Summary
Background
The E2F family of transcription factors play a crucial role in the development of various cancers. However, E2F members lack targetable binding pockets and are typically considered “undruggable”. Unlike canonical small-molecule therapeutics, molecular glues mediate new E3 ligase–protein interactions to induce selective proteasomal degradation, which represents an attractive option to overcome these limitations.
Methods
Human proteome microarray was utilized to identify a natural product-derived molecular glue for targeting E2F2 degradation. Co-IP analysis with stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative proteomics was carried out to further explore the E3 ligase for E2F2 degradation.
Findings
In this study, we identified a molecular glue bufalin, which significantly promoted E2F2 degradation. Unexpectedly, E2F2 underwent ubiquitination and proteasomal degradation via a previously undisclosed atypical E3 ligase, zinc finger protein 91 (ZFP91). In particular, we observed that bufalin markedly promoted E2F2-ZFP91 complex formation, thereby leading to E2F2 polyubiquitination via K48-linked ubiquitin chains for degradation. E2F2 degradation subsequently caused transcriptional suppression of multiple oncogenes including c-Myc, CCNE1, CCNE2, MCM5 and CDK1, and inhibited hepatocellular carcinoma growth in vitro and in vivo.
Interpretation
Collectively, our findings open up a new direction for transcription factors degradation by targeting atypical E3 ligase ZFP91. Meanwhile, the chemical knockdown strategy with molecular glue may promote innovative transcription factor degrader development in cancer therapy.
Funding
This work was financially supported by the National Key Research and Development Project of China (2022YFC3501601), National Natural Sciences Foundation of China (81973505, 82174008, 82030114), and China Postdoctoral Science Foundation (2019M650396), the Fundamental Research Funds for the Central Universities.
Keywords: E2F2 transcription factor, Proteasomal degradation, E3 ligase ZFP91, Molecular glue, Cancer therapy
Research in context.
Evidence before this study
E2F2 functions as a central transcription factor that is generally upregulated in advanced hepatocellular carcinoma, and the patients with a high expression of E2F2 have poor overall survival. However, available E2F2 targeting strategy for cancer therapy and drug development still remains largely unexplored.
Added value of this study
Molecular glue-based degradation of undruggable proteins, especially transcription factors, can serve as an unprecedented approach for E2F2 degradation. Our findings identify a natural-derived small-molecule bufalin, which promotes rapid degradation of E2F2 to inhibit hepatocellular carcinoma. Moreover, ZFP91 as an atypical E3 ligase is found to contribute to E2F2 degradation via the ubiquitin-proteasomal pathway.
Implications of all the available evidence
Our study provides that E2F2 serves as a targetable transcription factor for anti-cancer therapy with molecular glue bufalin. Meanwhile, the discovery of ZFP91 as a new E3 ligase for E2F2 degradation opens promising research opportunities into clinical agent development specially targeting transcription factors.
Introduction
The E2 factor (E2F) family of transcription factors perform key regulatory functions in the transcriptional control of cancer gene networks essential for cell cycle progression, proliferation, apoptosis, and differentiation.1,2 E2F2 is a crucial member of the E2F family, and its abnormal expression has been highly correlated with poor prognosis in various cancers,3, 4, 5, 6 which suggests a promising value of anti-cancer targets. For example, the E2F2 level was generally upregulated in advanced hepatocellular carcinoma, and the patients with a high expression of E2F2 have poor overall survival.7 Moreover, E2F2 expression is also positively correlated with the transcription in oncogenic pathway and immune cell infiltration during hepatocellular carcinoma development.8 However, as a transcription factor, E2F2 lacks a canonical ligand binding domain, and has been traditionally considered as 'undruggable' for therapeutics. Therefore, discovery of potential therapeutic strategy that selectively inhibit E2F2 function have attracted wide attention.
Molecular glues are a class of small-molecules that can induce protein–protein interactions in order to drive the degradation of previously undruggable proteins.9, 10, 11 Mechanistically, molecular glues promote protein–protein associations by enabling direct interaction between a disease-causative protein and components of the ubiquitin-proteasome system, thereby leading to selective degradation of the target. Unlike traditional enzyme inhibitors, molecular glues catalyze the rapid and substoichiometric degradation of previously inaccessible proteins through a proximity-induced interaction between the target protein and E3 ligases. Since molecular glues can exploit the protein–protein interactions required for a broad spectrum of biological functions, they have remarkable potential for therapeutic development against several human cancers, such as leukemia or colorectal cancer.12,13 However, up to now, the report of molecular glues with selective E2F2-targeting properties is still missing.
In order to explore E2F2 degradation strategy, in the present work, we identified small-molecule bufalin as a molecular glue to potently induce E2F2 degradation. Unexpectedly, we discovered a previously undisclosed atypical E3 ligase ZFP91 that recruited E2F2 to promote its proteasomal degradation, which is quite different from currently reported von Hippel-Lindau (VHL), SCFβ-TrCP, IAPs, and cereblon (CRBN) proteins. In particular, Cys349 was identified as a crucial covalent binding site for targeting ZFP91 with bufalin. Meanwhile, the formation of E2F2-ZFP91-bufalin trimer complex was observed in live cells, which induced significant inhibition of hepatocellular carcinoma growth in vitro and in vivo. Collectively, this study reveals ZFP91 as a distinct atypical E3 ligase for selective degrading E2F2, and also provides a proof-of-concept of molecular glue-based E2F2 targeting strategy for cancer therapy.
Methods
Cell lines
Human hepatoma cell lines HepG2 (Clone E6-1, TIB-152, derived from male) and human embryonic kidney 293T cells (HEK 293T) (CRL-2974, derived from female) was obtained from Peking union medical college, cell bank, China. Both cell lines were cultured in high glucose Dulbecco's Modified Eagle Medium (DMEM) (Corning, NY, USA) supplemented with 10% FBS, 1% sodium pyruvate, and 1% penicillin/streptomycin, 25 mM HEPES. Cells were grown at 37 °C in a humidified 5% CO2 atmosphere. All of the cells used were authenticated by short tandem repeat (STR) profiling and free of mycoplasma contamination before the experiments.
Compound preparation
Bufalin (>98% purity; Baoji Herbest Bio-Tech, Baoji, Shanxi, China) was dissolved in dimethylsulfoxide (DMSO) as stock solution at 10 mM and stored at −20 °C. Bufalin stock solution was freshly diluted with medium to the final concentration before each experiment in vitro. The final DMSO concentration did not exceed 0.1%.
