Abstract
Soil and rhizosphere microbiomes play important roles in suppression of plant pathogens through production of antagonistic secondary metabolites, yet mechanisms that determine the strength of pathogen control are not well understood. Many Pseudomonas species are associated with soil and rhizosphere microbiomes, and their ability to suppress pathogens is well documented. Here, we investigate how interactions within the Pseudomonas genus affect their production of antimicrobial metabolites. From a biosensor-based screen, we identify P. capeferrum species as capable of modulating secondary metabolite production in P. protegens. We show that P. capeferrum alters production of pyoluteorin and 2,4-diacetylphloroglucinol (DAPG) in P. protegens via two distinct and sequential mechanisms that depends on spatial proximity of the two species. Specifically, P. capeferrum secretes a diffusible signal that induce pyoluteorin production up to 100-fold in neighboring P. protegens colonies. In contrast, the interaction results in reduced DAPG production, but only within mixed-species colonies. Additionally, we found that increased pyoluteorin production and cell lysis of P. capeferrum is required for inhibition of DAPG production, suggesting that pyoluteorin-facilitated antibiosis of P. protegens on P. capeferrum leads to release of cell-associated metabolites and subsequent inhibition of DAPG production in P. protegens. As the interaction modulates in vitro bioactivity of the species, genus-specific interactions may assist in improving efficacy of biocontrol strains and consortia.
Subject terms: Microbial ecology, Bacteriology, Soil microbiology
Introduction
Soil- and rhizosphere-associated microbiomes play critical roles in plant development and health, for example by production of antimicrobial metabolites that suppress plant diseases by antagonizing growth of phytopathogens [1–3]. Understanding the processes that either potentiate or reduce this antagonism is essential for predicting the role and function of bacterial communities in controlling plant–pathogen interactions, and for enabling biological control applications in agriculture. Several studies have shown that antagonistic activity towards plant pathogens is related to bacterial diversity in ways that are not well understood [4–8]. For example, using defined consortia of Pseudomonas species it has been shown that increasing genotypic richness (i.e. increasing number of genotypically similar consortia members) correlate with increased disease suppressiveness and antagonistic activity towards phytopathogens [5, 7]. In contrast, another study has demonstrated that an increasing richness of distantly related Pseudomonas spp. is correlated with reduced pathogen inhibition [8]. These and other examples suggest that genus-specific interactions among Pseudomonas species can affect the strengths of their antagonistic activities and highlights the importance in understanding the underlying microbial interaction processes.
Pseudomonas spp. are a group of soil bacteria that play key roles in plant growth promotion and control of crop pathogens due to production of various antimicrobial metabolites [2, 4]. Among the fluorescent pseudomonads, Pseudomonas protegens has been studied intensively due to its potential as biocontrol agent and its array of antimicrobial secondary metabolites, including 2,4-diacetylphloroglucinol (DAPG), pyoluteorin, pyrrolnitrin, orfamide A and pyoverdine [9, 10]. This bacterium has been shown to exhibit antimicrobial activity against pathogenic bacteria, fungi and oomycetes [11–13]. At the mechanistic level, the biosynthesis and regulation of these metabolites has been well characterized in P. protegens, including that of DAPG and pyoluteorin. For the biosynthesis of DAPG, three molecules of malonyl-CoA are initially converted to phloroglucinol (PG) by the polyketide synthase, PhlD. PG is subsequently acetylated by the PhlACB enzyme complex once to mono-acetylphloroglucinol (MAPG) and twice to DAPG [14, 15]. The biosynthetic operon, phlACBDE, is under transcriptional control by the PhlF repressor. Additionally, Bottiglieri et al. found that a gene located upstream, phlG, encodes the PhlG enzyme, which degrades intracellular DAPG to MAPG [16]. The transcription of phlG is under control of the PhlH repressor [17]. The biosynthesis of pyoluteorin starts with L-proline, which is converted to a dichloro-pyrrole by a three-step enzymatic process by PltF, PltE and PltA. Pyoluteorin is formed by the addition of a resorcinol ring to the dichloro-pyrrole moiety catalyzed by the enzyme complex, PltBCG [18, 19]. Transcription of the biosynthetic operon, pltLABCDEFG, is activated by PltR. Recently, Yan et al. reported that the biosynthesis of DAPG and pyoluteorin is connected in P. protegens Pf-5, where the halogenase, PltM, converts the precursor, PG, from the DAPG pathway to PG-Cl and PG-Cl2 [10]. These molecules bind to the PltR activator, which in turn activates transcription of the pyoluteorin biosynthetic gene cluster.
The regulation of DAPG and pyoluteorin biosynthesis has been studied extensively in strains of P. protegens. It was demonstrated that these antimicrobial metabolites function in intra- and intercellular signaling circuits, as both molecules induced their own biosynthesis [20–22]. Auto-induction of DAPG was demonstrated in situ on wheat roots, as expression from the PphlA promoter was induced in two distinct Pseudomonas spp. by co-inoculation with a DAPG-producing strain [22]. Similarly, Brodhagen et al. reported that pyoluteorin functioned in positive autoregulation in P. protegens Pf-5 by inducing transcription of its respective biosynthetic gene cluster during growth on cucumber seedlings [21]. Recently, it was further reported that the intermediate PG in the biosynthesis of DAPG contributes to the regulation of production of both DAPG and pyoluteorin in a concentration-dependent manner [23, 24]. Additionally, production of both metabolites requires activation from the global regulatory system, Gac/Rsm [25]. Overall, previous work in the field has illuminated the complex regulatory intra- and intercellular mechanisms involved in fine-tuning the production of DAPG and pyoluteorin in strains of P. protegens. Additionally, Dubuis et al. demonstrated that interspecies interactions between distantly related Pseudomonas spp. stimulated antimicrobial activity of P. protegens CHA0 against Bacillus subtilis presumably via Gac/Rsm-induced production of DAPG [26]. However, the effects of microbial interactions and their underlying molecular mechanisms affecting the biosynthesis of DAPG and pyoluteorin has remained less characterized.
In this study we investigate the effects of interspecies interactions between the soil isolate, P. capeferrum F8, and a potential biocontrol agent, P. protegens DTU9.1, on the production of the two antimicrobial metabolites, DAPG and pyoluteorin. Our results show that cocultivation of the two bacteria in a biofilm on agar surfaces inhibits the production of DAPG, while significantly inducing the biosynthesis of pyoluteorin in P. protegens. We reveal that the two regulatory mechanisms occur from two independent signals – a cell-associated signal and an extracellular metabolite from P. capeferrum F8. Moreover, we demonstrate that the cocultivation of the two Pseudomonas species enhances the antimicrobial efficiency against two known bacterial phytopathogens. Lastly, we show that the antibiosis of P. protegens DTU9.1 on P. capeferrum F8 due to pyoluteorin production leads to cell lysis of F8 and release of an unknown metabolite inhibiting DAPG production. Overall, our study shows that bacterial secondary metabolites can be part of intricate interaction networks (even at dual species level) that may result in a multitude of changes including repression and induction of biosynthetic gene clusters. The dynamic, sequential interactions and resulting changes in chemical output (i.e. levels of secondary metabolites) from these interactions may help to further advance our understanding of e.g. microbial turnover in microbiomes and activities of biocontrol strains.
Results
P. protegens alters its secondary metabolism in coculture with a P. capeferrum soil isolate
In a previous screen for DAPG-producing Pseudomonas species from grassland soil samples, we identified a small fraction of isolates (9/864) unable to produce DAPG themselves, due to lack of the phlACBDE biosynthetic gene cluster (verified by PCR targeting phlD and chemical analysis). Yet the nine isolates were capable of eliciting a response in an engineered DAPG-responsive, PhlF-based whole-cell biosensor [27]. We hypothesized that these isolates had the potential to modulate DAPG production in Pseudomonas species known to produce DAPG (such as P. protegens) and to affect secondary metabolite production through unknown cell-cell interactions. Preliminary cocultivation experiments between the nine isolates and a DAPG producer, P. protegens DTU9.1, suggested that two isolates could inhibit DAPG production below the limit of detection. Based on their rpoD sequences, these two isolates were identified as P. capeferrum spp. and we selected one of them, P. capeferrum F8, for further studies of its ability to modulate secondary metabolite production in P. protegens DTU9.1.
