ABSTRACT
The SARS-CoV-2 pandemic resulted in a demand for highly specific and sensitive serological testing to evaluate seroprevalence and antiviral immune responses to infection and vaccines. Hence, there was an urgent need for a serology standard to harmonize results across different natural history and vaccine studies. The Frederick National Laboratory for Cancer Research (FNLCR) generated a U.S. serology standard for SARS-CoV-2 serology assays and subsequently calibrated it to the WHO international standard (National Institute for Biological Standards and Control [NIBSC] code 20/136) (WHO IS). The development included a collaborative study to evaluate the suitability of the U.S. serology standard as a calibrator for SARS-CoV-2 serology assays. The eight laboratories participating in the study tested a total of 17 assays, which included commercial and in-house-derived binding antibody assays, as well as neutralization assays. Notably, the use of the U.S. serology standard to normalize results led to a reduction in the inter-assay coefficient of variation (CV) for IgM levels (pre-normalization range, 370.6% to 1,026.7%, and post-normalization range, 52.8% to 242.3%) and a reduction in the inter-assay CV for IgG levels (pre-normalization range, 3,416.3% to 6,160.8%, and post-normalization range, 41.6% to 134.6%). The following results were assigned to the U.S. serology standard following calibration against the WHO IS: 246 binding antibody units (BAU)/mL for Spike IgM, 764 BAU/mL for Spike IgG, 1,037 BAU/mL for Nucleocapsid IgM, 681 BAU/mL for Nucleocapsid IgG assays, and 813 neutralizing international units (IU)/mL for neutralization assays. The U.S. serology standard has been made publicly available as a resource to the scientific community around the globe to help harmonize results between laboratories.
KEYWORDS: SARS-CoV-2, serology, standard
INTRODUCTION
In late 2019, the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was identified in Wuhan, China (1, 2), and by January of 2020, an international health emergency was declared by WHO, which was followed by a pandemic declaration in March 2020. The widespread testing and vaccine development efforts for SARS-CoV-2 have been unprecedented, enabled by recent technological advancements in molecular biology and vaccine development as well as the expansion of international collaborations and the rapid sharing of results.
Serology is a critical tool to evaluate rates of past infection and immune responses to vaccination. Given the crucial role of serology in the COVID-19 pandemic response, a very large number of serology assays have been developed by commercial and academic laboratories. However, most assays have not been standardized, precluding comparison of results between seroprevalence and vaccine studies.
To address these gaps and facilitate assay standardization across the United States, a U.S. serology standard was rapidly developed at the Frederick National Laboratory for Cancer Research (FNLCR), so serology laboratories in the United States could calibrate their assays and report results in the same units. The development process included the screening of large-volume plasma samples in October 2020 to identify suitable candidate samples, which were pooled to create the serology standard. A collaborative study (8 participating laboratories and 17 assays) was then conducted in November 2020 to determine the suitability for use of the standard, including detectability and repeatability of the standard in a wide range of assay platforms (commercial, in-house-developed, multiplex, and live virus neutralization assays). The serology standard was ready for distribution in December 2020, and the first vials were sent in January 2021 to laboratories in the United States. The standard was later calibrated to the WHO international standard as soon as received in February 2021. Even after development of the WHO international standard (National Institute for Biological Standards and Control [NIBSC] code 20/136) (WHO IS), the U.S. SARS-CoV-2 serology standard has remained a critical resource to enable assay calibration at the national and international level, especially following the recent depletion of the first primary WHO IS. Utilizing these calibrators globally on a routine basis, traced to the international standard, can increase the comparability of results from different studies and enable direct comparisons across different laboratories, as results can be expressed in the same units. These calibrators are key tools to compare data between independent clinical studies across the world using similar assays, enabling more efficient and better-informed decisions for public health. Moreover, assays that are standardized and harmonized across different laboratories facilitate review by regulators as the data are benchmarked and reported with the same units.
MATERIALS AND METHODS
Samples and participating laboratories.
A set of four laboratories participated in the initial screening of nine anticoagulant citrate dextrose (ACD) SARS-CoV-2 convalescent-phase plasma samples (nine individual donors) for neutralizing and binding antibody activity, to generate the candidate standard and to verify that different laboratories and assays had concordant findings regarding seropositivity/seronegativity of the candidate samples. The nine ACD SARS-CoV-2 convalescent-phase plasma samples (nine individual donors) were obtained through BEI Resources, NIAID, NIH, and by the Biomedical Advanced Research and Development Authority (BARDA), and the samples were collected under institutional review board (IRB)-approved protocols and informed consent. These SARS-CoV-2 convalescent-phase plasma samples were collected early on in the pandemic, likely infected with the original SARS-CoV-2 strain, but there is no information regarding PCR testing or hospitalization status. In the end, four donor plasma samples were selected from the initial nine donor samples to create the U.S. serology standard by pooling the four donor plasma samples. The pooled plasma was aliquoted at three different volumes (100 μL/vial, 250 μL/vial, or 500 μL/vial) and stored at −80°C from a liquid state.
Subsequently, a harmonization collaborative study was conducted to evaluate the performance of the candidate standard and the ability to normalize results across different assays and laboratories. That study included eight laboratories, and testing included either automated chemiluminescence assays, manufacturer-developed binding antibody assays, in-house-developed binding antibody assays, or live virus fluorescence reduction neutralization assay. Laboratories were deidentified, and exact assay methods are not disclosed, to maintain deidentification.
The WHO IS was provided as a freeze-dried product that was developed from pooling plasma from 11 donors. The plasma was further solvent-detergent treated to minimize the presence of any possible enveloped viruses.
Screening assays.
