Abstract
Staphylococcus aureus is a leading cause of prosthetic joint infections (PJI). Surface adhesins play an important role in the primary attachment to plasma proteins that coat the surface of prosthetic devices after implantation. Previous efforts to identify a genetic component of the bacterium that confers an enhanced capacity to cause PJI have focused on gene content, kmers, or single-nucleotide polymorphisms (SNPs) in coding sequences. Here, using a collection of S. aureus strains isolated from PJI and wounds, we investigated whether genetic variations in the regulatory region of genes encoding surface adhesins lead to differences in their expression levels and modulate the capacity of S. aureus to colonize implanted prosthetic devices. The data revealed that S. aureus isolates from the same clonal complex (CC) contain a specific pattern of SNPs in the regulatory region of genes encoding surface adhesins. As a consequence, each clonal lineage shows a specific profile of surface proteins expression. Co-infection experiments with representative isolates of the most prevalent CCs demonstrated that some lineages have a higher capacity to colonize implanted catheters in a murine infection model, which correlated with a greater ability to form a biofilm on coated surfaces with plasma proteins. Together, results indicate that differences in the expression level of surface adhesins may modulate the propensity of S. aureus strains to cause PJI. Given the high conservation of surface proteins among staphylococci, our work lays the framework for investigating how diversification at intergenic regulatory regions affects the capacity of S. aureus to colonize the surface of medical implants.
Keywords: Periprosthetic joint infection, LPXTG proteins, SNPs, Intergenic regions, Staphylococcus aureus, Biofilm
1. Introduction
A major challenge that microbiologists face in the post-genomic era is to identify those differences in bacterial genomes most likely related to phenotypic abilities to cause disease. Intraspecies variation in bacterial genomes occurs through differences in genome content (gene gain or loss) and point mutations caused by punctual insertions, deletions, and single nucleotide polymorphisms (SNPs) [[1], [2], [3], [4]]. Acquisition of new genes can promote more rapid adaptation to new environments than the accumulation of spontaneous mutations because they derive from strains already adapted to a specific niche [5]. The effect of point mutations on bacterial adaptation depends on whether mutations occur in protein-coding sequences or intergenic regions (IGRs). SNPs in protein-coding sequences can lead to frameshifts that interrupt protein translation or changes in the amino acid sequence that enhance or reduce the protein activity, and consequently, bacterial fitness to a particular niche [6]. When point mutations occur at IGRs, they may influence the expression of neighbouring genes or regulatory non-coding RNA molecules. In this case, the causal link between the SNPs and the host-adaptive trait is less recognizable, and functional validation assays are needed to support the relationship between the genotype and the phenotype [7,8].
Staphylococcus aureus is one of the most important Gram-positive bacterial pathogens that can form biofilms on medical devices such as catheters, valves and prostheses [9,10]. S. aureus from skin and mucous membranes from the patient or health care personnel can adhere to the surface of the implant via nonspecific interactions based on the physicochemical properties of the cell envelope or through specific binding between adhesins present on the cell surface and proteins of the host plasma that coat the surface of the implanted material soon after insertion [11,12]. Bacterial attachment is followed by bacterial division and synthesis of a matrix to form multicellular communities that protect bacteria from the immune system and the effects of antibiotics [13,14]. As a consequence, staphylococcal biofilm-associated infections are difficult to eradicate and, in most cases, contaminated implants need to be removed to cure the infection [15,16].
S. aureus produces several adhesins that promote initial attachment and cell-to-cell interactions during biofilm formation [17]. Proteinaceous adhesins include cell wall-anchored (CWA) proteins and non-covalently associated surface proteins [[17], [18], [19]]. The major group in the CWA proteins corresponds to the MSCRAMM (microbial surface components recognizing adhesive matrix molecules) family of proteins [20] and includes, among others, the clumping factor (Clf)-serine aspartate repeat (Sdr) family of proteins, the bone sialoprotein-binding protein (Bbp); the fibronectin-binding proteins (FnBPs); and the collagen-binding protein (Cna). Non-covalently associated surface adhesins include the SERAM (secretable expanded repertoire adhesive molecules) family with the extracellular fibrinogen binding protein (Efb), the extracellular matrix binding protein (Emp), and the extracellular adherence protein (Eap) as the most representative members [17,21]. Although the main function of surface adhesins is to promote adhesion to host extracellular matrix proteins, they can also contribute to other functions such as immune system evasion [10]. Another characteristic of surface adhesins is that they are functionally redundant and different surface adhesins can recognize the same ligand. This functional redundancy implies that the absence of an adhesin may be partially compensated by others [22]. The sequence of surface adhesins is under constant selective pressure to aid in the evasion of host defences. Sequence analysis of FnBPA and FnBPB of S. aureus strains isolated from patients with endocarditis showed polymorphisms in the amino acid sequence that might have been selected to increase the binding affinity to fibronectin (Fn) [23,24]. The level of surface adhesin expression can also vary between clinical S. aureus isolates [11], and within the same strain grown under different environmental conditions [11,25]. For instance, it is known that under iron-limiting conditions, S. aureus induces the expression of Isd proteins, as well as the secreted Eap and Emp proteins [26]. However, how important is genetic diversification of surface adhesins expression within S. aureus lineages that colonize medical implants in niche specialization remains unknown.
In this study, we investigated whether S. aureus clinical isolates might differ in their capacity to cause PJI depending on the presence of SNPs in the IGRs upstream of the translation start site of genes encoding surface adhesins. The results revealed that SNPs heterogeneity results in a specific pattern of surface adhesins expression in each genetic lineage that might confer a competitive advantage in the colonization of prostheses.
2. Materials and methods
2.1. Bacterial strains, plasmids, oligonucleotides, and culture conditions
Bacterial strains, plasmids, and oligonucleotides are listed in Supplementary tables S1, S2 and S3. The forty-five PJI S. aureus strains were previously isolated at Sahlgrenska Hospital (Sweden) from PJI of the hip or knee [27,28]. Twenty-six strains from wound infections, isolated between 1966 and 2010, were obtained from the Culture Collection University of Gothenburg (CCUG) [29,30]. Escherichia coli and S. aureus were grown in Luria-Bertani medium (LB; Conda-Pronadisa, Madrid, Spain) and Trypticase soy broth (TSB; Conda-Pronadisa, Madrid, Spain) at 37 °C. Bacteriological agar was used as a gelling agent (VWR, Radnor, PA, USA). When required, growth media were supplemented with antibiotics at the following concentrations: erythromycin (Ery), 10 μg/mL or 2.5 μg/mL; ampicillin (Amp), 100 μg/mL.
2.2. DNA manipulations
Unless specified otherwise, all the experiments were conducted in accordance with standard practices and manufacturer's instructions. Restriction enzymes (FastDigest), DNA polymerase (Phusion), and the quick DNA ligation kit from Thermo Scientific (Waltham, MA, USA) were used for cloning research. A Macherey-Nagel plasmid purification kit (Allentown, PA, USA) was used to purify the plasmids before they were electroporated into Escherichia coli (1 mm cuvette; 200 Ω, 25 μF, 1250 V) or S. aureus (1 mm cuvette; 100 Ω, 25 μF, 1250 V) with a Gene Pulser X-Cell electroporator. Gene Competent cells were prepared as previously stated [31]. The oligonucleotides were synthesized by STABVIDA Lda (Caparica, Portugal), and sanger sequencing was used to confirm the sequences of all generated plasmids.
