Abstract
New methods and technologies within the field of lung biology are beginning to shed new light into the microbial world of the respiratory tract. Long considered to be a sterile environment, it is now clear that the human lungs are frequently exposed to live microbes and their by-products. The nature of the lung microbiome is quite distinct from other microbial communities inhabiting our bodies such as those in the gut. Notably, the microbiome of the lung exhibits a low biomass and is dominated by dynamic fluxes of microbial immigration and clearance, resulting in a bacterial burden and microbiome composition that is fluid in nature rather than fixed. As our understanding of the microbial ecology of the lung improves, it is becoming increasingly apparent that certain disease states can disrupt the microbial–host interface and ultimately affect disease pathogenesis. In this Review, we provide an overview of lower airway microbial dynamics in health and disease and discuss future work that is required to uncover novel therapeutic targets to improve lung health.
Subject terms: Infectious diseases, Health care
Like the gut, the lung harbours a diverse, interacting assortment of microbiota. In this review, Natalini, Singh and Segal examine the role of the lung microbiome in health and disease and discuss future work that is needed to produce novel diagnostic and therapeutic tools aimed at improving lung health.
Introduction
Humans and microbes have symbiotically coevolved over hundreds of thousands of years, resulting in a human microbiome that is home to a wide range of microorganisms, including bacteria, yeasts, archaea, fungi, protozoa and viruses. These microbes, as well as their local environment, have a crucial role in immune system regulation and maturation, digestion, the production of certain vitamins, and the prevention of disease1. We now know that microbes occupy virtually all surfaces of the human body, and the respiratory tract is no exception.
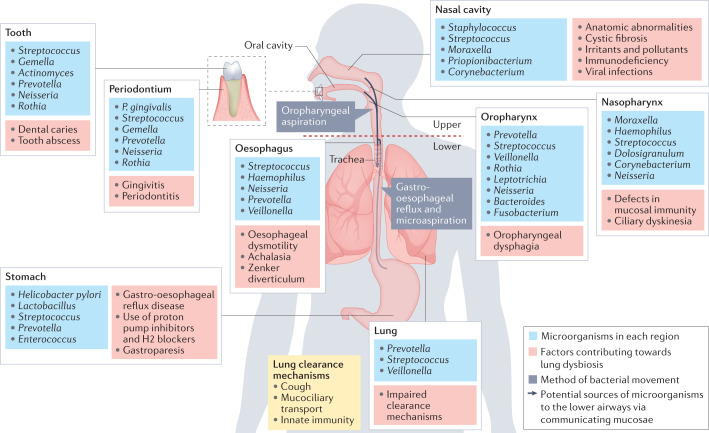
The upper and lower respiratory tracts differ in their microbial composition and biomass (Fig. 1). The upper respiratory tract, which includes the nasal cavity, paranasal sinuses, pharynx and supraglottic portion of the larynx, is largely colonized with bacteria. Moreover, within the upper respiratory tract, there are topographical differences in microbial composition. For example, the dominant taxa in the nasal cavity and nasopharynx include Moraxella, Staphylococcus, Corynebacterium, Haemophilus and Streptococcus species, whereas the oropharynx exhibits high abundance of Prevotella, Veillonella, Streptococcus, Leptotrichia, Rothia, Neisseria and Haemophilus species2 (Fig. 1). By contrast, the lower respiratory tract, comprised of the trachea and lungs, exhibits a relatively low biomass, with a major role in lower airway mucosal immunology (discussed below). Such low biomass is maintained by rapid microbial clearance via a number of physiological mechanisms (Fig. 1), enabling the lower respiratory tract to perform its most crucial function: the exchange of oxygen and carbon dioxide.
Fig. 1. Normal microbiota of the respiratory and proximal gastrointestinal tracts.
The upper respiratory tract, comprised of the nasal cavity, paranasal sinuses, nasopharynx, oropharynx and the supraglottic portion of the larynx, exhibits a relatively high biomass with topographically distinct microbiota within each section. By contrast, the lower respiratory tract, comprised of the infraglottic portion of the larynx, trachea and lungs, exhibits a relatively low biomass consisting mostly of oral commensals. Several factors can contribute to dysbiotic changes along the respiratory tract, as shown in red boxes. Aspiration of oropharyngeal or gastro-oesophageal contents is the predominant means by which bacteria reach the lower airways (arrows). Three major lung clearance mechanisms are coughing, mucociliary transport and the innate immune system.
