Abstract
Group A streptococci can be classified according to their tendency to cause either impetigo, pharyngitis, or both types of infection. Genotypic markers for tissue site preference lie within emm genes, which encode fibrillar surface proteins that play a key role in virulence. emm gene products (M and M-like proteins) display an extensive array of binding activities for tissue and plasma proteins of the human host. In a previous study, a high-affinity binding site for human plasmin(ogen) was mapped to the emm53 gene product. In this report, a structurally similar plasminogen-binding domain is found to be widely and selectively distributed among group A streptococci harboring the emm gene marker for the skin as the preferred tissue site for infection. The findings are highly suggestive of a central role for bacterial modulation of host plasmin(ogen) during localized infection at the epidermis.
Group A streptococci (GAS) are important human pathogens that can cause severe morbidity and mortality, as in cases of toxic shock syndrome and necrotizing fasciitis and during autoimmune sequelae such as rheumatic fever. However, the vast majority of GAS infections result in only mild disease, specifically pharyngitis and impetigo. The mucosal epithelium of the throat and the epidermal layer of the skin serve as the primary tissue reservoirs for the maintenance of this organism. GAS display a complex array of binding activities for human tissue and plasma components on their cell surfaces. For some host products, such as fibronectin and plasmin(ogen), there exists a multitude of distinct streptococcal structures capable of mediating these interactions. For example, human plasminogen can be bound by at least four distinct streptococcal cell surface proteins that differ in their binding affinities and are differentially expressed (4, 20, 25, 33). The precise reason for this redundancy is not well understood. However, GAS-bound plasminogen can be converted to plasmin—its active form—by secreted streptokinase (12, 27). Cell-bound plasmin functions as a broad-spectrum proteinase and therefore may act to modulate the bacterium’s microenvironment and movement through host tissue. Precisely how each of the distinct plasmin(ogen) binding activities function during streptococcal infection remains unclear.
Many host tissue and plasma proteins are specifically recognized by GAS through structurally discrete binding domains which comprise a major part of surface fibrils that are collectively known as M and M-like proteins. Included among the M-protein-bound host proteins are regulators of the complement cascade, major components of both the coagulation system and the fibrinolytic pathway, and immunoglobulins (reviewed in references 9 and 18). The different binding domain combinations give rise to mosaic-like arrays that impart a unique spectrum of biological activities to the streptococcal cell. The product of the emm gene of M serotype 53 streptococci binds both human plasminogen and plasmin with high affinity (designated PAM, for plasminogen-binding group A streptococcal M protein) (4). The plasmin(ogen) binding site of PAM has been localized to a 13-amino-acid repeated domain (11) (Fig. 1A).
FIG. 1.
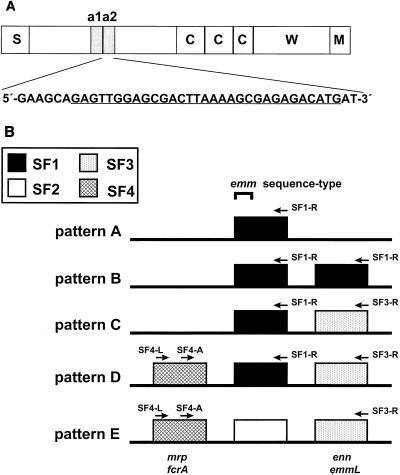
Structural organization of emm genes. (A) Location of the high-affinity plasminogen-binding site and corresponding nucleotide coding sequence within the Emm53 (PAM or M53) protein. Schematic organization of the Emm53 protein: S, signal sequence; C, conserved (C repeat region) repeats; W, cell wall (peptidoglycan)-spanning domain; M, membrane anchor. The a repeats (a1 and a2) are 13 amino acids in length, and each a repeat binds plasminogen with high affinity. The nucleotide sequence of the a2 repeat is given. An oligonucleotide (PAM-F) corresponding to the underlined sequence or its complimentary sequence (PAM-R) was paired with a SF-specific primer for PCR-based genotype mapping. (B) Arrangement of emm SF genes and position of SF-specific oligonucleotide hybridization sites. The emm and emm-like genes are represented by four emm gene SF forms that are based on nucleotide sequence differences in the 3′-end portion encoding for the peptidoglycan-spanning domain (8). Most GAS strains have one of the five emm patterns (designated A through E), which are defined by the number of emm genes, their SF content, and their relative arrangement on the chromosome. For strains with multiple emm or emm-like genes, each gene differs at its 5′ end, and genes are usually separated from one another by 0.2 to 0.3 kb. The centrally positioned gene is used for emm sequence typing (3). Alternative nomenclature for emm and emm-like genes is indicated (bottom). Arrows depict the hybridization sites for oligonucleotide primers. The emm53 gene, depicted in panel A, represents the central emm gene (SF1) of an emm pattern D strain.
