Abstract
Antibacterial resistance is a prominent issue with monotherapy often leading to treatment failure in serious infections. Many mechanisms can lead to antibacterial resistance including deactivation of antibacterial agents by bacterial enzymes. Enzymatic drug modification confers resistance to β-lactams, aminoglycosides, chloramphenicol, macrolides, isoniazid, rifamycins, fosfomycin and lincosamides. Novel enzyme inhibitor adjuvants have been developed in an attempt to overcome resistance to these agents, only a few of which have so far reached the market. This review discusses the different enzymatic processes that lead to deactivation of antibacterial agents and provides an update on the current and potential enzyme inhibitors that may restore bacterial susceptibility.
Enzymatic drug deactivation is an important contributor to bacterial resistance. Adjuvants which inhibit the β-lactamases help maintain the efficacy of the β-lactams, demonstrating the potential for this strategy for other antibacterial classes.
Introduction
The discovery of antibiotics has resulted in some of the most important innovations in healthcare. Many antibacterial agents were discovered in the decades following Fleming's discovery of penicillin but this era was also accompanied by the rapid emergence of resistance.1
The management of infectious diseases caused by bacteria relies on the continuing efficacy of antibacterials, but the rate of bacterial resistance has triggered global healthcare concerns of the advent of a post antibiotic era due to the decreasing availability of effective agents.2 Antimicrobial stewardship programs,3 which aim to ensure that antibacterial agents are used appropriately in order to retard the rate of development of resistance, are mostly restricted to developed nations and hospital rather than community usage.4 From 2017 to 2020, only eleven new antibacterial agents were approved, illustrating that the new antibacterial pipeline alone is insufficient to deal with the decrease in efficacy of current agents due to the steady emergence of drug-resistant infections.2
Antibacterial agents are gradually losing their effectiveness against bacteria (Table 1) and Mycobacterium tuberculosis, Pseudomonas aeruginosa, Acinetobacter baumannii, methicillin-resistant Staphylococcus aureus (MRSA), Enterococcus faecium, Helicobacter pylori, Streptococcus pneumoniae, Haemophilus influenzae, and Enterobacteriaceae have been defined by the World Health Organization (WHO) as priority pathogens.5 These bacteria are responsible for multiple nosocomial and community-acquired infections that can result in serious morbidities or mortality.6,7 Multi-drug resistant (MDR) bacteria are currently estimated to be the cause of 700 000 deaths globally every year and this is forecast to rise to 10 million by 2050.8
Examples of pathogenic bacterial species displaying resistance to antibacterials due to enzymatic drug modification.
| Pathogenic bacterium | Antibacterial resistance |
|---|---|
| S. aureus | β-Lactams9 |
| Aminoglycosides10 | |
| Fosfomycin11 | |
| S. pneumoniae | β-Lactams9 |
| C. difficile | β-Lactams9 |
| Aminoglycosides12 | |
| E. coli | Aminoglycosides13 |
| Chloramphenicol14 | |
| Macrolides15 | |
| Tetracyclines16 | |
| P. aeruginosa | Aminoglycosides17 |
| Macrolides18 | |
| Rifamycin19 | |
| A. baumannii | Aminoglycosides20 |
| Tetracyclines21 | |
| K. pneumoniae | β-Lactams9 |
| Aminoglycosides22 | |
| Rifamycin19 | |
| N. gonorrhoeae | β-Lactams9 |
| M. tuberculosis | Isoniazid23 |
Although resistance is a natural evolutionary process, its spread has been dramatically exacerbated by the overuse and misuse of antibacterial treatments.2 Resistance can be intrinsic (i.e. naturally present) or acquired through horizontal gene transfer or though mutations in chromosomal genes24 and can result in three main outcomes;
• The decreased accumulation of the drug as a result of decreased uptake or increased efflux,
• The modification of the drug target (i.e. enzymes or proteins), and
• Alterations to the drug structure making it inactive,24 for example deactivation by enzymatic hydrolysis (e.g. β-lactams) or enzymatic structural modification (e.g. aminoglycosides) (Table 2).25
Examples of resistance genes encoding for the enzymatic modification of antibacterials.
| Resistance mechanism | Resistance gene(s) | Enzyme | Resistance phenotype | Ref. |
|---|---|---|---|---|
| Hydrolytic modification | bla | β-Lactamases | β-Lactam antibiotics in Gram positive and Gram negative | 9 |
| ere | Esterases | Macrolides in Gram positive and Gram negative | 26 | |
| Chemical modification | aac | Acetyltransferases | Aminoglycosides in Gram positive and Gram negative | 27 |
| ant | Nucleotidyltransferases | |||
| aph | Phosphotransferases | |||
| fosA | Glutathionetransferase | Fosfomycin in Gram positive and Gram negative | 28 | |
| fosB | Bacillithioltransferase | |||
| fomA | Phosphotransferases | |||
| fomB | ||||
| cat | Acetyltransferases | Chloramphenicol in Gram positive and Gram negative | 29 | |
| orf2 | Phosphotransferase | |||
| nat | Acetyltransferase | Isoniazid in M. tuberculosis | 23 | |
| rgt | Glycosylation | Rifamycin Gram positive and mycobacteria | 30 | |
| rph | Phosphotransferase | |||
| arr | Ribosyltransferase | |||
| tetX | Oxidoreductases | Tetracyclines in Gram negative | 31 |
The discovery of new antibacterial agents is not the only means of overcoming the problems posed by resistance. The development and use of inhibitors of antibacterial deactivating enzymes, termed adjuncts or adjuvants, has had significant clinical effects through the restoration of antibacterial activity and thus improvements to treatment outcomes. Many enzymatic drug modification processes are still unknown or poorly studied, presenting a significant challenge to the development of novel inhibitors.2
The aims of this review are to investigate the range of enzymatic processes that can lead to resistance through antibacterial modification, describe the current and potential adjuvants that can be used to overcome resistance caused by enzymatic drug degradation, and highlight the areas that would benefit from further research.
This review will discuss current and potential inhibitors of enzymes which degrade the antibacterial classes listed in Table 2.
Antibacterial classes
β-Lactams
The β-lactams, consisting of the penicillins, cephalosporins, carbapenems and monobactams (Fig. 1), are among the most prescribed antibacterial agents, particularly those β-lactams which have a broad spectrum of activity and so are often the mainstays of empirical therapy.32
Fig. 1. The β-lactam classes of antibiotics.
Mechanism of action
β-Lactams target bacterial cell wall synthesis through covalent bond formation with penicillin binding proteins (PBPs) (Scheme 1), enzymes which catalyse the formation of the crosslinks needed to give the peptidoglycan layer its strength. During growth and division, the bacterial cell wall is a dynamic environment in which remodelling by hydrolysis (by murein (peptidoglycan) hydrolases, sometimes referred to as lysins) and synthesis (by PBPs) is carefully controlled. Thus, inhibition of PBPs by β-lactams can result in cell lysis and death.8,33
Scheme 1. Nucleophilic attack of the hydroxyl group of the PBP active site serine residue on the carbonyl of the β-lactam ring, forming a covalent bond.
Emergence of resistance
Resistance to the penicillins was discovered almost immediately following their discovery and, in his Nobel lecture in 1945, Fleming warned that exposing microbes to penicillin concentrations which were insufficient to kill them led to resistance.34 The clinical use of β-lactams has become increasingly challenging with the emergence of β-lactamase degrading enzymes. In 1980s, Gram negative bacteria producing extended-spectrum β-lactamases (ESBLs) were first identified which can hydrolyse penicillins, cephalosporins and monobactams.35 Carbapenems were considered the treatment of choice against such ESBL-producing organisms as they were able to resist rapid hydrolysis, but the emergence of carbapenemases has now created a significant new threat to the use of these ‘last resort’ agents.36
β-Lactamases
The β-Lactamase DataBase (BLDB)37 lists more than 7500 β-lactamases (see Table 3 for examples) and the Ambler system classifies them into classes A, B, C and D based upon similarities in sequence information and motifs.38 Although they differ structurally, class A, C and D all hydrolyse β-lactams through a distinct serine residue in the active site (Scheme 2), whereas class B utilises a metal ion to activate the water molecule that attacks the β-lactam amide bond.39
Common β-lactamase genes, examples of pathogenic hosts and the antibiotics that they hydrolyse39 (compiled with the aid of the Comprehensive Antibiotic Resistance Database; CARD; https://card.mcmaster.ca)40.
| β-Lactamase class | Examples of β-lactamase genes | Host pathogens | Substrates | Ref. |
|---|---|---|---|---|
| Class A | IMI | Enterobacter cloacae | Imipenem | 41 |
| CTX-M | Escherichia coli, K. pneumoniae | Cephalosporins (cefotaxime [CTX] activity greatest) | 42 | |
| Salmonella enterica, P. mirabilis | ||||
| KPC | K. pneumoniae, A. baumannii | Carbapenems, cephalosporins | 43–45 | |
| P. aeruginosa, S. enterica | ||||
| SME | Serratia marcescens | Carbapenems | 46 | |
| BlaZ | S. aureus | Penicillin, cephalosporin, carbapenem | 47 | |
| SHV | K. pneumonia | Penicillins, cephalosporins, carbapenem | 48 | |
| E. coli | ||||
| Class B (metallo-) | IMP | Enteric Gram negative bacteria | Carbapenem, cephalosporins | 49 |
| Pseudomonas spp., Acinetobacter spp. | ||||
| VIM | P. aeruginosa | Penicillins, cephalosporins, carbapenem | 49, 50 | |
| NDM-1 | E. coli, K. pneumoniae, E. cloacae | Carbapenems, cephalosporins, aztreonam, penicillins | 51 | |
| Proteus spp., Citrobacter freundii | ||||
| Class C | AmpC | Enterobacteriaceae | Cephalosporins, penicillin | 52 |
| Class D | OXA | Enterobacter spp., Serratia spp., C. freundii, Aeromonas spp., Proteus spp., Providencia spp., Morganella morganii (ESCAPPMs) | Penicillins, cephalosporins, carbapenems | 53 |
Scheme 2. Hydrolysis of cephalosporins by class A β-lactamases. Nucleophilic attack of the hydroxyl group of Ser70 on the amide group of β-lactams results in cleavage of the β-lactam ring.
The serine β-lactamase classes (A, C, D) share a common mechanism of action. The first step of deactivation mimics the first step of PBP inhibition by β-lactams, with the hydroxyl group of the serine residue undergoing nucleophilic attack on the amide bond of the lactam ring, forming a high-energy acyl–enzyme adduct. An activated water molecule then hydrolyses the adduct, breaking open the β-lactam ring and leading to deactivation of the drug. The amino acids which activate the serine residue and the water molecule form the basis of differentiation between the Ambler classes.54–56Scheme 2 illustrates this mechanism using a class A β-lactamase as an example.
Unlike the serine hydrolases, class B (metallo-)β-lactamases (MBLs) rely on Zn2+ ions in the active site to hydrolyse β-lactams and no amino acid residues are directly involved in the hydrolysis (Fig. 2).57
Fig. 2. Binding and hydrolysis of ampicillin by MBLs (PDB ID: 5O2F).58 The zinc ions facilitate the nucleophilic attack and help stabilise the tetrahedral intermediate and the hydrolysed product.
β-Lactamase inhibitors
Clinically approved β-lactamase inhibitors (Table 4) that are currently on the market include clavulanic acid, tazobactam and sulbactam (Fig. 3); as they contain a β-lactam ring, they act as suicide inhibitors, mimicking the structure of the β-lactams and forming a covalent enzyme–inhibitor bond (Fig. 4).32
Examples of β-lactamase inhibitors, their β-lactam adjuvant and the β-lactamase class inhibited59,60 (compiled with the aid of the Comprehensive Antibiotic Resistance Database; CARD; https://card.mcmaster.ca)40.
| Name of inhibitor | β-Lactam | β-Lactamase class inhibited | Ref. |
|---|---|---|---|
| Approved inhibitors | |||
| Clavulanic acid | Amoxicillin or ticarcillin | Class A, some class D | 61, 62 |
| Tazobactam | Piperacillin | Class A, some class D | 63 |
| Sulbactam | Ampicillin or cefoperazone | Class A | 64, 65 |
| Avibactam | Ceftazidime | Class A, C, D | 66 |
| Relebactam | Imipenem/cilastatin | Class A, C | 67, 68 |
| Vaborbactam | Meropenem | Class A, C | 67 |
| Developmental inhibitors in clinical trials | |||
| Nacubactam | Meropenem – phase I trial | Class A, C | 69, 70 |
| Zidebactam | Cefepime – phase III trial | Class A, C | 71 |
| ETX2514 | Sulbactam – phase II trial | Class A, C and some D | 72 |
| ETX0282 | Cefpodoxime – phase I trial | Class A, C | 73 |
| LN-1-255 | Ceftazidime – phase II trial | Class A, D (non-carbapenemases) | 74 |
| Enmetazobactam | Cefepime – phase III trial | Class A | 75, 76 |
| Taniborbactam | Cefepime – phase II trial | Class A, B, C, D | 77 |
| VNRX-7145 | Cefibutin – phase I trial | Class A, C, D | 78 |
| LK-157 | Cefuroxime – phase I trial | Class A, C | 78 |
| BLI-489 | Piperacillin – phase II trial | Class A, C, D | 79 |
| d-Captopril | Meropenem – phase I trial | Class B | 80 |
Fig. 3. Structures of the six currently approved β-lactamase inhibitors.
Fig. 4. Irreversible binding of clavulanic acid to serine β-lactamase (PDB ID: 6NVU).81 A The clavulanic acid has taken the place of a β-lactam antibiotic in the active site and the hydrogen bond between the carbonyl of the ring opened acyl–enzyme intermediate ii and serine237 is shown. B Schematic representation of the β-lactamase inhibition, involving nucleophilic attack by the hydroxyl group of serine70 on the carbonyl oxygen of the β-lactam ring, decarboxylation of the initially formed acyl–enzyme intermediate i, and hydrolysis of the imine ii, to form a high-energy acyl–enzyme adduct iii.