Dysbiosis, defined as deviation from a normal microbial composition, is associated with a number of adverse biological phenomena, sometimes with clinical consequences. In the lung, dysbiosis can have a significant impact on the development and progression of respiratory diseases, emphasizing the clinical need to understand the biology of the lung microbiome. Over the past decade, there has been a rapid expansion of studies applying culture-independent genomic methods to profile the lung microbial environment (Box 1). As we discuss below, several studies have now shown that certain microbial signals are associated with distinct lower airway inflammatory endotypes and disease processes.
The lung microbiome in health and disease is often conceptualized as a pendulum that swings between two static states — from a paucity of microbes in the lung airways to patterns of resilient microbial colonization. However, it is possible that these extremes are not mutually exclusive but rather snapshots of a dynamic microbial system. In this Review, we describe the evolutionary nature of the lung microbiome and the underlying mechanisms by which sustained dysbiosis might affect respiratory disease progression. We hope to challenge the conceptualization that the lung microbial environment is a static entity and emphasize the dynamic balance between microbial immigration and clearance. As discussed below, the dynamic nature of the lung microbiome has been documented by experimental mouse models of microaspiration3,4 and might be an important distinguishing feature from the microbial behaviour seen in other mucosae with high biomass, such as the oral cavity and gut, where microbial communities are highly resilient. We argue that focusing on elucidating the dynamic nature of the lung microbiome will be critical to understanding mechanisms of lung disease and for the development of novel therapeutic targets involving the microbial–host interface.
Box 1 Use of culture-independent methods in clinical practice.
The use of high-throughput sequencing methods is still in its infancy but not far from reaching clinical use. For example, culture-independent techniques are evolving as a promising approach to detect respiratory pathogens among patients suspected of having pneumonia. This approach promises not only a more rapid diagnosis than with traditional culture-based methods but also the identification of respiratory pathogens among patients with culture-negative pneumonia96,97. For example, a recently developed pneumonia panel using a multiplex PCR assay on sputum and bronchoalveolar lavage samples can provide diagnostic data within hours98 and has been shown to substantially increase the rate of microbiological diagnosis99,100. Targeted methods can also detect atypical bacterial and viral pathogens as well as certain antimicrobial resistance genes. Further, combining microbial and host genomic signals has the added potential benefit of accurately differentiating lower airway pulmonary infection from other pathological conditions101.
Targeted approaches are limited by the finite number of targets evaluated. With the development of nanopore sequencing technologies, such as the MinION, real-time evaluation of RNA or DNA can be performed on a clinical sample, thus providing immediate genomic detection of a pathogen97. Next-generation highly multiplexed methods may lead to an expanded ability to identify patients with bacterial, viral or fungal infections. Further, their use in combination with host transcriptome sequencing methods may provide important prognostic information among patients with pulmonary infections. For example, among critically ill patients with COVID-19, a combination of microbial and host genomic signals from the lower airways provides significant insights about the pathophysiological derangements occurring in these patients that may explain individual risk for poor clinical outcomes48. As the application of multi-omics becomes more widespread, it is possible that we will be better able to understand the phenotypic and endotypic characteristics that carry clinical significance across several lung conditions. For now, more studies employing longitudinal investigative approaches are needed before the adoption of culture-independent methods into clinical practice becomes a reality.
Culture-independent methods could also change our view of the effects of therapies commonly used in clinical practice. Characterization of the lower airway microbiome is challenging our current understanding of broad-spectrum and narrow antibiotic use, uncovering supposedly ‘off-target’ microbial effects. In cystic fibrosis, inhaled tobramycin, commonly given to target colonization with Pseudomonas, has broad effects on non-pathogenic microbes, which is an important consideration when interpreting results of antibiotic use102. For example, in the early stages of chronic obstructive pulmonary disease, chronic use of azithromycin leads to significant changes in the composition of the lower airway microbiome, which can be associated with the release of microbial metabolites with anti-inflammatory properties103. Thus, some of these off-target effects may lead to important mechanisms affecting disease progression. Further, azithromycin therapy in lung transplant recipients with chronic rejection has been shown to improve lung function, suggesting that host–microbial interaction may play a role in its pathogenesis104.
There are of course limitations to culture-independent methods that can severely limit their applicability in clinical practice. One example is that multiplex PCR assays cannot accurately distinguish DNA from viable versus non-viable organisms. Thus, initial detection does not necessarily indicate a need to treat nor does ongoing detection always necessitate prolonging antimicrobial therapy in a patient already being treated. Another limitation is that culture-independent methods applied on samples with a low microbial biomass are far more prone to sequencing noise and DNA contamination. In this regard, microbial RNA metatranscriptome sequencing methods may be particularly useful as detection of microbial RNA generally suggests the presence of viable microbes and given that RNA is rapidly degraded after cell lysis.
