Summary
Once naive B cells expressing germline VRC01-class B cell receptors become activated by germline-targeting immunogens, they enter germinal centers and undergo affinity maturation. Booster immunizations with heterologous Envs are required for the full maturation of VRC01-class antibodies. Here, we examined whether and how three adjuvants, Poly(I:C), GLA-LSQ, or Rehydragel, that activate different pathways of the innate immune system, influence the rate and type of somatic mutations accumulated by VRC01-class BCRs that become activated by the germline-targeting 426c.Mod.Core immunogen and the heterologous HxB2.WT.Core booster immunogen. We report that although the adjuvant used had no influence on the durability of plasma antibody responses after the prime, it influenced the plasma VRC01 antibody titers after the boost and the accumulation of somatic mutations on the elicited VRC01 antibodies.
Subject areas: Biological sciences, Immune response, Immunology, Virology
Graphical abstract
Highlights
-
•
VRC01 B cells can be activated in vivo by a germline-targeting HIV-1 Env immunogen
-
•
Adjuvants affect the magnitude of the initial VRC01 B cell response
-
•
Adjuvants affect plasma VRC01 antibody titers following heterologous boosting
-
•
Adjuvants affect the rate and type of mutations in maturing VRC01 BCRs
Biological sciences; Immune response; Immunology; Virology.
Introduction
Broadly neutralizing HIV-1 antibodies (bnAbs) have been isolated from HIV-1-infected individuals and the structures of diverse bnAbs, as well as those of their epitopes on the HIV-1 envelope glycoprotein trimeric spike (Env), have been well characterized (Burton and Hangartner, 2016; Mascola and Haynes, 2013; West et al., 2014). The epitopes targeted by bnAbs are located on multiple regions of Env, such as its apex (Bhiman et al., 2015; Doria-Rose et al., 2014; Gorman et al., 2016; McLellan et al., 2011; Pancera et al., 2010; Walker et al., 2009), the CD4-binding site (CD4-BS) (Barbas et al., 1992; Ditzel et al., 1995; Gristick et al., 2016; Huang et al., 2016; Kwong and Mascola, 2012; Sajadi et al., 2018; Scheid et al., 2011; Umotoy et al., 2019; Wu et al., 2011; Zhou et al., 2015), the interface between the gp120 and gp41 subunits (Blattner et al., 2014; Falkowska et al., 2014; Huang et al., 2012; Scharf et al., 2014, 2015), the silent face of gp120 (Schoofs et al., 2019; Zhou et al., 2018), and the membrane proximal regions (MPER) of gp41 (along with lipid moieties into which MPER is embedded) (Brunel et al., 2006; Cardoso et al., 2005; Huang et al., 2012; Lee et al., 2015; Muster et al., 1993, 1994; Stiegler et al., 2001; Zwick et al., 2001, 2005). In addition, some bnAbs recognize clusters of glycan moieties on the gp120 subunit (Calarese et al., 2003; Scanlan et al., 2002; Trkola et al., 1996), whereas other bnAbs recognize epitopes that contain both glycans and polypeptides (Bonsignori et al., 2017; Doores et al., 2015; Julien et al., 2013; Longo et al., 2016; Moore et al., 2012; Mouquet et al., 2012; Pancera et al., 2013; Pejchal et al., 2011; Walker et al., 2010, 2011a, 2011b).
bnAbs that recognize the same region of Env and share common genetic and structural features are grouped into ‘classes’ (Kwong and Mascola, 2012). The VRC01-class of antibodies recognize an epitope within the CD4-BS, and their heavy chains (HCs) are derived from the VH1-2∗02 allele whereas their light chains (LCs) express 5 amino acid (5 aa) long CDRL3 domains (Scharf et al., 2013, 2016; Scheid et al., 2011; Umotoy et al., 2019; West et al., 2012; Wu et al., 2011, 2015; Zhou et al., 2013, 2015). We note however that, not every Ab formed by the above-association of heavy and light chains targets the CD4-BS (Gray et al., 2021). They are among the most mutated bnAbs known (Klein et al., 2013) and can display up to 30% amino acid sequence divergence, yet they recognize their epitope on diverse Envs with similar angles of approach (Scheid et al., 2011; Wu et al., 2011; Zhou et al., 2015). VRC01-class bnAbs protect animals from experimental S/HIV infection (Balazs et al., 2014; Shingai et al., 2014) and one mAb of that class, VRC01, was recently shown to prevent HIV-1 acquisition from susceptible, circulating primary HIV-1 viruses, in large phase 3 clinical trials (Corey et al., 2021). Thus, it is expected that VRC01-class bnAbs should be a component of the immune responses elicited by an effective HIV-1 vaccine.
Although VRC01-class bnAbs isolated from HIV-1 infected individuals bind diverse recombinant (rec) Envs and potently neutralize HIV-1 viruses from different clades, their unmutated forms do not (Hoot et al., 2013; Jardine et al., 2013; McGuire et al., 2013, 2014b; Wu et al., 2011). So far, a natural Env (as expressed by a circulating virus) capable of binding the unmutated forms (‘germline’, gl) of VRC01-class antibodies has not been identified. It is believed that one reason for the lack of elicitation of VRC01-class antibodies through immunization with rec Envs is because of the failure of such proteins to activate naive B cells that express the unmutated B cell receptor (BCR) precursors of VRC01-class antibodies (Jardine et al., 2013; McGuire et al., 2013; Stamatatos et al., 2017).
We and others have designed Env-derived proteins capable of binding unmutated VRC01-class antibodies (Jardine et al., 2013, 2016; Lin et al., 2020; McGuire et al., 2013, 2016; Medina-Ramirez et al., 2017; Parks et al., 2019). Such constructs are commonly referred to as ‘germline-targeting’ (Stamatatos et al., 2017). Germline-targeting proteins have been designed on the backbone of the outer domain of gp120 (Jardine et al., 2013, 2016; Lin et al., 2020), the gp120 core (expressing both the inner and outer gp120 domains) (426c.Mod.Core) (McGuire et al., 2014a, 2016; Parks et al., 2019), and the entire extracellular portion of the viral Env (GT1.1) (Medina-Ramirez et al., 2017). A common feature of all such constructs is the elimination of the conserved N-linked glycosylation site at position N276 (Loop D of gp120), because glycans present at this position prevent glVRC01-antibodies/BCRs from binding to Env (Zhou et al., 2010). However, additional obstacles on the HIV-1 Envs, including the lengths of the V1-V3 regions, prevent the engagement of germline VRC01-class BCRs by rec Envs (Borst et al., 2018; McGuire et al., 2016).
Orthologs of the human VH1-2∗02 gene are not expressed in wild type animals such as mice, rats, rabbits, and non-human primates (West et al., 2012), thus the abilities of germline-targeting immunogens to activate naive B cells expressing glVRC01-class BCRs have so far been evaluated in specifically engineered mice. Indeed, germline-targeting immunogens activate B cells expressing glVRC01-class antibodies in vivo (Abbott et al., 2018; Briney et al., 2016; Chen et al., 2021; Dosenovic et al., 2018; Jardine et al., 2015; Lin et al., 2020; Medina-Ramirez et al., 2017; Parks et al., 2019; Sok et al., 2016; Tian et al., 2016), although by themselves are not capable of guiding the proper maturation of VRC01-class BCRs toward their broadly neutralizing forms, through the accumulation of specific somatic mutations (Chen et al., 2021; Dosenovic et al., 2018). It is hypothesized that following the activation of naive B cells expressing glVRC01-class BCRs by germline-targeting immunogens, sequential immunizations with diverse (and gradually more native-like rec Envs) will be necessary to achieve this goal (Chen et al., 2021; Tian et al., 2016).
Adjuvants alter the magnitude as well as the quality of the immune response (Lambrecht et al., 2009; Pulendran et al., 2021; Silva et al., 2021). However, the effect of adjuvant on the selection, expansion, and maturation of VRC01 B cell subclasses has not yet been investigated in detail, although there are some reports suggesting that different adjuvants may select for different HC/LC amino acids (Jardine et al., 2015). As immunogens targeting the unmutated forms of VRC01-class antibodies enter clinical evaluation, identifying the adjuvant that promotes high levels of the appropriate somatic mutations will be important.
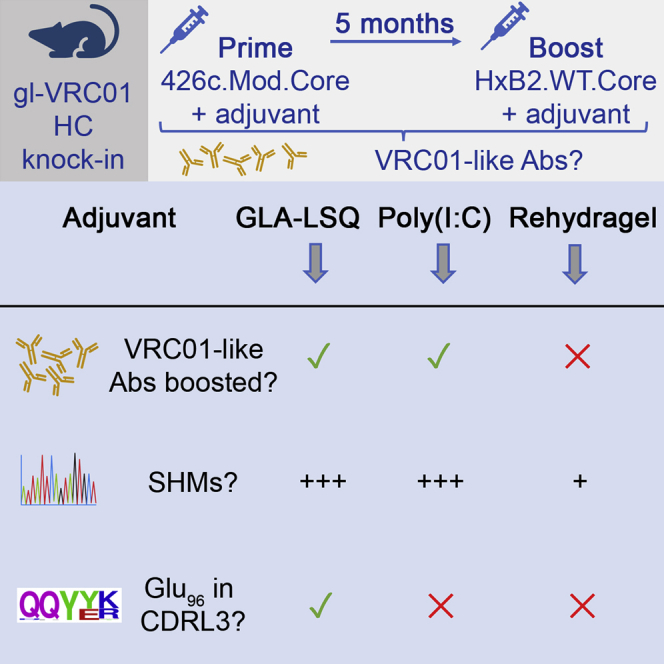
Here, we investigated how adjuvants affect the magnitude and duration of VRC01 antibody and B cell responses elicited by the 426c.Mod.Core germline-targeting immunogen (McGuire et al., 2014a, 2016) and whether these responses can be boosted by a heterologous Env that does not recognize glVRC01-class BCRs on its own. To this end, we compared the antibody and B cell responses elicited by 426c.Mod.Core when expressed on Ferritin nanoparticles and adjuvanted with polyinosinic-polycytidylic acid (Poly(I:C)), GLA-LSQ, or Rehydragel. Poly(I:C) is a double-stranded RNA analogue mimicking viral RNA that stimulates endosomal TLR3, GLA-LSQ consists of the TLR4 agonist glucopyranosyl lipid A (GLA) and the saponin QS21 in a lipid formulation, whereas Rehydragel, an aluminum hydroxide particulate formulation, does not activate known TLR pathways (Pulendran et al., 2021). We also investigated whether the adjuvants affect the maturation of VRC01-like B cells following a heterologous booster immunization with HxB2.WT.Core.
Our study indicates that long-lasting VRC01-like plasma antibody responses were generated irrespective of the adjuvant used during the prime immunization, but that the titers of plasma VRC01-like antibodies and the number of somatic mutations accumulated in VRC01-like antibodies were influenced by the adjuvant used during the heterologous boost immunization.
Results
High titers of anti-CD4-binding site antibodies are elicited by a single immunization with 426c.Mod.Core irrespective of the adjuvant used
The mouse model we employed here is heterozygous for the human inferred glHC of the VRC01 mAb (VRC01glHC) (Jardine et al., 2015). In this model, approximately 80% of B cells express the human transgene and all express mouse LCs (mLCs); ∼0.1% of which contain 5 aa-long CDRL3s. As a result, ∼ 0.08% of naive B cells in these mice express VRC01-like BCRs, as compared to ∼0.01% in humans (Abbott et al., 2018; Havenar-Daughton et al., 2018; Jardine et al., 2015). By VRC01-class BCRs, we mean antibodies expressing the knocked-in human glVRC01HC paired with mouse LCs expressing 5 amino acid long CDRL3.