Clavulanic acid and the other β-lactam containing β-lactamase inhibitors bind to the active site by forming a covalent bond with the hydroxyl group of the serine residue, the same residue that is involved in binding to the β-lactams. The leaving groups of the inhibitors allow the five-membered ring to open readily and form an imine intermediate that further rearranges itself to an enamine intermediate, eventually forming a stable acylated enzyme intermediate. The acyl group is very slowly hydrolysed compared to the acyl group formed by the attack of serine β-lactamases on β-lactams, making this bond quasi-irreversible.7 Avibactam and diazabicyclooctanes (DBOs) also form a covalent acyl–enzyme bond with the key serine residue.2,59 Avibactam is a non-β-lactam β-lactamase inhibitor which is used clinically in combination with ceftazidime and ceftaroline and the ring-opening acylation step is reversible. When bound to KPC-2, avibactam forms hydrogen bonds via its carbonyl oxygen with the nitrogen atoms of Ser70 and Cys237 and the amide of Ala132. The avibactam sulfate moiety forms hydrogen bonds with Asp130, Cys237, Gly235 (Fig. 5).82 Interestingly, the DBO nacubactam (Fig. 5) has been reported to have activity against class A, and B (weak) β-lactamases, as a result of its inhibition of β-lactamases and PBP-2, and a synergistic enhancer effect which is independent of its β-lactamase activity.83
Fig. 5. Taniborbactam, the first serine and metallo-β-lactamase inhibitor to enter clinical development, and nacubactam.
The spread of MBLs is increasing in pathogenic species such as Enterobacteriaceae, Pseudomonas and Acinetobacter species; while class B β-lactamases inhibitors are in trials or are the focus of ongoing research (Table 4), there are currently no approved MBL inhibitors, which is a growing concern due to the threat of class B carbapenemases.84
Taniborbactam (Fig. 5) is the first β-lactamase inhibitor active against both serine and metallo-β-lactamases. Taniborbactam is a cyclic boronate able to inhibit serine-β-lactamases as a reversible covalent inhibitor and metallo-β-lactamases as a competitive inhibitor. When used in combination susceptibility to cefepime was restored in all 34 E. coli strains expressing A, B, C, and D β-lactamases, revealing a potent broad-spectrum β-lactamase inhibitor, with early phase II trials demonstrating low toxicity.85
Aminoglycosides
Aminoglycosides are frequently employed in the empirical treatment of infections requiring hospital admission due to their bactericidal activity against Gram positive and negative organisms, as well as mycobacteria; the first, streptomycin, was isolated in 1943.86 Unfavourable side effects prompted the search for less toxic aminoglycosides with the newest agent, plazomicin, gaining FDA approval in 2018 for the treatment of complicated urinary tract infections in adults caused by E. coli, K. pneumoniae, P. mirabilis or E. cloacae.87,88
Mechanism of action
The hydroxyl and amino moieties of aminoglycosides (Fig. 6) are involved in binding to 16S rRNA of the ribosomal 30S subunit leading to misreading of mRNA, thus terminating protein synthesis.33,88
Fig. 6. General structure of some clinically used aminoglycosides consisting of free hydroxyl groups, two or more amino groups and an aminocyclitol ring linked to one or more aminosugars.33,88 As the hydroxyl and amino groups are involved in binding to rRNA, they are targeted by aminoglycoside modifying enzymes (AMEs).
Emergence of resistance to aminoglycosides
Resistance to the aminoglycosides has been spreading rapidly, mainly due to transmissible resistance genes which encode aminoglycoside modifying enzymes (AMEs); aminoglycoside N-acetyltransferases (AACs), nucleotidyltransferases (ANTs) or phosphotransferases (APTs).12 As there are many subgroups of each class of enzyme, this review will focus on the chemical reactions that lead to enzyme deactivation. Wright and Berghuis (2007) describe the structure and function of each of these enzymes.27
Aminoglycoside N-acetyltransferases (AACs)
AACs are the largest family of AMEs and due to their 3D structural similarities, they are thought to be a member of the GCN5 family of N-acyltransferases (GNAT),27 which catalyse the acetylation of a primary amino group in the substrate, using acetylCoA as the acetyl donor;89 AAC(1), AAC(2′), AAC(3) and AAC(6′) acetylate aminoglycosides at the 1, 2′, 3 and 6′ position, respectively, and can be further divided into subtypes (Table 5).12 The amino groups are all involved in binding to rRNA so acetylation at these sites, notably at the 6-position, leads to resistance.88 In 2007, a new acetyltransferase, enhanced intracellular survival (Eis) protein, was characterised in resistant M. tuberculosis strains, and was demonstrated to acetylate aminoglycosides at multiple positions.90,91
Examples of aminoglycoside N-acetyltransferases (AACs) and their substrates (compiled with the aid of the Comprehensive Antibiotic Resistance Database; CARD; https://card.mcmaster.ca)40.
| Enzyme | Gene coding enzyme | Pathogenic bacterial expression | Aminoglycoside substrate | Ref. |
|---|---|---|---|---|
| AAC(1)-I | aac(1) | Campylobacter fetus, E. coli | Paromomycin | 97 |
| Neomycin | ||||
| AAC(3)-I | aac(3)-Ib | E. coli, Enterobacter cloacae, K. pneumoniae, P. aeruginosa | Gentamicin | 98 |
| AAC(3)-II | aac(3)-II | A. baumannii, E. coli | Gentamicin | 99 |
| Tobramycin | ||||
| Kanamycin | ||||
| AAC(3)-III | aac(3)-III | P. aeruginosa | Tobramycin | 100 |
| Gentamicin | ||||
| Kanamycin | ||||
| Neomycin | ||||
| AAC(3)-IV | aac(3)-IV | E. coli, Campylobacter jejuni | Netilmicin | 101 |
| Dibekacin | ||||
| Apramycin | ||||
| AAC(3)-VI | aac(3)-VI | E. cloacae, Salmonella enterica | Apramycin | 102 |
| AAC(2′)-I | aac(2′)-I | A. baumannii | Tobramycin | 103 |
| Gentamicin | ||||
| Neomycin | ||||
| AAC(2′)-II | aac(2′)-II | Providencia stuartii | Gentamicin | 104 |
| Tobramycin | ||||
| Netilmicin | ||||
| AAC(6′)-I | aac(6′)-I | P. aeruginosa | Tobramycin | 105 |
| Amikacin | ||||
| Kanamycin | ||||
| AAC(6′)-II | aac(6′)-II | K. pneumoniae | Tobramycin | 106 |
| Gentamicin | ||||
| Kanamycin |
The active site of AACs is aspartate rich, forming a negatively charged area which can accommodate the positively charged aminoglycosides.27 The thioester group of acetyl CoA is subjected to nucleophilic attack by the charged amino group of the drug leading to an acetylated aminoglycoside.92 Each antibiotic binds to the enzyme differently however, the high-resolution X-ray structure of AAC found in M. abscessus shows that some residues are conserved in all subtypes.93
AAC(6′)-Ib is a plasmid mediated enzyme and is a cause of amikacin and kanamycin resistance, but has no reported effect on gentamicin (Fig. 7).94 Although the aac(6′)-Ib-cr gene is not associated with high levels of resistance, when combined with three or four chromosomal mutations it can cause ciprofloxacin and norfloxacin resistance, both in vitro and in vivo.95 Significant concerns arise from the fact that acc(6′)-Ib is carried by all the ESKAPE pathogens (E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter spp.) which are the cause of most hospital infections and have become increasingly resistant to aminoglycosides.96
Fig. 7. Kanamycin A (blue) and acetyl-CoA (orange-red) in the active site of AAC(6′)-Ib (PDB ID 1V0C).95.
AAC inhibitors
Although the search for AAC inhibitors has been ongoing for the last 40 years, no adjuvant has yet been approved for use. The search for an AAC inhibitor is based upon the hypothetical mechanism, in which both the aminoglycoside and the acetyl CoA donor are required to bind at the active site for the reaction to occur.107 Bisubstrate analogues have thus been developed to mimic the tetrahedral transition state geometry, containing aminoglycosides covalently liked to an acetyl CoA derivative through amide, phosphonate or sulphonamide groups (Fig. 8).108
Fig. 8. Structure of the a bisubstrate analogue which mimics the tetrahedral transition state of AAC-catalysed acetylation.108.
Aminoglycoside O-nucleotidyltransferases (ANTs)
ANTs can be divided into 5 classes (2′′,3′′,4′,6′ and 9) and catalyse the transfer of an adenosine monophosphate (AMP) group from adenosine triphosphate (ATP) to the hydroxyl group of an aminoglycoside (Table 6).88 ANTs are the smallest of the three groups of AMEs and most members show very little sequence similarity, with only the N-terminus having a conserved GSx10-12D/ExD/E motif.109
Substrates of the aminoglycoside nucleotidyltransferases (ANTs)12,110.
| Enzyme | Bacterial expression | Aminoglycoside substrates |
|---|---|---|
| ANT(2′′) | P. aeruginosa, E. coli, most ESCAPPM | Tobramycin, gentamicin, kanamycin |
| ANT(3′′)-I | A. baumannii, Pseudomonas spp., Vibrio spp., E. coli, Yersinia enterocolitica | Streptomycin, spectinomycin |
| ANT(4′)-Ia | S. epidermidis, S. aureus, Enterococcus spp., Bacillus spp. | Tobramycin, amikacin, kanamycin |
| ANT(4′)-IIa | P. aeruginosa, Enterobacteriaceae | Tobramycin, amikacin, kanamycin |
| ANT(6′)-I | C. fetus, E. faecium, S. aureus, E. faecalis, Streptococcus spp., Bacillus spp., C. jejuni | Streptomycin |
| ANT(9)-I | S. aureus, Enterococcus spp., Staphylococcus sciuri, E. faecalis | Spectinomycin |
Aminoglycoside O-nucleotidyltransferase inhibitors
7-Hydroxytropolone (7-HT) was identified as an ANT(2′′) and ANT(4′) inhibitor when screening of natural products was carried out.111 7-HT (Fig. 9) is a fermentation product, produced by some Pseudomonas spp. and Streptomyces spp., which does not inhibit aminoglycoside binding but instead acts as a competitive inhibitor of ATP.112E. coli strains expressing ANT(2′′) were unaffected by tobramycin, gentamicin, amikacin, or dibekacin yet became susceptible when the aminoglycosides were combined with 7-HT as their growth was inhibited. Other 7-HT derivatives which retain the 7-hydroxyl group as it is crucial for activity have been synthesised.113
Fig. 9. The fermentation product 7-hydroxytropolone.

Aminoglycoside O-phosphotransferases (APHs)
APHs consist of seven subclasses (Table 7) which can phosphorylate the hydroxyl group on positions 2′′, 3′, 3′′, 4, 6, 7′′ or 9 through direct attack on ATP. The enzyme facilitates this process by increasing the nucleophilicity of the hydroxyl group, with Asp190 acting as a catalytic base for proton abstraction.114
Substrates of aminoglycoside O-phosphotransferases (APHs) and their pathogenic hosts12,110.
| Enzyme | Subtype | Pathogenic host | Aminoglycoside substrates |
|---|---|---|---|
| APH(3′) | I | E. coli, S. enterica, K. pneumoniae, A. baumannii, Citrobacter spp. | Kanamycin, neomycin, paromomycin |
| II | E. coli, P. aeruginosa, S. maltophilia | Kanamycin, neomycin, paromomycin | |
| III | S. aureus, Enterococcus spp. | Kanamycin, neomycin, paromomycin, amikacin | |
| IV | B. circulans | Kanamycin, neomycin, paromomycin | |
| V | Streptomyces fradiae, Streptomyces ribosidificus | Neomycin, paromomycin | |
| VI | A. baumannii, K. pneumoniae, S. marcescens | Kanamycin, neomycin, paromomycin, amikacin | |
| VII | C. jejuni | Kanamycin, neomycin | |
| APH(2′′) | I–IV | S. aureus, C. difficile, E. faecium, E. coli, Enterococcus gallinarum | Kanamycin, gentamicin, tobramycin |
| APH(3′′) | Ia, Ib | S. griseus, Enterobacteriaceae, Pseudomonas spp., M. fortuitum | Streptomycin |
| APH(6) | Ia, Ib, Ic, Id | P. aeruginosa, E. coli, Salmonella spp., V. cholerae | Streptomycin |
| APH(9) | Ia, Ib | L. pneumophila | Spectinomycin |
APH inhibitors
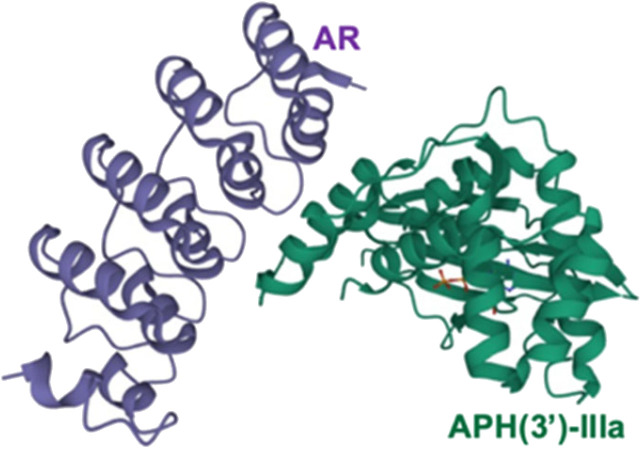
A specifically designed ankyrin repeat (AR) protein is an allosteric inhibitor of APH(3′)-IIIa as it binds to the APH C-terminal, stabilising the three α-helices that are required for aminoglycoside binding, therefore restricting its conformation and preventing it from binding to aminoglycosides (Fig. 10).115 However, AR is a relatively large molecule which could lead to issues with delivery and formulation. Other studies have identified isoquinoline sulphonamides, pyrido-pyrimidines and flavonoids as general inhibitors of APH but they were found to be non-selective to bacterial kinases.116,117 Leban et al. have investigated ways to disrupt normal functioning of the enzyme other than inhibition or competition at the active site and found a lead allosteric small kinase inhibitor, NL6, which could potentially inhibit both APH(3′)-IIIa and APH(2′′).118 As this study was only conducted in silico and in vitro, further investigation is needed.
Fig. 10. A designed AR in complex with APH(3′)-III (PDB ID 2BKK). The AR protein binds to the C-terminal outside the substrate active site, leading to an alteration in the active site so that the enzyme can no longer modify aminoglycosides115 (image created using Mol*119).
