Summary
Dusquetide is a next-generation IDR (innate defense regulator) targeting the major autophagy receptor protein SQSTM1/p62 and modulating the innate immune response. Here, we describe a protocol for determining dusquetide-binding sites of p62 by solution NMR spectroscopy. Step-by-step technique details were provided, including sample preparation, NMR experiment setup, data processing, and binding site analysis. This protocol could be applied to characterize other small molecules targeting the ZZ domain of p62 (9 kDa) or other proteins containing ZZ domains.
For complete details on the use and execution of this protocol, please refer to Zhang et al. (2022).
Subject areas: Molecular Biology, NMR, Protein Biochemistry, Protein expression and purification
Graphical abstract

Highlights
-
•
Optimized protocol to express and purify 15N/13C-labelled p62ZZ from bacteria cells
-
•
Methods for obtaining backbone assignments by solution NMR
-
•
Mapping of the drug-binding site onto p62ZZ sequence and structure
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Dusquetide is a next-generation IDR targeting the major autophagy receptor protein SQSTM1/p62 and modulating the innate immune response. Here, we describe a protocol for determining dusquetide-binding sites of p62 by solution NMR spectroscopy. Step-by-step technique details were provided, including sample preparation, NMR experiment set-up, data processing, and binding site analysis. This protocol could be applied to characterize other small molecules targeting the ZZ domain of p62 (9 kDa) or other proteins containing ZZ domains.
Before you begin
The protocol below describes the specific steps for determining dusquetide binding sites of p62 by solution NMR spectroscopy. However, this methodology could be applied for the characterization of other small molecules targeting the ZZ domain of p62 or other proteins containing ZZ domains.
In this protocol, we first described the procedure for expressing and purifying 13C/15N uniformly labeled p62ZZ protein. We then provided step-by-step instructions to set up triple-resonance experiments and analyze NMR spectra. Lastly, we described the procedure to perform backbone assignments of p62ZZ, trace cross-peak movement in ligand titration experiments to determine the interacting surface, and map the binding site onto 3D structure of p62ZZ.
Purification of PreScission protease
Timing: 4–5 days
-
1.
Transform the pGEX-4T plasmid encoding the GST-tagged PreScission protease into Escherichia coli BL21-CodonPlus (DE3) RIL chemically competent cells (Agilent Technologies) and grow colonies on LB agar plates containing 100 μg/mL ampicillin and 34 μg/mL chloramphenicol.
-
2.
Inoculate a single colony into 100 mL Luria-Bertani (LB) media supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol in a 250 mL PYREX® Round Media Storage Bottle (The Lab Depot). Grow the cells in a New Brunswick Scientific Model G25 Incubator Shaker at 37°C with shaking at 200 rpm, 14–18 h.
-
3.
Dilute 100 mL of cell culture into 2 L of LB media supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol in a 6 L Narrow-Mouth Erlenmeyer Flask (Thermo Fisher Scientific) and continue growing cells in a New Brunswick Excella E25 Incubator Shaker at 37°C with shaking at 200 rpm until the optical density of a 1 mL sample reaches 1.0 at an absorbance of 600 nm.
-
4.
Reduce the temperature of the shaker to 30°C and slow down the shaker speed to 180 rpm. Protein expression is induced by adding 2 mL of 1.0 M Isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubation at 30°C with shaking for 4 h.
-
5.
Harvest cells by centrifugation at 5,000 rpm (4,424 × g) using F10-6×500 Fixed Angle Rotor in Thermo Scientific Sorvall RC 6 Plus Centrifuge at 4°C for 10 min and discard supernatant.
-
6.
Resuspend cell pellet with 150 mL lysis buffer A. Lyse the cells on ice by sonication for 30 min at 60% power with intervals of 5 s on and 10 s off using Q700CA Sonicator (QSonica) attached to QSonica CL334 converter.
-
7.
Spin down the cell lysate in 50 mL high-speed PPCO centrifuge tubes (Nalgene Oak Ridge) at 15,000 rpm (26,964 × g) in a SS-34 Fixed-Angle Rotor (Thermo Fisher Scientific) at 4°C for 30 min to remove cell debris and insoluble fractions.
-
8.
Wash the 5 mL Glutathione Agarose Resin (Gold Biotechnology, stored at 4°C fridge) with 60 mL GST Resin wash buffer in an Econo-Pac chromatography gravity column (Bio-Rad).
-
9.
Incubate the supernatant with Glutathione Agarose Resin in a beaker at 25°C for 2 h with rocking on a Labnet GyroMini™ Nutating Mixer at 4°C.
-
10.
Elute the protein off the resin with 10 mL GST elution buffer and then dialyze the elution to 1 L storage buffer using a 10 kDa Regenerated Cellulose Dialysis Tubing (Thermo Fisher Scientific) at 4°C overnight.
-
11.
