Abstract
Traits shared among distantly related lineages are indicators of common evolutionary constraints, at the ecological, physiological, or molecular level. Here, we show that the vertebral stripe, a cryptic color pattern, has evolved hundreds of times in the evolutionary history of anurans (frogs and toads) and is favored in terrestrial habitats. Using a genome-wide association study, we demonstrate that variation near the Agouti signaling protein gene (ASIP) is responsible for the different vertebral stripe phenotypes in the African grass frog Ptychadena robeensis. RNAseq and real-time quantitative PCR revealed that differential expression of the gene and an adjacent long non-coding RNA is linked to patterning in this species. Surprisingly, and although the stripe phenotypes are shared with closely related species, we found that the P. robeensis alleles are private to the species and unlikely to evolve under long-term balancing selection, thus indicating that the vertebral stripe phenotypes result from parallel evolution within the group. Our findings demonstrate that this cryptic color pattern evolved rapidly and recurrently in terrestrial anurans, and therefore constitutes an ideal system to study repeated evolution.
Keywords: repeated evolution, adaptation, Agouti signaling protein, color polymorphism, Anura
Introduction
Animal color patterns are conspicuous hallmarks of selection. Color patterns may evolve because they are linked to a beneficial physiological trait (Bull 1977; Otaki 2008) or because they serve as sexual (Chen et al. 2012; Zhou et al. 2021) or warning signals (Briolat et al. 2019). Alternatively, color patterns can help avoid detection from visually oriented predators or prey by disrupting body shape recognition (Osorio et al. 1991; Stevens and Cuthill 2006; Cuthill and Székely 2009; Ruxton et al. 2018), masquerading as an object or animal (Skelhorn et al. 2010), countershading (Rowland et al. 2008), or substrate-matching (Godfrey et al. 1987; Woolbright and Stewart 2008; Rojas 2017). In many species, multiple color patterns coexist within or between populations. These polymorphisms can be maintained by divergent mating strategies (Hurtado-Gonzales and Uy 2009; Pérez i de Lanuza et al. 2013), apostatic selection (preference for the most common morph by predators), temporal, or spatial habitat heterogeneity (Briolat et al. 2019), or heterozygote advantage on correlated traits (Otaki 2008). This diversity of selective regimes makes color patterns an ideal system to investigate the evolutionary mechanisms underlying phenotypic evolution.
The vertebral stripe is a common color pattern element found across numerous distantly related anuran amphibians around the world (fig. 1A), and is believed to act as a substrate-matching and/or a disruptive camouflage (i.e., hindering the recognition of the animal's body shape by visually splitting it into two parts; Cott 1940; Ruxton et al. 2018). Phenotypes shared across such highly divergent lineages can result from ancestral alleles conserved over millions of years of evolution, or have evolved independently multiple times, perhaps driven by similar selective forces. While predator-mediated selection is a widely assumed mechanism for the evolution and maintenance of most color patterns in anurans (Hoffman and Blouin 2000), the link between the anuran vertebral stripe and survival is only empirically supported in a few species (Stewart 1974; Woolbright and Stewart 2008; McElroy 2016; but see Anderson et al. 2019). To gain a better understanding of the evolutionary history of the vertebral stripe in anurans, a broad-scale comparative analysis is necessary. Understanding the genomic architecture underlying phenotypes can also inform on the evolutionary mechanisms at play. Despite the commonness of the pattern, the genetic basis of the anuran vertebral stripe is largely unknown. In the few species investigated, the stripe is determined by a dominant allele at a single locus (e.g., Pyburn 1961; Browder et al. 1966; O’Neill and Beard 2010; reviewed in Hoffman and Blouin 2000). However, as these inferences were made solely from crossing experiments, the identity of the locus involved remains to be determined.
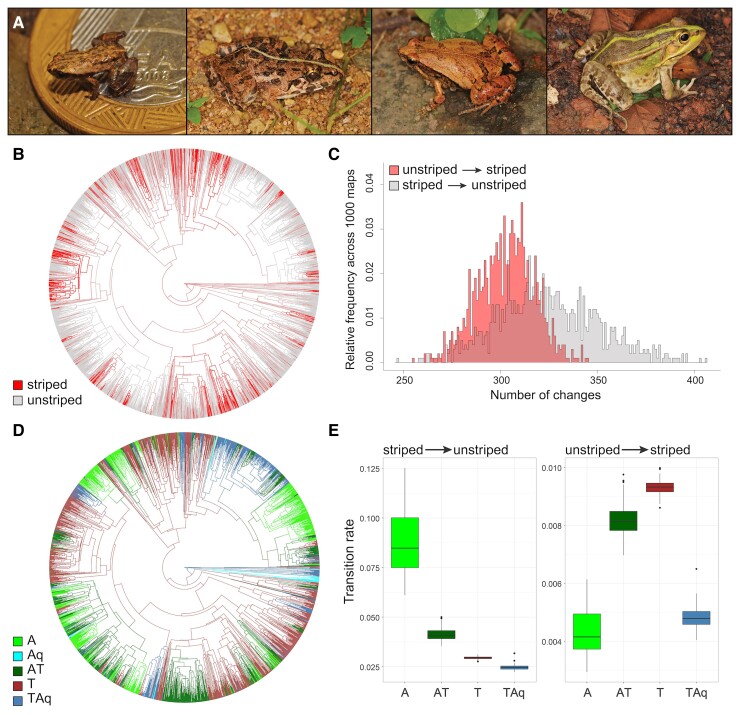
Fig. 1.
The evolution of the vertebral stripe in anurans. (A) Examples of the vertebral stripe in distantly related species: from left to right, Brachycephalus hermogenesi (family: Brachycephalidae), Fejervarya limnocharis (family: Dicroglossidae), Microhyla ornata (family: Microhylidae), Pelophylax nigromaculatus (family: Ranidae), (B) vertebral stripe morphs mapped on the phylogeny of Anura (n = 2,716 species; 1,000 stochastic maps), (C) estimated number of transitions between striped and unstriped morphs in the evolution of anurans based on 1,000 stochastic maps, (D) habitat use mapped on the phylogeny of Anura (n = 2,483 species; 100 stochastic maps), (E) transition rates between striped and unstriped phenotypes for each habitat, estimated for 100 stochastic maps. Habitat categories: A = arboreal, Aq = aquatic, AT = arboreal–terrestrial, T = terrestrial, TAq = terrestrial–aquatic.
Here we combine macro- and micro-evolutionary analyses with transcriptomic and histological data to investigate the evolutionary history and genomic architecture underlying the vertebral stripe in anurans. First, we ask how many times the vertebral stripe evolved during the evolutionary history of anurans and what ecological conditions may have favored its evolution. To do so, we create 1,000 stochastic maps of the trait on a molecular phylogeny including 2,716 species and we estimate the number of transitions between morphs. We then establish whether the vertebral stripe is differentially selected across habitats by comparing its rate of evolution in the five major anuran habitats. Second, we ask what is the genetic basis for the trait. Using genome- and transcriptome-wide analyses, we identify a single genomic region and differentially expressed (DE) transcripts associated with the color pattern in the African grass frog Ptychadena robeensis. Third, we ask what selective forces may maintain the polymorphism shared across the Ptychadena neumanni species complex, a radiation of 12 African grass frog species closely related to P. robeensis. We look for signatures of balancing selection in the genome of P. robeensis, and compare the genomic region of interest among the members of the radiation. By integrating results at three different scales (order, species group, and species), our study exemplifies how natural selection combined with rapidly evolving genomic regions may result in recurrent phenotypic evolution.
Results
Evolutionary History of the Vertebral Stripe in Anurans
To retrace the evolutionary history of the vertebral stripe in anurans, we examined the dorsal color pattern of 2,716 anuran species for which phylogenetic data was available (Jetz and Pyron 2018), representing 36.3% of species and all families currently recognized in Anura (supplementary table S1, Supplementary Material online; Frost 2021). A vertebral stripe morph was present in 17.9% of the 2,716 species included, and of those, 88.3% were lighter in color compared to the surrounding skin and 82.5% were polymorphic for the trait. We thus categorized species as either striped (including polymorphic taxa) or unstriped, and estimated the number of transitions between the two morphs in the evolutionary history of anurans based on 1,000 stochastic phylogenetic maps of the trait (fig. 1B and C). We estimated that the vertebral stripe pattern evolved independently 303 ± 15 times across the phylogeny and was lost 324 ± 26 times (fig. 1C). This result strongly supports the hypothesis of multiple origins of the anuran vertebral stripe.