Chloramphenicol
Mechanism of action
Chloramphenicol was first isolated from Streptomyces venezuelae in 1947 and has a wide spectrum of activity although, due to systemic toxicity issues, its use is normally limited to topical treatment of bacterial conjunctivitis and otitis externa. It inhibits protein synthesis by reversibly binding to the peptidyltransferase centre at the 50S ribosomal subunit, and the primary hydroxyl group is key in this binding to the ribosome.120,121
Chloramphenicol acetyltransferases (CATs)
The most common pathway of bacterial resistance to chloramphenicol involves enzymatic deactivation by chloramphenicol acetyltransferases (CATs) due to acetylation of the primary hydroxyl group, resulting in loss of antimicrobial activity. CATs are found in many bacterial strains including some of the WHO priority pathogens (Table 8).122
Mechanisms of chloramphenicol deactivation in pathogenic Gram positive and Gram negative hosts.
The specific genes coding for class A CATs (catA) have been extensively documented by Schwarz et al.29 The diversity in cat genes prevents the definition of a unified enzymatic model as only five of the seventeen residues are conserved between the different CAT active sites.122 Deactivation of chloramphenicol by the CATI subtype is shown in Scheme 3. CAT genes can be found on chromosomes, plasmids, integrons and transposons.126–128 There are currently no inhibitors available on the market but there have been several studies that have identified potential ligands.129–132
Scheme 3. Deactivation of chloramphenicol by CATI.133 Once bound, acetyl CoA reaches the enzyme active site through a tunnel.134 One of the most important residues for catalysis is His193, with the lone pair electrons of the imidazole deprotonating the primary alcohol of chloramphenicol, promoting the attack on the carbonyl of the thioester of acetyl CoA, yielding an acetylated chloramphenicol.134.
3′-O-Phosphotransferases
Mosher et al. identified 3′-O-phosphoryl ester as a metabolite of chloramphenicol, produced by 3′-O-phosphotransferase (CPT) enzymes.135 As mentioned previously, the primary hydroxyl group of chloramphenicol is important for its mechanism of action, so phosphorylation at this position leads to deactivation of the antibiotic (Scheme 4).135
Scheme 4. Other chloramphenicol enzymatic modifications.
The active site of CPT is lined with a hydrophobic and electron-dense region. The aromatic ring of chloramphenicol takes part in van der Waals interactions, the carbonyl group undergoes hydrogen bonding and the amide nitrogen undergoes hydrogen bonding with a sulphate anion bound to CPT. Asp37 is thought to deprotonate the primary hydroxyl group which attacks the γ-phosphate yielding a stabilised pentacovalent transition state, facilitating the transfer of a phosphoryl group to 3′-OH of chloramphenicol.123 There are no known inhibitors of CTP, which is mostly found in the chloramphenicol-producing actinomycete S. venezuelae.29
Hydrolases
As would be expected for a microorganism which produces an antibiotic, S. venezuelae has intrinsic resistance and produces chloramphenicol hydrolase, which catalyses the hydrolysis of chloramphenicol's amide bond to form the degradation products nitrophenylserinol and a dichloroacetyl moiety (Scheme 4).124,136 Chloramphenicol hydrolase is only produced by S. venezuelae, so there is no need for the development of an adjuvant molecule unless the enzymatic modification spreads to other bacterial species.
Nitroreductases
Nitroreductases reduce chloramphenicol activity through the catalysis of the reduction of the p-nitro group to form less active compounds such as the hydroxylamine and amino derivatives (Scheme 4).137 The nitro group is reduced by nicotinamide adenine dinucleotide (NADP) coenzyme and a water molecule ensures the modified products are stabilised.
Although there have been no studies investigating the potential of nitroreductase inhibitors as adjuvants, this mechanism of resistance has been found in P. aeruginosa, H. influenzae, and N. meningitidis so adjuvants may have potential in the treatment of infections caused by these MDR organisms.125,138
Macrolides
Due to their broad spectrum of activity, macrolides (Fig. 11) can be used in the treatment of a range of serious infections, and they are also an alternative for patients with penicillin allergies.139,140
Fig. 11. The structures of the macrolide antibiotics.
Mechanism of action33
Macrolides bind to the 50S subunit of bacterial ribosomes, adjacent to the peptidyl transferase centre made up of 23S rRNA.141 Macrolides bind through hydrophilic and hydrophobic interactions which block the lumen of the exit tunnel of the ribosome (Fig. 12) therefore terminating protein synthesis.142 Macrolides can exhibit bacteriostatic or bactericidal activities.143
Fig. 12. Interaction of erythromycin with the nucleotides in its binding site in the protein exit tunnel (PDB ID 1JZY). In ribosomal RNA nucleotide 2058 is an adenine to which the macrolide antibiotic binds strongly. Hydrogen bonds are formed between the 2′,6,11 and 12-hydroxyl groups of the macrolide and adenine bases 2058 and 2509. An electrostatic interaction exists between the conjugate acid of the tertiary amine of the macrolide and the guanine-2505 phosphate. All these interactions play a key role in macrolides binding to this site, with any modifications in structure disrupting the binding process.144.
Enzymatic modification
Resistance to macrolides can be the result of macrolide esterases (Ere) encoded by ere, macrolide 2′-phosphotransferases (MPH) encoded by gimA, ole, and mgt, and macrolide glycosyltransferases encoded by mph (Table 9). Resistance to macrolides has spread to S. pneumoniae and H. influenzae which, as highlighted previously, are on World Health Organisation's priority pathogens list.145,146
Macrolide deactivating enzymes, their hosts and the antibiotics that they modify (compiled with the aid of the Comprehensive Antibiotic Resistance Database; CARD; https://card.mcmaster.ca)40.
| Enzyme | Gene | Host | Substrate | Ref. |
|---|---|---|---|---|
| Esterase (Ere) | ereA | E. coli, K. pneumonia, P. aeruginosa, V. cholera | Erythromycin, roxithromycin, clarithromycin | 155 |
| ereB | E. coli, Salmonella spp. | Erythromycin, roxithromycin, clarithromycin, azithromycin | 148 | |
| ereC | K. pneumoniae | 14-Membered macrolactones (e.g. clarithromycin, erythromycin, oleandomycin, roxithromycin, solithromycin) except telithromycin | 156 | |
| ereD | Riemerella anatipestifer | Erythromycin | 157 | |
| Glycosyltransferase | gimA | Streptomyces ambofaciens | Oleandomycin | 158 |
| ole | Streptomyces antibioticus | Oleandomycin, tylosin | 159 | |
| mgt | S. lividans | Erythromycin, azithromycin | 150, 160 | |
| Phosphotransferase | mphA | E. coli, A. baumannii, ESCAPPM | 14- and 15-membered macrolactones (e.g. erythromycin, roxithromycin, clarithromycin, azithromycin) | 151, 161 |
| mphB | E. coli, K. pneumoniae, S. enterica, E. fergusonii | 14-, 15- and 16-membered macrolactones (e.g. erythromycin, roxithromycin, clarithromycin, azithromycin) | 151, 161 | |
| mphC | S. aureus | Erythromycin, roxithromycin, clarithromycin, azithromycin, oleandomycin | 151, 161, 162 |
Macrolide esterase (Ere)
These enzymes break the ester bond of the macrolide ring to give an inactive linear structure (Scheme 5).147 Macrolides bind to the esterase deep within the active site loop which contains a catalytic triad of His50, Glu78, and His289. His50 activates a water molecule which attacks the electrophilic carbon-1 of the ester group. There have been no macrolide esterase inhibitors tested, which is of growing concern as these enzymes lead to a high level of resistance, increasing the minimum inhibitory concentration (MIC) to greater than 1600 μg mL−1 in E. coli.148 The mode of binding of macrolides to the esterases has only been studied in silico so the mechanism requires further investigation.149
Scheme 5. Hydrolysis of macrolides by esterases. Macrolides, such as erythromycin, bind within a buried active site of EreC covered by a glycine and proline rich loop. When the loop is closed, it repositions the ester group of macrolides in close proximity to the catalytic triad, composed of His50, Glu78 and His289. The first step is deprotonation of a water molecule by His50, producing a reactive hydroxide ion which then attacks the carbonyl of the macrolide ester, forming a negatively charged transition-state intermediate which is stabilised by Arg261. The deprotonated hydroxyl group then retrieves the proton previously transferred to His50, releasing the antibiotic in an inactive hydrolysed form.149.
Macrolide glycosyltransferases
Macrolide glycosyltransferases are not actively involved in conferring antibiotic resistance as they are only present in macrolide producing bacteria, in which they contribute to inherent resistance.150
Macrolide phosphotransferase (MPHs)
MPHs are separated into different classes labelled Mph(2′)-I to Mph(2′)-XV each encoded by different genes but overall all have the same mechanism.151 All MPHs catalyse the attachment of a γ-phosphate from guanosine triphosphate (GTP) moiety to the 2′-hydroxyl group of the desosamine or cladinose sugar moiety, resulting in a reduction in the binding affinity of the macrolide for its ribosomal target (Fig. 13).
Fig. 13. 2′-Phosphorylation of erythromycin by an MPH(2′)-I (PDB ID: 5IGP).154 The nucleotide binds within the hydrophobic N-terminus and interacts with Tyr94 and aspartate residues (Asp200, Asp219, Asp234). The macrolide binding site consists of a hydrophobic region, a negatively charged region and an aspartate residue (Asp200), seen in all MPH subtypes, along with Tyr202, Ala234 and Phe280 residues. Asp200 is thought to position the target 2′-OH of macrolides in an orientation which allows proton transfer during catalysis. For MPHs which utilise GTP, a tyrosine residue (Tyr94) acts as a gatekeeper.154.
Macrolide phosphotransferases (MPHs) resemble aminoglycoside phosphotransferases but have a larger interdomain linker which widens the binding pocket. There are over fifteen mph gene subtypes and they can confer resistance to different antibiotics in both Gram positive and negative bacteria (Table 9).152,153 Although these genes are not widely spread, they are found on mobile genetic elements and some are also found to confer resistance to ketolides, an important class of antibiotics derived from macrolides.25,142
Macrolide phosphotransferases have a similar structure to eukaryotic protein kinases which are enzymes that have been studied extensively in drug discovery. One high throughput screening (HTS) study thus investigated the known eukaryotic ATP competitive kinase inhibitors, but none were found to inhibit MPH enzymes.154 There are currently no macrolide adjuvants despite the fact that these enzymes are responsible for conferring resistance in important pathogens (Table 9).147,163,164
Tetracyclines
Tetracyclines are a class of antibiotics that have been shown to bind to the 30S ribosomal subunit near the 16S subunit inhibiting protein synthesis. This occurs through competitive binding to the peptidyl transfer centre A site instead of the charged tRNA.165 Tetracycline binding is reversible, accounting for their bacteriostatic action.166,167
Tigecycline has a broad spectrum of activity and is approved for the treatment of serious infections in hospitalised adults, it is used as last resort against serious infections caused by carbapenem-resistant Enterobacteriaceae and Acinetobacter spp.168
Tetracyclines (Fig. 14) were first discovered in 1948, the newest tetracyclines, omadacycline and eravacycline, were approved in 2018 and three other tetracycline derivatives are currently in trials.169
Fig. 14. Examples of chemical structures of tetracyclines. Tetracyclines have a common linear fused tetracyclic nucleus; tigecycline is a third-generation tetracycline with an additional side chain on the D ring.
Mechanism of action
Tetracyclines bind to the 30S subunit most importantly at the A-site, by interacting with the sugar–phosphate backbone of helix-34 via an Mg2+ ion.170,171 The β-diketone region of tetracyclines is essential for activity as it forms a divalent metal chelation site, essential for binding and delivery into the cell.172 By complexing with a Mg2+ ion, the C and D rings of tetracyclines block the interaction of the first base of the tRNA anticodon with the third base of the mRNA codon, preventing the binding of aminoacyl tRNA to the A-site and therefore inhibiting protein synthesis.171,173
Tet(X)
Tet(X) is a flavin adenine dinucleotide (FAD) and NADP dependent monooxygenase (Fig. 15) that was first identified in Bacteroides fragilis.174 The tet(X) gene is transferrable by transposons and is found in many pathogenic MDR Gram negative bacteria, such as K. pneumonia, A. baumannii and P. aeruginosa.175–177
Fig. 15. X-ray crystal structure of Tet(X) with bound FAD (PDB ID: 2XDO).178 Tet(X) is composed of two domains stabilised by a C-terminal α-helix and another C-terminal α-helix involved in substrate recognition and binding.179 The X-ray crystal structure of Tet(X) show that the first domain contains a non-covalently bound FAD that adopts an IN (pointing towards the binding domain) or OUT (pointing away from the binding domain) conformation. The A ring is positioned near the bound FAD molecule and the D ring lies near the C-terminal α-helix. The OUT confirmation is required for the reduction of FAD to FADH2 by NADPH. The reduced form is then able to bind oxygen in the IN confirmation forming a hydroperoxide able to hydroxylate tetracycline at C11. The second domain covers the FAD-binding site while also containing the substrate recognition pocket178 (image created using Mol*119).

As the enzyme has a requirement for oxygen, this resistance mechanism is only found in aerobic bacteria. Tet(X) recognises the A and B ring of tetracyclines (Fig. 16); the D ring is not involved in binding to the active site which explains why third generation tetracyclines, such as tigecycline, are also susceptible to deactivation by Tet(X).180 The deactivation process can be divided into two steps—hydroxylation at C11 (weakening the binding to Mg2+ and therefore reducing the affinity of the drug for the ribosome) and decomposition of the hydroxylated tetracycline to an inactive polymer.172,173
Fig. 16. A Hydroxylation of 7-chlortetracycline by Tet(X). B The residues of Tet(X) recognise the A and B ring of tetracyclines and orient it for the hydroxylation at C-11a: FAD represented in green, tetracycline in purple (PDB ID: 2Y6R).178.
