To the Editor:
We read with great interest the recent articles by Wang et al. [1], Génin et al. [2], Sun et al. [3], and Németh et al. [4], in which the authors studied various genetic risk factors for chronic pancreatitis (CP). The ever increasing catalogue of risk genes and variants supports the long-held notion that CP is a complex genetic disorder. Thus, patients often carry multiple genetic alterations that interact in synergy and elicit disease onset and/or promote progression. To offer a conceptual framework for genetic risk in CP, we organized risk factors into mechanism-based schemes, which include the trypsin-dependent, misfolding-dependent, and ductal pathways [5].
More recently, novel mouse models harboring human pancreatitis-associated mutations emerged, providing strong evidence for the pathogenic nature of these risk variants [3, 6-10]. All mouse models of genetic CP published to date carry a single genetic alteration and none models CP as a complex genetic disease. We set out to fill this knowledge gap by generating a novel strain carrying both the T7K24R trypsinogen mutant and the Ctrb1-del chymotrypsin deletion alleles. The T7K24R mice have the p.K24R mutation in mouse cationic trypsinogen (isoform T7), which is analogous to the human hereditary-pancreatitis associated p.K23R PRSS1 mutation [9]. This mutation increases autoactivation of cationic trypsinogen and thus represents a gain-of-function variant. The Ctrb1-del mice are deficient in mouse chymotrypsins CTRB1 and CTRC [7, 8]. Chymotrypsins, primarily CTRC in humans and CTRB1 in mice, promote trypsinogen degradation and thereby protect against pancreatitis. The Ctrb1-del strain models the effect of loss-of-function chymotrypsin variants. Homozygous T7K24R or Ctrb1-del mice do not develop pancreatitis spontaneously, however, severity of cerulein-induced experimental pancreatitis is increased in both strains. Thus, the T7K24R and Ctrb1-del alleles do not cause pancreatitis but sensitize mice for the disease, in a manner that is reminiscent of the effects of human CP-associated risk variants.
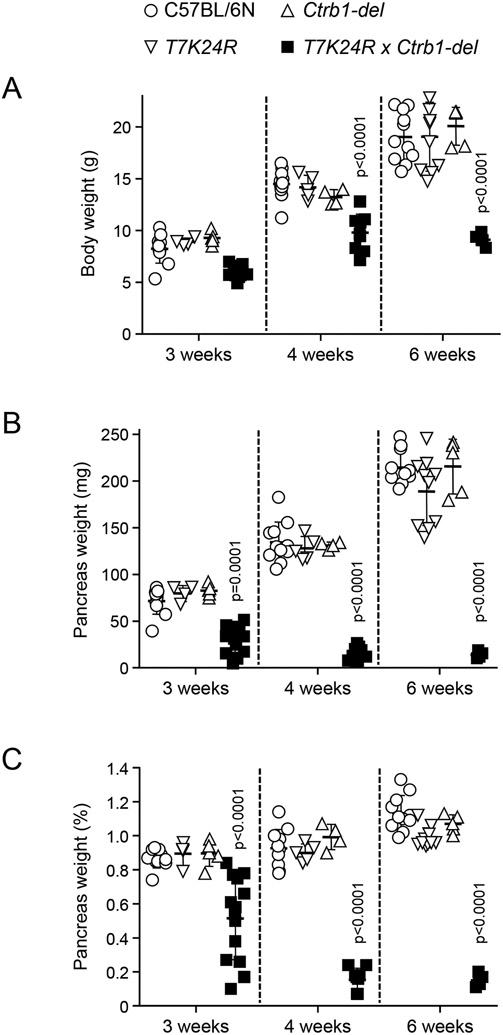
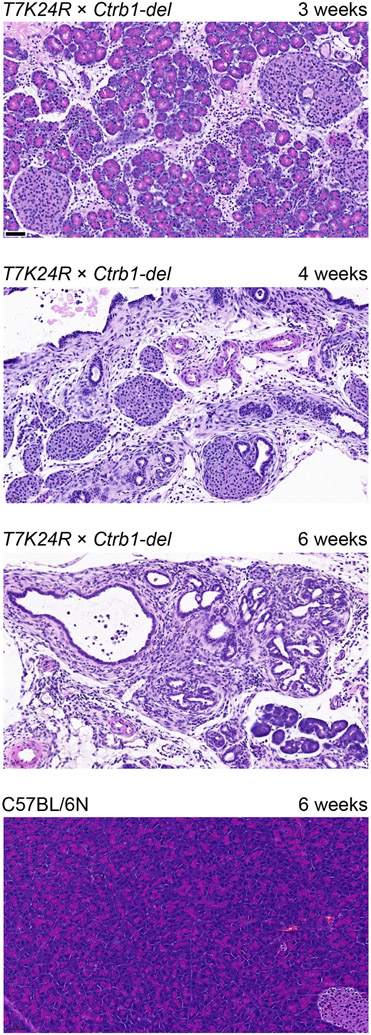
