Abstract
Objectives:
Studies have demonstrated immune dysfunction in adolescents with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS); however, evidence is varied. The current study used network analysis to examine relationships between cytokines among a sample of pediatric participants with ME/CFS.
Methods:
10,119 youth aged 5–17 in the Chicagoland area were screened for ME/CFS; 111 subjects and controls were brought in for a physician examination and completed a blood draw. Youth were classified as controls (Cs, N = 43), ME/CFS (N = 23) or severe (S-ME/CFS, N = 45). Patterns of plasma cytokine networks were analyzed.
Results:
All participant groups displayed a primary network of interconnected cytokines. In the ME/CFS group, inflammatory cytokines IL-12p70, IL-17A, and IFN-γ were connected and included in the primary membership, suggesting activation of inflammatory mechanisms. The S-ME/CFS group demonstrated a strong relationship between IL-17A and IL-23, a connection associated with chronic inflammation. The relationships of IL-6 and IL-8 in ME/CFS and S-ME/CFS participants also differed from Cs. Together, these results indicate pro-inflammatory responses in our illness populations.
Discussion:
Our data imply biological differences between our three participant groups, with ME/CFS and S-ME/CFS participants demonstrating an inflammatory profile. Examining co-expression of cytokines may aid in the identification of a biomarker for pediatric ME/CFS.
Keywords: Myalgic encephalomyelitis/chronic fatigue syndrome, pediatrics, cytokine networks, network analysis
Introduction
Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) is a complex illness that can cause significant impairment for adolescents, including severe exhaustion and disruption of social and academic activities.1,2 Several case definitions have been developed in attempts to recognize the illness in adults and some case definitions have been geared towards children and adolescents.3,4
There are multiple possible explanations regarding the cause of ME/CFS, and some studies have focused on immune dysfunction in adolescents with ME/CFS. For example, Sulheim et al. and Kristiansen et al. reported slightly elevated levels of C-reactive protein (CRP) in adolescents with ME/CFS, suggesting low-grade systemic inflammation.5,6 Additionally, ter Wolbeek et al. found increased expression of the anti-inflammatory cytokine IL-10, as well as reduced levels of pro-inflammatory cytokines IL-6 and TNF-α among adolescents with ME/CFS compared to controls and severely fatigued participants who did not meet criteria for ME/CFS.7 Others, however, have found no abnormalities in cytokine levels in adolescents with ME/CFS.8
While these studies have examined levels of individual cytokines, immune cells function as a distributed network.9 Several investigations have therefore used network analysis to examine cytokine relationships in adults with ME/CFS.10–12 With an adolescent sample, Wyller et al. found that plasma cytokine levels and cytokine network parameters were similar across ME/CFS and healthy control groups.13 However, participants with ME/CFS did show an upregulation of genes involved in inflammation compared to a healthy control group.14
Broderick et al. demonstrated the utility of cytokines as a biomarker to identify ME/CFS in a study analyzing the co-expression of cytokines in adolescents.15 Twenty-four months after infectious mononucleosis (IM), youth with ME/CFS showed higher levels of IL-2 and IL-6, lower levels of IL-23 and higher levels of IL-8 in those with ME/CFS than those without. The co-expression of these cytokines relative to the expression of IFN-γ was the basis for a classification model, allowing researchers to identify participants as cases or recovered controls with over 80% accuracy.
The current study used network analysis to examine the relationships between cytokines among a sample of pediatric patients with ME/CFS. Through network analysis, patterns of cytokine expression may be found to identify those with ME/CFS versus healthy controls and thus may serve as a biomarker for diagnosis or provide insight into specific therapeutic strategies.
Methods
Participants
Details of enrollment for this study have been previously published.16 Briefly, a sample of 10,119 youth aged 5–17 years from 5622 households in the Chicagoland area were screened for prolonged, unexplained fatigue and additional ME/CFS symptoms. Select participants were invited to the Ann & Robert H Lurie Children’s Hospital for a medical evaluation. Blood samples were taken from 68 screen-positive participants and 43 screen-negative, demographically matched controls. The average age of the sample was 13.75 years, 47.7% were white, and 57.7% were female. Serum and plasma were stored.
Cytokine analysis
For cytokine analysis, stored plasma was used. The following cytokines were evaluated using a multiple analyte platform and commercially customized kits from Millipore (Billerica, MA): IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12p70, IL-13, IL-15, IL-17A, IL-23, IFN-γ, TNF-α and TNF-β. Cytokine values below the detection limit were replaced with zero (792 of 1776 cytokine values; 44.6%). Cytokine concentrations were measured (pg/ml) and analyzed after logarithmic (x + 1) transformation. Two individuals had cytokine values outside of the limits of detection (2 of 1776 cytokine values; 0.1%) and these values were omitted from analyses.
ME/CFS diagnosis
Results from the DePaul Symptom Questionnaire (DSQ), the Child Health Questionnaire (CHQ), and a medical evaluation were used to determine whether participants met one or more of three case definitions of ME/CFS.17–21 The Fukuda et al. criteria is the least selective and brings in a wider number of patients.19,22–24 In addition, participants who met > 1 case definition for ME/CFS scored higher on the DSQ and CHQ than those who only met a single case definition. Participants who met no case definitions were labeled controls (Cs), those who met at least one case definition (almost always the Fukuda et al. criteria) were classified as having ME/CFS, and those who met more than one case definition (the Fukuda et al. criteria and at least either the Carruthers et al. criteria or the IOM criteria) were categorized as having severe ME/CFS (S-ME/CFS).19–21,25,26 There were 43 Cs, 23 with ME/CFS, and 45 participants with S-ME/CFS.
Statistical analysis
As cytokine data are non-normally distributed, all correlations were conducted using Spearman’s Rho, and we used network statistics to detect patterns among these correlations. Network statistics were dependent upon what correlation threshold value is selected. Generally a threshold between 0 and 1 is selected, where correlations above the selected threshold are used to detect patterns and correlations below the threshold are set to 0 (indicating no connection between cytokines).
