Abstract
The DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) allows efficient in vivo enrichment of transduced hematopoietic stem cells (HSC). Thus, linking this selection strategy to therapeutic gene expression offers the potential to reconstitute diseased hematopoietic tissue with gene-corrected cells. However, different dual-gene expression vector strategies are limited by poor expression of one or both transgenes. To evaluate different co-expression strategies in the context of MGMT-mediated HSC enrichment, we compared selection and expression efficacies in cells cotransduced with separate single-gene MGMT and GFP lentivectors to those obtained with dual-gene vectors employing either encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) or foot and mouth disease virus (FMDV) 2A elements for co-expression strategies. Each strategy was evaluated in vitro and in vivo using equivalent multiplicities of infection (MOI) to transduce 5-fluorouracil (5-FU) or Lin−Sca-1+c-kit+ (LSK)-enriched murine bone marrow cells (BMCs). The highest dual-gene expression (MGMT+GFP+) percentages were obtained with the FMDV-2A dual-gene vector, but half of the resulting gene products existed as fusion proteins. Following selection, dual-gene expression percentages in single-gene vector cotransduced and dual-gene vector transduced populations were similar. Equivalent MGMT expression levels were obtained with each strategy, but GFP expression levels derived from the IRES dual-gene vector were significantly lower. In mice, vector-insertion averages were similar among cells enriched after dual-gene vectors and those cotransduced with single-gene vectors. These data demonstrate the limitations and advantages of each strategy in the context of MGMT-mediated selection, and may provide insights into vector design with respect to a particular therapeutic gene or hematologic defect.
Keywords: O6-methylguanine-DNA methyltransferase, MGMT, P140K, IRES, F2A, lentivirus, gene therapy
1. Introduction
A number of drug resistance genes have been evaluated for selective enrichment of transduced stem cells in vivo1–4. However, the most robust stem cell selection has been achieved using specific point mutants of MGMT5,6. MGMT repairs O6-methylguanine and O6-chloroethylguanine DNA lesions induced by chemotherapeutics such as Temozolomide and 1,3-Bis(2-Chloroethyl)-Nitrosourea (BCNU), respectively. MGMT point mutants, including P140K and G156A, are resistant to the wild-type MGMT inhibitor, O6-benzylguanine (BG), but retain wild-type DNA repair activity7–9. Thus, the endogenous MGMT in untransduced BMCs is inactivated by BG treatment, sensitizing these cells to alkylating agent damage, while mutant MGMT-transduced cells are protected from the combined BG and alkylating agent insult10. With sequential treatments, this differential sensitivity allows transduced stem cells to survive and repopulate the hematopoietic compartment5,6,11–14. Therefore, dual-gene vectors that couple MGMT-mediated stem cell selection to therapeutic gene expression should allow diseased hematopoietic tissues to be reconstituted with functional, gene corrected cells. The feasibility of this approach has been demonstrated in animal models of β-thalassemia15,16, protoporphyria17, and HIV/AIDS18. However, higher MOIs are often required to compensate for the lower expression levels obtained with dual-gene vectors.
Several methods have been devised to co-express two genes from the same viral vector. Two of the most common strategies utilize either an IRES19,20, or ribosome slippage21,22 sequences for dual-gene expression. IRES elements form secondary RNA structures that act to initiate translation in a cap-independent fashion. Therefore, these elements can be used to express additional genes from the same transcript. Many endogenous and exogenous IRES sequences have been identified, but the sequence derived from the EMCV IRES is one of the most thoroughly characterized and widely used IRES elements in dual-gene expression vectors23,24. IRES-mediated translation efficiency is typically reduced by 20–50%, compared to the cap-dependent mechanism25,26. In addition, IRES activity can be influenced by the specific genes used, and their position with respect to the IRES sequence24,27.
Whereas IRES elements function to initiate translation, ribosome-slippage sites, such as the FMDV 2A element, are active during translation. The short nucleotide sequence encoding FMDV-2A is positioned between two genes, the first of which has the stop codon removed. Thus, both genes and the 2A element are joined as one open reading frame. After the first gene and 2A sequence are translated, cis-acting hydrolase activity within the 2A residues causes the ribosome to “skip” the last peptide bond in 2A. Thus, depending on the 2A nucleotide sequence employed the first gene product is released with 17–23 residues (with up to 32 residues demonstrating increased efficiency28) from the 2A element fused to its C-terminus. The ribosome then continues translating the second gene product, which contains an N-terminal proline from the 2A sequence29. Like IRES elements, the efficiency of ribosome slippage sequences appears to be sensitive to the specific gene combinations used30,31. Further, the activity of the first gene product can be perturbed by the 2A residues that remain fused to its C-terminus32.
Herein, we evaluated the use of two separate single-gene lentiviral vectors (one encoding MGMT-P140K and the other expressing GFP) to cotransduce and selectively enrich dual-gene expressing hematopoietic populations in vivo. Cotransduction was then compared to dual-gene vectors that utilize the EMCV-IRES or FMDV-2A elements for co-expression. Comparisons were made based on enrichment of dual-gene expressing cells, individual gene expression levels, and total vector insertion averages. Although each strategy may be more or less efficient, depending on the target cells and the specific genes used, this study demonstrates the advantages and limitations of each strategy in the context of MGMT-mediated hematopoietic progenitor cell enrichment.
2. Materials and Methods
2.1. Vectors
The self-inactivating lentiviral luciferase (Luc) vector, pCSO-rre-cppt-MCU3-LUC, was obtained from Donald Kohn (University of California, Los Angeles, CA). It contains the MND (myeloproliferative sarcoma virus enhancer, negative control region deleted, dl587rev primer-binding site substituted) promoter/enhancer sequence, the rev response element (RRE), and the central polypurine tract/central termination sequence (cPPT/CTS). Luc was removed by partial restriction with NcoI and EcoRI and replaced with a multiple cloning site linker. The MGMT-P140K and enhanced GFP coding sequences were separately inserted into the NcoI site (closest to the MND promoter) and a unique BamHI site. These constructs were then restricted at unique BamHI and EcoRI sites for insertion of the woodchuck hepatitis virus postranscriptional regulatory element (wPRE; provided by Thomas Hope, Northwestern University, Chicago, IL)33,34 to generate the pMND-MGMT and pMND-GFP single-gene vectors. The EMCV IRES element was obtained from the pCITE-2a vector (Clontech, Mountain View, CA). The IRES sequence was deleted from pCITE-2a by partial PvuII and MscI restriction and the resulting vector fragment was religated. 5′-NotI and 3′-EcoRI sites were added onto the deleted IRES sequence by PCR and the restricted PCR product was added back to the IRES-deleted pCITE-2a vector, generating an IRES vector with both 5′ and 3′ multiple cloning sites, termed pSOB. GFP was cloned into pSOB using NcoI and BamHI to position the GFP start codon at the site reported to maximize IRES-mediated translation24. The IRES-GFP cassette was then removed and placed 70 nt downstream of MGMT using a unique NotI site in pMND-MGMT, generating pMND-MIG. The FMDV-2A sequence was generated from oligos using overlap extension PCR. An MGMT-2A PCR fragment was generated using the oligos, MGMT-FOR (5′-GTGAGCAGGGTCTGCACGAA-3′) and MGMT-2A (5′-CTGCCAACTTGAGCAGGTCAAAGCTCAAAAGCTGTTTCACCGGTGCGTTTCGGCCAG CAG-3′). A separate 2A-GFP fragment was generated using the oligos 2A-GFP (5′-GGCAGGGGACGTCGAGTCCAACCCTGGGCC TATGGTGAGCAAGGGCGA-3′) and GFP-REV (5′-CTAGAGCGGCCGCTTTACTTGTACAGCTCGTCC-3′). The resulting MGMT-2A and 2A-GFP fragments were annealed, extended, and amplified with the MGMT-FOR and GFP-REV oligos. The resulting fragment was cloned into pMND-MIG using a unique SfiI site within MGMT and a NotI site 3′ of GFP, generating pMND-MAG. All constructs were verified by sequencing.
2.2. Virus preparation and transductions
Virus was generated as previously described35. In brief, 293T cells (ATCC, Manassas, VA) were transfected with the VSV-G pseudotyping vector (pMD.G)36, the packaging vector (pCMVΔR8.91)37, and the pMND transducing vectors at a 3:1:3 mass ratio, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. At 24–48 h after transfection, virus was harvested in Dulbecco’s modified Eagle medium (DMEM; Cellgro, Manassas, VA) containing 10% heat-inactivated (HI)-fetal bovine serum (FBS) (Cellgro) and 2 mM GlutaMAX (Invitrogen); then filtered through 0.45 μm syringe filter units (Millipore, Billerica, MA); and finally stored at −80 °C. Expression titers were determined on K562 cells (ATCC) using virus dilutions that resulted in less than 10% transduction. Titers ranged from 0.5–3.0 × 107 expression units (EU)/mL. Virus preps used for cotransductions were premixed at the specified MOI ratios prior to the addition of cells. K562 cells were transduced in Iscove’s medium (Cellgro) containing 10% HI-FBS, 2 mM GlutaMAX, and 8 μg/mL polybrene (Sigma, St. Louis, MO). BMCs (isolated from 5-FU treated animals as previously described35) and sorted Lin−Sca-1+c-kit+ (LSK) cell populations (isolated as previously described38) were transduced for 12 h in alpha-MEM containing 20% Hi-FBS, 2 mM GlutaMAX, 6 μg/mL polybrene, in the presence of murine IL-3 (20 ng/mL), murine IL-6 (50 ng/mL), and rat SCF (50 ng/mL) (R&D Systems Inc., Minneapolis, MN). MOIs defined for mouse cell transductions were derived from the expression titers established in K562 cells.
2.3. Animals and transplants
Transduced LSK cells (as above) were transplanted immediately after transduction. Six-week-old C57BL/6J recipient mice (Jackson Laboratories, Bar Harbor, ME) were lethally irradiated with 8.5 Gy using a Cs137 source and transplanted with 103 cells from each transduction culture, along with 2 × 106 Lin+ cells (for hematopoietic support) per mouse. The animals were sacrificed at 17 weeks for expression and vector insertion number analysis in BMCs and CFU, respectively. Fold-enrichment was calculated as follows: [(% GFP+ BMCs at 17 weeks) × (3 × 108 total BMCs in reconstituted mouse)] ÷ [(% GFP+ transplanted LSK cells) × (total # transplanted LSK cells)]. Analysis of vector insertions is described below.
