Summary
Archaeological research shows that the dispersal of the Neolithic took a more complex turn when reaching western Europe, painting a contrasted picture of interactions between autochthonous hunter-gatherers (HGs) and incoming farmers. In order to clarify the mode, the intensity, and the regional variability of biological exchanges implied in these processes, we report new palaeogenomic data from Occitanie, a key region in Southern France. Genomic data from 28 individuals originating from six sites spanning from c. 5,500 to c. 2,500 BCE allow us to characterize regional patterns of ancestries throughout the Neolithic period. Results highlight major differences between the Mediterranean and Continental Neolithic expansion routes regarding both migration and interaction processes. High proportions of HG ancestry in both Early and Late Neolithic groups in Southern France support multiple pulses of inter-group gene flow throughout time and space and confirm the need for regional studies to address the complexity of the processes involved.
Subject areas: Biological sciences, Paleobiology, Paleogenetics
Graphical abstract
Highlights
-
•
Genome-wide data from 28 individuals from Southern France (∼5,500–∼2,500 BCE)
-
•
Small groups associated with the Neolithic expansion along the Mediterranean
-
•
Early admixture between hunter-gatherers and Neolithic farmers in Southern France
-
•
Multiple pulses of HG legacy introgression in Western Europe throughout Neolithic
Biological sciences; Paleobiology; Paleogenetics
Introduction
Originating from the Middle East around 10,000 BCE, the Neolithic lifestyle has been shown to spread westward across Europe mainly during the seventh millennium, via two main routes: the Continental route, through the Struma and Vardar, and ultimately the Danube valleys and the Mediterranean route, through the Adriatic Region (Guilaine, 2001, 2003). Recent ancient DNA (aDNA) research significantly enriched our knowledge of the demographic processes taking place in Central and Western Europe following the arrival of the first farming communities and the subtle geographic structure between Neolithic groups (Brace et al., 2019; Brunel et al., 2020; Gamba et al., 2014; Haak et al., 2015; Hofmanová et al., 2016; Lipson et al., 2017; Mathieson et al., 2015, 2018; Skoglund et al., 2014; Rivollat et al., 2020; Valdiosera et al., 2018; ). Whereas the data gathered so far is unevenly distributed across Europe, genetic signals point to a differentiation between individuals from different regions and associated with different Neolithic expansion routes. These notably indicate diverse biological interaction processes between hunter-gatherer (HG) and farmer communities, both in terms of the number and intensity of gene flow events. Early farmer groups associated with the continental stream of expansion (Linearbandkeramik (LBK) and precursors like the Transdanubian and Alföld LBK, Starčevo, Körös, Criş cultures) are genetically well represented (N = 172), and the data at hand highlight very little initial gene flow during their expansion from southeastern to Central Europe, even though Neolithic settlers coexisted and exchanged material with neighboring HG (Haak et al., 2015; Lipson et al., 2017; Mathieson et al., 2018). The data from Mediterranean farmer communities related to the Impressed Wares or Impresso-Cardial cultural complex is smaller (ICC, N = 41), but the available Neolithic genomes from modern-day Italy, France, and Iberia have nonetheless displayed variable degrees of admixture between incoming farmers and local HG (Antonio et al., 2019; Rivollat et al., 2020; Olalde et al., 2019; Valdiosera et al., 2018; Villalba-Mouco et al., 2019). Indeed, despite low but noticeable levels of admixture between HG and farmer communities along the central Mediterranean, early farmers from Pendimoun and Les Bréguières in Southeastern France, dated to the second half of the 6th millennium BCE, showed unprecedentedly high proportions of HG ancestry (up to 56% in one Pendimoun individual; Rivollat et al., 2020). These observations demonstrated that admixture took place locally within only a few generations after the arrival of farming in this region, marking it as a contact zone between HG and farmer communities, and highlighting the need for more fine-grained regional genomic studies.
By 4,000 BCE, the agro-pastoral way of life was deeply anchored in human societies and had reached the far ends of Europe (Brace et al., 2019; Mittnik et al., 2018; Nordqvist and Kriiska, 2015). Whereas macro-regional genomic studies highlighted only minimal admixture during Early Neolithic (EN) between farmers and local HG in Central and Western Europe, HG ancestry increased over time in all farming communities as the Neolithic period progressed (Brunel et al., 2020; Haak et al., 2015; Lipson et al., 2017; Marcus et al., 2020; Papac et al., 2021; Olalde et al., 2019; Valdiosera et al., 2018).
The period between ∼3,500 and 2,000 BCE, corresponding to Late Neolithic (LN) and the transition into the Bronze Age in Western Europe, has been the focus of recent studies highlighting the demographic processes taking place with regard to the arrival of the Steppe-related ancestry (Haak et al., 2015; Furtwängler et al., 2020; Immel et al., 2021; Lösch et al., 2020; Olalde et al., 2019). For France, genomic data documenting LN groups’ ancestry is still scarce (Brunel et al., 2020; Seguin-Orlando et al., 2021). Recent studies provided unexpected insights into the HG legacy in LN farmer groups from Western Europe. Genetically heterogeneous communities with highly variable amounts of HG ancestry per individual were found at Mont-Aimé in the Paris Basin (France; Seguin-Orlando et al., 2021) and Niedertiefenbach in Hesse (Germany; Immel et al., 2021).
The pronounced heterogeneity of the HG legacy in Neolithic farmer communities clearly necessitates further time transects with a regional focus. Southern France is a key region for understanding the complex relationship between biological and cultural HG-farmer interactions. The rich archaeological record of Southern France indeed suggests various human groups entangled in a complex interplay of contact and exchange of elements attributed to both HG and Neolithic subsistence strategies (Binder, 2000; Guilaine and Manen, 2007; Perrin and Manen, 2021). We consequently analyzed genome-wide data recovered for 28 individuals from Occitanie, an archaeologically well-defined but still poorly genetically documented region that links the Iberian Peninsula and Southeastern France. The human remains originate from six sites, spanning from c. 5,500 to c. 2,500 BCE. We therefore aimed to assess a) whether the cultural variation was also reflected in the genetic heterogeneity, to discuss the connection between gene pools evolution and cultural transformations during the Neolithic, and b) whether sites like Pendimoun and Les Bréguières were localized exceptions or representing a wider regional phenomenon of intensified HG-farmer interactions in key regions of the Western Mediterranean area.
Results and discussion
Human remains and archaeological background
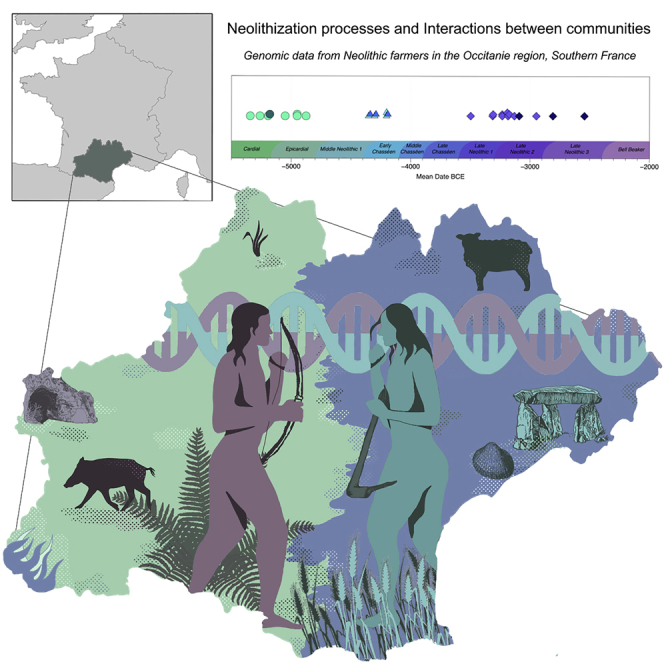
A total of 76 human remains were submitted to shallow shotgun sequencing to screen for the preservation of ancient human DNA molecules (see STAR Methods). All samples originate from six Neolithic archaeological sites located in Occitanie, Southern France (Figures 1C and S1): Baume Bourbon (BBB), Gazel (Gazel4), Le Crès (CRE), Champ du Poste (SP), the Dolmen des Fades (FAD) and the Aven de la Boucle (BOU).
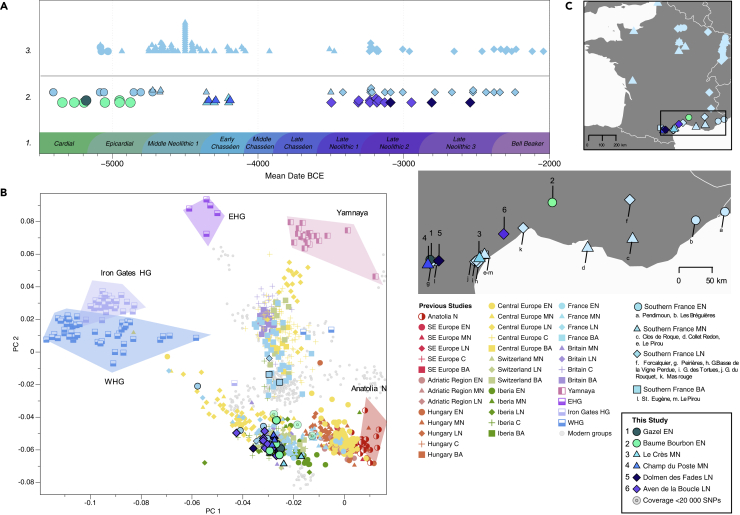
Figure 1.
Temporal and geographic distribution of the individuals included in this study
(A) Chronological timeline presenting (1) cultural chronology of Southern France, (2) mean radiocarbon dates obtained for the 23 samples in this study (lower) and for previously published samples from Southern France (upper), and (3) radiocarbon and context dates for previously published samples from the rest of France.
(B) Relevant published and 28 newly reported ancient individuals projected onto a principal component analysis (PCA) of 796 present-day West Eurasians (Lazaridis et al., 2017; Patterson et al., 2012). Individuals from this study presenting less than 20,000 SNPs are shown with a distinct symbol as the position of low-coverage genomes on the PCA must be discussed with caution (Günther and Jakobsson, 2019).
(C) Geographical distribution of archaeological sites and samples in today’s France with zoom in on Southern France.
DNA libraries from human remains presenting endogenous human DNA ranging between 5.4 and 72% were selected for whole-genome shotgun sequencing to obtain a total of 21 partial genomes with coverage between 0.078X and 0.68X (Tables S1 and S2). For individuals from the Baume Bourbon cave, who yielded lower endogenous DNA content (0.6–22.8%, with a median of 0.992%, Table S2), next-generation libraries were enriched for ∼1.2 million single nucleotide polymorphisms (SNPs) by using targeted in-solution capture (1240k SNP capture; Mathieson et al., 2018) at the Max Planck Institute for the Science of Human History in Jena, Germany. For this particular site, which provided some of the oldest Neolithic human remains in the region, the use of capture allowed us to obtain usable genomic data for a maximum number of individuals and to test the biological relatedness between the deceased in a cost-effective way.
Applying quality thresholds, we removed samples with >∼5% of contamination on the X chromosome (applicable on males) or coverage of fewer than 19,000 SNPs in the 1240k dataset (Mathieson et al., 2015). We also controlled contamination using mtDNA data (Table S3, STAR Methods) and the presence of DNA damage patterns consistent with ancient origin. As a result, 28 samples were kept for downstream analyses (Tables S1 and S3). To examine the genetic diversity, we co-analyzed all 28 newly typed genome-wide data with previously reported data from 1098 ancient individuals and the genotypes of 59 modern-day human populations from West Eurasia by conducting principal component analysis (PCA; on a reduced set of overlapping variants – Human origins [HO] dataset; Lazaridis et al., 2017; Patterson et al., 2012 Table S8). We explored the genetic affinities and legacies of ancient individuals using f-statistics, qpAdm, and DATES (Chintalapati et al., 2022; Harney et al., 2021; Narasimhan et al., 2019; Patterson et al., 2012; ). Using HapROH, we assessed the level of inbreeding and/or past limitations of the effective population’s size of newly reported and published ancient individuals (Ringbauer et al., 2021; Tables S8, S10, S11, S12, S13, S14, S15, S16, and S17; STAR Methods, ADMIXTOOLS). We also report direct 14C dates for 23 individuals, with dates ranging from 5,462 BCE to 2,470 BCE (Figure 1B, Tables S1 and S9).
Genome-wide data were recovered from eight individuals originating from two sepulchral contexts attributed to the second stage of the ICC complex: the Baume Bourbon cave (Cabrières, Gard; N = 7) and the contemporary Gazel cave (Sallèles-Cabardès, Aude; N = 1), with dates ranging between ∼5,400 and 4,800 BCE (Beyneix, 1997; Coste et al., 1987; Duday and Guilaine, 1980; Zemour, 2013). Direct dates obtained for these sites confirm their cultural attribution to Cardial-Epicardial cultures, which were followed by several centuries of the pioneer Neolithic settlements (∼5,850 BCE) in the Mediterranean Languedoc (Binder et al., 2018). Seven individuals come from two necropolises dated to the second half of the 5th millennium BCE: Champ du Poste (Convertini and Georjon, 2018; Carcassonne, Aude; N = 3) and Le Crès (Loison and Schmitt, 2009; Béziers, Hérault; N = 4). The respective 14C dates indicate that both necropolises were used over a few centuries and included individuals with dates between ∼4,500 and 4,050 BCE, attributed to the early Southern Chasséen culture (Ambert et al., 1989; Brunel et al., 2020; Loison and Schmitt, 2009; Vaquer, 1990, 1991, 1998) (STAR Methods). The remaining 13 low-coverage genomes come from individuals recovered in two collective burials used during LN and Bronze Age: the Aven de la Boucle (Duday, 1987; Jallet et al., 2010; Corconne, Gard; N = 10) and the Dolmen des Fades (Bonnery, 1991; Guilaine, 1998; Pépieux, Aude; N = 3). Individuals’ dates range between ∼3,600 and 2,800 BCE for the Aven de la Boucle and between ∼3,200 and 2,400 BCE for the Dolmen des Fades (Figure 1B, Tables S1 and S9, STAR Methods).