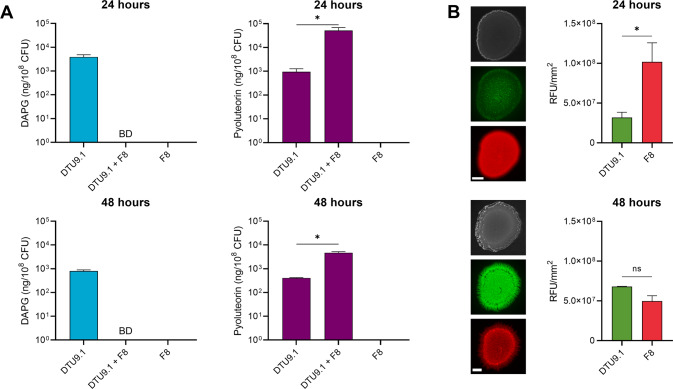
To investigate further, P. capeferrum F8 and P. protegens DTU9.1 were cocultivated in a mixed-species colony on a KBmalt agar surface. Quantitative analysis of DAPG and pyoluteorin was carried out using liquid chromatography-high resolution mass spectrometry (LC-HRMS) on agar plugs covering the entire microbial colony. The concentrations of DAPG and pyoluteorin were normalized to colony forming units (CFU) of the producing bacterium (Fig. 1A). Production of DAPG was inhibited below the limit of detection (LOD) in coculture with P. capeferrum F8 (Fig. 1A). Contrarily, the soil isolate significantly induced a 10–100 fold increase in pyoluteorin production in P. protegens DTU9.1 after 24 h (p = 4.8 × 10−2) and 48 h (p = 7.8 × 10−3). To examine the coculture dynamics, P. capeferrum F8 and P. protegens DTU9.1 were tagged with the Tn7 transposon from pBG42-mKate and pBG42-gfp, respectively. After 24 h of growth, P. capeferrum F8 constituted most of the bacterial colony (p = 1.1 × 10−2). On the second day of incubation, the ratio of the two bacteria had shifted towards an even distribution, as P. protegens DTU9.1 increased in abundance, while the abundance of P. capeferrum F8 decreased. However, despite this shift in coculture dynamics, DAPG production remained below the detection limit (Fig. 1A).
Fig. 1. P. protegens alters its secondary metabolism in coculture with P. capeferrum F8 on solid surface.
A Concentration of DAPG and pyoluteorin produced by P. protegens DTU9.1 after 24 and 48 h of growth on KBmalt agar. BD = Below detection (Student’s t-test: *p < 0.05). B Coculture dynamics of P. capeferrum F8 (red – mKate) and P. protegens DTU9.1 (green – gfp) on KBmalt. Top: Bright field image. Middle: gfp channel. Bottom mKate channel. Scale bar corresponds to 2 mm. Pictures are representative of three biological replicates with similar results. Fluorescence images were obtained with 500 ms exposure time. Relative fluorescence per colony for both channels was estimated with ImageJ.
P. capeferrum F8 affects metabolite production in P. protegens with two independent signals
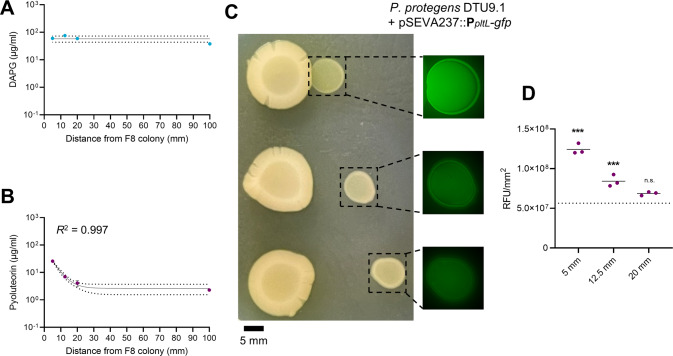
Based on the coculture interaction results above, we attempted to attribute the altered secondary metabolism in P. protegens DTU9.1 to secretion of a metabolite or signal from P. capeferrum using a distance assay on KBmalt agar. First, P. capeferrum F8 was grown for 48 h in bacterial colonies, followed by inoculating P. protegens DTU9.1 at a distance of 5, 12.5 and 20 mm from the edge of the F8 colony. Agar plugs covering the colony of P. protegens DTU9.1 were extracted after an additional 24 h of incubation. The concentrations of DAPG produced by P. protegens DTU9.1 was unaffected by diffusible signals from P. capeferrum F8, whereas pyoluteorin production was induced in a distance-dependent manner (Fig. 2A). P. protegens DTU9.1 was grown under axenic conditions as a control. To fit an overall non-linear regression model to the data, the control was assigned a 100 mm distance equivalent of the diameter of an entire petri dish. The induction of pyoluteorin as a function of distance is well described by a one phase decay function (R2 = 0.997) having a decay rate of −0.216 (p = 8.1 × 10−5). Closer inspection of the individual data points revealed that pyoluteorin production in the two closest colonies (5 and 12.5 mm) was induced significantly compared to the control as evidenced by ANOVA followed by Dunnett’s test (p = 2.1 × 10−5 and p = 3.2 × 10−3, respectively), whilst the 20 mm colony was not significantly induced.
Fig. 2. P. capeferrum F8 induces pyoluteorin biosynthesis in a distance-dependent manner.
Concentration of DAPG (A) and pyoluteorin (B) was measured in two independent experiments measured with HR-LCMS. The control represents P. protegens DTU9.1 grown in isolation on KBmalt agar (set to 100 mm distance to fit the model). Dotted lines represent the 95% confidence intervals of the mathematical model. C Distance assay with P. protegens DTU9.1 harboring pSEVA237::PpltL-gfp indicating that increased pyoluteorin production was due to induced transcription of the pyoluteorin biosynthetic gene cluster in a distance-dependent manner. Pictures are representative of three biological replicates with similar results. Fluorescence images were obtained with 500 ms exposure time. D Relative fluorescence per colony was estimated with ImageJ. The dotted line represents the average fluorescence measured in the control cultivated axenically (Dunnett’s test compared to control: ***p < 0.001).
The secreted signal from P. capeferrum F8 was shown to induce the transcription of the pyoluteorin biosynthetic gene cluster from the PpltL promoter, and this effect decreases with increasing distance between the bacterial colonies (Fig. 2C). Additionally, the relative fluorescence of P. protegens DTU9.1 harboring pSEVA::PpltL-gfp was measured with ImageJ after 24 h of cultivation (Fig. 2D). This showed that promoter activity was significantly induced at the 5 and 12.5 mm spots (p = 4.2 × 10−9 and p = 4.4 × 10−4, respectively), while the transcriptional activity was not significantly different from the control at the 20 mm spot. Overall, these findings suggests that DAPG inhibition and induction of pyoluteorin biosynthesis are affected by two independent molecular mechanisms. The signal inducing pyoluteorin biosynthesis is secreted by P. capeferrum F8, whereas direct cell-to-cell interaction may be required for the inhibition of DAPG as achieved in a mixed species colony.
Distance-dependent induction of pyoluteorin biosynthesis is observed among closely and distantly related bacteria
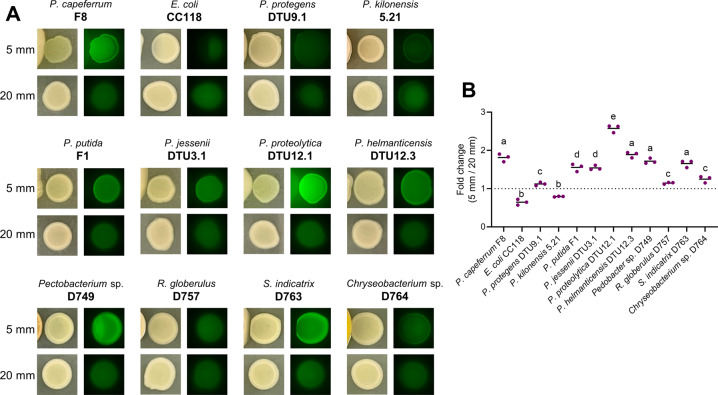
To explore the breadth of distance-dependent signaling from related Pseudomonas species, as well as unrelated bacteria, we tested six additional isolates of Pseudomonas and five phylogenetically distant bacteria for their ability to induce pyoluteorin biosynthesis in P. protegens DTU9.1. For the six Pseudomonas strains, P. putida F1 was included as a close relative to P. capeferrum F8, as part of the P. putida group. Moreover, we included five strains (P. helmanticensis DTU12.3, P. jessenii DTU3.1, P. kilonensis 5.21, P. protegens DTU9.1 and P. proteolytica DTU12.1) representing diverse soil Pseudomonas species of the P. fluorescens group belonging to five separate subgroups (P. koreensis, P. jessenii, P. corrugata, P. protegens and P. gessardii). The phylogenetic association between the Pseudomonas isolates in relation to known type strains [28] was evaluated based on part of the rpoD house-keeping gene (Figure S1). Four of the six tested Pseudomonas isolates induced a significant activation of the PpltL promoter in a distance-dependent manner (Fig. 3). However, two of the isolates (P. kilonensis 5.21 and P. protegens DTU9.1) did not induce promoter activity in P. protegens DTU9.1 harboring pSEVA237::PpltL-gfp. Additionally, we investigated if species outside the Pseudomonas genus could induce pyoluteorin biosynthesis in a distance-dependent manner. To this end, we tested a rapidly growing E. coli strain, as well as four commonly isolated soil bacteria, including Pectobacterium sp. D749, Rhodococcus globerulus D757, Stenotrophomonas indicatrix D763 and Chryseobacterium sp. D764 [29]. Both Pedobacter sp. D749 and S. indicatrix D763 secreted a signal capable of inducing pyoluteorin biosynthesis in P. protegens DTU9.1. These findings could indicate that P. protegens responds to a more general signal or cue from closely, as well as distantly related bacteria, by upregulating the production of pyoluteorin.