The M protein fibrils display extensive antigenic heterogeneity and provide the basis for a serological typing scheme, for which >80 serotypes have been defined. While decades of epidemiological studies have shown that some M serotypes have a strong tendency to be associated with only certain streptococcal diseases, more recent work has identified emm-related genetic markers for the so-called throat and skin types (8). In this report, by taking an epidemiological approach, we demonstrate a selective distribution of the PAM phenotype and genotype among a subpopulation of GAS strains that have a strong tendency to cause impetigo rather than pharyngitis.
MATERIALS AND METHODS
Bacterial strains.
Of the 83 GAS strains used in this study, all but three (AP52, AP53, and Manfredo) represent a broad selection from among those previously described (6, 7). Bacteria were isolated from infected humans between the years 1941 and 1989 at several locations throughout the world: 29 strains from New York; 16 from Trinidad; 7 from Alabama; 6 from the Czech Republic; 3 each from Illinois, Minnesota, Ohio, and Egypt; 1 each from Missouri, North Carolina, Nebraska, Utah, Chile, the former Yugoslavia, Japan, Kuwait, and United Kingdom; and 4 from unknown places. Serotyping was performed by the laboratory providing the strain. The emm sequence type is established based on 160 bp encoding for part of the leader peptide and NH2-terminal end of the mature M protein (Fig. 1B) (2, 3, 16a).
Measurement of the PAM genotype.
Chromosomal DNA purified from each of the streptococcal strains was used as a template in a PCR-based mapping strategy; emm chromosomal patterns A through E (Fig. 1B) were established by using emm subfamily (SF)-specific primers (6–8). To ascertain the presence or absence of the PAM genotype, PAM-specific oligonucleotide primers corresponding to the portion of the emm53 gene that encodes the binding site for human plasminogen (Fig. 1A) (11) were paired with emm SF-specific primers. The PAM primers are PAM-F (forward) (5′-GAGTTG[A/G]AACGACTTAAAA[A/G]CGAGAGACATG-3′) and its complement, PAM-R (reverse); they are degenerate in two positions in order to account for differences between the a1 and a2 repeat regions. For emm pattern A and B strains, PAM-F was paired with SF-specific primer SF1-R (5′-GTGCTTGACCTTTACCTGGAACAGCTT-3′). For emm pattern C strains, PAM-F was paired with SF-specific primers SF1-R and SF3-R (5′-GCTGTTTGAGCAGCTCTACC-3′). In emm pattern D and E strains, PAM-F was paired with SF3-R, whereas PAM-R was paired with SF-specific primer SF4-A (5′-CTCCTAGGTTCAGCTAAGCGTGAGTTG-3′) and/or SF4-L (5′-GAAATCCAAACAAGCACTACCTACTG-3′). For emm pattern D strains, PAM-F was also paired with SF1-R. All primer pairs were used at annealing temperatures of 55°C. Isolates giving PCRs that consistently provided moderate-to-high yields of a DNA fragment of the expected approximate size with one or more PAM-SF primer pair(s) were scored as positive for the PAM genotype.
Binding of radiolabeled plasminogen to streptococci.
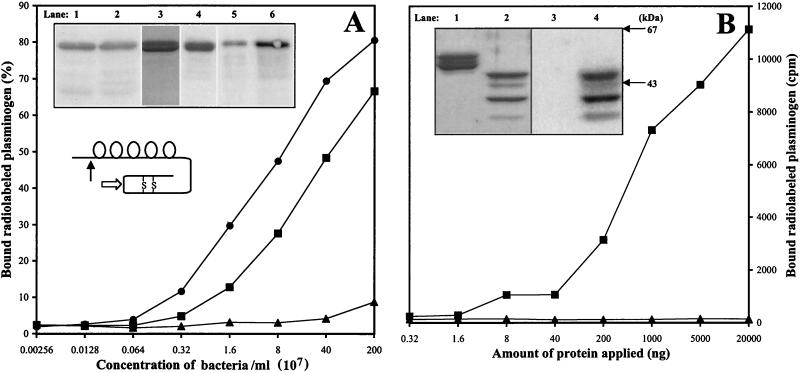
Human plasminogen was purified from human plasma by affinity chromatography using lysine-Sepharose 4B (Pharmacia). Bound material was eluted with 0.1 M glycine (pH 2.0), and fractions containing at least 95% pure plasminogen were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2A, inset). Using a monoclonal antibody (MAb) specific for the NH2-terminal peptide found only in the unprocessed Glu form of the zymogen (MAb n3641; American Diagnostica Inc.), Western blot analysis revealed that the purified preparation was largely the Glu form of plasminogen, and little or no plasmin was present (Fig. 2A, inset). Purified plasminogen was radiolabeled with 125I (Amersham) by using the chloramine-T method to a specific activity of 100,000 cpm/ng. For the whole-bacterial-cell adsorption assay, streptococci were cultured for 16 h on blood agar plates or in Todd-Hewitt broth (Difco) at 37°C with 5% CO2. Bacteria adjusted to the appropriate concentration were incubated with 125I-labeled plasminogen in a total volume of 250 μl of phosphate-buffered saline (PBS) containing 0.02% NaN3 and 0.1% Tween 20. Following incubation for 1 h at 20°C, an additional 2 ml of buffer was added, and the bacteria were centrifuged at 4,000 × g for 10 min. The supernatant was discarded, and radioactivity associated with the pellet was measured in a γ counter; measurements were performed in triplicate. Binding of ≥25% of 125I-labeled plasminogen is indicative of an “M53-like” plasminogen-binding activity (positive PAM phenotype).