Tet(X) inhibitors
Anhydrotetracycline, a shunt product involved in the biosynthesis of tetracycline, is structurally similar to tetracycline but dehydration at C6 alters its conformation and makes it more hydrophobic leading to differences in mechanisms of action.21,179 Tet(X) can hydroxylate anhydrotetracycline but at a much slower rate and specific modifications to the structure have been shown to improve stability and potency.21,181 Plumbagin is another potential inhibitor of a wide range of Tet(X) enzymes.182
Other tetracycline destructases were discussed in other studies but they will not be discussed in this review due to their clinical insignificance.172,183,184
Rifamycins
Alongside isoniazid, rifampicin is a first-line drug for the treatment of TB. When it was first used, the incidence and mortality due to TB was reduced by 25% and 50% respectively.185
Mechanism of action
Rifampicin is one of three rifamycins currently in use; rifampicin, rifabutin and rifapentine (Fig. 17). They are bactericidal antibiotics that inhibit the DNA-dependent RNA polymerase (RNAP) exit tunnel, leading to the termination of RNA elongation.186 Rifamycins specifically bind to the β subunit, a site encoded by the rpoB gene. Mutations to this gene represent the major resistance pathway.187 However, as the two free hydroxyl groups at C21 and C23 of the ansa chain as well as oxygen atoms at C1 and C8 of the naphthalene ring are required for binding, they are also targeted by drug modifying enzymes.188
Fig. 17. The structure of rifamycins is composed of a naphthalene core and an ansa bridge, which is a basket-like structure.30.
Emergence of resistance
In the 1980s and 1990s, TB cases started increasing again due to resistance, so derivatives were synthesised from the natural rifamycins, through modification at the C3 or C4 positions of the naphthalene ring, to expand their spectrum of activity.185 Most derivatives led to TB relapses except rifampicin, explaining why it is the treatment of choice.185
Rifamycin glycosyltransferases
The most prevalent mechanism is glycosylation of rifamycins by rifamycin glycosyltransferase (RGT) (Table 10), which uses UDP-glucose as the glycosyl donor, with the transfer occurring with inversion at the anomeric carbon. The enzyme glycosylates the drug at 23-OH of the ansa chain, thus preventing it from binding to RNAP.189 There are currently no studies describing the binding of the drug to the enzyme which thus restricts the search for inhibitors.
Rifamycin modifying enzymes, example of hosts and the antibiotics that they deactivate40.
| Enzyme | Host | Site modified | Substrate |
|---|---|---|---|
| Glycosyltransferase (RGT)188,189 | Nocardia brasiliensis, Streptomyces pseudogriseolus | 23-OH | Rifampicin, rifaximin, rifabutin, rifapentine |
| Phosphorylation (RPH)190 | Actinobacteria spp., Clostridia spp., Bacillus anthracis, E. faecalis, L. monocytogenes | 21-OH | Rifampicin, rifaximin, rifabutin, rifapentine |
| ADP-ribosylation (Arr)30,191 | M. smegmatis, P. aeruginosa, A. baumannii, some Enterobacteriaceae | 23-OH | Rifampicin, rifaximin, rifabutin, rifapentine |
| Monooxygenation + decomposition (Rox)192–194 | Nocardia farcinica | Naphthalene moiety | Rifampicin, rifaximin, rifabutin, rifapentine |
Rifamycin phosphotransferases
Rifamycin phosphotransferase (RPH) has a broad spectrum of activity as the R group, which distinguishes the rifamycins from one another, protrudes from the enzyme and does not bind internally. Phosphorylation (Scheme 6) reduces binding to RNAP as it may lead to a steric clash. There are currently no descriptions of RPH inhibitors, despite the fact that RPH causes resistance in lethal pathogens.
Scheme 6. Deactivation of rifamycins by RPH in L. monocytogenes. The enzyme has 3 domains: an N-terminal ATP-grasp domain, a swivel domain containing the catalytic His at the C-terminus and an intermediate domain responsible for binding rifampicin.195 The rifampicin binding domain contains three residues Val333, Met359 and Val368 forming a hydrophobic patch that stabilises rifampicin.196 The naphthol ring of rifampicin binds deep in the pocket through van der Waals interactions while the R groups point towards the opening of the pocket and binds to Pro356 and Phe479.196 The ATP-grasp domain positions the β-phosphate that is then transferred to His825 in the swivel domain. The Glu667–Arg666 residues found in the rifampicin-binding domain then facilitate the transfer of the phosphate group to 21-OH of the polyketide region.30.
Monooxygenases and decomposition
Decomposition of rifampicin by monooxygenases, encoded by iri or rox, resembles the mechanism of Tet(X) catalysed degradation. Rox is also a flavin-dependent monooxygenase and it leads to decomposition.199 The most accepted mechanism of action suggests that a rifamycins anion at C2 can attack the phenolate nucleophile on flavin, forming a hydroxyl intermediate which then opens the ansa ring, resulting in a 2-hydroxy-1,4-benzoquinone (Fig. 18). As the ring opening disrupts the basket-like structure required for binding to RNAP, the product of this reaction is inactive.198 There are currently no Rox or Iri inhibitors available.
Fig. 18. Deactivation of rifamycins by Rox in N. farcinica (5KOX).197 Rifampicin (purple) binds perpendicularly to FAD (light blue) with O4 of FAD undergoing hydrogen bonding with the drug.198 The exact binding interactions differ depending on the species expressing the Rox.198 For example, in S. venezuelae, Arg213 forms a hydrogen bond with 8-OH whereas, in N. farcinica, the 21-OH forms a hydrogen bond with the Arg43 residue.198 In N. farcinica, the drug binds to the enzyme through hydrophobic interactions mainly: the polyketide region interacts with the Val205 residue through hydrophobic interactions.198 In both examples, Rox transfers a hydroxyl group from FAD-OOH to RIF which leads to ring opening, disrupting the ansa bridge structure required for binding.
ADP-ribosyltransferases
ADP-ribosyltransferases (ARTs) are enzymes found in eukaryotic cells – for post-translation modifications and bacterial toxin inactivation – and prokaryotic organisms.200 The bacterial ADP-ribosyltransferase gene, Arr, can be found on the transposons of many bacteria and may confer resistance to rifamycins and also other antibiotics (Table 11).30,191 As Arr ribosylates the hydroxyl group which is involved in hydrogen bonding to the β-subunit of RNAP, Arr carriers are resistant to rifamycins.30
Examples of Arr enzymes and resistant gene location.
| Arr enzyme subtype | Gene coding enzyme | Pathogenic bacteria expression | Rifamycin substrate | Ref. |
|---|---|---|---|---|
| Arr-2 | arr-2 | P. aeruginosa, K. pneumoniae, A. baumannii | Rifaximin | 201 |
| Rifabutin | ||||
| Rifapentine | ||||
| Arr-3 | arr-3 | Vibrio fluvialis | Rifampicin | 202 |
| arr-6 | E. coli | Rifaximin | ||
| Arr-4 | arr-4 | P. aeruginosa, A. baumannii | Rifampicin | 203 |
| Rifaximin | ||||
| Rifapentine | ||||
| Arr-5 | arr-5 | P. aeruginosa, K. pneumoniae, S. marcescens | Rifampicin | 203 |
| Rifaximin | ||||
| Rifapentine | ||||
| Arr-7 | arr-7 | P. aeruginosa | Rifampicin | 204 |
| Arr-8 | arr-8 | Klebsiella oxytoca, K. pneumoniae, P. aeruginosa | Rifaximin | 205 |
| Rifabutin | ||||
| Rifapentine |
Arr inhibitors
Structural alignment showed that M. smegmatis Arr contains a catalytic His-Tyr-Asp triad which overlaps that of human polyADP ribose polymerase 1 (hPARP-1) (or His-Tyr-Glu), so hPARP inhibitors were screened for activity against M. smegmatis Arr. The hPARP inhibitors, such as olaparib led to 60% inhibition in the presence of rifampicin, as it binds to Arr at this NAD+ binding site. Upon expansion of the hydrophobic substituents attached to the non-terminal amide of olaparib, a lead compound, AZ9482, was found to be more potent in M. abscessus. As AZ9482 exhibited poor solubility and rapid clearance from the human body, further modifications and improvements need to be made before clinical testing.206
Isoniazid
Mechanism of action
Isoniazid is a synthetic antibacterial agent used as first line for the treatment of tuberculosis (TB),207 it is a prodrug activated by KatG, a catalase–peroxidase enzyme, to form reactive species and free radicals that damage cellular macromolecules such as nucleic acids, proteins and lipids.208 The activated form of isoniazid also inhibits the action of InhA, an enzyme involved in the synthesis of mycolic acid. Isoniazid is therefore bactericidal 24 hours after administration as a result of the inhibition of mycolic acid synthesis and a number of other metabolic processes that are essential for bacterial survival.209
M. tuberculosis is a top priority pathogen as TB is one of the leading causes of death in the world due to a single infectious agent. The standard treatment regimen for TB consists of an intensive phase (2 months) involving treatment with rifampicin, isoniazid, pyrazinamide and ethambutol (RIPE), followed by a continuation phase of 4–7 months of rifampicin or isoniazid. Drug-resistant TB often requires even more complex medication regimens and is responsible for 250 000 deaths every year.2 Although mutations in KatG and InhA are the major causes for isoniazid resistance, enzymatic modification also occurs in some mycobacterial species.209
N-Acetyltransferases
N-Acetyltransferase (NAT) and KATG compete for deactivation and activation of isoniazid, respectively. If isoniazid binds to NAT, acetylation of the terminal nitrogen of the hydrazide is induced by acetyl CoA which prevents the metabolism to its active form by KATG. Nat genes were found to confer some resistance in M. smegmatis and M. tuberculosis.210,211 The mode of binding of isoniazid to NAT was characterised in M. smegmatis as NATs found in M. tuberculosis are insoluble and therefore more difficult to study (Fig. 19) but there is a 60% identity between the nat sequence in M. smegmatis and M. tuberculosis with 10 of 13 active site amino acids being the same.210
Fig. 19. Deactivation of isoniazid by NAT (PDB ID: 1W6F).212 Isoniazid binds to a NAT catalytic triad composed of Cys70, His110 and Asp127. The terminal nitrogen of isoniazid is situated ∼2.4 Å from the γ-S atom of Cys70, making it the most important residue for this catalysis. Isoniazid also undergoes hydrophobic interactions and hydrogen bonding with other residues within the active site. The enzyme is initially acetylated by acetyl CoA and the acetyl group is then transferred onto the hydrazinyl group of isoniazid.212.
Not only does NAT deactivate isoniazid, but it has also been shown to be involved in essential cellular processes and in promoting the survival of M. tuberculosis inside macrophages.213 Inhibition of NAT can thus lead to M. tuberculosis death in different ways, which highlights the potential for inhibitors.209
A novel acetyltransferase, labelled Rv2170, is also capable of acylating the terminal nitrogen of isoniazid, leading to resistance.23
NAT inhibitors
There are currently no NAT inhibitors on the market. Westwood et al. described the characteristics of an ideal NAT inhibitor derived from 1,2,4-triazole-3-thiones and, based on their work, three nitrofurans were found to be potential NAT inhibitors and to have anti-mycobacterial activity of their own.213,214 The maximum N-acetyltransferase inhibition occurred when the substituent on a 1,2,4-triazole was an aliphatic or planar aromatic group. An increase in chain length meant more hydrophobic contacts with the active site occurred. The maximum chain length is 8 carbons, anything longer the binding energy is no longer favourable, and the inhibitory effect is lost.213
In addition, p-substituted piperidinols with a halide and an ethyl substituent on the nitrogen atom, were shown to increase potency; they can form an irreversible adduct with the cysteine residue of NAT in M. marinum, which is 74% identical in sequence to NAT in M. tuberculosis.215
11α-Hydroxycinnamosmolide from Warburgia salutaris inhibited N-acetyltransferase and weakened the M. bovis cell wall thus was able to inhibit mycobacterial growth (Fig. 20).216
Fig. 20. Chemical structure of 11α-hydroxycinnamosmolide an N-acetyltransferase inhibitor.216.

Lincosamides
Mechanism of action
Clindamycin and lincomycin are lincosamides217 which inhibit protein synthesis by binding to the 23S portion of the peptidyl transferase centre of the 50S subunit of the bacterial ribosome. Lincosamides block the protein tunnel and can cause early detachment of the peptidyl-tRNA meaning the peptide chain will grow until steric hindrance inhibits the translation process.218 Clindamycin is an alternative to the penicillins in patients who have allergies and is used in the treatment of dental infections and a range of other infections, including acne.
Enzymatic modification
There are two groups of O-adenylyltransferases which modify lincosamides and they are categorised based on sequence similarities.219
The first class is observed mostly in Staphylococci and is homologous to aminoglycoside nucleotidyltransferases. The class consists of LinA, LnuC and LnuD.220 LinA (161 amino acids) requires a nucleoside 5′-triphosphate (e.g. ATP, GTP, CTP, UTP) and Mg2+ as a cofactor. The enzyme modifies lincomycin at 3-OH of the sugar and clindamycin at the 4-OH group.221
The second class is found in Enterococci and consists of LinB and LnuF.220 Clindamycin undergoes hydrophobic interactions with Phe104, Tyr27 and Tyr44 of the enzyme, dipole–ion interactions between the 3-OH and Mg2+ and hydrogen bonds between 4-OH and Tyr27. After binding, the α-phosphate group is transferred, forming a pentacoordinate phosphate intermediate product which eventually leads to adenylation at 3-OH of clindamycin (Scheme 7).222
Scheme 7. Transfer of an AMP group from ATP to 3-OH of lincosamides by LinB.
These nucleotidyltransferase enzymes are encoded on mobile elements indicating that resistance can spread.223 As LinB and DNA polymerase exhibit many similarities, De Pascale and Wright suggested trialling DNA polymerase inhibitors on nucleotidyltransferases but no follow-up studies have been conducted.224
Fosfomycin
Fosfomycin is often used as an alternative treatment for osteomyelitis or urinary tract infections that are resistant to other antibacterial agents.225,226
Mechanism of action
Fosfomycin irreversibly inhibits cytosolic UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) through alkylation of Cys115 found in the enzyme active site (Fig. 21).227 MurA is utilised in the formation of N-acetylmuramic acid (NAM), an essential component of the peptidoglycan layer, thus its inhibition by fosfomycin leads to cell wall lysis, meaning that fosfomycin is bactericidal.226,227
Fig. 21. Fosfomycin binding to MurA, which is utilised in the formation of N-acetylmuramic acid, an essential component of the peptidoglycan layer, inhibition by fosfomycin thus leads to cell wall lysis. The catalytic domain of MurA is deep in the cavity of the enzyme's two domains and contains Cys115, which fosfomycin covalently attaches to. Three positively charged residues of MurA, Lys22, Arg120, and Arg397 surround the phosphonate group of fosfomycin creating electrostatic interactions and hydrogen bonds.230.