Fig. 3. Distantly related species induce pyoluteorin biosynthesis in P. protegens DTU9.1.
A Distance assay with P. protegens DTU9.1 harboring pSEVA237::PpltL-gfp. Pictures are representative of three biological replicates with similar results. Fluorescence images were obtained with 500 ms exposure time. B Fold change in relative fluorescence of P. protegens DTU9.1 harboring pSEVA237::PpltL-gfp cultivated 5 and 20 mm from the interacting bacteria. Relative fluorescence per colony was estimated with ImageJ from three biological replicates. The dotted line represents the control of P. protegens DTU9.1 cultivated alone. Letters above columns indicate treatments significantly different from one another, as determined by ANOVA and subsequent Tukey post-hoc analysis (p < 0.05).
DAPG and pyoluteorin biosynthesis are affected by two independent signals from P. capeferrum F8
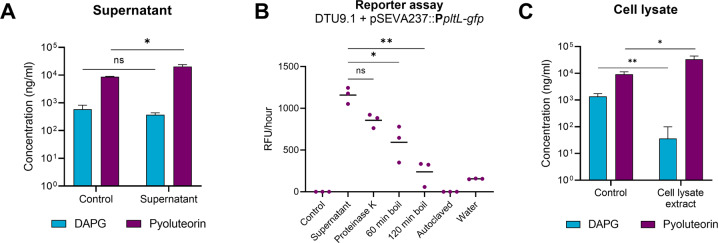
The results described above suggested the presence of two independent signals; a secreted signal inducing pyoluteorin biosynthesis and a signal requiring cell-to-cell contact between the two bacteria resulting in inhibition of DAPG production in P. protegens DTU9.1. Thus, we hypothesized that a supernatant of an outgrown P. capeferrum F8 culture would have the ability to induce pyoluteorin production in P. protegens DTU9.1, whereas molecules extracted from a lysate of the corresponding F8 cell-pellet would inhibit DAPG production. Exposing P. protegens DTU9.1 to 50% v/v fresh KBmalt broth and supernatant of a 48 h culture of P. capeferrum F8 significantly induced pyoluteorin production, while DAPG biosynthesis was unaffected (Fig. 4A). Moreover, the inducing molecule appeared to be heat sensitive and completely lost its activity following 15 min of autoclaving at 121 °C (Fig. 4B). However, treatment with proteinase K (30 U/ml) did not significantly reduce the activity of the supernatant, suggesting that the inducing molecule is not of proteinaceous nature. In contrast, exposing P. protegens DTU9.1 to 50% v/v fresh KBmalt broth and sterile water did not induce transcription from the PpltL promoter, which clearly indicated that the supernatant from P. capeferrum F8 contained a signal capable of inducing pyoluteorin production in P. protegens.
Fig. 4. DAPG and pyoluteorin biosynthesis are affected by two independent signals from P. capeferrum F8.
A Concentration of DAPG and pyoluteorin produced by P. protegens DTU9.1 cultured in KBmalt (control) or 50% v/v sterile-filtered supernatant from a 48 h culture of P. capeferrum F8. B Supernatant from a 48 h culture of P. capeferrum F8 was treated with proteinase K, boiled for 60 or 120 min at 100 °C or autoclaved for 15 min at 121 °C. P. protegens DTU9.1 harboring pSEVA237::PpltL-gfp was exposed to the treated supernatants, sterile water or fresh KBmalt media as control. Promoter activity was measured as the increase in relative fluorescence per h during stationary phase. C Concentration of DAPG and pyoluteorin produced by P. protegens DTU9.1 cultured in KBmalt supplemented with methanol as control or crude ethyl acetate extracted cell lysate of P. capeferrum F8. Significance levels were calculated with the Student’s t-test (*p < 0.05, **p < 0.005).
In order to test if cell-associated molecules could affect the secondary metabolism of P. protegens DTU9.1, we exposed P. protegens to molecules extracted from the lysate of a P. capeferrum F8 cell-pellet. To this end, P. protegens DTU9.1 was cultivated in three independent biological replicates in KBmalt broth supplemented with methanol as control or crude lysate extract. The supernatants were collected after 24 h and analyzed for the presence of DAPG and pyoluteorin with LC-HRMS (Fig. 4C). The crude cell lysate extract significantly inhibited DAPG production 38-fold (p = 3.3 × 10−3), where the DAPG concentration in two out of the three replicates was below the LOD. The production of pyoluteorin was induced 4-fold (p = 2.2 × 10−2). This suggests that cell lysis of P. capeferrum F8 leads to release of cell-associated metabolites inhibiting the production of DAPG. The minor induction of pyoluteorin biosynthesis is most likely due to small amounts of extracellular signaling molecules associated with P. capeferrum F8 cells during washing pre-lysis or that the inducing molecule is present intracellularly in low concentrations.
The antibiosis of P. Protegens on P. capeferrum F8 leads to inhibition of DAPG production
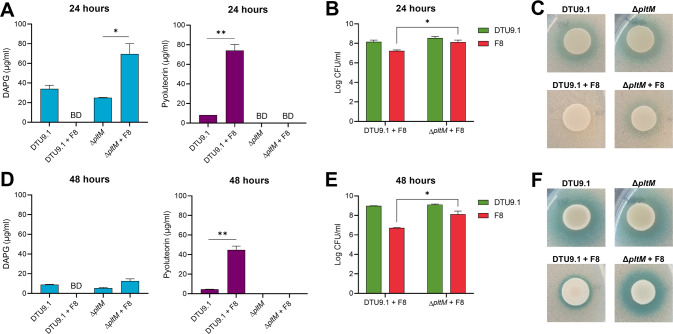
Based on these results, we hypothesized that antibiosis of P. protegens DTU9.1 on P. capeferrum F8 in mixed-species cocultures could facilitate the release of cell-associated metabolites from P. capeferrum leading to inhibition of DAPG production. We further hypothesized that the antimicrobial metabolite, pyoluteorin, was responsible for the antibiosis. Initially, we evaluated the minimal inhibitory concentration (MIC) of DAPG and pyoluteorin towards P. capeferrum F8 (Fig. S2). This revealed that pyoluteorin was toxic towards P. capeferrum F8, inhibiting growth at 64 µg/ml, whereas the MIC of DAPG exceeded the tested concentrations (> 128 µg/ml). To test the hypothesis that pyoluteorin caused antibiosis of P. capeferrum F8, we cultivated P. protegens DTU9.1 WT and a ΔpltM knock-out mutant in mono- and coculture colonies with P. capeferrum F8 on a KBmalt agar surface. The ΔpltM deletion mutant of P. protegens DTU9.1 was incapable of converting phloroglucinol (PG) to PG-Cl and PG-Cl2 [10], which in turn rendered the bacterium unable to initiate transcription of the pyoluteorin biosynthetic gene cluster. Cocultivation of P. capeferrum F8 and the ΔpltM deletion mutant of P. protegens DTU9.1 restored production of DAPG to levels comparable to or higher than monocultures of P. protegens DTU9.1 WT and ΔpltM mutant (Fig. 5A, D), demonstrating that pyoluteorin production is required for the observed suppression of DAPG production.
Fig. 5. Deletion of pltM restores DAPG production in coculture between P. protegens DTU9.1 and P. capeferrum F8.
A, D Concentration of DAPG and pyoluteorin produced by P. protegens DTU9.1 and the ΔpltM mutant in mono- and coculture with P. capeferrum F8 after 24 h (A) and 48 h (D). BD = Below detection limit B, E Abundance of P. protegens DTU9.1 or ΔpltM mutant and P. capeferrum F8 in mixed colonies on KBmalt agar after 24 h (B) and 48 h (E). Values represent means of two independent experiments. C, F P. protegens DTU9.1 and ΔpltM mutant in mono- and coculture with P. capeferrum F8 in bacterial colonies on top of a lawn of the biosensor harboring pSEVA225::DAPGlacZ on KBmalt + Xgal65 agar after 24 h (C) and 48 h (F). Significance levels were calculated with the Student’s t-test (*p < 0.05, **p < 0.005).