In order to distinguish between the three participant groups (Cs, ME/CFS, S-ME/CFS), we created a distribution of scores for each group, as opposed to calculating a single score, using the leave-one-out-cross-validation (LOOCV) technique.27 A distribution of correlational matrices was thus created for each group by removing a single individual from each iteration and correlating the remaining values for each participant group. This created a sample of correlational matrices identical in number to the number of participants in each group: 45 for the S-ME/CFS group, 23 for the ME/CFS group, and 43 for the Cs group.
Patterns of networks were classified according to the following characteristics: membership, modularity, Eigen centrality, total centrality and mean degree. Network statistics were previously described.28 Memberships are calculated by the Louvain method for indicating how many groups of cytokines exist within each of the three groups of participants (Cs, ME/CFS, S-ME/CFS). If a group of participants has only one membership, all cytokines within that group are inter-connected, whereas if there are several memberships, then there are several distinct patterns or groups of cytokines. Modularity provides evidence of the validity of the membership categories. Modularity can range from 0 to 1, and scores that are higher indicate more validity for the grouping of the cytokines; a modularity closer to 1 therefore indicates stronger connections within a group of cytokines but weaker connections between different cytokine groups. Eigen centrality, which is averaged for each cytokine, is a measure of the relative importance of a specific cytokine in connecting a network. Cytokines higher on Eigen centrality are those whose removal from the network would result in the most network fragmentation; in other words, such cytokines have a strong linking function in the network. For the network as a whole, Eigen centrality takes the value of 1.0 when one central cytokine is connected to all others, none of which are connected to each other, and zero when all cytokines serves as an intermediary for only one other cytokine. Total centrality, measuring the relative importance of a specific cytokine in connecting a network, is evaluated as a summation of the Eigen centralities of each cytokine in the overall network. Finally, mean degree indicates the average number of connections a specific cytokine forms with other cytokines. A cytokine with a mean degree of four indicates that the cytokine in question links to four other cytokines. These network statistics were compared across groups using a one-way analysis of variance (ANOVA) with post-hoc Bonferroni corrections.
Results
We used modularity statistics for each participant group in order to determine the most valid threshold value. As seen in Figure 1, correlations near 0.6 or 0.7 mark a change-in-slope for each of the three participant groups, indicating that the connection between the cytokines in the groups appears significant around these points. Given the visual representation of network graphs using thresholds within this range, 0.65 provided the best differentiation of cytokines for the current analysis and gave a clear depiction of relationships; therefore, this correlational value was utilized in further analyses for all participant groups. Thicker lines in subsequent figures indicate stronger correlations. Supplementary Tables 1, 2 and 3 present the correlations (strengths of the associations) between the cytokines for the Cs, ME/CFS and S-ME-CFS groups, respectively.
Figure 1.
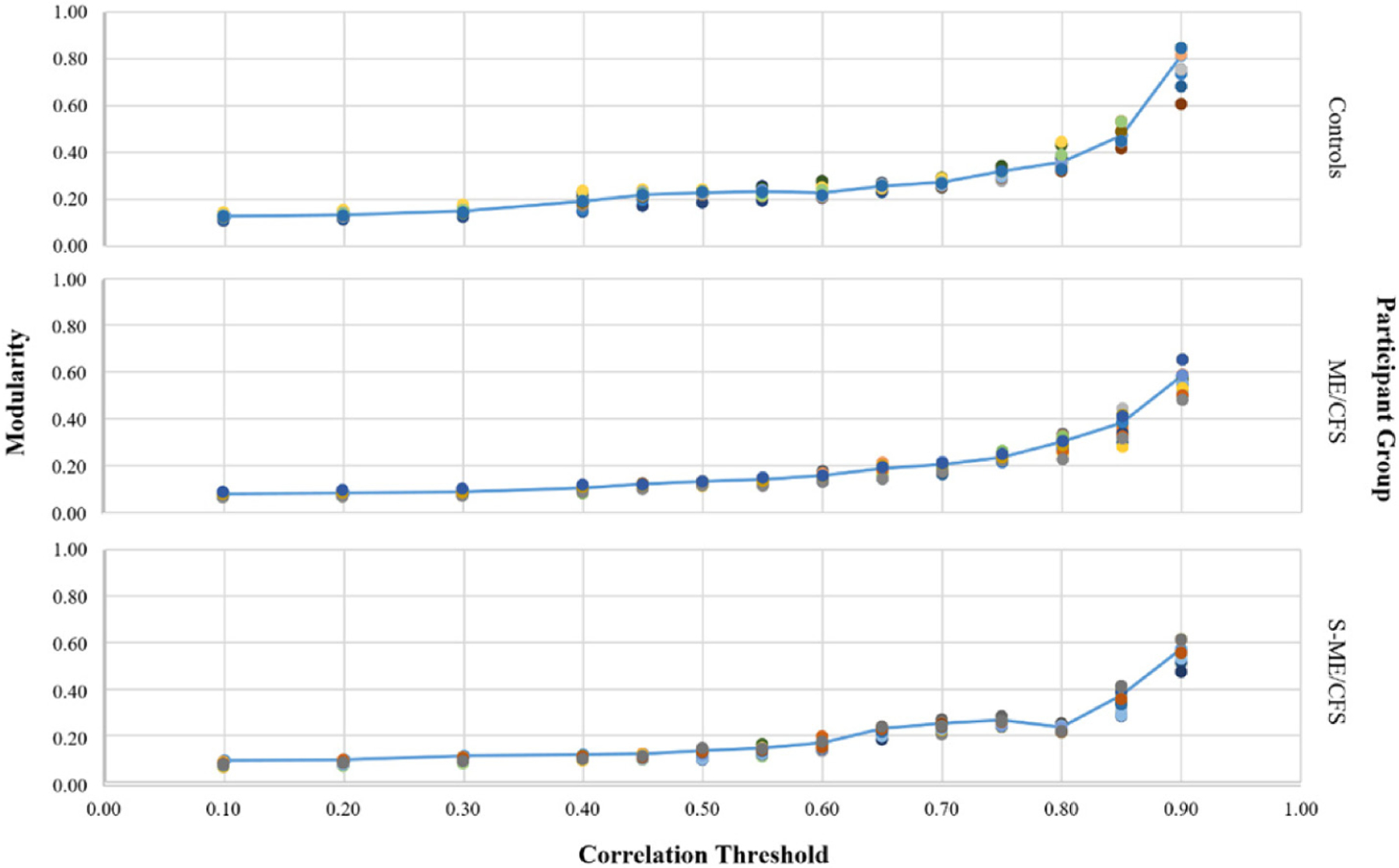
Modularity by threshold criterion for each participant group.