(i) SARS-CoV-2 Spike RBD ELISA (method from Florian Krammer's Lab). The SARS-CoV-2 Spike receptor binding domain (RBD) enzyme-linked immunosorbent assay (ELISA) was performed at the Frederick National Laboratory for Cancer Research (FNLCR) as previously described by Krammer’s lab (3, 4). Briefly, a 96-well plate was coated with SARS-CoV-2 RBD (2 μg/mL) and incubated overnight at 4°C. Following incubation, the plate was washed and blocked with a solution of 3% skim milk and 0.1% Tween 20 in phosphate-buffered saline (PBS) for 60 to 240 min at room temperature. The plate was washed, and heat-inactivated samples (diluted 1:50 in sample buffer [1% skim milk with 0.1% Tween 20 in PBS]) were then added in singlet to the plate and incubated at room temperature for 120 to 240 min. The plate was washed again, and diluted horseradish peroxidase (HRP) conjugate was added to the plate and incubated at room temperature for 50 to 65 min. After the plate was washed, SigmaFast ortho-phenylenediamine (OPD) solution was added to the plate and incubated for 10 min at room temperature. Finally, the reaction was stopped with 3 M HCl, and the plate was read at 490 nm. Data analyses were performed using SoftMax Pro GxP 7.0.3 and Microsoft Excel.
(ii) SARS-CoV-2 Spike ELISA (method from the CDC).
The SARS-CoV-2 Spike ELISA screen (pan-Ig) was performed at FNLCR as previously described by the CDC (4, 5). Briefly, a 96-well plate was coated with SARS-CoV-2 Spike protein (1 μg/mL) and incubated overnight (up to 1 week) at 4°C. Following incubation, the plate was washed and then blocked with 5% skim milk and 0.1% Tween 20 in PBS (blocking buffer) for 60 min at 37°C (humidified). Heat-inactivated samples (diluted 1:100 in blocking buffer [5% skim milk, 0.1% Tween 20 in PBS]) were then added in singlet to the plate and incubated at 37°C (humidified) for 60 min. Next, the plate was washed, and diluted goat anti-human IgG/IgA/IgM (pan-Ig) HRP conjugate was added to the plate and incubated at 37°C (humidified) for 60 min. After washing the plate, the 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) peroxidase substrate was added to the plate and incubated for 15 min at 37°C (humidified). Finally, ABTS peroxidase stop solution was added, and the plate was read at 405 nm and 490 nm. Data analyses were performed as previously described using SoftMax Pro GxP 7.0.3 and Microsoft Excel. All positive samples were further evaluated to determine anti-Spike IgG and IgM antibody titers. Briefly, rows 1 to 4 of a 96-well plate were coated with SARS-CoV-2 Spike protein (0.15 μg/mL) and incubated overnight (up to 1 week) at 4°C. Following incubation, the plate was washed and then blocking buffer was added to the plate and incubated for 60 min at 37°C (humidified). The plate was washed, and blocking buffer was added to all rows of the 96-well plate. Then, heat-inactivated sample (diluted 1:25 in blocking buffer) was added to the first and fifth rows of the plate. Next, 4-fold serial dilutions were performed for each sample. The plate was incubated at 37°C (humidified) for 60 min and washed, and diluted goat anti-human IgM HRP conjugate or diluted goat anti-human IgG HRP conjugate was added to each well and incubated at 37°C (humidified) for 60 min. The plate was washed, and the ABTS peroxidase substrate was added to the plate. Following a 15-min, 37°C (humidified) incubation, ABTS peroxidase stop solution was added to the plate, and the plate was read at 405 nm and 490 nm. Data analyses were performed using SoftMax Pro GxP 7.0.3 and Microsoft Excel.
(iii) FNLCR SARS-CoV-2 Spike IgG or IgM and Nucleocapsid IgG or IgM ELISA.
FNLCR developed four individual binding antibody assays, anti-SARS-CoV-2 Spike IgG, anti-SARS-CoV-2 Spike IgM, anti-SARS-CoV-2 Nucleocapsid IgG, and anti-SARS-CoV-2 Nucleocapsid IgM, to evaluate the antibody levels in the candidate samples. Each assay is conducted independently; however, the procedures for testing are similar between the four assays. Briefly, SARS-CoV-2 Spike protein (0.15 μg/mL) or Nucleocapsid protein (0.45 μg/mL) was applied as a coating onto a 96-well plate and incubated overnight (up to 1 week) at 4°C. Following incubation, the plate was washed and blocked with a solution of 4% skim milk and 0.2% Tween 20 in Dulbecco’s PBS (DPBS) for 90 min at room temperature. Next, the plate was washed, and samples (serially diluted 1:3) were added to the plate. The plate was incubated for 60 min at room temperature. After washing the plate, diluted goat anti-human IgM or IgG HRP conjugate was added to each well and incubated at room temperature for 60 min. The plate was washed, and tetramethylbenzidine (TMB) substrate solution was added to the plate. Following a 25-min, room-temperature incubation, 0.36 N sulfuric acid solution was added to the plate, and then the plate was read at 450 nm and 620 nm. Data analyses were performed using SoftMax Pro GxP 7.0.3 and Microsoft Excel. Antibody levels were calculated by interpolation of optical density (OD) values from the standard curve by averaging the calculated concentrations from all dilutions that fall within the working range of the reference curve. The standard curve was calculated from an internal daily assay reference sample (daily-use standard) that we generated by pooling serum from SARS-CoV-2-infected subjects. The daily-use standard is included in every plate and serially diluted to generate a standard curve.
Harmonization collaborative study protocol.
