Abstract
Plasmatic presepsin (PSP) is a novel biomarker reported to be useful for sepsis diagnosis and prognosis. During the pandemic, only few studies highlighted a possible correlation between PSP and COVID-19 severity, but results remain inconsistent. The present study aims to establish the correlation between PSP and COVID-19 severity. English-language papers assessing a correlation between COVID-19 and PSP from MEDLINE, PubMed, Google Scholar, Cochrane Library, MeSH, LitCovid NLM, EMBASE, CINAHL Plus and the World Health Organization (WHO) website, published from January 2020 were considered with no publication date limitations. Two independent reviewers performed data abstraction and quality assessment, and one reviewer resolved inconsistencies. The protocol was registered on PROSPERO (CRD42022325971).Fifteen articles met our eligibility criteria. The aggregate study population included 1373 COVID-19 patients who had undergone a PSP assessment. The random-effect meta-analysis was performed in 7 out of 15 selected studies, considering only those reporting the mean PSP levels in low- and high-severity cases (n = 707).The results showed that the pooled mean difference of PSP levels between high- and low-severity COVID-19 patients was 441.70 pg/ml (95%CI: 150.40–732.99 pg/ml).Our data show that presepsin is a promising biomarker that can express COVID-19 severity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-022-00936-8.
Keywords: COVID-19, Disease severity, Presepsin, SARS-CoV-2
Background
At the end of December 2019, a new zoonotic Coronavirus (SARS-CoV-2) was identified as the agent causing a cluster of pneumonia cases in Wuhan, China, and rapidly spreading throughout the world. Globally, data indicate that the COVID-19 pandemic involved over 530 millions of affected people with different clinical presentations and caused 6.3 millions of deaths [1, 2]. In February 2020, the World Health Organization designated the disease COVID-19 (coronavirus disease 2019), and different rates of mortality have been reported [3, 4]. Although many hypotheses have been proposed about its origin, the direct ancestral virus has not been identified yet [5, 6].
The clinical features of COVID-19 range from asymptomatic condition to severe/fatal lung injury and multi-organ failure due to an excessive immune response. Several risk factors for COVID-19 severity have been identified, namely a) “life-style factors” (e.g., obesity and smoking habit); b) demographic factors (e.g., age, male gender, post-menopausal status); and c) comorbidities (e.g., hypertension, coronary artery disease (CAD), diabetes, cerebrovascular disease (CVD), chronic kidney disease (CKD) and chronic obstructive pulmonary disease (COPD) [7]. Common complications of COVID-19 include acute respiratory distress syndrome (ARDS), acute kidney and liver dysfunctions, delirium/encephalopathy, thrombosis and cardiac damage (e.g., cardiomyopathy, arrhythmias and sudden cardiac death) [1].
Despite remarkable findings have been achieved since the beginning of the pandemic, an early identification and management of this novel coronavirus related disease is still limited. Since patients affected by COVID-19 may rapidly worsen and no effective antiviral therapy for SARS-CoV-2 infection has been found yet, an early identification of patients’ severity (through an effective and valuable biochemical marker) is key to guide the intensity of care and guarantee cardiorespiratory function [8]. In this regard, many efforts have been devoted to researching easily accessible biomarkers predicting COVID-19 severity.
Plasmatic presepsin (PSP) is a soluble N-terminal fragment of the cluster of differentiation marker protein 14 (CD14) reported to be a novel biomarker in sepsis [9, 10]. Indeed, as a glycoprotein expressed on monocytes and macrophages, CD14 is a receptor for the lipopolysaccharide (LPS)-LPS binding protein complexes, which is able to activate a series of signal transduction pathways leading to systemic inflammatory response. So far, two distinct forms of CD14 have been characterized, i.e., a membrane-bound (mCD14) and a soluble CD14 (sCD14). The sCD14 plays an essential role in mediating the immune responses to LPS of CD14-negative cells, such as endothelial and epithelial cells. During inflammatory stress, sCD14 is cleaved by plasmatic proteases which generate a truncated form of 64 aminoacidic residues of 13 kDa referred to as sCD14 subtype (sCD14-ST) or PSP [11, 12]. Since 2015, several studies have shown that PSP is not only useful for sepsis diagnosis [11–13], but also predicts the severity of this condition [14, 15]. A recent research highlighted that sepsis and SARS-CoV-2 infection share many immunopathological and pathophysiological similarities [16]. Therefore, it was recently postulated that elevated levels of PSP could predict the outcome of patients with SARS-CoV-2 infection [17, 18]. The relationship between PSP and COVID-19 severity is known, although not well detailed and comprehensively evaluated [17–31]. Thus, we conducted a systematic review and meta-analysis aimed at establishing the role of PSP in assessing SARS-CoV-2 infection severity.
Methods
Systematic review and meta-analysis
This paper has been performed following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [32] and Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines and checklists [33]. The protocol was registered on PROSPERO (CRD42022325971).
