Abstract
Biomarkers are crucial biological indicators in medical diagnostics and therapy. However, the process of biomarker discovery and validation is hindered by a lack of standardized protocols for analytical studies, storage and sample collection. Wearable chemical sensors provide a real-time, non-invasive alternative to typical laboratory blood analysis, and are an effective tool for exploring novel biomarkers in alternative body fluids, such as sweat, saliva, tears and interstitial fluid. These devices may enable remote at-home personalized health monitoring and substantially reduce the healthcare costs. This Review introduces criteria, strategies and technologies involved in biomarker discovery using wearable chemical sensors. Electrochemical and optical detection techniques are discussed, along with the materials and system-level considerations for wearable chemical sensors. Lastly, this Review describes how the large sets of temporal data collected by wearable sensors, coupled with modern data analysis approaches, would open the door for discovering new biomarkers towards precision medicine.
Subject terms: Sensors, Engineering, Bioanalytical chemistry, Biomarkers, Techniques and instrumentation
Wearable chemical sensors are effective tools for exploring novel non-invasive biomarkers in alternative body fluids. This Review introduces criteria, strategies and technologies involved in biomarker discovery using wearable chemical sensors coupled with data analysis towards precision medicine.
Introduction
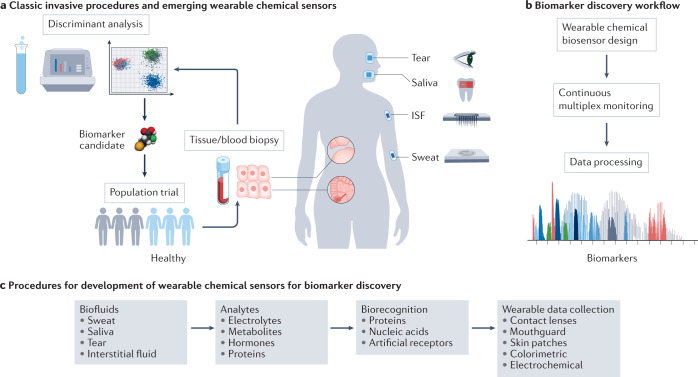
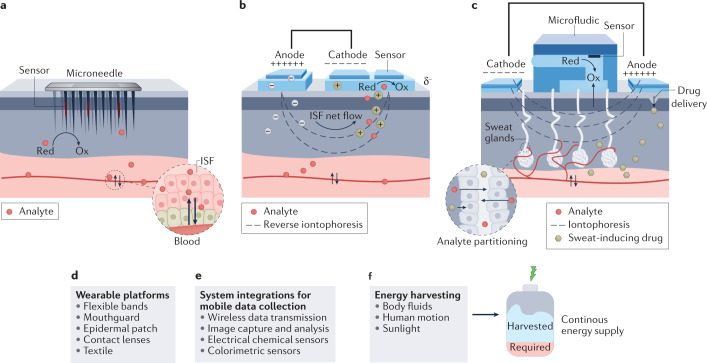
Biomarkers are important biophysical and biochemical parameters, detected as an indicator of certain biological, physiological and pathogenic processes or as pharmacological responses to therapeutic interventions1,2 (Box 1). The levels of certain biomarkers will fall above or below the norm in individuals who are sick. Advances in medicine related to the ability of precise diagnosis, treatment and prevention of diseases are directly related to the biomarkers. Therefore, the discovery of new biomarkers is very important to predict the onset and course of a disease and to develop effective therapeutic drugs. When identifying a potential biomarker, the lack of standardized protocols for sample collection, storage and analytical procedures makes it difficult to assess and verify the performance of the biomarker candidate in a specific health condition. The research for biomarker discovery usually requires participants to attend frequent hospital visits and tolerate invasive procedures (for example, blood draw or tissue biopsy), which increases the drop-out rate and hinders the study of biomarkers for diagnostics and prognostics of asymptomatic diseases1 (Fig. 1a). Because of this, when compared with episodic and invasive laboratory blood or tissue analyses, wearable chemical sensing is becoming an emerging technology for the screening of potential biomarkers in alternative body fluids, such as sweat, saliva, tears and interstitial fluid (ISF), as it can capture rich molecular information non-invasively and in real time3–6 (Fig. 1a). Discovering new non-invasively accessible biomarkers could be realized through continuous monitoring of the analytes using suitably designed wearable sensors coupled with data processing (Fig. 1b).
Fig. 1. Wearable chemical sensor-enabled biomarker discovery.
a, Biomarker discovery in precision medicine via classic tissue or blood biopsy procedures and via emerging wearable chemical sensors. b, General workflow for wearable chemical sensor-enabled biomarker discovery. c, Workflow procedures for the development of wearable chemical sensors towards biomarker discovery. ISF, interstitial fluid.
The emerging Internet of things, a network of connected objects (for example, sensors) that could exchange data over the Internet, promises to revolutionize traditional medical practices in centralized clinical settings. The Internet of things also presents a tremendous opportunity for developing wearable sensor technologies through integrating and miniaturizing analytical tools into a body-worn platform which may enable remote, wireless personalized health monitoring (Fig. 1c). Commercially available health monitors (for example, smartwatches) are already capable of autonomously monitoring several physiological parameters, such as heart rate, respiration rate, oxygen saturation, blood pressure and skin temperature. Wearable chemical sensors, such as continuous glucose monitors, convert the levels of various analytes in body fluids into measurable electrical, optical or piezoelectric signals. Recent advances in wearable biosensors have enabled continuous measurement of chemical biomarkers, including dynamic levels of electrolytes, metabolites, nutrients, drugs, hormones and proteins in blood surrogate biofluids4,6–10. This unique ability provides wearable chemical sensors with great value in the era of multi-omics (for example, genomics, proteomics, metabolomics, pharmacogenomics), enabling myriad molecular signatures to be monitored simultaneously and in real time, offering great advantages over conducting multiple single analyte assays11. Such wearable chemical sensors, in combination with information gathered from wearable physical sensors (that monitor biophysical parameters such as temperature, heart rate and motion), modern data processing and prediction algorithms, allow investigation into the roles of the distinct molecular signatures underlying disease prognosis. In this regard, wearable sensors could serve as a powerful tool for discovering novel biomarkers through identifying intricate correlations between analytes and disease status.
This Review provides an overview of wearable chemical sensor research and sheds insight into future directions of wearable sensor-based biomarker discovery. Firstly, the classification and chemical properties of potential biomarker candidates required for sensor design and fluid sampling are introduced. This is followed by a discussion of electrochemical and optical transduction techniques used by existing wearable chemical sensors as well as emerging methods for the continuous monitoring of a broad range of molecular signatures. Finally, a summary of the rich molecular data obtained continuously under normal and abnormal conditions with powerful computational tools for identifying new biomarkers is provided. The goal of this Review is to bring attention to the unique abilities of rapidly emerging wearable chemical sensors, in conjunction with modern data analysis tools, to expand our understanding of biomarkers from the bench to the body.
Box 1 Classification of biomarkers.
According to the definition by the US Food and Drug Administration (FDA)174, there are seven categories of biomarkers: susceptibility, prognostic, diagnostic, prediction, monitoring, response and safety. Understanding these definitions allows the implementation of standard protocols by matching a biomarker with its appropriate purpose and condition, increasing the efficiency in the development of precise diagnostic and therapeutic approaches. This, in turn, facilitates the introduction of new strategies and tools for novel biomarker discovery.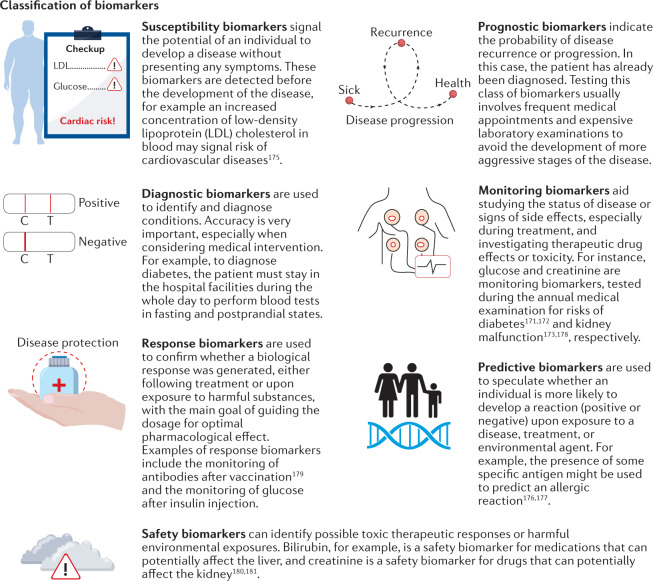
Wearable sensors in biomarker discovery
Despite the significance of the biomarkers in clinical assessment, it is challenging to discover new, specific, single-molecule biomarkers. Deep prior knowledge and understanding is required of a disease’s biological pathways and the pharmacology of new medications. However, most diseases are complex with intricate progressions depending on the patient’s underlying conditions, lifestyle and nutrition. Glucose, for example, can be used as a diagnostic and monitoring biomarker for diabetes, but can be also related to stress and other pathological conditions12. Moreover, electrolyte imbalance can be linked to dehydration, hyperkalaemia, kidney disorders and other pathologies13. The vast number of metabolites and small molecules linked to different medical conditions demonstrate the important role that these compounds play across processes of the human body14,15. Thus, the specificity of key biomarkers is a challenge, as it is likely that several known and unknown conditions might be related to any new compound. However, trial and error testing of the concentration of every biomarker for every disease is expensive and time-consuming.
Wearable chemical sensors offer an attractive approach to address these challenges by simultaneously monitoring a broad spectrum of molecular signatures towards multi-omics analysis7. The unique ability to perform continuous monitoring and identify temporal patterns can be a great asset in the discovery of biomarkers that vary rapidly within a short period of time. Examples of such important applications could include the use of wearable biosensors to monitor cardiac, epilepsy or other acute diseases, towards understanding the physiological changes in the body prior to such events. Currently, there are still no devices or tests able to predict life-threatening events such as heart attack, epilepsy episodes or heat stroke. Moreover, numerous health conditions, such as Alzheimer disease or long COVID-19, do not yet have standard alarming biomarkers. Continuous and real-time monitoring of multiple biomolecules during an individual’s daily activities could provide invaluable information about the chemical signature of the abnormal health conditions. Such an ability offers the fast screening of molecular signatures, and upon coupling to data-mining tools leads to a more effective biomarker discovery process which cannot be achieved with any other analytical tool.
To be suitable for wearable-based biomarker discovery, the molecule or molecular fragment candidates should be present in alternative biofluids such as sweat, saliva and ISF. Considering that they commonly originate from either blood or secretion glands, the biomarker candidates should be stable, preferably charged species, have small molecular weight for easy partition from blood and have potential links with their blood counterpart or targeted health condition8. These commonly include various metabolites, electrolytes, nutrients, hormones and therapeutic drugs7,8,16–25 (Table 1). The stability of these molecules could be impacted (directly or indirectly) by various parameters including the testing temperature and the pH and reactive oxygen species levels in the body fluids in situ. These parameters could vary with the activities and pathophysiological conditions. This can cause a shift in the chemical properties of the biomarker such as ionization states or surface charges, which may eventually impact the performance of wearable sensors. It should also be noted that most of the analytes in non-invasively accessible biofluids are not clinically approved biomarkers, although many of their counterparts in the blood are currently considered biomarkers. For example, sweat or saliva glucose are not used in a clinical context, whereas blood glucose is the well-recognized biomarker for diabetes.
Table 1.