FIG. 2.
Binding of Glu plasminogen to select strains and recombinant Emm proteins. (A) Inset, top: Human plasma (0.5 ml) was mixed with approximately 1010 CFUs of GAS (lane 1, strain AP53; lane 2, strain AP52), cells were washed, and bound proteins were eluted with 0.1 M glycine-HCl (pH 2.0). Blots of eluate were incubated with MAb directed to Glu plasminogen. Purified human plasminogen was analyzed by SDS-PAGE and Western blotting. The gel was stained with Coomassie brilliant blue (lane 3), and the blot was incubated with a MAb directed to the NH2-terminal peptide specific for Glu plasminogen (lane 4). Purified, 125I-labeled human plasminogen was analyzed by autoradiography (lane 5). Purified 125I-labeled human plasminogen (0.25 ng) was mixed with approximately 1010 CFUs of strain AP53 in 0.5 ml of PBS containing 2% bovine serum albumin. Following incubation, cells were washed extensively, and bound material was eluted with glycine and subjected to autoradiography (lane 6). (A) Inset, middle: A schematic representation of plasminogen. The five kringle domains and the activation cleavage sites for tissue-type plasminogen activator and urokinase in the serine protease domain (open arrow) are indicated. Also depicted is the cleavage site for plasmin, resulting in a 77-amino-acid residue NH2-terminal peptide and conversion of plasminogen from the Glu to the Lys form (closed arrow). (A) Bottom: Bacteria-bound 125I-labeled plasminogen was measured by an adsorption assay, except that 0.1 ng per 0.25 ml of 125I-labeled plasminogen was incubated with GAS present over a wide range of concentrations, expressed as CFU/ml. Shown are strain 1RP144 (M5, emm pattern A–C; ▴) and emm pattern D strains D617 (■) and AP53 (●) (Table 2). (B) Inset, top: Recombinant emm gene products (rM5, derived from M5 Manfredo, emm pattern A–C, lanes 1 and 3; rEmm52, derived from AP52, lanes 2 and 4) were separated by SDS-PAGE and stained with Coomassie blue (lanes 1 and 2); a replica of the gel was blotted and incubated with 125I-labeled human plasminogen (lanes 3 and 4). (B) Bottom: Purified rM5 (▴) and rEmm52 (■) proteins were immobilized in microtiter plate wells and incubated with 125I-labeled plasminogen. The amount of radio-iodinated material bound to various concentrations of recombinant proteins was measured in a γ counter.
Analysis of plasminogen binding to rEmm proteins.
The cloning and expression of recombinant Emm52 (rEmm52) from strain AP52 and rM5 from strain Manfredo has been previously described (11, 17). Western blot overlay with 125I-labeled plasminogen was used to detect binding activity by rEmm proteins. In addition, rEmm proteins were tested for plasminogen binding in a microtiter assay. Recombinant proteins were immobilized in microtiter plates (4°C for 16 h) over a range of concentrations, and wells were blocked with PBS containing 2% bovine serum albumin for 4 h at 20°C, washed with PBS containing 0.05% Tween 20, and incubated with 0.5 ng of 125I-labeled plasminogen. Following incubation (20°C for 2 h), plates were extensively washed, and bound radioactivity was measured in a γ counter.
Statistics.
Statistical significance was calculated by χ2 analysis with Yates’ correction for sample size (Epi Info version 6.04b; Centers for Disease Control and Prevention, Atlanta, Ga.).
RESULTS
Distribution of the PAM genotype among GAS.
The mapping of a plasminogen-binding motif to within the surface-exposed portion of M or M-like proteins (11) suggested that its distribution among members of the GAS species may be restricted. To ascertain whether PAM is tightly linked to genetic markers for principal tissue reservoir (i.e., emm patterns), PAM-specific oligonucleotides were designed for PCR-based mapping of the emm chromosomal region of 83 emm pattern-defined GAS strains (Fig. 1A and 1B). The GAS isolates under study represent all five of the emm patterns (A through E) and >40 distinct M serotypes or emm sequence types (Table 1). The wide range of M and emm types, combined with large spatial and temporal distances separating their isolation from human hosts, indicates that the GAS selected for this study are a biologically diverse sample set.
TABLE 1.