A panel of 16 samples (duplicate aliquots of the U.S. serology standard, duplicate aliquots of five selected positive samples, and duplicate aliquots of two negative samples) was developed to evaluate the candidate U.S. serology standard as well as within-day and between-day assay variability. Of note, the five selected positive samples came from the initial nine donor plasma samples that were screened prior to generating the U.S. serology standard. One of the two negative samples came from the initial nine donor plasma samples that were screened prior to generating the U.S. serology standard, and the other negative sample was selected based on being collected prior to December 2019. Each of the 16 samples had a unique coded identification number, COV1 to COV16. Table S1 in the supplemental material has a description of each of the 16 “COV” samples included in the collaborative study. In total, three panels (16 samples per panel) were distributed to eight laboratories. Each laboratory tested all 16 panel samples for SARS-CoV-2 antibody levels using each organization’s respective method(s) over the course of three separate days. Table 1 describes the participating laboratories, assay source (manufacturer or in-house), type of assay, the antigen tested, and the isotype used (IgG, IgM, or total).
TABLE 1.
Participating laboratory sites (deidentified) and their corresponding assay methodologies used for testing of the U.S. serology standard
| Site | Manufacturer | Assay | Antigen | Conjugate isotype |
|---|---|---|---|---|
| 1 | Abbott | AdviseDX SARS-CoV-2 | Spike | IgM |
| 1 | Abbott | Affinity I SARS-CoV-2 | Nucleocapsid | IgG |
| 1 | In-house | Anti-SARS-CoV-2 | Spike | IgG |
| 1 | In-house | Anti-SARS-CoV-2 orthogonal assay | RBD, then Spike | IgG |
| 1 | In-housea | Anti-SARS-CoV-2 orthogonal assay | RBD, then Spike | IgG |
| 2 | In-house | Anti-SARS-CoV-2 | Spike | IgG |
| 2 | In-house | Anti-SARS-CoV-2 | Spike | IgM |
| 3 | Roche | Elecys anti-SARS-CoV-2 | Nucleocapsid | Total |
| 4 | Euroimmun | Anti-SARS-CoV-2 | Spike S1 | IgG |
| 4 | Abbott | Architect SARS-CoV-2 | Nucleocapsid | IgG |
| 5 | In-house | Fluorescence reduction neutralization assay | Wild-type virus | Total |
| 6 | In-house | Anti-SARS-CoV-2 | Spike | IgG |
| 6 | In-house | RBD-ACE2-PE | RBD-ACE2 | Total |
| 7 | In-house | Anti-SARS-CoV-2 | Spike | IgG |
| 7 | In-house | Anti-SARS-CoV-2 | Spike | IgM |
| 7 | In-house | Anti-SARS-CoV-2 | Nucleocapsid | IgG |
| 7 | In-house | Anti-SARS-CoV-2 | Nucleocapsid | IgM |
| 8 | MSD | Anti-SARS-CoV-2 multiplex assay | RBD/Spike/Nucleocapsid | IgG |
As assay developed for commercial use.
The FNLCR Serology Laboratory personnel prepared the panel sample aliquots to which the collaborative study participants were blind to the sample identification on the same day as the U.S. serology standard was pooled and aliquoted. Samples were stored at −80°C. Participants were asked to complete and return an Excel spreadsheet with the raw data from each assay. The participants were also asked to record their interpretation of whether each sample was positive or negative for antibodies to SARS-CoV-2 based on the criteria in use in their laboratory and to indicate the cutoff value used.
Participants were requested to use the procedure for the detection of antibodies to SARS-CoV-2 in routine use in their laboratory. A diverse range of assay protocols was utilized in the harmonization collaborative study. Most of the immunoassays used either SARS-CoV-2 Spike or Nucleocapsid antigens. Across the eight participating laboratories, 11 assays utilized the SARS-CoV-2 Spike protein. Two laboratories detected neutralizing antibodies either with an in-house-developed live virus fluorescence reduction neutralization assay against wild-type SARS-CoV-2 or with a competitive RBD–angiotensin-converting enzyme 2 (ACE2)–phycoerythrin (PE) assay (measuring antibody-mediated inhibition of RBD binding to ACE2), six assays utilized Nucleocapsid, and four assays utilized the SARS-CoV-2 receptor binding domain of the Spike protein. Most assays utilized a direct ELISA protocol. The Meso Scale Discovery (MSD) assay is a multiplex assay of RBD, Spike, and Nucleocapsid utilizing IgG for detection. For laboratories that were included in the collaborative study and used the orthogonal approach, the samples were first screened in an RBD assay. If the sample tested positive in the RBD assay, then it was reflexed to the Spike assay for a concentration or titer. Hence, all positive results were initially positive in the RBD assay; however, the concentration or titer used for analysis came from the Spike assay, which also tested positive.
Calibration to WHO IS.
Following the receipt of the WHO IS in February 2021, both standards were tested using the four individual FNLCR Serology Laboratory in-house-developed binding antibody assays, anti-SARS-CoV-2 Spike IgG, anti-SARS-CoV-2 Spike IgM, anti-SARS-CoV-2 Nucleocapsid IgG, and anti-SARS-CoV-2 Nucleocapsid IgM, to determine the appropriate starting dilution to evaluate each sample or reagent within the assay linear range. Subsequently, for the calibration assay runs, the WHO IS and U.S. serology standard were tested in triplicate with 8 serial dilutions, on three separate days in each assay with the same conditions maintained across each day. Furthermore, the internal daily assay reference sample (sample with an arbitrarily assigned concentration of analyte tested within each plate to fit a concentration value to each unknown sample tested in the same plate) was run in quadruplicate each day, for a total of 12 replicates over the course of the study. For reference, the internal daily assay reference sample may be referred to as a “daily-use standard.” The daily-use standard is included in every assay plate and is serially diluted to generate a standard curve. A four- or five-parameter logistic regression curve is fit to the raw data generated from the serially diluted daily-use standard, and the raw data results from each sample tested on the plate are interpolated from the four- or five-parameter logistic regression curve fit to daily-use standard results to generate a concentration value for each sample. An unused vial of each sample/reagent was used for each day of testing to avoid variations from freeze-thaw events.