Cell counts of the bacteria residing in the mixed colonies further revealed that the absence of pyoluteorin in the ΔpltM mutant improved the ability of the two Pseudomonas species to co-exist (Fig. 5B, E). The abundance of P. capeferrum F8 was significantly higher in coculture with the ΔpltM mutant compared to wildtype P. protegens DTU9.1 after 24 h (p = 2.8 × 10−2) and 48 h (p = 2.5 × 10−2). Furthermore, the concentration of pyoluteorin in the coculture between the two bacteria reached levels comparable to the MIC (Fig. 5A), which supports the model that pyoluteorin causes antibiosis of P. capeferrum F8. This data indicates that the cell-associated, DAPG-inhibiting signal from P. capeferrum F8 is located intracellularly requiring cell lysis for its release. Additionally, the absence of pyoluteorin production in the ΔpltM mutant did not affect the ability of P. protegens DTU9.1 to proliferate in the mixed species coculture (Fig. 5B, E). Lastly, restoration of DAPG production in the cocultivation of P. capeferrum F8 and the ΔpltM deletion mutant of P. protegens DTU9.1 was visualized utilizing the whole-cell DAPG biosensor harboring pSEVA225::DAPGlacZ (Fig. 5C, F). Monocultures of P. protegens DTU9.1 WT and ΔpltM mutant yielded a strong response as evidenced by a clear blue halo surrounding the colony of Pseudomonas. Cocultivation of P. capeferrum F8 and the ΔpltM deletion mutant generated a similar response after 48 h of incubation but only a slight induction was observed after 24 h. This suggests that the biosensor response had an unexpected delay, since the LC-HRMS data showed restoration of DAPG production after 24 h (Fig. 5A). Taken together, these findings suggest that pyoluteorin facilitates lysis of P. capeferrum F8 leading to the release of one or more metabolites capable of inhibiting DAPG production in P. protegens DTU9.1.
The antimicrobial activity of P. protegens DTU9.1 against bacterial phytopathogens is increased in coculture with P. capeferrum F8
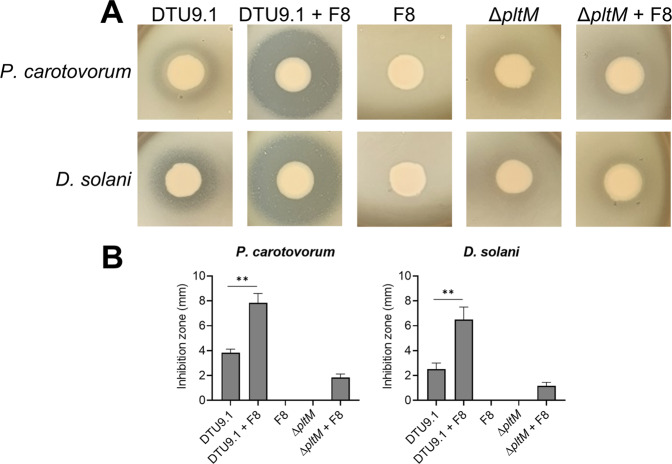
Previous research showed that using a diverse consortium of Pseudomonas species as biocontrol inoculum can increase disease suppressiveness [7]. Given the changes to secondary metabolite production in cocultures between P. protegens DTU9.1 and P. capeferrum F8, it could suggest that such cocultures would exhibit altered suppression of growth of DAPG and pyoluteorin sensitive microorganisms compared to monoculture. To test this, Pseudomonas species were cultivated in mono- and coculture colonies on KBmalt agar for 48 h to allow for metabolite production prior to chloroform-mediated cell inactivation to avoid undesirable swarming. Subsequently, the plates were covered with a soft agar layer inoculated with two known phytopathogenic bacteria, Pectobacterium carotovorum and Dickeya solani. Inhibition was evaluated based on the size and turbidity of the clearing zone arising after an additional 24 h of incubation (Fig. 6A). Cocultivation leads to a significant increase in antimicrobial effectiveness against both plant pathogens compared to both monocultures of Pseudomonas (p = 1.1 × 10−3 and p = 3.4 × 10−3 for P. carotovorum and D. solani, respectively) (Fig. 6B). To examine if the induced production of pyoluteorin was the cause of increased bioactivity, the pltM gene was knocked out in P. protegens DTU9.1 resulting in absence of pyoluteorin biosynthesis [10]. Bioactivity against both plant pathogens was lost in the ΔpltM mutant in monoculture and greatly reduced in coculture with P. capeferrum F8 compared to wildtype P. protegens DTU9.1 (Fig. 6).
Fig. 6. The antimicrobial effectiveness of P. protegens DTU9.1 against bacterial phytopathogens increases in coculture with P. capeferrum F8.
A The antimicrobial activity of P. protegens DTU9.1 towards two bacterial phytopathogens, P. carotovorum and D. solani, was enhanced in coculture with P. capeferrum F8. The ΔpltM mutant of P. protegens DTU9.1 was unable to produce pyoluteorin. Pictures are representative of three biological replicates with similar results. B Zones of inhibition were measured for three biological replicates (Student’s t-test: **p < 0.005).
Discussion
The biosynthesis of secondary metabolites has been extensively studied in strains of P. protegens, which has led to the characterization of complex intra- and intercellular regulatory mechanisms governing the fine-tuned production of specific metabolites [20–22, 24]. However, bacteria most often reside in microbial communities in which communication mediated through chemical signaling adds another layer of regulatory complexity. Additionally, previous studies have demonstrated conflicting results correlating Pseudomonas spp. richness and the effects on ecologically relevant mechanisms mediated by secondary metabolites (e.g. antimicrobial activity and plant protection) [5–8]. This supports the potential for uncharacterized, genus-specific interactions among Pseudomonas spp. affecting their secondary metabolism. Here we report on the effects of interspecies interactions between two fluorescent Pseudomonas on the production of DAPG and pyoluteorin in P. protegens DTU9.1. Our work sheds light on interspecies interactions between soil Pseudomonas that affect the biosynthesis of antimicrobial secondary metabolites. This further emphasizes the necessity of studying microbial interactions to broaden our current understanding of the natural role and biosynthesis of secondary metabolites, while bridging the anticipated in vitro and actual in situ efficacy of biocontrol agents.
In this study, the cocultivation of P. capeferrum F8 and a potential biocontrol agent, P. protegens DTU9.1, led to induced expression of the pyoluteorin biosynthetic gene cluster, while production of DAPG was inhibited (below LOD). We propose the sequential signaling model (Fig. 7) in which P. protegens initially responds to an extracellular, diffusible signal from P. capeferrum by inducing the biosynthesis of the antibacterial metabolite, pyoluteorin. The increased local concentration of pyoluteorin exceeds tolerable levels for P. capeferrum, leading to cell disruption. As a result of lysis within the P. capeferrum population, an intracellular signal (from P. capeferrum) is released, which subsequently inhibits production of DAPG in P. protegens DTU9.1. We speculate that the inhibition of DAPG production could be related to a yet uncharacterized ecological function, as the absence of DAPG has no apparent benefit to either organism in this dual species interaction. Previously, it was reported that P. aeruginosa releases an intracellular signal upon cell lysis, which in turn induces the biosynthesis of antibacterial metabolites in neighboring kin cells [30]. In the present study, however, we describe a dynamic, sequential interaction between two unrelated species of Pseudomonas affecting the biosynthesis of multiple secondary metabolites.
Fig. 7. Schematic overview of the altered secondary metabolism in P. protegens DTU9.1 in response to cocultivation with P. capeferrum F8.
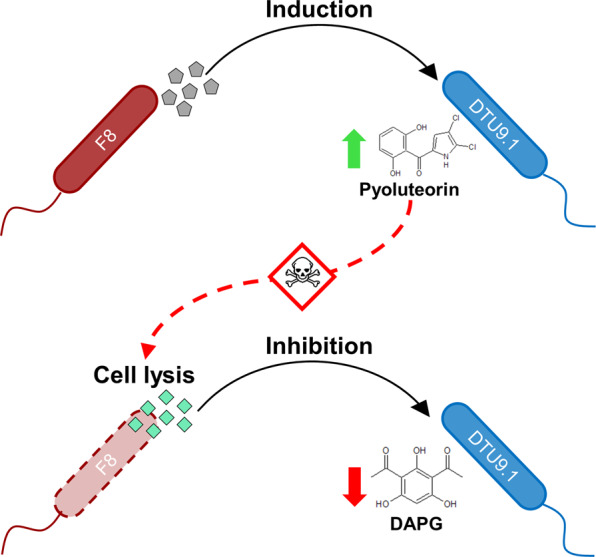
P. capeferrum F8 secretes a metabolite (grey pentagons), which induces biosynthesis of pyoluteorin in P. protegens DTU9.1. The increased concentration of pyoluteorin is toxic towards P. capeferrum F8 leading to cell lysis and release of an intracellular metabolite (cyan diamonds), which in turn inhibits production of DAPG in P. protegens.