Epidemiologic features of group A streptococcal strains under study
| emm pattern | No. of strains represented | No. of unique M or emm types | No. (%) isolated from:
|
||
|---|---|---|---|---|---|
| Nasopharynx | Impetigo | Unknown site | |||
| A, B, or C | 35 | 15 | 32 (91) | 2 (6) | 1 (3) |
| D | 28 | 17 | 5 (18) | 19 (68) | 4 (14) |
| E | 20 | 13 | 10 (50) | 10 (50) | 0 (0) |
Previous studies demonstrate that the majority of emm pattern A, B, and C (A–C) strains are oropharynx associated, whereas most pattern D strains are isolated from impetigo lesions; pattern E strains are readily found at both tissue sites (6, 8). Thus, emm patterns A–C and D can be regarded as genetic markers for the throat and skin, respectively, as the principal tissue reservoir in the human host. Of the 35 emm pattern A–C strains examined, representing 15 distinct M- or emm-sequence types, only four displayed the PAM genotype (11.4%; Fig. 3). Among the 20 emm pattern E isolates, representing 13 distinct M- or emm-sequence types, all tested negative for PCR-based detection of the PAM genotype.
FIG. 3.
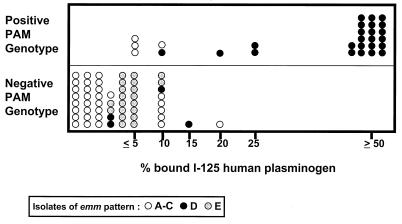
Binding of human plasminogen by intact GAS. Each symbol represents a different GAS isolate (n = 83) that is defined according to its emm pattern. Intact GAS were measured for the percentage of 125I-labeled human plasminogen bound in an adsorption assay. Approximately 2.0 × 109 CFUs were incubated with 125I-labeled plasminogen (∼10,000 cpm, corresponding to ∼1 ng of plasminogen) in a total reaction volume of 0.25 ml. The values for the percentage of plasminogen bound are rounded to the nearest 5% interval. Also, shown is the PAM genotype for each isolate, as determined by PCR amplification of GAS-derived DNA using PAM-specific oligonucleotide primers.
In sharp contrast to patterns A–C and E organisms, 24 of the 28 emm pattern D isolates tested positive with the PAM-specific oligonucleotides (85.7%; Fig. 3). Based on the generation of DNA products from multiple PCR amplifications, the PAM-specific priming site mapped to the central emm gene at a position that is upstream from the SF1 site (Fig. 1B). The difference between pattern D strains and either pattern A–C or E isolates, in terms of the PAM genotype, is highly significant (P < 0.001, df = 1). Of the 17 unique emm sequence types represented among the 28 pattern D strains, the positive PAM genotype was found among 13 emm types (76.5%), whereas the negative PAM genotype was limited to four emm types (23.5%) (Table 2). For emm types where multiple isolates were tested (emm33, emm52, emm53, emmpotter41, emmpt2110, and emmstD633), all were homogeneous in their display of the PAM genotype. The results indicate that the PAM genotype is largely restricted to emm pattern D strains.
TABLE 2.
Properties associated with plasminogen-binding activity among emm pattern D strains
| Strain | emm typea | Tissue site of isolationb | Isolation date | Placeb | PCR product (PAM primers) | % of bound 125I-labeled plasminogen |
|---|---|---|---|---|---|---|
| 13RS60 | 33 | NP | 1941 | New York | + | ≥50 |
| A910 | 33 | NP | 1966 | ND | + | ≥50 |
| A982 | 33 | Imp | 1968 | New York | + | ≥50 |
| 3732-S | 33 | Imp | 1969 | Alabama | + | ≥50 |
| 6010-S | 33 | Imp | 1971 | Alabama | + | ≥50 |
| D735 | 33 | Imp | 1973 | New York | + | ≥50 |
| 29487 | 33 | Imp | 1987 | Czech Republic | + | ≥50 |
| AP52c | 52 | Imp | 1967 | Minnesota | + | ≥50 |
| D617 | 52 | Imp | 1972 | Trinidad | + | ≥50 |
| AP53c | 53 | Imp | 1967 | Minnesota | + | ≥50 |
| ALAB49 | 53 | Imp | 1986 | Alabama | + | ≥50 |
| 29689 | pt2110 | Imp | 1988 | Czech Republic | + | ≥50 |
| ALAB55 | pt2110 | Imp | 1986 | Alabama | + | 25 |
| D964 | pt2631 | Imp | 1976 | Trinidad | + | ≥50 |
| D821 | pt5757 | Imp | 1975 | Trinidad | + | ≥50 |
| 29486 | st2370 | Imp | 1987 | Czech Republic | + | ≥50 |
| D432 | stD432 | ND | 1971 | Egypt | + | ≥50 |
| D631 | stD631 | Imp | 1972 | Trinidad | + | ≥50 |
| D633 | stD633 | Imp | 1972 | Trinidad | + | ≥50 |
| D680 | stD633 | NP | 1972 | Trinidad | + | ≥50 |
| D641 | stNS5 | Imp | 1972 | Trinidad | + | ≥50 |
| D466 | potter41 | ND | 1971 | Egypt | + | 25 |
| D502 | potter41 | Imp | 1971 | Trinidad | + | 10 |
| D998 | 70 | ND | 1976 | Japan | + | 20 |
| 10RS101 | 32 | NP | 1942 | New York | − | 15 |
| 1RS79 | 42 | NP | 1941 | New York | − | 10 |
| A457 | 36 | ND | 1961 | Former Yugoslavia | − | ≤5 |
| D626 | st88/31 | Imp | 1972 | Trinidad | − | ≤5 |
Binding activity for plasminogen among GAS.
