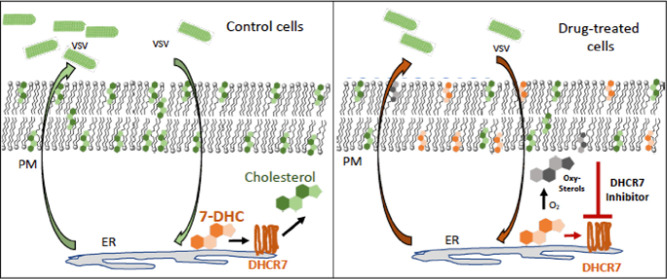
Abstract
Cholesterol is ubiquitous in cells; it plays a critical role in membrane structure and transport as well as in intracellular trafficking processes. There are suggestions that cholesterol metabolism is linked to innate immunity with inhibitors of DHCR7, the last enzyme in the cholesterol pathway, suggested to have potential as viral therapeutics nearly a decade ago. In fact, there are a number of highly prescribed pharmaceuticals that are off-target inhibitors of DHCR7, causing increased cellular levels of 7-dehydrodesmosterol (7-DHD) and 7-dehydrocholesterol (7-DHC). We report here dose–response studies of six such inhibitors on late-stage cholesterol biosynthesis in Neuro2a cells as well as their effect on infection of vesicular stomatitis virus (VSV). Four of the test compounds are FDA-approved drugs (cariprazine, trazodone, metoprolol, and tamoxifen), one (ifenprodil) has been the object of a recent Phase 2b COVID trial, and one (AY9944) is an experimental compound that has seen extensive use as a DHCR7 inhibitor. The three FDA-approved drugs inhibit replication of a GFP-tagged VSV with efficacies that mirror their effect on DHCR7. Ifenprodil and AY9944 have complex inhibitory profiles, acting on both DHCR7 and DHCR14, while tamoxifen does not inhibit DHCR7 and is toxic to Neuro2a at concentrations where it inhibits the Δ7−Δ8 isomerase of the cholesterol pathway. VSV itself affects the sterol profile in Neuro2a cells, showing a dose–response increase of dehydrolathosterol and lathosterol, the substrates for DHCR7, with a corresponding decrease in desmosterol and cholesterol. 7-DHD and 7-DHC are orders of magnitude more vulnerable to free radical chain oxidation than other sterols as well as polyunsaturated fatty esters, and the effect of these sterols on viral infection is likely a reflection of this fact of Nature.
Keywords: DHCR7, 7-dehydrocholesterol, 7-dehydrodesmosterol, vesicular stomatitis virus, oxidative stress
Cholesterol is a major constituent of the lipids present in viral envelopes, comprising up to 50% of the lipids present in some enveloped viruses. Cholesterol also plays an important role in the intracellular trafficking pathways commonly required by viruses for replication. Transport of viral cargo between intracellular compartments is a complex process that involves recruitment of cargo to membrane bilayers, curving donor membranes to form budding vesicles, and the sorting of vesicles in acceptor organelles. Cholesterol is critical to each of these steps, playing an important role in stabilizing membranes and in the formation of aggregate membrane structures involved in some of the transfers.
Interest in viral replication and late-stage cholesterol biosynthesis,1 shown in Figure 1, has also been stimulated by reports linking pharmacological inhibition of key steps in the transformations to a decrease in viral infections. 7-DHC, the enzymatic product of DHCR7, has recently been reported to inhibit various viral infections2 following Yang’s proposal a decade ago that DHCR7 is a potential target for antiviral development.3
Figure 1.
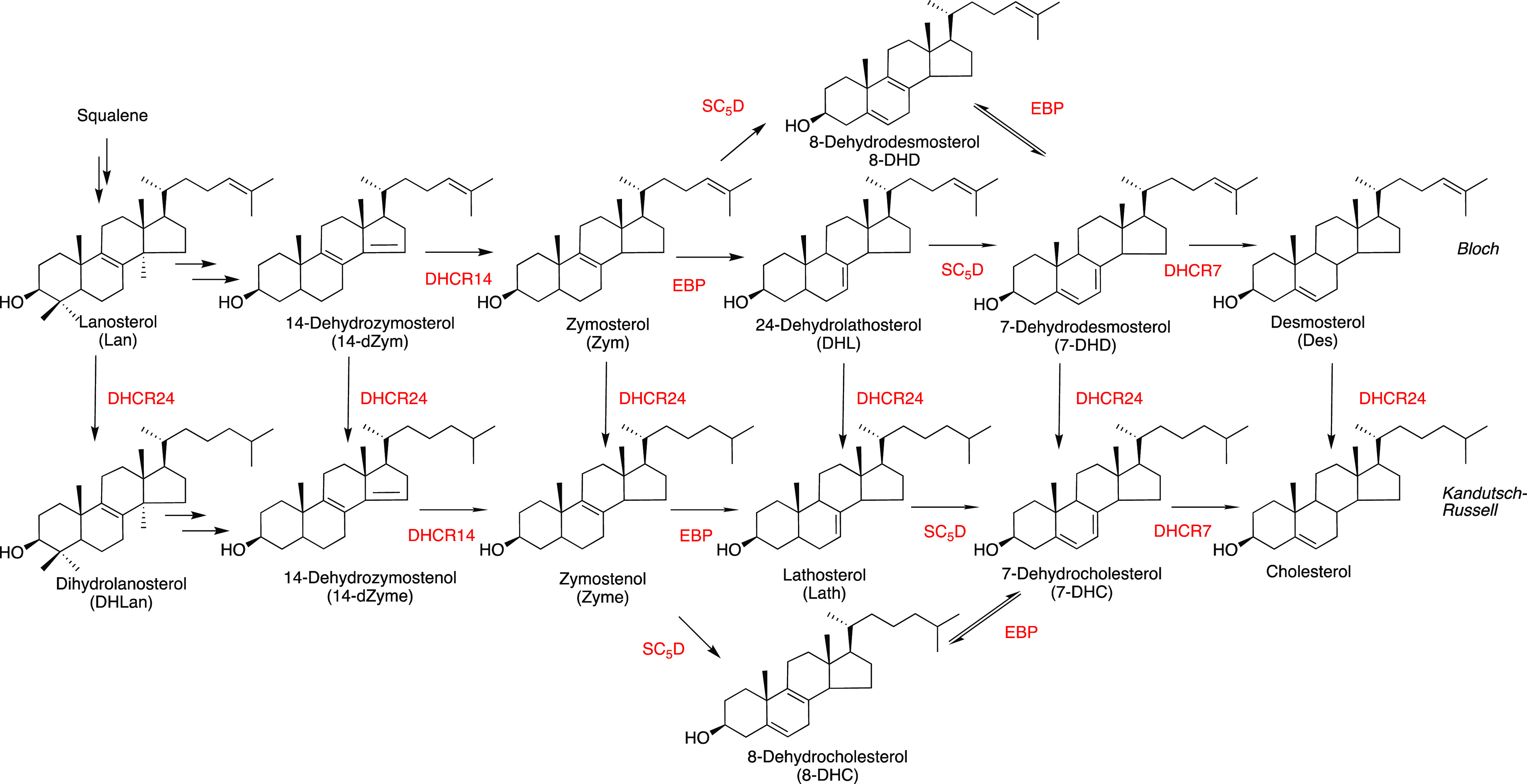
Post-lanosterol sterols and enzymes that catalyze the transformations (in red). The Bloch pathway from lanosterol to desmosterol is connected to the Kandutsch–Russell sterols (that include 7-DHC and cholesterol) via transformations promoted by DHCR24. The conjugated diene sterols,14-dZym, 14-dZyme, 7-DHD, and 7-DHC are particularly vulnerable to free radical peroxidation.
The structural differences between 7-DHC and cholesterol are minimal, the compounds differing by only 2 hydrogen atoms. They have the same physical properties and hydrophobic character, but the chemical difference between these two isosteric lipids could not be more stark. Cholesterol and 7-DHC both undergo free radical oxidation (lipid peroxidation),4−7 but 7-DHC is over 200 times more reactive than cholesterol in its reaction with membrane phospholipid-derived peroxyl radicals.8−10 7-DHC and its Bloch pathway analogue 7-DHD (Figure 1) are Nature’s most oxidizable lipids.10,11
Recent important studies have suggested that the exceptional reactivity of 7-DHC plays a role in mediating ferroptosis, a form of programmed cell death.12−15 7-DHC and 7-DHD, being orders of magnitude more vulnerable to free radical oxidation than polyunsaturated fatty (PUFA) ester phospholipids, apparently serve sacrificial roles by diverting membrane oxidative chains from PUFAs to sterols. The cellular protective response to a ferroptotic oxidative stress is upregulation of genes coding enzymes before 7-DHC and 7-DHD on the biosynthesis pathway and downregulation of DHCR7.16 The result of such a response is a change in the cellular sterol profile, with a presumed decrease in sterols biosynthesized after DHCR7 in the pathway (desmosterol and cholesterol) and an increase in levels of sterols before DHCR7 (7-DHC and 7-DHD).
Oxidative stress has been associated with viral infections in numerous studies, including evidence for the formation of peroxidation biomarkers from studies of VSV and RSV infections and in the analysis of plasma from COVID-19 patients. If ferroptosis is a consequence of viral oxidative stress,17−20 7-DHC and 7-DHD21 may provide a critical response to infection. Indeed, the identification of overabundant 7-DHC in replicating Hepatitis B cells by untargeted lipid profiling over a decade ago may have provided the first example of such a cellular response.22 From this perspective, the efficacy of DHCR7 inhibitors such as AY99442,3 and the FDA-approved drugs described here may come from the stabilizing effect of 7-DHD and 7-DHC on cellular membrane structures, their introduction by pharmacology providing sentries that protect membrane bilayers from radical assault.
There are a number of recent screening studies that have identified more than a handful of FDA-approved drugs that inhibit DHCR7 and increase cellular levels of 7-DHC at nanomolar concentrations.23−28 In one study of a 1000 compound library of biologically active compounds, including many FDA-approved drugs, over 5% of the compounds tested showed some activity as inhibitors of DHCR7,24 as measured by the Z-score for increase in 7-DHC over 24 h in Neuro2a cells at 1 μM drug.
