Abstract
DDX3X or DDX3, a member of the DEAD (asp, glu, ala, asp) box RNA helicase family of proteins, is a multifunctional protein, which is usurped by several viruses and is vital to their production. To date, 18 species of virus from 12 genera have been demonstrated to be dependent on DDX3 for virulence. In addition, DDX3 has been shown to function within 7 of 10 subcellular regions that are involved in the metabolism of viruses. As such, due to its direct interaction with viral components across most or all stages of viral life cycles, DDX3 can be considered an excellent host target for pan-antiviral drug therapy and has been reported to be a possible broad-spectrum antiviral target. Along these lines, it has been demonstrated that treatment of virally infected cells with small molecule inhibitors of DDX3 blunts virion productions. On the other hand, DDX3 bolsters an innate immune response and viruses have evolved capacities to sequester or block DDX3, which dampens an innate immune response. Thus, enhancing DDX3 production or co-targeting direct viral products that interfere with DDX3’s modulation of innate immunity would also diminish virion production. Here we review the evidence that supports the hypothesis that modulating DDX3’s agonistic and antagonistic functions during viral infections could have an important impact on safely and efficiently subduing a broad-spectrum of viral infections.
Keywords: DEAD-box RNA helicase, DDX3, antiviral, small molecule drug
1. Introduction
Antiviral therapies based on small molecule drugs have been developed into effective alternatives to vaccine resistance or for use in combination with vaccinations. Two broad categories of such therapeutics are direct-acting antiviral agents (DAAs) and those that are host-targeted antivirals (HTAs). An advantage to HTAs is the finding of host components that are required by a few or many viruses for cellular virion production and thus the potential of broad-spectrum antiviral agents may be achieved by targeting these components (Kumar et al., 2020). Given this, here we review a possible HTA therapy directed against a host DEAD (asp, glu, ala, asp)-box RNA helicase, DDX3X or DDX3, which has the potential of providing broad-spectrum antiviral therapy. We will focus on small molecule based HTA therapies directed against DDX3 including an inhibitor developed in our lab, RK-33, that has been designed to be a competitive inhibitor of DDX3’s ATPase site and has been demonstrated to abrogate its RNA helicase function (Bol et al., 2015; Heerma van Voss et al., 2018; Xie et al., 2016).
2. DDX3 is a multifunctional protein
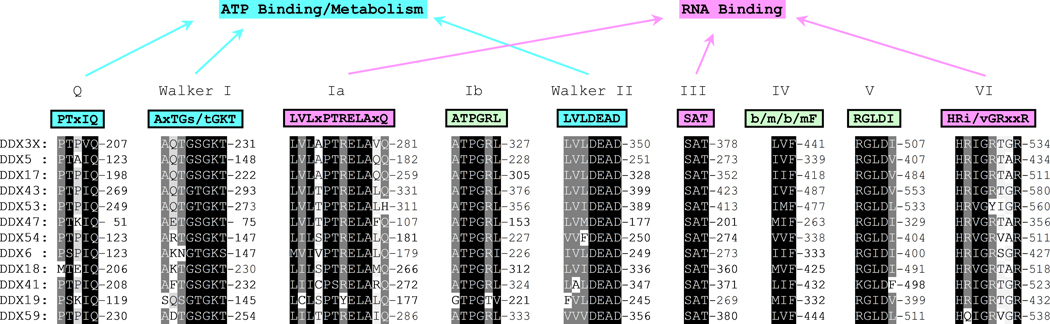
DDX3 belongs to the DEAD-box ATPase-dependent RNA helicase family of proteins (Song and Ji, 2019; Soto-Rifo and Ohlmann, 2013) that are characterized by two highly conserved Rec-A like core domains with the defining conserved motifs of this family of helicases: Q, Walker I, Ia, Ib, Walker II, III, IV, V, & VI, which are involved with ATPase/RNA binding helicase activity (Fig 1). Contrary to these domains are the amino and carboxy regions of the same proteins that vary greatly in length and sequence between proteins and exhibit a lack of any regions of homology or similarity (Fig 2). These latter domains imbues each member of this family with distinct functions that are independent of unwinding RNA (Bourgeois et al., 2016). DDX3 shuttles from the cytoplasm to the nucleus (Brennan et al., 2018) and has been demonstrated in be involved with a diversity of functions in both compartments (Bourgeois et al., 2016). Examples, in broad terms, of DDX3 functions are: translation activation of nuclear and mitochondrial mRNAs (Geissler et al., 2012; Heerma van Voss et al., 2018; Lee et al., 2008; Soto-Rifo et al., 2012; Waldron et al., 2019); transport, storage, and translation inhibition of mRNA (stress granules) (Gaete-Argel et al., 2019; Heaton et al., 2019; Shih et al., 2012); nuclear export of mRNA (Heaton et al., 2019); mRNA splicing (Bourgeois et al., 2016; Soto-Rifo and Ohlmann, 2013); modulation of transcription (Bourgeois et al., 2016; Fullam et al., 2018; Heaton et al., 2019; Khadivjam et al., 2017; Schroder, 2011); lipid hemostasis (Tsai et al., 2017); regulation of endoplasmic reticulum stress (Adjibade et al., 2017); modulation of epigenetic modifications (Chen et al., 2017); and innate immune system activation (Heaton et al., 2019; Oshiumi et al., 2010b; Taschuk and Cherry, 2020). Given this broad array of functions and the fact that viruses require the functions of cellular components to reproduce it should perhaps not be considered remarkable that a multifunctional protein such as DDX3 is essential for the replication and assembly of several species of viruses, as detailed below.
Fig 1. Illustration of the nine highly conserved protein motifs within the Rec-A like domains of DEAD box RNA helicases.
Protein sequences of DDX3, DDX5, DDX17, DDX43, DDX53, DDX47, DDX54, DDX6, DDX18, DDX41, DDX19, and DDX59 were obtained from the NCBI database as Fasta formats. In all cases of multiple isoforms, isoform 1 was used. Alignment was done in ClustalX2 with default settings (Gap opening: 10, Gap Extension: 0.2, and protein weight matrix: Gonnet series) and shading in GENEDOC. In all cases the sequences are represented by single letter amino acid code. The conserved consensus motif sequences above their respective shaded alignments are given with abbreviations: x = any amino acid and b = branched chain amino acids. Designations of each motif (Q, Walker I, Ia, Ib, Walker II, III, IV, V, & VI) are above the consensus sequences and those involved in ATP binding/metabolism and RNA binding are indicated by arrows.
Fig 2. Amino and carboxy termini sequences of the DEAD box RNA helicases represented in Fig 1.
(A) Amino-termini sequences and (B) carboxy-termini sequences. In either case, no homology or similar sequence motifs were found during the alignment of the sequences as described in Fig 1.
3. Viruses interact with multiple cellular pathways associated with DDX3
It is important to note that viral infections dramatically disrupt normal cellular functions and cellular homeostasis (Glingston et al., 2019) by diverting a cell’s functioning solely towards virion production. Table 1 presents a list, in broad terms, of the major subcellular regions and organelles that DDX3 is associated with and have been identified as regions that viruses are required to interact with throughout their life cycles (Glingston et al., 2019; Ji and Li, 2020; Kumar et al., 2020).
