Abstract
Advancements in cross-linking mass spectrometry (XL-MS) bridge the gap between purified systems and native tissue environments, allowing the detection of protein structural interactions in their native state. Here we use isobaric quantitative protein interaction reporter technology (iqPIR) to compare the mitochondria protein interactomes in healthy and hypertrophic murine hearts, 4 weeks post-transaortic constriction. The failing heart interactome includes 588 statistically significant cross-linked peptide pairs altered in the disease condition. We observed an increase in the assembly of ketone oxidation oligomers corresponding to an increase in ketone metabolic utilization; remodeling of NDUA4 interaction in Complex IV, likely contributing to impaired mitochondria respiration; and conformational enrichment of ADP/ATP carrier ADT1, which is non-functional for ADP/ATP translocation but likely possesses non-selective conductivity. Our application of quantitative cross-linking technology in cardiac tissue provides molecular-level insights into the complex mitochondria remodeling in heart failure while bringing forth new hypotheses for pathological mechanisms.
Keywords: quantitative cross-linking, heart failure, cardiac hypertrophy, mitochondria, protein interactions, interactome, mass spectrometry, systems structural biology
Introduction
Mitochondria contain over 1,100 proteins that orchestrate diverse cellular functions, from intermediary metabolism and energy production to signaling and cell death 1, 2. Extensive genomic and proteomic approaches have resulted in a curated compendium of mitochondrial proteins with their sub-organellar localization 1. Advancements in cryo-electron microscopy, X-ray crystallography, and NMR have greatly increased understanding of protein function and have enabled structure visualization of purified mitochondrial proteins and complexes with sizes unimaginable only a few years ago 3–7. However, within its native environment, protein function is dynamically regulated by the presence of other proteins, post-translational modifications, substrates, cofactors, and transient conformational changes that are not directly measured with conventional techniques.
Chemical cross-linking combined with mass spectrometry (XL-MS) methods have emerged to provide structural insight from complex samples 8. A cross-linker approach including MS cleavable features and affinity tags referred to as Protein Interaction Reporter (PIR) technology 9 has enabled interactome-level structural studies with isolated functional mitochondria 10, intact virus particles 11, live cells 12, 13, and even whole tissue samples 14. While identification of in vivo protein-protein interactions (PPIs) and conformational features from cross-linked peptides increase knowledge about how proteins function within cells, quantitation of cross-linked peptide level changes during perturbation reveals molecular features that confer functional changes at the systems level. For instance, quantitation of PIR cross-linked peptide levels in cells using SILAC 15 revealed functional differences relevant to acquired resistance to topoisomerase I inhibition therapy 16. Moreover, quantitative PIR applications to cellular pharmacological studies revealed interactome changes that are drug-concentration dependent and mechanism-of-action specific 13, 17. Thus, quantitative cross-linking technologies enable visualization of interactome changes in living systems relevant to functional changes that could be informative in pathological comparisons. Recently, PIR technologies were further advanced to include isobaric quantitative capabilities enabling quantitative interactome studies for systems without the need for SILAC 18, as is employed here for failing heart mitochondrial interactome studies.
Due to the extraordinary energy requirements of the heart, cardiomyocytes contain the highest concentration of mitochondria of any cell in the body across mammalian species 19, 20. Concurrently, mitochondrial dysfunction is a well-known maladaptive mechanism in the progression of heart failure. This study demonstrates the feasibility of applying isobaric quantitative PIR (iqPIR)18 XL-MS technology to mitochondria from healthy and failing mouse hearts. For the first time, integrated measurements of protein interactions, conformations, and surface accessibility allowing for comparative network analysis were enabled directly in tissue. These advancements provide the unique opportunity to study chronic, organ-level conditions where cell modeling does not holistically recapitulate the bewildering complexity of disease.
Results
Quantitation of mitochondrial interactome in failing hearts
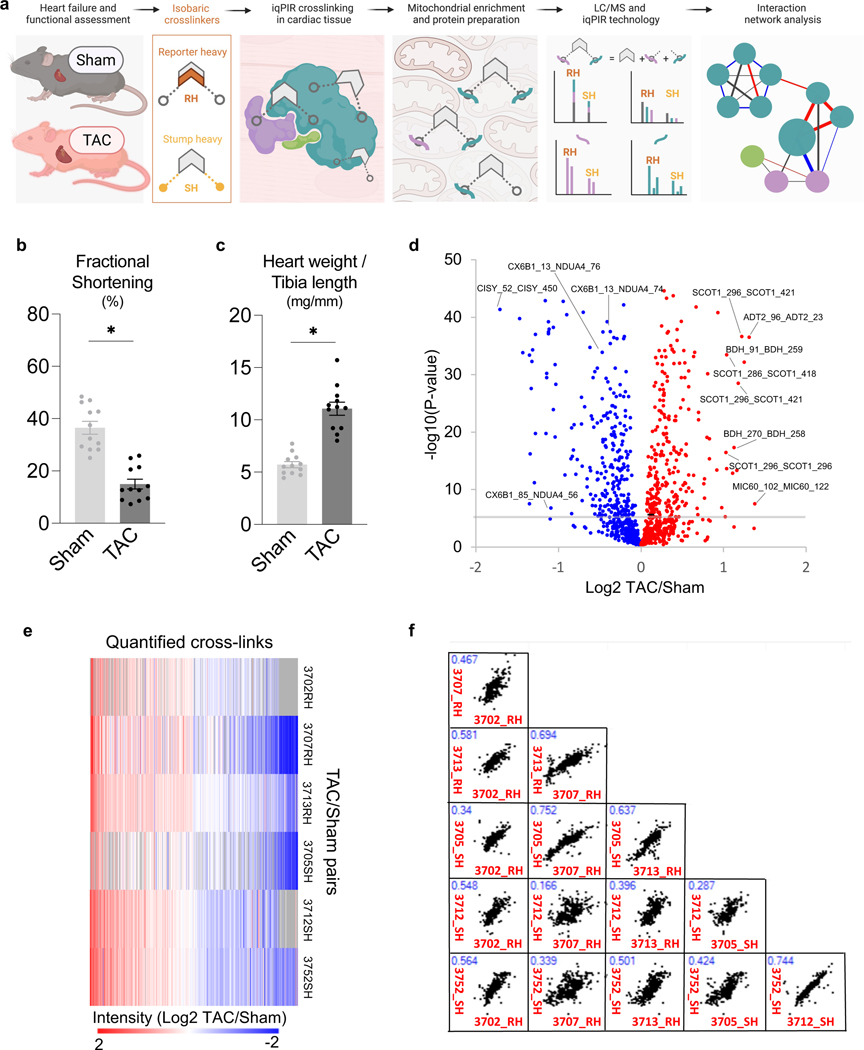
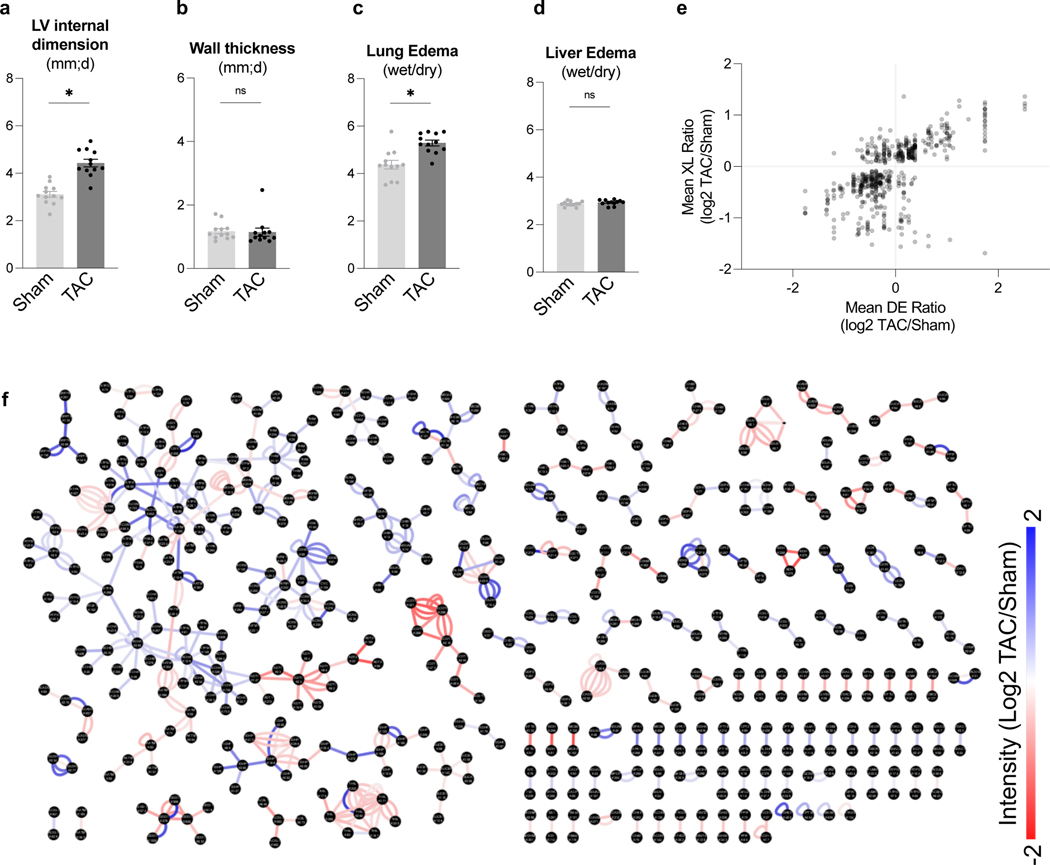
A workflow was developed to assess mitochondrial protein interactome in failing hearts using iqPIR technology (Fig. 1a). Heart failure was induced by transverse aortic constriction (TAC, n=12) surgery in mice. Sham-operated (Sham) animals were used as controls (n=12). TAC hearts demonstrated a decline in left ventricular fractional shortening (Fig. 1b) and increased chamber size (Extended Data Fig. 1a-b) compared to Sham. At harvest, TAC mice showed significant cardiac hypertrophy (Fig. 1c) and clear signs of pulmonary congestion (Extended Data Fig. 1c-d), indicating the development of heart failure.
Figure 1: Quantitation of mitochondrial protein interactome in failing hearts.
(a) Schematic of quantitative failing heart interactome pipeline. Briefly, TAC and Sham hearts were excised and subjected to stump-heavy and reporter-heavy iqPIR cross-linking reaction followed by mitochondrial enrichment. Samples were pooled for downstream processing and data acquisition by LC/MS. Identical cross-linked peptide pairs from TAC and Sham samples have identical masses yet produce distinct quantitative isotope signatures in MS2 spectra, and their intensities enable relative quantitation of cross-linked peptides. Illustration created with BioRender.com.
(b) Assessment of cardiac function measured by fractional shortening (%) using echocardiography in TAC and Sham groups four-weeks post-surgery.
(c) Cardiac hypertrophy (mg/mm) measured by heart weight normalized to tibia length.