Biological interactions between farmers and HGs on the Mediterranean coast
On the PCA plot, individuals from Baume Bourbon (BBB) and Gazel cluster closely with the earliest farmers from Southern France sites of Pendimoun (PEN) and Les Bréguières (LBR) (Figures 1B and 2A). Early farmers associated with the continental wave of Neolithization (e.g., LBK), located in Alsace and Eastern France (Brunel et al., 2020), and those linked to the Mediterranean wave from Southern France14 form two distinct clusters on the PCA plot, echoing the pan-European picture (Rivollat et al., 2020). Early farmers from Southern France (PEN, LBR, Gazel, BBB) are shifted toward the HG cluster on PC1, as opposed to other contemporaneous groups from Central Europe and LBK individuals from Alsace, and even further than Neolithic Iberia (Figure 2A). QpWave tests confirmed this clustering and revealed significant differences between the LBK individuals from France, the EN farmers from the Iberian Peninsula, and those from Southern France (LBR, Gazel, and BBB) (Table S17 and Figure S13). Genetic affinities of early farmers from Southern France with both farmers from the Mediterranean wave and HG are also seen in the results from uniparentally-inherited markers. The eight EN individuals from Southern France were indeed assigned to mitochondrial haplogroups J, H, K1, and U5 and male individuals carried Y chromosome haplogroup I2a1a2 (STAR Methods, Tables S5 and S6).
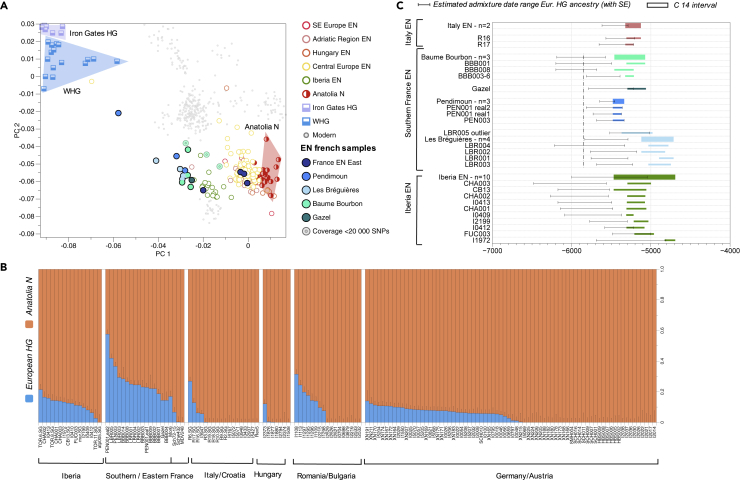
Figure 2.
Intensity and timeline of admixture between Early Neolithic (EN) farmers and hunter-gatherers in Europe
(A) PCA highlighting the genetic differentiation between European and Anatolia EN groups (∼8,300–4,800 BCE) regarding their affinity with individuals from Late Upper Palaeolithic/Mesolithic (∼16,000–5,600 BCE).
(B) QpAdm analysis. Western European individuals from EN contexts represented as a two-way model (Anatolia_N and European_HG (Loschbour, KO1, La Braña), qpAdm Model 1, Table S10).
(C) Admixture date estimates using DATES software, using the sources in (B). The conversion of generations to calendar years is using 28 years per generation and admixture time estimate is added to the oldest date of C14 interval (Table S15 and STAR Methods). The black dashed line materializes archaeologically attested farming establishment in Southern France, around 5,850 BCE.
To formally test for shared genetic drift between Neolithic individuals (test) and European HG we calculated outgroup-f3-statistics of the form f3(test, European HG; Mbuti) (Table S14 and Figure S6). For EN communities (6,100–4,700 BCE), we noted that individuals from Italy, the Iberian Peninsula, and Southern France exhibit higher f3-values, and thus higher amounts of shared genetic drift with HG compared to contemporaneous continental populations from Alsace, Germany, Austria, Hungary, and Romania (Table S14 and Figure S6). We conducted f4-statistics of the form f4(Mbuti, X; Y, Anatolia Neolithic) where X and Y correspond respectively to HG individuals and Neolithic individuals reported in this study to trace genetic affinities between Neolithic farmers and various HG communities. We were able to highlight that EN individuals from Baume Bourbon and Gazel share the most genetic affinities with HG specifically from Italy, France, Luxembourg, Germany, and the British Isles relatively to other HG groups (Table S18, SI4, Figure S9). We then used qpAdm (ADMIXTOOLS, STAR Methods) to explore European HG (Loschbour, KO1, La Braña) and Anatolian farmers (Anatolia_N) as potential sources of ancestry in European EN individuals and quantify the respective ancestry proportions (Figure 2B, qpAdm Model 1, Table S10). We were able to model Baume Bourbon and Gazel individuals as a two-way mixture of Anatolian farmers and European HG components with relatively high proportions of European HG ancestry ranging from 14.4 to 28.4% (qpAdm Model 1, Table S10). EN farmers from Southwestern Europe exhibit higher proportions of HG-related ancestry (18.5% on average) than LBK-associated individuals who either carry low amounts of HG ancestry or can be modeled with Anatolia_N ancestry, exclusively (7% on average; Lipson et al., 2017). In the Mediterranean region, the highest proportions of HG ancestry were found in individuals from Southern France (Pendimoun (22.9–57.6%), Les Bréguières (24.5–41.9%) and Baume Bourbon (14.4–28.4%) documenting substantial admixture in this region compared to contemporaneous, neighboring groups in Iberia and the Adriatic region. Whereas already identified for EN occupations of Pendimoun and Les Bréguières in Southeastern France, the high HG proportions found in individuals from Baume Bourbon and Gazel extend the geographic expanse of intensified HG-farmer interactions to the western part of the French Mediterranean, thus encompassing the entire Mediterranean zone of France.
We aimed to trace the presence of residual Magdalenian-associated ancestry (Villalba-Mouco et al., 2019), represented by ∼15,000-year-old Belgian GoyetQ2 (qpAdm model 3, Tables S12 and S14; Figures S7 and S11). Only BBB003_6 from Baume Bourbon could be modeled with GoyetQ2 as an additional source of HG ancestry (16.5%). The other individuals are best modeled as a two-way mixture of Villabruna-related ancestry and Anatolia farmers’ ancestry (Table S12 and Figure S11). Among contemporaneous EN farmers, high levels of GoyetQ2-like ancestry were reported in Iberians with the highest proportions identified in individuals from Andalusia (Cueva del Toro), and in lower proportions in Catalonia (Cueva de Chaves; Villalba-Mouco et al., 2019). These results argue in favor of networks connecting the populations of Languedoc and the Valencian region with a feedback effect from the Iberian Peninsula toward the East, a hypothesis that was put forward to explain the formation of the “Franco-Iberian Cardial” (Guilaine 2018a, 2018b; Guilaine and Manen, 2007). However, we have to consider that very little is known about the genetics of local Mesolithic groups, who could have equally contributed to the ancestry observed at Baume Bourbon. Among Mesolithic individuals, high proportions of GoyetQ2 ancestry were identified in French HGs from Les Perrats, on the West Atlantic façade during the early stage of Mesolithic, highlighting the late persistence of Madgalenian-associated genetic heritage outside of Iberia (Brunel et al., 2020). The presence and high percentage of GoyetQ2 ancestry at Baume Bourbon is all the more interesting as no EN individual from the Eastern part of the French Mediterranean appears to carry this type of ancestry. However, the distribution of this genetic component, which also manifests sporadically in Central Europe (Table S12), needs to be better characterized. This clearly highlights the need for further documentation of both Mesolithic and EN groups.
To document the processes of gene flow during the Mediterranean Neolithic expansion, we also estimated the timing of admixture events using the software DATES (Chintalapati et al., 2022; Narasimhan et al., 2019; DATES model 1, Table S14, Figure 2B, STAR Methods, Figure S14). All admixture estimations discussed here were calculated taking into account the interval between the maximum and minimum C14 dates of the individuals/groups to catch the most ancient and most recent possible admixture event as well as a generation time of 28 years (Fenner, 2005). However, estimations calculated for the median of C14 dates (and generation times of 25–30 years) can also be found in Table S15. For the individuals from Baume Bourbon (N = 3), admixture dates ranged between ∼6,200 and ∼5,600 BCE, and one individual yielded ∼5,900–5,300 BCE. For Gazel, Pendimoun, and Les Bréguières sites/individuals the estimated dates ranged between ∼6,200 and ∼5,000 BCE. When considering only the oldest C14 age for the Baume Bourbon group, DATES estimations (21.918 ± 4.037 generations before the age distribution of the group therefore between ∼6,200 and ∼5,950 BCE) place the admixture event at least four generations (∼111 years) before archaeologically attested farming establishment in the region, around 5,850 BCE (Binder, 2000). Nevertheless, when considering the minimum C14 age of the group, estimations place the admixture around 5,685 BCE, right after the farmers’ arrival. Pending estimates based on a more robust methodology, we cannot exclude that admixture between HG and incoming farmers took place prior to their arrival in the Occitanie region or right during their establishment in the region (Hofmanová et al., 2016). Of note, the other EN sites from Southern France returned date estimates suggesting that admixture could have occurred shortly after the arrival of farming in respective regions. Finally, admixture date ranges estimated for Italian and Iberian EN farmers were wider than those estimated for French sites. The variability of admixture dates measured between sites in the Western Mediterranean region and those measured within communities both suggest a complex and multi-phased history of admixture between HG and incoming farmers. Considering the whole date range of possible admixture events in France and Iberia, the earliest dates detected could place admixture events before attested farming establishment in both concerned regions. Consequently, these events could just as probably have taken place upstream of the Western Mediterranean expansion route, in regions that remain to be specified. Farming groups from Southern France therefore acquired specific HG ancestry, potentially from multiple origins. Only a few archaeological studies about Languedoc, Southeastern France, Central Italy have argued in favor of direct contacts between HG and farmer groups (Binder, 2000; Rigaud et al., 2018). In fact, geographical and chronological modeling of settlements in Italy rather highlighted that early farmers occupied regions that appeared to have been uninhabited, supporting a “no man’s land” model (Perrin and Manen, 2021). In Western Languedoc, shorter time gaps separate Mesolithic and Neolithic settlements, but still support a quick succession and reoccupation of a given area rather than the coexistence of HG and farmers (Perrin, 2010; Perrin and Manen, 2021). Both models imply a succession of HG and farmers either without or with very limited overlap. Therefore, our study shows how genomic data have the potential of adding new insights into complex interaction history during the Mediterranean expansion.
Small farming communities at the origin of the ICC complex
We estimated runs of homozygosity (ROH) to assess the level of inbreeding or past limitations of the effective population’s size for early farming groups by applying hapROH (Ringbauer et al., 2021; SI4, Table S16, Figure S15). Overall, we found few inbreeding signals among the early Western Mediterranean farmers, with the exception of individual Gazel4 from the Gazel cave, who surpassed the 50 cM threshold for the sum of ROH (sROH) over 20, with sROH>20 = 138.5 cM (Figures 3B and S15 and Table S16). This individual exhibits multiple long runs of homozygosity on different chromosomes, which can be interpreted as the offspring of a first-degree incestuous union (parent-offspring or full siblings, Figure 3B). Of note, we must acknowledge the difficulty of discussing specific funerary gestures surrounding Gazel4. The remains of this individual are very scarce and little archaeological arguments can be put forward to characterize this mortuary deposit. Therefore, there is nothing to support or contradict a potential social status awarded to this individual as what has been described for an inbred individual identified in Newgrange megalithic structure in Ireland (Cassidy et al., 2020).
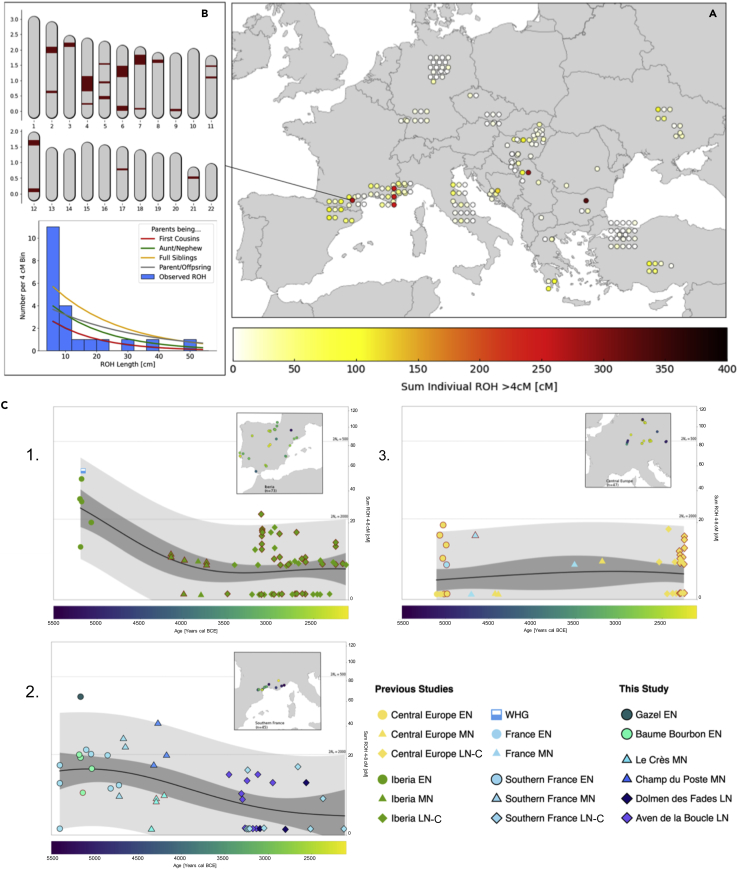
Figure 3.