Table 1:
Examples of DDX3’s functional association with cellular regions that are required for viral entry, processing, and egress from cells.
| Subcellular Region | Associated Function | References |
|---|---|---|
| Plasma Membrane | TRPV4 Mediated Ca2+ Influx Releases DDX3 to Nuclear Translocation & Inhibition of Viral Nuclear Export/Translation | (Donate-Macian et al., 2018; Ji and Li, 2020; Kumar et al., 2020) |
| Nucleus | Exportin/Viral mRNA Nuclear Export | (Bourgeois et al., 2016; Frohlich et al., 2016; Heaton et al., 2019; Ji and Li, 2020; Kumar et al., 2020) |
| Regulation of Epigenetic Modifiers | (Chen et al., 2017; Kumar et al., 2020) | |
| Transcription Viral Modulators | (Fullam et al., 2018; Schroder, 2011) | |
| Cytoplasm | Translation Viral mRNA | (Geissler et al., 2012; Han et al., 2020; Lee et al., 2008; Soto-Rifo et al., 2012; Su et al., 2018; Waldron et al., 2019) |
| Lipid | Synthesis/Metabolism | (Li et al., 2013; Tsai et al., 2017) |
| MLOs | mRNA Storage | (Gaete-Argel et al., 2019; Shih et al., 2012) |
| MIT | Translation of MIT Protein/Modulation of OXPOS/ROS Generation | (Heerma van Voss et al., 2018) |
| ER | Translation of ER Protein | (Adjibade et al., 2017) |
Abbreviations: ER – Endoplasmic Reticulum; MIT – Mitochondria; MLO – Membraneless Organelles (Stress Granules & Processing Bodies); TRPV4 – Transient Receptor Potential Vanilloid 4.
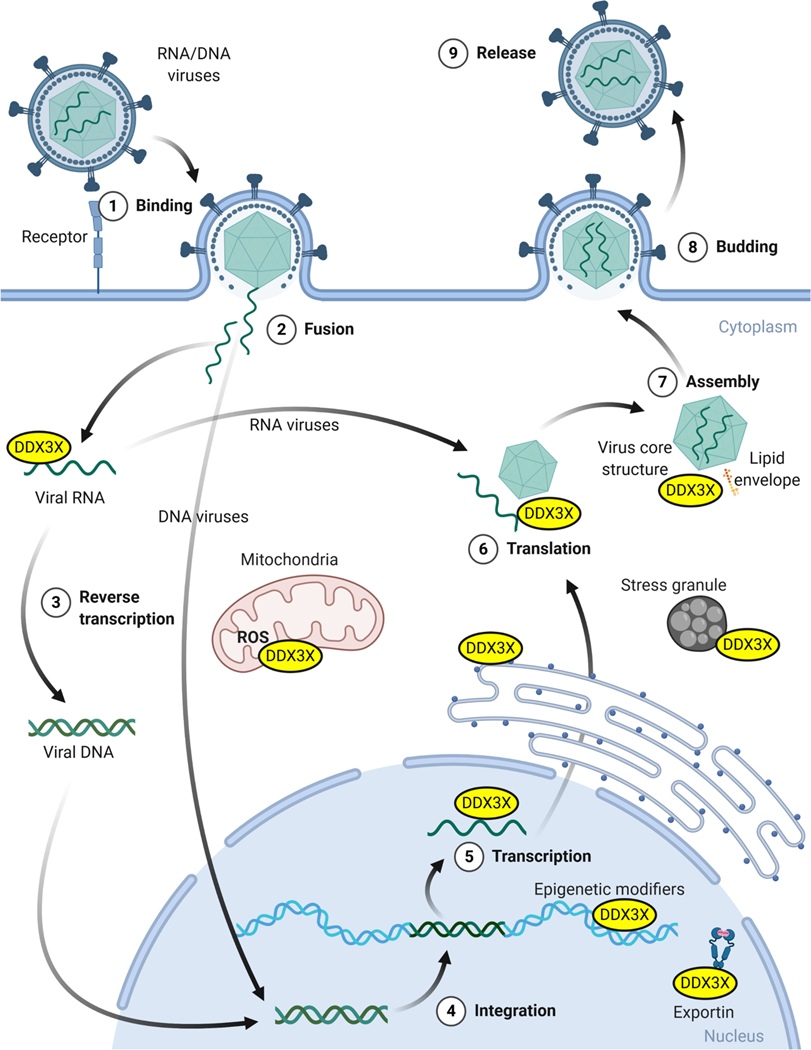
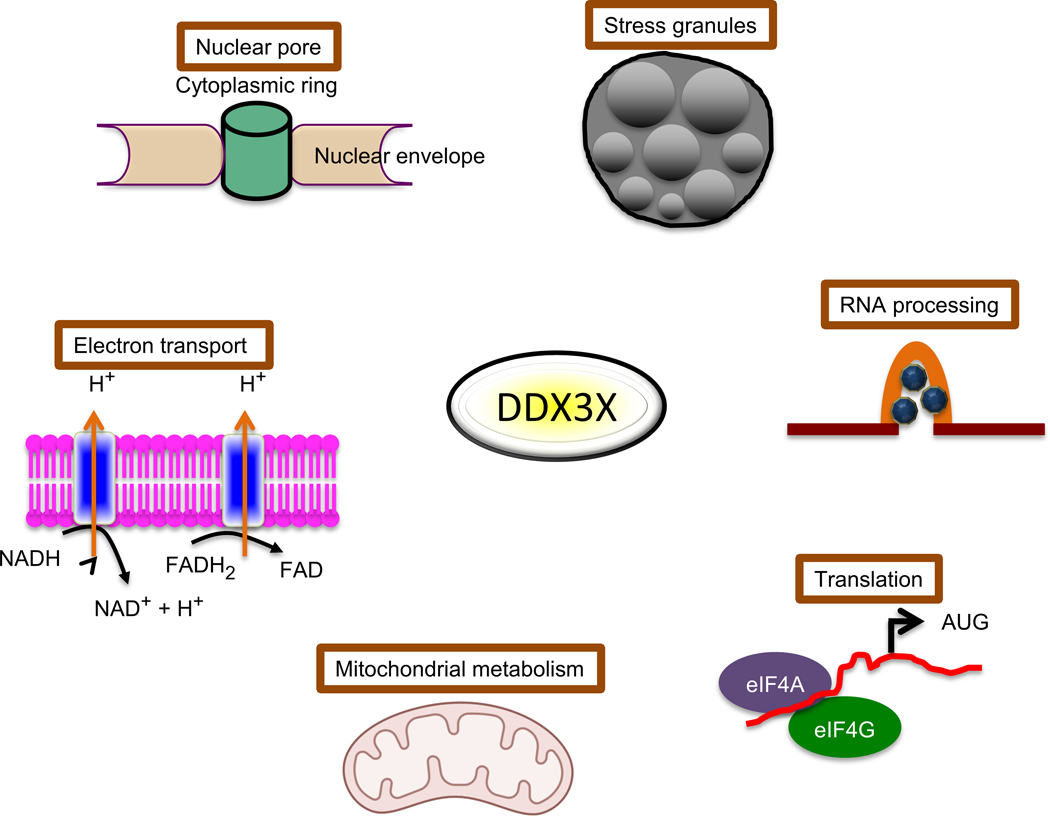
As noted (Table 1) and illustrated in Fig 3, in line with its multifaceted functions in cells, DDX3 has been found to be associated with 7 subcellular regions/organelles that are associated with a virus’ entry into a cell, nuclear/cytoplasmic processing/replication/translation/transcription, lipid envelope assembly, energy needs (mitochondrial), and egress from the cell. This is a very important finding as it indicates that DDX3 can be associated with many stages of viral life cycles, which increases the barrier of drug resistance, i.e., DDX3 is a multifunctional protein and viruses have evolved to utilize or block many of these functions making DDX3 essential for sustained virion production. Hence, if functions of DDX3 that are essential for multiple stages of virion production were abrogated (targeted) then the necessity of the co-evolution of multiple viral components that interact with DDX3 in a manner that allows a virus to remain fit for efficient propagation would be predicted to be slim, i.e., development of viral resistance to drugs targeting DDX3 would be improbable.