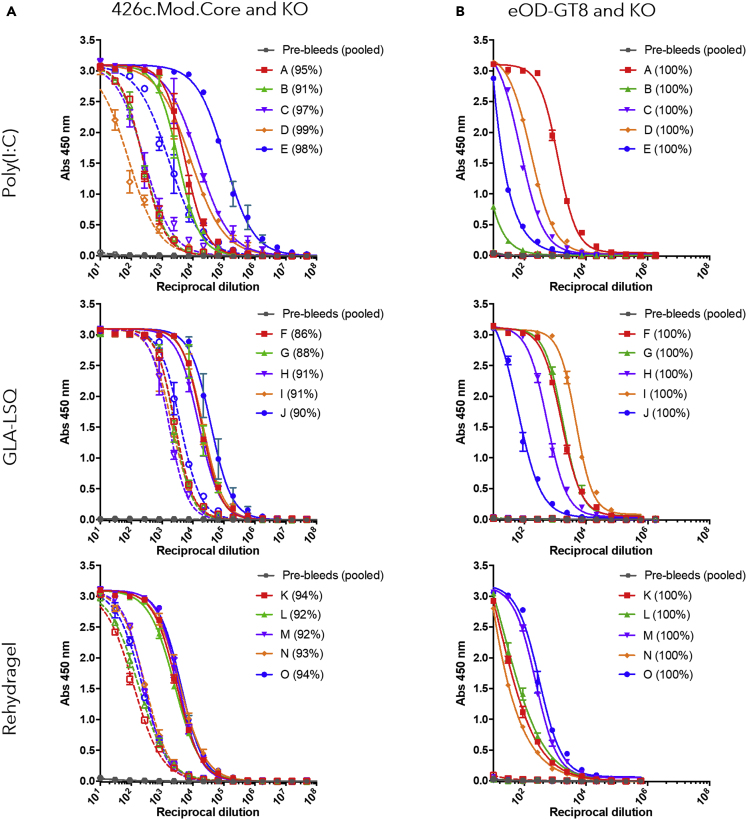
To test the effect of the adjuvant on the magnitude of the elicited antibody response, mice were immunized with 426c.Mod.Core Ferritin nanoparticles (24meric) adjuvanted with either Poly(I:C), GLA-LSQ, or Rehydragel. At the peak of the plasma antibody response (2 weeks after immunization), all animals, irrespective of the adjuvant, generated autologous plasma antibodies, the majority of which targeted the CD4-BS on 426c.Mod.Core (autologous anti-CD4-BS antibodies) (Figure 1A). At this point, the anti-426c.Mod.Core plasma antibody titers were significantly higher in the GLA-LSQ group than the Rehydragel group (p ≤ 0.05). However, no significant differences in the relative percentage of plasma antibodies against the CD4-BS of 426c.Mod.Core were observed at this time point. All animals, irrespective of the adjuvant used, also developed antibody responses against the heterologous germline-targeting immunogen, eOD-GT8 (Jardine et al., 2015), at this early time point after immunization (Figure 1B). The anti-eOD-GT8 plasma antibody responses exclusively targeted the VRC01 epitope on that protein, as they did not display reactivity to the version of the protein with a mutated VRC01 epitope (eOD-GT8 KO). We concluded that a single immunization with 426c.Mod.Core Ferritin nanoparticles, elicits high titers of autologous anti-CD4-BS antibodies, including antibodies that bind the VRC01 epitope, irrespective of the adjuvant used.
Figure 1.
Ab responses at 2 weeks following 426c.Mod.Core Ferritin immunization
Mice (n = 5 per group) were immunized with 426c.Mod.Core Ferritin with either Poly(I:C), GLA-LSQ, or Rehydragel. Mice were bled at 2 weeks after immunization and plasma was assayed by ELISA for binding against 426.Mod.Core (A) and eOD-GT8 (B) (full lines), as well as corresponding antigens with CD4-BS or VRC01 epitope knock-out (KO) (dotted lines). Figure legend indicates individual mouse, and percentage in parentheses indicates percent of response binding to CD4-BS, calculated based on the differences in areas under the curve. Pre-bleed samples from all animals (pool) was used as an internal control.
Sustained autologous and heterologous HIV-1 Env antibody responses following a single immunization with 426c.Mod.Core
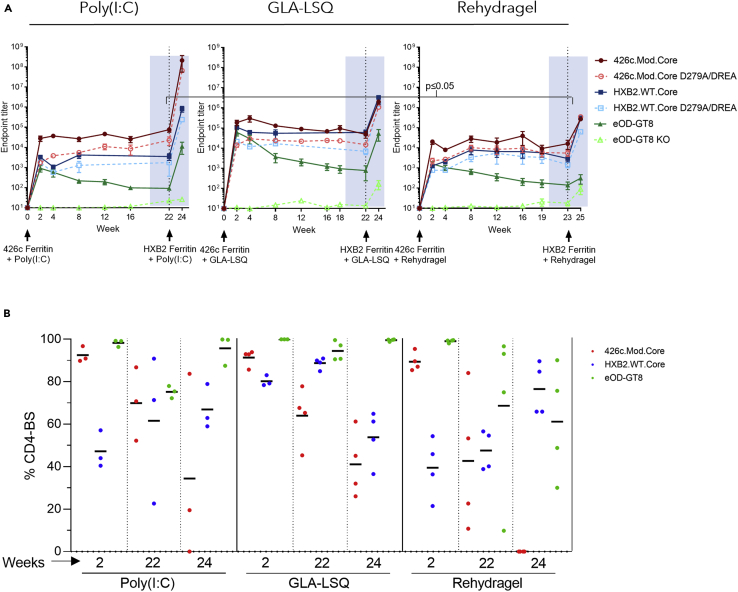
We next examined whether the longevity of the elicited antibody responses was affected by the adjuvant used. To this end, new groups of animals were immunized, and their responses were determined over a period of 22–23 weeks (Figure 2A). The autologous plasma IgG titers were maintained at high levels over the course of observation (22–23 weeks). At this late timepoint after immunization, the anti-426c.Mod.Core plasma antibody titers in the GLQ-LSQ and Poly(I:C) groups were not statistically different, although the antibody titers with Poly(I:C) were significantly higher than those in the Rehydragel group (p≤0.05).
Figure 2.
Ab responses following 426c.Mod.Core Ferritin prime and HXB2.WT.Core Ferritin boost immunization
Mice were primed with 426c.Mod.Core Ferritin at week 0 and boosted with HXB2.WT.Core Ferritin at week 22 (Poly(I:C) and GLA-LSQ groups), or week 23 (Rehydragel group). Mice were bled at the time points indicated in (A), and plasma was assayed for binding against the proteins listed in the legend by ELISA. Black dotted line indicates the time of the booster immunization.
(A) Mean endpoint titers against the indicated proteins over time. The anti-426c.Mod.Core plasma antibody titers in the Poly(I:C) at week 22 were significantly higher than those in the Rehydragel group at week 23 (p≤0.05)
(B) Percentage of anti-CD4-BS against 426c.Mod.Core (red), HxB2.WT.Core (blue) and VRC01 epitope on eOD-GT8 (green), calculated based on the differences in endpoint titer values, at the indicated times with the indicated adjuvants. Each dot represents an animal. See also Figure S1.
We also examined whether the fraction of autologous anti-CD4-BS antibody responses remained constant over time. These responses peaked early following prime immunization (2 weeks after immunization), but their relative proportions slowly decreased during the period of observation in all groups (Figure 2B). At 22–23 weeks after immunization, the autologous CD4-BS antibody responses represented ∼70% of the total autologous antibodies in the Poly(I:C) group (a drop of 24% from their peak value), 64% in the GLA-LSQ group (a drop of 30% from their peak value), and 43% in the Rehydragel group (a drop of 52% from their peak value) (Figure 2B).
At the peak of the response, the anti-eOD-GT8 plasma antibody titers were 500-1,500-fold lower than the anti-426c.Mod.Core antibody titers and although the anti-eOD-GT8 titers gradually declined over time, they were always detectable (Figure 2A). The majority of the anti-eOD-GT8 antibody responses targeted the VRC01 epitope on that protein for the duration of observation period as the reactivity to the eOD-GT8 KO was minimal (Figure 2A).
The VRC01-like antibodies elicited by 426c.Mod.Core in this mouse model recognize heterologous, fully glycosylated wild type gp120 Core proteins (Parks et al., 2019). These constructs (termed ‘WT Cores’ for simplicity) are not recognized by glVRC01-class antibodies, but once activated by the 426c.Mod.Core, the VRC01-expressing B cells enter the germinal centers where their BCRs accumulate somatic mutations that allow them to bypass N276- and V5- associated glycans on some heterologous WT Core proteins. One of these WT Cores is the HxB2.WT.Core. Indeed, all animals generated durable anti-HxB2.WT.Core antibody responses following a single immunization with the 426c.Mod.Core (Figure 2A), although the peak anti-HxB2.WT.Core responses were lower than those against 426c.Mod.Core. At 2 weeks after immunization, the anti-HxB2.WT.Core antibody titers were ∼88%, ∼15% and 92% lower than the anti-426c.Mod.Core titers for Poly(I:C), GLA-LSQ and Rehydragel, respectively. The anti-HxB2.WT.Core plasma antibody titers remained stable for the duration of the observation, irrespective of the adjuvant used, and a fraction of these Ab target the CD4-BS. At week 2 after immunization, the relative fraction of the plasma antibodies that recognized the CD4-BS on HxB2.WT.Core were ∼50% in the Poly(I:C) group, ∼ 80% in the GLA-LSQ group, and ∼40% in the Rehydragel group (Figure 2B). The relative proportion of these heterologous anti-CD4-BS plasma antibodies remained unchanged over the duration of the observation. Thus, at 23 weeks after immunization, the relative fraction was ∼60% in the Poly(I:C) group, ∼90% in the GLA-LSQ group, and ∼50% in the Rehydragel group. We conclude that a single immunization with 426c.Mod.Core Ferritin nanoparticles elicits long-lasting autologous and heterologous anti-CD4-BS antibodies, a fraction of which target the VRC01 epitope.
HXB2.WT.Core boosts autologous and heterologous anti-CD4-BS antibody responses primed by 426c.Mod.Core
We previously reported that immunization with a 7meric nanoparticle form of 426c.Mod.Core (426c.Mod.Core-C4b) adjuvanted with GLA-LSQ elicits VRC01-like antibody responses that are boosted by the heterologous HxB2.WT.Core (also in a 7meric nanoparticle form, HxB2.WT.Core-C4b), when administered 4 weeks after the prime immunogen, i.e., at the peak of the antibody response (Lin et al., 2020; Parks et al., 2019). As a result, the VRC01-like antibodies isolated after this prime-boost immunization schema, have more somatic mutations and enhanced Env-binding affinities than the VRC01-like antibodies isolated after the 426c.Mod.Core prime immunization alone (Parks et al., 2019). Here, we examined how the anti-CD4-BS antibody responses, and more specifically the VRC01-like antibody responses elicited by the Ferritin nanoparticle form of 426c.Mod.Core, were affected by the adjuvant and by delaying the timing of the boost immunization with HxB2.WT.core Ferritin nanoparticles. To this end, animals were immunized with HxB2.WT.core Ferritin nanoparticles 22–23 weeks after the prime immunization.
This boost immunization resulted in increases in the plasma antibody responses against 426c.Mod.Core and HxB2.WT.Core in all three groups (Figure 2A). The anti-426c.Mod.Core plasma antibody titers increased by ∼3,500-fold in the Poly(I:C) group, and by ∼1,500-fold in both the GLA-LSQ and the Rehydragel groups. The anti-HxB2.WT.Core titers increased by ∼3,000-fold in the Poly(I:C) group, by ∼2,000-fold in the GLA-LSQ group, and by ∼1,000-fold in the Rehydragel group. However, the relative proportions of anti-426c.Mod.Core and of anti-HxB2.WT.Core CD4-BS plasma antibodies were lower after the boost immunization than after the prime immunization (Figure 2B). Two weeks following the heterologous boost immunization, the proportions of plasma antibodies targeting the 426c.Mod.Core CD4-BS were ∼34% in the Poly(I:C) group and ∼41% in the GLA-LSQ group, and no longer apparent in the Rehydragel group. The proportion of antibodies targeting the CD4-BS of the booster antigen HxB2.WT.Core, was ∼67% in the Poly(I:C) group, ∼54% in the GLA-LSQ group, and ∼77% in the Rehydragel group. Of interest, the VRC01 plasma antibody titers, were boosted in the Poly(I:C) group by ∼2,000-fold and in the GLA-LSQ group by ∼1,500-fold, but not in the Rehydragel group. We concluded that Rehydragel may not be an optimal adjuvant to boost VRC01 plasma antibody responses elicited by the 426c.Mod.Core germline-targeting immunogen.