Table 1 shows the network statistics results for each group. Cs and those with S-ME/CFS had similar memberships, but both groups had significantly more memberships than those with ME/CFS. All groups were significantly different from others on remaining network statistics. Cs had the highest modularity, followed by S-ME/CFS and ME/CFS, demonstrating stronger division between cytokine communities. ME/CFS participants had a significantly greater degree of centrality and a greater average number of connections (mean connectedness) between cytokines compared to controls. The S-ME/CFS group had measures intermediate between Cs and those with ME/CFS.
Table 1.
Comparison of network statistics by condition.
| Controls | ME/CFS | S-ME/CFS | |
|---|---|---|---|
| Memberships | 6.16(0.37)a | 4.26(0.54)ab | 6.00(0.00)b |
| Modularity | 0.26(0.01)a | 0.19(0.02)a | 0.23(0.01)a |
| Average Eigen Centrality | 0.50(0.01)a | 0.62(0.02)a | 0.57(0.01)a |
| Total Eigen Centrality | 8.02(0.10)a | 9.93(0.39)a | 9.14(0.17)a |
| Mean Degree | 6.53(0.10)a | 9.01(0.44)a | 7.34(0.21)a |
Note. Rows with a similar letter superscript are significantly different at the p < 0.01 level.
Figures 2, 3, and 4 show representative network graphs of Cs, ME/CFS, and S-ME/CFS participants. Cytokines are color coded according to each cytokine’s main functioning: pro-inflammatory colored in red, and anti-inflammatory colored in blue; cytokines not colored are multi-purpose. In the Cs group (Figure 2), a primary association of 10 cytokines is present. A secondary membership is formed involving a weak relationship between IL12p70, IFN-γ, and IL-17A, while