We report here dose–response studies of three of these FDA-approved drugs, cariprazine, trazodone, and metoprolol, that cover a range of 7-DHC Z-scores, (Figure 2), along with investigations of these compounds as potential inhibitors of viral replication in cell culture. We used the enhanced green fluorescent protein (GFP)-encoding VSV (VSV-eGFP)29 to allow us to readily track the infection in cultured cells treated with various doses of the above drugs. We also investigated two compounds of interest, ifenprodil and AY9944, that are not approved for human use in the US. Ifenprodil was recently investigated in a Phase 2b study as a potential COVID-19 antiviral, and AY9944 has a history of use in cells and rodents as a DHCR7 inhibitor.30−32 In fact, earlier studies on DHCR7 inhibition as an antiviral strategy used only AY9944 to support DHCR7 efficacy,2,3 in one case showing increased survival for AY9944 treated mice exposed to VSV and H1N1.2
Figure 2.
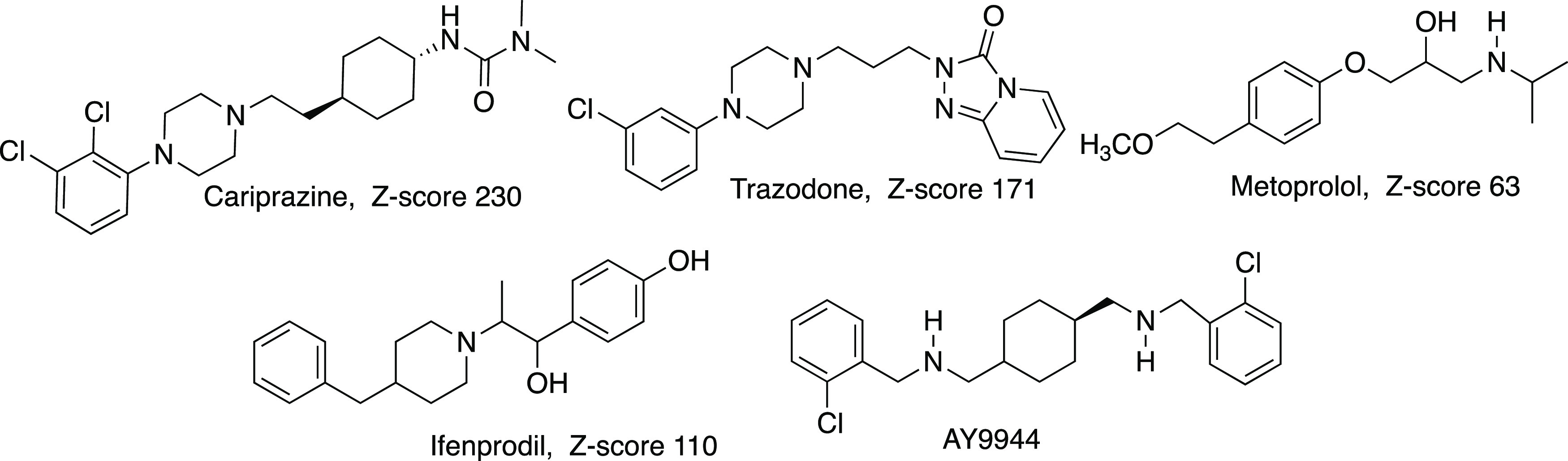
Structures and 7-DHC Z-scores for compounds studied. Cariprazine, trazodone, and metoprolol are FDA-approved drugs.
Results
Dose–Response Effects of Drugs on Cholesterol Biosynthesis
The effects of viral infections by VSV and the small molecules cariprazine, trazodone, metoprolol, ifenprodil, and AY9944 on cholesterol biosynthesis were studied in a neuroblastoma cell line, Neuro2a. Sterol levels in these cells are high enough that they can be conveniently measured in a 96-well format, and Neuro2a cellular sterol profiles correlate reasonably well with sterol profiles in the serum of individuals taking a drug, as do sterol profiles in tissues and fluids of exposed rodent.33,34 We have results similar to those reported here in Neuro2a and A549 cells, cultured cortical neurons, and human skin fibroblasts, but systematic studies in these cells have not yet been pursued.
An advantage found in the use of Neuro2a and the 96-well format was the fact that multiple conditions and exposures could be explored in a matter of days and weeks. Thus, the time course for changes in the level of the 12 sterols shown in Figure 1 was monitored by LC/MS (n > 6) for periods of time up to 24 h. The effect on the levels of the sterols shown in Figure 3 was observed after only 6 h of treatment, and perturbed sterol profiles were maintained throughout 24 h of drug exposure. The sterol levels shown in the figure are normalized with the cholesterol level set at 100% with no drug. The level of sterols in cells showed some variations from cell batch to batch (between 10 and 14 nmol/million cells), but the normalized sterol levels vs cholesterol/no drug were generally reproducible.
Figure 3.
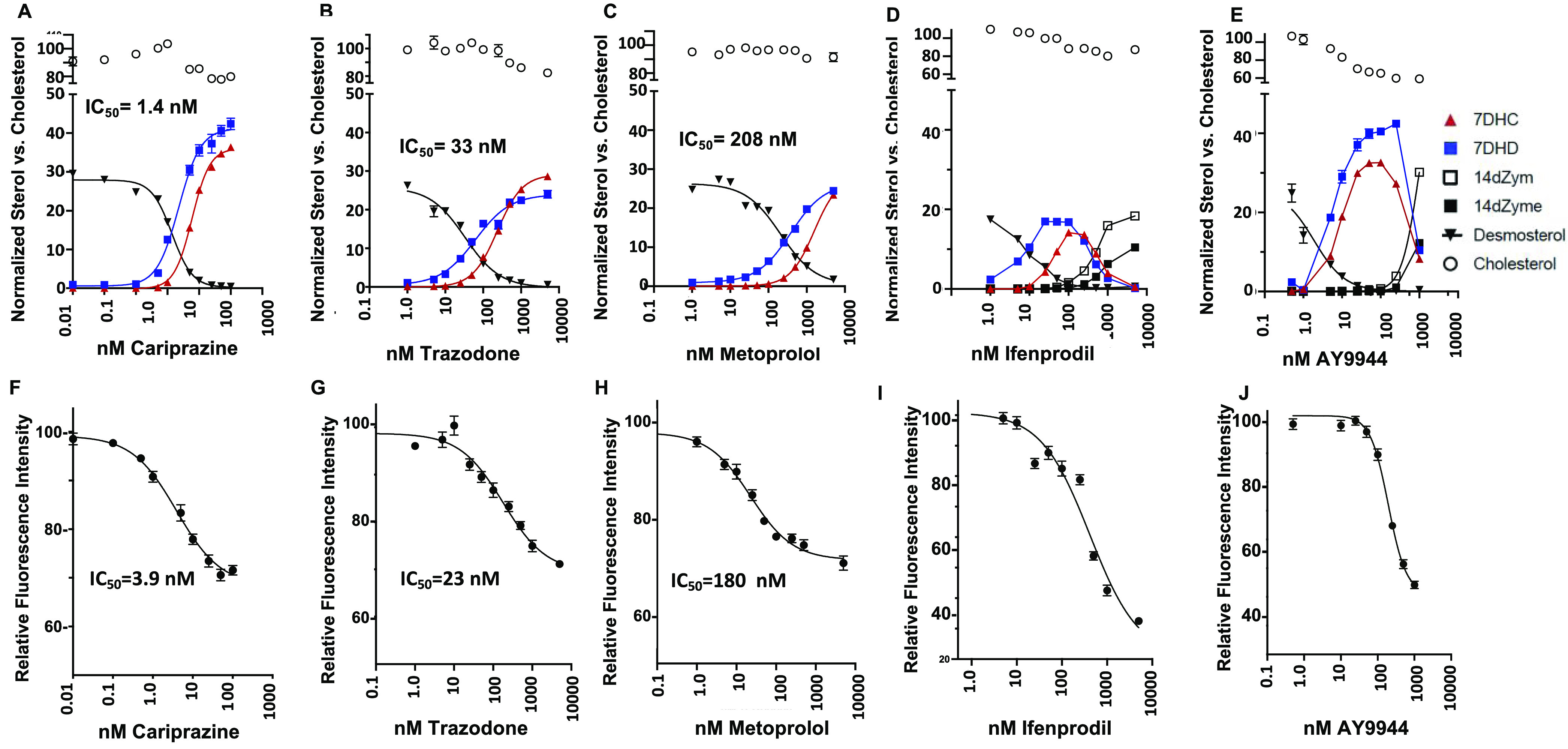
Dose–response of sterols and VSV-eGFP fluorescence after drug treatment of Neuro2a cells. (A–E) Sterols were assayed by LC/MS in nmol/million cells after a 24 h drug treatment and values presented here (n = 8) were normalized vs cholesterol, with the value for no drug set as 100%. Control levels of desmosterol in Neuro2a without drug are ∼30% of cholesterol. Only those sterols that undergo significant change as a result of drug treatment are shown. (F–J) Normalized fluorescence total integrated intensity of infected cells vs drug concentration. Cells were pretreated for 6 h with drug, followed by 0.1 MOI of VSV-eGFP for 1 h. After the removal of virus and further treatment with drug for 15 h, cells were fixed and assayed with an ImageXpress Pico instrument.
The three FDA-approved drugs, cariprazine, trazodone, and metoprolol, shown in Figure 3A–C, displayed relatively simple dose–responses that were confined to changes in desmosterol (Des), 7-dehydrodesmosterol (7-DHD), and 7-DHC. Good dose–response fits based on decreases in Des vs drug concentration showed potency of cariprazine (Z = 230, IC50 = 1.4 nM) that was greater than trazodone (Z = 171, IC50 = 33 nM), which was in turn greater than metoprolol (Z = 63, IC50 = 208 nM). Drugs generally affected levels of 7-DHD at lower concentrations than was the case for 7-DHC, and there were significant differences in the ratio of 7-DHD/7-DHC vs concentration for different drugs. This was likely due to differential effects of the drugs on DHCR7 and DHCR24, coupled with the dominant biosynthesis proceeding along the Bloch arm of the pathway in Neuro2a, see Figure 1. Decreases observed for desmosterol at low drug levels are consistent with the notion that the action of DHCR7 is dominant at these concentrations. DHCR24 still functions to convert desmosterol to cholesterol in these conditions, and steady-state levels of desmosterol decrease.