An independent laboratory performed the calibration testing of the U.S. serology standard to the WHO IS for neutralizing unitage. The WHO IS and U.S. serology standard were tested in quadruplicate with 12 serial dilutions, on three separate days with the same conditions maintained across each day. The calibration was conducted using a semiweighted sigmoid statistical assessment, rather than parallel line analysis, since the raw data lacked parallelism. An unused vial of each sample/reagent was used for each day of testing to avoid variations from freeze-thaw events.
Statistical methods.
Combistats (version 6.1, Council of Europe, France) was used to calculate intraday average and inter-day geometric mean values from parallel line analysis for each reagent, except when calibrating the U.S. serology standard to the WHO IS for neutralizing unitage. The calibration was conducted using a semiweighted sigmoid statistical assessment, rather than parallel line analysis, since the raw data lacked parallelism.
Data analysis of the harmonization collaborative study panel results was conducted as follows:
Results of replicates (laboratories were blind to sample identification) within a day were averaged (mean of day 1 results, mean of day 2 results, and mean of day 3 results).
The geometric mean (GM), standard deviation (SD), and coefficient of variation (%CV) were calculated from the 3-day results.
The inter-assay geometric mean was calculated from the geometric mean results calculated from the 3-day results for each assay.
The inter-assay geometric mean of the data was used to assign an arbitrary unit value to the U.S. serology standard for IgG Spike and Nucleocapsid binding antibody assays and IgM Spike and Nucleocapsid binding antibody assays. The arbitrary unit used was described as binding assay unit (BAU) per milliliter. For the neutralization assay (Table 1, described as fluorescence reduction neutralization assay), only the geometric mean of the results from the live virus wild-type assay was used to assign the arbitrary unit. The arbitrary unit used was described as neutralizing unit (NU) per milliliter. As these units are arbitrary, the goal was to assign units close to the inter-assay geometric mean results, with rounding to the nearest hundred, with minor adjustments. The assigned arbitrary values were used to normalize the replicate data using the following formula:
or
For the calibration of the U.S. serology standard to the WHO IS, the Combistats program calculated the geometric mean from the 3 days of testing for each assay relative to the WHO IS.
RESULTS
Sample screening and preparation of SARS-CoV-2 candidate U.S. serology standard.
A total of nine samples from human subjects who donated large volumes of ACD plasma were received. Samples were screened using SARS-CoV-2 enzyme-linked immunoassays (ELISAs) developed by the FNLCR Serology Laboratory. Samples were selected for pooling based on concordant results from four institutions involved in the preliminary serology screening. The plasma samples from four donors (subjects 2, 6, 7, and 9) were selected based on their strong IgG and IgM responses for Spike and strong IgG response for Nucleocapsid and consistency in response between assays and laboratories (Table 2). The U.S. serology standard was produced under the guidelines reported in previous WHO proceedings outlining guidance regarding the preparation, characterization, and calibration of reference materials (6). Specifically, the plasma samples from the four selected donors were pooled and aliquoted at three different volumes (100 μL/vial, 250 μL/vial, or 500 μL/vial) and stored at −80°C from a liquid state within the same day. Stability testing is ongoing as we receive feedback from users of the U.S. serology standard and from monitoring of the antibody response compared to the WHO IS.
TABLE 2.
Screening results of the nine U.S. serology standard candidate samples across seven different ELISAsc
| Subject IDb | Collaborative study panel IDs | Sample type | FNLCR |
Source of methoda |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Krammer, RBD IgG (OD) | CDC |
||||||||
| Spike IgM (AU/mL) | Spike IgG (AU/mL) | Nucleocapsid IgG (AU/mL) | Spike pan-Ig (OD) | Spike IgM (titer) | Spike IgG (titer) | ||||
| Subject 1 | Plasma | 230.2 | 923.3 | 6,240.1 | 2.4 | 2.7 | 100 | 1,600 | |
| Subject 2* | COV8, COV11 | Plasma | 201.6 | 4,971 | 15,838.9 | 2.8 | 2.8 | 100 | 6,400 |
| Subject 3 | COV13, COV15 | Plasma | NEG | NEG | NEG | 0.1 | 0.04 | NEG | NEG |
| Subject 4 | COV6, COV14 | Plasma | 610.8 | 12,281.5 | 10,813.7 | 2.8 | 2.7 | 1,600 | 6,400 |
| Subject 5 | Plasma | NEG | 295.8 | 647.1 | 1.6 | 2.8 | NEG | 400 | |
| Subject 6* | COV5, COV12 | Plasma | 473.1 | 5,319.1 | 12,356.2 | 2.7 | 2.7 | 100 | 6,400 |
| Subject 7* | COV9, COV16 | Plasma | 1,195 | 13,597.1 | 16,618.8 | 3.3 | 1.8 | 1,600 | 6,400 |
| Subject 8 | Plasma | 116.1 | 814.2 | 4,146.9 | 2.1 | 2.8 | 100 | 400 | |
| Subject 9* | COV2, COV10 | Plasma | 124.7 | 31,893.3 | 20,297.7 | 2.8 | 2.8 | 400 | 6,400 |
These assays were tested at FNLCR using protocols provided by Florian Krammer's lab and the CDC.
Asterisks indicate that samples 2, 6, 7, and 9 were pooled to develop the U.S. serology standard. For the collaborative study panel, COV1 and COV4 were assigned to the pooled U.S. serology standard, and COV3 and COV7 samples were assigned to an additional seronegative plasma sample collected prior to 2020, which was not included in the screening set of samples described in this table.
ID, identifier; AU, arbitrary units; NEG, negative; OD, optical density.
Harmonization collaborative study results and data harmonization.