Despite evolving data that supports use of mixed bacterial transfers, such as faecal microbiota transplantation, in treating gastrointestinal conditions like Clostridium difficile infection105, irritable bowel syndrome106 and ulcerative colitis107, much less is known about the impact of intentionally altering the lung microbiome in respiratory diseases. Interestingly, there is evolving literature to suggest that restoring the normal gastrointestinal microbiome with the use of prebiotics and probiotics has therapeutic benefit in a number of respiratory diseases such as lung cancer, asthma, tuberculosis and cystic fibrosis (the so-called ‘gut–lung axis’)108; however, therapeutic interventions directly targeting the lung microbiome, through prebiotics or probiotics, are lacking and will need to be explored further in future investigations.
The origins of the lung microbiome
Once thought to be a sterile environment, it is now understood that the lungs are frequently exposed to a variety of microorganisms. Human data support that microbial exposures start early in life, within days to weeks after birth, and have important implications for immune system maturation5. In a study of lower airway samples from infants, differences in the composition of the microbial community were associated with distinct changes in the host immune tone5. Within the first few weeks of life, domination of the lower airway microbial community by either Staphylococcus or Ureaplasma species represents two distinct signatures of the lung microbiome in association with the mode of neonate delivery. Lower airway enrichment with Ureaplasma, a common genital tract commensal, is associated with vaginal delivery, whereas enrichment with Staphylococcus, a common skin commensal, is associated with caesarean delivery, providing an early example of the role of the environment on the lung microbiome5. As neonates mature, their lung microbiome shifts towards enrichment with a more diverse mixture of oral commensals such as Streptococcus, Prevotella, Porphyromonas and Veillonella.
These longitudinal changes in the lung microbiome affect the regulation of immunoglobulins and innate immune responses, and are thus postulated to be one of the mechanisms by which lower airway immune maturation occurs5. Microbial changes in other parts of the respiratory tract may also affect the immune tone in early life. For example, microbial diversity and enrichment of the neonatal hypopharynx with microbes such as Prevotella and Veillonella species have been linked to the subsequent development of reactive airway diseases such as asthma, suggesting that early changes in the microbial environment, even if occurring among commensal taxa rather than among obvious respiratory pathogens, may predispose infants to certain respiratory conditions later in life6.
A dynamic microbial–host interface
As with any mucosal surface in the human body, microbial exposures are not invisible to the host immune system. Indeed, the microbial products found in the lower airways derive from a balancing act between microbial immigration and microbial clearance7. Unlike other mucosae, however, the lungs must maintain a lower bacterial burden in order to facilitate gas exchange, their main physiological purpose. Functionally, the conditions of the lower airways are distinct from those of other mucosae with regard to oxygen tension, protein content, presence of surfactants and other environmental factors. Further, in order to maintain a low bacterial burden, there is a significant reliance on mechanisms that facilitate the clearance of microbes that reach the lower airways, including mechanical mechanisms, such as mucociliary clearance and cough, as well as innate and adaptive host immune responses (Fig. 1).