Molecular signatures found in blood, interstitial fluid (ISF), saliva and sweat
| Target | Reported range | Molecular weight (Da) | Health association examples | Refs. | ||||
|---|---|---|---|---|---|---|---|---|
| Blood (μM) | ISF (μM) | Sweat (μM) | Saliva (μM) | |||||
| Metabolites | Glucose | 3.9 × 103–5.5 × 103 | 0.8 × 103–6 × 103 | 6–300 | 30–140 | 180 | Diabetes, metabolic syndrome | 3–10,12,15,22,24,34,38–41,49,50,86,102–105,109,111,116,124,140,144,171,172 |
| Lactate | 0.5 × 103–10 × 103 | 2.5 × 103–6.6 × 103 | 3.7 × 103–50 × 103 | <3.5 × 103 | 90 | Lactic acidosis | 3–10,15,28,29,33,116,124,131,134,140 | |
| Uric acid | 100–400 | 4–100 | 100–250 | 168 | Gout, cardiovascular diseases | 43,123 | ||
| Creatinine | 65–140 | 9–18 | 7–17 | 113 | Kidney diseases | 22,60,162,173 | ||
| Electrolytes | Na+ | 135 × 103–145 × 103 | 10 × 103–100 × 103 | 27 × 103–217 × 103 | 23 | Hydration, heart failure, liver disease | 3–10,18–20,33,42 | |
| Cl– | 96 × 103–106 × 103 | 10 × 103–100 × 103 | 7 × 103–18 × 103 | 35 | Cystic fibrosis | 3–10,18–20,33,42,106,107 | ||
| Hormones | Cortisol | 0.7 × 10–4–690 × 10–3 | Similar to blood plasma | 1 × 10–4–20 × 10–3 | 1 × 10–4–20 × 10–3 | 362 | Stress, anxiety, Cushing syndrome | 16–18,27 |
| Testosterone |
52 × 10–5–24 × 10–4 (females) 97 × 10–4–382 × 10–4 (males) |
Lack of report |
19 × 10–6–35 × 10–6 (females) 263 × 10–6–544 × 10–6 (males) |
288 | Heart muscle damage, fertility issues, irregular menstrual periods | 7,8 | ||
| Nutrients | Tyrosine | 200–400 | Similar to blood plasma | 66–300 | 0.07–205 | 181 | Metabolic disorders, gout | 7,43 |
| Tryptophan | 55–80 | 20–91 | Not reported | 204 | Anxiety, nerve damage | 7,22 | ||
| Vitamin C | 2.8–98 | <36 | 45–62 | 176 | Connective tissue defects, joint pain | 6,7,9,10,44 | ||
| Proteins | Cytokines | Picomolar to nanomolar | 15-42% of blood plasma | 1.97 × 10–5 (IL-1α) (mean) | 2.8 × 10–7–2.2 × 10–6 | >5,000–10,000 | Immunodeficiency disorders, cytokine storm | 8,21,158 |
| C-reactive protein | 7 × 10–3–29 × 10–3 | Lack of report | 4.2 × 10–6 – 2.5 × 10–4 | 3.3 × 10–6 – 1.8 × 10–4 | 120,000 | Coronary heart disease, obstructive pulmonary disease | 121 | |
| Drugs | Levodopa | 0.5–15 | Similar to blood plasma | Similar to plasma | 197 | Parkinson’s disease | 23 | |
| Acetaminophen | 66–132 | <50 | 151 | Fever, pain | ||||
Of all the criteria in biomarker discovery, the most critical is to confirm that the target molecule is present in the biofluid. Most analytes detected in alternative biofluids are small molecules as they can partition easily through tissue cells, whereas large macromolecules such as proteins may not be readily partitioned to these biofluids22,26. Once the presence of the molecule in the fluid is confirmed, the molecule needs sufficient stability to be detected. Although detecting radicals and unstable complexes is possible, it is not very accurate because of the short half-life of the analyte. Moreover, the molecule may react chemically with other compounds (for example, oxygen) in the body fluids, resulting in misleading lower concentrations5,8,10.
Assuming that a target is stable in the biofluid, its levels need to be correlated with its blood concentrations. For sweat, saliva and tears sensors, the molecule can either be partitioned from blood, where the correlation can usually be achieved successfully, or be excreted by their respective glands8,26 where the overall correlation with blood will be affected by the mixed molecules secreted from the glands. Even if a molecule’s secretion to these biofluids is primarily based on diffusion, there is still a time lag to be considered. For example, although blood glucose peaks within 10–15 min upon food ingestion, a wearable sensor can only detect the peak in an alternative biofluid after 15–20 min. Dilution effects are another major factor that needs to be considered as biofluid secretion rates could influence the final secreted molecular concentrations. Potentially, internal calibration methodologies27 can be introduced to obtain a normalized and accurate response.
To facilitate biomarker discovery, a wearable chemical sensor can be developed to analyse the selected molecular signature continuously in real time (Fig. 1c). To prove the effectiveness of a new chemical sensor, the biofluid is analysed simultaneously in situ and via external validation using benchtop laboratory analyses, to ensure similar trends are observed. The sensor operation range must cover the physiologically relevant concentrations, which usually requires a very low detection limit and high sensitivity along with a wide linear range. For wearable use, the desired sensor must be fully biocompatible and selective to the target molecule in the complex biofluid matrix without additional sample processing steps. This high selectivity is realized by using immobilized recognition receptors such as enzymes, aptamers or molecularly imprinted polymers (MIPs) that selectively bind to the target molecules. By following these criteria, and with the introduction of novel detection methodologies, it would be expected that a wearable sensor may be able to perform multi-omic analysis using non-invasive biofluids. Wearable sensors that can detect DNA and RNA molecules, fragments, hormones, proteins and whole small organisms, including viruses, will be of utmost importance for public health monitoring and surveillance.
Efficient biomarker discovery requires robust molecular collection in participants. The main challenges of wearable biosensors for collecting reliable data include the noise signal imposed by human motion, influences of the pH, temperature, conductivity and refreshing of the body fluids on sensing responses. Suitable filtering and calibration mechanisms are required, alongside selection of the platform/system for fluid extraction and sampling (Fig. 1c). To enable biomarker validation, longitudinal and cross-sectional studies need to be performed to collect large sets of data; modern data-mining techniques (such as machine learning) can facilitate biomarker identification and generate predictive algorithms for disease monitoring, diagnosis, treatment and prevention.
Wearable chemical sensing strategies
Wearable sensors have attracted tremendous attention over the past decade6,28–31 as they are able to perform real-time detection in situ directly on the body in a reagent-free and continuous manner. Protocols requiring additional washing or chemical addition steps are generally not suitable for on-body operation of wearable chemical sensors. Currently, wearable sensors are primarily electrochemical sensors21,32,33 or optical sensors34–37 based on signal transduction mechanisms.
Electrochemical biosensing strategies
Electrochemical sensors convert target analyte information into a measurable electrical signal in real time with high sensitivity. For this, bioreceptor (such as enzyme, antibody, DNA and aptamer) molecules are usually immobilized on the surface of the conductive electrode material to selectively interact with the target molecules. Amperometry, potentiometry and voltammetry are the most common electrochemical detection techniques32,33.
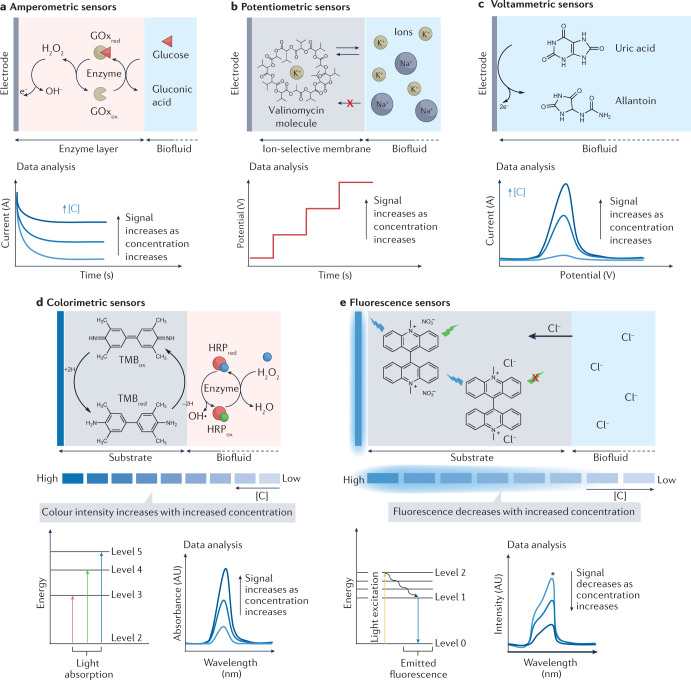
In the amperometric sensors, an applied potential facilitates the redox reaction of the electroactive molecule, and electron transfer between the molecule and the electrode is measured as the current signal. Enzymes are typically used in wearable amperometric sensors as the biocatalytic reaction can confer great selectivity and reversibility to the sensor system towards continuous analysis of target molecules (for example, glucose and lactate) in biofluids38,39. In Fig. 2a, the amperometric enzymatic detection mechanism is exemplified for the glucose sensor whose sensing electrode is modified with an enzyme (that is, glucose oxidase (GOx))-loaded biorecognition layer. Glucose molecules from the biofluid are oxidized to gluconic acid, catalysed by GOx. The detection of glucose is achieved by amperometric techniques. This is done by applying a fixed potential to detect the enzymatically generated hydrogen peroxide (H2O2) by-product (first-generation glucose biosensor) (Fig. 2a), to monitor the reaction of a mediator with low redox potential (second-generation glucose biosensor), or to monitor the direct electron transfer from the enzymatic reaction without any intermediate stage (third-generation glucose biosensor)38,39. The measured response in current is proportional to the concentration of the target substrate. Given the ability of performing continuous measurements with a fast response, amperometric enzymatic sensors have been successfully applied in commercial continuous glucose monitors40,41.
Fig. 2. The primary biorecognition and signal transduction strategies in wearable chemical sensors.
a–c, Recognition and signal transduction for wearable electrochemical biosensors. a, Amperometric enzymatic electrodes: for glucose sensing, the working electrode is modified with glucose oxidase (GOx) and the glucose molecule is oxidized to gluconic acid. Glucose is detected by applying a fixed potential to measure the hydrogen peroxide (H2O2) by-product or to monitor the reaction of a mediator with low redox potential. The generated current signal increases with the increase of glucose concentration. b, Potentiometric ion-selective sensors: the working electrode is modified with an ion-selective membrane containing the ionophore (for example, valinomycin for potassium ion sensing) molecules with cavities allowing only ions with specific charge and size to interact. The measured potential has a log-linear relationship with the target ion concentration. c, Voltammetric sensors: an electroactive molecule (for example, uric acid) can directly lose or donate electrons on the electrode surface when a redox potential is applied to the sensing electrode. The measured oxidation or reduction peak current height is directly proportional to the concentration of the target analyte. d,e, Recognition and transduction for wearable optical biosensors. d, Colorimetric sensors: the colour indicator molecule is immobilized on a substrate and upon contact with the analyte it changes colour. For hydrogen sensing based on horseradish peroxidase (HRP), the colour change is promoted by changes in the redox state of the chromophore molecule 3,3′,5,5′-tetramethylbenzidine (TMB) during the enzymatic degradation of H2O2; the measured light absorbance or colour intensity directly correlates with the concentration of target analyte. e, Fluorescence sensors: fluorometric sensing relies on a high-energy light source (for example, ultraviolet light or blue light) to excite the fluorophore molecule so it emits light with a longer wavelength through radiative electron transitions. For example, lucigenin can be used as a fluorescent indicator for chloride ion sensing; the chloride ion (Cl–) reacts with lucigenin by replacing NO3−, resulting in the quench of lucigenin fluorescence intensity through a collisional mechanism. The change of fluorescence intensity is directly proportional to the change in analyte concentration. red, reduced analyte; ox, oxidized analyte; [C], analyte concentration.
In contrast to amperometric sensors, there is no applied potential or current flow involved in potentiometric sensors. Instead, these sensors rely on the potential difference between a reference electrode and a working electrode. Solid-state ion-selective electrodes are typically employed in wearable biosensors to monitor the levels of electrolytes or ions (for example, Na+, K+ and Ca2+) in biofluids42 (Fig. 2b). The working electrode, or ion-selective electrode, is usually modified with an ion-selective recognition membrane composed of ionophore molecules that form cavities in the polymer film, allowing only ions with specific charge and size to interact. This selective recognition process creates an electrochemical phase boundary potential that is translated by the transducer into the voltage signal42. The potential signal measured between the ion-selective electrode and a solid-state reference electrode (for example, Ag/AgCl) has a log-linear relationship with the target ion concentration according to the Nernst equation3.
Another versatile electrochemical detection approach is voltammetric detection where the applied potential varies with controlled steps and speed, whereas the alternations in the resultant current signal are monitored. There are various approaches to sweep the potential including cyclic voltammetry, differential pulse voltammetry and square wave voltammetry. Voltammetric detection is often applied to monitor electroactive molecules (for example, uric acid and vitamin C) by detecting their direct oxidation/reduction reaction around a specific redox potential on the electrode surface under an applied potential waveform43,44 (Fig. 2c). Compared with amperometry or cyclic voltammetry, differential pulse voltammetry and square wave voltammetry are able to minimize the background charging current and achieve highly sensitive electrochemical measurements. Similarly, stripping voltammetry, involving an analyte preconcentration step followed by a voltammetric scan, is able to detect ultra-low levels of heavy metals in body fluids45–47. In the absence of specific bioreceptors (such as enzymes), the selectivity of voltammetric electroactive molecule detection is limited as different molecules may have a similar redox potential.