IL-2, IL-1β, and TNF-α are not a part of the network. In the ME/CFS group (Figure 3), the association between IL-12p70, IFN-γ, and IL-17A is stronger than in Cs and they are now weakly connected to the primary membership, along with IL-1β. Only IL-2 is removed from the network. Note also the stronger association of IL-8 and IL-23 with IL-17A and the other cytokines in the network in the ME/CFS and especially in the S-ME/CFS groups (Supplementary Tables 1–3). The pattern between IL-12p70 and TNF-α observed in ME/CFS is lost in the participants with S-ME/CFS, as they are separated from the primary association network (Figure 4), where a primary membership of 13 cytokines is present; IL-17A, however, remains and is more strongly attached to the primary network in the S-ME/CFS group; IL-1β is also attached to the network in the S-ME/CFS as well as in the ME/CFS groups. As noted in Supplemental Table 3, there was also a stronger positive relationship between IL-6 and IL-23 as well as IL-5 and IL-8 in the S-ME/CFS (r = .84 and 0.83, respectively) and ME/CFS (r = .79 and.84, respectively [Supplementary Table 2]) groups than Cs (r = .60 and.73, respectively [Supplementary Table 1]).
Figure 2.
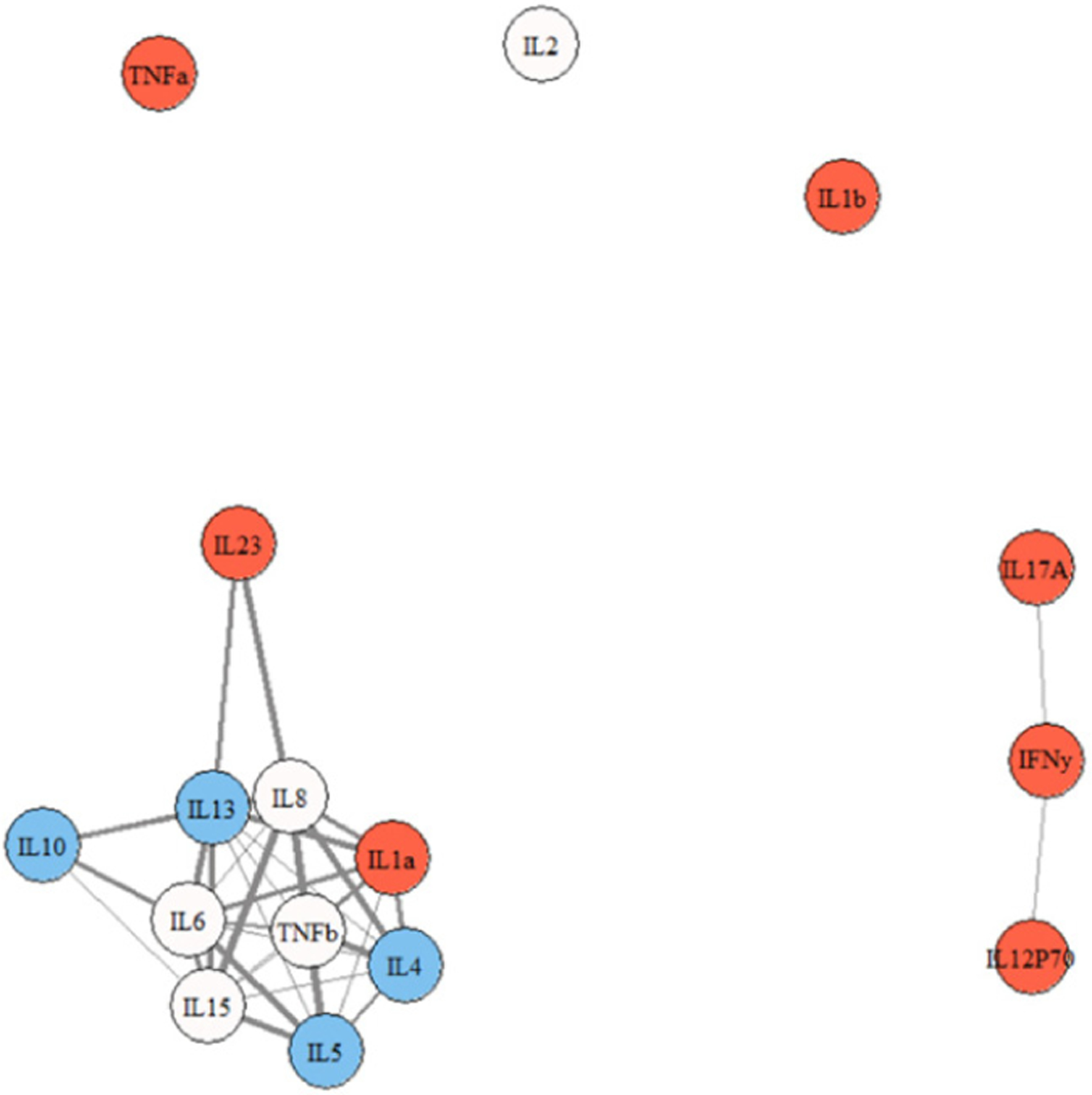
Cytokine network graph for Cs. Note. thicker lines indicate stronger correlations. Note. red = pro-inflammatory; blue = anti-inflammatory; not colored = multi-purpose.
Figure 3.
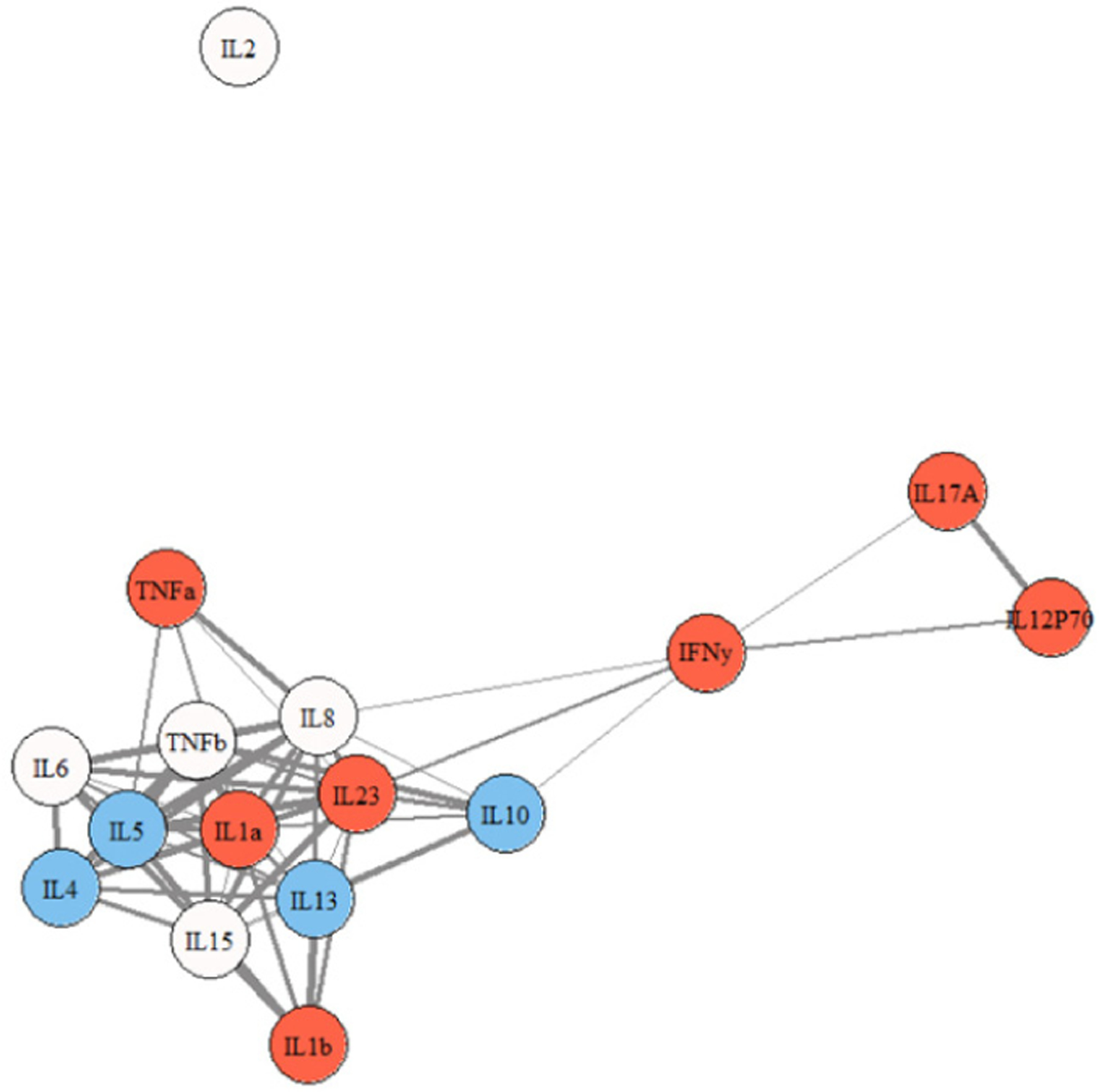
Cytokine network graph for participants with ME/CFS. Note. thicker lines indicate stronger correlations. Note. red = pro-inflammatory; blue = anti-inflammatory; not colored = multi-purpose.