2.4. Drug Selection
BG was synthesized by Robert Moschel at the Frederick Cancer Research Institute (Frederick, MD). BCNU was obtained from the Drug Synthesis and Chemistry Branch of the National Cancer Institute (NCI; Bethesda, MD). The in vitro drug treatment incubations were carried out in serum free media at 37 °C as previously described35. Briefly, cells were pretreated with BG for 1 h, then BCNU for 2 h. K562 cells were treated in Iscove’s media and then allowed to expand for 7 d. For in vitro selections, murine BMCs or LSK cells were treated with 25 μM BG and 0, 5, 15, or 25 μM BCNU in alpha-MEM containing 1.2% spleen cell conditioned media (Stem Cell Technologies, Vancouver, BC, Canada). Drug treated murine BMCs or LSK cells were expanded in liquid culture using the same media formulation as described for transduction without polybrene for 10 or 15 d, respectively. Progenitor (CFU) drug treatment survival assays were performed as previously described39. For in vivo selection, three rounds of selection, starting at 5 weeks post-transplant, were carried out allowing 3 weeks recovery between each treatment. Each round, mice received i.p. injection with 30 mg/kg BG (dissolved to 3 mg/mL in 40% polyethylene glycol (Union Carbide Corp., Danbury, CT) and 60% PBS, pH 8.0), followed by i.p. injection of 10 mg/kg BCNU (dissolved in ethanol and diluted to 1 mg/mL in PBS) an hour later.
2.5. Flow cytometry
Flow cytometry data were acquired with an LSR flow cytometer (Becton Dickinson, Franklin Lakes, NJ). GFP expression was measured in cells after a single wash in PBS. For MGMT detection, cells were fixed in 2% paraformaldehyde for 30 min at 4° C, permeablized in 1% Tween-20 for 30 or 60 min (K562 or murine cells, respectively) at 37 °C, and then blocked in 10% normal goat serum at room temperature for 15 min. Cells were then incubated with mouse anti-human MGMT monoclonal antibody (clone MT3.1; Kamiya Biomedical, Seattle, WA) and an APC-conjugated goat anti-mouse secondary antibody (Caltag Laboratories, Burlingame, CA). Dual detection of MGMT and GFP expression was carried out with the same staining protocol as that used for MGMT alone. Red blood cells were lysed in murine PB and BM samples prior to staining for flow cytometry. Monoclonal antibodies against Gr-1, CD11b, B220, CD19, CD3, CD4, CD8, Ter119, Sca-1, and c-kit were purchased from BioLegend (San Diego, CA) and BD Pharmingen (San Diego, CA).
2.6. Copy number analysis
The number of vector insertions per cell was evaluated using a LightCycler instrument and the LightCycler FastStart DNA Master SYBR Green I kit (Roche Diagnostics, Basel, Switzerland). MGMT and GFP copy numbers were determined using proviral-specific MGMT and GFP primers, as described previously35. Genomic copy numbers were determined using GAPDL4-specific primers, as previously described12. MGMT, GFP, and GAPDL4 copy numbers were assessed using a standard curve consisting of genomic DNA from murine PBLs spiked with known MAG vector copy numbers, and verified using K562 cell clones with single MAG vector insertion. One diploid copy of the mouse genome was assumed to contain two copies of GAPDL4 and 5.9 pg of DNA. MGMT, GFP, and GAPDL4 copy numbers were analyzed in 8–15 CFU per animal. The average number of vector insertions per marked CFU in cotransduced cells is the cumulative average of the total MGMT and GFP copies, whereas the average number of vector insertions per marked CFU in dual-gene vector transduced cells is the combined average of the individual MGMT and GFP copy numbers.
2.7. Cytospin and staining
At 72 h post-transduction, K562 cells were cytospun onto slides, fixed in 4% paraformaldehyde, and DAPI-stained (Vector Laboratories, Burlingame, CA) for nuclear visualization. GFP and DAPI fluorescence were visualized separately and the fluorescent images were superimposed using Image J software.
2.8. Western blotting
Following drug selection, K562 cells transduced with pMND.MIG, pMND.MAG, pMND.GAH (not subjected to drug selection), or cotransduced with pMND.GFP and pMND.MGMT, were harvested and prepared for Western blotting as described previously40.
2.9. Statistical analysis
The data are shown as means ± s.d. or s.e.m. Analysis of variance with Bonferroni’s post-test correction, for comparisons of three or more groups, was used to assess statistical differences using Prism 4.0 (GraphPad Software, La Jolla, CA, USA). Statistical significance was accepted at P<0.05.
3. Results
3.1. Dual-gene transfer strategies
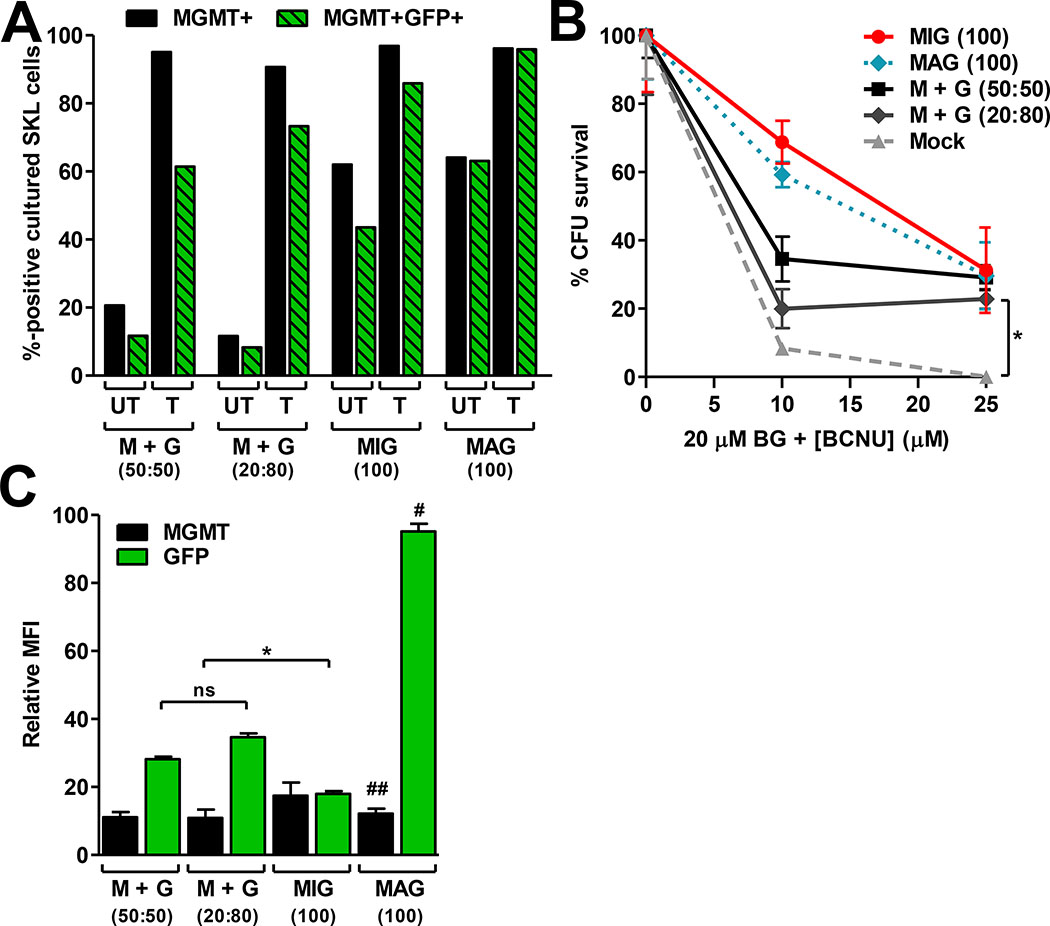
Four self-inactivating lentiviral vectors were constructed: two single-gene vectors containing MGMT-P140K (pMND-MGMT) or enhanced GFP (pMND-GFP), and two dual-gene vectors containing either the EMCV-IRES (pMND-MIG) or FMDV-2A (pMND-MAG) elements to allow co-expression of MGMT and GFP (Fig. 1). The MGMT-P140K gene was placed in the 5′ position of the dual-gene vectors to ensure that the level of drug resistance gene expression was equal to that obtained with the MGMT single-gene vector. Equivalent levels of expression and drug resistance were verified in K562 human erythroleukemia cells transduced with equal amounts of MGMT, MIG, or MAG lentiviruses (Fig. 2).
Fig. 1. Lentiviral vector constructs.
