Abstract
The pyrimidine nucleotide biosynthesis (pyr) operon in Bacillus subtilis is regulated by transcriptional attenuation. The PyrR protein binds in a uridine nucleotide-dependent manner to three attenuation sites at the 5′-end of pyr mRNA. PyrR binds an RNA-binding loop, allowing a terminator hairpin to form and repressing the downstream genes. The binding of PyrR to defined RNA molecules was characterized by a gel mobility shift assay. Titration indicated that PyrR binds RNA in an equimolar ratio. PyrR bound more tightly to the binding loops from the second (BL2 RNA) and third (BL3 RNA) attenuation sites than to the binding loop from the first (BL1 RNA) attenuation site. PyrR bound BL2 RNA 4–5-fold tighter in the presence of saturating UMP or UDP and 150- fold tighter with saturating UTP, suggesting that UTP is the more important co-regulator. The minimal RNA that bound tightly to PyrR was 28 nt long. Thirty-one structural variants of BL2 RNA were tested for PyrR binding affinity. Two highly conserved regions of the RNA, the terminal loop and top of the upper stem and a purine-rich internal bulge and the base pairs below it, were crucial for tight binding. Conserved elements of RNA secondary structure were also required for tight binding. PyrR protected conserved areas of the binding loop in hydroxyl radical footprinting experiments. PyrR likely recognizes conserved RNA sequences, but only if they are properly positioned in the correct secondary structure.
INTRODUCTION
The pyrimidine nucleotide biosynthesis (pyr) operon in Bacillus subtilis contains 10 cistrons. The first gene in the operon encodes PyrR, which has been shown to be the regulatory protein for the operon (1). The second gene in the operon encodes PyrP, which is a uracil permease. The remaining eight cistrons encode the six enzymes necessary for de novo biosynthesis of UMP (1–3). Regulation of the operon occurs through a transcriptional attenuation mechanism in which PyrR promotes transcriptional termination at three attenuation regions in the operon when uridine nucleotide levels are high (1,4). These attenuation regions are located in the 5′-leader region (binding loop 1, BL1), the pyrR–pyrP intercistronic region (BL2) and the pyrP–pyrB intercistronic region (BL3) of pyr mRNA.
Previous research led to a model in which high levels of UMP stimulate PyrR to bind to a conserved sequence and secondary structure in the mRNA in each attenuation region called the anti-antiterminator or the binding loop (5,6). Binding of PyrR to this sequence prevents the formation of an antiterminator stem–loop and allows a downstream ρ-independent transcription terminator to form, reducing expression of downstream genes. When UMP levels are low, the RNA-binding affinity of PyrR is reduced and the more stable antiterminator stem–loop is favored, leading to transcriptional read-through and expression of the downstream genes. This constitutes a feedback inhibition loop wherein accumulation of UMP, the direct product of the pathway, results in reduced expression of the genes responsible for its synthesis. Results presented in this work demonstrate that UDP and UTP also affect the RNA-binding affinity of PyrR and that UTP is likely the more physiologically important cofactor, rather than UMP as previously supposed. For a recent review of studies of attenuation control of the B.subtilis pyr operon see Switzer et al. (6).
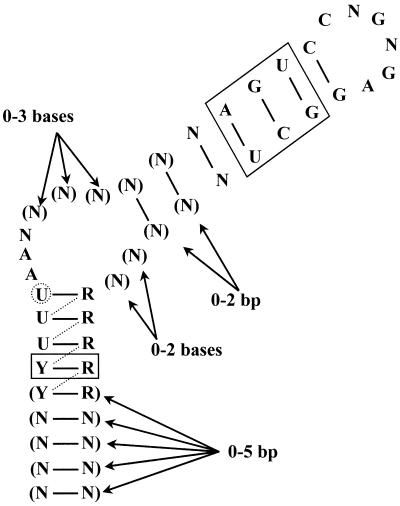
Bacillus subtilis is not the only organism to utilize this form of transcriptional attenuation to regulate its pyr genes. Homologous regulatory systems have been found in Bacillus caldolyticus (7), Enterococcus faecalis (8), Lactobacillus plantarum (9) and Lactococcus lactis (10–12). It appears from examination of genome sequences that PyrR-dependent regulation of pyr genes is found in many other bacterial species as well (6). All PyrR-binding loops from the various species contain the highly conserved sequence motif 5′-ARUCCNGNGAGGYU-3′. The computer program MFOLD v.3.1 (13,14) was used to predict possible secondary structures in the various binding loop sequences. Each binding loop could fold into a similar structure, comprised of a terminal 6 nt loop, an upper stem, a purine-rich internal bulge (with 3–6 nt on the 5′-side of the bulge and 0–2 nt on the 3′-side) and a lower stem (Fig. 1). The conserved sequence motif is always located in the same position, such that the nucleotides CNGNGA form the terminal hexaloop. We noticed that the terminal hexaloop could potentially fold into a GNRA tetraloop structure in which the first two bases of the terminal loop would be ‘flipped out’ of the loop. A precedent for this type of behavior was found by Legault et al. (15), who discovered that the λ phage boxB RNA hairpin, which binds to the transcriptional antitermination protein N, has a GAAGA pentaloop that is folded into a GNRA tetraloop with the fourth base of the pentaloop ‘flipped out’ of the structure. A second example was described by Cai et al. (16) in the boxB RNA hairpin of phage P22, in which a terminal pentaloop is converted to a GNRA-like tetraloop by ‘flipping out’ the second base of the pentaloop.
Figure 1.
Consensus PyrR-binding loop. The consensus sequence and secondary structure for the 20 known PyrR-binding loops from B.caldolyticus, B.subtilis, E.faecalis, L.lactis, Lactobacillus leichmanii, L.plantarum, Streptococcus pneumoniae, Streptococcus pyogenes and Thermus ZO5 are shown. R = A or G; Y = C or U; N = any nucleotide. Parentheses indicate bases that are present in some sequences. The secondary structure pictured is formed in 12 of the 20 known PyrR-binding loops; in the other eight, the uridine nucleotide in the dashed circle initiates the purine-rich internal bulge, with the resulting base pairs indicated by dashed lines. The base pairs shown in solid boxes are conserved in 18 of the 20 examples, except for the A-U base pair in the upper stem, which is replaced by other base pairs in seven of the 20 examples.
A second shorter conserved sequence motif was discovered when the folded binding loops were compared. This second motif, 5′-UUUAA-3′, is positioned so that one of the two underlined nucleotides initiates the purine-rich internal bulge. Additionally, the region of the lower stem immediately below the purine-rich internal bulge is pyrimidine-rich on the 5′-side and purine-rich on the 3′-side. The conservation of both primary and secondary structure in PyrR-binding mRNAs suggests that PyrR recognizes conserved RNA sequence motifs in the context of a particular RNA secondary/tertiary structure. To determine how PyrR recognizes its RNA target, RNA binding specificity and hydroxyl radical footprinting experiments were undertaken. It was found that the most highly conserved areas of the mRNA, which include the terminal loop, the top of the upper stem, the purine-rich internal bulge and the base pairs immediately below the purine-rich internal bulge, were critical for PyrR binding and were protected from hydroxyl radical cleavage in the presence of PyrR.
MATERIALS AND METHODS
Bacterial strains, plasmids, media and growth
Escherichia coli strains DH5αMCR (Life Technologies, Bethesda, MD) and TG1 (17) were used for cloning, propagation and purification of plasmids. LB broth and agar (18) were used for the growth of cultures. When cells harboring plasmids were grown, the medium was supplemented with 100 µg/ml ampicillin. All liquid cultures were grown aerobically at 37°C.
Recombinant DNA techniques and DNA sequencing
Restriction enzymes were from either Life Technologies or New England Biolabs (Beverly, MA). T4 DNA ligase was from Life Technologies. Klenow (exo–) fragment was from New England Biolabs. Plasmid DNA was isolated using the standard alkaline lysis technique (19) and purified using a QiaPrep Spin Kit or Qiagen Mini Kit (Qiagen, Valencia, CA). DNA was purified from agarose gel slices using gel extraction kits from Qiagen (either a QiaQuick or Qiaex II kit). DNA was sequenced at the W. M. Keck Center for Comparative and Functional Genomics, High Throughput Sequencing and Genotyping Unit at the University of Illinois.