2.3. Whole-genome sequencing and genomic analysis
Whole genome sequencing of PJI and wound isolates was previously performed [27,29]. The Sequence Read Archives (SRAs) including comprehensive data on the strains were submitted to the National Center for Biotechnology Information (NCBI) and given the BioProject accession number PRJNA765573.
Nodes containing the locus of twenty-three genes encoding adhesins, were localized in each strain using the blastn tool from the NCBI website. Analysis of the SNPs and comparison between the strains was performed with SnapGene v5.3.3. All sequences were aligned and compared against the S. aureus MW2 genome (GenBank accession number NC_003923). Genetic analysis was focused on the promoter region of adhesin-encoding genes fnbA, clfA, clfB, sdrC, spa, sasC, sasE (isdA), sasF, sasH (adsA), sasI (isdH), sasJ (isdB), eap, emp, vwb and efb, which were present in all strains. For PJI isolates, sequence typing (ST) was previously performed [27,28]. We used the MLST 2.0 tool (https://cge.cbs.dtu.dk/services/MLST/) and the Bacterial Isolate Genome Sequence Database (BIGSdb) (https://pubmlst.org/) to group the isolates from wounds.
2.4. Western blot analyses
To collect supernatant proteins, overnight cultures of one representative strain of CC15 (MIC 6935), CC8 (MIC 6947), CC45 (MIC 6981), and CC30 (MIC 6982) grown in 50 mL of TSB-glucose 0.25% (TSB-gluc) were pelleted by centrifugation at 4500 rpm for 30 min, at 4 °C and the supernatant was collected and centrifuged again. Culture supernatants were filtered and precipitated at 4 °C using 10% (v/v) trichloroacetic acid (Sigma-Aldrich, St Louis, MO, USA) for 1 h. The samples were centrifuged twice at 4000 rpm for 2 h at 4 °C and the supernatant was discarded. The pellets were resuspended in 800 μL ice-cold ethanol (96%) and then centrifuged at 14,000 rpm for 20 min at 4 °C. Resulting pellets were air dried at room temperature and proteins were solubilized with 100 μL of 8 M urea (Amresco, Dallas, TX, USA) for 30 min at room temperature. Protein concentration was quantified using a Bicinchoninic Acid Kit (BCA) (Sigma-Aldrich, St Louis, MO, USA).
CWA proteins from S. aureus clinical isolates were prepared as previously described [32]. In brief, overnight cultures of each of the above-selected strains grown in 5 mL of TSB-gluc were pelleted by centrifugation at 4500 rpm for 30 min, at 4 °C, washed with 1 mL of PBS and resuspended in 100 μL of isosmotic digestion buffer (phosphate-buffered saline (PBS) containing 26% [wt/vol] raffinose; Sigma-Aldrich, St Louis, MO, USA). CWA proteins were solubilized by adding 1.5 μL of a 1 mg/mL solution of lysostaphin (Sigma-Aldrich, St Louis, MO, USA) and incubation with shaking at 37 °C for 2 h. Samples were centrifuged at 8000×g for 30 min with slow deceleration and the supernatant was taken as the cell wall fraction. Protein concentration was quantified using a Bicinchoninic Acid Kit (BCA) (Sigma-Aldrich, St Louis, MO, USA).
For detection of cytoplasmic GFP, overnight cultures of S. aureus 132 containing pCN52:PxxCC plasmids were diluted 1:100 and cultured in 25 ml TSB-gluc at 37 °C under static conditions for 5 h. Cells were pelleted by centrifugation at 4500 rpm for 30 min at 4 °C, washed with 1 mL of PBS, suspended in 400 μL PBS and lysed using a FastPrep-24TM 5G homogenizer (MP Biomedicals, LLC, Irvine, CA, USA). The total amount of protein was quantified using a Bradford protein assay kit (Bio-Rad, Hercules, CA, USA).
For Western blot analysis, equal amounts of protein per sample were loaded on a 12% Stain-Free FastCastTM Acrylamide Kit, 12% (Bio-Rad, Hercules, CA, USA) after adding a volume of Laemmli buffer and boiling for 5 min. After the electrophoresis, the samples were transferred to AmershamTM ProtranTM Premium 0.45 μm nitrocellulose blotting membranes (Cytiva, Marlborough, MA, USA) by electroblotting. Membranes were blocked overnight in PBS containing 0.1% Tween 20 and 5% skimmed milk under shaking conditions and incubated with rabbit anti-S. aureus polyclonal antibody that reacts with soluble and structural antigens of the whole bacterium (Bio-Rad, Hercules, CA, USA) at 1 μg/mL in blocking solution for 1 h at room temperature. For GFP detection, the membranes were incubated with anti-GFP antibodies (Living Colors A.v. monoclonal antibody JL-8 (Clontech, Mountain View, CA, USA), diluted 1:2500 in blocking solution for 2 h at room temperature. Alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Thermo Scientific, Waltham, MA, USA) diluted 1:5000 in blocking solution was used as a secondary antibody and the subsequent chemiluminescence reaction was recorded with ECL Prime western blotting detection reagents (Cytiva, Marlborough, MA, USA) in the ChemidocTM MP Imaging System (Bio-Rad, Hercules, CA, USA).
2.5. Generation of posttranscriptional fusions of adhesin-encoding genes IGRs with gfp
To generate posttranscriptional fusions of the regulatory regions (promoter and 5’ UTR) of adhesins encoding genes and the gfpmut2 gene [33], the region comprising two hundred nucleotides immediately upstream of each gene coding sequence was amplified from one representative strain of CC15 (MIC 6935), CC8 (MIC 6947), CC45 (MIC 6981), and CC30 (MIC 6982), using primers listed in Supplementary Table S3 for each adhesin-encoding gene. The PCR products were cloned into the pJET 1.2 vector and then sub-cloned into the pCN52 plasmid [33] digested with SalI and KpnI, giving plasmids pCN52:PxxCC that were transformed into the S. aureus 132 strain.
2.6. Construction of sortase mutants
To construct the sortase A (srtA) mutants, two DNA fragments were amplified with the primer pairs BG_STAP79/LM97 and LM98/BG_STAP86 (Supplementary Table S3) from the MW2 wild type strain. The two PCR fragments were fused through overlapping PCR using primers BG_STAP79 and BG_STAP86, cloned into the pJET 1.2 vector and then subcloned into the pMAD vector [34] digested with NcoI and BamHI, generating plasmid pMAD:srtAAD. This plasmid was transformed into the MIC 6947, MIC 6981 (purified from S. aureus RN4220) [35], and MIC 6982 (purified from E. coli IM30B) wild-type strains by electroporation [36]. Homologous recombination experiments were performed as previously described [37]. Erythromycin-susceptible white colonies, which did not further contain the pMAD plasmid, were tested by PCR using the primers BG_STAP86 and BG_STAP87 and sanger sequencing was used to confirm the generated isogenic mutants.