Data sources and searches
A literature search for relevant documents was performed in the following sources: MEDLINE, PubMed, Google Scholar, Cochrane Library, MeSH, LitCovid NLM, EMBASE, CINAHL Plus, and the World Health Organization (WHO) website. Items published from January 2020 were considered. No publication date limitations have been established. The used search strategy included the following Medical Subject Heading terms and keywords: (“Coronavirus” OR “Coronaviridae” OR “nCoV” OR “Coronavirus Infections” OR “COVID-19” OR “severe acute respiratory syndrome coronavirus 2” AND (“human presepsin protein” OR “Presepsin” OR “Plasmatic presepsin” OR “PSP” OR “sCD14-ST”). Only studies that involved humans, and were written in English, were included.
Study selection
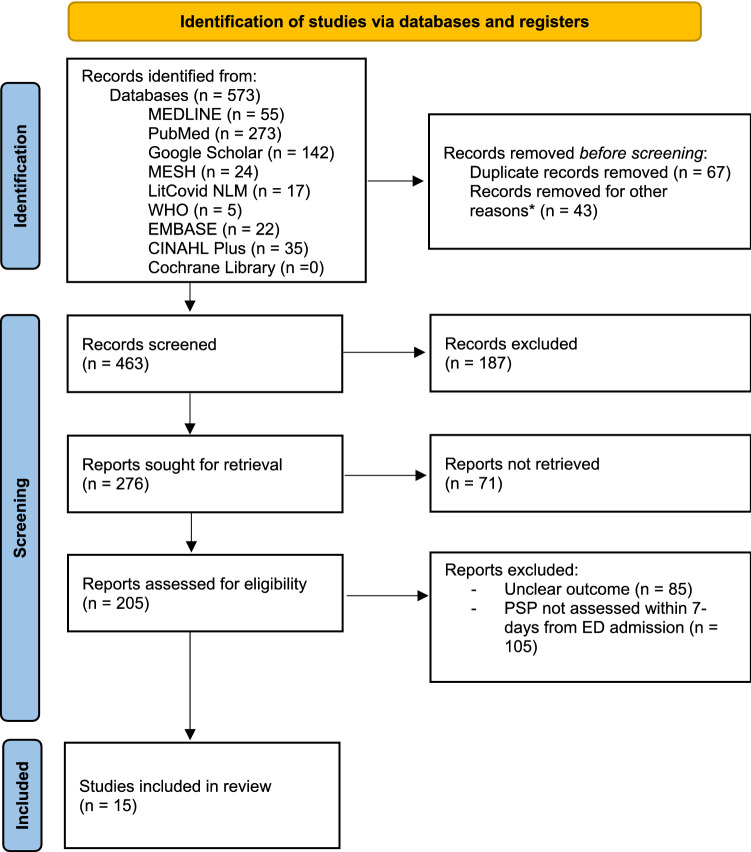
The systematic review was performed comprehending prospective and retrospective studies, pooled analysis, cross-sectional studies and case series. A study was eligible for inclusion in this review if: (a) participants were affected by SARS-CoV-2 infection, confirmed through polymerase chain reaction testing of nasopharyngeal swab; (b) PSP levels were assessed within 7 days from the admission to the Emergency Department; (c) severe COVID-19 was defined as follows: SpO2 < 94% on room air and/or PaO2/FiO2 < 300 and/or respiratory rate > 30 breaths/minute and/or lung infiltrates > 50% [8–34]; (d) the outcome was measured in terms of mechanical ventilation requirement or intensive care admission or mortality; (e) correlation between PSP levels and disease severity was assessed (see Fig. 1 for recruitment and exclusion criteria).
Fig. 1.
PRISMA 2020 flowchart for new systematic reviews that included only database and registry searches
The meta-analysis was performed comparing studies which expressed mean PSP levels, standard deviation (SD) and number of patients in two subgroups: experimental group (i.e., high-severity SARS-CoV-2 infection) vs. control group (i.e., low-severity disease).
Two independent reviewers (MG and BP) screened blindly the titles and abstracts of the identified documents and, for the record selected at this first step, retrieved and evaluated full manuscripts and appendices. Disagreements and inconsistencies were resolved by consensus and arbitration with a third reviewer (FR).
Data extraction and quality assessment
Two investigators (MG and BP) independently abstracted and recorded data, using standardized data abstraction form (Excel spreadsheet). The researchers were blinded to each other decisions. Extracted data included: study duration; study design; mean age; sex; sample size; numerosity of the two subgroups (when available); mean and SD of PSP levels in experimental and control group; time of PSP assessment and mortality. We did not contact study authors if data pertaining PSP levels or disease severity were not recorded. The quality assessment of the included studies has been performed following the NIH Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies [35]. Each study was evaluated according to a standardized set of predefined criteria consisting of 14 items, mainly exploring the following domains: study population, exposure and outcome (Table 2). Each item was rated as positive, negative or not available. Two independent reviewers scored each article for quality and any scoring inconsistencies were resolved by discussion and consensus between the two reviewers.
Table 2.