Ifenprodil and AY9944 treatments resulted in a more complex change in Neuro2a sterol profiles. Both of these compounds caused increases in 7-DHD and 7-DHC at concentrations between 1 and 10 nM, similar to the effect of cariprazine. In contrast to cariprazine, trazodone, and metoprolol, however, drug concentrations of ifenprodil and AY9944 above 100 nM caused a decrease in 7-DHD and 7-DHC, with a corresponding increase in two precursor sterols, 14-dehydrozymosterol (cholesta-8,14,24-trienol,14-dZym) and 14-dZyme (cholesta-8,14-dienol,14-dZym), see Figure 1 for structures. These results can be understood based upon the inhibition of DHCR7 at low drug concentrations with an effect on DHCR14 as concentrations exceed 100 nM.
Drug Effects on VSV Infections
We made use of VSV-eGFP29,35,36 to examine the effect of the drugs shown in Figure 2 on VSV infection in Neuro2a cells. These experiments were carried out in 96-well plates to explore a variety of conditions to find combinations of drug and virus that gave reproducible (n > 6) changes in VSV viral fluorescence and sterol profiles.
Neuro2a cells tolerated VSV-eGFP exposures when treated with different concentrations of drugs over extended time periods. In a typical experiment, cells were incubated with various drug concentrations for 6 h, followed by infection with 0.1 MOI of VSV-eGFP. After 1 h of VSV-eGFP exposure, the virus was replaced with medium containing drug, and incubations were continued for 15 h. Cells were then fixed, imaged for fluorescence, and counted (Hoechst) on an ImageXpress Pico instrument. Fluorescence levels and cell counts were monitored with virus alone or the combination of drug and virus, and the GFP total integrated fluorescence of infected cells was measured under these conditions as a function of drug concentration. Cell counts generally mirrored independent measures (alamarBlue) of cell viability.
Figure 3F–J shows the dose–responses of viral fluorescence for five of the compounds tested. The three FDA-approved drugs gave good dose–response fits with values of total integrated intensity that decreased at the highest drug concentrations to around 70% of the value for VSV-eGFP treatment alone. IC50s found for dose–response of viral fluorescence for these drugs were in the same order as the IC50s determined for changes in sterols. Thus, cariprazine had a (sterol IC50) = 1.4 nM vs (viral fluorescence IC50) = 3.9 nM, while the comparable values for trazodone were (sterol IC50) = 33 nM vs (viral IC50) = 23 nM, and for metoprolol, (sterol IC50) = 208 nM vs (viral IC50) = 180 nM. The viral response to treatments of infected cells with ifenprodil and AY9944, Figure 3I,J, showed a decrease of fluorescence values to 30–40% that of untreated cells, lower than the values found for the FDA-approved drugs (70%). Fitting the viral fluorescence data to a dose–response scheme gave results that were not as good for AY9944 and ifenprodil as those obtained for the FDA-approved drugs, a result that may be related to the complex sterol-concentration profiles for these compounds. Modest declines in cell counts were found for the highest concentrations of metoprolol, ifenprodil, and AY9944. Figures S2–S4 provide evidence that cell counts are a good read-out for viability. Figure 4A–F presents fluorescence images of Neuro2a cells infected with VSV-eGFP as described above, and Figure 4G,H shows immunoblots for GFP and the VSV-N viral protein as a function of cariprazine treatment.
Figure 4.
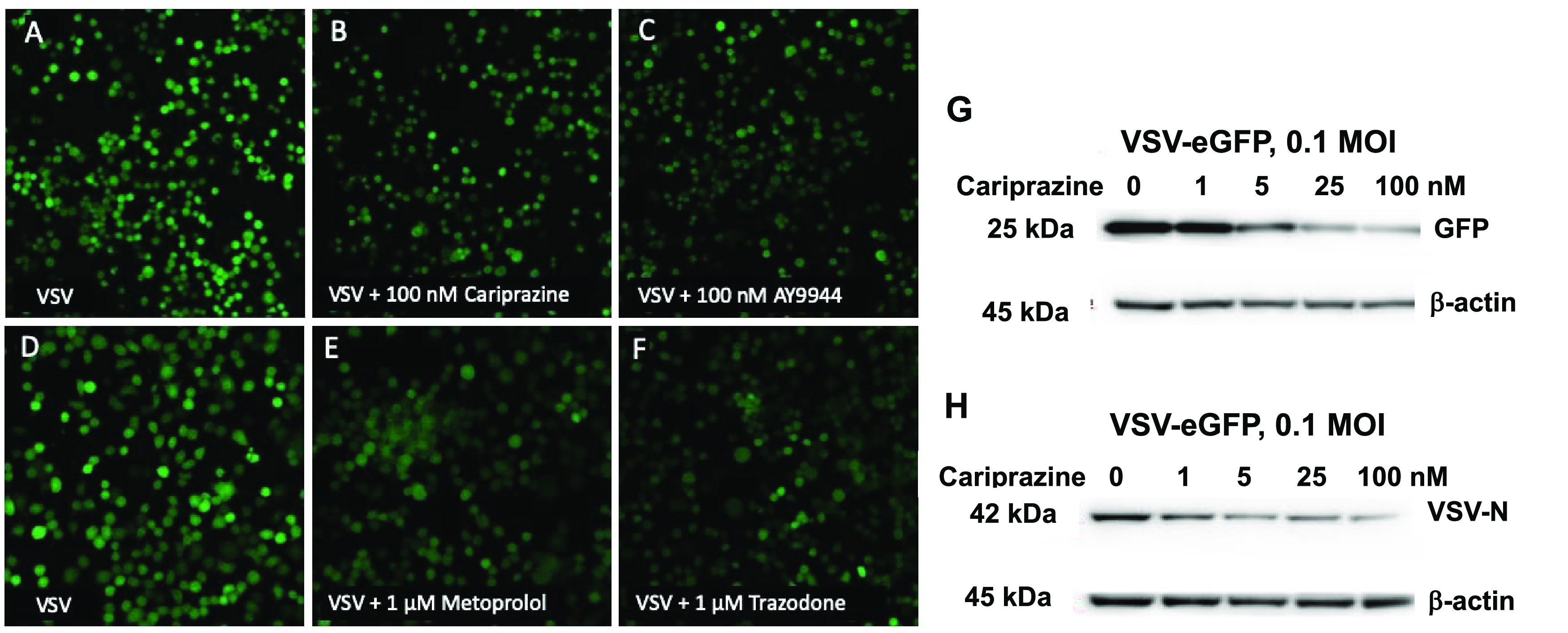
Fluorescent images and immunoblots of Neuro2a cells infected with VSV-eGFP in the presence of drugs. (A–C) Images (10×) of Neuro2a cells treated with 0.1 MOI VSV-eGFP, control vs VSV + 100 nM cariprazine or AY9944. (D–F) 20X images of Neuro2a treated with 0.1 MOI VSV-eGFP, control vs VSV + 1 μM metoprolol or trazodone. (G, H) Immunoblots of VSV-eGFP proteins (GFP and the VSV-N protein) vs β-actin isolated from cells with 0 to 100 nM cariprazine.
We note that Xiao et al. suggested that tamoxifen has antiviral properties similar to AY99442 and attributed this to its DHCR7 inhibitory activity. Other studies have indicated, however, that tamoxifen inhibits EBP (Δ7−Δ8 isomerase),37,38 and in our hands, tamoxifen did not affect 7-DHD and 7-DHC levels in Neuro2a, see Figure S4. Rather, levels of zymosterol and zymostenol increased at concentrations greater than 25 nM, supporting the notion that the main target of tamoxifen was EBP.39,40 The combination of tamoxifen and VSV-eGFP was also toxic to these cells under our standard conditions of incubation. Figure S4 shows that viral GFP fluorescence and cell counts both showed sharp declines at concentrations of tamoxifen over 100 nM, calling into question the antiviral activity suggested for this compound.2
Since the viral protein expression in infected cells was negatively affected by the drug treatments, we next examined if infectious viral progeny production was also similarly affected in the presence of the drugs. Following Neuro2a drug treatment of VSV-eGFP infected cells, supernatants were harvested, and VSV titers were determined by plaque assay. VSV titers decreased in a drug-dependent manner for all drugs, see Figure 5, with the effect of treatment on viral titers much more consequential than the fluorescence decreases shown for incubations of drugs and VSV-eGFP, Figure 3. Thus, the titer decreases at the highest doses were 8-fold for trazodone, 11-fold for cariprazine, and over 30-fold for both ifenprodil and AY9944. The viral titer response to drug concentration is also of interest, with potencies based on 50% titer reduction in the order trazodone (IC50 ∼ 150 nM) < AY9944 (IC50 ∼ 10 nM) = cariprazine (IC50 ∼ 10 nM) <ifenprodil (IC50 < 10 nM).
Figure 5.
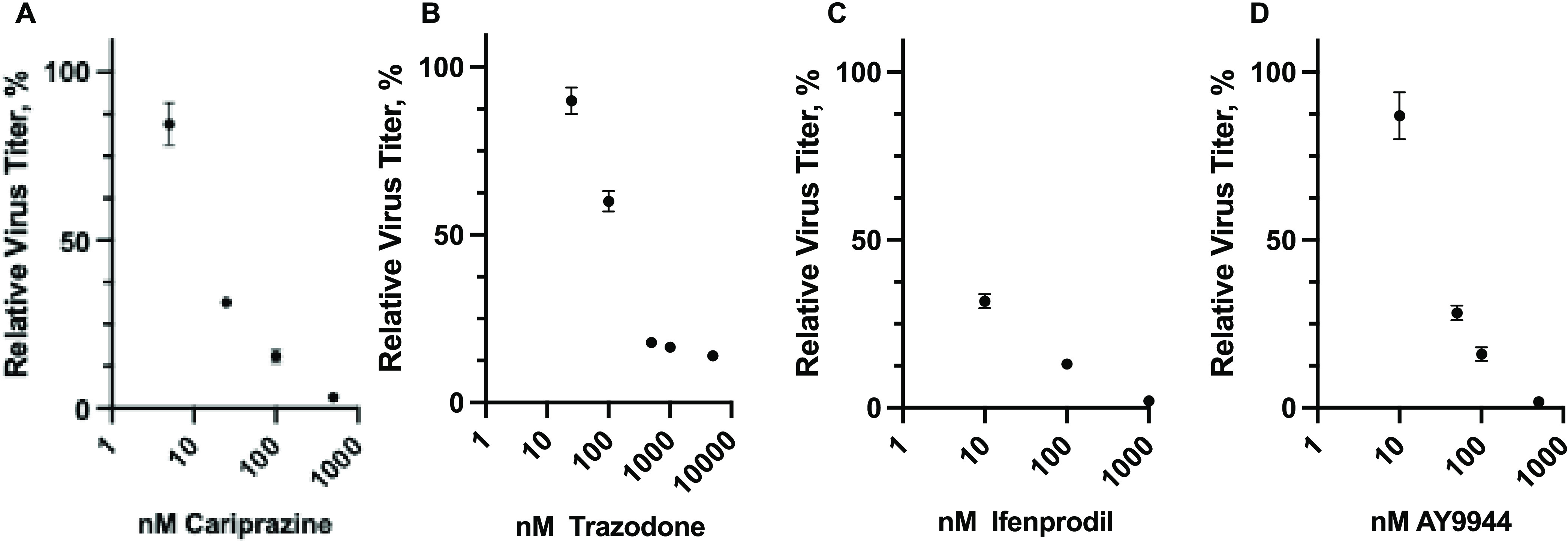
Neuro2a cells treated with drugs and infected with VSV-eGFP. Neuro2a cells were treated with drugs and 0.1 MOI VSV-eGFP, as described in Figure 5. Cell culture supernatants were collected, and viral titers (PFU/ml) were measured in BHK-21 cells. Viral titers (expressed as %) relative to the no-drug treatment control are shown for (A) Cariprazine, (B) Trazodone, (C) Ifenprodil, and (D) AY9944.