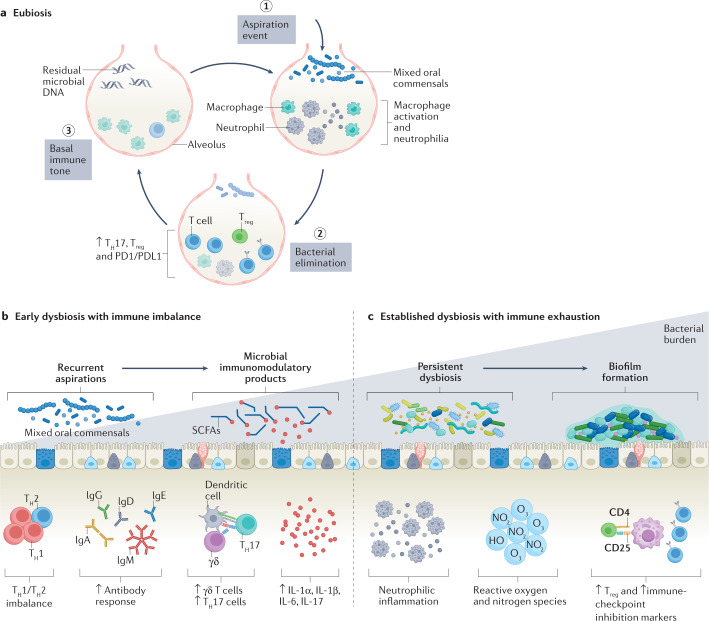
Unlike the oral cavity and gut lumen, both of which have a very high bacterial burden containing millions of viable and replicating microbes, the lower airways must maintain a thin bronchial epithelial fluid lining to facilitate effective gas transfer. Such unique conditions favour a more dynamic homeostatic state of the lung microbiome. Specifically, the microbiomes of the oral cavity and gastrointestinal tract tend to be much more static, dominated by reproducing site-specific microbial communities, changes in which are interpreted as a dysbiotic signature. However, the lower airway microbiome is frequently changing, with episodic exposure to microbes and their rapid clearance, resulting in a dynamic physiological process that defines eubiosis (Fig. 2). This is supported by several human cross-sectional investigations showing that traces of microbial DNA from oral commensals, despite a failure to isolate these bacteria by culture, are common in the healthy lung8–10. Genetic evaluation of the lower airway microbiota (by 16S rRNA gene sequencing, shotgun metagenomic sequencing and metatranscriptome sequencing) shows that a small proportion of individuals with detectable oral commensal microbial DNA also have detectable oral commensal microbial RNA3. As RNA is rapidly degraded after cell lysis, this suggests the transient presence of viable microbes after an aspiration event3. Experimental microaspiration with viable oral commensals in preclinical mouse models support these dynamic changes seen with culture-independent methods3.
Fig. 2. Lung microbial dynamics in eubiosis and dysbiosis.
a, A series of dynamic events promote eubiosis in the lower airways. Aspiration of a microorganism triggers the activation of leukocytes that orchestrate bacterial elimination by macrophage and neutrophil activity. After a transient upregulation of adaptive immunity, the immune tone returns to baseline with a low level of detectable residual microbial DNA. b, Steps commonly found in early dysbiotic events. Recurrent aspirations lead to changes in the lower airway microbial environment that can slowly become irreversible. c, Increased mucus production facilitates persistent colonization and increased bacterial burden and is associated with acute and chronic lower airway disease. Microbial immunomodulatory products, such as short-chain fatty acids (SCFAs), and host immune responses, such as a T helper 1 (TH1)/TH2 imbalance, an aggravated TH17 response and pro-inflammatory cytokines, can perpetuate neutrophilic inflammation, increased regulatory T (Treg) cell response and impaired immune surveillance. These changes can ultimately promote increased bacterial burden.
Lower airway immune tone: the lung–lung axis
Multiple investigations have shown the importance of gut microbial communities in setting the host immune tone, which affects lung immune responses as part of the gut–lung axis. Much less studied, lung microbial communities also affect lung immune responses. Indeed, existing cross-sectional human data, combined with data from murine models, suggest that there is frequent, episodic lung exposure to oral commensals that are subsequently rapidly cleared (Fig. 2). Models of microaspiration support that microbial clearance occurs at variable rates and is dependent on the species of the microbes aspirated4. Importantly, even with rapid lower airway clearance of aspirated oral commensals, such events lead to a durable and dynamic change in the lower airway immune tone4. For instance, mouse aspiration with a mixture of oral commensals (Prevotella melaninogenica, Veillonella parvula and Streptococcus mitis) leads to activation of CD4+ and CD8+ T cells, recruitment of IL-17-producing T cells (T helper 17 (TH17) and γδ T cells), and other counter-regulatory immunological responses such as an increase in regulatory T cells and immune-checkpoint inhibitor markers on T cells4. Importantly, this microbiome-dependent immunological priming is a durable phenomenon that persists beyond the transient presence of aspirated microbes and goes on to significantly impact innate immune defences against respiratory pathogens4. If such responses are representative of normal lung microbial dynamics, these dynamic changes may reflect true lung ‘eubiosis’.
The above experimental preclinical data is in line with multiple observations from cross-sectional studies, illustrating again that distinct lung microbial signatures are associated with lower airway immune tone10,11. For example, one study of mice with similar genetic backgrounds but heterogeneous microbial compositions found that the composition of the lower airway microbiome was associated with levels of several inflammatory markers in the lungs12. In humans, lower airway enrichment with supraglottic-predominant taxa, including Prevotella, Rothia and Veillonella, positively correlated with levels of multiple TH17 cytokines, such as IL-1α, IL-1β, IL-6, fractalkine and IL-17, as well as with the recruitment of both TH17 cells and neutrophils in the lung10. In another study of patients with severe asthma, Proteobacteria species were associated with activation of TH17-associated pathways11. Collectively, these data suggest that the lung microbiome, despite constantly being in flux with rapid clearance of microbes acquired via microaspiration or environmental exposures, leaves important signatures and imprints on the lower airway immune tone.