To establish the association between the PAM genotype and the plasminogen-binding phenotype, GAS were analyzed for their capacity to bind human plasminogen. A detailed analysis of plasminogen binding by a few select strains is summarized in Fig. 2. Using a human plasminogen source of either purified 125I-labeled plasminogen or unfractionated plasma, two emm pattern D strains (AP52 and AP53) were found to bind specifically to the unprocessed Glu form of the zymogen (Fig. 2A, inset). At concentrations of bacteria up to 2 × 109 CFU/ml, >50% of radiolabeled plasminogen was bound by two emm pattern D strains (D617 and AP53), whereas <10% of total plasminogen was bound by the emm pattern A–C strain (1RP144) (panel A). To demonstrate that the emm gene product represents the GAS component responsible for the observed binding of plasminogen, recombinant M proteins derived from two GAS strains were analyzed in direct binding experiments. As expected, the rEmm52 protein derived from strain AP52 bound Glu plasminogen efficiently, whereas the rM5 protein from emm pattern A–C strain Manfredo showed little, if any, affinity for the zymogen (Fig. 2B).
Using the whole-bacterial-cell adsorption assay, binding of human plasminogen was measured for all 83 GAS strains under study, at a single near-saturating concentration of bacteria (approximately 8 × 109 CFU/ml). Of the emm pattern D strains, 22 of 28 (78.6%) displayed high levels of plasminogen binding (ranging from 25 to >50% of total 125I-labeled plasminogen bound; Fig. 3). In contrast, the vast majority (83.6%) of emm patterns A–C and E strains bound ≤5% of 125I-labeled plasminogen; binding of >20% plasminogen was not observed for any of the patterns A–C and E strains tested.
Correlations of PAM phenotype, PAM genotype, and tissue site of isolation.
All 22 isolates binding ≥25% of human plasminogen in the whole-bacterial-cell adsorption assay also displayed the PAM genotype that is detected by PCR using PAM-specific oligonucleotide primers. Of the 22 strains that scored positive for plasminogen binding (i.e., those that had a positive PAM phenotype), 100% were emm pattern D (Fig. 3). Thus, the PAM phenotype positively correlates with the PAM genotype and furthermore, it is restricted to emm pattern D strains.
Of the 83 GAS isolates studied, 22 were positive for both the PAM phenotype and genotype, however, an additional six strains were positive for the PAM genotype only. Thus, six isolates hybridized with PAM oligonucleotides yet failed to exhibit strong plasminogen-binding activity. Of the four pattern A–C isolates that were positive for the PAM genotype but negative for the PAM phenotype (strains 87-214 [emm1], 1RP144 [emm5], 1GL217 [emm17], and 87-373 [emm18]), all were represented by emm types found among other strains that were clearly negative for the PAM genotype. Thus, it is possible that under the annealing conditions employed, the PAM-specific primers can hybridize to non-PAM-specific sequences whose distribution does not correlate well with emm type. For the two pattern D strains that were negative for the PAM phenotype (Table 2) but positive for the PAM genotype (strains D998 [emm70] and D502 [emmpotter41]), one displayed an emm type shared with a second isolate that is positive for the PAM phenotype (strain D466 [emmpotter41]). Since M and emm types are tightly linked to the 3′ end SF-specific regions that define emm pattern (Fig. 1B) (6), it seems likely that for strain D502, the PAM-specific oligonucleotides detected an emm gene whose product is not expressed at high levels or, alternatively, harbors a point mutation(s) that leads to a decrease in binding capacity.
The actual tissue site of isolation is known for 78 of the 83 GAS under study: 31 were isolated from impetigo lesions, whereas 47 were obtained from the nasopharyngeal mucosa and gave rise to either pharyngitis, clinically inapparent infections, or asymptomatic carriage. Of the 2 pattern A–C and 10 pattern E isolates derived from impetigo lesions, representing a total of nine distinct emm types, none displayed the PAM phenotype or genotype. In contrast, of the 18 pattern D impetigo isolates, representing 12 emm types, 16 scored positive for both the PAM phenotype and genotype, 1 had the PAM genotype only, and 1 was negative for both (Table 2). Of the five nasopharynx-derived pattern D organisms, three were positive for PAM (13RS60 [emm33], D680 [emmstD633], and A910 [emm33]), and two were negative for PAM (10RS101 [emm32] and 1RS79 [emm42]). Of the pattern D throat isolates, at least three (13RS60, D680, and 10RS101) were not known to be associated with clinical disease in their human hosts and most probably gave rise to an asymptomatic carrier state.