2.7. Co-infection experiments
To evaluate in vivo colonization, we used a murine model of catheter-associated biofilm. Bacteria grown overnight in TSB were suspended in PBS to an OD600nm of 0.2 (108 CFU/mL). A total of 24 five-week-old ICR female mice (Envigo, Indianapolis, IN, USA) were anesthetized with isoflurane, and two 15 mm intravenous catheters (24G; B. Braun Medical, Bethlehem, PA, USA) were implanted into the subcutaneous interscapular of each mouse, and co-infected by injection of 100 μL (a total of 107 CFU) of a bacterial mixture, containing the same proportion of each S. aureus strain (CC15 (MIC 6935), CC8 (MIC 6947), CC45 (MIC 6981), and CC30 (MIC 6982)). Five days after infection, animals were anesthetized by isoflurane inhalation and euthanatized by cervical dislocation. The catheters were removed aseptically and placed in Lysing Matrix E tubes (Thermo Scientific, Waltham, MA, USA) containing 1 mL of TES, and cells were lysed using a FastPrep-24™ 5G homogenizer (MP Biomedicals, LLC, Irvine, CA, USA) to isolate the DNA.
Extracted DNA was used for library preparation using Illumina Amplicon Library kit MS-102-3003 (Illumina, San Diego, CA, USA) followed by Illumina sequencing at Fisabio (Valencia, Spain) on a MiSeq System Illumina sequencer (San Diego, CA, USA) using a paired-end approach. The oligonucleotides LM63/LM64 and LM99/LM100 (Supplementary Table S3) were designed for PsasJ, and PclfB amplicon sequencing, respectively. DNA amplicon libraries were generated using a limited cycle PCR: initial denaturation at 95 °C for 3 min followed by 25 cycles of annealing (95 °C 30 s, 55 °C 30 s, 72 °C 30 s) and extension at 72 °C 5 min, using a KAPA HiFi HotStart ReadyMix (KK2602) (Sigma-Aldrich, St Louis, MO, USA). Then, Illumina sequencing adaptors and dual-index barcodes (Nextera XT index kit v2, FC-131-2001) were added to the amplicons. Libraries were normalized and pooled before sequencing. Quality assessment was performed by the use of the fastp program [38] applying the following parameters: min_length: 50; trim_qual_right: 30; trim_qual_type: mean; and trim_qual_window: 10. R1 and R2 from Illumina sequences were joined using the FLASH program applying default parameters [39]. Reports, graphs, and statistics were obtained using the ea-utils programs suite [40]. The MultiQC report was produced using the MultiQC program [41]. The Sequence Read Archives (SRAs) with detailed information of the samples were deposited in NCBI under the BioProject accession number PRJNA834761.
Sequence mapping was carried out with the software BWA v.0.7.17-r1188 and the variants within the reads were located with the software ivar v.1.3.1. For the feature counts, a bash script was written to count the number of reads that mapped to each specific sequence. The number of reads was used to determine the percentage of read abundance. Statistical analyses were performed with the GraphPad Prism v9.2.0 program. One-way ANOVA was used followed by Tukey's multiple comparisons. In all tests, p values of less than 0.05 were considered statistically significant: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
2.8. Biofilm formation phenotypes on protein precoated surfaces
To determine differences in initial adhesion between strains, a biofilm formation assay on protein pre-coated polystyrene plates was performed. First, Fn, Fibrinogen (Fg) and von Willebrand factor (VWF) were dissolved in buffer sodium carbonate 40 mM at a concentration of 5 μg/mL. Then, 100 μL of the corresponding coating was added to wells of a 48-well Nunclon™ Delta surface microtiter plate (Thermo Scientific, Waltham, MA, USA). After overnight incubation at 4 °C under shaking, the plate was rinsed once with water and the biofilm formation assays were performed. Briefly, overnight cultures were adjusted to an OD546nm of 0.13 (108 CFU/mL) in TSB-gluc and further diluted 1:1000. An inoculum consisting of 1 mL of each diluted suspension (105 CFU/mL) was added to the pre-coated wells. After 5 h of static incubation at 37 °C, wells were rinsed once with water, dried, and stained with 1 mL of crystal violet for 5 min (VWR, Radnor, PA, USA). To quantify stained cells, 1 mL of ethanol-acetone (80:20, vol/vol) was added to each well, 500 μL of the suspension were transferred to a new 48-well plate and the OD595nm was recorded in a FLUOstar Omega (BMG LABTECH, Ortenberg, Germany) microplate reader. Six biological replicates with two technical replicates were used.
2.9. Ethics statement
All animal studies were reviewed and approved by the Comité de Ética para la Experimentación Animal (CEEA) of the Universidad de Navarra (approved protocol 032-17). The experiments were performed at the Centro de Investigación Médica Aplicada (ES312010000132) under the principles and guidelines described in the European Directive 2010/63/EU for the protection of animals used for experimental purposes.
3. Results
3.1. Identification of genetic variations in the regulatory regions of genes encoding surface adhesins
Surface adhesins play an important role in the initial attachment to plasma proteins that cover the surface of prosthetic devices after implantation [19,20,42]. We explored if the differences in the presence and/or expression levels of genes encoding surface adhesins in clinical isolates of S. aureus might modulate the initial attachment and capacity of S. aureus to colonize implanted prosthetic devices. To explore this hypothesis, we examined a collection of clinical strains isolated from PJI (n = 45) and wounds (n = 26) [28] (Supplementary Fig. S1) for the presence/absence of twenty-three genes encoding CWA proteins and non-covalently associated surface proteins of the SERAM family. The results revealed that the genes fnbA, clfA, clfB, sdrC, spa, sasC, sasE (isdA), sasF, sasH (adsA), sasI (isdH), sasJ (isdB), eap, emp, vwb and efb were present in all strains, whereas the genes fnbB, cna, sdrD, sdrE, sasA (srAp), sasD, sasG and fmtB (sasB) were absent in some of them (Table 1). For instance, isolates from lineages CC15 and CC5 and most of the isolates from CC8 (93%) contained sasG, whereas none of the isolates from CC30 and CC45 contained this gene. On the contrary, the cna gene was present in all the isolates from CC45 and CC30 and was absent in CC15, CC5 and most of the CC8 (86%) isolates (Table 1). With respect to genes whose presence varied within the same lineage, no correlation was observed between their presence/absence and the source of infection (PJI or wounds).
Table 1.
Prevalence of genes encoding surface adhesins in PJI and wounds isolates.a.
