developers, and clinicians, whose collective efforts will be necessary to successfully deliver WRN inhibitors to the patients who would derive the most benefit (Fig. 2).
Fig. 2. Roadmap towards the use of WRN inhibitors in the clinic.
Future clinical success of WRN inhibitors hinges on three pillars: drug development, tumour profiling, and patient stratification. Development and refinement of WRN inhibitors that are potent, selective, bioavailable, and relatively safe is an absolute requirement. Improvements in genetic and epigenetic tumour profiling through deep sequencing, liquid biopsies, and other technologies would provide a clearer picture of intra-tumour heterogeneity. Moreover, MSI testing will need to be coupled with (TA)n repeat expansion profiling to predict the potential of WRN inhibition. Finally, risk analysis through cancer modelling and biomarker identification will invariably improve patient stratification. Figure was created with BioRender.com.
Acknowledgements
Research in the S.P.J. lab is funded by the Cancer Research UK Discovery grant (DRCPGM\100005); Wellcome Investigator Award (206388/Z/17/Z), and ERC Synergy grant DDREAMM (855741). D.A.M.J. is funded by a CONACYT-Cambridge scholarship and receives additional support from a gift from La Fondation ARC (to S.P.J.). We thank Dr Jessica Brown for her critical input and Dr Kate Dry for her help editing the manuscript.
Author contributions
D.A.M.J. conceived of and wrote the first draft of the manuscript, then worked closely with S.P.J. to evolve and refine the manuscript and associated figures.
Data availability
Not applicable to this article as no datasets were generated or analyzed.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
David A. Morales-Juarez, Email: dam71@cam.ac.uk
Stephen P. Jackson, Email: spj13@cam.ac.uk
References
- 1.Mateo J, et al. Delivering precision oncology to patients with cancer. Nat. Med. 2022;28:658–665. doi: 10.1038/s41591-022-01717-2. [DOI] [PubMed] [Google Scholar]
- 2.Meric-Bernstam F, Mills GB. Overcoming implementation challenges of personalized cancer therapy. Nat. Rev. Clin. Oncol. 2012;9:542–548. doi: 10.1038/nrclinonc.2012.127. [DOI] [PubMed] [Google Scholar]
- 3.Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80. doi: 10.1186/s13073-015-0203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mardis, E. R. The Impact of Next-Generation Sequencing on Cancer Genomics: From Discovery to Clinic. Cold Spring Harb Perspect Med9, a036269 (2019). [DOI] [PMC free article] [PubMed]
- 5.Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 7.Carden CP, Banerji U, Kaye SB, Workman P, de Bono JS. From darkness to light with biomarkers in early clinical trials of cancer drugs. Clin. Pharm. Ther. 2009;85:131–133. doi: 10.1038/clpt.2008.223. [DOI] [PubMed] [Google Scholar]
- 8.Prasad V, Mailankody S. Research and Development Spending to Bring a Single Cancer Drug to Market and Revenues After Approval. JAMA Intern Med. 2017;177:1569–1575. doi: 10.1001/jamainternmed.2017.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMasi JA, Grabowski HG. Economics of new oncology drug development. J. Clin. Oncol. 2007;25:209–216. doi: 10.1200/JCO.2006.09.0803. [DOI] [PubMed] [Google Scholar]
- 10.Kaelin WG., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 11.Mullard A. What’s next for the synthetic lethality drug discovery engine? Nat. Rev. Drug Disco. 2022;21:477–479. doi: 10.1038/d41573-022-00107-0. [DOI] [PubMed] [Google Scholar]
- 12.Lucchesi JC. Synthetic lethality and semi-lethality among functionally related mutants of Drosophila melanfgaster. Genetics. 1968;59:37–44. doi: 10.1093/genetics/59.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bridges CB. The origin of variations in sexual and sex-limited characters. Am. Nat. 1922;56:51–63. [Google Scholar]
- 14.O’Neil NJ, Bailey ML, Hieter P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017;18:613–623. doi: 10.1038/nrg.2017.47. [DOI] [PubMed] [Google Scholar]
- 15.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hua H, et al. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019;12:71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguirre, A. J. & Hahn, W. C. Synthetic Lethal Vulnerabilities in KRAS-Mutant Cancers. Cold Spring Harb. Perspect Med.8, a031518 (2018). [DOI] [PMC free article] [PubMed]
- 18.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 19.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018;15:564–576. doi: 10.1038/s41571-018-0055-6. [DOI] [PubMed] [Google Scholar]
- 21.Ricks TK, et al. Successes and Challenges of PARP Inhibitors in Cancer Therapy. Front Oncol. 2015;5:222. doi: 10.3389/fonc.2015.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boehm JS, et al. Cancer research needs a better map. Nature. 2021;589:514–516. doi: 10.1038/d41586-021-00182-0. [DOI] [PubMed] [Google Scholar]