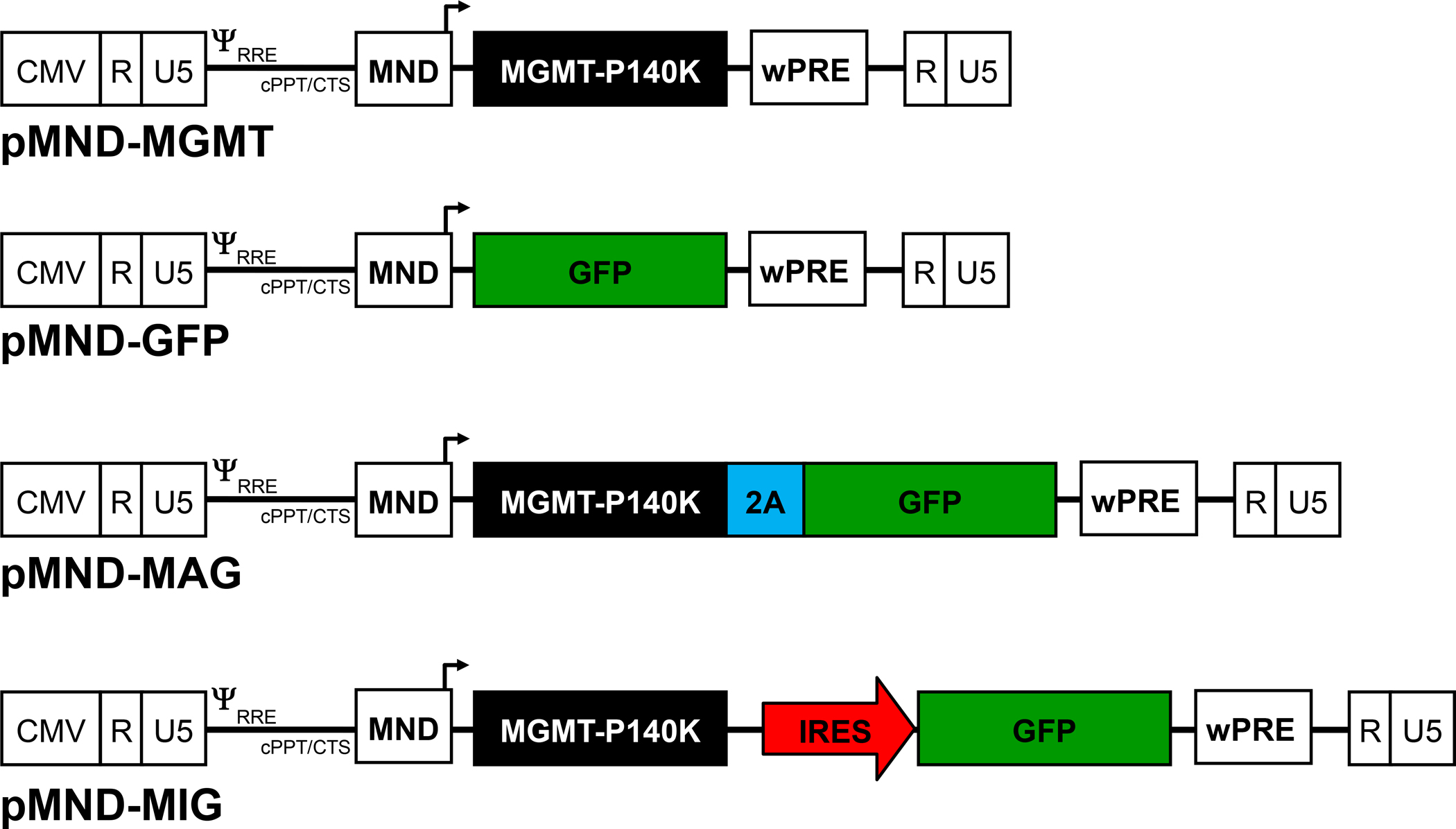
Schematic diagram of the self-inactivating lentiviral vectors used in this study. Each construct contains the MND promoter/enhancer (MND), central polypurine tract/central termination sequence (cPPT/CTS), as well as the woodchuck hepatitis virus post-transcriptional regulatory element (wPRE). The same 5′ restriction site was used in the construction of each vector to ensure that all transcripts have the same 5′-UTR sequences.
Fig. 2. MGMT expression vectors confer equivalent protection from drug treatment.
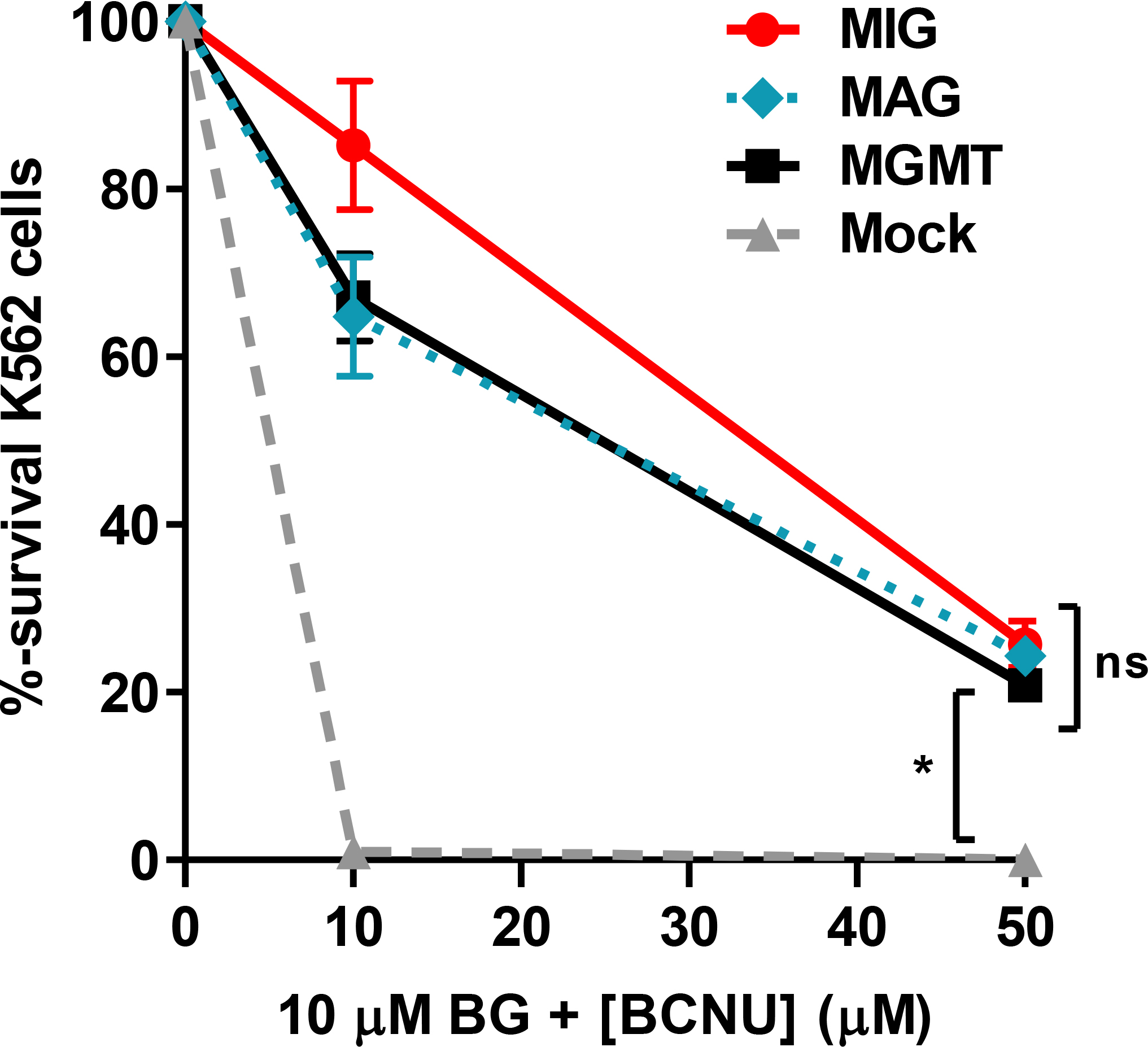
K562 cells were transduced (MOI = 0.5) with the MGMT single-gene, MIG, or MAG dual-gene vectors. At 72 h post-transduction the cultures were treated with 10 μM BG and 0, 10, or 50 μM BCNU and plated in methylcellulose for 10 d. Survival percentages are based on total colony-forming units (CFU), per number of cells plated, compared to untreated controls. Data are means ± s.e.m. of a representative experiment with triplicate samples. *, P<0.05; ns, not significant (P>0.05).
3.2. Dual-gene transfer and in vitro enrichment of K562 cells
As a first approach to comparing cotransduction with single-gene vectors to transduction with the MIG and MAG dual-gene vectors, K562 cells were transduced using the same total MOI for each method. The percentage of cells that expressed both MGMT and GFP (MGMT+GFP+) after cotransduction with pMND.MGMT and pMND.GFP (‘M+G’) was similar to that obtained with the dual-gene MIG vector (Fig. 3A). This similarity was observed for different total vector MOIs (1 or 5) for both sets of transductions and was maintained after selection with 10 μM BG and 25 μM BCNU. Since the total MOIs used for each transduction strategy were equivalent (1 or 5), the MGMT and GFP gene equivalents used for the cotransductions (0.5 each or 2.5 each, respectively) were a fraction of those used in the dual-gene vector transductions. Nevertheless, increased expression efficiency with the cotransduced vectors and the reduced GFP expression obtained with the IRES-based vector resulted in overall equivalent percentages of dual-gene expressing cells with either strategy. Poor IRES-mediated translation efficiency reduced the percentage of dual-gene expressing cells detected (i.e. expression of MGMT was consistently greater than GFP) (Fig. 3A). In contrast, MGMT and GFP were both efficiently expressed in K562 cells transduced with the dual-gene MAG vector and resulted in the highest percentage of dual-gene expressing cells in the untreated and drug-treated cultures (Fig. 3A). Interestingly, the GFP mean fluorescence intensity (MFI) in MAG-transduced cells was 3 to 8 times higher than in cotransduced or MIG-transduced cells, respectively (Fig. 3B). Western blotting revealed that approximately half of the MGMT protein expressed from the dual-gene MAG vector was still fused to GFP (Fig. 3C), suggesting the 2A hydrolase activity is not always effective31. However, the presence of fusion products did not affect the DNA repair activity (Fig. 2), as previously reported for MGMT-GFP fusion constructs41. Furthermore, the majority of GFP signal in K562 cells transduced with the MAG dual-gene vector localized to the nucleus (Supplementary Fig. S1), suggesting that most of the GFP signal arises from MGMT-GFP fusions that are directed to the nucleus via the nuclear localization signal (NLS) in MGMT. MFI values for GFP were much higher in cells transduced with the dual-gene MAG vector, compared to cells transduced with the other dual-gene delivery strategies, possibly due to nuclear localization. Alternatively, MGMT-P140K has been shown to be incredibly stable with a half-life of >18 h42. Therefore, it is also possible that the nuclear localized GFP fusion proteins resist degradation, or are sequestered in the nucleus through DNA-binding activity of MGMT binding activity, allowing the fluorescent potential to accumulate.
Fig. 3. Dual-gene transfer and expression in K562 cells.
