Abstract
In this paper we present a structurally-complex biomimetic scattering structure, fabricated with two-photon polymerization, and utilize this object in order to benchmark a computational imaging system. The phantom allows to tailor the scattering by modifying its degrees of freedom i.e. refractive index contrast and scattering layer dimensions and incorporates a 3D imaging quality test, representing a single cell within tissue. While the sample may be used with multiple 3D microscopy techniques, we demonstrate the impact of scattering on three tomographic phase microscopy (TPM) reconstruction methods. One of these methods assumes the sample to be weak-scattering, while the other two take multiple scattering into account. The study is performed at two wavelengths (visible and near-infrared), which serve as a scaling factor for the scattering phenomenon. We find that changing the wavelength from visible into near-infrared impacts the applicability of TPM reconstruction methods. As a result of reduced scattering in near-infrared region, the multiple-scattering-oriented techniques perform in fact worse than a method aimed for weak-scattering samples. This implies a necessity of selecting proper approach depending on sample’s scattering characteristics even in case of subtle changes in the object-light interaction.
Subject terms: Microscopy, Interference microscopy, Optical metrology
Introduction
One of the modern challenges in computational optics is to image scattering samples with high resolution1. This can be attributed to the fact that complex biological structures such as spheroids or organoids tend to be more relevant models than 2D cell cultures e.g. for drug discovery2. Also, most in vivo imaging techniques require the probing light to pass through the complex structure of a tissue which highly limits imaging depths due to multiple scattering. This demand stimulates the development of new methods1,3–5, however, it is difficult to select an appropriate one based on the scattering strength of the analyzed specimen. For this reason a versatile, repeatable and quantitative method for the evaluation of different imaging systems and algorithms is essential to determine their limits of applicability depending on the object’s scattering properties. One possibility is to use calibrated microphantoms as imaging targets. Unfortunately, existing microphantoms are typically either weak-scattering (e.g. index-matched microspheres) or overly simplistic (e.g. index-mismatched microspheres)6,7 compared to the types of heterogenously scattering multicellular samples that multiple-scattering methods are intended for. This is a critical limitation when characterizing computational imaging methods that utilize nonconvex solvers, where iterative convergence depends on the complexity of the energy landscape and directly associates with a sample’s 3D complexity8.
In this work we present a 3D-printed microphantom with multiple-scattering refractive index (RI) distribution. To do so, we leverage recent developments in 3D printing via direct laser writing9–12. Among multiple available 3D printing techniques13–18, we chose a two-photon polymerization that enables 3D printing of microphantom samples with known geometry and calibrated RI. When compared to other implementations of direct laser writing, it allows (1) to control the RI with relatively high modulation range, (2) to adjust the RI contrast or scattering strength post-fabrication using different immersion liquid and (3) to handle and measure the microphantom in the same way as biological specimens. Next, we present the application of the phantom in the field of tomographic phase microscopy (TPM), a technique which has demonstrated impressive biological imaging results in prior works. However, it is important to note that all computational imaging methods can be evaluated with the proposed procedure.
TPM is a quantitative, label-free imaging method that utilizes optical projections through a semi-transparent sample along various illumination angles to reconstruct the sample’s 3D RI. This method has found several applications in biological imaging, where RI is directly related to the dry mass distribution at the cellular and subcellular levels. Refractive index and dry mass are known to be crucial factors in analyzing the current stage of cell cycle19, cell structure20,21, photobiochemical effects on cells22, influence of external factors on cellular parameters23,24 and many others. Given the wealth of information provided by the dry mass at the single-cell level, there is significant demand to extend the capability to analyze dry mass to large multicellular clusters, thick tissue slices, or whole microorganisms. However, to reconstruct 3D RI, traditional TPM methods utilize critical assumptions in their computational reconstruction methodologies that rely on the sample being weakly scattering25. These assumptions limit samples to having thicknesses on the order of only tens of microns. For thick, complex samples, reconstruction frameworks that accommodate for multiple scattering must be utilized. To this end, numerous TPM approaches have been proposed in recent years that introduce new frameworks to accommodate multiple scattering26–31. Notably, these approaches utilize nonlinear and nonconvex solvers to iteratively solve for a sample’s 3D RI. Though these methods have demonstrated impressive results in reconstructing RI in multiple-scattering samples, their quantitative accuracy has not yet been robustly characterized experimentally, and the presented results usually do not allow comparison of different methods in order to select proper approach for a given scattering level in a sample. The general strategy to experimentally evaluate quantitative accuracy in TPM methods is to reconstruct 3D RI in samples with known RI distributions6,7,32. A multiple-scattering TPM method that outputs accurate 3D RI reconstructions of a weakly scattering or overly simplistic microphantom cannot be expected to output similarly accurate RI reconstructions for more complex multiple-scattering samples, where the probability of converging to local minima is drastically higher. To robustly characterize the quantitative accuracy of multiple-scattering TPM methods, it is imperative to use microphantoms with known 3D RI that mimic the structural complexity of the types of samples that the TPM methods are intended to image. To the best of our knowledge, these types of gold-standard multiple-scattering microphantoms do not exist.
Results
In this section we present the design of the 3D printed microphantom and its application in evaluation of three TPM reconstruction methods.