Fig 3. Illustrative summary of the multiple sub-compartments that DDX3 functions in and the association of viral components with the identical compartments.
A second arm of our rationale for selecting DDX3 for HTA therapy has been summarized in this section, i.e., once a cell has been infected by a virus it can no longer be considered as normal as it will have been converted into a virion factory with alterations to its gene expression repertoire and hence its transcriptome and proteome (Table 1 & (Claus and Liebert, 2014; El-Bacha and Da Poian, 2013; Gaete-Argel et al., 2019; Garcia-Sastre, 2017; Glingston et al., 2019; Hoffmann et al., 2017; Ji and Li, 2020; Kumar et al., 2020; Lange et al., 2019; Mohamed et al., 2020; Tanner et al., 2014). As such, targeting the functions DDX3, which has become an aberrantly required agent of virion production, may minimal impact the viability of normal uninfected cells. We hypothesize that a parallel with transformed cancer cells that present abnormal transcriptomes and proteomes can be considered. For example, there is convincing evidence to indicate that many viruses not only interact with various host proteins (Fung and Liu, 2019; Lim et al., 2016; van Hemert et al., 2008) but also replicate only in the S-phase of the cell cycle as viral replication proteins are regulated by cellular factors that are activated only in the S-phase (Schang, 2003). This has a direct association with DDX3, as DDX3 expression levels are important for the transition from G1 to S-phase of cell division (Ariumi, 2014) and a hallmark of cancer is that a large portion of cells in a tumor are rapidly dividing cells that have been tracked by scoring for the established division cycle protein Ki-67 (Pathmanathan and Balleine, 2013; Scholzen and Gerdes, 2000; Yang et al., 2018). Along these lines, we have demonstrated that in a number of cancer types, which have DDX3 is an essential component of the aggressiveness of these cancers and inhibition of DDX3 with a small molecule inhibitor, RK-33, kills cancer cells with no toxic effects on normal cells (Bol et al., 2015; Heerma van Voss et al., 2018; Xie et al., 2016).
4. DDX3 as a broad-spectrum antiviral target
To date, as listed in Table 2, 18 species of viruses grouped into 12 genera and 11 families have been reported as requiring DDX3 for efficient virion production and or sustained evasion of innate immunity (see references in Table 2). We searched for a few of the basic defining characteristics of these viruses in an attempt to identify common characteristics, such as type of genome (RNA or DNA), reliance on receptor entry into cells, enveloped or not, cytoplasmic or nuclear replication, 5’ 7-methylguanylate capped mRNA, and 3’ poly(A) appended mRNA, which might explain this broad reliance on DDX3. However, it is apparent that viruses with broadly different basic defining characteristics are represented in Table 2. For example, although most are single-stranded (ss) RNA viruses, double-stranded (ds) DNA viruses are also represented along with examples of segmented RNA viruses. The majority are packaged with a lipid envelop but three are not (Smith and Smith, 2019; Smyth and Martin, 2002), and 17 use protein specific receptor mediated cellular entry, but one does not (Edinger et al., 2014). For 13 of these, genome replication occurs exclusively in the cytoplasm, 3 replicate in the nucleus, and 2 replicate in a cytoplasmic stage coupled to a nuclear stage (Table 2). Obviously, the phylogenetic diversity represented in Table 2 explains the diversities enumerated here. In summary, the evidence presented in Table 1 & 2 indicates why DDX3 is an excellent candidate for HTA therapies across a broad and diverse spectrum of virus.
Table 2:
Viruses that require DDX3 for efficient sustained virion production.
| mRNA |
|||||||
|---|---|---|---|---|---|---|---|
| Species (Genus) | Genome | Env. | P.S.R. M.E. | Cyto. or Nucl. | 5′ Cap | 3′ poly-A | References |
| HIV (Lentivirus) | 2x +ssRNA | + | + | Cyto/Nucl | + | + | a |
| WNV (Flavivirus) | +ssRNA | + | + | Cyto | + | + | b |
| DENV (Flavivirus) | +ssRNA | + | + | Cyto | + | + | b |
| ZIKV (Flavivirus) | +ssRNA | + | + | Cyto | + | + | (Donate-Macian et al., 2018; Yang et al., 2020a) |
| JEV (Flavivirus) | +ssRNA | + | + | Cyto | + | + | (Li et al., 2014; Valiente-Echeverria et al., 2015) |
| HCV (Hepacivirus) | +ssRNA | + | + | Cyto | − | − | (Brai et al., 2016; Chatel-Chaix et al., 2013; Donate-Macian et al., 2018; Geissler et al., 2012; Li et al., 2013; Oshiumi et al., 2010a; Tsai et al., 2017; Valiente-Echeverria et al., 2015; Villareal et al., 2015) |
| EV-A71 (Enterovirus) | +ssRNA | − | + | Cyto | − | + | (Han et al., 2020; Su et al., 2018) |
| CV-B (Enterovirus) | +ssRNA | − | + | Cyto | − | + | (Quaranta et al., 2020) |
| NLV (Norovirus) | +ssRNA | − | + | Cyto | − | + | (Vashist et al., 2012) |
| HPIV-3 (Paramyxovirus) | -ssRNA | + | + | Cyto | + | + | (Heaton et al., 2019; Yang et al., 2020a) |
| RSV (Pneuovirus) | -ssRNA | + | + | Cyto | − | + | (Yang et al., 2020a) |
| FLUAV (α-Influenzavirus) | 8x -ssRNA | + | − | Nucl | + | + | (Thulasi Raman et al., 2016) |
| LASV (Mammarenavirus) | 2x +/−ssRNA | + | + | Cyto | + | − | (Loureiro et al., 2018) |
| JUNV (Mammarenavirus) | 2x +/−ssRNA | + | + | Cyto | + | − | (Loureiro et al., 2018) |
| LCMV (Mammarenavirus) | 2x +/−ssRNA | + | + | Cyto | + | − | (Loureiro et al., 2018) |
| HBV (Orthohepadnavirus) | dsDNA | + | + | Cyto/Nucl | + | + | (Megahed et al., 2020) |
| HSV-1 (Simplexvirus) | dsDNA | + | + | Nucl | + | + | (Khadivjam et al., 2017) |
| HCMV (Simplexivirus) | dsDNA | + | + | Nucl | + | + | (Cavignac et al., 2015; Lenarcic et al., 2015) |
| VACV (Orthopoxvirus) | dsDNA | + | + | Cyto | + | + | (Oda et al., 2009) |
(Brai et al., 2016; Brai et al., 2020b; de Bisschop et al., 2019; Frohlich et al., 2016; Gringhuis et al., 2017; Valiente-Echeverria et al., 2015; Yasuda-Inoue et al., 2013);
(Brai et al., 2016; Brai et al., 2019a; Geissler et al., 2012; Valiente-Echeverria et al., 2015; Yang et al., 2020a);
(Brai et al., 2016; Chatel-Chaix et al., 2013; Donate-Macian et al., 2018; Geissler et al., 2012; Li et al., 2013; Oshiumi et al., 2010a; Tsai et al., 2017; Valiente-Echeverria et al., 2015; Villareal et al., 2015);
Abbreviations: +ssRNA – positive sense single-stranded RNA; -ssRNA – negative sense single-stranded RNA; 2x – two genomic segments; 8x – eight genomic segments, +/−ssRNA – ambisense single-stranded RNA; HIV – Human Immunodeficiency virus; WNV – West Nile virus; DENV – Dengue virus; ZIKV – Zika virus; JEV – Japanese Encephalitis virus; HCV – Hepatitis C virus; EV-A71 – Enterovirus A71 virus; CV-B – Coxsackie virus group B; NLV – Norwalk virus; HPIV-3 – human Parainfluenza Type-3; RSV – Respiratory Syncytial virus; FLUAV – Influenza A virus; LASV – Lassa virus; JUNV – Junin virus; LCMV – Lymphocytic Choriomeningitis virus; HBV – Hepatitis B virus; HSV-1 – Herpes Simplex Type-1 virus; HCMV – Human Cytomegalovirus; VACV – Vaccinia virus; Env. – lipid envelope; P.S.R.M.E. – protein specific receptor mediated endocytosis; Cyto. or Nucl. – cytoplasmic or nuclear replication; 5’ Cap – 7-methylguanylate cap.