Once we established that the vertebral stripe evolved independently multiple times, we investigated the role of the environment in the evolution of this trait. We hypothesized that recurrent evolution of the vertebral stripe across anurans may be due to similar selective pressures in shared habitat types. To test this hypothesis, we assembled habitat data for 2,483 anuran species and categorized them as arboreal, aquatic, terrestrial, arboreal/terrestrial, or terrestrial/aquatic, based on the main habitat occupied by adults (supplementary table S1, Supplementary Material online). We built 100 stochastic maps of habitat data onto the anuran phylogeny, and for each map, we fitted a habitat-dependent model for the transition between the unstriped and striped morphs, and compared it to a habitat-independent model using a likelihood ratio test. Our analysis revealed that the rate of transition between morphs is correlated with habitat (χ2(8) = 74.8 ± 22.9, p = 1.5 × 10−10 ± 5.3 × 10−10; supplementary fig. S1, Supplementary Material online). We thus extracted and compared the estimated transition rates between the unstriped and striped morphs for each habitat across the 100 stochastic maps. The vertebral stripe evolved significantly more often in terrestrial clades compared to terrestrial–aquatic (Δ = 4.5 × 10−3, P < 0.001), arboreal (Δ = 4.7 × 10−3, P < 0.001), and terrestrial–arboreal linages, although the difference was smaller (Δ = 1.1 × 10−3, P < 0.001; fig. 1D and E; supplementary fig. S2, Supplementary Material online; table 1). Arboreal lineages also showed the lowest gain and highest loss rates for the color pattern. The vertebral stripe may thus be selected against in arboreal habitats and selected for in terrestrial habitats.
Table 1.
Tukey Honest Significant Differences Test of Morph Transition Rates Between Habitat Pairs.
| Striped -> unstriped | Unstriped -> striped | |||||||
|---|---|---|---|---|---|---|---|---|
| Δ | Lower | Upper | Adj. P-value | Δ | Lower | Upper | Adj. P-value | |
| AT - A | −46.28 | −49.68 | −42.89 | <0.001 | 3.86 | 3.64 | 4.09 | <0.001 |
| T - A | −58.03 | −61.42 | −54.63 | <0.001 | 4.96 | 4.73 | 5.18 | <0.001 |
| Taq - A | −62.75 | −66.15 | −59.35 | <0.001 | 0.46 | 0.23 | 0.68 | <0.001 |
| T - AT | −11.74 | −15.14 | −8.34 | <0.001 | 1.10 | 0.87 | 1.32 | <0.001 |
| TAq - AT | −16.47 | −19.87 | −13.07 | <0.001 | −3.41 | −3.63 | −3.18 | <0.001 |
| TAq - T | −4.73 | −8.12 | −1.33 | 0.002 | −4.50 | −4.73 | −4.28 | <0.001 |
Values were multiplied by 103 for readability. Average (Δ), lower, and upper values of the difference between rates based on 100 stochastic maps are given. Habitat categories: A = arboreal, Aq = aquatic, AT = arboreal–terrestrial, T = terrestrial, TAq = terrestrial–aquatic.
The Vertebral Stripe in a Grass Frog Radiation
To investigate the molecular and cellular mechanisms underlying the vertebral stripe in anurans, we focused on a radiation of grass frogs, the Ptychadena neumanni species complex. This monophyletic group encompasses 12 species (Goutte et al. 2021), which are all polymorphic for the vertebral stripe (the stripe could be thin, wide, or absent), except for two species: P. harenna, in which the vertebral stripe morph is always absent, and P. cooperi, for which all individuals present a thin vertebral stripe (fig. 2 and supplementary fig. S3, Supplementary Material online). In species other than P. harenna, the absence of vertebral stripe (hereafter referred to as the “unstriped” morph) is generally accompanied by an absence of melanization of the dorsum, and thus an overall lighter coloration (fig. 2B). While polymorphic species allow pinpointing the genomic region(s) linked to the polymorphic trait by comparing conspecifics of different morphs, shared polymorphism across radiation informs on the micro-evolutionary history of the region(s).
Fig. 2.
The vertebral stripe in Ethiopian Ptychadena. (A) Polymorphism of the vertebral stripe (wide or thin striped, or unstriped) in the Ptychadena neumanni species complex (phylogeny based on 500,000 genome-wide distributed SNPs, reproduced from Hariyani et al. in preparation). Presence of the morph is indicated in blue, absence in white. (B) Adult Ptychadena robeensis presenting the three possible vertebral stripe morphs. From left to right: wide striped, thin striped, unstriped. (C) Histological sections of the dorsal skin within (top) and outside (bottom) the vertebral stripe in a female Ptychadena erlangeri (SB548; live photograph on the left). Scale bar = 200 µm. Within the stripe (top), the few melanophores (blue arrows) are in a contracted state and do not entirely cover the xanthophores (green arrows), in contrast with the skin outside the stripe (bottom). Outside the stripe, numerous melanosomes (red arrows) are also present in the epidermal layer, creating a very dark coloration.
We examined the pigment cells organization in the dorsal skin of ten individuals of the P. neumanni species complex (P. robeensis, P. nana, P. erlangeri and P. amharensis) presenting different phenotypes (thin or wide striped, or unstriped; fig. 2B and supplementary fig. S3, Supplementary Material online). To do so, we produced histological sections of 4 µm thickness, which we stained with hematoxylin-eosin (HE), and examined chromatophore distribution within and outside the pattern. In all specimens examined, numerous melanophores with dispersed melanosomes covering other chromatophores, and melanosomes in the epidermal layer create a dark coloration outside the stripe (fig. 2C and supplementary fig. S4, Supplementary Material online). Within the stripe, melanophores with aggregated melanosomes (when present) and no or few epidermal melanosomes result in a lighter shade (fig. 2C and supplementary fig. S4, Supplementary Material online). The state (aggregated vs. dispersed) of the melanophores, as well as the concentration of epidermal melanosomes are thus the major determinants of the vertebral stripe pattern in the species examined.
Genomic Architecture of the Vertebral Stripe in Ptychadena robeensis
To identify the genomic region(s) involved in the vertebral stripe pattern, we conducted a genome-wide association study (GWAS) in one species of the P. neumanni complex, Ptychadena robeensis, which is polymorphic for the trait. We produced whole-genome resequencing data (4.84 × average coverage) for 52 individuals with either a wide (n = 25) or a thin (n = 27) vertebral stripe and aligned the reads on the recently assembled chromosome-level genome of the species (supplementary table S2, Supplementary Material online; Hariyani et al. in preparation), resulting in a dataset with 17,797,568 single-nucleotide polymorphisms (SNPs). The number of unstriped individuals collected being low (n = 5), we excluded this phenotype from the analysis. We identified a ∼88 kb region on chromosome 11, which included multiple significant SNPs (-log10(P) < 5 × 10−8) associated with the color pattern (fig. 3A and supplementary fig. S5, Supplementary Material online).
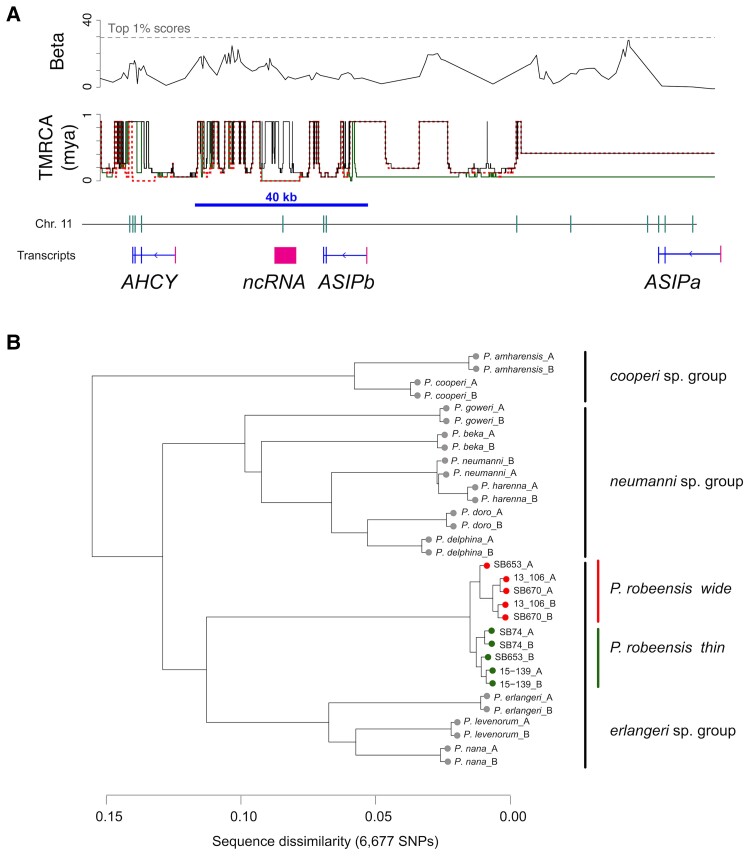
Fig. 3.