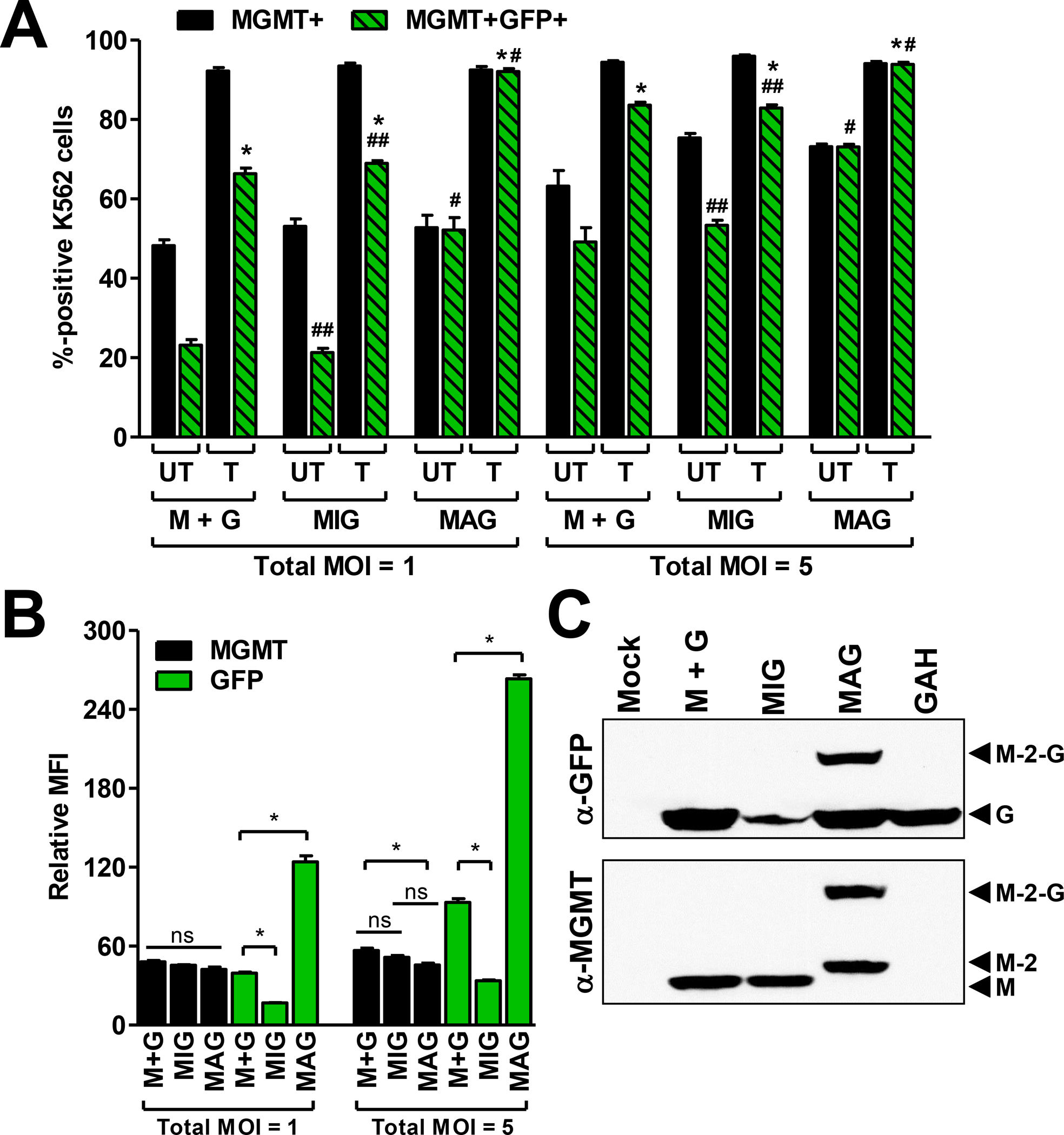
(A) Total MGMT+ or dual-positive (MGMT+GFP+) percentages obtained in K562 cells with each dual-gene transfer strategy. Equivalent total vector MOIs (1 or 5) were used for each transduction strategy. Cotransduction with single-gene vectors (M+G) was carried out using equivalent MGMT:GFP ratios (0.5:0.5 or 2.5:2.5). Transduced cultures were either mock treated (UT) or treated (T) with 10 μM BG and 25 μM BCNU and expanded in culture for 7 d prior to analysis. (B) Relative MFI of MGMT and GFP (measured separately) in cells from A after expansion in culture for 72 h. (C) Western blot of GFP (top) and MGMT (bottom) in protein extracts obtained from the drug-selected cultures in A (MOI = 1). The last lane in each blot is a control (K562 cells transduced with pMND-GFP-2A-HoxB4 not undergoing drug selection) demonstrating a lack of fusion product with a separate dual-gene vector with an identical 2A linker sequence. MGMT (M), GFP (G), MGMT with 2A peptide (M-2), and uncleaved MGMT-2A-GFP fusion product (M-2-G) are depicted by the black arrowheads. Data in A are means ± s.e.m. of a representative experiment with triplicate samples. *, P<0.05 compared to UT; #, P<0.05 compared to M+G or MIG; ##, P>0.05 compared to M+G. Data in B are means ± s.e.m. of a representative experiment with triplicate samples. *, P<0.05; ns, not significant (P>0.05).
3.3. Dual-gene transfer and in vitro enrichment of 5-FU-enriched BMCs
As a first approach to comparing these strategies in primary cells, 5-FU-enriched murine BMCs were transduced using each strategy. Higher MOIs were used to overcome the reduced transduction efficiency of primary murine cell populations17,43,44. Thus, BMC cultures were transduced with a total vector MOI of 20. Although the total MOI was held constant for each experiment, equivalent (10:10) or staggered (5:15) MGMT:GFP vector MOI ratios were used for cotransductions with the single-gene vectors. The total MGMT and dual-gene expression percentages in the cotransduced BMC cultures were lower, due to the lower MGMT vector MOIs used, and the interdependence of the individual MGMT and GFP vector transduction rates (Fig. 4A). Cotransduction with equal MGMT and GFP vector MOI ratios resulted in slightly higher percentages of dual-gene expressing cells compared to cotransduction with staggered MGMT:GFP vector MOI ratios (27% and 20%, respectively). However, 10 d after selection with 20 μM BG and 25 μM BCNU, the percentages of dual-gene expressing cells were highest in cultures cotransduced with the staggered (5:15) MGMT:GFP vector MOI ratios (67%) due to the greater selection pressure. This was similar to the dual-gene expression percentages observed in cultures transduced with the dual-gene MIG vector after drug treatment and culture expansion (62%). The same drug doses resulted in moderate enrichment of MGMT-expressing cells in cultures transduced with the dual-gene vectors (i.e. MIG or MAG) since these cultures had higher initial MGMT expression percentages (i.e. higher initial transduction resulted in less selection pressure).
Fig. 4. Dual-gene transfer and expression in 5-FU-enriched murine BMCs.
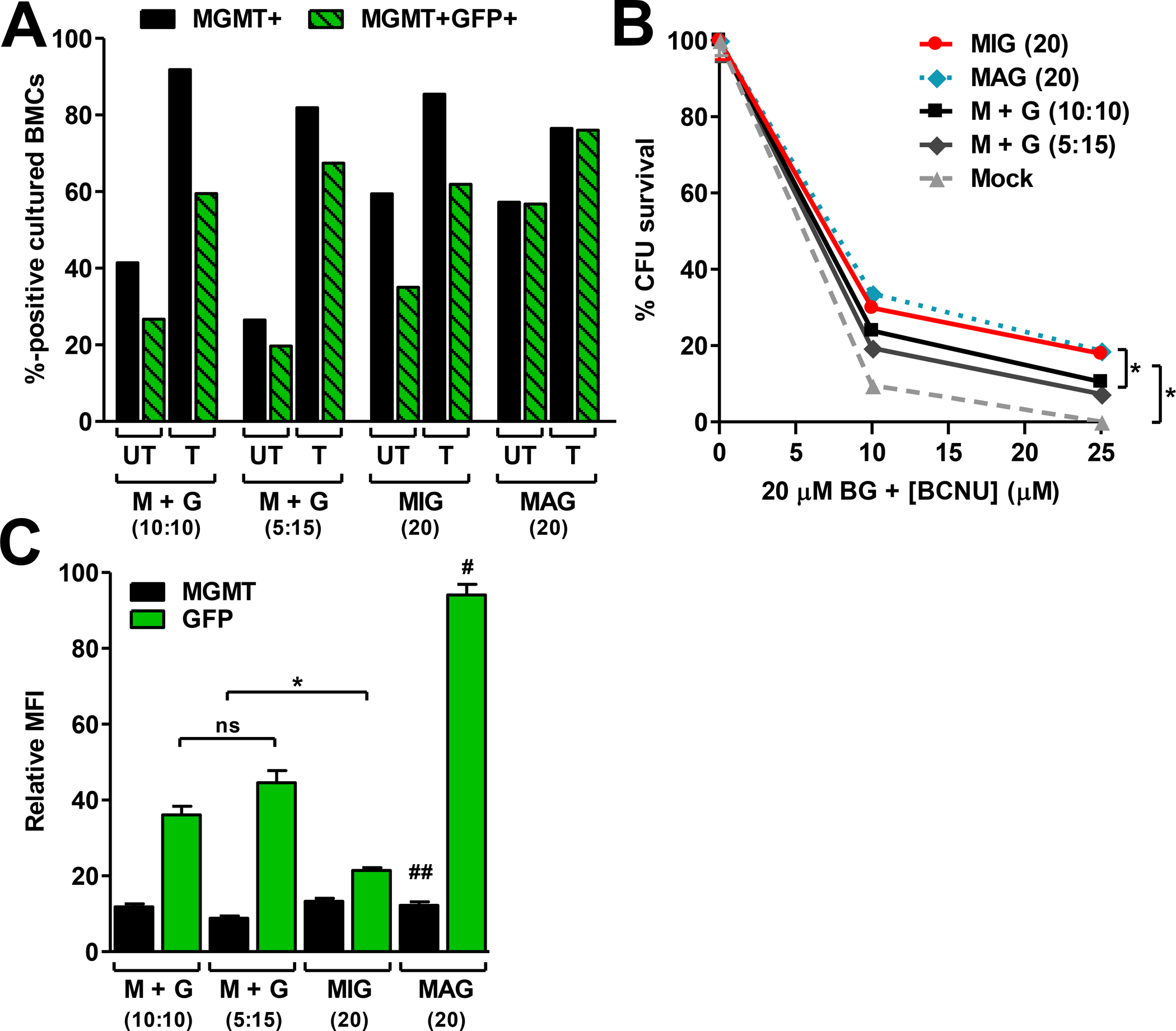
(A) Total MGMT+ or dual-positive (MGMT+GFP+) percentages obtained in BMCs with each dual-gene transfer strategy. Equivalent total vector MOIs of 20 were used for each strategy. Cotransductions were carried out using MGMT:GFP MOI ratios that were equivalent (10:10) or staggered (5:15). Transduced cultures were mock treated (UT) or treated (T) with 20 μM BG and 25 μM BCNU, and expanded in culture for 10 d prior to analysis. (B) CFU survival percentages in transduced BMC cultures following treatment with 20 μM BG and 0, 10, or 25 μM BCNU. (C) Relative MFI of MGMT or GFP expression in cells from A after expansion in culture for 10 d. Data in B and C are means ± s.e.m. of a representative experiment with triplicate samples. *, P<0.05; #, P<0.05 compared to M+G (10:10), M+G (5:15), and MIG; ##, P>0.05 compared to all other MGMT groups; ns, not significant (P>0.05).
