Abstract
The MCM proteins are a group of six proteins whose action is vital for DNA replication in eukaryotes. It has been suggested that they constitute the replicative helicase, with a subset of the proteins forming the catalytic helicase (MCM4,6,7) while the others have a loading or control function. In this paper we show that all six MCM proteins are present in equivalent amounts in soluble extracts and on chromatin. We have also analysed soluble and chromatin-associated MCM protein complexes under different conditions. This suggests that all six MCM proteins are always found in a complex with each other, although the interaction between the individual MCM proteins is not equivalent as stringent salt conditions are able to break the intact complex into a number of stable subcomplexes. These data contribute to the ongoing debate about the nature of MCM complexes, supporting the hypothesis that they act as a heterohexamer rather than as a number of different subcomplexes. Finally, using protein–protein cross-linking we have shown that MCM2 interacts directly with MCM5 and MCM6; MCM5 with MCM3 and MCM2; and MCM6 with MCM2 and MCM4. This provides the first direct information about specific subunit contacts in the MCM complex.
INTRODUCTION
An accumulation of evidence now suggests that DNA replication in eukaryotes is initiated by the formation of a constrained multiprotein complex on the chromatin, called the pre-replication complex (preRC) (1). The subsequent release of the constraints allows the initial synthesis to take place and the elongation phase of DNA replication to commence. Using a variety of genetic and biochemical methods a number of proteins involved in the assembly of the preRC have been defined and much work has been put into defining their order of action. This has resulted in a model for initiation where the origin of replication is initially recognised by the origin recognition complex (ORC). This promotes the binding of cdc6 and cdt1, which in turn are needed to recruit the MCM proteins. The subsequent loading of cdc45 and replication protein A enables the binding of DNA polymerase α, the enzyme that is presumed to be responsible for the synthesis of an RNA primer at the origin. For a recent review see (2) and (3). The inappropriate firing of origins is thought to be prevented by multiple redundant mechanisms. These act on both the formation and release of the preRC, and involve protein transport, proteolysis and both inhibitory and stimulatory post-translational modifications (3).
The MCM proteins constitute a group of six proteins that show regions of high similarity (the largest conserved domain is a region of ∼200 amino acids that encodes an NTP-binding motif) (4). Yeast genetics originally identified these proteins and suggested that they might have some role in initiation of DNA replication due to their requirement for the maintenance of mini-chromosomes (5). Many further studies in yeast and other organisms have confirmed the role of all six proteins in initiation, and defined the timing of their action in the initiation process. More recent genetic studies have also suggested that all of the MCM proteins are required throughout the elongation phase, and CHIP analysis has detected MCM4 and 7 moving with the elongating replication fork (6).
Although the exact function of the MCM proteins in replication and the relative roles of individual subunits have not been defined, recent studies have shed some light on their likely function. An oligomeric complex of the single MCM in archaebacteria has been shown to have helicase activity (7,8). In addition a complex of MCM4, 6 and 7 has been shown to possess helicase activity in humans (9) and Schizosaccharomyces pombe (10). This suggests that at least some of the MCM proteins could constitute the replicative helicase. A role for the other MCM proteins (2,3,5) as modulators of this activity has been suggested based on their suppression of the helicase activity of the 4/6/7 complex (11,12). Two other lines of evidence have also been used to suggest distinct functions for different MCM proteins. First, it is possible to isolate subcomplexes of the proteins from various organisms. The usual combinations that are found are MCM3/5, MCM4/6/7, MCM2/4/6/7 and MCM2/3/4/5/6/7. It should be noted, however, that all of the subcomplexes seem to be present at lower levels than the hexameric complex in both Xenopus and S.pombe extracts (12,13). In addition, in Xenopus, individual MCM proteins have been observed to associate with chromatin at different times (14).
An understanding of the physical arrangement of the various MCM complexes is important in helping to understand how the proteins interact together during their function. Of particular importance is how the subunits interact with each other when they are chromatin associated.
Most previous studies directed towards understanding the composition of complexes have been solution studies, and have not looked at the direct contacts made by the individual MCM proteins. Few studies have analysed MCM complexes occurring in the context of chromatin, and where this has been done complexes have been fixed using formaldehyde cross-linking (15). This agent produces long-range cross-linking and therefore again these studies do not provide information about direct contacts.
Since none of the previous studies has provided information about nearest neighbour contacts in MCM complexes we were interested in carrying out such an analysis for the Drosophila MCM proteins. In order to do this we chose to use a different cross-linking technique using a thiol-cleavable homobifunctional cross-linker dithiobis(succinimidylpropionate) (DSP) (16). This is a much less efficient cross-linker than formaldehyde, therefore allowing analysis of direct contacts and some distinction between primary and secondary contacts. It is also specific for protein–protein interactions, removing the complication of complexes held together through protein–DNA contacts (17).
In this paper we present our data quantifying the relative amounts of MCMs in different cellular compartments in Drosophila embryos. We also present an analysis of the components of MCM complexes observed both on the chromatin and in solution, and a nearest neighbour analysis of these interactions that provides the first direct information about specific subunit contacts.