Of the 21 isolates associated with cases of acute rheumatic fever, four strains displayed the PAM genotype but bound ≤10% of human plasminogen in the whole-bacterial-cell adsorption assay (all four are emm pattern A–C strains). Thus, rheumatic fever-associated strains tend to be deficient in PAM-mediated plasminogen binding activity. Of the seven GAS isolated from individuals with acute glomerulonephritis, three displayed both the PAM phenotype and genotype (all emm pattern D strains), whereas the remainder lacked both the PAM phenotype and genotype (emm pattern A–C and E strains).
Taken together, the data show a strong association between the PAM phenotype, PAM genotype and emm pattern D strains. All PAM-positive organisms segregate as a group with bacteria having a strong tendency to cause impetigo lesions. Furthermore, emm pattern E isolates and the occasional pattern A–C isolate that are obtained from impetigo lesions, are consistent in their lack of both the PAM phenotype and genotype, suggesting that alternative pathogenic mechanism(s) are used by these GAS subpopulations.
DISCUSSION
A recurring theme in the study of GAS is the existence of multiple gene products which bind various human tissue and plasma components. To better understand the biological role of bacterial-host binding activities during natural infection, one can test a hypothesis by comparing organisms that are genotypically and phenotypically well defined, using in vitro models consisting of human-derived components or alternatively, in vivo models that rely on animals. However, often the binding affinities are significantly higher for human components than for other mammalian forms, casting uncertainty on the true relevance of animal models for a disease that is uniquely human. For example, the molecule studied here (PAM) does not bind well to plasminogen of closely related species, such as the rhesus monkey, a nonhuman primate (10). An epidemiological approach provides a third general strategy and works by sorting a population of microorganisms according to their genotype, phenotype, and biological interactions with its natural host. Together, the three approaches can merge to form a more complete composite of the mechanisms underlying disease pathogenesis.
In the present investigation, we use a combined approach to further analyze the role of the GAS interaction with plasminogen, a zymogen found in high concentrations in plasma and tissue fluids. Cleavage of plasminogen can set off any one of several cascades of events that are part of important biological processes, such as dissolution of fibrin, wound healing, inflammatory cell migration, tumor cell metastasis, and the inhibition of angiogenesis (16, 28). Multiple pieces of evidence suggest that for some of these events, activation of plasminogen at the mammalian cell surface is required (14, 22, 24, 32). Therefore, the biological consequences of bacterial cell surface-bound plasmin activity are potentially quite numerous. GAS express surface proteins that allow for the capture of human plasmin(ogen) by at least four different mechanisms (4, 20, 25, 33). Furthermore, GAS produce at least two secreted proteins—streptokinase and cysteine protease—which can interact with plasmin(ogen) in a specific manner (26, 34). This report focuses on one of the plasminogen binding mechanisms exhibited by GAS, the high-affinity interaction that is mediated through PAM.
Recent evidence strongly suggests that surface-associated binding and activation of plasminogen enhances the invasion and dissemination of at least two bacterial pathogens, Yersinia pestis and Borrelia burgdorferi (13, 29). Invasion of normally sterile tissue by GAS is associated with high rates of morbidity and mortality; however, from an ecological standpoint, GAS invasive disease is a rare event. In contrast to the vector-borne pathogens Y. pestis and B. burgdorferi, which are transmitted to their mammalian hosts through insect bites, GAS are transmitted primarily by respiratory droplets or close contact. GAS colonize the mucosal epithelium of the upper respiratory tract or epidermis of the skin and, most often, cause only mild superficial infection. The strong epidemiological association demonstrated in this report narrows the focus for the most likely biological role of the PAM form of plasminogen-binding activity by GAS. Since emm pattern D strains display a strong tendency to cause impetigo and are much less often observed in association with pharyngitis (8), it seems most likely that PAM exerts its biological function within the epidermal tissue space.
The epidemiological associations uncovered in this study lead us to favor the hypothesis that PAM-directed plasmin(ogen) binding by GAS exerts its strongest biological effect during localized infection rather than during invasive disease. Population-based surveillance for invasive GAS disease during a 6-month time frame in Connecticut demonstrates that <2% of the isolates display emm pattern D (15). This finding is in further support of the idea that the key role of PAM is not linked to invasion of deep tissue. The concentration of plasminogen in tissue is highest in the circulation and interstitial fluids (1 × 10−6 to 2 × 10−6 M). However, the high affinity of PAM for human plasminogen (affinity constant, 8 × 10−8 M−1) (4) suggests that PAM has undergone adaptive evolution in order to function ideally under conditions whereby the concentration of plasminogen is low.
During the development of an impetigo vesicle, GAS are largely confined to the surface of the outermost layer of granular keratinocytes, located just below the cornified layer (stratum corneum) of the epidermis (1). As a continuing stream of neutrophils migrate from the dermal vessels through the differentiated keratinocyte layers of the epidermis and into the subcorneal space where adherent GAS persist, a mild spongiosis (intraepidermal intercellular edema) often develops and extracellular tissue fluid gradually accumulates within the epidermis. The vesicle itself contains coagulated serum and neutrophils in addition to bacteria. As the infection progresses and the vesicle ruptures, the stratum corneum is no longer present and the remaining epidermal layer is covered by a crust composed of fibrin and cellular debris. An expanding inflammatory response can eventually lead to a deeper erosion of the epidermal layer or penetration of the dermis as observed in ecthyma (19, 23, 30).