3D printed microphantom
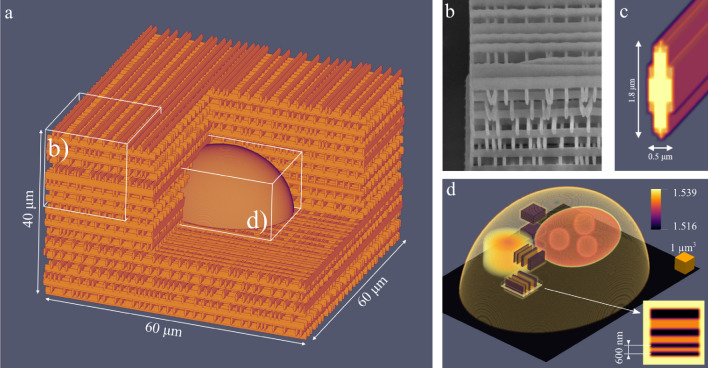
The scattering microphantom we designed and 3D printed consists of a cell-like target with internal test structures33 embedded within a pseudo-random distribution of rods that switch their orientation across various layers (Fig. 1a). The width and height of each rod is equal to 0.5 m and 1.8 m respectively. The lateral distance between the rods in each layer is randomized between 0.7 m to 3 m and the layers are stacked vertically every 1.4 m. The resulting structure is transparent (over 99% transmittance for the extinction coefficient of 0.1 mm34) and multiple scattering35. The final scattering region is a 60 m 60 m 40 m cube with a fill-factor of roughly 25% (fraction of volume occupied by the polymer).
Figure 1.
(a) Half-section view of the scattering microphantom design. (b) SEM image of a scattering layer made out of quasi-randomly distributed rods. (c) Individual rod that comprises a scattering layer. (d) Visualization of the 3D RI distribution of the imaging target—cell phantom. Subcellular features, such as resolution targets (shown in the inset) and cell nucleoli, are enclosed in truncated ellipsoid with the external dimensions of 30 m 25 m 12 m, which is then embedded in the center of the 60 m 60 m 40 m scattering cube.
The cell-like target embedded within the layers of randomly distributed rods mimic a single biological cell encased within a scattering volume, which represents e.g. tissue. The cell-target comprises of substructures that enable assessment of quantitative accuracy in TPM imaging systems36. Information about 3D printing procedure are given in Sect. “3D scattering microphantom fabrication”. The main features within the cell target include resolution test targets, nucleoli suspended in a nucleus, and a region of slow RI variation (see Fig. 1d). Notably, the resolution test target comprises of lines with increasing spatial frequency37 up to 1667 lp/mm. By assessing the maximum spatial frequency of lines that can be distinguished within the cell test target, the imaging resolution of a TPM method of choice can be characterized and potentially compared to different methods of assessing resolution described by other research groups38,39. The pseudo-random distribution of rods that compose the scattering portion of the whole microphantom are suppressed within 0.5 m of the cell target and do not intersect with any of the test structures. A model of the phantom is available in Dataset 140.
TPM systems evaluation
Three TPM reconstruction methods were implemented and evaluated through comparison of tomographic reconstructions of the proposed microphantom. The methods are: (1) Gerchberg-Paopulis with support constraint (GPSC)41, (2) multi-slice beam-propagation with electric-field measurements (MSBP-E)42 and (3) multi-slice beam-propagation with intensity-only measurements (MSBP-I)31. In order to perform the comparison, the phantom has been measured with the TPM device and two datasets were composed from the complex-valued scattered field measurements of the microphantom being illuminated at different angles, using both 633 nm and 835nm wavelengths. For GPSC and MSBP-E, these datasets were used directly. For MSBP-I, only the amplitude components were used. The measurements and their corresponding 3D RI reconstructions are available in Dataset 140.
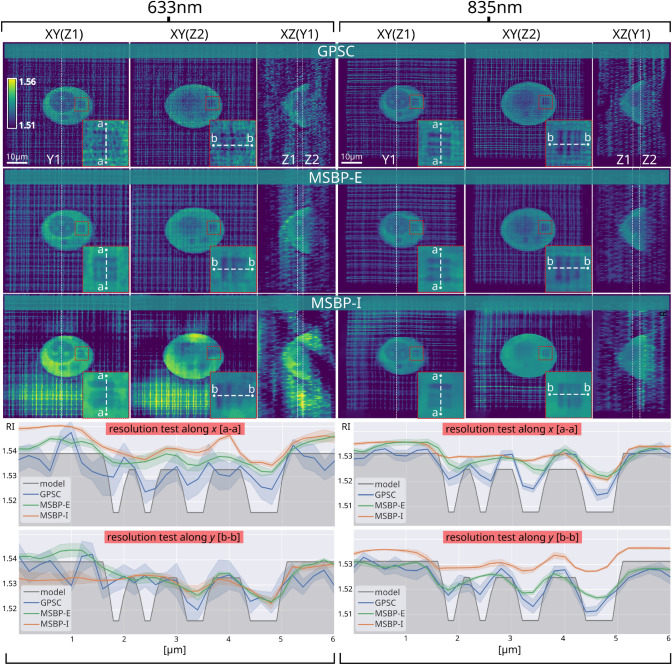
Figure 2 shows the reconstruction results for both wavelengths. We characterize lateral (x and y) resolution of the TPM reconstructions by visualizing the resolution test lines inside the microphantom. To quantitatively compare the three TPM reconstruction methods, horizontal and vertical cross-sectional plots across these resolution tests were generated by computing the average and standard-deviation of the pixel-values across rows or columns adjacent to the dashed white a-a and b-b lines, by pixels. These cross-sectional plots are shown below.