5. Small molecule inhibitors of DDX3 abrogate a broad-spectrum of viral infections.
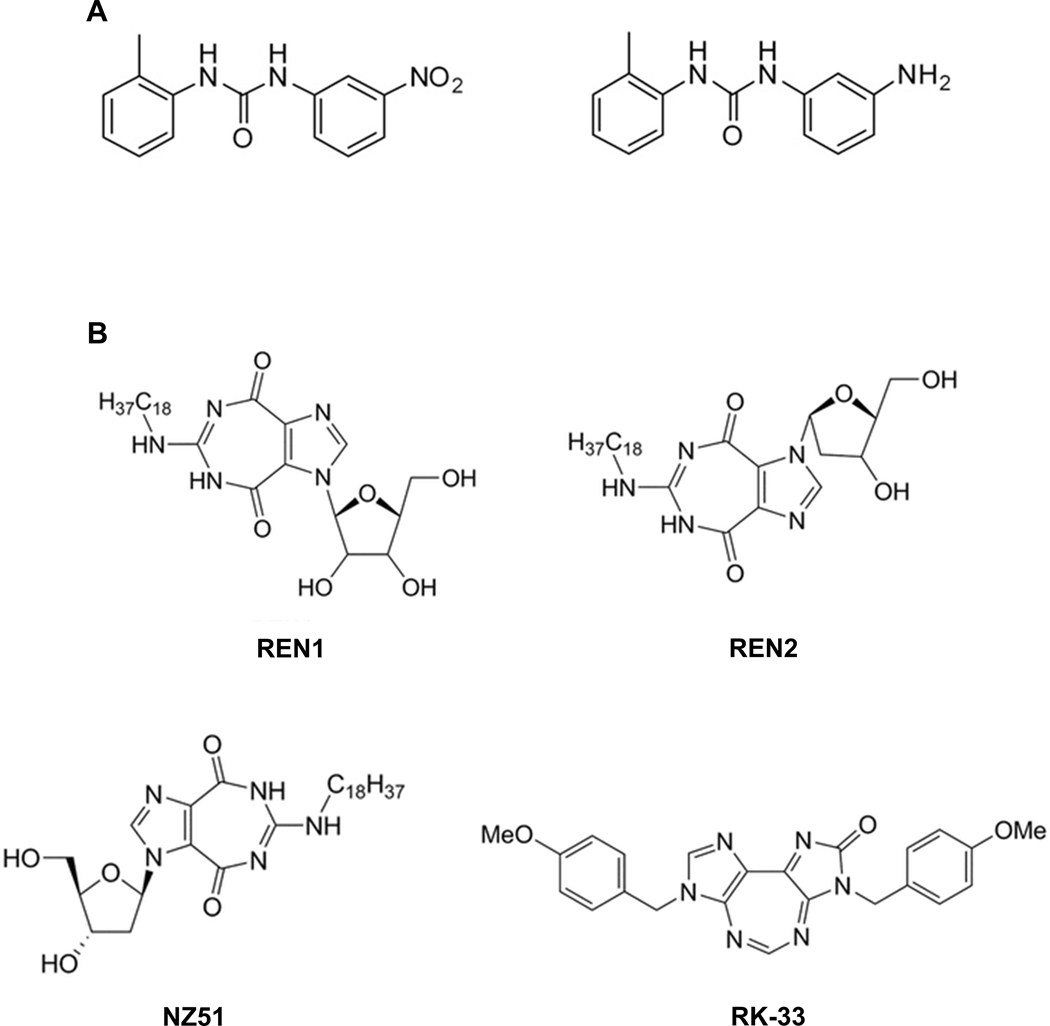
The viral species listed in Table 2 continue to have a devastating impact on human health by causing periodic deadly epidemics; e.g., West Nile virus (WNV), Dengue virus (DENV), Zika virus (ZIKA), Japanese Encephalitis virus (JEV), Influenza A virus (FLUAV), Lassa virus (LASV), Junin virus (JUNV), and Lymphocytic Choriomeningitis virus (LCMV), and persistent chronic illnesses; e.g., Human Immunodeficiency virus (HIV), Hepatitis C virus (HCV), and Human Cytomegalovirus (HCMV). As such, safe effective alternative antiviral treatments that overcome drug resistance as well as robust vaccines that control these infections are actively being sought (Brai et al., 2016; Ji and Li, 2020; Kumar et al., 2020). Along these lines, a goal is to develop safe HTA therapies as monotherapies or combination therapies, i.e., combinations of two or more HTA drugs or combinations of HTA therapeutics with DAA drugs, which can overcome drug resistance (Ji and Li, 2020; Kumar et al., 2020; Yang et al., 2020b). Given that DDX3 has been found as a common required host mediator for the efficient sustained production of the viruses listed in Table 2 and that vaccines or DAA antiviral therapies that are robust enough, i.e., are not limited by drug resistance, to effectively manage and ideally eliminate these viruses are lacking has brought many investigators to consider or actively pursue the targeting of DDX3 as a broad-spectrum antiviral strategy (Brai et al., 2020a; Brai et al., 2016; Brai et al., 2019a; Brai et al., 2020b; Brai et al., 2019b; Kukhanova et al., 2020; Quaranta et al., 2020; Schroder, 2011; Valiente-Echeverria et al., 2015; Yang et al., 2020a). For example, Botta and colleagues (Brai et al., 2020a; Brai et al., 2016; Brai et al., 2019a; Brai et al., 2020b; Brai et al., 2019b) have reported the development of several small molecule inhibitors that targets DDX3’s RNA binding site (Fig 4A). Using such inhibitors, these researchers have repeatedly demonstrated very effective anti-viral targeting of DDX3 in cells infected with HIV, HCV, DENV, and WNV (all positive-sense single stranded RNA viruses (+ssRNA – Table 2), which were non-toxic, i.e., in vitro half-maximum cytotoxic concentrations (CC50) of ~200 x the half-maximal effective concentrations (EC50), which were in the sub- to low-micromolar range as well as being safe in vivo with no brain, liver or kidney toxicity. Nevertheless, this strategy of targeting DDX3’s RNA binding site did not show anti-viral activity against negative-sense ssRNA (-ssRNA) viruses (Quaranta et al., 2020). However, a small molecule, originally developed as an anti-cancer therapeutic (Bol et al., 2015; Heerma van Voss et al., 2018; Xie et al., 2016), RK-33 (Fig 4B), that binds DDX3’s ATP binding site and inhibits its ATPase dependent RNA helicase activity, has been demonstrated to effectively (EC50 in the low micromolar range) abrogate virion production in both +ssRNA (DENV, WNV, and ZIKA) and -ssRNA (RSV and hPIV-3) infections (Yang et al., 2020a) (Table 2). Thus, both strategies: targeting RNA binding or ATPase activity, have produced similar results except that the RK-33 based targeting has the advantage of increasing the spectrum of viruses that can be suppressed. RK-33’s development was based on a rational design approach and has been demonstrated as being specific for DDX3 (Bol et al., 2015). For instance, RK-33 does not bind other members of the DEAD box RNA helicase family, despite the highly similar ATP binding sites across this family (Bourgeois et al., 2016). Moreover, the specificity of RK-33 to DDX3 was independently verified by Yang et al. (Yang et al., 2020a) using highly accurate physical biochemical methodologies: analytical ultracentrifugation and isothermal titration calorimetry, demonstrating that RK-33 binds DDX3 within the Walker I motif that contributes to ATP-binding (Fig 1), resulting in inhibition of DDX3’s ATPase and RNA unwinding activities. RK-33’s specificity is important to note because an objection to targeting the ATP binding pocket has been raised as it was argued such drugs would exhibit deleterious off target effects due to the similarity of ATP binding pockets across several classes of ATP binding proteins (Brai et al., 2016; Riva and Maga, 2019). Although, it has not been demonstrated that RK-33 will not bind any of the many DEAD box helicases or any of the ATP binding domain proteins, our animal studies have indicated that RK-33, even at significantly higher dose than the therapeutic window, had minimal or no toxicity, which indicates that it likely has minimal deleterious off target effects (Bol et al., 2015).