Bacterial communication mediated via chemical signaling is a concept that has been extensively studied. For instance, previous research has shown that dual-species interactions between culturable soil bacteria commonly result in increased antimicrobial activity, due to induced antibiotic production, compared to monocultures [31, 32]. An explanation for the distance-dependent induction of pyoluteorin observed in this study could be related to an interaction arising from bacterial competition sensing [33–35]. Such interactions often involve secretion of toxic metabolites proposed to have hormetic properties (e.g. stimulatory at subinhibitory concentrations and antagonistic at intolerable concentrations) [36]. At subinhibitory concentrations the toxic metabolites may act as cues of nearby microbial danger, inducing responses in the sensing bacterium, such as modifications to its own secondary metabolome [35–38]. As demonstrated recently, competitive interactions are commonly observed in interspecies interactions among closely related species, likely owing to similar nutrient preferences [39]. Dubuis et al. showed a Gac/Rsm-mediated induction of antimicrobial activity in P. protegens CHA0 resulting from interspecies interactions [26]. In this study we have shown that P. protegens DTU9.1 induces biosynthesis of pyoluteorin in response to a secreted signal from P. capeferrum F8 (Fig. 2), as well as diverse species of Pseudomonas and at least two other species of commonly isolated soil bacteria (Fig. 3). On the other hand, lack of induction was observed for several species, including other Pseudomonas strains and distantly related bacteria. Moreover, we could demonstrate that exposing P. protegens DTU9.1 to the supernatant of a P. capeferrum F8 culture induced biosynthesis of pyoluteorin (Fig. 4A). This supports the hypothesis that P. protegens DTU9.1 responds to a secreted signal rather than alterations in media composition caused by the interacting species. These findings suggest that P. protegens DTU9.1 responds to a more general bacterial cue, indicating the presence of a competing species. We could further show that the signal appeared heat sensitive, as boiling the supernatant significantly reduced the ability to induce transcription from the PpltL promoter, while autoclaving completely removed the ability to induce (Fig. 4B). Contrarily, treatment of the supernatant with proteinase K did not significantly reduce the ability to induce pyoluteorin biosynthesis, indicating that the secreted signal from P. capeferrum F8 is not of proteinaceous nature. Treatment of the supernatant with other enzymes, including α-amylase and lipase, could further aid in deducing the chemical nature of the secreted signal as recently demonstrated by Yang et al. [40]. In addition to enzymatic treatment, we also sequenced and de novo assembled the genome of P. capeferrum F8 (see Methods) to identify candidate genes required for biosynthesis of potential secreted signals. This analysis revealed the presence of the ppuI-rsaL-ppuR quorum sensing (QS) system [41] and an unknown polyketide synthase. Previous research has shown that QS molecules display varying levels of promiscuity, as they interact with both intra- and interspecies receptors [42]. As described above, secreted secondary metabolites with antimicrobial properties, including polyketide-derived metabolites, have been associated with danger signals sensed by neighboring microorganisms [33, 34, 36]. However, knock-out mutants either with a deficient QS system or lacking the polyketide synthase did not affect the ability of P. capeferrum F8 to induce pyoluteorin biosynthesis in P. protegens DTU9.1. Thus, further investigations are necessary to identify the secreted signal from P. capeferrum F8 capable of inducing pyoluteorin production in P. protegens.
The biosynthesis of DAPG in Pseudomonas is under tight intra- and intercellular regulation and studies have shown that metabolites from interacting microbes can affect its production [20, 43]. Schnider-Keel et al. showed that salicylate, a molecule with structural similarities to DAPG, could also inhibit biosynthesis of DAPG [20]. Interestingly, Li et al. found that P. putida RW10S1 produced promysalin, which is a salicylate-containing antibiotic selectively targeting other Pseudomonas species [44]. Since species of P. putida are closely related to P. capeferrum [28], we speculated if our soil isolate, P. capeferrum F8, possessed the genes required for salicylate and promysalin production. However, a tBLASTn analysis revealed the absence of this biosynthetic gene cluster. The signal inhibiting DAPG production in P. protegens DTU9.1 could be either intracellular or membrane-associated in P. capeferrum F8 (Fig. 4C). However, the abundance of the signaling bacterium (P. capeferrum F8) was stably maintained at approximately 108 CFU/ml in the coculture with the ΔpltM mutant compared to coculture with WT P. protegens DTU9.1, where the abundance of F8 declined 10-30 fold over the course of 48 h (Fig. 5B, E). This could indicate that the inhibiting signal is located intracellularly, requiring cell lysis to be released. Nevertheless, biosynthesis of DAPG was restored when the ΔpltM mutant was cultured with P. capeferrum F8. This indicates that cell lysis of P. capeferrum F8 caused by the elevated pyoluteorin concentration in coculture with WT P. protegens DTU9.1 is necessary for the release of an intracellular signal and inhibition of DAPG production. Further analyses are needed to identify the intracellular chemical signal from P. capeferrum F8 capable of inhibiting DAPG production. Additionally, we observed significantly higher levels of DAPG after 24 h in the coculture between P. capeferrum F8 and the ΔpltM mutant compared to monoculture of the mutant (Fig. 5A). This could indicate that P. capeferrum F8 additionally has the ability to induce DAPG production. This could further aid in explaining our initial observation utilizing the DAPG-responsive, PhlF-based whole-cell biosensor. Thus, the biosensor could have been responding to the presence of this inducing molecule, however, during cocultivation of P. protegens DTU9.1 and P. capeferrum F8 the release of the intracellular and inhibiting molecule overshadowed the effect of the inducing molecule, ultimately resulting in inhibition of DAPG production. Overall, our work and previous studies underline the importance of investigating microbial interactions to further elucidate the complex roles of secondary metabolites in natural communities.
In the recent decades, employing specific species of isolated bacteria as microbial biocontrol agents has gained increased attention for the suppression of phytopathogenic fungi and bacteria as a sustainable alternative to potentially harmful pesticides. The effectiveness of such biocontrol inoculants relies in part on antimicrobial activity against plant pathogens, which can typically be attributed to the production of antimicrobial secondary metabolites. However, despite the improvements in in vitro screening for novel candidate biocontrol microbes, less than 1% end up as commercial products [45]. The lack of efficacy in situ may in part be due to yet uncharacterized interactions between members of the natural soil microbiome and the introduced biocontrol agent. The bacterium, P. protegens, has been studied for decades for its biocontrol properties [2], and we show here that chemical interactions with another Pseudomonas species commonly found in soil and rhizosphere microbiomes [46] can radically alter its production of antimicrobial secondary metabolites (Fig. 1) and its activity towards bacterial phytopathogens (Fig. 6). We speculate that such phenotype-altering chemical interactions play an important role in determining the efficiency of biocontrol. Indeed, there are several recent demonstrations of improved plant growth promoting properties, as well as the overall bioactivity against plant-pathogenic microorganisms, by use of mixed species inoculants. For example, Niu et al. demonstrated that a synthetic microbial community consisting of seven species greatly reduced the severity of blight disease on maize seedling caused by the phytopathogen, Fusarium verticillioides [47]. Additionally, Zhuang et al. reported that a synthetic consortium consisting of six Pseudomonas spp. significantly affected plant growth promotion of radish seedlings [48]. Our results support that co-inoculation of two Pseudomonas strains; P. protegens DTU9.1 and P. capeferrum F8, augments the in vitro bioactivity against two known phytopathogens, P. carotovorum and D. solani, due to the increased production of pyoluteorin (Fig. 6). However, we also demonstrate a secondary microbial interaction leading to the inhibition of DAPG production. Natural conditions such as soil and the rhizosphere surrounding plant roots may facilitate spatial distribution of microorganisms, which favor distance-dependent interactions. As the inhibition of DAPG production requires cell-to-cell contact, it may only affect a minor fraction of P. protegens DTU9.1 cells, and hence the overall biocontrol activity against DAPG-sensitive microorganisms may be largely unaffected. This study therefore represents a defined mode of action for bacterial consortia applied as biocontrol inoculum in a natural setting and provides a stepping-stone into the much-needed mode of action studies in closer-to-field studies. An improved understanding of the underlying signalling events affecting microbial secondary metabolism may enable designs of signal-optimized microbial biocontrol consortia that potentiates production of relevant secondary metabolites to show improved efficiency in plant protection compared to single-species biocontrol.
Methods
Microorganisms and cultivation
Bacteria were routinely cultured in Luria-Bertani broth (LB, Merck) with appropriate antibiotics. E. coli was cultured by inoculating a single colony in 5 ml LB broth and incubating overnight at 37 °C with shaking (200 rpm). Pseudomonas were cultured by inoculating a single colony in 5 ml LB broth and incubating overnight at 30 °C with shaking (200 rpm). Plasmid cloning was performed in Escherichia coli CC118-λpir. Microorganisms used in this study are summarized in the Supplementary Table 1. Primers used for cloning and verification are found in the Supplementary Table 2.