The HXB2.WT.Core immunogen does not elicit VRC01-like plasma antibody responses
The HxB2.WT.Core does not bind glVRC01 antibodies (Parks et al., 2019), and it is thus expected that it will not activate naive B cells expressing glVRC01-like BCRs. Hence, we hypothesized that the increase in VRC01-like plasma antibody responses observed during the booster immunization with HxB2.WT.Core in the case of the Poly(I:C) and GLA-LSQ adjuvants was not due to the activation of naive VRC01-like B cells by the HxB2.WT.Core itself, but was due to a boosting of the memory VRC01-like B cells that transitioned to plasma cells secreting plasma antibodies at that stage. These memory VRC01-class B cells initially got activated by 426c.Mod.Core and their BCRs accumulated relevant somatic mutations that allowed them to bind HxB2.WT.Core.
To prove this point directly, we immunized mice with HxB2.WT.Core Ferritin nanoparticles adjuvanted with GLA-LSQ and examined the Env-recognition properties of the elicited plasma antibody. High autologous and anti-426c.Mod.Core plasma antibody titers were generated in all animals 2 weeks after immunization (Figure S1A). Between 20 and 70% of the anti-426c.Mod.Core, and between 60 and 80% of the anti-HxB2.WT.Core responses, targeted the corresponding CD4-BS (Figure S1B). Anti-eOD-GT8 plasma antibody responses were generated by 2 of 4 animals and were of lower magnitude than the anti-426c.Mod.Core or anti-HxB2.WT.Core plasma antibody responses (Figure S1A). However, the plasma antibody titers to eOD-GT8 and eOD-GT8 KO were either similar or the anti-eOD-GT8 KO titers were higher than the anti-eOD-GT8 titers (Figure S1A). These observations confirm that HxB2.WT.Core activates B cells that can target epitopes expressed on different Envs, some of which are located within the CD4-BS but is not capable of activating the B cells that produce VRC01 antibodies. This observation also supports the notion that HxB2.WT.Core can activate VRC01-like B cells only when they have accumulated relevant somatic mutations (Parks et al., 2019).
Characterizing the impact of adjuvants on the maturation of VRC01-like antibodies
To prove directly that the boost immunization with HxB2.WT.Core led to the expansion of memory B cells expressing VRC01-like BCRs, we isolated such cells at 2 weeks after the prime immunization and two weeks after the boost and sequenced their VH/VL genes.
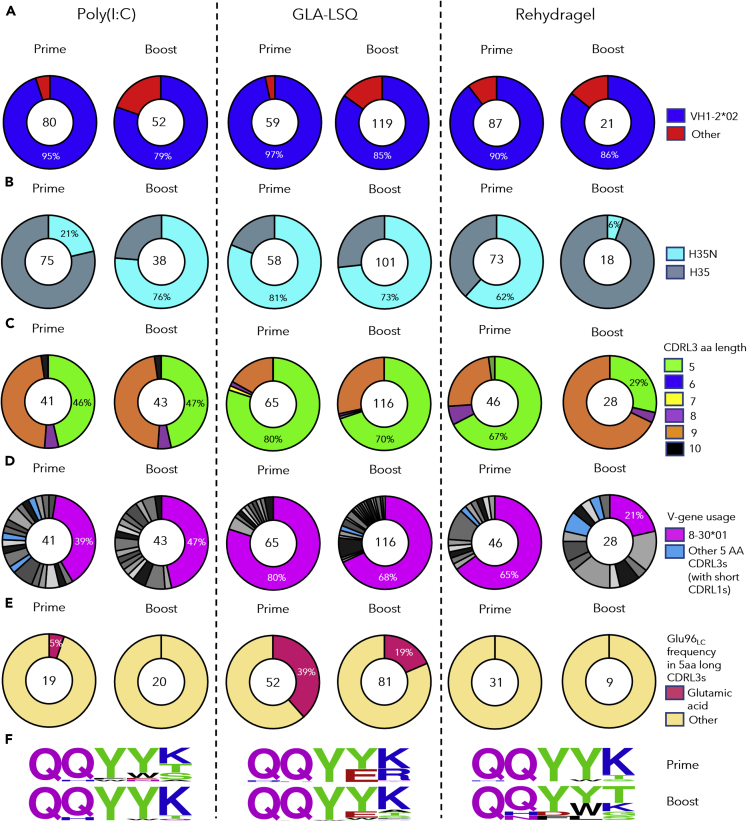
Two weeks after the prime immunization with 426c.Mod.Core Ferritin; 130, 206, and 172 class-switched Env+ B cells from pooled mouse sample, were individually isolated from the Poly(I:C), GLA-LSQ, and the Rehydragel group, respectively. 80 (62%), 59 (28%), and 87 (50%) of HCs were successfully sequenced from the Poly(I:C), GLA-LSQ, and Rehydragel groups (Table S2); of which 95%, 97%, and 90%, were derived from VH1-2∗02, respectively (Figure 3A). The H35N mutation in CDRH2, that stabilizes the interaction between CDRH1 and CDRH3 on VRC01-class antibodies (Jardine et al., 2015), was enriched by 21%, 81%, and 62%, of the VH1-2∗02 HCs isolated from the Poly(I:C), GLA-LSQ, and Rehydragel groups, respectively (Figure 3B). 41 (32%), 65 (32%), and 46 (27%) LCs, were successfully sequenced from the Poly(I:C), GLA-LSQ, and Rehydragel groups, respectively (Table S2). 19 (46%) in the Poly(I:C) group, 52 (80%) in the GLA-LSQ group, and 31 (67%) in the Rehydragel group, contained the characteristic 5 aa-long CDRL3s (Figure 3C). The majority of the 5 aa-long CDRL3s in all adjuvant groups were derived from the mouse 8-30∗01 κV gene (Figure 3D), as we previously reported (Parks et al., 2019). Antibodies expressing the VRC01 HC paired with other mouse LCs were also isolated (from the Poly(I:C) and Rehydragel groups) but only a few of these LCs expressed 5 aa-long CDRL3s (Figure 3D). Within the 8-30∗01 LCs, Glu96LC, a key CDRL3 feature of the mature VRC01-class antibodies, was detected in the Poly(I:C) group and more frequently in the GLA-LSQ group, but not in the Rehydragel group (Figures 3E and 3F). These results suggest that adjuvants may differentially affect the selection of VRC01-like BCRs with particular amino acid mutations in both their HC and LCs.
Figure 3.
VH/VL sequence analysis from antibodies developed after the prime and boost immunizations
Pie charts indicate VH (A and B) and VL (C–F) gene usage from individually sorted B cells from pooled mouse samples collected 2 weeks after immunization with 426c.Mod.Core (Prime) and 2 weeks after immunization with HxB2.WT.Core (Boost) with the indicated adjuvant. The number of VH and VL sequences analyzed is shown in the middle of each pie chart.
(A) VH-gene usage, (B) VHs with the H35N mutation.
(C) aa length of the CDRL3 domains in the VL, (D) VL-gene usage. Shades of gray/black slices represent non 5 aa-long CDRL3s, k8-30∗01 VLs are represented in pink whereas non k8-30∗01 VLs with 5 aa long CDRL3s are indicated in blue, (E) Presence or absence of Glu96LC within the LC sequences with 5 aa-long CDRL3 domains, (F) Logo plots represent 5 aa-long CDRL3 sequences in the 8-30∗01 VKs. See also Tables S1 and S2.
Two weeks following the boost immunization with HxB2.WT.Core, 124 B cells were isolated from a pool of Poly(I:C) group mouse sample, 248 B cells from a pool of GLA-LSQ group mouse sample, and 84 B cells from a pool of Rehydragel group mouse sample. 52 (42%) HCs and 43 (35%) LCs were successfully sequenced from the Poly(I:C) group, 119 (48%) HCs and 116 (47%) LCs from the GLA-LSQ group, and 21 (25%) HCs and 28 (33%) LCs from the Rehydragel group (Figures 3A and 3C; Table S2). Majority of the HC sequences (79% in the Poly(I:C) group, 85% in the GLA-LSQ group, and 86% in the Rehydragel group), expressed the VH1-2∗02 gene (Figure 3A). Of interest the fraction of HCs with the H35N mutation in the Poly(I:C) group increased from 21% after the prime to 76% after the boost, whereas it decreased from 61% after the prime to ∼6% after the boost in the Rehydragel group and remained at similar levels in the GLA-LSQ group (Figure 3B). 20 of 43 LCs (47%) in the Poly(I:C) group, 81 of 116 LCs (70%) in the GLA-LSQ group, and 8 of 28 (29%) LCs in the Rehydragel group, contained 5 aa-long CDRL3s (Figure 3C). These frequencies were comparable to those observed after the prime immunization. As expected, the majority of the 5-aa CDRL3s were derived from the mouse 8-30∗01 LC V gene (Figure 3D). Noticeably, only the GLA-LSQ group enriched for 5-aa-CDRL3s containing Glu96LC after the boost (19%; Figures 3E and 3F). These observations are in general agreement with those discussed above, in that the adjuvant influences which somatic mutations are selected by VRC01 BCRs.
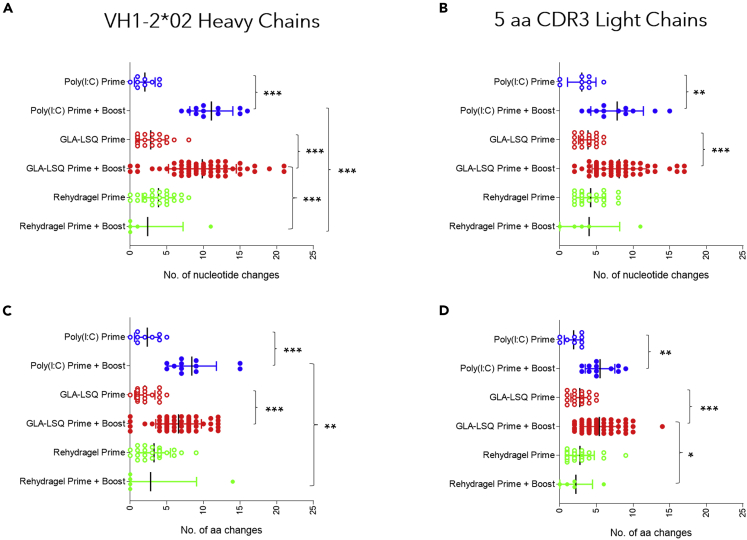
After the prime immunization, a range of somatic mutations resulting in amino acid changes were observed in both the HCs and LCs of paired sequences derived from pooled mouse sample (Figure 4) in all three adjuvant groups. The mean numbers of HC amino acid mutations were: 2.3, 2 and 3.3 (Figure 4C) and the mean numbers of LC amino acid mutations were: 1.9, 2.7 and 2.8 (Figure 4D) for the Poly(I:C), GLA-LSQ, and Rehydragel groups respectively, following prime immunization. After the boost immunization, statistically significant increases in both nucleotide (Figure 4A) and amino acid (Figure 4C) mutations in the HCs were observed in the Poly(I:C) and GLA-LSQ groups (p≤0.001), but not in the Rehydragel group. Similarly, statistically significant increases in both nucleotide (Figure 4B) and amino acid (Figure 4D) mutations in the LCs were observed in the Poly(I:C) (p<0.01) and GLA-LSQ (p≤0.001) groups but not in the Rehydragel group. In addition, the mean number of amino acid mutations in the three adjuvant groups differed significantly in the HCs (∼8.4, ∼6.6, and 2.8), and LCs (∼5.5, ∼5.4 and 2.2) for Poly(I:C), GLA-LSQ and Rehydragel, respectively. We concluded that Rehydragel may not be an optimal adjuvant for the maturation of VRC01 antibody responses.