Construction of templates for in vitro transcription
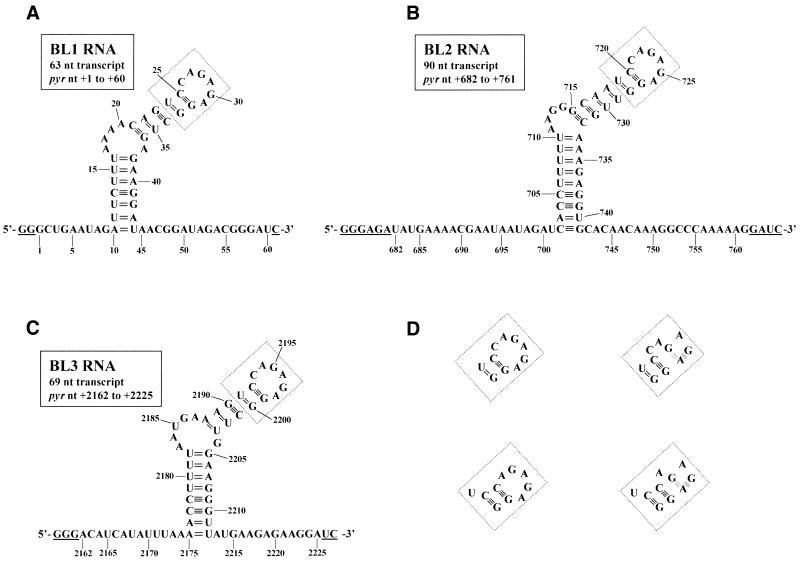
The construction of pBSBL2, which was used to transcribe BL2, a 90 nt RNA that contains the binding loop and some 5′- (20 pyr nt) and 3′-flanking (20 pyr nt) nucleotides from the second attenuation region in B.subtilis (nt +682 to +761, where +1 denotes the transcriptional start site) has been described previously (20). Templates pBSBL1 and pBSBL3, which were used to transcribe RNAs that contain the binding loops and some 5′- and 3′-flanking nucleotides from the first (BL1, 63 nt long RNA, nt +1 to +60 with 9 flanking pyr nt on the 5′-side and 15 flanking pyr nt on the 3′-side) and third (BL3, 69 nt long RNA, nt +2162 to +2225 with 13 flanking pyr nt on the 5′-side and, 13 flanking pyr nt on the 3′-side) attenuation regions, respectively, in B.subtilis were constructed as follows. For each template, two DNA oligonucleotides were annealed. One DNA oligonucleotide, called EcoT7, used for both templates, had the sequence 5′-CGGAATTCTAATACGACTCACTATAGGG-3′ (EcoRI site in bold, bacteriophage T7 RNA polymerase promoter underlined). For pBSBL1, the second DNA oligonucleotide had the sequence 5′-CGGGATCCCGTCTATCCGTTATCCTTCTCAGCCTCTCTGGACTGTTTTAAAGAATCTATTCAGCCCTATAGTGAGTCGTATTAGAATTCCG-3′ (BamHI site in bold, sequence complementary to EcoT7 oligonucleotide underlined). For pBSBL3, the second DNA oligonucleotide had the sequence 5′-CGGGATCCTTCTCTTCATAACCCTTCCAAGCCTCTCTGGACTTTCATTAAAAGGTTTTAAATATGATGTCCCTATAGTGAGTCGTATTAGAATTCCG-3′ (BamHI site in bold, sequence complementary to EcoT7 oligonucleotide underlined). Annealing was done by combining 100 pmol of each oligonucleotide and heating at 75°C for 15 min, then slow cooling. The overhanging regions were filled in using 5 U of Klenow (exo–) enzyme and 20 nmol each dNTP in a final volume of 25 µl. The full-length products were purified from an agarose gel, digested with EcoRI and BamHI and ligated into similarly digested pUC18 to create the desired vectors. The presence of the proper insert was confirmed by DNA sequencing. The vectors were linearized with BamHI prior to inclusion in a transcription reaction. The exact nucleotide sequences of BL1, BL2 and BL3 are shown in Figure 2.
Figure 2.
Nucleotide sequences and predicted secondary structure of pyr mRNA BL1, BL2 and BL3. The nucleotide sequences and MFOLD v.3.1 predicted secondary structures for the transcripts from pBSBL1/BamHI (A), pBSBL2/BamHI (B) and pBSBL3/BamHI (C) are shown. Numbers denote the pyr nucleotide number, where +1 is the transcriptional start site for the operon (2). The underlined nucleotides are not pyr sequences but are found in the transcript as a result of the method used to obtain T7 RNA polymerase transcripts of defined sequence. The boxed region in each panel could fold in several plausible ways, which are shown in (D). When the hexaloop/pentaloop is folded into the putative GNRA tetraloop structure, the double dashed line represents a reverse Hoogsteen G-A base pair and the shaded rectangle represents purine-adenosine stacking.
A similar strategy was used to create the various sequence variants of pBSBL2. For each construct, two DNA oligonucleotides were annealed, as described above for the construction of pBSBL1 and pBSBL3. One DNA oligonucleotide, called EcoT7pyr, was used for all templates, with the sequence 5′-CGGAATTCTAATACGACTCACTATAGGGTATGAAAACGAATAATAGAT-3′ (EcoRI site in bold, bacteriophage T7 RNA polymerase promoter underlined, and the sequence annealing to the second oligonucleotide in italic). The second oligonucleotide used for each sequence variant contained a BamHI site, followed by the sequence complementary to the desired RNA transcript. As an example, the second DNA oligonucleotide for the sequence variant U719C had the sequence 5′-CGGGATCCTTTTTGGGCCTTTGTTGTGCACCTCTTTGCAACCTCTCTGGGTTGCCCTTAAAAAGG-TGATCTATTATTCGTTTTCATACCC-3′ (BamHI site in bold, location of the single nucleotide substitution underlined, and the sequence annealing to the EcoT7pyr oligonucleotide in italic). For each sequence variant, the sequence of the second DNA oligonucleotide was altered to contain the desired changes. After annealing the oligonucleotides, the rest of the procedure was performed as noted above for the construction of pBSBL1 and pBSBL3. Each template was checked by DNA sequencing. MFOLD v.3.1 was used to ensure that each BL2 RNA sequence variant would have no unintended secondary structure changes.
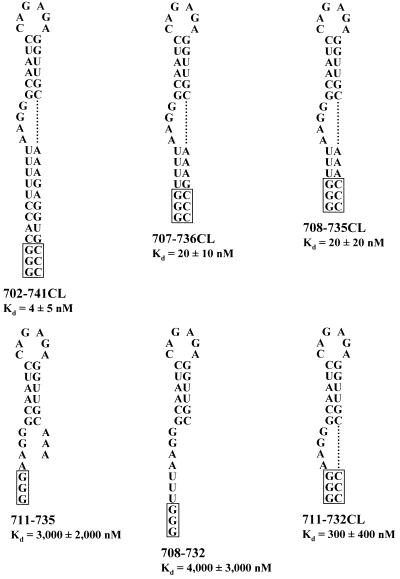
Templates for determination of the minimal RNA that binds to PyrR were constructed by annealing the ‘T7only’ oligonucleotide, 5′-TAATACGACTCACTATA-3′, to various ‘bottom strand’ oligonucleotides. Each bottom strand oligonucleotide contained the complement to the desired transcript at its 5′-end, followed by three C residues to allow for a 5′-GGG T7 RNA polymerase transcription start and, finally, the sequence complementary to the T7only oligonucleotide at its 3′-end. Many of the templates were designed with a ‘clamp’ in which the bottom strand oligonucleotide would originate with 5′-GGG, placing three C residues at the 3′-end of the transcript in the proper position to base pair to the three G residues at the 5′-end of the transcript. However, when the oligonucleotides were annealed and used directly as transcription templates as suggested by Milligan and Uhlenbeck (21), we observed an excessive amount of abortive transcription. We guessed that this was due to the single-stranded portion of the DNA template forming a hairpin-like secondary structure that could cause T7 RNA polymerase to stall during transcription, and eventually fall off the template due to the low concentration of ATP in the transcription reaction. To avoid this problem, Klenow (exo–) enzyme was used as previously described to fill in the single-stranded region and create a series of double-stranded DNA templates, which were less prone to abortive transcription. The RNAs used to determine the minimal sequence necessary for tight PyrR binding are shown in Figure 3.
Figure 3.
RNA sequences used to determine the minimal BL2 RNA that binds to PyrR. RNA sequences used to determine the minimal RNA necessary for PyrR binding are shown. Boxed regions denote nucleotides not found in the native pyr transcript. BL2 RNA from pBSBL2/BamHI has already been described (Fig. 2B). Shorter RNAs are 702–741CL, 707–736CL, 708–735CL, 711–735, 708–732 and 711–732CL, where CL indicates that three C nucleotides were added to the 3′-end of the transcript to base pair to the three G nucleotides at the 5′-end of the transcript.
RNA for hydroxyl radical footprinting and nuclease digestion experiments was prepared from templates pFIR1, pBSBL2 and pTHI1. The construction of pBSBL2 has already been described. pFIR1 and pTHI1 were constructed as follows. pFIR1 was constructed by cloning the 0.22 kb XmnI fragment from pTSE230 (R.J.Turner and R.L.Switzer, unpublished data) into the SmaI site of pSP72 (Promega, Madison, WI). pTHI1 was constructed by cloning the 0.31 kb DraI–BamHI fragment of pTSSP3 (R.J.Turner and R.L.Switzer, unpublished data) into SmaI + BamHI-digested pSP72. Prior to inclusion in in vitro transcription reactions, pFIR1 was linearized with BssHII to specify RNA corresponding to nt +9 to +75 and pTHI1 was linearized with HindIII to specify RNA corresponding to nt +2173 to +2229.
In vitro transcription and purification of RNA for gel mobility shift assays
RNA for electrophoretic gel mobility shift assays was produced by in vitro transcription and purified following denaturing acrylamide gel electrophoresis as described by Turner et al.(20), with the following exceptions. In some cases the ethanol precipitation step was replaced by dialyzing the RNA against RNase-free TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA) using Slide-A-Lyzer cassettes (Pierce Chemical Co., Rockford, IL), which gave a reproducibly higher yield of RNA. Another exception was for the titration experiment to determine the stoichiometry of the PyrR–pyr mRNA interaction, which required a much higher RNA concentration. This reaction was performed with undiluted unlabeled nucleotides to obtain a higher yield of RNA. This purified RNA was diluted to a stock concentration of 400 nM and used at a concentration of 100 nM RNA for gel mobility shift assays. A final exception was for the gel mobility shift assays to determine the dissociation constant for the BL2–PyrR interaction in the presence of saturating levels of UTP. As will be discussed later, this interaction was found to have a very tight binding constant, requiring PyrR concentrations so low that reaction concentrations of 1 pM RNA (instead of 50 pM) were needed in order to avoid using sub-stoichiometric levels of PyrR. To obtain a comparable number of counts per reaction at this lower RNA concentration, the transcription reactions for BL2 RNA used for these assays were modified to include 5 µl of [α-32P]ATP (3000 Ci/mmol; ICN, Costa Mesa, CA), 5 µl of [α-32P]ATP (800 Ci/mmol; ICN) and no unlabeled ATP.
The molar concentration of purified RNA was determined as follows in all cases. A 2 µl aliquot of the 20 µl transcription reaction was removed prior to electrophoresis, serially diluted and subjected to liquid scintillation counting to determine the total radioactivity present in the transcription reaction. Duplicate aliquots of purified RNA were also subjected to liquid scintillation counting; a ‘percent yield’ for the transcription reaction was calculated by comparing the radioactivity present in purified RNA to the radioactivity present in the transcription reaction. As the total amount of ATP in each transcription reaction was known, the ‘percent yield’ was used to determine the number of moles of ATP incorporated into purified RNA. The number of moles of purified RNA was determined by dividing the moles of ATP incorporated by the number of adenosine residues present in a given transcript; dividing this quantity by the volume of purified RNA gave the molar RNA concentration.