Fig 4. Examples of small molecule inhibitors of DDX3.
(A) Drugs that bind at the RNA binding site (Brai et al., 2016). (B) Drugs that are competitive inhibitors ATP binding at the ATP binding site (Bol et al., 2015).
Given the repeated evidence from independent laboratories that targeting DDX3’s RNA helicase activity has provided an effective safe abrogation of the production of a diversity of viruses indicates that further investigations aimed at developing drugs that block DDX3’s necessary interactions with viral propagation is warranted. As described above, to date, the focus has been on DDX3’s RNA helicase activity by either directly targeting the RNA binding site or its ATP binding site. However, the DEAD box family of proteins has diverse and generally non-overlapping functions, which are defined by their binding partners (Bourgeois et al., 2016; Shih et al., 2012; Soto-Rifo and Ohlmann, 2013). There is strong evidence that DDX3 interacts with several binding partners, such as chromosome region maintenance-1 (CRM1) (de Bisschop et al., 2019; Frohlich et al., 2016), poxvirus protein K7 (Oda et al., 2009), hepatocyte nuclear factor-4 (HNP4) (Tsai et al., 2017), small heterodimer partner (SHP) (Tsai et al., 2017), Tank-binding kinase1 (TBK1) (Khadivjam et al., 2017), calmodulin (CaM) (Khadivjam et al., 2017), transient receptor potential vanilloid-4 (TRPV-4) (Donate-Macian et al., 2018), ribosomal protein L13 (Han et al., 2020), dengue virus capsid protein (Kumar et al., 2017), eukaryotic translation initiation factor 4E (eIF4E) (Shih et al., 2012), eIF4F (Soto-Rifo et al., 2012), eIF3 (Geissler et al., 2012), 80s ribosomes (Geissler et al., 2012), p53 (Chen et al., 2017), retinoic acid inducible gene-1 (Oshiumi et al., 2010b), protein phosphatase 2A-C (PP2A-C) (Wang et al., 2017), and interferon-β promotor stimulator-1 (IPS-1) (Wang et al., 2017), through either its amino or carboxy termini, which thus, provides opportunities for the design of small molecule regents that block these interactions and inhibit those viruses that depend on the functions that such binding partners impart to their production.
6. DDX3’s function in innate immunity provides another anti-viral targeting strategy
DDX3 has been shown to be an integral component of an innate immune response to viral infections and, as such, is antiviral (Fullam et al., 2018; Heaton et al., 2019; Oshiumi et al., 2010b; Schroder, 2011; Wang et al., 2017); e.g., it contributes to the mediation of viral detection and functions in the induction of the type I interferon (IFN-I) pathway (Schroder, 2011). The necessity of an innate immune response to viral infections has led some investigators to caution against the targeting of DDX3 as an antiviral strategy as this might also comprise an innate immune response (Schroder, 2011). However, several counter points to this rationale can be made. First, viruses have evolved to block DDX3’s innate immunity functions or sequester DDX3 in subcellular virion producing regions, which provides an in situ evasion of an immune response (Gringhuis et al., 2017; Oda et al., 2009; Oshiumi et al., 2010a; Taschuk and Cherry, 2020). Second, the antiviral strategy targeting of DDX3 has been, as described above, based on the targeting of DDX3’s helicase activity, i.e., ATP or RNA binding sites, but these activities are independent and dispensable to DDX3’s innate immune antiviral functions (Khadivjam et al., 2017) and thus, these targeted strategies would be unlikely to affect innate immunity while simultaneously abrogating virion production. Third, during arenaviral infections DDX3 suppresses the IFN-I pathway (Loureiro et al., 2018) without being blocked or sequestered. However, the same report presented evidence that DDX3 promotes arenavirus replication and transcription in a helicase, i.e., ATPase/RNA binding, dependent manner. This latter finding is in line with all other reported evidence (as reviewed here) that DDX3 is generally directly utilized by viruses for virion production and in the case of arenaviruses the suppression of DDX3 mediated innate immunity is brought about in a manner other than the blocking or sequestering of DDX3 but with the same result.
It can be concluded that the most effective targeting of DDX3 would require drugs that do not solely target the ATP or RNA binding sites but also target domains outside of these with the aim of rectifying DDX3’s antiviral innate immunity activity. This concept of a dual drug approach can be extended to a combination drug approach where combinations of HTA antiviral agents might be used or DDX3 centered HTAs combined with DAAs could be advantageous (Kumar et al., 2020).