Runs of homozygosity of Western Europe Neolithic individuals
(A) Sum of Individual ROH >4cM in Early Farmers from Europe and Anatolia between ∼9000 and ∼6000 BP. (B) ROH in individual Gazel4 from Gazel, an adult female dated to 5,296–5,062 cal BCE. The upper panel shows the mapped length of ROH (in Morgan) on each chromosome. The lower panel shows a histogram of ROH length (in centimorgan; cM) with the distribution alongside projected curves of expected ROH patterns based on different scenarios of parental relatedness, as described in Ringbauer et al. (2021).
(C) sROH[4,8] for individuals belonging to three regional transects between 5,500 and 2,000 BCE, the Iberian peninsula (C1, n = 73), Southern France (C2, n = 44), and Central Europe (C3, n = 47), samples dated according to the archaeological context only are highlighted in red. The solid black line represents mean estimates, gray areas represent the 95% empirical confidence intervals for individuals (light gray) and for the estimated mean (dark gray), as described in Ringbauer et al. (2021). See Table S16 for further details.
When comparing Neolithic farmer communities in Europe, individuals associated with the Central and Western Mediterranean Neolithic show substantially higher sROH[4–8] values than those linked to the continental Neolithic (Figures 3A, 3C, and S15 and Table S16). Overall, the higher proportion of short ROH observed in Early Mediterranean farmers (mean sROH[4–8] = 15.542; median = 15.385) suggests smaller effective population sizes (Ne) for Early to Middle Neolithic communities in Southern France and Iberia, progressively increasing toward LN, whereas LBK groups were characterized by higher Ne from the EN (mean sROH[4–8] = 3.888; median = 4.541). The observed differences in effective population size between Continental and Mediterranean groups could be linked to migration modalities. The archaeological record indeed supports a fast dispersion following maritime routes of small ICC-associated groups westward from Southern Italy to Liguria and Southern France (Binder et al., 2018; Leppard, 2021; Robb, 2013; Zilhão, 1993, 2000). Expanding farmer groups likely faced various challenges along the coastline including those of seafaring (e.g., limited capacity of crafts or dependency on maritime currents), which could have reduced initial population size, and increased founder effects. Consequently, archaeological data and demographic estimates of population sizes suggested smaller pioneer populations sustained by long-distance networks (Binder et al., 2018; Leppard, 2021; Rowley-Conwy, 2011). During the second half of the 6th millennium BCE, the second stage of Mediterranean Neolithization is associated with the diversification of material cultures and increase of populations’ size (Leppard, 2021; Manen et al., 2019; Roberts et al., 2019). Our results provide more details about these demographic models. Population size estimates indeed argue in favor of smaller-sized human groups inhabiting Southern France during advanced phases of farming expansion, and to some extent characterized by a potential degree of isolation. This hypothesis could also be put forward to explain cases like the incestuous union observed at Gazel, although there is no evidence that such event was justified socially among small communities, and therefore incestuous mating could also be related to deviant behavior. Moreover, it is interesting to note that the reduced population size of early farmers groups in the Mediterranean could also have been a contributing factor in the higher observed intensity of HG admixture in these regions.
Genomic history of southern France farming communities throughout the Neolithic
The transition from Impressed Ware groups to the widespread Chasséen culture (4,400–3,600 BCE) in Occitanie corresponds to diverse cultural aspects provisionally grouped under the term meridional Middle Neolithic 1 (Ambert et al., 1989; Loison and Schmitt, 2009; Vaquer, 1990, 1991, 1998; MN1, 4,800–4,400 BCE; STAR Methods). This period was represented by five individuals from Southern France so far (Brunel et al., 2020; Olalde et al., 2018) (Table S8). We extended this dataset with seven new individuals, from Le Crès (N = 4) and Champ du Poste (N = 3), two necropolises attributed to Early Chasséen culture and dated to the second half of the 5th millennium BCE (Tables S1 and S8). The individuals carried maternal and paternal haplogroups falling within the variability of Western European Neolithic groups (STAR Methods; Tables S5 and S6).
All individuals attributed to the Early Chasséen culture fall within the variability of Western European MN populations (Figure 1B) and are indistinguishable from MN individuals from other French regions and the Iberian Peninsula on the PCA. These results are in line with a global genetic homogenization of contemporary Western European groups.
As the Neolithic period progressed, the Chasséen culture split into many smaller cultural entities in Southern France (Gutherz, 1894; Jedikian and Vaquer, 2001; Tarrête and Le Roux, 2008). In our study, the transition to LN (3,600–2,500 BCE) is represented by ten individuals from the Aven de la Boucle and three individuals from the Dolmen des Fades (Tables S1 and S2). These individuals also carried mtDNA and Y chromosomal haplogroups common to the Western European Neolithic (Tables S5 and S6; Figure S4 STAR Methods).
These individuals are genetically homogeneous and cluster with MN1 and Early Chasséen individuals from Southern France, suggesting a general continuity in the region throughout clear cultural transitions (Figure 1B). We then used qpWave (ADMIXTOOLS) to formally test shared genetic drift throughout the Neolithic period, including one outlier individual BOU6 from the LN Aven de la Boucle group (Table S17). This individual is further shifted on the PCA plot toward the HG cluster. This HG affinity was supported by qpWave through the clustering of BOU6 with ICC samples from Pendimoun and Les Bréguières (LBR5), known for their excess of HG ancestry, as well as qpAdm analyses (see below). The remaining early farmers from Southern France form a cluster in qpWave analysis comprising groups attributed to ICC (Baume Bourbon, Gazel, les Bréguières), NM1 and Early Chasséen (Le Pirou, Clos de Roque, Le Crès, and Champ du Poste), and LN cultures (Aven de la Boucle, Dolmen des Fades). This indicates a certain extent of genetic continuity throughout the Neolithic period in South of France, from the second phase of the Neolithization to the first half of the third millennium BCE, before the arrival of Steppe-related ancestry, despite substantial cultural transformations documented archaeologically. The situation is different in the North-East of France, where we observe a genetic discontinuity between the first stage of Neolithization and the post-LBK Neolithic in line with changes in the archaeological material culture (Table S17; Figure S8, S12, and S13).
We then quantified changes in European HG ancestry proportions in farming communities by running qpAdm on all individuals from this study and European Neolithic farmers dated between 6,300 and 2,400 cal BCE (qpAdm model 1, Table S10). Figure 4 highlights the special status of ICC farmers from Southern France, presenting the highest proportions of HG ancestry compared to any contemporaneous European farming groups. Moving through time, subsequent Neolithic groups (∼4,800 and ∼4,000 BCE) display a steady increase in European HG ancestry as already demonstrated for other Western European regions (Rivollat et al., 2020). The highest proportions of HG ancestry are observed in individuals from Clos de Roque in Southern France (Olalde et al., 2018; ∼4,800–4,550 BCE; 29–32%), one individual from Escalles site in North of France (Brunel et al., 2020; 31%) and the Obernai group in Northeast France (Rivollat et al., 2020; OBN B, ∼4,800–4,500 cal BCE, 46–41%), whereas most individuals from fifth millennium France range between ∼10 and 30%. This general trend is sustained in the subsequent period between ∼4,000 and ∼3,500 cal BCE (14–32%). All individuals from 5th to 3rd millennia BCE Neolithic contexts could be modeled as a two-way mixture of Villabruna HG and Western Anatolian Neolithic ancestries (qpAdm model 3, Table S12), and we found no further evidence of GoyetQ2-related ancestry. As with EN individuals, f4-statistics in the form f4(Mbuti, X; Y, Anatolia_N) where X and Y correspond respectively to previously reported HG individuals and Neolithic individuals discussed in this study, indicated that Southern France LN farmers are closest to HG originating from Italy, France, Luxembourg, Germany, and the British Isles compared with other HG individuals (Table S18; STAR Methods; Figure S9). This could indicate the preservation of this type of HG ancestry in the region throughout the Neolithic period.
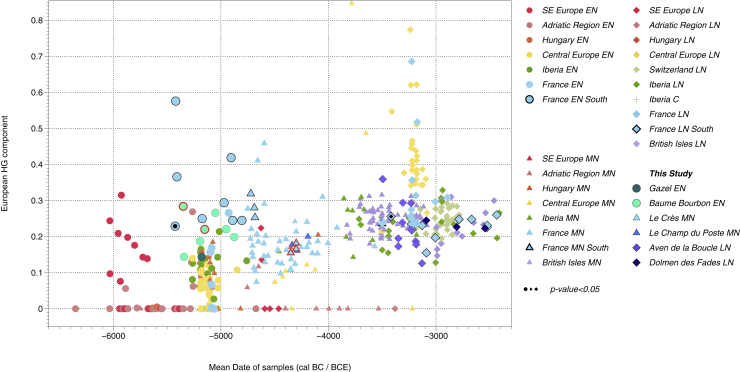
Figure 4.
Changes in European HG ancestry over time in Western Europe Neolithic communities
Results of 416 Neolithic individuals (plotted on the x-axis according to mean radiocarbon date, the five individuals dated by archaeological context among our corpus are highlighted in red), who were modeled as a two-way mixture of Anatolia_N and European_HG ancestry (represented by Loschbour, La Brana, and KO1; the y-axis) using qpAdm (Table S10).
The specific affinity of the la Boucle individual BOU6 to HG was also tested using qpAdm analysis (Model 1, Table S10). The HG component of BOU6 was estimated to be 36%, which exceeds the proportions found in other individuals from the same site (∼13–29%; Table S10 and Figure 4). This result mirrors recent findings of unexpectedly high HG proportions in late farmers from France and Germany: two individuals from the Mont-Aimé collective burial in the Paris Basin (∼3,300–3,100 BCE) carried 50.6 and 63.3% of Loschbour-related HG ancestry (Seguin-Orlando et al., 2021), MLN individuals from the Blätterhöhle cave (Lipson et al., 2017) carried ∼49 to 85%, the Wartberg-associated collective burial Niedertiefenbach (Immel et al., 2021) 34–58%, and one individual from the site of Tangermünde attributed to an Elb-Havel context carried ∼63.6% (Rivollat et al., 2020). We calculated admixture dates for Blätterhöhle MN individuals, Mont-Aimé individuals, the Niedertiefenbach community, and TGM009 (Table S15). Mont-Aimé individuals ranged between ∼4,600 and ∼3,600 cal BCE and the Niedertiefenbach community between ∼4,100 and ∼3,900 cal BCE. Blätterhöhle and TGM009 yielded similar results, ranging between ∼4,550 and ∼4,155 cal BCE and ∼4,300 and ∼3,400 cal BCE, respectively. Using the same methodology, the estimated admixture date for BOU6 falls between ∼5,000 and ∼4,200 cal BCE, whereas the rest of the la Boucle group was estimated between ∼5,800 and 5,100 cal BCE (Table S15).
The detection of high amounts of HG ancestry in LN farmers from different regions of France and Germany supports multi-phased gene flow between farmer groups (presenting less than 30% of HG component) and groups who had retained predominant HG ancestry. Indeed, the very recent admixture dates observed in some LN individuals would reinforce the hypothesis of continuous gene flow or multiple pulses of gene flow, implying recurrent genetic exchanges, over a long period of time (up until 3,600 BCE) and in different regions in Western Europe, as recently proposed by Chintalapati et al. (2022). This hypothesis evidently poses the challenge of identifying the groups contributing with substantial European HG ancestry during late periods. The French archaeological record clearly points out the absence of HG groups after 4,900 BCE, predating the admixture date estimated for BOU6 and the Paris Basin. The genomic data available from groups from the 5th to 3rd millennium BCE in Western Europe have not yet revealed communities carrying predominantly HG genomic legacy and therefore their geographical distribution remains elusive. The cultural attribution of these groups is also challenging as biological characteristics alone cannot be used to speculate on their cultural background. The invisibility of such groups from the archaeological and genomic record could either be linked to (i) conservation bias specific to their lifestyle and/or spatial distribution of the groups’ settlements in marginal areas, escaping current archaeological surveys (i.e., mountainous regions or coastal fringes) or (ii) the adoption of mortuary practices preventing the conservation of skeletal remains.
The restricted geographical area and wide time frame of this study allowed us to illustrate that individual variation in ancestries within a defined chronological and geographical frame can produce a more nuanced picture of contacts and mobility.
EN individuals from the 6th millennium BCE provide data showing that Neolithic groups attributed to the different waves of expansion experienced contrasting histories regarding both biological interactions with Mesolithic groups and dispersal modalities. The observed patterns therefore raise the question of the extent to which small population size and lower diversity of Early Mediterranean farmers may have been a contributing factor in biological exchanges between expanding EN farmers and autochthonous HG. The comparison of available data for EN farmers from Italy, Southern France, and the Iberian Peninsula however highlights slight differences in the intensity of admixture events between these regions. The observed differences could be connected to behavioral/cultural aspects or be tied to different densities of HG groups.
A remaining aspect also concerns the precise dynamics of admixture events between expanding farmers and local HGs, which seem to have been multiple, both in time and space. Additional genomic and radiocarbon data from Italy, the Aegean, and Central to Eastern Mediterranean Neolithic will be necessary to discuss the chronology and diversity of contacts along the expansion route, primordial for a holistic view of the Neolithization process in the Mediterranean. As we move into the Neolithic period, the new data also contributed valuable insights into modes of interaction throughout the Neolithic, illustrating both a genetic continuity despite cultural transformation and the presence of late and large HG ancestry in Southern France Neolithic groups.