Antibodies
The anti-E2F2 antibody (Cat# ab235837, RRID:AB_1140040) was obtained from Abcam (Cambridge, UK). Antibodies against GAPDH (Cat# 8884, RRID:AB_11129865), His (Cat# 9991, RRID:AB_2797714), Myc (Cat# 2276, RRID:AB_331783), Ubi (Cat# 3933, RRID:AB_2180538), HA (Cat# 3724, RRID:AB_1549585), CD4 (Cat# 25229, RRID:AB_2798898), CD8 (Cat# 98941, RRID:AB_2756376) and Flag (Cat# 14793, RRID:AB_2572291) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-C7orF31 (Cat# sc-515544) and anti-KCNAB1 (Cat# sc-377099) antibodies were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Anti-SDC4 (Cat# 11820-1-AP, RRID:AB_2877797) and anti-TRIM25 (Cat# 12573-1-AP, RRID:AB_2209732) antibodies were purchased from Proteintech (Chicago, IL, USA). Anti-ZFP91 antibody (Cat# EM1708-31) was obtained from HuaBio (Hangzhou, China) and anti-ZFP91 antibody (Cat# YT5823) was purchased from Immunoway (Plano, TX, USA). All antibodies were used in Western blot analysis at a dilution of 1:1000 unless otherwise specified.
SILAC media preparation and cell culture conditions
All standard stable isotope labeling with amino acids in cell culture (SILAC) media preparation and labeling steps were followed as previously described.14 Briefly, base media for DMEM (Macgene) was divided into two parts and to each added L-arginine (Arg0) and L-lysine (Lys0) (light) or 13C6 15N4-L-arginine (Arg10) and 13C6 15N2-L-Lysine (Lys8) (heavy) to generate the two SILAC labeling mediums. Each media with the full complement of amino acids at the standard concentration, was sterile filtered through a 0.22 μm filter (Milipore, Bedford, MA). Cells were grown in the corresponding labeling media, prepared as described above, supplemented with 2 mM l-glutamine (Gibco), and 10% dialyzed fetal bovine serum (Sigma) plus antibiotics (Gibco), in a humidified atmosphere with 5% CO2 at 37 °C. Cells were cultured in labeling media for at least six cell divisions.
Cell viability in 2D-adherent monolayers
MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed to measure the cell viability of monolayer cultured cells. Cells were seeded into 96-well plates at 5 × 103 cells/well and cultured under indicated treatment. MTT substrate (Sigma Aldrich) was added into each well and incubated for 4 h followed by careful removal of medium and addition of 100 μL of dimethylsulfoxide (DMSO). The plates were read with the absorbance at 570 nm.
Ultra-low adherent (ULA) 3D-spheroids assay
ULA 3D-spheroids assay was implemented to evaluate the cell viabilities as previously described.15 In brief, cells were plated in ULA 96-well plates (Corning, NY, USA) at a density of 3000 cells per well in 100 μL media and allowed to adhere or form spheroids overnight. After treatment with DMSO or bufalin at the indicated concentrations, cell viability was evaluated using CellTiter-Glo (Promega, no. G7570) and Viability/Cytotoxicity Assay Kit for Animal Live & Dead Cells (Calcein AM, EthD-1 Method). To normalize the differences of plating density, Day 0 plate of cells was inoculated at the beginning of the experiment and tested by CellTiter-Glo the day after plating. The final absorbance was then divided by the average Day 0 reading to normalize according to initial plating density.
Gene knockdown
The target siRNA sequences against ZFP91 (siZFP91, 5′-GAACUCAGAUAUACUCGGUTT -3′) and the scrambled siRNA (siControl) were synthesized by Gene Pharma (Shanghai, China). Cells were transfected with 100 nM siRNA using lipofectamine RNAiMAX (Invitrogen) in Opti-MEM medium according to the manufacturer's protocols. The siRNA-transfected cells were used for subsequent experiments after 48 h. Knockdown efficiency of siRNA was verified by western blotting.
Plasmids and transfections
The complete coding region of human E2F2 and ZFP91 cDNA was amplified by PCR. To generate E2F2 and ZFP91 plasmids for mammalian expression, the cDNA was subcloned into pcDNA3.1(+) containing a His tag sequence and pCMV-Tag2B containing a Flag tag at the N-terminal region, respectively. E2F2 deletion mutants were generated by standard molecular biology techniques. To create ZFP91 ubiqutin ligase-EGFP-HOTag3 fusions, the target protein was cloned into pcDNA3 containing EGFP. HOTag3 was then cloned into the pcDNA3 ZFP91 ligase-EGFP construct, resulting in pcDNA3 ZFP91 ligase-EGFP-HOTag3. Similar procedures were carried out to produce pcDNA3 E2F2 (125–310 amino acids) -EGFP-HOTag6. Myc-Ub and Myc-UbK48R expression constructs were obtained from Biogot technology (Nanjing, Jiangsu, China). HEK293T cells were transfected with the above plasmids using polyethylenimine (PEI) in Opti-MEM medium (Invitrogen).
Co-immunoprecipitation (Co-IP) assay
HEK293T cells, maintained in growth medium (SILAC) DMEM, were transiently transfected with His-E2F2 plasmid DNA and subjected to bufalin treatment as indicated. After washed twice with phosphate-buffered saline (PBS), cells were lysed in buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride and complete protease inhibitor cocktail (Roche) for 30 min. Cell lysates were centrifuged and supernatant was precleared by protein G agarose beads (Cell Signaling Technology), followed by incubation with Ni-beads (Cell Signaling Technology) for 4 h at 4 °C. The bound proteins were identified by mass spectrometry and subsequently confirmed by immunoblotting with indicated antibodies.
RNA isolation and quantitative RT-PCR
Total RNA was extracted using EasyPure RNA Kit (TransGen Biotech). The mRNA was reverse transcribed into cDNA by TransScript First-Strand cDNA Synthesis Super Mix (TransGen Biotech), according to manufacturer's protocol. 20 ng of total cDNA were subjected to real-time quantitative polymerase chain reaction using SYBR Green PCR Super Mix (TransGen Biotech). The Real-Time PCR amplification was then performed as 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s on Agilent Technologies Stratagene Mx3005P (CA, USA). GAPDH mRNAs were used as loading control to normalize mRNA expression. The threshold cycle (CT) values were provided at the end of PCR. The relative transcriptional level of target genes normalized to GAPDH was calculated by the comparative 2−ΔΔCT. Primer sequences are listed in Key Resources Table.
In vitro ubiquitination assay
For ubiquitination of E2F2 proteins in vitro, HEK293T cells were transfected with expression vectors for the individual His-E2F2 protein and Flag-ZFP91. The transfected cells were lysed and pre-cleared with protein A beads prior to incubation with Ni-beads for 4 h at 4 °C. Beads were washed twice with buffer B, twice with buffer A (25 mM Tris–HCl (pH 7.5), 10% (v/v) glycerol, 1 mM EDTA, 0.01% NP-40 and 0.1 M NaCl), and twice with reaction buffer (50 mM Tris–HCl (pH 7.5), 5 Mm MgCl2, 2 mM NaF, and 0.6 mM DTT). The pellets were incubated with ubiquitin (1 μg), E1 (50 nM), UbcH13/Mms2 (1 μg) and ATP (2 mM) in 1 × reaction buffer containing bufalin at indicated concentrations or a DMSO control for 1 h at 37 °C. Ubiquitin, E1, and UbcH13/Mms2 were purchased from Boston Biochem (MA, USA). The Ni-beads were centrifuged and resuspended in 2% SDS, 150 mM NaCl, 10 mM Tris–HCl (pH 8.0) and 1 mM DTT and boiled for 5 min to release bound proteins. Protein samples were subjected to immunoblot analysis with anti-ubiquitin antibodies.