In a normal response to damage at the dermal-epidermal junctional region, basal layer keratinocytes located at the margins of the “wound” will migrate laterally along a provisional matrix of fibrin and initiate the process of reepithelialization (21, 28). The migration of keratinocytes during wound healing, and possibly inflammatory cells responding to a chemotactic signal, is directed by pericellular proteolysis mediated by plasmin (16, 21). Conceivably, PAM acts to sequester plasmin(ogen) and thereby competes with the human cell receptors for its binding and, in doing so, circumvents the normal healing processes. Alternatively, plasminogen captured by PAM and subsequently activated by streptokinase might act directly on the fibrin network of either the overlying crust and/or dermal-epidermal junction, thereby allowing the bacteria to prolong their presence in superficial wounds. Finally, bacterial-surface-generated plasmin might function as a broad spectrum protease and modulate the bacterium’s microenvironment in an undefined manner that ultimately favors its reproduction and transmission to a new host. However, the failure of PAM to exhibit high-affinity binding for plasminogen of nonhuman origin may preclude the possibility of discerning among these possibilities by traditional in vivo models for infection.
ACKNOWLEDGMENTS
We thank Dominick Scaramuzzino and Jennifer McNiff for their feedback on the manuscript.
This work was supported by grants from the American Heart Association, the National Institutes of Health (AI-28944); the Swedish Medical Research Council (9926); and the Ax:son Johnson, Bergvall, Crafoord, Kock, and Österlund Foundations. M.D.S. is the recipient of a graduate scholarship from the Foundation for Strategic Research (Infection and Vaccinology program). D.E.B. is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Barnhill R L, editor. Textbook of dermatopathology. New York, N.Y: McGraw-Hill; 1998. [Google Scholar]
- 2.Beall B, Facklam R, Elliott J, Franklin A, Hoenes T, Jackson D, Laclaire L, Thompson T, Viswanathan R. Streptococcal emm types associated with T-agglutination types and the use of conserved emm gene restriction fragment patterns for subtyping group A streptococci. J Med Microbiol. 1998;47:1–6. doi: 10.1099/00222615-47-10-893. [DOI] [PubMed] [Google Scholar]
- 3.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge A, Sjobring U. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J Biol Chem. 1993;268:25417–25424. [PubMed] [Google Scholar]
- 5.Bessen D, Jones K F, Fischetti V A. Evidence for two distinct classes of streptococcal M protein and their relationship to rheumatic fever. J Exp Med. 1989;169:269–283. doi: 10.1084/jem.169.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessen D E, Izzo M W, Fiorentino T R, Caringal R M, Hollingshead S K, Beall B. Genetic linkage of exotoxin alleles and emm gene markers for tissue tropism in group A streptococci. J Infect Dis. 1999;179:627–636. doi: 10.1086/314631. [DOI] [PubMed] [Google Scholar]
- 7.Bessen D E, Izzo M W, McCabe E J, Sotir C M. Two-domain motif for IgG-binding activity by group A streptococcal emm gene products. Gene. 1997;196:75–82. doi: 10.1016/s0378-1119(97)00201-1. [DOI] [PubMed] [Google Scholar]
- 8.Bessen D E, Sotir C M, Readdy T L, Hollingshead S K. Genetic correlates of throat and skin isolates of group A streptococci. J Infect Dis. 1996;173:896–900. doi: 10.1093/infdis/173.4.896. [DOI] [PubMed] [Google Scholar]
- 9.Boyle M D P. Variation of multifunctional surface binding proteins: a virulence strategy for group A streptococci. J Theor Biol. 1995;173:415–426. doi: 10.1006/jtbi.1995.0073. [DOI] [PubMed] [Google Scholar]
- 10.Carlsson Wistedt A, Kotarsky H, Marti D, Ringdahl U, Castellino F J, Schaller J, Sjobring U. Kringle 2 mediates high affinity binding of plasminogen to an internal sequence in streptococcal surface protein PAM. J Biol Chem. 1998;273:24420–24424. doi: 10.1074/jbc.273.38.24420. [DOI] [PubMed] [Google Scholar]
- 11.Carlsson Wistedt A C, Ringdahl U, Muller-Esterl W, Sjobring U. Identification of a plasminogen-binding motif in PAM, a bacterial surface protein. Mol Microbiol. 1995;18:569–578. doi: 10.1111/j.1365-2958.1995.mmi_18030569.x. [DOI] [PubMed] [Google Scholar]
- 12.Christner R, Li Z, Raeder R, Podbielski A, Boyle M. Identification of key gene products required for acquisition of plasmin-like enzymatic activity by group A streptococci. J Infect Dis. 1997;175:1115–1120. doi: 10.1086/516450. [DOI] [PubMed] [Google Scholar]
- 13.Coleman J, Gebbia J, Piesman J, Degen J, Bugge T, Benach J. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell. 1997;89:1111–1119. doi: 10.1016/s0092-8674(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 14.Edwards D, Murphy G. Proteases: invasion and more. Nature. 1998;394:527–528. doi: 10.1038/28961. [DOI] [PubMed] [Google Scholar]
- 15.Fiorentino T R, Beall B, Mshar P, Bessen D E. A genetic-based evaluation of principal tissue reservoir for group A streptococci isolated from normally sterile sites. J Infect Dis. 1997;176:177–182. doi: 10.1086/514020. [DOI] [PubMed] [Google Scholar]
- 16.Heiple J M, Ossowski L. Human neutrophil plasminogen activator is localized in specific granules and is translocated to the cell surface by exocytosis. J Exp Med. 1986;164:826–840. doi: 10.1084/jem.164.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Infotech in the Biotechnology Core Facility Branch Website. 17 May 1999, revision date. [Online.] National Center for Infectious Diseases, Centers for Disease Control and Prevention. http://www.cdc.gov/ncidod/biotech/infotech_hp.html. [16 June 1999, last date accessed.]
