Abstract
Nicotinamide riboside (NR) is an effective precursor of nicotinamide adenine dinucleotide (NAD) in human and animal cells. NR supplementation can increase the level of NAD in various tissues and thereby improve physiological functions that are weakened or lost in experimental models of aging or various human pathologies. However, there are also reports questioning the efficacy of NR supplementation. Indeed, the mechanisms of its utilization by cells are not fully understood. Herein, we investigated the role of purine nucleoside phosphorylase (PNP) in NR metabolism in mammalian cells. Using both PNP overexpression and genetic knockout, we show that after being imported into cells by members of the equilibrative nucleoside transporter family, NR is predominantly metabolized by PNP, resulting in nicotinamide (Nam) accumulation. Intracellular cleavage of NR to Nam is prevented by the potent PNP inhibitor Immucillin H in various types of mammalian cells. In turn, suppression of PNP activity potentiates NAD synthesis from NR. Combining pharmacological inhibition of PNP with NR supplementation in mice, we demonstrate that the cleavage of the riboside to Nam is strongly diminished, maintaining high levels of NR in blood, kidney, and liver. Moreover, we show that PNP inhibition stimulates Nam mononucleotide and NAD+ synthesis from NR in vivo, in particular, in the kidney. Thus, we establish PNP as a major regulator of NR metabolism in mammals and provide evidence that the health benefits of NR supplementation could be greatly enhanced by concomitant downregulation of PNP activity.
Keywords: nicotinamide adenine dinucleotide (NAD), NAD biosynthesis, nicotinamide riboside, metabolism, purine nucleoside phosphorylase, human, mouse
Abbreviations: ENTs, equilibrative nucleoside transporters; FBS, fetal bovine serum; NA, nicotinic acid; mESC, mouse embryonic stem cells; NAD, nicotinamide adenine dinucleotide; Nam, nicotinamide; NAMN, NA mononucleotide; NAMPT, Nam phosphoribosyltransferase; NAPRT, NA phosphoribosyltransferase; NBTI, S-(4-nitrobenzyl)-6-thioinosine; NMN, Nam mononucleotide; NMNAT, NMN adenylyltransferase; NR, nicotinamide riboside; NRH, reduced form of nicotinamide riboside; NRK, NR kinase; PNP, purine nucleoside phosphorylase; RBCs, red blood cells
Nicotinamide adenine dinucleotide (NAD) metabolism has emerged as a major driver and key regulator of vital cellular and organismal processes. Recent research has established that dysregulation of NAD-dependent metabolism and signaling is associated with aging-related pathologies such as neurodegeneration (1, 2, 3), cancer (4), metabolic (5, 6) and cardiovascular (7, 8) diseases. In fact, a number of independent studies have established that NAD levels significantly decrease in the aging process of rodents (6, 9, 10, 11) and humans (12, 13). The mechanisms of this decline are not understood and could be caused by increased NAD consumption through NAD-dependent signaling or diminished NAD biosynthesis. Lowered NAD levels are considered to affect both metabolic, bioenergetic, and signaling pathways, thereby aggravating pathological alterations in aging. Therefore, so-called NAD supplementation strategies are under development aimed at re-adjusting NAD levels to normal values (14, 15). These strategies exploit alternative entry routes or downstream metabolites of NAD biosynthesis.
The cellular functions of NAD(P) include their well-known roles as major coenzymes of metabolic redox reactions as well as serving as substrates of several families of regulatory proteins such as protein deacylases (sirtuins), ADP-ribosyltransferases, and PARPs, which govern vital processes including gene expression, DNA repair, apoptosis, mitochondrial biogenesis, unfolded protein response, and many others (16, 17, 18). NAD(P) is also used to generate calcium mobilizing second messengers such as cyclic ADP-ribose and NAADP (19). Owing to the multiple functions of NAD and its phosphorylated form, NADP, and their continuous degradation in signaling reactions, the synthesis of these pyridine nucleotides is regarded as an important process which needs a permanent supply of precursors.
Human cells regulate their NAD supply through biosynthesis using various precursors delivered with the diet. The main NAD precursors are nicotinamide (Nam) and nicotinic acid known as vitamin B3 (20, 21). Nam is converted by the phosphoribosyltransferase NAMPT to the mononucleotide NMN, which in turn is adenylated by the adenylyl transferase of the NMNAT family to form NAD (Fig. S1). Nicotinic acid is converted to the corresponding mononucleotide NAMN by the phosphoribosyltransferase NAPRT. NAMN is adenylated by NMNATs to form the dinucleotide NAAD which is then amidated to NAD by NAD synthetase, (Fig. S1) (20, 21).
As shown in Fig. S1, the nucleoside form of Nam, nicotinamide riboside (NR) can enter NAD biosynthesis in an NAMPT-independent step to form NMN through phosphorylation by NR kinase (NRK) activity (22, 23). Since NAMPT represents the rate-limiting step in human NAD biosynthesis (24), NR can serve as an effective precursor to enhance NAD levels. Indeed, dietary supplementation with NR or other NAD biosynthetic intermediates, such as NMN, can increase intracellular NAD contents and thereby facilitates restoration of physiological functions that are weakened or lost in experimental models of aging and various pathologies (14, 15). In a previous work, we established that the import of NR into human cells is mediated by equilibrative nucleoside transporters (ENTs) (25, 26). However, the molecular mechanisms of NR utilization by human and animal cells are still not fully understood. In particular, there are studies that showed no beneficial effects of NR supplementation (27, 28, 29), whereas clear health benefits were observed in others (3, 5, 30, 31, 32). We showed recently that after being imported into cells, NR is readily converted to Nam (26, 33). While Nam generation can, in principle, be explained by degradation of NAD in signaling processes (Fig. S1), a plausible alternative would be the direct conversion of NR to Nam by purine nucleoside phosphorylase, PNP. This enzyme catalyzes the reversible phosphorolytic cleavage of a variety of nucleosides to the corresponding base and ribose-1-phosphate (34). It has been reported that NR can serve as a substrate for PNP (35, 36). Therefore, we reasoned that PNP may have a critical role in NR utilization in human cells. Moreover, modulation of PNP activity could potentially explain the observed differences in the efficiency of NR supplementation to boost NAD levels and improve health.
In the present study, we investigated the role of PNP in mammalian NAD metabolism driven by NR supplementation. Our results demonstrate that the utilization of NR as NAD precursor is considerably diminished by PNP activity present in mammalian cells. Of note, human red blood cells (RBCs) readily convert NR to Nam through PNP activity, thereby “deactivating” NR as an alternative to Nam as NAD precursor. On the other hand, inhibition of PNP activity strongly enhances the NAD boosting effect of NR in human cells and mouse tissues. These results indicate that the health benefits of NR supplementation could be greatly enhanced by concomitant downregulation of PNP activity.
Results
NR is intensively metabolized in mammalian cells by intracellular conversion to Nam
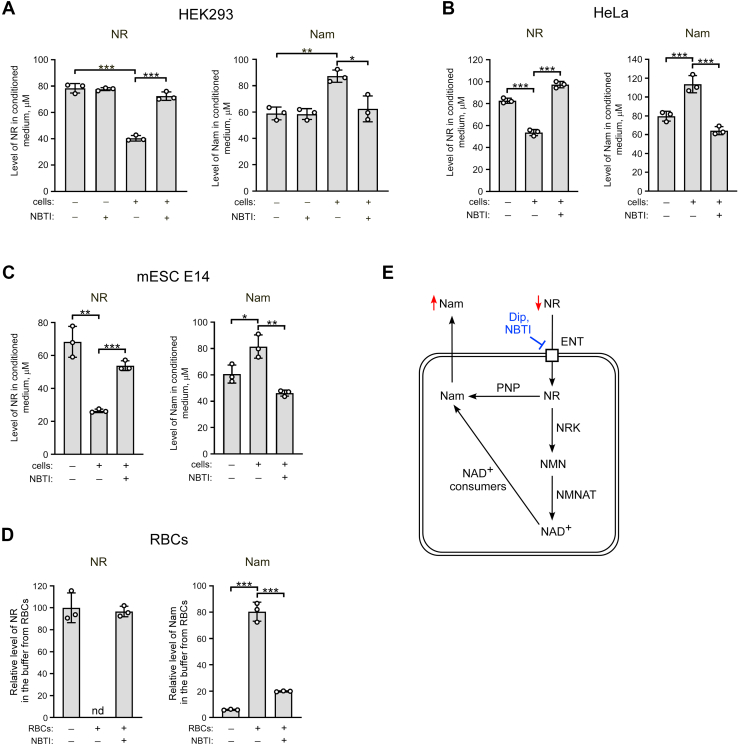
It was previously shown that members of the ENT family mediate the uptake of NR into human cells (25, 26). Moreover, in HEK293 cells, overexpression of ENT1 strongly enhanced the intracellular utilization of NR resulting in accumulation of Nam in the medium (26). Here, we first tested whether ENT-mediated uptake followed by intracellular conversion of NR to Nam is a general property of mammalian cells. To do this, we used an experimental approach developed previously, based on the quantitative assessment of NAD intermediates in conditioned culture medium using NMR spectroscopy (37). After incubation of cells with NR for 24 h at concentrations of 100 to 150 μM, we analyzed the culture medium for the contents of NR and Nam. As shown in Figures 1 and S2, ENT-specific inhibitors such as S-(4-nitrobenzyl)-6-thioinosine (NBTI) and dipyridamole strongly inhibited the disappearance of NR from the medium (left panels) and the concomitant accumulation of Nam (right panels) when incubated with mammalian cells including human HEK293, HeLa, A549 cells, and mouse embryonic stem cells (mESCs) E14 (Figs. 1, A–C and S2). The amount of NR in the medium dropped by ∼50 to 70% and was paralleled by a proportional increase of the Nam concentration. Since for NR supplementation the dynamics in circulating blood is of importance, we exposed isolated human RBCs (2 billion cells in 1 ml buffer) to NR at a final concentration 300 μM. After only 2 h of incubation, NR was undetectable in the buffer and converted into Nam (Fig. 1D). Again, inhibition of ENT-mediated uptake by NBTI nearly completely prevented the conversion of NR to Nam.