Western blotting
Whole cell lysates were prepared using RIPA lysis buffer containing complete protease inhibitor cocktail (Roche), homogenized and centrifuged at 12,000×g for 15 min at 4 °C. Protein concentration of cell lysates was determined by BCA protein assay reagent (TransGen). Cell lysates were incubated in SDS-PAGE sample loading buffer for 10 min at 98 °C, separated by 8%–12% SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membrane (Millipore), and incubated with primary antibodies specific for target proteins and horseradish peroxidase (HRP)-conjugated secondary antibodies. Super Signal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) was used for detection of protein of interest.
Cellular thermal shift assay (CETSA)
Cellular thermal shift assay was conducted as described previously.16,17 Briefly, Cells were treated with DMSO or 2 μM bufalin for 2 h, and then cells were collected and resuspended in PBS. Each aliquot were heated at indicated temperatures for 3 min on PCR instrument (Bio-Rad, CA, USA) and frozen twice in liquid nitrogen. The samples were centrifuged and the supernatants were subjected to immunoblotting analysis.
Pull-down assay
Cells were transfected with or without HA-E2F2, HA-E2F2 (1–310), HA-E2F2 (125–310), HA-E2F2 (125–438) plasmid DNA. After treatments, cells were washed with PBS and lysed for 1 h in the buffer (Tris–HCl at pH 8.0 12 mM; Glycerol 10%; NaCl 137 mM; EDTA 1 mM) with protease inhibitor cocktail (Roche) in the presence of 1% NP40. Cell lysates were then spun at 12,000×g for 10 min at 4 °C. Biotin-bufalin was coupled with streptavidin magnetic beads (Thermo) for 2 h at room temperature. 500 μg of cell lysate at 1 mg/mL was then used to incubate with the vehicle and bufalin-coupled beads in the absence or presence of bufalin (or α-pyrone, androsterone) cell lysates overnight at 4 °C. Agarose was then spun and washed for 6 times. Binding proteins were eluted by 2 × SDS-PAGE loading buffer and detected by western blotting.
SPPIER analysis
HEK293T cells were grown on laser confocal petri dishes. Cells were transiently transfected with plasmids using PEI and imaged 24 h after transfection. Time-lapse imaging was performed in an incubation chamber at 37 °C and at internal CO2 level of 5%. Chemical compounds were added to the incubation chamber as the time-lapse imaging was started. Fluorescence images were acquired every 6 min for a total of 1 h. Images were acquired and processed by the Zen-2 microscope imaging software v.14.0.18.201 (Zeiss).
Human proteome microarray fabrication analysis
Human proteome microarray fabrication analysis was carried out as previously described.18,19 In brief, proteome microarrays were blocked with blocking buffer (1% BSA in 0.1% Tween 20; TBST) for 1 h. Biotin-bufalin was diluted to 10 μM in blocking buffer and incubated on the proteome microarray. The microarrays were washed with TBST and incubated with Cy3-Streptavidin at 1:1000 dilution (Sigma) for 1 h at room temperature. To avoid false-positive detections, free biotin was incubated with the proteome microarray and used as a control. Finally, the microarrays were spun dry and scanned with a GenePix 4200A microarray scanner (Axon Instruments) to visualize the results. Data analysis was performed by GenePix Pro-6.0 software. Signal to noise ratio (SNR) which was defined as ratio of the median of foreground signal to the median of background signal, was calculated for each protein. The SNRs for microarrays incubated with and without Biotin-bufalin were set as SNR(+) and SNR(−), respectively. “Ratio” was defined as SNR (+)/SNR (−). The mean SNR was used to represent the signal of the protein. To screen the candidates, the cutoff was set as ratio ≥3.
Microscale thermophoresis analysis
The binding affinity of bufalin with E2F2 and ZFP91 were measured by microscale thermophoresis (MST). pcDNA3.1(+)-His-E2F2 and pEGFP-N1-ZFP91 were transiently transfected into HEK293T cells to express E2F2-His and ZFP91-GFP proteins. The cells were collected and then lysed in NP40 buffer. The His-tagged E2F2 was labelled with the Monolith His-Tag Labelling Kit RED-tris-NTA 2nd Generation (NanoTemper Technologies). Serially diluted bufalin compounds, with concentrations of 500 μM to 15 nM, were mixed with cell lysis solutions containing labelled E2F2, ZFP-GFP and GFP at room temperature and then loaded into Monolith standard-treated capillaries. Binding was measured by monitoring the samples at LED/excitation power of 20% and MST power of 40% on a Monolith NT.115 instrument (Nano Temper Technologies). The Kd values were determined using the MO.AffinityAnalysis software (Nano Temper Technologies).
Determination of bufalin-binding site on ZFP91 by LC-MS/MS
The pCMV-Tag2B-ZFP91 was transiently transfected into HEK293T cells to overexpress Flag-ZFP91 protein. After 48 h, cells were harvested and then lysised in NP40 buffer. Cell lysis solutions were incubated with anti-Flag magnetic beads for 4 h. Beads were washed 5–6 times with the IP buffer. The ZFP91 protein was enriched and then incubated with bufalin (4 μM) or DMSO with rotation overnight at 4 °C. The protein was further resolved by 12% SDS-PAGE and visualized by silver staining. The bands corresponding to ZFP91 were excised from the gel, digested in-gel with trypsin. The peptides were identified by nano-HPLC-tandem LTQ-Orbitrap Velos pro mass spectrometer (Thermo Fisher Scientific). The analytical column (75 μm, 10 cm) packed with 3 μm C18 reversed-phase material was used to separate the peptides by an EASY-nLC II system. The eluent was directly introduced into the mass spectrometer at a flow rate of 300 nL/min. Full scan MS spectra were acquired from 350 to 2000 m/z in the Orbitrap with resolution 60,000. The MS/MS data was analyzed through Proteome Discoverer (1.4) software.
Animal studies
Four-week-old male ICR mice were purchased from the VITAL RIVER Laboratories (Beijing, China). All ICR mice were maintained under pathogen-free and 12 h light/dark cycle conditions, and were free to get food and water. 200 μL of H22 suspension cells (3 × 106) was injected into the flanks of mice to generate xenografts. After subcutaneous inoculation of H22 cells for 3 days, tumors reached approximately 200 mm3 in size. The mice were randomly divided into 4 groups (6 mice in each group): a model group, a positive control group, and bufalin low- and high-dose groups. The mice were treated with bufalin (4 mg/kg), cyclophosphamide (25 mg/kg) and 0.9% saline solution respectively by intraperitoneal injection daily for 14 days. The body weights were recorded every day. The two perpendicular diameters of the tumors were measured and tumor volume was calculated by using the formula V = length (mm) × width (mm)2/2. After 14 days of treatment, mice were sacrificed via cervical dislocation. Then, tumors and organs (heart, liver, spleen, lung, and kidney) were weighted and fixed with 4% paraformaldehyde for further studies. All daily animal care and the experimental procedures were approved by Institutional Animal Care and Use Committee of Peking University (License No. LA2022505). All animal studies have followed the ARRIVE guidelines.