Figure 4.
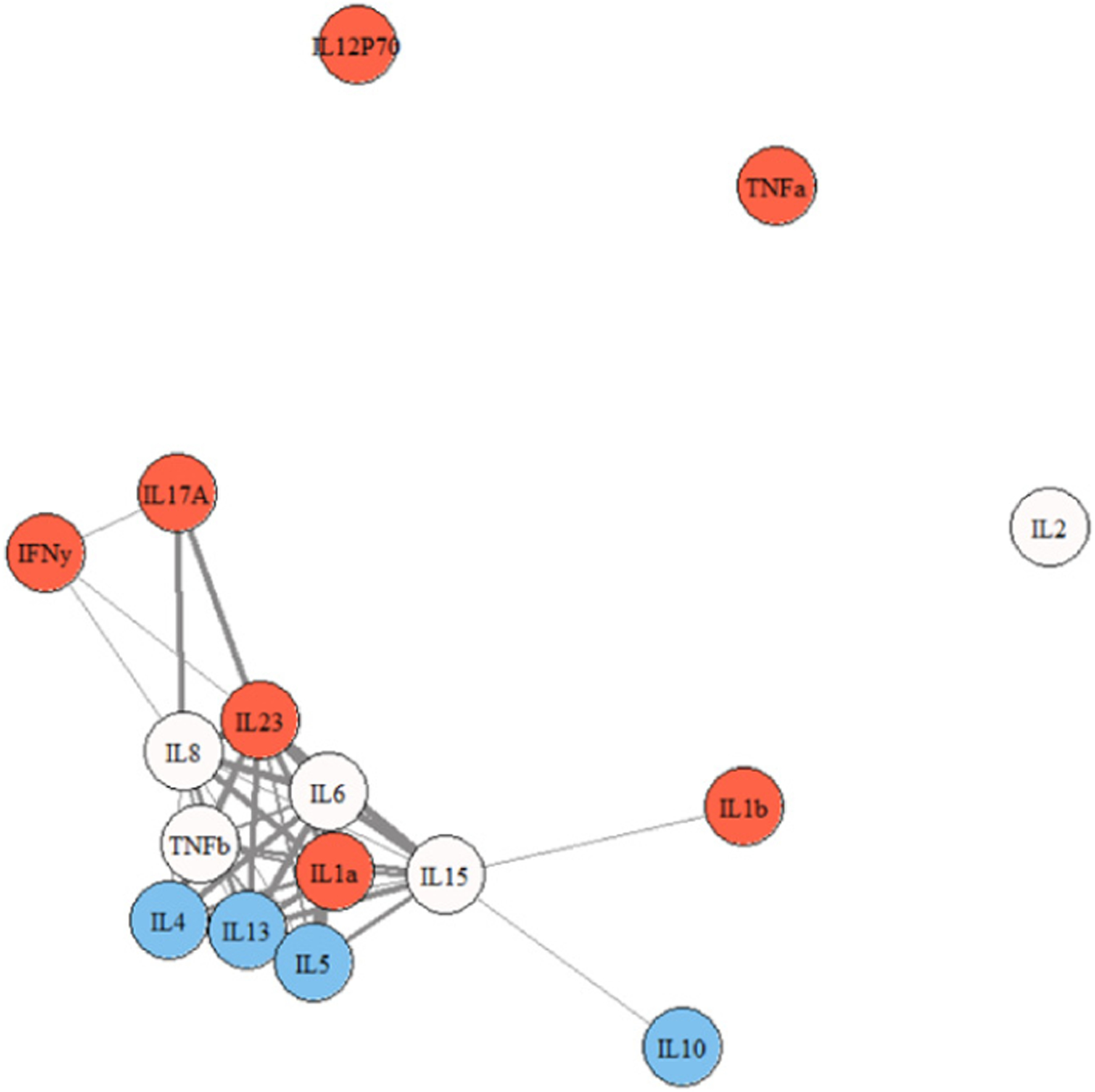
Cytokine network graph for participants with S-ME/CFS. Note. thicker lines indicate stronger correlations. Note. red = pro-inflammatory; blue = anti-inflammatory; not colored = multi-purpose.
Figure 5 shows the differences in the Eigen centrality values of each cytokine across groups, and Figure 6 shows the average number of connections a specific cytokine forms with other cytokines across participant groups. A value of 0 indicates no differences.
Figure 5.