In addition to these electrochemical signal transduction techniques, numerous other approaches involving the measurement of resistive, capacitive and impedance changes of the sensing electrode upon selective analyte biorecognition event could also be used for wearable electrochemical biosensing48. It should also be noted that several key challenges for wearable electrochemical sensors have yet to be fully addressed, including electrode biofouling, electrical noise, electronic backbone integration and power supply.
Optical biosensing strategies
Optical sensor systems rely on chemical or biological reactions capable of modifying optical detection elements, including changes in light absorbance (for example, colour) or emission (for example, fluorescence or luminescence)34–36.
In wearable colorimetric sensors, chromophore molecules are usually used as colour indicators. External stimuli can change the electron state of the chromophore molecule, resulting in the absorption of photons with different wavelengths (visualized as colour changes). Common examples of such stimuli for wearable colorimetric biosensors include gain or loss of electrons (electrochromism) or ions (ionochromism), or changes in pH (halochromism). A common colorimetric mechanism coupled with enzymatic reaction as the target biorecognition event is illustrated in Fig. 2d. The colour change of the chromophore 3,3′,5,5′-tetramethylbenzidine (TMB) is promoted by the reaction of the chromophore molecule with H2O2; the measured colour intensity directly correlates to the concentration of target analyte.
In wearable fluorescence sensors, fluorophores are commonly used for signal transduction. Under specific light excitation, the fluorophore molecules can absorb the light energy and re-emit at a longer wavelength. An ionochromism process that alters the fluorescent light signal intensity is demonstrated in Fig. 2e. The main difference between a colorimetric and a fluorescence sensor is the light source necessary to observe the phenomenon. Qualitative analysis of colorimetric sensors can be performed by the naked eye or directly using a phone camera, whereas additional optical accessories are often needed to accurately acquire fluorescence or luminescence outputs. As a result, fluorescence sensors are usually more sensitive than colorimetric sensors.
The main advantages of these optical biosensors are the simple fabrication process and system integration on the skin as no additional electronic components are needed on the body. However, high sensitivity, high long-term stability and high-frequency data collection are difficult to achieve with a miniaturized external wearable system (for example, cell phone or hand-held optical reader)49–51.
Emerging wearable sensing strategies
Despite the great promise for the existing wearable sensing strategies, enzymatic reactions, ionophore–ion interactions and direct molecular oxidation/reduction can only be used to directly monitor a limited number of targets4,5,8. The majority of body fluid analytes, such as circulating metabolites, nutrients, hormones and proteins, play important roles in clinical risk assessment and diagnosis (Table 1) but cannot be monitored using these wearable sensing approaches. For example, the concentrations of melatonin, prolactin and reproductive and growth hormones could be used for the prediction of seizures52, and troponin I and brain natriuretic peptides are closely related to the severity of heart failure53. Selective detection of analytes in biofluids usually requires bioaffinity receptors, such as antibodies, DNAs/RNAs, aptamers and MIPs. However, existing bioaffinity sensors are often one-time use, and require multiple incubations and washing processes for regenerating their receptor, making it challenging to apply such sensors towards continuous biosensing in situ48,54.
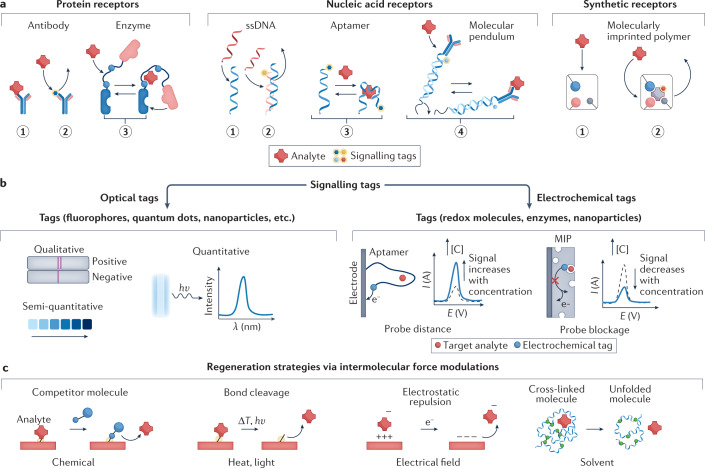
In this direction, a very promising approach for affinity sensors, towards detection of several classes of molecules, is the so-called ‘switch biosensor’55. Switch sensors mimic natural bioprocesses using artificial or modified biomolecules to undergo conformational changes upon binding the target molecules. Commonly used biomolecular switches include proteins56, oligonucleotides57,58 and synthetic receptors (MIPs)59–61 (Fig. 3a). The structural changes are based on a specific interaction such as hydrogen bonding, van der Waals force and hydrophilicity between the target and bioreporter, conferring selectivity and reversibility to the system. To achieve in situ reagent-free molecular detection, biomolecules can be coupled with either optical tags (for example, fluorophores or quantum dots) or electrochemical tags (for example, redox or enzyme molecules) for signal transduction62 (Fig. 3b).
Fig. 3. Emerging wearable sensing strategies for biosensing.
a, Target recognition based on bioaffinity receptors and molecular switches for wearable biosensing. Protein receptors include antibodies for target recognition through direct binding (panel 1) or displacement of a signalling probe (panel 2) and enzyme switches that change molecular configuration upon target binding (panel 3). Nucleic acid receptors include single-stranded DNA (ssDNA) used for target recognition through direct DNA hybridization (panel 1) or displacement of another DNA strand with a signal probe (panel 2), aptamer switch with shape change upon binding of the target molecule (panel 3) or a DNA-based molecular pendulum (coupled with a redox probe and a protein receptor) that changes the probe–electrode distance upon target binding (panel 4). Synthetic receptors such as molecularly imprinted polymers (MIPs) can perform target recognition through direct target binding (panel 1) or displacement of molecules with a signal probe (panel 2). b, Optical and electrochemical signalling tags used in bioaffinity sensors for in situ signal transduction. Optical tags such as fluorophores, quantum dots and nanoparticles can be used to transduce the target recognition to measurable optical signals. The resulting optical signal could offer qualitative analysis (yes/no response), semi-quantitative sensors based on colour change and quantitative analysis via absorbance or luminescence intensities. Electrochemical tags such as redox probe molecules, enzymes and nanoparticles are used to transduce the target recognition to measurable electrical signals; for example, the recognition of targets with aptamers decreases the distance of the tagged redox probe from the electrode, leading to increased redox signal; binding of target molecules with MIP sensors reduces the exposure of the redox probe to the biofluid, leading to a decreased redox signal. c, In situ sensor regeneration strategies based on intermolecular force modulations. Biosensors can be regenerated chemically, by introducing a competitor molecule that has a stronger binding affinity to the receptor than the target molecule; with heat and light, as external energy to cleave the bond between the receptor and target molecule; by electrical fields, which remove the target molecule by electrostatic repulsion; or by solvent effects, varying solution properties (for example, pH and ionic strength). λ, wavelength; E, potential; I, current; [C], analyte concentration.
Aptamers are molecules exhibiting target-induced conformational changes, and are highly attractive structures when analysing the presence of analytes by distance modulation between the electrode and the signalling redox indicator (for example, methylene blue)63–70 (Fig. 3b). A major advantage of aptamers is the ability to engineer their desired nucleic-base sequences specifically for the target analytes through an aptamer designing process called systematic evolution of ligands by exponential enrichment71. Electrochemical aptamer-based sensors have been exemplified in the continuous detection of multiple chemotherapeutic drugs, including doxorubicin, kanamycin and tobramycin in human whole blood and in live rats72. Binding of the target small molecules with the aptamer receptor triggered reversible conformational changes, resulting in an increased redox current correlating with the target concentration (Fig. 3b). Similarly, fluorometric switches are also commonly used in aptamer or protein biosensors in which the aptamer conformation modulates the quenching of the probe’s fluorescence signal73,74; the measured decrease in fluorescence intensity is proportional to the increased concentration.
In addition to aptamers, electrochemical DNA switches coupled with antibodies have recently been demonstrated for reagentless monitoring of proteins (for example, troponin I) in multiple biofluids (that is, saliva, sweat, tears and blood) based on the motion of an inverted molecular pendulum75. For the detection, a positive potential is applied to electrostatically attract the redox reporter-tagged DNA molecule to the surface, to decrease the probe–electrode distance; the binding of the target molecule increases the weight on the pendulum body, leading to a signal change that is proportional to the analyte concentration. In another example, wearable sensing of nucleic acids, such as SARS-CoV-2 guide RNA, was achieved in a breath sample via freeze-dried reactions in the face mask including lysis, reverse transcription–recombinase polymerase amplification and CRISPR–Cas12a-specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) sensing76. The presence of pieces of SARS-CoV-2-derived amplified RNAs — amplicons — leads to the cleavage of quenched single-stranded DNA (ssDNA) fluorophore probes which can be detected via a colorimetric lateral flow assay strip.
Although these emerging bioaffinity sensing technologies hold great promise in the detection of a broad spectrum of analytes in biofluids, there are many challenges related to stability and reversibility that need further attention. In particular, continuous detection is strongly dependent on the ability to regenerate the active sensor surface and realize reversible target bonding in situ without extra washing steps or chemical addition. Such sensor regeneration could be achieved by modulating the intermolecular forces between the bioreceptor and the target molecule (for example, hydrogen bond, van der Waals force and electrostatic interaction) through various stimuli such as heat, light, electrical field, ultrasound field, competitive binding and ionic strength or pH77–79 (Fig. 3c).
Materials and wearable system design
Flexible materials and miniaturized devices play an important role in developing wearable chemical sensors for enhanced, reliable and in situ molecular data collection. There are considerations both in materials and at the system level to effectively develop wearable sensors in biomarker discovery.
Material considerations
Electrode materials, the electrode surface and surface coatings, and skin conformability are key considerations for optimizing the sensitivity, stability and longevity of wearable sensors.
Electrode materials
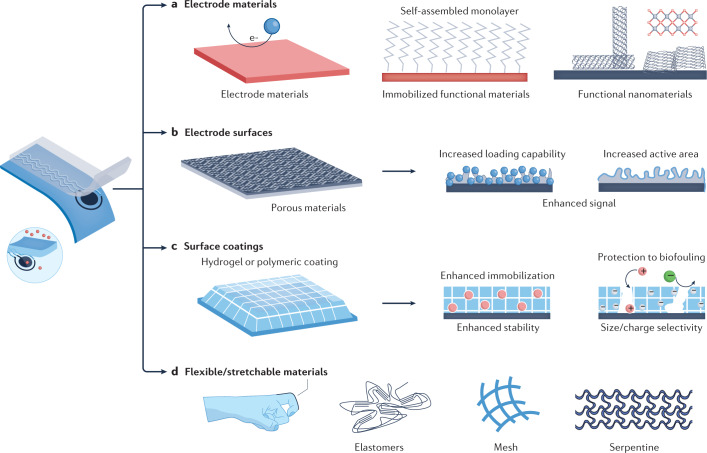
Common wearable electrochemical transducers rely on conducting carbon materials or gold surfaces owing to their high electrochemical stability, electrical conductivity80,81 and ability to anchor biorecognition elements efficiently using materials such as self-assembled monolayers82. Owing to their excellent electrochemical and optical properties, functional nanomaterials (for example, carbon nanotubes, graphene, MXenes, metal–organic framework) are also potential candidates for developing highly sensitive transducer surfaces83 (Fig. 4a). Moreover, carbon-based nanomaterials show great promise for wearable sensing electrodes as they can be mass produced at a low cost and have a wide operational potential window.
Fig. 4. Materials for enhanced wearable chemical sensing.
A sensor immobilized with functional materials could result in enhanced wearable sensing performance (left). a, Conducting and functional electrode materials for wearable chemical sensing. Gold transducer surfaces are suited for modification with self-assembled monolayers (for example, alkanethiol) for anchoring the biorecognition elements to introduce selectivity. Due to the excellent electrochemical and optical properties, functional nanomaterials (for example, carbon nanotubes, graphene, MXenes, metal–organic framework) are also used for highly sensitive transducer surfaces. b, Porous materials with high electrochemically active areas and high bioreceptor loading abilities towards enhanced in situ chemical sensing. c, Surface coatings (for example, hydrogel or polymeric coating) towards enhanced in situ chemical sensing. The properties of these coatings (for example, surface charge, hydrophilicity or porosity) can be tailored to increase receptor loading capabilities and resist the adhesion of fouling/interferent molecules. d, Soft flexible and biocompatible materials for enhanced wearable sensing. Flexible and stretchable materials can be used to realize enhanced skin conformability. The flexibility and stretchability of the sensors can be realized by using intrinsically stretchable materials (for example, elastomers) or through structure designs (for example, mesh or serpentine configuration designs).