Quantification and statistical analysis
All statistical analysis was performed using GraphPad prism 8.0 software. The data shown in the study were obtained from at least three independent experiments and all data in different experimental groups were expressed as mean ± SD (standard deviation). Statistical analyses were performed using one-way analysis of variance (ANOVA), with student’s t-test analysis. Details of each statistical analysis were provided in the figure legends. Differences with P-values < 0.05 were considered statistically significant.
Role of funders
The funders have no roles in the design of the research, the collection, analysis, interpretation of the data or writing of the report.
Results
Identification of a small-molecule E2F2 degrader
Bufalin is natural-derived small molecule that is previously reported to show anti-tumor potential.20 Here, we used bufalin as a chemical probe to determine the pharmacological effects in hepatocellular carcinoma (HCC). Then, we observed bufalin significantly inhibited HepG2 cells growth in two-dimensional (2D) cultures (Figure S1a) and three-dimensional (3D) sphere suspensions (Figs. 1a–c, S1b). Next, to identify the cellular target of bufalin, we synthesized a biotinylated bufalin probe (biotin-bufalin) as previously reported,21 and the chemical structure of biotin-bufalin was shown in Figure S2. Then we probed a human proteome microarray consisting of 20,000 purified recombinant proteins to detect potential bufalin-binding proteins via Cy-3-conjugated streptavidin labeling strategy (Fig. 1d). In total, 222 candidate bufalin-binding proteins were identified through these microarray assays (representative proteins shown in Fig. 1e). Of note, E2F2 produced the highest signal-to-noise ratio (SNR) of 30.28, which implied a crucial role in bufalin-mediated biological effect.
Fig. 1.
Bufalin A serves as a small-molecule E2F2 degrader. a, Bufalin inhibited HepG2 spheroids proliferation (Scale bars = 100 μm). 3D HepG2 spheroids were treated with bufalin for 48 h and the cell viability was evaluated via dual fluorescence staining (Calcein AM, EthD-I method). b, Day 0-normalized viability of HepG2 cells cultured as 3D spheroid suspensions with or without bufalin for 48 h. Cell viability of cultured 3D spheroids was measured by the CellTiter-glo assay. c, Day 0-normalized viability of HepG2 cells, treated with DMSO or bufalin (400 nM), with readings taken every 2 d. d, Schematic of the procedure for detecting binding events. e, Representative biotin-bufalin interacting proteins (left). Magnified image of biotin-bufalin binding proteins on the proteome microarray (right). Signal-to-noise ratio (SNR) > 2.45. f, Western blot analysis of biotin-bufalin pull-down experiments. g, Microscale thermophoresis analysis (MST) of bufalin binding to E2F2. A dissociation constant of 3.15 ± 1.43 μM was calculated from three independent replicates. h, Western blot analysis of E2F2 protein expression of HepG2 cells treated with various concentration bufalin 24 h. i, Western blot analysis of E2F2 protein expression of HepG2 cells treated with bufalin (400 nM) for the indicated time. j, Real-time qPCR analysis of E2F2 mRNA levels in HepG2 cells treated with bufalin for 6 h. k, Real-time qPCR analysis of E2F2 mRNA levels in HepG2 cells treated with bufalin (400 nM) or DMSO for the indicated time. Data were presented as mean ± SD. ∗∗P < 0.01 (Student's t-test analysis) vs. control group.
Next, pull-down assay validated the interaction of bufalin with E2F2 protein (Fig. 1f). Meanwhile, microscale thermophoresis (MST) experiments demonstrated that bufalin directly bound to E2F2 with a dissociation constant of 3.15 ± 1.43 μM (Fig. 1g). To determine which domain(s) in E2F2 could bind bufalin, we generated a series of E2F2 His-tagged deletion mutants. Pull-down assay showed that the segment containing amino acid residues 125–310, which harbors the dimerization and DNA binding domain of E2F2, was responsible for bufalin binding (Figure S3). In addition, to determine whether bufalin affected cellular levels of E2F2 protein accumulation, we treated HepG2 cells with a range of concentrations of bufalin. Compared with untreated cells, E2F2 protein levels significantly decreased in the presence of bufalin (Fig. 1h and i). Meanwhile, E2F2 mRNA levels moderately increased upon exposure to bufalin, suggesting that E2F2 protein downregulation was not resulted from genetic transcription inhibition (Fig. 1j and k). In addition, c7orf31, KCNAB1 and SDC4 proteins were also identified as potential candidate target proteins of bufalin. To verify whether bufalin affected these protein levels, we treated HepG2 cells with bufalin in a series of concentrations. The Western blot showed that the protein levels of c7orf31, KCNAB1 and SDC4 had no obvious changes, indicating that bufalin did not induce the degradation of c7orf31, KCNAB1 and SDC4 (Figure S4). Therefore, these observations indicate that bufalin serves as a small-molecule E2F2 degrader. Furthermore, we performed washout experiments to determine the reversibility of E2F2 degradation. After treatment with bufalin (400 nM) for 24 h, the cells were washed. Western blot assay suggested that E2F2 levels were completely recovered to baseline by 12 h after washout (Figure S5), indicating that bufalin-mediated E2F2 degradation in HepG2 cells was reversible.
Targeted E2F2 degradation via K48-linked ubiquitination mode
To further test that bufalin functioned as an E2F2 protein degrader, we performed cycloheximide (CHX) chase assay, which showed that a combination treatment of bufalin with CHX further accelerated the degradation rate of E2F2 (Fig. 2a). Next, we observed that treatment with proteasomal inhibitor MG-132 markedly blocked the bufalin-induced reduction in E2F2 protein levels (Fig. 2b). Moreover, lysosomal inhibitor bafilomycin A1 did not show similar effect (Fig. 2c), which excluded lysosomal degradation as a cause for E2F2 depletion. Further, co-immunoprecipitation (Co-IP) experiments indicated that ubiquitination of E2F2 was potentially increased in the presence of bufalin. As we known, all seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) in ubiquitin contribute to the synthesis of polyubiquitin chains on protein substrates. While, the lysine 48 (K48)- and lysine 63 (K63)-linked polyubiquitination are the two most abundant types. Here, we mainly focused on K48- and K63-linked chains which may be involved in E2F2 degradation. We found that K48R-linked ubiquitin (K48R Ub) completely abolished bufalin-mediated polyubiquitin chain formation on E2F2 (Fig. 2d) and K63R-linked ubiquitin (K63R Ub) showed no obvious effect on bufalin-mediated formation of polyubiquitin chain in E2F2 (Figure S6), suggesting that bufalin promoted E2F2 degradation via K48-linked polyubiquitin chains. Collectively, we speculated that K48 served as a major ubiquitin linkage for bufalin-induced E2F2 degradation.
Fig. 2.