Figure 4.
Number of nucleotide and amino acid changes in VRC01-like antibodies generated after the prime and after the boost immunizations
(A) number of nucleotide and (C) amino acid changes in the HC of paired sequences isolated from pooled mouse samples after the prime (week 2) or the boost administration (week 24 or 25), and (B) number of nucleotide and (D) amino acid changes in the LC of paired sequences isolated from pooled mouse samples isolated after the prime (week 2) or the boost administration (week 24 or 25) with the indicated adjuvants. Significance was calculated using one-way ANOVA (Tukey’s multiple comparisons test). (∗) ≤0.05; (∗∗) ≤0.01 and (∗∗∗) ≤0.001.
Neutralizing properties of the plasma antibody responses after the boost immunization
Plasma IgG was purified 2 weeks following the boost immunization with HxB2.WT.Core Ferritin from all three adjuvant groups and was first tested against the tier 2, WT 426c virus produced either in 293T or GnTI−/− cells, but neither version was neutralized by any of the samples (Table 1). We next tested these samples against a derivative of 426c that lacks 3 NLGS (N276, N460 and N463) (triple mutant, TM) expressed in GnTI−/− cells. The elimination of these NLGS renders the 426c susceptible to neutralization by some glVRC01-class antibodies (McGuire et al., 2013). Ten of eleven samples neutralized this variant. Sample M24 that did not display anti-TM neutralizing activity was derived from the Rehydragel group. To confirm that the neutralizing activity was due to VRC01-like antibodies present in these samples, we tested the neutralizing activities of each sample against a derivative of TM that contains the D279K mutation, which abrogates the neutralizing activity of VRC01-class antibodies (LaBranche et al., 2018). In the GLA-LSQ or Rehydragel groups, the neutralizing activities were exclusively due to VRC01-like antibodies. In Poly(I:C) group, the neutralizing activity in one of the three samples (M4) was entirely due to VRC01-like antibodies, whereas in the remaining two samples (M5 and 6), the anti-TM neutralizing activity was primarily but not exclusively due to VRC01-like antibodies. We concluded that, irrespective of the adjuvant used, this prime-boost immunization schema elicits plasma VRC01-like antibody responses that cannot yet bypass the glycans present in Loop D (N276) and V5 (N460 and N463) on the WT 426c virus but can neutralize this virus when the three NLGS are absent.
Table 1.
Neutralizing activities of plasma IgG and VRC01-like mAbs
| Mouse ID or mAb | Adjuvant | 426c.WT (293T) | 426c.WT (GnTI-) | 426c.TM (GnTI-) | 426c.TM.D279K (GnTI-) | 426c.SM (293T) | 426c.SM (GnTI-) |
|---|---|---|---|---|---|---|---|
| M4 | Poly(I:C) | >100 | >100 | 0.92 | >100 | NT | NT |
| M5 | >100 | >100 | 0.79 | 88.27 | NT | NT | |
| M6 | >100 | >100 | 0.84 | 88.09 | NT | NT | |
| M13 | GLA-LSQ | >100 | >100 | 3.53 | >100 | NT | NT |
| M14 | >100 | >100 | 1.83 | >100 | NT | NT | |
| M15 | >100 | >100 | 0.39 | >100 | NT | NT | |
| M16 | >100 | >100 | 0.14 | >100 | NT | NT | |
| M21 | Rehydragel | >98.5 | >98.5 | 0.84 | >98.5 | NT | NT |
| M22 | >84.5 | >84.5 | 0.48 | >84.5 | NT | NT | |
| M23 | >72 | >72 | 6.08 | >72 | NT | NT | |
| M24 | >81.9 | >81.9 | >81.9 | >81.9 | NT | NT | |
| mVRC01 | 2.47 | 0.18 | <0.005 | >10.00 | NT | NT | |
| MLK-002 | Poly(I:C) | >51.85 | >51.85 | <0.02 | NT | >17.33 | 1.724 |
| MLK-008 | GLA-LSQ | >50 | >50 | <0.02 | NT | 7.13 | 0.2 |
| MLK-009 | >50 | >50 | <0.02 | NT | NT | NT | |
| MLK-010 | >50 | >50 | <0.02 | NT | 2.748 | 0.13 | |
| MLK-014 | >50 | >50 | 0.05 | NT | >16.67 | 0.26 | |
| MLK-015 | >21 | >21 | <0.01 | NT | 2.89 | 0.06 | |
| MLK-016 | >50 | >50 | <0.02 | NT | 10.52 | 0.17 | |
| mVRC01 | 1.77 | 0.17 | <0.0023 | NT | 0.38 | 0.05 | |
| glVRC01 | >50 | >50 | 0.78 | NT | >33.33 | 0.81 |
See also Figures S2 and S3. IgG was isolated from plasma collected 2 weeks following the booster immunization with HxB2.WT.Core from the indicated mouse in the different adjuvant groups and was evaluated for the presence of neutralizing antibodies against the WT 426c virus and the 426c virus whose Env lacks three NLGS (N276, N460 and N463), TM, and against its variant with the D279K mutation that abrogates the neutralizing activity of VRC01 antibodies. Viruses were expressed in 293 GnTI- cells. The WT 426c virus was also expressed in 293T cells. Values represent IC50 neutralization values in μg/mL. Neutralization IC50 values of these same viruses with the mature VRC01 mAb are included. VRC01-like mAbs isolated after the boost immunization with HxB2.WT.Core from the Poly(I:C) and GLA-LSQ groups (see Figure 5 for binding information) were evaluated against the same viruses as the plasma samples as well as against the 426c variant whose Env only lacks the N276 NLGS (SM) expressed in 293T and 293 GnTI- cells. Values represent IC50 neutralization values in mg/mL. Neutralization IC50 values of these same viruses with the mature and germline VRC01 mAbs are included. Bold values indicate samples displaying neutralizing activity. NT indicates samples not tested.
Neutralizing properties of monoclonal VRC01-like antibodies isolated after the boost immunization
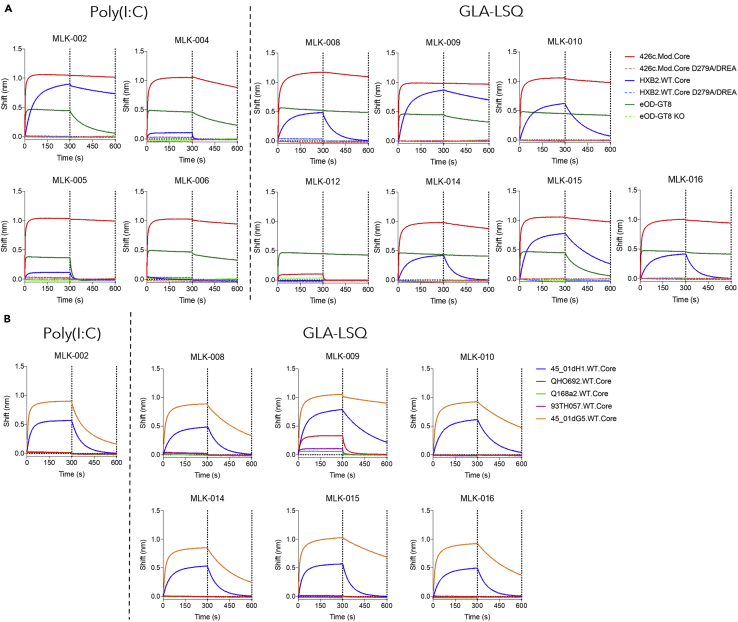
To prove directly that VRC01-like antibodies were responsible for the neutralizing activities of plasma-derived polyclonal IgGs, we generated eleven VRC01-like mAbs from mice immunized in the presence of Poly(I:C) or GLA-LSQ, two weeks after the booster immunization with HxB2.WT.Core Ferritin (Figure S2). The mAbs express the human glVRC01HC paired with mouse k8-30∗01 LCs expressing 5 aa-long CDRL3. With the exception of mAb MLK-012, they express amino acid mutations in both the HC and LCs (Table S3). All mAbs recognized 426c.Mod.Core and eOD-GT8, but not their KO versions (Figure 5A; comparable binding curves of mVRC01 and glVRC01 mAb are shown in Figure S3). With the exception of MLK-006 and MLK-012, all bound to HxB2.WT.Core but not its KO version. Among the mAbs that bound to HxB2.WT.Core, MLK-005 and MLK-006 displayed the weakest binding, whereas MLK-009 displayed the strongest binding.
Figure 5.
Binding properties of VRC01-like mAbs generated after the boost immunization
(A) Four VRC01-like mAb isolated from the Poly(I:C) group and seven VRC01-like mAbs isolated from the GLA-LSQ group were evaluated against the indicated soluble monomeric Envs.
(B) Those mAbs that displayed anti-HxB2.WT.Core reactivity were evaluated against the indicated five heterologous WT Cores. Dotted lines indicate end of association and dissociation phases. See also Figures S2 and S3; Table S3.
The seven mAbs that bound to HxB2.WT.Core efficiently were also tested for binding to heterologous WT Cores (Figure 5B; comparable binding curves of mVRC01 and glVRC01 mAb are shown in Figure S3). 45_01dG5 Env (clade B) is derived from a virus that circulated in patient 45, from which several VRC01-class antibodies have been isolated (including VRC01) (Lynch et al., 2015). It naturally lacks the 276 NLGS and expresses one NLGS in its V5 region (position 470). All seven mAbs bound to this protein, with MLK-009 displaying the slowest off-rate compared to the remaining six mAbs. 45_01dH1 is derived from a virus circulating at a later time point in patient 45 and expresses the N276 NLGS, and, in addition to the N460 NLGS it expresses a second NLGS in V5 (position 476). Although all seven mAbs bound to that Env, they all displayed lower maximum binding and faster off rates than for 45_01dG5. Only mAb MLK-009 bound to the QH0692-derived WT core protein (clade B), and minimally bound to the Q168a2-derived WT Core protein (clade A) and 93TH057-derived WT Core protein (clade A/E). The remaining six mAbs did not bind to these proteins.
In agreement with the above discussed neutralization results obtained with polyclonal plasma IgGs, none of the seven VRC01-like mAbs neutralized the 426c WT virus, irrespective of whether it was produced in 293T or 293 GnTI−/− cells (Table 1), but all neutralized the TM virus produced in 293 GnTI−/− cells. The mAbs also neutralized a 426c variant that only lacks the N276 NLGS (SM) when expressed in GnTI−/− cells and four of six mAbs neutralized this virus when expressed in 293T cells (mAbs MLK-002 and MLK-014 did not neutralize this virus). Importantly, glVRC01 mAb did not neutralize this virus. The fact that all mAbs neutralized the TM virus but only 4 neutralized the SM virus, indicate that the glycans in V5 (N460 and N463) are significant obstacles for the maturing VRC01-like antibodies.
Overall, the data strongly suggest that the above immunization schema activates and initiates the maturation of VRC01-like B cells, but that the maturation process is incomplete as the elicited VRC01-like antibodies have not yet accumulated mutations that allow them to accommodate the glycans present on N276. We also note that these mAbs did not neutralize heterologous viruses whose N276 glycosylation site was eliminated by mutagenesis (N276Q). Thus, in addition to the N276-associated glycans, additional steric obstacles are present on heterologous Env that prevented the binding of these immature VRC01-like antibodies.