Genes associated with the stripe phenotype in Ptychadena robeensis. (A) Genome-wide association study (GWAS) reveals a single locus governing the vertebral stripe phenotype on chromosome 11 of Ptychadena robeensis (top panel). Significant SNPs (indicated in blue in the bottom panel) are located in between and downstream ASIPa and ASIPb exons. (B) Position of ASIPa and ASIPb in the phylogeny of ASIP. Maximum-likelihood tree based on a protein alignment using MUSCLE (Edgar 2004). ASIPa and ASIPb are grouped with ASIP of Xenopus tropicalis, excluding the possibility for the two genes to correspond to fish ASIP1 and ASIP2. Examination of the protein alignment showed that ASIPa likely resulted from a duplication of ASIPb (see text and supplementary fig. S6, Supplementary Material online).
The genomic region encompasses three genes and one 4.6 kb long non-coding RNA (denoted hereafter ncRNA; Hariyani et al. in preparation). One of the genes was identified as AHCY, while the other two, 38 kb upstream of AHCY and 40 kb apart of each other, correspond to two copies of the Agouti signaling protein (ASIP). ASIP is a well-studied gene that is known to be involved in melanophore differentiation and melanin production in vertebrates (Sviderskaya et al. 2001; Cal et al. 2017). While a single ASIP gene is known in birds, mammals and in the African clawed frog Xenopus tropicalis, two genes, ASIP1 and ASIP2, are present in teleost fish (Braasch and Postlethwait 2011; Cortés et al. 2014; Cal et al. 2017; Liang et al. 2021). In order to determine whether the two duplicates in P. robeensis correspond to the fish ASIP1 and ASIP2, we translated the genomic sequences and produced a protein alignment and maximum-likelihood phylogeny of the ASIP gene family in vertebrates (fig. 3B). Both Ptychadena genes grouped together and with the ASIP of X. tropicalis, indicating that these duplicates arose independently from the fish ASIP genes. We thus named them ASIPa and ASIPb to avoid any confusion with ASIP1 and ASIP2 from teleost fish.
While vertebrates’ ASIP typically contains three coding exons, ASIPa, and ASIPb contain six and two predicted coding exons, respectively. The protein alignment revealed that ASIPa exons 2 & 3 correspond to ASIPb exons 1 & 2, and the two first exons of other vertebrates ASIP (supplementary fig. S6A, Supplementary Material online). Our analysis also revealed that the ncRNA, directly downstream of ASIPb, contains a region similar to the third exon of the ASIP gene present in other vertebrates but absent from ASIPa and ASIPb (supplementary fig. S6A, Supplementary Material online). Given the divergence between ASIPa and ASIPb, this pattern likely resulted from an ancient gene duplication, followed by the recruitment of four additional exons in ASIPa (supplementary fig. S6B, Supplementary Material online). Comparing the genomic region between the 12 species of the Ptychadena neumanni complex, we found that a ∼ 5 kb insertion between ASIPb exons 2 & 3 occurred in the common ancestor of the Ptychadena erlangeri species group (P. erlangeri, P. levenorum, P. nana, and P. robeensis) and may have caused the loss of functionality in the third exon of ASIPb (supplementary fig. S6B, Supplementary Material online).
The majority of the SNPs are in a 40 kb region overlapping with ASIPb and the ncRNA (fig. 3A), and all of them fall outside coding sequences. We thus hypothesize that they are located in a regulatory region and affect the expression of the gene. An up-regulation of ASIP is linked to lighter phenotypes in mammals (Jones et al. 2018; Liang et al. 2020) and fishes (Cal et al. 2017); we could thus expect an up-regulation of ASIPb in wide striped and unstriped individuals compared to thin striped frogs, as both morphs have reduced melanization (fig. 2C and supplementary fig. S4, Supplementary Material online).
Differential Gene Expression Associated With the Vertebral Stripe
To test the hypothesis that the stripe morphs are linked to differential expression of ASIPb, we sequenced the transcriptomes of the dorsal skin of wide striped (n = 4), thin striped (n = 4), and unstriped (n = 3) P. robeensis individuals, as well as the ventral skin (n = 2), as a control (as ventral skin lacks any melanization). Dorsal skin was sampled from the posterior half of the dorsum, and RNA was extracted within and outside the vertebral stripe separately for the four wide striped individuals in order to compare their respective gene expression levels (supplementary fig. S7, Supplementary Material online). Expression levels were significantly greater for ASIPb (t11 = 3.27, P < 0.01) and the ncRNA (t11 = 12.88, P < 0.01), and marginally greater for ASIPa (t11 = 2.05, P = 0.06) in ventral skin (n = 2) compared to dorsal skin (n = 11; fig. 4A). In dorsal skin, all three transcripts had the lowest concentration in thin striped (n = 4) compared to wide striped (n = 4) and unstriped individuals (n = 3), although the difference was not significant (ASIPa: F3,11 = 0.64, P = 0.61; ASIPb: F3,11 = 0.42, P = 0.74; ncRNA: F3,11 = 2.98, P = 0.08; fig. 4B). Interestingly, the ncRNA was expressed at significantly higher levels within versus outside the stripe in wide striped individuals (t6 = 4.10, P < 0.01), while neither ASIPa nor ASIPb were significantly DE (ASIPa: t6 = 0.25, P = 0.81; ASIPb: t6 = 0.16, P = 0.88). Additionally, expression levels of ASIPb and the ncRNA were not correlated in dorsal skin (t17 = 0.29, P = 0.77). To quantify more precisely ASIPb expression level in the dorsal skin, we conducted a quantitative real-time PCR experiment (qPCR), which confirmed that ASIPb expression level was higher in wide striped (n = 2; outside the stripe) compared to thin striped (n = 2) individuals (t = −2.28, P = 0.02; fig. 4C). Interestingly, ASIPb was expressed at higher levels outside (n = 2) versus within (n = 2) the vertebral stripe in wide striped individuals (t = 0.82, P = 0.04; fig. 4C). These results indicate that differential expression of both ASIPb and the ncRNA may play a role in the establishment of the stripe morphs in Ptychadena robeensis by regulating the differentiation of melanophores and/or the dispersion of melanosomes.
Fig. 4.
Differential gene expression associated with the vertebral stripe in Ptychadena robeensis. (A and B) Normalized expression levels of ASIPa, ASIPb and the ncRNA from RNAseq data are given in log count per million. (A) Expression levels of all three transcripts are greater in ventral skin (n = 2) than in dorsal skin (n = 15), (B) in dorsal skin, expression levels of ASIPa, ASIPb and the ncRNA are the lowest in thin striped (n = 4) compared with wide striped (n = 4) or unstriped individuals (n = 3), (C) relative expression levels of ASIPb in the dorsal skin of thin striped (n = 2) and wide striped (n = 2) individuals measured by qPCR. Each reaction was triplicated and average CT value for each individual was used. (D) Eight genes are differentially expressed between vertebral stripe phenotypes, and (E) 43 genes are down-regulated within versus outside the vertebral stripe in the same individuals.
To characterize further the genes interaction network associated with the vertebral stripe, we explored transcriptome-wide patterns of gene expression in the dorsal skin of P. robeensis (fig. 4D and E). Eight genes were up-regulated in wide striped individuals compared to thin striped frogs, and 43 genes were down-regulated within the stripe compared to outside the stripe (fig. 4D and E; supplementary table S3, Supplementary Material online). Interestingly, several of these genes have been shown to be involved in melanogenesis in mammals (ldha and aldoa; Slominski et al. 2014) and skin coloration in carp (aldoa; Ye et al. 2020), yet the majority had unknown functions. While morph-dependent DE genes are likely to be involved in the same pathways as ASIP, the genes DE between sections of the dorsal skin could be produced by the different mature chromatophores. Further gene expression and functional analyses on adult and developing individuals would be needed to elucidate the gene interaction network establishing the vertebral stripe in P. robeensis.