Figure 1.
NR is metabolized in mammalian cells predominantly by conversion to Nam. A–C, human HEK293, HeLa cells, or mouse embryonic stem cells (mESC) E14 were treated with nicotinamide riboside (NR) (150 μM, 150 μM, or 100 μM, respectively) in the presence or absence of inhibitor of equilibrative nucleoside transporters, S-(4-nitrobenzyl)-6-thioinosine (NBTI), as indicated. Twenty-four hours after the treatment, cell culture media were analyzed by quantitative NMR spectroscopy. Levels of NR and Nam in the medium are presented. D, 2 × 109 of isolated human red blood cells (RBCs) in 1 ml Hepes buffer were incubated with NR (300 μM) and NBTI for 2 h. The supernatant was then collected and analyzed by NMR as above. Relative levels of extracellular NR and Nam are presented. The concentration of NR in control buffer incubated without cells was taken as 100%. Data are presented as mean ± S.D. (n = 3). nd, not detected. Statistical analysis of differences between the groups was carried out by one-way ANOVA with post hoc comparisons using the Tukey test. ∗ indicates statistical significance at p < 0.05, ∗∗ indicates statistical significance at p < 0.01, ∗∗∗ indicates statistical significance at p < 0.001. E, schematic interpretation of the results shown in Panels A–D and possible mechanisms of NR to Nam conversion. Dip, dipyridamole; ENT, equilibrative nucleoside transporter; Nam, nicotinamide; NMN, Nam mononucleotide; NMNAT, NMN adenylyltransferase; NRK, NR kinase; PNP, purine nucleoside phosphorylase.
These observations confirm that mammalian cells import NR through ENTs followed by active conversion to Nam. Nam is then released from the cells into the culture medium. In principle, there are two plausible mechanisms of intracellular conversion of NR to Nam (Fig. 1E). First, NR is metabolized into NAD+, which is then converted to Nam by NAD+-consuming enzymes. Second, direct cleavage of NR to Nam by PNP (Fig. 1E). The massive conversion of NR to Nam in erythrocytes indicated a strong contribution of the latter mechanism, because these cells lack both a nucleus and mitochondria, the compartments in which most NAD+-degrading signaling processes take place in human cells. Therefore, we next wished to establish the role of PNP in NR metabolism in human cells.
Overexpression of PNP increases NR conversion to Nam in HEK293 cells
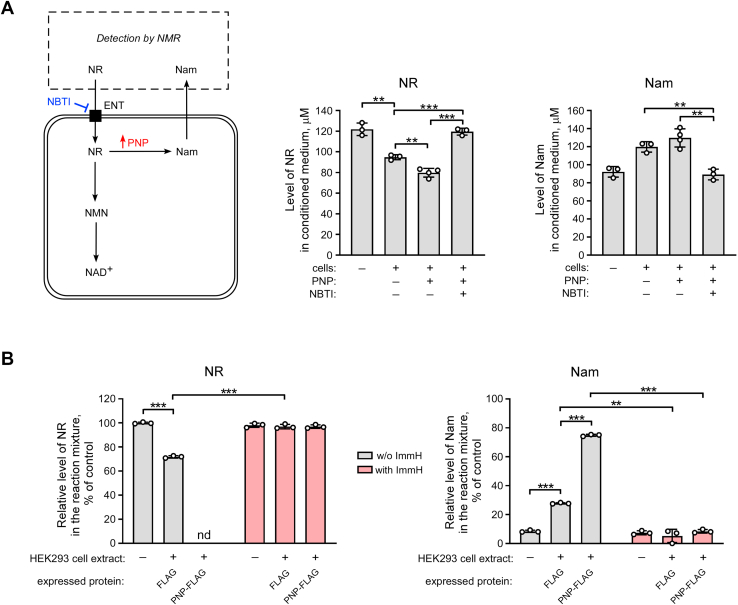
To explore the role of PNP, we first transiently overexpressed FLAG-tagged PNP in HEK293 cells and evaluated changes in the rate of utilization of extracellular NR. The level of overexpression of the FLAG-fusion protein was assessed by immunoblotting using antibodies recognizing the FLAG epitope (Fig. S3A). Twenty-four hours after transfection of the FLAG-PNP-encoding vector, NR was added to the culture medium at a concentration of 200 μM. After another 24 h, the conditioned culture medium was analyzed by NMR spectroscopy (Fig. 2A, left panel). Overexpression of PNP-FLAG led to a moderate increase of NR conversion to Nam, whereas NBTI efficiently blocked NR consumption and Nam accumulation in the medium. A similar effect was observed under conditions when NR uptake into HEK293 cells was stimulated by ENT4 overexpression (26). When ENT4 and PNP were co-expressed, the NR level in the medium dropped by 50% compared to the decrease obtained from cells expressing ENT4 alone (Fig. S3B), indicating that both NR uptake and PNP control the intracellular conversion of NR to Nam.
Figure 2.
Overexpression of purine nucleoside phosphorylase (PNP) increases NR conversion to Nam in HEK293 cells. A, HEK293 cells were treated with 200 μM NR and NBTI, as indicated. Twenty-four hours before the treatment, cells were transiently transfected with a vector encoding FLAG-tagged PNP or the FLAG peptide only (control). Twenty-four hours after the treatment, culture media were analyzed by NMR spectroscopy. Levels of NR and Nam in culture medium are presented. Left panel, schematic representation of the experimental approach. B, 24 h after the transfection of НЕК293 cells with vectors encoding FLAG-tagged PNP or the FLAG peptide, cell extracts were incubated with NR (1 mM) and an inhibitor of purine nucleoside phosphorylase, Immucillin H (ImmH), for 40 min. NR cleavage and the formation of Nam in these reactions were detected by NMR as above. Relative levels of NR and Nam in the reaction mixture are presented. The concentration of NR in control reaction mixture incubated without cell extract was taken as 100%. Data are presented as mean ± S.D. (n = 3–4). nd, not detected. Statistical analysis of differences between the groups was carried out by one-way ANOVA with post hoc comparisons using the Tukey test. ∗∗ indicates statistical significance at p < 0.01, ∗∗∗ indicates statistical significance at p < 0.001. ENT, equilibrative nucleoside transporter; Nam, nicotinamide; NBTI, S-(4-nitrobenzyl)-6-thioinosine; NMN, Nam mononucleotide; NR, nicotinamide riboside.
To further analyze the role of PNP, we characterized the efficiency of NR cleavage in cell extracts obtained from control cells or from cells overexpressing the PNP-FLAG protein. Twenty-four hours after transfection of НЕК293 cells with the vector encoding FLAG-tagged PNP or the FLAG peptide (control), cell extracts were incubated with 1 mM NR for 40 min at 37 °C and then analyzed by NMR. After incubation with extracts derived from cells overexpressing FLAG, the amount of NR in the reaction dropped to ∼70% (Fig. 2B, left panel). At the same time, the Nam concentration increased proportionally (Fig. 2B, right panel). In cell extracts derived from HEK293 cells overexpressing FLAG-PNP, NR cleaving activity was dramatically increased. In fact, after the incubation, NR was undetectable and converted to Nam (Fig. 2B). Notably, neither NR depletion nor Nam formation was observed when the potent pharmacological inhibitor of human PNP, Immucillin H (38), was added to the reaction (Fig. 2B). Consequently, under these conditions, PNP accounted for most, if not all activity converting NR to Nam.