The samples were tested to measure the SARS-CoV-2 IgM and IgG antibody levels, and the data were normalized as described in Materials and Methods. Tables 3 and 4 show the IgM levels and CVs before and after normalization, respectively, while Fig. 1 is a graphic display of the results. In Fig. 1A, the pre-normalization IgM values for the five seropositive samples, excluding the U.S. serology standard, had a 25- to 193-fold range in antibody responses between the minimum and maximum geometric mean values (also Table 3), and in post-normalization (Fig. 1B), the samples had only a 2- to 15-fold range in antibody responses between the minimum and maximum geometric mean values (also Table 4). The highest single-sample inter-assay CV pre-normalization was 1,026.7%, reduced to 145.7% post-normalization. This was an 86% reduction of inter-assay CV.
TABLE 3.
IgM pre-normalization assay resultsb
| ID | Inter-assay GM | Inter-assay SDa | Inter-assay CV (%) | Max GM | Min GM |
|---|---|---|---|---|---|
| Subject 4 | 228.9 | 0.8 | 516.7 | 1,406.6 | 18.6 |
| Subject 2 | 100.5 | 0.7 | 370.6 | 345.0 | 13.7 |
| Subject 6 | 82.5 | 0.9 | 718.7 | 371.4 | 4.0 |
| Subject 7 | 170.2 | 0.9 | 609.8 | 1,605.7 | 18.1 |
| Subject 9 | 115.5 | 1.1 | 1,026.7 | 618.3 | 3.2 |
| U.S. serology standard | 111.4 | 0.7 | 423.0 | 409.8 | 10.3 |
Standard deviation calculated from the log10GM results from each assay.
GM, geometric mean; SD, standard deviation; CV, coefficient of variation.
TABLE 4.
IgM post-normalization assay resultsb
| ID | Inter-assay GM | Inter-assay SDa | Inter-assay CV (%) | Max GM | Min GM |
|---|---|---|---|---|---|
| Subject 4 | 215.7 | 0.3 | 82.3 | 351.5 | 99.4 |
| Subject 2 | 95.7 | 0.2 | 52.8 | 133.9 | 53.9 |
| Subject 6 | 77.9 | 0.2 | 62.0 | 118.2 | 38.9 |
| Subject 7 | 160.2 | 0.5 | 242.3 | 398.7 | 27.3 |
| Subject 9 | 109.4 | 0.4 | 145.7 | 238.4 | 31.6 |
| U.S. serology standard | 100.0 |
Standard deviation calculated from the log10GM results from each assay.
GM, geometric mean; SD, standard deviation; CV, coefficient of variation.
FIG 1.
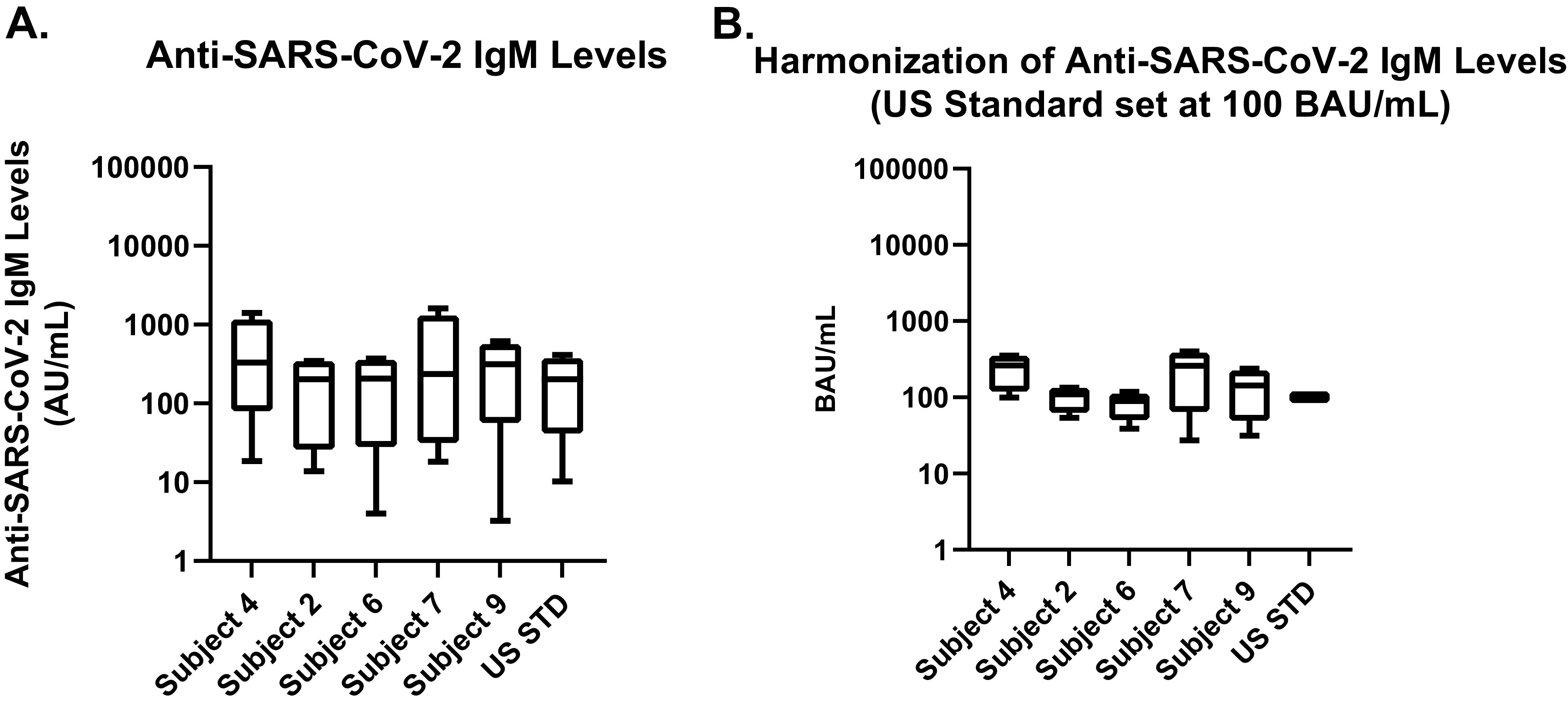
Pre- and post-normalization values for sample test results across all assays and antigens for IgM. (A) Pre-normalization values utilize different units and unit scales. (B) Serology results following normalization using the U.S. serology standard.