Tagging Pseudomonas with fluorescent transposons for visualization of coculture dynamics
To visualize the development of bacterial cocultivation on solid agar surfaces, P. protegens DTU9.1 and P. capeferrum F8 were chromosomally tagged with Tn7 transposons carrying a constitutively expressed gfp or mKate gene, respectively. The plasmid, pBG42-gfp, was a kind gift from Victor de Lorenzo [49]. The mKate-version was cloned by replacing gfp with a PCR-amplified mKate gene from phymKate [50] with BciI/HindIII overhangs. The cloned plasmid, pBG42-mKate, was verified by Sanger sequencing at Eurofins Genomics.
The plasmids were mobilized into Pseudomonas by quad-parental mating and the Tn7 transposon was subsequently integrated chromosomally downstream of glmS in a transposase-dependent manner. For the quad-parental mating the optical density at OD600 was normalized to 1.0 for E. coli HB101 harboring pRK600, E. coli CC118 λpir harboring pTNS2, E. coli CC118 λpir harboring pBG42-gfp/-mKate and the recipient Pseudomonas. Then, the bacterial suspensions were mixed in a 1:1:1:1 ratio, concentrated 50 times and finally spotted on an LB agar plate incubated at 30 °C O/N. Transconjugants were selected on Pseudomonas Isolation Agar (PIA, Merck) supplemented with 50 µg ml−1 gentamycin and confirmed to be expressing gfp or mKate constitutively by examination under a fluorescence microscope.
Cocultivation of P. protegens DTU9.1 and P. capeferrum F8 on solid surface
For bacterial cocultivations, 1 ml samples of Pseudomonas O/N cultures were washed twice in 0.9% NaCl by centrifuging at 10,000 rpm for 1 min, followed by discarding the supernatant and resuspending the pellet in fresh liquid. Subsequently, the optical density at OD600 was normalized to 1.0. For cocultivations on agar surfaces, the bacterial suspensions of P. protegens DTU9.1 and P. capeferrum F8 were mixed in a 1:1 ratio, followed by spotting on KBmalt agar (20 g l−1 Proteose peptone No. 3, 1.5 g l−1 K2HPO4, 1.5 g l−1 MgSO4, 7.5 g l−1 malt extract, 10 ml l−1 glycerol and 20 g l−1 Bacto agar).
Visualization and analysis of fluorescent colonies
Fluorescent colonies were imaged on a Zeiss Axio Zoom V16 stereomicroscope with a Zeiss CL 9000 LED Light source, an AxioCam 503 Monochrome camera and a PlanApo Z 0.5x/0.125 FWD 114 mm objective. The filter sets used were: Zeiss 38 HE eGFP (Ex: 470/40 nm, Em: 525/50 nm) and Zeiss 63 HE (Ex: 572/25 nm, Em: 629/62 nm). Exposure time and focus were manually optimized for each experiment, but kept identical for every sample in a given experiment. Images were acquired with Zeiss Zen and analyzed in ImageJ v1.53 [51]. Initially, the colony of interest was segmented by automatic thresholding with the Triangle method on the bright field channel with a Gaussian blur (radius = 2.00 pixels). Relative fluorescence intensity per area was measured within the resulting region of interest outlining the entire colony.
Construction of pSEVA237::PpltL-gfp reporter fusion
To investigate the effect of the secreted signal from P. capeferrum F8 on the transcription of the pyoluteorin biosynthetic gene cluster, a PpltL reporter fusion was constructed in pSEVA237RBS30 with a strong ribosomal binding site (RBS30). The promoter region was PCR-amplified with BamHI/HindIII overhangs and ligated with pSEVA237RBS30. The cloned reporter fusion was verified by Sanger sequencing at Eurofins Genomics.
The plasmid was subsequently mobilized into P. protegens DTU9.1 by tri-parental mating. Initially, the optical density at OD600 was normalized to 1.0 for E. coli HB101 harboring pRK600, E. coli CC118 λpir harboring pSEVA237::PpltL-gfp and the recipient Pseudomonas. Then, the bacterial suspensions were mixed in a 1:1:1 ratio, concentrated 50 times and finally spotted on an LB agar plate incubated at 30 °C O/N. A transconjugant was selected on PIA supplemented with 50 µg ml−1 kanamycin and confirmed to be expressing gfp by examination under a fluorescence microscope.
Distance assay
A distance assay was carried out to determine if P. capeferrum F8 produced a single molecule affecting both DAPG and pyoluteorin production in P. protegens DTU9.1. Initially, P. capeferrum F8 was normalized to OD600 = 1.0 and spotted in 20 µl on KBmalt agar plates. The plates were incubated at 30 °C for 48 h. Then, a culture of P. protegens DTU9.1 was normalized to OD600 = 1.0 and spotted in 5 µl with varying distance to the edge of the P. capeferrum F8 colony (5, 12.5 and 20 mm). As a control, P. protegens DTU9.1 was spotted in isolation on a separate plate. The plates were incubated at 30 °C for 24 h, followed by agar plug extraction for HR-LCMS. The decay of pyoluteorin production as a function of distance to the edge of the P. capeferrum F8 colony was modelled with a one phase decay function in GraphPad Prism v8.3.0, where Y0 represents the intercept and K represents the decay rate.
Additionally, distance assays were conducted to evaluate the effect of secreted molecules on the transcription of the pyoluteorin biosynthetic genes measured as PpltL promoter activity. Similar to above, a culture of P. protegens DTU9.1 harboring pSEVA237::PpltL-gfp normalized to OD600 = 1.0 was spotted in 5 µl with varying distance to the edge of a pre-cultured colony of an interacting bacterium. After 24 h of incubation at 30 °C, colonies were analyzed by fluorescent microscopy on a Zeiss Axio Zoom V16 stereomicroscope as described above.
Supernatant assay
To evaluate if P. capeferrum F8 secreted a signal capable of inducing transcription of the pyoluteorin biosynthetic genes in P. protegens DTU9.1, a supernatant assay was conducted. Initially, P. capeferrum F8 was cultivated for 48 h in 100 ml KBmalt broth at 30 °C with shaking (200 rpm). Supernatant was harvested by centrifuging the culture at 8,000 rpm for 5 min followed by sterile-filtration. Aliquots of the supernatant were either boiled for 60 or 120 min at 100 °C, autoclaved for 15 min at 121 °C or treated with Proteinase K (New England Biolabs) at a final concentration of 30 U/ml for 1 h at 56 °C. Then, P. protegens DTU9.1 harboring pSEVA237::PpltL-gfp was inoculated to an initial OD600 = 0.001 in 50% v/v fresh KBmalt broth and supernatant (treated or untreated). Growth and fluorescence was measured every 30 min during incubation at 30 °C and continuous shake for 24 h in a Cytation5 microplate reader.
Additionally, P. protegens DTU9.1 was inoculated to an initial OD600 = 0.001 in 50% v/v fresh KBmalt and sterile-filtered supernatant of P. capeferrum F8 or 100% fresh KBmalt (control). Tubes were incubated at 30 °C with shaking (200 rpm) for 24 h, followed by sterile filtration of the supernatant. Levels of DAPG and pyoluteorin were analyzed with HR-LCMS.
Cell lysate assay
To investigate the effect of cell-associated metabolites from P. capeferrum F8 on the metabolite production in P. protegens DTU9.1 a cell lysate assay was performed. Firstly, the cell lysate was prepared by lysis of cell pellet from 4 ml O/N culture of P. capeferrum F8 with NEB gDNA Tissue Lysis Buffer (New England Biolabs) and lysozyme from chicken egg-white (Sigma-Aldrich). Cell-associated metabolites were extracted with ethyl:acetate (EtOAc) extraction and resuspended in methanol to a concentration of 1 mg/ml. Then, P. protegens DTU9.1 was inoculated to an initial OD600 = 0.001 in 10 ml KBmalt broth supplemented with 50 µl methanol (control) or 50 µl cell lysate extract. Tubes were incubated at 30 °C with shaking (200 rpm) for 24 h, followed by sterile filtration of the supernatant. Levels of DAPG and pyoluteorin were analyzed with HR-LCMS.