Discussion
Sterol Dose–Responses
There are over two dozen steps required to produce cholesterol from acetate, and drugs that modulate many of these steps and perturb sterol homeostasis are known. Statins target cholesterol formation at the first committed step of biosynthesis, essentially shutting down the formation of any isoprenoid products, but a number of drugs have been found that inhibit late-stage transformations in the process. Inhibitors of DHCR7, the last enzyme in the post-lanosterol stage, have been of interest since DHCR7 is linked to a neurodevelopmental disorder, Smith–Lemli–Opitz syndrome (SLOS).41,42 Over 200 pathogenic mutations of the gene that encodes DHCR7 have been reported,43−45 and a carrier frequency of up to 3% has been suggested.46 Individuals who inherit two pathogenic DHCR7 alleles have elevated levels of 7-DHC in tissues and fluids, a diagnostic feature of SLOS. A rodent model of SLOS, based on the treatment of pregnant rats with AY9944,31,32,47−49 has been developed with elevated levels of 7-DHC found in tissues and fluids in the pups from treated animals.
The suggestion2,3 that targeting DHCR7 with AY9944 eliminated infection of a number of viruses called attention to the potential involvement of DHCR7 in viral replication, and 7-DHC itself has been shown to protect cultured cells from VSV infection.2 This, and the fact that dozens of FDA-approved drugs cause increases in cellular levels of 7-DHC23−28 prompted us to re-examine the effects of a selection of “DHCR7 active” compounds in an established viral model.2,29,35,36 We selected five compounds for further study, three of which are FDA-approved drugs, cariprazine, trazodone, and metoprolol. Cariprazine, having one of the highest DHCR7 Z-scores of compounds tested, is marketed in the US to treat schizophrenia and bipolar depression. Trazodone is also a potent DHCR7 inhibitor. It is a serotonin receptor antagonist and reuptake inhibitor, prescribed for depression and off-label as a sleep aid over 20 million times a year. Metoprolol is a β-blocker, prescribed over 80 million times a year in the US for hypertension and angina. Based on our initial screening studies, metoprolol showed significant activity, although its potency (Z = 63) was significantly lower than any of the other compounds studied here.
The results of the study of the three FDA-approved drugs gave good dose–response fits that showed the same order of activities found in the sterol screening study (Figure 3A–C), found in viral fluorescence IC50s, with decreasing activity in the order metoprolol >trazodone >cariprazine (Figure 3F–H). In contrast, ifenprodil and AY9944 gave results consistent with inhibition of both DHCR7 and DHCR14 (Figure 3D–E), a reductase enzyme earlier in the biosynthetic cascade, Figure 1. The reductase enzymes DHCR7 and DHCR14 show a high level of sequence homology,50 along with lamin-B receptor (LBR). DHCR14 and LBR both reduce the sterol Δ14 double bond, the former being a tunable protein, the latter constitutively,50 so the fact that a molecule can inhibit the transformations of each of these reductases is not surprising. Indeed, AY9944, haloperidol, and ziprasidone have been previously reported to inhibit both DHCR7 and DHCR14 in HL-60 cells23,51,52 and cortical neurons.53 As shown in Figure 3, the two-stage inhibition of DHCR7 and DHCR14 by AY9944 and ifenprodil give dose–response maxima for 7-DHC and 7-DHD at approximately 100 nM in Neuro2a cells. Both of these compounds affect DHCR7 at concentrations similar to cariprazine, so 1–10 nM of the three compounds give similar sterol profiles. But above 100 nM of ifenprodil or AY9944, cellular levels of 14-dZym and 14-dZyme increase at the expense of 7-DHC and 7-DHD.
Neuro2a-VSV Infections
Cariprazine, trazodone, and metoprolol caused a decrease of the VSV-eGFP total integrated fluorescence of infected cells to about 70% of the value of untreated cells after 15 h of exposure to 0.1 MOI VSV-eGFP. Cell morphology appeared to be unchanged after treatment as well, showing no evidence of significant toxicity. AY9944 and ifenprodil also reduced the GFP fluorescence of infected cells compared to untreated cells, with fluorescence levels decreasing to roughly 40% of the values found for untreated cells.
Plaque-forming assays gave results that were consistent with reduced VSV infection at concentrations above the IC50 for cariprazine, trazodone, ifenprodil, and AY9944, as were immunoblots of GFP and the viral N protein in treated cells, see Figures 4 and 5. The fact that the PFU studies showed a reduction in virion production up to 30-fold for ifenprodil and AY9944 during a 15 h incubation is noteworthy since this is the standard commonly used to determine drug efficacy. In this regard, it is notable that both the fluorescence assay, Figure 3I,J, and PFU studies, Figure 5C,D, show greater antiviral activity for ifenprodil and AY9944, the dual enzyme inhibitors.
Ifenprodil is of particular interest because of the disclosure of topline findings of a Phase 2b COVID-19 trial (04/26/2021, Algernon Pharmaceuticals). The findings for hospitalized patients of this trial included improved all-cause mortality, return to normal oxygen levels, and time spent in ICU in the 20 mg dose arm of the trial. While these data showed promise, the efficacy of the drug was called into question by the disclosure that higher dose ifenprodil treatments showed no change in clinical outcomes. A number of other reports of small molecule antivirals have been made that may now be interpreted based on their effect on sterol homeostasis. A recent screen for antiviral activity of Niemann–Pick-C disease mimetics notably identified several active compounds that are common to our screens.54 Claims of antiviral properties have also been made for amiodarone55,56 and clomiphene57 compounds that affect steps in biosynthesis other than DHCR7 and DHCR14. Recent disclosures of antiviral effects of the psychotropic approved drug fluvoxamine prompted us to examine this compound in our sterol and VSV-eGFP fluorescence screens, but we found no activity of the compound on GFP fluorescence or sterol levels at concentrations below 1000 nM, with a very small increase in zymosterol, zymostenol, and 7-DHD at 5000 nM.
Sterols and Viral Replication
The results of our efforts support proposals that link sterol biosynthesis inhibition with antiviral activity.2,3 We emphasize that the DHCR7 inhibitors studied here affect cellular levels of 7-DHD, 7-DHC, desmosterol, and cholesterol, and it may be premature to assign “activity” at this juncture. Indeed, the fact that DHCR14 is also a target for some of the compounds studied is an additional complication, with 14-dZym and 14-dZyme becoming prominent cellular constituents at higher concentrations of ifenprodil and AY9944. It is nevertheless notable that 7-DHC itself has been observed to have antiviral effects against VSV in cell culture.2 It therefore seems reasonable to ask how the introduction of sterols like 7-DHD, 7-DHC, 14-dZym, and 14-dZyme into the cellular lipidome may affect viral infections. The lipid compositions of VSV58−62 closely resemble that of its infected host plasma membranes, with cholesterol making up over 50% of the envelope lipids.58,63 The pairing of cell and virus may be an important determinant in drug efficacy, and VSV-Neuro2a may be a good pair since Neuro2a has active cholesterol biosynthesis and VSV has high sterol levels in its envelope. At any rate, virion particle stability, survival, and infectivity may be compromised if the lipid envelope is derived from “DHCR7 and DHCR14 compromised” cells that are producing oxidatively vulnerable 7-DHC, 7-DHD, 14-dZym, and 14-dZyme instead of cholesterol and desmosterol.
Most enveloped viruses enter cells by direct fusion of their envelope with the plasma membrane or through endocytic entry and exit cells by exocytosis or budding from the plasma membrane. Cholesterol plays an important role in all of these steps since membrane bilayer structure and stability are critically important to these processes.64−69 Likewise, many viruses use cholesterol-rich plasma membrane lipid raft microdomains to bud from the infected host cells,69−71 and cholesterol is important for intracellular trafficking processes for the assembly of replication complexes and for viral genome replication.72,73
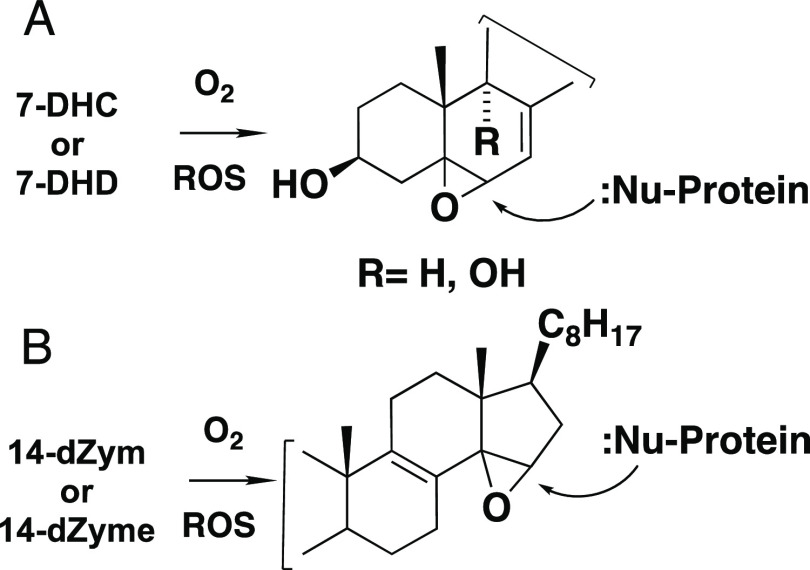
Conditions that alter homeostasis3,74,75 and introduce oxidatively reactive sterols into the cellular lipidome have the potential to perturb critical steps in the infection process. Indeed, there is evidence that cellular 7-DHC gives highly reactive electrophilic oxysterols upon peroxidation, see Figure 6A.76−79 These species adduct to several lipid synthesis and transport proteins,80 prominent among these proteins being constituents of COPI, the transport complex81−83 involved in traffic between the Golgi and endoplasmic reticulum and ACSL4, an acyl-CoA synthetase. ACSL4 enriches cellular membranes with long-chain PUFA fatty esters84,85 and shapes cellular lipid composition, influencing cellular sensitivity to ferroptosis, a form of regulated cell death.12−15 7-DHC-derived adducts of FASD2 and LPCAT3, other lipid synthesis proteins linked to ferroptosis,20,84 have also been observed.80 There is reason to believe that 14-dehydrosterols like 14-dZym and 14-dZyme would also form reactive sterol electrophiles under peroxidation conditions, although the free radical chemistry of these sterols has not been explored in detail. Conjugated dienes like those present in these structures typically undergo peroxyl radical addition reactions, giving reactive allylic epoxide electrophiles, as shown in Figure 6B.86,87
Figure 6.