Electrode surfaces
To further enhance the sensitivity of wearable chemical sensing, porous surfaces can be incorporated into the transducer — for example, nanoporous graphene84,85, graphitic nanocarbon structures86 or nanoporous gold87 — as they can increase the electron transfer area or bioreceptor loading capability resulting in an enhanced signal (Fig. 4b).
Surface coatings
Prolonged on-body monitoring of different biofluids is often hindered by gradual surface passivation in protein-rich biofluids. Anti-biofouling protective hydrogels or polymeric coatings, such as chitosan88, polyvinyl chlorides89, Nafion90 or serum albumin91, are widely used for extending the operational stability towards continuous reliable molecular profiling (Fig. 4c). The use of these surface-coating materials can tailor the surface charge, surface hydrophilicity or porosity to resist the adhesion of fouling/interferent molecules onto the electrode. For example, Nafion is a negatively charged polymer used as an anti-fouling coating against negatively charged proteins whereas some hydrogels consist of porous networks that allow only analytes with smaller sizes to reach the electrode surface.
Skin conformability
Conformal contact between wearable chemical sensors and the human body is realized by matching the mechanical properties of the device with those of the human tissues, through use of skin-friendly, soft and stretchable elastomers such as polyimide, polydimethylsiloxane (PDMS) or poly(styrene–butadiene–styrene) (SBS)92 (Fig. 4d). In addition, carbon nanomaterials, metal–carbon nanocomposites or metallic nanomaterials are commonly employed for developing stress-enduring carbon inks along with silver-based interconnect circuitry. Compared with highly elastic substrate materials, the conducting materials usually have low stretchability. A trade-off between the conductivity and stretchability is achieved by adopting an optimal ratio of conductive and elastomeric materials and design-based techniques (for example, mesh-like or serpentine-like structures), resulting in an attractive sensor performance along with comfort81,92 (Fig. 4d).
It should be noted that, in addition to the above factors, biocompatibility of the transducer and substrate materials is always crucial for wearable chemical sensors; biocompatible polymers, carbon materials and noble metals are preferred for meeting this requirement93–96.
System-level considerations
To ensure high performance for on-body analyte monitoring, the integrated wearable system must be carefully designed to allow efficient fluid sampling, reliable in situ sensing, wireless data collection and communication, and can be powered sustainably.
Fluid extraction and sampling
During blood circulation, the transcapillary filtration of the blood forms ISF, which has similar composition and levels of biomarkers to blood97,98. For efficient ISF profiling of disease-related biomarkers, microneedle-based electrochemical sensing systems have received considerable attention owing to their painless, minimally invasive, transdermal access97,99–101 (Fig. 5a). ISF can also be accessed non-invasively using reverse iontophoresis, a technique that facilitates the transport of charged molecules by applying an electric current. This approach allows the extraction of the ions and neutral molecules from the ISF to the skin surface102–105 (Fig. 5b). Potential challenges here are interpersonal variations due to differences in the skin properties. Direct iontophoresis can also initiate localized sweating without the need for heat or vigorous exercise, particularly in connection with transdermal delivery of muscarinic agonists (for example, pilocarpine, acetylcholine, carbachol) to induce localized sweating106–111. The sweat-stimulating electrodes (cathode and anode) are thus interfaced with the skin by a separating thin layer of the drug-loaded hydrogel (Fig. 5c). In addition, the microfluidics-based sample handling modules can be used for in situ intermittent storage and controlled handling of the secreted sweat without dilution, mixing or cross-contamination110,112–115 (Fig. 5c).
Fig. 5. System development for in situ wearable chemical sensing.
a, Minimally invasive microneedle-based interstitial fluid (ISF) extraction and chemical sensing. b, Non-invasive reverse iontophoresis-based ISF extraction and chemical sensing via a wearable device. c, Non-invasive iontophoresis-based sweat extraction, microfluidic sweat sampling and chemical sensing via iontophoresis using a wearable device. The sweat-stimulating electrodes are interfaced with the skin via a thin layer of the hydrogel loaded with agonists (for example, pilocarpine or carbachol). A small current is applied to transdermally deliver agonist molecules that stimulate the sweat gland to trigger local sweating. The microfluidics-based sample handling modules can be used for in situ intermittent storage and controlled handling of the secreted sweat. d, General wearable platforms. e, Wireless wearable system integration for mobile data collection. f, Wearable energy harvesting systems. These systems convert chemical energy in body fluids, mechanical energy from human motion and light into electricity to continuously power wearable sensors. Red, reduced analyte; Ox, oxidized analyte.
Wearable sensor platforms
Various skin-worn wearable sensors have been developed over the past decade for detecting and assessing the sweat-based biomarkers non-invasively, including epidermal tattoos98, patches116, flexible smart bands3,117 or smart textiles86 (Fig. 5d). Microfluidic wearable sensors can be developed to achieve fully autonomous sweat extraction, sampling and multiplexed sensing61. Such microfluidic wearables could minimize sweat evaporation to enable rapid sweat analysis. Tears and saliva are other important biofluids that contain a plethora of biomarkers of clinical importance118–121. Wearable biosensors based on contact lenses49, dental chips122 and mouthguard-based123 devices have thus been recently developed but need more evaluation in participants. Wearable microneedle arrays show considerable promise for realizing continuous monitoring of multiple biomarkers beyond glucose in ISF124. Owing to the close analyte correlation between blood and ISF, minimally invasive wearable ISF monitors are expected to provide the most accurate metabolic profiling compared with other wearables.
Wireless and continuous data collection
Wearable devices continuously collect biomarker profiles from different biofluids of the wearer, accompanied by real-time wireless signal transmission to mobile devices (Fig. 5e). Pre-calibrated wearable sensor arrays may accurately profile the level of multiple chemical biomarkers from different biofluids to create specific signature patterns. Similarly, in colorimetric-based optical devices, the signal can be directly seen in the chromogenic bands and these colour changes can be quantified for the patches using a pre-calibrated app-based data processor for finding the biomarker levels88 (Fig. 5e). Combining rich biomarker temporal profiles based on wearable electrochemical and optical sensing, and relating these profiles to abnormal body conditions, would serve to pinpoint the symptom-specific physicochemical fluctuation of the individual towards creating biomarker signatures.
Power
Complex wearable biosensing systems require suitable power sources to reach their full ability. Existing systems are commonly powered by batteries or through near-field communication but suffer from limited long-term continuous usability. Self-powered wearables, harvesting energy from body fluids, human motion or sunlight, are attractive candidates for next-generation wearable biosensors for biomarker discovery125–130.
Data-driven biomarker discovery
The emergent multiplexed wearable chemical sensors have broadened the spectrum of biomarkers available to diagnose, monitor and treat diseases. The continuous monitoring of multiple analytes via wearable biosensors has resulted in the collection of large sets of molecular data able to facilitate the assessment of a patient’s health status131,132. Alongside developing novel approaches to enhance the sensors’ sensitivity, selectivity and stability and creating smart bioelectronics for in situ measurements, systematic analysis of the chemical data provides insightful information and broader data sets of biomarkers related to a specific health condition43. In this regard, implementing modern data analysis tools such as machine learning algorithms to interpret and predict patterns in vast amounts of measurements can aid in the identification of robust and accurate biomarkers133.
Multiplexed multimodal wearable sensing
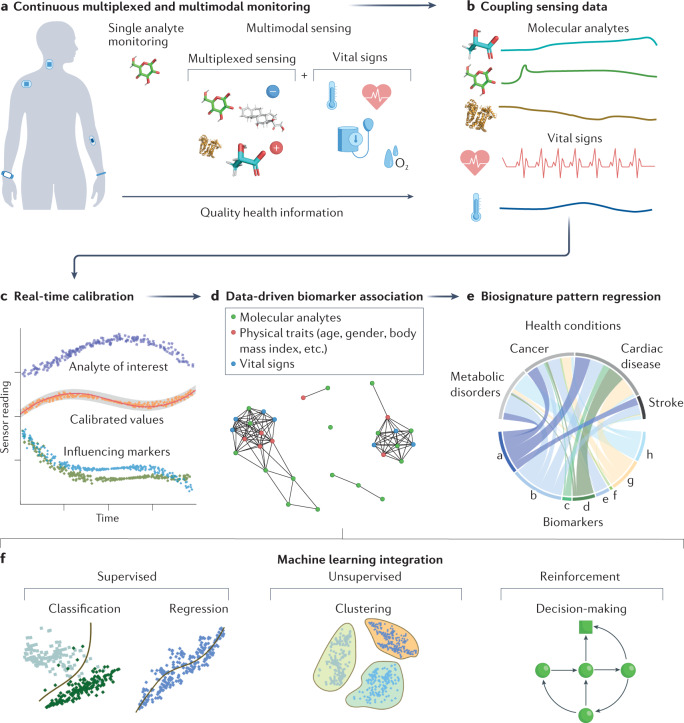
Physiological monitoring of a single chemical analyte has been proven ineffective for disease diagnostics as, in most cases, they are affected by a combination of other chemical and physical markers134. Although there are several models proposed for realizing multi-omics analysis135–137, they usually rely on molecular data from different techniques collected at different times, which increases the errors and challenges the implementation of a precise health condition prediction/correlation. Advances in the miniaturization of wearable bioelectronic devices, through modern fabrication protocols, have allowed the integration of several sensors in a confined space. This has allowed the development of microsensor arrays able to perform detection of different analytes from the same biofluid sample86,134,138–140 (Fig. 6a). Multiplexed wearable sensors can monitor multiple analytes simultaneously and in real time, giving them a unique potential in chemical biomarker discovery. Moreover, other physical biomarkers (that is, vital signs) contain rich information which can be included in the diagnosis and prognosis of numerous diseases (Fig. 6a).
Fig. 6. Wearable biosensor-enabled data-driven biomarker discovery.
a, Multiplexed and multimodal wearable sensors continuously collect chemical and physical signatures from people’s daily activities. b, Simultaneous collection and coupling of sensing data from multiplexed and multimodal sensors is imperative for robust post-processing. c, With coupled sensor information, real-time calibrated values of chemical analytes can be generated. d, After obtaining calibrated sensor data, multiplexed associations between other physical traits (age, gender, body mass index and so on) and markers (vital signs) can be used for improved diagnostics. e, From multiplexed biomarker associations, molecular signatures corresponding to a biomarker’s significance for determination of various diseases can be discovered. f, General classification of machine learning algorithms and models that can be implemented for biomarker discovery using wearable chemical sensors.
Recently, continuous wearable monitoring of cardiac and respiratory-related signals such as coughing, body temperature and patterns of activity (movements) was successfully demonstrated for detecting early symptoms and evolution of COVID-19 (ref.141). Moreover, a multiplex physical sensor was demonstrated for neonatal and paediatric critical care applications where physiological sensors (for example, blood pressure, heart rate and oxygenation) were integrated with movement, orientation and vocal sensors for tracking the tonality and temporal characteristics of crying142.
Within this context, multimodal wearable sensors that simultaneously monitor chemical and physical health features could offer more powerful and comprehensive information of an individual’s health state (Fig. 6a,b) than single-modal wearables, increasing diagnostic accuracy89. Integrated wearable chemical and physical sensors have been demonstrated for the simultaneous monitoring of chemical analytes and blood pressure: a wearable skin patch capable of monitoring sweat caffeine, alcohol, lactate and ISF glucose was used to deconvolute the additive effects on blood pressure and heart rate throughout daily activities116. This ability is crucial to improve the reliability during daily tracking and avoid erratic interpretation of biomarker behaviour. Such combinations are significantly important when applied to the early detection of sepsis, characterized by a surge in lactate levels and a drop in blood pressure143, and for the daily supervision of biomarkers in patients with diabetes who are obese and hypertensive.
Owing to the unique complexities of biofluid secretion and the uncontrolled operational conditions of wearable sensors, it is also important to create multiplexed and multimodal sensors that can re-calibrate their analyte readings in real time based on other chemical and physical marker concentrations (Fig. 6c). Although informative of various diseases, fluctuations in skin temperature have been demonstrated to cause substantial overestimation of sweat metabolites. To resolve this, a fully integrated wearable metabolite, electrolyte and skin temperature sensing array was created to accurately measure sweat glucose and lactate levels by calibrating sensor values based on skin temperature3,144. In parallel, multiplexed sensors have also used measurements of sweat pH and NH4+ as calibration to normalize enzyme activity and improve the detection accuracy of sweat urea and glucose concentrations, reducing miscalculation of the actual analyte levels125.
