Abstract
In this study, we sought to investigate the various characteristics of Salmonella spp. isolated from raw chicken meats available in Korean markets. The data collected, such as food source of isolation, sampling information, serotype, virulence, and genetic profile including sequence type, were registered in the database for further comparative analysis of the strains isolated from the traceback investigation samples. To characterize serotype, virulence and gene sequences, we examined 113 domestically distributed chicken meat samples for contamination with Salmonella spp. Phylogenetic analysis was conducted on 24 strains (21.2%) of Salmonella isolated from 113 commercially available chicken meats and by-products, using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). Serotyping of the isolated Salmonella spp. revealed S. Enteritidis in 11 strains (45.8%), S. Virchow in 6 strains (25%), S. Montevideo in 2 strains (8.3%), S. Bsilla in 2 strains (8.3%), S. Bareilly in 1 strain (4.2%), S. Dessau in 1 strain (4.2%), and S. Albany in 1 strain (4.2%). The genetic correlation indicated that 24 isolated strains were classified into 18 clusters with a genetic similarity of 64.4-100% between them. Eleven isolated S. Enteritidis strains were classified into 9 genotypes with a sequence identity of 74.4%, whereas the most distantly related S. Virchow was divided into five genotypes with 85.9% identity. Here, the MLST analysis indicated that the major Sequence Type (ST) of the Salmonella spp. isolated from domestic chicken sold in Chungcheong Province belongs to the ST 11 and 16, which differs from the genotype of Salmonella isolated from imported chicken. The differential sequence characteristics can be a genetic marker for identifying causative bacteria for epidemiological investigations of food poisoning.
Keywords: Salmonella, serotype, sequence type, chicken, foodborne pathogen
Introduction
Many foodborne illness outbreaks of harmful bacteria in foods are reported each year, and these human health-threatening incidences have had various patterns [1]. Across the globe, outbreaks of food poisoning tend to occur in groups and become larger while the key pathogenic organisms causing food poisoning include Salmonella spp., Escherichia coli, Staphylococcus aureus, Clostridium perfringens, and Vibrio parahaemolyticus [2, 3]. Among them, food poisoning from Salmonella has become more frequent worldwide, making up a higher share of the number of food poisoning cases reported in the Republic of Korea [2-4].
The average number of individuals in a Salmonella outbreak has varied significantly. For instance, from 2009 to 2020, an average of 857 cases were reported each year of individuals becoming sick with Salmonella, which is the third-highest, behind 1,777 people infected with pathogenic E. coli and 1,161 with norovirus, according to the statistics report of the Ministry of Food and Drug Administration (MFDS), Republic of Korea [4]. In particular, as seen in the case of the nationwide foodborne outbreak of 2018, which was linked to chocolate cake distributed to 190 schools and facilities, food poisoning attributed to egg white contaminated with Salmonella is likely to spread easily, leading to soaring numbers of patients.
In addition, according to data from the Korea Meteorological Administration, the year 2018 was one of the hottest ever, with a total of 27.8 heat wave days that had a daily maximum apparent temperature of 33°C or higher [5]. In terms of temperature-induced proliferation of Salmonella in eggs, it is reported that the maximum specific growth rate (log CFU/h) of Salmonella contamination caused by high temperatures (35°C) during heat waves increases from 0.39 to 0.86 compared to normal temperatures (25°C) [6], which means that the growth rate of each germ per hour under 35°C is up to three times faster than under 25°C, indicating that extra care must be taken not only to prevent Salmonella contamination during the manufacturing process but to maintain cold chains to prevent proliferation in the distribution and consumption stages.
Salmonella species are a group of bacteria that can live in the intestinal tract of animals. These bacteria are widely distributed in nature, particularly in livestock such as chickens, pigs, and cattle, as well as soil and water [3, 7]. They consist of two species: Salmonella enterica and S. bongori, and so far, more than 2,500 serotypes have been reported. Salmonella Enteritidis and Salmonella Typhimurium are two major serotypes that cause food poisoning [3, 8, 9, 10]. In addition, these strains cause food poisoning in both humans and animals as they have no specificity in terms of who can be infected with them and develop illness; in most cases, contaminated food, improper handling, and distribution of meat and inadequate cooking cause illness in humans [3, 11, 12]. Known causes of Salmonella food poisoning include diverse foods such as poultry, eggs, meat, fish, and dairy products. In particular, Salmonella is a pathogenic bacterium that causes diseases in humans, making it a representative, hazardous factor that poses a threat to the safety of agricultural foods [13, 14]. According to the data from the Centers for Disease Control and Prevention (CDC), it was chicken that caused the most foodborne illness in the United States between 2009 and 2015; about 3,000 people were found to have suffered from food poisoning from eating contaminated chickens, and 64 out of 149 mass food poisoning cases were attributed to Salmonella enterica [15]. Salmonella species are present in the intestinal tract of such animals as ducks, chickens, cattle, and pigs and are transmitted to humans through contaminated food, causing symptoms in the gastrointestinal tract and food poisoning, even with a small amount of 100 to 1,000 CFU [15]. Salmonella species are disease-causing bacteria that proliferate in livestock products including chicken, and it is crucial that we properly manage food hygiene and public health to prevent outbreaks of foodborne illness [3].