FosA
FosA is a homodimeric Mn2+- and K+-dependent glutathione S-transferase made up of two active sites and a central cavity.228,229 FosA deactivates fosfomycin by opening the epoxide ring, a critical structure for binding to MurA, via nucleophilic attack (Fig. 22).
Fig. 22. Fosfomycin binding to FosA from K. pneumoniae (PDB ID 5V3D). FosA key residues involved in the binding of fosfomycin include Lys93, Ser97, Ser101, Tyr103, and Arg122. The Mn2+ ion acts as a Lewis acid during the nucleophilic attack by glutathione. A K+ at the active site does not take part in the catalysis reaction but enhances the electrophilicity of the Mn2+ ion, increasing the rate of reaction.229.
FosA genes are found in plasmids (e.g. ESBL E. coli and carbapenemase producing K. pneumonia) or chromosomes (e.g. Klebsiella spp., Enterobacter spp., and P. aeruginosa), highlighting the importance and urgency of developing strategies to overcome fosfomycin resistance.226
Although there are many subtypes of FosA enzymes (Table 12), their active site has been shown to be conserved which makes the development of broad-spectrum FosA inhibitors possible.229 For a long time, fosfomycin resistance was not of concern as it did not significantly affect bacteria which were susceptible to fosfomycin however it has now reached E. coli and other Gram negative pathogens.231
Examples of FosA enzymes and pathogenic bacteria which express them.
Several phosphonates, such as phosphonoformate, have been shown to be inhibitors of FosA.238 Phosphonoformate can form a five-coordinate complex with the Mn2+ centre with a geometry similar to that of the transition state of the reaction. Phosphonoformate can greatly increase the susceptibility K. pneumoniae, E. cloacae, P. aeruginosa, and to a lesser extent E. coli, to fosfomycin. One disadvantage is the association of phosphonoformate with nephrotoxicity and seizures, thus limiting its use in practice.239
One study showed that compound 19 583 672 from the Zinc database was a potent inhibitor of FosA variant 7 from K. pneumoniae and could form a stable complex with the enzyme.240 ANY1 (3-bromo-6-[3-(3-bromo-2-oxo-1H-pyrazolo[1,5-a]pyrimidin-6-yl)-4-nitro-1H-pyrazol-5-yl]-1H-pyrazolo[1,5-a]pyrimidin-2-one) is a small-molecule active site non-competitive inhibitor of FosA which exhibits antibacterial activity against K. pneumoniae in combination with fosfomycin (Fig. 23).
Fig. 23. Crystal structure of FosA3 with inhibitor ANY1 bound (PDB ID 5WEP). ANY1 binds to the active site of FosA by interacting with Lys93, Ser97 and Tyr103.239.
These beneficial effects were not seen when ANY1 was given alone or in combination with gentamicin, demonstrating its potential as a fosfomycin adjuvant.239
FosB
The FosB enzyme was discovered first in Staphylococcus epidermidis and has now also been found in the chromosome of S. aureus which is a leading cause of global morbidity and mortality (Table 13).241 FosB is a Mg2+-dependent enzyme unique to Gram positive Firmicutes as they do not produce glutathione.226,242 Fosfomycin is deactivated by FosB through the nucleophilic addition of l-cysteine (l-Cys) or bacillithiol (BSH) to C1. The fosB gene is found in the plasmids of Enterobacterales and both the plasmids and chromosomes of S. epidermidis and Bacillus subtilis.226
Examples of FosB enzymes and the pathogenic bacteria that express resistance.
FosB inhibitors
FosB activity is inhibited by Zn2+ ions as they have a full valence orbital and do not accept electrons from fosfomycin thus, preventing the formation of the partial positive charge needed for the nucleophilic attack by l-Cys or BSH.245 This mechanism can be used as a basis for the development of other adjuvants, using Zn2+ itself is unfavourable as the effect is lower when BSH is the donor proposing a problem as BSH is the preferred substrate.241,246 Disulfides, including disulfiram and S-octyl disulfides with either N,N-dimethyl- or N-pyrrolidinodithiocarbamate substituents, have been analysed for their ability to inhibit FosB. The results showed that both disulfiram and octyl pyrrolidine-1-carbo(dithioperoxo)thioate could sensitise S. aureus to the effects of fosfomycin. Mechanistic studies have shown these compounds decrease intracellular levels of FosB cofactors such as BSH through a thiol disulfide exchange reaction. This exchange results in the formation of a stable adduct, depleting the co-factor stores and preventing the enzyme from degrading fosfomycin.246
Susceptibility testing showed that the antibiotic activity of fosfomycin was potentiated by the addition of disulfiram (Fig. 24) or octyl pyrrolidine-1 carbo(dithioperoxo)thioate.
Fig. 24. Chemical structure of disulfiram. Disulfiram is able to react with BSH, forming a stable adduct and depleting BSH stores.247.

FosX
FosX is an Mn2+ dependent epoxide hydrolase, the active site of the enzyme contains Glu44, which acts as a general base in the catalysis of the hydration of fosfomycin. This reaction causes inversion of configuration at carbon-1 in the formation of the inactive vicinal diol.248
FosX inhibitors
Virtual screening of the indofine natural-product database and FDA-approved drugs was used to identify potential FosX inhibitors. The results showed that phenolic acid compounds, caffeic acid and chlorogenic acid could inhibit FosX with high affinity. Caffeic acid and chlorogenic acid were tested at 1.5 mg mL−1 and 3 mg mL−1 respectively with 50 mg L−1 fosfomycin and were shown to inhibit the growth of Listeria monocytogenes expressing FosX.249
Conclusions
Enzymatic drug deactivation is an important contributor to the resistance of priority pathogens to many classes of antibacterial agents. The aim of this review has been to describe the enzymatic processes involved in antibacterial deactivation to highlight opportunities for adjuvant development. Serine β-lactamase inhibitors are currently the only adjuvants in clinical use, although other resistant enzymes are encoded on mobile genetic elements, which contribute to a serious spread in resistance. Soon, infections such as sexually transmitted diseases, urinary tract infections and tuberculosis may become impossible to treat without the development of novel drugs or drug-adjuvant combinations, making a post-antibiotic era worryingly possible.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the Gadigal people of the Eora Nation as traditional custodians of the land upon where we work, and where this research was conducted.
Notes and references
- Aminov R. I. Front. Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W. H. O. (WHO), 2020 antibacterial agents in clinical and preclinical development: an overview and analysis, https://www.who.int/publications/i/item/9789240021303, (accessed May, 2022)
- Leuthner K. D. Doern G. V. J. Clin. Microbiol. 2013;51:3916–3920. doi: 10.1128/JCM.01751-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathwani D. Varghese D. Stephens J. Ansari W. Martin S. Charbonneau C. Antimicrob. Resist. Infect. Control. 2019;8:35. doi: 10.1186/s13756-019-0471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W. H. O. (WHO), Antimicrobial resistance, https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance#:~:text=What%20is%20antimicrobial%20resistance%3F,spread%2C%20severe%20illness%20and%20death, (accessed May, 2022)
- Raoult D. Leone M. Roussel Y. Rolain J. Lancet Infect. Dis. 2019;19:128–129. doi: 10.1016/S1473-3099(18)30800-4. [DOI] [PubMed] [Google Scholar]
- Lim C. Takahashi E. Hongsuwan M. Wuthiekanun V. Thamlikitkul V. Hinjoy S. Day N. P. J. Peacock S. J. Limmathurotsakul D. eLife. 2016;5:180–182. doi: 10.7554/eLife.18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W. H. O. (WHO), New report calls for urgent action to avert antimicrobial resistance crisis, https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis, (accessed May, 2022)
- Bush K. Antimicrob. Agents Chemother. 2018;62:e01076-18. doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. Leclercq S. O. Tian J. Wang C. Yahara K. Ai G. Liu S. Feng J. PLoS Genet. 2017;13:e1006602. doi: 10.1371/journal.pgen.1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Liu Y. Chen C. Guo Y. Ma Y. Yang Y. Hu F. Xu X. Wang M. PLoS One. 2016;11:e0154829. doi: 10.1371/journal.pone.0154829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M. S. Tolmasky M. E. Drug Resistance Updates. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwocho L. R. Schaffner C. P. Miller G. H. Hare R. S. Shaw K. J. Antimicrob. Agents Chemother. 1995;39:1790–1796. doi: 10.1128/AAC.39.8.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisha B. G. Steyn L. Plasmid. 1991;25:96–104. doi: 10.1016/0147-619X(91)90020-W. [DOI] [PubMed] [Google Scholar]
- Plante I. Centrón D. Roy P. H. J. Antimicrob. Chemother. 2003;51:787–790. doi: 10.1093/jac/dkg169. [DOI] [PubMed] [Google Scholar]
- Sun J. Chen C. Cui C.-Y. Zhang Y. Liu X. Cui Z.-H. Ma X.-Y. Feng Y. Fang L.-X. Lian X.-L. Zhang R.-M. Tang Y.-Z. Zhang K.-X. Liu H.-M. Zhuang Z.-H. Zhou S.-D. Lv J.-N. Du H. Huang B. Yu F.-Y. Mathema B. Kreiswirth B. N. Liao X.-P. Chen L. Liu Y.-H. Nat. Microbiol. 2019;4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seupt A. Schniederjans M. Tomasch J. Häussler S. Antimicrob. Agents Chemother. 2021;65:e01166-20. doi: 10.1128/AAC.01166-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D. Toleman M. A. Giske C. G. Cho H. S. Sundman K. Lee K. Walsh T. R. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlet G. Nadjar D. Herrmann J. L. Donay J. L. Lagrange P. H. Philippon A. Antimicrob. Agents Chemother. 2001;45:2971–2972. doi: 10.1128/AAC.45.10.2971-2972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghazadeh M. Rezaee M. A. Nahaei M. R. Mahdian R. Pajand O. Saffari F. Hassan M. Hojabri Z. Microb. Drug Resist. 2013;19:282–288. doi: 10.1089/mdr.2012.0223. [DOI] [PubMed] [Google Scholar]
- Markley J. L. Wencewicz T. A. Front. Microbiol. 2018;9:1058. doi: 10.3389/fmicb.2018.01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D. A. Rana A. P. Krapp F. Patel S. R. Huang Y. Q. Ozer E. A. Hauser A. R. Bulman Z. P. J. Antimicrob. Chemother. 2021;76:671–679. doi: 10.1093/jac/dkaa480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun K. B. Madhavan A. Abraham B. Balaji M. Sivakumar K. C. Nisha P. Kumar R. A. Antimicrob. Agents Chemother. 2020;65:e00456-20. doi: 10.1128/AAC.00456-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reygaert W. C. AIMS Microbiol. 2018;4:482–501. doi: 10.3934/microbiol.2018.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D. Adv. Drug Delivery Rev. 2005;57:1451–1470. doi: 10.1016/j.addr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Miklasińska-Majdanik M. Antibiotics. 2021;10:1406. doi: 10.3390/antibiotics10111406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D. and Berghuis A. M., in Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition, 2007, pp. 21–33, 10.1128/9781555815615.ch3 [DOI] [Google Scholar]
- Falagas M. E. Athanasaki F. Voulgaris G. L. Triarides N. A. Vardakas K. Z. Int. J. Antimicrob. Agents. 2019;53:22–28. doi: 10.1016/j.ijantimicag.2018.09.013. [DOI] [PubMed] [Google Scholar]
- Schwarz S. Kehrenberg C. Doublet B. Cloeckaert A. FEMS Microbiol. Rev. 2004;28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Surette M. D. Spanogiannopoulos P. Wright G. D. Acc. Chem. Res. 2021;54:2065–2075. doi: 10.1021/acs.accounts.1c00048. [DOI] [PubMed] [Google Scholar]
- Linkevicius M. Sandegren L. Andersson D. I. Antimicrob. Agents Chemother. 2016;60:789–796. doi: 10.1128/AAC.02465-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Bradford P. A. Cold Spring Harbor Perspect. Med. 2016;6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R., Groundwater P. W., Todd A. and Worsley A., Antibacterial Agents: Chemistry, Mode of Action, Mechanisms of Resistance and Clinical Applications, John Wiley & Sons, Incorporated, New York, 2012 [Google Scholar]
- Fleming A., Penicillin - Nobel Lecture, https://www.nobelprize.org/uploads/2018/06/fleming-lecture.pdf, (accessed May, 2022)
- Paterson D. L. Bonomo R. A. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S. K. Lee N. R. Ko J.-H. Choi J. K. Moon S.-Y. Joo E. J. Peck K. R. Park D. A. J. Antimicrob. Chemother. 2018;73:2631–2642. doi: 10.1093/jac/dky168. [DOI] [PubMed] [Google Scholar]