Pharmacological inhibition of PNP blocks NR conversion to Nam in various types of mammalian cells
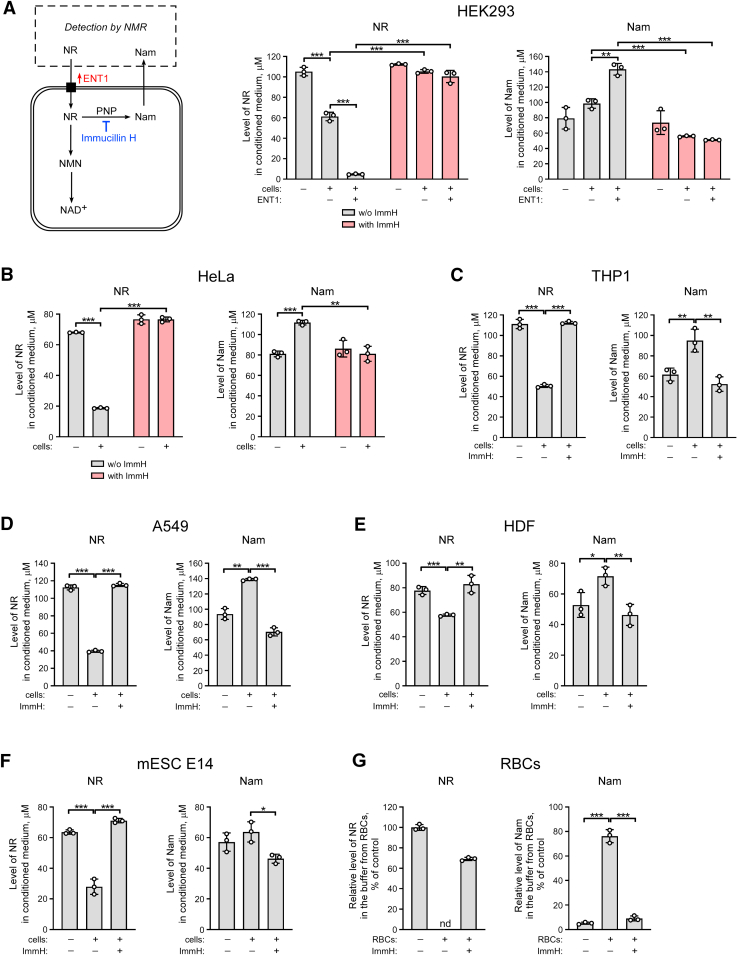
To test whether suppression of PNP activity with Immucillin H can modulate NR conversion to Nam in cultured cells, we used an experimental approach which is schematically presented in Figure 3A, left panel. As shown in Figure 3, A–G, incubation of various human cell lines, including HEK293, HeLa, THP1, A549, dermal fibroblasts (HDF),and RBCs as well as mESCs, with high concentrations (100–150 μM) of NR led to considerable conversion of the nucleoside to Nam. This conversion was nearly completely prevented when the PNP inhibitor Immucillin H was present during the incubation. Using Western blot analysis, we verified that PNP is expressed in all cell lines tested. Moreover, the treatment with NR did not change PNP expression (Fig. S4A).
Figure 3.
Pharmacological inhibition of PNP blocks NR conversion to Nam in various types of mammalian cells. A–F, HEK293, HeLa, THP1, A549 cells, or HDF (human dermal fibroblasts) were treated with 150 μM NR and Immucillin H (ImmH). mESC E14 cells were treated with 100 μM NR and ImmH. A, 24 h before the treatment, HEK293 cells were transiently transfected with a vector encoding FLAG-tagged ENT1. Left panel, schematic representation of the experimental approach. Twenty-four hours after the treatment, culture media were analyzed by NMR spectroscopy. Levels of NR and Nam in culture medium are presented. G, 2 × 109 of isolated human red blood cells (RBCs) in 1 ml Hepes buffer were incubated with NR (300 μM) and ImmH for 2 h. The supernatant was then collected and analyzed by NMR as above. Relative levels of extracellular NR and Nam are presented. The concentration of NR in control buffer incubated without cells was taken as 100%. Data are presented as mean ± S.D. (n = 3). nd, not detected. Statistical analysis of differences between the groups was carried out by one-way ANOVA with post hoc comparisons using the Tukey test. ∗ indicates statistical significance at p < 0.05, ∗∗ indicates statistical significance at p < 0.01, ∗∗∗ indicates statistical significance at p < 0.001. ENT, equilibrative nucleoside transporter; Nam, nicotinamide; NBTI, S-(4-nitrobenzyl)-6-thioinosine; NMN, Nam mononucleotide; NR, nicotinamide riboside; PNP, purine nucleoside phosphorylase.
When FLAG-tagged ENT1 was overexpressed in HEK293 cells, the amount of NR in the medium dramatically decreased compared to samples obtained from mock-transfected cells (Fig. 3A, middle panel), which was accompanied by a proportional accumulation of Nam (Fig. 3A, right panel). Addition of Immucillin H still potently inhibited the degradation of NR and accumulation of Nam. These results demonstrate that PNP significantly contributes to NR metabolism in various types of mammalian cells by mediating the cleavage to Nam.
PNP knockout HEK293 cells lack NR cleaving activity
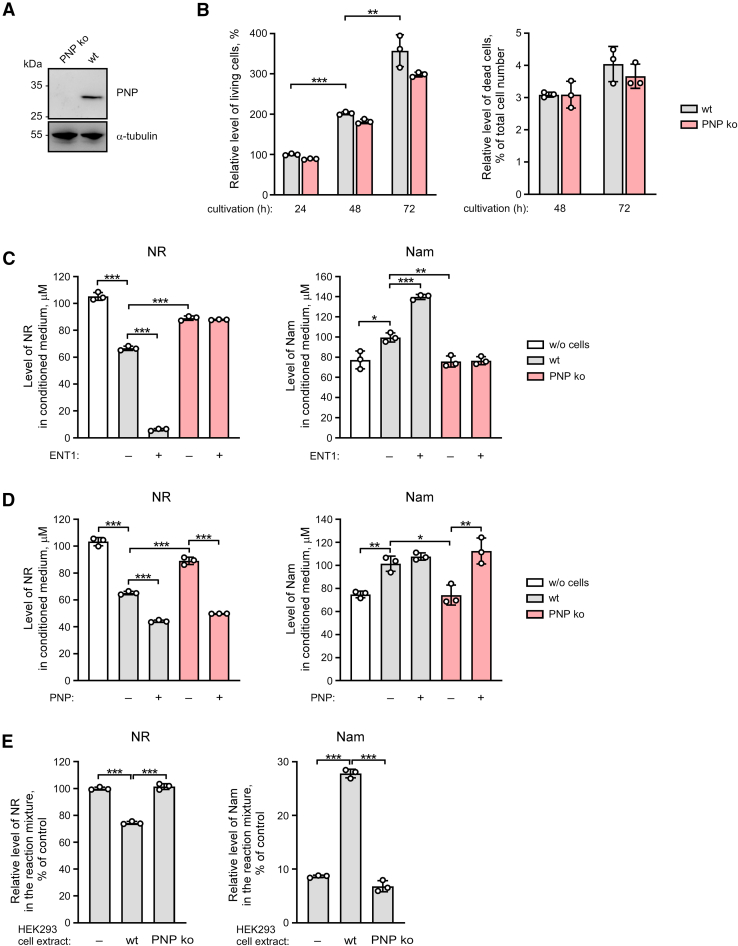
To further substantiate the role of PNP in NR metabolism, we exploited the CRISPR-Cas9 system to generate PNP knockout HEK293 cells. As expected, expression of the endogenous PNP protein was detected by immunoblotting only in wildtype, but not in PNP knockout HEK293 cells (Fig. 4A). The absence of PNP did not noticeably affect cell viability or proliferation as established by propidium iodide staining and FACS analysis (Fig. 4B).
Figure 4.
PNP knockout HEK293 cells lack NR cleaving activity. A purine nucleoside phosphorylase knockout (ko) HEK293 cell line was generated using the CRISPR-Cas9 system. A, expression of the PNP protein was detected in wildtype (wt) but not in PNP ko HEK293 cells as revealed by immunoblotting using an antibody to PNP. B, knocking out of PNP did not noticeably affect cell proliferation and did not lead to increased cell death, as established by flow cytometry. C and D, wt or PNP ko HEK293 cells were treated with NR (150 μM). Twenty-four hours before the treatment with NR, cells were transiently transfected with a vector encoding FLAG-tagged ENT1 (panel C) or FLAG-tagged PNP (panel D). Twenty-four hours after the treatment, culture media were analyzed by NMR spectroscopy. Levels of NR and Nam in culture medium are presented. E, HEK293 cell extracts derived from wt or PNP ko cells were incubated with NR (1 mM) for 40 min. NR cleavage and the formation of Nam in these reactions were detected by NMR as above. Relative levels of NR and Nam in the reaction mixture are presented. The concentration of NR in control reaction mixture incubated without cell extract was taken as 100%. Data are presented as mean ± S.D. (n = 3). Statistical analysis of differences between the groups was carried out by one-way ANOVA with post hoc comparisons using the Tukey test. ∗ indicates statistical significance at p < 0.05, ∗∗ indicates statistical significance at p < 0.01, ∗∗∗ indicates statistical significance at p < 0.001. ENT, equilibrative nucleoside transporter; Nam, nicotinamide; NR, nicotinamide riboside; PNP, purine nucleoside phosphorylase.