Limitations of the study
The Mediterranean is a key geographical location to gain access to a more refined understanding of Neolithization processes. However, its genomic documentation faces two major challenges. First, the scarcity of human remains originating from Late Mesolithic and earliest Neolithic contexts introduces bias in studying the pioneer establishment of farmers and exchanges with local groups in the region. Second, in this region, palaeogenomic studies have to tackle the highly variable DNA preservation among sites. As exemplified in this study by the results from Baume Bourbon, the mediocre conservation of aDNA can drastically reduce the number of accessible data.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Osteological remain | This study | Gazel4 |
| Osteological remain | This study | BBB001 |
| Osteological remain | This study | BBB002 |
| Osteological remain | This study | BBB003_6 |
| Osteological remain | This study | BBB004 |
| Osteological remain | This study | BBB008 |
| Osteological remain | This study | BBB009 |
| Osteological remain | This study | BBB014 |
| Osteological remain | This study | SP193 |
| Osteological remain | This study | SP237 |
| Osteological remain | This study | SP249 |
| Osteological remain | This study | CRE11C |
| Osteological remain | This study | CRE14 |
| Osteological remain | This study | CRE20B |
| Osteological remain | This study | CRE20D |
| Osteological remain | This study | BOU1 |
| Osteological remain | This study | BOU3 |
| Osteological remain | This study | BOU4 |
| Osteological remain | This study | BOU5 |
| Osteological remain | This study | BOU6 |
| Osteological remain | This study | BOU7 |
| Osteological remain | This study | BOU9 |
| Osteological remain | This study | BOU10 |
| Osteological remain | This study | BOU11 |
| Osteological remain | This study | BOU14 |
| Osteological remain | This study | FAD1 |
| Osteological remain | This study | FAD3 |
| Osteological remain | This study | FAD10 |
| Chemicals, peptides, and recombinant proteins | ||
| Proteinase K 100MG | Sigma Aldrich | P2308-100MG |
| EDTA | Invitrogen | 10135423 |
| Guanidinium chloride | Sigma Aldrich | G3272-500g |
| Sodium Acetate | Invitrogen | AM9740 |
| H2O | Dutscher | 33311 |
| Ethanol Absolute | Vwr | 8187602500 |
| Tween 20 | Sigma Aldrich | 11332465001 |
| dNTP Set | Dutscher | 755086 |
| BSA, Molecular Biology Grade | New England Biolabs Cat# B9000S | B9000 S |
| Adenosine 5′-Triphosphate (ATP) | New England Biolabs | P0756 S |
| Buffer PE | Qiagen | 19065 |
| Buffer EBT | Qiagen | 19086 |
| Buffer QG | Qiagen | 19063 |
| iso-propanol | Vwr | 1009952500 |
| 5M Sodium Chloride | Sigma Aldrich | S7899-500ML |
| NEBNext® End Repair Module | New England Biolabs | E6050L |
| NEBNext® Quick Ligation Module | New England Biolabs | E6056L |
| OneTaq 2X Master Mix with Standard Buffer | New England Biolabs | M0484L |
| Silica magnetic beads | Invitrogen | 10099482 |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs | M0530L |
| USER Enzyme | New England Biolabs | M5505L |
| Critical commercial assays | ||
| MinElute PCR Purification kit | Qiagen | 28004 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32851 |
| NextSeq 500/550 High Output Kit v2.5 (150 Cycles) | Illumina | 20024907 |
| Agilent High Sensitivity DNA Kit | Agilent | 5067-4626 |
| Deposited data | ||
| Raw and analyzed data | This Study | ENA project : PRJEB50995 |
| Software and algorithms | ||
| READ | Monroy Kuhn et al. (2018) | https://bitbucket.org/tguenther/read/src/master/ |
| TKGWV2 | Fernandes et al. (2021) | https://github.com/danimfernandes/tkgwv2 |
| Admixtools | Patterson et al. (2012) | https://github.com/DReichLab/AdmixTools |
| DATES | Narasimhan et al. (2019); Chintalapati et al. (2022) | https://github.com/priyamoorjani/DATES |
| EAGER | Peltzer et al., 2016 | https://github.com/nf-core/eager |
| ANGSD | Korneliussen et al. (2014) | https://github.com/ANGSD/angsd |
| ContamMix | Fu et al. (2013) | |
| Haplogrep | Weissensteiner et al. (2016) | https://github.com/seppinho/haplogrep-cmd |
| Yleaf | Ralf et al. (2018) | https://github.com/genid/Yleaf |
| smartPCA | Patterson et al. (2006) | https://github.com/chrchang/eigensoft |
| HapROH | Ringbauer et al. (2021) | https://github.com/hringbauer/hapROH |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Ana Arzelier (ana.arzelier@u-bordeaux.fr).
Materials availability
Raw sequence data and alignments are available at the European Nucleotide Archive (ENA project) under accession number ENA PRJEB50995. All other previously published genomic data discussed in this study is reported in Table S8.
Experimental model and subject details
Archaeological and Anthropological information
Authorisation to perform destructive sampling protocols for genetic analysis and radiocarbon dating was granted by the Service Régional de l’Archéologie Occitanie as well as the Institut National de Recherches Archéologiques Préventives through the PAS ANCESTRA (Coord. MP. Pruvost and F. Maziere), the project ANR ANCESTRA (coord M. Pruvost, ANR15-CE27-0001) and the project ANR-DFG INTERACT (coord M.-F. Deguilloux and W. Haak, Grant ANR 17-FRAL-0010, DFG-HA-5407/4-1). Human remains were provided to the laboratory PACEA by F. Convertini (Champ du Poste), H. Duday (Baume Bourbon, Aven de la Boucle), J. Guilaine (Gazel, Dolmen des Fades) and M. Gandelin (Le Crès).
Both samples denominations used in this study and corresponding excavation inventory labels are reported in Table S1 for the 28 samples sequenced on the genomic scale. For the 48 samples that were not sequenced further at this stage we also report archaeological inventory IDs and initial screening results in Table S2.
The 28 samples discussed here include four petrous bones, two teeth and two long bones (Femur and metatarsal bone) from Baume Bourbon, one tooth from Gazel, three petrous bones from Champ du Poste, four petrous bones from Le Crès, ten petrous bones from the Aven de la Boucle, three petrous bones from the Dolmen des Fades.
General information on archaeological context, funerary context and osteological observations are provided in Table S1 and are discussed hereafter.
Baume Bourbon, Cabrières, Gard – (Contact: H. Duday)
The Baume Bourbon cave is located on the side of a small combe carved into the urganian limestone of the Nîmes garrigue, 3km north-east of Cabrières (Gard). The occupation spans from Early Neolithic to modern era. The cave is divided into three areas, an entry area, known as the porch, as well as two other areas further down (S2 and S3). These later areas, located deeper inside the cavity revealed human remains belonging to a minimum number of 15 individuals.
This archaeological layer containing human remains also contained Cardial pottery, faunal remains, quartz and flint flakes and burnt cereals. Human remains mainly belong to the superior part of this layer, without anatomical connections. The stratigraphic position of these remains places the funerary deposits at the end of the Early Neolithic occupation of the cave. Shortly after, a colluvium closed the access between the porch and the further down areas S2 and S3, therefore making the Early Neolithic occupation the last one in this sector of the cavity. Moreover, the layer containing Early Neolithic human remains was fossilized by deposits of calcite and the formation of a flowstone. An isotopic analysis of charcoal pieces dated the Early Neolithic funerary layer around 6080 ± 100 BP (MC-794) (Coste et al., 1987; Le Bras-Goude, 2007; Zemour, 2013).
Of this layer, genome-wide data was recovered from seven individuals attributed to Cardial and Epicardial occupations of the Baume Bourbon cave (Coste et al., 1987; Zemour, 2013) dated between ∼5,400 and 4,800 BCE (N = 7; BBB samples; Table S1).
Gazel, Sallèles-Cabardès, Aude – (Contact: J. Guilaine)
The Gazel cave (Duday and Guilaine, 1980; Beyneix, 1997) in Sallèles-Cabardès (Aude) was frequented from the Upper Palaeolithic until the Middle Ages and includes a series of stratified occupations of the late Mesolithic and the early Neolithic. Over 1.80 m of stratigraphy, the deposits of the ancient Neolithic correspond to four stages (Gazel I to Gazel IV), the first correspond to Cardial, the other three to an original Languedoc Epicardial. Human remains corresponding to four Epicardial burials have been identified, two complete and two partial individuals. The genome recovered from Gazel Cave corresponds to an individual associated with the Cardial stage deposits, identified in the C2 sector and dated to 5,296-5,062 BCE (N = 1, Gazel4 sample; Table S1).
Champ du Poste, Carcassonne, Aude – (Contact: F. Convertini)
The site of the Champ du Poste (Carcassonne, France) was excavated in 2005 and 2006 (Convertini and Georjon, 2018). It is an important site for several archaeological periods that span from Neolithic to Antiquity. The Middle Neolithic is the period best represented. The duration of the occupations is known by a series of radiocarbon dates. During the first half of the 5th millennium BCE, the first settlers' installations leave few traces (heated stone hearths) and some burial pits. Then, during the Early Chasséen (second half of the 5th millennium BCE), occupations become more numerous and the site yields abundant artifacts (ceramics, flints, bones). Domestic structures (pits, silos, hearths) as well as burial pits are present. At the end of the Middle Neolithic, few structures and remains are present on the site. It was reoccupied at the end of the Neolithic (domestic structures and remains), then at the beginning and the end of the Bronze Age and finally again during Antiquity. The three individuals from Champ du Poste necropolis characterized on the genomic level are attributed to the second half of the 5th millennium BCE, ranging between ∼4,400 and 4,050 BCE (N = 3; SP individuals; Table S1).
Le Crès, Béziers, Hérault – (Contact: M. Gandelin)
The site of Le Crès in Béziers was excavated in 2001 by the Inrap (Loison and Schmitt, 2009). This deposit was explored on an area of 5,000 m2 that does not correspond to the entire archaeological site. Only 190 structures remain preserved and of these 30 yielded single or multiple primary burials. A total of 49 individuals has been discovered, which makes it one of the most important burial complexes known for the southern Chasséen. Some of the deceased, men or women, are installed in clear funerary graves, oblong plan and often arranged with large blocks of stones, whereas others are deposited, sometimes without particular care and most often without grave goods, in domestic pits, including silos. The latter type of pits can contain several individuals, including children. The grouping of burials testifies to a true spatial organization of the tombs with a separate burial area located close to the settlement. This grave group is dated to the Early Chasséen, between 4,350 and 4,100 BCE. Indeed, the site of Le Crès offers very homogeneous archaeological material attributed to the Early Chasséen and distinctive practices, which, combine to several radiocarbon dates, allow a reliable chrono-cultural attribution (Loison and Schmitt, 2009). For instance the disposition of corpses inside plural burials such as structures 20 and 11 is characteristic of Early Chasséen traditions from the Western Hérault region. This is also exemplified by grave goods associated with the individuals buried inside these pit graves (cups and ceramics with “funicular” handles, strip handles and bedoulian flint). The four low-coverage genomes recovered from Le Crès belong to one single adult burial (CRE14) and two distinct plural burials, Burial 11 (CRE11C) and burial 20 (CRE20B and CRE20D). Burial 11 is a relatively shallow and small pit, which comprised three individuals, one adult and two immature individuals deposited simultaneously. Burial 20 is a larger silo-type pit, which comprised five immature individuals. Given the clear association of these individuals with specific archaeological material and direct or indirect radiocarbon dates, all four individuals from Le Crès discussed in this study fit within the time frame of Early Chasséen, with dates ranging between ∼4,500 and 4,050 BCE (Loison and Schmitt, 2009; Brunel et al., 2020), (N = 4, CRE samples; Table S1).
Dolmen des Fades, Pépieux, Aude – (Contact: J. Guilaine)
The Dolmen des Fades in Pépieux (Aude), also called "Palet de Roland", is the longest dolmenic tomb in the South of France (Bonnery, 1991; Guilaine, 1998). It is a megalithic gallery of 24 m, excluding the tumulus. It is split into a corridor of 12 m, an antechamber and a room, both 6 m in length. The passage between these three parts was done through “oven doors”. After early disorganized excavations, a survey was carried out in 1946 by O. and J. Taffanel and J. Arnal in its terminal room, a sector that had in fact been disturbed in the Middle Ages. From 1962, J. Guilaine has excavated the corridor and the antechamber. The artifacts revealed a long use beginning in the late Neolithic and continuing until the Bronze Age. Several radiocarbon dates confirm this long occupation during the third millennium BCE. The three individuals providing genomic data and originating from this dolmen are attributed to Late Neolithic, ranging dates between ∼3,200 and 2,400 cal BCE (N = 3, FAD samples; Table S1).
Aven de la Boucle, Corconne, Gard – (Contact: H. Duday)
The aven de la Boucle at Corconne (Gard) is a natural cavity that served as a collective burial from the recent Neolithic, in the second half of the fourth millennium BCE (Duday, 1987; Jallet et al., 2010). It continued into the late Neolithic (Ferrières culture), at the beginning of the third millennium. The group of deceased consists of roughly 75 individuals, with a marked selection according to the age of individuals (massive exclusion of children). Complex layouts were highlighted, particularly at the level of access: a natural diaclase had been covered in the manner of a megalithic corridor, with a large removable horizontal slab equivalent to the "entrance cap" of covered walkways or dolmens; steps were intended to facilitate the descent of the bodies to the deep part of the cavity, located more than 10 m below the surface. This passage was condemned after its use for funerary purposes: a wall of slabs closed the inner part of the diaclase, the roof of the passage was tilted and the outer part of the diaclase was completely filled with stones.
The ten individuals analyzed on the genomic scale from this collective burial are all attributed to the second half of the fourth and beginning of the third millennium BCE, with dates ranging between ∼3,600 and 2,800 BCE (N = 10, BOU samples; Table S1).
Method details
Sample selection and preparation
A total of 76 samples was processed and analyzed from the 6 different sites located in Modern-day France Occitanie region illustrated by Figure S1.
Human remains were sampled, DNA extracted and prepared for next-generation sequencing in a clean room at a dedicated ancient DNA (aDNA) facility in the Laboratory of PACEA, University of Bordeaux, France.
We privileged sampling of cranial remains (petrous bones and tooth). For Baume Bourbon (BBB) sampling included 13 cranial and 13 non-cranial remains (long bones and coxal bones).
First, skeletal remains were irradiated with UV light during 30 min on each side. A layer of bone surface was then abraded around the sampled area before low-speed drilling into the remains to retrieve between 20 and 100mg of bone powder. For petrous bones, the powder was drilled from the cochlea (Pinhasi et al., 2015). Teeth were cleaned with bleach solution to remove surface contaminants before being sawn in half along the cementum-enamel junction, then powder was drilled from the pulp cavity.