7. Future research
7.1. Rationale for a potential role for DDX3 in SARS-CoV-2 pathogenesis
Evolutionary studies have revealed that SARS-CoV-2 is closely related to SARS-CoV with a nearly identical genome size and organization as well as replication (Coronaviridae Study Group of the International Committee on Taxonomy of, 2020; Petrosillo et al., 2020; Stammler et al., 2011; van den Born et al., 2005; Yang and Leibowitz, 2015; Yoshimoto, 2020). SARS-CoV has been extensively studied and it is known that its replication initially proceeds through a 5’ cap-dependent translation using the host’s translation complexes (Cencic et al., 2011). As such, SARS-CoV-2 also has a 5’ capped mRNA genome that requires cap-dependent translation (Gordon et al., 2020). As pointed out above, there is strong evidence that DDX3 forms functional complexes with several eukaryotic translation initiation co-factors including eIF4E, eIF4E, eIF4G and eIF3 (Geissler et al., 2012; Lee et al., 2008; Shih et al., 2012; Soto-Rifo et al., 2012) and has been demonstrated as contributing a required function in promoting translational as a part of the 80S translation initiation complex (Geissler et al., 2012). Moreover, this is not the only example of a potential overlap between DDX3’s function and subcellular locations with those of SARS-CoV-2 virion processing/production. A recent detailed study (Gordon et al., 2020) of SARS-CoV-2 resulted in a comprehensive depiction of the SARS-CoV-2 interactome along with the associated subcellular compartments and organelles that fits the general schema laid out in Table 1 and illustrated in Fig 3. Thus, SARS-CoV-2 interactome maps reveal signaling pathways and organelles, which, as noted above, have been demonstrated as being involved with the functional and regulatory activities of DDX3 (Table 1 and Fig 5). Moreover, the same study found that the SARS-CoV-2 interactome was highly similar to that of WNV along with a high similarity with Zika and HIV and these three viruses usurp DDX3 during their infection cycles (Table 2). It is also noteworthy, in this context, that two members of the DEAD family, DDX1 and DDX5, have been shown to participate in viral replication of SARS-CoV ((Chen et al., 2009; Wu et al., 2014; Xu et al., 2010)) and further, that DDX3 has been demonstrated as binding with DDX5 forming a complex that functions in shuttling mRNP export from the nucleus to the cytoplasm (Choi and Lee, 2012; Riva and Maga, 2019). Consequently, although not yet demonstrated, given the structural similarity of DEAD box family members, it is not unreasonable to hypothesize that an antiviral strategy in SARS-CoV-2 infections might be achieved by targeting DDX3 to diminish or abolish cellular virion production of SARS-CoV-2.
Fig 5. Association of DDX3 in SARS-Cov2 host protein-protein interactome map.
7.2. Rationale for the consideration that DDX3 centered HTA strategy might be extended to other viral infections
The viruses listed in Table 2 cover a broad diversity of characteristics and some of these will be common to several other human viral infections. This is important to note because the rationale discussed in this review need not be limited to only the viruses listed in Table 2. We suggest that continued molecular characterization of the motifs/domains of DDX3 and its viral partners (Oda et al., 2009) will provide the capability to use rational design strategies to develop safe highly specific drugs that block the advantages that DDX3 supplies to virion production. Given DDX3’s critical function in inducing an innate immune response (Fullam et al., 2018; Meier-Stephenson et al., 2018; Schroder, 2011; Taschuk and Cherry, 2020), the development of reagents that bolster this activity and subvert the viral programs that thwart it is warranted.
Human host DEAD box RNA helicase, DDX3, has been demonstrated as a necessary component for the replication of several viruses.
Specific targeting of multifunctional DDX3 has the potential of providing a broad-spectrum antiviral therapy strategy.
DDX3 associated sub-cellular organelles have been identified in SARS-CoV-2 interactome maps.
Functional identification of antiviral and proviral activities of DDX3 can be exploited for defined virus treatment.
Small molecule inhibitors of DDX3 have been demonstrated to reduce viral titers and could have clinical applications as potential host-targeted antivirals.
Funding
This research was supported by R01CA207208 to VR.
Abbreviations:
- b
branched chain amino acid
- x
any amino acid
References
- Adjibade P, Grenier St-Sauveur V, Bergeman J, Huot ME, Khandjian EW, Mazroui R, 2017. DDX3 regulates endoplasmic reticulum stress-induced ATF4 expression. Sci Rep 7, 13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y, 2014. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection. Front Genet 5, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol GM, Vesuna F, Xie M, Zeng J, Aziz K, Gandhi N, Levine A, Irving A, Korz D, Tantravedi S, Heerma van Voss MR, Gabrielson K, Bordt EA, Polster BM, Cope L, van der Groep P, Kondaskar A, Rudek MA, Hosmane RS, van der Wall E, van Diest PJ, Tran PT, Raman V, 2015. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med 7, 648–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CF, Mortreux F, Auboeuf D, 2016. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Biol 17, 426–438. [DOI] [PubMed] [Google Scholar]
- Brai A, Boccuto A, Monti M, Marchi S, Vicenti I, Saladini F, Trivisani CI, Pollutri A, Trombetta CM, Montomoli E, Riva V, Garbelli A, Nola EM, Zazzi M, Maga G, Dreassi E, Botta M, 2020a. Exploring the Implication of DDX3X in DENV Infection: Discovery of the First-in-Class DDX3X Fluorescent Inhibitor. ACS Med Chem Lett 11, 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brai A, Fazi R, Tintori C, Zamperini C, Bugli F, Sanguinetti M, Stigliano E, Este J, Badia R, Franco S, Martinez MA, Martinez JP, Meyerhans A, Saladini F, Zazzi M, Garbelli A, Maga G, Botta M, 2016. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc Natl Acad Sci U S A 113, 5388–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brai A, Martelli F, Riva V, Garbelli A, Fazi R, Zamperini C, Pollutri A, Falsitta L, Ronzini S, Maccari L, Maga G, Giannecchini S, Botta M, 2019a. DDX3X Helicase Inhibitors as a New Strategy To Fight the West Nile Virus Infection. Journal of medicinal chemistry 62, 2333–2347. [DOI] [PubMed] [Google Scholar]
- Brai A, Riva V, Saladini F, Zamperini C, Trivisani CI, Garbelli A, Pennisi C, Giannini A, Boccuto A, Bugli F, Martini M, Sanguinetti M, Zazzi M, Dreassi E, Botta M, Maga G, 2020b. DDX3X inhibitors, an effective way to overcome HIV-1 resistance targeting host proteins. Eur J Med Chem 200, 112319. [DOI] [PubMed] [Google Scholar]
- Brai A, Ronzini S, Riva V, Botta L, Zamperini C, Borgini M, Trivisani CI, Garbelli A, Pennisi C, Boccuto A, Saladini F, Zazzi M, Maga G, Botta M, 2019b. Synthesis and Antiviral Activity of Novel 1,3,4-Thiadiazole Inhibitors of DDX3X. Molecules 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R, Haap-Hoff A, Gu L, Gautier V, Long A, Schroder M, 2018. Investigating nucleo-cytoplasmic shuttling of the human DEAD-box helicase DDX3. Eur J Cell Biol 97, 501–511. [DOI] [PubMed] [Google Scholar]