Next, we incubated wildtype or PNP knockout cells with NR (150 μM) for 24 h. Genetic suppression of PNP resembled the effect of pharmacological inhibition by Immucillin H. Neither a drop in the NR concentration nor an increase in the Nam level was observed in the culture medium obtained from PNP knockout cells. Even after stimulation of NR uptake by overexpression of ENT1, the absence of PNP virtually precluded the conversion of NR to Nam (Figs. 4C and S5A). Importantly, overexpression of ectopic PNP in knockout cells restored the conversion of NR to Nam (Figs. 4D and S5B). These data validate that PNP is required for NR cleavage to Nam in НЕК293 cells. Another argument supporting this conclusion is that no cleavage of NR was observed in in vitro reaction in the presence of cell extract derived from PNP-knockout HEK293 cells (Fig. 4E).
Pharmacological or genetic suppression of PNP potentiates NAD synthesis from NR in human cells
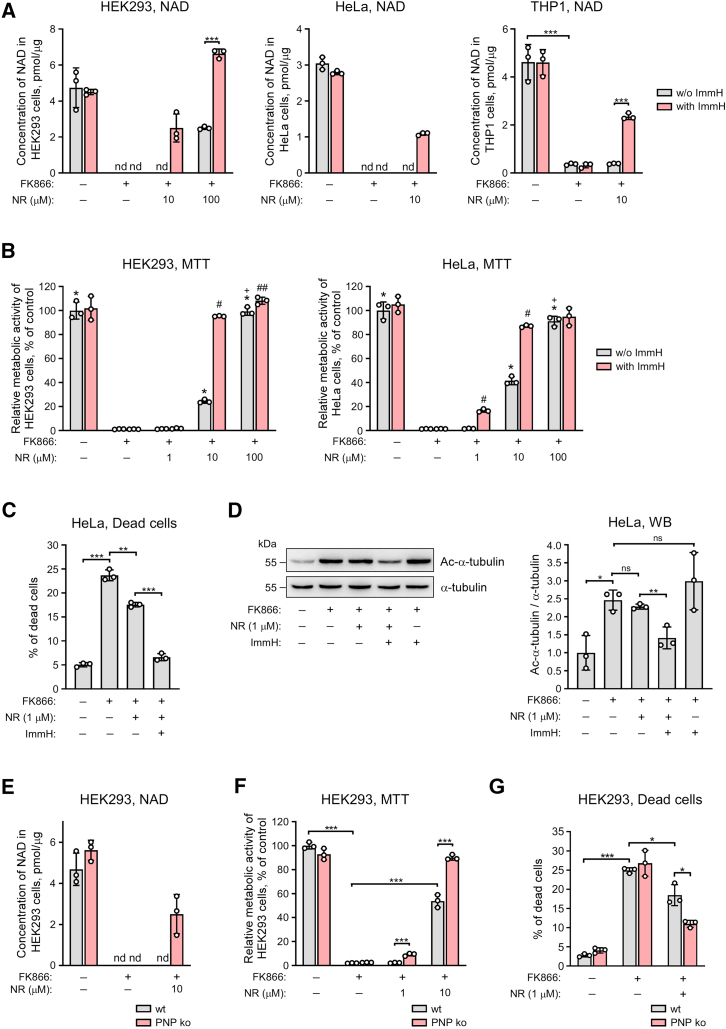
We reasoned that the highly active cleavage of NR by PNP might affect the utilization of the nucleoside for NAD synthesis. Therefore, we tested whether PNP suppression could improve the efficiency of NR as precursor for NAD biosynthesis. First of all, using NMR spectroscopy, we analyzed changes in NAD and its intermediates in HEK293 cell extracts prepared 24 h after treatment of cells with 150 μM NR. Following incubation with NR, the level of intracellular NR was below the limit of detection, as in control cells. Nevertheless, we observed a significant increase in the NAD concentration. As expected, cotreatment with Immucillin H led to considerable accumulation of intracellular NR; however, the NAD level remained the same as in cells treated with NR alone Fig. S6A). Similar effects were observed when comparing wildtype and PNP knockout HEK293 cells (Fig. S6B). The failure of Immucillin H to further increase NAD accumulation may possibly reflect the activation of a safety mechanism to prevent excessive NAD accumulation. In addition, the increase of the NAD concentration will kinetically activate NAD consuming enzymes. That is, to maintain higher NAD levels, biosynthesis needs also to counterbalance the accelerated consumption.
To evaluate the utility of NR supplementation and PNP suppression at lowered NAD levels, we inhibited NAD synthesis from Nam by inhibiting NAMPT (Fig. S1). Cells were treated with NAMPT inhibitor, FK866, which causes a rapid drop in NAD levels and cell death (39). NR was added to the medium at a concentration of 1, 10, or 100 μM. To inhibit PNP, cells were additionally treated with Immucillin H. Twenty-four hours after the treatment, we estimated the concentration of intracellular NAD using NMR spectroscopy. Treatment with FK866 depleted NAD to undetectable levels in HEK293, HeLa, and THP1 cells (Fig. 5A). NAD in extracts obtained from cells additionally treated with 10 μM NR was still below the detection limit, while cotreatment with Immucillin H was able to recover NAD to up to 50% of the control level. Consequently, PNP inhibition considerably improved the ability of cells to synthesize NAD from NR. When NR was added to HEK293 cells at a concentration of 100 μM, the cellular NAD concentration could be maintained at normal levels, even in the presence of FK866, but only when PNP was inhibited (Fig. 5A, left panel). We additionally assessed the NAD(P)H-dependent cell metabolic activity using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide test (Fig. 5B). FK866 led to a significant decrease in the metabolic activity of HEK293 and HeLa cells. NR, at a concentration of 100 μM, completely restored metabolic activity of the cells to control level. However, NR at a concentration of 10 μM was less effective and restored metabolic activity only up to 30 to 40%. Cotreatment with NR (10 μM) and Immucillin H led to complete recovery of the metabolic activity of NAD-depleted cells (Fig. 5B).
Figure 5.
Pharmacological or genetic suppression of PNP potentiates the NAD synthesis from NR in human cells. A–D, HEK293, HeLa or THP1 cells were treated with NR and the inhibitor of PNP, Immucillin H (ImmH) as indicated. To inhibit NAD synthesis from Nam, cells were treated with FK866. E and F, wildtype (wt) or PNP knockout (ko) HEK293 cells were treated with NR and FK866, as indicated. Twenty-four hours after the treatment, intracellular NAD levels were measured by NMR spectroscopy (A and E), and relative metabolic activity was assessed using the MTT-assay (B and F). Forty-eight hours after the treatment the fraction of dead cells was determined by flow cytometry (C and G). Twenty-four hours after the treatment, the extent of α-tubulin (K40) acetylation was estimated by immunoblotting (D); right panel, densitometric quantification of immunoblots as in left panel. Data are presented as mean ± S.D. (n = 3). nd, not detected. ns, not significant. Statistical analysis of differences between the groups was carried out by one-way ANOVA with post hoc comparisons using the Tukey test. Panels A, and C–G, ∗ indicates statistical significance at p < 0.05, ∗∗ indicates statistical significance at p < 0.01, ∗∗∗ indicates statistical significance at p < 0.001. Panel B, ∗ indicates statistical difference at p < 0.001 versus the FK866-treated cells, # indicates statistical difference at p < 0.001 and ## - statistical difference at p < 0.05 versus the treatment with the corresponding concentration of NR, but without Immucillin H, + indicates statistical difference at p < 0.001 versus the treatment with 10 μM NR, but without Immucillin H. MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; NR, nicotinamide riboside; PNP, purine nucleoside phosphorylase.
Next, we analyzed how pharmacological inhibition of PNP may affect the viability of HeLa cell in the presence of FK866 and NR using flow cytometry. Following treatment with FK866 for 48 h, the number of dead cells, as evaluated by staining with propidium iodide, increased significantly in comparison with the control untreated cells (Fig. 5C). Cotreatment with 1 μM NR led to a partial recovery of cell viability. Importantly, when cells were treated with both NR and Immucillin H, cell viability increased almost up to control level observed in population of cells grown in standard medium (Fig. 5C).
We also used these experimental conditions to characterize the possible functional consequences of the suppression of PNP-dependent cleavage of NR to Nam. As shown in Figure 5D, the treatment of HeLa cells with FK866 for 24 h led to the accumulation of acetylated α-tubulin K40—the target of NAD+-dependent cytoplasmic protein deacetylase SIRT2 (40, 41). Cotreatment with 1 μM NR had no effect on the acetylation status of this lysine; however, when Immucillin H was simultaneously added to cells, we observed a significant decrease in the level of acetylated α-tubulin K40 (Fig. D) indicating that, under this condition, more cytosolic NAD+ is available.
When using PNP knockout cells under similar experimental conditions, NR at a concentration of 10 μM was sufficient to maintain NAD synthesis in PNP knockout cells in the presence of FK866 (Fig. 5E). Likewise, knocking out the PNP gene increased NAD(P)H-dependent metabolic activity in cells treated with FK866 and NR (Fig. 5F). Moreover, NR supplementation counteracted FK866-induced cell death more efficiently in PNP knockout in comparison to wild type HEK293 cells (Fig. 5G).