Based on this background, we aimed to isolate Salmonella spp. from raw chicken meat distributed domestically, and the isolated bacteria were characterized by serotyping, virulence gene-targeted PCR, and use of PFGE and MLST.
Materials and Methods
Sampling
From February to April 2018, 113 samples of chicken and by-products sold in department stores, large discount stores and traditional markets in Chungcheong Province were collected. After purchase, the samples were kept in a cooling box for refrigeration while being transported to the laboratory for testing.
Isolation and Identification of Salmonella spp.
In accordance with the testing method of the Korean Food Code laid out by the MFDS, a 25 g sample and 225 ml of Buffered Peptone Water (Oxoid, UK) were mixed thoroughly and enriched in the incubator at 37°C for around 24 h. The enriched culture solution was added to two enrichment media, 1 ml to 10 ml of teterathionate medium (Biomeriux Inc., Spain) and 0.1 ml to 10 ml of Rappaport-Vassiliadis medium (Oxid, UK), which underwent secondary enrichment at 37°C (tetrathionate) and 42°C (RV medium) for 20-24 h. The secondary enrichment culture solution was smeared on the selective media of XLD agar (Oxoid) and Brilliant green sulfa agar (Remel, UK), and cultured at 37°C for 18-24 h, and then a typical colony was selected and subcultured in the nutrient medium, which was identified by Vitek MS (Biomeriux Inc., France).
Pathogenic Gene Analysis Using Polymerase Chain Reaction (PCR)
Genes subject to genetic characterization of Salmonella were invA, his, stn, sefA, spvC, and hin for pathogenic and serotype identification. To extract purely isolated strain DNA, a single colony was taken and DNA was extracted using automated equipment (EZ1 Advance XL, Qiagen, UK) according to the manufacturer’s methods, and this was then used as the DNA template.
For his, invA, and stn gene detection from Salmonella spp., 5 μl of the template DNA was put into the mixture using a detection kit according to the method proposed by the Korean manufacturer (Kogenbiotech Co., Ltd., Korea). This brought the total to 20 μl, with which real-time PCR (7500 Fast Real-Time PCR, Applied Biosystems, USA) was performed.
For spvC, sefA, and hin gene identification, real-time PCR and conventional PCR were used, referencing the methods of Bugarel et al. [16], Seo et al. [17] and Kim et al. [18], and the primer/probe PCR conditions used are shown in Table 1.
Table 1.
Primers/probe and PCR conditions used in the present study.
| Target gene | Sequence (5'-3') | Size (bp) | PCR cycling conditions |
|---|---|---|---|
| spvC | F: AATGAACTACGAAGTGGGCG R: TCAAACGATAAAACGGTTCCTC P: FAM-ATGGTGGCGAAATGCAGAGACAGGC-BHQ1 |
112 | 50°C, 2 m→95°C, 10 m→95°C, 15 s→60°C, 1 m: 40 cycles |
| sefA | F: GGCTTCGGTATCTGGTGGTGTA R: GGTCATTAATATTGGCCCTGAATA P:Cy5-CCACTGTCCCGTTCGTTGATGGACA-BHQ2 |
98 | 50°C, 2 m→95°C, 10 m→95°C, 15 s→60°C, 1 m: 40 cycles |
| hin | F: TCCATGAGAAAAGCGACTAAAAT R: AGCCGACTAATCTGTTCCTGTTC |
572 | 95°C, 3 m→95°C, 30 s→57°C, 30 s→72°C, 1 m: 30 cycles→72°C, 2 m |
For spvC, and sefA genes, 5 μl of the extracted DNA, 1 μl and 1.5 μl of forward and reverse primer (10 pmole/μl), respectively, 0.5 μl of probe (10 pmole/μl) and PCR mastermix (Kogenbiotech Co., Ltd., Korea) were used, bringing the total to 20 μl, after which real-time PCR (7500 Fast Real-Time PCR, Applied Biosystems) was performed.
For hin gene, 5 μl of the extracted DNA, 1 μl of forward and reverse primer (10 pmole/μl) each, and PCR mastermix (Bioneer, Korea) were used to make a total of 20 μl, and then real-time PCR (C1000 Touch Thermal Cycler, Bio-Rad, USA) was performed, and with the resulting product, a specific band was verified through electrophoresis with a 2% agarose gel.
Serology Testing
Tests were conducted in accordance with the method provided by the MFDS [4] to verify serotypes of isolated strains. Difco Antisera by somatic (O) antigen (A, B, C, D, E, Vi) and by flagellar (H) antigens (a, b, c, d, e, h, i, k, r, y, z) were used to perform slide and tube agglutination tests for identification of serotypes.
Pulsed-Field Gel Electrophoresis (PFGE)
PFGE analysis of Salmonella spp. was performed in accordance with PFGE Standard Testing published by the MFDS. Pure-isolated strains were put into cell suspension TE buffer (100 mM Tris, 100 mM EDTA, pH 8.0) and suspended at suspened to 0.8-1.0 O.D. at 610nm using a spectrophotometer. Then, 200 μl of 1.2% Seakem Gold agarose was added to the strain suspension, mixed gently, and immediately solidified in the plug mold. The solidified plug was transferred to 1.5 ml cell lysis buffer (50 mM Tris, 50 mM EDTA, pH 8.0; 1% sodium-lauroyl sarcosine) to which 50 μl of Proteinase K was added, and after reaction in a 55-L shaking water bath for 1.5-2 h, the plug was washed five times with plug wash TE buffer (10 mM Tris, 1mM EDTA, pH 8.0) for 20 min.