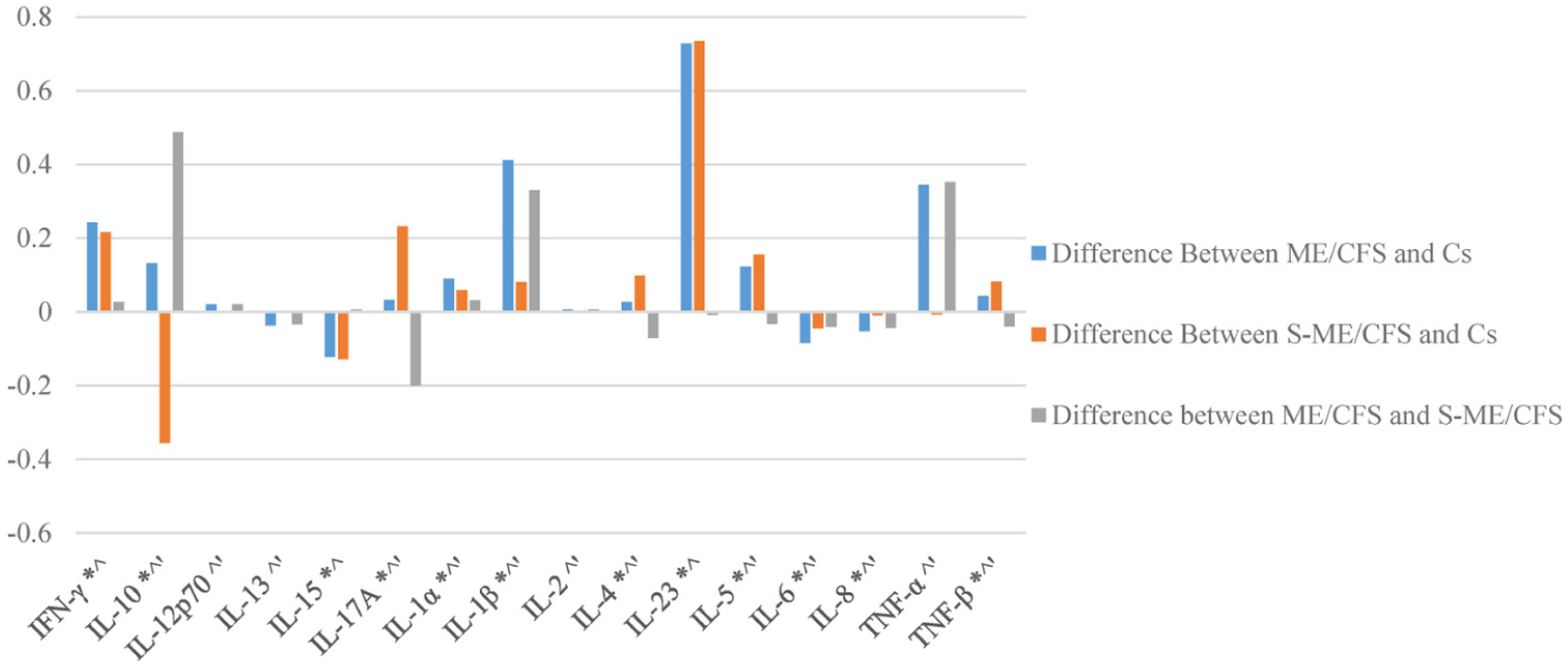
Differences in eigen centrality values for each cytokine among patient and control groups. * S-ME/CFS is significantly different from Cs. ^ ME/CFS is significantly different from Cs. ‘ ME/CFS is significantly different from S-ME/CFS.
Figure 6.
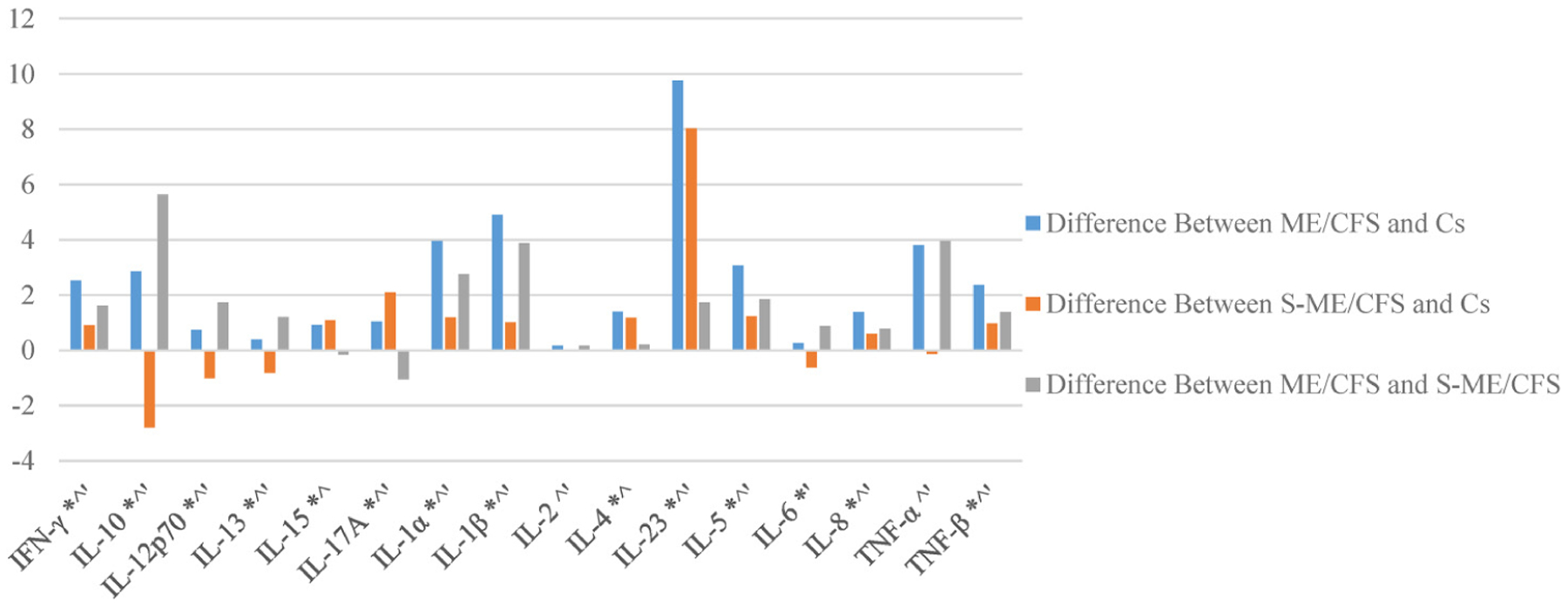
Differences in mean connections within cytokine groups for each cytokine among patient and control groups. * S-ME/CFS is significantly different from Cs. ^ ME/CFS is significantly different from Cs. ‘ ME/CFS is significantly different from S-ME/CFS.