Discussion
The VRC01 antibody maturation process can be initiated by a single immunization with specifically designed Env-derived germline-targeting immunogens (Briney et al., 2016; Dosenovic et al., 2015; Jardine et al., 2015; Lin et al., 2020; Parks et al., 2019; Sok et al., 2016). The completion of the maturation process, however, will require booster immunizations with distinct heterologous Envs (Chen et al., 2021; Tian et al., 2016). In the transgenic animal model used here, that expresses the inferred human glHC of VRC01 paired with mLCs expressing 5 aa-long CDRL3 at a frequency of ∼0.08%, we and others reported on the incremental, but still incomplete, maturation of VRC01-like antibodies initiated by a prime immunization with a germline-targeting immunogen (426c.Mod.Core or eOD-GT8) followed by booster immunizations with 1–2 heterologous Env-derived proteins (Briney et al., 2016; Parks et al., 2019). In a separate animal model, expressing both the inferred human glHC and human glLC of VRC01, Chen et al., reported on a more extensive maturation of VRC01-like antibodies using nine distinct immunogens administered sequentially that resulted in the isolation of VRC01-like antibodies displaying ∼50% neutralizing breadth (Chen et al., 2021). These observations support the overall ‘germline-targeting’ immunization approach (Dimitrov, 2010) for the elicitation of VRC01 bnAbs and validate the potential of the utilized germline-targeting immunogens to initiate the antibody maturation process (Stamatatos et al., 2017). It is however important to identify ways to optimize and accelerate this process. Here, we examined whether and how adjuvants may affect the maturation of VRC01-like antibody responses. The three adjuvants evaluated here were chosen because they activate different pathways of the innate response Poly(I:C) via TLR3 (Alexopoulou et al., 2001; Salem et al., 2005), GLA-LSQ via TLR4 (Coler et al., 2011; Radtke et al., 2017; Reed et al., 2018), whereas Rehydragel acts independently of the TLR pathways (Gavin et al., 2006).
Our results suggest that long-lived populations of plasma cells, including ones producing VRC01-like antibodies, can be generated by a single administration of the germline-targeting immunogen 426c.Mod.Core, irrespective of the adjuvant used. The fact that immunization with 426c.Mod.Core also elicited high and sustained plasma antibody responses against the heterologous HxB2.WT.Core suggests that this germline-targeting immunogen activates B cells that recognize conserved epitopes between these two proteins. A high fraction of these cross-reactive plasma antibodies targeted conserved elements of the CD4-BS. Of interest, the anti-CD4-BS plasma antibody titers (against 426c.Mod.Core or HxB2.WT.Core) decreased over time in all three adjuvant groups, whereas the total anti-426c.Mod.Core and anti-HxB2.WT.Core plasma antibody titers remained stable. This suggests that the numbers of plasma cells that produce anti-CD4-BS antibodies gradually decreased during the observation period whereas the numbers of plasma cells that target epitopes outside the CD4-BS gradually increased. A fraction of the anti-CD4-BS plasma antibody responses recognize the VRC01 epitope (expressed on eOD-GT8), and their titers also gradually decreased over the period of observation, in agreement with the overall decrease of the anti-CD4-BS plasma antibody responses.
Although the plasma antibody responses to epitopes outside the CD4-BS increased by the boost immunization with HxB2.WT.Core, the anti-CD4-BS responses (to HxB2.WT.Core itself and to the prime immunogen 426c.Mod.Core) did not increase. This is not because the CD4-BS is not immunogenic on the HxB2.WT.Core. One possibility is that pre-existing anti-CD4-BS antibodies (elicited by the prime immunogen), bind the CD4-BS on the HxB2.WT.Core immunogen and prevent its recognition by naive, or memory anti-CD4-BS B cells. That possibility however does not appear to apply to epitopes outside the CD4-BS. Importantly, however, in the case of Poly(I:C) and GLA-LSQ, an increase in plasma VRC01-like antibodies was observed after the boost immunization. This ‘boosting’ of VRC01-like plasma antibody responses suggests that an increase in the number of VRC01-like producing plasma cells took place by the heterologous immunization. The HxB2.WT.Core does not activate germline VRC01 B cells, and thus the observed increase in plasma VRC01 antibodies is due to the binding of HxB2.WT Core to B cells expressing VRC01-like BCRs that had been activated by the 426c.Mod.Core and had accumulated somatic mutations during the intervening period. The corresponding partially mutated plasma VRC01 antibodies represent a small fraction of the total antibodies in circulation at the time of the HxB2.WT.Core boost and thus do not prevent that protein from binding to VRC01-like B cells. Thus, despite a predominance of B cell responses to epitopes other than the VRC01 epitope on HxB2.WT.Core, boosting of VRC01-like B cells responses was possible in the presence of Poly(I:C) and GLA-LSQ, but not in the presence of Rehydragel. Presently, we do not know why the plasma VRC01-like B cell response were not boosted in the Rehydragel group, but we speculate that is related to the activation of different innate pathways.
Importantly, the adjuvant used affected the rate at which activated VRC01-class B cells accumulated somatic mutations in their VH/VL genes. Significantly more nucleotide and amino acid mutations were present in the VRC01-like antibodies isolated after the boost from animals that received immunogens adjuvanted with Poly(I:C) or GLA-LSQ than Rehydragel. It is possible that adjuvants affect the activation of CD4+T cells that assist B cells in GCs, and/or the adjuvants directly affect these B cells. We expect the above effect to not be specific for VRC01 B cells, but to all B cells that became activated by the two immunogens used here.
As a result of the different rates of somatic mutations observed in the Rehydragel group and the GLA-LSQ or Ploy(I:C) groups, B cells expressing VRC01-like BCRs with particular amino acids at key positions were selected. Thus, the Glu96LC amino acid was present in VRC01-like B cells found in the Poly(I:C) and GLA-LSQ groups following the prime immunization with 426c.Mod.Core Ferritin, but not in the Rehydragel group. Glu96LC is found in all presently known human VRC01-class bnAbs and is the product of affinity maturation (West et al., 2012). It forms hydrogen bonds with the side chain of Asn280 in Loop D and with the backbone amide of Gly459 in V5 and contributes to increased antibody-Env affinity (West et al., 2012; Zhou et al., 2010). Glu96LC was only present in VRC01-like antibodies generated from the GLA-LSQ group after the boost immunization with HxB2.WT.Core Ferritin. Similarly, a higher frequency of VRC01-like antibodies isolated after the boost immunization expressed N35HC (rather than H) in the case of Poly(I:C) and GLA-LSQ than Rehydragel. The H35N mutation allows for a hydrogen bond formation with N100a in CDRH3, thus improving the HC/LC interaction (Jardine et al., 2015).
In sum, our study provides direct evidence that adjuvants influence the activation and maturation of B cells expressing VRC01-like BCRs. As the elicitation of fully matured VRC01-like antibodies requires the accumulation of an extensive number of mutations in both the VH and VL genes (Klein et al., 2013) and several immunogens have to be administered in a particular sequence to select VRC01 BCRs with particular mutations at each immunization step (Chen et al., 2021), our study highlights the importance that adjuvants have in the selection of the appropriate mutations.
Limitations of the study
Immunizations were performed in transgenic mice expressing unphysiologically high precursor frequencies of VRC01-like BCRs. In future studies, the immunizations will be performed in animal models expressing VRC01-like precursor frequencies similar to those present in humans. Although the rate and type of somatic mutations accumulated by VRC01-like B cells following the prime immunization with the germline-targeting immunogen 426c.Mod.core were affected by the adjuvant employed, the corresponding antibodies have not yet accumulated all the necessary mutations to accommodate complex glycans, both in Loop D and V5. For this, additional booster immunizations with more native-like Envs, which are presently unknown, will most likely be required.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC Rat Anti-Mouse IgG1 (clone A85-1) | BD Biosciences | Cat# 553443; RRID:AB_394862 |
| FITC Rat Anti-Mouse IgG2b (clone R12-3) | BD Biosciences | Cat# 553395; RRID:AB_394833 |
| Goat antiMouse IgG2c:FITC | Bio-Rad | Cat# STAR135F; RRID:AB_1102667 |
| FITC Rat Anti-Mouse IgG3 (clone R40-82) | BD Biosciences | Cat# 553403; RRID:AB_394840 |
| PerCP/Cy5.5 anti-mouse IgD Antibody | Biolegend | Cat# 405710; RRID:AB_1575113 |
| eBioscience Fixable Viability Dye eFluor 506 | ThermoFisher Scientific | 65-0866-14 |
| BV510 Hamster Anti-Mouse CD3e (clone 145-2c11) | BD Biosciences | Cat# 563024; RRID:AB_2737959 |
| BV510 Rat Anti-Mouse CD4 (clone RM4-5) | BD Biosciences | Cat# 563106; RRID:AB_2687550 |
| Brilliant Violent 510 anti-mouse F4/80 Antibody | Biolegend | Cat# 123135; RRID:AB_2562622 |
| Brilliant Violet 605 anti-mouse IgM Antibody | Biolegend | Cat# 406523; RRID:AB_2563358 |
| BV786 Rat Anti-Mouse CD45R/B220 | BD Bioscience | Cat# 563894; RRID:AB_2738472 |
| BV650 Rat Anti-Mouse CD19 | BD Biosciences | Cat# 563235; RRID:AB_2738085 |
| 6x-His Tag Monoclonal Antibody (HIS.H8) | ThermoFisher Scientific | Cat# MA1-21315; RRID:AB_557403 |
| HRP Goat anti-mouse IgG | Biolegend | Cat# 405306; RRID:AB_315009 |
| Bacterial and virus strains | ||
| NEB® 5-alpha Competent E. coli (High Efficiency) | New England BioLabs | C2987H |
| HIV-1 Env-pseudotyped viruses | D. Montefiori and C. LaBranche, Duke University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Poly (I:C) | InvivoGene | tlrl-pic-5 |
| GLA-LSQ | Infectious Disease Reseach Institute (IDRI) | IDRI-LS130 |
| Rehydragel | NIAID | N/A |
| Steptavidin-R-Phycoerythirin | Prozyme | PJRS25 |
| Streptavidin-Allophycocyanin | Prozyme | PJ27S |
| RNaseOUT™ Recombinant Ribonuclease Inhibitor | ThermoFisher Scientific | 10777019 |
| HotStar Taq Plus | Qiagen | 203607 |
| Gel Red Nucleic Acid Stain | Biotium | 41002 |
| ExoSap-IT | Affymetrix | 78201 |
| SureBlue Reserve TMB Microwell Peroxidase Substrate | KPL | 53-00-02 |
| 293-Free Transfection Reagent | Millipore Sigma | 72181 |
| 5x In-Fusion enzyme | Takara Bio | 1805251A |
| SuperScript IV RT | ThermoFisher Scientific | 18090200 |
| Critical commercial assays | ||
| EasySep Mouse B Cell Isolation Kit | Stemcell Technologies | 19854 |
| InFusion HD Cloning Kit | Takara Bio | 639649 |
| QIAprep Spin Miniprep Kit | Qiagen | 27106 |
| britelite plus Reporter Gene Assay System | PerkinElmer | 6066766 |
| Thrombin, Restriction Grade | EMD Millipore | 69671-3 |
| Deposited data | ||
| Antibody Sequence | This paper | Table S2 |
| Experimental models: Organisms/strains | ||
| Mouse: glH-VRC01 | Jardine et al., 2015 | N/A |
| Oligonucleotides | ||
| Random Primers | ThermoFisher Scientific | 48190011 |
| GeneAmp dNTP Blend | ThermoFisher Scientific | N8080261 |
| Primers for PCR | This paper; Jardine et al., 2015; Tiller et al., 2008 | Table S1 |
| Recombinant DNA | ||
| Ptt3 k vector | Snijder et al., 2018 | N/A |
| PMN 4-341 vector | Mouquet et al., 2012 | N/A |
| Software and algorithms | ||
| Geneious (Version 8.1.9) | Biomatters Ltd. | https://www.geneious.com |
| IMGT/V-QUEST | Brochet et al., 2008; Giudicelli et al., 2011 | http://www.imgt.org/IMGT_vquest/input |
| Prism 7 | GraphPad | https://www.graphpad.com |
| FlowJo_V10 | BD | https://www.flowjo.com |
| Other | ||
| Anti-PE microbeads | Miltenyi Biotech | 130-048-801 |
| Anti-APC microbeads | Miltenyi Biotech | 130-090-855 |
| LS Columns | Miltenyi Biotech | 130-042-401 |
| Pierce Protein A agarose beads | ThermoFisher Scientific | 18-5063 |
| Anti-human IgG Fc Capture (AHC) Biosensors | ForteBio | 18-5126 |
| MyOne Tosylactivated Dynabeads | ThermoFisher Scientific | 65501 |
| His60 Ni-Superflow Resin | Takara Bio | 636660 |
| HiLoad 16/600 Superdex 200 pg (GE) column | GE Healthcare | 28989335 |
| Superose 6 10/300 GL | GE Healthcare | 17-6172-01 |
| Agarose Bound Galanthus Nivalis Lectin | Vector Labs | AL-1243-5 |
| Step-Tactin SuperFlow Plus | Qiagen | 30002 |
| HisTrap FF Column | GE Healthcare | 17525501 |
Resource availability
Lead contact
Further information and requests for reagents should be directed to and will be fulfilled by Leonidas Stamatatos (lstamata@fredhutch.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Knock-in mice expressing the inferred germline HC of the human VRC01 Ab (VRC01glH) and endogenous mouse LCs (Jardine et al., 2015) were bred and kept at the Fred Hutchinson Cancer Research Center. Mice (both male and female) were 6–12 weeks old at the initiation of experiments. Proteins and adjuvants were diluted in PBS and administered intramuscularly with 50 μL in each hind leg in the gastrocnemius muscle (total volume 100 μL/mouse). Env antigens were administered at 50 (GLA-LSQ groups) or 60 μg (Poly(I:C) and Rehydragel groups), and adjuvants at 60 μg for Poly(I:C), 50 μL GLA-LSQ (containing 5 μg TLR4 agonist and 2 μg Saponin for GLA-LSQ), and 100 μg for Rehydragel. Blood was collected by the retroorbital route into tubes containing 25 μL citrate-phosphate-dextrose solution (Sigma-Aldrich). Terminal bleeds were collected by cardiac puncture into tubes containing 100 μL citrate-phosphate-dextrose solution. Organs were harvested into cold IMDM media (Gibco). All experiments conform to relevant regulatory standards. Mouse experiments were approved and carried out in accordance with Fred Hutchinson Cancer Center’s IACUC protocol number 50879.