Moving forward, skin-interfaced wearables should continue to expand their detection repertoire of target analytes and integrate continuous chemical sensing with biophysical markers (that is, blood pressure, heart rate). There are great opportunities for new categories of multimodal wearables; for example, more research is expected in combining brain activity monitoring (electroencephalogram) sensors with chemical sensors, which can open invaluable possibilities, including early detection of epileptic seizures, stress, fatigue and depression145. These smart multiplexed, multimodal chemical sensors will become more reliable and create personalized data for more accurate prediction of an individual’s physiological response to specific health conditions. At the same time, compilation of these extended data will yield new complex information about known diseases and medications, which will consequently lead to the discovery of new effective biomarkers that can augment the selectivity and robustness of disease detection and prevention (Fig. 6d). By using various data analysis tools, the association between the molecular signatures and health conditions can be decoded and the resulted pattern regression could enable disease prediction, forecasting and diagnosis (Fig. 6e).
Machine learning-driven biomarker discovery
Technological innovations in the wearable multimodal and multiplexed sensors have allowed significant leaps in the continuous acquisition of high-resolution multi-omics data related to patient health from the same biofluid at the same time. These complex data are composed of multivariable, non-linear physiological patterns that cannot be readily identified by conventional processing methods. In the medical field, machine learning has already been proven an effective tool for interpreting large volumes of intricate data sets. For example, for insulin dosage recommendations for adults with type 1 diabetes, continuous glucose monitor sensors could predict the insulin bolus and perform self-calibration and correction. A similar approach was presented for other feedback sensors such as the levodopa closed loop system for Parkinson disease146, feedback systems for cancer diagnosis and systems for the detection of cardiovascular diseases, pulmonary diseases and conditions related to patterns of movement, gait and posture147–154. As many possible variations, such as ionic strength, temperature, pH, humidity or body locations, could affect the readings of wearable biosensors during real-life on-body usage, machine learning can be applied for multiplexed data processing towards more accurate prediction of target analyte levels.
In generalized terms, the umbrella of machine learning algorithms can be divided into three classes: supervised, unsupervised and reinforced learning algorithms (Fig. 6f). Within each algorithm class, specific models (regression and classification models from supervised learning algorithms, clustering models from unsupervised learning algorithms and decision-making models from reinforced learning models) can be implemented based on the behaviour of the data set and the information or conclusion to be recovered from the study. Each specific machine learning model has a distinctive approach of interpreting and associating features from a data set and the adequate selection of them has been comprehensively reviewed in the literature1,133. A number of these machine learning models, such as the support vector machine (SVM), k-nearest neighbours, neural networks, k-means clustering and the Markov decision process, can be applied for supervised and unsupervised data mining and decision-making based on big data collected by wearable biosensors (Fig. 6f).
Enhanced target sensitivity and selectivity through advances in chemical sensing have enabled new understanding of the correlations between chemical biomarkers and specific diseases155,156. Out of the three general categorizations of machine learning algorithms, supervised learning models have been used to establish input–output correlations based on molecular analytes and vital signs. Focusing on classification models, an SVM algorithm can be applied to establish relationships between volatile organic compound footprints from breath and blood glucose levels157. Using published diabetic breath analyses as training data, the SVM algorithm categorized artificial breath samples to corresponding blood glucose levels with 97.1% accuracy. With increased focus on generating more durable wearable chemical sensors, implementing such SVM models will uncover new real-time correlations between validated biomarkers and the analyte monitored through non-invasively accessible bodily fluids. Furthermore, through cytokine and chemokine profiling using a single plasma sample, other machine learning classification and regression models (for example, naïve Bayes and decision tree algorithms) helped predict the sepsis host-immune response with high accuracy up to 96.64%158. Using machine learning, wearable chemical sensing systems could continuously monitor various target candidates, and an individual’s chemical profile of ISF, sweat or saliva will serve as feedback information on the patient’s metabolic or even immune response. Additionally, multiplexed analysis of biomarkers in different body fluids will also yield connections to clinically relevant disorders. Recent studies demonstrated this by sampling armpit sweat and salivary cortisol levels during exercise; integrating a k-nearest neighbours algorithm facilitated estimation of the score on the Kessler 10 Psychological Distress Scale (a globally recognized questionnaire for determination of psychological distress with the scores range from 10 to 50) with an error rate of 3.6 (ref.18).
Looking at regression models, long short-term memory (a neural network algorithm capable of learning sequenced dependencies) was used to predict future blood glucose levels based on continuous monitoring data159. Biophysical markers recorded from wearable sensors (heart rate, blood pressure), physical characteristics (weight, age, gender) and blood glucose data measured from a glucometer over 6 h were combined to predict blood glucose concentration with a 15-min horizon (predicting the concentration 15 min ahead of the actual measurement) and an average error of 15.43 mg dl–1 (ref.160). Using this regression model and establishing associations between blood glucose and the partitioned alternatives measured with wearable sensors, personalized ISF, sweat and saliva glucose–insulin dynamics could become a standardized method for managing individuals with diabetes. In another application, by analysing neuropsychiatric signatures in blood coupled with mental health questionnaires as training information, researchers developed a decision tree algorithm to reduce the misdiagnosis of bipolar disorder from major depressive disorder161. Supplementing these studies with chemical signatures provided by the wearable sensors could enhance the accuracy and efficiency of those diagnostic pathways. However, this will predominantly be achieved by generating machine learning models that can produce associations between multiple biomarker candidates in different body fluids. This was recently demonstrated by applying linear regression and neural network algorithms for learning from clinical data obtained from serum levels of creatinine in patients with renal disease, which helped correlate the serum creatinine concentrations with tear creatinine levels monitored by a wearable chemical sensor 162.
As wearable chemical sensors become more robust, machine learning algorithms are more able to classify chemical and biophysical data corresponding to unique health patterns and provide accurate predictions of medical outcomes based on molecular signatures for different health conditions163. Machine learning can be applied not only for the prediction of event but also to recognize when one has already happened; such a system can be applied in the detection of convulsion for patients with epilepsy, where galvanic skin response data, heart rate, temperature and movement sensors can detect when an event is happening and send an alert to the caregiver164. Besides supervised regression and classification models, unsupervised machine learning algorithms can be implemented to identify key unrecognized characteristics165 from unlabelled molecular sensing data. This can be further used for dimensionality-reduction representations of the sensor information166 and detection of anomalies within clusters of data (through models such as k-means clustering) (Fig. 6f). After comprehensive processing of the wearable biosensing data, reinforced learning algorithms can be integrated for assisted medical recommendations and timely interventions167 (Fig. 6f).
Overall, the addition of multiplexed, multimodal real-time chemical sensors to the big data industry will open the gate to new correlations between existing biomarkers and new chemical signatures that can be monitored continuously through easily accessible bodily fluids. Combining these big data analytics, streaming from wearable sensing devices that monitor chemical and biophysical markers, machine learning techniques should have a profound impact upon the discovery of biomarkers and upon the healthcare field in general168. A platform that could integrate chemical and biophysical data from different wearables is imperative to test and validate newly discovered biomarkers and their relationship with relevant diseases169. Ultimately, integrating new chemical data from alternative body fluids provided by wearable chemical sensors will considerably facilitate the diagnostic and prevention of numerous diseases.
Conclusions
In the era of omics and digital medicine, wearable chemical sensing technology is emerging as a powerful tool to aid the biomarker discovery process. Biomarker discovery can be facilitated by employing wearable multiplexed sensing systems that can perform continuous monitoring of potential molecular signatures. The data collected by these wearables can be correlated with the studied physiological and biological conditions using suitable data analytic tools. Compared with traditional laboratory tests based on blood draws and tissue biopsy, which are invasive, costly, require physical clinic visits and provide only discrete analyte information, the wearable sensor approach could substantially increase the efficiency and lower the costs of the process. Wearable chemical sensors may open a new era of medical tools by offering means of remote, continuous and non-invasive real-time monitoring of potential multi-omics biomarkers during people’s daily activities.
Despite great promise, the scope of molecular analytes able to be continuously screened on the body is currently limited to a small number of metabolites and electrolytes. There is an urgent need to develop next-generation wearable chemical sensors that can monitor a broader spectrum of biomarker candidates. In this regard, new bioaffinity sensors based on molecular switches coupled with suitable signal tags and in situ regeneration strategies are highly desired. Another critical limitation is that the catalogue of accessible and detectable analytes is limited to small molecules that can easily partition from blood or be secreted by exocrine/endocrine glands. Moreover, the time lag related to the molecular secretion into alternative body fluids from blood vessels needs to be carefully considered. Compared with gene sequencing or metabolomics analysis that can quantify thousands of molecules at a time, the number of biosensors that can be integrated into a single miniaturized wearable system is limited as each sensor requires the dimensions to reach desired selectivity, stability and sensitivity. Furthermore, the successful realization of wearable sensors with suitable sensitivity, selectivity and stability requires the careful selection of electrode materials, bioreceptor immobilization strategies and seamless wearable system integration that allows efficient fluid extraction and reliable, robust in situ data collection. Moreover, more studies are needed to verify the accuracy, precision and repeatability of the wearable sensors in participants. Such validation studies should be realized using existing gold standard tests to consolidate wearable biosensors in the omics era towards the widespread acceptance and credibility of these new non-invasive monitoring platforms170.
Overall, wearable sensor-based biomarker discovery is a highly interdisciplinary field. Chemists, biologists, engineers and physicians need to work closely together to develop high-performance integrated multiplexed wearable chemical sensor systems with the power to predict and prevent the inevitable. We envisage that such evolution of wearable sensing technologies would lead to continuous real-time analyses of metabolomics, proteomics, genomics and other omics. The tremendous volume of data collected continuously from large-scale human studies, coupled with efficient data-fusion and data-mining methods, would enable early disease prediction, diagnosis and timely interventions.
Acknowledgements
This project was supported by National Institutes of Health (NIH) grant R01HL155815; National Science Foundation (NSF) grant 2145802; Office of Naval Research grants N00014-21-1-2483 and N00014-21-1-2845; the Translational Research Institute for Space Health through NASA NNX16AO69A; American Cancer Society Research Scholar Grant RSG-21-181-01-CTPS; High Impact Pilot Research Award T31IP1666 from the Tobacco-Related Disease Research Program; and the Center of Wearable Sensors at University of California San Diego.
Author contributions
All authors contributed to researching data for the article, and writing and review/editing of the manuscript before submission.
Peer review
Peer review information
Nature Reviews Chemistry thanks H. Tom Soh, Koji Sode, Atul Sharma and Zong-Hong Lin for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Joseph Wang, Email: josephwang@ucsd.edu.
Wei Gao, Email: weigao@caltech.edu.