The initially low MGMT expression percentages in the cotransduced cultures resulted in fewer BM progenitor CFU surviving drug treatment, compared to cultures transduced with the dual-gene vectors (Fig. 4B). Nevertheless, dual-gene expressing cells enriched from the single-gene vector cotransduction cultures expressed both genes at high levels (Fig. 4C). Equivalent MGMT expression levels were obtained in all of the BMC cultures after drug treatment, but GFP expression levels in each cohort were significantly different (MAG > cotransduction > MIG; Fig. 4C). As in K562 cells, the GFP fluorescent intensities in BMCs transduced with the MAG dual-gene vector were significantly higher (Fig. 4C), possibly due to nuclear localization of MGMT-GFP fusion products. The level of GFP expression in single-gene vector cotransduced cells was 50% higher than in cells transduced with the MIG dual-gene vector. Thus, in primary hematopoietic cells, cotransduction is limited by lower initial dual-gene expression percentages, while IRES vectors yield poor second gene expression levels, and 2A vectors are potentially limited by inefficient protein processing.
3.4. Dual-gene transfer and in vitro enrichment of LSK cells
Transduction efficiencies are reduced in more primitive hematopoietic cell populations. To determine whether the dual-gene expression percentages and the GFP expression levels differed in more primitive populations, similar comparisons were carried out on Lin−Sca-1+c-kit+ (LSK) cells. LSK cell cultures were transduced with each strategy using a total MOI of 100, which is typical for murine HSC transduction studies17,35,43. However, the total vector MOI was again divided into equal (50:50) or staggered (20:80) MGMT:GFP vector MOI mixtures for cotransduction with the single-gene vectors. The initial GFP expression percentages in the single-gene vector cotransduced LSK cultures had a greater impact on the dual-gene expression percentages after drug treatment than MGMT expression percentages, since limiting numbers of drug resistance cells were efficiently enriched (Fig. 5A). Therefore, the staggered MGMT:GFP vector MOI ratio was further skewed in favor of GFP expression in the single-gene vector cotransduced LSK cells. As an indicator of the overall lower transduction rate of LSK cells, the 5-fold higher dual-gene vector MOIs used to transduce LSK cells resulted in dual-gene expressing cell percentages that were similar to those obtained in BMCs transduced with lower MOIs (Figs. 4A and 5A).
Fig. 5. Dual-gene transfer and expression in LSK-enriched murine BMCs.
(A) Total MGMT+ or dual-positive (MGMT+GFP+) LSK cells obtained with each dual-gene transfer strategy. Equivalent total MOIs of 100 were used for each strategy. Cotransductions were carried out using MGMT:GFP MOI ratios that were equivalent (50:50) or staggered (20:80). Transduced cultures were mock treated (UT) or treated (T) with 20 μM BG and 25 μM BCNU, and expanded in culture 10 d prior to analysis. (B) CFU survival percentages in transduced LSK cultures following treatment with 20 μM BG and 0, 10, or 25 μM BCNU. (C) Relative MFI of MGMT or GFP expression in LSK cells from A after expansion in culture for 15 d. Data in B and C are means ± s.e.m. of a representative experiment with triplicate samples. *, P<0.05; #, P<0.05 compared to M+G (50:50), M+G (20:80), and MIG; ##, P>0.05 compared to all other MGMT groups; ns, not significant (P>0.05).
Transduction with the dual-gene MAG vector resulted in the highest percentage of dual-gene expressing cells, in both untreated and drug treated cultures. After drug treatment, the highest percentage of dual-gene expressing cells among the single-gene vector cotransduction cultures was obtained using the staggered (20:80) MGMT:GFP vector MOI mixture, which was likely due to the greater selection pressure (Fig. 5A). However, a slightly higher percentage of dual-gene expressing cells was obtained in cultures transduced with the dual-gene MIG vector after drug selection, compared to the maximum achieved with cotransduction (86% and 73%, respectively). Since independent transduction events are required for cotransduction, the reduced transduction efficiency of LSK cells likely affected cotransduction to a greater extent than that in the dual-gene vector transduced cultures.
MGMT expression percentages correlated with drug resistant CFU survival in each of the transduced LSK cultures (Fig. 5A, B). As expected, the percentage of drug-resistant CFU in MIG and MAG transduced cultures were equivalent at both BCNU doses evaluated, while CFU survival rates in cotransduced cultures were proportionately lower, since these cultures were exposed to lower MOIs of MGMT and were subjected to greater selective pressure (Fig. 5B). Nevertheless, this greater degree of selective pressure favored gene expression, as post drug-selected liquid cultures of cotransduced progenitors expressed GFP at levels twice as high as those transduced with the dual-gene MIG vector (Fig. 5C).
3.5. Enrichment of dual-gene expressing HSCs in vivo
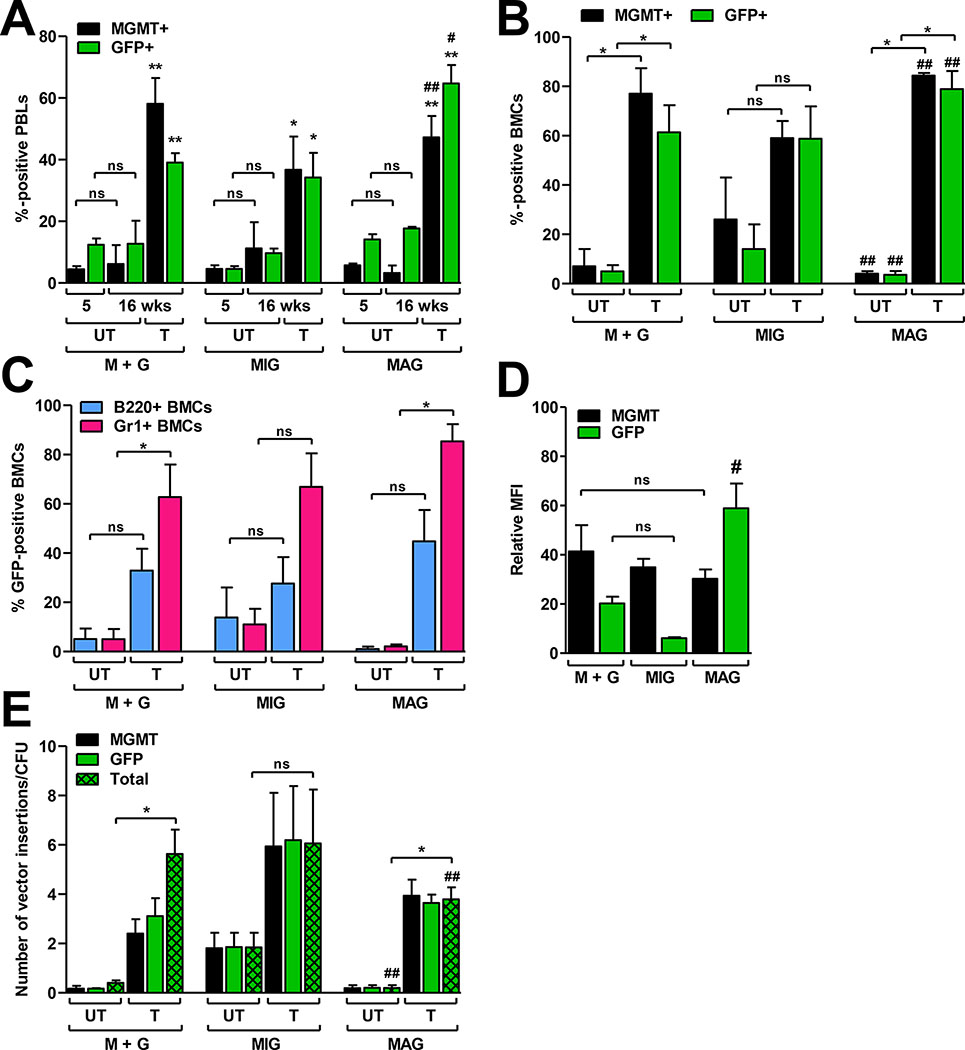
To evaluate dual-gene expression efficiencies over a longer time frame and more robust expansion conditions, we next compared each strategy in vivo. Donor LSK cell populations were transduced using a total MOI of 100 for each strategy. The staggered (20:80) MGMT:GFP vector MOI ratio was used for cotransduction, based on the superior dual-gene expression obtained with this ratio in LSK cultures treated in vitro. Lethally irradiated recipients were transplanted with 103 LSK cells from each transduction culture and 2 × 106 lineage-positive (Lin+) BMCs for hematopoietic support. At 5 weeks post-transplant the mice were mock treated or treated with three rounds of 30 mg/kg BG and 10 mg/kg BCNU, allowing 3 weeks recovery between each treatment. MGMT+ and GFP+ percentages were measured separately in peripheral blood leukocytes (PBL) 1 d prior to each treatment. MGMT and GFP measurements were carried out individually due to loss of GFP fluorescence intensity in primary cells when staining for MGMT (intranuclear), which made it difficult to resolve single and dual-gene expressing cell populations. Therefore, total GFP or total MGMT expression percentages are reported.
At 5 weeks post-transplant (before drug selection), the percentage of MGMT+ PBL were equivalent (5 ± 4%) in each of the three cohorts (Fig. 6A). GFP+ PBL percentages in cotransduced and MAG-transduced cell recipients were similar (14 ± 5% and 13 ± 4%, respectively), while MIG-transduced cell recipients had a lower percentage of GFP+ cells (5 ± 3%). After drug treatment and recovery (16 weeks post-transplant), the average MGMT+ and GFP+ PBL percentages were enriched to a similar degree in each cohort; the average MGMT+ PBL percentage in cotransduced recipient animals increased 11-fold (from 5 ± 3% to 58 ± 22%), compared to 8-fold increases (from 5 ± 4% to 37 ± 24%) in MIG-transduced cell recipients, and 9-fold increases (from 5 ± 2% to 47 ± 17%) in MAG-transduced cell recipients (Fig. 6A). Average GFP+ PBL percentages increased by 7-fold (from 5 ± 3% to 34 ± 18%) in MIG-transduced cell recipients and by 5-fold (from 13 ± 4% to 65 ± 15%) in MAG-transduced cell recipients. Enrichment for GFP+ cells was lower in the cohort cotransduced with the single-gene vectors (3-fold; from 14 ± 5% to 39 ± 8%) due to expansion of MGMT singly-transduced cells and loss of the drug sensitive GFP singly-transduced cells (Fig. 6A). One animal in the MIG-transduced cohort had to be euthanized after the third treatment. MGMT+ PBLs in this animal declined after the first and second treatments, indicating a lack of engraftment with drug-resistant cells. Mock treated animals in all three transduction cohorts did not show enrichment of either MGMT+ or GFP+ PBLs at 16 weeks post-transplant (p > 0.05).