(d) Volcano plot of quantified cross-link lysine ratios (Log2 TAC/Sham) versus statistical significance. Blue circles indicate cross-linked peptide ratios with decreasing quantitation, while red circles indicate cross-linked peptide ratios with increasing quantitation (relative to TAC). The cross-link Log2 ratio p-value is calculated based on an unpaired, two-tailed t-test using the Log2 ratio mean value, standard deviation, and a number of contributing quantified ions. Significance threshold set to p=2.5×10−6 (-log10=5.6).
(e) Heat map illustration of quantified cross-linked peptide pairs showing significant changes between TAC and Sham groups, filtered for values present in at least 4/6 biological replicates. 3 pairs of replicates were forward-labeled samples (Reporter Heavy (RH) crosslinked TAC combined with Stump Heavy (SH) crosslinked Sham) and 3 pairs were reverse samples (SH labeled TAC combined with RH labeled Sham).
(f) Pair-wise scatter plots of 6 pairs of biological replicates. Correlation of Log2 ratios of two biological replicates as shown in X-axis and Y-axis was evaluated with linear regression. R-squared value marked in blue font on top left of each figure.
For (b-c), all data are n=12 animals, AVG+/−SEM, * denotes p<0.05 by unpaired, two-tailed Student’s t test.
Dissected TAC or Sham cardiac tissue was cross-linked with a 2-plex iqPIR cross-linker with either Reporter Heavy (RH) or Stump Heavy (SH) iqPIR versions. TAC and Sham cross-linked samples were then mixed pairwise in a 1:1 ratio. Thus, a total of 6 pairs of biological replicates were generated, including 3 pairs of forward samples and 3 pairs of reverse samples. The forward/reverse labeling strategy evaluated if the labeling direction caused any quantitation bias.
In total, 3,792 non-redundant cross-linked peptide pairs were identified from 6 pairs of TAC/Sham heart samples at an estimated FDR of 1% or less, corresponding to 2,734 lysine residue pairs and 507 protein pairs. The failing heart interactome network is available online in XLinkDB 21 (xlinkdb.gs.washington.edu, network table name = Caudal_iqPIR_TACsham_Bruce). All quantified cross-link ratios were plotted against the p-value of each ratio (Fig. 1d), which determined statistical outliers (significance threshold p-value= 2.5×10−6). The data were further processed by applying a statistical t-test and Bonferroni correction to all quantified cross-linked peptides, then filtered to a maximum of 2 missing ratios out of 6 pairs of biological replicates. A final number of 588 non-redundant crosslinked peptide pairs showed statistically significant differences between TAC and Sham hearts (Supplementary Data 1). The heat map containing 588 cross-linked peptides was generated using NG-CHM BUILDER 22 (Fig. 1e). A total of 297 cross-linked peptide pairs were decreased, while 291 cross-linked peptide pairs were increased in TAC.
iqPIR cross-linkers employ reactive esters to react with surface accessible lysine residues covalently. Dead-end (DE) labeled peptides are generated when only one ester reacts with a lysine residue while the other ester is hydrolyzed. This feature is useful as a read-out for the surface accessibility of proteins and an indirect survey of relative protein level changes. Since PPI levels can be affected by endogenous protein levels, DE-labeled peptide quantitation provided estimated protein abundance for each cross-linked peptide pair as demonstrated previously by Chavez et al. 18. For each non-redundant cross-link pair, the mean Log2 ratios for Protein A and Protein B were calculated from all DE peptides (Supplementary Data 1). In parallel, label-free quantitation (LFQ) of mitochondrial proteins from 6 biological replicates of TAC and sham samples and combined Log2 ratios of TAC and sham are reported in Supplementary Data 2. In addition, the reproducibility of the data was evaluated by applying linear regression to 6 pairs of biological replicates on a pair-wise basis. R-squared values were calculated and shown in Fig. 1f. Despite the known individual variations in the TAC model, strikingly high data correlation and consistency were observed across all 15 comparisons of 6 TAC/Sham pairs. It is worth noting that each sample is a 1:1 mix of cross-linked proteins from two different animals; thus, regression fitting of two samples involved 4 different animals. The correlations of forward and forward labeling samples (average R2=0.53) are close to those of forward and reverse labeling samples (average R2=0.47) (Supplementary Data 3), which further confirmed the consistency and reproducibility of iqPIR quantitation results. The subset of cross-linked peptide pairs displaying statistically different ratios in TAC were further analyzed by interaction network analysis (Extended Data Fig. 1f). Comprehensive views of pathways and proteins exhibiting large statistical changes (Log2 ratios are generally ≥1 or ≤−1) led to key highlights of TAC-induced structural changes that will be further discussed in greater detail in the following sections.
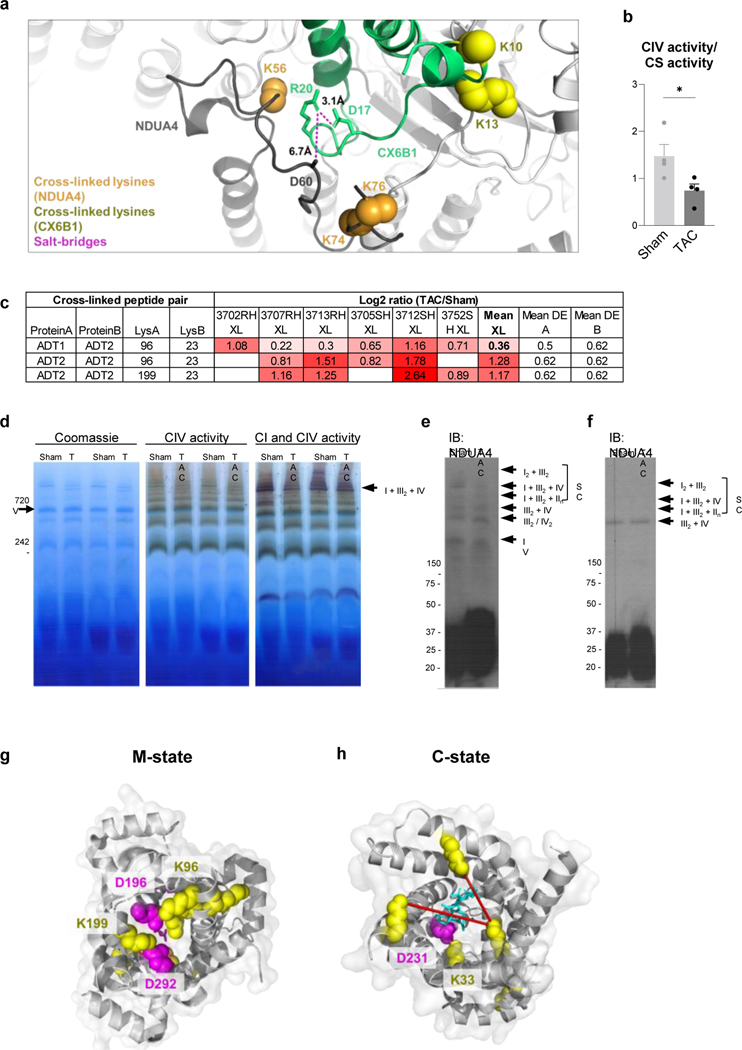
Altered NDUA4-C6XB1 interaction and reduced CIV activity in TAC
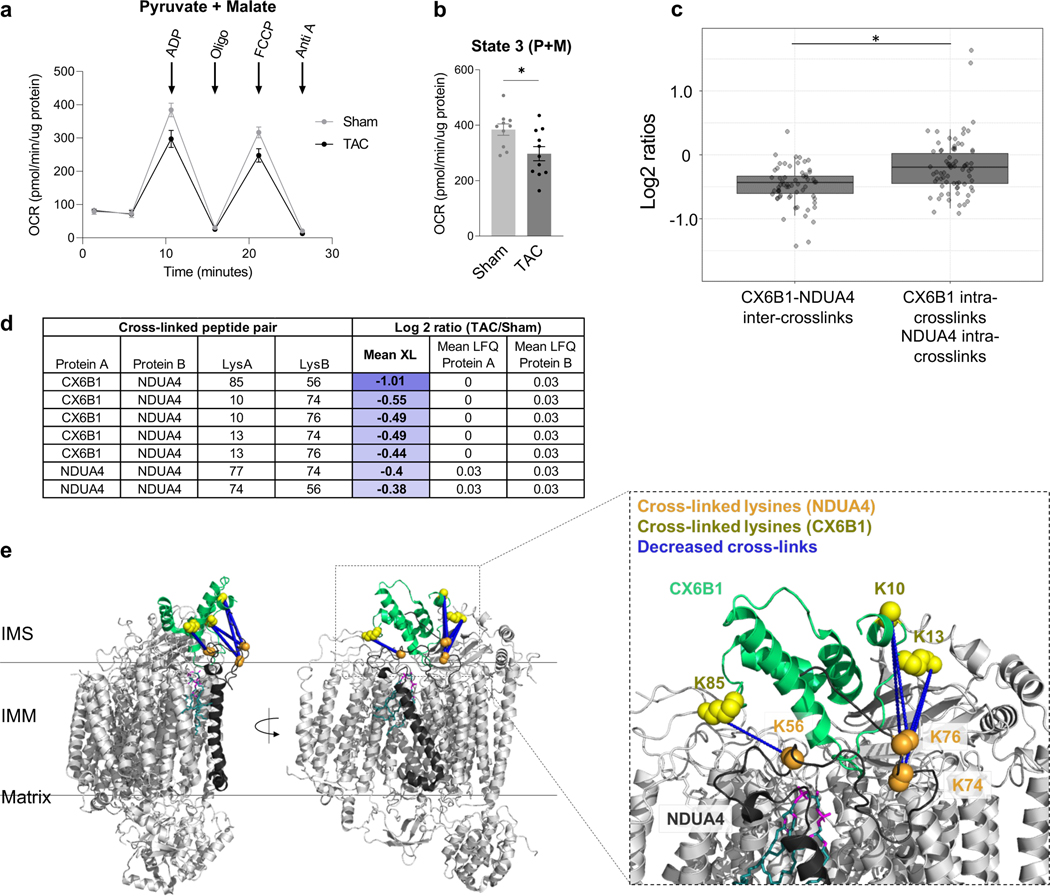
Mitochondrial respiration, assessed by oxygen consumption rate (OCR), was reduced in TAC hearts (Fig. 2a-b). Multiple mechanisms have been proposed for impaired OCR, among which altered function of electron transport chain (ETC) has been proposed 19. Quantitative mitochondrial interactome data analysis revealed five cross-linked lysine pairs between NDUA4 and CX6B1 (human COX6B isoform 1) in Complex IV that were significantly decreased in TAC (Fig. 2c-d, Supplementary Data 1). Complex IV (Cytochrome C oxidase, CIV) is the terminal end of the electron transport chain, which accepts two electrons from Cytochrome C (CYC) to reduce oxygen into water. Due to its high sensitivity to detergent conditions 23, 24 the interaction between NDUA4 and other subunits of Complex IV has been difficult to study. Since iqPIR cross-linkers were applied directly to heart tissue to secure PPIs with covalent bonds, the disruption of detergent-sensitive interactions such as those of NDUA4 was avoided with this approach. Identification of multiple NDUA4 inter-protein cross-linked peptide pairs provides direct and definitive evidence that NDUA4 exists as a component of CIV in cardiac mitochondria and provides insight into the NDUA4-CX6B1 interaction. Cross-linked peptides containing residues K10, K13, and K85 of CX6B1 and K56, K74, and K76 of NDUA4 were mapped to disordered regions of both proteins residing in the IMS (Fig. 2e, PDB: 5Z62), making further structural analysis challenging. Mutations in CX6B1 R20 which disrupt a salt-bridge with D17 25 are hypothesized to cause instability of CIV, leading to encephalomyopathy and hypertrophic cardiomyopathy in patients 26. R20 is likely stabilized by a salt-bridge with nearby NDUA4 D60, although this side-chain is only partially resolved in the structure (Extended Data Fig. 2a, PDB: 5Z62). However, Complex IV activity was significantly decreased in TAC (Extended Data Fig. 2b), raising the possibility that the NDUA4-CX6B1 interaction is a regulator of CIV function in heart tissues.