Spun down the dialyzed protein for 2 min at 12,000 rpm (13,000 × g) in falcon tubes to remove any precipitates. Concentrate the 10 mL protein to 10–15 mg/mL using a 10 kDa MWCO centrifugal filter (Millipore) at 3,000 ×g on Eppendorf 5810 R Centrifuge with an A-4-81 rotor and store the protein at −80°C.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli BL21 (DE3) RIL | Zhang et al., 2018 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dithiothreitol | Gold Biotechnology | 27565-41-9 |
| Duesquetide | Synpeptide | N/A |
| Centrum Adults Multivitamins | GlaxoSmithKline | N/A |
| 15NH4Cl | Sigma-Aldrich | 299251 |
| IPTG | Gold Biotechnology | I2481C100 |
| Glutathione agarose resin | Gold Biotechnology | G-250 |
| PreScission proteases | Home expressed | N/A |
| L-Arginine (98+%) | Acros Organics | AC104991000 |
| Disodium hydrogen phosphate heptahydrate | Sigma-Aldrich | 7782-85-6 |
| Magnesium sulfate anhydrous | Fisher Chemical | 7487-88-9 |
| Calcium chloride dihydrate | Fisher Chemical | 10035-04-8 |
| Potassium dihydrogen phosphate | Fisher Chemical | 7778-77-0 |
| Sodium chloride | Fisher Chemical | J21618.A1 |
| Tris base | Sigma-Aldrich | 77-86-1 |
| Zinc chloride | Thermo Fisher Scientific | AAA1628136 |
| TritionX-100 | Sigma-Aldrich | 9036-19-5 |
| Glutathione reduced | Fisher Bio Reagents | 70-18-8 |
| Glycerol | Fisher Chemical | BP2294 |
| Deposited data | ||
| p62-ZZ: dusquetide complex | This study | PDB: 7R1O |
| Recombinant DNA | ||
| Plasmid: modified pGEX6p-1 | Zhang et al., 2018 | N/A |
| Software and algorithms | ||
| Topspin 4.1.4 | Bruker | |
| NMRPIPE | https://www.ibbr.umd.edu/nmrpipe/ | |
| CCPN 3.1.0 | https://ccpn.ac.uk | |
| NMRBOX | https://nmrbox.nmrhub.org | |
| Other | ||
| Econo-Pac® Chromatography Columns | Bio-Rad | 7321010 |
| PierceTM Protein Concentrator PES, 3K MWCO | Thermo Scientific | XB344046 |
| Shigemi NMR tubes (BMS-005TB) | Sigma-Aldrich | Z529451 |
| Nalgene Oak Ridge High-Speed PPCO centrifuge tubes | Thermo Scientific | 3119-0050 |
| SnakeSkin Dialysis Tubing | Thermo Scientific | 68100 |
| CellPro 1,000 mL, 0.22-μm bottle top filter, PES filter material | Alakli Scientific | VH100022 |
| Beckman Coulter centrifuge bottle (polypropylene bottle with Noryl cap) | Beckman Coulter | 50-163-1991 |
| Sterile Polystyrene Disposable Serological Pipets | Thermo Fisher Scientific | 13-678-11E |
| pH buffer sachets, METTLER TOLEDO | METTLER TOLEDO | 01-911-289 |
Materials and equipment
M9 minimal media
20× Stock preparation
| Reagent | Final concentration | Amount |
|---|---|---|
| Na2HPO4·7H2O | 954 mM | 256 g |
| KH2PO4 | 440 mM | 60 g |
| NaCl | 171.2 mM | 10 g |
Prepare the following chemical stocks as well in 0.1 L Milli-Q water.
| Stocks | Reagents | Final concentration | Amount |
|---|---|---|---|
| 1000× | MgSO4 | 1,000 mM | 12 g |
| 10000× | CaCl2 | 1,000 mM | 11.1 g |
| 2000× | ZnCl2 | 100 mM | 1.363 g |
Dilute the following stocks to 1× in 1 L Milli-Q water and autoclave the media at 121°C for 30 min.
| Stock concentration | Reagent | Final concentration | Amount |
|---|---|---|---|
| 20× | Na2HPO4·7H2O | 47.7 mM | 50 mL |
| KH2PO4 | 22 mM | ||
| NaCl | 8.56 mM | ||
| 15NH4Cl | 18.4 mM | 1 g | |
| 1000× | MgSO4 | 2 mM | 2 mL |
| 10000× | CaCl2 | 100 μM | 100 μL |
| 2000× | ZnCl2 | 50 μM | 0.5 mL |
Add the following reagents before applying to cell culture:
| Reagent | Amount |
|---|---|
| vitamin power (Centrum Adults Multivitamins) dissolved in 40 mL Milli-Q water, mixed and filtered | 0.1 g |
| 13C-labeled glucose dissolved in 30 mL PBS buffer, mixed and filtered | 4 g |
The M9 minimal media can be stored at 25°C for 1 week.
Lysis buffer A
Lysis buffer A contains 50 mM Tris (pH 7.5), 500 mM NaCl, 2 mM DTT and 0.5% Triton x-100. To prepare lysis buffer A, add the reagents below to 800 mL Milli-Q water.
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-base | 50 mM | 6.06 g |
| NaCl | 500 mM | 29.25 g |
| DTT | 2 mM | 0.31 g |
| Triton x-100 | 0.5% | 5 mL |
Adjust the pH of the buffer by adding 12 N HCl while stirring until the pH reaches 7.5 monitored by a METLLER TOLEDO pH-meter at 25°C. Add Milli-Q water up to 1 L and filter the buffer using a 0.22 μm bottle top filter (CellPro). The buffer can be stored at 4°C for 1 week.