Method details
Recombinant HIV-1 envelope proteins
Recombinant HIV-1 envelope proteins (rec Envs) were expressed by transient transfection in HEK 293-F cells and then purified directly from conditioned media as we previously described (McGuire et al., 2016). Briefly, cell supernatants were purified by lectin affinity chromatography (Galanthus nivalis, Vector Labs), then subjected to Superdex 200 size exclusion chromatography (GE Healthcare). The “CD4-BS knockout” (KO) versions of rec Envs contain the D279A, D368R and E370A mutations (D279A/DREA). In the case of eOD-GT8, the KO version contains the D368R mutation and the amino acids DWRD at positions 276–279 were substituted by NFTA. Purified Env proteins were aliquoted in PBS and stored frozen in −20°C until further use. Ferritin nanoparticles expressing 426c.Mod.Core and HxB2.WT.Core were produced and purified as previously described (Parks et al., 2019). Specifically, ferritin nanoparticles underwent two rounds of size exclusion chromatography, first on a Superose 6 10/300GL column and then on a HiLoad 16/600 Superdex 200pg column. They were stored at 4°C. Tetramers of Avi-tagged eOD-GT8 and of eOD-GT8 KO were generated as previously reported (Parks et al., 2019).
Adjuvants
Polyinosinic-polycytidylic acid (Poly(I:C)) was obtained from InvivoGene. GLA-LSQ was provided by the Infectious Disease Research Institute (IDRI) and liposomal formulations of GLA were prepared as previously published (Baldwin et al., 2021). Rehydragel was provided by the NIAID.
Organ processing
Spleens and lymph nodes were first mashed through 70-μm-pore-size nylon cell strainers (Falcon) to obtain a single cell suspension. Cells from lymph nodes were then washed twice with PBS, while splenocytes were first treated with Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich) for 1.5–2 min followed by two washes with PBS. Cells were resuspended in FBS supplemented with 10% DMSO and frozen in a Mr. Frosty Freezing Container (Thermo Fisher Scientific) at −80°C overnight, then moved to liquid nitrogen for storage until use.
Single B cell sorting
Splenocytes or lymph node cells were thawed and first stained with Fc-block (2.4G2; BD Biosciences), 1pmolGb phycoerythrin (PE)-DyLight (DL)650 (PE-Decoy) and 3pmolGb allophycocyanin (APC)-DL755 (APC-Decoy) diluted in fluorescence-activated cell sorting (FACS) buffer (2% FBS, 1 mM EDTA in PBS). PE-Decoy and APC-Decoy were made by first combining streptavidin-PE (SA-PE) or streptavidin APC (SA-APC) (Prozyme) with a DL NHS ester (650 or 755; Thermo Fisher Scientific). 1 pmol eOD-GT8-PE tetramer and 3 pmol eOD-GT8 KO-APC tetramers were added to the cells and incubated on ice for 25 mins. Samples were then washed and incubated with anti-PE and anti-APC MicroBeads (both Miltenyi Biotec) after which decoy- and tetramer-positive cells were enriched by putting the samples through magnetic LS columns positioned on a QuadroMACS separator (all from Miltenyi Biotec) (19). Non-bound cells present in the flow-through were collected to use as controls. After a wash, samples (both bound and non-bound fractions) were incubated with Fixable Viability Dye eFluor 506 (eBioscience, ThermoFisher Scientific) and the following Abs diluted in Brilliant Stain Buffer (BD Biosciences): Anti-IgG1-fluorescein isothiocyanate (FITC; A85-1), anti-IgG2b-FITC (R12-3; both from BD Biosciences), anti-IgG2c-FITC (polyclonal; Bio-Rad), anti-IgG3-FITC (R40-82; BD Biosciences), anti-IgD-PerCP-Cy5.5 (11-26c.2a; BioLegend), anti-CD3-Brilliant Violet (BV)510 (145-2C11), anti-CD4-BV510 (RM4-5), anti-Gr-1-BV510 (RB6-8C5; all from BD Biosciences), anti-F4/80-BV510 (BM8), anti-IgM-BV605 (RMM-1; both from BioLegend), anti-CD19-BV650 (1D3), anti-B220-BV786 (RA3-6B2; both BD Biosciences). Samples were washed and resuspended in FACS buffer, AccuCheck Counting Beads (Thermo Fisher Scientific) were added, and the samples were then evaluated on a FACSAria II (BD Biosciences). Stained UltraComp eBeads (Thermo Fisher Scientific) were used for compensating voltages. Non-bound cells collected in the enrichment step described above were used for setting up gates. Samples were single cell-sorted into 96-well skirted plates (Eppendorf) and stored at −80°C until further processing. 20 μL lysis buffer per well (20 U Rnase out, SSIV buffer, 6.25 μM DTT (all from Thermo Fisher Scientific), 0.3% Igepal) was added either immediately before sorting cells, or after thawing plates for further processing.
VH/VL gene sequencing
Amplification and sequencing of the antibody VH/VL genes was performed as we previously described (Jardine et al., 2015; Lin et al., 2020; Parks et al., 2019). Briefly, cDNA was generated by adding 6 μL per well of a mix containing Random Primers, 3.3 mM dNTPs, 200 U Superscript IV Reverse Transcriptase (all from Thermo Fisher Scientific) and with PCR program 42°C 10 min, 25°C 10 min, 50°C 60 min, 94°C 5 min. cDNA was amplified and sequenced by two rounds of PCR using primers listed in Table S1 using PCR program: 94°C 5 min, 50 × (94°C 30 sec; XoC for 30 sec (X being the corresponding annealing temperature of the primers) and 72°C for 55 sec), and 72°C 10 min. All reactions were performed in a 40 μL volume with 2.4 U HotStarTaq Plus DNA Polymerase (Qiagen), 0.24 μM of a 5′ primers pool, 0.24 μM of a 3′ primer pool (all from Integrated DNA Technologies), 0.35 mM dNTPs. Aliquots from each well were subjected to agarose gel electrophoresis, which were then treated with ExoSAP-IT PCR Product Cleanup Reagent (Applied Biosystems, Thermo Fisher Scientific) and sequenced by Sanger sequencing using the primers indicated in Table S1, as previously reported (Jardine et al., 2015; Tiller et al., 2008).VH and VL sequences were analyzed using the Geneious software (Biomatters, Ltd.) and the online IMGT/V-QUEST tool (Brochet et al., 2008). To calculate numbers of nucleotide and amino acid mutations, sequenced HC and LC pairs were aligned against the VH regions of the VRC01 gH knocked-in sequence and LC reference sequences obtained from IMGT/V-QUEST, respectively, using sequences starting from CDR1 to CDR3. VH/VL sequences are provided in Table S2.
VH/VL cloning and antibody expression
DNA products from the 1st round of PCR were used as templates for gene-specific PCR to amplify the gene of interest and add ligation sites to allow for insertion of the DNA fragment into the human IgG1 vectors: ptt3 for κ light chain (Snijder et al., 2018) and PMN 4-341 for γ heavy chain (23). Each gene-specific PCR reaction consisted of 0.5 μL of each 10 μM 5′ and -3′ primer, 22.5 μL Accuprime Pfx Supermix (Thermo Fisher Scientific) and 1.5 μL of 1st round PCR product. The gene-specific PCR product was then infused into cut IgG1 vector in a 2.5 μL volume reaction containing 12.5 ng of cut vector, 50 ng of insert, 0.5 μL of 5× Infusion enzyme (Takara Bio). Competent E. coli cells were transformed with the entire reaction and plated onto ampicillin agar plates. Colonies were picked and grown in LB broth containing ampicillin, and DNA was extracted and purified using QIAprep Spin Miniprep Kit (Qiagen). 293E cells were then transfected with equal amounts of HC and LC DNA as well as 293F transfection reagent (Millipore Sigma) and grown for 5–7 days, at which time Abs were purified from cell supernatants using Pierce Protein A agarose beads (Thermo Fisher Scientific). Abs were eluted with 0.1 M Citric Acid into 1 M Tris buffer followed by buffer exchange into PBS using an Amicon centrifugal filter (Millipore Sigma).
Purification of IgG from mouse plasma
IgGs were purified from mouse plasma using Protein G HP-Ab Spin Trap columns (GE Healthcare Life Sciences) according to the manufacturer’s protocol. Eluted antibodies were buffer-exchanged into PBS using Amicon Ultra-4 centrifugal filter units (30K, Merck Millipore Ltd.).
ELISA
384 well ELISA plates (Thermo Fisher Scientific) were coated with 0.1 μM his/avi-tagged protein (426c.Mod.Core, 426c.Mod.Core KO, HXB2.WT.Core, HXB2.WT.Core KO, eOD-GT8, eOD-GT8-KO) diluted in 0.1 M sodium bicarbonate, at room temperature (RT) overnight. Plates were then washed four times with wash buffer (PBS plus 0.02% Tween20) using a microplate washer (BioTek) and incubated with block buffer (10% milk, 0.03% Tween20 in PBS) for 1-2h at 37°C. Plates were washed, mouse plasma added, and serially diluted (1:3) in block buffer. After 1h incubation at 37°C, plates were washed, and horse radish peroxidase-conjugated goat anti-mouse IgG (BioLegend) was added and incubated for 1h at 37°C. After a final wash, SureBlue Reserve TMB Microwell Peroxidase Substrate (KPL Inc.) was added to the plates for 5 mins. The reaction was stopped with 1 N H2SO4, and the optical density (OD) was read at 450 nm with a SpectraMax M2 Microplate reader (Molecular Devices). The average OD of blank wells from the same plate were subtracted from all wells before analysis.