Recent Evolution of the Thin and Wide Alleles
As the vertebral stripe is under selective pressure in terrestrial habitats, we hypothesized that the stripe polymorphism present across the Ethiopian Ptychadena radiation is maintained by predator-driven balancing selection across the group. In order to test this hypothesis, we looked for signatures of long-term balancing selection in our genomic data. First, we computed β, a summary statistic measuring the degree of shared allele frequencies between neighboring loci (Siewert and Voight 2017). The value of β increases in regions under long-term balancing selection, and loci in the top 1% β values in a given genome can be hypothesized as under ancient balancing selection (Siewert and Voight 2020). We computed β on our region of interest across the 56 low-coverage Ptychadena robeensis samples on a 4 kb sliding window using BetaScan (Siewert and Voight 2017). All values in the region were below the top 1% β values threshold for the genome (fig. 5A), indicating that the alleles are unlikely to be under long-term balancing selection.
Fig. 5.
Recent evolution of the thin and wide alleles in Ptychadena robeensis. (A) β scores and TMRCA across the region of interest computed using BetaScan on low-coverage data (n = 56) and ARGweaver on high-coverage data (n = 4), respectively. Red dashed and green solid lines indicate wide and thin haplotypes, respectively. Black solid lines indicate the overall population. The positions of ASIPa, ASIPb, AHCY and the ncRNA are indicated below and the 40 kb region containing most significant SNPs in the GWAS analysis is indicated by a horizontal blue bar. In the region of interest, haplotypes have β scores below the top 1% values for the genome (top panel), and an equal or more recent TMRCA than the overall population and surrounding region (bottom panel), indicating a lack of long-term balancing selection of the alleles. (B) Dendrogram based on dissimilarity of sequences in the 40 kb region of interest (6,677 SNPs) in the Ptychadena neumanni species complex (8.29 × average coverage). Haplotypes are denoted by A or B after the species or sample name. For P. robeensis, leaves are color-coded by haplotypes (green for thin and red for wide). Within P. robeensis, haplotypes are grouped by color pattern, while across the Ptychadena neumanni complex, they are grouped according to species relatedness and form the three species groups previously described (fig. 2A; Goutte et al. 2021).
Second, we estimated the Times to the Most Recent Common Ancestor (TMRCA) of the thin and wide alleles using Ancestral Recombination Graphs (ARG) analyses implemented in ARGweaver (Hubisz and Siepel 2020). Regions under ancient balancing selection should show older TMRCA than neutral genomic regions and an equal TMRCA between the haplotypes and the overall population. Positive selection or recent balancing selection, on the other hand, should have an overall TMRCA similar to neutral regions but more recent haplotype TMRCAs than the overall TMRCA. We ran ARGweaver on the high-coverage P. robeensis samples (n = 4; 12.97 × on average) and categorized the phased haplotypes as wide or thin, thereby considering haplotypes rather than individuals in subsequent analyses. TMRCA for the thin and/or wide alleles were more recent than the total population TMRCA in our region of interest, and notably at the level of the ncRNA where both alleles were younger than the overall population (fig. 5A). This result is consistent with a partial selective sweep and not ancient balancing selection. We estimated the wide and thin alleles to have diverged recently, less than a million years ago (fig. 5A). These alleles thus arose long after the divergence between Ptychadena robeensis and its closest relatives, estimated at 4.3–6.0 million years ago (Freilich et al. 2014).
To confirm this result, we compared the region of interest between all 12 species of the P. neumanni complex (8.29 × average coverage). We phased each genome and built a phylogeny of the haplotypes in the 40 kb region of interest (6,677 SNPs). Haplotypes grouped according to clades within the radiation, and not phenotypes (except within P. robeensis), confirming that the thin and wide alleles were indeed private to Ptychadena robeensis and not shared with the other Ptychadena species (fig. 5B; supplementary fig. S9, Supplementary Material online). Additional GWAS analyses on two other members of the P. neumanni complex, P. amharensis (n = 32; 1.95 × average coverage; 82,580,376 SNPs) and P. beka (n = 42; 4.58 × average coverage; 33,430,567 SNPs), failed to detect a single locus responsible for the dorsal pattern for either species. Additionally, none of the significant SNPs found in the GWAS conducted on P. robeensis were shared with P. amharensis or P. beka. These results indicate that the alleles found in P. robeensis have evolved recently and other alleles are responsible for similar dorsal patterns in closely related species.
Discussion
In this study, we show that the anuran vertebral stripe evolved multiple times, and significantly more often in terrestrial lineages compared to terrestrial–aquatic, arboreal, and terrestrial–arboreal linages. The vertebral stripe might increase concealment from visually oriented predators coming from above, such as birds or mammals which are more prevalent in terrestrial habitats, and thus confer a fitness advantage. A similar pattern is observed in several polymorphic species, where the incidence of the striped morph correlates with geographical or habitat features, with a greater proportion of striped individuals in more open habitats (e.g., Fishbeck and Underhill 1971; Stewart 1974; Schueler and Cook 1980; Tarkhnishvili and Gokhelashvili 1996). Interestingly, the vertebral stripe was lost significantly more often in arboreal lineages than in other groups, indicating a potential fitness cost of the pattern in this habitat, or that other color patterns are selected for and the vertebral stripe lost as a side-effect. Although the molecular and evolutionary mechanisms may vary across lineages, similar selective pressures may have favored the presence of striped morphs in terrestrial anurans.
In the grass frog Ptychadena robeensis, we identified a single genomic region associated with the thin and wide vertebral stripe morphs. The difference in melanophore proliferation and melanosome dispersion leading to these morphs is likely linked to differential expression of ASIPb and of the adjacent ncRNA. Indeed, the identified haplotypes differ in non-protein-coding regions near the gene, and both ASIPb and the ncRNA are DE between the morphs and within versus outside the pattern. The ncRNA could have a regulatory role on ASIPb, although we did not observe any significant ncRNA/ASIPb co-expression pattern in adult skin.
Although we found morph-dependent differential expression of ASIPb, the gene's expression levels were very low in all adult dorsal skin. The role of ASIPb in skin pigmentation may be more significant at an earlier stage of the animal's development, when the pattern first appears. In zebrafish, ASIP1's expression level varies dynamically during development and is lower in adults (60 days post fertilization) than in metamorphic individuals (30 days post fertilization; Ceinos et al. 2015). In the Ptychadena neumanni species complex, the vertebral stripe emerges at the final stages of metamorphosis, or soon after (observation based on 92 individuals identified through barcoding at different developmental stages). Investigating ASIPb's expression levels in the dorsal skin of metamorphic and juvenile individuals will be necessary to determine the gene's role in the establishment of the color pattern during development.
When compared to dorsal skin, the expression levels of ASIPb and adjacent ncRNA were extremely high in the ventral skin, which, as in many other anurans, is uniformly white. This dorso-ventral differential expression is comparable to expression patterns found in several species of fish which also present a white ventrum (Ceinos et al. 2015; Jones et al. 2018). ASIP thus seems to play a determining role in both the dorsal color pattern and the lack of melanization in ventral skin. Our results suggest that ASIP could correspond to the melanization inhibiting factor (MIF) identified in cultures of Xenopus laevis’ ventral skin cells, as was previously suggested (Fukuzawa and Ide 1988; Fukuzawa et al. 1995; Mills and Patterson 2009). Although the involvement of ASIP in melanophore differentiation and melanin production has been extensively studied in mammals (Girardot et al. 2006; Norris and Whan 2008; Almathen et al. 2018; Liang et al. 2020), and particularly in rodents (Galbraith 1964; Siracusa 1994; Dolinoy 2008), its role in amphibians had not previously been demonstrated. This study provides a strong line of evidence for a causal link between ASIP and the establishment of color patterns in anurans.