Figure 2: Decreased interaction between NDUA4 and C6XB1 affects CIV activity in TAC.
(a) Oxygen consumption rate of mitochondria isolated from TAC (n=11 animals) and Sham (n=10 animals) hearts given pyruvate/malate substrates followed by sequential injections of ADP, Oligomycin, FCCP, and Antimycin A, measured by Seahorse XFe24 Analyzer.
(b) State III-driven respiration of isolated mitochondria (after addition of ADP).
(c) Box plot illustrates a comparison of Log2 ratios of inter-links versus intra-links of NDUA4 and CX6B1 (n=66 inter-protein crosslinks, n=84 intra-protein crosslinks, visualized as median and 25th and 75th percentiles, with whiskers indicating minima and maxima excluding outliers). Welch’s two-sample, two-sided t-test was used to determine statistical significance.
(d) Table summarizing the mean cross-linking ratio and LFQ ratio for cross-linked peptide pairs exhibiting quantitative changes in NDUA4 and CX6B1 subunits of CIV.
(e) Monomeric Complex IV (grey, PDB: 5Z62). Interactions between NDUA4 (black) and CX6B1 (green) subunits are shown. Cross-linked lysine sidechains are shown as yellow (CX6B1) or orange (NDUA4) spheres. Cross-links exhibit a structural scaffold on the IMS-facing interface of CIV, which are decreased in TAC. Cardiolipin bound to CIV is shown in teal.
For (a-b), TAC (n=11 animals) and Sham (n=10 animals) data is represented as AVG+/−SEM, *denotes p<0.05 by unpaired, two-tailed Student’s t test.
Furthermore, as NDUA4 blocks the CIV dimerization interface, its presence maintains CIV primarily as a monomer, thus facilitating Complex I-III2-IV respirasome assembly 23. Alterations in the interaction between NDUA4 and other subunits of CIV could therefore affect respirasome formation. Decreased CI-CIII2-CIV respirasome has been previously reported in heart failure 27. Consistently, BN-PAGE showed decreased association of NDUA4 with Complex I-III2-IV and reduced formation of I-III2-IV respirasome in TAC (Extended Data Fig. 2c-e). Thus, quantitative mitochondrial interactome analysis reveals remodeling of Complex IV structure as a new contributor to impaired mitochondrial respiration within the failing heart.
Active ketone-oxidation enzyme conformations increased in TAC
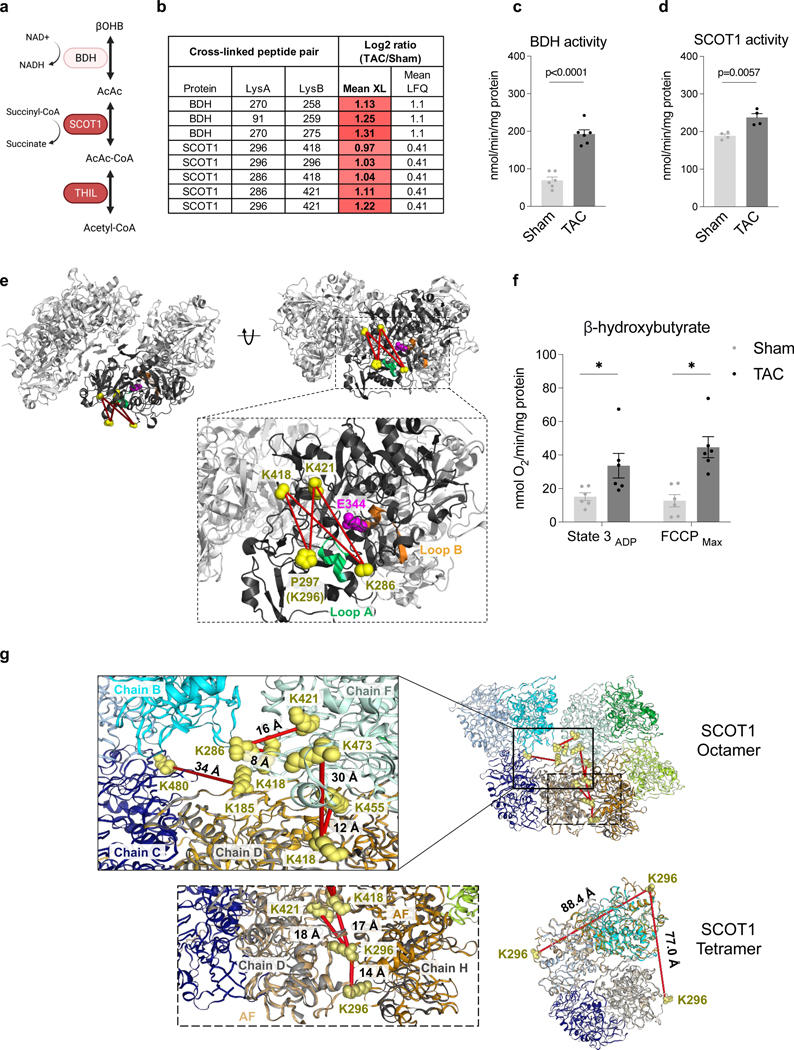
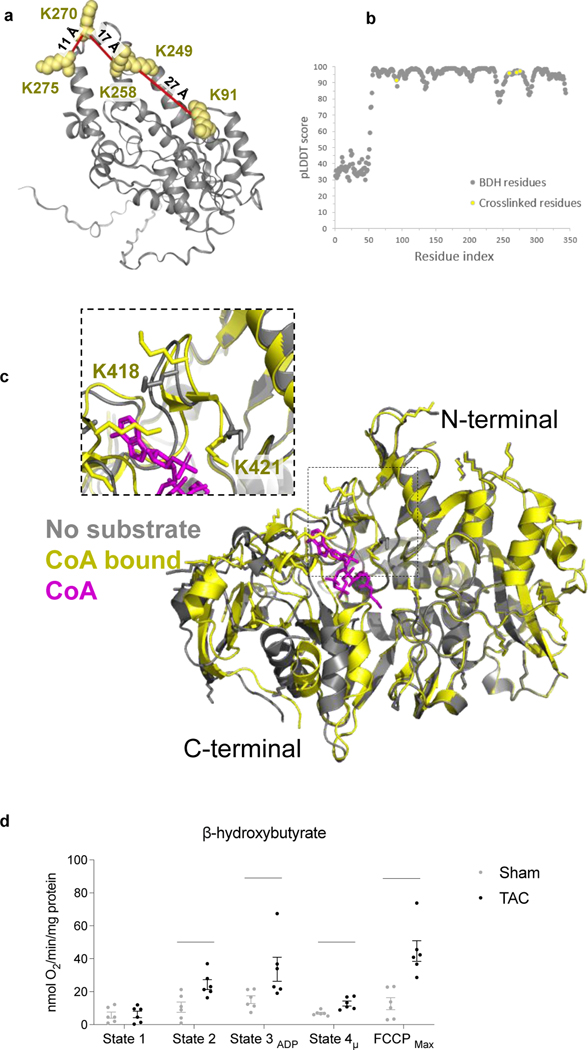
Recent studies demonstrated increased ketone oxidation as part of mitochondrial metabolic reprogramming in failing hearts 28–30. Enzymes for ketone oxidation, D-β-hydroxybutyrate dehydrogenase (BDH), and Succinyl-CoA:3-oxoacid-CoA transferase (SCOT1) were reported to be upregulated in the failing heart. Still, mechanisms driving the flux of ketone oxidation are not fully understood 29–31. Circulating ketones, primarily generated by the liver, are taken up by cardiomyocytes and metabolized via three consecutive reactions catalyzed by BDH, SCOT1, and Acetyl-CoA acetyltransferase (THIL) to produce acetyl-CoA for further oxidation in the TCA cycle (Fig. 3a). Consistent with increased expression of ketone metabolism proteins in failing myocardium 29, 30, BDH levels were increased significantly (over 2-fold) in the TAC mitochondria, which agreed well with the results reported by Uchihashi et al., who used iTRAQ relative quantitation of TAC and sham heart 32 (Fig. 3b). Consistently, BDH activity was increased in TAC mitochondria (Fig. 3c). The structure of BDH is unavailable; thus, AlphaFold (AF), a newly developed computational software tool by DeepMind, was used to predict BDH_mouse structure 33. The identified crosslinks of BDH mapped to the AF-generated model were within the spatial distance constraints of the PIR crosslinker (< 40 Å) (Extended Data Fig. 3a-b), supporting the Alphafold model with empirical data.
Figure 3: Active conformational states of ketone oxidation proteins enriched in TAC.
(a) Schematic of ketone oxidation pathway. Illustration created with BioRender.com.
(b) Table summarizing the mean cross-linking ratio and LFQ ratio for cross-linked peptide pairs exhibiting quantitative changes in ketone oxidation machinery.
(c) Enzymatic activity of BDH1 in mitochondria isolated from TAC and Sham hearts, normalized to total protein. N=6 animals, AVG+/−SEM, *denotes p<0.05 by unpaired, two-tailed Student’s t-test.
(d) Enzymatic activity of SCOT1 from heart lysates from TAC and Sham, normalized to total protein. N=4 animals, AVG+/−SEM, *denotes p<0.05 by unpaired, two-tailed Student’s t-test.
(e) Human SCOT1 tetrameric structure (PDB: 3DLX) shown in grey. Monomeric unit is shown in black centered around the catalytic E344 (magenta). Cross-linked lysine side-chains (K286, K296/P297, K418, and K421, shown as yellow spheres) surround Loop A and together form four cross-links found to increase in TAC. Loop A (CoA binding site, residues 321–329, chain shown in green) is a dynamic region that must undergo conformational change to prevent steric clashing of N- and C-termini during catalysis. Loop B is Succinate/Acetoacetate binding site (residues 374–386 chain shown in orange). P297 is shown due to missing density of K296.
(f) Oxygen consumption rate (OCR) of mitochondria isolated from TAC and Sham hearts given β-hydroxybutyrate/malate and stimulated with ADP stimulation (State 3) and FCCP (FCCPmax). N=6 animals, AVG+/−SEM, *denotes p<0.05 by unpaired, two-tailed Student’s t-test.