Biolayer interferometry
Biolayer interferometry (BLI) assays were performed on the Octet Red instrument (ForteBio) as previously described (Lin et al., 2020; Parks et al., 2019). Briefly, anti-human IgG FC capture biosensors (ForteBio) were used to immobilize mAbs (20 μg/μL in PBS) for 5 min, followed by baseline interference reading for 60 s in kinetics buffer (PBS, 0.01% BSA, 0.02% Tween-20, 0.005% NaN3). Sensors were then immersed into wells containing Env Core monomers (2 μM) diluted in kinetics buffer for 300 s (association phase) and another 300 s (dissociation phase). mVRC01 and glVRC01 mAbs were used as internal controls for comparison. All measurements were corrected by subtracting the signal obtained from simultaneous tracing of the corresponding Env using an irrelevant IgG Abs in place of the mAbs tested. Curve fitting was performed using the Data analysis software (ForteBio).
TZM-bl neutralization assay
Plasma IgG and individual mAbs were tested for neutralization against a panel of selected HIV-1 pseudoviruses using TZM-bl target cells, as previously described (LaBranche et al., 1999). Briefly, neutralizing antibody activity was measured in 96-well culture plates by using Tat-regulated luciferase (Luc) reporter gene expression to quantify reductions in virus infection in TZM-bl cells. For the assays, mAbs were used at highest concentration possible, diluted over 3-fold dilutions and pre-incubated with virus (∼150,000 relative light unit equivalents) for 1hr at 37°C before addition of cells. Following a 48 hr incubation, cells were lysed and Luc activity determined using a microtiter plate luminometer and BriteLite Plus Reagent (PerkinElmer, Cat#: 6066766). Germline and mature VRC01 mAbs were used as controls in every assay.
Quantification and statistical analysis
For pairwise comparisons between two groups, the unpaired t-test was used. For comparisons between three or more groups, the ANOVA test was used for a priori analyses, followed by Tukey’s or Sidak’s multiple comparison’s test for post hoc analyses. A p value of ≤0.05 was considered statistically significant. Statistical analyses were carried out using the GraphPad Prism software (GraphPad Software).
Acknowledgments
This work was supported by grants P01 AI138212, 2R01 AI104384 and contract #HHSN272201800004C from the National Institutes of Health.
Author contributions
Conceptualization: L.S., M.L.K., and P.A.; Methodology: L.S., M.L.K., and P.A.; Validation: L.S., M.L.K., and P.A.; Formal analysis: M.L.K., P.A., and D.M.; Investigation: M.L.K., P.A., A.M., K.R.P., M.D.G., B.N.T., A.K., K.R.S., and R.N.C., Writing – Original Draft: L.S., M.L.K., and P.A., Writing – Review Editing: All; Visualization: M.L.K. and P.A., Supervision: L.S., Project Administration: L.S.; Funding Acquisition: L.S.
Declarations of interests
The authors declare no competing interests. Patent US 2018/0117140 ‘Engineered and Multimerized Human Immunodeficiency Virus Envelope Glycoproteins and Uses Thereof’’ was awarded to LS.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105473.
Supplemental information
Amino acid sequences are aligned to the V gene from which they are derived. For sequences where the PCR product did not contain the 5’ end, the sequence is shown beginning at CDR1.
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This paper does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abbott R.K., Lee J.H., Menis S., Skog P., Rossi M., Ota T., Kulp D.W., Bhullar D., Kalyuzhniy O., Havenar-Daughton C., et al. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity. 2018;48:133–146.e36. doi: 10.1016/j.immuni.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Balazs A.B., Ouyang Y., Hong C.M., Chen J., Nguyen S.M., Rao D.S., An D.S., Baltimore D. Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 2014;20:296–300. doi: 10.1038/nm.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin S.L., Reese V.A., Larsen S.E., Beebe E., Guderian J., Orr M.T., Fox C.B., Reed S.G., Coler R.N. Prophylactic efficacy against Mycobacterium tuberculosis using ID93 and lipid-based adjuvant formulations in the mouse model. PLoS One. 2021;16:e0247990. doi: 10.1371/journal.pone.0247990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas C.F., 3rd, Björling E., Chiodi F., Dunlop N., Cababa D., Jones T.M., Zebedee S.L., Persson M.A., Nara P.L., Norrby E., Burton D. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA. 1992;89:9339–9343. doi: 10.1073/pnas.89.19.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhiman J.N., Anthony C., Doria-Rose N.A., Karimanzira O., Schramm C.A., Khoza T., Kitchin D., Botha G., Gorman J., Garrett N.J., et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat. Med. 2015;21:1332–1336. doi: 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C., Lee J.H., Sliepen K., Derking R., Falkowska E., de la Peña A.T., Cupo A., Julien J.P., van Gils M., Lee P.S., et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M., Kreider E.F., Fera D., Meyerhoff R.R., Bradley T., Wiehe K., Alam S.M., Aussedat B., Walkowicz W.E., Hwang K.K., et al. Staged induction of HIV-1 glycan-dependent broadly neutralizing antibodies. Sci. Transl. Med. 2017;9:eaai7514. doi: 10.1126/scitranslmed.aai7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A.J., Weidle C.E., Gray M.D., Frenz B., Snijder J., Joyce M.G., Georgiev I.S., Stewart-Jones G.B., Kwong P.D., McGuire A.T., et al. Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core. Elife. 2018;7:e37688. doi: 10.7554/eLife.37688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B., Sok D., Jardine J.G., Kulp D.W., Skog P., Menis S., Jacak R., Kalyuzhniy O., de Val N., Sesterhenn F., et al. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell. 2016;166:1459–1470.e11. doi: 10.1016/j.cell.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel F.M., Zwick M.B., Cardoso R.M.F., Nelson J.D., Wilson I.A., Burton D.R., Dawson P.E. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J. Virol. 2006;80:1680–1687. doi: 10.1128/JVI.80.4.1680-1687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D.R., Hangartner L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese D.A., Scanlan C.N., Zwick M.B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M.R., Stanfield R.L., Roux K.H., et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Cardoso R.M.F., Zwick M.B., Stanfield R.L., Kunert R., Binley J.M., Katinger H., Burton D.R., Wilson I.A. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhou T., Schmidt S.D., Duan H., Cheng C., Chuang G.Y., Gu Y., Louder M.K., Lin B.C., Shen C.H., et al. Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth. Immunity. 2021;54:324–339.e8. doi: 10.1016/j.immuni.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coler R.N., Bertholet S., Moutaftsi M., Guderian J.A., Windish H.P., Baldwin S.L., Laughlin E.M., Duthie M.S., Fox C.B., Carter D., et al. Development and characterization of synthetic glucopyranosyl lipid adjuvant system as a vaccine adjuvant. PLoS One. 2011;6:e16333. doi: 10.1371/journal.pone.0016333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey L., Gilbert P.B., Juraska M., Montefiori D.C., Morris L., Karuna S.T., Edupuganti S., Mgodi N.M., deCamp A.C., Rudnicki E., et al. HVTN 704/HPTN 085 and HVTN 703/HPTN 081 Study Teams Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N. Engl. J. Med. 2021;384:1003–1014. doi: 10.1056/NEJMoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. Therapeutic antibodies, vaccines and antibodyomes. mAbs. 2010;2:347–356. doi: 10.4161/mabs.2.3.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel H.J., Binley J.M., Moore J.P., Sodroski J., Sullivan N., Sawyer L.S., Hendry R.M., Yang W.-P., Barbas C.F., III, Burton D.R. Neutralizing recombinant human antibodies to a conformational V2- and CD4-binding site-sensitive epitope of HIV-1 gp120 isolated by using an epitope-masking procedure. J. Immunol. 1995;154:893–906. [PubMed] [Google Scholar]
- Doores K.J., Kong L., Krumm S.A., Le K.M., Sok D., Laserson U., Garces F., Poignard P., Wilson I.A., Burton D.R. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J. Virol. 2015;89:1105–1118. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Schramm C.A., Gorman J., Moore P.L., Bhiman J.N., DeKosky B.J., Ernandes M.J., Georgiev I.S., Kim H.J., Pancera M., et al. NISC Comparative Sequencing Program Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P., Kara E.E., Pettersson A.K., McGuire A.T., Gray M., Hartweger H., Thientosapol E.S., Stamatatos L., Nussenzweig M.C. Anti-HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl. Acad. Sci. USA. 2018;115:4743–4748. doi: 10.1073/pnas.1803457115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P., von Boehmer L., Escolano A., Jardine J., Freund N.T., Gitlin A.D., McGuire A.T., Kulp D.W., Oliveira T., Scharf L., et al. Immunization for HIV-1 broadly neutralizing antibodies in human Ig knockin mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E., Le K.M., Ramos A., Doores K.J., Lee J.H., Blattner C., Ramirez A., Derking R., van Gils M.J., Liang C.H., et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.L., Hoebe K., Duong B., Ota T., Martin C., Beutler B., Nemazee D. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 2006;314:1936–1938. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli, V. Brochet, X. Lefranc, M. P. 2011. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb Protoc. Vol 2011, pg 695-715. [DOI] [PubMed]
- Gorman J., Soto C., Yang M.M., Davenport T.M., Guttman M., Bailer R.T., Chambers M., Chuang G.Y., DeKosky B.J., Doria-Rose N.A., et al. NISC Comparative Sequencing Program Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat. Struct. Mol. Biol. 2016;23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.D., Feng J., Weidle C.E., Cohen K.W., Ballweber-Fleming L., MacCamy A.J., Huynh C.N., Trichka J.J., Montefiori D., Ferrari G., et al. Characterization of a vaccine-elicited human antibody with sequence homology to VRC01-class antibodies that binds the C1C2 gp120 domain. bioRxiv. 2021 doi: 10.1101/2021.08.21.457217. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick H.B., von Boehmer L., West A.P., Jr., Schamber M., Gazumyan A., Golijanin J., Seaman M.S., Fätkenheuer G., Klein F., Nussenzweig M.C., Bjorkman P.J. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Sarkar A., Kulp D.W., Toy L., Hu X., Deresa I., Kalyuzhniy O., Kaushik K., Upadhyay A.A., Menis S., et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci. Transl. Med. 2018;10:eaat0381. doi: 10.1126/scitranslmed.aat0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoot S., McGuire A.T., Cohen K.W., Strong R.K., Hangartner L., Klein F., Diskin R., Scheid J.F., Sather D.N., Burton D.R., Stamatatos L. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Kang B.H., Ishida E., Zhou T., Griesman T., Sheng Z., Wu F., Doria-Rose N.A., Zhang B., McKee K., et al. Identification of a CD4-binding-site antibody to HIV that evolved near-Pan neutralization breadth. Immunity. 2016;45:1108–1121. doi: 10.1016/j.immuni.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Ofek G., Laub L., Louder M.K., Doria-Rose N.A., Longo N.S., Imamichi H., Bailer R.T., Chakrabarti B., Sharma S.K., et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J., Julien J.P., Menis S., Ota T., Kalyuzhniy O., McGuire A., Sok D., Huang P.S., MacPherson S., Jones M., et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J.G., Kulp D.W., Havenar-Daughton C., Sarkar A., Briney B., Sok D., Sesterhenn F., Ereño-Orbea J., Kalyuzhniy O., Deresa I., et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J.G., Ota T., Sok D., Pauthner M., Kulp D.W., Kalyuzhniy O., Skog P.D., Thinnes T.C., Bhullar D., Briney B., et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J.P., Sok D., Khayat R., Lee J.H., Doores K.J., Walker L.M., Ramos A., Diwanji D.C., Pejchal R., Cupo A., et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Diskin R., Scheid J.F., Gaebler C., Mouquet H., Georgiev I.S., Pancera M., Zhou T., Incesu R.B., Fu B.Z., et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong P.D., Mascola J.R. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche C.C., Hoffman T.L., Romano J., Haggarty B.S., Edwards T.G., Matthews T.J., Doms R.W., Hoxie J.A. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBranche C.C., McGuire A.T., Gray M.D., Behrens S., Kwong P.D., Chen X., Zhou T., Sattentau Q.J., Peacock J., Eaton A., et al. HIV-1 envelope glycan modifications that permit neutralization by germline-reverted VRC01-class broadly neutralizing antibodies. PLoS Pathog. 