Radiocarbon dating
Direct radiocarbon dating was conducted for 25 samples. Dating was performed using standard protocole at the C14 laboratory of CEDAD in Lecce or through ARTEMIS program (MMC) in the C14 dedicated laboratory of Lyon.
Bone fragments between 0.9 and 2g were prepared in a dedicated clean room at the PACEA lab. To ensure the exact correspondence of genomic and radiocarbon data, sampling was conducted on the same remains sampled for ancient DNA or on contiguous bones, i.e. cranial vault for sub-complete skulls and in the case of individuals BBB003-6 and BBB004, radiocarbon sampling was conducted on the mandibles holding the teeth sampled for DNA.
Radiocarbon dates were obtained for 23 samples and failed for samples BBB002 and BBB014 due to lack of collagen. The “conventional radiocarbon age” was calculated with a 13C correction based on the 13C/12C ratio measured directly with the accelerator. For the estimation of the measurement uncertainty (standard deviation) both the radioisotope counting statistics and the scattering of the data have been taken into account. Calibration was carried out using OxCal online (OxCal 4.494) and the IntCal20 calibration curve (Table S9).
Dates are reported in Tables S1, S8, and S9 according to two formats: dates provided by direct radiocarbon dating on the 23 analyzed samples that provided positive results, given in calibrated 2-sigmas interval, and associated date in BP of the form “5,309-5,077 BCE (6248 ± 30 BP)”. In the absence of direct dating for the BBB002 and BBB014 samples from Baume Bourbon cave (that failed to provide radiocarbon dating due to the lack of collagen) the interval provided relies on archaeological context and is given in the form “4,500-4,100 BCE”.
Of the 23 directly dated samples, seven individuals originate from two collective sepulchral caves attributed to Early Neolithic period: the Baume Bourbon cave (Cabrières, Gard) dated between ∼5,400 and 4,800 BCE (N = 6; BBB samples) and the contemporary Gazel cave (Sallèles-Cabardès, Aude), with one individual dated to 5,296-5,062 BCE. Direct dates obtained for these sites confirm their cultural attribution to Cardial-Epicardial cultures and the Impresso-cardial complex (ICC), following by a few centuries the pioneer Neolithic settlements reported in Mediterranean Languedoc ∼5,850 BCE (Binder et al., 2018).
We were able to date four individuals originating from two necropolises dated to Middle Neolithic: Champ du Poste (Convertini and Georjon, 2018; Carcassonne, Aude; N = 3; SP individuals) and Le Crès (Loison and Schmitt, 2009; Béziers, Hérault; N = 1, CRE samples). The individual from Le Crès necropolis directly dated ranged between 4,442 and 4,261 BCE (CRE14), perfectly fitting with the dates obtained for other individuals from the site between ∼4,500 and 4,050 BCE (Loison and Schmitt, 2009; Brunel et al., 2020). The three other individuals were not directly dated; however, several arguments allow their reliable chrono-cultural attribution. Both burial 11 (CRE11C) and 20 (CRE20B and CRE20D) reflect a distinctive funerary expression, that can be attributed to Early Chasséen and fit the homogeneity of the archaeological material displayed on the site. Inside burial 11, individual CRE11C was associated with several objects such as bedoulian flint or ceramic with a strip handle, which are typically encountered in Early Chasséen contexts. Inside grave 20, individuals CRE20B and CRE20D were buried simultaneously alongside three other individuals, among which one is directly dated to 4,335-4,058 cal BCE (5380 ± 35 BP) (Loison and Schmitt, 2009). These individuals were also associated with ceramics typical of Early Chasséen contexts.
Therefore, radiocarbon dating conducted on human remains can be put in perspective with the typology of archaeological material as well as the chronology of the pits to place the occupation of the site and utilization of the necropolis within a restricted time range (Loison and Schmitt, 2009; Brunel et al., 2020).
The three individuals from Champ du Poste necropolis yielded similar dates ranging between ∼4,400 and 4,050 BCE. All dates in hand confirm that both necropolises were used over a few centuries and included individuals who can be attributed to Early Southern Chasséen culture (Loison and Schmitt, 2009; Ambert et al., 1989; Vaquer, 1990, 1991, 1998).
Finally, direct dates were obtained for 13 individuals recovered from two collective burials used during Late Neolithic and Bronze Age: the Aven de la Boucle cave (Duday, 1987; Jallet et al., 2010; Corconne, Gard; N = 10, BOU samples) and the Dolmen des Fades (Bonnery, 1991; Guilaine, 1998; Pépieux, Aude; N = 3, FAD samples). Radiocarbon dates obtained on the 10 individuals sequenced for the Aven de la Boucle confirmed that this collective burial cave was mainly active during the second half of the 4th and beginning of the 3rd millennium BCE (Ferrières culture), with dates ranging between ∼3,600 and 2,800 BCE. Finally, as initially demonstrated by the artifacts, radiocarbon dates confirmed a long occupation history for the Dolmen des Fades collective burial, beginning during Late Neolithic and continuing into the Bronze Age. Three individuals providing genomic data and originating from this dolmen were dated between ∼3,200 and 2,400 BCE (FAD1, FAD3 and FAD10).
DNA extraction and sequencing
All samples were processed for DNA extraction at the Laboratory of PACEA, University of Bordeaux.
Samples from Baume Bourbon were processed following the procedure described in Dabney et al. (2013), (dx.doi.org/10.17504/protocols.io.baksicwe) whereas other samples were processed according to a two-step extraction procedure described in Brunel et al. (Damgaard et al., 2015; Brunel et al., 2020).
Purification of samples was conducted according to a modified version of the method described in Dabney et al. published in Brunel et al. (Dabney et al., 2013; Brunel et al., 2020).
For Baume Bourbon samples, double-stranded libraries were built from 20μL of DNA template, following the protocol proposed by Meyer and Kircher (2010) and using unique index pairs (Kircher, 2012). A partial uracil-DNA-glycosylase (UDG half) treatment was applied to remove deaminated cytosines except for the final nucleotides at the 5′ and-3′ reads ends to preserve part of the damage pattern characteristic for ancient DNA (Rohland et al., 2015) (dx.doi.org/10.17504/protocols.io.bmh6k39e).
For other samples, double stranded libraries were constructed according to a protocol adapted from Gorgé et al. (2016; Brunel et al., 2020).
We first screened all indexed libraries via shotgun sequencing targeting 1 million reads. Libraries were pooled and sequenced on an Illumina NextSeq 500 at Institut de Recherches Biomédicales des Armées, using a NextSeq 500/550 High Output Kit v2.5 (150 Cycles).
For Baume Bourbon samples, we enriched next-generation ancient DNA libraries for ∼1.2 million single-nucleotide polymorphisms (SNPs) using targeted in-solution capture (Mathieson et al., 2015), as well as an independent capture array for the complete mitogenome (Maricic et al., 2010), and sequenced these on Illumina platforms to an average depth per site of 0.14896 for 1240K and 1.42010 for mitochondrial capture.
Read processing, alignment and postmortem damage
After demultiplexing, we processed raw sequence data using EAGER (Peltzer et al., 2016). Steps included trimming for adaptors sequences and processing into single reads with Clip&Merge, trimmed sequences were then mapped to Human Reference Genome Hs37d5 using BWA v.0.7.12 (Li and Durbin, 2009; Schubert et al., 2016). Duplicates were removed with De-Dup and mapDamage v.2.06 (Ginolhac et al., 2011) was used to assess DNA damage patterns consistent with ancient origin and remove reads with a mapping quality<30. Sequencing results are presented in Table S2.
Genotyping
All our bam files were trimmed for 2 bases on each side using trimBam function from BamUtil package and were genotyped by using PileupCaller. Considering the human genome as pseudo-haploïd, alleles were randomly called for each position of the 1,233,013 SNPs and ∼600,000 SNPs HO panels (Mathieson et al., 2015; Patterson et al., 2012; Lazaridis et al., 2016). Individuals having at least 19,000 SNPs on the 1240k panel were considered for further analysis.
Quantification and statistical analysis
Genetic sex determination
We determined genetic sex using the methods described in Skoglund et al. (2013), based on the estimations of reads mapping to X and Y chromosomes compared to reads mapping to the autosomes. Females are expected to have a ratio of 1 on the X and 0 on the Y, as opposed to males, who are expected to have a ratio of 0.5 on X and Y chromosomes. We determined a threshold of Y ratio based on the method published by Skoglund et al. (2013). We used an upper threshold of 0.016 of the ratio of sequence mapping to the Y chromosome for females and a lower bound of 0.077 for males (Table S4, Figures S2 and S3). Using this threshold, among the 26 newly reported individuals, twelve individuals could be identified as females and 14 as males. Two individuals are undetermined, one (BBB002) is most likely to be female and one (BOU3) is most likely to be male (Table S4, Figures S2 and S3). The results are highlighted in Table S4.
Contamination estimations
We used ANGSD (Analysis of Next Generation Sequencing Data) package to test the degree of heterozygosity on the X chromosome and estimate contamination levels in all male individuals (Korneliussen et al., 2014). Considering a contamination threshold of ∼5%, we excluded one individual from Baume Bourbon (BBB7) of downstream analyses. We used the likelihood-based method ContamMix (Fu et al., 2013) to estimate contaminations in sequences aligning to the human mitochondrial genome. To build consensus sequences we used Geneious v.11.1.5, mitochondrial captured samples from Baume Bourbon were aligned to the mitochondrial genome NC_012920.1 (HG19) and other samples were aligned to the mitochondrial genome MT (hs35d7). Alignment of the samples to 311 reference genomes was performed using mafft v.7.453. (Katoh et al., 2002). Each fastQ was aligned to its consensus sequence using BWA v.0.5.10. (Li and Durbin, 2009). Results for contamination estimations are reported in Table S3, displaying Number of Reads on mitochondrial consensus sequence, estimated error rate, estimated maximum posterior proportion authentic, 2.5% credible quantile for proportion authentic, 97.5% credible quantile for proportion authentic, Gelman and Rubin diagnostic point estimate, Gelman and Rubin diagnostic upper confidence limit. The relative contamination rate was calculated by subtracting the proportion authentic to 1. We have to acknowledge that four samples displayed high contamination signals with this method (BBB002, BBB004, BBB009, BBB014). Of note, these samples as well as 8 other samples provided less than 2,000 reads on mitochondrial consensus sequences. This low coverage can affect the reliability of contamination estimations with ContamMix, therefore, BAM files were also inspected manually for possible contamination on mitochondrial reads using the Integrative Genomic Viewer. This inspection permitted to detect very limited contamination patterns, allowing us to proceed with downstream analyses for these samples.
Therefore, samples BBB002, BBB004, BBB009 and BBB014 displaying the highest contaminations signals, were cautiously integrated to this study. The results obtained on the population genetic level for these samples is very consistent with authentic data, as it is very coherent with samples displaying low contamination signals from the same contexts. In order to process these results with caution, we did not include these individuals with lowest quality data in more sensitive analysis (DATES, ROH). As a result, the low number of reads and the difficulty of providing reliable contamination estimation for these individuals does not affect our discussion and interpretations.
Uniparental markers analysis
Reads mapped on the revised Cambridge reference mitogenome were converted into VCF files and mitochondrial haplotypes were inferred using HaploGrep (Weissensteiner et al., 2016). Detailed haplogroup attribution results are provided in Table S5.
The most represented mtDNA haplogroups in our samples are haplogroups H and K1 (35,5 and 29%), what is consistent with previous reports regarding the genetic makeup of Southwestern Neolithic communities (Olalde et al., 2015; Hofmanová et al., 2016; Lipson et al., 2017; Lazaridis et al., 2017; Szécsényi-Nagy et al., 2017; Fregel et al., 2018; Valdiosera et al., 2018; Mathieson et al., 2018; Olalde et al., 2019; Villalba-Mouco et al., 2019; Antonio et al., 2019; Rivollat et al., 2020; Brunel et al., 2020). The 8 newly typed Early Neolithic individuals from Southern France are assigned to mitochondrial haplogroups J, H, K1 and U5. These haplogroups are typically found among Early farmers and are consistent with mtDNA data reported for Mediterranean Early farming communities (Bramanti et al., 2009; Mathieson et al., 2015, 2018; Olalde et al., 2015; Hofmanová et al., 2016; Lipson et al., 2017; Lazaridis et al., 2017; Szécsényi-Nagy et al., 2017; Fregel et al., 2018; Valdiosera et al., 2018; Olalde et al., 2019; Villalba-Mouco et al., 2019; Antonio et al., 2019; Rivollat et al., 2020). Haplogroup U5, carried by individual BB009 from La Baume Bourbon sepulchral cave is however predominantly found in WHG individuals and was previously identified in individual LBR002 from ICC Southern France site Les Bréguières (Rivollat et al., 2020). The presence of this haplogroup is consistent with admixture with WHG.
Individuals dated the second half of the 5th millennium cal BCE from Le Crès and Le Champ du Poste carry mtDNA haplogroups H2, K1, J2, V and X2, which fall into the variability of Western Europe Neolithic groups. Few ancient farmers carry haplogroup V, it was identified at Le Pirou (Pir4; Brunel et al., 2020), among western Europe Neolithic and Bronze Age individuals, and also in Poland and Hungary for Bell Beaker groups (Olalde et al., 2015; Hofmanová et al., 2016; Lipson et al., 2017; Lazaridis et al., 2017; Szécsényi-Nagy et al., 2017; Fregel et al., 2018; Valdiosera et al., 2018; Mathieson et al., 2018; Olalde et al., 2019; Villalba-Mouco et al., 2019; Antonio et al., 2019; Rivollat et al., 2020).
Characteristic mtDNA haplogroups associated with Neolithic farmers were also identified for the Aven de la Boucle and the Dolmen des Fades. The 4th and 3rd millennium cal BCE individuals from these collective burials carry haplogroups H, K1, J1, T2 and X2, U5 and U2. These lineages are commonly found among Neolithic European farmers groups, with the exception of haplogroup U5 (BOU3 and BOU6) and U2 (BOU1), found in high frequency in HG populations and manifesting a potential HG related ancestry at the Aven de la Boucle.