The studies described above give some insight into how the human lung might respond to sporadic exposure to certain microbes. However, numerous unanswered questions remain. It is unclear what happens when the frequency of microbial exposures increases. For example, it is unknown whether the balance between pro-inflammatory effects and counter-regulatory mechanisms becomes altered as aspiration events become more frequent. Whether recurrent exposure leads to chronic inflammation is also unclear. Despite a paucity of answers, data suggest that persistent exposure to certain microbes can trigger mechanisms leading not only to increased inflammation but also to immune exhaustion (Fig. 2). For example, in individuals where the lower airway microbiome is dominated by oral commensals, there is a blunting of the Toll-like receptor 4 (TLR4) response of alveolar macrophages10. In another human study, microbial products, such as short-chain fatty acids (SCFAs), had significant immunomodulatory properties and blunted IFNγ and IL-17 responses to pathogen-associated molecular patterns13. Finally, in mice, experimental exposure of the lower airways to oral commensals not only triggers an increase in inflammatory cytokines but also an increase in immune-checkpoint inhibitor markers, such as PD1, among T cells and recruitment of regulatory T cells4. These findings collectively suggest that the dynamic nature of microbial–host interactions in the lung likely changes as the frequency of exposure to microbes, rate of elimination and type of immune response change over time.
Dysbiosis in respiratory inflammatory diseases
Microbial dysbiosis is present in various lung diseases, such as cystic fibrosis, chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF), in which a reduction in the diversity of bacterial composition has been linked to disease progression14–16. However, whether microbial dysbiosis itself is the cause of disease or a result of the disease process is not clear. For example, in lung disease, pathophysiological changes in the lung architecture and impaired mucus clearance mechanisms might result in microbial dysbiosis rather than be caused by it. Alternatively, dysbiosis may play a causative role in disease through upregulation of inflammatory signals (such as NF-κB, Ras, IL-17 and PI3K) or blunting of TNF and IFNγ production in response to pathogens in the lower airways10,13,17,18. In this section, we review some examples of dysbiotic signatures identified among pulmonary diseases commonly described as inflammatory and discuss the potential role of lower airway microbes in their pathogenesis.
Chronic obstructive pulmonary disease
COPD serves as a useful example of the evolutionary nature of the lung microbiome and its impact on respiratory disease progression. Initial microbiome studies in patients with stable COPD demonstrated a clear association between the presence of potentially pathogenic microbes (PPMs), such as Haemophilus species, Streptococcus pneumoniae, Moraxella catarrhalis, Staphylococcus aureus, Pseudomonas aeruginosa and gram-negative enteric bacteria, and the presence of neutrophilic inflammation and increased expression of cytokines, such as IL-8 and TNF, in the lower airways19–22. Lower airway colonization with PPMs has been correlated with increased severity of daily symptoms in COPD and progressive radiographic changes23. Nonetheless, precisely how these microbial events contribute to early stages of disease onset is poorly understood. Smoking alone does not seem to be associated with a significant change in the lower airway microbiome24,25; however, this situation changes as smokers start to develop early pathophysiological evidence of COPD. In a recent study of the lower airway microbiome in patients with early COPD, enrichment with Streptococcus, Staphylococcus, Prevotella and Gemella (strains that are part of the normal oral microbiota) was associated with lung function abnormalities, including an impaired response to bronchodilator therapy26. In support of a causative role of the microbiome in COPD, longitudinal enrichment with oral commensals in repeated lower airway samples results in the development of COPD-like changes in a macaque preclinical model27.