Figure 2.
Comparison of tomographic reconstructions of the microphantom measured with 633 nm and 835 nm wavelength and calculated with 3 algorithms. The shaded colored regions surrounding each of the 1D plots at the bottom represent the standard deviation.
Discussion
The known complex geometry and RI distribution of the developed microphantom allows to show that changing the wavelength from visible into near-infrared impacts the applicability of TPM reconstruction methods43,44. The magnified views of the resolution test region after reconstructing the microphantom using the GPSC algorithm with 633 nm wavelength light reveals significant grainy artifacts which occlude the line features within the microphantom’s test region. We note that these artifacts are heavily decreased when reconstructing with 835 nm wavelength light. This suggests that, at 633 nm wavelength, the microphantom is too highly-scattering for GPSC to be applicable. The decreased GPSC reconstruction artifacts for 835 nm matches conventional knowledge that longer wavelengths of light are more resistant to scattering than shorter wavelengths. In order to confirm that this observation is due to reduced scattering and not different noise characteristics between the two light sources, we analyzed the standard deviation of phase-noise in an object-free region within the input datasets. For 633 nm wavelength light, a phase-noise standard deviation of radians was observed, while for 835 nm wavelength light, a phase-noise standard deviation of radians was observed. Given such a small variation between these noise characteristics, we conclude that the major factor in the GPSC reconstruction quality that affects its ability to visualize the microphantom’s test lines is the scattering strength of the microphantom at the two different wavelengths.
Notably, both MSBP-E and MSBP-I use total-variation (TV) regularization in order to stabilize the convergence of the nonconvex iterative solver in the presence of noise45. Especially in the case of using 633 nm wavelength light, TV regularization results in 3D RI reconstructions with less noise compared to the 3D reconstructions computed via GPSC, which does not use TV regularization. This can be directly visualized in the 2D cross-sections in Fig. 2, and is confirmed by the bounds of standard deviation shown in the 1D cross-sectional plots. However, the drawback of regularization is that it has a blurring effect on high-resolution features. Because the microphantom is less scattering in 835 nm wavelength light, only GPSC managed to reconstruct the high spatial-frequency test lines within the microphantom. Typically, the strength of TV regularization is manually tuned to fit experimental factors and balance the tradeoff between achieving iterative stability versus high imaging resolution.
In terms of the average RI, the knowledge about the ground truth RI distribution of the phantom makes it possible to quantitatively compare reconstruction results with GPSC, MSBP-E, and MSBP-I. We see that all methods successfully capture the bulk characteristics of the microphantom. As described above, the 3D reconstruction via GPSC exhibits grainy artifacts when using 633 nm wavelength light, likely due to the microphantom being multiple-scattering at that wavelength. Furthermore, MSBP-I outputs slightly overestimated RI values, and also suffers from low-frequency spatial artifacts (which have been observed in other intensity-only phase-imaging techniques46,47). Other works have shown that MSBP-I demonstrates higher accuracy when using partially-coherent illumination, which drastically reduces coherent noise31. Future work may include repeating this analysis across a larger range of TPM reconstruction techniques with more complex scattering microphantoms.
With the presented results we show that changing the illumination wavelength affects the scattering nature of the microphantom. Specifically, though the microphantom is multiple scattering with 633 nm wavelength light, it is weak scattering with 835 nm wavelength light. This naturally indicates that the optimum choice for illumination wavelength must balance between resolution ( for single projection) and scattering strength. As has been shown, there are cases when instead of applying multiple-scattering methods, it is advantageous to increase the illumination wavelength (thus decreasing the scattering strength of the sample) and apply a method based on the Fourier Diffraction Theorem that does not utilize the total-variation constraint. More fundamentally however, we showed that 3D printed microphantoms enable quantitative assessment of 3D RI reconstruction accuracy across various TPM methodologies. This capability is important when choosing a TPM method optimized for specific classes of samples and imaging conditions.
Future works will focus on developing methods to quantify scattering strengths of phantoms and relate these quantities with the scattering properties of different tissue types. If successful, this would enable us to design and fabricate (using the presented methods) 3D microphantom structures to mimic a wide range of biological specimens ranging from multicellular clusters to bulk tissues and small organisms. Another possible direction is to tune scattering parameters in the phantom to characterize imaging performance for various techniques used to image into scattering tissue, such as optical coherence tomography or confocal reflectance microscopy. Finally, we envision that one can exploit the flexibility of two-photon polymerization to fabricate microphantoms on different substrates (e.g. at the end-face of the optical fibre for sample rotation tomography), using biocompatible resins (to combine test targets with the living cells in the single measurement volume48) or modify the resin with functional particles18 (e.g. to accommodate systems that also measure absorption49–53).
Materials and methods
We present below the (1) methodology with which we 3D-print multiple scattering microphantoms; (2) the optical design of the TPM imaging systems that we use to experimentally collect scattering electric-field measurements of the microphantom; and (3) short theoretical descriptions of three tomographic algorithms that were used to reconstruct 3D RI from the measured data.