A 1 mm-thick slice was cut from the washed plug and reacted at 37°C for 2 h using 40 U/μl XbaI (Roche, Switzerland). Electrophoresis was performed with the plug gel treated with the restriction enzyme using the electrophoresis equipment at 14°C for 18 h under an initial time of 2.16 s, final time of 63.8 s, a voltage gradient of 6 V/cm, and an included angle of 120°.
S. enterica serovar Braenderup BAA-664 standards were used as the size marker, and the testing was carried out in the same way as for isolated strains. Once electrophoresis was completed, the gel was put into the SYBR gold stain (Invitrogen, USA) and dyed for 30 min, and after decoloring, UV was used for identification. Identified pictures were analyzed using the program BioNumerics (Applied Maths, Belgium).
Multilocus Sequence Typing (MLST)
With MLST, the sequence type was identified by analyzing the sequences of seven house-keeping genes (thrA, purE, sucA, hisD, aroC, hemD and dnaN) (Table 2). present amplification cycles pre-denaturation was first performed at 94°C for 10 min, followed by denaturation at 94°C for 1 min at 35 cycles annealing at 55°C for 1 min, elongation at 72°C for 1 min, and a final extension at 72°C for 5 min.
Table 2.
PCR and sequencing primer for MLST used in this study.
| Gene | Primer sequence (5'-3') | Product size (bp) |
|---|---|---|
| thrA | F: GTCACGGTGATCGATCCGGT R: CACGATATTGATATTAGCCCG |
852 |
| purE | F: GACACCTCAAAAGCAGCGT' R: AGACGGCGATACCCAGCGG |
635 |
| sucA | F: CGCGCTCAAACAGACCTAC R: GACGTGGAAAATCGGCGCC |
793 |
| hisD | F: GAAACGTTCCATTCCGCGC R: GCGGATTCCGGCGACCAG |
788 |
| aroC | F: CCTGGCACCTCGCGCTATAC R: CCACACACGGATCGTGGCG |
826 |
| hemD | F: GAAGCGTTAGTGAGCCGTCTGCG R: ATCAGCGACCTTAATATCTTGCCA |
666 |
| dnaN | F: ATGAAATTTACCGTTGAACGTGA R: AATTTCTCATTCGAGAGGATTGC |
833 |
Sequences were assembled and analyzed using Lasergene 7.2.1 software (DNAStar). Sequence type (ST) numbers were assigned by submitting the sequences and strain information to the Salmonella MLST website (http://www.pubmlst.org/organisms/salmonella-spp). The phylogenetic analysis with MEGA6 (version 6.05) confirmed their homology [19].
Results
Prevalence of Salmonella spp. from Raw Chicken Meat
Among the 113 samples of chicken purchased in retail stores in Chungcheong Province, 24 chicken samples (21.2%) were determined to be positive for Salmonella spp. (data not shown).
Distribution of Salmonella Serotypes
The identified Salmonella serotypes are provided in Table 3. As a result of serotyping on 24 isolates of Salmonella bacteria, the group of O-antigens, in most cases, consisted of bacterial strains belonging to C - E. Various types were isolated, including S. Enteritidis in 11 strains (45.8%), S. Virchow in 6 strains (25%), S. Montevideo in 2 strains (8.3%), S. Bsilla in 2 strains (8.3%), S. Bareilly in 1 strain (4.2%), S. Dessau in 1 strain (4.2%), and S. Albany in 1 strain (4.2%).
Table 3.
Serotypes of Salmonella spp. isolated from raw chicken meat.
| Sample No. | Source of isolates | Somatic antigens | Flagellar antigens | Serovar | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Group I | Group II | O-antigen | H phase 1 | H phase 2 | |||
| 1 | Meat | C | Group O:8 (C2-C3) | 6, 8 | r | 1, 2 | S. Bsilla |
| 2 | Meat | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 3 | Meat | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 4 | Gizzard | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 5 | Meat | C | Group O:7 (C1) | 61,2, 7 | g, m, s | [1, 2, 7] | S. Montevideo |
| 6 | Meat | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 7 | Gizzard | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 8 | Meat | C | Group O:7 (C1) | 61,2, 7 | r | 1, 2 | S. Virchow |
| 9 | Meat | C | Group O:8 (C2-C3) | 6, 8 | r | 1, 2 | S. Bsilla |
| 10 | Meat | C | Group O:7 (C1) | 61,2, 7 | r | 1, 2 | S. Virchow |
| 11 | Meat | C | Group O:7 (C1) | 61, 7 | r | 1, 2 | S. Virchow |
| 12 | Meat | C | Group O:7 (C1) | 61,2, 7 | r | 1, 2 | S. Virchow |
| 13 | Meat | C | Group O:7 (C1) | 61,2, 7, 14 | y | 1, 5 | S. Bareilly |
| 14 | Heart | D | Group O:9 (D1) | 1, 9, 12 | g, m | - | S. Enteritidis |
| 15 | Meat | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 16 | Feet | C | Group O:7 (C1) | 61,2, 7 | r | 1, 2 | S. Virchow |
| 17 | Gizzard | C | Group O:7 (C1) | 61,2, 7 | r | 1, 2 | S. Virchow |
| 18 | Meat | E | Group O:1,3,19 (E4) | 1, 3, 19 | g, s ,t | - | S. Dessau |
| 19 | Meat | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 20 | Meat | C | Group O:8 (C2-C3) | 8, 20 | z4, z24 | - | S. Albany |
| 21 | Meat | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 22 | Meat | C | Group O:7 (C1) | 61,2, 7, 14 | g, m, s | [1, 2, 7] | S. Montevideo |
| 23 | Meat | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
| 24 | Gizzard | D | Group O:9 (D1) | 9, 12 | g, m | - | S. Enteritidis |
PCR Targeted to Pathogenic Genes
The results from gene detection of the 24 isolates of Salmonella spp. are summarized in Table 4. The results reveal that all isolated Salmonella spp. from raw chicken meat have invA, his, and stn genes. On the other hand, the detection rate of sefA, spvC, and hin genes was 45.8% (11/24), 41.7% (10/24), and 37.5% (9/24), respectively.