Fig. 6. Dual-gene transfer, selection, and expression in vivo.
Three different cohorts of mice (n = 7–9 animals per cohort) were transplanted with cotransduced (M+G), MIG-transduced, or MAG-transduced LSK cells. (A) Total MGMT+ and GFP+ percentages in murine PBLs at 5 and 16 weeks after transplant. At 5 weeks post-transplant animals were mock treated or selected with 3 rounds of 30 mg/kg BG and 10 mg/kg BCNU, with 3 weeks between each round of selection. Values at 5 weeks are the expression percentages prior to the first treatment. Values depicted at 16 weeks are expression percentages in mock treated (UT; n = 2 animals per transduction cohort) or thrice-treated (T; n = 5–7 animals per transduction cohort) animals. (B–E) Recipient animals in A were sacrificed at 17 weeks post-transplant for BMC analysis. (B) Total MGMT+ and GFP+ expression percentages in total BMCs. (C) Total GFP+ expression percentages in lymphoid (B220+) and granulocytic (Gr-1+) BMCs. (D) Relative MFI of MGMT or GFP expression (measured separately) in BMCs. (E) The average number of vector insertions per cell, calculated from individual CFU. Values were determined using primers for either MGMT or GFP on individual CFU (n = 8–15 CFU per animal). Mean MGMT and GFP values were calculated from all of the CFU analyzed in each animal. Mean MGMT and GFP values were totaled (cotransduced CFU), or averaged (MIG and MAG transduced CFU) to get the total number of vector insertion per cell. For individual animal data, please see Supplementary Fig. S2. Data in A are means ± s.e.m. *, P<0.05 compared to 5 wks UT; **, P<0.05 compared to 5 or 16 wks UT; #, P<0.05 for MAG compared to either M+G or MIG; ##, P>0.05 for MAG compared to either M+G or MIG; ns, not significant (P>0.05). Data in B–E are means ± s.e.m. *, P<0.05 (T compared to UT); #, P<0.05 for MAG compared to either M+G or MIG; ##, P>0.05 for MAG compared to either M+G or MIG; ns, not significant (P>0.05).
At 17 weeks post-transplant, the animals were sacrificed and BMCs were harvested. At this point, stable hematopoiesis post-drug selection allowed analysis of progenitor selection, as impacted by the different transduction strategies. Drug selection increased the proportion of gene-expressing cells (Fig. 6B). MGMT+ BMC percentages in cotransduced cell recipient animals increased from 0–14% in the untreated controls to an average of 77 ± 27% in the drug treated animals. In the same cohort, GFP+ BMC percentages increased from 5 ± 4% in untreated animals, to 61 ± 29% in the drug treated animals (Fig. 6B). Among the dual-gene vector cohorts, the average MGMT+ and GFP+ BMC percentages in MIG-transduced recipients were not significantly different between drug treated and untreated control animals (p > 0.05) since one of the untreated animals engrafted with a high percentage of dual-gene expressing cells in the absence of selection (see Supplementary Fig. S2 for individual mouse data). In contrast, among the MAG-transduced cell recipients, MGMT+ BMC percentages increased from 4 ± 2% in untreated controls to 84 ± 3% in the drug treated animals. Likewise, GFP+ BMC percentages increased from 4 ± 2% to 79 ± 18% after drug treatment. Furthermore, multilineage selection was observed in each of the three drug treated animal cohorts, with increased GFP expression in B220+ and Gr-1+ BMC fractions (Fig. 6C). As with the in vitro experiments, the level of MGMT expressed in the MGMT+ BMCs was not significantly different among the cohorts, while the GFP expression levels were (Fig. 6D). As expected, GFP expression was localized to the nuclei of MAG-transduced cells (data not shown), which may artificially inflate MFI-based assessment of GFP expression levels. Interestingly, the GFP expression levels in cotransduced recipient BMCs was over 3-fold higher than that in cells transduced with the MIG vector.
3.6. Enrichment of transduced HSCs does not increase copy number
Real-time PCR analysis was then used to determine the number of total vector insertions per cell in individual CFU recovered from each cohort (Fig. 6E). While the proportion of transduced CFU increased after drug treatment, the average number of insertions in marked CFU was not significantly different among the different cohorts of drug treated animals (P>0.05). In drug-treated animals, the number of vector insertions per marked cell averaged 5 ± 3 (range 1.7 ± 0.2 to 10 ± 1) in cotransduced recipients, compared to 6 ± 5 (range 1.3 ± 0.5 to 14.1 ± 0.1) and 4 ± 1 (range 2.6 ± 0.1 to 6 ± 1) in MIG-transduced and MAG-transduced recipients, respectively. These data indicate that selection does not skew towards cells containing high copy numbers and, compared to dual-gene vector strategies, cotransduction with separate single-gene vectors does not result in an overall increase in vector insertions.
Numerical expansion levels were then estimated based on the engrafted cell transduction and expression efficiencies, as well as the total cellular reconstitution levels. On this basis, and assuming the normal mouse bone marrow compartment consists of ~3 × 108 BMCs45,46, dual-gene expressing cells in cotransduced recipient animals were enriched by an average of 1.8 ± 0.5 × 107-fold, compared to 1.8 ± 0.9 × 107-fold in MIG-transduced recipients and 1.18 ± 0.08 × 106-fold in MAG-transduced recipients. Additionally, studies by Colvin et al. suggest the bone marrow compartment is comprised of closer to 4.7 × 108 BMCs, which would result in further gains (~155% increase) in the enrichment of dual-gene expressing cells47. The lower level of MAG-transduced cell expansion was due to the lower selection pressure exerted on initially greater numbers of dual-gene expressing cells in the transduced donor cell populations. Thus, the overall level of dual-positive cell enrichment in the cotransduced and MIG-transduced recipients was equivalent.
These data indicate that similar levels of vector insertions are obtained with each strategy before and after drug selection. In addition, despite initially lower levels of MGMT+ cells, the cotransduced populations still contained sufficient numbers for engraftment and multilineage repopulation with cells expressing high levels of both. Thus, cotransduction with single-gene vectors may be an efficient alternative to dual-gene vectors for coupling hematopoietic progenitor selection to therapeutic gene expression, particularly as a first approach to establishing feasibility.
4. Discussion
Studies aimed at coupling selective stem cell enrichment to therapeutic gene expression have focused on the use of dual-gene vectors15–18. Many strategies have been employed to co-express two genes from the same vector, including the use of alternative splice sites, internal promoters, gene fusions, IRES elements, and ribosome slippage sites19–22. However, higher MOIs are often needed for dual-gene vectors to compensate for low expression levels. Recently, we demonstrated that cotransduction, coupled with MGMT-mediated selection, allows enrichment of dual-positive cells in vivo35. In the present study we aimed to directly compare cotransduction with single-gene vectors to transduction with dual-gene vectors in the context of MGMT-mediated hematopoietic progenitor cell enrichment. The work reported herein supports our previous work in that MGMT-mediated selection can be used to efficiently enrich hematopoietic progenitor cells cotransduced with separate single-gene lentivectors. Although the initial dual-gene transfer rates are reduced with this strategy, the percentage of dual-expressing cells that are enriched after drug selection are similar to percentages obtained after transduction with dual-gene vectors. In addition, the populations cotransduced with the single-gene vectors expressed both gene products at high levels with vector insertion averages equivalent to those obtained with dual-gene vectors.
Advantages and disadvantages exist for each dual-gene vector strategy that are specific to the particular application or gene combinations used. Strong selective pressure applied to alternative splicing or internal promoter strategies can lead to preferential splice site recognition or promoter interference, favoring drug resistance gene expression at the cost of therapeutic gene expression48. Gene fusion is likely the most efficient strategy, but requires that both gene products are functional as a fusion and active in the same cellular compartment. Previously, Amendola et al. reported the creation of dual-gene lentivectors using bidirectional promoters49. These promoters co-express two genes that are positioned in opposite orientations to one another in the same vector. However, the efficiency of this strategy in the setting of stringent drug selection remains to be determined.
The most common dual-gene expression strategies utilize IRES or ribosome slippage sequences for co-expression. Therefore, we compared cotransduction of single-gene MGMT and GFP vectors to dual-gene vectors that utilized IRES or 2A ribosome slippage sites to co-express MGMT and GFP. The EMCV IRES and FMDV-2A elements were specifically chosen, since these sequences have been thoroughly characterized and are routinely used in dual-gene expression vectors. The dual-gene MIG vector used in these studies was constructed to maximize IRES-mediated GFP expression. IRES activity has been reported to be most efficient when the IRES is placed less than 100 nucleotides downstream of the first gene50. Therefore, the IRES in the dual-gene MIG vector was inserted 70 nucleotides downstream of MGMT. The position of the second gene’s start codon with respect to the EMCV-IRES sequence is also critical, since a specific AUG site is preferentially used for IRES-initiated translation24. Thus, GFP was cloned into an NcoI site engineered to position the GFP start codon at the optimal site for IRES-initiated translation. Although the same genes had to be maintained for a comparative study, cis-acting complications have also been attributed to the specific genes used in bicistronic constructs27. In the studies reported here, MIG-transduced cells efficiently expressed MGMT, but the GFP expression levels were 2 to 3-fold lower than levels obtained in cells cotransduced with the single-gene vectors, despite similar vector insertion averages. Low IRES-mediated translation efficiencies have been reported using similar gene combinations (i.e. MGMT and GFP)51 as well as other gene combinations, and IRES elements27,31,52. IRES-mediated expression appears to be particularly low in lentiviral vectors. This may be due to the lower transcription rates obtained with internal lentivector promoters, compared to LTR-mediated transcription in retroviral vectors.