In vitro transcription and purification of RNA for hydroxyl radical footprinting
RNA for use in hydroxyl radical footprinting experiments was synthesized using the MAXIscript T7 kit from Ambion (Austin, TX) with the linearized DNA templates described above. Plasmids were transcribed using T7 RNA polymerase and a 5:1 ratio of guanosine to GTP, allowing most transcripts to possess a free 5′-OH group. Transcription products were 5′-end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (7000 Ci/mmol; ICN) for 30 min at 37°C. Reaction products were separated by electrophoresis at 200 V on a 5% polyacrylamide, 7 M urea gel. Following autoradiography, full-length mRNA was excised from the gel, passively eluted into elution buffer (0.1% SDS, 1 mM EDTA, 5 mM ammonium acetate), extracted twice with acid phenol:chloroform (5:1, pH 4.7), precipitated with 0.1 vol 5 M ammonium acetate and 3 vol absolute ethanol and resuspended in 20 µl of sterile H2O. The radioactivity of the mRNA was measured by liquid scintillation counting and the mRNA was diluted to an activity of 100 000 c.p.m./µl.
Electrophoretic gel mobility shift assay to measure PyrR–pyr mRNA interaction
This assay was performed using purified PyrR protein, essentially as described in Turner et al. (20), with several significant modifications for this study. The concentration of PyrR was determined using the Bradford protein assay (22) with reagents purchased from Pierce. As previously described in Turner et al. (20), this method agreed well with the method of Gill and von Hippel (23), in which protein concentration was determined based on a molar extinction coefficient calculated based on the amino acid content of the protein. To minimize any potential problems with protein stability (highly diluted B.subtilis PyrR is necessary for the gel mobility shift assay), freshly thawed aliquots of PyrR were immediately diluted and incorporated into the binding reaction mix. The final step in the PyrR purification protocol was changed so that PyrR was stored in 100 mM Tris–acetate pH 7.5, 10 mM potassium acetate, 20% glycerol buffer. The PyrR dilution buffer for gel mobility shift assays was identical to the new PyrR storage buffer except that 1 mg/ml RNase-free acetylated bovine serum albumin (Ambion) was also included. PyrR purified and diluted in this way gave very reproducible binding data and no fraction detectable on immunoblots was unable to bind RNA. Each binding reaction was set up using 10 µl of ‘binding mix’, 5 µl of appropriately diluted PyrR and 5 µl of RNA to give final reaction concentrations of 25 mM Tris–acetate pH 7.5, 2.5 mM Tris–HCl pH 8.0, 50 mM potassium acetate, 1 mM magnesium acetate, 0.1 mg/ml yeast RNA, 5 µg/ml heparin, 0.01% Igepal CA-630, 0.08 U/µl RNase inhibitor (Ambion), 0.25 mM EDTA, 5% glycerol and appropriate amounts of the desired co-effector molecules.
All gel mobility shift experiments included a lane in which PyrR was omitted. This ‘RNA-only’ lane was used to determine the cut-off between what was counted as free RNA and what was counted as PyrR–RNA complex. All species with a slower mobility than the RNA-only species (normally shifted species plus ‘smeared’ and ‘super-shifted’ species) were counted as PyrR–RNA complex for quantitation. Data were fitted to regular hyperbolic binding curves using the Solver function of Microsoft Excel 97. The apparent dissociation constant was defined as the PyrR concentration required to cause half-maximal PyrR–RNA complex formation.
‘Titration’ gel mobility shift experiments and western blotting
To determine the stoichiometry of the PyrR–RNA interaction, gel mobility shift experiments were done in which the BL2 RNA concentration was held constant at 100 nM, well above the observed dissociation constant, and the PyrR concentration was varied to determine the [PyrR]/[RNA] ratio at which binding became saturated. Concentrations of PyrR and RNA were determined as previously described. To test if all of the PyrR was active and able to bind RNA, these experiments were done in duplicate, meaning that identical reactions were loaded on both sides of the gel. After electrophoresis, the gel was cut to separate the two halves. One half was subjected to autoradiography and phosphorimager analysis as usual, while the other half was used to detect PyrR using western blotting as follows. Protein was transferred to polyvinylidene difluoride (PVDF) membrane (Midwest Scientific, Valley Park, MO) using a BioRad (Hercules, CA) TransBlot SD semi-dry electroblotting unit at 25 V, 0.6 A initial current for 1 h. A transfer stack consisting of three sheets of filter paper, PVDF membrane, gel and three sheets of filter paper was used; all components were equilibrated in transfer buffer (48 mM Tris buffer, 39 mM glycine, 20% methanol). The membrane was equilibrated in blocking solution (2% non-fat dry milk in phosphate-buffered saline) for 1 h and a 1/5000 dilution of primary antibody (rabbit anti-PyrR, prepared at the Immunological Resource Center at the University of Illinois, Urbana-Champaign, IL) was added for 1 h. The membrane was washed three times with phosphate-buffered saline containing 0.05% Tween-20, followed by a 1 h incubation in blocking solution containing a 1/20 000 dilution of secondary antibody (goat anti-rabbit IgG conjugated to horseradish peroxidase; Pierce). The membrane was washed three times with phosphate-buffered saline containing 0.05% Tween-20, followed by three washes in phosphate-buffered saline. PyrR was detected using a SuperSignal kit (Pierce), according to the manufacturer’s suggested protocol. The membrane was placed in a plastic sheet protector and exposed to Hyperfilm ECL film (Amersham International, Little Chalfont, UK) for 1 min.
Hydroxyl radical footprinting
Hydroxyl radical footprinting followed the procedure of Tullius and Dombroski (24), with modifications. Diluted PyrR (25 pmol in 1 µl; PyrR dilution buffer was 10 mM Tris–acetate, pH 7.5, 1 mg/ml RNase-free acetylated bovine serum albumin) was mixed with 18 µl of binding solution (10 mM Tris–acetate, pH 7.5, 10 mM MgCl2, 100 mM potassium glutamate) and diluted radiolabeled RNA (5 pmol in a volume of 1 µl) was added. When necessary, UMP was included as part of the binding solution to give a reaction concentration of 560 µM. The reagents to produce hydroxyl radicals [1 µl each of 50 mM Fe(NH4)2(SO4)2, 0.1 M EDTA, 1% H2O2 and 0.25 M DTT] were added to the side of the microcentrifuge tube, but not in contact with the binding reaction. The reaction was incubated at 4°C for 15 min and then the tube was centrifuged to collect the fluid at the bottom. The Fe–EDTA reactions were allowed to proceed for 1 min, then 100 µl of 0.1 M thiourea was added to stop the reactions. The reactions were extracted with acid phenol:chloroform and precipitated with 0.1 vol of 5 M ammonium acetate and 3 vol of absolute ethanol. The precipitate was resuspended in 3 µl of gel loading buffer (95% formamide, 0.5 mM EDTA, 0.025% SDS, 0.025% xylene cyanol, 0.025% bromophenol blue), loaded on an 8% acrylamide, 7 M urea sequencing gel and electrophoresed at 40 mA constant current.
Nuclease digestion and partial alkaline hydrolysis
RNase T1 digestion and partial alkaline hydrolysis of the mRNA were performed as described by Donis-Keller et al. (25). The method used for RNase I and RNase V1 digestion of the mRNA was as follows. Diluted PyrR (25 pmol in 1 µl), or PyrR dilution buffer for controls without PyrR, was added to 18 µl of binding solution (as for the hydroxyl radical footprinting experiments) and radiolabeled RNA (5 pmol in 1 µl). The reaction was incubated at room temperature for 10 min, after which appropriately diluted nuclease was added, followed by another 10 min incubation at room temperature. The mRNA was extracted with acid phenol:chloroform, precipitated with ethanol, resuspended, and analyzed by electrophoresis as previously described for hydroxyl radical footprinting experiments.
RESULTS
Determination of apparent dissociation constants for PyrR–pyr mRNA interaction
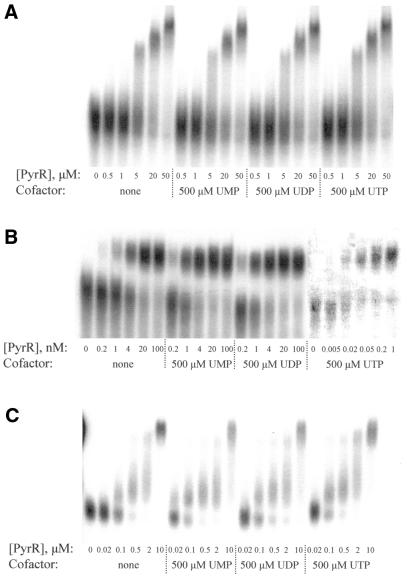
The relative affinity of B.subtilis PyrR protein for pyr mRNA from each of the three attenuation regions was determined by electrophoretic gel mobility shift assays. 32P-labeled RNA corresponding to the three pyr binding loop sequences (BL1, BL2 and BL3; Fig. 2) was held at a constant concentration of 50 pM (or 1 pM to determine the apparent dissociation constant for the BL2–PyrR interaction in the presence of saturating UTP; see Materials and Methods) and the percentage of RNA bound to increasing amounts of PyrR was determined. Apparent dissociation constants for BL1, BL2 and BL3 binding to PyrR in the absence of nucleotides and in the presence of saturating levels of UMP, UDP and UTP are listed in Table 1. Representative examples of gel mobility shift experiments for BL1, BL2 and BL3 binding to PyrR are shown in Figure 4.