Table 4.
Pathogenic gene-targeted PCR results of Salmonella serovars.
| Sample No. | Serovar | Serological type | Real-time PCR | PCR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| O-antigen Group | H Phase 1 | H Phase 2 | his | invA | stn | sefA | spvC | hin | ||
| 1 | S. Bsilla | C | r | 1, 2 | + | + | + | - | - | + |
| 2 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 3 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 4 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 5 | S. Montevideo | C | g, m, s | [1, 2, 7] | + | + | + | - | - | - |
| 6 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 7 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 8 | S. Virchow | C | r | 1, 2 | + | + | + | - | - | + |
| 9 | S. Bsilla | C | r | 1, 2 | + | + | + | - | - | + |
| 10 | S. Virchow | C | r | 1, 2 | + | + | + | - | - | + |
| 11 | S. Virchow | C | r | 1, 2 | + | + | + | - | - | + |
| 12 | S. Virchow | C | r | 1, 2 | + | + | + | - | - | + |
| 13 | S. Bareilly | C | y | 1, 5 | + | + | + | - | - | + |
| 14 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 15 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 16 | S. Virchow | C | r | 1, 2 | + | + | + | - | - | + |
| 17 | S. Virchow | C | r | 1, 2 | + | + | + | - | - | + |
| 18 | S. Dessau | E | g, s ,t | - | + | + | + | - | - | - |
| 19 | S. Enteritidis | D | g, m | - | + | + | + | + | - / - | - |
| 20 | S. Albany | C | z4, z24 | - | + | + | + | - | - | - |
| 21 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 22 | S. Montevideo | C | g, m, s | [1, 2, 7] | + | + | + | - | - | - |
| 23 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
| 24 | S. Enteritidis | D | g, m | - | + | + | + | + | + | - |
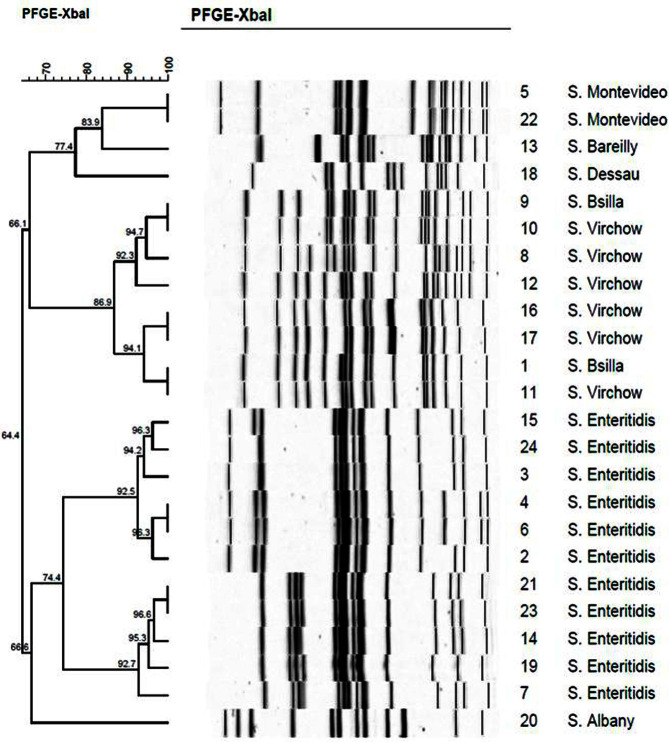
Comparison of Isolates of Salmonella Bacteria Using PFGE
The PFGE results on the 24 Salmonella isolates, which were classified into 18 clusters indicating a genetic similarity of 64.4-100%, is shown in Fig. 1. S. Enteritidis in 11 strains, isolated the most, was classified into 9 genotypes with a homology of 74.4%, followed by S. Virchow which was classified into 5 genotypes with an 85.9%homology.
Fig. 1. Relatedness of Salmonella spp. isolated from raw chicken meat by PFGE analysis with XbaI.