Lysis buffer B
Lysis buffer B contains 50 mM Tris (pH 7.0), 100 mM NaCl, 5 mM DTT. To prepare lysis buffer B, add the reagents below to 800 mL Milli-Q water.
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-base | 50 mM | 6.06 g |
| NaCl | 100 mM | 5.85 g |
| DTT | 5 mM | 0.77 g |
Adjust the pH of the buffer to 7.0 at 25°C. Add Milli-Q water up to 1 L and filter the buffer using a 0.22 μm filter. The buffer can be stored at 4°C for 1 week.
GST resin wash buffer
Wash buffer contains 50 mM Tris (pH 7.5), 500 mM NaCl and 2 mM DTT. To prepare wash buffer, add the reagents below to 800 mL Milli-Q water:
| Reagents | Final concentration | Amount |
|---|---|---|
| Tris-base | 50 mM | 6.06 g |
| NaCl | 500 mM | 29.25 g |
| DTT | 2 mM | 0.31 g |
Adjust pH of the buffer to 7.5 at 25°C. Add Milli-Q water up to 1 L and filter the buffer using a 0.22 μm filter. The buffer can be stored at 4°C for 14 days.
GST elution buffer
GST elution buffer contains 50 mM Tris (pH 7.5), 500 mM NaCl, 2 mM DTT and 50 mM reduced glutathione. To prepare GST elution buffer, add the reagents below to 800 mL Milli-Q water.
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-base | 50 mM | 6.06 g |
| NaCl | 500 mM | 29.25 g |
| DTT | 2 mM | 0.31 g |
| Glutathione | 50 mM | 15.35 g |
Adjust the pH of the buffer to 7.5 at 25°C. Add Milli-Q water up to 1 L and filter the buffer using a 0.22 μm filter. The buffer can be stored at 4°C for 1 day.
Storage buffer
Storage buffer contains 50 mM Tris (pH 7.5), 500 mM NaCl, 2 mM DTT and 5% glycerol. To prepare GST storage buffer, add the reagents below to 800 mL Milli-Q water.
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-base | 50 mM | 6.06 g |
| NaCl | 500 mM | 29.25 g |
| DTT | 2 mM | 0.31 g |
| Glycerol | 5% | 50 mL |
Adjust the pH of the buffer to 7.5 at 25°C. Add Milli-Q water up to 1 L and filter the buffer using a 0.22 μm filter. The buffer can be stored at 4°C for 1 day.
Gel-filtration buffer
Gel-filtration buffer contains 50 mM Tris (pH 7.0), 100 mM NaCl and 5 mM DTT. To prepare gel-filtration buffer, add the reagents below to 800 mL Milli-Q water:
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-base | 50 mM | 6.06 g |
| NaCl | 100 mM | 5.85 g |
| DTT | 5 mM | 0.77 g |
Adjust the pH of the buffer to 7.0 at 25°C. Add Milli-Q water up to 1 L and filter the buffer using a 0.22 μm filter. The buffer can be stored at 4°C for 1 day.
Step-by-step method details
p62ZZ expression and purification
Timing: 4–5 days
Prepare the protein samples for NMR experiments.
-
1.
The human p62 ZZ domain (aa 115–190) was cloned into a pGEX 6p-1 vector.
-
2.
Transform the expression plasmid that encodes p62ZZ fused with GST tag into BL21 (DE3) RIL chemically competent cells (Agilent Technologies) according to the manufacturer’s transformation protocol. Plate the cells onto LB agar plates supplemented with 34 μg/mL chloramphenicol and 100 μg/mL ampicillin.
Note: The detailed transformation protocol could be found from Agilent Technologies website (https://www.agilent.com/cs/library/usermanuals/public/200133.pdf).
-
3.
Inoculate single colony into 150 mL LB media. Culture the cells in a New Brunswick Scientific Model G25 Incubator Shaker at 37°C and 200 rpm for 14–18 h.
-
4.
Dilute the overnight culture into the 3 L LB media by adding 50 mL cells into 1 liter of LB media. Culture the cells at 37°C in a New Brunswick Excella E25 Incubator Shaker with an agitation speed of 200 rpm until the optical density of a 1 mL sample reaches 0.3–0.4 at an absorbance of 600 nm.
-
5.
Reduce the shaker temperature to 16°C.
-
6.
Spin down cells using Beckman Coulter 500 mL polypropylene sterile bottles for centrifuge in the Thermo Scientific Sorvall RC 6 Plus Centrifuge with a speed of 3,500 rpm for 5 min using F10-6×500 Fixed Angle Rotor (Thermo Fisher Scientific).
-
7.
Resuspend the cells with 50 mL M9 minimal media using a 50 mL pipette (Globe) and transfer the cells back to 1 L M9 minimal media.
-
8.
Weigh out 4 g 13C-labeled glucose (Sigma) and 1 g 15N-labeled NH4Cl (Sigma), dissolve in 30 mL M9 minimal media, and transfer to the M9 minimal media.
-
9.
Weight out 0.1 g vitamin power Centrum, dissolve in 40 mL Milli-Q water. Remove the undissolved solids by centrifuge. Filter the supernatant using a Ezflow syringe filter 0.22 μM. Transfer the vitamin solution to the M9 minimal media.
-
10.
Add 0.2 mM IPTG in to the M9 minimal media. Protein expression was induced at 16°C for 20 h.