In multiple organisms, ASIP alleles evolved rapidly leading to parallel evolution of similar phenotypes within species (Jones et al. 2020) or groups of closely related species (Liang et al. 2020). In the P. neumanni species complex, species presenting the same color patterns did not share alleles with P. robeensis. Indeed, we estimated P. robeensis’ alleles to have evolved after the species divergence from its closest known relatives. Recurrent evolution in the regulatory region of ASIP could thus have led to the same color patterns in this group of closely related species, similarly to the horizontal stripes in African cichlids caused by repeated evolution at agrp2 regulatory region (Kratochwil et al. 2018). However, mutations impacting the expression of genes interacting with ASIP (e.g., MC1R;Hoekstra et al. 2006; Cal et al. 2017) may also be responsible for the vertebral stripe in Ptychadena spp. and other terrestrial anurans. The lability of cryptic color pattern elements such as the anuran vertebral stripe may allow species to adapt to rapid environmental changes or variable local conditions. Intense predation or other strong selection pressures can then favor a rapid spreading of advantageous alleles. By demonstrating the involvement of ASIP in a widespread trait, our study opens new research avenues on color patterns in anurans. Mutations in the regulatory regions of ASIP or interacting genes causing the appearance or loss of the vertebral stripe are likely occurring at a high rate in anurans, making this trait an ideal system to study parallel evolution.
Methods
Comparative Analysis of Vertebral Stripe Evolution in Anurans
We conducted a comparative analysis across the Anura using the largest dated molecular phylogeny published to date (Jetz and Pyron 2018). This phylogeny comprises 3,449 anuran species (=46% of currently recognized species; Frost 2021), representing all families (Jetz and Pyron 2018). We collected data on dorsal color pattern for 2,716 of these species by examining all photographs available for each species on Amphibiaweb (https://amphibiaweb.org). When no or few photos were available, we searched additional sources such as original species descriptions and the number of photographs examined for each species was systematically recorded (supplementary table S1, Supplementary Material online). Dorsal color patterns were classified in the following categories: thin, medium or wide striped, and unstriped. If a species had at least one individual counted in the unstriped and one of the striped categories, the species was considered polymorphic for the trait.
Habitat use data were collected independently for 2,483 species based on multiple large studies, and species accounts in Amphibiaweb and the IUCN red list websites (https://amphibiaweb.org; https://www.iucnredlist.org; supplementary table S1, Supplementary Material online). Anuran habitats were categorized as arboreal, aquatic, terrestrial, arboreal/terrestrial, and terrestrial/aquatic, based on the main habitat occupied by adult individuals (when reproductive and general habitats were available). We searched for precise microhabitat use descriptions such as “Individuals are found on roots, leaf litter and crevices” or “Adults are found in phytotelmas up to four meters height”, and considered insufficient general habitat description such as “This species inhabits riparian forests” or “This frog is linked to streams” as they do not provide sufficient details on whether adults spend their time on vegetation, on the ground or in the water.
Ancestral State Reconstruction of Dorsal Color Patterns
To reconstruct the evolutionary history of the vertebral stripe, we first estimated the rate of evolution between the striped, unstriped, and polymorphic states using the function fitpolyMk in the R package phytools (Revell 2014) and a model allowing only transition from striped to unstriped through a polymorphic state. We compared the goodness of fit of models with all transition rates equal (ER; 1 rate), all rate different (ARD; 4 rates), rates from and to the polymorphic state equal, but different for each fixed state (SYM; 2 rates), rates from polymorphic to either fixed state equal, and from either fixed state to the polymorphic state equal (transient; 2 rates). The model in which all rates were allowed to be different best fitted the data (supplementary table S4, Supplementary Material online). The rate of transitions between the striped and polymorphic states was the highest, and the transition from unstriped to the polymorphic state had the lowest rate (supplementary fig. S8, Supplementary Material online).
Because most of the striped species were polymorphic for the trait (82.5%), and as erroneous categorization was more likely for the fixed than for the polymorphic categories (if only few photos were available), we recategorized species as either having at least some individuals presenting a vertebral stripe, or without any striped individuals. We thus used two categories, striped (including polymorphic species) and unstriped with a model allowing transition rates between the two morphs to be different (ARD; 2 rates), and estimated the number of transitions during the evolutionary history of anurans. We then created 1,000 stochastic maps of the trait onto the phylogeny using the function make.simmap.
Dorsal Pattern Evolution in Different Habitats
To test the hypothesis that the dorsal stripe might be selected for in particular habitats, we first built 100 stochastic maps of habitat data for the 2,483 species categorized on the phylogeny. For each of the stochastic trees, we fitted a model for which transition rates between dorsal color patterns differed for each habitat using the fitmultiMk function from the R package phytools, and compared them to a model for which transition rates were independent of habitat using a likelihood ratio test (supplementary fig. S1A, Supplementary Material online). The habitat-dependent model of evolution for the vertebral stripe was systematically better fitted than the independent model. We then compared the fits of habitat-dependent models with equal rates (ER; 5 rates) and all rates different (ARD; 10 rates) across the 100 stochastic maps (supplementary fig. S1B and C, Supplementary Material online); for each map, the ARD model was favored. Finally, we extracted the estimated transition rates between striped and unstriped phenotypes for each habitat and each of the 100 stochastic maps. Because few species were categorized as aquatic (1.6% of included taxa, i.e., 40 species), the variance in transition rate estimates was much greater than for the other habitats (supplementary fig. S2, Supplementary Material online), so we excluded aquatic lineages from further analyses. We compared the transition rates between pairs of habitats using a Tukey honest significant differences test (table 1).
Sampling of Ethiopian Ptychadena
Individuals of the Ptychadena neumanni species complex were collected in the highlands of Ethiopia between 2011 and 2019. Our study was approved by the relevant Institutional Animal Care and Use Committee at Queens College and New York University School of Medicine (IACUC; Animal Welfare Assurance Number A32721–01 and laboratory animal protocol 19–0003). Frogs were sampled according to permits DA31/305/05, DA5/442/13, DA31/454/07, DA31/192/2010, DA31/230/2010, DA31/7/2011, and DA31/02/11 provided by the Ethiopian Wildlife Conservation Authority. We photographed individuals in life and euthanized them by ventral application of 20% benzocaine gel. We extracted tissue samples and stored them in RNAlater or 95% ethanol. Adult individuals were fixed in 10% formalin for 24–48 h, and then transferred to 70% ethanol. After preservation, we took additional photographs of all individuals. All specimens were deposited at the Natural History Collection of the University of Addis Ababa, Ethiopia. Tissue samples are deposited at the Vertebrate Tissue Collection, New York University Abu Dhabi (NYUAD).
Histological Skin Sections
Dorsal and ventral skin sections were extracted from ten preserved adult specimens: two thin striped (SB81, SB82) and one unstriped (SB69) Ptychadena robeensis specimens, two thin striped (SB493, SB510) and one wide striped (SB494) P. nana, two wide striped (SB552, SB548) P. erlangeri, and two wide striped (SB584, SB593) P. amharensis. The skin samples were embedded in paraffin blocks and sections of 4 µm thickness were produced. The sections were stained with hematoxylin-eosin (HE) and chromatophores were examined using a Leica DMI 6000 B microscope. We used the following staining protocol: xylene 2 × 2 min, absolute ethanol 2 × 2 min, 95% ethanol 1 min, 70% ethanol 1 min, water for 2 × 30 s, hematoxylin 5 min, water for 2 × 30 s, bluing reagents 30 s, water 30 s, 95% ethanol 30 s, Eosin 3 min, 95% ethanol 30 s, absolute ethanol 2 × 30 s, xylene for 2 × 30 s.
Ptychadena robeensis Reference Genome
The assembly and annotation of the Ptychadena robeensis genome is described in details elsewhere (Hariyani et al. in preparation). Briefly, a de novo assembly was first constructed using two paired end libraries of mean insert size ∼350 bp and one paired end library of ∼550 bp insert size. The shotgun reads were incorporated with low-coverage PacBio to generate a hybrid assembly. To achieve longer scaffolds, two Chicago libraries were sequenced as described in Putnam et al. (2016). The input hybrid assembly and Chicago library reads were then used as input data for HiRise, a software pipeline designed specifically for using proximity ligation data to scaffold genome assembly (Putnam et al. 2016). Finally, an Omni-C library was sequenced. The Omni-C reads and the input de novo assembly from the previous step were used as input data for HiRise. The resulting genome is 1.59 Gb in length and consists of 12 scaffolds, which corresponds to the haploid chromosome number reported in the genus Ptychadena, with an N50 of 157 Mb.