Y chromosome haplotypes were called using Yleaf statistical software (Ralf et al., 2018), with standard settings. Output results are described in Table S6 and Figure S4.
Among the four Early Neolithic male individuals dated to the 6th millennium cal BCE, we were able to identify Y chromosome haplogroup I2a1a2 for two individuals, whereas too low coverage data prevented to assign Y chromosome haplogroup for remaining individuals. The two male individuals dated from the 5th millennium cal BCE and originating from Champ du Poste site (SP249 and SP193) carry paternal haplogroups G2a2a1a2a1 and H2m (Rohrlach et al., 2021; Figure S4; Table S6). The male individuals dated to the 4th and 3rd millennium cal BCE and originating from Aven de la Boucle and Dolmen des Fades were assigned to haplogroups I2 and G2a. Haplogroup I2 has been reported in several Mesolithic male individuals from Western Europe before the arrival of the Neolithic lifestyle as well as among various Western Europe Farmers groups (Mathieson et al., 2015, 2018).
Biological relatedness analysis
Degrees of genetic relatedness between all individuals included in this study were estimated by applying Relationship Estimation from Ancient DNA (READ) to infer pairwise relationships up to the second degree (Monroy Kuhn et al., 2018). We used standard parameters, ie the median of all average P0s, for normalization. We were able to identify two samples from Baume Bourbon corresponding to one same individual (BBB003 and BBB006), and consequently merged the corresponding bam files in a unique file for downstream analysis (BBB003_6). We identified one first-degree related pair of individuals among the Aven de la Boucle samples (BOU9 and BOU10).
To complement biological relatedness analysis, we used the TKGWV2 pipeline that can be used to infer pairwise relatedness for individuals presenting very low coverage (Fernandes et al., 2021). We used default parameters, i.e. 30 for minimum mapping and base quality, default 1 for setting the threshold for the minimum number of SNPs allowed to estimate relatedness. Combining the two methods, we kept positive results when based on a minimum of 5000 SNPs shared between individuals or when positive results from TKGWV2 based on lower number of shared SNPs were backed up by READ results. This allowed us to identify three second-degree kinship pairs at Baume Bourbon (BBB004-BBB014, BBB001-BBB002, BBB001-BBB009), some kinship being reinforced by shared maternal lineages (Tables S5 and S7). This could be consistent with the collective use of this natural cavity for funerary purposes by a small community during the second part of the 6th millennium cal BCE. At the Aven de la Boucle, we confirmed the first-degree kinship between BOU9 and BOU10. No other biological relatedness could be inferred for other sites. Biological relatedness results for Baume Bourbon are provided in Table S7. From each relative pair, we removed the individual presenting the lowest coverage, keeping only “unrelated” individuals for population genetic analyses.
Population size and inbreeding estimates
We assessed levels of inbreeding in our samples using the 1000 Genome as a reference panel, calling SNPs for each individual chromosome by chromosome, through hapROH (Ringbauer et al., 2021). We were able to retrieve ROH data for all our samples except 3 (BBB004, BBB009, BBB014) and confronted ROH data for our 25 new samples to a subset of 322 ancient individuals from the Near-East, Central Europe, and Western Europe (results are presented in Table S15 and illustrated by Figure S15).
Merging datasets
Genomic data of all 28 individuals included in this study were merged with previously published genotypes of modern and ancient individuals on two different datasets used for genome-wide analysis. New data were merged to the HO dataset panel for PCA construction, whereas they were merged to the 1240k SNPs dataset panel provided on David Reich’s website v42.5. We also included individuals reported in Rivollat et al. (2020), Brunel et al. (2020), Seguin-Orlando et al. (2021) and Immel et al. (2021) for all other population genetic analyses. For these recent studies, data were subjected through the same procedure as the individuals reported in this study from the fastq files, which we downloaded from the ENA project site. For individuals reported in Immel et al. (2021), the procedure started from the individual BAM files, also downloaded from the ENA project site. The reprocessing of raw previously published data for population genetic analysis produced highly similar results to the original studies. Therefore, we chose to proceed with population genetic analysis for both the HO and 1240k datasets without reprocessing all raw data through the same pipeline. The newly obtained genomic data were specifically co-analyzed with ancient human groups from the periods considered, such as Neolithic individuals from Anatolia (Anatolia_N); Bulgaria, Romania, Serbia (SE Europe) Hungary; Greece, Italy (Adriatic Region); Germany, Austria, Poland, Czech Republic (Central Europe); Switzerland; France; Spain, Portugal (Iberia); Ireland and Great Britain (Britain).
Principal Component Analysis
PCA was constructed after merging our new data to the HO dataset panel for 592,998 autosomal genotypes in 796 modern individuals from western Eurasia, using the smartpca program (Patterson et al., 2006; See Figures 1, 2, and S5).
Genotypes were downloaded from David Reich’s website (v42.5 https://reichdata.hms.harvard.edu/pub/datasets/amh_repo/curated_releases/index_v42.4.html). We projected only individuals genotyped for more than 10,000 SNPs, using option modes lsqproject: YES, shrinkmode: YES.
The distribution of European Neolithic individuals follows genetic variation between European HGs and Anatolian farmers and can be split in two regionally distinct clusters, i.e. (i) a first cluster, closer to Anatolian farmers, comprising samples from Central and South-eastern Europe and (ii) a second cluster slightly shifting toward WHGs and comprising individuals from Western and South-western Europe (Figure S5). All newly reported Neolithic samples from Occitanie cluster with previously published Western Europe Neolithic individuals from the Iberian Peninsula, France, and the British Isles, and share all the same cluster from Early to Late Neolithic.
Early Neolithic farmers from Baume Bourbon and Gazel cluster with individuals reported for Southern France sites Pendimoun and Les Bréguières, but not with individuals from the Adriatic region and attributed to Impresso-Cardial. However, three individuals from Baume Bourbon appear to be slightly shifted toward the centroid, which could be explained partly by low coverage on the HO reference panel (<20,000 SNPs; Table S1). Middle Neolithic individuals from Champ du Poste group closely together on the PCA as more variation is observed for the three individuals from Le Crès necropolis. Late Neolithic individuals from the Aven de la Boucle and the Dolmen des Fades collective burials form a homogeneous cluster with the exception of the individual BOU6 from the Aven de la Boucle that is shifted further toward the HG cluster, suggesting higher affinities with this group.
F-statistics
We used ADMIXTOOLS to perform f-statistics analyses, applying qp3Pop to calculate outgroup f3-statistics to measure shared genetic drift in ancient individuals relative to Mbuti population used as outgroup (Patterson et al., 2012).
We performed f3-statistics of the form f3(test1, test2; outgroup) to investigate population level similarities within Early Neolithic groups from the Mediterranean and Continental currents, combining all the possible pairs of Early Neolithic Western Europe individuals as test. We then attempted to convert f3-statistics into a pairwise distance matrix by subtracting all f3 values from 1 (detailed results are provided in Table S13).
To measure shared genetic drift between Western Europe Neolithic individuals and European hunter-gatherers we performed f3-outgroup statistics of the form f3(test, European_HG; Mbuti). The f3-value estimates the shared genetic drift between European HG (Loschbour, La Braña, KO1) and each of the 680 Western Europe individuals tested, dated between ∼6,300 and 2,000 BCE. Detailed results are available in Table S14 and described by Figure S6.
We also performed outgroup-f3 statistics of the form f3(Mbuti; GoyetQ2, Test) with Test representing samples from Baume Bourbon, to test whether these individuals displayed specific affinities to GoyetQ2-like ancestry. Detailed results are available in Table S14 and described by Figure S7.
To test the affinities between Early Farmers and subsequent Neolithic groups from France we conducted outgroup statistics of the form f3(Mbuti; France_EN_LBK, Test) and f3(Mbuti; France_EN_Occitanie, Test), where test representing Neolithic groups from different regions and periods. The results for both tests are provided in Table S14 and illustrated by Figure S8.
We performed f4-Statistics using qpDstat with f4mode: YES, printSE: YES, in the form of f4(Mbuti, X; Y, Anatolia Neolithic) where X and Y correspond respectively to previously reported HG and Neolithic individuals. This analysis aimed to trace genetic proximity and gene flow between X and Y, reflected by negative f4 values significantly supported by Z-scores inferior or equal to |3|. Figure S9 and Table S18 describe the results of this analysis.
Population modeling and clustering
We used qpWave from ADMIXTOOLS (Patterson et al., 2012; Haak et al., 2015) in order to cluster individuals belonging to coherent archaeological contexts and investigate the evolution of the genetic structure of Southern France communities throughout the Neolithic. We used parameter allsnps: YES and a set of “right” populations comprising Mbuti.DG, Han.DG, Karitiana.DG, Papuan.DG, Ethiopia_4500BP_published.SG, CHG, Russia_Ust_Ishim_HG_published.DG, Czech_Vestonice, Russia_MA1_HG.SG, Israel_Natufian, Italy_North_Villabruna_HG, Jordan_PPNB_published, Russia_EBA_Yamnaya_Samara, Anatolia_N, European_HG. We based pairwise clustering on a p value>0.01 threshold as described in Fernandes et al. (2020). Below 0.01, clustering of individuals/groups was rejected. Results provided in Table S17 were used to create a heatmap using the heatmap3 R-package (Zhao et al., 2014; Warnes et al., 2014; Figures S12 and S13; R package version 2.4.1).
We used the 1240k SNPs panel to proceed to qpAdm modeling in order to estimate ancestry proportions in our samples and previously published ancient individuals with ADMIXTOOLS (Patterson et al., 2012; Haak et al., 2015; Harney et al., 2021; setting details: YES, allSNPs: YES). The following populations were used as outgroup “right” populations: Mbuti, Papuan, Han, Karitiana, Ust-Ishim, Malta, Ethiopia_4500BP, Vestonice, Israel_Raqefet_M_Natufian, Italy_North_Villabruna_HG. We included different outgroup populations depending on the model and populations tested as ancestral sources. Models results and details of individuals considered for each left and right populations are available in Tables 10, S11, and S12.
In a first model (Model 1) we tried to characterize the HG component detected in the EN farmers by testing different models with qpAdm using different HG sources that could have been encountered in Southern France as well as in the eastern part of the expansion route (KO1, La Brana, Loschbour). We tested Anatolia_N and European_HG as left populations, providing Anatolia farmers and European hunter-gatherers ancestry proportions estimates in each ancient individual considered, and testing whether individuals were best explained by both or either one of the two ancestries (Table S10, Figures 2B and 4).
In a second three-way model (Model 2) we tried to differentiate between different central and western HG contributions, respectively represented by KO1 and Loschbour-related ancestries in Neolithic individuals from France. We used “right” populations Mbuti.DG Papuan.DG Han.DG Karitiana.DG CHG Italy_North_Villabruna_HG Ethiopia_4500BP.SG Russia_Ust_Ishim.DG Czech_Vestonice Russia_MA1_HG.SG Israel_Raqefet_M_Natufian Jordan_PPNB_published Russia_EHG and “left” populations KO1, Anatolia_N and Loschbour. In several cases both Loschbour and KO1 were supported by the analysis, the results are detailed in Table S11.
In a third three-way model (Model 3), we aimed to test whether an excess of GoyetQ2-like ancestry could be detected in newly reported Neolithic individuals. We considered a model identical to the one described in Rivollat et al. (2020) (Model D). We used “right” populations Mbuti.DG, Papuan.DG, Han.DG, Karitiana.DG, CHG, Belgium_UP_GoyetQ116_1_published_all, Ethiopia_4500BP.SG, Hungary_KO1, Russia_Ust_Ishim.DG, Czech_Vestonice, Russia_MA1_HG.SG, Israel_Raqefet_M_Natufian, and “left” populations Anatolia_N, GoyetQ2, Villabruna (Table S12 and Figure S11).
Admixture dating
To estimate the dates of admixture events between Anatolia_N and European_HG components we used linkage disequilibrium (LD)-decay method DATES described in Narasimhan et al. (2019). We used the same ancestral populations defined as Anatolia_N and European_HG in our first qpAdm model.
The analysis models ancestry of admixed individual as a combined likelihood of ancestries from two sources. First, it calculates the global admixture proportions with the genotype vector of each target individual, described as a linear combination of the allele frequency vectors of the two ancestral source populations.
The second step is the estimation of local ancestry, to estimate whether a genomic block descends from one source or the other, by multiplying the genotype residual to the allele frequency difference between the two ancestral sources. The correlation between markers across an individual’s genome is expected to exponentially decay in accordance to genetic distance between markers and recombination processes and thus the number of generations since admixture. For each individual DATES (Narasimhan et al., 2019; Chintalapati et al., 2022) will estimate an admixture date, given in generations, by welding genetic distance and exponential decay.
To convert the estimated times of admixture given by DATES we assumed a number of 25, 28 and 30 years per generation (Fenner, 2005). Dates were estimated for both individuals and coherent groups to evaluate inter-individuals and inter-groups variability in admixture histories (See Table S15 and Figure S14).
Parameters binsize: 0.001; maxdis: 1; seed: 77; jackknife: YES; qbin: 10; runfit: YES; afffit: YES; lovalfit: 0.45 and minparentcount: 1 were used.
Admixture dates estimations are provided in Table S15 for individuals exhibiting clear admixture signal with qpAdm Model 1.
DATES were treated as irrelevant considering several arguments:
-
(1)
When the individual considered lacked indices of admixture (<5% European HG in qpAdm Model 1 or infeasible Model).
-
(2)
When aberrant estimation was provided by the software, that is for instance when a negative number of generations was estimated or when the estimated admixture date predated 10th millennium BCE.
-
(3)
When estimated std. Err was superior to the estimated number of generations.