To test whether the T7K24R and Ctrb1-del alleles would synergize and elicit spontaneous pancreatitis, we created mice homozygous for both alleles (T7K24R × Ctrb1-del). Remarkably, mice with the compound genotype developed severe, early-onset pancreatitis (Figures 1 and 2). Body weight of T7K24R × Ctrb1-del mice was noticeably reduced at 3, 4 and 6 weeks of age compared to the parent strains or C57BL/6N controls (Figure 1A), suggesting pancreatic insufficiency. Consistent with atrophic CP, the pancreas weight was markedly diminished in T7K24R × Ctrb1-del mice (Figure 1B), even after normalization to body weight (Figure 1C). Hematoxylin-eosin staining of pancreas sections confirmed end-stage CP in 4-6-week-old T7K24R × Ctrb1-del mice while pancreas histology of the parent strains (not shown) was normal, indistinguishable from C57BL/6N controls (Figure 2). From the 3-week-old T7K24R × Ctrb1-del mice (n=8) analyzed histologically, 3 were still unaffected, 2 had acute pancreatitis, and 3 had CP of varying severity. In contrast, histology of the 7 mice evaluated at 4 weeks (n=4) or 6 weeks (n=3) of age showed severe CP in all cases, suggesting that in this model the age of pancreatitis onset is around 3 weeks of age and a brief period of acute pancreatitis might precede chronic progression.
Figure 1.
Body weight and pancreas weight of T7K24R × Ctrb1-del mice at 3, 4, and 6 weeks of age. For comparison, age-matched data for C57BL/6N mice and the parent strains T7K24R and Ctrb1-del are also shown. A, Body weight. B, Pancreas weight in mg units. C, Pancreas weight as percent of body weight. Individual data points were graphed with the mean and SD indicated. The differences of means between the groups were analyzed by one-way ANOVA followed by Tukey’s post-hoc test. P < 0.05 was considered statistically significant.
Figure 2.
Histology of the pancreas from T7K24R × Ctrb1-del mice at 3, 4, and 6 weeks of age. For comparison, pancreas section from a 6-week-old C57BL/6N mouse is shown. Representative hematoxylin-eosin stained pancreas sections are shown. The scale bars correspond to 50 μm. Note that the phenotype at 3 weeks of age was variable, see text for details. Although not shown, pancreas histology of 6-week-old T7K24R and Ctrb1-del mice was normal, indistinguishable from that of C57BL/6N controls.
When the disease phenotype of T7K24R × Ctrb1-del mice is compared to those of the parent strains, the difference is striking and demonstrates how the pathogenic alleles amplify each other’s effect. These observations offer the first line of experimental evidence that independent risk factors for CP, particularly those within the same mechanistic pathway, can synergize and provoke overt pancreatic disease. The experiments also demonstrate that modeling CP in mice as a complex genetic disorder is feasible and facilitates the study of interactions between various CP risk alleles.