| CC (n) | ST (n) | Prevalence (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| fnbB | cna | sdrD | sdrE | sasA | sasD | sasG | fmtB | |||
| PJI | CC45 (12) | ST45 (12) | 75 | 100 | 83 | 83 | 100 | 100 | 0 | 100 |
| CC30 (8) | ST30 (8) | 88 | 100 | 100 | 88 | 100 | 100 | 0 | 100 | |
| CC8 (6) | ST8 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | |
| ST630 (4) | 75 | 0 | 100 | 75 | 0 | 100 | 100 | 100 | ||
| ST789 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CC15 (6) | ST15 (5) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | |
| ST3457 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CC5 (3) | ST5 (3) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | |
| CC22 (1) | ST22 (1) | 0 | 100 | 100 | 0 | 0 | 0 | 100 | 100 | |
| CC1 (1) | ST1 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 0 | 100 | |
| ST50 (2) | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 100 | ||
| ST20 (2) | 50 | 0 | 50 | 100 | 100 | 100 | 100 | 100 | ||
| ST25 (1) | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 100 | ||
| ST80 (1) | 100 | 0 | 100 | 0 | 100 | 100 | 0 | 100 | ||
| ST50 (1) | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | ||
| ST182 (1) | 100 | 100 | 0 | 0 | 100 | 0 | 100 | 0 | ||
| Wounds | CC8 (8) | ST8 (2) | 100 | 0 | 100 | 100 | 100 | 100 | 50 | 100 |
| ST254 (1) | 100 | 0 | 100 | 0 | 100 | 100 | 100 | 100 | ||
| ST247 (3) | 33 | 0 | 100 | 33 | 100 | 100 | 100 | 100 | ||
| ST239 (2) | 100 | 100 | 100 | 50 | 100 | 100 | 100 | 100 | ||
| CC30 (3) | ST30 (3) | 33 | 100 | 100 | 100 | 100 | 100 | 0 | 100 | |
| CC5 (3) | CC5/ST5 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | |
| CC5/ST225 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CC5/ST1649 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| CC45 (1) | ST45 (1) | 0 | 100 | 100 | 100 | 100 | 100 | 0 | 100 | |
| CC15 (1) | ST15 (1) | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | |
| ST25 (3) | 100 | 0 | 100 | 100 | 100 | 100 | 0 | 100 | ||
| ST80 (3) | 33 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | ||
| ST182 (2) | 100 | 100 | 0 | 0 | 100 | 0 | 100 | 0 | ||
| ST121 (1) | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 100 | ||
| ST1693 (1) | 0 | 100 | 100 | 100 | 100 | 0 | 0 | 100 | ||
The fnbA, clfA, clfB, sdrC, spa, sasC, sasE, sasF, sasH, sasI, sasJ, eap, emp, vwb and efb genes are not included because they are present in all strains.
We next explored the possibility that genetic variations between isolates might be located in the regulatory regions (romoter and 5’ UTR) controlling the expression of adhesin proteins. Thus, we compared a sequence of 200 nt upstream the first codon of each of the 15 genes encoding surface adhesins present in all the strains with the corresponding sequence in the reference genome of S. aureus MW2 strain (CC1). The conservation rate of the sequence in the regulatory regions was highly variable depending on the gene (Fig. 1 and Supplementary Fig. S2). The sequence of the regulatory region of sasE and fnbA genes was highly conserved with variation rates below 5%. In contrast, the sequence of the regulatory region of sdrC showed a variation rate of 28% (Supplementary Table S4). Notably, the SNPs were highly conserved between isolates of the same sequence type, regardless of whether the strain was isolated from PJI or wounds. Taken together, these results indicated that SNPs variations in the regulatory region of surface adhesin genes are characteristic of each S. aureus ST lineage.
Fig. 1.
Analysis of genetic variations in the regulatory region controlling the expression of surface adhesins. The region controlling sasF expression is shown as an example. PJI and wound isolates were grouped into twelve different clusters according to the SNPs present in the region comprising 200 nt upstream of the sasF gene compared to the sequence of the reference strain MW2. The most representative sequence variant for the most prevalent clonal complexes is shown. The black lines show the SNPs or indels found. All sequence variations found in CC8 are depicted (nucleotide changes from a black to a red nucleotide in the upper strand). In the rest of CCs, only sequence variations that are different from the ones found above are detailed. Eleven SNPs were identified among the representative sequence variants; none of the strains contained the same sequence as the MW2 reference strain.
3.2. Contribution of SNPs to surface adhesins expression
We next wondered whether the SNPs present in IGRs may have an impact on surface adhesins expression levels. For that, we first used a commercial polyclonal antibody (Bio-Rad, Hercules, CA, USA) raised against soluble and structural antigens of the whole bacterium to compare the overall expression levels of CWA and secreted proteins between selected strains from different CC: CC15 (MIC 6935), CC8 (MIC 6947), CC45 (MIC 6981), and CC30 (MIC 6982). To identify the CWA proteins, we constructed sortase A (srtA) mutants for each strain. Our reasoning was that those bands that disappear in the absence of the SrtA activity would correspond to CWA proteins; regarding secreted proteins, no differences between wild-type and isogenic srtA mutants should be observed, therefore secreted proteins profiles are shown as a control. We generated srtA mutants for CC18, CC45 and CC30 strains. However, we were unable to genetically manipulate and generate the srtA mutant in the CC15 strain. Western Blot analysis revealed differences in the levels of CWA proteins between isolates of different CCs (Supplementary Fig. S3).
To specifically investigate the contribution of the SNPs in IGRs to the expression of surface adhesins without the interference of particular regulators that control the expression of surface adhesins in each strain, the region encompassing 200 nt upstream of the first codon of each adhesin-encoding gene present in the representative PJI isolates of CC15, CC8, CC45 and CC30 was fused with the gfp gene in the pCN52 plasmid. Then, the resulting plasmids were inserted in the S. aureus 132 strain, which is able to produce a proteinaceous-biofilm matrix when grown in TSB-glucose medium [43]. The effect of the genetic variations in IGRs on GFP expression levels was analyzed by Western blot. The results revealed large differences in the expression level of each surface adhesin. Of note, in general, expression from the regulatory region of spa, clfA, clfB, vwb and efb genes was high when compared to other regions such as the ones controlling expression of sdrC, sasC, sasE, sasI, sasJ, eap and emp (Fig. 2).
Fig. 2.
Comparison of surface adhesins expression levels within the same strain. Western blots show GFP protein levels in S. aureus 132 strain transformed with plasmids containing reporter fusions of 200 nt upstream the initial codon of 15 surface adhesin-encoding genes, amplified from the same isolate, with the gfp gene. One representative S. aureus PJI isolate of each of the four most abundant clonal complexes CC15, CC8, CC45 and CC30 was used for amplification of regulatory sequences. Bacteria were grown until the exponential phase and proteins were transferred to nitrocellulose membranes, incubated with anti-GFP monoclonal antibodies and developed using peroxidase-conjugated goat anti-mouse antibodies and a bioluminescence kit. Stain-free gels are shown as loading controls (LD).
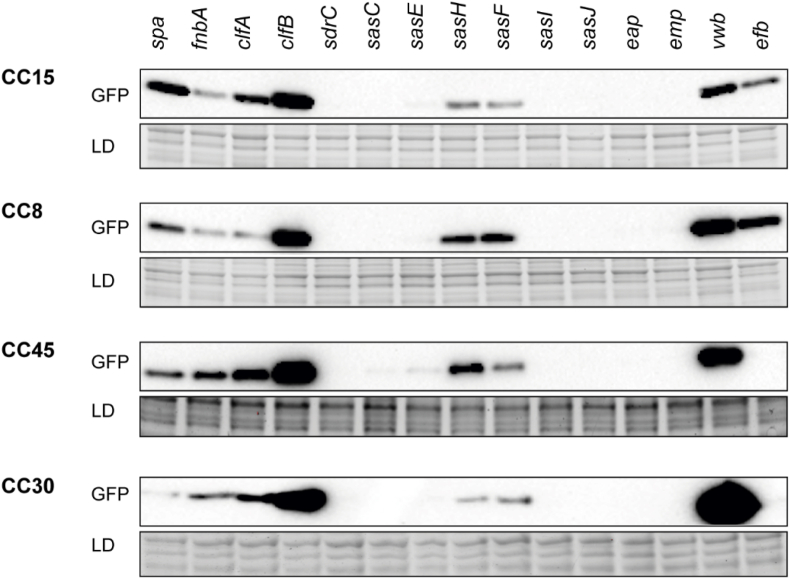
Next, we compared the expression of gfp under the regulatory region of each surface adhesin-encoding gene amongst the four different CCs. The results showed variability in the expression pattern of spa, sdrC, sasC, sasF, sasH, sasI, sasJ, emp, eap, vwb and efb genes, indicating that the SNPs present in the regulatory regions of the genes encoding for surface adhesins affect their expression levels (Fig. 3). The level of expression of fnbA, clfA, clfB and sasE genes remained unchanged among the isolates of different CCs, indicating that the SNPs present in the regulatory region of these genes were not involved in transcriptional and/or post-transcriptional regulation of such genes expression. Collectively, these results provided robust evidence that surface adhesin expression levels vary among S. aureus ST lineages due to sequence variations in IGRs of the corresponding genes.