- 23.Pecina-Slaus N, Kafka A, Salamon I, Bukovac A. Mismatch Repair Pathway, Genome Stability and Cancer. Front Mol. Biosci. 2020;7:122. doi: 10.3389/fmolb.2020.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vernole P, et al. Common fragile sites in colon cancer cell lines: role of mismatch repair, RAD51 and poly(ADP-ribose) polymerase-1. Mutat. Res. 2011;712:40–48. doi: 10.1016/j.mrfmmm.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 27.Lothe RA. Microsatellite instability in human solid tumors. Mol. Med Today. 1997;3:61–68. doi: 10.1016/S1357-4310(96)10055-1. [DOI] [PubMed] [Google Scholar]
- 28.Kang S, et al. The significance of microsatellite instability in colorectal cancer after controlling for clinicopathological factors. Med. (Baltim.) 2018;97:e0019. doi: 10.1097/MD.0000000000010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang QX, et al. The degree of microsatellite instability predicts response to PD-1 blockade immunotherapy in mismatch repair-deficient/microsatellite instability-high colorectal cancers. Exp. Hematol. Oncol. 2021;10:2. doi: 10.1186/s40164-020-00193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shenoy, S. Mismatch repair mutations: Biomarker for immunotherapy in colorectal cancers. Indian J. Cancer, 10.4103/ijc.IJC_548_20 (2021). [DOI] [PubMed]
- 31.Lizardo DY, et al. Immunotherapy efficacy on mismatch repair-deficient colorectal cancer: From bench to bedside. Biochim Biophys. Acta Rev. Cancer. 2020;1874:188447. doi: 10.1016/j.bbcan.2020.188447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corcoran RB, Grothey A. Efficacy of Immunotherapy in Microsatellite-Stable or Mismatch Repair Proficient Colorectal Cancer-Fact or Fiction? JAMA Oncol. 2020;6:823–824. doi: 10.1001/jamaoncol.2020.0504. [DOI] [PubMed] [Google Scholar]
- 33.Le DT, et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J. Clin. Oncol. 2020;38:11–19. doi: 10.1200/JCO.19.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overman MJ, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuca, G. et al. Ascites and resistance to immune checkpoint inhibition in dMMR/MSI-H metastatic colorectal and gastric cancers. J. Immunother. Cancer10, 4001 (2022). [DOI] [PMC free article] [PubMed]
- 36.Behan FM, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 37.Lieb, S. et al. Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. Elife8, e43333 (2019). [DOI] [PMC free article] [PubMed]
- 38.Kategaya L, Perumal SK, Hager JH, Belmont LD. Werner Syndrome Helicase Is Required for the Survival of Cancer Cells with Microsatellite Instability. iScience. 2019;13:488–497. doi: 10.1016/j.isci.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan EM, et al. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature. 2019;568:551–556. doi: 10.1038/s41586-019-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh DK, Ahn B, Bohr VA. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology. 2009;10:235–252. doi: 10.1007/s10522-008-9205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 43.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst.) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol. Carcinog. 2009;48:379–388. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Wietmarschen N, et al. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature. 2020;586:292–298. doi: 10.1038/s41586-020-2769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dayn A, et al. Formation of (dA-dT)n cruciforms in Escherichia coli cells under different environmental conditions. J. Bacteriol. 1991;173:2658–2664. doi: 10.1128/jb.173.8.2658-2664.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowater R, Aboul-ela F, Lilley DM. Large-scale stable opening of supercoiled DNA in response to temperature and supercoiling in (A + T)-rich regions that promote low-salt cruciform extrusion. Biochemistry. 1991;30:11495–11506. doi: 10.1021/bi00113a003. [DOI] [PubMed] [Google Scholar]
- 48.Zlotorynski E, et al. Molecular basis for expression of common and rare fragile sites. Mol. Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaushal S, et al. Sequence and Nuclease Requirements for Breakage and Healing of a Structure-Forming (AT)n Sequence within Fragile Site FRA16D. Cell Rep. 2019;27:1151–1164.e5. doi: 10.1016/j.celrep.2019.03.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orren DK, Theodore S, Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- 51.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.GlaxoSmithKline. How GSK is using synthetic lethality to develop cancer therapies. Nat. Sponsor Feature. https://www.nature.com/articles/d42473-021-00591-9.