References
- 1.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat. Rev. Drug Discov. 2003;2:566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 2.Firestein GS. A biomarker by any other name. Nat. Clin. Pract. Rheumatol. 2006;2:635–635. doi: 10.1038/ncprheum0347. [DOI] [PubMed] [Google Scholar]
- 3.Gao W, et al. Fully integrated wearable sensor arrays for multiplexed in situ perspiration analysis. Nature. 2016;529:509–514. doi: 10.1038/nature16521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J, Campbell AS, de Ávila BE-F, Wang J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019;37:389–406. doi: 10.1038/s41587-019-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J, Guo H, Li J, Bandodkar AJ, Rogers JA. Body-interfaced chemical sensors for noninvasive monitoring and analysis of biofluids. Trends Chem. 2019;1:559–571. doi: 10.1016/j.trechm.2019.07.001. [DOI] [Google Scholar]
- 6.Yang Y, Gao W. Wearable and flexible electronics for continuous molecular monitoring. Chem. Soc. Rev. 2019;48:1465–1491. doi: 10.1039/C7CS00730B. [DOI] [PubMed] [Google Scholar]
- 7.Broza YY, et al. Disease detection with molecular biomarkers: from chemistry of body fluids to nature-inspired chemical sensors. Chem. Rev. 2019;119:11761–11817. doi: 10.1021/acs.chemrev.9b00437. [DOI] [PubMed] [Google Scholar]
- 8.Heikenfeld J, et al. Accessing analytes in biofluids for peripheral biochemical monitoring. Nat. Biotechnol. 2019;37:407–419. doi: 10.1038/s41587-019-0040-3. [DOI] [PubMed] [Google Scholar]
- 9.Ray TR, et al. Bio-integrated wearable systems: a comprehensive review. Chem. Rev. 2019;119:5461–5533. doi: 10.1021/acs.chemrev.8b00573. [DOI] [PubMed] [Google Scholar]
- 10.Bariya M, Nyein HYY, Javey A. Wearable sweat sensors. Nat. Electron. 2018;1:160–171. doi: 10.1038/s41928-018-0043-y. [DOI] [Google Scholar]
- 11.Picard M, Scott-Boyer M-P, Bodein A, Périn O, Droit A. Integration strategies of multi-omics data for machine learning analysis. Comput. Struct. Biotechnol. J. 2021;19:3735–3746. doi: 10.1016/j.csbj.2021.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butterfield DA, Halliwell B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019;20:148–160. doi: 10.1038/s41583-019-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nahkuri S, Becker T, Schueller V, Massberg S, Bauer-Mehren A. Prior fluid and electrolyte imbalance is associated with COVID-19 mortality. Commun. Med. 2021;1:51. doi: 10.1038/s43856-021-00051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcantonio ER, et al. Serum biomarkers for delirium. J. Gerontol. A Biol. Sci. Med. Sci. 2006;61:1281–1286. doi: 10.1093/gerona/61.12.1281. [DOI] [PubMed] [Google Scholar]
- 15.Ernst J, et al. Increased pregenual anterior cingulate glucose and lactate concentrations in major depressive disorder. Mol. Psychiatry. 2017;22:113–119. doi: 10.1038/mp.2016.73. [DOI] [PubMed] [Google Scholar]
- 16.Torrente-Rodríguez, et al. Investigation of cortisol dynamics in human sweat using a graphene-based wireless mHealth system. Matter. 2020;2:921–937. doi: 10.1016/j.matt.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearlmutter P, et al. Sweat and saliva cortisol response to stress and nutrition factors. Sci. Rep. 2020;10:19050. doi: 10.1038/s41598-020-75871-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallapur B, et al. Quantitative estimation of sodium, potassium and total protein in saliva of diabetic smokers and nonsmokers: a novel study. J. Nat. Sci. Biol. Med. 2013;4:341–345. doi: 10.4103/0976-9668.117006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohan AMV, Rajendran V, Mishra RK, Jayaraman M. Recent advances and perspectives in sweat based wearable electrochemical sensors. TrAC. Trends Anal. Chem. 2020;131:116024. doi: 10.1016/j.trac.2020.116024. [DOI] [Google Scholar]
- 20.Sonner Z, et al. The microfluidics of the eccrine sweat gland, including biomarker partitioning, transport, and biosensing implications. Biomicrofluidics. 2015;9:031301. doi: 10.1063/1.4921039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katchman BA, Zhu M, Blain Christen J, Anderson KS. Eccrine sweat as a biofluid for profiling immune biomarkers. Proteom. Clin. Appl. 2018;12:1800010. doi: 10.1002/prca.201800010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey CJ, LeBouf RF, Stefaniak AB. Formulation and stability of a novel artificial human sweat under conditions of storage and use. Toxicol. Vitr. 2010;24:1790–1796. doi: 10.1016/j.tiv.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Lin S, et al. Noninvasive wearable electroactive pharmaceutical monitoring for personalized therapeutics. Proc. Natl Acad. Sci. USA. 2020;117:19017–19025. doi: 10.1073/pnas.2009979117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abikshyeet P, Ramesh V, Oza N. Glucose estimation in the salivary secretion of diabetes mellitus patients. Diabetes, Metab. Syndr. Obes. Targets Ther. 2012;5:149–154. doi: 10.2147/DMSO.S32112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng R, et al. A new oil/membrane approach for integrated sweat sampling and sensing: sample volumes reduced from μl’s to nL’s and reduction of analyte contamination from skin. Lab Chip. 2016;16:4415–4423. doi: 10.1039/C6LC01013J. [DOI] [PubMed] [Google Scholar]
- 26.Baker LB, Wolfe AS. Physiological mechanisms determining eccrine sweat composition. Eur. J. Appl. Physiol. 2020;120:719–752. doi: 10.1007/s00421-020-04323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang B, et al. Wearable aptamer-field-effect transistor sensing system for noninvasive cortisol monitoring. Sci. Adv. 2022;8:eabk0967. doi: 10.1126/sciadv.abk0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandodkar AJ, Jeerapan I, Wang J. Wearable chemical sensors: present challenges and future prospects. ACS Sens. 2016;1:464–482. doi: 10.1021/acssensors.6b00250. [DOI] [Google Scholar]
- 29.Sempionatto JR, Jeerapan I, Krishnan S, Wang J. Wearable chemical sensors: emerging systems for on-body. Anal. Chem. 2019;92:378–396. doi: 10.1021/acs.analchem.9b04668. [DOI] [PubMed] [Google Scholar]
- 30.Xu C, Yang Y, Gao W. Skin-interfaced sensors in digital medicine: from materials to applications. Matter. 2020;2:1414–1445. doi: 10.1016/j.matt.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandodkar AJ, Wang J. Non-invasive wearable electrochemical sensors: a review. Trends Biotechnol. 2014;32:363–371. doi: 10.1016/j.tibtech.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Zhao C. Wearable electrochemical sensors for noninvasive monitoring of health — a perspective. Curr. Opin. Electrochem. 2020;23:42–46. doi: 10.1016/j.coelec.2020.03.008. [DOI] [Google Scholar]
- 33.Min J, Sempionatto JR, Teymourian H, Wang J, Gao W. Wearable electrochemical biosensors in North America. Biosens. Bioelectron. 2021;172:112750. doi: 10.1016/j.bios.2020.112750. [DOI] [PubMed] [Google Scholar]
- 34.Xiao J, et al. Microfluidic chip-based wearable colorimetric sensor for simple and facile detection of sweat glucose. Anal. Chem. 2019;91:14803–14807. doi: 10.1021/acs.analchem.9b03110. [DOI] [PubMed] [Google Scholar]
- 35.Choe A, et al. Stretchable and wearable colorimetric patches based on thermoresponsive plasmonic microgels embedded in a hydrogel film. NPG Asia Mater. 2018;10:912–922. doi: 10.1038/s41427-018-0086-6. [DOI] [Google Scholar]
- 36.Koh A, et al. A soft, wearable microfluidic device for the capture, storage, and colorimetric sensing of sweat. Sci. Transl. Med. 2016;8:366ra165. doi: 10.1126/scitranslmed.aaf2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghaffari R, et al. Soft wearable systems for colorimetric and electrochemical analysis of biofluids. Adv. Funct. Mater. 2019;30:1907269. doi: 10.1002/adfm.201907269. [DOI] [Google Scholar]
- 38.Kim J, Campbell AS, Wang J. Wearable non-invasive epidermal glucose sensors: a review. Talanta. 2018;177:163–170. doi: 10.1016/j.talanta.2017.08.077. [DOI] [PubMed] [Google Scholar]
- 39.Lee H, Hong YJ, Baik S, Hyeon T, Kim DH. Enzyme-based glucose sensor: from invasive to wearable device. Adv. Healthc. Mater. 2018;7:1701150. doi: 10.1002/adhm.201701150. [DOI] [PubMed] [Google Scholar]
- 40.Wolkowicz KL, et al. A review of biomarkers in the context of type 1 diabetes: biological sensing for enhanced glucose control. Bioeng. Transl. Med. 2021;6:e10201. doi: 10.1002/btm2.10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teymourian H, Barfidokht A, Wang J. Electrochemical glucose sensors in diabetes management: an updated review (2010–2020) Chem. Soc. Rev. 2020;49:7671–7709. doi: 10.1039/D0CS00304B. [DOI] [PubMed] [Google Scholar]
- 42.Parrilla M, Cuartero M, Crespo GA. Wearable potentiometric ion sensors. TrAC. Trends Anal. Chem. 2019;110:303–320. doi: 10.1016/j.trac.2018.11.024. [DOI] [Google Scholar]
- 43.Yang Y, et al. A laser-engraved wearable sensor for sensitive detection of uric acid and tyrosine in sweat. Nat. Biotechnol. 2020;38:217–224. doi: 10.1038/s41587-019-0321-x. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J, et al. A wearable nutrition tracker. Adv. Mater. 2021;33:2006444. doi: 10.1002/adma.202006444. [DOI] [PubMed] [Google Scholar]
- 45.Ariño C, et al. Electrochemical stripping analysis. Nat. Rev. Methods Prim. 2022;2:62. doi: 10.1038/s43586-022-00143-5. [DOI] [Google Scholar]
- 46.Gao W, et al. Wearable microsensor array for multiplexed heavy metal monitoring of body fluids. ACS Sens. 2016;1:866–874. doi: 10.1021/acssensors.6b00287. [DOI] [Google Scholar]
- 47.Kim J, et al. Wearable temporary tattoo sensor for real-time trace metal monitoring in human sweat. Electrochem. Commun. 2015;51:41–45. doi: 10.1016/j.elecom.2014.11.024. [DOI] [Google Scholar]
- 48.Takaloo S, Moghimi Zand M. Wearable electrochemical flexible biosensors: with the focus on affinity biosensors. Sens. Bio-Sens. Res. 2021;32:100403. doi: 10.1016/j.sbsr.2021.100403. [DOI] [Google Scholar]
- 49.Elsherif M, Hassan MU, Yetisen AK, Butt H. Wearable contact lens biosensors for continuous glucose monitoring using smartphones. ACS Nano. 2018;12:5452–5462. doi: 10.1021/acsnano.8b00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng M, et al. Wearable fluorescent contact lenses for monitoring glucose via a smartphone. Sens. Actuator B Chem. 2022;352:131067. doi: 10.1016/j.snb.2021.131067. [DOI] [Google Scholar]
- 51.Sekine Y, et al. A fluorometric skin-interfaced microfluidic device and smartphone imaging module for in situ quantitative analysis of sweat chemistry. Lab Chip. 2018;18:2178–2186. doi: 10.1039/C8LC00530C. [DOI] [PubMed] [Google Scholar]
- 52.Brinkmann BH, et al. Seizure diaries and forecasting with wearables: epilepsy monitoring outside the clinic. Front. Neurol. 2021;12:1128. doi: 10.3389/fneur.2021.690404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia X, et al. High-sensitivity troponin I and incident coronary events, stroke, heart failure hospitalization, and mortality in the aric study. Circulation. 2019;139:2642–2653. doi: 10.1161/CIRCULATIONAHA.118.038772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu J, Torrente-Rodríguez RM, Wang M, Gao W. The era of digital health: a review of portable and wearable affinity biosensors. Adv. Funct. Mater. 2019;29:1906713. [Google Scholar]
- 55.Plaxco KW, Soh HT. Switch-based biosensors: a new approach towards real-time, in vivo molecular detection. Trends Biotechnol. 2011;29:1–5. doi: 10.1016/j.tibtech.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quijano-Rubio A, et al. De novo design of modular and tunable protein biosensors. Nature. 2021;591:482–487. doi: 10.1038/s41586-021-03258-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Modi S, et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 2009;4:325–330. doi: 10.1038/nnano.2009.83. [DOI] [PubMed] [Google Scholar]
- 58.Liu S, et al. DNA nanotweezers for biosensing applications: recent advances and future prospects. ACS Sens. 2022;7:3–20. doi: 10.1021/acssensors.1c01647. [DOI] [PubMed] [Google Scholar]
- 59.Ahmad OS, Bedwell TS, Esen C, Garcia-Cruz A, Piletsky SA. Molecularly imprinted polymers in electrochemical and optical sensors. Trends Biotechnol. 2019;37:294–309. doi: 10.1016/j.tibtech.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 60.Subrahmanyam S, et al. ‘Bite-and-switch’ approach using computationally designed molecularly imprinted polymers for sensing of creatinine. Biosens. Bioelectron. 2001;16:631–637. doi: 10.1016/S0956-5663(01)00191-9. [DOI] [PubMed] [Google Scholar]