3D scattering microphantom fabrication
The phantom is fabricated using two-photon laser lithography, in which a focused laser beam is scanned within liquid resin. The resin within the laser’s focal volume is locally polymerized. Adjusting the scanning trajectory and the exposure time of the laser beam enables simultaneous control over the 3D printed geometry (accuracy at the order of 100 nm) and RI (accuracy at the order of , maximal RI = 0.03 within the structure) in three dimensions. We used Photonic Professional GT (Nanoscribe GmbH) equipped with a 1.3 NA 100 microscope objective and piezo scanning stage. The phantom is fabricated in the IP-Dip resin (Nanoscribe GmbH) on top of a #1.5H coverslip (dip-in configuration54). After fabrication the structure was developed in PGMEA (Propylene glycol monomethyl ether acetate; 12 min), followed by isopropyl alcohol (10 min) and then blow-dried. The full methodology for fabrication and validation of the features can be found in our previous work33.
To conduct our TPM imaging experiments, the microphantom was immersed in Zeiss Immersol 518F oil (RI nm = 1.5123), which provides similar RI contrast as in the case of cells immersed in culture medium. By using immersion oils with varying RI, it is possible to adjust the scattering properties of the microphantom post-fabrication.
Measurement system
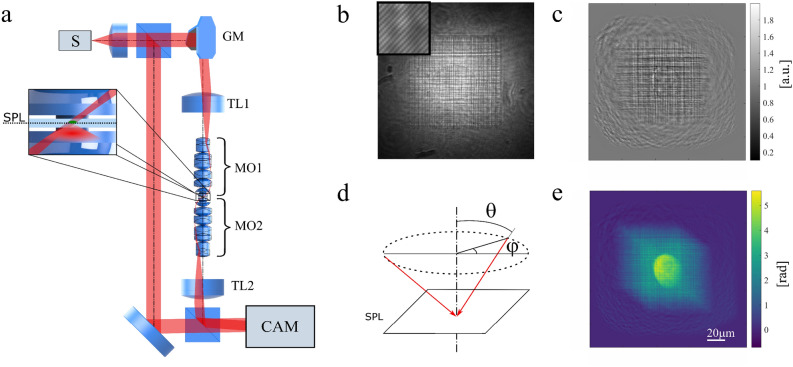
In this work, an optical system, as shown in Fig. 3a) was used in order to study the scattering phantom. The system is a Mach-Zehnder-based TPM microscope55, working in a limited-angle configuration with stationary sample and illumination rotated with a galvo mirror (Thorlabs GVS212/M)56. The research was performed with two wavelengths and thus there were two modified versions of the presented tomographic microscope. First version, works with wavelength 633 nm and the second, with 835 nm. The input beam (S in Fig.3a) is delivered with an optical fiber, collimated and then split into object and reference arm. In the system the light source was a volume Bragg grating laser (Necsel NovaTru Chroma 633 SLM), , providing a single longitudinal mode and offering long coherence length. The system utilized a swept source (Superlum Broadsweeper BS-840-2-HP, = 800–870 nm), set at 835 nm. Due to difference in coherence length, an additional delay module was placed in the reference beam for measurements. The beam-splitting cubes in this work were either coated for 400–700 nm or 700–1100 nm depending on the wavelength used. The focal length of the tube lens TL1 was 150 mm and 200 mm respectively. Both microscope objectives (MO1 and MO2) in Fig. 3 were 100 NA 1.3 Semi plan-apochromatic, infinity-corrected objectives. The second tube lens TL2 used was either 200 mm or 300 mm. This provided magnifications and . The camera used in the system was a CMOS sensor in both cases, with 3.45 pixel size (JAI BM500GE) in case of and 5.5 pixel size (Basler acA2040-180km) in case of . The minimum magnification, which is imposed by pixel size and wavelength in order to assure correct hologram recording for each projection57 is and , which is satisfied in both cases. A sample hologram is presented in Fig. 3b). Both systems were set to illuminate the sample with a circular scanning scenario (see Fig. 3d) at zenith angle and provided 180 projections spaced at . A sample of the phase and amplitude provided by the system at 835nm is presented in Fig. 3c and e.
Figure 3.
(a) Mach-Zehnder-based TPM measurement system. S, light source; GM, galvo system; TL1 and TL2, tube lenses; MO1 and MO2; microscope objective; SPL, sample plane; CAM, camera; (b) hologram acquired at axial illumination of the sample; (c) amplitude of a projection at ; (d) circular scanning scenario used in the measurement. Rotation angle: , zenith angle: ; (e) phase of the projection.
Reconstruction algorithms
There were three different TPM reconstruction methods for computing 3D RI of the microphantom used for comparison. The data for each method was acquired through the multi-angle scattering measurements captured as described in Sect. “Measurement system”. We provide a short description of these methods below. Complete descriptions of these methods is given in respective references.