Table 1. Apparent dissociation constants for PyrR–RNA interactions in the presence or absence of uridine nucleotide cofactors.
| RNA | Cofactor | Kd (nM) |
|---|---|---|
| BL1 | None | 10 000 ± 2000 (n = 3)a |
| BL1 | 500 µM UMP | 16 000 ± 1000 (n = 2) |
| BL1 | 500 µM UDP | 13 000 ± 2000 (n = 3) |
| BL1 | 500 µM UTP | 12 000 ± 1000 (n = 2) |
| BL2 | None | 3 ± 1 (n = 3) |
| BL2 | 500 µM UMP | 0.7 ± 0.4 (n = 16) |
| BL2 | 500 µM UDP | 0.6 ± 0.1 (n = 3) |
| BL2 | 500 µM UTP | 0.02 ± 0.006 (n = 3) |
| BL3 | None | 200 ± 100 (n = 4) |
| BL3 | 500 µM UMP | 100 ± 50 (n = 4) |
| BL3 | 500 µM UDP | 70 ± 30 (n = 4) |
| BL3 | 500 µM UTP | 60 ± 30 (n = 4) |
an, number of determinations.
Figure 4.
Apparent dissociation constants for the interaction of PyrR with BL1, BL2 and BL3 RNA were determined in the presence of no cofactor, 500 µM UMP, 500 µM UDP or 500 µM UTP; these constants are listed in Table 1. Representative gel mobility shift experiments for BL1 (A), BL2 (B) and BL3 (C) are shown here. All reactions contained 50 pM of the indicated RNA, except for the PyrR–BL2 binding reactions containing 500 µM UTP; these reactions used 1 pM BL2 RNA due to the extremely low PyrR concentrations needed to determine the apparent dissociation constant. As these reactions contained much less radioactivity than all other BL2 reactions, they were scanned separately to improve visibility. ‘Super-shifted’ bands were observed with all RNA species when the PyrR concentration was ≥5 µM (evident here in the BL1 and BL3 reactions); this phenomenon is discussed in the text. ‘Smeared’ RNA between the free RNA and PyrR–RNA species was counted as bound RNA, as was ‘super-shifted’ RNA, for calculation of apparent dissociation constants.
The binding of BL1 to PyrR appeared to be much weaker than for BL2 and BL3 in gel mobility shift analysis and did not respond to uridine nucleotide levels. The binding of BL3 to PyrR was weaker than for BL2, but stronger than BL1, and showed an intermediate response to uridine nucleotide levels, lacking the strong response of BL2 to UTP. These results are probably due to a more rapid dissociation of the BL1–PyrR and BL3–PyrR complexes than the BL2–PyrR complex, leading to dissociation of the complex during electrophoresis. Evidence for this suggestion is given by a ‘smeared’ appearance between the free and bound RNA species in gel mobility shift assays for both BL1 and BL3, whereas for BL2 the two species are well separated with no smearing. Dissociation during electrophoresis would mask the true strength of these interactions, as well as possibly obscuring any potential effect of uridine nucleotide ligands. ‘Smeared’ RNA was counted as bound RNA in the analysis, but RNA that dissociates very quickly relative to the time of electrophoresis (3 h) would not be counted. However, for the well-behaved BL2–PyrR interaction, UMP and UDP increase binding affinity by 4–5-fold and UTP by 150-fold. The large effect of UTP on binding suggests that UTP, rather than UMP as previously thought (6), is the more physiologically important effector of pyr operon regulation. ‘Super-shifted’ species were observed when the PyrR concentration was ≥5 µM, as shown in Figure 4 for the PyrR–BL1 and PyrR–BL3 interactions. This phenomenon was also observed with BL2 RNA, but is not shown in Figure 4 due to the lower concentrations of PyrR used in these assays. The reason for this phenomenon is unclear, but it is likely that the extremely high concentrations of PyrR in these assays led to extra molecules of PyrR associating with the PyrR–RNA complex, either through protein–protein or protein–RNA interactions.
PyrR binds to pyr mRNA as a monomer
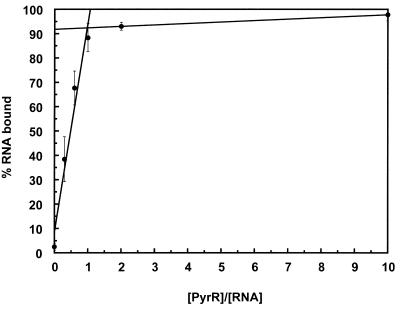
To determine the stoichiometry of the PyrR–pyr mRNA interaction, we performed a gel mobility shift ‘titration’ experiment, suggested to us by the work of Batey and Williamson(26), using BL2 RNA, which showed the tightest binding to PyrR. The RNA concentration was held constant at 100 nM, well above the observed apparent dissociation constant for the BL2–PyrR interaction. Therefore, PyrR was the limiting reagent at lower concentrations, and binding would only be saturated when the concentration of the functional binding unit of PyrR (whether monomer, dimer, etc.) was equal to the RNA concentration. The results of three independent determinations were averaged. The amount of RNA bound by PyrR increased linearly with increasing PyrR concentration and reached saturation when ∼92% of the total RNA was bound (Fig. 5). A sharp inflection indicated the point at which the PyrR concentration was high enough to saturate binding. The molar ratio of PyrR subunits to RNA at this point was 1.0. Two of the determinations were done with saturating UTP and one with saturating UMP; the results of these experiments agreed well.
Figure 5.
Titration of BL2 mRNA with PyrR. Gel mobility shift assays were performed with BL2 RNA held at 100 nM, which was over 100 times greater than the observed apparent dissociation constant for the BL2–PyrR interaction. Increasing amounts of PyrR were added. The binding data were fitted to two straight lines using linear least squares methods. The lines intersect at [PyrR]/[RNA] = 1.0 and RNA bound = 92%, indicating that one PyrR subunit binds to one molecule of pyr mRNA. Data averaged from three independent experiments are shown, two with 500 µM UTP and one with 500 µM UMP.
To ensure that all of the PyrR protein was active and able to bind RNA, each of these gel mobility shift experiments was done in duplicate; identical reactions were run on the right and left sides of the gel. After electrophoresis, the two sides of the gel were cut apart and one side was dried and subjected to autoradiography and phosphorimager detection as usual. The other side of the gel was used for PyrR detection by western immunoblotting. When PyrR that had been purified and stored in 100 mM Tris–acetate pH 7.5, 10 mM potassium acetate, 20% glycerol was used, all of the protein was able to bind RNA, as demonstrated by the absence of any free PyrR until all available RNA had also been bound by PyrR (27). PyrR that was purified as previously described(20) and stored in 10 mM Tris–acetate pH 7.5 showed different behavior, in that the number of PyrR subunits per RNA molecule varied significantly from experiment to experiment and some free PyrR was found in reactions in which there was still free RNA available to be bound (27).
Determination of the minimal RNA sequence needed for PyrR binding
The BL2 transcript, which contains a 40 nt anti-antiterminator stem–loop and 20 flanking single-stranded nucleotides on both the 5′- and 3′-sides, bound PyrR very tightly (Kd = 0.7 nM) in the presence of saturating levels of UMP. To determine the minimal RNA sequence necessary for tight PyrR binding, DNA templates were constructed to serve as transcription templates for progressively shorter RNA species derived from BL2, which are shown in Figure 3. Gel mobility shift experiments were performed with 500 µM UMP to measure the dissociation constant for each RNA–PyrR interaction (Fig. 3). Deleting all 5′ and 3′ single-stranded nucleotides in BL2, as well as the bottom 6 bp of the lower stem, had only a small effect on the strength of binding, as evidenced by the 20 nM dissociation constant for the 708–735CL RNA species. All shorter RNA species tested showed much weaker binding to PyrR. Of these three shorter species, 711–735 and 708–732 RNA bound more weakly than 711–732CL RNA; this suggests that the ability to form a lower stem is necessary for tight binding by PyrR.
Hydroxyl radical footprinting
RNA was synthesized by in vitro transcription using T7 RNA polymerase and the linearized DNA template pBSBL2, then end-labeled with [γ-32P]ATP for use in hydroxyl radical footprinting experiments. End-labeled RNA was reacted with hydroxyl radicals after preincubation with PyrR, PyrR and UMP or buffer only. The reaction products were separated by denaturing PAGE and visualized by autoradiography (Fig. 6). Partial alkaline hydrolysis of the end-labeled RNA gave a complete ladder of bands (Fig. 6, lanes 4, 8 and 13). When PyrR was bound to the RNA from the second attenuation region, a total of 13 nt were protected from cleavage by hydroxyl radicals (Fig. 6, lanes 6 and 7). Nucleotides U710–G714 were weakly protected, A724 was moderately protected and A722 and nt A726–G728 and A734–G736 were strongly protected (see also Fig. 7).
Figure 6.
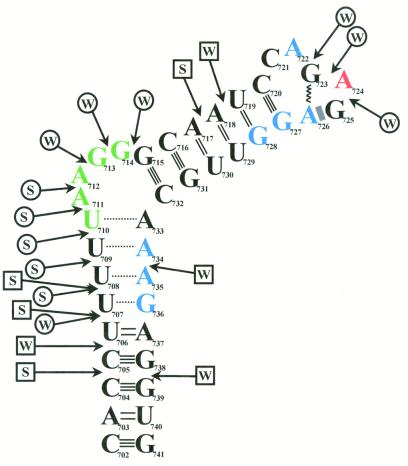
Autoradiogram of hydroxyl radical footprinting and nuclease digestion experiments. End-labeled BL2 RNA was checked for degradation or radiolysis both in the absence of any other factors (lane 1) and also in the presence of PyrR and UMP (lane 2). BL2 RNA was reacted with RNase T1 (lane 3) to produce a ladder of fragments that terminate at each G in the transcript and subjected to partial alkaline hydrolysis (lanes 4, 8 and 13) to produce a ladder of fragments that terminate at every position in the transcript. BL2 RNA was reacted with hydroxyl radicals in the absence of any other factors (lane 5), in the presence of PyrR alone (lane 6) or in the presence of PyrR and UMP (lane 7). BL2 RNA was reacted with RNase I in the absence of any other factors (lane 9) or in the presence of PyrR and UMP (lane 10). BL2 RNA was reacted with RNase V1 in the absence of any other factors (lane 11) or in the presence of PyrR and UMP (lane 12). The sequence diagram on the right has every fifth nucleotide numbered and predicted secondary structure features of the RNA noted. The predicted single-stranded regions of RNA that flank the BL2 stem–loop are denoted by 5′-SS and 3′-SS.