- 61.Wang M, et al. A wearable electrochemical biosensor for the monitoring of metabolites and nutrients. Nat. Biomed. Eng. 2022 doi: 10.1038/s41551-022-00916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vallée-Bélisle A, Plaxco KW. Structure-switching biosensors: inspired by nature. Curr. Opin. Struct. Biol. 2010;20:518–526. doi: 10.1016/j.sbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gotrik MR, Feagin TA, Csordas AT, Nakamoto MA, Soh HT. Advancements in aptamer discovery technologies. Acc. Chem. Res. 2016;49:1903–1910. doi: 10.1021/acs.accounts.6b00283. [DOI] [PubMed] [Google Scholar]
- 64.Ahmad KM, et al. Probing the limits of aptamer affinity with a microfluidic selex platform. PLoS ONE. 2011;6:e27051. doi: 10.1371/journal.pone.0027051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao Y, Lai RY, Plaxco KW. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protoc. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 66.Dauphin-ducharme P, Yang K, Plaxco KW. Electrochemical aptamer-based sensors for improved therapeutic drug monitoring and high-precision, feedback-controlled drug delivery. ACS Sens. 2019;4:2832–2837. doi: 10.1021/acssensors.9b01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White RJ, Phares N, Lubin AA, Xiao Y, Plaxco KW. Optimization of electrochemical aptamer-based sensors via optimization of probe packing density and surface chemistry. Langmuir. 2008;24:10513–10518. doi: 10.1021/la800801v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao Y, et al. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem. Int. Ed. 2005;117:5592–5595. doi: 10.1002/ange.200500989. [DOI] [PubMed] [Google Scholar]
- 69.Chamorro-Garcia A, et al. Switching the aptamer attachment geometry can dramatically alter the signalling and performance of electrochemical aptamer-based sensors. Chem. Commun. 2021;57:11693–11696. doi: 10.1039/D1CC04557A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leung KK, Downs AM, Ortega G, Kurnik M, Plaxco KW. Elucidating the mechanisms underlying the signal drift of electrochemical aptamer-based sensors in whole blood. ACS Sens. 2021;6:3340–3347. doi: 10.1021/acssensors.1c01183. [DOI] [PubMed] [Google Scholar]
- 71.Röthlisberger P, Hollenstein M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018;134:3–21. doi: 10.1016/j.addr.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 72.Ferguson BS, et al. Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci. Transl. Med. 2013;5:213ra165. doi: 10.1126/scitranslmed.3007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poudineh M, et al. A fluorescence sandwich immunoassay for the real-time continuous detection of glucose and insulin in live animals. Nat. Biomed. Eng. 2021;5:53–63. doi: 10.1038/s41551-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson BD, Hariri AA, Thompson IAP, Eisenstein M, Soh HT. Independent control of the thermodynamic and kinetic properties of aptamer switches. Nat. Commun. 2019;10:5079. doi: 10.1038/s41467-019-13137-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das J, et al. Reagentless biomolecular analysis using a molecular pendulum. Nat. Chem. 2021;13:428–434. doi: 10.1038/s41557-021-00644-y. [DOI] [PubMed] [Google Scholar]
- 76.Nguyen PQ, et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat. Biotechnol. 2021;39:1366–1374. doi: 10.1038/s41587-021-00950-3. [DOI] [PubMed] [Google Scholar]
- 77.Morales MA, Halpern JM. Guide to selecting a biorecognition element for biosensors. Bioconjug. Chem. 2018;29:3231–3239. doi: 10.1021/acs.bioconjchem.8b00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Satishkumar BC, et al. Reversible fluorescence quenching in carbon nanotubes for biomolecular sensing. Nat. Nanotechnol. 2007;2:560–564. doi: 10.1038/nnano.2007.261. [DOI] [PubMed] [Google Scholar]
- 79.Aleman J, Kilic T, Mille LS, Shin SR, Zhang YS. Microfluidic integration of regeneratable electrochemical affinity-based biosensors for continual monitoring of organ-on-a-chip devices. Nat. Protoc. 2021;16:2564–2593. doi: 10.1038/s41596-021-00511-7. [DOI] [PubMed] [Google Scholar]
- 80.Ambaye AD, Kefeni KK, Mishra SB, Nxumalo EN, Ntsendwana B. Recent developments in nanotechnology-based printing electrode systems for electrochemical sensors. Talanta. 2021;225:121951. doi: 10.1016/j.talanta.2020.121951. [DOI] [PubMed] [Google Scholar]
- 81.Mamun MAAl, Yuce MR. Recent progress in nanomaterial enabled chemical sensors for wearable environmental monitoring applications. Adv. Funct. Mater. 2020;30:2005703. doi: 10.1002/adfm.202005703. [DOI] [Google Scholar]
- 82.Ulman A. Formation and structure of self-assembled monolayers. Chem. Rev. 1996;96:1533–1554. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 83.Joshi P, Mishra R, Narayan RJ. Biosensing applications of carbon-based materials. Curr. Opin. Biomed. Eng. 2021;18:100274. doi: 10.1016/j.cobme.2021.100274. [DOI] [Google Scholar]
- 84.Wang M, Yang Y, Gao W. Laser-engraved graphene for flexible and wearable electronics. Trends Chem. 2021;3:969–981. doi: 10.1016/j.trechm.2021.09.001. [DOI] [Google Scholar]
- 85.Basu J, RoyChaudhuri C. Graphene nanoporous FET biosensor: influence of pore dimension on sensing performance in complex analyte. IEEE Sens. J. 2018;18:5627–5634. doi: 10.1109/JSEN.2018.2841060. [DOI] [Google Scholar]
- 86.He W, et al. Integrated textile sensor patch for real-time and multiplex sweat analysis. Sci. Adv. 2019;5:eaax0649. doi: 10.1126/sciadv.aax0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Downs AM, et al. Nanoporous gold for the miniaturization of in vivo electrochemical aptamer-based sensors. ACS Sens. 2021;6:2299–2306. doi: 10.1021/acssensors.1c00354. [DOI] [PubMed] [Google Scholar]
- 88.Bandodkar AJ, et al. Battery-free, skin-interfaced microfluidic/electronic systems for simultaneous electrochemical, colorimetric, and volumetric analysis of sweat. Sci. Adv. 2019;5:eaav3294. doi: 10.1126/sciadv.aav3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Imani S, et al. A wearable chemical–electrophysiological hybrid biosensing system for real-time health and fitness monitoring. Nat. Commun. 2016;7:11650. doi: 10.1038/ncomms11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bariya M, et al. Glove-based sensors for multimodal monitoring of natural sweat. Sci. Adv. 2020;6:8308–8336. doi: 10.1126/sciadv.abb8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sabaté del Río J, Henry OYF, Jolly P, Ingber DE. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 2019;14:1143–1149. doi: 10.1038/s41565-019-0566-z. [DOI] [PubMed] [Google Scholar]
- 92.Xu K, Lu Y, Takei K. Multifunctional skin-inspired flexible sensor systems for wearable electronics. Adv. Mater. Technol. 2019;4:1800628. doi: 10.1002/admt.201800628. [DOI] [Google Scholar]
- 93.Zhang S, Wright G, Yang Y. Materials and techniques for electrochemical biosensor design and construction. Biosens. Bioelectron. 2000;15:273–282. doi: 10.1016/S0956-5663(00)00076-2. [DOI] [PubMed] [Google Scholar]
- 94.Feron K, et al. Organic bioelectronics: materials and biocompatibility. Int. J. Mol. Sci. 2018;19:2382. doi: 10.3390/ijms19082382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwon YT, et al. All-printed nanomembrane wireless bioelectronics using a biocompatible solderable graphene for multimodal human–machine interfaces. Nat. Commun. 2020;11:3450. doi: 10.1038/s41467-020-17288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cui C, et al. Recent progress in natural biopolymers conductive hydrogels for flexible wearable sensors and energy devices: materials, structures, and performance. ACS Appl. Bio Mater. 2021;4:85–121. doi: 10.1021/acsabm.0c00807. [DOI] [PubMed] [Google Scholar]
- 97.Kim Y, Prausnitz MR. Sensitive sensing of biomarkers in interstitial fluid. Nat. Biomed. Eng. 2021;5:3–5. doi: 10.1038/s41551-020-00679-5. [DOI] [PubMed] [Google Scholar]
- 98.Shi T, et al. Modeling and measurement of correlation between blood and interstitial glucose changes. J. Diabetes Res. 2016;2016:4596316. doi: 10.1155/2016/4596316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee H-R, Kim C-C, Sun J-Y. Stretchable ionics — a promising candidate for upcoming wearable devices. Adv. Mater. 2018;30:1704403. doi: 10.1002/adma.201704403. [DOI] [PubMed] [Google Scholar]
- 100.Wang Z, et al. Microneedle patch for the ultrasensitive quantification of protein biomarkers in interstitial fluid. Nat. Biomed. Eng. 2021;5:64–76. doi: 10.1038/s41551-020-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Samant PP, Prausnitz MR. Mechanisms of sampling interstitial fluid from skin using a microneedle patch. Proc. Natl Acad. Sci. USA. 2018;115:4583–4588. doi: 10.1073/pnas.1716772115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lipani L, et al. Non-invasive, transdermal, path-selective and specific glucose monitoring via a graphene-based platform. Nat. Nanotechnol. 2018;13:504–511. doi: 10.1038/s41565-018-0112-4. [DOI] [PubMed] [Google Scholar]
- 103.Chen Y, et al. Skin-like biosensor system via electrochemical channels for noninvasive blood glucose monitoring. Sci. Adv. 2017;3:e1701629. doi: 10.1126/sciadv.1701629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pu Z, et al. A thermal activated and differential self-calibrated flexible epidermal biomicrofluidic device for wearable accurate blood glucose monitoring. Sci. Adv. 2021;7:199–226. doi: 10.1126/sciadv.abd0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim J, et al. Simultaneous monitoring of sweat and interstitial fluid using a single wearable biosensor platform. Adv. Sci. 2018;5:1800880. doi: 10.1002/advs.201800880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Emaminejad S, et al. Autonomous sweat extraction and analysis applied to cystic fibrosis and glucose monitoring using a fully integrated wearable platform. Proc. Natl Acad. Sci. USA. 2017;114:4625–4630. doi: 10.1073/pnas.1701740114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ray TR, et al. Soft, skin-interfaced sweat stickers for cystic fibrosis diagnosis and management. Sci. Transl. Med. 2021;13:eabd8109. doi: 10.1126/scitranslmed.abd8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hauke A, et al. Complete validation of a continuous and blood-correlated sweat biosensing device with integrated sweat stimulation. Lab Chip. 2018;18:3750–3759. doi: 10.1039/C8LC01082J. [DOI] [PubMed] [Google Scholar]
- 109.Karpova EV, et al. Noninvasive diabetes monitoring through continuous analysis of sweat using flow-through glucose biosensor. Anal. Chem. 2019;91:3778–3783. doi: 10.1021/acs.analchem.8b05928. [DOI] [PubMed] [Google Scholar]
- 110.Paul B, Demuru S, Lafaye C, Saubade M, Briand D. Printed iontophoretic-integrated wearable microfluidic sweat-sensing patch for on-demand point-of-care sweat analysis. Adv. Mater. Technol. 2021;6:2000910. doi: 10.1002/admt.202000910. [DOI] [Google Scholar]
- 111.Bolat G, et al. Wearable soft electrochemical microfluidic device integrated with iontophoresis for sweat biosensing. Anal. Bioanal. Chem. 2022;414:5411–5421. doi: 10.1007/s00216-021-03865-9. [DOI] [PubMed] [Google Scholar]
- 112.Nyein HYY, et al. A wearable patch for continuous analysis of thermoregulatory sweat at rest. Nat. Commun. 2021;12:1823. doi: 10.1038/s41467-021-22109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bandodkar AJ, et al. Sweat-activated biocompatible batteries for epidermal electronic and microfluidic systems. Nat. Electron. 2020;3:554–562. doi: 10.1038/s41928-020-0443-7. [DOI] [Google Scholar]
- 114.Liu C, Xu T, Wang D, Zhang X. The role of sampling in wearable sweat sensors. Talanta. 2020;212:120801. doi: 10.1016/j.talanta.2020.120801. [DOI] [PubMed] [Google Scholar]
- 115.Nyein HYY, et al. Regional and correlative sweat analysis using high-throughput microfluidic sensing patches toward decoding sweat. Sci. Adv. 2019;5:eaaw9906. doi: 10.1126/sciadv.aaw9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sempionatto JR, et al. An epidermal patch for the simultaneous monitoring of haemodynamic and metabolic biomarkers. Nat. Biomed. Eng. 2021;5:737–748. doi: 10.1038/s41551-021-00685-1. [DOI] [PubMed] [Google Scholar]
- 117.Tai L-C, et al. Methylxanthine drug monitoring with wearable sweat sensors. Adv. Mater. 2018;30:1707442. doi: 10.1002/adma.201707442. [DOI] [PubMed] [Google Scholar]
- 118.Salvisberg C, et al. Exploring the human tear fluid: discovery of new biomarkers in multiple sclerosis. Proteom. Clin. Appl. 2014;8:185–194. doi: 10.1002/prca.201300053. [DOI] [PubMed] [Google Scholar]
- 119.Bachhuber F, Huss A, Senel M, Tumani H. Diagnostic biomarkers in tear fluid: from sampling to preanalytical processing. Sci. Rep. 2021;11:10064. doi: 10.1038/s41598-021-89514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yoshizawa JM, et al. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013;26:781–791. doi: 10.1128/CMR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Iyengar A, et al. Detection and potential utility of C-reactive protein in saliva of neonates. Front. Pediatr. 2014;2:131. doi: 10.3389/fped.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mannoor MS, et al. Graphene-based wireless bacteria detection on tooth enamel. Nat. Commun. 2012;3:763. doi: 10.1038/ncomms1767. [DOI] [PubMed] [Google Scholar]