- Naas T. Oueslati S. Bonnin R. A. Dabos M. L. Zavala A. Dortet L. Retailleau P. Iorga B. I. J. Enzyme Inhib. Med. Chem. 2017;32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. Philos. Trans. R. Soc., B. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Tooke C. L. Hinchliffe P. Bragginton E. C. Colenso C. K. Hirvonen V. H. A. Takebayashi Y. Spencer J. J. Mol. Biol. 2019;431:3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock B. P. Raphenya A. R. Lau T. T. Y. Tsang K. K. Bouchard M. Edalatmand A. Huynh W. Nguyen A.-L. V. Cheng A. A. Liu S. Min S. Y. Miroshnichenko A. Tran H. Werfalli R. E. Nasir J. A. Oloni M. Speicher D. J. Florescu A. Singh B. Faltyn M. Hernandez-Koutoucheva A. Sharma A. N. Bordeleau E. Pawlowski A. C. Zubyk H. L. Dooley D. Griffiths E. Maguire F. Winsor G. L. Beiko R. G. Brinkman F. S. L. Hsiao W. W. L. Domselaar G. V. McArthur A. G. Nucleic Acids Res. 2020;8:517–525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen B. A. Bush K. Keeney D. Yang Y. Hare R. O'Gara C. Medeiros A. A. Antimicrob. Agents Chemother. 1996;40:2080–2086. doi: 10.1128/AAC.40.9.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind A. Stemplinger I. Jungwirth R. Ernst S. Casellas J. M. Antimicrob. Agents Chemother. 1996;40:509–513. doi: 10.1128/AAC.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo I. E. Aquino E. E. Vázquez G. J. Antimicrob. Agents Chemother. 2011;55:2968–2970. doi: 10.1128/AAC.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jure M. A. Duprilot M. Musa H. E. López C. Castillo M. C. D. Weill F. X. Arlet G. Decré D. Antimicrob. Agents Chemother. 2014;58:6335–6336. doi: 10.1128/AAC.03322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokri D. Mobasherizadeh S. Baruq M. N. Yaran M. Maj. Danishkadah-i Pizishki-i Isfahan. 2013;31:1247–1256. [Google Scholar]
- Queenan A. M. Torres-Viera C. Gold H. S. Carmeli Y. Eliopoulos G. M. Moellering, Jr. R. C. Quinn J. P. Hindler J. Medeiros A. A. Bush K. Antimicrob. Agents Chemother. 2000;44:3035–3039. doi: 10.1128/AAC.44.11.3035-3039.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R. J. Biol. Chem. 1981;256:11283–11291. doi: 10.1016/S0021-9258(19)68589-3. [DOI] [PubMed] [Google Scholar]
- Bradford P. A. Antimicrob. Agents Chemother. 1999;43:2960–2963. doi: 10.1128/AAC.43.12.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger P. Ai N. DuPrez K. T. Welsh W. J. Toney J. H. J. Med. Chem. 2010;53:3013–3027. doi: 10.1021/jm9012938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Saez I. Docquier J. D. Rossolini M. Dideberg O. J. Mol. Biol. 2007;375:604–611. doi: 10.1016/j.jmb.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Kumarasamy K. K. Toleman M. A. Walsh T. R. Bagaria J. Butt F. Balakrishnan R. Chaudhary U. Doumith M. Giske C. G. Irfan S. Krishnan P. Kumar A. V. Maharjan S. Mushtaq S. Noorie T. Paterson D. L. Pearson A. Perry C. Pike R. Rao B. Ray U. Sarma J. B. Sharma M. Sheridan E. Thirunarayan M. A. Turton J. Upadhyay S. Warner M. Welfare W. Livermore D. M. Woodford N. Lancet Infect. Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterly J. S. Richardson C. L. Eltoukhy N. S. Qi C. Scheetz M. H. Ann. Pharmacother. 2011;45:218–228. doi: 10.1345/aph.1P084. [DOI] [PubMed] [Google Scholar]
- Evans B. A. Amyes S. G. B. Clin. Microbiol. Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes N. T. Fisher J. F. Antibiotics. 2014;3:398–434. doi: 10.3390/antibiotics3030398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambler R. P. Coulson A. F. W. Frère J. M. Ghuysen J. M. Joris B. Forsman M. Levesque R. C. Tiraby G. Waley S. G. Biochem. J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J. F. Meroueh S. O. Mobashery S. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- Rotondo C. M. Wright G. D. Curr. Opin. Microbiol. 2017;39:96–105. doi: 10.1016/j.mib.2017.10.026. [DOI] [PubMed] [Google Scholar]
- Raczynskaa J. E. Shabalin I. G. Minor W. Wlodawer A. Jaskolski M. Drug Resistance Updates. 2018;40:1–12. doi: 10.1016/j.drup.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Int. J. Antimicrob. Agents. 2015;46:483–493. doi: 10.1016/j.ijantimicag.2015.08.011. [DOI] [PubMed] [Google Scholar]
- Drawz S. M. Bonomo R. A. Clin. Microbiol. Rev. 2010;23:160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu H. C. Fu K. P. Antimicrob. Agents Chemother. 1978;14:650–655. doi: 10.1128/AAC.14.5.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlaes D. M. Ann. N. Y. Acad. Sci. 2013;1277:105–114. doi: 10.1111/nyas.12010. [DOI] [PubMed] [Google Scholar]
- van Duin D. Bonomo R. A. Clin. Infect. Dis. 2016;63:234–241. doi: 10.1093/cid/ciw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akova M. Clin. Microbiol. Infect. 2008;14:185–188. doi: 10.1111/j.1469-0691.2007.01847.x. [DOI] [PubMed] [Google Scholar]
- Shapiro A. B. Antimicrob. Agents Chemother. 2017;61:e01612-17. doi: 10.1128/AAC.01612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann D. E. Jahić H. Ross P. L. Gu R.-F. Hu J. Durand-Réville T. F. Lahiri S. Thresher J. Livchak S. Gao N. Palmer T. Walkup G. K. Fisher S. L. J. Biol. Chem. 2013;288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G. G. Lawrence C. K. Adam H. Schweizer F. Zelenitsky S. Zhanel M. Lagacé-Wiens P. R. S. Walkty A. Denisuik A. Golden A. Gin A. S. Hoban D. J. Lynch J. P. Karlowsky J. A. Drugs. 2017;78:65–98. doi: 10.1007/s40265-017-0851-9. [DOI] [PubMed] [Google Scholar]
- Heo Y.-A. Drugs. 2021;81:377–388. doi: 10.1007/s40265-021-01471-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallalieu N. L. Winter E. Fettner S. Patel K. Zwanziger E. Attley G. Rodriguez I. Kano A. Salama S. M. Bentley D. Geretti A. M. Antimicrob. Agents Chemother. 2020;64:e02229-19. doi: 10.1128/AAC.02229-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M. Warner M. Mushtaq S. Woodford N. Antimicrob. Agents Chemother. 2016;60:554–560. doi: 10.1128/AAC.02184-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J. A. Hackel M. A. Bouchillon S. K. Sahm D. F. Antimicrob. Agents Chemother. 2020;64:e01432-20. doi: 10.1128/AAC.01432-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes M. D. Kumar V. Bethel C. R. Moussa S. H. O'Donnell J. Rutter J. D. Good C. E. Hujer K. M. Hujer A. M. Marshall S. H. Kreiswirth B. N. Richter S. S. Rather P. N. Jacobs M. R. Papp-Wallace K. M. van den Akker F. Bonomo R. A. MBio. 2019;10:e00159-19. doi: 10.1128/mBio.00159-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Réville T. F. Comita-Prevoir J. Zhang J. Wu X. May-Dracka T. L. Romero J. A. C. Wu F. Chen A. Shapiro A. B. Carter N. M. McLeod S. M. Giacobbe R. A. Verheijen J. C. Lahiri S. D. Sacco M. D. Chen Y. O'Donnell J. P. Miller A. A. Mueller J. P. Tommasi R. N. A. J. Med. Chem. 2020;63:12511–12525. doi: 10.1021/acs.jmedchem.0c00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Ucha J. C. Maneiro M. Martínez-Guitián M. Buynak J. Bethel C. R. Bonomo R. A. Bou G. Poza M. González-Bello C. Beceiro A. Antimicrob. Agents Chemother. 2017;61:e01172-17. doi: 10.1128/AAC.01172-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang P. A. Raj R. Tumber A. Lohans C. T. Rabe P. Robinson C. V. Brem J. Schofield C. J. Proc. Natl. Acad. Sci. U. S. A. 2022;119:e2117310119. doi: 10.1073/pnas.2117310119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard F. Odedra R. Sordello S. Cardin R. Franzoni S. Charrier C. Belley A. Warn P. Machacek M. Knechtle P. Antimicrob. Agents Chemother. 2020;64:e00078-20. doi: 10.1128/AAC.00078-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. Trout R. E. L. Chu G.-H. McGarry D. Jackson R. W. Hamrick J. C. Daigle D. M. Cusick S. M. Pozzi C. De Luca F. Benvenuti M. Mangani S. Docquier J.-D. Weiss W. J. Pevear D. C. Xerri L. Burns C. J. J. Med. Chem. 2020;63:2789–2801. doi: 10.1021/acs.jmedchem.9b01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trout R. E. Zulli A. Mesaros E. Jackson R. W. Boyd S. Liu B. Hamrick J. Daigle D. Chatwin C. L. John K. McLaughlin L. Cusick S. M. Weiss W. J. Pulse M. E. Pevear D. C. Moeck G. Xerri L. Burns C. J. J. Med. Chem. 2021;64:10155–10166. doi: 10.1021/acs.jmedchem.1c00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen P. J. Hal Jones C. Venkatesan A. M. Bradford P. A. Antimicrob. Agents Chemother. 2009;53:1698–1700. doi: 10.1128/AAC.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D. Li H. Yue C. Sun K. Dai Y. Zhang H. Liu Y. Gao Y. Li J. J. Inorg. Biochem. 2021;218:111381. doi: 10.1016/j.jinorgbio.2021.111381. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Castro V. Rodriguez-Almazan C. Silva-Sanchez J. Rudino-Pinera E. Biochem. Biophys. Res. Commun. 2020;522:545–551. doi: 10.1016/j.bbrc.2019.11.138. [DOI] [PubMed] [Google Scholar]
- Krishnan N. P. Nguyen N. Q. Papp-Wallace K. M. Bonomo R. A. van den Akker F. PLoS One. 2015;10:e0136813. doi: 10.1371/journal.pone.0136813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaka A. Tsutsumi Y. Yamada M. Suzuki K. Watanabe T. Abe T. Furuuchi T. Inamura S. Sakamaki Y. Mitsuhashi N. Ida T. Livermore D. M. J. Antimicrob. Chemother. 2015;70:2779–2786. doi: 10.1093/jac/dkv166. [DOI] [PubMed] [Google Scholar]
- Bedenić B. Sardelić S. Luxner J. Bošnjak Z. Varda-Brkić D. Lukić-Grlić A. Mareković I. Frančula-Zaninović S. Krilanović M. Šijak D. Grisold A. Zarfel G. Infect., Genet. Evol. 2016;43:74–82. doi: 10.1016/j.meegid.2016.05.011. [DOI] [PubMed] [Google Scholar]
- Hamrick J. C. Docquier J.-D. Uehara T. Myers C. L. Six D. A. Chatwin C. L. John K. J. Vernacchio S. F. Cusick S. M. Trout R. E. Antimicrob. Agents Chemother. 2020;64:e01963-19. doi: 10.1128/AAC.01963-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz A. Bugie E. Waksman S. A. Proc. Soc. Exp. Biol. Med. 1944;55:66–69. doi: 10.3181/00379727-55-14461. [DOI] [Google Scholar]
- Clark J. A. Burgess D. S. Ther. Adv. Infect. Dis. 2020;7:1–15. doi: 10.1177/2049936120952604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B. Cooper M. A. ACS Chem. Biol. 2013;8:105–115. doi: 10.1021/cb3005116. [DOI] [PubMed] [Google Scholar]
- Neuwald A. F. Landsman D. Trends Biochem. Sci. 1997;22:154–155. doi: 10.1016/S0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- Samuel L. P. Song C.-H. Wei J. Roberts E. A. Dahl J. L. Barry C. E. Jo E.-K. Friedman R. L. Microbiology. 2007;153:529–540. doi: 10.1099/mic.0.2006/002642-0. [DOI] [PubMed] [Google Scholar]
- Chen W. Biswas T. Porter V. R. Tsodikov O. V. Garneau-Tsodikova S. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9804–9808. doi: 10.1073/pnas.1105379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S. Deb J. K. Appl. Microbiol. Biotechnol. 2006;70:140–150. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- Ung K. L. Alsarraf H. M. A. B. Olieric V. Kremer L. Blaise M. FEBS J. 2019;286:4342–4355. doi: 10.1111/febs.14975. [DOI] [PubMed] [Google Scholar]
- Centron D. Roy P. H. Antimicrob. Agents Chemother. 2002;46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting M. W. Park C. H. Hegde S. S. Jacoby G. A. Hooper D. C. Blanchard J. S. Biochemistry. 2008;47:9825–9835. doi: 10.1021/bi800664x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira D. M. P. D. Forde B. M. Kidd T. J. Harris P. N. A. Schembri M. A. Beatson S. A. Paterson D. L. Walker M. J. Clin. Microbiol. Rev. 2020;33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunada A. Nakajima M. Ikeda Y. Kondo S. Hotta K. J. Antibiot. 1999;52:809–814. doi: 10.7164/antibiotics.52.809. [DOI] [PubMed] [Google Scholar]
- Schwocho L. R. Schaffner C. P. Miller G. H. Hare R. S. Shaw K. J. Antimicrob. Agents Chemother. 1995;39:1790–1796. doi: 10.1128/AAC.39.8.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P.-L. Wong R. C. Lo S. W. Chow K.-H. Wong S. S. Que T.-L. J. Med. Microbiol. 2010;59:702–707. doi: 10.1099/jmm.0.015032-0. [DOI] [PubMed] [Google Scholar]
- Vliegenthart J. S. Ketelaar-Van Gaalen P. A. G. Van De Klundert J. A. M. Antimicrob. Agents Chemother. 1991;35:892–897. doi: 10.1128/AAC.35.5.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H. Krögerrecklenfort E. Wellington E. M. H. Egan S. van Elsas J. D. van Overbeek L. Collard J. M. Guillaume G. Karagouni A. D. Nikolakopoulou T. L. Smalla K. FEMS Microbiol. Ecol. 2002;42:289–302. doi: 10.1111/j.1574-6941.2002.tb01019.x. [DOI] [PubMed] [Google Scholar]