Discussion
Children and adolescents in our ME/CFS and S-ME/CFS groups showed cytokine signatures indicative of inflammation that was not present in Cs. In the relatively healthy C group, the main markers of pro-inflammatory differentiation, IL-2 and IL-12p70, were not linked to the primary cytokine network. A major protein involved in viral defense, IFN-γ, was also missing, as was IL-17A, a cytokine important for innate immunity, and TNF-α, another important pro-inflammatory cytokine; this would support the notion of an immune system that is not activated in Cs.
The patterns seen in the ME/CFS and S-ME/CFS groups, on the other hand, indicate linking and activation of pro-inflammatory pathways. In the ME/CFS group, the relationship between IL-12p70, IFN-γ, and IL-17A was strong and weakly associated with the primary grouping. This could reflect activation of inflammation, given the role of IL-12p70 and IL-17A in the regulation of pro-inflammatory T cell lineage commitment. The pro-inflammatory cytokine, TNF-α, was also more intrinsically linked in the ME/CFS group, again likely reflecting a greater inflammatory environment. Of note also were the new relationships between IL-17A and cytokines IL-8 and IL-23 seen in the S-ME/CFS group. IL-17A and IL-23 both have significantly greater centrality and significantly more connections in these illness group networks than in controls (Figures 5 and 6). The production of IL-17A can result in activation of a subset of T cells that then produce IL-23. IL-23 in turn can lead to the differentiation of naïve T cells into IL-17A producing cells. This IL-17A/IL-23 axis is associated with chronic inflammation, suggesting a persistent inflammatory response in the S-ME/CFS group.29
Interestingly, TNF-α and IL-12p70 were separated from the primary membership among S-ME/CFS participants, similar to the controls, implying that these major differentiation proteins are no longer tied to the immunologic response network in this group of participants. The disassociation of IL-12p70 is unexpected given the ability of this protein to activate Th1 cells in a manner that facilitates release of IFN-γ. IFN-γ and IL-17A, however, remained in the primary grouping of cytokines, with IL-17A being even more strongly connected. This provides additional evidence for disruption of immune-regulatory processes in those with S-ME/CFS.
The network associations within the primary ME/CFS and S-ME/CFS groups were also reflective of a pattern of ongoing inflammation. Of note were the stronger associations of IL-8 and IL-6, cytokines both linked to inflammation and dysregulated immune responses in the groups with ME/CFS and S-ME/CFS.30,31 A recent study by Russell and colleagues suggested that these cytokines, along with IL-1α, adjusted for illness duration, may serve as biomarkers for pediatric ME/CFS.32 The association of IL-8 with IL-5 was stronger in the ME/CFS and S-ME/CFS populations than in Cs. IL-5 is a Th2 associated protein, and this increased relationship is congruent with might be expected as part of a physiologic response to increased inflammation in the ME/CFS group. Similarly, there was a stronger positive relationship among IL-6 and IL-23 in the ME/CFS and S-ME/CFS populations than Cs. This new connection with an inflammatory cytokine may provide further evidence for an inflammatory involvement in ME/CFS and S-ME/CFS.
Research using network analyses to examine cytokine relationships in pediatric ME/CFS is scarce and has produced mixed results. Our results are consistent with prior research on individual cytokine levels indicating a pro-inflammatory environment in pediatric ME/CFS.5,6,14,33 These results are also comparable to our previous study on cytokines networks among college students who developed ME/CFS following IM.28 In that study, young adults in the illness groups also showed distinct, more highly interconnected protein networks than observed in Cs even before the development of IM. While these two studies observed participants in different stages of illness (at the time of ME/CFS six months following IM in college students versus a random time point in younger adolescents with ME/CFS in this study), pro-inflammatory states were observed in both illness groups compared to healthy controls.
In the current study, using network analysis, we found noticeable shifts in cytokine connections and network structure between groups that are not observable when solely considering individual cytokine expression levels. Evaluating the co-expression of cytokines in this way implicates pro-inflammatory responses in youth with ME/CFS. Identifying biological components involved in the pathogenesis of pediatric ME/CFS, such as these patterns of cytokine expression and immune functioning, may lead to objective criteria for diagnosis and the identification of a potential therapeutic target.
Supplementary Material
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, (grant number R01 HD072208).
Guarantor
LAJ
Footnotes
Trial registration
Not applicable, because this article does not contain any clinical trials.
Informed consent
Written informed consent was attained from all study participants.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval
Ethical approval for this study was obtained from DePaul University, Office of Research Services, Institutional Review Board, Research Protocol #LJ071012PSY.
Supplemental material
Supplemental material for this article is available online.