- 123.Kim J, et al. Wearable salivary uric acid mouthguard biosensor with integrated wireless electronics. Biosens. Bioelectron. 2015;74:1061–1068. doi: 10.1016/j.bios.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tehrani F, et al. An integrated wearable microneedle array for the continuous monitoring of multiple biomarkers in interstitial fluid. Nat. Biomed. Eng. 2022 doi: 10.1038/s41551-022-00887-1. [DOI] [PubMed] [Google Scholar]
- 125.Yu Y, et al. Biofuel-powered soft electronic skin with multiplexed and wireless sensing for human–machine interfaces. Sci. Robot. 2020;5:eaaz7946. doi: 10.1126/scirobotics.aaz7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Song Y, et al. Wireless battery-free wearable sweat sensor powered by human motion. Sci. Adv. 2020;6:eaay9842. doi: 10.1126/sciadv.aay9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Song Y, Mukasa D, Zhang H, Gao W. Self-powered wearable biosensors. Acc. Mater. Res. 2021;2:184–197. doi: 10.1021/accountsmr.1c00002. [DOI] [Google Scholar]
- 128.Yin L, et al. A passive perspiration biofuel cell: high energy return on investment. Joule. 2021;5:1888–1904. doi: 10.1016/j.joule.2021.06.004. [DOI] [Google Scholar]
- 129.Yin L, et al. A self-sustainable wearable multi-modular e-textile bioenergy microgrid system. Nat. Commun. 2021;12:1542. doi: 10.1038/s41467-021-21701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhao J, et al. A fully integrated and self-powered smartwatch for continuous sweat glucose monitoring. ACS Sens. 2019;4:1925–1933. doi: 10.1021/acssensors.9b00891. [DOI] [PubMed] [Google Scholar]
- 131.Terse-Thakoor T, et al. Thread-based multiplexed sensor patch for real-time sweat monitoring. npj Flex. Electron. 2020;4:18. doi: 10.1038/s41528-020-00081-w. [DOI] [Google Scholar]
- 132.Torrente-Rodríguez RM, et al. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;6:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu K-H, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018;2:719–731. doi: 10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- 134.Yokus MA, Songkakul T, Pozdin VA, Bozkurt A, Daniele MA. Wearable multiplexed biosensor system toward continuous monitoring of metabolites. Biosens. Bioelectron. 2020;153:112038. doi: 10.1016/j.bios.2020.112038. [DOI] [PubMed] [Google Scholar]
- 135.Trejo Banos D, et al. Bayesian reassessment of the epigenetic architecture of complex traits. Nat. Commun. 2020;11:2865. doi: 10.1038/s41467-020-16520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Conesa A, Beck S. Making multi-omics data accessible to researchers. Sci. Data. 2019;6:251. doi: 10.1038/s41597-019-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Eren AM, et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021;6:3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Heifler O, et al. Clinic-on-a-needle array toward future minimally invasive wearable artificial pancreas applications. ACS Nano. 2021;15:12019–12033. doi: 10.1021/acsnano.1c03310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Teymourian H, Tehrani F, Mahato K, Wang J. Lab under the skin: microneedle based wearable devices. Adv. Healthc. Mater. 2021;17:2002255. doi: 10.1002/adhm.202002255. [DOI] [PubMed] [Google Scholar]
- 140.Teymourian H, et al. Microneedle-based detection of ketone bodies along with glucose and lactate: toward real-time continuous interstitial fluid monitoring of diabetic ketosis and ketoacidosis. Anal. Chem. 2020;92:2291–2300. doi: 10.1021/acs.analchem.9b05109. [DOI] [PubMed] [Google Scholar]
- 141.Jeong H, Rogers JA, Xu S. Continuous on-body sensing for the COVID-19 pandemic: gaps and opportunities. Sci. Adv. 2020;6:eabd4794. doi: 10.1126/sciadv.abd4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chung HU, et al. Skin-interfaced biosensors for advanced wireless physiological monitoring in neonatal and pediatric intensive-care units. Nat. Med. 2020;26:418–429. doi: 10.1038/s41591-020-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Reddy B, et al. Point-of-care sensors for the management of sepsis. Nat. Biomed. Eng. 2018;2:640–648. doi: 10.1038/s41551-018-0288-9. [DOI] [PubMed] [Google Scholar]
- 144.Lee H, et al. A graphene-based electrochemical device with thermoresponsive microneedles for diabetes monitoring and therapy. Nat. Nanotechnol. 2016;11:566–572. doi: 10.1038/nnano.2016.38. [DOI] [PubMed] [Google Scholar]
- 145.Iqbal SMA, Mahgoub I, Du E, Leavitt MA, Asghar W. Advances in healthcare wearable devices. npj Flex. Electron. 2021;5:9. doi: 10.1038/s41528-021-00107-x. [DOI] [Google Scholar]
- 146.Teymourian H, et al. Closing the loop for patients with Parkinson disease: where are we? Nat. Rev. Neurol. 2022;18:497–507. doi: 10.1038/s41582-022-00674-1. [DOI] [PubMed] [Google Scholar]
- 147.Huang L, et al. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 2020;11:3556. doi: 10.1038/s41467-020-17347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Krittanawong C, et al. Integration of novel monitoring devices with machine learning technology for scalable cardiovascular management. Nat. Rev. Cardiol. 2021;18:75–91. doi: 10.1038/s41569-020-00445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Odish OFF, Johnsen K, van Someren P, Roos RAC, van Dijk JG. EEG may serve as a biomarker in Huntington’s disease using machine learning automatic classification. Sci. Rep. 2018;8:16090. doi: 10.1038/s41598-018-34269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Green EM, et al. Machine learning detection of obstructive hypertrophic cardiomyopathy using a wearable biosensor. npj Digit. Med. 2019;2:57. doi: 10.1038/s41746-019-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kehoe ER, et al. Biomarker selection and a prospective metabolite-based machine learning diagnostic for lyme disease. Sci. Rep. 2022;12:1478. doi: 10.1038/s41598-022-05451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Islam MT, Xing L. A data-driven dimensionality-reduction algorithm for the exploration of patterns in biomedical data. Nat. Biomed. Eng. 2021;5:624–635. doi: 10.1038/s41551-020-00635-3. [DOI] [PubMed] [Google Scholar]
- 153.Tyler NS, et al. An artificial intelligence decision support system for the management of type 1 diabetes. Nat. Metab. 2020;2:612–619. doi: 10.1038/s42255-020-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Strain T, et al. Wearable-device-measured physical activity and future health risk. Nat. Med. 2020;26:1385–1391. doi: 10.1038/s41591-020-1012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jin X, Liu C, Xu T, Su L, Zhang X. Artificial intelligence biosensors: challenges and prospects. Biosens. Bioelectron. 2020;165:112412. doi: 10.1016/j.bios.2020.112412. [DOI] [PubMed] [Google Scholar]
- 156.Bashir A, et al. Machine learning guided aptamer refinement and discovery. Nat. Commun. 2021;12:2366. doi: 10.1038/s41467-021-22555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Boubin M, Shrestha S. Microcontroller implementation of support vector machine for detecting blood glucose levels using breath volatile organic compounds. Sensors. 2019;19:2283. doi: 10.3390/s19102283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sardesai AU, et al. An approach to rapidly assess sepsis through multi-biomarker host response using machine learning algorithm. Sci. Rep. 2021;11:16905. doi: 10.1038/s41598-021-96081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.van Doorn WPTM, et al. Machine learning-based glucose prediction with use of continuous glucose and physical activity monitoring data: the Maastricht study. PLoS ONE. 2021;16:e0253125. doi: 10.1371/journal.pone.0253125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rodríguez-Rodríguez I, et al. Utility of big data in predicting short-term blood glucose levels in type 1 diabetes mellitus through machine learning techniques. Sensors. 2019;19:4482. doi: 10.3390/s19204482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tomasik J, et al. A machine learning algorithm to differentiate bipolar disorder from major depressive disorder using an online mental health questionnaire and blood biomarker data. Transl. Psychiatry. 2021;11:41. doi: 10.1038/s41398-020-01181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kalasin S, Sangnuang P, Surareungchai W. Lab-on-eyeglasses to monitor kidneys and strengthen vulnerable populations in pandemics: machine learning in predicting serum creatinine using tear creatinine. Anal. Chem. 2021;93:10661–10671. doi: 10.1021/acs.analchem.1c02085. [DOI] [PubMed] [Google Scholar]
- 163.Cui F, Yue Y, Zhang Y, Zhang Z, Zhou HS. Advancing biosensors with machine learning. ACS Sens. 2020;5:3346–3364. doi: 10.1021/acssensors.0c01424. [DOI] [PubMed] [Google Scholar]
- 164.Bruno E, Viana PF, Sperling MR, Richardson MP. Seizure detection at home: do devices on the market match the needs of people living with epilepsy and their caregivers? Epilepsia. 2020;61:S11–S24. doi: 10.1111/epi.16521. [DOI] [PubMed] [Google Scholar]
- 165.Waldstein SM, et al. Unbiased identification of novel subclinical imaging biomarkers using unsupervised deep learning. Sci. Rep. 2020;10:12954. doi: 10.1038/s41598-020-69814-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fortino V, et al. Machine-learning-driven biomarker discovery for the discrimination between allergic and irritant contact dermatitis. Proc. Natl Acad. Sci. USA. 2020;117:33474–33485. doi: 10.1073/pnas.2009192117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Oh SH, Lee SJ, Noh J, Mo J. Optimal treatment recommendations for diabetes patients using the Markov decision process along with the South Korean electronic health records. Sci. Rep. 2021;11:6920. doi: 10.1038/s41598-021-86419-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dunn J, et al. Wearable sensors enable personalized predictions of clinical laboratory measurements. Nat. Med. 2021;27:1105–1112. doi: 10.1038/s41591-021-01339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Quer G, et al. Wearable sensor data and self-reported symptoms for COVID-19 detection. Nat. Med. 2021;27:73–77. doi: 10.1038/s41591-020-1123-x. [DOI] [PubMed] [Google Scholar]
- 170.Tu J, Gao W. Ethical considerations of wearable technologies in human research. Adv. Healthc. Mater. 2021;10:2100127. doi: 10.1002/adhm.202100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Harris MI, Hadden WC, Knowler WC, Bennett PH. International criteria for the diagnosis of diabetes and impaired glucose tolerance. Diabetes Care. 1985;8:562–567. doi: 10.2337/diacare.8.6.562. [DOI] [PubMed] [Google Scholar]
- 172.Inzucchi SE. Clinical practice. Diagnosis of diabetes. N. Engl. J. Med. 2012;367:542–550. doi: 10.1056/NEJMcp1103643. [DOI] [PubMed] [Google Scholar]
- 173.Tonelli M, Manns B. Supplementing creatinine-based estimates of risk in chronic kidney disease: is it time? JAMA. 2011;305:1593–1595. doi: 10.1001/jama.2011.502. [DOI] [PubMed] [Google Scholar]
- 174.FDA-NIH Biomarker Working Group. BEST (biomarkers, endpoints, and other tools) Resource (FDA, 2016). This publication aims to harmonize terms, such as biomarkers, used in translational science and medicine.
- 175.Miksenas H, Januzzi JL, Natarajan P. Lipoprotein(a) and cardiovascular diseases. JAMA. 2021;326:352–353. doi: 10.1001/jama.2021.3632. [DOI] [PubMed] [Google Scholar]
- 176.Minton K. Maternal sensitization to neonatal allergy. Nat. Rev. Immunol. 2020;21:2–3. doi: 10.1038/s41577-020-00484-w. [DOI] [PubMed] [Google Scholar]
- 177.Valenta R. The future of antigen-specific immunotherapy of allergy. Nat. Rev. Immunol. 2002;2:446–453. doi: 10.1038/nri824. [DOI] [PubMed] [Google Scholar]
- 178.Levey AS, Inker LA, Coresh J. Chronic kidney disease in older people. JAMA. 2015;314:557–558. doi: 10.1001/jama.2015.6753. [DOI] [PubMed] [Google Scholar]
- 179.Rubin R. COVID-19 vaccines vs variants-determining how much immunity is enough. JAMA. 2021;325:1241–1243. doi: 10.1001/jama.2021.3370. [DOI] [PubMed] [Google Scholar]
- 180.Dieterle F, et al. Urinary clusterin, cystatin C, β2-microglobulin and total protein as markers to detect drug-induced kidney injury. Nat. Biotechnol. 2010;28:463–469. doi: 10.1038/nbt.1622. [DOI] [PubMed] [Google Scholar]
- 181.Chertow GM, et al. Guided medication dosing for inpatients with renal insufficiency. JAMA. 2001;286:2839–2844. doi: 10.1001/jama.286.22.2839. [DOI] [PubMed] [Google Scholar]