- 53.Picco G, et al. Werner Helicase Is a Synthetic-Lethal Vulnerability in Mismatch Repair-Deficient Colorectal Cancer Refractory to Targeted Therapies, Chemotherapy, and Immunotherapy. Cancer Disco. 2021;11:1923–1937. doi: 10.1158/2159-8290.CD-20-1508. [DOI] [PubMed] [Google Scholar]
- 54.Black JRM, McGranahan N. Genetic and non-genetic clonal diversity in cancer evolution. Nat. Rev. Cancer. 2021;21:379–392. doi: 10.1038/s41568-021-00336-2. [DOI] [PubMed] [Google Scholar]
- 55.van Lier MG, et al. A review on the molecular diagnostics of Lynch syndrome: a central role for the pathology laboratory. J. Cell Mol. Med. 2010;14:181–197. doi: 10.1111/j.1582-4934.2009.00977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N. Engl. J. Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 57.Moller P, et al. Cancer incidence and survival in Lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective Lynch syndrome database. Gut. 2017;66:464–472. doi: 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaffrelot M, et al. An unusual phenotype occurs in 15% of mismatch repair-deficient tumors and is associated with non-colorectal cancers and genetic syndromes. Mod. Pathol. 2022;35:427–437. doi: 10.1038/s41379-021-00918-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baxter, J. S., Zatreanu, D., Pettitt, S. J. & Lord, C. J. Resistance to DNA repair inhibitors in cancer. Mol. Oncol.10.1002/1878-0261.13224 (2022). [DOI] [PMC free article] [PubMed]
- 60.Datta A, Brosh RM., Jr. New Insights Into DNA Helicases as Druggable Targets for Cancer Therapy. Front Mol. Biosci. 2018;5:59. doi: 10.3389/fmolb.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben-David U, Beroukhim R, Golub TR. Genomic evolution of cancer models: perils and opportunities. Nat. Rev. Cancer. 2019;19:97–109. doi: 10.1038/s41568-018-0095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020;11:346. doi: 10.1038/s41419-020-2531-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida K, Sanada M, Ogawa S. Deep sequencing in cancer research. Jpn J. Clin. Oncol. 2013;43:110–115. doi: 10.1093/jjco/hys206. [DOI] [PubMed] [Google Scholar]
- 64.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum. Genomics. 2019;13:34. doi: 10.1186/s40246-019-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Metzcar J, Wang Y, Heiland R, Macklin P. A Review of Cell-Based Computational Modeling in Cancer Biology. JCO Clin. Cancer Inf. 2019;3:1–13. doi: 10.1200/CCI.18.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berg KD, et al. Detection of microsatellite instability by fluorescence multiplex polymerase chain reaction. J. Mol. Diagn. 2000;2:20–28. doi: 10.1016/S1525-1578(10)60611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hempelmann JA, et al. Microsatellite instability in prostate cancer by PCR or next-generation sequencing. J. Immunother. Cancer. 2018;6:29. doi: 10.1186/s40425-018-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waalkes A, et al. Accurate Pan-Cancer Molecular Diagnosis of Microsatellite Instability by Single-Molecule Molecular Inversion Probe Capture and High-Throughput Sequencing. Clin. Chem. 2018;64:950–958. doi: 10.1373/clinchem.2017.285981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheah PL, et al. Screening for microsatellite instability in colorectal carcinoma: Practical utility of immunohistochemistry and PCR with fragment analysis in a diagnostic histopathology setting. Malays. J. Pathol. 2019;41:91–100. [PubMed] [Google Scholar]
- 70.Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J. Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 71.de la Chapelle A. The incidence of Lynch syndrome. Fam. Cancer. 2005;4:233–237. doi: 10.1007/s10689-004-5811-3. [DOI] [PubMed] [Google Scholar]
- 72.Bhattacharya, P. & McHugh, T. W. in StatPearls (2022).
- 73.Stoffel EM, Mangu PB, Limburg PJ, American Society of Clinical, O.European Society for Medical, O. Hereditary colorectal cancer syndromes: American Society of Clinical Oncology clinical practice guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology clinical practice guidelines. J. Oncol. Pr. 2015;11:e437–e441. doi: 10.1200/JOP.2015.003665. [DOI] [PubMed] [Google Scholar]
- 74.Boland CR, Shike M. Report from the Jerusalem workshop on Lynch syndrome-hereditary nonpolyposis colorectal cancer. Gastroenterology. 2010;138:2197.e1–7. doi: 10.1053/j.gastro.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bakry D, et al. Genetic and clinical determinants of constitutional mismatch repair deficiency syndrome: report from the constitutional mismatch repair deficiency consortium. Eur. J. Cancer. 2014;50:987–996. doi: 10.1016/j.ejca.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 76.Barrow E, et al. Colorectal cancer in HNPCC: cumulative lifetime incidence, survival and tumour distribution. A report of 121 families with proven mutations. Clin. Genet. 2008;74:233–242. doi: 10.1111/j.1399-0004.2008.01035.x. [DOI] [PubMed] [Google Scholar]
- 77.Wimmer K, Kratz CP. Constitutional mismatch repair-deficiency syndrome. Haematologica. 2010;95:699–701. doi: 10.3324/haematol.2009.021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durno CA, et al. Phenotypic and genotypic characterisation of biallelic mismatch repair deficiency (BMMR-D) syndrome. Eur. J. Cancer. 2015;51:977–983. doi: 10.1016/j.ejca.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 79.Bonneville, R. et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol2017, 10.1200/PO.17.00073 (2017). [DOI] [PMC free article] [PubMed]
- 80.Hampel H, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer. N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 81.Ward R, et al. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48:821–829. doi: 10.1136/gut.48.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gryfe R, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable to this article as no datasets were generated or analyzed.