Figure 7.
Map of hydroxyl radical footprinting and nuclease digestion results. End-labeled BL2 RNA was exposed to hydroxyl radicals either in the presence or absence of PyrR. In the presence of PyrR, nucleotides in blue were strongly protected, nucleotides in red were moderately protected and nucleotides in green were weakly protected from hydroxyl radical cleavage (see Fig. 6, lanes 6 and 7). The secondary structure of the RNA was mapped using end-labeled RNA, RNase I (specific for single-stranded RNA) and RNase V1 (specific for double-stranded RNA). A speculative model for the secondary structure of BL2 RNA is shown, based on the structure predicted by MFOLD v.3.1, as well as the nuclease digestion pattern that was observed. Locations of nuclease cuts are shown by arrows, with circle-headed arrows representing single-strand-specific cuts and square-headed arrows representing double-strand-specific cuts (see Fig. 6, lanes 9 and 11). S and W represent strong and weak cleavages, respectively. The dashed lines in the lower stem represent base pairs that are predicted to form transiently or not at all, due to strong cleavage by RNase I in this region. The hexaloop is folded into a GNRA tetraloop structure with the first two bases of the hexaloop extruded. The zigzag line represents a reverse Hoogsteen G-A base pair and the shaded rectangle represents purine-adenosine stacking. No direct evidence exists for a GNRA tetraloop structure actually forming in vivo or in vitro.
When the ionic strength of the binding buffer was lowered by omitting potassium glutamate (binding buffer was usually 10 mM Tris–acetate, pH 7.5, 10 mM MgCl2, 100 mM potassium glutamate), a similar protection pattern was seen, although A724 was not protected under these conditions (data not shown). PyrR protection of the mRNA from the second attenuation region required a divalent cation, such as Mg2+ or Ca2+, at a minimum concentration of 2 mM. Spermidine was insufficient for protection in the absence of divalent cations (data not shown). Hydroxyl radical footprinting was attempted with RNAs from the first and third attenuation regions, produced from templates pFIR1 and pTHI1. These experiments were unsuccessful, probably because of the weaker interaction between these RNAs and PyrR in vitro, as demonstrated in the gel mobility shift assay.
Nuclease digestion studies of pyr mRNA
We tested the computer predicted secondary structures of BL1, BL2 and BL3 using RNase I, which specifically degrades single-stranded RNA (28,29), and RNase V1, which specifically degrades double-stranded RNA (30–32). End-labeled RNAs were produced by transcription in vitro from templates pFIR1, pBSBL2 and pTHI1, digested with appropriately diluted nuclease, separated by denaturing PAGE and the gel subjected to autoradiography. The results are interpreted in Figure 7. Only data obtained with BL2 are shown in Figure 6 (lanes 9–12) and discussed here. End-labeled RNAs from the first and third attenuation regions showed essentially the same nuclease digestion patterns as BL2.
RNase I cleaved the region upstream of the predicted BL2 stem–loop (5′ SS in Fig. 6) extensively and cleaved the region downstream of the predicted stem–loop (3′ SS in Fig. 6) well except for a small area of essentially no cleavage (Fig. 6, lane 9). Nucleotides 698–701 were cleaved more weakly by RNase I, perhaps due to the proximity of this region to the double-stranded lower stem. RNase I cleaved extensively in the purine-rich internal bulge and the nucleotides immediately below it, with strong cuts after nt 707–711 and weak cuts after nt 706, 712, 713 and 714. The region immediately below the purine-rich internal bulge is predicted to be composed of weak U-A and U-G base pairs by MFOLD v.3.1, so cleavage after nt 706–709 indicates that these base pairs form transiently, at best, and may not form at all, as shown in Figure 7. The entire sequence from G715 to G741 was in a structure that protected it from RNase I cleavage, except for three very weak cuts in the terminal hexaloop, after nt 722–724. The weakness of these cleavages, as well as the absence of other cuts in the terminal loop, suggests that the hexaloop possesses some compact structure (such as a GNRA tetraloop) that is not recognized as single-stranded by RNase I. Figure 7 shows the terminal hexaloop folded into a GNRA tetraloop structure with the first two bases of the hexaloop extruded, but there is no direct evidence that this particular structure actually forms. It is interesting that the terminal hexaloop is closed by a conserved C-G base pair, as it is known that C-G closing base pairs provide extra stability to UNCG tetraloops (33) and UUA triloops (34). The role of C-G closing base pairs in GNRA tetraloop stabilization is unknown, however.
RNase V1 did not cleave the region upstream of the predicted stem–loop, but did cleave the region downstream of the predicted stem–loop in four places (Fig. 6, lane 11); interestingly, these cleavages became more prominent in the presence of PyrR and UMP (Fig. 6, lane 12). As two of these cleaved species are present in the RNA in the absence of any other factors (Fig. 6, lane 1), they likely result from RNA degradation. RNase V1 cleaved strongly after nt 704, 706 and 707 of the lower stem and nt 717 of the upper stem, all regions predicted by MFOLD v.3.1 to be base paired. RNase V1 cleaved weakly after nt 705, 734 and 738 of the lower stem and nt 718 of the upper stem; MFOLD v.3.1 also predicts these regions to be base paired. Altogether, the results of nuclease cleavage are consistent with the secondary structure of BL2 shown in Figure 7. Our data do indicate the possibility of a small hairpin in the single-stranded 3′-region that was not predicted by MFOLD v.3.1. All nuclease cleavages were either absent or much less prominent (see Fig. 6, lanes 10 and 12) when the RNA was preincubated with PyrR and UMP. This extends the protection pattern over a much larger area than that observed when hydroxyl radicals were used as the cleavage agent. This is likely due to the relative size difference between the hydroxyl radical and a RNase enzyme.
Effects of RNA sequence on PyrR binding
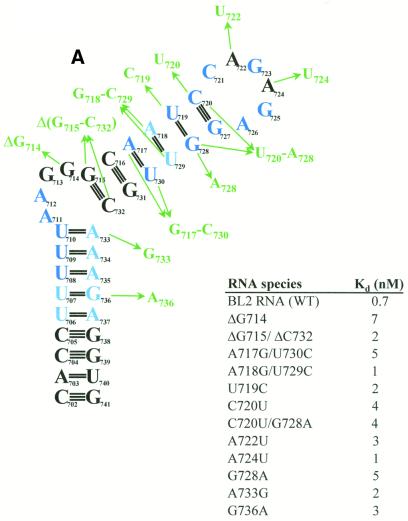
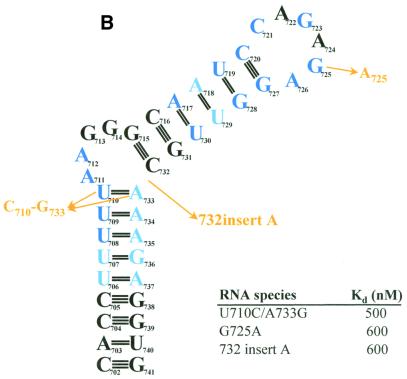
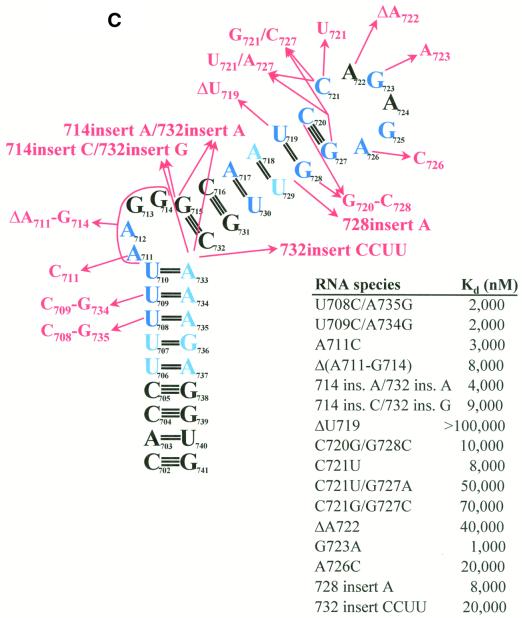
To determine the structural requirements for RNA binding by PyrR, the binding of a series of BL2 sequence variants was studied using gel mobility shift assays (Fig. 8A–C). Of the 31 BL2 variants, 12 were essentially unaffected in binding (defined as having an apparent Kd ≤ 10 nM in the presence of saturating UMP; Fig. 8A), three showed moderate defects in binding (defined as having an apparent Kd between 10 and 1000 nM in the presence of saturating UMP; Fig. 8B) and 16 showed very poor or undetectable binding (defined as having an apparent Kd > 1000 nM in the presence of saturating UMP; Fig. 8C).
Figure 8.
Effects of structural variants of BL2 on binding to PyrR. The effects of various changes in BL2 RNA sequence on the BL2–PyrR interaction were measured using the gel mobility shift assay (500 µM UMP was used) and are shown as apparent dissociation constants for each RNA. (A) Sequence changes in green had little effect on binding (apparent Kd < 10 nM; apparent Kd for BL2 = 0.7 nM). (B) Sequence changes in yellow had a moderate effect on binding (10 nM < apparent Kd < 1000 nM). (C) Sequence changes in red had a severe effect on binding (apparent Kd > 1000 nM). Nucleotides in dark blue are highly conserved among pyr anti-antiterminator sequences from various bacteria, while nucleotides in light blue denote positions of pyrimidine or purine conservation.