Sterol-derived electrophiles. (A) Epoxide formed from 7-DHC and 7-DHD peroxidation. (B) Predicted 14-dZyme-derived electrophile. ROS = reactive oxygen species.
It is of interest that changes in the Neuro2a sterol profile occurred after VSV treatment alone, see Figures S5 and S6. The effect on desmosterol was dramatic, with levels falling over 50% after 24 h of exposure to 0.1 MOI VSV. The decrease of cholesterol to 60% of control levels was also significant under these conditions. But the increases in cellular lathosterol and dehydrolathosterol found as a result of infection indicate a response to viral exposure that is consistent with disruption at a late step in the biosynthesis, in this case, at SC5D. 7-DHD and 7-DHC levels are very low in Neuro2 cells, and their steady-state levels may not reflect any changes of DHCR7 activity that result from infection.
Finally, we emphasize the importance of the earlier AY9944 antiviral studies.2,3 Our work shows that with VSV in Neuro2a cells, the effect can be generalized beyond AY9944 and underlines the fact that multiple sterol biosynthesis enzymes may be affected by the inhibitors. We also emphasize the unusual vulnerability of sterols that are substrates for DHCR7 and DHCR14 to free radical chain peroxidation and oxysterol formation. The effect of these conjugated diene sterols on virion stability, intracellular trafficking processes, and the protection of membrane phospholipids from peroxidation all rely on the fact that if present in the lipidome, they will dominate radical chain reactions.
Materials and Methods
Chemicals
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich Co (St. Louis, MO). HPLC grade solvents were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). The derivatizing reagents 2-methyl-6-nitrobenzoic anhydride and N,N-dimethylglycine were purchased from Combi-Blocks (San Diego, CA). Trazodone, cariprazine, metoprolol, and ifenprodil were obtained from Selleck Chemicals (Houston, TX). AY9944 was synthesized by the Vanderbilt University Synthesis Core. All deuterated sterol standards were synthesized as previously described (34) and are available from Kerafast, Inc. (Boston, MA).
Cell Cultures, VSV Infection, ImageXpress, and Preparation for LC-MS/MS
Mouse neuroblastoma cell line Neuro2a was purchased from ATCC (Rockville, MD). The Neuro2a cells were maintained in EMEM supplemented with 10% FBS and kept at 37 °C in a 5% CO2 cell culture incubator. Cells were subcultured once a week, and the culture medium was changed every 2 days. Most experiments were done with the cells plated in 96-well plates (for cell viability, sterol analysis, VSV infection). To analyze the effects of compounds on cellular sterol synthesis, the cultures were grown in a defined medium without cholesterol and without lipids using EMEM with N2 supplement (Gibco, Thermo Fisher Scientific). At the end point of the incubation, Hoechst dye was added to all wells in the 96-well plate, and the total number of cells was counted using an ImageXpress Pico and cell counting algorithm in CellReporterXpress. After removing the medium, wells were rinsed with 1× PBS, and 10 μL of butylated hydroxytoluene-/triphenylphosphine (BHT/PPh3) was added to each well, and the plates were stored at −80 °C for sterol analysis. All samples were analyzed within 2 weeks of freezing. Sterol levels were analyzed in individual wells, and, for most experiments, cellular levels correspond to the average of 8–12 technical replicates. Sterols in individual wells were normalized to the total cell count within the well. VSV infections: VSV-eGFP was prepared as previously described.29 For measuring the effects of VSV on sterol synthesis in cells, Neuro2a cells were plated in a 96-well plate in EMEM plus 10% FBS. Several hours after cells adhered to the plate, the medium was changed into EMEM plus N2 supplement, and cells were kept in the cell culture incubator overnight. For each experiment, several 96-well plates were prepared. One plate was used to count cells just before VSV infection to calculate the multiplicity of infection (MOI). Cells were infected with different MOI (0.001, 0.01, 0.1) of VSV for 1 h, at which time the medium with VSV was removed, fresh medium was added, and the cultures were incubated for different times, as shown in the Results section and figures. At the end of the experiment, Hoechst was added to the plates, followed by imaging in ImageXpress Pico. Medium was removed, plates were rinsed with 1× PBS and 70% EtOH with butylated hydroxytoluene, and triphenylphosphine (BHT/ PPh3) was added to the plates, followed by incubation at 60 °C for 2 min. The plates were stored at −80 °C for sterol analysis. Cell Viability. In addition to cell counting, alamarBlue was used to monitor cell viability. At the end point, alamarBlue was added to individual wells in a 96-well plate, and the plates were incubated for 3 h at 37 °C in the 5% CO2 cell culture incubator. Fluorescence was read on a SpectraMax fluorescence microplate reader (Molecular Devices, San Jose, CA) with excitation/emission 530/590 nm.
For the analysis of drug effects on VSV infections, cells were plated in a 96-well plate in EMEM plus 10% FBS. After overnight culture, the medium was replaced with EMEM plus N2 supplement plus/minus different concentrations of drugs and cultures incubated for 6 h at 37 °C in a 5% CO2 cell culture incubator. One extra plate was used to count the total number of cells for accurate calculation of VSV MOI. The cultures were infected for 1 h with VSV MOI 0.1. After 1 h, the medium was completely replaced with fresh EMEM plus N2 supplement plus/minus pharmaceuticals, and cultures were kept for additional 14 h at 37 °C in a 5% CO2 cell culture incubator. At the end of the experiment, Hoechst dye was added to the plates, and plates were imaged on ImageXpress Pico using 10X and 20X Plan Fluor objective and GFP and DAPI channels. VSV-eGFP fluorescence and nuclei were imaged using the Multiwavelength Cell Scoring application within CellReporterXpress. Total integrated intensity measurement was used for analyzing the effects of compounds on VSV infection. The effects of compounds on VSV infection were analyzed in Neuro2a grown for different lengths of time: 15, 18, and 24 h. The results were similar for all time points. Each experiment was repeated at least three times. Data are shown for one representative experiment.
Plaque Assays
BHK-21 cells were grown in 12- or 6-well plates to 90% confluency. Cells were infected in duplicates, with serial 10-fold dilutions of cell culture supernatants. After an initial adsorption of 1 h, the inoculum was aspirated out, and cells were overlaid with 1.5 mL of medium containing 1% low-melting-point agarose and incubated for 15–18 h. After the incubation period, cells were fixed with 1 mL of 10% formaldehyde for 1 h at room temperature. The agarose plugs were removed, and the cells were stained for 10–15 min with 0.01% crystal violet in 30% methanol. Cells were washed with water and air-dried. The plaques were counted, and titers were determined.
Western Blotting
For western blotting, Neuro2a cells were grown in 60 mm dishes in a defined medium, as described above. The cells were infected with VSV-eGFP as above. At the end of the experiment, cells were rinsed twice with 1× PBS, scraped, and pelleted by centrifugation at 4 °C. The cell pellets were homogenized by sonication in ice-cold RIPA lysis buffer (VWR International, Radnor, PA) plus phosphatase inhibitors (Sigma-Aldrich) and protease inhibitors (Thermo Fisher Scientific) and incubated on ice for 30 min. To clear the lysates, the samples were spun at 14,000g at 4 °C for 5 min to pellet the debris. The total protein of the supernatant was quantified with Pierce BCA assay (Thermo Fisher Scientific), reading absorbance at 562 nm using a SpectraMax Plus 384 (Molecular Devices). Equal amounts of protein from each sample were mixed with reducing reagent and loading buffer and heated to 70 °C for 10 min. Proteins were separated on NuPAGE 4–12% Bis-Tris protein gels (Thermo Fisher Scientific). Prestained protein was used to evaluate the molecular weight. The Bio-Rad Mini Trans-Blot Electrophoretic Transfer Cell was used for the electrophoretic transfer using the polyvinylidene difluoride membranes (Immobilon-P PVDF Membrane, Sigma-Aldrich) and transfer buffer (25 mM Tris-Cl, 192 mM glycine and 20% (v/v) methanol (pH 8.3)). Following transfer, PVDF membranes were blocked in 5% milk in TBS (50 mM Tris-Cl, 150 mM NaCl, pH 7.5) with 0.05% Igepal (Spectrum Chemical, New Brunswick, NJ) and incubated in primary antibodies (anti-VSV antibody) or anti-GFP (Santa Cruz) antibody overnight at +4 °C and secondary antibodies at room temperature for 1 h. Western blots were developed using Azure’s Radiance substrate, imaged on Azure C300 with the cSeries Capture software, and saved as TIFF images (Azure Biosystems).