E2F2 undergoes degradation via K48-linked ubiquitination. a, Western blot and quantification analysis of E2F2 levels. The protein stability of E2F2 was analyzed by incubating bufalin-treated HepG2 cells and control cells for 0, 4, 6, 8, and 12 h with cycloheximide (CHX). b, Proteasome inhibitor rescued the observed reduction in E2F2. HepG2 cells were treated with bufalin, alone or in combination with MG-132 (10 μM) for 24 h. c, Lysosome inhibitor did not reverse the down-regulation of E2F2. HepG2 cells were treated with bufalin, alone or in combination with Bafilomycin A1 (Baf-A1) (1 μM) for 24 h. d, Co-IP assay showing K48-type ubiquitination of E2F2 induced by bufalin in cells. Data were presented as mean ± SD. ∗∗P < 0.01 (Student's t-test analysis) vs. control group.
Atypical E3 ligase ZFP91 contributes to proteasomal degradation of E2F2
To further identify which specific E3 ligase targets E2F2 for proteasomal degradation, we used Co-IP analysis with stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative proteomics (Fig. 3a). Whole protein extracts of HEK293T cells expressing His-E2F2 were subjected to immunoprecipitation (IP) and determined by LC-MS/MS. A total of 706 proteins were identified from the UniProt database, and four proteins were associated with ubiquitin-proteasome pathway. Particularly, two E3 ligases, ZFP91 and TRIM25, were identified to suggest that E2F2 degradation may be potentially mediated by them (Fig. 3b). Since E2F2 predominantly functions as a nuclear transcription factor, we surmised that physiologically relevant interactions with other proteins were more likely to occur in the nucleus. Indeed, we found that E2F2 protein tended to co-immunoprecipitate with the nuclear localized ZFP91, but not with TRIM25 (Fig. 3c and d). Western blots also showed greater precipitation by ZFP91 compared to that in the control group (Figure S7). These results strongly suggested that ZFP91 served as the E3 ligase responsible for E2F2 protein degradation. Besides, the biotin linkage of bufalin did not influence the interaction between E2F2 and ZFP91 (Figure S8). To verify whether ZFP91 was involved in E2F2 ubiquitination and degradation, we next performed experiments to show that E2F2 polyubiquitination levels induced by bufalin were substantially decreased by small interfering RNA (siRNA) silencing of ZFP91 in the presence of MG-132 (Fig. 3e). In addition, the silencing of ZFP91 also reversed the degradation of E2F2 induced by bufalin (Figure S9). Furthermore, in vitro ubiquitination assays confirmed that both bufalin and ZFP91 were required for robust ubiquitination of E2F2 (Fig. 3f). Taken together, these results show that ZFP91 serves as a specific E3 ligase in E2F2 protein polyubiquitination and proteasomal degradation.
Fig. 3.
ZFP91 serves as an E3 ligase responsible for E2F2 polyubiquitination and degradation. a, Schematic diagram for SILAC-based proteomic assay. b, The proteins that interacted with E2F2 were identified by combining Co-IP and mass spectrometry assay. c, The interaction of ZFP91 and E2F2 was examined by Co-IP assay. HEK293T cells overexpressing His-E2F2 were treated with bufalin (400 nM) or DMSO for 3 h. d, The interaction of TRIM25 and E2F2 was examined by Co-IP assay. HEK293T cells overexpressing His-E2F2 were treated with bufalin (400 nM) or DMSO for 3 h. e, Bufalin promoted the ubiquitination of E2F2 through E3 ubiquitin ligase ZFP91. HEK293T cells were cotransfected with the indicated plasmids and siRNAs for 36 h and then treated with MG-132 (10 μM) and bufalin (400 nM) for 4 h. f, In vitro analysis of ZFP91-mediated E2F2 ubiquitination. His-E2F2 was enriched on Ni-NTA beads and then incubated with E1, UbcH13/Mms2, ZFP91-Flag, ubiquitin, ATP and bufalin.
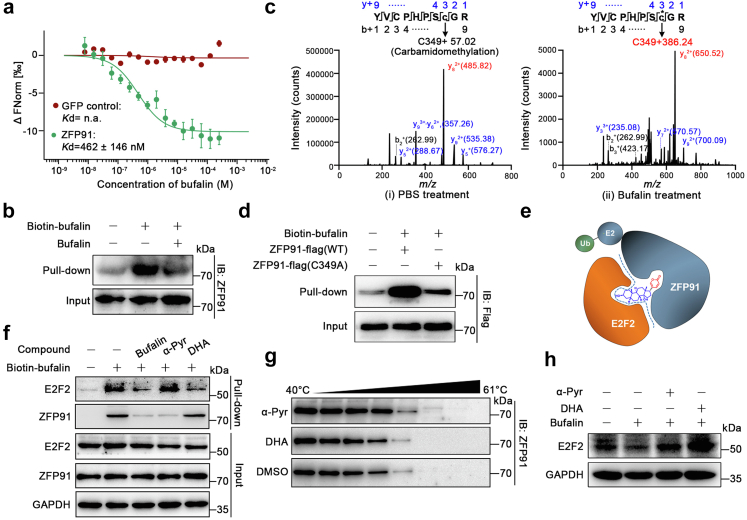
Cys349 serves as a druggable site in ZFP91
To deeply explore the mechanism that bufalin promoted the interaction between E2F2 and ZFP91, we speculated that bufalin may act as a molecular glue to stabilize their binding. To test this hypothesis, we performed pull-down assays to characterize the mechanisms underlying interaction between bufalin and ZFP91. The MST binding assay revealed that bufalin bound to ZFP91 with a dissociation constant (Kd) of 462 ± 146 nM (Fig. 4a). Moreover, we found that biotin-bufalin successfully enriched ZFP91 from cell lysates, which was blocked by adding excess amount of bufalin for competition (Fig. 4b). Given bufalin contains a conjugated unsaturated system which can react with Cys via a Michael addition reaction, we speculated whether bufalin could irreversibly covalently bind to Cys of ZFP91. To this end, ZFP91 was incubated with bufalin, followed by LC-MS/MS analysis. A search for these modifications revealed that a mass shift (386.24 Da) consistent with the addition of molecular weight of a bufalin molecule, and Cys349 was identified as a potential binding site for bufalin modification on ZFP91 (Fig. 4c). Then, we also mutated Cys349 residue of ZFP91 into Ala. As shown in Fig. 4d, C349A mutation weakened the binding of ZFP91 with bufalin, revealing that Cys349 was a direct covalent binding site of bufalin on ZFP91. Besides, we carried out MST experiment. The results revealed that α-Pyr directly bound to ZFP91 protein (Kd = 900 ± 322 nM), but not the C349A mutation of ZFP91 (Kd not detected), indicating a crucial role of the covalent modification site Cys349 in α-Pyr binding to ZFP91 (Figure S10). Therefore, we proposed that bufalin could act as a molecular glue to mediate formation of E2F2-ZFP91 complexes, further promoting the ubiquitination and degradation of E2F2.
Fig. 4.