DNA and RNA Extractions and Sequencing
Genomic DNA of 61 Ptychadena robeensis individuals was extracted from liver tissue using the DNeasy blood and tissue kit (Qiagen, Valencia, CA). RNA was extracted from the skin of 13 individuals using a RNeasy mini kit (five wide striped, five thin striped, three unstriped; Qiagen, Valencia, CA). For four wide striped individuals, RNA was extracted from dorsal skin within and outside the vertebral stripe separately, and for two individuals (one wide and one thin striped), RNA from ventral skin was extracted. For all other individuals, RNA was extracted from dorsal skin (see supplementary fig. S7, Supplementary Material online). We quantified extracted DNA and RNA using a Qubit fluorometer (Life Technologies). Libraries were prepared using a NEB library prep kit and sequenced on Illumina NextSeq 550 flow cells at the Genome Core Facility of New York University Abu Dhabi. After quality filtering, reads were aligned to the de novo assembly Ptychadena robeensis reference genome (Hariyani et al. in preparation). The average coverage of the genomic data was 4.84X, except for four individuals which we sequenced at an average of 12.97X. Variants were called using the function HaplotypeCaller from gatk v3.5 (McKenna et al. 2010). The low-coverage and higher-coverage samples were then combined and genotyped in two separate datasets using CombineGVCF and GenotypeGVCFs functions from gatk.
Genome-wide Association Study on Ptychadena robeensis
After examination of the low-coverage genomic dataset (n = 61 individuals; SNPs PCA and visual examination in IGV), we realized that five individuals were likely hybrids resulting from the crossing of P. robeensis and P. levenorum, a closely related species with a partially overlapping distribution range (Goutte et al. 2021). We thus ran an admixture analysis including all the individuals from our low-coverage dataset and 12 additional P. levenorum individuals sequenced at the same average depth. To do so, we excluded linked sites (pairs of SNPs with r2 > 0.1 within a 50 SNP window), as well as sites with >25% missing data, minor allele frequency <0.05, Phred quality score <30, or that were multiallelic or monomorphic, using PLINK 1.9 (Purcell et al. 2007). The final dataset comprised 615,561 unlinked SNPs. We then ran ADMIXTURE 1.3 (Alexander et al. 2009) with K = 2–5. Cross validation error was lowest for K = 2, with five individuals showing admixture between the two species (supplementary fig. S10, Supplementary Material online).
After removing the hybrids, the dataset comprised 17,797,568 SNPs. We further excluded four individuals, which presented no dorsal melanization and could not be categorized as thin or wide striped, and our final dataset contained 52 individuals (25 wide striped and 27 thin striped). Stripe phenotype (thin, wide or unstriped) and coloration (brown or green) were not correlated. Quality checks and the genome-wide association study (GWAS) were done using PLINK 1.9 (Purcell et al. 2007). We checked for individual relatedness as well as major discrepancies between samples in data missingness and minor allele frequency. However, because of the low-coverage nature of our data, we did not apply any stringent quality filtering. We extracted and visualized the result of the GWAS using the R package qqman (Turner 2014).
Test for Balancing Selection in Ptychadena robeensis
We searched for signatures of balancing selection in the regions linked to dorsal stripe pattern and aimed to determine the age of the alleles using multiple approaches. First, we used the summary statistic β of BetaScan (Siewert and Voight 2017) to search specifically for signatures of long-term balancing selection. In short, β scores are higher when neighboring loci share similar allele frequencies, which is indicative of long-term balancing selection. We computed β across the entire genome on 4 kb windows in order to determine a threshold based on the top 1% β values. Loci with β values above the threshold can be considered as likely under long-term balancing selection. We then compared the β values in our region of interest to the threshold value. We ran BetaScan on the 56 samples low-coverage Ptychadena robeensis dataset, as the program requires a relatively large sample size.
Second, we estimated the time to the most recent common ancestor (TMRCA) of our alleles using ARGweaver (Hubisz and Siepel 2020). In short, ARGweaver reconstructs a set of ARGs for every non-recombining interval in the genomic region of interest. The program then samples from the posterior distribution of ARGs given an evolutionary model. Regions under positive selection should display a reduced coalescence time whereas regions under ancient balancing selection should have older coalescence time compared to neutral regions.
We ran ARGweaver on the high-coverage Ptychadena robeensis dataset (12.97 × average coverage) comprising four individuals. As priors, we used a mutation rate map and a recombination rate map computed for the species on a 12 kb window and with a 2-year generation time using VCFtools (Danecek et al. 2011) and LDhat (McVean et al. 2002). The effective population size was estimated as a function of time in SMC++ (Terhorst et al. 2017) elsewhere (Hariyani et al. in preparation), and we used a maximum coalescence time of two million generations, around ten times the harmonic mean of the estimated effective population size over time. ARGweaver was run for 5,000 iterations and ARGs were sampled every 100 iterations. Convergence of the chain was monitored visually by plotting multiple ARG statistics (priors, likelihood, number of recombination events, total branch length, number of variant sites not explained by a single mutation) against iteration number. All statistics were stationary after the first 1,000 iterations, so we discarded these 1,000 first iterations as burn-in for further analyses.
We inspected the phased haplotypes and categorized them in wide or thin haplotypes, thereby considering haplotypes rather than individuals in the subsequent analyses. We extracted times to the most recent common ancestor (TMRCA) for the wide and thin haplotypes, as well as for the whole population and compared the three curves across our region of interest. Ancient balancing selection should show TMRCA older than neutral genomic regions and an equal TMRCA between the haplotypes and the overall population. Positive selection or recent balancing selection, on the other hand, should have and overall TMRCA similar to neutral regions but haplotype TMRCAs more recent than the overall TMRCA.
Phylogeny of Haplotypes
In order to compare the region of interest across the Ptychadena neumanni species complex, we reconstructed a haplotype phylogeny. The genomes of the 12 species (one individual per species, except for P. robeensis for which the genomes of five individuals were included) were phased using Beagle 5.1 (Browning et al. 2021). We then built a dissimilarity matrix based on variable sites and including all individuals (40 kb region; 6,677 SNPs) using the R package SNPRelate (Zheng et al. 2012). Finally, we created a dendrogram of the haplotypes in the region of interest using the cluster analysis function snpgdsHCluster of the same R packaged and the UPGMA algorithm (fig. 5B). In order to compare with the results of the BetaScan analysis, we also reconstructed ten haplotype phylogenies on 4 kb windows in the same manner (supplementary fig. S9, Supplementary Material online). With some variation in topology, haplotypes of P. robeensis systematically grouped separately from other species of the radiation, indicating that the haplotypes are not shared among species of the Ptychadena neumanni complex.
Gene Expression Analysis
Reads were aligned to the annotated reference genome using HISAT2 (Kim et al. 2019) and StringTie2 (Kovaka et al. 2019). A transcriptome-wide gene count matrix was then created using the script prepDE.py3 provided on the StringTie website. Subsequent analyses were conducted in the R environment (R Core Team 2020). We used the R package edgeR (Robinson et al. 2010) to filter and normalized our data prior analysis. We filtered out genes which had a count inferior to 1 count-per-million (cpm) in at least 16 samples (>50% of the 21 samples in total) and applied a trimmed mean of M-values (TMM) normalization of the data. We then calculated a contrast matrix and corrected for Poisson count noise using the makeContrast and voom functions of the R package limma (Ritchie et al. 2015), respectively. Finally, we identified DE genes between wide and thin striped individuals as well as between sections of dorsal skin within and outside the vertebral stripe using the eBayes function while correcting for multiple testing using the topTreat function and the Benjamini–Hockberg adjustment method.
Annotation of the Region of Interest
In order to determine the type of variant linked with the vertebral stripe pattern in Ptychadena robeensis, we annotated the region of chromosome 11 containing significant SNPs in the GWAS. We visually examined the transcripts obtained from our RNAseq data against the annotation for protein-coding sequences predicted by Augustus (Keller et al. 2011; Hariyani et al. in preparation) in the region using IGV (Robinson et al. 2011). Significant SNPs outputted by the GWAS were all located outside protein-coding regions, in between three genes: two predicted genes 40 kb apart were identified as ASIP and a third, 38 kb downstream, was identified as AHCY using MegaBlast (Johnson et al. 2008).
While a single ASIP gene is known in birds and mammals, two genes, ASIP1 and ASIP2 are present in teleost fish (Cortés et al. 2014; Cal et al. 2017). In Xenopus tropicalis, a single ASIP gene is predicted, but no focal study in amphibians has been conducted to date. In order to determine whether the two genes in P. robeensis correspond to ASIP, ASIP1, or ASIP2, we translated the genomic sequences and produced a protein alignment and maximum-likelihood phylogeny with the ASIP gene family in vertebrates using seaview (Galtier et al. 1996; fig. 3B and supplementary fig. S6A, Supplementary Material online).