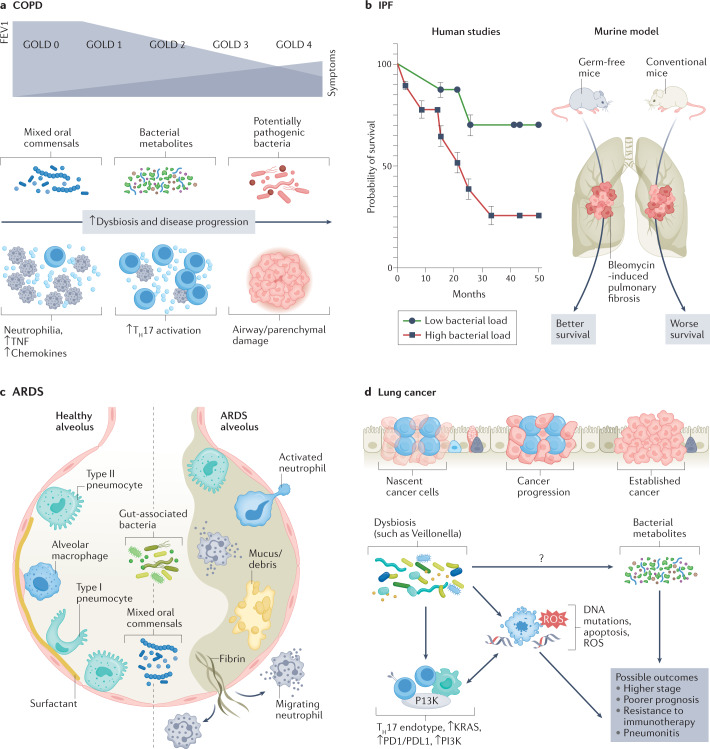
As COPD progresses, chronic inflammation results in impairment of the lung innate immune defences, which in turn paves the way for increased bacterial burden of both new and existing pathogens. Multiple investigations have shown that, in moderate-stage to advanced-stage COPD, there is enrichment with Gammaproteobacteria (which includes many PPMs such as Haemophilus and Moraxella species)14,28,29. In another study of patients with COPD, the presence of Proteobacteria-dominated microbes in sputum was associated with poorer lung function, worse scores on the Medical Research Council dyspnoea scale, and more frequent exacerbations30. In a multicentre trial involving longitudinal sampling from three large cohorts of patients with moderate to severe COPD, the airway microbiome (as assessed in sputum samples) showed clear associations between its composition and the inflammatory endotype31. Specifically, the Haemophilus-dominated sputum microbiota behaved rather stably over time and was associated with elevated IL-1β and TNF levels. These findings are supported by another study of patients with stable COPD, which similarly showed that colonization with Haemophilus influenzae was associated with increased airway inflammation32. It is therefore possible that these patients might benefit from targeted antimicrobial therapies to mitigate chronic inflammation. In contrast, patients with a ‘balanced’ microbiome profile, characterized by enrichment with several oral commensals such as Veillonella and Prevotella, exhibited a more dynamic microbiome over time and had elevations in serum and sputum IL-17A31. This subgroup was more susceptible to greater microbiome shifts during exacerbations. Thus, it is possible that these patients may benefit from a different, perhaps distinctly targeted, anti-inflammatory therapeutic approach or closer monitoring of airway ecology and pathogen acquisition, particularly at times of acute exacerbations. Importantly, increases in the relative abundance of some oral commensals, such as Campylobacter and Granulicatella, were associated with intra-patient switches from neutrophilic to eosinophilic inflammation31. Taken together, these findings are consistent with a model in which recurrent microaspiration of oral secretions starts occurring at a higher rate in early stages of disease, which contributes to lower airway inflammation; subsequently, further impairment of microbial clearance results in immune exhaustion and in eventual lower airway colonization with respiratory pathogens, a phenomenon that has been well documented in later stages of disease through both genomic-based and culture-based methods28,33–35. Thus, multiple forms of dysbiosis can be seen in patients with inflammatory airway diseases such as COPD (Fig. 3a).
Fig. 3. Disease-specific dysbiotic signals.
Different forms of dysbiosis coexist with different immune endotypes and pathological phenotypes in various disease states. a, In chronic obstructive pulmonary disease (COPD), progression to advanced stages, characterized by decrease in forced expiratory volume in one second (FEV1) and higher Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage, is hallmarked by the presence of potentially pathogenic microbes, neutrophilia, and increased inflammatory cytokines and chemokines. Newer investigations are identifying early dysbiotic events involving some oral commensals and bacterial metabolites that might precede the development of advanced-stage COPD. b, Increased bacterial burden has been shown to be associated with a poorer prognosis in patients with idiopathic pulmonary fibrosis (IPF). In animal studies, germ-free mice had a survival benefit after the induction of bleomycin-induced pulmonary fibrosis as compared to conventional mice with normal bacterial burden. c, In acute respiratory distress syndrome (ARDS), there is a relationship between gut-associated bacteria and mixed oral commensals with alveolar filling, increased mucus or cellular debris, and immune activation. d, Mixed oral commensals can play a role in the progression of lung cancer by perpetuating a T helper 17 (TH17) endotype, increased PI3K signalling and immune-checkpoint inhibition. These events, in conjunction with the more direct carcinogenesis precursors (such as DNA mutations and cell apoptosis, which further contribute to a pro-inflammatory tumour microenvironment), can have serious effects on the prognosis of lung cancer. It is likely that metabolites regulated by microbial metabolism play a significant role in cancer pathogenesis, which deserves further investigation. ROS, reactive oxygen species.