References
- 1.Kennedy G, Underwood C, and Belch JJF. Physical and functional impact of chronic fatigue syndrome/myalgic encephalomyelitis in childhood. Pediatr 2010; 125: e1324–e1330. [DOI] [PubMed] [Google Scholar]
- 2.Nijhof SL, Maijer K, Bleijenberg G, et al. Adolescent chronic fatigue syndrome: prevalence, incidence, and morbidity. Pediatr 2011; 127: e1169–e1175. [DOI] [PubMed] [Google Scholar]
- 3.Brurberg KG, Fønhus MS, Larun L, et al. Case definitions for chronic fatigue syndrome/myalgic encephalomyelitis (CFS/ME): a systematic review. BMJ open 2014; 4: e003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jason LA, Jordan K, Miike T, et al. A pediatric case definition for myalgic encephalomyelitis and chronic fatigue syndrome. J Chronic Fatigue Syndr 2006; 13: 1–44. [Google Scholar]
- 5.Sulheim D, Fagermoen E, Winger A, et al. Disease mechanisms and clonidine treatment in adolescent chronic fatigue syndrome: a combined cross-sectional and randomized clinical trial. JAMA Pediatr 2014; 168: 351–360. [DOI] [PubMed] [Google Scholar]
- 6.Kristiansen MS, Stabursvik J, O’Leary EC, et al. Clinical symptoms and markers of disease mechanisms in adolescent chronic fatigue following Epstein-Barr virus infection: an exploratory cross-sectional study. Brain Behav Immun 2019; 80: 551–563. [DOI] [PubMed] [Google Scholar]
- 7.Wolbeek MT, Van Doornen LJ, Kavelaars A, et al. Longitudinal analysis of pro- and anti-inflammatory cytokine production in severely fatigued adolescents. Brain Behav Immun 2007; 21: 1063–1074. [DOI] [PubMed] [Google Scholar]
- 8.Wyller VB, Nguyen CB, Ludviksen JA, et al. Transforming growth factor beta (TGF-β) in adolescent chronic fatigue syndrome. J Transl Med 2017; 15: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang JM and An J. Cytokines, inflammation, and pain. Int Anesthesiol Clin 2007; 45: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broderick G, Fuite J, Kreitz A, et al. A formal analysis of cytokine networks in chronic fatigue syndrome. Brain Behav Immun 2010; 24: 1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornig M, Gottschalk G, Peterson DL, et al. Cytokine network analysis of cerebrospinal fluid in myalgic encephalomyelitis/chronic fatigue syndrome. Mol Psychiatry 2016; 21: 261–269. [DOI] [PubMed] [Google Scholar]
- 12.Sorenson M, Furst J, Matthews H, et al. Dysregulation of cytokine pathways in chronic fatigue syndrome and multiple sclerosis. Fatigue 2017; 5: 145–158. [Google Scholar]
- 13.Wyller BV, Sorenson O, Sulheim D, et al. Plasma cytokine expression in adolescent chronic fatigue syndrome. Brain Behav Immun 2015; 46: 80–86. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen CB, Alsøe L, Lindvall JM, et al. Whole blood gene expression in adolescent chronic fatigue syndrome: an exploratory cross-sectional study suggesting altered B cell differentiation and survival. J Transl Med 2017; 15: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broderick G, Katz BZ, Fernandes H, et al. Cytokine expression profiles of immune imbalance in post-mononucleosis chronic fatigue. J Transl Med 2012; 10: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jason LA, Katz BZ, Sunnquist M, et al. The prevalence of pediatric myalgic encephalomyelitis/chronic fatigue syndrome in a community-based sample. Child Youth Care Forum 2020; 49: 563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jason LA and Sunnquist M. The development of the DePaul Symptom Questionnaire: original, expanded, brief, and pediatric versions. Front Pediatr 2018; 6: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landgraf JM, Abetz L, and Ware JE. The child health questionnaire (CHQ): a user’s manual. Boston: The Health Institute, New England Medical Center, 1996. [Google Scholar]
- 19.Fukuda K, Straus SE, Hickie I, et al. The chronic fatigue syndrome: a comprehensive approach to its definition and study. Ann Intern Med 1994; 121: 953–959. [DOI] [PubMed] [Google Scholar]
- 20.Carruthers BM, Jain AK, De Meirleir KL, et al. Myalgic encephalomyelitis/chronic fatigue syndrome: clinical working case definition, diagnostic and treatment protocols. J Chronic Fatigue Syndr 2003; 11: 7–115. [Google Scholar]
- 21.Institute of Medicine. Beyond myalgic encephalomyelitis/chronic fatigue syndrome: redefining an illness. Washington, DC: National Academic Press, 2015. [Google Scholar]
- 22.Jason LA, Sunnquist M, Brown A, et al. Reflections on the Institute of Medicine’s systemic exertion intolerance disease. Pol Arch Intern Med 2015; 125: 576–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McManimen SL, Jason LA, and Williams YJ. Variability in symptoms complicates utility of case definitions. Fatigue 2015; 3: 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jason LA, Fox PA, and Gleason KD. The importance of a research case definition. Fatigue 2018; 6: 52–58. [Google Scholar]
- 25.Katz BZ, Reuter C, Lupovitch Y, et al. A validated scale for assessing the severity of acute infectious mononucleosis. J Pediatr 2019; 209: 130–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jason LA, Cotler J, Islam MF, et al. Risks for developing ME/CFS in college students following infectious mononucleosis: a prospective cohort study. Clin Infect Dis 2020; 73: e3740–e3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gelfand AE, Dey DK, Chang H. Model determination using predictive distributions with implementation via sampling-based methods. In: Bernardo JM, Berger JO, Dawid AP, et al. (eds) Bayesian Statistics. Oxford: Oxford University Press, 1992, pp.147–167. [Google Scholar]
- 28.Jason LA, Cotler J, Islam MF, et al. Cytokine networks analysis unconvers further differences between those who develop myalgic encephalomyelitis/chronic fatigue syndrome following infectious mononucliousis. Fatigue 2021; 9: 45–57. [Google Scholar]
- 29.Cătană CS, Berindan-Neagoe I, Cozma V, et al. Contribution of the IL-17/IL-23 axis to the pathogenesis of inflammatory bowel disease. World J Gastroenterol 2015; 21: 5823–5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorenson M, Jason L, Lerch A, et al. The production of interleukin-8 is increased in plasma and peripheral blood mononuclear cells of patients with fatigue. Neurosci Med 2012; 3: 47–53. [Google Scholar]
- 31.Papanicolaou DA, Wilder RL, Manolagas SC, et al. The pathophysiologic roles of interleukin-6 in human disease. Ann Intern Med 1998; 128: 127–137. [DOI] [PubMed] [Google Scholar]
- 32.Russell L, Broderick G, Taylor R, et al. Illness progression in chronic fatigue syndrome: a shifting immune baseline. BMC Immunol 2016; 17: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kavelaars A, Kuis W, Knook L, et al. Disturbed neuroendocrine-immune interactions in chronic fatigue syndrome. J Clin Endocrinol Metab 2000; 85: 692–696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