(g) SCOT1 octameric (PDB: 3OXO) and tetrameric (PDB: 3DLX) structure. Crosslinked residues were indicated in yellow spheres and crosslinks indicated in red lines mean they were increased in TAC samples. AlphaFold (AF) was used to construct SCOT1_MOUSE monomeric structure (in orange cartoon) and aligned with chain D (light grey) and H (dark grey) in 3OXO to cover unresolved region containing residues 287–300 in 3OXO. D299 were used in 3DLX due to missing density of K296.
Interestingly, SCOT1 activity increased significantly in TAC (Fig. 3d), although changes in its protein level were very modest (Fig. 3b), suggesting potential structural changes in SCOT1.
SCOT1 is a tetramer (Fig. 3e), and it transfers a Coenzyme A (CoA) moiety from succinyl-CoA to acetoacetate to form acetoacetyl-CoA and succinate (Fig. 3a). The reaction involves E344 in the active site, which sits between two binding pockets for succinate/acetoacetate (residues 321–329, Loop A) and CoA (residues 374–386, Loop B), respectively 34. Five non-redundant cross-linked peptide pairs in SCOT1 were increased significantly more than DE with TAC operation (Fig. 3b). Cross-linked peptide pairs containing residue K286, K296, K418, and K421 were mapped onto the SCOT1 structure (human SCOT1 PDB: 3DLX) and pinpointed a critical surface near Loop A (Fig. 3e). The C-terminal undergoes a 17-degree angle domain rotation during catalysis where Loop A must conform to alleviate steric clashing with the static N-terminal 34. We hypothesize that the rotation renders the cross-linking of the four peptide pairs possible. Thus, increases in the cross-linked peptide pairs suggest that a greater fraction of SCOT1 assumes an active conformational state in TAC. As K286 and K296 reside in a relatively disordered region of SCOT1, detailed structural information is unavailable. However, a large conformational change in the C-terminal domain, where the four cross-linked peptide pairs reside, is observed when apo (PDB: 3OXO, chain A) and substrate-bound (PDB: 3OXO, chain E) porcine SCOT1 monomers are aligned (Extended Data Fig. 3c) 35, 36 supporting the notion that the active enzyme assumes a distinct conformation in the region. In humans, naturally occurring mutations in Loop A, such as G324E and L327P, result in SCOT1 loss-of-function 37, indicating the importance of flexible conformations observed in this region.
The homodimer link between SCOT1 K296-K296 increased ∼2-fold in TAC (p-value=3.98E−17), indicating increased interaction between two monomeric units of SCOT1 (Fig. 3b). However, the K296-K296 distance mapped onto the human tetramer structure (PDB: 3DLX) (Fig. 3g, labeled SCOT1 tetramer) is over 65 Å, which exceeds the span possible by iqPIR cross-linkers. These results suggested that SCOT1 exists in an alternative conformation or higher-order assembly that increases in TAC. The structure 3OXO is a SCOT1 octamer; however, regions of residues 287–300 (containing K296 residue) were not resolved in this octamer. To determine if the K296-K296 link can be mapped in the octamer structure, we first constructed the SCOT1_MOUSE structure using AlphaFold 33. Next, we applied the TM-align software tool 38 to align the AF-generated structure with two adjacent subunits (Chain D and H) of 3OXO (Supplementary Data 4). As shown in the bottom inset of Fig. 3g, the distance of residue pair K296-K296 is approximately 14 Å, indicating this link is consistent with octamer but not tetramer SCOT1 oligomers. Along with 7 other inter-chain crosslinked residue pairs (Fig. 3g), the increased quantitation of these crosslinks indicates enrichment of SCOT1 octamer assemblies in TAC hearts.
This evidence suggests that a higher-order assembly could account for increased SCOT1 activity in the absence of large changes in protein amount. If this is the case, enrichment of SCOT1 octamer should be accompanied by increased ketone body oxidation as SCOT1 catalyzes the rate-limiting step of this pathway. We thus assessed ketone body oxidation by measuring β-hydroxybutyrate supported respiration (OCR) in mitochondria isolated from sham and TAC hearts. Although ketone-supported respiration is relatively low in isolated mitochondria due to the lack of anaplerotic reactions, the OCR was 2–3 folds higher in TAC than the sham group under this condition (Fig. 3f, Extended Data Fig. 3d). Thus, these data substantiate the notion that ketone oxidation increases in TAC and functionally correlates with the cross-linking discovery of higher-order oligomers.
Taken together, quantitative iqPIR analysis confirmed the upregulation of ketone metabolism proteins as previously reported and suggests possible mechanisms for active protein conformations and higher-order assembly in the failing hearts. These findings provide greater insight into how ketone metabolism is regulated and adapted in the failing heart, enabling future therapeutic developments.
Enrichment of ADP/ATP carrier P-state detected in TAC
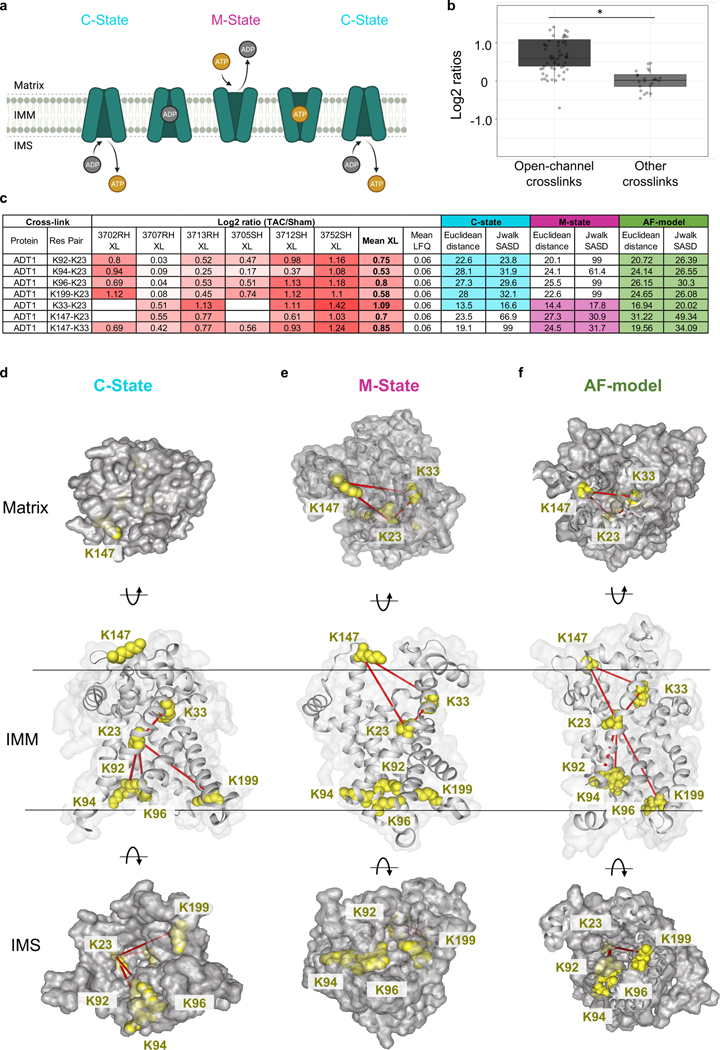
The exchange of ATP and ADP between mitochondria and cytosol is achieved by the mitochondrial ADP/ATP carrier (ADT), a highly conserved, abundant, and extensively studied translocase 39. ADT is a dual gated transporter that interconverts between at least two known distinct structural conformations (Fig. 4a). During a normal cycle, the cytoplasm-open state (C-state) faces the IMS. The gate to the mitochondrial matrix is closed via a series of salt-bridges, allowing for the release of a bound ATP and the binding of a new cytosolic ADP 39. The binding of ADP induces a conformational shift to the matrix-open state (M-state), in which the gate to the IMS is closed by the formation of new salt-bridges and a hydrophobic plug 40. The M-state releases the ADP from the cytosol into the matrix in exchange for a newly charged ATP, and the cycle repeats. These conformational changes permit the transport of large nucleotide solutes across small protein carriers while preventing proton leak throughout the exchange process 41. Mammalian ADT exists in four isoforms which exhibit tissue-dependent but overlapping expression patterns.
Figure 4: Enrichment of an intermediate state of ADP/ATP carrier detected in TAC.
(a) Schematic of ADT1 cycling between two dominant conformational states. The cytoplasmic-facing state (C-state) delivers ATP to the cytosol and binds ADP. Binding of ADP induces a conformational change to the matrix-facing state (M-state), which returns ADP to the mitochondria and binds a newly charged ATP. Illustration created with BioRender.com.
(b) Box plot illustrates a comparison of Log2 ratios of ADT1 open-channel crosslinks versus other crosslinks (n=60 open-channel crosslinks, n=24 other crosslinks, visualized as median and 25th and 75th percentiles, with whiskers indicating minima and maxima excluding outliers. Welch’s two-sample, two-sided t-test was used to determine statistical significance.
(c) Table summarizing mean cross-linking ratio and LFQ ratio for each cross-linked peptide pair detected in ADT1 exhibiting changes in TAC. Euclidean distance (EI) and Solvent Accessible Surface distance (Jwalk SASD) are listed for each cross-linked peptide pair when mapped onto both C-state and M-state of ADT1. Highlighted values satisfy iqPIR molecular distance constraint (EI<42Å, Jwalk SASD<51Å).
(d) Cross-linked peptide pairs (yellow lysine side chains) mapped onto the C-state conformation of bovine ADT1 (PDB: 1OKC). In this conformation, K23, K92, K94, K96, and K199 are available and can satisfy cross-linking distance constraints. K147 and K33 are not accessible in the M-state
(e) Cross-linked peptide pairs (yellow lysine side chains) mapped onto the M-state of ADT1 (PDB: 6GCI) where residues K92, K94, K96 and K199 form the gate closed to the matrix, and are inaccessible for cross-linking. Three cross-links are possible and shown between K23-K33, K23-K147, and K33-K147.
(f) Cross-linked peptide pairs (yellow lysine side chains) mapped onto the AF-model with an open channel. All 7 crosslinks in (b) satisfied PIR crosslinker distance constraints.
Quantitative iqPIR analysis revealed increased levels of seven non-redundant cross-linked peptide pairs of ADT1 in TAC (Fig. 4b). It should be noted that other ADT isoforms and multimeric ADT structures may also be present that could explain increases in these observed links (Extended Data Fig. 4a), but ADT1 is the predominant isoform in cardiac and skeletal muscle 42. Cross-links between K23-K92, K23-K94, K23-K96, and K23-K199 are compatible with the C-state 39 of ADT1 because all lysine residues are solvent-exposed in this conformation (Fig. 4b-4c, bovine ADT1 PDB: 1OKC, Extended Data Video 1). However, these links are not compatible with the M-state because links to K92, K94, K96, and K199 must penetrate occluded protein volume in this conformation. These links also exceed Jwalk43 SASD distance allowed with the PIR crosslinker (51Å). Moreover, K96 and K199 form salt bridge interactions with D196 and D292 that contribute to the ADT1 gate closure to the IMS (Fig. 4d, ADT1 PDB: 6GCI, Extended Data Video 1). Although the K23 and K33 cross-link are compatible with distance constraints, it is known that K33 partakes in the hydrogen-bond network needed to stabilize the C-state by forming a salt-bridge with D232 39 (Extended Data Fig. 4b-c). On the other hand, cross-links between K23-K33, K23-K147, and K33-K147 are compatible only with the M-state (Fig. 4d). Thus, increases of all seven links would indicate that both C-state and M-state have increased in TAC; however, this is unlikely given the mutual exclusivity of M- and C-states. Functionally, increased enrichment of the C-state in failing hearts, in which ATP production is known to be impaired, is also counter-intuitive. Collectively, increases in these seven cross-linked peptide pairs and ADT1 protein level measurements in TAC hearts cannot be explained by either a shift between the C-state and M-state of ADT1 in heart failure.