ACKNOWLEDMENT
The authors thank Andrea Geisz for critical reading of the manuscript.
FUNDING
This study was supported by the National Institutes of Health (NIH) grants R01 DK058088, R01 DK117809, and R01 DK082412 to MST, and the Department of Defense grant W81XWH2010134 (PR192583) to ZJ and MST.
Footnotes
COMPETING INTERESTS
The authors do not have competing interests.
ETHICAL APPROVAL
Animal experiments were performed at the University of California Los Angeles (UCLA) and at Boston University (BU) with the approval and oversight of the Animal Research Committee and the Institutional Animal Care and Use Committee, respectively, including protocol review and post-approval monitoring. The animal care programs at these institutions are managed in full compliance with the US Animal Welfare Act, the United States Department of Agriculture Animal Welfare Regulations, the US Public Health Service Policy on Humane Care and Use of Laboratory Animals and the National Research Council's Guide for the Care and Use of Laboratory Animals. UCLA and BU have approved Animal Welfare Assurance statements (A3196-01 and A3316-01, respectively) on file with the US Public Health Service, National Institutes of Health, Office of Laboratory Animal Welfare. Both institutions are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
REFERENCES
- 1.Wang YC, Mao XT, Yu D, Mao SH, Li ZS, Zou WB, Liao Z. Alcohol amplifies the association between common variants at PRSS1-PRSS2 locus and chronic pancreatitis in a dose-dependent manner. Gut 2022. Jan 7. gutjnl-2021-326670. [DOI] [PubMed] [Google Scholar]
- 2.Génin E, Cooper DN, Masson E, Férec C, Chen JM. NGS mismapping confounds the clinical interpretation of the PRSS1 p.Ala16Val (c.47C>T) variant in chronic pancreatitis. Gut 2022, 71:841–842 [DOI] [PubMed] [Google Scholar]
- 3.Sun C, Liu M, An W, Mao X, Jiang H, Zou W, Wu H, Liao Z, Li Z. Heterozygous Spink1 c.194+2T>C mutant mice spontaneously develop chronic pancreatitis. Gut 2020, 69:967–968 [DOI] [PubMed] [Google Scholar]
- 4.Németh BC, Orekhova A, Zhang W, Nortman SA, Thompson T, Hegyi P, Abu-El-Haija M. Novel p.K374E variant of CPA1 causes misfolding-induced hereditary pancreatitis with autosomal dominant inheritance. Gut 2020, 69:790–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayerle J, Sendler M, Hegyi E, Beyer G, Lerch MM, Sahin-Tóth M. Genetics, cell biology, and pathophysiology of pancreatitis. Gastroenterology 2019, 156:1951–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisz A, Sahin-Tóth M. A preclinical model of chronic pancreatitis driven by trypsinogen autoactivation. Nat Commun 2018, 9:5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jancsó Z, Hegyi E, Sahin-Tóth M. Chymotrypsin reduces the severity of secretagogue-induced pancreatitis in mice. Gastroenterology 2018, 155:1017–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisz A, Jancsó Z, Németh BC, Hegyi E, Sahin-Tóth M. Natural single-nucleotide deletion in chymotrypsinogen C gene increases severity of secretagogue-induced pancreatitis in C57BL/6 mice. JCI Insight 2019, 4:e129717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jancsó Z, Sahin-Tóth M. Mutation that promotes activation of trypsinogen increases severity of secretagogue-induced pancreatitis in mice. Gastroenterology 2020, 158:1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gui F, Zhang Y, Wan J, Zhan X, Yao Y, Li Y, Haddock AN, Shi J, Guo J, Chen J, Zhu X, Edenfield BH, Zhuang L, Hu C, Wang Y, Mukhopadhyay D, Radisky ES, Zhang L, Lugea A, Pandol SJ, Bi Y, Ji B. Trypsin activity governs increased susceptibility to pancreatitis in mice expressing human PRSS1R122H. J Clin Invest 2020, 130:189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]