Asthma
Several investigations have focused on the airway microbiome in asthma. Although studies in early childhood with lower airway samples are lacking, multiple studies have shown associations between changes in the microbiome and development of wheezing and asthma diagnosis36–38. Even among children with an established asthma diagnosis, compositional changes in the upper airway microbiome are associated with disease control39. Evaluation of the lower airway microbiome of patients with asthma has been performed more extensively in adults. Several studies have now shown that the composition of the lower airway microbiome is associated with distinct clinical features among patients with asthma, including airway bronchial responsiveness and severity11,40. It is now understood that asthma comprises a group of very heterogeneous endotypes with a large proportion of patients with eosinophilic or type 2 (T2)-high inflammation that is more susceptible to steroids and targeted therapies. In contrast, treatment is less effective in patients with a non-T2-high inflammatory endotype (that is, patients with mostly neutrophilic airway inflammation) for reasons that remain unknown41. One intriguing hypothesis is that the lung microbiome may have a major role specifically among patients with a non-T2-high inflammatory neutrophilic endotype. For example, evaluation of the lower airway microbiota using airway brushings obtained from asthmatics showed that bacterial composition was not associated eosinophilic inflammation. In contrast, significant associations were found between the enrichment with Proteobacteria and other taxa and TH17 inflammatory markers, likely resulting in neutrophilic recruitment11. Thus, better understanding of the molecular mechanism affecting these associations may allow for the discovery of novel treatable traits among patients with asthma presenting with more refractory forms of the disease.
Idiopathic pulmonary fibrosis
The lung microbiome has important implications for disease progression and survival in various other pulmonary diseases. There are now several studies showing an association between lower airway bacterial burden and disease progression-free survival among patients with IPF42–44 (Fig. 3b). In particular, one study reported a correlation between alveolar inflammatory and fibrotic cytokines and lung microbiota diversity and composition, and lower Shannon diversity indices (within-sample diversity) were associated with higher concentrations of IL-1Ra, IL-1β, CXCL8, MIP1α, G-CSF and EGF42. In addition, alveolar concentrations of IL-6 were positively correlated with relative abundance of the lung Firmicutes phylum, whereas alveolar IL-12p70 was negatively correlated with relative abundance of the lung Proteobacteria phylum42. Other IPF studies have also shown clear associations between lung dysbiosis and both peripheral blood mononuclear cell transcriptomic expression of immune pathways and progression-free survival, suggesting that microbial–host interactions have an important role in the pathogenesis of pulmonary fibrosis44,45. However, a major limitation of these human studies is the inability to test the directionality of observed associations. In a bleomycin murine model of pulmonary fibrosis, microbial dysbiosis preceded peak lung injury and persisted until the development of fibrosis42. In contrast, germ-free mice exposed to bleomycin-induced pulmonary fibrosis had a mortality benefit compared to conventional mice42 (Fig. 3b). In another preclinical murine model, Bacteroides and Prevotella species were associated with increased fibrotic pathogenesis through IL-17R signalling46. Taken together, these data suggest that certain microbial exposures may act as persistent stimuli for repetitive alveolar injury, ultimately contributing to pulmonary fibrosis and continual inflammatory injury in this disease.
Acute respiratory distress syndrome
Both in preclinical models of sepsis and in humans with established acute respiratory distress syndrome, the lung microbiome likely plays a major role in the pathogenesis of the disease (Fig. 3c). Using culture-independent methods, a study from 2016 demonstrated that enrichment of the lower airways with gut-associated bacteria correlated with concentrations of alveolar TNF, a key mediator of alveolar inflammation in acute respiratory distress syndrome47. In addition, a recent study of patients with COVID-19 on mechanical ventilation demonstrated worse clinical outcomes among patients whose lower airway microbiome was enriched with Mycoplasma salivarium, another oral commensal48. Additionally, a recent large study of over 300 patients on mechanical ventilation demonstrated that enrichment of the lower airways with Staphylococcus or Pseudomonadaceae was associated with increased lower airway inflammation and worse clinical outcomes, including poorer 30-day survival and longer time to liberation from mechanical ventilation49. Taken together, these data suggest that microbial dysbiosis may have an important role in the dysregulation of local and systemic inflammation that consequently impacts clinical outcomes in acute respiratory failure.