Perhaps a more likely explanation is the existence of an alternative state of ADT1, with which all quantified cross-links are compatible, and this new conformation has increased in TAC. To satisfy all the cross-links in one single state, lysine residues involved in gating at either C- or M-state must be accessible to cross-linkers. This new conformation could be consistent with the P-state, which was recently proposed to be simultaneously open to both the IMS and the matrix, synonymous with non-selective mPTP conductance40, 41. Although there is currently no structural evidence, the non-selective P-state is hypothesized to provide an open channel where the lysine residues responsible for gating are not involved in salt-bridge formation and thus, may be available for cross-linking. To this end, the modified version of Alphafold, ColabFold using shallow multiple sequence alignment44, was applied to generate ADT1_MOUSE structural models with diversity.
On the other hand, it was reported that the conformation of UCP2 protein (resolved NMR structure PDB, 2LCK) closely resembles ADT1 structure and 2LCK has an open channel. Thus, AF-generated models were compared against 2LCK using TM-align38. The model with the highest TM-score was proposed as an alternate conformer of ADT1 with an open channel, as shown in Fig. 4f, which satisfies all 7 crosslinks with SASD distance less than 51 Å (Fig. 4b) (Supplementary Data 5). Therefore, our results provide structural evidence that heart failure is associated with increased conformational enrichment of ADT1 that is non-functional for ADP/ATP translocation but likely possesses non-selective conductivity 45.
Discussion
Using iqPIR technology as a discovery-based approach, conformational and interaction changes in mitochondrial proteins and complexes were observed that possess direct functional relevance to metabolic changes in heart failure. In the present study, the identification of SCOT1 oligomer metabolons, increased active conformational states, and higher-order assembly of SCOT1 is corroborated by previously reported metabolic changes in failing hearts. Therefore, this agreement affirms the utility of quantitative in vivo cross-linking and mass spectrometry to gain greater molecular-level insight into changes in heart failure. Importantly, changes in ketone metabolism have been thus far attributed to substrate and enzyme abundance in heart failure with no information on regulatory mechanisms 31. Our results provide insight into metabolic remodeling, suggesting that active conformations exist beyond mere increases in protein levels.
The first direct evidence for the NDUA4 existence as a CIV subunit within cardiac tissue was previously provided by PIR technology 14. The present iqPIR data confirms that interaction and enables the first quantitation of the NDUA-CX6B1 interaction in failing and control heart tissues with five non-redundant Lys-Lys residue pairs between NDUA4 and CX6B1. The decrease in NDUA4-CX6B1 interactions and the concomitant decrease in CIV activity in heart failure samples suggest that this interaction could potentially modulate CIV activity. NDUA4 is thought to maintain CIV monomeric population by occupying the CIV dimerization interface. Decreased NDUA4-CX6B1 cross-links, coincident with reduced CIV activity in TAC, support the hypothesis that monomer and respirasome populations are functionally active populations23. It remains unclear whether destabilization of NDUA4 during TAC results in consequential alterations of CIV dimeric pools and what role this population may have in disease pathology 46. Nonetheless, the present observation that NDUA4-CX6B1 interactions decrease in heart failure presents a previously unrecognized target for future studies and possible therapies that stabilize this interaction as a treatment to restore or prevent CIV functional decline in heart failure.
Mitochondrial ADP/ATP carrier (ADT) has long been proposed to contribute a pore-forming component of the mitochondrial permeability transition pore (mPTP) 41, 47. However, several decades of genetics and physiological studies have yielded ambiguous evidence. Transient opening of mPTP has physiological functions such as regulating mitochondrial calcium homeostasis or reactive oxygen species signaling. In contrast, prolonged mPTP contributes to cell death under stress conditions, including heart failure 42. Our results provide critical structural information consistent with the recently proposed P-state of ADT, which opens to both matrix and inter-membrane space 42. It remains unclear whether the P-state is static or results from rapid interconversion between two incompletely gated structures.
Nonetheless, an increased presence of such a state compromises nucleotide translocation function and increases mitochondrial proton leaks leading to impaired energetics. In support of this notion, a mitochondrial-targeted tetrapeptide, elamipretide (SS-31), is shown to bind K23 of ADT1 and reduces proton leak in mitochondria of aging animals 48. Thus, changes in the conformational state of ADT warrant further investigation both as a disease mechanism and a therapeutic target.
In summary, these findings advance mitochondrial and heart failure research in several ways. First, the data elucidate structural information on key players in mitochondrial function, providing a basis for developing future therapies. Second, the results enable the generation of novel hypotheses for mechanisms of mitochondrial maladaptation in disease. The quantitative interactome dataset provides a valuable resource for exploring and visualizing changes in many other proteins not discussed here that can further advance improved understanding of mitochondrial function and heart failure pathology.
METHODS
Animal Model
All protocols concerning animal use were approved by the Institutional Animal Care and Use Committee at the University of Washington (Protocol 4214–01). This study utilized 24 wild-type mice, strain C67BL6/NCrl (IMSR_CRL:27). Female and male mice have different responses to pressure overload. TAC surgery causes a mild and variable phenotype in females. Thus, only male mice were used in this study. Adult (10–12 weeks old, weighing 24–26g) male mice were chosen randomly into experimental groups. Mice were housed in a vivarium with a 12 hr light/dark cycle at 25°C. Mice were maintained on ad libitum standard rodent diet and water.
Transverse aortic constriction (TAC) surgery
Male mice (n=12 animals per group) aged 10–12 weeks, weighing 24–28g, underwent TAC or Sham surgery 1. Mice were anesthetized with 4% isoflurane and intubated with a 20-gauge cannula. Mice were ventilated at 2.5% isoflurane at 135 breaths per minute by a small animal TOPO ventilator (Kent Scientific, Torrington, CT). The aortic arch was exposed and separated from the thymus via left thoracotomy. A 27-gauge blunt needle was held near the aorta (between the brachiocephalic and left common carotid arteries). A constriction of the transverse aorta was generated by tying a 6–0 Ethilon ligature against the blunt needle. The needle was then promptly removed. The lungs were inflated, and the chest was closed by a 5–0 polypropylene suture. The animal was removed from the ventilation system and given SR buprenorphine (subcutaneous, 0.5mg/kg) analgesic and 0.9% saline (intraperitoneal, 0.2ml) for hydration after the mice regained consciousness (2 hrs after ventilation). Sham-operated mice underwent all the same procedures as TAC, excluding ligature of the aorta. Mice were randomly assigned to TAC and Sham procedures, and the researcher was blind to operation until after echocardiogram analysis was performed. Combined mortality (acute and chronic) was less than 25%.
Transthoracic echocardiography
Mice from TAC and Sham groups were anesthetized and maintained with 0.8%−2% isofluorane in 95% oxygen at 550–600 beats per minute 2. Trans-thoracic echocardiography was conducted 4 weeks post-TAC and Sham surgery with Vevo 2100 high-frequency, high-resolution digital imaging system (VisualSonics) with an MS400 Microscan Transducer. A parasternal long-axis view was used to collect M-mode images to analyze fractional shortening, ejection fraction, and other functional parameters.
Cross-linking of cubed heart tissue
Mice from TAC and Sham groups were euthanized by cervical dislocation. The chest cavity was opened, and the heart was rapidly excised and placed into an ice-cold mitochondria isolation medium (MIM: 70mM sucrose, 220mM mannitol, 5mM MOPS, 1.6mM carnitine hydrochloride, 1mM EDTA, 0.025% fatty acid-free BSA, pH 7.4 with 5M KOH) to flesh out blood. The aorta and atria were removed. The myocardium was weighed (heart weight). The heart was transferred to a pre-chilled 60mm dry petri dish and maintained on ice. Heart tissue was finely minced with a razor blade to a homogenous tissue size distribution of approximately 1mm2 cubes.
Heart tissue was centrifuged at 1500 x g for 5 min at 4°C, and MIM was replaced with cross-linking buffer (170 mM Na2HPO4, pH 8). iqPIR 3 is a novel technology recently developed for quantitation of cross-linked peptide pairs using isobaric stable isotopes selectively incorporated into the PIR cross-linker, which enables MS2-based interactome quantitation in a way analogous to how TMT 4 or iTRAQ 5 is used for proteome quantitation. Briefly, reporter heavy (RH) or stump heavy (SH) iqPIR cross-linkers were applied to different samples individually first, then TAC/Sham cross-linked sample pairs were pooled together for further downstream sample processing and data acquisition. This strategy greatly decreases sample handling errors and reduces instrument time for data acquisition. Identical cross-linked peptide pairs from TAC and sham samples have identical masses and LC retention times yet produce different and distinct quantitative isotope signatures in MS2 spectra. The intensities of TAC- or Sham-distinct isotope-labeled peptide and fragment ions in each MS2 spectrum enable relative quantitation of cross-linked peptides in TAC and Sham samples. The samples were mixed for 30 min at room temperature on a Thermomixer at 800 rpm. The tissue was centrifuged at 1500 x g for 5 min at 4°C, and the supernatant was removed. Cross-linked tissue was then further subjected to modified mitochondrial isolation as described below.
Mitochondrial isolation from cross-linked heart tissue
Cross-linked heart tissue was washed in fresh MIM and centrifuged at 1500 x g for 5 min at 4°C. Subsequent steps are the same as the regular mitochondrial isolation protocol below. However, all resulting fractions and supernatants were collected and frozen at −80°C until processing.
Protein extraction from cross-linked mitochondria
Frozen fractions were transferred to a stainless-steel cryogrinding jar cooled to −196°C with liquid nitrogen in 0.1M NH4HCO3. The samples were cryoground for five 3 min cycles at 30 Hz using a Retch MM 400 mixer mill. Between cycles, the cryogrinder was cooled with liquid nitrogen. The resulting frozen powder was transferred to a falcon tube where 8M urea (in 0.1M Tris, pH 8.0) was added. Samples were sonicated using a GE-130 ultrasonic processor, followed by reduction of cysteine residues by incubation with 5mM Tris (2-carboxyethyl) phosphine (TCEP, Fisher Scientific) for 30 min, followed by alkylation with 45 min incubation with 10mM iodoacetamide (Fisher Scientific). Urea concentration was reduced to less than 1M by diluting the samples by a factor of 10 with fresh 0.1M Tris buffer (pH 8.0). The protein concentration was measured using the Pierce Coomassie protein assay (Thermo Scientific).