Extraction of Sterols
A stock solution of deuterated standards was made containing 300 μM d7-Chol, 30 μM d6-Lan, d7-dHLan, and 3 μM d7–7-DHC, d7-8-DHC, d6-Des, d6-7-DHD, d6-8-DHD, d7-Lath, d6-DHL, d7-Zyme, d6-Zym, d7-14d-Zyme, d6-14d-Zym in MeOH. The stock solution of standards contained 1% (v/v) Et3N and an antioxidant mixture (BHT and PPh3) to prevent isomerization and/or oxidation. The antioxidant mixture contained BHT (2.5 mg/mL) and PPh3 (1 mg/mL) in EtOH. N.B. 7-DHC and 7-DHD undergo air oxidation readily when concentrated in the atmosphere. Appropriate care should be taken, or samples and isotopically labeled standards will be contaminated with oxidation products.
At the conclusion of the experiment, the plates were treated with the antioxidant mixture (10 μL/well) and stored at −80 °C for sterol analysis. Plates containing VSV were treated with 70% EtOH with BHT/PPh3 (100 μL), followed by incubation at 60 °C for 2 min to kill the virus. The plates were stored at −80 °C for sterol analysis. To each well of the 96-well plate was added the standard mixture (10 μL) and MeOH (100 μL). The plate was agitated on a shaker for 20 min and then allowed to rest to settle cell debris. The MeOH was transferred to a 96-well MS analysis plate, dried under vacuum, and derivatized as described below.
Derivatization and LC-MS/MS Analysis of Sterols
The derivatizing reagent was freshly prepared with 2-methyl-6-nitrobenzoic anhydride (20 mg), N,N-dimethylglycine, DMG (14 mg), dimethylaminopyridine, DMAP (6 mg), and triethylamine, Et3N (0.1 mL) in anhydrous CHCl3 (0.9 mL). To each sample was added derivatizing reagent (100 μL) and the mixture was allowed to react at room temperature for 30 min. The samples were dried under vacuum and subsequently dissolved in MeOH (100 μL) for LC-MS/MS analysis. Samples were analyzed on an Acquity UPLC system equipped with ANSI-compliant well plate holder. The sterols (10 μL injection) were analyzed on an Agilent Poroshell EC-C18 (10 cm × 2.1 mm, 1.9 μm) with CH3CN/MeOH/H2O, 70:25:5 (0.01% (v) formic acid, 1 mM NH4OAc) mobile phase at a column temperature of 40 °C. The flow rate was 400 μL/min for 11.5 min and then ramped to 600 μL/min at 11.6 min with a total run time of 16 min. A TSQ Quantum Ultra tandem mass spectrometer (Thermo Fisher) was used for MS detections, and data were acquired with a Finnigan Xcalibur software package. Selected reaction monitoring (SRM) of the DMG derivatives was acquired in the positive ion mode using electrospray ionization (ESI). MS parameters were optimized using DMG-Chol and were as follows: spray voltage at 4500 V, capillary temperature at 300 °C, auxiliary nitrogen gas pressure at 55 psi, and sheath gas pressure at 60 psi. Collision energy (CE) was optimized for each sterol under a collision gas pressure of 1.5 mTorr. The monitored transitions are listed in the Supporting Information (Table S1). Endogenous sterol levels were quantified based on the known matched deuterated standard amount and then normalized to cell count.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 8 for Windows. Unpaired two-tailed t-tests were performed for individual comparisons between two groups. Welch’s correction was employed when the variance between the two groups was significantly different. One-Way ANOVA analyses were performed for comparisons between three or more groups. The p-values for statistically significant differences are highlighted in the figure legend.
Acknowledgments
This work was supported by The National Institutes of Health NIMH R01 MH110636 (KM and NAP), R01 MH067234 (KM), and R01 HD064727 (NAP).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.2c00051.
MS parameters for sterol analysis (Table S1); infection protocol for drug and VSV-eGFP treatments in Neuro2a cells (Figure S1); comparison of viability assay and cell counts for drug treatments (Figure S2); viability assay after drug treatment and VSV infection (Figure S3); comparison of viability assay and cell counts of VSV infections (Figure S4); effects VSV-eGFP on Sterol levels (Figure S5); time course of sterol profiles during Neuro2a infection with VSV-eGFP (Figure S6); effects of tamoxifen on Sterols and VSV (Figure S7); fluorescent images for ifenprodil treatments and VSV-eGFP infection (Figure S8); and compound purity information (Figure S9) (PDF)
Author Present Address
# Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, United States
Author Contributions
∇ Z.K. and K.A. contributed equally to this work. N.A.P. proposed experiments, first draft, edits, and the final version of the manuscript; multiple versions of figures and statistics; K.A.T. performed the development and verification of DMG method; synthesis of sterol and oxysterol standards, LC-MS for sterols in Neuro2a cells (control vs drug treated vs VSV infected); Z.K. performed cell cultures, VSV infection, ImageXpress analysis; KM performed strategy, edits, GraphPad analysis; TG-M performed LC-MS analysis in Neuro2a cells; MB performed western blotting, PFU; A.P. and A.P. helped with VSV, PFU; H.-Y.K. performed western blotting; GraphPad analysis. The final version was read and approved by all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Nes W. D. Biosynthesis of cholesterol and other sterols. Chem. Rev. 2011, 111, 6423–6451. 10.1021/cr200021m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J.; Li W.; Zheng X.; Qi L.; Wang H.; Zhang C.; Wan X.; Zheng Y.; Zhong R.; Zhou X.; Lu Y.; Li Z.; Qiu Y.; Liu C.; Zhang F.; Zhang Y.; Xu X.; Yang Z.; Chen H.; Zhai Q.; Wei B.; Wang H. Targeting 7-Dehydrocholesterol Reductase Integrates Cholesterol Metabolism and IRF3 Activation to Eliminate Infection. Immunity 2020, 52, 109–122. 10.1016/j.immuni.2019.11.015. [DOI] [PubMed] [Google Scholar]
- Rodgers M. A.; Villareal V. A.; Schaefer E. A.; Peng L. F.; Corey K. E.; Chung R. T.; Yang P. L. Lipid Metabolite Profiling Identifies Desmosterol Metabolism as a New Antiviral Target for Hepatitis C Virus. J. Am.Chem. Soc. 2012, 134, 6896–6899. 10.1021/ja207391q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti A. W.; Korytowski W. Cholesterol as a natural probe for free radical-mediated lipid peroxidation in biological membranes and lipoproteins. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2016, 1019, 202–209. 10.1016/j.jchromb.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti A. W.; Korytowski W. Cholesterol Hydroperoxide Generation, Translocation, and Reductive Turnover in Biological Systems. Cell Biochem. Biophys. 2017, 75, 413–419. 10.1007/s12013-017-0799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotti A. W. Mechanisms of lipid peroxidation. J. Free Radicals Biol. Med. 1985, 1, 87–95. 10.1016/0748-5514(85)90011-x. [DOI] [PubMed] [Google Scholar]
- Girotti A. W. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res. 1998, 39, 1529–1542. 10.1016/S0022-2275(20)32182-9. [DOI] [PubMed] [Google Scholar]
- Xu L.; Davis T. A.; Porter N. A. Rate Constants for Peroxidation of Polyunsaturated Fatty Acids and Sterols in Solution and in Liposomes. J. Am. Chem. Soc. 2009, 131, 13037–13044. 10.1021/ja9029076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Xu L.; Shelton R.; Porter N. A. Biological activities of 7-dehydrocholesterol-derived oxysterols: implications for Smith-Lemli-Opitz syndrome. J. Lipid Res. 2010, 51, 3259–3269. 10.1194/jlr.M009365. [DOI] [PMC free article] [PubMed] [Google Scholar]; Epub 2010/08/13.