Collectively, these results demonstrated that the utilization of NR for NAD biosynthesis in mammalian cells is severely affected by the presence of PNP activity. Suppression of this activity strongly increases the conversion of NR into NAD which, in turn, supports vital processes under conditions of NAD deficiency.
PNP inhibition suppresses NR cleavage to Nam and potentiates NAD+ synthesis from NR in mice
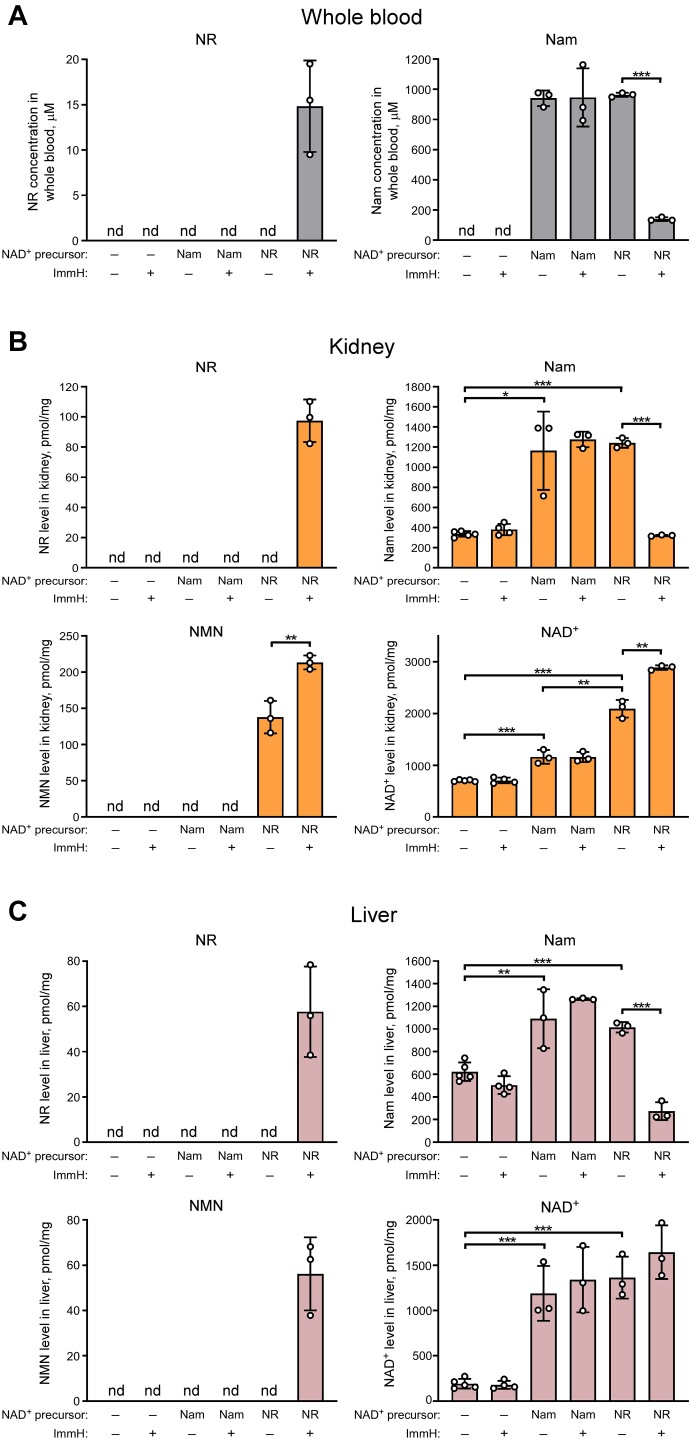
Given the strong effect of PNP activity on NR metabolism in cultured cells, we next asked the question whether a similar interrelationship between NR cleavage and utilization for NAD synthesis is also relevant in vivo. First, we tested whether PNP-dependent cleavage of NR to Nam occurs in vivo by intraperitoneally injecting six groups (n = 3–5) of young male C57BL/6J mice with phosphate-buffered saline (PBS), NR (at a dose of 400 mg/kg) or Nam (at equimolar dose) with or without Immucillin H. Two hours after injection, whole blood, kidney, and liver were collected and analyzed by NMR spectroscopy. Surprisingly, we did not detect any traces of NR in whole blood, kidney, and liver when mice were injected with NR only. Instead, we observed high concentrations of Nam in these samples, which were comparable to Nam levels in whole blood, kidney, and liver detected after injection of Nam itself (Fig. 6, A–С, upper panels). Conversely, cotreatment of mice with Immucillin H led to accumulation of NR in blood, kidney, and liver, while Nam accumulation was strongly diminished. At the same time, co-injection with Immucillin H had no effect on the level of Nam in whole blood, kidney, and liver after the treatment of mice with Nam itself. We also observed considerable accumulation of NMN in the kidney when mice were injected with NR but not after Nam injection. Accordingly, the treatment with NR increased the NAD+ levels in the kidney far more strongly than the treatment with Nam (Fig. 6B, lower panels). When mice were injected with NR under conditions of PNP inhibition, NR accumulation in the kidney resulted in the stimulation of NMN and NAD+ synthesis. The concentrations of NMN and NAD+ in the kidney increased by 40% and 60%, respectively, compared to samples obtained from animals treated with NR only. Importantly, Immucillin H did not affect the levels of these nucleotides in control or Nam-treated mice (Fig. 6B, lower panels). These results demonstrated that PNP inhibition suppresses NR cleavage to Nam and stimulates NAD+ synthesis from NR in vivo.
Figure 6.
PNP inhibition suppresses NR cleavage to Nam and potentiates NAD+synthesis from NR in mice. A–C, C57BL/6 mice were intraperitoneally injected with PBS, NR (400 mg/kg) or Nam (molar equivalent) and Immucillin H (2.5 mg/kg) as indicated. Two hours after injection, whole blood, kidney, and liver were collected and analyzed by NMR spectroscopy. Concentrations of NAD+ intermediates are presented as mean ± S.D. (n = 3–5 in each group). nd, not detected. Statistical analysis of differences between the groups was carried out by one-way ANOVA with post hoc comparisons using the Tukey test. ∗ indicates statistical significance at p < 0.05, ∗∗ indicates statistical significance at p < 0.01, ∗∗∗ indicates statistical significance at p < 0.001. NR, nicotinamide riboside; Nam, nicotinamide; NAD, nicotinamide adenine dinucleotide; NMN, Nam mononucleotide; PNP, purine nucleoside phosphorylase.
Interestingly, unlike in the kidney, treatment with NR did not lead to the accumulation of NMN in the liver. In addition, the extent to which NAD+ levels were increased in this organ was similar when using Nam or NR (Fig. 6C, lower panels). These observations can probably be explained, at least in part, by higher expression of PNP in liver compared to kidney (Fig. S4B. Indeed, when PNP was inhibited by Immucillin H, NR treatment also resulted in the accumulation of NMN in the liver.
Discussion
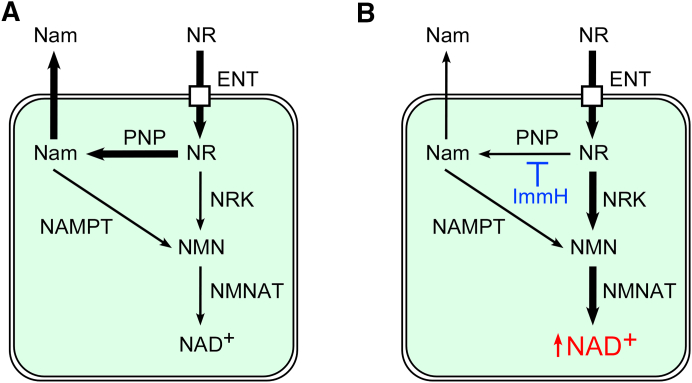
The results from the present study support two major conclusions. First, PNP has emerged as a key contributor to NR metabolism whose role has so far been largely underestimated. Once NR has entered the cells, the enzyme cleaves the nucleoside to generate Nam (Fig. 7A). Thereby, much of the NR meant to increase NAD synthesis is diverted. In view of the considerably elevated NAD concentration in the liver of mice following Nam treatment, it appears plausible that some of the positive organismal effects ascribed to NR are mediated by the accumulating Nam, which could stimulate NAD synthesis through NAMPT activity. This mechanism may be particularly relevant in circulating blood, because RBCs exhibited a high PNP activity. Indeed, treatment of Nrk1-deficient mice with NR still led to considerable elevation of tissue NAD levels, albeit to somewhat lesser extent than in wildtype animals (42). The second major conclusion relates to the possibility of downregulation of PNP to boost NAD synthesis from NR (Fig. 7B). Both genetic and pharmacological suppression of PNP activity resulted in re-directing NR utilization toward NAD generation through NRK activity. Moreover, inhibition of PNP by Immucillin H maintained high concentrations of NR in blood, kidney, and liver of mice (Fig. 6). Consequently, the NAD boosting effect of dietary NR supplementation could be synergistically potentiated by concomitant inhibition of PNP.
Figure 7.