- Cavignac Y, Lieber D, Laib Sampaio K, Madlung J, Lamkemeyer T, Jahn G, Nordheim A, Sinzger C, 2015. The Cellular Proteins Grb2 and DDX3 Are Increased upon Human Cytomegalovirus Infection and Act in a Proviral Fashion. PLoS One 10, e0131614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ, Pelletier J, 2011. Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication. J Virol 85, 6381–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Germain MA, Motorina A, Bonneil E, Thibault P, Baril M, Lamarre D, 2013. A host YB-1 ribonucleoprotein complex is hijacked by hepatitis C virus for the control of NS3-dependent particle production. J Virol 87, 11704–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Chen WN, Poon KM, Zheng BJ, Lin X, Wang YX, Wen YM, 2009. Interaction between SARS-CoV helicase and a multifunctional cellular protein (Ddx5) revealed by yeast and mammalian cell two-hybrid systems. Arch Virol 154, 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Wang WT, Tsai TY, Li HK, Lee YW, 2017. DDX3 localizes to the centrosome and prevents multipolar mitosis by epigenetically and translationally modulating p53 expression. Sci Rep 7, 9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lee SG, 2012. The DEAD-box RNA helicase DDX3 interacts with DDX5, co-localizes with it in the cytoplasm during the G2/M phase of the cycle, and affects its shuttling during mRNP export. J Cell Biochem 113, 985–996. [DOI] [PubMed] [Google Scholar]
- Claus C, Liebert UG, 2014. A renewed focus on the interplay between viruses and mitochondrial metabolism. Arch Virol 159, 1267–1277. [DOI] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of, V., 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bisschop G, Ameur M, Ulryck N, Benattia F, Ponchon L, Sargueil B, Chamond N, 2019. HIV-1 gRNA, a biological substrate, uncovers the potency of DDX3X biochemical activity. Biochimie 164, 83–94. [DOI] [PubMed] [Google Scholar]
- Donate-Macian P, Jungfleisch J, Perez-Vilaro G, Rubio-Moscardo F, Peralvarez-Marin A, Diez J, Valverde MA, 2018. The TRPV4 channel links calcium influx to DDX3X activity and viral infectivity. Nature communications 9, 2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger TO, Pohl MO, Stertz S, 2014. Entry of influenza A virus: host factors and antiviral targets. J Gen Virol 95, 263–277. [DOI] [PubMed] [Google Scholar]
- El-Bacha T, Da Poian AT, 2013. Virus-induced changes in mitochondrial bioenergetics as potential targets for therapy. Int J Biochem Cell Biol 45, 41–46. [DOI] [PubMed] [Google Scholar]
- Frohlich A, Rojas-Araya B, Pereira-Montecinos C, Dellarossa A, Toro-Ascuy D, Prades-Perez Y, Garcia-de-Gracia F, Garces-Alday A, Rubilar PS, Valiente-Echeverria F, Ohlmann T, Soto-Rifo R, 2016. DEAD-box RNA helicase DDX3 connects CRM1-dependent nuclear export and translation of the HIV-1 unspliced mRNA through its N-terminal domain. Biochim Biophys Acta 1859, 719–730. [DOI] [PubMed] [Google Scholar]
- Fullam A, Gu L, Hohn Y, Schroder M, 2018. DDX3 directly facilitates IKKalpha activation and regulates downstream signalling pathways. Biochem J 475, 3595–3607. [DOI] [PubMed] [Google Scholar]
- Fung TS, Liu DX, 2019. Human Coronavirus: Host-Pathogen Interaction. Annu Rev Microbiol 73, 529–557. [DOI] [PubMed] [Google Scholar]
- Gaete-Argel A, Marquez CL, Barriga GP, Soto-Rifo R, Valiente-Echeverria F, 2019. Strategies for Success. Viral Infections and Membraneless Organelles. Front Cell Infect Microbiol 9, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, 2017. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 22, 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R, Golbik RP, Behrens SE, 2012. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res 40, 4998–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glingston RS, Deb R, Kumar S, Nagotu S, 2019. Organelle dynamics and viral infections: at cross roads. Microbes Infect 21, 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Huttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O’Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d’Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, Garcia-Sastre A, Shokat KM, Shoichet BK, Krogan NJ, 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis SI, Hertoghs N, Kaptein TM, Zijlstra-Willems EM, Sarrami-Forooshani R, Sprokholt JK, van Teijlingen NH, Kootstra NA, Booiman T, van Dort KA, Ribeiro CM, Drewniak A, Geijtenbeek TB, 2017. HIV-1 blocks the signaling adaptor MAVS to evade antiviral host defense after sensing of abortive HIV-1 RNA by the host helicase DDX3. Nat Immunol 18, 225–235. [DOI] [PubMed] [Google Scholar]
- Han S, Sun S, Li P, Liu Q, Zhang Z, Dong H, Sun M, Wu W, Wang X, Guo H, 2020. Ribosomal Protein L13 Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus in a Helicase DDX3-Dependent Manner. J Virol 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton SM, Atkinson SC, Sweeney MN, Yang SNY, Jans DA, Borg NA, 2019. Exportin-1-Dependent Nuclear Export of DEAD-box Helicase DDX3X is Central to its Role in Antiviral Immunity. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerma van Voss MR, Vesuna F, Bol GM, Afzal J, Tantravedi S, Bergman Y, Kammers K, Lehar M, Malek R, Ballew M, Ter Hoeve N, Abou D, Thorek D, Berlinicke C, Yazdankhah M, Sinha D, Le A, Abrahams R, Tran PT, van Diest PJ, Raman V, 2018. Targeting mitochondrial translation by inhibiting DDX3: a novel radiosensitization strategy for cancer treatment. Oncogene 37, 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HH, Schneider WM, Blomen VA, Scull MA, Hovnanian A, Brummelkamp TR, Rice CM, 2017. Diverse Viruses Require the Calcium Transporter SPCA1 for Maturation and Spread. Cell Host Microbe 22, 460–470 e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Li Z, 2020. Medicinal chemistry strategies toward host targeting antiviral agents. Med Res Rev. [DOI] [PMC free article] [PubMed]
- Khadivjam B, Stegen C, Hogue-Racine MA, El Bilali N, Dohner K, Sodeik B, Lippe R, 2017. The ATP-Dependent RNA Helicase DDX3X Modulates Herpes Simplex Virus 1 Gene Expression. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukhanova MK, Karpenko IL, Ivanov AV, 2020. DEAD-box RNA Helicase DDX3: Functional Properties and Development of DDX3 Inhibitors as Antiviral and Anticancer Drugs. Molecules 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Sharma S, Kumar R, Tripathi BN, Barua S, Ly H, Rouse BT, 2020. Host-Directed Antiviral Therapy. Clin Microbiol Rev 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Singh N, Abdin MZ, Patel AH, Medigeshi GR, 2017. Dengue Virus Capsid Interacts with DDX3X-A Potential Mechanism for Suppression of Antiviral Functions in Dengue Infection. Front Cell Infect Microbiol 7, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PT, Lagunoff M, Tarakanova VL, 2019. Chewing the Fat: The Conserved Ability of DNA Viruses to Hijack Cellular Lipid Metabolism. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R, 2008. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res 36, 4708–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic EM, Ziehr BJ, Moorman NJ, 2015. An unbiased proteomics approach to identify human cytomegalovirus RNA-associated proteins. Virology 481, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Ge LL, Li PP, Wang Y, Dai JJ, Sun MX, Huang L, Shen ZQ, Hu XC, Ishag H, Mao X, 2014. Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions. Virology 449, 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ, 2013. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med 19, 722–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YX, Ng YL, Tam JP, Liu DX, 2016. Human Coronaviruses: A Review of Virus-Host Interactions. Diseases 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro ME, Zorzetto-Fernandes AL, Radoshitzky S, Chi X, Dallari S, Marooki N, Leger P, Foscaldi S, Harjono V, Sharma S, Zid BM, Lopez N, de la Torre JC, Bavari S, Zuniga E, 2018. DDX3 suppresses type I interferons and favors viral replication during Arenavirus infection. PLoS pathogens 14, e1007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megahed FAK, Zhou X, Sun P, 2020. The Interactions between HBV and the Innate Immunity of Hepatocytes. Viruses 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Stephenson V, Mrozowich T, Pham M, Patel TR, 2018. DEAD-box helicases: the Yin and Yang roles in viral infections. Biotechnol Genet Eng Rev 34, 3–32. [DOI] [PubMed] [Google Scholar]