- Porter N. A.; Xu L.; Pratt D. A. Reactive Sterol Electrophiles: Mechanisms of Formation and Reactions with Proteins and Amino Acid Nucleophiles. Chemistry 2020, 2, 390–417. 10.3390/chemistry2020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberson C. R.; Muchalski H.; McDuffee K. B.; Tallman K. A.; Xu L.; Porter N. A. Propagation rate constants for the peroxidation of sterols on the biosynthetic pathway to cholesterol. Chem. Phys. Lipids 2017, 207, 51–58. 10.1016/j.chemphyslip.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J.; Lemberg K. M.; Lamprecht M. R.; Skouta R.; Zaitsev E. M.; Gleason C. E.; Patel D. N.; Bauer A. J.; Cantley A. M.; Yang W. S.; Morrison B.; Stockwell B. R. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. S.; SriRamaratnam R.; Welsch M. E.; Shimada K.; Skouta R.; Viswanathan V. S.; Cheah J. H.; Clemons P. A.; Shamji A. F.; Clish C. B.; Brown L. M.; Girotti A. W.; Cornish V. W.; Schreiber S. L.; Stockwell B. R. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell 2014, 156, 317–331. 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. S.; Kim K. J.; Gaschler M. M.; Patel M.; Shchepinov M. S.; Stockwell B. R. Peroxidation of Polyunsaturated Fatty Acids by Lipoxygenases Drives Ferroptosis. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E4966–E4975. 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.; Pratt D. A. The chemical basis of ferroptosis. Nat. Chem. Biol. 2019, 15, 1137–1147. 10.1038/s41589-019-0408-1. [DOI] [PubMed] [Google Scholar]
- Angeli J. P. F.; Freitas F. P.; Nepachalovich P.; Puentes L.; Zilka O.; Inague A.; Lorenz S.; Kunz V.; Nehring H.; da Silva T. N. X.; Chen Z.; Doll S.; Schmitz W.; Imming P.; Miyamoto S.; Klein-Seetharaman J.; Kumar L.; Genaro-Mattos T. C.; Mirnics K.; Meierjohann S.; Kroiss M.; Weigand I.; Bommert K.; Bargou R.; Garcia-Saez A.; Pratt D.; Fedorova M.; Wehmann A.; Horling A.; Bornkamm G.; Conrad M.. 7-Dehydrocholesterol is an endogenous suppressor of ferroptosis Res. Square 2021, (Oct 6), 10.21203/rs.3.rs-943221/v1. [DOI]
- Hosakote Y. M.; Liu T. F.; Castro S. M.; Garofalo R. P.; Casola A. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am. J. Respir. Cell Mol. Biol. 2009, 41, 348–357. 10.1165/rcmb.2008-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva D. A.; de Molina M. C. R.; Rocchetta I.; Gerhardt E.; Coulombié F. C.; Mersich S. E. Oxidative Stress in Vero Cells Infected with Vesicular Stomatitis Virus. Intervirology 2006, 49, 294–298. 10.1159/000094245. [DOI] [PubMed] [Google Scholar]
- Martín-Fernández M.; Aller R.; Heredia-Rodríguez M.; Gómez-Sánchez E.; Martínez-Paz P.; Gonzalo-Benito H.; Prada LS.-d.; Gorgojo Ó.; Carnicero-Frutos I.; Tamayo E.; Tamayo-Velasco Á. Lipid peroxidation as a hallmark of severity in COVID-19 patients. Redox Biol. 2021, 48, 102181 10.1016/j.redox.2021.102181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane D.; Hayashi Y.; Matsumoto M.; Nakanishi H.; Imagawa H.; Kohara M.; Lemon S. M.; Ichi I. FADS2-dependent fatty acid desaturation dictates cellular sensitivity to ferroptosis and permissiveness for hepatitis C virus replication. Cell Chem. Biol. 2021, 29, 799–810.e4. 10.1016/j.chembiol.2021.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.-p.; Joshua B.; Jin N.-y.; Du S.-w.; Li C. Ferroptosis in viral infection: the unexplored possibility. Acta Pharmacol Sin. 2021, 43, 1905–1915. 10.1038/s41401-021-00814-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers M. A.; Saghatelian A.; Yang P. L. Identification of an Overabundant Cholesterol Precursor in Hepatitis B Virus Replicating Cells by Untargeted Lipid Metabolite Profiling. J. Am. Chem. Soc. 2009, 131, 5030–5031. 10.1021/ja809949r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfrán-Duque A.; Casado M. E.; Pastor O.; Sanchez-Wandelmer J.; Pena Gdl.; Lerma M.; Mariscal P.; Bracher F.; Lasuncion M. A.; Busto R. Atypical antipsychotics alter cholesterol and fatty acid metabolism in vitro. J. Lipid Res. 2013, 54, 310–324. 10.1194/jlr.m026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. Y.; Korade Z.; Tallman K. A.; Liu W.; Weaver C. D.; Mirnics K.; Porter N. A. Inhibitors of 7-Dehydrocholesterol Reductase: Screening of a Collection of Pharmacologically Active Compounds in Neuro2a Cells. Chem. Res. Toxcol. 2016, 29, 892–900. 10.1021/acs.chemrestox.6b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korade Z.; Kim H.-YH.; Tallman K. A.; Liu W.; Koczok K.; Balogh I.; Xu L.; Mirnics K.; Porter N. A. The Effect of Small Molecules on Sterol Homeostasis: Measuring 7-Dehydrocholesterol in Dhcr7-Deficient Neuro2a Cells and Human Fibroblasts. J. Med. Chem. 2016, 59, 1102–1115. 10.1021/acs.jmedchem.5b01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Tallman K. A.; Allen L. B.; Anderson A.; Mirnics K.; Korade Z.; Porter N. A. Dichlorophenyl piperazines, including a recently-approved atypical antipsychotic, are potent inhibitors of DHCR7, the last enzyme in cholesterol biosynthesis. Toxicol. Appl. Pharmacol. 2018, 349, 21–28. 10.1016/j.taap.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wages P. A.; Kim H. H.; Korade Z.; Porter N. A. Identification and characterization of prescription drugs that change levels of 7-dehydrocholesterol and desmosterol. J Lipid Res. 2018, 59, 1916–1926. 10.1194/jlr.M086991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horling A.; Müller C.; Barthel R.; Bracher F.; Imming P. A new class of selective and potent 7-dehydrocholesterol reductase inhibitors. J. Med. Chem. 2012, 55, 7614–7622. 10.1021/jm3006096. [DOI] [PubMed] [Google Scholar]
- Das S. C.; Nayak D.; Zhou Y.; Pattnaik A. K. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 2006, 80, 6368–6377. 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux C.; Wolf C.; Mulliez N.; Gaoua W.; Cormier V.; Chevy F.; Citadelle D. Role of cholesterol in embryonic development. Am. J. Clin. Nutr. 2000, 71, 1270s–9s. 10.1093/ajcn/71.5.1270s. [DOI] [PubMed] [Google Scholar]; Epub 2000/05/09
- Fliesler S. J.; Bretillon L. The ins and outs of cholesterol in the vertebrate retina. J. Lipid Res. 2010, 51, 3399–3413. 10.1194/jlr.R010538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliesler S. J.; Peachey N. S.; Herath K. B.; Hines K. M.; Weinstock N. I.; Rao R. S.; Xu L. Prevention of Retinal Degeneration in a Rat Model of Smith-Lemli-Opitz Syndrome. Sci. Rep. 2018, 8, 1286 10.1038/s41598-018-19592-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Klingelsmith K. B.; Allen L. B.; Anderson A.; Tallman K. A.; Porter N. A.; Korade Z.; Mirnics K. Sterol Biosynthesis Inhibition in Pregnant Women Taking Prescription Medications. ACS Pharmacol. Transl. Sci. 2021, 4, 848–857. 10.1021/acsptsci.1c00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genaro-Mattos T. C.; Anderson A.; Allen L. B.; Tallman K. A.; Porter N. A.; Korade Z.; Mirnics K. Maternal cariprazine exposure inhibits embryonic and postnatal brain cholesterol biosynthesis. Mol. Psych. 2020, 25, 2685–2694. 10.1038/s41380-020-0801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D.; Das A.; Dinh P. X.; Subramaniam S.; Nayak D.; Barrows N. J.; Pearson J. L.; Thompson J.; Kelly D. L.; Ladunga I.; Pattnaik A. K. RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 19036–19041. 10.1073/pnas.1113643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.; Panda D.; Nayak D.; Pattnaik A. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged m protein allows dynamic imaging of m protein and virus uncoating in infected cell. J. Virol. 2009, 83, 2611–2622. 10.1128/JVI.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T.; Hassan A.; Thompson B. M.; McDonald J. G.; Wang J.; Li X. Structural basis for human sterol isomerase in cholesterol biosynthesis and multidrug recognition. Nat. Commun. 2019, 10, 2452 10.1038/s41467-019-10279-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Medina P.; Paillasse M. R.; Segala G.; Poirot M.; Silvente-Poirot S. Identification and pharmacological characterization of cholesterol-5,6-epoxide hydrolase as a target for tamoxifen and AEBS ligands. Proc. Natl. Acad. Sci. U.S A. 2010, 107, 13520–13525. 10.1073/pnas.1002922107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allimuthu D.; Hubler Z.; Najm F. J.; Tang H.; Bederman I.; Seibel W.; Tesar P. J.; Adams D. J. Diverse Chemical Scaffolds Enhance Oligodendrocyte Formation by Inhibiting CYP51, TM7SF2, or EBP. Cell Chem. Biol. 2019, 26, 593–599. 10.1016/j.chembiol.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubler Z.; Allimuthu D.; Bederman I.; Elitt M. S.; Madhavan M.; Allan K. C.; Shick H. E.; Garrison E.; Karl M.; Factor D. C.; Nevin Z. S.; Sax J. L.; Thompson M. A.; Fedorov Y.; Jin J.; Wilson W. K.; Giera M.; Bracher F.; Miller R. H.; Tesar P. J.; Adams D. J. Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature 2018, 560, 372–376. 10.1038/s41586-018-0360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F. D. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 2008, 16, 535–541. 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- Porter F. D.; Herman G. E. Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 2011, 52, 6–34. 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham H. R.; Hennekam R. C. M. Mutational spectrum of Smith-Lemli-Opitz syndrome. Am. J. Med. Genet. 2012, 160C, 263–284. 10.1002/ajmg.c.31346. [DOI] [PubMed] [Google Scholar]