Tables 5 and 6 and Fig. 2 are organized similarly for the IgG antibody levels detected among the assays evaluated. In Fig. 2A, the pre-normalization IgG values for the five seropositive samples, excluding the U.S. serology standard, had a 17,000- to 101,000-fold range in antibody responses between the minimum and maximum geometric mean values (also Table 5), and in post-normalization (Fig. 2B), the samples had a 3- to 19-fold range in antibody responses between the minimum and maximum geometric mean values (also Table 6). The highest single-sample inter-assay CV pre-normalization was 6,160.8%, reduced to 90.9% post-normalization. This was a 99% reduction of inter-assay CV. Although 10 of the 13 IgG assays (includes only assays detecting IgG, not total Ig assays, and the multiplex assay accounts for three IgG assays) ranked subject 9 as having the highest antibody response, the antibody responses for the other subjects were more varied, such as subject 4 being ranked as having the lowest antibody response in 6 of the 13 IgG assays tested. Even when grouping the assays by antigen, there was variability with the low- to intermediate-antibody-response samples. However, it is notable that there were a few inter-assay antibody response rankings that were perfectly matched, so overall the agreement was low to moderate between the assays.
TABLE 5.
IgG pre-normalization assay resultsb
| ID | Inter-assay GM | Inter-assay SDa | Inter-assay CV (%) | Max GM | Min GM |
|---|---|---|---|---|---|
| Subject 4 | 822.3 | 1.7 | 4,393.8 | 80,929.4 | 4.9 |
| Subject 2 | 844.8 | 1.5 | 3,416.3 | 137,642.1 | 6.4 |
| Subject 6 | 1,001.3 | 1.7 | 4,703.3 | 614,402.9 | 6.1 |
| Subject 7 | 1,498.9 | 1.7 | 5,223.8 | 197,620.6 | 5.9 |
| Subject 9 | 2,096.9 | 1.8 | 6,160.8 | 321,956.8 | 6.5 |
| U.S. serology standard | 1,241.1 | 1.6 | 4,334.8 | 303,158.3 | 6.4 |
Standard deviation calculated from the log10GM results from each assay.
GM, geometric mean; SD, standard deviation; CV, coefficient of variation.
TABLE 6.
IgG post-normalization assay resultsb
| ID | Inter-assay GM | Inter-assay SDa | Inter-assay CV (%) | Max GM | Min GM |
|---|---|---|---|---|---|
| Subject 4 | 807.5 | 0.4 | 134.6 | 3,428.8 | 182.1 |
| Subject 2 | 841.8 | 0.2 | 41.6 | 1,547.5 | 547.8 |
| Subject 6 | 998.5 | 0.2 | 43.3 | 2,449.3 | 576.9 |
| Subject 7 | 1,490.5 | 0.2 | 60.6 | 3,508.8 | 571.7 |
| Subject 9 | 2,097.3 | 0.3 | 90.9 | 5,192.1 | 901.1 |
| U.S. serology standard | 1,200.0 |
Standard deviation calculated from the log10GM results from each assay.
GM, geometric mean; SD, standard deviation; CV, coefficient of variation.
FIG 2.
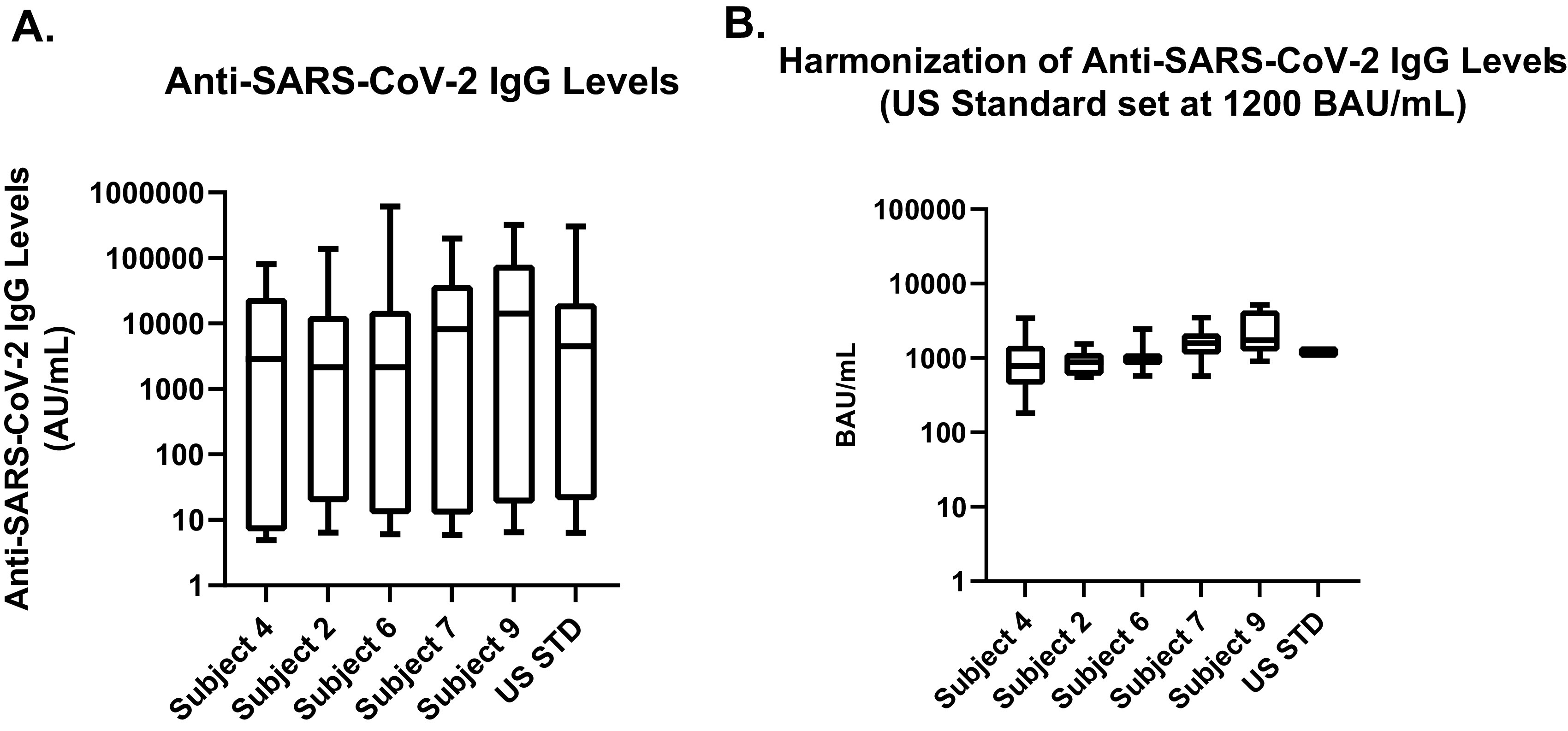
Pre- and post-normalization values for sample test results across all assays and antigens for IgG. (A) Pre-normalization values utilize different units and unit scales. (B) Serology results are normalized using the U.S. serology standard.
Pre-normalization results from different assays span several orders of magnitude, making direct comparison of results difficult without the application of a common calibration factor. The common calibration factor was determined to be 1,200 binding assay units (BAU)/mL for IgG and 100 BAU/mL for IgM, based on the overall geometric mean response of the U.S. serology standard prior to the normalized results. As only one live virus neutralization assay was tested in the harmonization collaborative study, an arbitrary calibration value of 200 neutralizing units (NU)/mL was assigned, based on the geometric mean of the titers from the 3 days of testing, and then normalized.
Calibration to the WHO IS.