MLST Analysis
Six Sequence Types (STs) from Salmonella spp. based on allele type for seven loci sequences are defined in Table 5.
Table 5.
ST definitions based on allele type for each of seven loci sequenced and assigned by the Salmonella enterica database.
| ST | Allele type | No. isolates | % of total | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| thrA | purE | sucA | hisD | aroC | hemD | dnaN | |||
| 11 | 11 | 6 | 6 | 7 | 5 | 3 | 2 | 11 | 45.8 |
| 16 | 14 | 8 | 10 | 10 | 6 | 10 | 7 | 8 | 33.3 |
| 4 | 4 | 34 | 13 | 13 | 43 | 16 | 41 | 2 | 8.3 |
| 203 | 17 | 68 | 12 | 12 | 81 | 36 | 69 | 1 | 4.2 |
| 14 | 13 | 7 | 8 | 8 | 7 | 8 | 6 | 1 | 4.2 |
| 292 | 48 | 104 | 9 | 78 | 104 | 54 | 100 | 1 | 4.2 |
After being classified through the PubMLST program to verify the diversity of clones, 6 STs belonged to ST11 and 16. Of the 6 STs, ST11 (11 strains) and ST16 (8 strains) were the most common types. The rest of the strains were ST4 (2 strains), and ST203, 14, 292 (1 strain). In addition, in most cases, ST11, ST16, ST4, ST203, ST14, and ST292 appeared in isolates of bacterial species in 2001 but did not appear in those thereafter.
Discussion
In this study, Salmonella spp. were isolated from 24 domestic raw chicken meat samples out of a total 113 samples for a 21.2% detection rate. According to Pilar et al. [20], Salmonella was detected in chicken collected from retail stores at a rate of 42%, and from supermarkets at 36%; a detection rate of 17.41% was found by a study conducted by Rodriguez et al. [21], and 8.3% from one by Anisa et al. [22], showing the difference in detection rate with this study. Salmonella bacteria infecting humans through contaminated eggs, poultry meats, and by-products [23, 24, 25] can cause cross-contamination through diverse routes in the process of distribution.
The serotypes of Salmonella spp. isolated from chicken meat were identified as S. Enteritidis (11/24, 45.8%), S. Virchow (6/24, 25%), S. Montevideo (2/24, 8.3%), S. Bsilla (2/24, 8.3%), S. Bareilly (1/24, 4.2%), S. Dessau (1/24, 4.2%), and S. Albany (1/24, 4.2%). Lee et al. [26] reported that, of isolates from 24 Salmonella bacterial strains, S. Enteritidis was isolated the most, at 70.8%; according to a study conducted by Kim et al. [27] on serotypes of Salmonella bacteria isolated from chicken meat, S. Enteritidis and S. Montevideo were found to be widely distributed. As a result of a study by Jung et al. [28] on serotypes of Salmonella bacteria in chickens from 2003 to 2004, S. Enteritidis was reported to be present in about 52% (39/75). A study by Yang et al. [29] also revealed that of five serotypes, S. Enteritidis, S. Newport, S. Typhimurium, S. Derby, and S. Galinarum, S. Enteritidis was detected the most, at 46%. Based on earlier studies, the serotype isolated the most from chicken produced locally was S. Enteritidis, showing the same pattern as in the past. In terms of distribution of Salmonella bacteria in foreign countries from a Canadian study, S. Typhimurium (44.4%, 123/277) showed the highest rate of frequency, with Kentucky (32%, 120/382), Heidelberg (20%, 78/382) and Enteritidis (16%, 62/382), showing a difference from cases in South Korea in terms of the distribution pattern. The major serotypes causing Salmonella-derived food poisoning were reported to be S. Typhimurium, S. Heidelberg, S. Enteritidis, S. Thompson, and S. Montevideo [30]. In particular, Salmonella Enteritidis and Salmonella Typhimurium are bacterial strains that are most frequently related to food poisoning both worldwide and in South Korea. Various serotypes are isolated from source foods infected with Salmonella, including chicken and by-products, of which S. Enteritidis is found to be the most prevalent.
With respect to genotypes of isolated Salmonella spp. in this study, as described in the results, the invA, his, and stn genes were detected in all isolated Salmonella spp. Conversely, the detection rate of sefA, spvC, and hin genes was 45.8% (11/24), 41.7% (10/24), and 37.5% (9/24), respectively. Of genes related to Salmonella bacteria, those identified were: inv, related to adhesion and invasion into epithelial cells [31], his, involved in regulating histidine transport [32], sefA, which encodes fimbria and specifically detects S. Enteritidis [33], spv, that causes cytotoxicity by moving into the host cell [11], Salmonella enterotoxin (stn), which causes diarrhea by Salmonella invading the intestines, and hin, expressing flagella corresponding to the two flagellar antigens phase 1 and 2. PCR is used to specifically detect Salmonella by identifying Salmonella-related genes [34]. In this study, all the Salmonella spp. isolated from monitoring had genes such as invA, stn, and his, showing the same trend of earlier studies that Salmonella bacteria carried the invA gene [35, 36]. The sefA gene that encodes thin filamentous fimbria of S. Enteritidis was detected in 11 Salmonella serogroup D isolates from this study, and there is a report that it is observed specifically in serogroup D1 [34, 37]. In the case of spv, a gene that expresses pathogenicity, derived from a plasmid that can specifically detect S. Enteritidis, studies conducted by Araque and Chaudhary et al. [38, 39] reported that spvC was not detected in any isolates of S. Enteritidis. On the other hand, Soto et al. [40] reported that spvC was detected in all 60 strains of S. Enteritidis, indicating that plasmid-derived genes show different results depending on the bacterial strains used; it was found that all S. Enteritidis isolated in this study, had spvC. In addition, PCR with hin could identify a hin-specific product of 572 bp in 9 bacterial strains of 24 Salmonella. According to Kim et al. [18], it was found that Salmonella strains with monophasic flagella do not have the hin gene and that all monophasic Salmonella were expressed as phase 1. Furthermore, the study also reported that in the case of Salmonella bacteria with no hin gene, the composition of O-antigens and phase 1 of H antigens could identify serotypes of Salmonella bacteria without conducting a phase 2 test, similar to this study.