-
11.
Harvest cells using 500 mL bottles (Fisherbrand) by centrifugation at 5,000 rpm (4,424 × g) using a F10-6×500 Fixed Angle Rotor in Thermo Scientific Sorvall RC 6 Plus Centrifuge for 10 min at 4°C and discard the supernatant.
Pause point: Pellets can be flash frozen in liquid nitrogen and stored at −80°C for up to 3 months.
-
12.
Resuspend the cell pellets in 50 mL lysis buffer B by pipetting up and down using 10 mL transfer pipette.
-
13.
Lyse the resuspended cells on ice by sonication for 10 min at 60% power with intervals of 5 s on and 10 s off using Q700CA SONICATOR (QSonica) attached to QSonica CL334 converter.
-
14.
Spin down the cell lysate using 50 mL Nalgene Oak Ridge centrifuge tubes (LPS) at 14,000 g using a SS-34 Fixed-Angle Rotor in Thermo Scientific Sorvall RC 6 Plus Centrifuge for 15 min at 4°C to remove cell debris and insoluble fractions.
-
15.
Pass the supernatant to a 0.45 μM Syringe Filter (PALL) to remove the residual cell debris completely.
-
16.
Incubate the filtered solution with 2 mL Glutathione Agarose Resin (Gold Biotechnology) in a 50 mL Falcon tube for 2 h rocking on a Roto-Mini™ Rotator test tube rocker at 4°C.
-
17.
Apply the suspension to an Econo-Pac® Chromatography Column (Bio-Rad) to collect the protein bound resin.
-
18.
To remove impurities, wash the resin with 50 mL lysis buffer B supplemented with 100 mM arginine.
-
19.
Resuspend the resin with 15 mL lysis buffer B, add the home-made PreScission enzyme, and incubate the mixture rocking on a Labnet GyroMini™ Nutating Mixer for 4–24 h at 4°C to cleave the GST tag.
-
20.Analyze a 10 μL sample of the PreScission cleavage reaction by polyacrylamide gel electrophoresis (PAGE). Once the desirable extent of the cleavage reaction is achieved (> 80%), collect and concentrate the cleaved protein.
-
a.Apply the reaction mixture to an Econo-Pac gravity column (Bio-rad) to separate the beads. Collect the flow-through, which contains the cleaved protein.
-
b.Centrifuge to remove the potential precipitate and then concentrate the protein to 2 mL using a 3 kDa MWCO centrifugal filter (Millipore) at 2,800 g in an Eppendorf 5810 R Centrifuge with an A-4-81 rotor at 4°C.
-
a.
-
21.
Inject the concentrated protein onto a PierceTM Protein Concentrator PES, 3 kDa (Thermo-Scientific) pre-equilibrated in gel-filtration buffer supplemented with 100 mM arginine. Run the sample at a flowrate of 0.4 mL/min.
-
22.
Perform the polyacrylamide gel electrophoresis using a 15-well homemade 15% gel to check the purity of the fractions. The gel was prepared according to standard Bio-rad gel making protocol (https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6201.pdf).
-
23.
Pool protein-containing fractions and concentrate to ∼10 mg/mL using a 3 kDa MWCO centrifugal filter (Millipore) at 3,000 g in an Eppendorf 5810 R Centrifuge with an A-4-81 rotor (Eppendorf) at 4°C.
Backbone NMR experiments using topspin
Timing: 3–5 days
Acquiring NMR data needed for backbone assignments.
-
24.
Prepare the 13C/15N-labelled protein sample using above protocol.
-
25.
Add 8%–10% D2O to protein sample (250 μL at 1.1 mM) prepared in 20 mM Tris-HCl (pH 7.0) buffer, supplemented with 100 mM NaCl, 100 mM free arginine and loaded the sample Shigemi NMR-tube using the NMR pipette.
Note: Concentration of the protein was quantified by measuring 1.5 μL of the protein sample at 280 nm using a Nanodrop spectrometer. The NMR tube was sealed with parafilm to prevent evaporation during NMR experiments.
Note: The Shigemi NMR microtubes (BMS-005TB) require less sample and provide better signal to noise ratio then regular 5 mm symmetric tubes.
-
26.
Execute the “topspin” command in the terminal or click topspin icon on NMR instrument and open the topspin-gui.
-
27.
Enter “ej” command in topspin-gui terminal and load the sample and enter the ij to load the NMR sample on to the probe.
-
28.
Create a new dataset and using the command “iexpno” and load the pulse sequence zg. Set up the sample temperature of the sample to 298K using edte panel (command: edte) on topspin-gui and equilibrate the temperature of the sample for 15 min.
-
29.
Perform lock, tune and match, and auto-shim using commands “lock”, “atma”, and “topshim” as directed in the Bruker guide and/or local NMR facility guide.
-
30.
Calibrate the 90° pulse width for 1H (90° hard pulse) for the sample using standard calibration experiment.
Optional: Optimize the N15 and C13 pulse widths per instructions at local NMR facility.
-
31.
Calibrate the water resonance frequency to set the o1p value.
-
32.
Load the Bruker pulse sequences for 2D 1H-15N HSQC and 1H-13C HSQC experiments using the following commands i.e., “rpar hsqcetf3gpsi” and “rpar hsqcetgp” from Bruker library.
-
33.