- Call D. R. Singer R. S. Meng D. Broschat S. L. Orfe L. H. Anderson J. M. Herndon D. R. Kappmeyer L. S. Daniels J. B. Besser T. E. Antimicrob. Agents Chemother. 2010;54:590–596. doi: 10.1128/AAC.00055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather P. N. Orosz E. Shaw K. J. Hare R. Miller G. J. Bacteriol. 1993;175:6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii A. Moriyama H. Fukuhara T. Appl. Environ. Microbiol. 2012;78:5555–5564. doi: 10.1128/AEM.01155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Cabrera M. Pérez-González J. A. Heinzel P. Piepersberg W. Jiménez A. J. Bacteriol. 1989;171:321–328. doi: 10.1128/jb.171.1.321-328.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casin I. Hanau-Berçot B. Podglajen I. Vahaboglu H. Collatz E. Antimicrob. Agents Chemother. 2003;47:697–703. doi: 10.1128/AAC.47.2.697-703.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. W. Northrop D. B. J. Biol. Chem. 1978;253:5908–5914. doi: 10.1016/S0021-9258(17)34554-4. [DOI] [PubMed] [Google Scholar]
- Vong K. Auclair K. MedChemComm. 2012;3:397–407. doi: 10.1039/C2MD00253A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A. Baker E. N. Curr. Drug Targets: Infect. Disord. 2002;2:143–160. doi: 10.2174/1568005023342533. [DOI] [PubMed] [Google Scholar]
- Vakulenko S. B. Mobashery S. Clin. Microbiol. Rev. 2003;16:430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch D. R. Cox G. D'Erasmo M. P. Shakya T. Meck C. Mohd N. Wright G. D. Murelli R. P. Bioorg. Med. Chem. Lett. 2014;24:4943–4947. doi: 10.1016/j.bmcl.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst H. A. Marconi G. G. Counter F. T. Ensminger P. W. Jones N. D. Chaney M. O. Toth J. E. Allen N. E. J. Antibiot. 1982;35:1651–1657. doi: 10.7164/antibiotics.35.1651. [DOI] [PubMed] [Google Scholar]
- Allen N. E. Alborn, Jr. W. E. Hobbs Jr. J. N. Kirst H. A. Antimicrob. Agents Chemother. 1982;22:824–831. doi: 10.1128/AAC.22.5.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K. Caldwell S. J. Fong D. H. Berghuis A. M. Front. Cell. Infect. Microbiol. 2013;3:22. doi: 10.3389/fcimb.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl A. Amstutz P. Parizek P. Binz H. K. Briand C. Capitani G. Forrer P. Plückthun A. Grütter M. G. Structure. 2005;13:1131–1141. doi: 10.1016/j.str.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Daigle D. M. McKay G. A. Wright G. D. J. Biol. Chem. 1997;272:24755–24758. doi: 10.1074/jbc.272.40.24755. [DOI] [PubMed] [Google Scholar]
- Shakya T. Stogios P. J. Waglechner N. Evdokimova E. Ejim L. Blanchard J. E. McArthur A. G. Savchenko A. Wright G. D. Chem. Biol. 2011;18:1591–1601. doi: 10.1016/j.chembiol.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Leban N. Kaplan E. Chaloin L. Godreuil S. Lionne C. Biochim. Biophys. Acta, Gen. Subj. 2017;1861:3464–3473. doi: 10.1016/j.bbagen.2016.09.012. [DOI] [PubMed] [Google Scholar]
- Sehnal D. Bittrich S. Deshpande M. Svobodová R. Berka K. Bazgier V. Velankar S. Burley S. K. Koča J. Rose A. S. Nucleic Acids Res. 2021;49:W431–W437. doi: 10.1093/nar/gkab314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrilich J. Bartz Q. R. Smith R. M. Joslyn D. A. Burkholder P. R. Science. 1947;106:417. doi: 10.1126/science.106.2757.417. [DOI] [PubMed] [Google Scholar]
- Dinos G. P. Athanassopoulos C. M. Missiri D. A. Giannopoulou P. C. Vlachogiannis I. A. Papadopoulos G. E. Papaioannou D. Kalpaxis D. L. Antibiotics. 2016;5:20. doi: 10.3390/antibiotics5020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Leslie A. G. W. Annu. Rev. Biophys. Biophys. Chem. 1991;20:363–386. doi: 10.1146/annurev.bb.20.060191.002051. [DOI] [PubMed] [Google Scholar]
- Izard T. Protein Sci. 2008;10:1508–1513. doi: 10.1002/pro.101508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik V. S. Vining L. C. Can. J. Microbiol. 1970;16:173–179. doi: 10.1139/m70-030. [DOI] [PubMed] [Google Scholar]
- Smith A. L. Erwin A. L. Kline T. Unrath W. C. T. Nelson K. Weber A. Howald W. N. Antimicrob. Agents Chemother. 2007;51:2820–2829. doi: 10.1128/AAC.00087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alton N. K. Vapnek D. Nature. 1979;282:864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Parent R. Roy P. H. J. Bacteriol. 1992;174:2891–2897. doi: 10.1128/jb.174.9.2891-2897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe-Magnus D. A. Mazel D. Int. J. Med. Microbiol. 2002;292:115–125. doi: 10.1078/1438-4221-00197. [DOI] [PubMed] [Google Scholar]
- Tanaka H. Izaki K. Takahashi H. J. Biochem. 1974;76:1009–1019. [PubMed] [Google Scholar]
- Miyamura S. Koizumi K. Nakagawa Y. J. Antibiot. 1979;32:1217–1218. doi: 10.7164/antibiotics.32.1217. [DOI] [PubMed] [Google Scholar]
- Fitton J. E. Shaw W. V. Biochemistry. 1979;177:575–582. doi: 10.1042/bj1770575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous C. Cullis P. M. Shaw W. V. Biochemistry. 1985;24:5307–5313. doi: 10.1021/bi00341a006. [DOI] [PubMed] [Google Scholar]
- Biswas T. Houghton J. L. Garneau-Tsodikova S. Tsodikov O. V. Protein Sci. 2012;21:520–530. doi: 10.1002/pro.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G. W. Moody P. C. E. Shaw W. V. Proc. Natl. Acad. Sci. U. S. A. 1988;85:4133–4137. doi: 10.1073/pnas.85.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R. H. Camp D. J. Yang K. Brown M. P. Shaw W. V. Vining L. C. J. Biol. Chem. 1995;270:27000–270006. doi: 10.1074/jbc.270.45.27000. [DOI] [PubMed] [Google Scholar]
- Cundliffe E. Demain A. L. J. Ind. Microbiol. Biotechnol. 2010;37:643–672. doi: 10.1007/s10295-010-0721-x. [DOI] [PubMed] [Google Scholar]
- Corbett M. D. Chipko B. R. Antimicrob. Agents Chemother. 1978;13:193–196. doi: 10.1128/AAC.13.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis A. A. Annu. Rev. Pharmacol. Toxicol. 1988;28:83–100. doi: 10.1146/annurev.pa.28.040188.000503. [DOI] [PubMed] [Google Scholar]
- Guay D. R. P. Drugs. 1996;51:515–536. doi: 10.2165/00003495-199651040-00002. [DOI] [PubMed] [Google Scholar]
- MacLaughlin E. J. Saseen J. J. Malone D. C. Arch. Fam. Med. 2000;9:722–726. doi: 10.1001/archfami.9.8.722. [DOI] [PubMed] [Google Scholar]
- Douthwaite S. Clin. Microbiol. Infect. 2008;7:11–17. doi: 10.1111/j.1469-0691.2001.00049.x. [DOI] [PubMed] [Google Scholar]
- Golkar T. Zielinski M. Berghuis A. M. Front. Microbiol. 2018;9:1942. doi: 10.3389/fmicb.2018.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov M. S. Vázquez-Laslop N. Mankin A. S. Proc. Natl. Acad. Sci. U. S. A. 2017;114:13673–13678. doi: 10.1073/pnas.1717168115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlünzen F. Zarivach R. Harms J. Bashan A. Tocilj A. Albrecht R. Yonath A. Franceschi F. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- Bergman M. Huikko S. Huovinen P. Paakkari P. Seppälä H. Antimicrob. Agents Chemother. 2006;50:3646–3650. doi: 10.1128/AAC.00234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein R. A. Al-Ouqaili M. T. S. Majeed Y. H. Saudi J. Biol. Sci. 2022;29:513–520. doi: 10.1016/j.sjbs.2021.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wondrack L. Massa M. Yang B. V. Sutcliffe J. Antimicrob. Agents Chemother. 1996;40:992–998. doi: 10.1128/AAC.40.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A. Nakazawa K. Miyakozawa I. Mizukoshi S. Tsurubuchi K. Nakagawa M. O'hara K. Sawai T. J. Antibiot. 2000;53:516–524. doi: 10.7164/antibiotics.53.516. [DOI] [PubMed] [Google Scholar]
- Zieliński M. Park J. Sleno B. Berghuis A. M. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-020-20314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundliffe E. Antimicrob. Agents Chemother. 1992;36:348–352. doi: 10.1128/AAC.36.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski A. C. Stogios P. J. Koteva K. Skarina T. Evdokimova E. Savchenko A. Wright G. D. Nat. Commun. 2018;9:112. doi: 10.1038/s41467-017-02680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka M. Endou K. Kobayashi H. Inoue M. Nakajima Y. FEMS Microbiol. Lett. 1998;167:221–227. doi: 10.1111/j.1574-6968.1998.tb13232.x. [DOI] [PubMed] [Google Scholar]
- Nakamura A. Miyakozawa I. Nakazawa K. Sawai O. H. K. T. Antimicrob. Agents Chemother. 2000;44:3241–3242. doi: 10.1128/AAC.44.11.3241-3242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong D. H. Burk D. L. Blanchet J. Yan A. Y. Berghuis A. M. Structure. 2017;25:750–761. doi: 10.1016/j.str.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Morar M. Pengelly K. Koteva K. Wright G. D. Biochemistry. 2012;51:1740–1751. doi: 10.1021/bi201790u. [DOI] [PubMed] [Google Scholar]
- Yong D. Toleman M. A. Giske C. G. Cho H. S. Sundman K. Lee K. Walsh T. R. Antimicrob. Agents Chemother. 2009;53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L. Yu H. Qi J. Jiang P. Sun B. Cui J. Ou C. Chang W. Hu Q. PLoS One. 2015;10:e0131078. doi: 10.1371/journal.pone.0131078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourmelen A. Blondelet-Rouault M.-H. L. N. Pernodet J.-L. Antimicrob. Agents Chemother. 1998;42:2612–2619. doi: 10.1128/AAC.42.10.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós L. M. Aguirrezabalaga I. Olano C. Méndez C. Salas J. A. Mol. Microbiol. 2002;28:1177–1185. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- Sasaki J. Mizoue K. Morimoto S. Omura S. J. Antibiot. 1996;49:1110–1118. doi: 10.7164/antibiotics.49.1110. [DOI] [PubMed] [Google Scholar]
- Chesneau O. Tsvetkova K. Courvalin P. FEMS Microbiol. Lett. 2007;269:317–322. doi: 10.1111/j.1574-6968.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- Matsuoka M. Inoue M. Endo Y. Nakajima Y. FEMS Microbiol. Lett. 2003;220:287–293. doi: 10.1016/S0378-1097(03)00134-4. [DOI] [PubMed] [Google Scholar]
- Kim Y. H. Cha C. J. Cerniglia C. E. FEMS Microbiol. Lett. 2002;210:239–244. doi: 10.1111/j.1574-6968.2002.tb11187.x. [DOI] [PubMed] [Google Scholar]
- Thungapathra M. Amita Sinha K. K. Chaudhuri S. R. Garg P. Ramamurthy T. Nair G. B. Ghosh A. Antimicrob. Agents Chemother. 2002;46:2948–2955. doi: 10.1128/AAC.46.9.2948-2955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epe B. Woolley P. Hornig H. FEBS Lett. 1987;213:443–447. doi: 10.1016/0014-5793(87)81539-9. [DOI] [PubMed] [Google Scholar]
- Chopra I. Roberts M. Microbiol. Mol. Biol. Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G. G. Homenuik K. Nichol K. Noreddin A. Vercaigne L. Embil J. Gin A. Karlowsky J. A. Hoban D. J. Drugs. 2004;64:63–88. doi: 10.2165/00003495-200464010-00005. [DOI] [PubMed] [Google Scholar]
- Tucker H. Wible M. Gandhi A. Quintana A. Infect. Drug Resist. 2017;10:401–417. doi: 10.2147/IDR.S143306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G. G. Esquivel J. Zelenitsky S. Lawrence C. K. Adam H. J. Golden A. Hink R. Berry L. Schweizer F. Zhanel M. A. Bay D. Lagacé-Wiens P. R. S. Walkty A. J. Lynch III J. P. Karlowsky J. A. Int. J. Emerg. Med. 2020;80:285–313. doi: 10.1007/s40265-020-01257-4. [DOI] [PubMed] [Google Scholar]
- Pioletti M. Schlünzen F. Harms J. Zarivach R. Glühmann M. Avila H. Bashan A. Bartels H. Auerbach T. Jacobi C. Hartsch T. Yonath A. Franceschi F. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen D. E. Clemons W. M. Carter A. P. Morgan-Warren R. J. Wimberly B. T. Ramakrishnan V. Cell. 2000;103:1143–1154. doi: 10.1016/S0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Thaker M. Spanogiannopoulos P. Wright G. D. Cell. Mol. Life Sci. 2010;67:419–431. doi: 10.1007/s00018-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen F. Starosta A. L. Arenz S. Sohmen D. Dönhöfer A. Wilson D. N. Biol. Chem. 2014;395:559–575. doi: 10.1515/hsz-2013-0292. [DOI] [PubMed] [Google Scholar]
- Yang W. Moore I. F. Koteva K. P. Bareich D. C. Hughes D. W. Wright G. D. J. Biol. Chem. 2004;279:52346–52352. doi: 10.1074/jbc.M409573200. [DOI] [PubMed] [Google Scholar]
- Speer B. S. Bedzyk L. Salyers A. A. J. Bacteriol. 1991;173:176–183. doi: 10.1128/jb.173.1.176-183.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. Liu L. Sun J. Feng Y. Sci. Bull. 2019;64:1478–1481. doi: 10.1016/j.scib.2019.08.024. [DOI] [PubMed] [Google Scholar]
- Leski T. A. Bangura U. Jimmy D. H. Ansumana R. Lizewski S. E. Stenger D. A. RoweTaitt C. Vora G. J. Int. J. Antimicrob. Agents. 2013;42:83–86. doi: 10.1016/j.ijantimicag.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Volkers G. Palma G. J. Weiss M. S. Wright G. D. Hinrichs W. FEBS Lett. 2011;585:1061–1066. doi: 10.1016/j.febslet.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Park J. Gasparrini A. J. Reck M. R. Symister C. T. Elliott J. L. Vogel J. P. Wencewicz T. A. Dantas G. Tolia N. H. Nat. Chem. Biol. 2017;13:730–736. doi: 10.1038/nchembio.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkers G. Damas J. M. Palm G. J. Panjikar S. Soares C. M. Hinrichs W. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013;69:1758–1767. doi: 10.1107/S0907444913013802. [DOI] [PubMed] [Google Scholar]