NIH Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies
| Authors | Year | Questions | Quality Rating | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |||
| Fukada et al.[17] | 2020 | Y | Y | N/A | Y | N | Y | N/A | N/A | Y | Y | N | N | N | N | Poor |
| Zaninotto et al. [18] | 2020 | Y | Y | N/A | Y | N | Y | Y | N/A | N | Y | Y | N | Y | N | Fair |
| Dell'Aquila et al.[19] | 2020 | Y | Y | Y | Y | Y | Y | Y | N/A | Y | Y | Y | N | Y | Y | Good |
| Ducastel et al.[20] | 2020 | Y | Y | Y | Y | N | Y | Y | N/A | Y | N | Y | N | Y | N | Good |
| Keskinidou et al.[21] | 2020 | Y | Y | Y | Y | N | Y | Y | N/A | Y | N | Y | N | Y | N | Fair |
| Schirinzi et al.[22] | 2020 | Y | Y | Y | Y | Y | Y | Y | N/A | Y | N | Y | N | Y | N | Good |
| Kocyigit et al.[23] | 2020 | Y | Y | Y | Y | Y | Y | Y | N/A | Y | N | Y | N | Y | Y | Good |
| Hasegawa et al.[24] | 2021 | Y | Y | Y | Y | N | Y | Y | N/A | Y | N | Y | N | Y | N | Fair |
| Domi et al.[25] | 2021 | Y | Y | Y | Y | Y | Y | Y | N/A | Y | N | Y | N | Y | N | Good |
| Dewi et al.[26] | 2021 | Y | Y | N | Y | N | Y | N/A | N/A | Y | Y | Y | N | Y | N | Fair |
| Mirza et al.[27] | 2021 | Y | Y | Y | Y | Y | Y | Y | N/A | Y | N | N | N | Y | N | Fair |
| Farag et al.[28] | 2021 | Y | Y | Y | Y | Y | Y | Y | N/A | N | Y | N | N | Y | Y | Good |
| Kim et al.[29] | 2021 | Y | Y | Y | Y | Y | Y | N/A | N/A | N | Y | Y | N | N/A | N | Fair |
| Çaglar et al.[30] | 2022 | Y | Y | Y | Y | Y | Y | Y | N/A | Y | N | Y | N | Y | Y | Good |
| Morales-Cely et al.[31] | 2022 | Y | Y | N/A | Y | Y | Y | N/A | N/A | N | N | Y | N | N/A | N | Poor |
Note: N/A = Not applicable; Y = Yes; N = No
Questions
Was the research question or objective in this paper clearly stated?
Was the study population clearly specified and defined?
Was the participation rate of eligible persons at least 50%?
Were all the subjects selected or recruited from the same or similar populations (including the same time period)? Were inclusion and exclusion criteria for being in the study prespecified and applied uniformly to all participants?
Was a sample size justification, power description, or variance and effect estimates provided?
For the analyses in this paper, were the exposure(s) of interest measured prior to the outcome(s) being measured?
Was the timeframe sufficient so that one could reasonably expect to see an association between exposure and outcome if it existed?
For exposures that can vary in amount or level, did the study examine different levels of the exposure as related to the outcome (e.g., categories of exposure, or exposure measured as continuous variable)?
Were the exposure measures (independent variables) clearly defined, valid, reliable, and implemented consistently across all study participants?
Was the exposure(s) assessed more than once over time?
Were the outcome measures (dependent variables) clearly defined, valid, reliable, and implemented consistently across all study participants?
Were the outcome assessors blinded to the exposure status of participants? Was loss to follow-up after baseline 20% or less?
Were key potential confounding variables measured and adjusted statistically for their impact on the relationship between exposure(s) and outcome(s)?
Data synthesis and analysis
The meta-analysis was performed using the meta package of R statistical program (version 4.0.5) [36]. Mean serum levels of PSP and Standard Deviation (SD) in high- and low-severity patient groups were collected from the 7 out of 15 selected studies according to the data availability. Through a random-effect meta-analysis was performed to estimate the pooled mean difference and 95% confidence interval (95%CI) of serum levels of PSP between the high and low severity patients. Statistical heterogeneity was evaluated through Chi-squared test and expressed as I2 statistic of the proportion of total variation. A p value < 0.10 was considered statistically significant, and an I2 statistic > 75% indicated a high grade of heterogeneity. The publication bias of the selected studies was assessed both graphically and quantitatively, through test for asymmetry of funnel plots and Egger’s regression test, respectively. As a sensitivity analysis, we repeated our analysis after excluding those studies that could determine publication bias in light of the graphical evaluation of the funnel plot.
Role of the funding source
No funding sources have been used to produce this manuscript.
Results
Literature search results
A total of 573 studies were identified through database searching (273 from PubMed, 55 from MEDLINE, 24 from MESH, 22 from EMBASE, 35 from CINAHL, 142 from Google Scholar, 17 from LitCovid NLM, and 5 from WHO website). The flowchart of the studies’ selection is illustrated in Fig. 1. After the title-abstract and full-text screenings, 15 documents were identified, and their main characteristics are reported in Table 1. Among the selected studies, 14 involved adult (n = 1373) and one pediatric (n = 20) patients. All studies highlighted a possible relationship between PSP levels and COVID-19 severity, but this correlation was statistically significant in 10 works (1005 patients over 1373) [18, 19, 22–24, 27–31]. Among studies involving adult patients [17–25, 27–31], the mean age of the pooled sample was 62.7 ± 5.7 years. Mortality in the included studies ranged between 8 and 45%.