The BL2 RNA will be divided into four sections for the purposes of analysis (see Fig. 2B). The terminal loop (nt 721–726) contains 6 nt and may or may not be folded into a GNRA tetraloop with nucleotides C721 and A722 bulged out. The upper stem is comprised of nt 715–720 base paired to nt 727–732. An alternative fold for the upper stem is possible (see Fig. 2D), in which U719 is bulged out of the upper stem, C720 is base paired with G728 and C721 is base paired with G727, creating a terminal pentaloop instead of a hexaloop. This alternative fold is predicted to be slightly less stable by MFOLD v.3.1. The purine-rich internal bulge consists of nt 711–714. The lower stem consists of nt 702–710 and 733–741, of which nt 702–706 are base paired to nt 737–741, but nt 707–710 are more weakly base paired (or not at all) to nt 733–736.
In the terminal loop, the G723A and A726C substitutions, which alter consensus positions and would disrupt formation of the putative GNRA tetraloop structure, strongly disrupt binding. The G725A replacement alters a consensus position, but should still be able to form a GNRA tetraloop; this change caused a moderate defect in binding. As expected, either of the two non-conserved A residues (A722 and A724) can be replaced by U with little effect. The C721U replacement, which binds PyrR poorly, would likely cause a U721-A726 base pair to form, lengthening the upper stem by 1 bp and changing the terminal hexaloop to a tetraloop. The size of the terminal loop is critical, as deletion of the non-conserved residue A722 strongly disrupted binding.
In the upper stem, several structural variants were designed to test whether the predicted fold (continuous upper stem with a terminal hexaloop) or the alternative fold (U719 bulged out of the upper stem with a terminal pentaloop) of the RNA actually forms. The C721U/G727A and C721G/G727C double replacements both bound weakly to PyrR. If the predicted fold is correct, both of these changes would disrupt the base pair below the terminal hexaloop. However, in the alternative fold, both of these variants should be able to form a base pair (U-A or G-C) below the terminal pentaloop. The C720U and C720U/G728A variants bound tightly to PyrR, whereas the C720G/G728C double replacement bound very poorly. In the predicted fold, the first two changes allow 2 bp to form below the terminal hexaloop, while the final change disrupts both base pairs. In the alternative fold, all three variants should be able to form a base pair (U-G, U-A or G-C) in this position. If the RNA folds with a continuous upper stem and a terminal hexaloop, as predicted by MFOLD v.3.1, and this structure is important for PyrR recognition, then replacements that disrupt upper stem base pairs would be expected to disrupt binding, while replacements which permit base pairing should allow tight PyrR binding. This was observed in our experiments. If the RNA folds with U719 bulged out and a terminal pentaloop, the explanation of the binding of these variants is much more difficult. In the alternate fold, the two C-G base pairs below the terminal pentaloop are completely conserved; yet these results imply that the C721-G727 base pair cannot be altered at all, whereas the C720-G728 base pair can be replaced with any pyrimidine-purine base pair. We believe that the results with this set of structural variants favors the predicted hexaloop fold of the RNA as the structure required for PyrR binding.
The ΔU719 and 728insertA variants both bound poorly. In the predicted fold these changes would disrupt a base pair or would insert a bulged nucleotide on the 3′-side of the upper stem, respectively, and are predicted by MFOLD v.3.1 to favor the alternate fold of the RNA (terminal pentaloop with two C-G base pairs immediately below it). However, in the alternate pentaloop fold, the bulged U719 nucleotide would be deleted or would become base paired. In either RNA fold these variants might disrupt binding to PyrR, as was observed.
The U719C and G728A replacements both bound tightly to PyrR. In the predicted fold, both of these mutations alter an existing base pair, but do not abolish it. The U719C replacement should strongly favor formation of two C-G base pairs below a terminal hexaloop. The G728A replacement would almost certainly force U719 to base pair with A728, as A728 would be unable to base pair to C720, and would make a pentaloop fold of the RNA impossible; this strongly suggests that PyrR is able to bind tightly to an RNA with a continuous upper stem and terminal hexaloop. Altogether, we conclude that this set of RNA variants demonstrates that PyrR can bind to RNA with the predicted hexaloop fold, but cannot prove that RNA with the alternate pentaloop fold would not also be bound by PyrR.
Replacing the A718-U729 base pair with a G-C base pair (which is more common at this position among PyrR-binding loops from various bacteria) had no effect on binding. More surprisingly, replacing the conserved A717-U730 base pair with a G-C base pair did not disrupt binding. However, 7 of the 20 known PyrR-binding loop mRNAs do not have an A-U base pair in this position. Shortening the intermediate stem by 1 bp by deleting the G715-C732 base pair had little effect on binding. However, lengthening the intermediate stem by inserting a C-G base pair above the purine-rich internal bulge disrupted binding significantly.
The purine-rich internal bulge appears to be critical for tight PyrR binding. Deletion of the bulge or insertion of complementary nucleotides on the 3′-side of the stem that would cause it to become base paired, strongly disrupted binding. Inserting a single nucleotide on the opposite side of the bulge (as found in BL1, BL3 and several other PyrR-binding loops) caused a significant decrease in binding. Inserting a single nucleotide on both sides of the purine-rich bulge (714insertA/732insertA) causes the purine-rich bulge of BL2 to closely resemble the purine-rich bulge of BL3; this substitution bound PyrR poorly. However, deleting G714 had little effect on binding. The A711C substitution does not affect the structure of the bulge, but does disrupt the sequence of the conserved 5′-UUUAA-3′ motif. This substitution causes a defect in binding, suggesting that A711 may be involved in base-specific interactions with PyrR. The data indicate that an internal bulge is required on the 5′-side of the RNA and that the exact structure of the loop can have a profound effect on the strength of the binding interaction.
The identity of the nucleotides in the lower stem is important; nuclease digestion experiments suggest that the weak U-purine base pairs below the purine-rich internal bulge form transiently, at best, and may not form at all (Fig. 7). Changing any of the three weak U-purine base pairs immediately below the purine-rich internal bulge to strong C-G base pairs caused a moderate to severe defect in binding. However, substituting a U-G base pair for the U-A base pair immediately below the purine-rich internal bulge (as found in BL1, BL3 and several other PyrR-binding loops) had little effect on binding. Also, 8 of the 20 known PyrR-binding loop mRNAs contain a U-G base pair somewhere in their lower stem. Changing the single U-G base pair in the lower stem of BL2 to a U-A base pair had no effect on binding.
Effects of small molecule cofactors on PyrR–pyr mRNA interaction
The effects of various concentrations of several possible metabolite effectors were examined in gel mobility shift experiments using 50 pM BL2 RNA and 1 nM PyrR. The concentrations of uridine nucleotides that yielded half-maximal effects on increasing the amount of PyrR–RNA complex formed under these conditions were 9 ± 7 µM (n = 7) for UMP, 160 ± 90 µM (n = 7) for UDP and 130 ± 70 µM (n = 7) for UTP. Uracil, uridine, CTP, AMP, ATP and GTP showed no effect on the PyrR–BL2 RNA interaction at any concentration up to 1 mM. CMP and GMP both showed a systematic increase in the percentage of bound RNA with increasing ligand concentration (data not shown). However, the magnitude of the effect was small (both showed a 10–15% increase in the percent of total RNA bound with saturating ligand, as opposed to a 35% increase for UMP and a 70% increase for UTP) and required a fairly high ligand concentration (half-maximal effective concentrations were ∼40 µM for CMP and 80 µM for GMP).
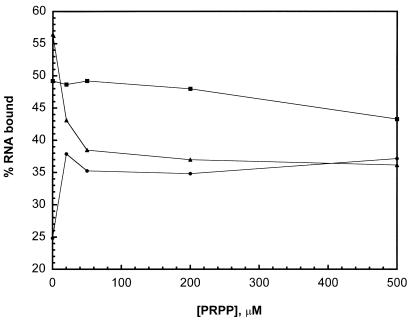
In addition to its regulatory function, PyrR is also an enzyme, possessing uracil phosphoribosyltransferase (UPRTase) activity (20). It has been shown that 5′-phosphoribosyl-α-1′-pyrophosphate (PRPP), a substrate for the UPRTase reaction, can antagonize the stimulatory effect of UMP on attenuation by PyrR in an in vitro transcription assay (35). We tested the effect of PRPP on the PyrR–BL2 RNA interaction directly using the gel mobility shift assay. Holding the PyrR (1 nM) and BL2 RNA (50 pM) concentrations constant, the concentration of PRPP was varied from 0 to 500 µM, in the presence of either no cofactor, 200 µM UMP or 200 µM UTP (Fig. 9). PRPP caused a modest increase in the amount of RNA bound in the absence of other cofactors and antagonized the effects of UTP. At 50 µM PRPP (PRPP:UTP ratio 1:4), the amount of RNA bound was reduced to the level present with PRPP only. However, PRPP antagonized the effects of UMP only weakly, requiring high concentrations to achieve even a modest decrease in the amount of RNA bound. These results are consistent with the relative concentrations of UMP and UTP necessary to affect transcription attenuation by PyrR (35). Since UMP has been shown to be a competitive inhibitor of PRPP saturation in kinetic experiments on the UPRTase reaction of PyrR (G.Grabner, unpublished results), we suggest that PRPP antagonizes the effects of UMP and UTP by competing for binding at the same site.
Figure 9.
Effect of PRPP on PyrR–BL2 RNA interaction. PyrR (1 nM) and BL2 RNA (50 pM) concentrations were held constant. The concentration of PRPP was increased from 0 to 500 µM in the presence of no cofactor (filled circles), 200 µM UMP (filled squares) or 200 µM UTP (filled triangles).