Bufalin serves as a molecular glue promoting E2F2-ZFP91 interaction. a, MST demonstrated direct interaction between bufalin and GFP-tagged ZFP91 in lysates from GFP-ZFP91 expressing HEK293T cells. GFP protein was used as negative control. b, Biotin-bufalin pull-down experiments. c, LC-MS/MS analysis showed modification of ZFP91 by bufalin at Cys349 residue. Recombinant ZFP91 protein was incubated with (left) or without (right) bufalin overnight at 4 °C. d, Bufalin interacts with Cys349 of ZFP91. Cell lysates were incubated with bufalin-coupled beads at 4 °C for 4 h, and the proteins bound to bufalin beads were detected by Western blot. e, Schematic diagram of bufalin as a molecular glue to mediate the interactions between ZFP91 and E2F2. Bufalin consists of two parts, structure A (blue) and B (red). f, Pull-down assay of α-Pyr binding to ZFP91 and DHA binding to E2F2. HepG2 cell lysates were incubated with vehicle and bufalin-coupled beads in the presence of bufalin, α-Pyr or DHA, respectively. g, CETSA assays confirmed the binding of α-Pyr to ZFP91 in cells. HepG2 cells were treated with α-Pyr (10 μM), DHA (10 μM) or DMSO for 2 h. h, Western blot analysis of E2F2 in HepG2 cells treated with bufalin (400 nM), α-Pyr (10 μM), DHA (10 μM) or DMSO for 24 h.
As shown in Fig. 4e, bufalin consists of two parts, structure A (red) and B (blue). To explore the potential structure-binding relationship, we utilized two compounds (Figure S11), androsterone (DHA) (analogue of structure A) and α-Pyrone (α-Pyr) (analogue of structure B), as competitive binding elements in pull-down experiments. The results showed that DHA significantly limited the binding of biotin-bufalin to E2F2, and α-Pyr significantly blocked the binding of biotin-bufalin to ZFP91 (Fig. 4f). The CETSA assays further demonstrated that α-Pyr improved the thermal stability of ZFP91, instead of DHA (Fig. 4g). Moreover, both α-Pyr and DHA suppressed bufalin-induced E2F2 degradation in HepG2 cells (Fig. 4h). These findings suggested that structure A was responsible for binding to E2F2 and structure B mainly interacted with ZFP91. In particular, E2F2 was significantly degraded at a concentration of bufalin as low as 0.8 μM and the degradation of E2F2 was reversed at around 1.2 μM (Figure S12).
In situ E2F2-ZFP91 complex formation is driven by bufalin in live cells
To visualize E2F2-bufalin-ZFP91 trimer complex formation, we applied a fluorophore phase transition-based strategy to design a separation of phases-based protein interaction reporter (SPPIER) system.22, 23, 24 The SPPIER protein was comprised of three domains, including a protein-of-interest, an enhanced GFP (EGFP), and a homo-oligomeric tag (HOTag). Upon small molecule-induced protein–protein interactions (PPIs) between two proteins-of-interest, multivalent PPIs from HOTags drive EGFP phase separation, resulting in the formation of fluorescent droplets. Here, to detect bufalin-induced PPI between E2F2 and ZFP91, we engineered the DNA binding and dimerization domains (DD; residues 125–310) of E2F2 into SPPIER to produce the E2F2DD-EGFP-HOTag3 construct, which forms hexamers in cells (Fig. 5a). Similarly, the ZFP91-EGFP-HOTag6 fusion construct, which forms tetramers in cells, was used as the E3 ligase SPPIER (Fig. 5a). Thus, when bufalin promotes E2F2-ZFP91 interaction in cells, the E2F2DD-EGFP-HOTag3 hexamers crosslink with ZFP91-EGFP-HOTag6 tetramers to induce EGFP phase separation, which is then observed by fluorescence microscopy.
Fig. 5.
Formation of E2F2-ZFP91-bufalin trimer complex in live cells. a, Schematic diagram showing the design of the SPPIER assay. b, GFP-fluorescence images of HEK293T cells expressing ZFP91-EGFP-HOTag6 and E2F2DD-EGFP-HOTag3 at the indicated time point after treatment with bufalin (400 nM), MG-132 (10 μM), α-Pyr (10 μM), DHA (10 μM) or equivalent volume of DMSO (Scale bars = 5 μm). c, Fluorescence intensity analysis of the red line across the HEK293T cells treated with bufalin (400 nM) and MG-132 (10 μM). d, Analysis of E2F2-Bufalin-ZFP91 ternary complex formation in HEK293T cells by quantifying the fluorescent foci numbers. Data are represented as mean ± SD. ∗∗P < 0.01 vs. control group, by one-way ANOVA.
In our study, live cell fluorescence imaging showed that treatment with bufalin resulted in the formation of numerous green fluorescent puncta, which were undetectable in control group (Figs. 5b and S13). The green fluorescent signal accumulated to a higher intensity following addition of MG-132 (Fig. 5b and c). Moreover, we found that treatment with either α-Pyr or DHA attenuated green fluorescent puncta formation in cells (Fig. 5b, quantified in 5d). These SPPIER data thus illustrate the formation of an E2F2-bufalin-ZFP91 trimer complex in live cells.
E2F2 degradation regulates the transcription of oncogenic genes
To investigate the transcription of E2F2-dependent genes, RNA sequencing was carried out in this research. As shown in Fig. 6a, 3000 genes were upregulated, while 2450 genes were downregulated after bufalin treatment. In addition, TRRUST database (Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining) was used to predict the target genes of E2F2. In order to reveal the relation of bufalin-regulated transcription and E2F2-regulated genes, venn diagram was used to show numerical distribution of differential genes regulated by bufalin (Fig. 6b). Then, we screened out a panel of E2F2 target genes including c-Myc, Polr2a, GMNN, CDK1, CCNE1, CCNE2 and MCM5 (Fig. 6c). We next conducted qPCR-based relative expression analysis of these genes, and results showed that bufalin significantly promoted the down-regulation of c-Myc, Polr2a, GMNN, CDK1, CCNE1, CCNE2 and MCM5 gene expression (Fig. 6d). Collectively, our proposed that the natural product bufalin may function as a molecular glue that induces E2F2 degradation via recruitment of the E3 ligase ZFP91, resulting in down-regulation of target genes to inhibit hepatocellular carcinoma growth (Fig. 6e).
Fig. 6.
Bufalin inhibits the transcription of E2F2-dependent oncogenic genes. a, The volcano plot represents differentially expressed genes. HepG2 cells were treated with vehicle (control group) or bufalin (400 nM) for 12 h, respectively. b, Venn diagram showing numerical distribution of differential genes and E2F2 target genes. E2F2-regulated genes were obtained from TRRUST (Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining). c, The heatmap analysis of down-regulated genes. d, The mRNA expression levels of E2F2 target genes were measured by qPCR. Data were presented as mean ± SD. ∗∗P < 0.01, by independent Student's t test. e, Mechanism of bufalin-induced E2F2-ZFP91 complex to promote E2F2 protein degradation.