Gerchberg-Papoulis with object support
To provide a baseline standard to compare to TPM reconstruction algorithms utilizing multiple-scattering models, we first reconstruct the microphantom’s 3D RI using a weak-scattering TPM method. We specifically use the Gerchberg-Papoulis algorithm enhanced with additional finite object support constraint (GPSC)41. Complex-valued electric-fields measured by our TPM systems are used as inputs. The procedure is performed in two steps. First, an initial tomographic 3D RI distribution is reconstructed from the electric-field measurements with strong total-variation regularization45. This is performed through the Chambolle-Pock58 optimization method and implemented with ASTRA tomography toolbox59. The result undergoes binarization and a finite object support is generated. Secondly, a classic Gerchberg-Papoulis algorithm is used which is an iterative version of Direct Inversion method (also known as the Wolf transform)60. This iterative procedure is based on the Fourier Diffraction Theorem61 and utilizes first-order scattering approximation. Here, the reconstruction and its Fourier transform are calculated alternately and constraints are applied: nonnegativity and finite object support in the signal domain, and replenishment of original projections in the frequency domain.
Multi-slice beam-propagation with electric-field measurements
Our main method to model multiple-scattering is the multi-slice beam-propagation method (MSBP)62, which has recently shown promising results for biological imaging42,63. In our first implementation of MSBP, we use the same exact electric-field dataset utilized by GPSC from above. An initial guess of the sample’s 3D RI is selected to start off the iterative procedure. Afterwards, the MSBP method is used to simulate scattering measurements resulting from plane waves propagating through the sample’s initial estimated 3D RI. The scattered fields resulting from this simulation are compared with those obtained experimentally with our TPM systems. The error computed between the simulated and experimental measurements is back-propagated through each layer of the 3D sample estimate to incrementally modify the RI value of each voxel. Continued iterations repeating these steps eventually result in the 3D sample estimate converging to a stable steady-state solution. We implemented the MSBP with electric-field measurements (MSBP-E) through the Learning Tomography algorithm (LT)42. The LT procedure is an iterative optimization algorithm with additional weak total-variation (TV) regularization applied in each iteration to ensure convergence. We found that the method works best when an initial-guess is chosen as a starting point for the iterative process. In this paper, we use the Direct Inversion method to provide the initial guess.
Multi-slice beam-propagation with intensity-only measurements
Recent works have demonstrated that the gradient-update step within the MSBP method can be reformulated to reconstruct 3D RI from only non-interferometric intensity measurements31. The key advantages of this method include the use of a non-interferometric imaging system, which are resistant to mechanical instabilities that often limit long-term use of dual-arm interferometers without realignment. Furthermore, the light source can be partially coherent, to avoid coherent speckle artifacts in the measurements, while retaining sufficient coherence necessary for RI reconstruction. For the purposes of demonstrating 3D RI reconstruction using this intensity-only variant of MSBP (which we refer to here as MSBP-I), we simply use the amplitude component of the electric-field measurements used for the GPSC and MSBP-E reconstructions, described above. Similarly to MSBP-E, total-variation regularization is applied at every iteration. The starting point for MSBP-I is a matrix of zeros.
Stopping criterion
All described TPM reconstruction methods are iterative procedures that use the same stopping criterion to automatically terminate the computations. This criterion is a modification of a method presented earlier64, and is described in Alg. 1 below. The general intuition behind this procedure is to terminate the iterative computation process when the dynamics of the change between 3D sample estimates outputted from consecutive iterations drops below a certain saturation level . In order to be less dependent on outliers, the median value of the dynamics from the last 10 iterations is calculated. The value for is chosen empirically for each algorithm. For GPSC , for LT and MS . Values of were empirically chosen to balance between incomplete convergence and reconstruction speed.
Acknowledgements
The authors want to acknowledge prof. Maciej Szkulmowski from Nicolaus Copernicus University for providing access to the near-infrared light source used in this work. The authors want to thank Demetri Psaltis and Joowon Lim from École Polytechnique Fédérale de Lausanne for providing the code for Learning Tomography algorithm.
Author contributions
CRediT. W.K.: conceptualization, formal analysis, methodology, software, visualization, writing–original draft. A.K.: conceptualization, funding acquisition, investigation, methodology, project administration, visualization, writing–original draft. M.Z.: data curation, investigation, methodology, resources, visualization, writing–original draft. M.H.: software, writing–review & editing. S.C.: funding acquisition, software, writing–review & editing. M.K.: funding acquisition, project administration, writing–review & editing.
Funding
The research leading to the described results was carried out within the program TEAM TECH/2016-1/4 of Foundation for Polish Science, co-financed by the European Union under the European Regional Development Fund. Calibration object development was funded by FOTECH-1 project granted by Warsaw University of Technology under the program Excellence Initiative: Research University (ID-UB). We also gratefully acknowledge support from the University of Texas at Austin, Cockrell School of Engineering, and the Chan Zuckerberg Initiative.