Deletion of pltM in P. protegens DTU9.1
To obtain a strain of P. protegens DTU9.1 incapable of synthesizing pyoluteorin, pltM was deleted by allelic replacement according to Hmelo et al. [52]. In short, DNA fragments directly upstream and directly downstream of the gene of interest were PCR amplified. The fragments were joined by splicing-by-overlap extension PCR with XbaI/SacI overhangs. The purified PCR product was restriction-digested and inserted in pNJ1 [53]. The resulting plasmid, pNJ1-pltM-del, was mobilized into P. protegens DTU9.1 via triparental mating with E. coli HB101 harboring the helper plasmid pRK600. Merodiploid transconjugants were initially selected on PIA supplemented with 50 µg ml−1 tetracycline. A second selection was performed on NSLB agar (10 g l−1 tryptone, 5 g l−1 yeast extract, 15 g l−1 Bacto agar) with 15% v/v sucrose. Candidates for successful deletion were confirmed by PCR and verified by Sanger sequencing at Eurofins Genomics.
Detection of DAPG with a whole-cell biosensor
For biosensor-guided detection of DAPG production, 1 ml O/N culture of E. coli CC118 λpir harboring pSEVA225::DAPGlacZ was washed twice in 0.9% NaCl and its optical density at OD600 set to 0.5. Then, 100 µl of the biosensor suspension was spread on KBmalt plates supplemented with 65 µg ml−1 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Xgal, Thermo Fisher Scientific). Pseudomonas mono- and cocultures normalized to OD600 = 1.0 were spotted on top as 20 µl colonies. All agar plates were incubated at 30 °C.
Detection of secondary metabolites with HR-LCMS
To extract secondary metabolites of mono- and cocultivated Pseudomonas, an agar plug covering the bacterial colony (6 mm diameter) was transferred to a vial and extracted with 1 ml of isopropanol:ethyl acetate (1:3, v/v), containing 1% formic acid, under ultrasonication for 60 min. Extracts were subsequently evaporated under N2 and re-dissolved in 200 µl of methanol for further sonication for 15 min. Lastly, the samples were centrifuged at 13400 rpm for 3 min and supernatants transferred to HPLC vials and subjected to ultra high-performance liquid chromatography-high resolution electrospray ionization mass spectrometry analysis (abbreviated as HR-LCMS).
HR-LCMS was performed on an Agilent Infinity 1290 UHPLC system. Liquid chromatography of 1 µL or 5 µL extract was performed using an Agilent Poroshell 120 phenyl-C6column (2.1 × 150 mm, 1.9 μm) at 60 oC using CH3CN and H2O, both containing 20 mM FA. Initially, a linear gradient of 10% CH3CN/H2O to 100% CH3CN over 10 min was employed, followed by isocratic elution of 100% CH3CN for 2 min. Then, the gradient was returned to 10% CH3CN/H2O in 0.1 min and finally isocratic condition of 10% CH3CN/H2O for 1.9 min, all at a flow rate of 0.35 min ml−1. HRMS data was recorded in positive ionization on an Agilent 6545 QTOF MS equipped with an Agilent Dual Jet Stream electrospray ion (ESI) source with a drying gas temperature of 250 °C, drying gas flow of 8 min l−1, sheath gas temperature of 300 °C and sheath gas flow of 12 min l−1. Capillary voltage was 4000 V and nozzle voltage was set to 500 V. HRMS data was processed and analyzed using Agilent MassHunter Qualitative Analysis B.07.00. HPLC grade solvents (VWR Chemicals) were used for extractions while LCMS grade solvents (VWR Chemicals) were used for LCMS.
Inhibition assay
The effect of cocultivating P. protegens DTU9.1 and P. capeferrum F8 on bioactivity towards known phytopathogenic bacteria was investigated in an inhibition assay. Cultures of P. protegens DTU9.1 wild type and ΔpltM mutant, as well as P. capeferrum F8 were normalized to an OD600 = 1.0. Mono- and cocultures were spotted in 10 µl on KBmalt agar plates and incubated for 48 h at 30 °C, followed by bacterial inactivation by exposure to chloroform vapors for 30 min. Then, 6 ml 0.5% soft KBmalt agar was inoculated with the phytopathogenic bacteria, Pectobacterium carotovorum ATCC 15713 and Dickeya solani LMG 25993, to an OD600 = 1.0 and added to the plates with inactivated Pseudomonas colonies. Plates were incubated for another 24 h at 30 °C, followed by measuring the size of inhibition zones surrounding each Pseudomonas colony.
Genome sequencing
Genomic DNA from P. capeferrum F8 was purified with the Monarch HMW DNA Extraction Kit for Tissue (New England Biolabs) according to the manufacturer’s instructions. Long reads were generated with a Nanopore MinION instrument using the R9/FLO-MIN106 flow cell, while short paired-end reads were generated on the NovaSeq 6000 platform with 2×150-bp reads (Illumina). The long reads were used to de novo assemble the genome with Flye v2.8.1. The assembled genome was subsequently polished with the forward Illumina reads using Racon v1.4.13. The assembled genome was uploaded to and annotated by NCBI Genbank (Accession number: CP099575).
Supplementary information
Acknowledgements
We thank professor Victor de Lorenzo for providing the SEVA plasmids used in this study. Additionally, we thank Mikael Lenz Strube for aiding with statistical data analysis, as well as Mikkel Anbo and members of the Centre for Microbial Secondary Metabolites (CeMiSt) for general scientific discussions.
Author contributions
MLH and LJ conceived and designed the study. MLH carried out microbiological, molecular and genetic assays. MW and SAJ carried out metabolite extractions and chemical analyses. MW, SAJ and TOL supervised interpretation of LCMS data. MLH and LJ wrote the manuscript, with contributions and approval of all authors.
Funding
This study was carried out as part of the Center of Excellence for Microbial Secondary Metabolites funded by The Danish National Research Foundation (DNRF137).
Data availability
All data are included in the article and the supplemental material will be made available upon request to the corresponding author. Additionally, the de novo assembled genome sequence of P. capeferrum F8 generated in this study is available in the NCBI GenBank under the accession number CP099575.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01322-8.
References
- 1.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–86. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–19. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 3.Whipps JM. Microbial interactions and biocontrol in the rhizosphere. J Exp Bot. 2001;52:487–511. doi: 10.1093/jexbot/52.suppl_1.487. [DOI] [PubMed] [Google Scholar]
- 4.Mendes R, Kruijt M, De Bruijn I, Dekkers E, Van Der Voort M, Schneider J, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 5.Jousset A, Becker J, Chatterjee S, Karlovsky P, Scheu S, Eisenhauer N. Biodiversity and species identity shape the antifungal activity of bacterial communities. Ecology. 2014;95:1184–90. doi: 10.1890/13-1215.1. [DOI] [PubMed] [Google Scholar]
- 6.Becker J, Eisenhauer N, Scheu S, Jousset A. Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecol Lett. 2012;15:468–74. doi: 10.1111/j.1461-0248.2012.01759.x. [DOI] [PubMed] [Google Scholar]
- 7.Hu J, Wei Z, Friman VP, Gu SH, Wang XF, Eisenhauer N, et al. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio. 2016;7:e01790–16. doi: 10.1128/mBio.01790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehrabi Z, McMillan VE, Clark IM, Canning G, Hammond-Kosack KE, Preston G, et al. Pseudomonas spp. diversity is negatively associated with suppression of the wheat take-all pathogen. Sci Rep. 2016;6:1–10. doi: 10.1038/srep29905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Z, Geudens N, Kieu NP, Sinnaeve D, Ongena M, Martins JC, et al. Biosynthesis, chemical structure, and structure-activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Front Microbiol. 2016;7:1–16. doi: 10.3389/fmicb.2016.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Q, Philmus B, Chang JH, Loper JE. Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. Elife. 2017;6:e22835. doi: 10.7554/eLife.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramette A, Moënne-Loccoz Y, Défago G. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol Ecol. 2003;44:35–43. doi: 10.1111/j.1574-6941.2003.tb01088.x. [DOI] [PubMed] [Google Scholar]
- 12.Raaijmakers JM, Weller DM. Natural Plant Protection by 2,4-Diacetylphloroglucinol-Producing Pseudomonas spp. in Take-All Decline Soils. Mol Plant-Microbe Interact. 1998;11:144–52. [Google Scholar]
- 13.Murata K, Suenaga M, Kai K. Genome Mining Discovery of Protegenins A–D, Bacterial Polyynes Involved in the Antioomycete and Biocontrol Activities of Pseudomonas protegens. ACS Chem Biol. 2021. https://pubs.acs.org/doi/10.1021/acschembio.1c00276. Online ahead of print. [DOI] [PubMed]
- 14.Achkar J, Xian M, Zhao H, Frost JW. Biosynthesis of Phloroglucinol. J Am Chem Soc. 2005;127:5332–3. doi: 10.1021/ja042340g. [DOI] [PubMed] [Google Scholar]