Adjust the carrier frequency using o2p (117 ppm), spectral width SW (36 ppm) for 1H-15N HSQC, o3p (43 ppm), spectral width 84 ppm for 1H-13C HSQC, and number of scans (ns) and time domain points (TD) and other default parameters.
-
34.
Acquire the 1H-15N HSQC and 1H-13C HSQC data and process using “xfb” command in the topspin terminal. Use the phase correction tab to adjust the phase in F1 and F2 dimensions.
-
35.Acquire a set of 3D-NMR experiments [HNCACB, CBCA(CO)NH and HNCA] using the following pulse sequence from Bruker library. Adjust the parameters used as per in the given table and adjust the carrier frequency and spectral widths using the 1H-13C HSQC and 1H-15N HSQC experiments.
-
a.hncacbgp3d.
-
b.cbcaconhgp3d.
-
c.hncagp3d.
-
a.
CRITICAL: Before collecting 3D spectra, 2D NMR must be performed to confirm that the target protein is folded and stable without aggregation or visible precipitation in the NMR tube. Typically, a well-dispersed 1H-15N HSQC spectrum with cross peaks displaying uniform intensity is a good start.
CRITICAL: Please consult the local NMR facility and confirm NMR parameters such as pulse width and power level are up to date. For protein backbone assignments, other triple resonance experiments can also be used depending on the size and property of the target protein.
-
36.
Adjust the receiver gain using “rga” command in the topspin terminal.
-
37.
Use the “zg” command for individual experiment or “multizg” command to run all above experiments.
Note: Please consult the local NMR facility to set up the triple resonance experiments using non-uniform sampling (NUS) methods, which can reduce data acquisition time to 25%. With NUS, the data acquisition times for the HNCACB, CBCA(co)NH, and HNCA experiments used in this protocol were 13 h (64 scans), 5 h (32 scans), and 4 h (16 scans), respectively.
Note: Prior or during data acquisition, the sample stability should be monitored by running N15-HSQCs every day (or experiment). If the overlay of the HSQC spectra shows extra peaks in the ∼8 ppm [H]/∼126 ppm [N] region, the protein sample is undergoing degradation or denaturing. See also problem 4.
NMR data processing
Timing: 1–2 days
Processing NMR data for backbone assignments (Figure 1).
Figure 1.
The 1H-13C planes projected from 3D HNCACB and CBCA(co)NH spectra
Prerequisite: Topspin, NMRPipe (Delaglio et al., 1995), and CCPNMR/Assign (Skinner et al., 2016) on your Linux workstation or NMRbox account (Maciejewski et al., 2017).
-
38.
Open 3D NMR data in the topspin using menu tab (left of the topspin panel) or drag the NMR data into the topspin main GUI.
-
39.
Use the “xfb” command in the topspin terminal to process the data into 13 (CH) plane and 23 (NH) plane and correct the phase correction. Note down phase values in 1H, N15 and C13 planes.
-
40.
Use the “convert2pipe” command in the topspin terminal and topspin opens a window to chemical shift reference window, select the option depending on the mode of data collection.
-
41.
Topspin opens another window to generate the NMRPipe script, chose option 1 to generate the simple NMRPipe script for 3D NMR data. Open another window to display the fid.com file and click “ok” option. Open the text file to modify the acquisition parameters i.e., spectral parameters, mode of acquisition parameters, phasing parameters.
-
42.
Enter the phasing values that noted while performing the phase correction and save the file using save button.
-
43.
Open the terminal command under 3D NMR data folder on the Linux OS or NMRbox virtual machine. Change to c-shell using “csh” command (default shell is bash shell on Linux OS).
-
44.
Enter the “fid.com” command in the 3D-NMR folder (i.e., HNCACB, CBCACONH, HNCA).
-
45.
Use steps 38–44 to process all 3D-NMR data.
Backbone NMR assignments using CCPNmr
Timing: 5–7 days
Peak picking and backbone chemical shift assignment.
-
46.
Convert the nmr data from pipe format into sparky using the below command or use pipe-format files (xxxx.pipe) in CCPN assign module.
pipe2ucsf xxxxxx.ft2 xxxx.ucsf.
-
47.
Open CCPNmr Analysis Assign from terminal using “assign” command.
-
48.
Select all ucsf or pipe files from the NMR data folder and drag them into the sidebar or load the spectrums using “ls” command and opens a tab to navigate the folder that contains the NMR data files.
-
49.
Drag and drop the 15N-HSQC, CBCAcoNH, HNCACB & HNCA spectra anywhere on the purple box in the drop box area. Spectra with the same frequencies can be shown in the same module, which allows to toggle and overlay between multiple spectra in on/off display mode.
-
50.
Right-click on each spectrum and click on contours or use the command CO, then uncheck the negative contours except for HNCACB spectra. Adjust the contour levels to minimize the noise levels for all spectra.
-
51.
Click on the 15N-HSQC spectrum and use Ctrl (or Cmd for Mac) + Shift + Left-drag on the contour to create the blue box in the region. Release the button and keys to select the peaks or perform auto peak picking using shortcut PP. Define spectral regions for N & H to select the positive peaks in 15N-HSQC spectra. Inspect the peaks in 15N-HSQC spectrum and remove the noise peaks and sidechain peaks.