Data availability
Data underlying the results presented in this paper are available in Dataset 140.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yoon S, Kim M, Jang M, Choi Y, Choi W, Kang S, Choi W. Deep optical imaging within complex scattering media. Nature Rev. Phys. 2020;2:141–158. [Google Scholar]
- 2.Chatzinikolaidou M. Cell spheroids: The new frontiers in in vitro models for cancer drug validation. Drug Discov. Today. 2016;21:1553–1560. doi: 10.1016/j.drudis.2016.06.024. [DOI] [PubMed] [Google Scholar]
- 3.Pierré W, Hervé L, Paviolo C, Mandula O, Remondiere V, Morales S, Grudinin S, Ray PF, Dhellemmes M, Arnoult C, Allier C. 3D time-lapse imaging of a mouse embryo using intensity diffraction tomography embedded inside a deep learning framework. Appl. Opt. 2022;61:3337. doi: 10.1364/AO.453910. [DOI] [PubMed] [Google Scholar]
- 4.Rios AC, Clevers H. Imaging organoids: A bright future ahead. Nature Methods. 2018;15:24–26. doi: 10.1038/nmeth.4537. [DOI] [PubMed] [Google Scholar]
- 5.Dekkers JF, Alieva M, Wellens LM, Ariese HC, Jamieson PR, Vonk AM, Amatngalim GD, Hu H, Oost KC, Snippert HJ, Beekman JM, Wehrens EJ, Visvader JE, Clevers H, Rios AC. High-resolution 3D imaging of fixed and cleared organoids. Nature Protocols. 2019;14:1756–1771. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- 6.Lim J, Lee K, Jin KH, Shin S, Lee S, Park Y, Ye JC. Comparative study of iterative reconstruction algorithms for missing cone problems in optical diffraction tomography. Opt. Express. 2015;23:16933–16948. doi: 10.1364/OE.23.016933. [DOI] [PubMed] [Google Scholar]
- 7.Sung Y, Dasari RR. Deterministic regularization of three-dimensional optical diffraction tomography. JOSA A. 2011;28:1554–1561. doi: 10.1364/JOSAA.28.001554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindi, H. A tutorial on convex optimization. In Proceedings of the 2004 American Control Conference, vol. 4 3252–3265 (IEEE, 2004).
- 9.Lamont AC, Restaino MA, Alsharhan AT, Liu Z, Hammer DX, Sochol RD, Agrawal A. Direct laser writing of a titanium dioxide-laden retinal cone phantom for adaptive optics-optical coherence tomography. Opt. Mater. Express. 2020;10:2757–2767. [Google Scholar]
- 10.LaFratta CN, Baldacchini T. Two-photon polymerization metrology: Characterization methods of mechanisms and microstructures. Micromachines. 2017;8:101. [Google Scholar]
- 11.Horng H, O’Brien K, Lamont A, Sochol RD, Pfefer TJ, Chen Y. 3d printed vascular phantoms for high-resolution biophotonic image quality assessment via direct laser writing. Opt. Lett. 2021;46:1987–1990. doi: 10.1364/OL.412849. [DOI] [PubMed] [Google Scholar]
- 12.Eifler M, Hering J, Von Freymann G, Seewig J. Calibration sample for arbitrary metrological characteristics of optical topography measuring instruments. Opt. Express. 2018;26:16609–16623. doi: 10.1364/OE.26.016609. [DOI] [PubMed] [Google Scholar]
- 13.Vora HD, Sanyal S. A comprehensive review: Metrology in additive manufacturing and 3D printing technology. Progress Addit. Manufact. 2020;5:319–353. [Google Scholar]
- 14.Bellec M, Royon A, Bousquet B, Bourhis K, Treguer M, Cardinal T, Richardson M, Canioni L. Beat the diffraction limit in 3D direct laser writing in photosensitive glass. Opt. Express. 2009;17:10304. doi: 10.1364/oe.17.010304. [DOI] [PubMed] [Google Scholar]
- 15.Kotz F, Arnold K, Bauer W, Schild D, Keller N, Sachsenheimer K, Nargang TM, Richter C, Helmer D, Rapp BE. Three-dimensional printing of transparent fused silica glass. Nature. 2017;544:337–339. doi: 10.1038/nature22061. [DOI] [PubMed] [Google Scholar]
- 16.Jonušauskas L, Rekštytė S, Buividas R, Butkus S, Gadonas R, Juodkazis S, Malinauskas M. Hybrid subtractive-additive-welding microfabrication for lab-on-chip applications via single amplified femtosecond laser source. Opt. Eng. 2017;56:1. [Google Scholar]
- 17.Varapnickas, S. & Malinauskas, M. Processes of laser direct writing 3D nanolithography. In Handbook of Laser Micro- and Nano-Engineering Vol. 12, 1–31 (Springer International Publishing, 2020).
- 18.Yang L, Mayer F, Bunz UHF, Blasco E, Wegener M. Multi-material multi-photon 3D laser micro- and nanoprinting. Light: Adv. Manufact. 2021;2:1. [Google Scholar]
- 19.Girshovitz P, Shaked NT. Generalized cell morphological parameters based on interferometric phase microscopy and their application to cell life cycle characterization. Biomed. Opt. Express. 2012;3:1757–1773. doi: 10.1364/BOE.3.001757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y, Shim H, Kim K, Park H, Heo JH, Yoon J, Choi C, Jang S, Park Y. Common-path diffraction optical tomography for investigation of three-dimensional structures and dynamics of biological cells. Opt. Express. 2014;22:10398–10407. doi: 10.1364/OE.22.010398. [DOI] [PubMed] [Google Scholar]
- 21.Stanly TA, Suman R, Rani GF, O’Toole PJ, Kaye PM, Hitchcock IS. Quantitative optical diffraction tomography imaging of mouse platelets. Front. Physiol. 2020;11:568087. doi: 10.3389/fphys.2020.568087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baczewska M, Stȩpień P, Mazur M, Krauze W, Nowak N, Szymański J, Kujawińska M. Method to analyze effects of low-level laser therapy on biological cells with a digital holographic microscope. Appl. Opt. 2022;61:B297–B306. doi: 10.1364/AO.445337. [DOI] [PubMed] [Google Scholar]
- 23.Baczewska M, Eder K, Ketelhut S, Kemper B, Kujawińska M. Refractive index changes of cells and cellular compartments upon paraformaldehyde fixation acquired by tomographic phase microscopy. Cytometry Part A. 2021;99:388–398. doi: 10.1002/cyto.a.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eder, K., Marzi, A., Barroso, Á., Kemper, B., & Schnekenburger, J. Medical nanoparticle impact on macrophage temporal dry mass development quantified in vitro by digital holographic microscopy. In Quantitative Phase Imaging VII, vol. 11653 132–138 (SPIE, 2021).