MATERIALS AND METHODS
Quantitation methods
Overexpressed GST–MCM6, GST–MCM3 (18), His6-MCM2 and His6-MCM5 concentrations were determined by Coomassie staining and comparison with known amounts of standards using the Alpha Innotech Alphaimager.
Concentrations of MCM2, MCM3, MCM5 and MCM6 in total extract and formaldehyde cross-linked chromatin were determined by quantitative western blots comparing the levels in crude extracts with known amounts of overexpressed protein using the Alpha Innotech Alphaimager.
Preparation of extracts, cross-linking methods
Total extract. Drosophila embryos were homogenised in 1 vol buffer A [15 mM HEPES pH 7.9, 15 mM NaCl, 60 mM KCl, 2 mM EDTA, 0.34 M sucrose and complete™ protease inhibitors (Roche)] using a loose pestle. The homogenate was centrifuged for 5 min at 5000 g at 4°C in a benchtop centrifuge. The middle layer was collected, aliquoted and flash frozen in liquid nitrogen.
For DSP cross-linking of crude extract, the extract was homogenised as above, filtered through four layers of Miracloth, then centrifuged for 20 min at 25 000 r.p.m. at 4°C in a TL100.3 (Beckman) rotor. The middle layer was re-centrifuged for 30 min at 100 000 r.p.m. (Beckman TL100.3 rotor) at 4°C. To the clear supernatant, DSP (Sigma) was added to a final concentration of 0.5 mg/ml (freshly prepared in dimethylsulphoxide at 10 mg/ml). After 30 min on ice the reaction was stopped by the addition of Tris pH 8 (final concentration 25 mM). Cross-linked material was denatured by adjusting to 1% SDS and incubating for 5 min at 100°C. The extract was centrifuged for 30 min at 100 000 r.p.m. (Beckman TL100.3 rotor) at 20°C and adjusted to 0.1% SDS and 1% Triton X-100 by the addition of 9 vol phosphate-buffered saline (PBS), 1% Triton X-100 plus complete™ protease inhibitors (Roche). Prior to immunoprecipitation the extract was filtered through a 0.22 µm filter.
Chromatin. Drosophila embryos were homogenised with 1 vol buffer A with a loose pestle. The homogenate was filtered through Miracloth, centrifuged for 20 min at 4000 r.p.m. (SS34, Sorvall) and washed once with the same buffer. The nuclei-enriched pellet was resuspended in 2 vol buffer A.
DSP was added to a final concentration of 0.5 mg/ml and the incubation carried out on ice for 20 min. The reaction was stopped by the addition of Tris pH 8 to 25 mM and the nuclei pelleted by centrifugation for 10 min at 7000 r.p.m. (SS34, Sorvall). To remove the nuclear membrane and nucleoplasm the pellet was resuspended in 2 vol buffer A, adjusted to 1% Triton X-100 and centrifuged for 10 min at 7000 r.p.m. (SS34, Sorvall). This was repeated twice.
The final pellet was resuspended in 0.1 vol PBS and denatured by the addition of SDS to 1%. The viscosity of the homogenate was reduced by sonication and particulate material removed by centrifugation for 30 min at 100 000 r.p.m. (TL100.3). The clear supernatant was adjusted to 0.1% SDS and 1% Triton X-100 in PBS and filtered through a 0.22 µm filter prior to the immunoprecipitation.
Micrococcal nuclease-released chromatin. The micrococcal nuclease-released chromatin was prepared as described (19) with the following modifications: Drosophila embryos were homogenised in 1 vol 0.25 mM EDTA, 1 mM HEPES pH 7.9, 1 mM ATP plus protease inhibitors, using a loose pestle to reduce nuclei disruption. The homogenate was filtered through Miracloth and the filtrate centrifuged for 10 min at 4000 r.p.m. (SS34). The pellet was washed twice by resuspension in 0.5 vol of the same buffer and centrifugation for 10 min at 4000 r.p.m. The final pellet was resuspended in 0.4 vol of the same buffer adjusted to 0.5% NP-40, layered on top of 0.1 M sucrose, 1 mM Tris pH 8, 1 mM ATP plus protease inhibitors and centrifuged for 10 min at 7000 r.p.m. (SS34, Sorvall). This wash was repeated. The pellet was resuspended in 0.5 vol 0.25 mM EDTA, 1 mM HEPES pH 7.9, 1 mM ATP plus protease inhibitors and adjusted to 10 mM Tris pH 7.5 and 2 mM CaCl2. Micrococcal nuclease (Sigma) was added to a final concentration of 2 U/ml and the reaction incubated on ice for 2 h. The reaction was stopped by the addition of 8 mM EDTA and centrifuged for 15 min at 100 000 r.p.m. (TL100). This treatment reduces the DNA in the supernatant to the size of mononucleosomes (data not shown). The supernatant was filtered through a 0.22 µm filter before being used for immunoprecipitation.
Immunoprecipitations
Antibodies. Anti-MCM2/5/3/6 and the control antibody (anti-hsp90) were made by immunisation of rabbits with the relevant overproduced MCM protein. Anti-MCM4 was a kind gift of T. Su and P. O’Farrell.
DMP cross-linking of the antibodies. Antibodies (anti-MCM2, anti-MCM5, anti-MCM6 and anti-Hsp90) were cross-linked to protein A–Sepharose using dimethyl pimelimidate (DMP) according to Harlow and Lane (20).
Immunoprecipitations. For immunoprecipitation from crude extracts the antibody beads were incubated with the extract for 1 h. For the immunoprecipitation from micrococcal nuclease-released chromatin, the antibody beads were incubated with the extract overnight at 4°C. The beads were washed 10 times with 20 vol of either PBS or RIPA buffer (PBS containing 0.1% SDS and 1% Triton X-100) as indicated in the text (protease inhibitors are included in all buffers).
For immunoprecipitation from denatured extract, cross-linked extract or DSP cross-linked chromatin, the antibody beads were incubated with extracts overnight at 4°C. The beads were washed 10 times with 20 vol RIPA buffer (adjusted to 650 mM NaCl).
In all cases immunoprecipitated proteins were eluted using 2% SDS in PBS.
RESULTS
Quantitation of MCMs in crude extract and on chromatin
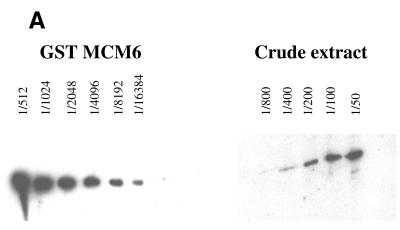
Although all MCMs seem to be required for both initiation and elongation phases of replication, the presence of identifiable subcomplexes and the apparent sequential loading of individual MCM proteins onto chromatin in Xenopus might suggest some independence of function. We were interested to see whether this was reflected in the relative levels of the MCM proteins in the cytoplasm or on chromatin. For this analysis we chose to use antibodies against MCM2, 3, 5 and 6, thus including at least one MCM from each subgrouping. We prepared crude extracts from Drosophila 0–5 h embryos and quantified the amount of each MCM present in the fraction by comparison of a titration of the crude extract with a titration of the overexpressed fragment that the antibody was raised against. An example of this analysis is shown for MCM6 in Figure 1A.
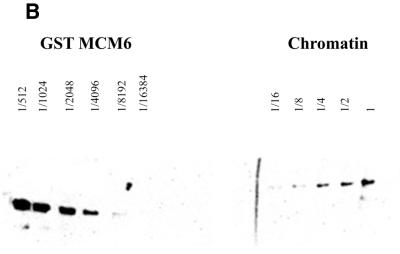
Figure 1.
Titration of MCM6 in cytoplasmic extract (A) and on chromatin (B). (A) To make crude cytoplasmic extract Drosophila embryos were homogenised in 1 vol buffer A [15 mM HEPES pH 7.9, 15 mM NaCl, 60 mM KCl, 2 mM EDTA, 0.34 M sucrose and complete™ protease inhibitors (Roche)] using a loose pestle. The homogenate was centrifuged for 5 min at 5000 g at 4°C in a benchtop centrifuge. The middle layer was collected and serial dilutions of it were subjected to PAGE and western blotting with anti-MCM6 antibody. A serial dilution of the peptide against which the MCM6 antibody was raised was run on the same gel and scanned to generate a titration curve. The level of MCM6 present in the crude extract was calculated by comparison of the signal obtained from the crude extract with the titration curve. (B) Chromatin was prepared by the method of Hancock and the associated proteins were fixed onto it using formaldehyde cross-linking. The chromatin was purified on caesium chloride gradients, the cross-linking reversed and serial dilutions of the extract analysed by western blotting. Again quantitation was performed by titration against known standard concentrations of the overproduced protein as described above.
In crude extract all four MCMs were present in nearly equimolar amounts (variation in amounts between individual MCM proteins was less than a factor of 2). Calculation of the absolute amount present gave a figure of between 20 and 40 fg per nucleus (equivalent to one MCM per 80–100 bp chromatin). For quantitation of chromatin-bound MCMs, chromatin was prepared by the method of Hancock, and associated proteins were fixed using formaldehyde cross-linking. The chromatin was purified on caesium chloride gradients, and again quantitation was performed by titration against known standard concentrations of the overproduced protein. An example for MCM6 is shown in Figure 1B. Again we found that the four MCM proteins tested were present in similar amounts equivalent to one molecule of each MCM for every 8–16 kb.
From these data we conclude that MCM2, 3, 5 and 6 are present in stoichiometric amounts with each other both in the cytoplasm and on chromatin. Although we were not able to test this for MCM4 and 7 the close association seen between these proteins and MCM6 in other systems makes it likely that these will also be present at the same stoichiometry.
Analysis of cytoplasmic complexes of MCM proteins
A complex of all six MCMs can be seen under non-denaturing conditions. In other systems heterohexameric complexes containing all six MCM proteins have been observed in low salt conditions (13). However, previous analyses in Drosophila extracts did not reveal the presence of such complexes (21). Our initial experiments were therefore aimed at defining whether there were any conditions where heterohexameric MCM complexes could be detected in Drosophila extracts.
We therefore subjected our extracts to immunoprecipitation using antibodies against MCM2, 5 and 6 (again chosen as representatives of each of the previously identified subcomplexes of MCM proteins). The precipitates were washed extensively in PBS (150 mM NaCl) before elution of the beads, and the bead eluates were analysed for the presence of MCM2, MCM3, MCM4, MCM5 and MCM6 by SDS–PAGE and western blotting. The results of this analysis are shown in Figure 2.
Figure 2.
Co-immunoprecipitation of the MCM proteins under non-denaturing conditions. Crude cytoplasmic extracts were generated as described above and were then subjected to immunoprecipitation with specific anti-MCM antibodies as shown. The pellets were washed extensively with PBS before elution and analysis by western blotting. CE, crude extract control; IP, SDS eluate from the beads after extensive washing with PBS. In some cases proteolytic cleavage products of individual MCM proteins can also be seen on the gel.
If anti-MCM2 antibodies were used for the immunoprecipitation our bead eluates contained MCM2, MCM3, MCM4, MCM5 and MCM6. Using anti-MCM6 antibodies we could identify MCM2, MCM3, MCM4, MCM5 and MCM6. Analysis of the levels of each of the proteins present in the pellet suggested that all of the MCMs were present in these precipitates in approximately equal amounts. These co-immunoprecipitations are not due to non-specific binding to antibody beads, as they are not immunoprecipitated by an unrelated antibody (data not shown).
Under the same conditions the MCM5 antibody was only able to co-precipitate MCM3 in near equimolar amounts. The other MCMs could also be detected in the immunoprecipitate, but in much lower amounts. Since we have already shown that MCM5 appears to be an equimolar constituent in the MCM complexes, and that the amounts of MCM5 protein are not drastically different from the other MCMs, the most likely explanation for this is that the MCM5 antibody partially disrupts the complex. In further analyses of situations where the proteins have not been covalently linked together prior to immunoprecipitation we have therefore taken this into account when interpreting our results.
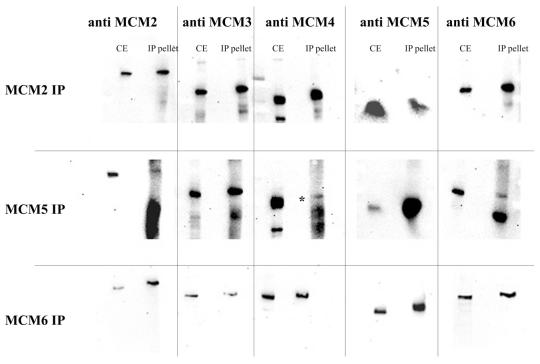
Subcomplexes of MCMs can be detected under mildly disruptive conditions. In order to determine whether the subcomplexes of MCM proteins that we detect in cytoplasmic fractions were similar to those reported by others, the immuno-adsorbed complexes were washed in RIPA buffer containing 0.1% SDS and 1% Triton X-100 prior to bead elution. This should retain only the more tightly bound partners. The results for this analysis are shown in Figure 3.
Figure 3.
Co-immunoprecipitation of the MCM proteins under stringent conditions. Crude cytoplasmic extracts were generated as described above and were then subjected to immunoprecipitation with specific anti-MCM antibodies as shown. The pellets were washed extensively with RIPA buffer (PBS containing 0.1% SDS and 1% Triton X-100) before elution and analysis by western blotting. CE, crude extract control; IP, SDS eluate from the beads after extensive washing with RIPA.
In this case only MCM6 was detected precipitating with MCM2 in near equimolar amounts. MCM3, MCM4 and MCM5 were not detected even after long exposures.
Using the anti-MCM5 antibody, only MCM3 is seen to co-precipitate in roughly equimolar amounts to MCM5. MCM2, MCM4 and MCM6 were not detected under these conditions.
Using anti-MCM6 antibody both MCM4 and MCM2 were detected in addition to MCM6. In this case MCM6 is more abundant than MCM2 and MCM4. MCM3 and MCM5 were never detected.
From these data it therefore seems likely that our cytoplasmic Drosophila extracts contain complexes comparable to those observed in other systems.
Nearest neighbour contacts can be identified by DSP cross-linking. The methods used above do not tell us whether the contacts we are observing are direct. In order to address this question we generated covalent linkages between the proteins in crude Drosophila embryo extracts by treating them with DSP. DSP is a thiol-cleavable homobifunctional N-hydroxysuccinimide ester causing linkages principally through primary amines (17). To ensure that we were only looking at covalent interactions SDS was added to a final concentration of 1% and the solutions were incubated at 100°C after cross-linking to break any non-covalent bonds.
Under the cross-linking conditions used we determined by glycerol gradient (data not shown) that 15–20% of the MCM subunits are found migrating at about twice their monomeric size. This indicates that 15–20% of each individual MCM is cross-linked. At the levels of detection that we were using we did not detect trimers on our gradients. These conditions are therefore appropriate for a nearest neighbour analysis since they only allow the detection of direct interactions. The results from this analysis are shown in Figure 4.
Figure 4.
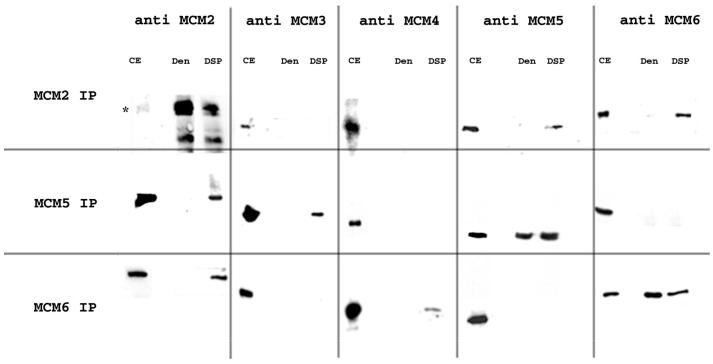
Co-immunoprecipitation of the MCM proteins under denaturing conditions after cross-linking with DSP. Crude cytoplasmic extracts were generated as described above and cross-linked by the addition of DSP. Non-covalent interactions between proteins were disrupted by addition of SDS to a final concentration of 1%. Immunoprecipitations were then carried out in RIPA buffer as described above. Cross-links were broken by the addition of dithiothreitol (DTT) and the proteins present were analysed by western blotting. CE, crude extract control; Den, analysis of proteins present in immunoprecipitate after denaturation with SDS but without DSP cross-linking; IP, proteins released from immunoprecipitate pellets of DSP cross-linked extracts by SDS and treated with DTT to break cross-links.
Using the anti-MCM2 antibody as expected MCM2 is the most efficiently immunoprecipitated. It should be noted that in this case DSP cross-linking leads to a reduced efficiency of immunoprecipitation (Fig. 4, compare the Den and DSP lanes on the anti-MCM2 column). MCM5 and MCM6 are detected in the cross-linked complex in roughly similar amounts but much lower than MCM2. MCM3 and MCM4 are not detected.
Using the anti-MCM5 antibody for the immunoprecipitation, MCM5 is efficiently immunoprecipitated and here DSP cross-linking does not seem to affect the efficiency. MCM3 and MCM2 are clearly identified in the cross-linked complex; MCM4 and MCM6 are not detected.
Using the anti-MCM6 antibody, MCM6 is clearly detected but again DSP cross-linking affects the efficiency of immunoprecipitation. MCM2 is detected in the cross-linked complex as well as MCM4, although in lower amounts. MCM3 and MCM5 are not detected.
In all cases in the absence of cross-linker (Fig. 4, Den lanes) only the MCM subunit corresponding to the antibody used for the immunoprecipitation is detected.
These data therefore suggest that in cytoplasmic fractions direct contacts occur between MCM2 and MCM5, MCM2 and MCM6, MCM5 and MCM3, and MCM6 and MCM4.
Analysis of chromatin-associated complexes of MCM proteins
All our analyses so far have been carried out in cytoplasmic extracts; however, the likely point of action of the MCM proteins is on chromatin. We were interested to ask whether the pattern of contacts that we had observed in the cytoplasm were maintained once the proteins had been loaded onto chromatin. We therefore performed similar experiments to those described above except that in this case chromatin was substituted for the cytoplasmic extract.
Chromatin-associated MCM proteins are more tightly associated with each other than those in the soluble fraction. For analysis of interactions under non-disruptive and mildly disruptive conditions we purified chromatin and solubilised chromatin-bound proteins using micrococcal nuclease digestion. Conditions were adjusted so that the DNA was mostly reduced to the size of mononucleosomes (data not shown). The released proteins were used for immunoprecipitations using anti-MCM2, 5 and 6 antibodies as described above.
Under non-disruptive conditions (PBS), the results for the MCM2 and MCM6 antibodies were the same as those obtained from cytoplasmic extracts, that is, in both cases all the MCMs tested for were present (data not shown). On chromatin, in contrast to cytoplasmic extracts, a similar result is also obtained for the MCM5 antibody.
In the immunoprecipitation under more stringent conditions (after washing pellets in RIPA buffer) (Fig. 5) the complexes identified show some differences from those observed in the cytoplasm.
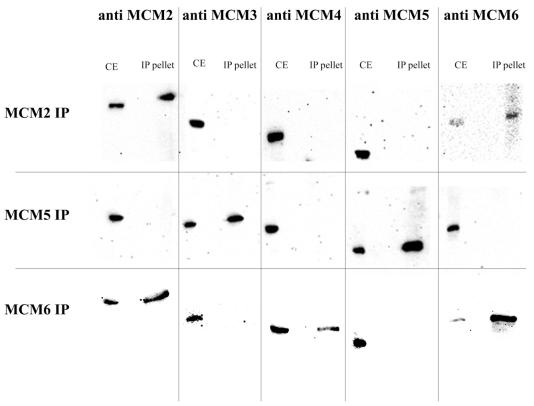
Figure 5.
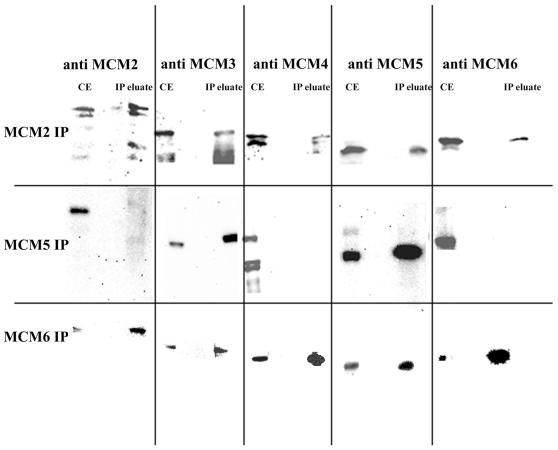
Co-immunoprecipitation of the MCM proteins on chromatin under stringent conditions. Chromatin was generated as described above and reduced to the size of mononucleosomes using micrococcal nuclease. It was then subjected to immunoprecipitation with specific anti-MCM antibodies as shown. The pellets were washed extensively with RIPA buffer before elution and analysis by western blotting. CE, crude extract control; IP eluate, SDS eluate from the beads after extensive washing with RIPA.
The MCM2 antibody is able to co-immunoprecipitate all of the other MCMs tested. MCM2 is more abundant and MCM3, MCM4, MCM5 and MCM6 are in near equimolar amounts.
Using anti-MCM5 antibody only MCM3 could be co-precipitated in near equimolar amounts, the other MCMs could not be detected even at long exposures.
With anti-MCM6 antibody, MCM6 is much more abundant, but MCM2, MCM3, MCM4 and MCM5 were all identified in near equimolar amounts in the immunopellet.
These results suggest that on chromatin the interactions between MCM proteins are stronger than those observed in soluble extracts.
Nearest neighbour interactions can also be detected on chromatin using DSP cross-linking. The differences in co-precipitation of MCM proteins seen from the analysis of complexes present under non-cross-linked conditions suggested that complexes formed on chromatin may differ from those observed in solution. We were therefore interested in determining whether there were any measurable changes in nearest neighbours in chromatin-bound complexes. Chromatin was therefore prepared and cross-linked as indicated in Materials and Methods and immunoprecipitated with the anti-MCM antibodies as described above.
As determined by glycerol gradient under denaturing conditions, the proportion of MCM that is cross-linked is between 5 and 10% and the only high molecular form detected is the size of a dimer (data not shown). Therefore, again we are confident that we are looking at direct contacts between subunits. The results of this analysis are shown in Figure 6.
Figure 6.
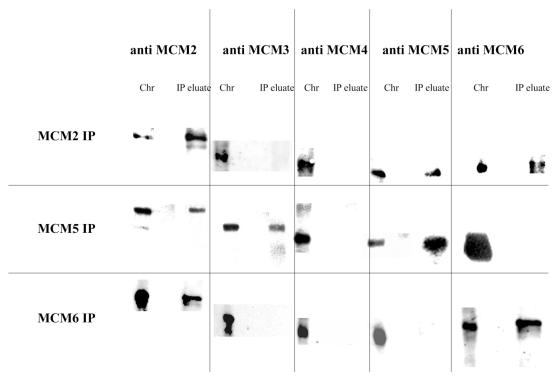
Co-immunoprecipitation of the MCM proteins on chromatin under denaturing conditions after cross-linking with DSP. Chromatin was generated as described above and cross-linked by the addition of DSP. Non-covalent interactions between proteins and DNA were then disrupted by addition of SDS to a final concentration of 1%. Immunoprecipitations were then carried out in RIPA buffer as described above. Cross-links were broken by the addition of DTT and the proteins present were analysed by western blotting. Chr, chromatin extract; IP eluate, proteins released from immunoprecipitate pellets of DSP cross-linked extracts by SDS and treated with DTT to break cross-links.
The results for MCM2 and MCM5 immunoprecipitations are as seen for cytoplasmic extracts, specifically both MCM5 and MCM6 can be seen to co-immunoprecipitate with MCM2, and both MCM2 and MCM3 are seen to co-precipitate with MCM5. When precipitations were carried out with MCM6 antibody a significant decrease in the efficiency of precipitation was seen (at least 5-fold). Analysis of the co-precipitating proteins also revealed a difference from what was observed in cytoplasmic extracts. On chromatin we can only detect an MCM6 interaction with MCM2; the interaction with MCM4 is not observed in this case even at long exposures.
DISCUSSION
It has been known for some time that the MCM proteins are necessary for DNA replication in higher eukaryotes (5); however, for much of that time the nature of their function has been unclear. More recent data have suggested that some combination of MCM proteins may function as a helicase (9); however, the exact nature of the helicase complex and how the MCM proteins interact with each other in both functional and non-functional complexes have been a subject of some controversy.
Some of the information presented in this paper is pertinent to the ongoing debate regarding the structure of MCM protein complexes. First, we have confirmed that in Drosophila, as in other systems, we can detect interactions of MCM proteins under non-denaturing conditions in cytoplasmic extracts consistent with the existence of a heterohexameric complex. Furthermore, we have also observed that under more harsh conditions we are able to see the apparent breakdown of these hexameric complexes into subcomplexes, also consistent with observations in other systems (13).
The observation that subcomplexes can be purified as separate entities, and the differential effects of these subcomplexes with respect to helicase activity in vitro, have led to suggestions that there may be multiple MCM complexes with different functionality (11,12). If this were the case it might be expected that different levels of the proteins involved in the individual subcomplexes might be seen associated with chromatin. The data presented here, however, suggest that this is not the case and that the MCM proteins are present in approximately stoichiometric amounts to each other both in the cytoplasm and on chromatin. The total amounts of MCM proteins in Drosophila extracts appear to be higher than those in other systems. This is likely to be due to the large maternal contribution present in the stages used for analysis.
The observation of comparable stoichiometry levels alone does not show that the MCM proteins function as a heterohexameric complex. It is also consistent with a situation where there are the same amounts of separate subcomplexes that perform independent functions on the chromatin. Our analysis of the complexes that are released from chromatin by micrococcal nuclease digestion does, however, suggest that any subcomplexes that are generated must remain within close proximity to each other while associated with chromatin, since, regardless of which MCM antibody is used for the precipitation, comparable amounts of each MCM tested are present. Additional evidence that would favour the hypothesis that the MCM complex retains its heterohexameric nature at all times during its action in the replication of DNA is that the proportions of MCM5 and MCM6 co-precipitating with MCM2 are the same in DSP cross-linked extracts and DSP cross-linked chromatin. This situation would not be expected if the hexameric complex split into subcomplexes during its action on the chromatin.
Our results further suggest that the predominant form of the MCM complex found in the cytoplasm also contains comparable amounts of each of the MCM proteins. This is consistent with the proposal that the MCM proteins assemble in solution and are loaded as a complex onto the chromatin rather than being loaded separately (13). Although the cytoplasmic and chromatin-bound complexes have the same proportions of each MCM we find that the hexameric nature of the complex is more stable when loaded on chromatin (hexameric complexes are detected upon RIPA washing of chromatin but not crude extract immunopellets). However, at this point we cannot distinguish whether this is due to a conformational change in the complex resulting in stronger interactions between the individual MCM proteins, or whether the apparent increase in stability is due to bridging interactions with DNA or other chromatin-associated proteins.
Based on the arguments presented above we therefore suggest that our data support a model where the MCM proteins function throughout as a heterohexameric complex or multiple thereof, and that alterations in the activity of the complex are achieved by modification of subunits rather than removal of subunits from the complex. In this case the appearance of subcomplexes may simply reflect the relative strengths of the interactions between the MCMs in the complex rather than true subcomplexes. This result is consistent with observations that suggest that all six subunits are needed throughout the initiation and elongation phases of replication (22). It is apparently at odds with the observation in Xenopus (14) that there are differences in the time of chromatin loading of individual MCMs. This could represent a species difference between Drosophila and Xenopus. However, it could also be explained by the differential stability of MCM subunit interactions and their susceptibility to various preparative techniques.
In addition to contributing data to the discussion of whether the MCM proteins function as a heterohexamer on chromatin, our data provide the first analysis of nearest neighbour partners in the MCM complex and therefore some insight into the architecture of the MCM complex (Table 1).
Table 1. Summary of MCM interactions.
| MCM2 IP | MCM5 IP | MCM6 IP | |
|---|---|---|---|
| Crude extracts | |||
| PBS | 2, 3, 4, 5, 6 | 3, 5 | 2, 3, 4, 5, 6 |
| RIPA | 2, 6 | 3, 5 | 2, 4, 6 |
| DSP | 2, 5, 6 | 2, 3, 5 | 2, 4, 6 |
| Chromatin | |||
| PBS | 2, 3, 4, 5, 6 | 2, 3, 4, 5, 6 | 2, 3, 4, 5, 6 |
| RIPA | 2, 3, 4, 5, 6 | 3, 5 | 2, 3, 4, 5, 6 |
| DSP | 2, 5, 6 | 2, 3, 5 | 2, 6 |
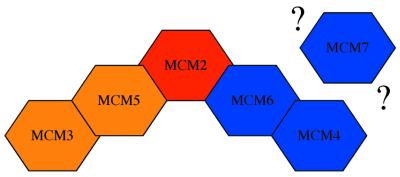
Putting our results together with what is known about subunit interactions from the known subcomplexes we suggest that in soluble extracts MCM2 is the link between the MCM3/MCM5 complex and the MCM4/MCM6/MCM7 complex by binding directly to MCM5 on the one hand and MCM6 on the other. We have expressed this in a schematic way in Figure 7. It should be noted that at present this only represents an approximation to the situation since we do not know about the contacts made by MCM7 against which we do not have antibodies.
Figure 7.
Schematic of MCM nearest neighbour contacts. This arrangement of proteins is proposed based on the analysis of interactions under non-denaturing conditions from this study and earlier studies, and the interactions seen at high stringency and by DSP cross-linking from this study. From other studies MCM7 is likely to be associated with MCM4 and 6, but the direct contacts made between this protein and the other members of the MCM group have not yet been established. This figure is only intended to show the contacts made by the proteins; it is not intended to portray the final overall structure of the MCM heterocomplex.
When on the chromatin, the nearest neighbour interactions that we detect are largely unchanged with the exception of the apparent loss of direct contact between MCM4 and MCM6. This, taken together with the huge decrease in efficiency with which the MCM6 immunoprecipitate can be carried out under these conditions, might be an indicator of a conformational change that has taken place. Since both MCM4 and MCM6 are part of the complex implicated in helicase activity this could represent a functionally significant alteration in the configuration of the complex related to its putative helicase function.
REFERENCES
- 1.Diffley J.F., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- 2.Lei M. and Tye,B.K. (2001) Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci., 114, 1447–1454. [DOI] [PubMed] [Google Scholar]
- 3.Blow J.J. (2001) Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J., 20, 3293–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kearsey S.E. and Labib,K. (1998) MCM proteins: evolution, properties and role in DNA replication. Biochim. Biophys. Acta, 1398, 113–136. [DOI] [PubMed] [Google Scholar]
- 5.Sinha P., Chang,V. and Tye,B.K. (1986) A mutant that affects the function of autonomously replicating sequences in yeast. J. Mol. Biol., 192, 805–814. [DOI] [PubMed] [Google Scholar]
- 6.Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- 7.Chong J.P., Hayashi,M.K., Simon,M.N., Xu,R.M. and Stillman,B. (2000) A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA, 97, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shechter D.F., Ying,C.Y. and Gautier,J. (2000) The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum delta H minichromosome maintenance protein. J. Biol. Chem., 275, 15049–15059. [DOI] [PubMed] [Google Scholar]
- 9.Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.-K. and Hurwitz,J. (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6 and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA, 98, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishimi Y., Komamura,Y., You,Z. and Kimura,H. (1998) Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem., 273, 8369–8375. [DOI] [PubMed] [Google Scholar]
- 12.Lee J.K. and Hurwitz,J. (2000) Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem., 275, 18871–18878. [DOI] [PubMed] [Google Scholar]
- 13.Prokhorova T.A. and Blow,J.J. (2000) Sequential MCM/P1 subcomplex assembly is required to form a heterohexamer with replication licensing activity. J. Biol. Chem., 275, 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maiorano D., Lemaitre,J.M. and Mechali,M. (2000) Stepwise regulated chromatin assembly of MCM2-7 proteins. J. Biol. Chem., 275, 8426–8431. [DOI] [PubMed] [Google Scholar]
- 15.Ritzi M., Baack,M., Musahl,C., Romanowski,P., Laskey,R.A. and Knippers,R. (1998) Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem., 273, 24543–24549. [DOI] [PubMed] [Google Scholar]
- 16.Liu H.T. and Yung,B.Y. (1999) In vivo interaction of nucleophosmin/B23 and protein C23 during cell cycle progression in HeLa cells. Cancer Lett., 144, 45–54. [DOI] [PubMed] [Google Scholar]
- 17.Lomant A.J. and Fairbanks,G. (1976) Chemical probes of extended biological structures: synthesis and properties of the cleavable protein cross-linking reagent [35S]dithiobis(succinimidyl propionate). J. Mol. Biol., 104, 243–261. [DOI] [PubMed] [Google Scholar]
- 18.Ohno K., Hirose,F., Inoue,Y.H., Takisawa,H., Mimura,S., Hashimoto,Y., Kiyono,T., Nishida,Y. and Matsukage,A. (1998) cDNA cloning and expression during development of Drosophila melanogaster MCM3, MCM6 and MCM7. Gene, 217, 177–185. [DOI] [PubMed] [Google Scholar]
- 19.Holthoff H.P., Baack,M., Richter,A., Ritzi,M. and Knippers,R. (1998) Human protein MCM6 on HeLa cell chromatin. J. Biol. Chem., 273, 7320–7325. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Su T.T., Feger,G. and O’Farrell,P.H. (1996) Drosophila MCM protein complexes. Mol. Biol. Cell, 7, 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labib K., Tercero,J.A. and Diffley,J.F. (2000) Uninterrupted MCM2-7 function required for DNA replication fork progression. Science, 288, 1643–1647. [DOI] [PubMed] [Google Scholar]