Translational study of small-molecule E2F2 degrader in hepatocellular carcinoma
We next explored the potential translational medical value of targeted E2F2 degradation strategy. We determined the inhibitory effect of bufalin on the growth of hepatoma H22 cell-derived xenograft allograft in ICR mice (Fig. 7a). Following intraperitoneal injection of bufalin (4 mg/kg) once daily for 14 days, we found that bufalin significantly decreased the tumor volume in ICR mice compared with control group (Fig. 7b and c). Meanwhile, bufalin obviously reduced the tumor weight (Fig. 7d). In addition, there was no significant change in body and organ weights, suggesting that bufalin showed no obvious side effects (Fig. 7e and f). Pathological examination by H&E staining showed that xenograft tumor cells were loosely packed with small nuclei compared with vehicle group (Fig. 7g). Next, protein expression levels of E2F2 were measured in tumors using immunohistochemical (IHC) staining. As expected, we found that bufalin administration significantly decreased E2F2 expression (Fig. 7h), which quite differed from positive drug CTX. Therefore, these results indicate that bufalin serving as an available molecular glue small-molecule E2F2 degrader exhibits a potent inhibitory effect on the growth of hepatocellular carcinoma in vivo. In addition, to further investigate whether bufalin was involved in antitumor immune regulation, we then established an ICR mice model of tumor transplantation. In particular, we analyzed the immune cells (especially T cells) infiltration. The results showed that bufalin greatly enhanced the amounts of CD4+ and CD8+ T cells in tumor tissues, indicating that bufalin enhanced antitumor immune response (Figure S14).
Fig. 7.
Translational study of E2F2 degrader in hepatocellular carcinoma growth in vivo. a, Schematic diagram of the administration procedure of bufalin in the established H22 tumor-bearing mice. Cyclophosphamide (CTX) was used as a positive control. b, The dynamic change of tumor volume in subcutaneous models were displayed. c, Bufalin decreased tumor volume in ICR mice transplanted with H22 cells (n = 6). d, Bufalin obviously reduced tumor weight in H22 xenograft mice compared with vehicle group (n = 6). e, bufalin treatment had no significant effect on the body weight of H22-xenograft mice. f, bufalin treatment did not significantly change the weight of heart, liver, spleen, lung, and kidney in H22-xenograft mice (n = 6). g, Representative histological analysis of tumor specimen stained by H&E (Scale bars = 100 μm). h, Representative images of IHC staining for E2F2 in tumor tissues. Data were presented as mean ± SD. ∗∗∗P < 0.001, by one-way ANOVA. N.S. not significant.
Discussion
Zinc finger protein 91 (ZFP91) is reported to be involved in several biological processes. However, its potential application as a target of cancer therapeutics remains largely unexplored. Previous studies have shown that ZFP91 serves as an atypical E3 ligase responsible for ubiquitin-mediated degradation of a wide range of substrate proteins in a proteasome-dependent manner, including NF-κB inducing kinase (NIK),25 forkhead box A1 (FOXA1),26 hnRNPA1,27 and hnRNPAB.28 Here, we provide the evidence that E2F2 degradation is selectively regulated in a ZFP91-dependent manner. These findings further expand the range of possible future applications of a previously undisclosed E3 ligase ZFP91 in depleting undruggable proteins particularly for transcription factors in a wide range of human diseases. Moreover, so far, the number of available E3 ligases being explored by molecular gules or proteolysis-targeting chimera (PROTAC) still remains limited (typically, CRBN, VHL, IAPs, MDM2, DCAF15, and RNF114). Therefore, expansion of the E3 ligase toolbox will be crucial to accelerate the research and development small-molecule protein degraders.
Currently, the application of molecular glues that recruit E3 ligases is very limited. Our research reveals bufalin as a unique natural-derived molecular glue that targets an as-of-yet undescribed E3 enzyme, thus expanding our understanding of the diversity of molecular glues. Moreover, its pyrone structure may be a potential structural group for targeting ZFP91. Of note, the carbonyl-conjugated structure may possess electrophilic addition capability to covalently bind to the cysteine in ZFP91. Thus, more study will enable the use of this structural group in the discovery of novel drugs with molecular glue properties. Furthermore, we noticed that the degradation range of E2F2 by bufalin was relatively moderate. In fact, different from traditional "occupancy-driven" mode, molecular glue only provides binding activity, which is "event-driven" reusable and then possesses a relative long half-life. Thus, bufalin can promote the target protein degradation in a persistent and moderate manner.
Meanwhile, we observed that ZFP91 did not show obvious binding SNR value to bufalin in the microarray. A possible reason was considered that the specific binding spatial conformation of ZFP91 on the microarray surface led to the insufficient exposure to bufalin, which resulted in a false negative result. Therefore, we performed MST and pull-down experiments to further confirm the interaction between ZFP91 and bufalin in this study. While, we noted that SDC4 was another potential binding protein in this array, which was consistent with our previous report that SDC4 functioned as a direct cellular target of bufalin for anti-HCC metastasis and invasion.21 Besides, bufalin was also reported to bind to ATPase (ATP1B1).29 Nonetheless, we found that ATPase (ATP1B1) showed a low SNR value (1.26) in our study. In fact, previous report obtained the crystal of ATPase-bufalin in the specific condition with potassium ion; meanwhile, the ATPase in that study was isolated from pig kidney with genetic encoding difference from human. Thus, the interaction between bufalin and ATPase remain to be further determined.
As a special type of molecular glue, bufalin also exhibits a specific binding capacity with the target protein E2F2, which is different from the classical molecular glue (only targeting E3 ligase). However, the binding capacity of bufalin to ZFP91 is much higher than that of E2F2. Therefore, this property provides a key biochemical fundamental for efficient degradation of substrate protein via molecular glue-mediated mechanism.
Although the crystal structure has not been solved for bufalin in complex with E2F2 and ZFP91 due to high E2F2 instability under crystallization conditions, dynamic, in situ, real-time analysis with fluorescent sensors confirmed the bufalin-mediated interplay between E2F2 and ZFP91, as well as formation of a complex in the cytoplasm. Anticancer drug thalidomide is representative of molecular glue, which can redirect the E3 ubiquitin ligase CRBN to polyubiquitinate the transcription factors IKZF1 and IKZF3 for proteasomal degradation.30 However, our research reveals bufalin as a unique molecular glue skeleton that targets ZFP91 as an as-of-yet undescribed E3 enzyme, thus expanding our understanding of the targeted protein degradation strategy.
Collectively, our findings demonstrate a new atypical E3 ligase as a druggable target to degrade E2F2 transcription factor. Moreover, bufalin functions as a molecular glue, significantly inducing E2F2 degradation via ZFP91, thereby resulting in promising medicinal value in cancer therapy.
Contributors
K.Z. and P.T. conceived and supervised the project. T.L. and H.Y. did most of the pharmacologic experiments. F.Z, Z.Y., Y.L., M.Z., and Q.G. assisted in part of the biochemical experiments. D.L. performed LC-MS/MS analysis. K.Z., H.Y., and T.L. wrote the manuscript, have accessed and verified the data. All authors have read and approved the final version of the manuscript.