- 25.Chen B, Stamnes JJ. Validity of diffraction tomography based on the first Born and the first Rytov approximations. Appl. Opt. 1998;37:2996–3006. doi: 10.1364/ao.37.002996. [DOI] [PubMed] [Google Scholar]
- 26.Lee M, Hugonnet H, Park Y. Inverse problem solver for multiple light scattering using modified Born series. Optica. 2022;9:177–182. [Google Scholar]
- 27.Liu H-Y, Liu D, Mansour H, Boufounos PT, Waller L, Kamilov US. Seagle: Sparsity-driven image reconstruction under multiple scattering. IEEE Trans. Comput. Imaging. 2017;4:73–86. [Google Scholar]
- 28.Sun Y, Xia Z, Kamilov US. Efficient and accurate inversion of multiple scattering with deep learning. Opt. Express. 2018;26:14678–14688. doi: 10.1364/OE.26.014678. [DOI] [PubMed] [Google Scholar]
- 29.Soubies E, Pham T-A, Unser M. Efficient inversion of multiple-scattering model for optical diffraction tomography. Opt. Express. 2017;25:21786–21800. doi: 10.1364/OE.25.021786. [DOI] [PubMed] [Google Scholar]
- 30.Chen M, Ren D, Liu H-Y, Chowdhury S, Waller L. Multi-layer Born multiple-scattering model for 3d phase microscopy. Optica. 2020;7:394–403. [Google Scholar]
- 31.Chowdhury S, Chen M, Eckert R, Ren D, Wu F, Repina N, Waller L. High-resolution 3D refractive index microscopy of multiple-scattering samples from intensity images. Optica. 2019;6:1211. doi: 10.1364/optica.6.001211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim J, Wahab A, Park G, Lee K, Park Y, Ye JC. Beyond Born-Rytov limit for super-resolution optical diffraction tomography. Opt. Express. 2017;25:30445–30458. doi: 10.1364/OE.25.030445. [DOI] [PubMed] [Google Scholar]
- 33.Ziemczonok M, Kuś A, Wasylczyk P, Kujawińska M. 3D-printed biological cell phantom for testing 3D quantitative phase imaging systems. Sci. Rep. 2019;9:18872. doi: 10.1038/s41598-019-55330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid M, Ludescher D, Giessen H. Optical properties of photoresists for femtosecond 3D printing: Refractive index, extinction, luminescence-dose dependence, aging, heat treatment and comparison between 1-photon and 2-photon exposure. Opt. Mater. Express. 2019;9:4564–4577. [Google Scholar]
- 35.Marakis E, Uppu R, Meretska ML, Gorter K, Vos WL, Pinkse PWH. Deterministic and controllable photonic scattering media via direct laser writing. Adv. Opt. Mater. 2020;8:2001438. [Google Scholar]
- 36.Ziemczonok M, Kuś A, Kujawińska M. Optical diffraction tomography meets metrology—measurement accuracy on cellular and subcellular level. Measurement. 2022;195:111106. [Google Scholar]
- 37.Horstmeyer R, Heintzmann R, Popescu G, Waller L, Yang C. Standardizing the resolution claims for coherent microscopy. Nature Photon. 2016;10:68–71. [Google Scholar]
- 38.Cotte Y, Toy F, Jourdain P, Pavillon N, Boss D, Magistretti P, Marquet P, Depeursinge C. Marker-free phase nanoscopy. Nature Photon. 2013;7:113–117. [Google Scholar]
- 39.Debailleul M, Georges V, Simon B, Morin R, Haeberlé O. High-resolution three-dimensional tomographic diffractive microscopy of transparent inorganic and biological samples. Opt. Lett. 2009;34:79–81. doi: 10.1364/ol.34.000079. [DOI] [PubMed] [Google Scholar]
- 40.Krauze, W., Kuś, A., Ziemczonok, M., Haimowitz, M., Chowdhury, S., & Kujawińska, M. Optical tomography measurements and reconstructions of a multiple-scattering 3d-printed microphantom. Zenodo (2022). 10.5281/zenodo.7233563.