2018;14:e1007431. doi: 10.1371/journal.ppat.1007431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht B.N., Kool M., Willart M.A.M., Hammad H. Mechanism of action of clinically approved adjuvants. Curr. Opin. Immunol. 2009;21:23–29. doi: 10.1016/j.coi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Leaman D.P., Kim A.S., Torrents de la Pena A., Sliepen K., Yasmeen A., Derking R., Ramos A., de Taeye S.W., Ozorowski G., et al. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat. Commun. 2015;6:8167. doi: 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.R., Parks K.R., Weidle C., Naidu A.S., Khechaduri A., Riker A.O., Takushi B., Chun J.H., Borst A.J., Veesler D., et al. HIV-1 VRC01 germline-targeting immunogens select distinct epitope-specific B cell receptors. Immunity. 2020;53:840–851.e6. doi: 10.1016/j.immuni.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo N.S., Sutton M.S., Shiakolas A.R., Guenaga J., Jarosinski M.C., Georgiev I.S., McKee K., Bailer R.T., Louder M.K., O'Dell S., et al. Multiple antibody lineages in one donor target the glycan-V3supersite of the HIV-1 envelope glycoprotein and display a preference for quaternary binding. J. Virol. 2016;90:10574–10586. doi: 10.1128/JVI.01012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch R.M., Wong P., Tran L., O'Dell S., Nason M.C., Li Y., Wu X., Mascola J.R. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J. Virol. 2015;89:4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Haynes B.F. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Dreyer A.M., Carbonetti S., Lippy A., Glenn J., Scheid J.F., Mouquet H., Stamatatos L. HIV antibodies. Antigen modification regulates competition of broad and narrow neutralizing HIV antibodies. Science. 2014;346:1380–1383. doi: 10.1126/science.1259206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Glenn J.A., Lippy A., Stamatatos L. Diverse recombinant HIV-1 Envs fail to activate B cells expressing the germline B cell receptors of the broadly neutralizing anti-HIV-1 antibodies PG9 and 447-52D. J. Virol. 2014;88:2645–2657. doi: 10.1128/JVI.03228-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Gray M.D., Dosenovic P., Gitlin A.D., Freund N.T., Petersen J., Correnti C., Johnsen W., Kegel R., Stuart A.B., et al. Specifically modified Env immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat. Commun. 2016;7:10618. doi: 10.1038/ncomms10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Hoot S., Dreyer A.M., Lippy A., Stuart A., Cohen K.W., Jardine J., Menis S., Scheid J.F., West A.P., et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Pancera M., Carrico C., Gorman J., Julien J.P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Ramirez M., Garces F., Escolano A., Skog P., de Taeye S.W., Del Moral-Sanchez I., McGuire A.T., Yasmeen A., Behrens A.J., Ozorowski G., et al. Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo. J. Exp. Med. 2017;214:2573–2590. doi: 10.1084/jem.20161160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore P.L., Gray E.S., Wibmer C.K., Bhiman J.N., Nonyane M., Sheward D.J., Hermanus T., Bajimaya S., Tumba N.L., Abrahams M.R., et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat. Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J.F., Halper-Stromberg A., Gnanapragasam P.N.P., Spencer D.I.R., Seaman M.S., et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T., Guinea R., Trkola A., Purtscher M., Klima A., Steindl F., Palese P., Katinger H. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T., Steindl F., Purtscher M., Trkola A., Klima A., Himmler G., Rüker F., Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., McLellan J.S., Wu X., Zhu J., Changela A., Schmidt S.D., Yang Y., Zhou T., Phogat S., Mascola J.R., Kwong P.D. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M., Yang Y., Louder M.K., Gorman J., Lu G., McLellan J.S., Stuckey J., Zhu J., Burton D.R., Koff W.C., et al. N332-Directed broadly neutralizing antibodies use diverse modes of HIV-1 recognition: inferences from heavy-light chain complementation of function. PLoS One. 2013;8:e55701. doi: 10.1371/journal.pone.0055701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks K.R., MacCamy A.J., Trichka J., Gray M., Weidle C., Borst A.J., Khechaduri A., Takushi B., Agrawal P., Guenaga J., et al. Overcoming steric restrictions of VRC01 HIV-1 neutralizing antibodies through immunization. Cell Rep. 2019;29:3060–3072.e7. doi: 10.1016/j.celrep.2019.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.S., Wang S.K., Stanfield R.L., Julien J.P., Ramos A., Crispin M., et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., Arunachalam P.S., O'Hagan D.T. Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 2021;20:454–475. doi: 10.1038/s41573-021-00163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke A.J., Anderson C.F., Riteau N., Rausch K., Scaria P., Kelnhofer E.R., Howard R.F., Sher A., Germain R.N., Duffy P. Adjuvant and carrier protein-dependent T-cell priming promotes a robust antibody response against the Plasmodium falciparum Pfs25 vaccine candidate. Sci. Rep. 2017;7:40312. doi: 10.1038/srep40312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S.G., Carter D., Casper C., Duthie M.S., Fox C.B. Correlates of GLA family adjuvants' activities. Semin. Immunol. 2018;39:22–29. doi: 10.1016/j.smim.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadi M.M., Dashti A., Rikhtegaran Tehrani Z., Tolbert W.D., Seaman M.S., Ouyang X., Gohain N., Pazgier M., Kim D., Cavet G., et al. Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell. 2018;173:1783–1795.e14. doi: 10.1016/j.cell.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem M.L., Kadima A.N., Cole D.J., Gillanders W.E. Defining the antigen-specific T-cell response to vaccination and poly(I: C)/TLR3 signaling: evidence of enhanced primary and memory CD8 T-cell responses and antitumor immunity. J. Immunother. 2005;28:220–228. doi: 10.1097/01.cji.0000156828.75196.0d. [DOI] [PubMed] [Google Scholar]
- Scanlan C.N., Pantophlet R., Wormald M.R., Ollmann Saphire E., Stanfield R., Wilson I.A., Katinger H., Dwek R.A., Rudd P.M., Burton D.R. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J. Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L., Scheid J.F., Lee J.H., West A.P., Jr., Chen C., Gao H., Gnanapragasam P.N.P., Mares R., Seaman M.S., Ward A.B., et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 2014;7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L., Wang H., Gao H., Chen S., McDowall A.W., Bjorkman P.J. Broadly neutralizing antibody 8ANC195 recognizes closed and open states of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L., West A.P., Jr., Gao H., Lee T., Scheid J.F., Nussenzweig M.C., Bjorkman P.J., Diskin R. Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc. Natl. Acad. Sci. USA. 2013;110:6049–6054. doi: 10.1073/pnas.1303682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L., West A.P., Sievers S.A., Chen C., Jiang S., Gao H., Gray M.D., McGuire A.T., Scheid J.F., Nussenzweig M.C., et al. Structural basis for germline antibody recognition of HIV-1 immunogens. Elife. 2016;5:e13783. doi: 10.7554/eLife.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y.K., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs T., Barnes C.O., Suh-Toma N., Golijanin J., Schommers P., Gruell H., West A.P., Jr., Bach F., Lee Y.E., Nogueira L., et al. Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity. 2019;50:1513–1529.e9. doi: 10.1016/j.immuni.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M., Donau O.K., Plishka R.J., Buckler-White A., Mascola J.R., Nabel G.J., Nason M.C., Montefiori D., Moldt B., Poignard P., et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J. Exp. Med. 2014;211:2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M., Kato Y., Melo M.B., Phung I., Freeman B.L., Li Z., Roh K., Van Wijnbergen J.W., Watkins H., Enemuo C.A., et al. A particulate saponin/TLR agonist vaccine adjuvant alters lymph flow and modulates adaptive immunity. Sci. Immunol. 2021;6:eabf1152. doi: 10.1126/sciimmunol.abf1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J., Ortego M.S., Weidle C., Stuart A.B., Gray M.D., McElrath M.J., Pancera M., Veesler D., McGuire A.T. An antibody targeting the fusion machinery neutralizes dual-tropic infection and defines a site of vulnerability on epstein-Barr virus. Immunity. 2018;48:799–811.e9. doi: 10.1016/j.immuni.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D., Briney B., Jardine J.G., Kulp D.W., Menis S., Pauthner M., Wood A., Lee E.C., Le K.M., Jones M., et al. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science. 2016;353:1557–1560. doi: 10.1126/science.aah3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Pancera M., McGuire A.T. Germline-targeting immunogens. Immunol. Rev. 2017;275:203–216. doi: 10.1111/imr.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler G., Kunert R., Purtscher M., Wolbank S., Voglauer R., Steindl F., Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Tian M., Cheng C., Chen X., Duan H., Cheng H.L., Dao M., Sheng Z., Kimble M., Wang L., Lin S., et al. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell. 2016;166:1471–1484.e18. doi: 10.1016/j.cell.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J.P., Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umotoy J., Bagaya B.S., Joyce C., Schiffner T., Menis S., Saye-Francisco K.L., Biddle T., Mohan S., Vollbrecht T., Kalyuzhniy O., et al. IAVI Protocol C Investigators. IAVI African HIV Research Network Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity. 2019;51:141–154.e6. doi: 10.1016/j.immuni.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.P., Wang S.K., Ramos A., Chan-Hui P.Y., Moyle M., et al. Protocol G Principal Investigators Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L., et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Simek M.D., Priddy F., Gach J.S., Wagner D., Zwick M.B., Phogat S.K., Poignard P., Burton D.R. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Sok D., Nishimura Y., Donau O., Sadjadpour R., Gautam R., Shingai M., Pejchal R., Ramos A., Simek M.D., et al. Rapid development of glycan-specific, broad, and potent anti-HIV-1 gp120 neutralizing antibodies in an R5 SIV/HIV chimeric virus infected macaque. Proc. Natl. Acad. Sci. USA. 2011;108:20125–20129. doi: 10.1073/pnas.1117531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Jr., Diskin R., Nussenzweig M.C., Bjorkman P.J. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc. Natl. Acad. Sci. USA. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Jr., Scharf L., Scheid J.F., Klein F., Bjorkman P.J., Nussenzweig M.C. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhang Z., Schramm C.A., Joyce M.G., Kwon Y.D., Zhou T., Sheng Z., Zhang B., O'Dell S., McKee K., et al. NISC Comparative Sequencing Program Maturation and diversity of the VRC01-antibody lineage over 15 Years of chronic HIV-1 infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K., et al. NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Lynch R.M., Chen L., Acharya P., Wu X., Doria-Rose N.A., Joyce M.G., Lingwood D., Soto C., Bailer R.T., et al. NISC Comparative Sequencing Program Structural repertoire of HIV-1-Neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Zheng A., Baxa U., Chuang G.Y., Georgiev I.S., Kong R., O'Dell S., Shahzad-Ul-Hussan S., Shen C.H., Tsybovsky Y., et al. NISC Comparative Sequencing Program A neutralizing antibody recognizing primarily N-linked glycan targets the silent face of the HIV envelope. Immunity. 2018;48:500–513.e6. doi: 10.1016/j.immuni.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I.S., Altae-Tran H.R., Chuang G.Y., Joyce M.G., et al. NISC Comparative Sequencing Program Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class Antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M.B., Jensen R., Church S., Wang M., Stiegler G., Kunert R., Katinger H., Burton D.R. Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J. Virol. 2005;79:1252–1261. doi: 10.1128/JVI.79.2.1252-1261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick M.B., Labrijn A.F., Wang M., Spenlehauer C., Saphire E.O., Binley J.M., Moore J.P., Stiegler G., Katinger H., Burton D.R., Parren P.W. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequences are aligned to the V gene from which they are derived. For sequences where the PCR product did not contain the 5’ end, the sequence is shown beginning at CDR1.
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This paper does not report any original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.