Fig. 3.
Comparison of surface adhesins expression levels in strains from different CCs. Western blots show GFP protein levels in S. aureus 132 strain transformed with plasmids containing reporter fusions of 200 nt upstream the initial codon of each surface adhesin-encoding genes, amplified from representative S. aureus PJI isolates of the four most abundant CCs, with the gfp gene. Bacteria were grown until the exponential phase and proteins were transferred to nitrocellulose membranes, incubated with anti-GFP monoclonal antibodies and developed using peroxidase-conjugated goat anti-mouse antibodies and a bioluminescence kit. Membrane exposure was adjusted in each case to allow band visualization in all cases. Stain-free gels are shown as loading controls.
3.3. Comparison of the in vivo colonization capacity of S. aureus isolates of different CCs
Taking into account the above results demonstrating that the strength of the promoters encoding for surface adhesins varies between S. aureus isolates of different CCs, we hypothesized that differences in the capacity to colonize implanted prostheses might occur. To investigate this possibility, we carried out an in vivo catheter colonization assay with a mixture containing four strains representative of the four CCs that most frequently caused prosthesis infections in our PJI collection. An intrinsic difficulty in co-infection experiments is that a selective analytical method is required in order to distinguish and quantify each strain in the mixture. To address this methodological challenge, we amplified a 200 nt region upstream of the sasJ and clfB genes that contain the specific SNPs of each CC (Fig. 4A) [44]. To validate the selected DNA signatures, we prepared in vitro mock mixtures containing different proportions of the four isolates, total DNA was purified, PCR amplified and sequenced. The reads were processed and mapped to sasJ and clfB regions. The total number of reads that passed a quality filter was higher than 99% for both PsasJ and PclfB, respectively. The results confirmed that the number of mapped reads obtained corresponding to each DNA signature was proportional to the amount of each bacterium in the original mixture (Fig. 4B). We next used this strategy to determine the proportion of each isolate on the surface of catheters implanted in the subscapular space of mice. A mixture of equal numbers (107 CFU) of the four representative strains was used to colonize the implanted catheters (Supplementary Fig. S4) and after five days, catheters were recovered, DNA was purified, and the amount of each bacterium was quantified based on the abundance of reads for each DNA signature. The results revealed that read abundance corresponding to strains from CC15 and CC8 was significantly higher than the reads corresponding to strains from CC45 and CC30 (Fig. 4C). No significant differences were found between the number of reads corresponding to CC15 and CC8 strains and CC30 and CC45 strains. Together, these results evidenced that S. aureus isolates show CC dependent variation of the capacity to colonize an implanted catheter.
Fig. 4.
DNA signatures for quantifying isolates from different clonal lineages during co-infection experiments of catheters in a mouse model. (A) Natural sequence variants of the regulatory regions of sasJ and clfB genes from representative strains ascribed to CC15, CC8, CC45, and CC30. The red lines show the SNPs or indels found compared with the CC15 sequence (nucleotide changes from a black to a red nucleotide in the upper strand). (B) Relative proportion of each isolate based on the number of reads corresponding to sequence variants. The results represent the mean of three independent in vitro experiments (n = 3). M1: Mixture one (equal proportion of each of the four strains); M2: Mixture two (double proportion of CC15 strain than CC8 and CC45); M3: Mixture three (triple proportion of CC15 strain than CC8). (C) Differences in colonization capacity of S. aureus isolates from different clonal complexes in a murine model of catheter colonization. Percentage of reading abundance after 5 days of infection according to the number of reads of sequence variants of sasJ and clfB regulatory regions. Catheters were coinfected with equal amounts (107 CFU) of representative S. aureus PJI isolates ascribed to the most prevalent clonal complexes. CC15: MIC 6935; CC8: MIC 6947; CC45: MIC 6981; CC30: MIC 6982. The boxes indicate the range between the first and third quartiles (25th and 75th percentiles). The horizontal line inside the box is the median. The size of the box represents the interquartile range (IQR). The whiskers indicate the spread of data outside the box at up to 1.5 times the IQR from the edge of box (1.5 times the size of the box below the 25th percentile or above the 75th percentile). Data further than 1.5 times the IQR from the box are designated outliers and plotted individually. Statistically significant differences were determined using one-way ANOVA, followed by Tukey's multiple comparison (**, p < 0.01), (***, p < 0.001) (****, p < 0.0001).
If differences in the ability of the strains from the different clonal groups to adhere to implanted catheters were due to differences in their adhesion to plasma proteins coating the catheter surface, similar variances in their capacity to adhere to abiotic surfaces covered with plasma proteins in vitro could be expected. To explore this hypothesis, we performed primary attachment assays on polystyrene pre-coated with Fn, Fg and VWF (Fig. 5). The results revealed that the representative strains from CC15 and CC8 showed a significantly higher capacity than isolates from CC30 and CC45 to adhere to polystyrene surfaces coated with Fn and Fg, whereas in the case of surfaces coated with VWF, all isolates showed a similar primary adhesion capacity. Interestingly, isolates from CC15 and CC8 lineages also showed a higher capacity to adhere to uncoated polystyrene surfaces than isolates from CC30 and CC45. Notably, adhesion of CC15 and CC8 isolates to the uncoated surface was lower than to Fn and Fg coated surfaces. Together, these results demonstrated that the strains winning the race for colonization of the surface of the implant in vivo also displayed a higher capacity to colonize abiotic surfaces coated with plasma proteins in vitro.
Fig. 5.
Biofilm biomass formed by representative S. aureus PJI isolates from CC15, CC8, CC45 and CC30 on protein coated surfaces. The wells of polystyrene plates were pre-coated with 5 μg/mL of fibronectin, fibrinogen, and the von Willebrand factor and kept overnight at 4 °C. Static biofilms were grown in TSB-gluc on the pre-coated 48-well plates for 5 h. Biofilm formation was quantified by crystal violet staining, followed by 80:20 ethanol: acetone elution and OD595nm measurement. The data represent the mean of six biological replicates (n = 6) with two technical replicates; error bars represent ± SD. Statistically significant differences were determined using one-way ANOVA, followed by Tukey's multiple comparisons (*, p < 0.05), (**, p < 0.01)), (***, p < 0.001).
4. Discussion
PJI is the most severe complication following arthroplasty [45]. S. aureus is known as the main bacterium causing PJI due to its ability to adhere to and establish biofilms on abiotic surfaces [13,46]. Epidemiologic studies have shown that several S. aureus lineages can cause PJI [47,48]. However, the fact that strains from different clonal complexes are isolated from PJI does not necessarily mean that isolates from different CCs have the same ability to adhere irreversibly to the implant surface and cause an infection.