Quantitative Real-time PCR Experiment
Quantitative real-time PCR (qPCR) was conducted on RNA extracted from dorsal skin within (n = 2) and outside (n = 2) the vertebral stripe in wide striped individuals, outside the vertebral stripe in thin-striped individuals (n = 2), and ventral skin (n = 1). Each reaction was triplicated to minimize the impact of experimental error. Two reference genes were selected using our RNAseq dataset with the following criteria: a minimum of 50 count per million in all samples, the lowest possible variance in expression level among samples and a minimum of two exons. Candidate reference genes were then checked for functional independence and compared to genes typically used for qPCR in Xenopus laevis. We retained rpl27 and abce1 as candidate reference genes.
Primers for ASIPb, rpl27, and abce1 were designed based on the Ptychadena robeensis reference genome and annotation using Primer3Plus (Untergasser et al. 2007; supplementary table S5, Supplementary Material online). We targeted PCR products spanning an exon–intron junction in order to avoid amplification of genomic DNA during the qPCR. The experiment was run using a StepOnePlus real-time PCR system and a Power SYBR Green RNA-to-CT 1-step kit (Applied Biosystems) on a volume of 20 µl. Results were analyzed using the R package pcr (Ahmed and Kim 2018). We compared the expression levels of rpl27 and abce1 across samples and retained rpl27 as reference gene as its expression level was more stable across the samples. Relative expression levels of ASIPb between our samples were calculated using the delta CT method and rpl27 as reference gene and dorsal skin outside the stripe as reference group as it should have the lowest level of ASIPb.
Supplementary Material
Acknowledgments
We would like to thank the Ethiopian Wildlife Conservation Authority and the Ethiopian Biodiversity Institute for providing us with collecting and export permits for the samples. Fieldwork in Ethiopia would not have been possible if not for the invaluable assistance of Megersa Kelbessa, Itbarek Kibret, and Samuel Woldeyes of Rock Hewn Tours. We also thank the important number of students and postdocs who collected Ptychadena specimens and samples over the years, and in particular Xenia Freilich, Jacobo Reyes-Velasco, Justin Wilcox, Sebastian Kirchhof and Marcin Falis. We are very thankful for the help from Marc Arnoux and Nizar Drou, from the Genome Core Facility and the Bioinformatics group at NYUAD. This research was carried out on the High-Performance Computing resources at New York University Abu Dhabi. We also thank David Howse and Sayel Daoud of The National Reference Laboratory and Rachid Rezgui from the Microscopy Core Facility at NYUAD for their help in producing and visualizing histological sections. This project was funded by NYUAD Grant AD180 to S.B. The NYUAD Sequencing Core is supported by NYUAD Research Institute grant G1205A to the NYUAD Center for Genomics and Systems Biology.
Contributor Information
Sandra Goutte, Division of Science, New York University Abu Dhabi, Saadiyat Island, Abu Dhabi, United Arab Emirates.
Imtiyaz Hariyani, Division of Science, New York University Abu Dhabi, Saadiyat Island, Abu Dhabi, United Arab Emirates.
Kole Deroy Utzinger, Division of Science, New York University Abu Dhabi, Saadiyat Island, Abu Dhabi, United Arab Emirates.
Yann Bourgeois, School of Biological Sciences, University of Portsmouth, Portsmouth, United Kingdom.
Stéphane Boissinot, Division of Science, New York University Abu Dhabi, Saadiyat Island, Abu Dhabi, United Arab Emirates.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
S.G. and S.B. designed the study. K.D.U. and S.G. collected color pattern and habitat data. S.G., Y.B., and S.B. collected Ptychadena spp. specimens and samples. S.G. extracted DNA and RNA from Ptychadena spp. samples and ran the qPCR. S.G. produced and interpreted histological section photographs and conducted comparative, genomic and gene expression analyses. I.H. and Y.B. provided help and advice on genomic and transcriptomic analyses. All authors read and contributed to the manuscript.
Competing Interests
The authors declare no competing interest.
Data Availability Statement
All sequences are publicly available in GenBank (accession numbers are given supplementary table S2, Supplementary Material online).
References
- Ahmed M, Kim DR. 2018. Pcr: an R package for quality assessment, analysis and testing of qPCR data. PeerJ. 6:e4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almathen F, Elbir H, Bahbahani H, Mwacharo J, Hanotte O. 2018. Polymorphisms in MC1R and ASIP genes are associated with coat color variation in the Arabian Camel. J Hered. 109:700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NK, Gutierrez SO, Bernal XE. 2019. From forest to city: urbanization modulates relative abundance of anti-predator coloration. J Urban Ecol. 5(1):1–8. [Google Scholar]
- R Core Team . 2020. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: http://www.R-project.org/. [Google Scholar]
- Braasch I, Postlethwait JH. 2011. The teleost agouti-related protein 2 gene is an ohnolog gone missing from the tetrapod genome. Proc Natl Acad Sci U S A. 108:E47–E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briolat ES, Burdfield-Steel ER, Paul SC, Rönkä KH, Seymoure BM, Stankowich T, Stuckert AMM. 2019. Diversity in warning coloration: selective paradox or the norm? Biol Rev. 94:388–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browder LW, Underhill JC, Merrell DJ. 1966. Mid-dorsal stripe in the wood frog. J Hered. 57:65–67. [DOI] [PubMed] [Google Scholar]
- Browning BL, Tian X, Zhou Y, Browning SR. 2021. Fast two-stage phasing of large-scale sequence data. Am J Hum Genet. 108:1880–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull CM. 1977. Black pattern polymorphism and tadpole growth rate in two Western Australian frogs. Aust J Zool. 25:243–248. [Google Scholar]
- Cal L, Suarez-Bregua P, Cerdá-Reverter JM, Braasch I, Rotllant J. 2017. Fish pigmentation and the melanocortin system. Comp Biochem Physiol A Mol Integr Physiol. 211:26–33. [DOI] [PubMed] [Google Scholar]
- Ceinos RM, Guillot R, Kelsh RN, Cerdá-Reverter JM, Rotllant J. 2015. Pigment patterns in adult fish result from superimposition of two largely independent pigmentation mechanisms. Pigment Cell Melanoma Res. 28:196–209. [DOI] [PubMed] [Google Scholar]
- Chen I-P, Stuart-Fox D, Hugall AF, Symonds MRE. 2012. Sexual selection and the evolution of complex color patterns in dragon lizards. Evolution 66:3605–3614. [DOI] [PubMed] [Google Scholar]
- Cortés R, Navarro S, Agulleiro MJ, Guillot R, García-Herranz V, Sánchez E, Cerdá-Reverter JM. 2014. Evolution of the melanocortin system. Gen Comp Endocrinol. 209:3–10. [DOI] [PubMed] [Google Scholar]
- Cott HB. 1940. Adaptive coloration in animals. London: Methuen & Co. [Google Scholar]
- Cuthill IC, Székely A. 2009. Coincident disruptive coloration. Philos Trans R Soc B Biol Sci. 364:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC. 2008. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr Rev. 66:S7–S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbeck DW, Underhill JC. 1971. Distribution of stripe polymorphism in wood frogs, Rana sylvatica LeConte, from Minnesota. Copeia. 1971:253–259. [Google Scholar]
- Freilich X, Tollis M, Boissinot S. 2014. Hiding in the highlands: evolution of a frog species complex of the genus Ptychadena in the Ethiopian highlands. Mol Phylogenet Evol. 71:157–169. [DOI] [PubMed] [Google Scholar]
- Frost DR. 2021. Amphibian Species of the World: An Online Reference. Version 6.1. Available from: https://amphibiansoftheworld.amnh.org/index.php.