Table 1.
Synopsis of the main features described in papers (n = 15) correlating PSP and COVD-19
| Author | Year | Duration (months) | Study design | Patients (n) | Mean age (ys) | Male (n) | Level of PSP in low-severity patients (pg/ml) | Level of PSP in high-severity patients (pg/ml) | p | Time of PSP assessing | Mortality (%) | Conclusion |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fukada et al.[17] | 2020 | 2 | Case series | 6 | N/A | N/A | N/A | N/A | N/A | Admission | 16.0 | PSP has potential as a biomarker for severe COVID-19 pneumonia |
| Zaninotto et al.[18] | 2020 | 3 | Retrospective study | 75 | 67.0 | 56 | 408 | 1069 | < 0.001 | Day 2–7 | 8.0 | PSP seems to have a role in providing prognostic information in COVID-19 pts in the early phase |
| Dell'Aquila et al.[19] | 2020 | 2 | Prospective study | 143 | 73.0 | 86 | 518 | 892 | < 0.001 | Admission | 45.0 | PSP is a very specific predictor (92%) of 30-day mortality in COVID-19 patients |
| Ducastel et al.[20] | 2020 | 2 | Retrospective study | 160 | 60.0 | 92 | N/A | N/A | N/A | Admission | 10.6 | PSP was associated with worse outcome |
| Keskinidou et al.[21] | 2020 | 8 | Observational study | 66 | 64.0 | 51 | N/A | 1300 | N/A | Within 24 h post ICU admission | 34.9 | PSP could differentiate patients who did not survive, independently of dexamethasone administration |
| Schirinzi et al.[22] | 2020 | 2 | Observational study | 86 | 67.0 | 58 | 737 | 1234 | < 0.0001 | Admission (and every 24 h) | 22.0 | PSP reflects the clinical course of the disease and might be used to predict the evolution of COVID-19 disease |
| Kocyigit et al.[23] | 2020 | 2 | Observational study | 88 | 51.0 | 41 | 590 | 3500 | < 0.001 | Within 24 h post admission | N/A | There was a significant correlation between PSP and disease severity |
| Hasegawa et al. [24] | 2021 | 1 | Observational study | 57 | 59.0 | 33 | 563 | 1217 | 0.007 | Admission | N/A | PSP is significantly higher in COVID-19 pts with ARDS |
| Domi et al.[25] | 2021 | 11 | Retrospective study | 97 | 68.0 | 67 | 433 | 579 | 0.183 | Admission | 14.4 | The complication of bacterial superinfection might be associated with PSP elevation |
| Dewi et al.[26] | 2021 | 8 | Cross-sectional study | 20 | 10.0 | 10 | N/A | N/A | N/A | Admission | 40.0 | High levels of PSP were related to higher mortality rate |
| Mirza et al.[27] | 2021 | 10 | Cross-sectional study | 80 | 67.5 | 54 | 17 | 56 | < 0.001 | Admission | N/A | PSP is the most useful tool in predicting the severity of COVID-19 infection |
| Farag et al.[28] | 2021 | 4 | Observational study | 42 | 59.6 | 26 | 390 | 950 | 0.008 | Admission | 26.2 | Potential utility of PSP as a predictive indicator of severity in COVID-19 patients |
| Kim et al.[29] | 2021 | N/A | Retrospective study | 42 | 59.5 | 20 | N/A | N/A | 0.007 | Admission | N/A | PSP has the potential to be a useful severity marker in patients with COVID-19 |
| Çaglar et al.[30] | 2022 | 3 | Observational study | 259 | 58.1 | 146 | 40.17 | 55.40 | 0.013 | Admission | 14 | Presepsin may be of value for risk stratification of COVID-19 patients |
| Morales-Cely et al. [31] | 2022 | N/A | Prospective study | 152 | N/A | N/A | 570 | 1358 | < 0.0001 | Admission | N/A | Median level of PSP was higher in patients deceased by COVID-19 than in survived |
N/A: not available; PSP: Plasmatic presepsin
Concerning the study design, we selected 5 observational, 4 retrospective, and 2 prospective studies, 2 cross-sectional works, one case series and one pooled analysis. The quality assessment is reported in Table 2. No study was excluded because of a quality score less than 8 (< 50%).
Study characteristics
The characteristics of the studies with main clinical features eligible for our paper are summarized in Table 1. The quality assessment has been performed following NIH criteria (Table 2). No study was excluded solely because of low-quality scores less than 8 (< 50%). No randomized, controlled trials met our selection criteria. This manuscript involved observational (n = 6), retrospective (n = 4) and prospective (n = 2) studies, cross-sectional analysis (n = 2) and case series (n = 1).
Publication bias and heterogeneity
The evaluation of the funnel plot suggested a possible publication bias across the selected studies (Supplementary Fig. 1), which was not confirmed at the Egger’s regression test (Egger regression intercept = 3.558, 95%CI: -0.376, 7.492, p = 0.130). In order to establish the level of consistency among involved studies, the heterogeneity has been calculated, resulting quite high (I2 = 93%, p < 0.01).