In this study, the serotype of Salmonella isolated from raw chicken and by-products was determined as S. Enteritidis, a representative serotype that causes food poisoning in humans. As shown here, Salmonella spp. isolated from domestic chicken sold in Chungcheong Province showed specific sequence types as the ST of Salmonella spp., which several studies reported were isolated from Brazilian poultry. Salmonella Typhimurium isolated from poultry revealed ST-19 [41, 42] and most of the Salmonella Dublin isolates (n = 112) from human and animal presented ST-10 (n = 68), ST-3734 (n = 28), and ST-4030 (n = 9) [43].
The Sequence Type determined by the MLST can be used as an important clue for traceback investigation particularly when multiple outbreaks of foodborne illness derived from the same Salmonella spp. occur. For example, useful information can be obtained relatively fast when we analyze the suspected source of contamination between two or more independent outbreak cases. To be a meaningful clue, species identification, serotyping, and pathogenic gene-targeted PCR are carried out in advance according to the traceback investigation manual by the National Institute of Food and Drug Safety (NIFDS). If all the test results are decided to be identical between the strains, the sequence type determined by MLST is a useful marker for the final confirmation. Considering that the aim of an outbreak investigation is to find its source through comparing many strains isolated from a specimen, whether that may come from a sample of ingested food or from the environment, MLST can provide evidence for the coincidence of strains within the same outbreak. To that end, an accumulation of data on Salmonella spp. as a food source of isolation, sampling information, serotype, virulence, and genetic data including sequence type, has been registered in the Integrated Foodborne Pathogen Data System operated by the NIFDS for further comparative analysis between strains. This study is in line with the data construction of Salmonella spp. with its virulence characteristics and sequence type of isolated from domestic poultry.
Along with the rapid growth of the global food trade, the consumption of food or ingredients has become highly dependent on importation, and the possibility of food poisoning sources from imported food is increasing. In the case of an outbreak suspected to be caused by an imported food source, a traceback investigation is conducted by authorities in the importing and exporting countries and the investigating country requests the gene sequence data of isolated pathogens from the suspected source, which becomes important scientific evidence for the investigation. So, it is crucial to monitor the prevalence and gene sequence profile of pathogens isolated from domestic products through sustainable national surveillance programs in response to a foodborne illness outbreak investigation as well as to protect the health of people and the agricultural industry.
In this study, we attempted to investigate the various characteristics of Salmonella including the prevalence of serotype, and gene sequence profile isolated from domestic raw chicken meats. As Salmonella food poisoning repeatedly occurs along with the consumption of poultry products worldwide, these gene sequence characteristics can be used as important clues to identifying causative bacteria for epidemiological investigations and traceback studies. Additionally, sustainable monitoring programs at the national level are necessary to establish gene sequence profiles of Salmonella isolates from various conditions, such as domestic and imported products, regional data, food type, and seasonal data.
Acknowledgment
This research was supported by a grant (18161MFDS033) from the Ministry of Food and Drug Safety in 2018.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Lim TH, Lee HJ, Kim MS, Kim BY, Yang SY, Song CS. Evaluation of efficacy of bacteriophage CJø07 against Salmonella enteritidis infection in the SPF chicks. Korean J. Poult. Sci. 2010;37:283–287. doi: 10.5536/KJPS.2010.37.3.283. [DOI] [Google Scholar]
- 2.Hong SH, Park NY, JO HJ, Ro EY, Ko YM, Na YJ, et al. Risk ranking determination of combination of foodborne pathogens and livestock or livestock products. J. Food Hyg. Saf. 2015;30:1–12. doi: 10.13103/JFHS.2015.30.1.1. [DOI] [Google Scholar]
- 3.Kim SH, Lee YS, Joo IS, Kwak HS, Chung GT, Kim SH. Rapid detection for Salmonella spp. by ultrafast real-time PCR assay. J. Food Hyg. Saf. 2018;33:50–57. doi: 10.13103/JFHS.2018.33.1.50. [DOI] [Google Scholar]
- 4.Ministry of Food and Drug Safety (MFDS), author 2022. www.foodsafetykorea.go.kr.