Data sharing statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Declaration of interests
All authors have declared no competing interests.
Acknowledgements
The authors are grateful to Dr. Xia Yuan, Qian Wang and Jing Wang for the helpful discussions. This work was financially supported by the National Key Research and Development Project of China (2022YFC3501601), National Natural Science Foundation of China of China (81973505, 82174008, 82030114), and China Postdoctoral Science Foundation (2019M650396), the Fundamental Research Funds for the Central Universities.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104353.
Contributor Information
Ke-Wu Zeng, Email: ZKW@bjmu.edu.cn.
Peng-Fei Tu, Email: pengfeitu@bjmu.edu.cn.
Appendix A. Supplementary data
References
- 1.Bertoli C., Skotheim J.M., de Bruin R.A. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14(8):518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H.Z., Tsai S.Y., Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y., Hu C., Du P., Huang G. E2F1 maintains gastric cancer stemness properties by regulating stemness-associated genes. J Oncol. 2021;2021 doi: 10.1155/2021/6611327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun C.C., Li S.J., Hu W., et al. Comprehensive analysis of the expression and prognosis for E2Fs in human breast cancer. Mol Ther. 2019;27(6):1153–1165. doi: 10.1016/j.ymthe.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Li Y., Sturgis E.M., Zhu L., et al. E2F transcription factor 2 variants as predictive biomarkers for recurrence risk in patients with squamous cell carcinoma of the oropharynx. Mol Carcinog. 2017;56(4):1335–1343. doi: 10.1002/mc.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng Z., Cao Z., Tang Y. Increased E2F2 predicts poor prognosis in patients with HCC based on TCGA data. BMC Cancer. 2020;20(1):1037. doi: 10.1186/s12885-020-07529-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen S., Wang Y.F. Expression and prognostic role E2F2 in hepatocellular carcinoma. Int J Gen Med. 2021;14:8463–8472. doi: 10.2147/IJGM.S334033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González-Romero F., Mestre D., Aurrekoetxea I., et al. E2F1 and E2F2-mediated repression of CPT2 establishes a lipid-rich tumor-promoting environment. Cancer Res. 2021;81(11):2874–2887. doi: 10.1158/0008-5472.CAN-20-2052. [DOI] [PubMed] [Google Scholar]
- 9.Schreiber S.L. The rise of molecular glues. Cell. 2021;184(1):3–9. doi: 10.1016/j.cell.2020.12.020. [DOI] [PubMed] [Google Scholar]
- 10.Isobe Y., Okumura M., McGregor L.M., et al. Manumycin polyketides act as molecular glues between UBR7 and P53. Nat Chem Biol. 2009;16(11):1189–1198. doi: 10.1038/s41589-020-0557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Słabicki M., Kozicka Z., Petzold G., et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature. 2020;585(7824):293–297. doi: 10.1038/s41586-020-2374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surka C., Jin L., Mbong N., et al. CC-90009, a novelcereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells. Blood. 2021;137(5):661–677. doi: 10.1182/blood.2020008676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bussiere D.E., Xie L., Srinivas H., et al. Structuralbasis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex. Nat Chem Biol. 2020;16(1):15–23. doi: 10.1038/s41589-019-0411-6. [DOI] [PubMed] [Google Scholar]
- 14.Liao L.X., Song X.M., Wang L.C., et al. Highly selective inhibition of IMPDH2 provides the basis of antineuroinflammation therapy. Proc Natl Acad Sci U S A. 2017;114(29):E5986–E5994. doi: 10.1073/pnas.1706778114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinci M., Box C., Eccles S.A. Three-dimensional (3D) tumor spheroid invasion assay. J Vis Exp. 2015;99 doi: 10.3791/52686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jafari R., Almqvist H., Axelsson H., et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc. 2014;9(9):2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 17.Martinez Molina D., Jafari R., Ignatushchenko M., et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341(6141):84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 18.Jeong J.S., Jiang L., Albino E., et al. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol Cell Proteomics. 2012;11(6) doi: 10.1074/mcp.O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox E., Uzoma I., Guzzo C., et al. Identification of SUMO E3 ligase-specific substrates using the HuProt human proteome microarray. Methods Mol Biol. 2015;1295:455–463. doi: 10.1007/978-1-4939-2550-6_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takai N., Kira N., Ishii T., et al. Bufalin, a traditional oriental medicine, induces apoptosis in human cancer cells. Asian Pac J Cancer Prev. 2012;13(1):399–402. doi: 10.7314/apjcp.2012.13.1.399. [DOI] [PubMed] [Google Scholar]
- 21.Yang H., Liu Y., Zhao M.M., et al. Therapeutic potential of targeting membrane-spanning proteoglycan SDC4 in hepatocellular carcinoma. Cell Death Dis. 2021;12(5):492. doi: 10.1038/s41419-021-03780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung C.I., Zhang Q., Shu X. Dynamic imaging of small molecule induced protein-protein interactions in living cells with a fluorophore phase transition based approach. Anal Chem. 2018;90(24):14287–14293. doi: 10.1021/acs.analchem.8b03476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posternak G., Tang X., Maisonneuve P., et al. Functional characterization of a PROTAC directed against BRAF mutant V600E. Nat Chem Biol. 2020;16(11):1170–1178. doi: 10.1038/s41589-020-0609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q., Huang H., Zhang L., et al. Visualizing dynamics of cell signaling in vivo with a phase separation-based kinase reporter. Mol Cell. 2018;69(2):334–346. doi: 10.1016/j.molcel.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin X., Jin H.R., Jung H.S., et al. An atypical E3 ligase zinc finger protein 91 stabilizes and activates NF-κB-inducing kinase via lys63-linked ubiquitination. J Biol Chem. 2010;285(40):30539–30547. doi: 10.1074/jbc.M110.129551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang D.E., Dai Y., Xu Y., et al. The ubiquitinase ZFP91 promotes tumor cell survival and confers chemoresistance through FOXA1 destabilization. Carcinogenesis. 2020;41(1):56–66. doi: 10.1093/carcin/bgz085. [DOI] [PubMed] [Google Scholar]
- 27.Chen D., Wang Y., Lu R., et al. E3 ligase ZFP91 inhibits hepatocellular carcinoma metabolism reprogramming by regulating PKM splicing. Theranostics. 2020;10(19):8558–8572. doi: 10.7150/thno.44873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan T., Yu Z., Jin Z., et al. Tumor suppressor lnc-CTSLP4 inhibits EMT and metastasis of gastric cancer by attenuating HNRNPAB-dependent snail transcription. Mol Ther Nucleic Acids. 2021;23:1288–1303. doi: 10.1016/j.omtn.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laursen M., Gregersen J.L., Yatime L., Nissen P., Fedosova N.U. Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex. Proc Natl Acad Sci U S A. 2015;112(6):1755–1760. doi: 10.1073/pnas.1422997112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer E.S., Böhm K., Lydeard J.R., et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512(7512):49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.