A simple mechanism to generate variability in the capacity to colonize implants between different isolates may be due to variations in the presence/absence of adhesin-encoding genes [49,50]. For instance, SasG, a protein important in promoting biofilm formation during the accumulation phase, is present in strains of CC15 and CC8 whereas it is absent in strains from CC30 and CC45 [51,52]. On the contrary, the presence of the collagen-binding protein (Cna) has been mainly associated with strains from CC30 and CC45 [53]. The analysis of the genome sequences of our collection of PJI isolates confirmed the presence of fnbA, clfA, clfB, sdrC, spa, sasC, sasE, sasF, sasH, sasI, sasJ, eap, emp, vwb and efb in all the isolates. In agreement with previous studies, the sasG gene was found in all PJI strains from CC8 and CC15 and was not detected in any CC30 and CC45 isolate, whereas cna was present in all strains from CC30 and CC45 and absent in all CC15 and CC8 PJI isolates. Our results revealed that the selected strains from CC8 and CC15 showed a higher capacity to colonize catheters in vivo and a higher ability to form a biofilm on protein precoated surfaces than isolates from CC30 and CC45, and therefore we cannot exclude that the presence of SasG might contribute to an increased S. aureus propensity to colonize and accumulate on the surface of implanted prostheses. Following the same reasoning, our findings suggest that the presence of Cna may not be as important when competing to colonize a surface.
A second source of variability in the ability of S. aureus to colonize implants can be generated by changes in the sequence of adhesin proteins. It is well known that the exposure of the bacteria to the pressure of the host immune system contributes to the accumulation of polymorphisms within surface proteins. Studies on S. aureus have revealed that clinical isolates accumulate SNPs in their surface proteins as the infection progresses [48,54]. Ma et al. showed that increasing genotypic variation in adhesin-encoding genes between the first and later isolates from PJI outcomes with changes in the ability to bind to plasma proteins. These changes might confer advantages to successfully colonizing surfaces and/or evading the immune system. Similarly, several studies have evidenced that polymorphisms in FnBPA-binding repeats in isolates causing infection of cardiovascular devices are associated with an enhanced capacity to adhere to Fn [23,55,56].
A third level of variability can be generated through mutations at the IGRs of certain genes that cause changes in the expression levels of compounds important for implant colonization. Most studies dedicated to investigating how bacteria adapt to the host environment have focused on changes that occur within coding regions, whereas the role of intergenic mutations has remained mostly disregarded. Importantly, several evolution studies have shown that mutations in the regulatory elements upstream of transcriptional start sites cause changes in the transcription levels of genes important for evolution of pathogenic phenotypes, including essential genes that are less permissive to accumulate mutations in the coding sequence [[57], [58], [59]]. In the case of S. aureus, SNPs accumulation in the IGRs of adhesin-encoding genes present in strains isolated from later points of infection has been described [48]. On the contrary, regarding the production of the main exopolysaccharide of the S. aureus biofilm matrix, SNPs in the highly conserved IGRs of the icaADBCR operon were not associated with changes in icaADBC expression, PIA/PNAG production and adaptation to PJI [29].
A fourth level of variability can be generated by changes in the expression levels of global transcriptional regulators controlling the expression of surface adhesins. Several regulatory elements, such as agrAC, saeRS, arlRS, mgrA, and sarA are known to directly or indirectly regulate the expression of staphylococcal adhesins in response to different environmental conditions [26]. For instance, the agr system represses adhesins expression and its activity is inhibited by proteins found in human serum and blood [60]. Thus, the agr-dependent expression of adhesins is inhibited in plasma or human serum. Similarly, SaeRS, Agr, and SarA, upregulate the expression of some SERAMs such as eap and emp under low-iron conditions [26,61]. SarA also responds to O2 or CO2 levels and can induce (fnbA and fnbB) or repress (spa) the expression of some adhesins either in an agr-dependent or independent way [62]. Moreover, the repression of the protease activity by SarA can affect the accumulation of some staphylococcal proteins [63].
In the present study, the comparison of the IGRs upstream 15 adhesin-encoding genes of the 71 isolates under study revealed strong differences in conservation rates. In particular, the IGR upstream the fnbA gene was highly conserved, only comprising seven SNPs, whereas the IGR upstream sdrC accumulated 56 SNPs that resulted in a 28% variation rate. The high degree of conservation in the IGR upstream fnbA was somehow unexpected because the FnBPA amino acid sequence varies considerably between S. aureus lineages [64] and polymorphisms associated with binding mechanisms have been described in isolates from cardiovascular devices infections [55]. A significant difficulty for inferring the functional value of SNPs in IGRs compared to those in coding regions is that the synonymous/non-synonymous distinction does not apply for IGRs. Thus, experimental evaluation of the impact of each set of mutations becomes necessary in order to reach a conclusion on its function and relevance. Here, the potential functional relevance of the SNPs in the IGRs of the 15 adhesins encoding genes to their expression was determined using posttranscriptional fusions with the gfp reporter gene. The results showed that each CC shows a characteristic profile of adhesins expression. The Spa, vWbp and Efb reporters showed a higher level of expression in CC15 and CC8 isolates compared to CC30 and CC45 strains. Protein A (Spa) has been shown to promote catheter-associated infection [65], and the secreted VWF-binding protein (vWbp) promotes ClfA-mediated adhesion to VWF [66]. The Efb is a very large secreted protein that binds host Fg and complement C3 and plays an important role in evasion of the immune system [67]. On the other hand, the presence of SNPs in the IGRs upstream of genes encoding for FnBPA, ClfA, and ClfB did not affect expression levels, suggesting that the SNPs were silent, at least in the conditions tested.
Based on the notion that S. aureus colonization capacity of prosthetic implants is very likely due to the expression profile of the whole family of surface adhesins, we explored if S. aureus isolates from different CCs, with a particular profile in the presence and SNPs of adhesins genes, might show an advantage in colonizing an implant, using a catheter infection model in mice. At the same time, aiming to apply the reduction principle for ethical use of animals in scientific research [68], we coinfected each catheter with a mixture containing equal numbers of representative strains of four CCs. Because the four isolates were genetically very closely related, it was necessary to design oligonucleotides that amplify DNA regions containing enough number of SNPs to unambiguously distinguish and quantify the amount of bacteria corresponding to each isolate on the surface of the catheter after the infection process. Once we selected the regions comprising IGRs of sasJ and clfB genes, we applied the well-established methodology used to sequence and characterize microbiomes using 16S amplification products. Interestingly, isolates from CC15 and CC8 showed a higher capacity to colonize the catheters surface than isolates from CC30 and CC45. These results corresponded to the ability of each isolate to adhere to abiotic surfaces coated with individual plasma proteins, strongly suggesting that differences in the capacity to colonize implanted devices might result from the sum of adhesion properties of each individual adhesin.
We anticipate that this experimental approach for quantifying and detecting closely related bacterial isolates will be very helpful to identify competitive advantages for certain clinical bacterial isolates in specific stages of the infectious process or in polymicrobial biofilm research.