Serum level of PSP and clinical outcomes
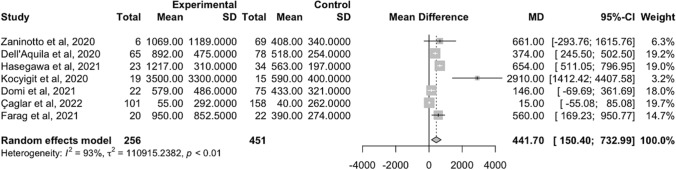
Among the selected studies, higher serum levels of PSP at hospital admission in patients with COVID-19 disease were related to worse clinical outcomes (i.e., higher severity of COVID-19 and necessity of respiratory support). The random-effect meta-analysis on 707 individuals (Fig. 2) showed a significant pooled mean difference in serum PSP levels between patients with high- and low-severity of COVID-19 disease was 441.70 pg/ml (95%CI: 150.40–732.99 pg/ml).
Fig. 2.
Forest plot on the mean difference of PSP levels between the high- and low-severity patient groups from the random-effect meta-analysis
Sensitivity analysis
The results were confirmed in the sensitivity analysis after excluding the study of Kocyigit et al. [23]. Indeed, we excluded the analysis with more graphical distance of the effect size from the polled one in the forest plot of meta-analysis and with the smallest sample size. From the additional meta-analysis, including 673 patients, we found a pooled mean difference of PSP between the high- and low-severity patient groups of 350.02 pg/ml (95%CI: 115.15–584.89 pg/ml) (Supplementary Fig. 2).
Discussion
Early prediction of COVID-19 severity is still challenging although it represents a crucial step in defining the risk of fatal outcomes and the most appropriate recovery setting for adequate treatment. As reported by the last update of the Surviving Sepsis Campaign Guidelines [8], disease severity, which is currently assessed mainly by clinical parameters, has a key role in managing COVID-19 patients. However, since these criteria do not predict the risk of clinical worsening, a tool able to assess COVID-19 evolution would be helpful for physicians. Furthermore, it is not currently possible to define the severity of the disease relying upon the viral load [34].
Different biochemical markers have been proposed to integrate the WHO criteria in predicting COVID-19 severity. In particular, a recent research performed on diabetic patients confirmed that C-reactive protein (CRP) is a valuable predictor of COVID-19 progression and severity. Furthermore, serum levels of inflammation-related (e.g., interleukin-6 or serum ferritin) and coagulation parameter (D-dimer) were higher in patients with SARS-CoV-2 infection and diabetes mellitus vs. those without, suggesting that diabetic patients could be more susceptible to the cytokine storm that leads to ARDS and fatal outcome [37]. However, different factors can alter levels of these markers (e.g., tumors, autoimmune diseases) making them less specific in the diagnosis and risk stratification of patients with COVID-19 [38].
Recently, different studies highlighted the role of a novel biochemical marker (i.e., PSP), which seems to have better sensitivity and specificity in the diagnosis and severity assessment of sepsis [9–15, 37]. Considering that sepsis and SARS-CoV-2 infection share immunopathogenetic and pathophysiological similarities, we believe that PSP may help in risk stratification [16]. In the last two years of pandemic, the interest in the possible correlation between COVID-19 severity and levels of PSP has increased and several studies have been published [17–31]. In 2021, Amhed et al. proposed a review on this correlation [39] highlighting that PSP levels predicted the aggravation of COVID-19 infection. However, the limited number of pertinent manuscripts hampered this analysis as only three articles [17, 18, 22] were considered eligible for the review.
In 2021, Lippi et al. proposed a pooled analysis on this topic concluding that PSP values were significantly higher in COVID-19 patients with severe/critical illness vs. those without [24]. In our opinion, this result was interesting but the sample size was small (n = 420). Moreover, this paper presented an unclear definition of disease severity (i.e., death, need for tracheostomy, mechanical ventilation, respiratory distress, ICU recovery).
Our work considered studies that assessed PSP levels in the first 7 days of hospitalization. This choice allowed us to consider, not only patients identified as critical because of their clinical manifestations, but also those ones who showed a rapid worsening in the first days from admission. Indeed, Faes et al. reported that an average time of 5 to 7 days to progress from the first manifestations to ARDS [40]. As indicated by the pooled results, PSP can be considered a valuable biomarker of COVID-19 severity. Indeed, higher PSP levels might help physicians in recognizing potentially critical patients, even when clinical condition are not alarming yet.
We would like to acknowledge some limitations of our study: First, there is a complete lack of multicenter randomized clinical trials, which are fundamental to confirm the effective usefulness of this biomarker to stratify COVID-19 severity. Second, the heterogeneity among the involved studies resulted very high (> 90%). This result was expected since PSP has only recently been proposed as a biomarker for COVID-19 severity with few published studies. Third, all the included studies in the present meta-analysis considered the disease severity as a primary outcome. The definition of this condition is often heterogeneous; therefore, the choice of other outcomes (e.g., mortality) might be preferable. Fourth, the included studies used different PSP assessment methods (e.g., PATHFAST Presepsin–Mitsubishi Chemical Europe GmbH, Düsseldorf, Germany or STACIA Presepsin–LSI Medience Corporation, Tokyo, Japan), which can be a further source of heterogeneity.