Protein digestion and enrichment of cross-linked peptides
The procedures for protein digestion, desalting, and enrichment of cross-linked peptides by strong cation exchange chromatography (SCX) and biotin-capture were followed as described in a previous publication 6. Briefly, extracted proteins were digested with trypsin (200:1 in weight ratio) overnight at 37°C and then quenched with acidification of digest to pH ∼3 by adding TFA. The digest was then desalted using Sep-Pak cartridges and fractionated on SCX using a flow rate of 1.5 mL/min and 97.5-min gradient with an increasing percentage of SCX solvent B (solvent A: 7 mM KH2PO4, pH 2.6, 30% (vol/vol) acetonitrile and solvent B: 7 mM KH2PO4, pH 2.6, 350 mM KCl, 30% (vol/vol) acetonitrile). The SCX fractions were adjusted to pH 8.0 and then subjected to biotin capture by adding monomeric avidin UltraLink resin (Thermo Scientific) to each pooled fraction. After 3 times of washing the avidin beads with 0.1 M NH4HCO3, pH 8.0, the cross-linked peptides were eluted with 70% (vol/vol) acetonitrile, 1% (vol/vol) formic acid (FA), and then dried in a speed vacuum. The dried samples were reconstituted in 0.1% FA and ready for LC-MS/MS analysis.
LC-MS/MS
Cross-linked peptide samples were analyzed in technical duplicate by liquid chromatography mass spectrometry using an Easy-nLC (Thermo Scientific) coupled to a Q Exactive Plus mass spectrometer (Thermo Scientific). Each analysis used a 3 μL injection of sample onto a 3 cm x 100 μm inner diameter fused silica trap column packed with a stationary phase consisting of 5 μm Reprosil C8 particles with 120 Å pores (Dr. Maisch GmbH) with a flow rate of 2 μL/ min of mobile phase consisting of solvent A (H2O containing 0.1% formic acid) for 10 minutes. Peptides were then fractionated over a 60 cm x 75 μm inner diameter fused silica analytical column packed with 5 μm Reprosil C8 particles with 120 Å pores by applying a linear gradient from 95% solvent A, 5% solvent B (acetonitrile containing 0.1% formic acid) to 60% solvent A, 40% solvent B over either 120 or 240 minutes at a flow rate of 300 nL/min. Eluting peptide ions were ionized by electrospray ionization by applying a positive 2.2 kV potential to a laser pulled spray tip at the end of the analytical column. The mass spectrometer was operated using a top five data-dependent acquisition method with a resolving power setting of 70,000 for MS1 and MS2 scans. Additional settings include an AGC target value of 1e6 with a maximum ion time of 100 ms for the MS1 scans and an AGC value of 5e4 with a maximum ion time of 300 ms for the MS2 scans. Charge state exclusion parameters were set to only allow ions with charge states from 4+ to 7+ to be selected for MS2. Ions selected for MS2 were isolated with a 3 m/z window and fragmented by HCD using a normalized collision energy setting of 30. Ions for which MS2 was performed were then dynamically excluded from further selection for MS2 for 30 s.
Heat Map Analysis
Perseus (version 1.6.15.0) 7 was used for statistical data analysis of cross-link quantitation ratios. Data was first filtered based on 70% of total valid values across all biological replicates and then applied with a T-test using Benjamini-Hochberg FDR at 0.05. T-test q-values less than 0.05 were considered significant. Heatmaps were generated using NG-CHM GUI software (version 2.20.0) 8 with Euclidean distance and complete linkage selected for both row and column hierarchical clustering.
Mitochondrial Isolation from cardiac tissue
Hearts were excised from mice, and the aortas and atria were removed. Heart tissues were rinsed briefly in ice-cold mitochondria isolation medium (70mM sucrose, 220mM mannitol, 5mM MOPS, 1.6mM carnitine hydrochloride, 1mM EDTA, 0.025% fatty acid-free BSA, pH 7.4 with 5M KOH) with the addition of 2 mM taurine, to remove residual blood. Tissues were minced on ice and resuspended in fresh MIM, followed by trypsin digestion (10μg/ml) and incubated on ice for 10 min. Trypsin digestion was stopped by adding trypsin inhibitor (0.5mg/ml) and additional BSA (1 mg/ml) to MIM. The suspension was centrifuged for 1 min at 1,500 x g at 4°C, and the supernatant was discarded. The tissue pellets were resuspended in fresh MIM containing 1mg/ml BSA, transferred to a Teflon-glass tube, and homogenized on ice with a Teflon pestle. The homogenates were centrifuged for 10 min at 800 x g at 4°C. The supernatants were collected and centrifuged for 10 min at 8,000 x g at 4°C. The supernatant was discarded, and the mitochondrial pellets were resuspended in MIM to wash. The resuspension was centrifuged for 10 min at 8,000 x g at 4°C, and the supernatant was discarded. The mitochondrial pellet was resuspended to a concentrated volume for protein quantification by BCA Assay (Pierce).
Mitochondrial respiration using Seahorse XFe24 Analyzer
The oxygen consumption rate (OCR) of isolated cardiac mitochondria was measured with an XF24e Flux Analyzer (Seahorse Bioscience, Agilent) 9. The Seahorse utility plate was hydrated with calibration buffer and incubated overnight at 37°C prior to respiration experiments. The mitochondria were seeded in 24-well microplates at 1–2 μg protein per well in 1x mitochondrial assay solution (70mM sucrose, 220mM mannitol, 10mM KH2PO4, 5mM MgCl2, 2mM HEPES, 1mM EGTA, 0.2% BSA, pH 7.35 at room temperature) containing substrates and centrifuged for 2,000 x g for 20 min at 4°C. Respiration buffer was added to each well to a final volume of 500ul; then, the plate was placed in a non-CO2 incubator for 10 min at 37°C. The plate was incubated in the Seahorse Analyzer, and a baseline oxygen consumption rate (OCR) was measured. Sequential injections of inhibitors (final concentration 4mM ADP, 2.5 μg/mL oligomycin, 4uM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone, and 4uM antimycin A) were added into each well and changes in OCR were measured. For Complex I-supported respiration, 10mM pyruvate and 2mM malate were given to mitochondria seeded at 2 μg/well.
Mitochondrial Respiration using β-hydroxybutyrate
Isolated mitochondria respiration experiments were performed in a Clark-type oxygen electrode Oxygraph System (Hansatech Instruments) at 30°C. Briefly, isolated mitochondria were added to respiration buffer (125mM KCl, 20mM HEPES, 3mM MgCl2, 400µM EGTA, 5mM KH2PO4, 0.2% BSA (fatty acid-free), pH 7.2) and placed in the respiration chamber to measure baseline respiration (State 1). The change in oxygen levels following the immediate addition of 10mM β-hydroxybutyrate and 2mM malate (State 2) followed by the sequential addition of 2.5 mM ADP (State 3), 1mg/ml Oligomycin (State 4μ), and 1µM FCCP (FCCP Max). Antimycin A (2.5µM) was added last to measure the residual oxygen consumption not accounted for by the electron transfer chain, and it was subtracted from the previous measurements.
BDH Enzyme Activity
BDH1 activity was determined in isolated mitochondria homogenates using an adapted method 10. Mitochondrial homogenates were preincubated in assay buffer (50mM Tris-HCl (pH 8.0), 2.0 mM NAD+, 0.3 μg of rotenone, 0.4mg of BSA) for 10 min at 30°C. The reaction was initiated by the 50ul addition of 0.2M β-hydroxybutyrate. The rate of NAD+ reduction was calculated from the increase in 340nm for 2 minutes. Enzyme activity is expressed as nmol of NAD+ reduced/min/mg of protein.
SCOT1 Enzyme Activity
SCOT activity was determined in heart tissue homogenates at 37°C using an adapted protocol 11, 12. Briefly, heart tissue was homogenized with phosphate buffer saline (pH 7.2) containing protease inhibitors, followed by centrifugation at 20,000 x g at 4C for 20 minutes. The assay buffer contained 50mmol/L Tris HCl, pH 8.5, 10mmol/L MgCl2, 4mmol/L iodoacetamide, 1mmol/L succinyl-CoA, 10mmol/L lithium acetoacetate, and 50μg of the heart lysate. SCOT activity was measured by following the formation of acetoacetyl-CoA at 313nm for 2 minutes.
Structural modeling
Sequences of BDH_MOUSE and SCOT1_MOUSE were subjected to structural modeling using AlphaFold v2.0 13. TM-align software tool 14 was applied to align the AF-generated SCOT1_MOUSE structure with two adjacent subunits (Chain D and H) of the octameric structure of SCOT1 (PDB: 3OXO). Modeling of ADT1 conformers was performed using Alphafold v2.0 13, with a shallow multiple sequence alignment1 (MSA) using a modified version (https://github.com/delalamo/af2_conformations) of ColabFold 15. Conformers were modeled using the ADT1 mouse sequence (Uniprot: P48962) as input without templates and setting max_extra_msa = 16 and max_msa_clusters = 8 to perform modeling using MSAs with 16 sequences. A total of 50 models were generated from 50 random seeds. These models were then compared against the structure of the mitochondrial uncoupling protein 2 (UCP2, PDB:2LCK 16) using TM-align 14. UCP2 was selected for comparison due to its structural similarity to ADT1 while having an open channel 16. The model with the highest TM-score was imported into XLinkDB for further analysis. Cross-links were mapped to structures based on sequence homology 17. In the case of structure 2LCK, conservation of amino acids in the structure to which cross-linked residues were mapped were not required to be conserved with lysine.
Cytochrome C Oxidase and Citrate Synthase activity from heart tissue homogenate
Citrate synthase (CS) and Cytochrome C Oxidase (COX) activities were determined in heart tissue homogenates at 30°C using an adapted protocol 18. Briefly, frozen cardiac tissue (25mg) for CS activity was homogenized at 4°C in CelLyticTM MT Cell Lysis Reagent (C3228) and then incubated on ice for 30 minutes. The samples were then centrifuged at 10,000 x g for 10 minutes at 4°C, and protein concentration was determined. CS activity was analyzed in 100mM Tris buffer containing 0.1% Triton X-100, pH 8.0 18.
For COX activity, 25mg of cardiac tissue was homogenized at 4°C in 50 mM potassium phosphate buffer containing 1 mM EDTA, and 0.1%Triton X-100 (pH 7.4; final concentration, 5 mg tissue/mL). The samples were incubated on ice for 30 minutes, followed by centrifugation at 10,000 x g for 10 minutes at 4°C. COX activity was analyzed in 100mM potassium phosphate buffer 18.