Alternative to PP shortcut: Main Menu ⟶ Spectrum ⟶ Pick Peaks ⟶ Pick ND Peaks.
-
52.
To create the default peak labels for picked peaks in the previous step, use the shortcut SN command. This creates a default index label for each contour in the 15N-HSQC spectrum. Use the shortcut PL to toggle between different ways to display the peak labels.
Source PeakList: PL:hsqc.x.
NmrChain: NC:@-.
Click Setup NMR Residues.
Alternative to SN shortcut: go to Main Menu ⟶ Assign ⟶ Setup NMR Residues.
-
53.
To generate the chain, go to Main Menu ⟶ Molecules ⟶ Chain from the fasta option or copy and paste the fasta sequence into the New Chain option.
-
54.
Select all 3D spectrums, i.e., HNCA, HNCACB and CBCAcoNH, in the side panel and open all spectrums as a module. To pick peaks in the 3D spectra based on the 15N-HSQC spectrum, use the Pick & Assign module, and use the restricted peak picking tab to pick the peaks. Noise peaks should be removed while using the restricted peak picking tab and then select the following option in the tab NmrChain: NC:@-.
Alternative to PA shortcut: go to Main Menu ⟶ Assign ⟶ Pick and Assign.
CRITICAL: Accurate peak picking is critical for backbone chemical shift assignment protocol, particularly when using automatic programs. Noise, artifacts and peak overlap strongly affect subsequence steps and should be inspected carefully.
-
55.
To define the atom nomenclature in the 3D spectrum, Select the NMR atom assigner. Using peak and assign tab or short cut PA, select the peak with index 0 and select the contour using NA tab to define the CA(i), CA(i-1) and CA(i), CA(i-1), CB(i), CB(i-1) and CB(i), CB(i-1) in the HNCA, HNCACB, and CBCAcoNH spectrum respectively. Use shortcut AN and select the following option in the tabs. NmrChain: NC:@- and Assign by axis: C.
For example, define the following atom names in the 3D spectra as follows.
HNCA: CA (i, i-1),
HNCACB: CA (i, i-1) and CB (i, i-1).
CBCAcoNH: CA (i-1) and CB(i-1).
Alternative to AN shortcut: go to Main Menu ⟶ Assign ⟶ NmrAtom Assigner.
-
56.
Similarly, select peak index (N=1,2,3,4 …) in the Peak and Assign tab and use NMR Atom Assigner (command AN) to define the CA(i) an CB(i) or CA (i-1) CB(i-1) using HNCA, HNCACB and CBCAcoNH spectrum for each peak N(i)-H(i) in 15N-HSQC spectrum.
-
57.
To monitor the default connectivity while defining the CA, CB, or CA (i-1), CB(i-1) atoms to N(i)-H(i), The default sequence graph allows the probability of each peak N(i)-H(i) as amino acid type depending on the CA/CB chemical shifts.
Alternative to SG shortcut: go to Main Menu ⟶ view ⟶ Sequence Graph.
-
58.
Continue the above steps until all assignments of CA an CB and/or CA (i-1) CB(i-1) are done for each peak in 15N-HSQC spectrum.
-
59.
To link the connectivity of i-1, i, i+1 residues of the spectra, use the BB command that opens a tab with a list of index number as a table for 15N-HSQC default Peak list.
Alternative to BB shortcut: go to Main Menu ⟶ Assign ⟶ Backbone Assignment.
-
60.
Double click on the index number that makes changes in module tab which allows to connect the CA (i-1), CB(i-1) and CA (i), CB(i) of i-1, i, i+1 residues. The best matched residues of the CA and CB of linking residues indicated on percentage basis on top of the module tab in both i+1 and i-1 directions and allows to make sequential connection by drag and drop of that strip to left/right depends on the i+1/i-1 direction, respectively.
Note: Many other NMR software and online server such as SPARKY (Lee et al., 2015) and PINE (Lee et al., 2019) are also frequently used for analyzing NMR spectra, peak picking, and chemical shift assignment. Alternative to CCPNmr, PINE-SPARKY extension (Lee et al., 2009) also supports interactive assignments.
-
61.
Use the Spin Graph assigner to inspect the sequential connection in the given sequence and continue the assignments until all sequential assignments of residues are completed.
-
62.
Open the Assignment Inspector module to check the connectivity of all residues in the given protein sequence.
Main Menu ⟶ Assign ⟶ Assignment Inspector or shortcut AI.
Note: Video tutorials and manual for using CcpNmr Analysis V3 can be found at https://ccpn.ac.uk/manual/v3/index.html.
Chemical shift perturbations (CSPs) calculation and histogram plotting
Timing: 1 day
Calculate CSPs in NMR titration experiments (Figure 2) and plot as a histogram. Ligands that specifically bind to the target protein typically induce significant but un-uniform CSPs on multiple crosspeaks, whereas the rest of crosspeaks were little perturbed. Alterations in the solvent environment due to the addition of ligands, such as pH or crowding effect, typically induce uniform changes in all perturbed crosspeaks. In addition, CSPs induced by specific ligands will become saturated as the ligand concentration increases.
-
63.
Input the processed NMR spectra of apo-formed p62ZZ and p62ZZ bound to dusquetide peptide, arginine amide and free arginine at different molar ratios into the CCPNmr or Sparky software.
-
64.