- 41.Krauze W. Optical diffraction tomography with finite object support for the minimization of missing cone artifacts. Biomed. Opt. Express. 2020;11:1919–1926. doi: 10.1364/BOE.386507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamilov US, Papadopoulos IN, Shoreh MH, Goy A, Vonesch C, Unser M, Psaltis D. Learning approach to optical tomography. Optica. 2015;2:517. [Google Scholar]
- 43.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nature Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 44.Rubart M. Two-photon microscopy of cells and tissue. Circul. Res. 2004;95:1154–1166. doi: 10.1161/01.RES.0000150593.30324.42. [DOI] [PubMed] [Google Scholar]
- 45.Strong D, Chan T. Edge-preserving and scale-dependent properties of total variation regularization. Inverse Problems. 2003;19:S165. [Google Scholar]
- 46.Wang Z, Millet L, Mir M, Ding H, Unarunotai S, Rogers J, Gillette MU, Popescu G. Spatial light interference microscopy (slim) Opt. Express. 2011;19:1016–1026. doi: 10.1364/OE.19.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian L, Waller L. Quantitative differential phase contrast imaging in an led array microscope. Opt. Express. 2015;23:11394–11403. doi: 10.1364/OE.23.011394. [DOI] [PubMed] [Google Scholar]
- 48.Liao C, Wuethrich A, Trau M. A material odyssey for 3D nano/microstructures: Two photon polymerization based nanolithography in bioapplications. Appl. Mater. Today. 2020;19:100635. [Google Scholar]
- 49.Simon B, Debailleul M, Beghin A, Tourneur Y, Haeberlé O. High-resolution tomographic diffractive microscopy of biological samples. J. Biophoton. 2010;3:462–467. doi: 10.1002/jbio.200900094. [DOI] [PubMed] [Google Scholar]
- 50.Simon B, Debailleul M, Houkal M, Ecoffet C, Bailleul J, Lambert J, Spangenberg A, Liu H, Soppera O, Haeberlé O. Tomographic diffractive microscopy with isotropic resolution. Optica. 2017;4:460–463. [Google Scholar]
- 51.Lauer V. New approach to optical diffraction tomography yielding a vector equation of diffraction tomography and a novel tomographic microscope. J. Microscopy. 2002;205:165–176. doi: 10.1046/j.0022-2720.2001.00980.x. [DOI] [PubMed] [Google Scholar]
- 52.Kim K, Yoon H, Diez-Silva M, Dao M, Dasari RR, Park Y. High-resolution three-dimensional imaging of red blood cells parasitized by plasmodium falciparum and in situ hemozoin crystals using optical diffraction tomography. J. Biomed. Opt. 2013;19:011005. doi: 10.1117/1.JBO.19.1.011005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wedberg T, Wedberg W. Tomographic reconstruction of the cross-sectional refractive index distribution in semi-transparent, birefringent fibres. J. Microsc. 1995;177:53–67. [Google Scholar]
- 54.Bunea, A.-I., del Castillo Iniesta, N., Droumpali, A., Wetzel, A. E., Engay, E., & Taboryski, R.Micro 3D printing by two-photon polymerization: Configurations and parameters for the nanoscribe system. Micro 1, 164–180 (2021).
- 55.Kuś A. Illumination-related errors in limited-angle optical diffraction tomography. Appl. Opt. 2017;56:9247–9256. doi: 10.1364/AO.56.009247. [DOI] [PubMed] [Google Scholar]
- 56.Kuś A, Krauze W, Makowski PL, Kujawińska M. Holographic tomography: Hardware and software solutions for 3D quantitative biomedical imaging (Invited paper) ETRI J. 2019;41:61–72. [Google Scholar]
- 57.Sánchez-Ortiga E, Doblas A, Saavedra G, Martínez-Corral M, Garcia-Sucerquia J. Off-axis digital holographic microscopy: Practical design parameters for operating at diffraction limit. Appl. Opt. 2014;53:2058–2066. doi: 10.1364/AO.53.002058. [DOI] [PubMed] [Google Scholar]
- 58.Chambolle A, Pock T. A first-order primal-dual algorithm for convex problems with applications to imaging. J. Math. Imaging Vis. 2011;40:120–145. [Google Scholar]
- 59.Van Aarle W, Palenstijn WJ, De Beenhouwer J, Altantzis T, Bals S, Batenburg KJ, Sijbers J. The ASTRA Toolbox: A platform for advanced algorithm development in electron tomography. Ultramicroscopy. 2015;157:35–47. doi: 10.1016/j.ultramic.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Wolf E. Three-dimensional structure determination of semi-transparent objects from holographic data. Opt. Commun. 1969;1:153–156. [Google Scholar]
- 61.Haeberlé O, Belkebir K, Giovaninni H, Sentenac A. Tomographic diffractive microscopy: Basics, techniques and perspectives. J. Modern Opt. 2010;57:686–699. [Google Scholar]
- 62.Van Roey J, Van der Donk J, Lagasse P. Beam-propagation method: Analysis and assessment. JOSA. 1981;71:803–810. [Google Scholar]
- 63.Kamilov US, Papadopoulos IN, Shoreh MH, Goy A, Vonesch C, Unser M, Psaltis D. Optical tomographic image reconstruction based on beam propagation and sparse regularization. IEEE Trans. Comput. Imaging. 2016;2:59–70. [Google Scholar]
- 64.Makowski PL, Ziemczonok M. Projection extrapolation routine for tight-frame limited-angle optical diffraction tomography. Opt. Lett. 2019;44:3442–3445. doi: 10.1364/OL.44.003442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data underlying the results presented in this paper are available in Dataset 140.