- Markley J. L. Fang L. Gasparrini A. J. Symister C. T. Kumar H. Tolia N. H. Dantas G. Wencewicz T. A. ACS Infect. Dis. 2019;5:618–633. doi: 10.1021/acsinfecdis.8b00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. Zhou Y. Niu S. Liu Z. Zou Y. Yang Y. Feng H. Liu D. Niu X. Deng X. Wang Y. Wang J. Lancet. 2022;78:103943. doi: 10.1016/j.ebiom.2022.103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka L. Suzuki S. Antimicrob. Agents Chemother. 2002;46:1550–1552. doi: 10.1128/AAC.46.5.1550-1552.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C. FEMS Microbiol. Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Adams R. A. Leon G. Miller N. M. Reyes S. P. Thantrong C. H. Thokkadam A. M. Lemma A. S. Sivaloganathan D. M. Wan X. Brynildsen M. P. J. Antibiot. 2021;74:786–798. doi: 10.1038/s41429-021-00462-x. [DOI] [PubMed] [Google Scholar]
- Campbell E. A. Korzheva N. Mustaev A. Murakami K. Nair S. Goldfarb A. Darst S. A. Cell. 2001;104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Floss H. G. Yu T.-W. Chem. Rev. 2005;105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- Yazawa K. Mikami Y. Maeda A. Akao M. Morisaki N. Iwasaki S. Antimicrob. Agents Chemother. 1993;37:1313–1317. doi: 10.1128/AAC.37.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanogiannopoulos P. Thaker M. Koteva K. Waglechner N. Wright G. D. Antimicrob. Agents Chemother. 2012;56:5061–5069. doi: 10.1128/AAC.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski A. C. Wang W. Koteva K. Barton H. A. McArthur A. G. Wright G. D. Nat. Commun. 2016;7:1–10. doi: 10.1038/ncomms13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysarowich J. Koteva K. Hughes D. W. Ejim L. Griffiths E. Zhang K. Junop M. Wright G. D. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4886–4891. doi: 10.1073/pnas.0711939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbs E. R. FEMS Microbiol. Lett. 1987;44:395–399. doi: 10.1111/j.1574-6968.1987.tb02320.x. [DOI] [Google Scholar]
- Dabbs E. R. Yazawa K. Tanaka Y. Mikami Y. Miyaji M. M. Andersen S. J. Morisaki N. Iwasaki S. Shida O. Takagi H. J. Antibiot. 1995;48:815–819. doi: 10.7164/antibiotics.48.815. [DOI] [PubMed] [Google Scholar]
- Andersen S. J. Quan S. Gowan B. Dabbs E. R. Antimicrob. Agents Chemother. 1997;41:218–221. doi: 10.1128/AAC.41.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios P. J. Cox G. Spanogiannopoulos P. Pillon M. C. Waglechner N. Skarina T. Koteva K. Guarné A. Savchenko A. Wright G. D. Nat. Commun. 2016;7:1–12. doi: 10.1038/ncomms11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qia X. Lin W. Ma M. Wang C. He Y. He N. Gao J. Zhou H. Xiao Y. Wang Y. Zhang P. Proc. Natl. Acad. Sci. U. S. A. 2015;113:3803–3808. doi: 10.1073/pnas.1523614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Abdelwahab H. Campo J. S. M. D. Mehra-Chaudhary R. Sobrado P. Tanner J. J. J. Biol. Chem. 2016;291:21553–21562. doi: 10.1074/jbc.M116.745315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koteva K. Cox G. Kelso J. K. Surette M. D. Zubyk H. L. Ejim L. Stogios P. Savchenko A. Sorensen D. Wright G. D. Cell Chem. Biol. 2018;25:403–412. doi: 10.1016/j.chembiol.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Hoshino Y. Fujii S. Shinonaga H. Arai K. Saito F. Fukai T. Satoh H. Miyazaki Y. Ishikawa J. J. Antibiot. 2010;63:23–28. doi: 10.1038/ja.2009.116. [DOI] [PubMed] [Google Scholar]
- Lüscher B. Bütepage M. Eckei L. Krieg S. Verheugd P. Shilton B. H. Chem. Rev. 2018;118:1092–1136. doi: 10.1021/acs.chemrev.7b00122. [DOI] [PubMed] [Google Scholar]
- Tribuddharat C. Fennewald M. Antimicrob. Agents Chemother. 1999;43:960–962. doi: 10.1128/AAC.43.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury G. Pazhani G. P. Nair G. B. Ghosh A. Ramamurthy T. Int. J. Antimicrob. Agents. 2011;38:169–173. doi: 10.1016/j.ijantimicag.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Lourenco Da Fonseca E. Dos Santos Freitas F. De Amorim J. C. Vicente A. C. P. Antimicrob. Agents Chemother. 2008;52:1865–1867. doi: 10.1128/AAC.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsen Ø. Toleman M. A. Walsh T. R. Giske C. G. Sundsfjord A. Rydberg J. Leegaard T. M. Walder M. Lia A. Ranheim T. E. Rajendra Y. Hermansen N. O. Antimicrob. Agents Chemother. 2010;54:346–352. doi: 10.1128/AAC.00824-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A. C. S. Cavalcanti F. L. S. Martins W. M. B. Vilela M. A. Gales A. C. Morais Junior M. A. Morais M. M. C. Antimicrob. Agents Chemother. 2013;57:4077–4078. doi: 10.1128/AAC.02376-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M. Lupoli T. J. ACS Infect. Dis. 2021;7:2604–2611. doi: 10.1021/acsinfecdis.1c00297. [DOI] [PubMed] [Google Scholar]
- McDermott W. J. Infect. Dis. 1969;119:678–693. doi: 10.1093/infdis/119.6.678. [DOI] [PubMed] [Google Scholar]
- Timmins G. S. Mol. Microbiol. 2006;62:1220–1227. doi: 10.1111/j.1365-2958.2006.05467.x. [DOI] [PubMed] [Google Scholar]
- Unissa A. N. Subbian S. Hanna L. E. Selvakumar N. Infect., Genet. Evol. 2016;45:474–492. doi: 10.1016/j.meegid.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Payton M. Auty R. Delgoda R. Everett M. Sim E. J. Bacteriol. 1999;181:1343–1347. doi: 10.1128/JB.181.4.1343-1347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton A. M. Mushtaq A. Victor T. C. Sampson S. L. Sandy J. Smith D.-M. Helden P. V. V. Sim E. Mol. Microbiol. 2008;42:309–317. doi: 10.1046/j.1365-2958.2001.02648.x. [DOI] [PubMed] [Google Scholar]
- Sandy J. Holton S. Fullam E. Sim E. Noble M. Protein Sci. 2005;14:775–782. doi: 10.1110/ps.041163505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood I. M. Bhakta S. Russell A. J. Fullam E. Anderton M. C. Kawamura A. Mulvaney A. W. Vickers R. J. Bhowruth V. Besra G. S. Lalvani A. Davies S. G. Sim E. Protein Cell. 2010;1:82–95. doi: 10.1007/s13238-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agre N. Tawari N. Maitra A. Gupta A. Munshi T. Degani M. Bhakta S. Antibiotics. 2020;9:368. doi: 10.3390/antibiotics9070368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuhammad A. Fullam E. Bhakta S. Russell A. J. Morris G. M. Finn P. W. Sim E. Molecules. 2014;19:16274–16290. doi: 10.3390/molecules191016274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madikane V. E. Bhakta S. Russell A. J. Campbell W. E. Claridge T. D. W. Elisha B. G. Davies S. G. Smith P. Sim E. Bioorg. Med. Chem. 2007;15:3579–3586. doi: 10.1016/j.bmc.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Hoeksema H. Bannister B. Birkenmeyer R. D. Kagan F. Magerlein B. J. MacKellar F. A. Schroeder W. Slomp G. Herr R. R. J. Am. Chem. Soc. 1964;86:4223–4224. doi: 10.1021/ja01073a083. [DOI] [Google Scholar]
- Böttger E. C. Springer B. Prammananan T. Kidan Y. Sander P. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitcheltree M. J., PhD thesis, Harvard University, 2018 [Google Scholar]
- Petinaki E. Guérin-Faublée V. Pichereau V. Villers C. Achard A. Malbruny B. Leclercq R. Antimicrob. Agents Chemother. 2008;52:626–630. doi: 10.1128/AAC.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson-Noël A. Delrieu P. Samain D. Courvalin P. J. Biol. Chem. 1988;263:15880–15887. doi: 10.1016/S0021-9258(18)37532-X. [DOI] [PubMed] [Google Scholar]
- Morar M. Bhullar K. Hughes D. Junop M. Wright G. Structure. 2009;17:1649–1659. doi: 10.1016/j.str.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Egorov A. M. Ulyashova M. M. Rubtsova M. Y. Acta Naturae. 2018;10:33–48. doi: 10.32607/20758251-2018-10-4-33-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascale G. Wright G. D. ChemBioChem. 2010;11:1325–1334. doi: 10.1002/cbic.201000067. [DOI] [PubMed] [Google Scholar]
- Falagas M. E. Giannopoulou K. P. Kokolakis G. N. Rafailidis P. I. Clin. Infect. Dis. 2008;46:1069–1077. doi: 10.1086/527442. [DOI] [PubMed] [Google Scholar]
- Diez-Aguilar M. Canton R. Rev. Esp. Quimioter. 2019;32:8–18. [PMC free article] [PubMed] [Google Scholar]
- Kahan F. M. Kahan J. S. Cassidy P. J. Kropp H. Ann. N. Y. Acad. Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- Pakhomova S. Rife C. L. Armstrong R. N. Newcomer M. E. Protein Sci. 2009;13:1260–1265. doi: 10.1110/ps.03585004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klontz E. H. Tomich A. D. Günther S. Lemkul J. A. Deredge D. Silverstein Z. Shaw J. F. McElheny C. Doi Y. Wintrode P. L. Antimicrob. Agents Chemother. 2017;61:e01572-17. doi: 10.1128/AAC.01572-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijong Han Y. Y. Olesen S. H. Becker A. Betzi S. Schönbrunn E. Biochemistry. 2010;49:4276–4282. doi: 10.1021/bi100365b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Zheng B. Li Y. Zhu S. Xue F. Liu J. PLoS One. 2015;10:e0135269. doi: 10.1371/journal.pone.0135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. Miao V. Kwong W. Xia R. Davies J. Lett. Appl. Microbiol. 2011;52:427–429. doi: 10.1111/j.1472-765X.2011.03016.x. [DOI] [PubMed] [Google Scholar]
- Alrowais H. McElheny C. L. Spychala C. N. Sastry S. Guo Q. Butt A. A. Doi Y. Emerging Infect. Dis. 2015;21:2045. doi: 10.3201/eid2111.150750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura G. Wachino J.-i. Sato N. Kimura K. Yamada K. Jin W. Shibayama K. Yagi T. Kawamura K. Arakawa Y. J. Clin. Microbiol. 2014;52:3175–3179. doi: 10.1128/JCM.01094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y. Xu X. Guo Q. Wang P. Wang W. Wang M. Lett. Appl. Microbiol. 2015;60:259–264. doi: 10.1111/lam.12366. [DOI] [PubMed] [Google Scholar]
- Guo Q. Tomich A. D. McElheny C. L. Cooper V. S. Stoesser N. Wang M. Sluis-Cremer N. Doi Y. J. Antimicrob. Chemother. 2016;71:2460–2465. doi: 10.1093/jac/dkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman M. A. Yin X. Persaud-Lachhman M. G. Diarra M. S. Antimicrob. Agents Chemother. 2017;61:e00410–e00417. doi: 10.1128/AAC.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigsby R. E. Rife C. L. Fillgrove K. L. Newcomer M. E. Armstrong R. N. Biochemistry. 2004;43:13666–13673. doi: 10.1021/bi048767h. [DOI] [PubMed] [Google Scholar]
- Tomich A. D. Klontz E. H. Deredge D. Barnard J. P. McElheny C. L. Eshbach M. L. Weisz O. A. Wintrode P. Doi Y. Sundberg E. J. Sluis-Cremer N. Antimicrob. Agents Chemother. 2019;63:e01524-18. doi: 10.1128/AAC.01524-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D. T. Lavanya P. Doss C. G. P. Tayubi I. A. Kumar R. N. Yesurajan I. F. Siva R. Balaji V. J. Cell. Biochem. 2017;118:4088–4094. doi: 10.1002/jcb.26064. [DOI] [PubMed] [Google Scholar]
- Thompson M. K. Goodman M. Keithly M. Hammer N. Cook P. Jagessar K. Harp J. Skaar E. Armstrong R. N. Biochemistry. 2014;106:45a. doi: 10.1021/bi4015852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M. Bernat B. A. Wang Z. Armstrong R. N. Helmann J. D. J. Bacteriol. 2000;183:2380–2383. doi: 10.1128/JB.183.7.2380-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read T. D. Peterson S. N. Tourasse N. Baillie L. W. Paulsen I. T. Nelson K. E. Tettelin H. Fouts D. E. Eisen J. A. Gill S. R. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- Xu X. Chen C. Lin D. Guo Q. Hu F. Zhu D. Li G. Wang M. PLoS One. 2013;8:e78106. doi: 10.1371/journal.pone.0078106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. K. Keithly M. E. Sulikowski G. A. Armstrong R. N. Perspect. Sci. 2015;4:17–23. doi: 10.1016/j.pisc.2014.12.004. [DOI] [Google Scholar]
- Lamers A. P. Keithly M. E. Kim K. Cook P. D. Stec D. F. Hines K. M. Sulikowski G. A. Armstrong R. N. Org. Lett. 2012;14:5207–5209. doi: 10.1021/ol302327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A. D. Riedel T. M. Kesler M. B. A. Varney M. E. Long T. E. J. Antibiot. 2022;75:146–154. doi: 10.1038/s41429-022-00500-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillgrove K. L. Pakhomova S. Schaab M. R. Newcomer M. E. Armstrong R. N. Biochemistry. 2007;46:8110–8120. doi: 10.1021/bi700625p. [DOI] [PubMed] [Google Scholar]
- Zhang F. Zhai T. Haider S. Liu Y. Huang Z. J. J. Am. Chem. Soc. 2020;5:7537–7544. doi: 10.1021/acsomega.0c00352. [DOI] [PMC free article] [PubMed] [Google Scholar]