The FMDV-2A element has also been thoroughly characterized and successfully used in multi-gene vectors21,30–32,51,53–56. The 2A consensus sequence has been used in our lab to co-express other gene combinations. While, the C-terminal 2A fusion residues perturbed gene function in one such construct, no evidence of fusion products were previously detected. Similar complications have been reported in the literature by others. Lengler et al. found that the C-terminal 2A residues specifically destabilized some gene products and not others32. Milsom et al. and Chinnasamy et al. reported the use of dual-gene vectors, containing an MGMT-2A-GFP cassette similar to that of the dual-gene MAG vector reported here30,51. Although slightly different linker sequences may have been used, similar levels of MGMT-GFP fusion products were reported. Thus, the efficiency of 2A processing appears to be more related to the specific gene combinations31 or length of the 2A element28,57 used, rather than a cell type-specific phenomenon. Furthermore, other 2A sequence elements in addition to the FMDV-2A have been identified and evaluated in various systems, with data indicating that 2A processing efficiency also varies among these different elements55. Nevertheless, despite that as much as half of detected MGMT protein in MAG transduced cells was actually uncleaved MGMT-2A-GFP, MGMT repair activity was unaffected by this fusion with GFP. In this regard, the most efficient dual gene expression strategy in the studies reported here was achieved with the dual-gene MAG vector, similar to others51.
The selection stringency imparted through the BG + BCNU selection strategy used in this report impacts the level of enrichment which is primarily dependent on the level of MGMT expressed. In our hands there appears to be a threshold MOI required to efficiently transduce LSK cells, as determined by expression-based readouts. However, even limiting amounts of MGMT affords protection, but multiple insertions may be required to achieve even limiting expression. Nevertheless, we have previously demonstrated that more stringent selection does not necessarily translate to increased copy numbers58. Unlike the dual-gene vector strategies, which were limited by lower expression levels or inefficient processing, cells cotransduced with the single-gene vectors expressed both genes at high levels. Cotransduction is limited by the reduced dual-gene transfer rates inherent to this strategy. However, MGMT-mediated selection sufficiently enriched limiting numbers of cotransduced progenitor cells. Lower MGMT gene equivalents were used in cotransduction experiments to keep the total MOIs equivalent. This led to lower initial percentages of drug-resistant cells, compared to the percentages obtained with the dual-gene vectors. However, after selection the percentage of MGMT+ cells in the cotransduced cultures were equivalent to the percentages obtained with the dual-gene vectors. Thus, the level of expansion did not appear to be a limiting factor for cells cotransduced with the single-gene vectors. The initial proportion of dual-expressing cells to MGMT-singly transduced cells determined the dual-expressing cell percentage after selection. While the cotransduction studies reported here relied on the individual vector transduction rates, strategies that physically couple the separate vectors should improve the efficiency of cotransduction. Zhang et al. reported that low molecular weight polylysine polymers allow low-speed concentration of VSVG-pseudotyped lentivectors59. The virus-polylysine precipitates in these studies resembled “beads on a string.” Thus, a similar strategy might be used to co-precipitate separate viruses used for cotransduction, using vector MOI ratios that are optimized for each application.
5. Conclusions
These studies demonstrate that cotransduction with single-gene vectors is an efficient alternative to dual-gene vectors for selective expansion of dual-gene expressing hematopoietic progenitor cells in vivo. Although higher levels of expansion are required with this strategy, MGMT-mediated enrichment was not limiting. Further, the single-gene vectors were less prone to the cis-acting complications in the dual-gene vectors that reduced protein expression or processing. Since the degree of selective expansion and expression obtained using the single-gene MGMT vector is independent from that of another cotransduced vector, previously-established MGMT vector MOIs and drug-selection conditions could be applied to a wide range of therapeutic vectors as a first approach to establishing feasibility in HSC gene therapy models.
Supplementary Material
Supplementary Fig. S1. Nuclear visualization of GFP in K562 cells 72 h after transduction by each dual-gene transfer strategy (total vector MOI = 5 for each). Nuclei were stained with DAPI (blue) for comparison with GFP (green).
Supplementary Fig. S2. Dual-gene transfer, selection, and expression in vivo. Individual animal data corresponding to the pooled cohort data presented in Fig. 6. Three different cohorts of mice (n = 7–9 animals per cohort) were transplanted with cotransduced (M+G), MIG-transduced, or MAG-transduced LSK cells. At 5 weeks post-transplant animals were mock treated (UT; n = 2 animals per transduction cohort) or selected with 3 rounds of 30 mg/kg BG and 10 mg/kg BCNU, with 3 weeks between each round of selection (T; n = 5–7 animals per transduction cohort). Each panel represents data from a single mouse over the course of time at 5, 16, and 17 weeks post-transplant. Total MGMT+ (black bars) and GFP+ (green bars) percentages in murine PBLs are shown at 5 and 16 weeks after transplant. Values at 5 weeks are the expression percentages prior to the first treatment in treated animals. Animals were then sacrificed at 17 weeks post-transplant for BMC analysis. Total MGMT+ (black bars) and GFP+ (green bars) expression percentages in total BMCs are shown. The average number of vector insertions per cell, calculated from individual CFU is also depicted. Values were determined using primers for either MGMT or GFP on individual CFU (n = 8–15 CFU per animal). Mean MGMT (filled black circles) and GFP (filled green circles) values were calculated and plotted from all of the CFU analyzed in each animal. Mean MGMT and GFP values were totalled (cotransduced CFU), or averaged (MIG and MAG transduced CFU) to get the total number of vector insertion per cell (filled black and green diamonds).
HIGHLIGHTS.
Single-gene vector cotransduction was compared to transduction with dual-gene vectors
Equivalent MGMT expression levels were obtained with each strategy
Second-gene expression levels were lower in cells transduced with IRES dual-gene vector
Cotransduction is equivalent to dual-gene vectors for MGMT-mediated HSC selection in vivo
Vector insertions were similar for dual-gene vectors and single-gene vector cotransduction
Acknowledgements
This research was supported by the National Institutes of Health (NIH) grant R01CA073062 (SLG). Additional support was provided through the Flow Cytometry Core Facility of the Comprehensive Cancer Center of Case Western Reserve University and University Hospitals of Cleveland (NIH grant P30CA43703). JCR was supported in part by NIH training grants T32CA059366 (Research Oncology) and T32HL07147 (Hematology). The authors wish to thank Donald Kohn (University of California, Los Angeles, CA) for providing the pCSO-rre-cppt-MCU3-LUC plasmid and Thomas Hope (Northwestern University, Chicago, IL) for the woodchuck hepatitis virus posttranscriptional regulatory element (wPRE) template DNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Corey CA, DeSilva AD, Holland CA, Williams DA. Serial transplantation of methotrexate-resistant bone marrow: protection of murine recipients from drug toxicity by progeny of transduced stem cells. Blood. 1990;75(2):337–343. [PubMed] [Google Scholar]
- 2.Allay JA, Persons DA, Galipeau J, et al. In vivo selection of retrovirally transduced hematopoietic stem cells. Nat Med. 1998;4(10):1136–1143. [DOI] [PubMed] [Google Scholar]
- 3.Allay JA, Davis BM, Gerson SL. Human alkyltransferase-transduced murine myeloid progenitors are enriched in vivo by BCNU treatment of transplanted mice. Exp Hematol. 1997;25(10):1069–1076. [PubMed] [Google Scholar]
- 4.Sorrentino BP, Brandt SJ, Bodine D, et al. Selection of drug-resistant bone marrow cells in vivo after retroviral transfer of human MDR1. Science. 1992;257(5066):99–103. [DOI] [PubMed] [Google Scholar]
- 5.Davis BM, Koc ON, Gerson SL. Limiting numbers of G156A O(6)-methylguanine-DNA methyltransferase-transduced marrow progenitors repopulate nonmyeloablated mice after drug selection. Blood. 2000;95(10):3078–3084. [PubMed] [Google Scholar]
- 6.Bowman JE, Reese JS, Lingas KT, Gerson SL. Myeloablation is not required to select and maintain expression of the drug-resistance gene, mutant MGMT, in primary and secondary recipients. Mol Ther. 2003;8(1):42–50. [DOI] [PubMed] [Google Scholar]
- 7.Crone TM, Pegg AE. A single amino acid change in human O6-alkylguanine-DNA alkyltransferase decreasing sensitivity to inactivation by O6-benzylguanine. Cancer Res. 1993;53(20):4750–4753. [PubMed] [Google Scholar]
- 8.Crone TM, Goodtzova K, Edara S, Pegg AE. Mutations in human O6-alkylguanine-DNA alkyltransferase imparting resistance to O6-benzylguanine. Cancer Res. 1994;54(23):6221–6227. [PubMed] [Google Scholar]
- 9.Loktionova NA, Pegg AE. Point mutations in human O6-alkylguanine-DNA alkyltransferase prevent the sensitization by O6-benzylguanine to killing by N,N’-bis (2-chloroethyl)-N-nitrosourea. Cancer Res. 1996;56(7):1578–1583. [PubMed] [Google Scholar]
- 10.Reese JS, Koc ON, Lee KM, et al. Retroviral transduction of a mutant methylguanine DNA methyltransferase gene into human CD34 cells confers resistance to O6-benzylguanine plus 1,3-bis(2-chloroethyl)-1-nitrosourea. Proc Natl Acad Sci U S A. 1996;93(24):14088–14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollok KE, Hartwell JR, Braber A, et al. In vivo selection of human hematopoietic cells in a xenograft model using combined pharmacologic and genetic manipulations. Hum Gene Ther. 2003;14(18):1703–1714. [DOI] [PubMed] [Google Scholar]
- 12.Zielske SP, Reese JS, Lingas KT, Donze JR, Gerson SL. In vivo selection of MGMT(P140K) lentivirus-transduced human NOD/SCID repopulating cells without pretransplant irradiation conditioning. J Clin Invest. 2003;112(10):1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beard BC, Sud R, Keyser KA, et al. Long-term polyclonal and multilineage engraftment of methylguanine methyltransferase P140K gene-modified dog hematopoietic cells in primary and secondary recipients. Blood. 2009;113(21):5094–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beard BC, Trobridge GD, Ironside C, McCune JS, Adair JE, Kiem HP. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J Clin Invest. 2010;120(7):2345–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persons DA, Allay ER, Sawai N, et al. Successful treatment of murine beta-thalassemia using in vivo selection of genetically modified, drug-resistant hematopoietic stem cells. Blood. 2003;102(2):506–513. [DOI] [PubMed] [Google Scholar]