- 17.Johnsson E, Thern A, Dahlback B, Heden L O, Wikstrom M, Lindahl G. A highly variable region in members of the streptococcal M protein family binds the human complement regulator C4BP. J Immunol. 1996;157:3021–3029. [PubMed] [Google Scholar]
- 18.Kehoe M A. Cell wall-associated proteins in Gram-positive bacteria. New Compr Biochem. 1995;27:217–261. [Google Scholar]
- 19.Lever W, Schaumberg-Lever G. Histopathology of the skin. 7th ed. Philadelphia, Pa: J. B. Lippincott Co.; 1990. [Google Scholar]
- 20.Lottenberg R, Broder C C, Boyle M D P, Kain S J, Schroeder B L, Curtiss R I. Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. J Bacteriol. 1992;174:5204–5210. doi: 10.1128/jb.174.16.5204-5210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin P. Wound healing: aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 22.Mignatti P, Robbins E, Rifkin D. Tumor invasion through the human amniotic membrane: requirement for a protein cascade. Cell. 1986;47:487–498. doi: 10.1016/0092-8674(86)90613-6. [DOI] [PubMed] [Google Scholar]
- 23.Montgomery H. Dermatopathology. New York, N.Y: Harper & Row; 1967. [Google Scholar]
- 24.O’Reilly M, Holmgren L, Shing Y, Chen C, Rosenthal R, Moses M, Lane W, Cao Y, Sage E, Folkman J. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 25.Pancholi V, Fischetti V A. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273:14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 26.Poon-King R, Bannan J, Viteri A, Cu G, Zabriskie J. Identification of an extracellular plasmin binding protein from nephritogenic streptococci. J Exp Med. 1993;178:759–763. doi: 10.1084/jem.178.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ringdahl U, Svensson M, Wistedt A, Renné T, Kellner R, Müller Esterl W, Sjöbring U. Molecular co-operation between protein PAM and streptokinase for plasmin acquisition by Streptococcus pyogenes. J Biol Chem. 1998;273:6424–6430. doi: 10.1074/jbc.273.11.6424. [DOI] [PubMed] [Google Scholar]
- 28.Romer J, Bugge T H, Pyke C, Lund L R, Flick M J, Degen J L, Dano K. Impaired wound healing in mice with a disrupted plasminogen gene. Nat Med. 1996;2:287–292. doi: 10.1038/nm0396-287. [DOI] [PubMed] [Google Scholar]
- 29.Sodeinde O, Subrahmanyam Y, Stark K, Quan T, Bao Y, Goguen J. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 30.Swartz M. Cellulitis and subcutaneous tissue infections. In: Mandell G, Bennett J, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1995. pp. 909–929. [Google Scholar]
- 31.Top F H, Wannamaker L W, Maxted W R, Anthony B F. M antigens among group A streptococci isolated from skin lesions. J Exp Med. 1967;126:667–685. doi: 10.1084/jem.126.4.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassalli J-D, Sappino A-P, Belin D. The plasminogen activator/plasmin system. J Clin Investig. 1991;88:1067–1072. doi: 10.1172/JCI115405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Lottenberg R, Boyle M D. A role for fibrinogen in the streptokinase-dependent acquisition of plasmin(ogen) by group A streptococci. J Infect Dis. 1995;171:85–92. doi: 10.1093/infdis/171.1.85. [DOI] [PubMed] [Google Scholar]
- 34.Young K, Shi G, Wu D, Chang L, Chang B, Ou C, Wu H. Plasminogen activation by streptokinase by a unique mechanism. J Biol Chem. 1998;173:3110–3116. doi: 10.1074/jbc.273.5.3110. [DOI] [PubMed] [Google Scholar]