Main strengths of this study include: First, we performed a systematic review on the role of PSP in COVID-19 severity according to specific guidelines. Second, we examined a significant number of scientific databases leading to a consistent number of eligible papers (resulting in a large sample size). Third, this study allowed for a new quantitative analysis on this topic.
Conclusion
Our results show that PSP alone is a reliable tool to assess COVID-19 severity. The possible integration of this biomarker with clinical criteria might be useful to improve the accuracy of risk stratification in COVID-19 patients. Furthermore, since SARS-COV-2 infection and sepsis share similar immunopathological manifestations and PSP showed its intrinsic value in predicting the severity of both diseases, we can hypothesize that other conditions with similar immunopathological features, a biomarker as PSP might help in the risk stratification of affected patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to thank Dr. Donato Bragatto, Dr. Claudia Righini, Mrs. Manuela Zappaterra (Biblioteca Interaziendale di Scienza della Salute, Hospital of Ferrara), Mrs. Egizia Zironi and Mrs. Silvia Bellotti (Unità Servizi Interbibliotecari, University of Ferrara) for their valuable and precious collaboration. Also, the authors state that they abide by the “Requirements for Ethical Publishing in Biomedical Journals”.
Abbreviations
- ARDS
Acute respiratory distress syndrome
- CAD
Coronary artery disease
- CD14
Cluster of differentiation marker protein 14
- CKD
Chronic kidney disease
- COPD
Chronic obstructive pulmonary disease
- COVID-19
Coronavirus disease 2019
- CVD
Cerebrovascular disease
- FiO2
Inspired fraction of oxygen
- ICU
Intensive care unit
- LPS
Lipopolysaccharide
- mCD14
Membrane-bound CD14
- MOOSE
Meta-analysis of Observational Studies in Epidemiology
- PaO2
Arterial partial pressure of oxygen
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- PSP
Plasmatic presepsin
- sCD14
Soluble CD14
- SD
Standard deviation
- SpO2
Peripheral oxygen saturation
- WHO
World health organization
Author’s contributions
MG and BP designed the project and wrote the paper. MG, BP and FR built the database. FR, CT and SV analyzed the database. MG, MM, MDS, AC, CC and RDG critically reviewed the paper. All authors have read and agreed to the published version of the manuscript.
Funding
Open access funding provided by Università degli Studi di Ferrara within the CRUI-CARE Agreement.
Availability of data and materials
The dataset is available for reviewers on reasonable request.
Declarations
Conflict of interest
Authors have no conflicts of interest to disclose.
Ethical approval
This research does not directly involve patients so an ethical approval is not deemed necessary.
Reproducible research statement
The study protocol was registered on PROSPERO (CRD42022325971).
Statistical code and data set
All data and statistical analyses codes are available upon request to the corresponding author.
Footnotes
Matteo Guarino and Benedetta Perna share co-first authorship.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organisation. https://covid19.who.int/ (accessed 09 June 2022).
- 3.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vahidy FS, Drews AL, Masud FN, Schwartz RL, Askary BB, Boom ML, Phillips RA. Characteristics and outcomes of COVID-19 patients during initial peak and resurgence in the Houston metropolitan area. JAMA. 2020;324:998–1000. doi: 10.1001/jama.2020.15301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotondo JC, Martini F, Maritati M, Mazziotta C, Di Mauro G, Lanzillotti C, Barp N, Gallerani A, Tognon M, Contini C. SARS-CoV2 infection: new molecula, phylogenetic and pathogenetic insight efficacy of current vaccines and the potential risk of variants. Viruses. 2021;13:1687. doi: 10.3390/v13091687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contini C, Di Nuzzo M, Barp N, Bonazza A, De Giorgio R, Tognon M, Rubino S. The novel zoonotic COVID-19 pandemic: an expected global health concern. J Infect Dev Ctries. 2020;14:254–264. doi: 10.3855/jidc.12671. [DOI] [PubMed] [Google Scholar]
- 7.Wolff D, Nee S, Hickey NS, Marschollek M. Risk factors for Covid-19 severity and fatality: a structured literature review. Infection. 2021;49:15–28. doi: 10.1007/s15010-020-01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alhazzani W, Evans L, Alshamsi F, Møller MH, Ostermann M, Prescott HC, Arabi YM, Loeb M, Ng Gong M, Fan E, Oczkowski S, Levy MM, Derde L, Dzierba A, Du B, Machado F, Wunsch H, Crowther M, Cecconi M, Koh Y, Burry L, Chertow DS, Szczeklik W, Belley-Cote E, Greco M, Bala M, Zarychanski R, Kesecioglu J, McGeer A, Mermel L, Mammen MJ, Nainan Myatra S, Arrington A, Kleinpell R, Citerio G, Lewis K, Bridges E, Memish ZA, Hammond N, Hayden FG, Alshahrani M, Al Duhailib Z, Martin GS, Kaplan LJ, Coopersmith CM, Antonelli M, Rhodes A. Surviving Sepsis Campaign Guidelines on the Management of Adults With Coronavirus Disease 2019 (COVID-19) in the ICU: First Update. Crit Care Med. 2021;49:e219–e234. doi: 10.1097/CCM.0000000000004899. [DOI] [PubMed] [Google Scholar]