CRediT authorship contribution statement
Liliana Morales-Laverde: Investigation, Visualization, Methodology, Validation, Writing – original draft. Margarita Trobos: Methodology, Writing – review & editing, Resources, Funding acquisition. Maite Echeverz: Investigation. Cristina Solano: Writing – review & editing, Supervision. Iñigo Lasa: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Prof. Ola Rolfson (Department of Orthopaedics, Institute of Clinical Sciences, University of Gothenburg, Sweden) and Assoc. Prof. Bodil Jönsson (Department of Infectious Diseases, Institute for Biomedicine, University of Gothenburg, Sweden) for the collection of the strains from periprosthetic joint infections. We thank Prof. Edward Moore (Culture Collection at the University of Gothenburg, CCGU, Sweden) for kindly providing the S. aureus strains from wounds. This work was financially supported by the Spanish Ministry of Science and Innovation grant PID2020-113494RB-I00 to I.L. (Agencia Española de Investigación/Fondo Europeo de Desarrollo Regional, European Union), the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 754412 [MoRE2020 - Region Väs-tra Götaland], and the IngaBritt and Arne Lundberg Foundation (LU2021-0048). L.M.L was supported by the European Union's H2020 research and innovation programme under Marie Sklodowska-Curie grant agreement No 801586 (IberusTalent).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2022.100093.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Schürch A.C., Arredondo-Alonso S., Willems R.J.L., Goering R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin Microbiol Infect. 2018;24:350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 2.Raskin D.M., Seshadri R., Pukatzki S.U., Mekalanos J.J. Bacterial genomics and pathogen evolution. Cell. 2006;124:703–714. doi: 10.1016/j.cell.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Sheppard S.K., Guttman D.S., Fitzgerald J.R. Population genomics of bacterial host adaptation. Nat Rev Genet. 2018;19:549–565. doi: 10.1038/s41576-018-0032-z. [DOI] [PubMed] [Google Scholar]
- 4.Toft C., Andersson S.G.E. Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet. 2010;11:465–475. doi: 10.1038/nrg2798. [DOI] [PubMed] [Google Scholar]
- 5.Du X., Larsen J., Li M., Walter A., Slavetinsky C., Both A., et al. Staphylococcus epidermidis clones express Staphylococcus aureus-type wall teichoic acid to shift from a commensal to pathogen lifestyle. Nat Microbiol. 2021;6:757–768. doi: 10.1038/s41564-021-00913-z. [DOI] [PubMed] [Google Scholar]
- 6.Viana D., Comos M., McAdam P.R., Ward M.J., Selva L., Guinane C.M., et al. A single natural nucleotide mutation alters bacterial pathogen host tropism. Nat Genet. 2015;47:361–366. doi: 10.1038/ng.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheppard S.K., Didelot X., Meric G., Torralbo A., Jolley K.A., Kelly D.J., et al. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci USA. 2013;110:11923–11927. doi: 10.1073/pnas.1305559110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pascoe B., Méric G., Murray S., Yahara K., Mageiros L., Bowen R., et al. Enhanced biofilm formation and multi-host transmission evolve from divergent genetic backgrounds in Campylobacter jejuni. Environ Microbiol. 2015;17:4779–4789. doi: 10.1111/1462-2920.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arciola C.R., Campoccia D., Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol. 2018;16:397–409. doi: 10.1038/s41579-018-0019-y. [DOI] [PubMed] [Google Scholar]
- 10.Raafat D., Otto M., Reppschläger K., Iqbal J., Holtfreter S. Fighting Staphylococcus aureus biofilms with monoclonal antibodies. Trends Microbiol. 2019;27:303–322. doi: 10.1016/j.tim.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speziale P., Pietrocola G., Foster T.J., Geoghegan J.A. Protein-based biofilm matrices in Staphylococci. Front Cell Infect Microbiol. 2014;4:171. doi: 10.3389/fcimb.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster T.J. Surface proteins of Staphylococcus epidermidis. Front Microbiol. 2020;11:1829. doi: 10.3389/fmicb.2020.01829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otto M. Staphylococcal biofilms. Microbiol Spectr. 2018;6 doi: 10.1128/microbiolspec.gpp3-0023-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schilcher K., Horswill A.R. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol Mol Biol Rev. 2020;84 doi: 10.1128/mmbr.00026-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montanaro L., Speziale P., Campoccia D., Ravaioli S., Cangini I., Pietrocola G., et al. Scenery of Staphylococcus implant infections in orthopedics. Future Microbiol. 2011;6:1329–1349. doi: 10.2217/fmb.11.117. [DOI] [PubMed] [Google Scholar]
- 16.Puhto T., Puhto A.-P., Vielma M., Syrjälä H. Infection triples the cost of a primary joint arthroplasty. Infect Dis Newsl. 2019;51:348–355. doi: 10.1080/23744235.2019.1572219. [DOI] [PubMed] [Google Scholar]
- 17.Heilmann C. vol. 715. Springer Netherlands; Dordrecht: 2011. pp. 105–123. (Adhesion mechanisms of staphylococci). [DOI] [PubMed] [Google Scholar]
- 18.Geoghegan J.A., Foster T.J. Staphylococcus aureus, microbiology, pathology, immunology, therapy and prophylaxis. Curr Top Microbiol. 2015:95–120. doi: 10.1007/82_2015_5002. [DOI] [PubMed] [Google Scholar]
- 19.Clarke S.R., Foster S.J. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol. 2006;51:187–224. doi: 10.1016/s0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
- 20.Foster T.J. The MSCRAMM family of cell-wall-anchored surface proteins of gram-positive cocci. Trends Microbiol. 2019;27:927–941. doi: 10.1016/j.tim.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Chavakis T., Wiechmann K., Preissner K.T., Herrmann M. Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thromb Haemostasis. 2005;94:278–285. doi: 10.1160/th05-05-0306. [DOI] [PubMed] [Google Scholar]
- 22.Foster T.J., Geoghegan J.A., Ganesh V.K., Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casillas-Ituarte N.N., Lower B.H., Lamlertthon S., Fowler V.G., Lower S.K. Dissociation rate constants of human fibronectin binding to fibronectin-binding proteins on living Staphylococcus aureus isolated from clinical patients. J Biol Chem. 2012;287:6693–6701. doi: 10.1074/jbc.m111.285692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Speziale P., Pietrocola G. The multivalent role of fibronectin-binding proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in host infections. Front Microbiol. 2020;11:2054. doi: 10.3389/fmicb.2020.02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roche F., Massey R., Peacock S., Day N., Visai L., Speziale P., et al. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology. 2003;149:643–654. doi: 10.1099/mic.0.25996-0. [DOI] [PubMed] [Google Scholar]
- 26.Paharik A.E., Horswill A.R. The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.vmbf-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malchau K.S., Tillander J., Zaborowska M., Hoffman M., Lasa I., Thomsen P., et al. Biofilm properties in relation to treatment outcome in patients with first-time periprosthetic hip or knee joint infection. J Orthop Transl. 2021;30:31–40. doi: 10.1016/j.jot.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trobos M., Firdaus R., Malchau K.S., Tillander J., Arnellos D., Rolfson O., et al. Genomics of Staphylococcus aureus and Staphylococcus epidermidis from periprosthetic joint infections and correlation to clinical outcome. Microbiol Spectr. 2022 doi: 10.1128/spectrum.02181-21. e02181-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morales-Laverde L., Echeverz M., Trobos M., Solano C., Lasa I. Experimental Polymorphism survey in intergenic regions of the icaADBCR Locus in Staphylococcus aureus isolates from periprosthetic joint infections. Microorganisms. 2022;10:600. doi: 10.3390/microorganisms10030600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner A.B., Gerner E., Firdaus R., Echeverz M., Werthén M., Thomsen P., et al. Role of sodium salicylate in Staphylococcus aureus quorum sensing, virulence, biofilm formation and antimicrobial susceptibility. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.931839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenk S., Laddaga R. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 32.Arrizubieta M.J., Toledo-Arana A., Amorena B., Penadés J.R., Lasa I. Calcium inhibits bap-dependent multicellular behavior in Staphylococcus aureus. J Bacteriol. 2004;186:7490–7498. doi: 10.1128/jb.186.22.7490-7498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charpentier E., Anton A.I., Barry P., Alfonso B., Fang Y., Novick R.P. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol. 2004;70:6076–6085. doi: 10.1128/aem.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnaud M., Chastanet A., Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/aem.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreiswirth B., Lofdahl S., Betley M., O’Reilly M., Schlievert P., Bergdoll M., et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 36.Monk I.R., Tree J.J., Howden B.P., Stinear T.P., Foster T.J. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio. 2015;6:e00308–e00315. doi: 10.1128/mbio.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valle J., Toledo‐Arana A., Berasain C., Ghigo J., Amorena B., Penadés J.R., et al. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aronesty E. Comparison of sequencing utility programs. Open Bioinf J. 2013;7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 41.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson J.M., Rodriguez A., Chang D.T. Foreign body reaction to biomaterials. Semin Immunol. 2008;20:86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vergara-Irigaray M., Valle J., Merino N., Latasa C., García B., Mozos IR de los, et al. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun. 2009;77:3978–3991. doi: 10.1128/iai.00616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zahariev M., Chen W., Visagie C.M., Lévesque C.A. Cluster oligonucleotide signatures for rapid identification by sequencing. BMC Bioinf. 2018;19:395. doi: 10.1186/s12859-018-2363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klug A., Gramlich Y., Rudert M., Drees P., Hoffmann R., Weißenberger M., et al. The projected volume of primary and revision total knee arthroplasty will place an immense burden on future health care systems over the next 30 years. Knee Surg Sports Traumatol Arthrosc. 2020:1–12. doi: 10.1007/s00167-020-06154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosteius T., Jansen O., Fehmer T., Baecker H., Citak M., Schildhauer T.A., et al. Evaluating the microbial pattern of periprosthetic joint infections of the hip and knee. J Med Microbiol. 2018;67:1608–1613. doi: 10.1099/jmm.0.000835. [DOI] [PubMed] [Google Scholar]
- 47.Wildeman P., Tevell S., Eriksson C., Lagos A.C., Söderquist B., Stenmark B. Genomic characterization and outcome of prosthetic joint infections caused by Staphylococcus aureus. Sci Rep-Uk. 2020;10:5938. doi: 10.1038/s41598-020-62751-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma D., Brothers K.M., Maher P.L., Phillips N.J., Simonetti D., Pasculle A.W., et al. Staphylococcus aureus genotype variation among and within periprosthetic joint infections. J Orthop Res. 2021 doi: 10.1002/jor.25031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy A.J., Lindsay J.A. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 2010;10:173. doi: 10.1186/1471-2180-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasmussen G., Monecke S., Ehricht R., Söderquist B. Prevalence of clonal complexes and virulence genes among commensal and invasive Staphylococcus aureus isolates in Sweden. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corrigan R.M., Rigby D., Handley P., Foster T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology (Read) 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 52.Geoghegan J.A., Corrigan R.M., Gruszka D.T., Speziale P., O'Gara J.P., Potts J.R., et al. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol. 2010;192:5663–5673. doi: 10.1128/jb.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arciola C., Campoccia D., Gamberini S., Baldassarri L., Montanaro L. Prevalence of cna, fnbA and fnbB adhesin genes among Staphylococcus aureus isolates from orthopedic infections associated to different types of implant. FEMS Microbiol Lett. 2005;246:81–86. doi: 10.1016/j.femsle.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 54.Young B.C., Wu C.-H., Gordon N.C., Cole K., Price J.R., Liu E., et al. Severe infections emerge from commensal bacteria by adaptive evolution. Elife. 2017;6 doi: 10.7554/elife.30637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lower S.K., Lamlertthon S., Casillas-Ituarte N.N., Lins R.D., Yongsunthon R., Taylor E.S., et al. Polymorphisms in fibronectin binding protein A of Staphylococcus aureus are associated with infection of cardiovascular devices. Proc Natl Acad Sci USA. 2011;108:18372–18377. doi: 10.1073/pnas.1109071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piroth L., Que Y.-A., Widmer E., Panchaud A., Piu S., Entenza J.M., et al. The fibrinogen- and fibronectin-binding domains of Staphylococcus aureus fibronectin-binding protein A synergistically promote endothelial invasion and experimental endocarditis. Infect Immun. 2008;76:3824–3831. doi: 10.1128/iai.00405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khademi S.M.H., Sazinas P., Jelsbak L. Within-host adaptation mediated by intergenic evolution in Pseudomonas aeruginosa. Genome Biol Evol. 2019;11:1385–1397. doi: 10.1093/gbe/evz083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blanka A., Düvel J., Dötsch A., Klinkert B., Abraham W.-R., Kaever V., et al. Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Sci Signal. 2015;8 doi: 10.1126/scisignal.2005943. ra36–ra36. [DOI] [PubMed] [Google Scholar]
- 59.Cui Z., Li Y., Cheng S., Yang H., Lu J., Hu Z., et al. Mutations in the embC-embA intergenic region contribute to Mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother. 2014;58:6837–6843. doi: 10.1128/aac.03285-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenul C., Horswill A.R. Regulation of Staphylococcus aureus Virulence. Gram-Positive Pathogens. Third Edition. 2019 doi: 10.1128/9781683670131.ch41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson M., Cockayne A., Morrissey J.A. Iron-regulated biofilm formation in Staphylococcus aureus newman requires ica and the secreted protein emp. Infect Immun. 2008;76:1756–1765. doi: 10.1128/iai.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan P., Foster S. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/JB.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenul C., Horswill A.R. Regulation of Staphylococcus aureus virulence. Microbiol Spectr. 2019;7:669–686. doi: 10.1128/microbiolspec.gpp3-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clarke N., Loughman A., Meade G., Foster T. Molecular basis for Staphylococcus aureus-mediated platelet aggregate formation under arterial shear in vitro. Arterioscler Thromb Vasc Biol. 2008;28:335–340. doi: 10.1161/ATVBAHA.107.152058. [DOI] [PubMed] [Google Scholar]
- 65.Merino N., Toledo-Arana A., Vergara-Irigaray M., Valle J., Solano C., Calvo E., et al. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol. 2009;191:832–843. doi: 10.1128/jb.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Viljoen A., Viela F., Mathelié-Guinlet M., Missiakas D., Pietrocola G., Speziale P., et al. Staphylococcus aureus vWF-binding protein triggers a strong interaction between clumping factor A and host vWF. Commun Biology. 2021;4:453. doi: 10.1038/s42003-021-01986-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ko Y.-P., Liang X., Smith C.W., Degen J.L., Höök M. Binding of efb from Staphylococcus aureus to fibrinogen blocks neutrophil adherence. J Biol Chem. 2011;286:9865–9874. doi: 10.1074/jbc.m110.199687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burch WMSR and RL, Burch Rl. The principles of humane experimental technique. vol. 1, 1959, 10.5694/j.1326-5377.1960.tb73127.x. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.