PNP controls NR metabolism in mammals. A, after being imported into cells by members of the ENT family, NR is intensively metabolized by PNP-dependent cleavage to Nam which is then released from cells. B, suppression of PNP activity by Immucillin H (ImmH) blocks NR conversion to Nam and thereby redirects NR toward NAD+ synthesis via NRK and NMNAT activities. ENTs, equilibrative nucleoside transporters; Nam, nicotinamide; NAMPT, Nam phosphoribosyltransferase; NMN, Nam mononucleotide; NMNAT, NMN adenylyltransferase; NR, nicotinamide riboside; NRK, NR kinase; PNP, purine nucleoside phosphorylase.
Providing several lines of evidence, we have established that PNP represents the by far strongest activity cleaving NR in mammalian cells. Overexpression of cytosolic PNP increased, whereas PNP inhibition by Immucillin H blocked NR conversion to Nam in various types of mammalian cells. Moreover, we observed no NR cleaving activity in PNP knockout HEK293 cells. This activity was restored by PNP overexpression. Finally, we established that when NAD synthesis from Nam is diminished pharmacological or genetic suppression of PNP potentiates the NAD synthesis from NR. In turn, this leads to an increase in metabolic activity and cell survival as well as stimulates deacetylation of α-tubulin at lysine 40—the target of NAD-dependent cytoplasmic protein deacetylase SIRT2. Taken together, these results indicate that PNP controls NR metabolism in mammals.
Phosphorolytic cleavage of NR to Nam was already observed more than half a century ago in extracts from mammalian RBCs and liver (43, 44). However, the biochemical characterization of the purified bovine enzyme was conducted more recently, and it suggested that NR represents a less preferred substrate compared to other nucleosides such as inosine (35, 36). PNP activity toward NR has been established in yeast, where it also represents an alternative mechanism to the utilization for NAD synthesis through Nrk1 (36, 45). Moreover, human PNP can functionally substitute for its homolog in yeast (Pnp1) to mediate NAD synthesis from Nam generated by NR cleavage (36).
Our data are consistent with the observations of other researchers, according to which intraperitoneal injections of NR into mice at different doses (from 50 to 500 mg/kg) caused a significant accumulation of Nam in plasma and liver, while no significant accumulation of NR was observed (42, 46, 47). Similarly, when mice were intravenously injected with NR isotopically labeled on both the nicotinamide and ribose moieties, NR was readily degraded. Even though a fraction of the uncleaved nucleoside was delivered to liver, kidney, and muscle, as deduced from the presence of both labels in NAD, the main circulating product of the administered NR was Nam (48). According to our observations, the high activity of PNP in RBCs may be a major contributor to this effect.
Recent studies have discovered a new pathway for the synthesis of NAD from the reduced form of NR, NRH, which requires adenosine kinase activity (47, 49). In various cell and mouse models, it was shown that NRH is a more efficient precursor of NAD compared to NR (47, 50). NRH is far more stable than NR after administration to animals (47). Presumably, the higher efficiency of NRH as an NAD booster can be explained by the different configuration of the chemical bond between the pyridine base and the ribose in the reduced form, NRH, which is unlikely to be cleaved by PNP (51).
In this study, we found that intraperitoneal administration of Nam and NR to mice evokes somewhat different changes in the NAD metabolome of the kidney and the liver. NR treatment led to accumulation of NMN and significantly increased the NAD+ levels in the kidney, although NR itself was undetectable. In contrast, when mice were injected with Nam, no NMN was detected and only a moderate increase of NAD+ was observed in the kidney. Notably, administration of either Nam or NR led to a comparable accumulation of Nam in this organ. These observations support the conclusion that following intraperitoneal administration, part of NR is effectively delivered to the kidney, despite extensive cleavage to Nam in the blood. Moreover, it would appear that in the kidney, NAD+ is synthesized more efficiently from NR than from Nam. In line with this suggestion, inhibition of PNP-dependent cleavage of NR by Immucillin H potentiates NR-induced accumulation of NMN and, in turn, NAD+ in the kidney. In the liver, administration of Nam or NR resulted in a comparable increase in the level of NAD+, while the amount of NMN remained at levels below the limit of detection. Cotreatment of mice with NR and Immucillin H led to NR accumulation in the liver, which stimulated the synthesis of NMN. It is noteworthy that the significant elevation of the NMN concentration resulted in only a small increase in NAD+ levels. This observation could point to a regulatory mechanism that limits liver NAD+ levels that can be produced from NMN. The possibility of the existence of such a mechanism in mammals is also indirectly evidenced by the results obtained on NR-treated HEK293 cells, showing that a significant accumulation of intracellular NR after PNP suppression does not lead to a further increase in NAD+ accumulation compared to cells treated with NR only (Fig. S6). Possibly, excessive NAD+ accumulation might be prevented by activation of NAD+ consuming enzymes such as PARPs, sirtuins, or CD38/SARM1 that cleave NAD+ into Nam and ADP-ribose. Alternatively, intracellular NAD+ levels can be regulated by members of the NUDIX hydrolase family, which cleave NAD+ to NMN and AMP.
Collectively, the present study has revealed PNP as a major component of NR metabolism in humans and animals. We speculate that the partially contradicting observations regarding health benefits upon NR supplementation might be due to individual variations in PNP activity in the model systems or human subjects. The possibility of pharmacological PNP inhibition could potentially be exploited to effectively boost NAD synthesis and thereby the beneficial effects of NR supplementation. However, we suppose that health benefits from PNP suppression leading to NR accumulation would be cell type specific and would depend not only on the level of PNP expression but also on the activity of NAMPT and NRK, generating NMN from Nam and NR, respectively, in each target organ.
Experimental procedures
Materials
Unless otherwise specified, all chemicals and reagents were of analytical grade and were purchased from Sigma and Amresco. Cell culture reagents were from Gibco, Greiner Bio-One, and Orange Scientific. HPLC-grade methanol and acetonitrile were obtained from Merck. The ultrapure water was obtained from a Milli-Q Synthesis purification system (Millipore). NR was synthesized as reported previously (52). DNA-modifying and restriction enzymes were purchased from Thermo Fisher Scientific. The following antibodies were used: rabbit anti-PNP (Sigma), mouse anti-FLAG (Sigma), mouse anti-β-tubulin (Sigma), rabbit anti-α-tubulin (Abcam), mouse anti-α-acetylated (K40) tubulin (Sigma), HRP-conjugated rabbit anti-mouse (Sigma), and HRP-conjugated goat anti-rabbit (Sigma). Enhanced chemiluminescence (ECL) reagents were from GE Healthcare.
Cell culture
Human cell lines HEK293, HeLa, A549, and THP1 were obtained from the shared research facility “Vertebrate cell culture collection” (Institute of Cytology of the Russian Academy of Sciences). HEK293, HeLa, and A549 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine. THP1 cells were cultured in RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine. Human dermal fibroblasts (purchased from Pokrovsky Stem Cell Bank) were cultured in MEM supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM glutamine. mESCs E14 Tg2α (obtained from Bay Genomics) were cultured in knockout Dulbecco's modified Eagle's medium (Gibco) supplemented with 15% HyClone FBS (Cytiva), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM glutamine, 100 mM 2-mercaptoethanol, minimum essential medium non-essential amino acids (Gibco), and leukemia inhibitory factor (Millipore). The cells were cultured at 37 °C in a humidified atmosphere of 5% CO2. NR, S-(4-nitrobenzyl)-6-thioinosine (10 μM), dipyridamole (2 μM), Immucillin H (5 μM), and FK866 (2 μM) were added to the culture medium as indicated. Transient transfection of cells was performed using Effectene reagent (Qiagen) or the calcium phosphate precipitation method. Metabolic activity was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
RBCs isolation from the human whole blood
Human blood was collected from healthy volunteers in S-monovette tubes (9NC, Sarstedt) with the addition of 2 mM EGTA. RBCs were prepared by centrifugation of whole blood at 400g in Hepes buffer with EGTA for 3 min at room temperature. Washed RBCs were resuspended in Hepes buffer and adjusted to 2 × 109 cells/ml (corresponding to Hematocrit 4.0–4.5%). The main blood parameters (RBC count and mean cell volume) were controlled by the hematological counter Medonic-M20 (Boule Medical A.B.). Blood was collected from healthy volunteers after obtaining informed consent and according to the Declaration of Helsinki. Our studies with human blood were approved and reconfirmed (Protocol #2–02; 26.02.2021) by the local Ethical Committee of the Sechenov Institute of Evolutionary Physiology and Biochemistry.
Generation of eukaryotic expression vector
For transient expression of FLAG-tagged PNP in human cells, its corresponding open reading frame was amplified from HEK293 cDNA and inserted into the pFLAG-CMV-4 plasmid (Sigma). Cloned DNA fragment was verified by DNA sequencing.