Adjust the counter level of the spectra to get an appropriate signal to noise ratio for further peak picking process.
-
65.
Perform the automatic peak picking. Inspect all the selected peaks and manually pick the missed ones and delete those that were picked out incorrectly.
-
66.
Label the peaks with the assigned residues on the spectrum of arginine-bound p62ZZ based on the previous assignments.
-
67.
Copy the assignments to the spectrum of apo-p62ZZ and adjust peak positions by tracing arginine-induced movement of crosspeaks.
CRITICAL: Assigning apo-p62ZZ based on the assignment obtained using arginine bound form is highly accurate in this case as the affinity of ZZ/Arg interaction is estimated to be in the millimolar range. All amide crosspeaks in p62ZZ are in the fast-exchange regime and were confidently mapped.
-
68.
Label the peaks with the assigned residues on the spectrum of apo-formed p62ZZ based on the mapped assignments.
-
69.
Copy the assignments to the spectrum of p62ZZ bound to dusquetide peptide or arginine amide at a molar ratio of 1:4 and adjust peak positions based on titration experiments.
-
70.
Output the peak lists containing the 1H and 15N coordinates of individual peaks on the spectra.
-
71.
Copy the coordinates of the peaks in apo form and substrate-bound p62ZZ NMR spectra to a Microsoft Excel table.
-
72.
Calculate the observed CSPs in p62ZZ bound to substrates at a protein/ligand ratio of 1:4 relative to free state using the following formula: , where δH and δN are the chemical shift changes in ppm for the 1H and 15N dimension respectively, and the values are the differences of the corresponding coordinates of individual peaks between apo-formed and substrate-bound p62ZZ. The coefficient α equals 0.2.
Note: Calculation of CSPs can be performed automatically and raw histogram can be generated in the software. For record keeping purpose, this protocol exports the peaklists and performed the analysis in Microsoft Excel.
-
73.
Plot the histogram using the residue numbers and the CSPs in Excel. The p62ZZ domain comprised of amino acid 115–190 are included in the plotting except for the proline residues that are not visible in HSQC spectrum. The missing prolines are indicated by filled dots.
-
74.
Treat the CSP value as zero for unassigned residues and indicates them with filled squares. CSPs for amino acids 174–190 of p62ZZ are less than 0.01 ppm and not shown.
-
75.
Mark the threshold with a dash line. Residues with the CSPs above the threshold are considered to be significantly perturbed and involved in the interaction with ligands.
Note: The threshold is empirically based on each experiment. There is no standard rule to define significantly perturbed crosspeaks across different protein-ligand pairs. In general, a value at one or two times the standard deviation can be used as an initial threshold.
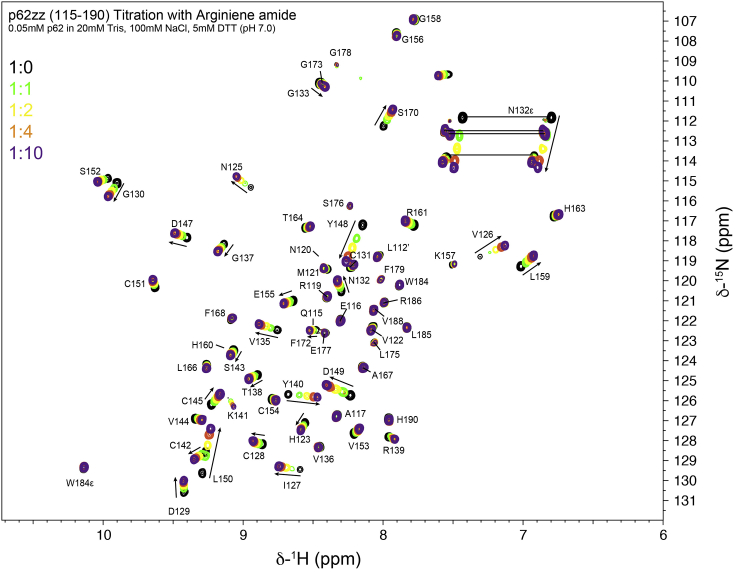
Figure 2.
Assignments of amide resonances and analysis of the arginine amide titration experiment
Superimposed 1H,15N HSQC spectra of p62ZZ(115-190) collected while arginine amide was titrated in. Assignment of backbone amides were shown.
Mapping the perturbed residues onto p62ZZ surface
Timing: 1 h
Map the CSPs onto known target protein structure.
-
76.
Open the PDB file of p62ZZ in complex with dusquetide peptide in Pymol software (DeLano, 2020).
-
77.
Display the background in white color.
-
78.
Present the p62ZZ protein as surface in white and the dusquetide peptide as sticks in pink. Set the surface quality value to 2.
-
79.
Select the most perturbed residues with CSPs above the threshold in the histogram and show these residues as surface in pink.
-
80.
Label the surface-exposed residues on the surface of p62ZZ.
-
81.
Output the graph for figure making.