- Balogh I.; Koczok K.; Szabo G. P.; Torok O.; Hadzsiev K.; Csabi G.; Balogh L.; Dzsudzsak E.; Ajzner E.; Szabo L.; Csakvary V.; Olah A. V. Mutational spectrum of Smith-Lemli-Opitz syndrome patients in Hungary. Mol. Syndromol. 2012, 3, 215–222. 10.1159/000343923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingson M. S.; Wick M. J.; White W. M.; Raymond K. M.; Saenger A. K.; Pichurin P. N.; Wassif C. A.; Porter F. D.; Babovic-Vuksanovic D. Pregnancy in an individual with mild Smith-Lemli-Opitz syndrome. Clin Genet. 2014, 85, 495–497. 10.1111/cge.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross J. L.; Iben J.; Simpson C. L.; Thurm A.; Swedo S.; Tierney E.; Bailey-Wilson J. E.; Biesecker L. G.; Porter F. D.; Wassif C. A. Determination of the allelic frequency in Smith-Lemli-Opitz syndrome by analysis of massively parallel sequencing data sets. Clin. Genet. 2015, 87, 570–575. 10.1111/cge.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.; Liu W.; Sheflin L. G.; Fliesler S. J.; Porter N. A. Novel oxysterols observed in tissues and fluids of AY9944-treated rats - a model for Smith-Lemli-Opitz Syndrome. J. Lipid Res. 2011, 52, 1810–1820. 10.1194/jlr.M018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapphahn R. J.; Richards M. J.; Ferrington D. A.; Fliesler S. J. Lipid-derived and other oxidative modifications of retinal proteins in a rat model of Smith-Lemli-Opitz syndrome. Exp. Eye Res. 2019, 178, 247–254. 10.1016/j.exer.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer B. A.; Xu L.; Porter N.; Rao S.; Fliesler S. Differential cytotoxic effects of 7-dehydrocholesterol derived oxysterols on cultured retina derived cells: Dependence on sterol structure, cell type, and density. Exp. Eye Res. 2016, 145, 297–316. 10.1016/j.exer.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell-Hattam I. M.; Sharpe L. J.; Qian L.; Hart-Smith G.; Prabhu A. V.; Brown A. J. Twin enzymes, divergent control: The cholesterogenic enzymes DHCR14 and LBR are differentially regulated transcriptionally and post-translationally. J. Biol. Chem. 2020, 295, 2850–2865. 10.1074/jbc.ra119.011323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giera M.; Plössl F.; Bracher F. Fast and easy in vitro screening assay for cholesterol biosynthesis inhibitors in the post-squalene pathway. Steroids 2007, 72, 633–642. 10.1016/j.steroids.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Müller C.; Junker J.; Bracher F.; Giera M. A gas chromatography–mass spectrometry-based whole-cell screening assay for target identification in distal cholesterol biosynthesis. Nat. Protoc. 2019, 14, 2546–2570. 10.1038/s41596-019-0193-z. [DOI] [PubMed] [Google Scholar]
- Tallman K. A.; Allen L. B.; Klingelsmith K.; Anderson A.; Genaro-Mattos T. C.; Mirnics K.; Porter N. A.; Korade Z. Prescription Medications Alter Neuronal and Glial Cholesterol Synthesis. ACS Chem. Neurosci. 2021, 12, 735–745. 10.1021/acschemneuro.0c00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturley S. L.; Rajakumar T.; Hammond N.; Higaki K.; Márka Z.; Márka S.; Munkacsi A. B. Potential COVID-19 therapeutics from a rare disease: weaponizing lipid dysregulation to combat viral infectivity. J. Lipid Res. 2020, 61, 972–982. 10.1194/jlr.R120000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimo A.; Baritussio A.; Emdin M.; Tascini C. Amiodarone as a Possible Therapy for Coronavirus Infection. Eur. J. Prev. Cardiol. 2021, 28, e16–e8. 10.1177/2047487320919233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler K.; Ha H. R.; Ciminale V.; Spirli C.; Saletti G.; Schiavon M.; Bruttomesso D.; Bigler L.; Follath F.; Pettenazzo A.; Baritussio A. Amiodarone Alters Late Endosomes and Inhibits SARS Coronavirus Infection at a Post-Endosomal Level. Am. J. Respir. Cell Mol. Biol. 2008, 39, 142–149. 10.1165/rcmb.2007-0217OC. [DOI] [PubMed] [Google Scholar]
- Nelson E. A.; Barnes A. B.; Wiehle R. D.; Fontenot G. K.; Hoenen T.; White J. M. Clomiphene and Its Isomers Block Ebola Virus Particle Entry and Infection with Similar Potency: Potential Therapeutic Implications. Viruses 2016, 8, 206. 10.3390/v8080206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvodova L.; Sampaio J. L.; Cordo S.; Ejsing C. S.; Shevchenko A.; Simons K. The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol. 2009, 83, 7996–8003. 10.1128/JVI.00635-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova P. T.; Myers D. S.; Milne S. B.; McClaren J. L.; Thomas P. G.; Brown H. A. Lipid composition of viral envelope of three strains of influenza virus - not all viruses are created equal. ACS Infect. Dis. 2015, 1, 399–452. 10.1021/acsinfecdis.5b00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSharry J. J.; Wagner R. R. Lipid composition of purified vesicular stomatitis viruses. J. Virol. 1971, 7, 59–70. 10.1128/jvi.7.1.59-70.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley J. P.; Rifkin D. B.; Reich E. Phospholipid composition of Rous sarcoma virus, host cell membranes and other enveloped RNA viruses. Virology 1971, 46, 106–116. 10.1016/0042-6822(71)90010-9. [DOI] [PubMed] [Google Scholar]
- Rawat S. S.; Viard M.; Gallo S. A.; Rein A.; Blumenthal R.; Puri A. Modulation of entry of enveloped viruses by cholesterol and sphingolipids. Mol. Membr. Biol. 2003, 20, 243–254. 10.1080/0968768031000104944. [DOI] [PubMed] [Google Scholar]
- Gerl M. J.; Sampaio J. L.; Urban S.; Kalvodova L.; Verbavatz J.-M.; Binnington B.; Lindemann D.; Lingwood C. A.; Shevchenko A.; Schroeder C.; Simons K. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J. Cell Biol. 2012, 196, 213–221. 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro A. C.; Damonte E. B. Requirement of cholesterol in the viral envelope for dengue virus infection. Virus Res. 2013, 174, 78–87. 10.1016/j.virusres.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Campbell S. M.; Crowe S. M.; Mak J. Virion-associated cholesterol is critical for the maintenance of HIV-1 structure and infectivity. AIDS 2002, 16, 2253–2261. 10.1097/00002030-200211220-00004. [DOI] [PubMed] [Google Scholar]
- Graham D. R. M.; Chertova E.; Hilburn J. M.; Arthur L. O.; Hildreth J. E. K. Cholesterol Depletion of Human Immunodeficiency Virus Type 1 and Simian Immunodeficiency Virus with β-Cyclodextrin Inactivates and Permeabilizes the Virions: Evidence for Virion-Associated Lipid Rafts. J. Virol. 2003, 77, 8237–8248. 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna-Ramos J. F.; Reyes-Ruiz J. M.; del Ángel R. M. The Role of Host Cholesterol During Flavivirus Infection. Front. Cell. Infect. Microbiol. 2018, 8, 388. 10.3389/fcimb.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R.; Case J. B.; Yutuc E.; Ma X.; Shen S.; Gomez Castro M. F.; Liu Z.; Zeng Q.; Zhao H.; Son J.; Rothlauf P. W.; Kreutzberger A. J. B.; Hou G.; Zhang H.; Bose S.; Wang X.; Vahey M. D.; Mani K.; Griffiths W. J.; Kirchhausen T.; Fremont D. H.; Guo H.; Diwan A.; Wang Y.; Diamond M. S.; Whelan S. P. J.; Ding S. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 32105–32113. 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa I. P.; Carvalho C. A. M.; Gomes A. M. O. Current Understanding of the Role of Cholesterol in the Life Cycle of Alphaviruses. Viruses 2021, 13, 35. 10.3390/v13010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazal N.; Gerlier D. Virus Entry, Assembly, Budding, and Membrane Rafts. Microbiol. Mol. Biol. Rev. 2003, 67, 226–237. 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrero T.; Hagan M. F. Simulations show that virus assembly and budding are facilitated by membrane microdomains. Biophys. J. 2015, 108, 585–595. 10.1016/j.bpj.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng C. G.; Coppens I.; Govindarajan D.; Pisciotta J.; Shulaev V.; Griffin D. E. Effect of host cell lipid metabolism on alphavirus replication, virion morphogenesis, and infectivity. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 16326–16331. 10.1073/pnas.0808720105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichit S.; Hamel R.; Bernard E.; Talignani L.; Diop F.; Ferraris P.; Liegeois F.; Ekchariyawat P.; Luplertlop N.; Surasombatpattana P.; Thomas F.; Merits A.; Choumet V.; Roques P.; Yssel H.; Briant L.; Missé D. Imipramine Inhibits Chikungunya Virus Replication in Human Skin Fibroblasts through Interference with Intracellular Cholesterol Trafficking. Sci. Rep. 2017, 7, 3145 10.1038/s41598-017-03316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello D. A.; Villareal V. A.; Yang P. L. Desmosterol Increases Lipid Bilayer Fluidity during Hepatitis C Virus Infection. ACS Infect. Dis. 2016, 2, 852–862. 10.1021/acsinfecdis.6b00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallorin L.; Villareal V. A.; Hsia C.-Y.; Rodgers M. A.; Burri D. J.; Pfeil M.-P.; Llopis P. M.; Lindenbach B. D.; Yang P. L. Hepatitis C virus NS3-4A protease regulates the lipid environment for RNA replication by cleaving host enzyme 24-dehydrocholesterol reductase. J. Biol. Chem. 2020, 295, 12426–12436. 10.1074/jbc.ra120.013455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillasse M. R.; Saffon N.; Gornitzka H.; Silvente-Poirot S.; Poirot M.; de Medina P. Surprising unreactivity of cholesterol-5,6-epoxides towards nucleophiles. J. Lipid Res. 2012, 53, 718–725. 10.1194/jlr.M023689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot M.; Silvente-Poirot S. Oxysterols and related sterols: implications in pharmacology and pathophysiology. Biochem. Pharmacol. 2013, 86, 1–2. 10.1016/j.bcp.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Poirot M.; Silvente-Poirot S. Cholesterol-5,6-epoxides: chemistry, biochemistry, metabolic fate and cancer. Biochimie 2013, 95, 622–631. 10.1016/j.biochi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- Poirot M.; Silvente-Poirot S. When cholesterol meets histamine, it gives rise to dendrogenin A: a tumour suppressor metabolite. Biochem. Soc. Trans. 2016, 44, 631–637. 10.1042/bst20150232. [DOI] [PubMed] [Google Scholar]
- Tallman K. A.; Kim H. H.; Korade Z.; Genaro-Mattos T. C.; Wages P. A.; Liu W.; Porter N. A. Probes for protein adduction in cholesterol biosynthesis disorders: Alkynyl lanosterol as a viable sterol precursor. Redox Biol. 2017, 12, 182–190. 10.1016/j.redox.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cureton D. K.; Burdeinick-Kerr R.; Whelan S. P. J. Genetic inactivation of COPI coatomer separately inhibits vesicular stomatitis virus entry and gene expression. J. Virol. 2012, 86, 655–666. 10.1128/jvi.05810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R.; Ravet M.; Wieland F. T.; Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009, 583, 2701–2709. 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Dodonova S. O.; Diestelkoetter-Bachert P.; Appen Av.; Hagen W. J. H.; Beck R.; Beck M.; Wieland F.; Briggs J. A. G. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 2015, 349, 195–198. 10.1126/science.aab1121. [DOI] [PubMed] [Google Scholar]
- Dixon S. J.; Winter G. E.; Musavi L. S.; Lee E. D.; Snijder B.; Rebsamen M.; Superti-Furga G.; Stockwell B. R. Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem. Biol. 2015, 10, 1604–1609. 10.1021/acschembio.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll S.; Proneth B.; Tyurina Y. Y.; Panzilius E.; Kobayashi S.; Ingold I.; Irmler M.; Beckers J.; Aichler M.; Walch A.; Prokisch H.; Trümbach D.; Mao G.; Qu F.; Bayir H.; Füllekrug J.; Scheel C. H.; Wurst W.; Schick J. A.; Kagan V. E.; Angeli J. P. F.; Conrad M. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do Q.; Lee D. D.; Dinh A. N.; Seguin R. P.; Zhang R.; Xu L. Development and Application of a Peroxyl Radical Clock Approach for Measuring Both Hydrogen-Atom Transfer and Peroxyl Radical Addition Rate Constants. J. Org. Chem. 2020, 86, 153–168. 10.1021/acs.joc.0c01920. [DOI] [PubMed] [Google Scholar]
- Zielinski Z. A. M.; Pratt D. A. Lipid Peroxidation: Kinetics, Mechanisms, and Products. J. Org. Chem. 2017, 82, 2817–2825. 10.1021/acs.joc.7b00152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.