DISCUSSION
Reliability of RNA binding studies using the gel mobility shift assay
In analyzing our RNA binding studies, it is necessary to consider the limitations imposed by use of the electrophoretic gel mobility shift assay. The gel mobility shift assay of RNA binding to PyrR was used because PyrR does not bind quantitatively to nitrocellulose or PVDF filters (J.N.D’Elia and R.L.Switzer, unpublished results). Since dissociation of nucleic acid–protein complexes during electrophoresis is known to occur (36), the values listed here as apparent ‘dissociation constants’ may well not be thermodynamically valid. The magnitude of the deviation of the observed apparent Kd values from true dissociation constants is probably greatest for those RNAs that exhibited the weakest apparent binding, as these RNAs likely have the fastest rate of dissociation from PyrR. In fact, we generally observed that RNAs that bound weakly to PyrR usually showed a significant fraction of total radioactivity smeared between the PyrR–RNA band and the free RNA band after electrophoresis. Even though this smeared radioactivity was included as PyrR–RNA complex in the analysis of the data, we believe that the amount of RNA bound to PyrR is still underestimated in many cases where the PyrR–RNA complex dissociates rapidly.
These considerations limit our ability to interpret these values quantitatively. Nonetheless, we believe that our results provide an adequate qualitative picture of the RNA nucleotide sequence and secondary structural requirements exhibited by PyrR for RNA binding. We believe that comparisons of the relative affinity of the various RNA species for PyrR are reliable, although the reported apparent Kd values for RNAs that bind weakly (apparent Kd > 1000 nM) are beyond the range that the gel mobility shift assay can accurately determine. Our confidence in the qualitative conclusions drawn in this work is strengthened by the fact that they are well supported by hydroxyl radical footprinting experiments. Furthermore, a series of eight mutations in BL1 were isolated by Ghim and Switzer (37) by screening in vivo for strains with cis-acting defects in pyr–lacZ regulation by pyrimidines. All but one of these mutants would be predicted from our generalizations about requirements for tight binding of PyrR to RNA to have reduced affinity for PyrR (27). Given that the structural variants we studied were in BL2, not BL1, this is a remarkable agreement.
Binding of BL1, BL2 and BL3 to PyrR
The three anti-antiterminator RNAs had surprisingly different apparent affinities for PyrR in the gel mobility shift assay. BL2 bound PyrR most tightly, BL3 bound PyrR less well, and BL1 bound PyrR much more weakly (Table 1). We were surprised that the binding of BL1 to PyrR appeared so weak in the gel mobility shift assay, compared to BL2 and BL3. While the three binding loops from B.subtilis are similar, there are several regions that have significant differences. Several of the BL2 structural variants studied were designed to test why BL2 binds PyrR more tightly than BL1 or BL3. BL2 has an A-U base pair (A718-U729) immediately below the U-G base pair in the upper stem, while BL1 and BL3 (as well as most other binding loops from various bacteria) have a G-C base pair in this position. Changing this base pair to a G-C base pair (A718G/U729C) had no effect on binding. The upper stem in BL2 is 1 bp longer than in BL1 or BL3, but shortening this stem by 1 bp (ΔG715/ΔC732) had little effect on binding. BL2 has a U-A base pair in the position immediately below the purine-rich internal bulge, which would presumably be more stable than the U-G base pair found in the same position in BL1 and BL3. However, changing this base pair to a U-G (A733G) had little effect on binding. The only change in BL2 structure to make it resemble BL1 and BL3 more closely that caused a significant change in binding was 732insertA, in which a single base was inserted on the 3′-side of the purine-rich internal bulge, as found in BL1 and BL3. This mutation reduced binding affinity by ∼500-fold. This difference, when combined with the small differences caused by the other structural variants described, might account for the several thousand-fold difference in apparent binding affinity between BL1 and BL2 in the presence of saturating levels of UMP. However, BL3 contains a single nucleotide on the 3′-side of the purine-rich internal bulge, but only binds ∼100-fold more weakly than BL2 in the presence of saturating levels of UMP. Since BL1 and BL3 have 4 and 5 nt, respectively, on the 5′-side of the purine-rich internal bulge, it is possible that the exact configuration of this region is critical. BL1 also has the least stable lower stem (7 bp, only one C-G base pair) compared to BL2 (9 bp, three C-G) or BL3 (8 bp, two C-G). It is possible that in the in vitro binding assays the folding of BL1 is less favorable, leading to quicker dissociation of the PyrR–BL1 complex and a higher apparent dissociation constant.
While BL1 appeared to bind PyrR weakly compared to BL2 and BL3 in the gel mobility shift assay, each attenuation region showed a similar capability to be regulated by pyrimidine levels when pyr–lacZ fusions containing them were compared in vivo (38). The relationship between direct studies of RNA binding by gel mobility shift analysis and the ability of PyrR to regulate an attenuator containing the same binding loop RNA sequence in vivo requires a much more systematic analysis. To this end, our laboratory is currently constructing a series of pyr–lacZ fusions based on the second pyr (BL2-containing) attenuation region in which the set of RNA sequence variants in Figure 8 replace the normal BL2 sequence, but which are otherwise identical to the native sequence. The ability of these variants to be regulated by pyrimidines in vivo will be tested by the methods of Lu et al. (38). Such a study should give a better picture of the degree to which regulation of pyr genes can be predicted from measurements of the apparent affinity of PyrR for a given binding loop sequence. It would be surprising if the correlation were a close one. As noted above, the gel mobility shift measurements require a tight, long-lived PyrR–RNA complex to be formed, but only transient binding of PyrR may be required to cause attenuation. Furthermore, attenuation is undoubtedly a more complex process than binding of PyrR to RNA, involving rates of transcriptional elongation and the relative stability of the downstream antiterminator and terminator stem–loops, as well as possible direct interactions between PyrR and RNA polymerase.
Effects of metabolites on the PyrR–RNA interaction
Of the metabolites tested for their effects on the PyrR–BL2 interaction, only UMP, UDP and UTP had significant effects. Effects of these nucleotides on the termination of pyr transcription in vitro were previously demonstrated by Lu et al. (38). As in those studies, UMP exerted its effects on RNA binding at 10–15-fold lower concentration than UTP, but we were surprised to find that at saturating concentrations UTP increased the apparent affinity of PyrR for BL2 RNA much more than did UMP. PRPP antagonized the effects of UTP on RNA binding much more effectively than it antagonized the effects of UMP. However, PRPP was able to antagonize the effects of UMP on pyr transcription termination in vitro (38). For reasons that are not clear, the effects of uridine nucleotides on the binding of other RNA species were much less dramatic than were seen with BL2.
The ability of uridine nucleotides to increase transcription termination by increasing the affinity of PyrR for anti-antiterminator regions of pyr mRNA provides an obvious mechanism for the regulation of transcription of the pyr operon by the end products of the pathway. Antagonism of the effects of uridine nucleotides by PRPP is also readily rationalized, and has been discussed by Turner et al. (1). The results of the current work lead us to suggest that UTP plays a more important role in this regulation than we had previously recognized. While accurate measurements of the intracellular nucleotide concentrations in B.subtilis are few, it has been shown that the intracellular level of ATP in growing B.subtilis cells is 3.3 nmol/mg dry weight and that PRPP is about one-third of this level (39). Setlow (40) reported that the total uridine nucleotide pool in growing Bacillus megaterium cells was ∼56% of the ATP pool and that at least 70% of that was present as UTP. If one assumes that the ATP pool corresponds to an intracellular concentration of 3 mM and that the UMP and UDP pools are equal, these data lead to values of 1 mM for PRPP, 1.2 mM UTP and 0.25 mM UDP and UMP in growing B.subtilis cells. Even if these estimates are high by 2-fold, the concentrations of uridine nucleotides and PRPP that affected BL2 RNA binding in our studies are within physiologically meaningful ranges.
PyrR and pyr mRNA form a one-to-one complex
The stoichiometry of the Pyr–pyr mRNA interaction was studied using the PyrR–BL2 interaction as a model. Our results demonstrated that the stoichiometry of binding is one molecule of PyrR per molecule of BL2 RNA. While BL1 and BL3 were not tested, we assume that the stoichiometry for these interactions is the same. The three-dimensional structure of PyrR has been determined by X-ray crystallography by Tomchick et al. (41); both hexameric and dimeric crystal forms of PyrR were obtained. Gel filtration experiments reported by Turner et al. (20) suggested that in solution, a concentration-dependent equilibrium exists between dimeric and hexameric forms of PyrR. At physiologically relevant PyrR concentrations it seems most likely that a PyrR monomer binds to one molecule of pyr mRNA, but other equimolar combinations (e.g. a PyrR dimer binding to two molecules of pyr mRNA) are certainly possible. It is unknown which form or forms of PyrR possess UPRTase activity, although all Type I phosphoribosyltransferases studied to date have crystallized as dimers (42–47) with the exception of the E.coli xanthine phosphoribosyltransferase, which crystallized as a tetramer (48). In some cases dimerization is required for phosphoribosyltransferase catalysis (44–47).
Structural specificity of the binding of BL2 RNA to PyrR
The binding of BL2 RNA structural variants to PyrR has been interpreted in the context of the RNA secondary structure shown in Figure 7. The present studies cannot establish that this exact structure is formed in solution or when the RNA is bound to PyrR, but there is good reason to postulate that the RNA that binds to PyrR is folded into a stem–loop structure. First, such a structure is consistently predicted by computer-based RNA folding programs such as MFOLD v.3.1 that search out the most energetically favored base paired structures for single-stranded RNA molecules. Second, the experiments of Lu et al. (5) on the effects of single-stranded deoxyoligonucleotides on the attenuation of pyr transcription in vitro demonstrated that interfering with formation of the anti-antiterminator stem–loop decreased termination at a downstream terminator, providing strong evidence that the predicted secondary structure does form in vitro and is functional in affecting attenuation. Finally, and most convincingly, digestion of end-labeled BL2 RNA with single- and double-strand-specific RNases in this study demonstrates that BL2 RNA folds into a secondary structure very similar, if not identical, to the one shown in Figure 7.