Blue Native polyacrylamide gel electrophoresis (BN-PAGE) of isolated cardiac mitochondria
Frozen isolated mitochondrial pellet was thawed and gently resuspended in permeabilization medium (1x NativePage Sample Buffer, 1% digitonin or 1% n-Dodecyl β-D-maltoside (DDM), and distilled water) and incubated on ice for 20 min. Samples were centrifuged at 20,000 x g for 20 min at 4°C. The supernatants were collected and mixed with Coomassie G-250 additive. Samples were loaded into pre-cast Native gradient gels (4%−16% Bis-Tris protein gel). Protein was run at 150V for 30 mins in dark blue cathode buffer (2X NativePAGE Dark blue cathode running buffer). Running buffer was replaced with light blue running buffer (0.2X NativePAGE light blue cathode running buffer) and run at 200V for 1.5 hrs. Gels were washed in ice cold water immediately after electrophoresis. Gels were stained with 0.25% Coomassie Brilliant Blue R-250 for 1 hr with constant agitation. Gels were destained (40% methanol, 10% acetic acid) for 1 hr followed by washing for one hour with water. For Complex IV staining, gels were incubated in freshly prepared in-gel activity substrate (for 100ml of substrate 50 mg Diaminobenzidine (DAB), 100 mg Cytochrome C, 90ml 50 mM phosphate buffer pH 7.4 to 10ml water) for 1 hr at 37°C. For Complex I staining, gels were incubated in freshly prepared in-gel activity substrate (2mM Tris-HCl pH 7.4., 0.1mg/ml NADH, 2.5mg/ml Nitrotetrazolium Blue chloride) for 1 hour at 37°C.
For immunoblotting, Native gels were incubated in transfer buffer (20% methanol, 10% Tris-glycine) supplemented with 2% SDS for 15 mins. Protein was transferred to PVDF membrane in transfer buffer at 120V for 1 hour at 4°C. After transfer, membranes were immediately washed in methanol for three times for 5 minutes. Membranes were then washed three times in water for 5 minutes then incubated in blocking buffer (5% BSA in Tris-buffered saline with 1% Tween 20) for 1 hr. Membranes were incubated in primary antibody (1:1000 in blocking buffer, anti-Ndua4 #ab129752) overnight at 4°C. Membranes were washed three times with TBST to remove primary antibody. Secondary antibodies raised against primary species (1:5,000 goat anti-rabbit #HAF008) were diluted in TBST and membranes were incubated for 2 hr at room temperature. Membranes were washed four times with TBST. HRP substrate was added to membranes and protein bands were detected with film.
Discrepancies in quantitation of cross-linked residue pairs from fully-cleaved and missed-cleaved tryptic peptide pairs
It is not unusual that the same crosslinked residue pair is detected multiple times from different forms of non-redundant crosslinked peptide pairs, such as peptides containing missed trypsin cleavage sites and modified residues (e.g., oxidized methionine), as observed in Supplementary Data 6. The consistent quantitation over multiple distinct peptide pairs containing the same residue pair is confirmatory for the quantitation of the cross-linked residue pair. It was recommended to consolidate the quantitation of various forms of crosslinked peptide pairs into a single data point for the same residue pair using the median value of all its supporting peptide pairs 19, 20. With iqPIR quantitation, the final Log2 ratio of the cross-linked residue pair was generated using the mean value of all contributing ions containing the quantitative iqPIR stumps from all distinct peptide pairs. A total of 2717 crosslinked residue pairs were quantified in at least one of the six pairs of biological replicates.
Further filtering of the data with maximum 2 missing values across 6 biological replicates reduced the number to 1292 residue pairs, of which 454 residue pairs were quantified with 2 or more non-redundant peptide pairs and 451 residue pairs have mean Log2 ratios with 95% confidence level < 0.5, indicating highly consistent quantitation across biological replicates and multiple distinct peptide pairs (Supplementary Data 6). A total number of 108 residue pairs (filtered with a maximum of 2 missing Log2 values across 6 biological replicates) were generated from both fully-cleaved tryptic peptide pairs, and corresponding missed-cleaved peptide pairs were evaluated with an ANOVA test to analyze the Log2 ratio difference between the two groups. 92 (85%) residue pairs showed consistent quantitation, while 16 residue pairs had a significant deviation between the fully-cleaved peptide pairs and missed-cleaved peptide pairs (Supplementary Data 7). Interestingly, 14 cross-linked peptide pairs with missed cleavage significantly decreased across multiple TAC/sham heart comparisons, and 10 pairs showed over a 2-fold decrease in TAC samples (Supplementary Data 7). For comparison, the ANOVA test was also applied to a previously published iqPIR dataset which used HEK293 cells 3. Among 102 residue pairs generated from fully-cleaved and corresponding missed-cleaved peptide pairs, only 2 residue pairs showed significant quantitation difference between full- and missed-cleavage peptide pairs, and the residue pairs from missed-cleavage peptide pairs were not decreased significantly. The consistent decreases observed in missed-cleaved peptide pairs in TAC samples indicate systematic changes in TAC-induced processes. The underlying cause is unknown; however, one hypothesis could be that the preceding missed cleavage lysine or arginine sites were somehow modified more in TAC samples, leading to fewer missed cleavage peptides in TAC. Several groups have reported that mitochondrial protein acetylation levels increased in heart failure, and these modified residues could prevent tryptic cleavage in TAC samples, leading to reduced crosslinked peptide pairs with missed cleavage sites.
QUANTIFICATION AND STATISTICAL ANALYSIS
Assessment of cardiac and mitochondrial function
The number of independent experiments are specified in the relevant figure legends. Data are expressed as the mean ± standard error of the mean (SEM). Statistical comparisons between two groups were conducted by unpaired, two-tailed Student’s t-test. P<0.05 was considered to be statistically significant. Statistical analysis was performed with Prism 9.0 software (GraphPad).
LC-MS/MS data analysis
The LC-MS/MS results were analyzed to identify and quantify iqPIR cross-linked peptide pairs 3. LC-MS/MS data raw files were converted to .mzXML format using the ReADW tool in the Trans Proteomic Pipeline software suite 21. Mango 22 was used to search for PIR mass relationships, and Comet 23 was used to search the mzXML files against the mouse Mitocarta 2 database 24 consisted of both forward and reverse protein sequences (2,084 total sequences). The resulting pepXML files were analyzed with XLinkProphet 25 and filtered for an estimated 1% FDR at the non-redundant cross-link level. For dead-end peptides, PeptideProphet 26 was applied to filter Comet search results at < 1% FDR. All cross-links passing the threshold were used for quantitation. Light (SH) and heavy (RH) isotope peptide precursors and their fragment peaks containing the stump group were deconvoluted, and their peak intensities were extracted to calculate Log2 ratios. The final ratio for each cross-linked peptide pair is the normalized mean value from all contributing ion ratios to the same cross-link from different charge states, different scans, and separate replicate runs with outliers removed, where normalization is achieved by subtracting from each cross-link Log2 ratio the median value of all cross-link Log2 ratios. Finally, the cross-link Log2 ratio p-value reflecting its likelihood of being 0 is calculated based on a statistical t-test using the Log2 ratio mean value, standard deviation, and a number of contributing quantified ions. Protein quantitation was estimated by combining normalized Log2 ratios of all intra-protein cross-links and dead-end peptides corresponding to the protein. The mean Log2 ratio of all contributing ratios (intra-protein cross-link and dead-end) was used after excluding outliers.
Data availability
Cross-linking data has been deposited at XlinkDB (http://xlinkdb.gs.washington.edu/xlinkdb/index.php) and is publicly available (Network Name: Caudal_iqPIR_TACsham_Bruce). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE 49 partner repository with the dataset identifiers: PXD027757 and PXD035622. The following publicly available files were included: PDB 5Z62, PDB 3DLX, PDB 3OXO, PDB 1OKC, PDB 6GCI, PDB 2LCK, and Uniprot P48962. Any additional information required to reanalyze the data reported in this paper is available from the lead contacts upon request. All other data supporting the findings in this study are included in the main article and associated files.
Software availability statement
This work did not employ any custom, or previously unreported, computer code or algorithms.
Extended Data
Extended Data Fig. 1.
(a-b) Left ventricle internal dimension and wall thickness in TAC and Sham groups determined by echocardiography four weeks post-surgery.
(c-d) Lung and liver edema (wet weight/dry weight in mg) measured at tissue harvest.
(e) Quantitation of mean cross-link (XL) ratio vs mean dead-end (DE) ratio for each cross-linked peptide pair statistically changed in TAC in at least 4/6 biological pairs (Log2 TAC/Sham). The sum of the Mean DE ratio for Protein A and Protein B is shown to account for cross-links between two different proteins.
(f) Interaction network of lysine residues (black nodes) connected by observed cross-links (edges). Edges are colored according to increasing (red) and decreasing (blue) quantitation of statistically significant subset of cross-links (Log2 TAC/Sham) shown by color scale.
For (a-d), all data are n=12 animals, AVG+/−SEM, *denotes p<0.05 by unpaired, two-tailed Student’s t-test.
Extended Data Fig. 2.
(a) Structural insight into CX6B1 R20 forming salt-bridges at NDUA4-CX6B1 interface. R20 side chain (green) forms a salt bridge with CX6B1 D16 (green) and NDUA4 D60 (black), which pinpoints an interface necessary for the stability of CIV. Side-chains of R20, D16, and NDUA4 D60 (partially resolved) are depicted in stick representation. Cross-linked lysine sidechains are shown as yellow (CX6B1) or orange (NDUA4) spheres.
(b) Cytochrome C oxidase enzymatic activity assay in tissue homogenates from TAC and Sham hearts, normalized to Citrate Synthase activity. N=4 animals, AVG+/−SEM, *denotes p<0.05 by unpaired, two-tailed Student’s t-test.
(c) Representative Blue Native-PAGE (BN-PAGE) analysis of mitochondria isolated from Sham and TAC groups. Coomassie stain (left) for total protein loading, In-gel CIV activity stain (middle), with CI activity stain overlay (right). Gels were run in duplicates.
(d) Representative BN-PAGE immunoblot of NDUA4 containing SCs from digitonin solubilized isolated mitochondria. Blots were run in duplicates.
(e) Representative BN-PAGE immunoblot of NDUA4 containing SCs from DDM solubilized isolated mitochondria. Blots were run in duplicates.
Extended Data Fig. 3.
(a) AlphaFold-predicted BDH1_MOUSE structure. Crosslinked residues were indicated in yellow spheres, and crosslinks shown in red lines mean they were increased in TAC samples.
(b) The pLDDT plot of the predicted BDH_mouse structure.
(c) Alignment of apo (grey, PDB: 3OXO chain A) and substrate-bound (yellow, PDB: 3OXO chain E) monomers of porcine SCOT1. CoA is colored in magenta and bound to the active site. Lysine sidechains are shown in stick representation. Alignment depicts the structural differences between the dynamic C-terminal domain and the static N-terminal domain during substrate-binding. A close-up view specifies cross-linked lysines (K418 and K421).
(d) Ketone-driven oxygen consumption rate (OCR) of mitochondria isolated from TAC and Sham hearts. Baseline OCR (State 1) was measured, followed by sequential injections of β-hydroxybutyrate/malate (State 2), ADP (State 3), Oligomycin (State 4μ), and FCCP (FCCPmax). N=6 animals, AVG+/−SEM, *denotes p<0.05 by unpaired, two-tailed Student’s t-test.
Extended Data Video 1.
Supplementary Material
Acknowledgments
We thank all Tian and Bruce lab members for their thoughtful discussion and support. We thank the University of Washington Proteomics Resource for advice and helpful discussions. We thank Dr. Yun-Wei A. Hsu for assistance with the animal models. We thank Dr. Julia Ritterhoff and Dr. Frauke Drees for their technical support and guidance.