The lung microbiome and immune surveillance
Although much research has focused on how the lung microbiome promotes inflammatory states, its effects on immune surveillance are much less understood but are likely very important. Among patients with cystic fibrosis, a reduction in alpha diversity, which quantifies microbial diversity within a sample, is directly correlated with disease severity and colonization with bacterial pathogens50. In many cases, this is likely due to the emergence of a dominant pathogen that increases in relative abundance. In addition, decreased microbial diversity sometimes precedes the development of cystic fibrosis exacerbations51. These data suggest that a reduction in biodiversity over time can render the lower airways more susceptible to colonization with a dominant pathogen such as Pseudomonas or Burkholderia species.
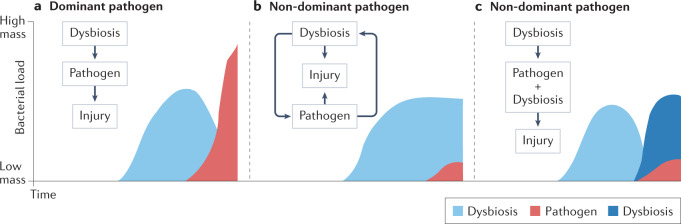
However, there are different possible scenarios in which dysbiosis might affect host susceptibility to pathogens. Figure 4 depicts some of these scenarios in which pathogens become either dominant or non-dominant in the lower airway microbiome. For example, in patients with bronchiectasis, those with Pseudomonas aeruginosa lower airway colonization experience more frequent exacerbations and often have more severe disease. Colonization with Pseudomonas, which, when present, tends to dominate the airway microbiota, has been associated with a distinct lower airway host immune profile characterized by severe neutrophilic inflammation, including high levels of neutrophil elastase52. However, the microbial events that occur prior to acquisition of Pseudomonas are unknown (Fig. 4a). Moreover, it remains to be determined whether we can identify the lower airway dysbiosis that precedes Pseudomonas colonization. Such investigations would require longitudinal sampling of the lower airways prior to the emergence of a dominant pathogen. In contrast, there are other respiratory pathogens that, when present, frequently do not dominate the lower airway microbial community (Fig. 4b,c). For example, non-tuberculous mycobacterium (NTM) is frequently a non-dominant infectious pathogen, thus potentially allowing for the identification of dysbiotic signatures beyond the presence of the pathogen itself53. In that case, it is possible that an initial lower airway dysbiosis that favours host susceptibility to NTM persists after acquisition of this pathogen, possibly contributing to the inflammatory injury in this disease (Fig. 4b). Interestingly, in a small case series of patients with NTM disease, levels of inflammatory cytokines measured in the lower airways correlated with relative abundances of several oral commensals in the lower airways rather than with the abundance of Mycobacterium53. Alternatively, in the case of non-dominant pathogens, a pre-pathogen lower airway dysbiosis may lead not only to acquisition of the non-dominant pathogen but also to a replacement by another dysbiotic signature, which may be important for the survival of the pathogen itself (microorganism–microorganism interaction) and affect the inflammatory injury (Fig. 4c).
Fig. 4. Dysbiotic events rendering increased host susceptibility to pathogens.
a, In cases of acquisition of a dominant pathogen, dysbiotic changes occurring prior to pathogen seeding may promote host susceptibility. b, In cases of non-dominant respiratory pathogens, changes in the lower airway microbiome might continue as a dysbiotic signature once pathogens are established in the lower airways causing lung injury. c, Alternatively, non-dominant pathogens could lead to a new dysbiotic signature that favours pathogen survival and contributes to lung injury.
HIV
As another example of dysbiosis that might result in decreased host immune surveillance, people living with HIV continue to experience higher rates of respiratory infections and lung cancer despite widespread use of antiretroviral therapy (ART). It is hypothesized that residual levels of HIV in deep tissues leads to chronic immune activation and lung inflammation. This longstanding pro-inflammatory state can eventually contribute to immune cell exhaustion and pulmonary immune dysregulation, thereby making people living with HIV be at higher risk for pulmonary infections and malignancies54. