Abstract
Identification of specific leukemia subtypes is a key to successful risk-directed therapy in childhood acute lymphoblastic leukemia (ALL). Although RNA sequencing (RNA-seq) is the best approach to identify virtually all specific leukemia subtypes, the routine use of this method is too costly for patients in resource-limited countries. This study enrolled 295 patients with pediatric ALL from 2010 to 2020. Routine screening could identify major cytogenetic alterations in approximately 69% of B-cell ALL (B-ALL) cases by RT-PCR, DNA index, and multiplex ligation–dependent probe amplification. STIL-TAL1 was present in 33% of T-cell ALL (T-ALL) cases. The remaining samples were submitted for RNA-seq. More than 96% of B-ALL cases and 74% of T-ALL cases could be identified based on the current molecular classification using this sequential approach. Patients with Philadelphia chromosome–like ALL constituted only 2.4% of the entire cohort, a rate even lower than those with ZNF384-rearranged (4.8%), DUX4-rearranged (6%), and Philadelphia chromosome–positive (4.4%) ALL. Patients with ETV6-RUNX1, high hyperdiploidy, PAX5 alteration, and DUX4 rearrangement had favorable prognosis, whereas those with hypodiploid and KMT2A and MEF2D rearrangement ALL had unfavorable outcomes. With the use of multiplex ligation–dependent probe amplification, DNA index, and RT-PCR in B-ALL and RT-PCR in T-ALL followed by RNA-seq, childhood ALL can be better classified to improve clinical assessments.
Childhood acute lymphoblastic leukemia (ALL) is one of the most curable cancers, with a 5-year event-free survival (EFS) and a 5-year overall survival (OS) exceeding 80% and 90%, respectively, in many developed countries.1, 2, 3, 4, 5, 6, 7 This remarkable achievement is partly attributed to the more precise risk stratification of patients based on the identification of genetic abnormalities with prognostic and therapeutic implications and the evaluation of early treatment response using the minimal residual disease (MRD) test.8,9 The mapping of the human genome and subsequent advances in DNA and RNA sequencing (RNA-seq) techniques and bioinformatic analysis pipelines have revolutionized understanding of the genomic landscape of ALL.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22 Using RNA-seq, molecular classification can be performed for nearly 97% of childhood B-cell ALL (B-ALL) cases into 23 subtypes, some with prognostic significance and others with targetable lesions.23,24 In T-cell ALL (T-ALL), RNA-seq can provide provisional classifications according to transcriptional factor gene expression profiles or dysregulation of targetable functional pathways.25, 26, 27
In childhood B-ALL, conventional genetic analyses can identify ETV6-RUNX1 and high hyperdiploidy, the two most common subtypes, in 40% to 45% of patients. Other common fusions, such as BCR-ABL1 and TCF3-PBX1, account for 5% to 7% of patients.28,29 Several novel subtypes of ALL that are not evident through conventional genetic analysis but can be identified by transcriptome analysis include Philadelphia chromosome (Ph)–like ALL; DUX4-, MEF2D-, and ZNF384-rearranged B-ALL; and KMT2A-like and ETV6–RUNX1–like B-ALL.13,16,17,19,20,22 These new ALL subtypes have distinct clinical and biological characteristics. Unlike common recurrent ALL subtypes, such as ETV6-RUNX1, TCF3-PBX1, and BCR-ABL1, these subtypes have diverse genetic alterations that involve different partner genes, breakpoints, or signaling pathways, making routine screening by simple RT-PCR almost impractical. Some have no fusions but similar gene expression to known subtypes, which requires gene expression profiles for a precise diagnosis.24 Several molecular methods, such as RT-PCR, DNA index (DI), and multiplex ligation–dependent probe amplification (MLPA), can identify common genetic subtypes of B-ALL, including ETV6-RUNX1, high hyperdiploidy, hypodiploidy, BCR-ABL1, and TCF3-PBX1. This approach can identify approximately 60% to 70% of subtypes of B-ALL30; RNA-seq can be applied to the remaining samples with negative findings to determine subtypes.
In the case of T-ALL, half have aberrant transcriptional factor expression, and whole genome sequencing (WGS) may be required to identify the precise breaking point that led to aberrant expression.25 Gene expression defined by RNA-seq can identify some important genetic translocations and classify the basic T-ALL subtype by transcriptional factors. Although transcriptome analysis is vital to the comprehensive and precise classification of ALL, cost is a major hurdle to its widespread clinical use in a resource-limited country. In this study, a sequential approach was designed to identify childhood ALL genetic profiles using RT-PCR, DI, and RNA-seq to improve molecular classification while saving costs.
Materials and Methods
Patients and Protocols
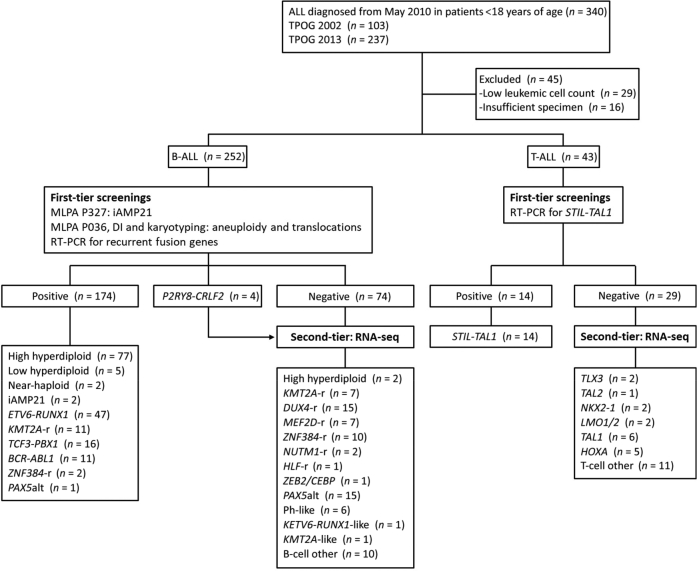
Between May 2010 and December 2020, a total of 340 children with newly diagnosed ALL treated at the National Taiwan University Hospital were enrolled in two consecutive Taiwan Pediatric Oncology Group (TPOG) protocols: 2002 protocol (n = 103) and 2013 protocol (n = 237) (Figure 1).31, 32, 33 Diagnostic bone marrow or peripheral blood samples were available for 295 children, and clinical features are described in Supplemental Table S1. Cases with insufficient material or a leukocyte count <70% were excluded from further analysis. The diagnosis of ALL was based on bone marrow morphology, and the immunophenotype of leukemic cells was determined using flow cytometry. Conventional cytogenetic analysis was performed using G-banding as part of the routine workup. Although both protocols used clinical and biological characteristics and remission induction response for risk assignment, TPOG 2013 incorporated the level of MRD on day 15 and the end of remission induction (between day 35 and day 42) to direct the intensity of treatment. The institutional review board of the National Taiwan University Hospital approved the study (201510016RIND), and written informed consent was obtained from parents, guardians, or patients in accordance with the Declaration of Helsinki.
Figure 1.
A flowchart of patients with the genetic diagnosis of acute lymphoblastic leukemia (ALL) enrolled in this study. ALL, acute lymphoblastic leukemia; B-ALL, B-cell acute lymphoblastic leukemia; DI, DNA index; MLPA, multiplex ligation–dependent probe amplification; T-ALL, T-cell acute lymphoblastic leukemia; TPOG, Taiwan Pediatric Oncology Group.
RT-PCR for Fusion Genes
Total RNA was isolated from bone marrow or blood samples using NucleoZOL (Macherey-Nagel, Düren, Germany). cDNA was synthesized using the Maxima First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). A total of 1 μg of RNA was used for cDNA synthesis according to the manufacturer's instructions. The prepared reaction mix was incubated at 25°C for 10 minutes, followed by 60°C for 30 minutes; then the reaction was terminated by heating at 85°C for 5 minutes. PCR was performed using MyTaq HS Mix (Bioline, London, UK), and thermocycling was performed as follows: 95°C for 60 seconds, followed by 38 cycles at 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 30 seconds, followed by final extension at 72°C for 5 minutes. Primers used for RT-PCR are listed in Supplemental Table S2. The PCR products were visualized using agarose gel electrophoresis. Suspected bands were purified using the FavorPrep GEL/PCR Purification Kit (Favorgen, Ping-Tung, Taiwan). Sanger sequencing was performed using an ABI 3730XL DNA analyzer (Thermo Fisher Scientific). Sequencing results were analyzed with SnapGene software version 4.1.3 (GSL Biotech, San Diego, CA) (https://www.ncbi.nlm.nih.gov/nuccore; accession number NM_000546.6 and https://www.ncbi.nlm.nih.gov/nuccore; accession number NM_001987).
Ploidy Status Analysis
The ploidy status was evaluated using SALSA MLPA Probemix P036 Subtelomeres Mix 1 (MLPA P036) (MRC-Holland, Amsterdam, the Netherlands) and DI, as previously reported.30 Briefly, DI was used to detect DNA aneuploidy, and MLPA P036 was used to identify individual chromosome gain or loss. The cases were suspected of masked hypodiploidy based on a hypodiploidy-like chromosome pattern.34 These cases were further analyzed by single-nucleotide polymorphism array or short tandem repeat typing to confirm chromosomal loss of heterozygosity.
Genomic DNA Extraction
Lymphoblasts were purified from bone marrow or peripheral blood specimens using the Ficoll-Paque centrifugation method, according to the manufacturer's instructions (GE Healthcare, Piscataway, NJ). Genomic DNA was extracted from leukemic cells using standard phenol/chloroform-based methods. Briefly, 1 × 106 cells were lysed in 10 mmol/L Tris hydrochloride, 10 mmol/L sodium chloride, 10 mmol/L EDTA, 20 μg proteinase K, and 0.5% SDS by incubating at 37°C for 16 hours. Total RNA was further removed by adding 500 μg PureLink RNase A (Thermo Fisher Scientific) and incubating for 10 minutes at 37°C. An equal volume of phenol-chloroform-isopropanol (25:24:1) was added to the lysates and mixed by vigorous shaking, followed by centrifugation at 16,100 × g at 4°C for 5 minutes. The upper aqueous phase was transferred to a fresh tube; genomic DNA was then precipitated by adding 2× 100% ethanol stored at −80°C. The DNA pellet was washed with 75% ethanol and rehydrated with Tris-EDTA buffer. The concentration of DNA was determined using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific).
MLPA Analysis
Genomic DNA was analyzed using the SALSA MLPA kit (MRC-Holland), according to the manufacturer's instructions, as described in a previous study.30 SALSA MLPA Probemix P335 ALL-IKZF1 was used for the detection in the alterations of the IKZF1, PAX5, and ETV6 genes. SALSA MLPA Probemix P327 iAMP21-ERG was used for detecting alterations in ERG and the iAMP21 subtype.
Mutation Analysis of TP53 and ETV6
Genomic DNA was used for mutation analysis. For sequencing analysis, the coding regions were amplified using Phusion Hot Start II High-Fidelity PCR Master Mix (Thermo Fisher Scientific). The primers are listed in Supplemental Table S2. Thermocycling was performed as follows: 98°C for 30 seconds, then 38 cycles of 98°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, followed by a final extension at 72°C for 5 minutes. The PCR products were confirmed using 2% agarose gel electrophoresis. DNA bands were purified using the FavorPrep GEL/PCR Purification Kit. Sanger sequencing was performed using an ABI 3730XL DNA analyzer. The sequencing results were aligned to the National Center for Biotechnology Information (NCBI) GenBank (http://www.ncbi.nlm.nih.gov; GenBank accession number NM_000546 or NM_001987) using SnapGene software version 4.1.3.
Transcriptome Sequencing and Bioinformatic Analysis
RNA-seq was performed using the TruSeq Library Prep Kit and the HiSeq 2000 Sequencing System (Illumina, San Diego, CA). All sequence reads were paired end and analyzed using total RNA-seq (100–base pair reads). Default parameters were used for all software programs in a high-performance computing environment, and fastq files were mapped to the GRCh37 human genome reference using STAR version 2.5.3a (https://github.com/alexdobin/STAR/releases). Gene annotation software downloaded from the Ensembl website (http://www.ensembl.org, last accessed January 1, 2021) was used for STAR mapping and the read count evaluation.35 The bioinformatic tools used included STAR-Fusion version 1.8.1 (https://github.com/STAR-Fusion/STAR-Fusion), FusionCatcher version 1.2.0 (https://github.com/ndaniel/fusioncatcher), Squid version 1.5 (https://github.com/Kingsford-Group/squid), Pizzly version 0.37 (https://github.com/pmelsted/pizzly), arriba version 1.2.0 (https://github.com/suhrig/arriba), and Pindel version 0.2.0 (https://github.com/genome/pindel), which specifically search for internal tandem duplication.36 The primers used for RNA-seq validation are listed in Supplemental Table S2.
Evaluation of Gene Expression Level from RNA-Seq Data
The HTSeq package version 0.11.2 (https://htseq.readthedocs.io/en/release_0.11.1/count.html) was used to evaluate gene expression levels, and the DESeq2 version 1.32.0 Bioconductor R package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to perform gene expression level normalization and differential expression analysis. A regularized log-transformed value was calculated using DESeq2 to evaluate the digital gene expression levels. To correct the batch effect introduced by different library preparation strategies and sequencing lengths, the ComBat function in the sva R package version 3.40.0 (https://bioconductor.org/packages/release/bioc/html/sva.html) was used.RNA-seq from 1268 reference samples with clear ALL subtypes predefined was included in the analysis to increase accuracy.24 With the log-transformed gene expression level, the R package Rtsne version 0.15 (https://cran.r-project.org/web/packages/Rtsne/index.html) was used to map the samples to a 2-dimensional t-distributed stochastic neighbor embedding (tSNE) plot with the 1000 most variable genes (based on the median absolute deviation), and the tSNE perplexity parameter was set to 30. Different gene numbers (200, 500, 1000, and 2000) and tSNE parameters (perplexity of 20, 30, 40, and 50) were explored and stable clusters were observed.24 The R package umap version 0.2.7.0 (https://cran.r-project.org/web/packages/umap/index.html) was used to map the T-ALL samples to a two-dimensional Uniform Manifold Approximation and Projection (UMAP) plot with the top 1000 median absolute deviation genes, and the number of the nearest neighbors was set to 15. Gene signature analysis was also performed using DESeq2 with default parameters to evaluate differentially expressed genes.
Statistical Analysis
The EFS was calculated from the date of diagnosis to the first major adverse event, including induction failure, relapse, development of a second malignancy, or death from any cause. The OS was calculated from the date of diagnosis to death of any cause. The time was censored at the date of last contact if no event occurred. The EFS and OS were estimated according to the Kaplan-Meier method and compared using the log-rank test. Univariate and multivariate Cox regression analyses were performed to evaluate hazard ratios and 95% CIs of risk factors. t-tests were performed for surface marker expression. P < 0.05 was considered statistically significant. All statistical analyses were performed using the SAS software version 9.4 (SAS Institute, Cary, NC).
Availability of Data and Materials
The data sets used and/or analyzed during the current study are publicly available and have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo; accession number GSE207057).
Results
RT-PCR and DI to Identify Samples with Recurrent Fusions and the Status of Ploidy of B-ALL and STIL-TAL1 to Screen T-ALL
In this study, 252 patients with B-ALL were recruited (Figure 1). Because most B-ALL subtypes were mutually exclusive, RT-PCR was used to screen the samples for ETV6-RUNX1, BCR-ABL1, KMT2A-AFF1, TCF3-PBX1, and P2RY8-CRLF2 and DI to verify ploidy. Four cases of P2RY8-CRLF2 and one case with hyperdiploidy and BCR-ABL1 were further analyzed using RNA-seq. Two cases with iAMP21 were diagnosed using MLPA.30 Two cases with ZNF384-r were identified by karyotyping and validated by RT-PCR. These approaches could identify specific subtypes in 173 of 252 patients (69%) with B-ALL. For T-ALL, only RT-PCR was used to screen for STIL-TAL1 fusion and identified 14 cases (32.6%) among 43 patients studied. The results of RT-PCR, DI, and MLPA are available within 5 working days after diagnosis.
Integrative Genetic and Genomic Classification of B-ALL by RNA-Seq
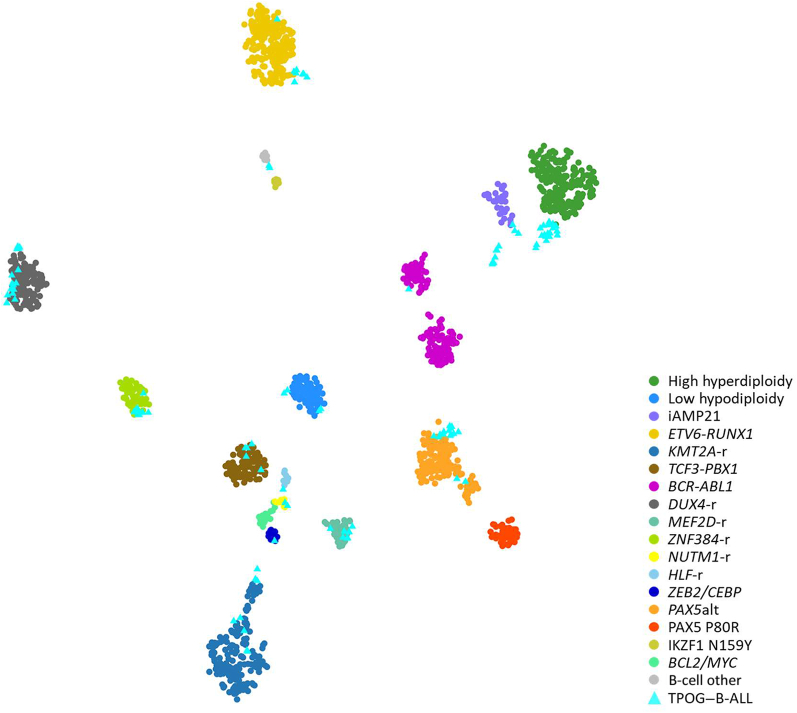
The remaining cases without specific leukemia subtypes identified by conventional genetic analyses were analyzed using RNA-seq (Figure 1) and classified according to the criteria proposed by Jeha et al.37 RNA-seq data were analyzed from 111 individuals with B-ALL (including 33 B-ALL samples with known subtypes, four samples with P2RY8-CRLF2, and one sample with hyperdiploidy status and BCR-ABL1) to obtain gene expression profiles analyzed using hierarchical clustering, tSNE analysis, prediction analysis of microarrays, and predictive modeling 17 using cases of known subtypes (Figure 2).24 Of these, 10 patients harbored ZNF384-r, 15 PAX5alt, 7 MEF2D-r, 6 Ph-like, 15 DUX4-r, 7 KMT2A-r, 1 BCR-ABL1, 2 NUTM1 fusion, 1 TCF3-HLF, and 1 ZEB2/CEBP. With the use of this approach, molecular classification could not be performed for only 10 cases (4%). The heatmap of the entire B-ALL cohort is shown in Supplemental Figure S1, and the final genetic subtypes of all patients are listed in Table 1.
Figure 2.
Integrative acute lymphoblastic leukemia (B-ALL) subtypes. Gene expression profiling of reference samples with clear B-ALL subtype predefined and Taiwan Pediatric Oncology Group (TPOG)–B-ALL samples shown in a two-dimensional t-distributed stochastic neighbor embedding plot. Each dot represents a sample. Major subtypes of reference samples are represented as circles in different colors and TPOG–B-ALL samples are represented as light blue triangles.
Table 1.
Details of the Entire Cohort
| Subtype | No. (%) |
|---|---|
| B-cell acute lymphoblastic leukemia (n = 252) | |
| High hyperdiploidy | 79 (31.3) |
| Low hypodiploidy | 5 (2.0) |
| Near-haploid | 2 (0.8) |
| iAMP21 | 2 (0.8) |
| ETV6-RUNX1 | 47 (18.7) |
| KMT2A-r | 18 (7.1) |
| TCF3-PBX1 | 16 (6.3) |
| BCR-ABL1 | 11 (4.4) |
| DUX4-r | 15 (6.0) |
| MEF2D-r | 7 (2.8) |
| ZNF384-r | 12 (4.8) |
| NUTM1-r | 2 (0.8) |
| HLF-r | 1 (0.4) |
| ZEB2/CEBP | 1 (0.4) |
| PAX5alt | 16 (6.3) |
| Ph-like | 6 (2.4) |
| ETV6-RUNX1–like | 1 (0.4) |
| KMT2A-like | 1 (0.4) |
| B-cell other | 10 (4.0) |
| Total | 252 (100) |
| T-cell acute lymphoblastic leukemia (n = 43) | |
| TAL1 | 20 (46.5) |
| TAL2 | 1 (2.3) |
| TLX3 | 2 (4.7) |
| LMO1/2 | 2 (4.7) |
| NKX2-1 | 2 (4.7) |
| HOXA | 5 (11.6) |
| T-cell other | 11 (25.6) |
| Total | 43 (100) |
IKZF1 Deletions in Patients with DUX4 Rearrangements
Of the 15 cases with DUX4 rearrangements, 14 had sufficient genomic material for further analysis. Two samples had somatic TP53 mutations. Seven cases had IKZF1 deletions [7 of 14 cases (50%) that could be evaluated], a rate higher than that in other reports (Table 2).16,18,22 In contrast, the ERG deletions detected in 2 of 14 cases (14.3%) were less common compared with other studies. In line with previous studies,16, 37 CD2 expression was higher in patients with DUX4-r (Supplemental Figure S2). IKZF1 deletions were not associated with inferior clinical outcomes in patients with DUX4-r (Supplemental Figure S3).
Table 2.
IKZF1, ERG, and TP53 Alterations in DUX4-r ALL
| Alteration | TPOG, no. (%) (n = 252) | Zhang et al16 (n = 1743) |
Lilljebjörn et al22 (n = 195) |
Liu et al18 (n = 94) |
|||
|---|---|---|---|---|---|---|---|
| No. (%) | P∗ | No. (%) | P∗ | No. (%) | P∗ | ||
| DUX4-r ALL | 15† (6.0) | 134 (7.7) | 0.37 | 8 (4.1) | 0.52 | 6 (6.4) | >0.99 |
| IKZF1 | |||||||
| Deletion | 7 (50) | 27 (23.3) | 0.02 | 1 (12.5) | 0.07 | 3 (50.0) | >0.99 |
| Wild type | 7 (50) | 89 (76.7) | 7 (87.5) | 3 (50.0) | |||
| ERG | |||||||
| Deletion | 3 (21.4) | 68 (59.1) | 0.002 | 6 (75.0) | 0.008 | 4 (66.7) | >0.99 |
| Wild type | 11 (88.6) | 47 (40.9) | 2 (25.0) | 2 (33.3) | |||
| TP53 | |||||||
| Mutation | 2 (14.3) | 2 (3.3) | 0.04 | 0 (0.0) | 0.27 | 0 (0.0) | 0.52 |
| Wild type | 12 (85.7) | 59 (96.7) | 8 (100.0) | 6 (100.0) | |||
ETV6 Deletion May Be Enriched in the ZNF384 Subtype
Several novel fusion partners in the ZNF384 fusions were identified (Supplemental Figure S4). Like previous reports,20,38,39 ZNF384 fusion is associated with loss of CD10 and expression of CD33 and CD13 compared with other subtypes (Supplemental Figure S4, Supplemental Table S3). Similar to cases with ETV6-RUNX1, the ETV6 alteration was also enriched in this cohort, a finding not identified in previous studies (Table 3).20, 38, 39 Two patients with somatic ETV6 mutations were identified. The Sanger sequencing of these two samples is shown in Supplemental Figure S5.
Table 3.
ETV6 Alterations Were Enriched in ZNF384-r ALL
Pediatric Cases of B-ALL with Multiple Primary Alterations
Three patients with two primary alterations were identified in this cohort (Table 4). All samples were submitted for the screening of fusion genes, DI, and MLPA P036 to determine the status of the ploidy, and the analysis was stopped at this step if the samples had positive results. The three patients had hyperdiploidy status besides BCR-ABL1 (case 1098) or P2RY8-CRLF2 (cases 829 and 1019). To determine the exact molecular subtype, RNA-seq was performed for these cases, and they were classified as BCR-ABL1 or high hyperdiploidy based on gene expression clustering.
Table 4.
Pediatric B-Cell Acute Lymphoblastic Leukemia Cases with Multiple Primary Alterations
| ID | Molecular subtype | Fusion gene | Cytogenetics |
|---|---|---|---|
| 1098 | BCR-ABL1 | BCR-ABL1 | 58,XX,+X,+4,+4,+5,+6,+9,t(9;22)(q34;q11),+10,+14,+18,+20,+21,+21,der(22)t(9;22) |
| 829 | High hyperdiploidy | P2RY8-CRLF2 | 57,XY,+X,+X,+4,+8,+9,+10,+11,+14,+14,+21,+21 |
| 1019 | High hyperdiploidy | P2RY8-CRLF2 | 52,XY,del(3)(q13q2?7),+X,+8,+10,der(16)t(7;16)(q11;p11),add(16)(p11),+21,+21,+22 |
Genomic Alterations in T-ALL
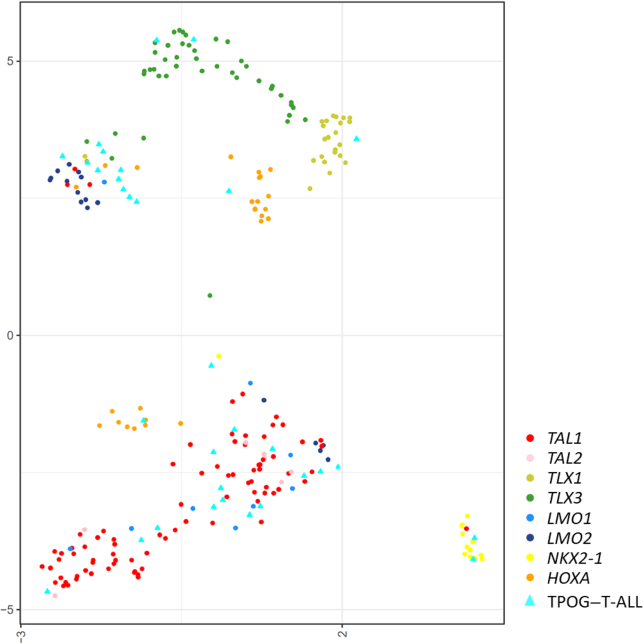
A total of 32 T-ALL samples were submitted for RNA-seq, including three samples with STIL-TAL1. An ensemble approach was used to identify the fusion transcripts in these samples. The gene expression profiles of the RNA-seq data were analyzed using hierarchical clustering (UMAP analysis), which revealed seven clusters of tumors, indicative of different subtypes of T-ALL (Figure 3). The most common subgroup was TAL1 [20 of 43 (46.5%)]. Six samples were identified to have fusions, including BCR-BAL1, KMT2A-r, ETV6-NCOA2, SFPQ-ZFP36L1, and PICALM-MLLT10. The heatmap of T-ALL is presented in Supplemental Figure S6. Complete genetic alterations and molecular classification by RNA-seq and gene expression of the entire cohort are listed in Supplemental Tables S4 and S5.
Figure 3.
Integrative T-cell acute lymphoblastic leukemia (T-ALL) subtypes. Gene expression profiling of reference samples with clear T-ALL subtype predefined and Taiwan Pediatric Oncology Group (TPOG)–T-ALL samples shown in a two-dimensional uniform manifold approximation and projection plot. Each dot represents a sample. Major subtypes of reference samples are represented as circles in different colors and TPOG–T-ALL samples are represented as light blue triangles.
Outcome Analysis by Genotyping
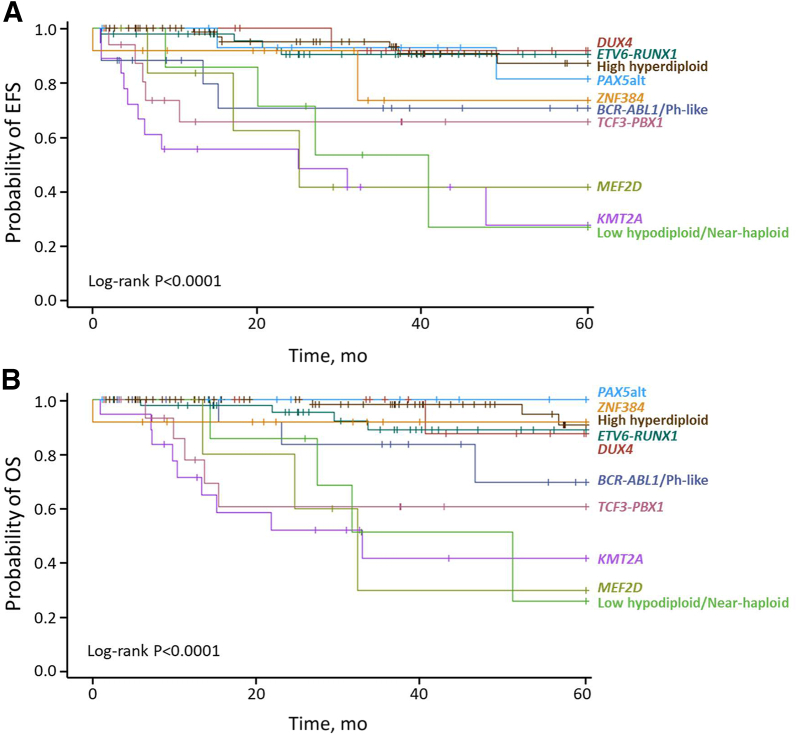
Patients with ETV6-RUNX1, high hyperdiploidy, PAX5alt, and DUX4-r had favorable outcomes, whereas those with hypodiploidy and KMT2A and MEF2D fusions had unfavorable outcomes based on EFS and OS (Figure 4, Supplemental Table S6).
Figure 4.
The 5-year event-free survival (EFS) (A) and 5-year overall survival (OS) (B) according to the risk groups by genetic subtypes. The EFS and OS were estimated using the Kaplan-Meier method, and P values were calculated using the log-rank test.
Discussion
In developing countries with limited resources, a major hurdle to the comprehensive use of RNA-seq for childhood ALL is the cost associated with it. With the use of conventional molecular techniques, such as RT-PCR, DI, and MLPA, approximately 70% of childhood B-ALL can be genetically classified. RT-PCR can aid in the identification of approximately 30% of patients with STIL-TAL1 in the T-ALL cohort. The results of these molecular methods used in this study can be available within 5 working days, and the sequential approach decreases the number of samples required for RNA-seq analysis. RNA-seq can provide information on targetable genetic fusions and molecular classification of transcriptional factors in T-ALL. The data showed that the prevalence rates of genetic alterations in childhood B-ALL in Taiwan differ from those reported in the United States and Europe, and the incidence of Ph-like ALL in Taiwan is much lower than that reported in White and Hispanic populations.12,13,16,22,24,40,41
Until now, most reports on the application of RNA-seq in childhood ALL are from White populations, whereas only a few RNA-seq studies have focused on Asian populations.18,42,43 Although all leukemia subtypes occur in all racial or ancestral groups, there are substantial differences in the incidence rates of several subtypes of B-ALL. Ph-like ALL is a high-risk subtype of B-ALL, with a frequency of 10% to 25% in different age populations among White and Hispanic populations. However, in Japan, Imamura et al42 identified 29 pediatric patients with Ph-like ALL by transcriptome and multiplex RT-PCR analyses among 373 Ph-negative B-ALL patients without recurrent genetic abnormalities. Another study in China that used RNA-seq showed that only 5% of children with ALL exhibited Ph-like subtype.18 Using RNA-seq among children with ALL in Singapore and Malaysia, Ni Chin et al43 found that the frequency of Ph-like ALL was only 2%, a rate similar to the rate [6 of 252 (2.4%)] in this report. When more sequencing studies are conducted in Asian populations, the true distribution of these genetic subtypes could become clearer in the future.
Some differences in genetic alterations have been observed between subtypes. In this cohort, 14.3% of patients with DUX4-r had TP53 somatic mutations, which have not been previously reported.16,22 One DUX4-r case with the TP53 mutation had tetraploidy (Supplemental Table S4), which might have resulted from somatic TP53 mutations.44 The significance of somatic TP53 mutations was unclear because of the low sample number in this study. Besides samples with ETV6-RUNX1 fusion, ETV6 alterations were enriched in patients with ZNF384 fusions, which were not previously identified.20,38,39
The prevalence rates of these novel subtypes might affect the diagnosis strategy. Unlike the higher incidence rate of Ph-like ALL in White and Hispanic populations,13,40,45,46 the incidence rate in this cohort is <3%. Several methods are used to screen for Ph-like ALL because of the availability of possible targeted treatment options after its diagnosis.13,40,45,47 The very low prevalence rate of Ph-like ALL may make its diagnosis by RNA-seq easier in Asian populations, especially for patients who receive risk-directed therapy. RNA-seq alone can identify all cases with targetable lesions. However, DUX4-r accounts for many of the patients in this study, and its diagnosis requires RNA-seq.16,22,48 Some subtypes, such as the ETV6-RUNX1–like and KMT2A-like subtypes, had no specific fusions but had gene expression profiles similar to those of known subtypes.22,24 In such cases, RNA-seq can provide the required information and be helpful for risk-directed therapy. Several samples using gene expression profiles were identified in this cohort.
Liu et al25 used WGS and RNA-seq to identify genetic alterations in T-ALL. So far, this is the largest T-ALL cohort with comprehensive genome sequencing data. Unlike B-ALL, only 50% of the samples had in-frame fusions in T-ALL; the remaining samples could be classified using gene expression profiles, although specific breaking points that result in overexpression should be identified using WGS instead of RNA-seq.49, 50, 51 However, RNA-seq can identify targetable kinase fusions suitable for treatment with tyrosine kinase inhibitors.25 A patient with T-ALL with BCR-ABL1 using RNA-seq was identified, although the t(9;22)(q34;q11) translocation could not be identified by cytogenetic analysis. However, other translocations can be identified using gene expression analysis.
The gene expression profile may be another important determinant of subtype classification, especially in B-ALL, because it may be the only way to identify leukemia subtypes in some patients. Although MRD is the most powerful prognostic marker in childhood ALL, genotyping is also an important tool to determine whether patients are at high risk of relapse. In this sense, negative MRD could not preclude a high incidence of relapse among patients with certain unfavorable leukemia subtypes.9,37,52 Furthermore, it is important to genotype all patients not only for prognostic value but also for possible targeted or novel therapy. With the development of next-generation sequencing, it will be ideal to apply tools such as whole exome sequencing and/or WGS and RNA-seq to all samples. However, application of these tools is difficult to achieve in a developing country. RNA-seq is currently the best approach to identify unknown subtypes and provides useful information for targeted therapy to augment traditional chemotherapy. In countries with limited resources, MLPA and DI also provide important information on genetic alterations of B-ALL, if interpreted accurately.
This study had limitations. Although a comprehensive genome analysis requires RNA-seq, the prevalence rates for some B-ALL subtypes, such as PAX5 P80R and IKZF1 N159Y, are very low. This cohort comprised only 340 patients, and all rare subtypes could not be identified. The hypothesis of the sequential approach that the genetic subtypes in B-ALL are mutually exclusive may not be completely accurate. In this cohort, three patient samples had two genetic alterations, which had high hyperdiploidy. However, with the use of DI and fusion gene tests, samples with high hyperdiploidy and fusion genes could be identified. In addition, not all patients with CRLF2 rearrangements were Ph-like. For patients with P2RY8-CRLF2, RNA-seq might be indicated to confirm Ph-like status. In the largest cohort in the St. Jude's report, 77 of 1261 patients (6.1%) had more than one genetic alteration. These subtypes included KMT2A-r, PAX5alt, CRLF2-r, and iAMP21. The sequential approach might have missed iAMP21, high hyperdiploidy, and CRLF2-r if these cases had more than one genetic alteration and had not been submitted for the RNA-seq.
In conclusion, RNA-seq, RT-PCR, MLPA, and DI could classify 96% of patients with B-ALL in this study. This approach might improve the risk-directed classification system in childhood ALL and help identify targetable and prognostic genetic lesions in patients with childhood ALL. RNA-seq could also provide information about gene expression classification and a few targetable fusions in T-ALL. This study showed that the distribution of genetic alterations in childhood B-ALL is different among various racial populations. More sequencing efforts are required to obtain a clearer picture of the genetic distribution of childhood ALL in Asian populations. The most important hurdle to the comprehensive use of RNA-seq is high cost. The approach described in this study may decrease the size of samples required for RNA-seq analysis, which can be saved for patients for whom molecular classification cannot be performed with traditional methods. Therefore, genetic classification of childhood ALL could be performed in an economical manner without compromising precision medicine in countries with limited resources.
Acknowledgments
We thank all the patients who participated in this study as well as their parents, the TPOG and the Childhood Cancer Foundation in Taiwan, the Pharmacogenomics Laboratory of the National Core Facility for Biopharmaceuticals, and the Next-Generation Sequencing and Microarray Core Facility of the National Taiwan University Centers of Genomic and Precision Medicine for technical support.
Footnotes
Supported by Ministry of Science and Technology, Taiwan grants MOST-107-2314-B-002-173-MY2 (Y.-L.Y.), 110-2314-B-002-091-MY3 (Y.-L.Y.), and MOST-108-2319-B-002-001 (S.-L.Y.), National Taiwan University Hospital grant 110-L1007 (Y.-L.Y.), the Rasing Foundation, the Center of Precision Medicine from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan, NIH grant CA21765, and the American Lebanese Syrian Associates Charities. The bioinformatics analysis was supported in part by National Cancer Institute grant P30 CA021765.
C.-H.Y., G.W., and C.-C.C. contributed equally to this work.
S.-L.Y. and Y.-L.Y. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2022.08.001.
Contributor Information
Sung-Liang Yu, Email: slyu@ntu.edu.tw.
Yung-Li Yang, Email: yangyl92@ntu.edu.tw.
Author Contributions
C.-H.Y., G.W., J.J.Y., C.-H.P., S.-L.Y., and Y.-L.Y. conceived and designed the study; S.-L.Y. and Y.-L.Y. obtained funding; Y.-L.N. and S.-W.L. provided administrative support; S.-T.J., M.-Y.L., K.-H.L., S.-H.C., K.-H.W., F.-L.H., C.-N.C., H.-H.C., J.-L.W., H.-J.Y., M.-J.L., S.-W.C., D.-T.L., and Y.-L.Y. provided study material or patients; C.-T.H., Z.-S.L., and Y.-C.H. performed sequencing experiments; C.-H.Y., G.W., C.-C.C., and Y.-C.H. collected data; G.W., C.-C.C., D.H., Y.-C.H., C.-Y.L., and H.-Y.C. analyzed and interpreted data; all authors wrote, revised, and approved the manuscript for publication. All authors are accountable for all aspects of the work.
Supplemental Data
Heatmap of B-cell ALL.
A:ERGalt transcript was identified by RNA-sequencing in DUX4-r cases. B: CD2 expression was higher in DUX4-r cases than in non–DUX4-r cases. ∗∗∗∗P < 0.0001 (t-test).
IKZF1 deletions are not in association with inferior 5-year event-free survival (A) and 5-year overall survival (B) in DUX4-r patients, although the P value is not significant because of small case numbers.
A:ZNF384 novel fusions. B: CD10 expression is lower and CD33 and CD13 expression is higher in the ZNF384 rearrangements patients. ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 (t-test).
A and B:ETV6 mutations were detected in two patients with ZNF384 fusions.
Heatmap of T-cell ALL.
References
- 1.Pui C.H., Yang J.J., Hunger S.P., Pieters R., Schrappe M., Biondi A., Vora A., Baruchel A., Silverman L.B., Schmiegelow K., Escherich G., Horibe K., Benoit Y.C., Izraeli S., Yeoh A.E., Liang D.C., Downing J.R., Evans W.E., Relling M.V., Mullighan C.G. Childhood acute lymphoblastic leukemia: progress through collaboration. J Clin Oncol. 2015;33:2938–2948. doi: 10.1200/JCO.2014.59.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vora A., Goulden N., Mitchell C., Hancock J., Hough R., Rowntree C., Moorman A.V., Wade R. Augmented post-remission therapy for a minimal residual disease-defined high-risk subgroup of children and young people with clinical standard-risk and intermediate-risk acute lymphoblastic leukaemia (UKALL 2003): a randomised controlled trial. Lancet Oncol. 2014;15:809–818. doi: 10.1016/S1470-2045(14)70243-8. [DOI] [PubMed] [Google Scholar]
- 3.Yeoh A.E., Ariffin H., Chai E.L., Kwok C.S., Chan Y.H., Ponnudurai K., Campana D., Tan P.L., Chan M.Y., Kham S.K., Chong L.A., Tan A.M., Lin H.P., Quah T.C. Minimal residual disease-guided treatment deintensification for children with acute lymphoblastic leukemia: results from the Malaysia-Singapore Acute Lymphoblastic Leukemia 2003 Study. J Clin Oncol. 2012;30:2384–2392. doi: 10.1200/JCO.2011.40.5936. [DOI] [PubMed] [Google Scholar]
- 4.Conter V., Arico M., Basso G., Biondi A., Barisone E., Messina C., Parasole R., De Rossi G., Locatelli F., Pession A., Santoro N., Micalizzi C., Citterio M., Rizzari C., Silvestri D., Rondelli R., Nigro L.L., Ziino O., Testi A.M., Masera G., Valsecchi M.G. Long-term results of the Italian Association of Pediatric Hematology and Oncology (AIEOP) Studies 82:87, 88, 91 and 95 for childhood acute lymphoblastic leukemia. Leukemia. 2009;24:255–264. doi: 10.1038/leu.2009.250. [DOI] [PubMed] [Google Scholar]
- 5.Conter V., Bartram C.R., Valsecchi M.G., Schrauder A., Panzer-Grümayer R., Möricke A., Aricò M., Zimmermann M., Mann G., De Rossi G., Stanulla M., Locatelli F., Basso G., Niggli F., Barisone E., Henze G., Ludwig W.D., Haas O.A., Cazzaniga G., Koehler R., Silvestri D., Bradtke J., Parasole R., Beier R., van Dongen J.J., Biondi A., Schrappe M. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 Study. Blood. 2010;115:3206–3214. doi: 10.1182/blood-2009-10-248146. [DOI] [PubMed] [Google Scholar]
- 6.Hunger S.P., Mullighan C.G. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 7.Veerman A.J., Kamps W.A., van den Berg H., van den Berg E., Bökkerink J.P., Bruin M.C., van den Heuvel-Eibrink M.M., Korbijn C.M., Korthof E.T., van der Pal K., Stijnen T., van Weel Sipman M.H., van Weerden J.F., van Wering E.R., van der Does-van den Berg A., Dutch Childhood Oncology Group Dexamethasone-based therapy for childhood acute lymphoblastic leukaemia: results of the prospective Dutch Childhood Oncology Group (DCOG) protocol ALL-9 (1997–2004) Lancet Oncol. 2009;10:957–966. doi: 10.1016/S1470-2045(09)70228-1. [DOI] [PubMed] [Google Scholar]
- 8.Pui C.H., Pei D., Campana D., Cheng C., Sandlund J.T., Bowman W.P., Hudson M.M., Ribeiro R.C., Raimondi S.C., Jeha S., Howard S.C., Bhojwani D., Inaba H., Rubnitz J.E., Metzger M.L., Gruber T.A., Coustan-Smith E., Downing J.R., Leung W.H., Relling M.V., Evans W.E. A revised definition for cure of childhood acute lymphoblastic leukemia. Leukemia. 2014;28:2336–2343. doi: 10.1038/leu.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pui C.H., Pei D., Raimondi S.C., Coustan-Smith E., Jeha S., Cheng C., Bowman W.P., Sandlund J.T., Ribeiro R.C., Rubnitz J.E., Inaba H., Gruber T.A., Leung W.H., Yang J.J., Downing J.R., Evans W.E., Relling M.V., Campana D. Clinical impact of minimal residual disease in children with different subtypes of acute lymphoblastic leukemia treated with response-adapted therapy. Leukemia. 2017;31:333–339. doi: 10.1038/leu.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson A.K., Ma J., Wang J., Chen X., Gedman A.L., Dang J., et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47:330–337. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmfeldt L., Wei L., Diaz-Flores E., Walsh M., Zhang J., Ding L., et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander T.B., Gu Z., Iacobucci I., Dickerson K., Choi J.K., Xu B., et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018;562:373–379. doi: 10.1038/s41586-018-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts K.G., Li Y., Payne-Turner D., Harvey R.C., Yang Y.L., Pei D., et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts K.G., Morin R.D., Zhang J., Hirst M., Zhao Y., Su X., et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J., McCastlain K., Yoshihara H., Xu B., Chang Y., Churchman M.L., et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48:1481–1489. doi: 10.1038/ng.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu Z., Churchman M., Roberts K., Li Y., Liu Y., Harvey R.C., et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7:13331. doi: 10.1038/ncomms13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y.F., Wang B.Y., Zhang W.N., Huang J.Y., Li B.S., Zhang M., et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBiomedicine. 2016;8:173–183. doi: 10.1016/j.ebiom.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki K., Okuno Y., Kawashima N., Muramatsu H., Okuno T., Wang X., Kataoka S., Sekiya Y., Hamada M., Murakami N., Kojima D., Narita K., Narita A., Sakaguchi H., Sakaguchi K., Yoshida N., Nishio N., Hama A., Takahashi Y., Kudo K., Kato K., Kojima S. MEF2D-BCL9 fusion gene is associated with high-risk acute B-cell precursor lymphoblastic leukemia in adolescents. J Clin Oncol. 2016;34:3451–3459. doi: 10.1200/JCO.2016.66.5547. [DOI] [PubMed] [Google Scholar]
- 20.Qian M., Zhang H., Kham S.K., Liu S., Jiang C., Zhao X., Lu Y., Goodings C., Lin T.N., Zhang R., Moriyama T., Yin Z., Li Z., Quah T.C., Ariffin H., Tan A.M., Shen S., Bhojwani D., Hu S., Chen S., Zheng H., Pui C.H., Yeoh A.E., Yang J.J. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and Crebbp. Genome Res. 2017;27:185–195. doi: 10.1101/gr.209163.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulsson K., Lilljebjörn H., Biloglav A., Olsson L., Rissler M., Castor A., Barbany G., Fogelstrand L., Nordgren A., Sjögren H., Fioretos T., Johansson B. The genomic landscape of high hyperdiploid childhood acute lymphoblastic leukemia. Nat Genet. 2015;47:672–676. doi: 10.1038/ng.3301. [DOI] [PubMed] [Google Scholar]
- 22.Lilljebjörn H., Henningsson R., Hyrenius-Wittsten A., Olsson L., Orsmark-Pietras C., von Palffy S., Askmyr M., Rissler M., Schrappe M., Cario G., Castor A., Pronk C.J., Behrendtz M., Mitelman F., Johansson B., Paulsson K., Andersson A.K., Fontes M., Fioretos T. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat Commun. 2016;7:11790. doi: 10.1038/ncomms11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J.F., Dai Y.T., Lilljebjörn H., Shen S.H., Cui B.W., Bai L., Liu Y.F., Qian M.X., Kubota Y., Kiyoi H., Matsumura I., Miyazaki Y., Olsson L., Tan A.M., Ariffin H., Chen J., Takita J., Yasuda T., Mano H., Johansson B., Yang J.J., Yeoh A.E., Hayakawa F., Chen Z., Pui C.H., Fioretos T., Chen S.J., Huang J.Y. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A. 2018;115:E11711–E11720. doi: 10.1073/pnas.1814397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Z., Churchman M.L., Roberts K.G., Moore I., Zhou X., Nakitandwe J., et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y., Easton J., Shao Y., Maciaszek J., Wang Z., Wilkinson M.R., et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017;49:1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teachey D.T., Pui C.H. Comparative features and outcomes between paediatric T-cell and B-cell acute lymphoblastic leukaemia. Lancet Oncol. 2019;20:e142–e154. doi: 10.1016/S1470-2045(19)30031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gocho Y., Liu J., Hu J., Yang W., Dharia N.V., Zhang J., et al. Network-based systems pharmacology reveals heterogeneity in LCK and BCL2 signaling and therapeutic sensitivity of T-cell acute lymphoblastic leukemia. Nat Cancer. 2021;2:284–299. doi: 10.1038/s43018-020-00167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pui C.H., Nichols K.E., Yang J.J. Somatic and germline genomics in paediatric acute lymphoblastic leukaemia. Nat Rev Clin Oncol. 2019;16:227–240. doi: 10.1038/s41571-018-0136-6. [DOI] [PubMed] [Google Scholar]
- 29.Iacobucci I., Mullighan C.G. Genetic basis of acute lymphoblastic leukemia. J Clin Oncol. 2017;35:975–983. doi: 10.1200/JCO.2016.70.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu C.H., Lin T.K., Jou S.T., Lin C.Y., Lin K.H., Lu M.Y., Chen S.H., Cheng C.N., Wu K.H., Wang S.C., Chang H.H., Li M.J., Ni Y.L., Su Y.N., Lin D.T., Chen H.Y., Harrison C.J., Hung C.C., Lin S.W., Yang Y.L. MLPA and DNA index improve the molecular diagnosis of childhood B-cell acute lymphoblastic leukemia. Sci Rep. 2020;10:11501. doi: 10.1038/s41598-020-68311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M.J., Liu H.C., Yen H.J., Jaing T.H., Lin D.T., Yang C.P., et al. Treatment for childhood acute lymphoblastic leukemia in Taiwan: Taiwan Pediatric Oncology Group ALL-2002 study emphasizing optimal reinduction therapy and central nervous system preventive therapy without cranial radiation. Pediatr Blood Cancer. 2017;64:234–241. doi: 10.1002/pbc.26142. [DOI] [PubMed] [Google Scholar]
- 32.Liang D.C., Yang C.P., Lin D.T., Hung I.J., Lin K.H., Chen J.S., et al. Long-term results of Taiwan Pediatric Oncology Group studies 1997 and 2002 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:397–405. doi: 10.1038/leu.2009.248. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y.L., Hung C.C., Chen J.S., Lin K.H., Jou S.T., Hsiao C.C., Sheen J.M., Cheng C.N., Wu K.H., Lin S.R., Yu S.L., Chen H.Y., Lu M.Y., Wang S.C., Chang H.H., Lin S.W., Su Y.N., Lin D.T. IKZF1 deletions predict a poor prognosis in children with B-cell progenitor acute lymphoblastic leukemia: a multicenter analysis in Taiwan. Cancer Sci. 2011;102:1874–1881. doi: 10.1111/j.1349-7006.2011.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safavi S., Paulsson K. Near-haploid and low-hypodiploid acute lymphoblastic leukemia: two distinct subtypes with consistently poor prognosis. Blood. 2017;129:420–423. doi: 10.1182/blood-2016-10-743765. [DOI] [PubMed] [Google Scholar]
- 35.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR, ultrafast universal RNA-Seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLeod C., Gout A.M., Zhou X., Thrasher A., Rahbarinia D., Brady S.W., et al. St. Jude Cloud: a pediatric cancer genomic data-sharing ecosystem. Cancer Discov. 2021;11:1082–1099. doi: 10.1158/2159-8290.CD-20-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeha S., Choi J., Roberts K.G., Pei D., Coustan-Smith E., Inaba H., Rubnitz J.E., Ribeiro R.C., Gruber T.A., Raimondi S.C., Karol S.E., Qu C., Brady S.W., Gu Z., Yang J.J., Cheng C., Downing J.R., Evans W.E., Relling M.V., Campana D., Mullighan C.G., Pui C.H. Clinical Significance of novel subtypes of acute lymphoblastic leukemia in the context of minimal residual disease-directed therapy. Blood Cancer Discov. 2021;2:326–337. doi: 10.1158/2643-3230.BCD-20-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirabayashi S., Ohki K., Nakabayashi K., Ichikawa H., Momozawa Y., Okamura K., et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102:118–129. doi: 10.3324/haematol.2016.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shago M., Abla O., Hitzler J., Weitzman S., Abdelhaleem M. Frequency and outcome of pediatric acute lymphoblastic leukemia with ZNF384 gene rearrangements including a novel translocation resulting in an ARID1B/ZNF384 gene fusion. Pediatr Blood Cancer. 2016;63:1915–1921. doi: 10.1002/pbc.26116. [DOI] [PubMed] [Google Scholar]
- 40.Reshmi S.C., Harvey R.C., Roberts K.G., Stonerock E., Smith A., Jenkins H., Chen I.M., Valentine M., Liu Y., Li Y., Shao Y., Easton J., Payne-Turner D., Gu Z., Tran T.H., Nguyen J.V., Devidas M., Dai Y., Heerema N.A., Carroll A.J., Raetz E.A., Borowitz M.J., Wood B.L., Angiolillo A.L., Burke M.J., Salzer W.L., Zweidler-McKay P.A., Rabin K.R., Carroll W.L., Zhang J., Loh M.L., Mullighan C.G., Willman C.L., Gastier-Foster J.M., Hunger S.P. Targetable kinase gene fusions in high-risk B-ALL: a study from the Children's Oncology Group. Blood. 2017;129:3352–3361. doi: 10.1182/blood-2016-12-758979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herold T., Schneider S., Metzeler K.H., Neumann M., Hartmann L., Roberts K.G., Konstandin N.P., Greif P.A., Bräundl K., Ksienzyk B., Huk N., Schneider I., Zellmeier E., Jurinovic V., Mansmann U., Hiddemann W., Mullighan C.G., Bohlander S.K., Spiekermann K., Hoelzer D., Brüggemann M., Baldus C.D., Dreyling M., Gökbuget N. Adults with Philadelphia chromosome-like acute lymphoblastic leukemia frequently have IGH-CRLF2 and JAK2 mutations, persistence of minimal residual disease and poor prognosis. Haematologica. 2017;102:130–138. doi: 10.3324/haematol.2015.136366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imamura T., Kiyokawa N., Kato M., Imai C., Okamoto Y., Yano M., Ohki K., Yamashita Y., Kodama Y., Saito A., Mori M., Ishimaru S., Deguchi T., Hashii Y., Shimomura Y., Hori T., Kato K., Goto H., Ogawa C., Koh K., Taki T., Manabe A., Sato A., Kikuta A., Adachi S., Horibe K., Ohara A., Watanabe A., Kawano Y., Ishii E., Shimada H. Characterization of pediatric Philadelphia-negative B-cell precursor acute lymphoblastic leukemia with kinase fusions in Japan. Blood Cancer J. 2016;6:e419. doi: 10.1038/bcj.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni Chin W.H., Li Z., Jiang N., Lim E.H., Suang Lim J.Y., Lu Y., Chiew K.H., Yin Kham S.K., Zhi Oh B.L., Tan A.M., Ariffin H., Yang J.J., Eng-Juh Yeoh A. Practical considerations for using RNA sequencing in management of B-lymphoblastic leukemia: Malaysia-Singapore acute lymphoblastic leukemia 2020 implementation strategy. J Mol Diagn. 2021;23:1359–1372. doi: 10.1016/j.jmoldx.2021.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Galipeau P.C., Cowan D.S., Sanchez C.A., Barrett M.T., Emond M.J., Levine D.S., Rabinovitch P.S., Reid B.J. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci U S A. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiaretti S., Gianfelici V., O'Brien S.M., Mullighan C.G. Advances in the genetics and therapy of acute lymphoblastic leukemia. Am Soc Clin Oncol Educ. 2016;35:e314–e322. doi: 10.1200/EDBK_156628. [DOI] [PubMed] [Google Scholar]
- 46.Roberts K.G., Reshmi S.C., Harvey R.C., Chen I.M., Patel K., Stonerock E., Jenkins H., Dai Y., Valentine M., Gu Z., Zhao Y., Zhang J., Payne-Turner D., Devidas M., Heerema N.A., Carroll A.J., Raetz E.A., Borowitz M.J., Wood B.L., Mattano L.A., Maloney K.W., Carroll W.L., Loh M.L., Willman C.L., Gastier-Foster J.M., Mullighan C.G., Hunger S.P. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: a report from the Children's Oncology Group. Blood. 2018;132:815–824. doi: 10.1182/blood-2018-04-841676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegele B.J., Nardi V. Laboratory testing in BCR-ABL1-like (Philadelphia-like) B-lymphoblastic leukemia/lymphoma. Am J Hematol. 2018;93:971–977. doi: 10.1002/ajh.25126. [DOI] [PubMed] [Google Scholar]
- 48.Yasuda T., Tsuzuki S., Kawazu M., Hayakawa F., Kojima S., Ueno T., Imoto N., Kohsaka S., Kunita A., Doi K., Sakura T., Yujiri T., Kondo E., Fujimaki K., Ueda Y., Aoyama Y., Ohtake S., Takita J., Sai E., Taniwaki M., Kurokawa M., Morishita S., Fukayama M., Kiyoi H., Miyazaki Y., Naoe T., Mano H. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat Genet. 2016;48:569–574. doi: 10.1038/ng.3535. [DOI] [PubMed] [Google Scholar]
- 49.Mansour M.R., Abraham B.J., Anders L., Berezovskaya A., Gutierrez A., Durbin A.D., Etchin J., Lawton L., Sallan S.E., Silverman L.B., Loh M.L., Hunger S.P., Sanda T., Young R.A., Look A.T. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu S., Qian M., Zhang H., Guo Y., Yang J., Zhao X., He H., Lu J., Pan J., Chang M., Du G., Lin T.N., Kham S.K., Quah T.C., Ariffin H., Tan A.M., Cheng Y., Li C., Yeoh A.E., Pui C.H., Skanderup A.J., Yang J.J. Whole-genome noncoding sequence analysis in T-cell acute lymphoblastic leukemia identifies oncogene enhancer mutations. Blood. 2017;129:3264–3268. doi: 10.1182/blood-2017-03-771162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z., Abraham B.J., Berezovskaya A., Farah N., Liu Y., Leon T., Fielding A., Tan S.H., Sanda T., Weintraub A.S., Li B., Shen S., Zhang J., Mansour M.R., Young R.A., Look A.T. APOBEC signature mutation generates an oncogenic enhancer that drives LMO1 expression in T-ALL. Leukemia. 2017;31:2057–2064. doi: 10.1038/leu.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Connor D., Enshaei A., Bartram J., Hancock J., Harrison C.J., Hough R., Samarasinghe S., Schwab C., Vora A., Wade R., Moppett J., Moorman A.V., Goulden N. Genotype-specific minimal residual disease interpretation improves stratification in pediatric acute lymphoblastic leukemia. J Clin Oncol. 2018;36:34–43. doi: 10.1200/JCO.2017.74.0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heatmap of B-cell ALL.
A:ERGalt transcript was identified by RNA-sequencing in DUX4-r cases. B: CD2 expression was higher in DUX4-r cases than in non–DUX4-r cases. ∗∗∗∗P < 0.0001 (t-test).
IKZF1 deletions are not in association with inferior 5-year event-free survival (A) and 5-year overall survival (B) in DUX4-r patients, although the P value is not significant because of small case numbers.
A:ZNF384 novel fusions. B: CD10 expression is lower and CD33 and CD13 expression is higher in the ZNF384 rearrangements patients. ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 (t-test).
A and B:ETV6 mutations were detected in two patients with ZNF384 fusions.
Heatmap of T-cell ALL.
Data Availability Statement
The data sets used and/or analyzed during the current study are publicly available and have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo; accession number GSE207057).