Acknowledgments
We thank Lab technicians at MPI SHH for processing captures on Baume Bourbon samples in the aDNA lab facility of Jena, as well as Olivier Gorgé and the Molecular Biology team of the Département des Plateformes et Recherche Technologique at Institut de Recherche Biomédicale des Armées for processing samples for Next-generation sequencing. We are grateful to the genotoul bioinformatics platform Toulouse Occitanie (Bioinfo Genotoul, https://doi.org/10.15454/1.5572369328961167E12) for providing help, computing, and storage resources.
We thank Detelf Gronenborn for discussions regarding archaeological data and cultural background. We thank Etienne Patin at the Institut Pasteur for discussion and insights into computational analysis. The authors are grateful to Dominique Castex, Fanny Mendisco, Diego Lopez-Onaindia, Frédéric Santos, as well as other members from PACEA lab for discussions.
The authors thank the Service Régional de l’Archéologie Occitanie as well as the Institut National de Recherches Archéologiques Préventives through the PAS ANCESTRA (Coord. MP. Pruvost and F. Maziere) for granting access to archaeological material, as well as the CEDAD for processing samples for radiocarbon dating.
This project is a part of a PhD Research project carried out at the University of Bordeaux, supervised by Marie-France Deguilloux and Mélanie Pruvost and funded by the Ministère de l'Enseignement Supérieur et de la Recherche. Research was conducted in the PACEA lab (UMR 5199 CNRS). This project has received funding from the French National Research Agency (ANR) and Deutsche Forschungsgemeinschaft (DFG) under both the ANR project ANCESTRA, Grant ANR15-CE27-0001 coordinated by Mélanie Pruvost and the Franco-German Call in Humanities and Social Sciences project ANR-DFG INTERACT, grant ANR-17-FRAL-0010, DFG-HA-5407/4-1, 2018-2023, coordinated by Marie-France Deguilloux and Wolfgang Haak. This study received financial support from the French government in the framework of the University of Bordeaux’s IdEx “Investments for the Future” program/GPR “Human Past.”
Author contributions
A.A., M.F.D., and M.P. conceived the project and carried out research. D.B., F.C., H.D., M.G., and J.G. provided samples and/or information about archaeological contexts and cultural aspects. M.H.P, H.D.B., and M.P. carried out aDNA laboratory work with input from A.A. D.B. and M.P. supervised radiocarbon dating. A.A. carried out computational analysis with input from M.F.D., M.P., and M.R. A.A. wrote the paper and STAR Methods with input from D.B., W.H., M.F.D., M.P., and M.R as well as insights notably regarding archaeological background from all co-authors.
Declaration of interests
The authors declare no competing interests.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105387.
Contributor Information
Ana Arzelier, Email: ana.arzelier@u-bordeaux.fr.
Marie-France Deguilloux, Email: marie-france.deguilloux@u-bordeaux.fr.
Mélanie Pruvost, Email: melanie.pruvost@u-bordeaux.fr.
Supplemental information
(A) f3-outgroup test results of the form f3(Mbuti; Test, European HG), exploring the genetic affinity between Western Europe Neolithic individuals (Test) and European HG (La Brana, Loschbour, KO1) (B) outgroup-f3 test results of the form f3(Mbuti; EN_LBK_FR, Test) and (C) outgroup-f3 test results of the form f3(Mbuti; France_EN_Occitanie, Test) exploring the genetic affinities between Early Neolithicfarmers and subsequent Neolithic groups from France, related to STAR Methods.
Data and code availability
Raw sequence data and alignments have been deposited at the European Nucleotide Archive (ENA project) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ambert P., Genna A., Taffanel O. Contribution à l’étude du Chasséen du Minervois. Le Chasséen en Languedoc oriental, Préhistoire, Université Paul Valéry, n°. 1989;1:25–36. [Google Scholar]
- Antonio M.L., Gao Z., Moots H.M., Lucci M., Candilio F., Sawyer S., Pritchard J.K. Ancient Rome: a genetic crossroads of Europe and the Mediterranean. Science. 2019;366:708–714. doi: 10.1126/science.aay6826. PMID: 31699931; PMCID: PMC7093155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyneix A. Les sépultures cardiales et épicardiales de France méridionale. Bulletin de la Société préhistorique française. 1997;94-2:191–197. doi: 10.3406/bspf.1997.10877. [DOI] [Google Scholar]
- Binder D. Europe’s first farmers. 2000. Mesolithic and Neolithic interaction in southern France and northern Italy: new data and current hypotheses; pp. 117–143. [DOI] [Google Scholar]
- Binder D., Lanos P., Angeli L., Gomart L., Guilaine J., Manen C., Maggi R., Muntoni I.M., Panelli C., Radi G., et al. Modelling the earliest north-western dispersal of Mediterranean Impressed Wares: new dates and Bayesian chronological model. Doc. praeh. 2018;44:54–77. doi: 10.4312/dp.44.4. [DOI] [Google Scholar]
- Bonnery B. L'allée couverte mégalithique de Pépieux. Bulletin de la Société Scientifique, Historique et Archéologique de la Corrèze. 1991;113:31–46. [Google Scholar]
- Brace S., Diekmann Y., Booth T.J., van Dorp L., Faltyskova Z., Rohland N., Mallick S., Olalde I., Ferry M., Michel M., Barnes I. Ancient genomes indicate population replacement in Early Neolithic Britain. Nat. Ecol. Evol. 2019;3:765–771. doi: 10.1038/s41559-019-0871-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramanti B., Thomas M.G., Haak W., Unterlaender M., Jores P., Tambets K., Antanaitis-Jacobs I., Haidle M.N., Jankauskas R., Kind C.J., Burger J. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326:137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- Brunel S., Bennett E.A., Cardin L., Garraud D., Barrand Emam H., Beylier A., Boulestin B., Chenal F., Ciesielski E., Convertini F., Pruvost M. Ancient genomes from present-day France unveil 7, 000 years of its demographic history. Proc. Natl. Acad. Sci. USA. 2020;117:12791–12798. doi: 10.1073/pnas.1918034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy L.M., Maoldúin R.Ó., Kador T., Lynch A., Jones C., Woodman P.C., Murphy E., Ramsey G., Dowd M., Noonan A., Bradley D.G. A dynastic elite in monumental Neolithic society. Nature. 2020;582:384–388. doi: 10.1038/s41586-020-2378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapati M., Patterson N., Moorjani P. The spatiotemporal patterns of major human admixture events during the European Holocene. Elife. 2022;11 doi: 10.7554/eLife.77625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Convertini F., Georjon C. Archives d'Ecologie Préhistorique; 2018. Le Champ du Poste (Carcassonne, Aude). Une succession d'occupations du début du Néolithique moyen à l'âge du Bronze ancien. Catalogue des structures; p. 133. [Google Scholar]
- Coste A., Duday H., Gutherz X., Roudil J.L. Premières communautés paysannes en Méditerranée occidentale. 1987. Les sépultures de la baume Bourbon à Cabrières (Gard) pp. 531–535. [Google Scholar]
- Dabney J., Knapp M., Glocke I., Gansauge M.T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J.L., Meyer M. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard P.B., Margaryan A., Schroeder H., Orlando L., Willerslev E., Allentoft M.E. Improving access to endogenous DNA in ancient bones and teeth. Sci. Rep. 2015;5:11184. doi: 10.1038/srep11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duday H., Guilaine J. Deux sépultures à la grotte Gazel. (Les) Dossiers de l’Archéologie Dijon. 1980;44:88–89. [Google Scholar]
- Duday H. Anthropologie physique et archéologie: méthodes d’étude des sépultures: actes du colloque de Toulouse, 4, 5 et 6 novembre 1982. 1987. Organisation et fonctionnement d’une sépulture collective néolithique. L’aven de la Boucle à Corconne (Gard) pp. 89–104. [Google Scholar]
- Fenner J.N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- Fernandes D.M., Cheronet O., Gelabert P., Pinhasi R. TKGWV2: an ancient DNA relatedness pipeline for ultra-low coverage whole genome shotgun data. Sci. Rep. 2021;11:21262–21269. doi: 10.1038/s41598-021-00581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes D.M., Mittnik A., Olalde I., Lazaridis I., Cheronet O., Rohland N., Mallick S., Bernardos R., Broomandkhoshbacht N., Carlsson J., et al. The spread of steppe and Iranian-related ancestry in the islands of the western Mediterranean. Nat. Ecol. Evol. 2020;4:334–345. doi: 10.1038/s41559-020-1102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregel R., Méndez F.L., Bokbot Y., Martín-Socas D., Camalich-Massieu M.D., Santana J., Morales J., Ávila-Arcos M.C., Underhill P.A., Shapiro B., et al. Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6774–6779. doi: 10.1073/pnas.1800851115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q., Mittnik A., Johnson P.L.F., Bos K., Lari M., Bollongino R., Sun C., Giemsch L., Schmitz R., Burger J., Krause J. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtwängler A., Rohrlach A.B., Lamnidis T.C., Papac L., Neumann G.U., Siebke I., Reiter E., Steuri N., Hald J., Denaire A., Krause J. Ancient genomes reveal social and genetic structure of Late Neolithic Switzerland. Nat. Commun. 2020;11:1915. doi: 10.1038/s41467-020-15560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba C., Jones E.R., Teasdale M.D., McLaughlin R.L., Gonzalez-Fortes G., Mattiangeli V., Domboróczki L., Kővári I., Pap I., Anders A., Pinhasi R. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 2014;5:5257–5259. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginolhac A., Rasmussen M., Gilbert M.T.P., Willerslev E., Orlando L. mapDamage: testing for damage patterns in ancient DNA sequences. Bioinformatics. 2011;27:2153–2155. doi: 10.1093/bioinformatics/btr347. [DOI] [PubMed] [Google Scholar]
- Gorgé O., Bennett E.A., Massilani D., Daligault J., Pruvost M., Geigl E.M., Grange T. Microbial Environmental Genomics (MEG) Humana Press; 2016. Analysis of ancient DNA in microbial ecology; pp. 289–315. [DOI] [PubMed] [Google Scholar]
- Guilaine J. Privat, FeniXX; 1998. Au temps des dolmens: mégalithes et vie quotidienne en France méditerranéenne il y a 5000 ans; p. 315. [Google Scholar]
- Guilaine J. La diffusion de l’agriculture en Europe: une hypothèse arythmique. La difusión de L’agricultura en Europa: una hipótesis aritmética. Zephyrus. 2001;53:54. [Google Scholar]
- Guilaine J. The widening harvest. The Neolithic transition in Europe: Looking back, looking forward. 2003. Aspects de la Néolithisation en Méditerranée et en France; pp. 189–206. [Google Scholar]
- Guilaine J., Manen C. In: Pont de Roque-Haute. Nouveaux regards sur la Néolithisation de la France méditerranéenne, Toulouse. Guilaine J., Manen et C., Vigne J.-D., editors. Archives d'écologie préhistorique; 2007. Du Mésolithique au Néolithique en Méditerranée de l'Ouest: aspects culturels; pp. 303–322. [Google Scholar]
- Guilaine J. A personal view of the neolithisation of the Western Mediterranean. Quat. Int. 2018;470:211–225. doi: 10.1016/j.quaint.2017.06.019. [DOI] [Google Scholar]
- Guilaine J. In: Multas per Gentes et Multa per Secula, Mélanges J.K. Kozlowski. ValdeNowak P., Sobczyk K., Nowak M., Zralka J., editors. Krakow; 2018. A new hypothesis on the emergence of the early neolithic cardial culture; pp. 295–300. [Google Scholar]
- Günther T., Jakobsson M. Handbook of Statistical Genomics: Two Volume Set. 2019. Population genomic analyses of DNA from ancient remains; pp. 295–340. [DOI] [Google Scholar]
- Gutherz X. Unknown; 1984. Les cultures du Néolithique récent et final en Languedoc oriental (Doctoral dissertation) [Google Scholar]
- Haak W., Lazaridis I., Patterson N., Rohland N., Mallick S., Llamas B., Brandt G., Nordenfelt S., Harney E., Stewardson K., Reich D. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney É., Patterson N., Reich D., Wakeley J. Assessing the performance of qpAdm: a statistical tool for studying population admixture. Genetics. 2021;217:iyaa045. doi: 10.1093/genetics/iyaa045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmanová Z., Kreutzer S., Hellenthal G., Sell C., Diekmann Y., Díez-Del-Molino D., van Dorp L., López S., Kousathanas A., Link V., Burger J. Early farmers from across Europe directly descended from neolithic aegeans. Proc. Natl. Acad. Sci. USA. 2016;113:6886–6891. doi: 10.1073/pnas.152395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immel A., Pierini F., Rinne C., Meadows J., Barquera R., Szolek A., Susat J., Böhme L., Dose J., Bonczarowska J., Krause-Kyora B. Genome-wide study of a Neolithic Wartberg grave community reveals distinct HLA variation and hunter-gatherer ancestry. Commun. Biol. 2021;4:113. doi: 10.1038/s42003-020-01627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallet F., Duday H., Cours S. Vol. 1. Société Préhistorique Française; 2010. Néolithique récent et Néolithique final de l’aven de la Bouche (Corconne, Gard), regards d’archéologues; pp. 243–256. (Transitions, Ruptures et Continuité en Préhistoire: XXVIIème Congrès Préhistorique de France, Bordeaux-Les Eyzies 31 mai-5 juin 2010). [Google Scholar]
- Jedikian G., Vaquer J. Il declino del mondo neolitico: ricerche in Italia centro-settentrionale fra aspetti peninsulari, occidentali e nord-alpini: atti del Convegno. 2001. Repères pour les changements culturels et sociaux dans le Néolithique du Midi de la France au IVème millénaire avant J.-C; pp. 5–7. [Google Scholar]
- Katoh K., Misawa K., Kuma K.I., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M. Ancient DNA. Humana Press; 2012. Analysis of high-throughput ancient DNA sequencing data; pp. 197–228. [DOI] [PubMed] [Google Scholar]
- Korneliussen T.S., Albrechtsen A., Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinf. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I., Nadel D., Rollefson G., Merrett D.C., Rohland N., Mallick S., Fernandes D., Novak M., Gamarra B., Sirak K., Reich D. Genomic insights into the origin of farming in the ancient Near East. Nature. 2016;536:419–424. doi: 10.1038/nature19310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I., Mittnik A., Patterson N., Mallick S., Rohland N., Pfrengle S., Furtwängler A., Peltzer A., Posth C., Vasilakis A., Stamatoyannopoulos G. Genetic origins of the minoans and mycenaeans. Nature. 2017;548:214–218. doi: 10.1038/nature23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras-Goude G. Doctoral dissertation, Université Sciences et Technologies-Bordeaux I; Universität Leipzig); 2007. Étude des modes de subsistance de populations néolithiques (VIe-IVe millénaires av. J.-C.) dans le nord-ouest de la Méditerranée. Approche par l'utilisation des isotopes stables (d13C et d15N) du collagène; p. 416. [Google Scholar]
- Leppard T.P. Process and dynamics of Mediterranean neolithization (7000–5500 bc) J. Archaeol. Res. 2021;30:231–283. doi: 10.1007/s10814-021-09161-5. [DOI] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M., Szécsényi-Nagy A., Mallick S., Pósa A., Stégmár B., Keerl V., Rohland N., Stewardson K., Ferry M., Michel M., Reich D. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 2017;551:368–372. doi: 10.1038/nature24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loison G., Schmitt A. Diversité des pratiques funéraires et espaces sépulcraux sectorisés au Chasséen ancien sur le site du Crès à Béziers (Hérault): croisements de données archéologiques et anthropologiques. Gall. Prehist. 2009;51:245–272. doi: 10.3406/galip.2009.2480. [DOI] [Google Scholar]
- Lösch S., Siebke I.K.E., Furtwängler A., Steuri N., Hafner A., Ramstein M., Krause J. 89th Annual Meeting of the American Association of Physical Anthropologists. 2020. Bioarchaeological analysis of Late Neolithic inhumations from a dolmen in Switzerland. [Google Scholar]
- Manen C., Perrin T., Guilaine J., Bouby L., Bréhard S., Briois F., Durand F., Marinval P., Vigne J.D. The Neolithic transition in the western Mediterranean: a complex and non-linear diffusion process—the radiocarbon record revisited. Radiocarbon. 2019;61:531–571. doi: 10.1017/RDC.2018.98. [DOI] [Google Scholar]
- Marcus J.H., Posth C., Ringbauer H., Lai L., Skeates R., Sidore C., Beckett J., Furtwängler A., Olivieri A., Chiang C.W.K., Novembre J. Genetic history from the Middle neolithic to present on the Mediterranean island of Sardinia. Nat. Commun. 2020;11:939. doi: 10.1038/s41467-020-14523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic T., Whitten M., Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I., Lazaridis I., Rohland N., Mallick S., Patterson N., Roodenberg S.A., Harney E., Stewardson K., Fernandes D., Novak M., Reich D. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I., Alpaslan-Roodenberg S., Posth C., Szécsényi-Nagy A., Rohland N., Mallick S., Olalde I., Broomandkhoshbacht N., Candilio F., Cheronet O., Reich D. The genomic history of southeastern Europe. Nature. 2018;555:197–203. doi: 10.1038/nature25778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Mittnik A., Wang C.C., Pfrengle S., Daubaras M., Zariņa G., Hallgren F., Allmäe R., Khartanovich V., Moiseyev V., Tõrv M., Krause J. The genetic prehistory of the Baltic Sea region. Nat. Commun. 2018;9:442. doi: 10.1038/s41467-018-02825-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy Kuhn J.M., Jakobsson M., Günther T. Estimating genetic kin relationships in prehistoric populations. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan V.M., Patterson N., Moorjani P., Rohland N., Bernardos R., Mallick S., Lazaridis I., Nakatsuka N., Olalde I., Lipson M., Reich D. The formation of human populations in South and Central Asia. Science. 2019;365:eaat7487. doi: 10.1126/science.aat7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordqvist K., Kriiska A. The Dąbki Site in Pomerania and the Neolithisation of the North European Lowlands (c. 5000–3000 calBC) Vol. 8. 2015. Towards Neolithisation: the Mesolithic-Neolithic transition in the central area of the eastern part of the Baltic Sea; pp. 537–556. [Google Scholar]
- Olalde I., Brace S., Allentoft M.E., Armit I., Kristiansen K., Booth T., Rohland N., Mallick S., Szécsényi-Nagy A., Mittnik A., Reich D. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 2018;555:190–196. doi: 10.1038/nature25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalde I., Mallick S., Patterson N., Rohland N., Villalba-Mouco V., Silva M., Dulias K., Edwards C.J., Gandini F., Pala M., Reich D. The genomic history of the Iberian Peninsula over the past 8000 years. Science. 2019;363:1230–1234. doi: 10.1126/science.aav4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalde I., Schroeder H., Sandoval-Velasco M., Vinner L., Lobón I., Ramirez O., Civit S., García Borja P., Salazar-García D.C., Talamo S., et al. A Common Genetic Origin for Early Farmers from Mediterranean Cardial and Central European LBK Cultures. Mol. Biol. Evol. 2015;32:3132–3142. doi: 10.1093/molbev/msv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papac L., Ernée M., Dobeš M., Langová M., Rohrlach A.B., Aron F., Neumann G.U., Spyrou M.A., Rohland N., Velemínský P., Haak W. Dynamic changes in genomic and social structures in third millennium BCE central Europe. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abi6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin T. In: Transitions, ruptures et continuité durant la Préhistoire, actes du XXVIIe Congrès préhistorique de France (Bordeaux - Les Eyzies, 2010) Perrin T., Manen C., Marchand G., Allard P., Binder D., Ilett M., editors. Société préhistorique française; 2010. Potentialités de contacts entre mésolithiques et néolithiques dans le sud de la France; pp. 357–372. [Google Scholar]
- Perrin T., Manen C. Potential interactions between Mesolithic hunter-gatherers and Neolithic farmers in the Western Mediterranean: the geochronological data revisited. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltzer A., Jäger G., Herbig A., Seitz A., Kniep C., Krause J., Nieselt K. EAGER: efficient ancient genome reconstruction. Genome Biol. 2016;17:60. doi: 10.1186/s13059-016-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R., Fernandes D., Sirak K., Novak M., Connell S., Alpaslan-Roodenberg S., Gerritsen F., Moiseyev V., Gromov A., Raczky P., Hofreiter M. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf A., Montiel González D., Zhong K., Kayser M. Yleaf: software for human Y-chromosomal haplogroup inference from next-generation sequencing data. Mol. Biol. Evol. 2018;35:1291–1294. doi: 10.1093/molbev/msy032. [DOI] [PubMed] [Google Scholar]
- Rigaud S., Manen C., García-Martínez de Lagrán I. Symbols in motion: flexible cultural boundaries and the fast spread of the Neolithic in the western Mediterranean. PLoS One. 2018;13 doi: 10.1371/journal.pone.0196488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringbauer H., Novembre J., Steinrücken M. Parental relatedness through time revealed by runs of homozygosity in ancient DNA. Nat. Commun. 2021;12:5425. doi: 10.1038/s41467-021-25289-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivollat M., Jeong C., Schiffels S., Küçükkalıpçı İ., Pemonge M.H., Rohrlach A.B., Alt K.W., Binder D., Friederich S., Ghesquière E., Haak W. Ancient genome-wide DNA from France highlights the complexity of interactions between Mesolithic hunter-gatherers and Neolithic farmers. Sci. Adv. 2020;6:eaaz5344. doi: 10.1126/sciadv.aaz5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb J. Material culture, landscapes of action, and emergent causation: a new model for the origins of the European Neolithic. Curr. Anthropol. 2013;54:657–683. doi: 10.1086/673859. [DOI] [Google Scholar]
- Roberts C.N., Woodbridge J., Palmisano A., Bevan A., Fyfe R., Shennan S. Mediterranean landscape change during the Holocene: synthesis, comparison and regional trends in population, land cover and climate. Holocene. 2019;29:923–937. doi: 10.1177/0959683619826697. [DOI] [Google Scholar]
- Rohland N., Harney E., Mallick S., Nordenfelt S., Reich D. Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2013.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrlach A.B., Papac L., Childebayeva A., Rivollat M., Villalba-Mouco V., Neumann G.U., Penske S., Skourtanioti E., van de Loosdrecht M., Akar M., Haak W. Using Y-chromosome capture enrichment to resolve haplogroup H2 shows new evidence for a two-Path Neolithic expansion to Western Europe. Sci. Rep. 2021;11:15005. doi: 10.1038/s41598-021-94491-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley-Conwy P. Westward Ho! The spread of agriculture from central Europe to the atlantic. Curr. Anthropol. 2011;52:S431–S451. doi: 10.1086/658368. [DOI] [Google Scholar]
- Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin-Orlando A., Donat R., Der Sarkissian C., Southon J., Thèves C., Manen C., Tchérémissinoff Y., Crubézy E., Shapiro B., Deleuze J.F., Orlando L. Heterogeneous hunter-gatherer and steppe-related ancestries in late neolithic and Bell beaker genomes from present-day France. Curr. Biol. 2021;31:1072–1083.e10. doi: 10.1016/j.cub.2020.12.015. [DOI] [PubMed] [Google Scholar]
- Skoglund P., Malmström H., Omrak A., Raghavan M., Valdiosera C., Günther T., Hall P., Tambets K., Parik J., Sjögren K.G., Jakobsson M. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344:747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- Skoglund P., Storå J., Götherström A., Jakobsson M. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 2013;40:4477–4482. doi: 10.1016/j.jas.2013.07.004. [DOI] [Google Scholar]
- Szécsényi-Nagy A., Roth C., Brandt G., Rihuete-Herrada C., Tejedor-Rodríguez C., Held P., García-Martínez-de-Lagrán Í., Arcusa Magallón H., Zesch S., Knipper C., et al. The maternal genetic make-up of the Iberian Peninsula between the Neolithic and the Early Bronze Age. Sci Rep. 2017;7:15644. doi: 10.1038/s41598-017-15480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrête J., Le Roux C.T. Revue archéologique de l'Ouest; 2008. J. Tarrête et C.-T. Le Roux (dir.), 2008–Archéologie de la France. Le Néolithique; p. 393. [Google Scholar]
- Valdiosera C., Günther T., Vera-Rodríguez J.C., Ureña I., Iriarte E., Rodríguez-Varela R., Simões L.G., Martínez-Sánchez R.M., Svensson E.M., Malmström H., Jakobsson M. Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc. Natl. Acad. Sci. USA. 2018;115:3428–3433. doi: 10.1073/pnas.1717762115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquer J. In: Autour de Jean Arnal, Montpellier, Recherches sur les premières communautés paysannes en Méditerranée occidentale. Guilaine J., Gutherz X., editors. Laboratoire de Paleobotanique. Université des Sciences et techniques du Languedoc; 1990. - L'évolution du Chasséen méridional: essai dans le bassin de l'Aude; pp. 177–190. [Google Scholar]
- Vaquer J. In: Beeching A., Binder D., Blanchet J.-C., et al., editors. Vol. 4. 1991. Aspects du Chasséen en Languedoc oriental. Habitat et culture matérielle; pp. 27–37. (Identité du chasséen, Nemours, APRAIF (Mémoires du Musée de Préhistoire d'Ile de France). [Google Scholar]
- Vaquer J. In: Sépultures d’occident et genèse des mégalithismes (9500–3500) Guilaine J., editor. Errance; 1998. Les sépultures du Néolithique moyen en France méditerranéenne; pp. 167–186. [Google Scholar]
- Villalba-Mouco V., van de Loosdrecht M.S., Posth C., Mora R., Martínez-Moreno J., Rojo-Guerra M., Salazar-García D.C., Royo-Guillén J.I., Kunst M., Rougier H., Haak W. Survival of late pleistocene hunter-gatherer ancestry in the Iberian peninsula. Curr. Biol. 2019;29:1169–1177.e7. doi: 10.1016/j.cub.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Weissensteiner H., Pacher D., Kloss-Brandstätter A., Forer L., Specht G., Bandelt H.-J., Kronenberg F., Salas A., Schönherr S. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–W63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes G.R., Bolker B., Gorjanc G., Grothendieck G., Korosec A., Lumley T., Rogers J. gdata: various R programming tools for data manipulation. R package version. 2014;2:35. [Google Scholar]
- Zemour A. Université Nice Sophia Antipolis & Università di Roma - La Sapienza; 2013. Gestes, espaces et temps funéraires au début du Néolithique (6e millénaire et 1ère moitié du 5e millénaire cal-BC) en Italie et en France méridionale. Reconnaissance des témoins archéologiques de l'après-mort; p. 1115. [Google Scholar]
- Zhao S., Guo Y., Sheng Q., Shyr Y. Heatmap3: an improved heatmap package with more powerful and convenient features. BMC Bioinf. 2014;15:166. doi: 10.1186/1471-2105-15-S10-P16. [DOI] [Google Scholar]
- Zilhão J. The spread of agro-pastoral economies across Mediterranean Europe: a view from the far west. J. Mediterr. Archaeol. 1993;6:5–63. doi: 10.1558/jmea.v6i1.5. [DOI] [Google Scholar]
- Zilhão J. In: Europe’s First Farmers. Price T.D., editor. Cambridge University Press; 2000. From the mesolithic to the neolithic in the Iberian Peninsula; pp. 144–182. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) f3-outgroup test results of the form f3(Mbuti; Test, European HG), exploring the genetic affinity between Western Europe Neolithic individuals (Test) and European HG (La Brana, Loschbour, KO1) (B) outgroup-f3 test results of the form f3(Mbuti; EN_LBK_FR, Test) and (C) outgroup-f3 test results of the form f3(Mbuti; France_EN_Occitanie, Test) exploring the genetic affinities between Early Neolithicfarmers and subsequent Neolithic groups from France, related to STAR Methods.
Data Availability Statement
Raw sequence data and alignments have been deposited at the European Nucleotide Archive (ENA project) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.