- 9.Shozushima T, Takahashi G, Matsumoto N, Kojika M, Okamura Y, Endo S. Usefulness of presepsin (sCD14-ST) measurements as a marker for the diagnosis and severity of sepsis that satisfied diagnostic criteria of systemic inflammatory response syndrome. J Infect Chemother. 2011;17:764–769. doi: 10.1007/s10156-011-0254-x. [DOI] [PubMed] [Google Scholar]
- 10.Velissaris D, Zareifopoulos N, Karamouzos V, Karanikolas E, Pierrakos C, Koniari I, Karanikolas M. Presepsin as a diagnostic and prognostic biomarker in sepsis. Cureus. 2021;13:e15019. doi: 10.7759/cureus.15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Hu L, Zhang G, Wu F, He T. Accuracy of presepsin in sepsis diagnosis: a systematic review and meta-analysis. PLoS ONE. 2015;10:e0133057. doi: 10.1371/journal.pone.0133057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Hu ZD, Song J, Shao J. Diagnostic value of presepsin for sepsis: a systematic review and meta-analysis. Medicine. 2015;94:e2158. doi: 10.1097/MD.0000000000002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Liu D, Liu YN, Wang R, Xie LX. The accuracy of presepsin (sCD14-ST) for the diagnosis of sepsis in adults: a meta-analysis. Crit Care. 2015;19:323. doi: 10.1186/s13054-015-1032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endo S, Suzuki Y, Takahashi G, Shozushima T, Ishikura H, Murai A, Nishida T, Irie Y, Miura M, Iguchi H, Fukui Y, Tanaka K, Nojima T, Okamura Y. Usefulness of presepsin in the diagnosis of sepsis in a multicenter prospective study. J Infect Chemother. 2012;18:891–897. doi: 10.1007/s10156-012-0435-2. [DOI] [PubMed] [Google Scholar]
- 15.Masson S, Caironi P, Spanuth E, Thomae R, Panigada M, Sangiorgi G, Fumagalli R, Mauri T, Isgrò S, Fanizza C, Romero M, Tognoni G, Latini R, Gattinoni L2014; ALBIOS Study Investigators. Presepsin (soluble CD14 subtype) and procalcitonin levels for mortality prediction in sepsis: data from the Albumin Italian Outcome Sepsis trial. Crit Care 18:R6. [DOI] [PMC free article] [PubMed]
- 16.Olwal CO, Nganyewo NN, Tapela K, Djomkam Zune AL, Owoicho O, Bediako Y, Duodu S. Parallels in sepsis and COVID-19 conditions: implications for managing severe COVID-19. Front Immunol. 2021;3(12):602848. doi: 10.3389/fimmu.2021.602848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukada A, Kitagawa Y, Matsuoka M, Sakai J, Imai K, Tarumoto N, Orihara Y, Kawamura R, Takeuchi S, Maesaki S, Maeda T. Presepsin as a predictive biomarker of severity in COVID-19: A case series. J Med Virol. 2021;93:99–101. doi: 10.1002/jmv.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaninotto M, Mion MM, Cosma C, Rinaldi D, Plebani M. Presepsin in risk stratification of SARS-CoV-2 patients. Clin Chim Acta. 2020;507:161–163. doi: 10.1016/j.cca.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dell'Aquila P, Raimondo P, Orso D, De Luca P, Pozzessere P, Parisi CV, Bove T, Vetrugno L, Grasso S, Procacci V. A simple prognostic score based on troponin and presepsin for COVID-19 patients admitted to the emergency department: a single-center pilot study. Acta Biomed. 2021;92:e2021233. doi: 10.23750/abm.v92i4.11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ducastel M, Chenevier-Gobeaux C, Ballaa Y, Meritet JF, Brack M, Chapuis N, Pene F, Carlier N, Szwebel TA, Roche N, Terrier B, Borderie D. Oxidative stress and inflammatory biomarkers for the prediction of severity and ICU admission in unselected patients hospitalized with COVID-19. Int J Mol Sci. 2021;22:7462. doi: 10.3390/ijms22147462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keskinidou C, Vassiliou AG, Zacharis A, Jahaj E, Gallos P, Dimopoulou I, Orfanos SE, Kotanidou A. Endothelial, immunothrombotic, and Inflammatory biomarkers in the risk of mortality in critically Ill COVID-19 patients: the role of Dexamethasone. Diagnostics (Basel) 2021;11:1249. doi: 10.3390/diagnostics11071249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schirinzi A, Cazzolla AP, Lovero R, Lo Muzio L, Testa NF, Ciavarella D, Palmieri G, Pozzessere P, Procacci V, Di Serio F, Santacroce L. New insights in laboratory testing for COVID-19 patients: looking for the role and predictive value of human epididymis secretory protein 4 (HE4) and the innate immunity of the oral cavity and respiratory tract. Microorganisms. 2020;8:1718. doi: 10.3390/microorganisms8111718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kocyigit A, Sogut O, Durmus E, Kanimdan E, Guler EM, Kaplan O, Yasar O. Circulating furin, IL-6, and presepsin levels and disease severity in SARS-CoV-2–infected patients. Sci Prog. 2021;104(2-suppl):00368504211026119. doi: 10.1177/00368504211026119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippi G, Sanchis-Gomar F, Henry BM. Presepsin value predicts the risk of developing severe/critical COVID-19 illness: results of a pooled analysis. Clin Chem Lab Med. 2021;60:e1–e3. doi: 10.1515/cclm-2021-0848. [DOI] [PubMed] [Google Scholar]