The development and implementation of the U.S. serology standard coincided with the release of the WHO IS in December 2020; however, the U.S. serology standard was not calibrated to the WHO IS until its receipt by the FNLCR Serology Laboratory in February 2021. The protocol used to calibrate our assays to the WHO IS is described in Materials and Methods, and the results are summarized in Table 7. Based on the Combistats results, the calibrated results for the U.S. serology standard were assigned as follows: 246 BAU/mL and 764 BAU/mL for the anti-SARS-CoV-2 Spike IgM and IgG, respectively, and 1,037 BAU/mL and 681 BAU/mL for the Nucleocapsid IgM and IgG, respectively. A live virus wild-type neutralization assay was performed to determine the calibration of the U.S. serology standard to the WHO IS, and the neutralizing international unit (IU) per milliliter assigned based on the results from the calibration study was 813 IU/mL.
TABLE 7.
Calibrated concentrations to the WHO IS, reported in binding antibody unit per milliliter
| Standard | Spike IgG (BAU/mL) | Nucleocapsid IgG (BAU/mL) | Spike IgM (BAU/mL) | Nucleocapsid IgM (BAU/mL) | Neutralization assay (IU/mL) |
|---|---|---|---|---|---|
| WHO IS | 1,000 | 1,000 | 1,000 | 1,000 | 1,000 |
| U.S. serology standard | 764 | 681 | 246 | 1,037 | 813 |
| Internal daily assay reference | 94 | 74 | 911 | 10,707 | NAa |
NA, not available.
DISCUSSION
A serological standard that can be used for virtually all serology assays is intended for calibration and harmonization of assay data. It is important that a serological standard is used as early as possible when evaluating serology data resulting from a natural infection in serosurveillance studies or receiving a candidate vaccine. Here, we have presented the development of a U.S. serology standard, the collaborative inter-assay testing results, and the calibration of the U.S. serology standard to the WHO IS.
We developed the U.S. serology standard, following WHO and NIBSC general guidelines for evaluation of suitability for a broad range of different assays and reflecting how a test sample would behave on those assays. The U.S. serology standard is a pool of plasma samples from four donors that were shown to be broadly reactive to SARS-CoV-2 Spike and Nucleocapsid antigens in a collaborative harmonization study that included 8 laboratories that between them utilized 17 different assays. The candidate U.S. serology standard was initially assigned arbitrary units in December 2020 until the WHO IS was received in February 2021 in the FNLCR laboratory. At that time, the U.S. serology standard was finally calibrated to the WHO IS. It must be noted that the calibration of an assay with a secondary standard and WHO IS is specific to the assay antigen and class of immunoglobulin. Also, when harmonizing assays to the WHO IS, only results from assays with the same specificity and isotype should be compiled. The main goal for producing the U.S. serology standard was to ensure that a serology standard would be available to the scientific serology community as rapidly as possible, so results from different assays could be reported in the same units and use the same benchmark for both seroprevalence and vaccine studies. The U.S. serology standard is available free of charge, following request at the Frederick National Laboratory for Cancer Research website, and upon material transfer agreement (MTA) execution. As of June 2022, we have distributed more than 1,000 aliquots of the U.S. serology standard to over 155 organizations inside and outside the United States, including government, academia, and industry.