Our analysis of the binding of BL2 RNA structural variants led us to conclude that the requirements for tight PyrR binding are as follows. The RNA must have a terminal loop with the sequence (C)NGNGA. The terminal loop may contain either 5 or 6 nt, but we believe the evidence fits a hexaloop structure better. Studies of the specificity of binding to PyrR and phylogenetic conservation of PyrR-binding loop sequences are consistent with but do not prove the existence of a GNRA tetraloop structure with one or two preceding bases ‘flipped out’ of the loop. The upper stem of the binding loop varies in length from 5 to 7 bp in various bacteria. Despite the high degree of sequence conservation in the upper part of the upper stem, the sequence requirements for PyrR binding seem to be lenient; the only changes in this region that adversely affected binding were those which destroyed base pairs. A purine-rich internal bulge on the 5′-side of the stem–loop is required immediately below the upper stem. The exact configuration of this bulge is likely to affect the relative positioning of the upper and lower stems and has a large effect on binding affinity. The lower stem must have weak U-purine base pairs immediately below the purine-rich internal bulge, followed by one or two pyrimidine-purine base pairs. Additional base pairs are often found in the lower stem, but these are probably for stabilization of the RNA stem–loop structure and are probably not involved in direct binding to PyrR.
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge Dr Robert J. Turner for assistance in developing the RNA gel mobility shift assay and Heather K. Savacool for assistance with protein purification techniques, as well as critical reading of this manuscript. This work was supported by United States Public Health Service Grant GM47112.
REFERENCES
- 1.Turner R.J., Lu,Y. and Switzer,R.L. (1994) Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol., 176, 3708–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinn C.L., Stephenson,B.T. and Switzer,R.L. (1991) Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon. J. Biol. Chem., 266, 9113–9127. [PubMed] [Google Scholar]
- 3.Kahler A.E. and Switzer,R.L. (1996) Identification of a novel gene of pyrimidine nucleotide biosynthesis, pyrDII, that is required for dihydroorotate dehydrogenase activity in Bacillus subtilis. J. Bacteriol., 178, 5013–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Y. and Switzer,R.L. (1996) Evidence that the Bacillus subtilis pyrimidine regulatory protein PyrR acts by binding to pyr mRNA at three sites in vivo. J. Bacteriol., 178, 5806–5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Y., Turner,R.J. and Switzer,R.L. (1996) Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon. Proc. Natl Acad. Sci. USA, 93, 14462–14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Switzer R.L., Turner,R.J. and Lu,Y. (1999) Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acid Res. Mol. Biol., 62, 329–367. [DOI] [PubMed] [Google Scholar]
- 7.Ghim S.-Y. and Neuhard,J. (1994) The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J. Bacteriol., 176, 3698–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghim S.-Y., Kim,C.C., Bonner,E.R., D’Elia,J.N., Grabner,G.K. and Switzer,R.L. (1999) The Enterococcus faecalis pyr operon is regulated by autogenous transcriptional attenuation at a single site in the 5′ leader. J. Bacteriol., 181, 1324–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elagöz A., Abdi,A., Hubert,J.-C. and Kammerer,B. (1996) Structure and organization of the pyrimidine biosynthesis pathway genes in Lactobacillus plantarum: a PCR strategy for sequencing without cloning. Gene, 182, 37–43. [DOI] [PubMed] [Google Scholar]
- 10.Andersen P.S., Martinussen,J. and Hammer,K. (1996) Sequence analysis and identification of the pyrKDbF operon from Lactococcus lactis including a novel gene, pyrK, involved in pyrimidine biosynthesis. J. Bacteriol., 178, 5005–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinussen J. and Hammer,K. (1998) The carB gene encoding the large subunit of carbamoyl phosphate synthetase from Lactococcus lactis is transcribed monocistronically. J. Bacteriol., 180, 4380–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinussen J., Schallert,J., Andersen,B. and Hammer,K. (2001) The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol., 183, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical approach. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology, NATO ASI Series. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 11–43.
- 14.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 15.Legault P., Li,J., Mogridge,J., Kay,L.E. and Greenblatt,J. (1998) NMR structure of the bacteriophage λ N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell, 93, 289–299. [DOI] [PubMed] [Google Scholar]
- 16.Cai Z., Gorin,A., Frederick,R., Ye,X., Hu,W., Majumdar,A., Kettani,A. and Patel,D. (1998) Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex. Nature Struct. Biol., 5, 203–212. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, p. 4.14.
- 18.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidmann,J.G., Smith,J.A. and Struhl,K. (eds) (2000) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY, pp. 1.1.2 and 1.1.4.
- 19.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidmann,J.G., Smith,J.A. and Struhl,K. (eds) (2000) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY, pp. 1.6.1–1.6.2.
- 20.Turner R.J., Bonner,E.R., Grabner,G.K. and Switzer,R.L. (1998) Purification and characterization of Bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyltransferase. J. Biol. Chem., 273, 5932–5938. [DOI] [PubMed] [Google Scholar]
- 21.Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 22.Bradford M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 23.Gill S.C. and von Hippel,P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 24.Tullius T.D. and Dombroski,B.A. (1985) Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science, 230, 679–681. [DOI] [PubMed] [Google Scholar]
- 25.Donis-Keller H., Maxam,A.M. and Gilbert,W. (1977) Mapping adenines, guanines and pyrimidines in RNA. Nucleic Acids Res., 4, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Batey R.T. and Williamson,J.R. (1996) Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16 S rRNA: I. Defining the minimal RNA site. J. Mol. Biol. 261, 536–549. [DOI] [PubMed] [Google Scholar]
- 27.Bonner E.R. (2001) Investigation of the molecular interaction between pyr mRNA and the Bacillus subtilis attenuation protein, PyrR. PhD thesis, University of Illinois at Urbana-Champaign, Urbana-Champaign, IL.
- 28.Spahr P.F. and Hollingsworth,B.R. (1961) Purification and mechanism of action of ribonuclease from Escherichia coli ribosomes. J. Biol. Chem., 236, 823–831. [Google Scholar]
- 29.Meador J., Cannon,B., Cannistraro,V.J. and Kennel,D. (1990) Purification and characterization of Escherichia coli RNase I. Comparisons with RNase M. Eur. J. Biochem., 187, 549–553. [DOI] [PubMed] [Google Scholar]
- 30.Favorova O.O., Fasiolo,F., Keith,G., Vassilenko,S.K. and Ebel,J.-P. (1981) Partial digestion of transfer RNA-aminoacyl transfer RNA synthetase complexes with cobra venom RNase. Biochemistry, 20, 1006–1011. [DOI] [PubMed] [Google Scholar]
- 31.Lockard R.E. and Kumar,A. (1981) Mapping tRNA structure in solution using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res., 9, 5125–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lowman H.B. and Draper,D.E. (1986) On the recognition of helical RNA by cobra venom V-1 nuclease. J. Biol. Chem., 261, 5396–5403. [PubMed] [Google Scholar]
- 33.Woese C.R., Winker,S. and Gutell,R.R. (1990) Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc. Natl Acad. Sci. USA, 87, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shu Z. and Bevilacqua,P.C. (1999) Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry, 38, 15369–15379. [DOI] [PubMed] [Google Scholar]
- 35.Lu Y. and Switzer,R.L. (1996) Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J. Bacteriol., 178, 7206–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidmann,J.G., Smith,J.A. and Struhl,K. (eds) (2000) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY, p. 12.2.8.
- 37.Ghim S.-Y. and Switzer,R.L. (1996) Characterization of cis-acting mutations in the first attenuator region of the Bacillus subtilis pyr operon that are defective in pyrimidine-mediated regulation of expression. J. Bacteriol., 178, 2351–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y., Turner,R.J. and Switzer,R.L. (1995) Roles of the three transcriptional attenuators of the Bacillus subtilis pyrimidine biosynthetic operon in the regulation of its expression. J. Bacteriol., 177, 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxild H.H. and Nygaard,P. (1991) Regulation of levels of purine biosynthetic enzymes in Bacillus subtilis: effects of changing purine nucleotide pools. J. Gen. Microbiol., 137, 2387–2394. [DOI] [PubMed] [Google Scholar]
- 40.Setlow P. (1975) Energy and small-molecule metabolism during germination of Bacillus spores. In Gerhardt,P., Costilow,R.W. and Sadoff,H.C. (eds), Spores VI. American Society for Microbiology, Washington, DC, pp. 443–450.
- 41.Tomchick D.R., Turner,R.J., Switzer,R.L. and Smith,J.L. (1998) Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase. Structure, 6, 337–350. [DOI] [PubMed] [Google Scholar]
- 42.Somoza J.R., Chin,M.S., Focia,P.J., Wang,C.C. and Fletterick,R.J. (1996) Crystal structure of the hypoxanthine-guanine-xanthine phosphoribosyltransferase from the protozoan parasite Tritrichomonas foetus. Biochemistry, 35, 7032–7040. [DOI] [PubMed] [Google Scholar]
- 43.Heroux A., White,E.L., Ross,L.J., Davis,R.L. and Borhani,D.W. (1999) Crystal structure of Toxoplasma gondii hypoxanthine-guanine phosphoribosyltransferase with XMP, pyrophosphate and two Mg2+ ions bound: insights into the catalytic mechanism. Biochemistry, 38, 14495–14506. [DOI] [PubMed] [Google Scholar]
- 44.Scapin G., Grubmeyer,C. and Sacchettini,J.C. (1994) Crystal structure of orotate phosphoribosyltransferase. Biochemistry, 33, 1287–1294. [DOI] [PubMed] [Google Scholar]
- 45.Focia P.J., Craig,S.P., Nieves,A.R., Fletterick,R.J. and Eakin,A.E. (1998) A 1.4 Å crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi. Biochemistry, 37, 15066–15075. [DOI] [PubMed] [Google Scholar]
- 46.Schumacher M.A., Carter,D., Scott,D.M., Roos,D.S., Ullman,B. and Brennan,R.G. (1998) Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding. EMBO J., 17, 3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips C.L., Ullman,B., Brennan,R.G. and Hill,C.P. (1999) Crystal structures of adenine phosphoribosyltransferase from Leishmania donovani. EMBO J., 18, 3533–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vos S., De-Jersey,J. and Martin,J.L. (1997) Crystal structure of Escherichia coli xanthine phosphoribosyltransferase. Biochemistry , 36, 4125–4134. [DOI] [PubMed] [Google Scholar]