- 5.KMA Weather Data Service, Open MET Data Portal, Korea Meteorological Administration, Republic of Korea, author. (https://data.kma.go.kr. )
- 6.Moon HJ, Lim JG, Yoon KS. Comparative study of change in Salmonella Enteritidis and Salmonella Typhimurium populations in Egg white and yolk. J. Food Hyg. Safety. 2016;31:342–348. doi: 10.13103/JFHS.2016.31.5.342. [DOI] [Google Scholar]
- 7.Na DH, Hong YH, Yoon MY, Park EJ, Lim TH, Jang JH, et al. Prevalence of Salmonella species isolated from old hen delivery trucks in Korea and application of disinfectant for the reduction of Salmonella contamination. Korean J. Poult. Sci. 2013;40:11–16. doi: 10.5536/KJPS.2013.40.1.011. [DOI] [Google Scholar]
- 8.Kweon OG, Jin SK, Kim GO, Lee CI, Jeong KH, Kim JY. Characterization of Salmonella spp. clinical isolates in Gyeongsangbuk-do province, 2012 to 2013. Ann. Clin. Microbiol. 2014;17:50–56. doi: 10.5145/ACM.2014.17.2.50. [DOI] [Google Scholar]
- 9.Lee HJ. Salmonellosis. Korean J. Clin. Microbiol. 2001;4:5–10. [Google Scholar]
- 10.Yang B, Qu D, Zhang X, Shen J, Cui S, Shi Y, et al. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 2010;141:63–72. doi: 10.1016/j.ijfoodmicro.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Ha DY, Cha HG, Han KS, Jang EH, Park HY, Bae MJ, et al. Analysis of virulence gene profiles of Salmonella spp. and Enterococcus faecalis isolated from the freshly slaughtered poultry meats produced in Gyeong-Nam province. Korean J. Vet. Serv. 2018;41:157–163. [Google Scholar]
- 12.Wang X, Jothikumar N, Griffiths MW. Enrichment and DNA extraction protocols for the simultaneous detection of Salmonella and Listeria monocytogenes in raw sausage meat with multiplex real-time PCR. J. Food Prot. 2004;67:189–192. doi: 10.4315/0362-028X-67.1.189. [DOI] [PubMed] [Google Scholar]
- 13.Shin WS, Kim YS, Lee JS, Kim MH. Analysis of Salmonella species from eggs using immunoliposomes and comparison with a commercial test kit. Korean J. Food Sci. Ani. Resour. 2009;29:533–538. doi: 10.5851/kosfa.2009.29.4.533. [DOI] [Google Scholar]
- 14.Kim GY, Yang GM, Park SB, Kim YH, Lee KJ, Son JY, et al. Rapid detection kit got Salmonella Typhimurium. J. Biosyst. Eng. 2011;36:140–146. doi: 10.5307/JBE.2011.36.2.140. [DOI] [Google Scholar]
- 15.Lee JB, Yoon JW. Hazard analysis of Salmonella contamination in the broiler and duck farms and their slaughtering processing. Safe Food. 2018;13:3–10. [Google Scholar]
- 16.Bugarel M, Granier SA, Weill FX, Fach P, Brisabois A. A multiplex real-time PCR assay targeting virulence and resistance genes in Salmonella enterica serotype Typhimurium. BMC Microbiol. 2011;11:151. doi: 10.1186/1471-2180-11-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo KH, Valentin-Bon IE, Brackett RE, Holt PS. Rapid, specific detection of Salmonella Enteritidis in pooled eggs by Real-Time PCR. J. Food Prot. 2004;67:864–869. doi: 10.4315/0362-028X-67.5.864. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Kim SH, Lee SW, Kang YH, Lee BK. Rapid serological identification for monophasic Salmonella serovars with a hin gene-specific polymerase chain reaction. J. Bacteriol. Virol. 2005;35:291–297. [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilar DG, Clavijo V, Tafur MA, Gonzales S, Hume M, LEO' N M, et al. Prevalence of Salmonella on retail broiler chicken meat carcasses in Colombia. J. Food Prot. 2012;75:1134–1138. doi: 10.4315/0362-028X.JFP-11-513. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez JM, Rondon IS, Verjan N. Serotypes of Salmonella in broiler carcasses marketed at Ibague, Colombia. Brazilian J. Poultry Sci. 2015;17:545–552. doi: 10.1590/1516-635X1704545-552. [DOI] [Google Scholar]
- 22.Khan AS, Georges1 K, Rahaman S, Abdela W, Adesiyun AA. Prevalence and serotypes of Salmonella spp. on chickens sold at retail outlets in Trinidad. PLoS One. 2018;13:e0202108. doi: 10.1371/journal.pone.0202108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorns CJ. Bacterial food-borne zoonoses. Rev. Sci. Tech. 2000;19:226–239. doi: 10.20506/rst.19.1.1219. [DOI] [PubMed] [Google Scholar]
- 24.Wilson IG. Salmonella and Campylobacter contamination of raw retail chickens from different producers: a six year survey. Epidemiol. Infect. 2002;129:635–645. doi: 10.1017/S0950268802007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang MS, Lee SJ, Shin YS. Prevalence and antimicrobial resistance of Salmonella isolated in poultry farms. Korean J. Vet. Serv. 2015;38:95–100. doi: 10.7853/kjvs.2015.38.2.95. [DOI] [Google Scholar]