One of the limitations of our study is that the assigned units for the neutralization assays were based on the results from the single live virus neutralization assay used, as the competitive RBD-ACE2-PE assay was a surrogate for the live virus neutralization assay and, therefore, the results from the two assays were not combined. Considering the need for a biosafety level 3 (BSL-3) facility to perform the testing and the need for rapid development of the secondary standard material for the scientific community, it was reasonable to assess neutralization with the one available assay, which had demonstrated great specificity and sensitivity results (data not shown). Also, we only had limited assay data from assays measuring total Ig. As these assays can detect several isotypes, we could not group the results with either IgG or IgM analysis alone, so these assays were utilized only to assess general antibody response levels or lack of response to the two seronegative samples included in the collaborative study panel.
The supply of the international standard is limited and intended to last several years, given the complexity of the process and timelines associated with production and the formal WHO approval by the Expert Committee on Biological Standardization (ECBS), prior to release of an international standard for public distribution. Thus, production of the U.S. serology standard was critical to serve as a tool for calibration and more regular use by serology laboratories, particularly prior to the release of the first WHO IS or when the International Standard was exhausted later in 2021.
With the collaborative study, we were able to demonstrate the utility of the U.S. serology standard, as its use enabled normalization of assay data from the various laboratories, which reduced by orders of magnitude the differences between assay results. The problem with assay comparability in the absence of a standard when utilizing any calculated arbitrary unit measure(s) is that the differences in results may be extreme between laboratories, which does not allow for direct comparison of study results in the absence of a common reference. As we respond to each new variant as it arises, the need to share data quickly, effectively, and calibrated to the same standard can help support faster decisions to slow the spread of SARS-CoV-2 while enabling us to further explore the population-level dynamics of the pandemic.
Overall, serology is instrumental in identifying individuals who may have been exposed to the SARS-CoV-2 virus and may have developed an adaptive immune response. It can be also used to determine the immune response to vaccines and potency of convalescent-phase plasma. However, the current emergency use authorization (EUA)-authorized SARS-CoV-2 serology assays are not recommended to assess protection against COVID-19 (7, 8). Studies are ongoing to determine correlates of protection and minimal levels of antibodies required for protection against infection.
A key aspect related to the U.S. serology standard implementation was global access and correct utilization. As more organizations report data calibrated to the international serology standard for SARS-CoV-2, the easier it will be to compare results and advance understanding in the areas of seroprevalence and vaccine immunogenicity. We highly encourage laboratories conducting serosurveillance and evaluation of serological responses to vaccines to use a serology standard and report data in international units. To ensure correct utilization, WHO and we have provided several seminars in this area (https://cdn.who.int/media/docs/default-source/biologicals/covid-19/agenda-ppt-q-a-ik.pdf?sfvrsn=89c3ddfa_5) and worked with laboratories individually to provide guidance on the use of the standard, as often laboratories have been unclear on how to use it. The FNLCR Serology Laboratory-generated U.S. serology standard and the NIBSC-generated WHO IS are powerful tools available to laboratories worldwide to improve data comparability for ongoing and future SARS-CoV-2 serology studies, which may facilitate use of data to inform public health measures.
ACKNOWLEDGMENTS
We thank the anonymous plasma donors for their generosity during the SARS-CoV-2 pandemic. Without their commitment to biomedical research, developing and qualifying this material would not be possible for SARS-CoV-2 serological assays. Furthermore, we thank the collaborative study laboratories for their continued support for evaluating this material as well as support for harmonizing SARS-CoV-2 serology assays across different organizations. Critical samples and materials used in the evaluation were provided by the NIH National Institute of Allergy and Infectious Diseases-supported BEI Resources Repository (beiresources.org). Furthermore, we thank all the analysts at the Serology Laboratory at the FNLCR for their strong dedication and time to prepare sample aliquots and conduct testing.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. 75N91019D00024 and HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supplemental material is available online only.
Contributor Information
Ligia A. Pinto, Email: pintol@mail.nih.gov.
Melissa B. Miller, UNC School of Medicine
REFERENCES
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C, Kirkpatrick E, Meade P, Brito RN, Teo C, McMahon M, Simon V, Krammer F. 2020. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinto LA, Shawar RM, O’Leary B, Kemp TJ, Cherry J, Thornburg N, Miller CN, Gallagher PS, Stenzel T, Schuck B, Owen SM, Kondratovich M, Satheshkumar PS, Schuh A, Lester S, Cassetti MC, Sharpless NE, Gitterman S, Lowy DR. 2022. A trans-governmental collaboration to independently evaluate SARS-CoV-2 serology assays. Microbiol Spectr 10:e0156421. doi: 10.1128/spectrum.01564-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman B, Lester S, Mills L, Rasheed MAU, Moye S, Abiona O, Hutchinson GB, Morales-Betoulle M, Krapinunaya I, Gibbons A, Chiang CF, Cannon D, Klena J, Johnson JA, Owen SM, Graham BS, Corbett KS, Thornburg NJ. 2020. Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and serosurveillance. bioRxiv. doi: 10.1101/2020.04.24.057323. [DOI]
- 6.World Health Organization. 2006. WHO Expert Committee on Biological Standardization. World Health Organ Tech Rep Ser 932:1–137. [PubMed] [Google Scholar]
- 7.CDC. 2022. Interim guidelines for COVID-19 antibody testing. CDC, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html.
- 8.FDA. 2022. Antibody testing is not currently recommended to assess immunity after COVID-19 vaccination: FDA safety communication. FDA, Silver Spring, MD. https://www.fda.gov/medical-devices/safety-communications/antibody-testing-not-currently-recommended-assess-immunity-after-covid-19-vaccination-fda-safety.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jcm.00995-22-s0001.pdf, PDF file, 0.1 MB (13KB, pdf)