- 25.Domi H, Matsuura H, Kuroda M, Yoshida M, Yamamura H. Simple prognostic factors and change of inflammatory markers in patients with severe coronavirus disease 2019: a single-center observational study. Acute Med Surg. 2021;8:e683. doi: 10.1002/ams2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewi R, Kaswandani N, Karyanti MR, Setyanto DB, Pudjiadi AH, Hendarto A, Djer MM, Prayitno A, Yuniar I, Indawati W, Prawira Y, Handryastuti S, Sjakti HA, Hidayati EL, Muktiarti D, Soebadi A, Puspaningtyas NW, Muhaimin R, Rahmadhany A, Octavius GS, Puspitasari HA, Jasin MR, Tartila T, Putri ND. Mortality in children with positive SARS-CoV-2 polymerase chain reaction test: Lessons learned from a tertiary referral hospital in Indonesia. Int J Infect Dis. 2021;107:78–85. doi: 10.1016/j.ijid.2021.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirza FH, Baig FA, Syed S. Diagnostic value of novel presepsin and inflammatory biomarkers in predicting the clinical course of COVID-19. Med Bulletin of Haseki/Haseki Tip Bulteni. 2021;59:5. [Google Scholar]
- 28.Farag SM, Nasr RA, El Sheikh NG, Khattab MA. Presepsin as a predictive indicator of severity in Coronavirus disease-2019 (COVID-19) Novel Research in Microbiology Journal. 2021;5:1325–1337. doi: 10.21608/nrmj.2021.190249. [DOI] [Google Scholar]
- 29.Kim SW. Presepsin and monocyte distribution width as a useful early biomarker of severity in patients with covid-19. Chest. 2021;160:A1074. doi: 10.1016/j.chest.2021.07.994. [DOI] [Google Scholar]
- 30.Çaglar FN, Yildiz C, Korkusuz R, Yasar KK, Isiksacan N. Serum presepsin levels among patients with COVID-19. Indian J Med Spec. 2022;13:17–22. doi: 10.4103/injms.injms_77_21. [DOI] [Google Scholar]
- 31.Morales-Cely L, Bravo-Castelo LA, Bustos-Moya IG, Fuentes Y, Lozada-Arciniegas J, Ibañez-Prada ED, Narváez – Ramírez O, Ramirez P, Parra-Tanoux D, Gomez-Duque D, Gamboa-Silva E, Caceres E, Reyes LF, 319 Presepsin as a prognostic biomarker for mortality in COVID-19 patients vs community-adquired pneumonia CAP patients in open forum infectious diseases. Oxford Univ Press. 2021;8:S265–S265. [Google Scholar]
- 32.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology MOOSE group meta- analysis of observational studies in epidemiology: a proposal for reporting. JAMA. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 34.Rotondo JC, Martini F, Maritati M, Caselli E, Gallenga CE, Tramarin ML, Guarino M, De Giorgio R, Mazziotta C, Badiale G, Mauro Tognon M, Contini C. Advanced molecular and immunological diagnostic methods to detect SARS-CoV-2 infection. Microorganisms. 2022;10:1193. doi: 10.3390/microorganisms10061193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Institutes of Health (2014). Quality assessment tool for observational cohort and cross-sectional studies.
- 36.R Development Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 37.Lisco G, De Tullio A, Stragapede A, Solimando AG, Albanese F, Capobianco M, Giagulli VA, Guastamacchia E, De Pergola G, Vacca A, Racanelli V, Triggiani V. COVID-19 and the endocrine system: a comprehensive review on the theme. J Clin Med. 2021;10:2920. doi: 10.3390/jcm10132920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bivona G, Agnello L, Ciaccio M. Biomarkers for prognosis and treatment response in COVID-19 patients. Ann Lab Med. 2021;41:540–548. doi: 10.3343/alm.2021.41.6.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed S, Mansoor M, Shaikh MS, Siddiqui I. Presepsin as a predictive biomarker of severity in COVID-19: a systematic review. Indian J Crit Care Med. 2021;25:1051–1054. doi: 10.5005/jp-journals-10071-23967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faes C, Abrams S, Van Beckhoven D, Meyfroidt G, Vlieghe E, Hens N, Belgian, Collaborative group on COVID-19 hospital surveillance time between symptom onset hospitalisation and recovery or Death: statistical analysis of Belgian COVID-19 patients. Int J Environ Res Public Health. 2020;17:7560. doi: 10.3390/ijerph17207560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset is available for reviewers on reasonable request.