- 15.Bangera MG, Thomashow LS. Identification and Characterization of a Gene Cluster for Synthesis of the Polyketide Antibiotic 2,4-Diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J Bacteriol. 1999;181:3155–63. doi: 10.1128/jb.181.10.3155-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bottiglieri M, Keel C. Characterization of PhlG, a hydrolase that specifically degrades the antifungal compound 2,4-diacetylphloroglucinol in the biocontrol agent Pseudomonas fluorescens CHA0. Appl Environ Microbiol. 2006;72:418–27. doi: 10.1128/AEM.72.1.418-427.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan X, Yang R, Zhao R-X, Han J-T, Jia W-J, Li D-Y, et al. Transcriptional Regulator PhlH Modulates 2,4-Diacetylphloroglucinol Biosynthesis in Response to the Biosynthetic Intermediate and End Product. Appl Environ Microbiol. 2017;83:e01419–17. doi: 10.1128/AEM.01419-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. Dichlorination of a pyrrolyl-S-carrier protein by FADH2- dependent halogenase PltA during pyoluteorin biosynthesis. Proc Natl Acad Sci USA. 2005;102:13843–8. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas MG, Burkart MD, Walsh CT. Conversion of L-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem Biol. 2002;9:171–84. doi: 10.1016/s1074-5521(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 20.Schnider-Keel U, Seematter A, Maurhofer M, Blumer C, Duffy B, Gigot-Bonnefoy C, et al. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J Bacteriol. 2000;182:1215–25. doi: 10.1128/jb.182.5.1215-1225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brodhagen M, Henkels MD, Loper JE. Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5. Appl Environ Microbiol. 2004;70:1758–66. doi: 10.1128/AEM.70.3.1758-1766.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maurhofer M, Baehler E, Notz R, Martinez V, Keel C. Cross Talk between 2,4-Diacetylphloroglucinol-Producing Biocontrol Pseudomonads on Wheat Roots. Appl Environ Microbiol. 2004;70:1990–8. doi: 10.1128/AEM.70.4.1990-1998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clifford JC, Buchanan A, Vining O, Kidarsa TA, Chang JH, McPhail KL, et al. Phloroglucinol functions as an intracellular and intercellular chemical messenger influencing gene expression in Pseudomonas protegens. Environ Microbiol. 2016;18:3296–308. doi: 10.1111/1462-2920.13043. [DOI] [PubMed] [Google Scholar]
- 24.Kidarsa TA, Goebel NC, Zabriskie TM, Loper JE. Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol Microbiol. 2011;81:395–414. doi: 10.1111/j.1365-2958.2011.07697.x. [DOI] [PubMed] [Google Scholar]
- 25.Hassan KA, Johnson A, Shaffer BT, Ren Q, Kidarsa TA, Elbourne LDH, et al. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ Microbiol. 2010;12:899–915. doi: 10.1111/j.1462-2920.2009.02134.x. [DOI] [PubMed] [Google Scholar]
- 26.Dubuis C, Haas D. Cross-species GacA-controlled induction of antibiosis in pseudomonads. Appl Environ Microbiol. 2007;73:650–4. doi: 10.1128/AEM.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen ML, He Z, Wibowo M, Jelsbak L. A Whole-Cell Biosensor for Detection of 2,4- Diacetylphloroglucinol (DAPG)-Producing Bacteria from Grassland Soil. Appl Environ Microbiol. 2021;87:e01400–e01420. doi: 10.1128/AEM.01400-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hesse C, Schulz F, Bull CT, Shaffer BT, Yan Q, Shapiro N, et al. Genome‐based evolutionary history of Pseudomonas spp. Environ Microbiol. 2018;20:2142–59. doi: 10.1111/1462-2920.14130. [DOI] [PubMed] [Google Scholar]
- 29.Lozano-Andrade CN, Strube ML, Kovács ÁT. Complete genome sequences of four soil-derived isolates for studying synthetic bacterial community assembly. Microbiol Resour Announc. 2021;10:e00848–21. doi: 10.1128/MRA.00848-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Roux M, Kirkpatrick RL, Montauti EI, Tran BQ, Brook Peterson S, Harding BN, et al. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife. 2015;2015:1–65. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyc O, van den Berg M, Gerards S, van Veen JA, Raaijmakers JM, de Boer W, et al. Impact of interspecific interactions on antimicrobial activity among soil bacteria. Front Microbiol. 2014;5:1–10. doi: 10.3389/fmicb.2014.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi SS, Bogdanov A, Cnockaert M, Acar T, Ranty-Roby S, Coenye T, et al. Induction of antibiotic specialized metabolism by co-culturing in a collection of phyllosphere bacteria. Environ Microbiol. 2021;23:2132–51. doi: 10.1111/1462-2920.15382. [DOI] [PubMed] [Google Scholar]
- 33.Cornforth DM, Foster KR. Competition sensing: The social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–93. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 34.LeRoux M, Peterson SB, Mougous JD. Bacterial danger sensing. J Mol Biol. 2015;427:3744–53. doi: 10.1016/j.jmb.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westhoff S, van Wezel GP, Rozen DE. Distance-dependent danger responses in bacteria. Curr Opin Microbiol. 2017;36:95–101. doi: 10.1016/j.mib.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–53. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Garbeva P, Silby MW, Raaijmakers JM, Levy SB, Boer WDE. Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J. 2011;5:973–85. doi: 10.1038/ismej.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrudan MI, Smakman F, Grimbergen AJ, Westhoff S, Miller EL, Van Wezel GP, et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA. 2015;112:11054–9. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kehe J, Ortiz A, Kulesa A, Gore J, Blainey PC, Friedman J. Positive interactions are common among culturable bacteria. Sci Adv. 2021;7:1–10. doi: 10.1126/sciadv.abi7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang KM, Kim JS, Kim HS, Kim YY, Oh JK, Jung HW, et al. Lactobacillus reuteri AN417 cell-free culture supernatant as a novel antibacterial agent targeting oral pathogenic bacteria. Sci Rep. 2021;11:1–16. doi: 10.1038/s41598-020-80921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubern JF, Lugtenberg BJJ, Bloemberg GV. The ppuI-rsaL-ppuR quorum-sensing system regulates biofilm formation of Pseudomonas putida PCL1445 by controlling biosynthesis of the cyclic lipopeptides putisolvins I and II. J Bacteriol. 2006;188:2898–906. doi: 10.1128/JB.188.8.2898-2906.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wellington S, Peter Greenberg E. Quorum sensing signal selectivity and the potential for interspecies cross talk. mBio. 2019;10:e00146–19. doi: 10.1128/mBio.00146-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duffy BK, Défago G. Zinc Improves Biocontrol of Fusarium Crown and Root Rot of Tomato by Pseudomonas fluorescens and Represses the Production of Pathogen Metabolites Inhibitory to Bacterial Antibiotic Biosynthesis. Phytopathology. 1997;87:1250–7. doi: 10.1094/PHYTO.1997.87.12.1250. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Estrada-de los Santos P, Matthijs S, Xie G-L, Busson R, Cornelis P, et al. Promysalin, a Salicylate-Containing Pseudomonas putida Antibiotic, Promotes Surface Colonization and Selectively Targets Other Pseudomonas. Chem Biol. 2011;18:1320–30. doi: 10.1016/j.chembiol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Parnell JJ, Berka R, Young HA, Sturino JM, Kang Y, Barnhart DM, et al. From the lab to the farm: An industrial perspective of plant beneficial microorganisms. Front Plant Sci. 2016;7:1–12. doi: 10.3389/fpls.2016.01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berendsen RL, van Verk MC, Stringlis IA, Zamioudis C, Tommassen J, Pieterse CMJ, et al. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics. 2015;16:1–23. doi: 10.1186/s12864-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu B, Paulson JN, Zheng X, Kolter R. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci USA. 2017;114:E2450–E2459. doi: 10.1073/pnas.1616148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhuang L, Li Y, Wang Z, Yu Y, Zhang N, Yang C, et al. Synthetic community with six Pseudomonas strains screened from garlic rhizosphere microbiome promotes plant growth. Micro Biotechnol. 2021;14:488–502. doi: 10.1111/1751-7915.13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zobel S, Benedetti I, Eisenbach L, De Lorenzo V, Wierckx N, Blank LM. Tn7-Based Device for Calibrated Heterologous Gene Expression in Pseudomonas putida. ACS Synth Biol. 2015;4:1341–51. doi: 10.1021/acssynbio.5b00058. [DOI] [PubMed] [Google Scholar]
- 50.Van Gestel J, Weissing FJ, Kuipers OP, Kovács ÁT. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 2014;8:2069–79. doi: 10.1038/ismej.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10:1820–41. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Hengzhuang W, Wu H, Damkiær S, Jochumsen N, Song Z, et al. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2012;65:366–76. doi: 10.1111/j.1574-695X.2012.00936.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article and the supplemental material will be made available upon request to the corresponding author. Additionally, the de novo assembled genome sequence of P. capeferrum F8 generated in this study is available in the NCBI GenBank under the accession number CP099575.