- Fukuzawa T, Ide H. 1988. A ventrally localized inhibitor of melanization in Xenopus laevis skin. Dev Biol. 129:25–36. [DOI] [PubMed] [Google Scholar]
- Fukuzawa T, Samaraweera P, Mangano FT, Law JH, Bagnara JT. 1995. Evidence that MIF plays a role in the development of pigmentation patterns in the frog. Dev Biol. 167:148–158. [DOI] [PubMed] [Google Scholar]
- Galbraith DB. 1964. The agouti pigment pattern of the mouse: a quantitative and experimental study. J Exp Zool. 155:71–89. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. 1996. SEAVIEW And PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci CABIOS. 12:543–548. [DOI] [PubMed] [Google Scholar]
- Girardot M, Guibert S, Laforet M-P, Gallard Y, Larroque H, Oulmouden A. 2006. The insertion of a full-length Bos taurus LINE element is responsible for a transcriptional deregulation of the Normande Agouti gene. Pigment Cell Res. 19:346–355. [DOI] [PubMed] [Google Scholar]
- Godfrey D, Lythgoe JN, Rumball DA. 1987. Zebra stripes and tiger stripes: the spatial frequency distribution of the pattern compared to that of the background is significant in display and crypsis. Biol J Linn Soc. 32:427–433. [Google Scholar]
- Goutte S, Reyes-Velasco J, Freilich X, Kassie A, Boissinot S. 2021. Taxonomic revision of grass frogs (Ptychadenidae, Ptychadena) endemic to the Ethiopian highlands. ZooKeys 1016:77–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariyani I, Reyes-Velasco J, Goutte S, Boissinot S. in preparation. A chromosome-scale genome assembly for the Ethiopian Highland frog Ptychadena robeensis.
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313:101–104. [DOI] [PubMed] [Google Scholar]
- Hoffman EA, Blouin MS. 2000. A review of colour and pattern polymorphisms in anurans. Biol J Linn Soc. 70:633–665. [Google Scholar]
- Hubisz M, Siepel A. 2020. Inference of ancestral recombination graphs using ARGweaver. In: Dutheil JY, editor. Statistical population genomics. Methods in molecular biology. New York, NY: Springer US. p. 231–266. [DOI] [PubMed] [Google Scholar]
- Hurtado-Gonzales JL, Uy JAC. 2009. Alternative mating strategies may favour the persistence of a genetically based colour polymorphism in a pentamorphic fish. Anim Behav. 77:1187–1194. [Google Scholar]
- Jetz W, Pyron RA. 2018. The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nat Ecol Evol. 2:850–858. [DOI] [PubMed] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Mills LS, Alves PC, Callahan CM, Alves JM, Lafferty DJR, Jiggins FM, Jensen JD, Melo-Ferreira J, Good JM. 2018. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360(6395):1355–1358. [DOI] [PubMed] [Google Scholar]
- Jones MR, Mills LS, Jensen JD, Good JM. 2020. Convergent evolution of seasonal camouflage in response to reduced snow cover across the snowshoe hare range. Evolution 74:2033–2045. [DOI] [PubMed] [Google Scholar]
- Keller O, Kollmar M, Stanke M, Waack S. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27:757–763. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil CF, Liang Y, Gerwin J, Woltering JM, Urban S, Henning F, Machado-Schiaffino G, Hulsey CD, Meyer A. 2018. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362:457–460. [DOI] [PubMed] [Google Scholar]
- Liang Y, Grauvogl M, Meyer A, Kratochwil CF. 2021. Functional conservation and divergence of color-pattern-related agouti family genes in teleost fishes. J Exp Zool B Mol Dev Evol. 336:443–450. [DOI] [PubMed] [Google Scholar]
- Liang D, Zhao P, Si J, Fang L, Pairo-Castineira E, Hu X, Xu Q, Hou Y, Gong Y, Liang Z, et al. 2020. Genomic analysis revealed a convergent evolution of LINE-1 in coat color: a case study in water buffaloes (Bubalus bubalis). Mol Biol Evol. 38:1122–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy MT. 2016. Teasing apart crypsis and aposematism – evidence that disruptive coloration reduces predation on a noxious toad. Biol J Linn Soc. 117:285–294. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160:1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills MG, Patterson LB. 2009. Not just black and white: pigment pattern development and evolution in vertebrates. Semin Cell Dev Biol. 20:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris BJ, Whan VA. 2008. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res. 18:1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EM, Beard KH. 2010. Genetic basis of a color pattern polymorphism in the coqui frog Eleutherodactylus coqui. J Hered. 101:703–709. [DOI] [PubMed] [Google Scholar]
- Osorio D, Srinivasan MV, Pettigrew JD. 1991. Camouflage by edge enhancement in animal coloration patterns and its implications for visual mechanisms. Proc R Soc Lond B Biol Sci. 244:81–85. [DOI] [PubMed] [Google Scholar]
- Otaki JM. 2008. Physiological side-effect model for diversification of non-functional or neutral traits: a possible evolutionary history of Vanessa butterflies (Lepidoptera, Nymphalidae). Lepidoptera Sci. 59:87–102. [Google Scholar]
- Pérez i de Lanuza G, Font E, Carazo P. 2013. Color-assortative mating in a color-polymorphic lacertid lizard. Behav Ecol. 24:273–279. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O’Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyburn WF. 1961. Inheritance of the green vertebral stripe in Acris crepitans. Southwest Nat. 6:164–167. [Google Scholar]
- Revell LJ. 2014. Ancestral character estimation under the threshold model from quantitative genetics. Evolution 68:743–759. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas B. 2017. Behavioural, ecological, and evolutionary aspects of diversity in frog colour patterns. Biol Rev. 92:1059–1080. [DOI] [PubMed] [Google Scholar]
- Rowland HM, Cuthill IC, Harvey IF, Speed MP, Ruxton GD. 2008. Can’t tell the caterpillars from the trees: countershading enhances survival in a woodland. Proc R Soc B Biol Sci. 275:2539–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruxton GD, Allen WL, Sherratt TN, Speed MP. 2018. Avoiding attack: the evolutionary ecology of crypsis, aposematism, and mimicry. Oxford: Oxford University Press. [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler FW, Cook FR. 1980. Distribution of the middorsal stripe dimorphism in the wood frog, Rana sylvatica, in eastern North America. Can J Zool. 58:1643–1651. [Google Scholar]
- Siewert KM, Voight BF. 2017. Detecting long-term balancing selection using allele frequency correlation. Mol Biol Evol. 34:2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert KM, Voight BF. 2020. Betascan2: standardized statistics to detect balancing selection utilizing substitution data. Genome Biol Evol. 12:3873–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siracusa LD. 1994. The agouti gene: turned on to yellow. Trends Genet. 10:423–428. [DOI] [PubMed] [Google Scholar]
- Skelhorn J, Rowland HM, Speed MP, Ruxton GD. 2010. Masquerade: camouflage without crypsis. Science 327:51–51. [DOI] [PubMed] [Google Scholar]
- Slominski A, Kim T-K, Brożyna AA, Janjetovic Z, Brooks DLP, Schwab LP, Skobowiat C, Jóźwicki W, Seagroves TN. 2014. The role of melanogenesis in regulation of melanoma behavior: melanogenesis leads to stimulation of HIF-1α expression and HIF-dependent attendant pathways. Arch Biochem Biophys. 563:79–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M, Cuthill IC. 2006. Disruptive coloration, crypsis and edge detection in early visual processing. Proc R Soc B Biol Sci. 273:2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MM. 1974. Parallel pattern polymorphism in the genus Phrynobatrachus (Amphibia: Ranidae). Copeia 1974:823–832. [Google Scholar]
- Sviderskaya EV, Hill SP, Balachandar D, Barsh GS, Bennett DC. 2001. Agouti signaling protein and other factors modulating differentiation and proliferation of immortal melanoblasts. Dev Dyn Off Publ Am Assoc Anat. 221:373–379. [DOI] [PubMed] [Google Scholar]
- Tarkhnishvili D, Gokhelashvili R. 1996. A contribution to the ecological genetics of frogs: age structure and frequency of striped specimens in some Caucasian populations of the Rana macrocnemis complex. Alytes 14:27–41. [Google Scholar]
- Terhorst J, Kamm JA, Song YS. 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 49:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD. 2014. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv:005165.
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolbright LL, Stewart MM. 2008. Spatial and temporal variation in color pattern morphology in the tropical frog, Eleutherodactylus coqui. Copeia 2008:431–437. [Google Scholar]
- Ye X, Zhou L, Jia J, Wei L, Wen Y, Yan X, Huang J, Gan B, Liu K, Lv Y, et al. 2020. ITRAQ Proteomic analysis of yellow and black skin in Jinbian Carp (Cyprinus carpio). Life 10(10):226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Gogarten S, Laurie C, Weir B. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Yu L, Kwek BZW, Jin G, Zeng H, Li D. 2021. Sexual selection on jumping spider color pattern: investigation with a new quantitative approach. Behav Ecol. 32(4):695–706. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences are publicly available in GenBank (accession numbers are given supplementary table S2, Supplementary Material online).