- Mohamed B, Mazeaud C, Baril M, Poirier D, Sow AA, Chatel-Chaix L, Titorenko V, Lamarre D, 2020. Very-long-chain fatty acid metabolic capacity of 17-beta-hydroxysteroid dehydrogenase type 12 (HSD17B12) promotes replication of hepatitis C virus and related flaviviruses. Sci Rep 10, 4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda S, Schroder M, Khan AR, 2009. Structural basis for targeting of human RNA helicase DDX3 by poxvirus protein K7. Structure 17, 1528–1537. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Ikeda M, Matsumoto M, Watanabe A, Takeuchi O, Akira S, Kato N, Shimotohno K, Seya T, 2010a. Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS-1-mediated IFN-beta induction. PLoS One 5, e14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Sakai K, Matsumoto M, Seya T, 2010b. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. European journal of immunology 40, 940–948. [DOI] [PubMed] [Google Scholar]
- Pathmanathan N, Balleine RL, 2013. Ki67 and proliferation in breast cancer. J Clin Pathol 66, 512–516. [DOI] [PubMed] [Google Scholar]
- Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E, 2020. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect 26, 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaranta P, Lottini G, Chesi G, Contrafatto F, Russotto R, Macera L, Lai M, Spezia PG, Brai A, Botta M, Freer G, Pistello M, 2020. DDX3 inhibitors show antiviral activity against positive-sense single-stranded RNA viruses but not against negative-sense single-stranded RNA viruses: The coxsackie B model. Antiviral Res 178, 104750. [DOI] [PubMed] [Google Scholar]
- Riva V, Maga G, 2019. From the magic bullet to the magic target: exploiting the diverse roles of DDX3X in viral infections and tumorigenesis. Future Med Chem 11, 1357–1381. [DOI] [PubMed] [Google Scholar]
- Schang LM, 2003. The cell cycle, cyclin-dependent kinases, and viral infections: new horizons and unexpected connections. Prog Cell Cycle Res 5, 103–124. [PubMed] [Google Scholar]
- Scholzen T, Gerdes J, 2000. The Ki-67 protein: from the known and the unknown. J Cell Physiol 182, 311–322. [DOI] [PubMed] [Google Scholar]
- Schroder M, 2011. Viruses and the human DEAD-box helicase DDX3: inhibition or exploitation? Biochem Soc Trans 39, 679–683. [DOI] [PubMed] [Google Scholar]
- Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH, 2012. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J 441, 119–129. [DOI] [PubMed] [Google Scholar]
- Smith HQ, Smith TJ, 2019. The Dynamic Capsid Structures of the Noroviruses. Viruses 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth MS, Martin JH, 2002. Picornavirus uncoating. Mol Pathol 55, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Ji X, 2019. The mechanism of RNA duplex recognition and unwinding by DEAD-box helicase DDX3X. Nature communications 10, 3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Rifo R, Ohlmann T, 2013. The role of the DEAD-box RNA helicase DDX3 in mRNA metabolism. Wiley interdisciplinary reviews. RNA 4, 369–385. [DOI] [PubMed] [Google Scholar]
- Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T, 2012. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. Embo J 31, 3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stammler SN, Cao S, Chen SJ, Giedroc DP, 2011. A conserved RNA pseudoknot in a putative molecular switch domain of the 3’-untranslated region of coronaviruses is only marginally stable. RNA 17, 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YS, Tsai AH, Ho YF, Huang SY, Liu YC, Hwang LH, 2018. Stimulation of the Internal Ribosome Entry Site (IRES)-Dependent Translation of Enterovirus 71 by DDX3X RNA Helicase and Viral 2A and 3C Proteases. Front Microbiol 9, 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner LB, Chng C, Guan XL, Lei Z, Rozen SG, Wenk MR, 2014. Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J Lipid Res 55, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschuk F, Cherry S, 2020. DEAD-Box Helicases: Sensors, Regulators, and Effectors for Antiviral Defense. Viruses 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasi Raman SN, Liu G, Pyo HM, Cui YC, Xu F, Ayalew LE, Tikoo SK, Zhou Y, 2016. DDX3 Interacts with Influenza A Virus NS1 and NP Proteins and Exerts Antiviral Function through Regulation of Stress Granule Formation. J Virol 90, 3661–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TY, Wang WT, Li HK, Chen WJ, Tsai YH, Chao CH, Wu Lee YH, 2017. RNA helicase DDX3 maintains lipid homeostasis through upregulation of the microsomal triglyceride transfer protein by interacting with HNF4 and SHP. Sci Rep 7, 41452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente-Echeverria F, Hermoso MA, Soto-Rifo R, 2015. RNA helicase DDX3: at the crossroad of viral replication and antiviral immunity. Rev Med Virol 25, 286–299. [DOI] [PubMed] [Google Scholar]
- van den Born E, Posthuma CC, Gultyaev AP, Snijder EJ, 2005. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J Virol 79, 6312–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ, 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS pathogens 4, e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Urena L, Chaudhry Y, Goodfellow I, 2012. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J Virol 86, 11977–11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal VA, Rodgers MA, Costello DA, Yang PL, 2015. Targeting host lipid synthesis and metabolism to inhibit dengue and hepatitis C viruses. Antiviral Res 124, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron JA, Tack DC, Ritchey LE, Gillen SL, Wilczynska A, Turro E, Bevilacqua PC, Assmann SM, Bushell M, Le Quesne J, 2019. mRNA structural elements immediately upstream of the start codon dictate dependence upon eIF4A helicase activity. Genome Biol 20, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang R, Luo M, Li C, Wang HX, Huan CC, Qu YR, Liao Y, Mao X, 2017. (DEAD)-box RNA helicase 3 modulates NF-kappaB signal pathway by controlling the phosphorylation of PP2A-C subunit. Oncotarget 8, 33197–33213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Chen PJ, Yeh SH, 2014. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe 16, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Vesuna F, Tantravedi S, Bol GM, Heerma van Voss MR, Nugent K, Malek R, Gabrielson K, van Diest PJ, Tran PT, Raman V, 2016. RK-33 Radiosensitizes Prostate Cancer Cells by Blocking the RNA Helicase DDX3. Cancer Res 76, 6340–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX, 2010. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol 84, 8571–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Zhang J, Ding M, Xu K, Li L, Mao L, Zheng J, 2018. Ki67 targeted strategies for cancer therapy. Clin Transl Oncol 20, 570–575. [DOI] [PubMed] [Google Scholar]
- Yang D, Leibowitz JL, 2015. The structure and functions of coronavirus genomic 3’ and 5’ ends. Virus Res 206, 120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SNY, Atkinson SC, Audsley MD, Heaton SM, Jans DA, Borg NA, 2020a. RK-33 Is a Broad-Spectrum Antiviral Agent That Targets DEAD-Box RNA Helicase DDX3X. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SNY, Atkinson SC, Wang C, Lee A, Bogoyevitch MA, Borg NA, Jans DA, 2020b. The broad spectrum antiviral ivermectin targets the host nuclear transport importin alpha/beta1 heterodimer. Antiviral Res 177, 104760. [DOI] [PubMed] [Google Scholar]
- Yoshimoto FK, 2020. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19. Protein J 39, 198–216. [DOI] [PMC free article] [PubMed] [Google Scholar]