CRISPR-CAS9
The sequence of the variable part of the RNA guide (GTGGGTACCCTTCATACATG) was developed using the Benchling online resource (https://benchling.com/). Oligonucleotides encoding this sequence were cloned into the pX330-GFP plasmid, which also encodes Cas9 and GFP, under the U6 promoter for RNA polymerase III. HEK293 cells were transiently transfected with these plasmids using Lipofectamine-2000. GFP-labeled cells were selected using FACS and subcloned. Genomic DNA was isolated from the clones, and the corresponding fragments of the PNP gene were amplified and sequenced. RNA was isolated from 2 HEK293-PNP ko clones, with confirmed deletions in genomic DNA, and the corresponding PNP gene fragments were amplified, cloned into an AT vector, and sequenced. The lack of PNP gene expression in these clones was also confirmed by Western blotting.
Western blotting
Cells were washed with PBS and lysed in 50 mM Tris/HCl, pH 6.8, 4% (w/v) SDS, 5M urea, 10% (v/v) glycerol, 5% β-mercaptoethanol, and 0.01% (w/v) bromophenol blue for 30 min at 37 °C. Gel electrophoresis and immunoblotting were carried out according to standard procedures. ECL was used for immunodetection. Pictures were taken using the ChemiDoc Imaging System (Bio-Rad). Densitometric quantification of bands of acetylated α-tubulin at lysine 40 and total α-tubulin was performed using Image Lab program (Version 6.0.0 build 25, Bio-Rad). For each experimental point, three independently prepared cell extracts were analyzed. Acetylated (K40) α-tubulin band intensities were normalized to intensities of the corresponding total α-tubulin bands.
Metabolite measurements
For metabolite extraction, HEK293, HeLa, or THP1 cells (2 × 107) grown on 100 mm cell culture plates were washed twice with ice-cold PBS and put on ice. Following addition of 80% methanol, the cells were kept on ice for 30 min. Thereafter, the cells were scraped off and centrifuged at 150,00g for 30 min at 4 °C. The obtained pellets were used for protein determination using BCA Protein Assay kit (Thermo Fisher Scientific). Cell extracts were lyophilized and then resuspended in DBP buffer, D2O-based buffer containing 50 mM NaPi (pH 6.5), and 1 mM sucrose as a chemical shift reference (d(1H), 5.42 ppm) and internal standard for quantification. To remove oxygen, the samples were kept under vacuum (80 mm Hg) for 10 min with occasional agitation. Samples were stored at −80 °C until NMR analysis. Culture medium was collected and stored at −80 °C. To precipitate proteins, the samples were incubated on ice with 2 volumes of acetonitrile for 30 min and then centrifuged at 150,00g for 30 min at 4 °C. Supernatants were then treated in the same way as the cell extracts. All NMR experiments were performed using a Varian DirectDrive NMR System 700-MHz spectrometer equipped with a 5-mm z-gradient salt-tolerant as described in (37). Briefly, the one-pulse sequence with the suppression of solvent signal by presaturation was used for acquisition of 1H spectra. The following acquisition parameters were used: relaxation delay, 2.0 s; acquisition time, 3.0 s; and number of scans, 256 to 1536. Data were acquired using VNMRJ 4.2 (Agilent Technologies) and then analyzed by Mestrelab Mnova (version 12; Mestrelab). The concentrations of NAD+, NR, and Nam were determined by integration of the corresponding nonoverlapping proton signals with the following chemical shifts: 9.34 ppm, 9.15 ppm, 8.84 ppm and 8.44 ppm for NAD+, 9.62 ppm for NR, and 8.72 ppm and 7.60 ppm for Nam.
NR cleavage to Nam in cell extracts
Wildtype, PNP knockout, or PNP-FLAG overexpressing HEK293 cells were grown to confluence on a 60 mm plate and were collected and lysed by Dounce homogenizer in 50 mM Na phosphate buffer, pH 8.0, containing 100 mM NaCl and protease inhibitor cocktail (Roche). Cell homogenates were then centrifuged for 20 min at 21,000g and 4 °C, and the cell extracts were collected. 250 μg of cell extract was incubated with NR (2 mM) with or without Immucillin H (5 μM) in 100 μl reaction mixture containing 50 mM Na phosphate buffer, pH 8.0, 100 mM NaCl at 37 °C for 40 min. Reaction was quenched by adding 400 μl of ice-cold methanol. Samples were then incubated on ice for 20 min and centrifuged for 20 min at 21,000g and 4 °C. Supernatants were lyophilized and analyzed by NMR spectroscopy as described above.
NR cleavage to Nam in vivo
Five- to six-month-old male C57BL/6J mice were purchased from the Stolbovaya Nursery. Mice were divided into six groups: (1) control, (2) Immucillin H, (3) Nam, (4) Nam + Immucillin H, (5) NR, and (6) NR + Immucillin H, with three to five mice per treatment group. Mice were intraperitoneally injected with 400 mg per kg (body weight) NR, equimolar dose of Nam, or 2.5 mg per kg Immucillin H. The injection volume was 100 μl/10 g. Animals from control group were injected with PBS. Mice from group 2, 4, and 6 were also preinjected (2 h before the main injection) with 2.5 mg per kg Immucillin H. Two hours after the injection, mice were euthanized, and the whole blood, kidney, and liver samples were immediately collected. Kidney and liver samples were frozen in liquid nitrogen and stored at −80 °C until further analysis. Metabolites from 200 μl of whole blood were extracted by adding 800 μl of ice-cold methanol. Then, the mixture was kept on ice with occasional vortexing for 20 min, centrifuged for 20 min at 21,000g and 4 °C. The supernatant was lyophilized and analyzed by NMR spectroscopy. To determine the amount of NAD+ and its intermediates in kidney and liver, frozen samples were placed on ice for 10 min. Then, 0.7 ml of cooled glass beads (diameter 1.7 mm) and 1 ml of 80% methanol cooled to –80 °C were added into the tubes. Then samples were homogenized under cryogenic conditions using a FastPrep-24 homogenizer (MP Biomedicals) equipped with a CoolPrep adapter filled with dry ice. Five cycles of homogenization were carried out for 60 s at a rate of 6.5 m/s with 5 min breaks, when the tubes were kept on dry ice. Further, the extraction was continued by stirring at 4 °C for 30 min. Then, the resulting extracts were centrifuged for 20 min at 21,000g and 4 °C. The supernatant was lyophilized and analyzed by NMR spectroscopy. All experiments with animals were approved by the Animal Care and Use Committee of the Sechenov Institute of Evolutionary Physiology and Biochemistry (Protocol #1/2021; 28.01.2021).
Cell proliferation and cell death analysis
2.5 × 105 wildtype or PNP knockout HEK293 cells were plated on 6-well plates. After 24, 48, and 72 h, cells were trypsinized and stained with 50 μg/ml propidium iodide. Flow cytometry was performed using the CytoFLEX instrument (Beckman Coulter Inc). Analysis was carried out using CytExpert Software.
Statistical analysis
Statistical analysis was performed using the SigmaPlot 12.0 (Systat Software Inc). Differences between groups were analyzed using one-way or two-way ANOVA with Tukey’s post-hoc test. p-values < 0.05 were considered to be significant.
Data availability
All data are contained within the article and the Supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
A. K., V. K., L. S., A. Y., K. N., M. S., J. S., A. P., M. A., S. G., and A. N. investigation; A. K., A. Y., K. N., and M. E. M. methodology; A. K. validation; A. K., V. K., L. S., and A. N. visualization; A. K., M. Z., and A. N. writing-original draft; V. K. and L. S. data curation; A. N., V. K. and M. Z. funding acquisition; L. S., M. Z., and A. N. writing-review & editing; A. Y. software; A. Y. formal analysis; M. K., M. E. M., S. G., and M. Z. resources; A. N. conceptualization; A. N. supervision; A. N. project administration.
Funding and additional information
This work was partially supported by the Russian Foundation for Basic Research (grant № 19–34–60039). NMR-based metabolite measurements and experiments with mouse embryonic stem cells were supported by the grant from the Russian Science Foundation (grant № 21–14–00319). M. Z. and M. E. M. are supported by a network program (VitaCross) from the Research Council of Norway (grant № 309567). Human cell lines used in this study were obtained from the shared research facility “Vertebrate cell culture collection” supported by the Ministry of Science and Higher Education of the Russian Federation (Agreement № 075–15–2021–683). Part of the work was carried out using scientific equipment of the Center of Shared Usage “The analytical center of nano- and biotechnologies of SPbPU” and was supported by the Program ‘Priority 2030’ (Agreement 075–15–2021–1333).
Edited by Ruma Banerjee
Contributor Information
Mathias Ziegler, Email: mathias.ziegler@uib.no.
Andrey Nikiforov, Email: andrey.nikiforov@gmail.com.