Sources of funding
This work was supported in part by U.S. National Institutes of Health (NIH) grants HL110349, HL129510, HL142628 (to R.T.), HL144778, GM097112, GM086688, R35GM136255 (to J.E.B), American Heart Association (AHA) Predoctoral Fellowship 20PRE35120126 (to A.C.), AHA Postdoctoral Fellowship 18POST33990352 (to B.Z.), China Scholarship Council Fellowship 202006320416 (to H.C.), NIH 2T32DK007247–41 and AHA Career Development Award 930223 to (M.A.W).
Footnotes
Competing Interests Statement The authors declare no competing interests.
References
- 1.Rath S et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Research 49, D1541–D1547 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pagliarini DJ et al. A Mitochondrial Protein Compendium Elucidates Complex I Disease Biology. Cell 134, 112–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qi L et al. Cryo-EM structure of the human mitochondrial translocase TIM22 complex. Cell Research 31, 369–372 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridges HR et al. Structure of inhibitor-bound mammalian complex I. Nature Communications 11, 5261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu J et al. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science 364, 1068 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Spikes TE, Montgomery MG & Walker JE. Structure of the dimeric ATP synthase from bovine mitochondria. Proceedings of the National Academy of Sciences 117, 23519 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker K & Park E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nature Structural & Molecular Biology 26, 1158–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinz A. Crosslinking Mass Spectrometry Goes In-Tissue. Cell Systems 6, 10–12 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Tang X, Munske GR, Siems WF & Bruce JE. Mass Spectrometry Identifiable Cross-Linking Strategy for Studying Protein−Protein Interactions. Analytical Chemistry 77, 311–318 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Schweppe DK et al. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proceedings of the National Academy of Sciences 114, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavez JD et al. Cross-linking measurements of the Potato leafroll virus reveal protein interaction topologies required for virion stability, aphid transmission, and virus-plant interactions. Journal of proteome research 11, 2968–2981 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisbrod CR et al. In Vivo Protein Interaction Network Identified with a Novel Real-Time Cross-Linked Peptide Identification Strategy. Journal of Proteome Research 12, 1569–1579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chavez, Juan D, Schweppe, Devin K, Eng, Jimmy & Bruce, James E. In Vivo Conformational Dynamics of Hsp90 and Its Interactors. Cell Chemical Biology 23, 716–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez JD et al. Chemical Crosslinking Mass Spectrometry Analysis of Protein Conformations and Supercomplexes in Heart Tissue. Cell Systems 6, 136–141.e135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong S-E et al. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics*. Molecular & Cellular Proteomics 1, 376–386 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Chavez JD et al. Quantitative interactome analysis reveals a chemoresistant edgotype. 6, 7928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chavez JD, Keller A, Zhou B, Tian R & Bruce JE. Cellular Interactome Dynamics during Paclitaxel Treatment. Cell Reports 29, 2371–2383.e2375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chavez JD, Keller A, Mohr JP & Bruce JE. Isobaric Quantitative Protein Interaction Reporter Technology for Comparative Interactome Studies. Analytical Chemistry 92, 14094–14102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown DA et al. Mitochondrial function as a therapeutic target in heart failure. Nature Reviews Cardiology 14, 238–250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barth E, Stämmler G, Speiser B & Schaper J. Ultrastructural quantitation of mitochondria and myofilaments in cardiac muscle from 10 different animal species including man. Journal of Molecular and Cellular Cardiology 24, 669–681 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Schweppe DK et al. XLinkDB 2.0: integrated, large-scale structural analysis of protein crosslinking data. Bioinformatics 32, 2716–2718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broom BM et al. A Galaxy Implementation of Next-Generation Clustered Heatmaps for Interactive Exploration of Molecular Profiling Data. Cancer Research 77, e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zong S et al. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Research 28, 1026–1034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balsa E et al. NDUFA4 Is a Subunit of Complex IV of the Mammalian Electron Transport Chain. Cell Metabolism 16, 378–386 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Massa V et al. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. American journal of human genetics 82, 1281–1289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdulhag UN et al. Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. European Journal of Human Genetics 23, 159–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosca MG et al. Cardiac mitochondria in heart failure: decrease in respirasomes and oxidative phosphorylation. Cardiovascular Research 80, 30–39 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murashige D et al. Comprehensive quantification of fuel use by the failing and nonfailing human heart. Science 370, 364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aubert G et al. The Failing Heart Relies on Ketone Bodies as a Fuel. Circulation 133, 698–705 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedi KC et al. Evidence for Intramyocardial Disruption of Lipid Metabolism and Increased Myocardial Ketone Utilization in Advanced Human Heart Failure. Circulation 133, 706–716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolwicz SC, Airhart S & Tian R. Ketones Step to the Plate. Circulation 133, 689–691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchihashi M et al. Cardiac-Specific Bdh1 Overexpression Ameliorates Oxidative Stress and Cardiac Remodeling in Pressure Overload-Induced Heart Failure. Circ Heart Fail 10 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tammam SD, Rochet J-C & Fraser ME. Identification of the Cysteine Residue Exposed by the Conformational Change in Pig Heart Succinyl-CoA:3-Ketoacid Coenzyme A Transferase on Binding Coenzyme A. Biochemistry 46, 10852–10863 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Fraser ME, Hayakawa K & Brown WD. Catalytic Role of the Conformational Change in Succinyl-CoA:3-Oxoacid CoA Transferase on Binding CoA. Biochemistry 49, 10319–10328 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Coker S-F et al. The high-resolution structure of pig heart succinyl-CoA:3-oxoacid coenzyme A transferase. Acta Crystallographica Section D 66, 797–805 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Shafqat N et al. A structural mapping of mutations causing succinyl-CoA:3-ketoacid CoA transferase (SCOT) deficiency. Journal of inherited metabolic disease 36, 983–987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y & Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic acids research 33, 2302–2309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pebay-Peyroula E et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Ruprecht JJ et al. The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier. Cell 176, 435–447.e415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karch J et al. Inhibition of mitochondrial permeability transition by deletion of the ANT family and CypD. Science Advances 5, eaaw4597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bround MJ, Bers DM & Molkentin JDA 20/20 view of ANT function in mitochondrial biology and necrotic cell death. Journal of Molecular and Cellular Cardiology 144, A3–A13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bullock JMA, Thalassinos K & Topf M. Jwalk and MNXL web server: model validation using restraints from crosslinking mass spectrometry. Bioinformatics 34, 3584–3585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirdita M et al. ColabFold - Making protein folding accessible to all. bioRxiv, 2021.2008.2015.456425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertholet AM et al. H+ transport is an integral function of the mitochondrial ADP/ATP carrier. Nature 571, 515–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramzan R, Rhiel A, Weber P, Kadenbach B & Vogt S. Reversible dimerization of cytochrome c oxidase regulates mitochondrial respiration. Mitochondrion 49, 149–155 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Kokoszka JE et al. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature 427, 461–465 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chavez JD et al. Mitochondrial protein interaction landscape of SS-31. Proceedings of the National Academy of Sciences 117, 15363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Riverol Y et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research 47, D442–D450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
References (Methods)
- 1.Tarnavski O et al. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiological Genomics 16, 349–360 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Ritterhoff J et al. Metabolic Remodeling Promotes Cardiac Hypertrophy by Directing Glucose to Aspartate Biosynthesis. Circulation Research 126, 182–196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chavez JD, Keller A, Mohr JP & Bruce JE. Isobaric Quantitative Protein Interaction Reporter Technology for Comparative Interactome Studies. Analytical Chemistry 92, 14094–14102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson A et al. Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Analytical Chemistry 75, 1895–1904 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Ross PL et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. [DOI] [PubMed] [Google Scholar]
- 6.Chavez JD et al. Systems structural biology measurements by in vivo cross-linking with mass spectrometry. Nature Protocols 14, 2318–2343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyanova S et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Broom BM et al. A Galaxy Implementation of Next-Generation Clustered Heatmaps for Interactive Exploration of Molecular Profiling Data. Cancer Research 77, e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers GW et al. High Throughput Microplate Respiratory Measurements Using Minimal Quantities Of Isolated Mitochondria. PLOS ONE 6, e21746 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grinblat L, Pacheco Bolaños LF & Stoppani AO. Decreased rate of ketone-body oxidation and decreased activity of d-3-hydroxybutyrate dehydrogenase and succinyl-CoA:3-oxo-acid CoA-transferase in heart mitochondria of diabetic rats. Biochemical Journal 240, 49–56 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson DH, Bates MW, Page MA & Krebs HA. Activities of enzymes involved in acetoacetate utilization in adult mammalian tissues. Biochemical Journal 121, 41–47 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahma MK et al. Increased Glucose Availability Attenuates Myocardial Ketone Body Utilization. Journal of the American Heart Association 9, e013039 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y & Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic acids research 33, 2302–2309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirdita M et al. ColabFold - Making protein folding accessible to all. bioRxiv, 2021.2008.2015.456425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berardi MJ, Shih WM, Harrison SC & Chou JJ. Mitochondrial uncoupling protein 2 structure determined by NMR molecular fragment searching. Nature 476, 109–113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller A, Chavez JD, Tang X & Bruce JE. Leveraging the Entirety of the Protein Data Bank to Enable Improved Structure Prediction Based on Cross-Link Data. J Proteome Res 20, 1087–1095 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spinazzi M, Casarin A, Pertegato V, Salviati L & Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nature Protocols 7, 1235–1246 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Chen Z et al. Quantitative cross-linking/mass spectrometry reveals subtle protein conformational changes. Wellcome Open Res 1, 5-5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen ZA & Rappsilber J. Protein Dynamics in Solution by Quantitative Crosslinking/Mass Spectrometry. Trends in Biochemical Sciences 43, 908–920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller A, Eng J, Zhang N, Li X. j. & Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular Systems Biology 1, 2005.0017 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohr JP, Perumalla P, Chavez JD, Eng JK & Bruce JE. Mango: A General Tool for Collision Induced Dissociation-Cleavable Cross-Linked Peptide Identification. Analytical Chemistry 90, 6028–6034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chavez JD, Weisbrod CR, Zheng C, Eng JK & Bruce JE. Protein Interactions, Post-translational Modifications and Topologies in Human Cells. Molecular & Cellular Proteomics 12, 1451–1467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo SE, Clauser KR & Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Research 44, D1251–D1257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller A, Chavez JD & Bruce JE. Increased sensitivity with automated validation of XL-MS cleavable peptide crosslinks. Bioinformatics 35, 895–897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller A, Nesvizhskii AI, Kolker E & Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Analytical Chemistry 74, 5383–5392 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cross-linking data has been deposited at XlinkDB (http://xlinkdb.gs.washington.edu/xlinkdb/index.php) and is publicly available (Network Name: Caudal_iqPIR_TACsham_Bruce). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE 49 partner repository with the dataset identifiers: PXD027757 and PXD035622. The following publicly available files were included: PDB 5Z62, PDB 3DLX, PDB 3OXO, PDB 1OKC, PDB 6GCI, PDB 2LCK, and Uniprot P48962. Any additional information required to reanalyze the data reported in this paper is available from the lead contacts upon request. All other data supporting the findings in this study are included in the main article and associated files.