Expected outcomes
Solution NMR spectroscopy is a unique technique that can provide target protein and drug interaction with amino acid level accuracy without solving the atomic resolution structures (Ziarek et al., 2011). It directly examines protein-drug interaction in solution without the need for co-crystallization and is suited for proteins below the size limit of cryo-EM. Titration-based chemical shift perturbation mapping is exceptionally powerful at the early stage of drug or lead molecules assessment where high-affinity ligands (Kd < 1 μM) were yet to be discovered. While this protocol was developed to study the ligand binding to the ZZ domain of p62, it can be broadly used to produce and characterize other ZZ domain-containing proteins or other small molecule ligands in general. In our study, 1 L cell culture grown in M9 media yields 5–10 mg of p62ZZ. This protocol yielded near complete backbone assignment and allowed the determination of binding sites for three different ligands.
Limitations
While the isotope-labeling, NMR data acquisition, backbone assignments, ligand-induced CSP calculation, and structure mapping steps described here are suitable for all target proteins, this protocol will not work for proteins with apparent molecular weight higher than 25 kDa. In addition, protein expression conditions, such as temperature, duration, IPTG concentration, and the expressing cell line often vary for individual proteins and need to be optimized to increase the yield. Proteins with a tendency to oligomerize or aggregate are also not suitable for characterization using the above protocol. The instrument used to collect NMR data is a Bruker 600 MHz spectrometer equipped with a TCI cryo probe. It also remains challenging to collect NMR data affordably using spectrometers without a cryogenic probe or below 500 MHz.
Troubleshooting
Problem 1
The yield of the labeled protein is low (steps 4–11).
Potential solution
Optimize the concentration of IPTG. Increase IPTG concentration might improve the protein expression. However, if the concentration is too high, it may also cause potential aggregation of protein. Alternatively, increase the amount of starting culture will also increase the yield. For toxic proteins, leaky expression should be checked and avoided to increase cell viability.
Problem 2
Protein cleavage is not complete (step 20).
Potential solution
Extend cleavage time, add more PreScission enzyme, or cleave the protein at room temperature.
Problem 3
Protein amide signals in 15N-NHSQC are heterogenous in intensity (step 32).
Potential solution
Optimize protein concentration, buffer system, salt concentration, additives, and data collection temperature. Electrically conductive buffers and salts are typically required for protein stability; however, they reduce the signal-to-noise ratio in NMR experiments. A guide for evaluating buffer conditions was described (Kelly et al., 2002).
Problem 4
Protein is degraded during NMR data collection (step 35).
Potential solution
Add 0.1% sodium azide to the NMR sample or increase protein concentration to shorten the data acquisition time.
Problem 5
The signal noise ratio in triple resonance spectra is low (step 35).
Potential solution
Increase NMR sample concentration or increase the number of scans and data collection time.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yi Zhang (yi.zhang26@case.edu).
Materials availability
All reagents generated in this study will be made available on reasonable request.
Acknowledgments
This work was supported by a grant from NIH R00 CA241301 to Y.Z. and R01 AG067664 to T.G.K.
Author contributions
All authors contributed to writing the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
No coordinates or structure factors were generated in this protocol.
References
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. Schrödinger LLC; 2020. The PyMOL Molecular Graphics System. Version 2.3. [Google Scholar]
- Lee W., Westler W.M., Bahrami A., Eghbalnia H.R., Markley J.L. PINE-SPARKY: graphical interface for evaluating automated probabilistic peak assignments in protein NMR spectroscopy. Bioinformatics. 2009;25:2085–2087. doi: 10.1093/BIOINFORMATICS/BTP345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/BIOINFORMATICS/BTU830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Ou H.D., Withers R., Dötsch V. Low-conductivity buffers for high-sensitivity NMR measurements. J Am Chem Soc. 2002;124:12013–12019. doi: 10.1021/ja026121b. [DOI] [PubMed] [Google Scholar]
- Lee W., Bahrami A., Dashti H.T., Eghbalnia H.R., Tonelli M., Westler W.M., Markley J.L. I-PINE web server: an integrative probabilistic NMR assignment system for proteins. J. Biomol. NMR. 2019;73:213–222. doi: 10.1007/S10858-019-00255-3/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewski M.W., Schuyler A.D., Gryk M.R., Moraru I.I., Romero P.R., Ulrich E.L., Eghbalnia H.R., Livny M., Delaglio F., Hoch J.C. NMRbox: a resource for biomolecular NMR computation. Biophys. J. 2017;112:1529–1534. doi: 10.1016/J.BPJ.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner S.P., Fogh R.H., Boucher W., Ragan T.J., Mureddu L.G., Vuister G.W. CcpNmr AnalysisAssign: a flexible platform for integrated NMR analysis. J. Biomol. NMR. 2016;66:111–124. doi: 10.1007/S10858-016-0060-Y/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mun S.R., Linares J.F., Ahn J., Towers C.G., Ji C.H., Fitzwalter B.E., Holden M.R., Mi W., Shi X., Moscat J., et al. ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat Commun. 2018;9:4373. doi: 10.1038/s41467-018-06878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Towers C.G., Singh U.K., Liu J., Håkansson M., Logan D.T., Donini O., Kutateladze T.G. Dusquetide modulates innate immune response through binding to p62. Structure. 2022;30:1055–1061.e7. doi: 10.1016/j.str.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziarek J.J., Peterson F.C., Lytle B.L., Volkman B.F. In: Methods in Enzymology. Kuo K.C., editor. Vol. 493. Academic Press; 2011. Binding site identification and structure determination of ProteinLigand complexes by NMR: a semiautomated approach; pp. 241–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No coordinates or structure factors were generated in this protocol.