Supporting information
References
- 1.Fang E.F., Kassahun H., Croteau D.L., Scheibye-Knudsen M., Marosi K., Lu H., et al. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016;24:566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J., Zhai Q., Chen Y., Lin E., Gu W., McBurney M.W., et al. A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 2005;170:349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown K.D., Maqsood S., Huang J.Y., Pan Y., Harkcom W., Li W., et al. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014;20:1059–1068. doi: 10.1016/j.cmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiarugi A., Dolle C., Felici R., Ziegler M. The NAD metabolome--a key determinant of cancer cell biology. Nat. Rev. Cancer. 2012;12:741–752. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- 5.Trammell S.A., Weidemann B.J., Chadda A., Yorek M.S., Holmes A., Coppey L.J., et al. Nicotinamide riboside opposes type 2 diabetes and neuropathy in mice. Sci. Rep. 2016;6 doi: 10.1038/srep26933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshino J., Mills K.F., Yoon M.J., Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu C.P., Oka S., Shao D., Hariharan N., Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ. Res. 2009;105:481–491. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diguet N., Trammell S.A.J., Tannous C., Deloux R., Piquereau J., Mougenot N., et al. Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation. 2018;137:2256–2273. doi: 10.1161/CIRCULATIONAHA.116.026099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braidy N., Guillemin G.J., Mansour H., Chan-Ling T., Poljak A., Grant R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Gomes A.P., Price N.L., Ling A.J., Moslehi J.J., Montgomery M.K., Rajman L., et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Canto C., et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massudi H., Grant R., Braidy N., Guest J., Farnsworth B., Guillemin G.J. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu X.H., Lu M., Lee B.Y., Ugurbil K., Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2876–2881. doi: 10.1073/pnas.1417921112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsyuba E., Romani M., Hofer D., Auwerx J. NAD(+) homeostasis in health and disease. Nat. Metab. 2020;2:9–31. doi: 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- 15.Covarrubias A.J., Perrone R., Grozio A., Verdin E. NAD(+) metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021;22:119–141. doi: 10.1038/s41580-020-00313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen M.S., Chang P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat. Chem. Biol. 2018;14:236–243. doi: 10.1038/nchembio.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupte R., Liu Z., Kraus W.L. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31:101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guse A.H. Calcium mobilizing second messengers derived from NAD. Biochim. Biophys. Acta. 2015;1854:1132–1137. doi: 10.1016/j.bbapap.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Nikiforov A., Kulikova V., Ziegler M. The human NAD metabolome: functions, metabolism and compartmentalization. Crit. Rev. Biochem. Mol. Biol. 2015;50:284–297. doi: 10.3109/10409238.2015.1028612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y., Sauve A.A. NAD(+) metabolism: bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta. 2016;1864:1787–1800. doi: 10.1016/j.bbapap.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bieganowski P., Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 23.Tempel W., Rabeh W.M., Bogan K.L., Belenky P., Wojcik M., Seidle H.F., et al. Nicotinamide riboside kinase structures reveal new pathways to NAD+ PLoS Biol. 2007;5:e263. doi: 10.1371/journal.pbio.0050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Revollo J.R., Grimm A.A., Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 25.Nikiforov A., Dolle C., Niere M., Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kropotov A., Kulikova V., Nerinovski K., Yakimov A., Svetlova M., Solovjeva L., et al. Equilibrative nucleoside transporters mediate the import of nicotinamide riboside and nicotinic acid riboside into human cells. Int. J. Mol. Sci. 2021;22:1391. doi: 10.3390/ijms22031391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purhonen J., Rajendran J., Tegelberg S., Smolander O.P., Pirinen E., Kallijarvi J., et al. NAD(+) repletion produces no therapeutic effect in mice with respiratory chain complex III deficiency and chronic energy deprivation. FASEB J. 2018;32:5913–5926. doi: 10.1096/fj.201800090R. [DOI] [PubMed] [Google Scholar]
- 28.Dall M., Trammell S.A.J., Asping M., Hassing A.S., Agerholm M., Vienberg S.G., et al. Mitochondrial function in liver cells is resistant to perturbations in NAD(+) salvage capacity. J. Biol. Chem. 2019;294:13304–13326. doi: 10.1074/jbc.RA118.006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frederick D.W., McDougal A.V., Semenas M., Vappiani J., Nuzzo A., Ulrich J.C., et al. Complementary NAD(+) replacement strategies fail to functionally protect dystrophin-deficient muscle. Skelet Muscle. 2020;10:30. doi: 10.1186/s13395-020-00249-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong B., Pan Y., Vempati P., Zhao W., Knable L., Ho L., et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer's mouse models. Neurobiol. Aging. 2013;34:1581–1588. doi: 10.1016/j.neurobiolaging.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canto C., Houtkooper R.H., Pirinen E., Youn D.Y., Oosterveer M.H., Cen Y., et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H., Ryu D., Wu Y., Gariani K., Wang X., Luan P., et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 33.Kulikova V., Shabalin K., Nerinovski K., Yakimov A., Svetlova M., Solovjeva L., et al. Degradation of extracellular NAD(+) intermediates in cultures of human HEK293 cells. Metabolites. 2019;9:293. doi: 10.3390/metabo9120293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bzowska A., Kulikowska E., Shugar D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Ther. 2000;88:349–425. doi: 10.1016/s0163-7258(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 35.Wielgus-Kutrowska B., Kulikowska E., Wierzchowski J., Bzowska A., Shugar D. Nicotinamide riboside, an unusual, non-typical, substrate of purified purine-nucleoside phosphorylases. Eur. J. Biochem. 1997;243:408–414. doi: 10.1111/j.1432-1033.1997.0408a.x. [DOI] [PubMed] [Google Scholar]
- 36.Belenky P., Christensen K.C., Gazzaniga F., Pletnev A.A., Brenner C. Nicotinamide riboside and nicotinic acid riboside salvage in fungi and mammals. Quantitative basis for Urh1 and purine nucleoside phosphorylase function in NAD+ metabolism. J. Biol. Chem. 2009;284:158–164. doi: 10.1074/jbc.M807976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shabalin K., Nerinovski K., Yakimov A., Kulikova V., Svetlova M., Solovjeva L., et al. NAD metabolome analysis in human cells using (1)H NMR spectroscopy. Int. J. Mol. Sci. 2018;19:3906. doi: 10.3390/ijms19123906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bantia S., Miller P.J., Parker C.D., Ananth S.L., Horn L.L., Kilpatrick J.M., et al. Purine nucleoside phosphorylase inhibitor BCX-1777 (Immucillin-H)--a novel potent and orally active immunosuppressive agent. Int. Immunopharmacol. 2001;1:1199–1210. doi: 10.1016/s1567-5769(01)00056-x. [DOI] [PubMed] [Google Scholar]
- 39.Hasmann M., Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 40.North B.J., Marshall B.L., Borra M.T., Denu J.M., Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 41.Skoge R.H., Dolle C., Ziegler M. Regulation of SIRT2-dependent alpha-tubulin deacetylation by cellular NAD levels. DNA Repair (Amst) 2014;23:33–38. doi: 10.1016/j.dnarep.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Ratajczak J., Joffraud M., Trammell S.A., Ras R., Canela N., Boutant M., et al. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 2016;7 doi: 10.1038/ncomms13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grossman L., Kaplan N.O. Nicotinamide riboside phosphorylase from human erythrocytes. II. Nicotinamide sensitivity. J. Biol. Chem. 1958;231:727–740. [PubMed] [Google Scholar]
- 44.Rowen J.W., Kornberg A. The phosphorolysis of nicotinamide riboside. J. Biol. Chem. 1951;193:497–507. [PubMed] [Google Scholar]
- 45.Belenky P., Racette F.G., Bogan K.L., McClure J.M., Smith J.S., Brenner C. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+ Cell. 2007;129:473–484. doi: 10.1016/j.cell.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 46.Trammell S.A., Schmidt M.S., Weidemann B.J., Redpath P., Jaksch F., Dellinger R.W., et al. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 2016;7 doi: 10.1038/ncomms12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giroud-Gerbetant J., Joffraud M., Giner M.P., Cercillieux A., Bartova S., Makarov M.V., et al. A reduced form of nicotinamide riboside defines a new path for NAD(+) biosynthesis and acts as an orally bioavailable NAD(+) precursor. Mol. Metab. 2019;30:192–202. doi: 10.1016/j.molmet.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L., Su X., Quinn W.J., 3rd, Hui S., Krukenberg K., Frederick D.W., et al. Quantitative analysis of NAD synthesis-Breakdown fluxes. Cell Metab. 2018;27:1067–1080.e1065. doi: 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y., Zhang N., Zhang G., Sauve A.A. NRH salvage and conversion to NAD(+) requires NRH kinase activity by adenosine kinase. Nat. Metab. 2020;2:364–379. doi: 10.1038/s42255-020-0194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y., Mohammed F.S., Zhang N., Sauve A.A. Dihydronicotinamide riboside is a potent NAD(+) concentration enhancer in vitro and in vivo. J. Biol. Chem. 2019;294:9295–9307. doi: 10.1074/jbc.RA118.005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayat F., Migaud M.E. Nicotinamide riboside-amino acid conjugates that are stable to purine nucleoside phosphorylase. Org. Biomol. Chem. 2020;18:2877–2885. doi: 10.1039/d0ob00134a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makarov M.V., Harris N.W., Rodrigues M., Migaud M.E. Scalable syntheses of traceable ribosylated NAD(+) precursors. Org. Biomol. Chem. 2019;17:8716–8720. doi: 10.1039/c9ob01981b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the Supporting information.