- 26.Lee HW, Hong CH, Jung BY. Characteristics of Salmonella spp isolated from poultry carcasses. Kor. J. Vet. Serv. 2007;30:339–351. [Google Scholar]
- 27.Kim SR, Nam HM, Jang GC, Kim AR, Kang MS, Chae MH, et al. Antimicrobial resistance in Salmonella isolates from food animals and raw meats in Korea during 2010. Kor. J. Vet. Publ. Health. 2011;35:246–254. [Google Scholar]
- 28.Jung SC, Song SW, Kim SI, Jung ME, Kim KH, Lee JY, et al. Antimicrobial susceptibility of Salmonella spp. isolated from carcasses in slaughterhouse. Kor. J. Vet. Publ. Health. 2007;31:51–56. [Google Scholar]
- 29.Yang HY, Lee SM, Park EJ, Kim JH, Lee JG. Analysis of antimicrobial resistance and PFGE patterns of Salmonella spp. isolated from chickens at slaughterhouse in Incheon area. Korean J. Vet. Serv. 2009;32:325–334. [Google Scholar]
- 30.Lim SY, Ryu SR. Prevalence of Salmonella Enterotoxin gene(stn) among clinical strains isolated in Korea and regulation of stn expression. Kor. J. Appl. Microbiol. Biotechnol. 2000;28:316–321. [Google Scholar]
- 31.Galan JE, Ginocchio C, Costeas P. Molecular and functional characterization of Salmonella the invasion gene invA: homology of invA to members of a new protein family. J. Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziemer CJ, Steadham SR. Evaluation of the specificity of Salmonella PCR primers using various intestinal bacterial species. Lett. Appl. Microbiol. 2003;37:463–469. doi: 10.1046/j.1472-765X.2003.01430.x. [DOI] [PubMed] [Google Scholar]
- 33.Jeon MH, Kim TJ, Jang GS, Kang GI, Kim GH, Kim GS, et al. Specific detection of Salmonella serogroup D1 by Polymerase Chain Reaction (PCR) for sefA gene. Korean J. Vet. Res. 1999;39:523–530. [Google Scholar]
- 34.Lee WW, Lee SM, Lee GR, Lee DS, Park HK. Identification of Salmonella Enteritidis and S. Typhimurium by multiplex polymerase chain reaction. Korean J. Vet. Serv. 2009;32:147–153. [Google Scholar]
- 35.Swamy SC, Barnhart HM, Lee MD, Dreesen DW. Virulence determinants invA and spvC in Salmonellae isolated from poultry products, wastewater, and human sources. Appl. Environ. Microbiol. 1996;62:3768–3771. doi: 10.1128/aem.62.10.3768-3771.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira SD, Rodenbusch CR, Michae GB, Cardoso MI, Canal CW, Brandelli A. Detection of virulence genes in Salmonella Enteritidis isolated from different sources. Braz. J. Microbiol. 2003;34:123–124. doi: 10.1590/S1517-83822003000500042. [DOI] [Google Scholar]
- 37.Woodward MJ, Kirwan SES. Detection of Salmonella Enteritidis in eggs by the polymerase chain reaction. Vet. Rec. 1996;138:411–413. doi: 10.1136/vr.138.17.411. [DOI] [PubMed] [Google Scholar]
- 38.Araque M. Nontyphoid Salmonella gastroenteritis in pediatric patients from urban areas in the city of Mérida, Venezuela. J. Infect. Dev. Ctries. 2009;3:28–34. doi: 10.3855/jidc.102. [DOI] [PubMed] [Google Scholar]
- 39.Chaudhary JH, Nayak JB, Brahmbhatt MN, Makwana PP. Virulence genes detection of Salmonella serovars isolated from pork and slaughterhouse environment in Ahmedabad, Gujarat. Vet. World. 2015;8:121–124. doi: 10.14202/vetworld.2015.121-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soto SM, Rodríguez I, Rodicio MR, Vila J, Mendoza MC. Detection of virulence determinants in clinical strains of Salmonella enterica serovar Enteritidis and mapping on macrorestriction profiles. J. Med. Microbiol. 2006;55:365–373. doi: 10.1099/jmm.0.46257-0. [DOI] [PubMed] [Google Scholar]
- 41.Almeida F, Silva PD, Medeiros MIC, Rodrigues DDP, Moreira CG, Allard MW, et al. Multilocus sequence typing of Salmonella Typhimurium reveals the presence of the highly invasive ST313 in Brazil. Infect. Genet. Evol. 2017;51:41–44. doi: 10.1016/j.meegid.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Amanda Ap, Seribelli AA, Silva P, Cruz1 MF, Almeida F, Frazao MR, et al. Insights about the epidemiology of Salmonella Typhimurium isolates from different sources in Brazil using comparative genomics. Gut Pathog. 2021;13:27. doi: 10.1186/s13099-021-00423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campioni F, Vilela FP, Cao G, Kastanis G, Rodrigues RP, Costa RG, et al. Whole genome sequencing analyses revealed that Salmonella enterica serovar Dublin strains from Brazil belonged to two predominant clades. Sci. Rep. 2022;12:10555. doi: 10.1038/s41598-022-14492-4. [DOI] [PMC free article] [PubMed] [Google Scholar]