- 16.Falahati R, Zhang J, Flebbe-Rehwaldt L, Shi Y, Gerson SL, Gaensler KM. Chemoselection of allogeneic HSC after murine neonatal transplantation without myeloablation or post-transplant immunosuppression. Mol Ther. 2012;20(11):2180–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard E, Robert E, Cario-Andre M, et al. Hematopoietic stem cell gene therapy of murine protoporphyria by methylguanine-DNA-methyltransferase-mediated in vivo drug selection. Gene Ther. 2004;11(22):1638–1647. [DOI] [PubMed] [Google Scholar]
- 18.Trobridge GD, Wu RA, Beard BC, et al. Protection of stem cell-derived lymphocytes in a primate AIDS gene therapy model after in vivo selection. PLoS One. 2009;4(11):e7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan RA, Couture L, Elroy-Stein O, Ragheb J, Moss B, Anderson WF. Retroviral vectors containing putative internal ribosome entry sites: development of a polycistronic gene transfer system and applications to human gene therapy. Nucleic Acids Res. 1992;20(6):1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto Y, Aksentijevich I, Gottesman MM, Pastan I. Efficient expression of drug-selectable genes in retroviral vectors under control of an internal ribosome entry site. Biotechnology (N Y) 1994;12(7):694–698. [DOI] [PubMed] [Google Scholar]
- 21.de Felipe P, Martin V, Cortes ML, Ryan M, Izquierdo M. Use of the 2A sequence from foot-and-mouth disease virus in the generation of retroviral vectors for gene therapy. Gene Ther. 1999;6(2):198–208. [DOI] [PubMed] [Google Scholar]
- 22.Klump H, Schiedlmeier B, Vogt B, Ryan M, Ostertag W, Baum C. Retroviral vector-mediated expression of HoxB4 in hematopoietic cells using a novel coexpression strategy. Gene Ther. 2001;8(10):811–817. [DOI] [PubMed] [Google Scholar]
- 23.Jang SK, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4(9):1560–1572. [DOI] [PubMed] [Google Scholar]
- 24.Davies MV, Kaufman RJ. The sequence context of the initiation codon in the encephalomyocarditis virus leader modulates efficiency of internal translation initiation. J Virol. 1992;66(4):1924–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuguchi H, Xu Z, Ishii-Watabe A, Uchida E, Hayakawa T. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol Ther. 2000;1(4):376–382. [DOI] [PubMed] [Google Scholar]
- 26.Yu X, Zhan X, D’Costa J, et al. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol Ther. 2003;7(6):827–838. [DOI] [PubMed] [Google Scholar]
- 27.Hennecke M, Kwissa M, Metzger K, et al. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;29(16):3327–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnelly ML, Hughes LE, Luke G, et al. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol. 2001;82(Pt 5):1027–1041. [DOI] [PubMed] [Google Scholar]
- 29.de Felipe P Skipping the co-expression problem: the new 2A “CHYSEL” technology. Genet Vaccines Ther. Vol 22004:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milsom MD, Woolford LB, Margison GP, Humphries RK, Fairbairn LJ. Enhanced in vivo selection of bone marrow cells by retroviral-mediated coexpression of mutant O6-methylguanine-DNA-methyltransferase and HOXB4. Mol Ther. 2004;10(5):862–873. [DOI] [PubMed] [Google Scholar]
- 31.Chan HY, S V, Xing X, et al. Comparison of IRES and F2A-based locus-specific multicistronic expression in stable mouse lines. PLoS One. 2011;6(12):e28885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lengler J, Holzmuller H, Salmons B, Gunzburg WH, Renner M. FMDV-2A sequence and protein arrangement contribute to functionality of CYP2B1-reporter fusion protein. Anal Biochem. 2005;343(1):116–124. [DOI] [PubMed] [Google Scholar]
- 33.Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73(4):2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hlavaty J, Schittmayer M, Stracke A, et al. Effect of posttranscriptional regulatory elements on transgene expression and virus production in the context of retrovirus vectors. Virology. 2005;341(1):1–11. [DOI] [PubMed] [Google Scholar]
- 35.Roth JC, Ismail M, Reese JS, Lingas KT, Ferrari G, Gerson SL. Cotransduction with MGMT and Ubiquitous or Erythroid-Specific GFP Lentiviruses Allows Enrichment of Dual-Positive Hematopoietic Progenitor Cells In Vivo. ISRN Hematol. 2012;2012:212586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. [DOI] [PubMed] [Google Scholar]
- 37.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15(9):871–875. [DOI] [PubMed] [Google Scholar]
- 38.Reese JS, Liu L, Gerson SL. Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood. 2003;102(5):1626–1633. [DOI] [PubMed] [Google Scholar]
- 39.Davis BM, Reese JS, Koc ON, Lee K, Schupp JE, Gerson SL. Selection for G156A O6-methylguanine DNA methyltransferase gene-transduced hematopoietic progenitors and protection from lethality in mice treated with O6-benzylguanine and 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 1997;57(22):5093–5099. [PubMed] [Google Scholar]
- 40.Allay JA, Dumenco LL, Koc ON, Liu L, Gerson SL. Retroviral transduction and expression of the human alkyltransferase cDNA provides nitrosourea resistance to hematopoietic cells. Blood. 1995;85(11):3342–3351. [PubMed] [Google Scholar]
- 41.Choi U, DeRavin SS, Yamashita K, et al. Nuclear-localizing O6-benzylguanine-resistant GFP-MGMT fusion protein as a novel in vivo selection marker. Exp Hematol. 2004;32(8):709–719. [DOI] [PubMed] [Google Scholar]
- 42.Davis BM, Roth JC, Liu L, Xu-Welliver M, Pegg AE, Gerson SL. Characterization of the P140K, PVP(138–140)MLK, and G156A O6-methylguanine-DNA methyltransferase mutants: implications for drug resistance gene therapy. Hum Gene Ther. 1999;10(17):2769–2778. [DOI] [PubMed] [Google Scholar]
- 43.Visigalli I, Delai S, Politi LS, et al. Gene therapy augments the efficacy of hematopoietic cell transplantation and fully corrects mucopolysaccharidosis type I phenotype in the mouse model. Blood. 2010;116(24):5130–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestina TI, Hargrove PW, Jay D, Gray JT, Boyd KM, Persons DA. Correction of murine sickle cell disease using gamma-globin lentiviral vectors to mediate high-level expression of fetal hemoglobin. Mol Ther. 2009;17(2):245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boggs DR. The total marrow mass of the mouse: a simplified method of measurement. Am J Hematol. 1984;16(3):277–286. [DOI] [PubMed] [Google Scholar]
- 46.Reese JS, Qin X, Ballas CB, Sekiguchi M, Gerson SL. MGMT expression in murine bone marrow is a major determinant of animal survival after alkylating agent exposure. J Hematother Stem Cell Res. 2001;10(1):115–123. [DOI] [PubMed] [Google Scholar]
- 47.Colvin GA, Lambert JF, Abedi M, et al. Murine marrow cellularity and the concept of stem cell competition: geographic and quantitative determinants in stem cell biology. Leukemia. 2004;18(3):575–583. [DOI] [PubMed] [Google Scholar]
- 48.Emerman M, Temin HM. Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell. 1984;39(3 Pt 2):449–467. [PubMed] [Google Scholar]
- 49.Amendola M, Venneri MA, Biffi A, Vigna E, Naldini L. Coordinate dual-gene transgenesis by lentiviral vectors carrying synthetic bidirectional promoters. Nat Biotechnol. 2005;23(1):108–116. [DOI] [PubMed] [Google Scholar]
- 50.Attal J, Theron MC, Houdebine LM. The optimal use of IRES (internal ribosome entry site) in expression vectors. Genet Anal. 1999;15(3–5):161–165. [DOI] [PubMed] [Google Scholar]
- 51.Chinnasamy D, Milsom MD, Shaffer J, et al. Multicistronic lentiviral vectors containing the FMDV 2A cleavage factor demonstrate robust expression of encoded genes at limiting MOI. Virol J. 2006;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ibrahimi A, Vande Velde G, Reumers V, et al. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther. 2009;20(8):845–860. [DOI] [PubMed] [Google Scholar]
- 53.Furler S, Paterna JC, Weibel M, Bueler H. Recombinant AAV vectors containing the foot and mouth disease virus 2A sequence confer efficient bicistronic gene expression in cultured cells and rat substantia nigra neurons. Gene Ther. 2001;8(11):864–873. [DOI] [PubMed] [Google Scholar]
- 54.Ryan MD, Drew J. Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 1994;13(4):928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JH, Lee SR, Li LH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One. 2011;6(4):e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szymczak AL, Workman CJ, Wang Y, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589–594. [DOI] [PubMed] [Google Scholar]
- 57.Minskaia E, Ryan MD. Protein coexpression using FMDV 2A: effect of “linker” residues. Biomed Res Int. 2013;2013:291730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zielske SP, Lingas KT, Li Y, Gerson SL. Limited lentiviral transgene expression with increasing copy number in an MGMT selection model: lack of copy number selection by drug treatment. Mol Ther. 2004;9(6):923–931. [DOI] [PubMed] [Google Scholar]
- 59.Zhang B, Xia HQ, Cleghorn G, Gobe G, West M, Wei MQ. A highly efficient and consistent method for harvesting large volumes of high-titre lentiviral vectors. Gene Ther. 2001;8(22):1745–1751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Nuclear visualization of GFP in K562 cells 72 h after transduction by each dual-gene transfer strategy (total vector MOI = 5 for each). Nuclei were stained with DAPI (blue) for comparison with GFP (green).
Supplementary Fig. S2. Dual-gene transfer, selection, and expression in vivo. Individual animal data corresponding to the pooled cohort data presented in Fig. 6. Three different cohorts of mice (n = 7–9 animals per cohort) were transplanted with cotransduced (M+G), MIG-transduced, or MAG-transduced LSK cells. At 5 weeks post-transplant animals were mock treated (UT; n = 2 animals per transduction cohort) or selected with 3 rounds of 30 mg/kg BG and 10 mg/kg BCNU, with 3 weeks between each round of selection (T; n = 5–7 animals per transduction cohort). Each panel represents data from a single mouse over the course of time at 5, 16, and 17 weeks post-transplant. Total MGMT+ (black bars) and GFP+ (green bars) percentages in murine PBLs are shown at 5 and 16 weeks after transplant. Values at 5 weeks are the expression percentages prior to the first treatment in treated animals. Animals were then sacrificed at 17 weeks post-transplant for BMC analysis. Total MGMT+ (black bars) and GFP+ (green bars) expression percentages in total BMCs are shown. The average number of vector insertions per cell, calculated from individual CFU is also depicted. Values were determined using primers for either MGMT or GFP on individual CFU (n = 8–15 CFU per animal). Mean MGMT (filled black circles) and GFP (filled green circles) values were calculated and plotted from all of the CFU analyzed in each animal. Mean MGMT and GFP values were totalled (cotransduced CFU), or averaged (MIG and MAG transduced CFU) to get the total number of vector insertion per cell (filled black and green diamonds).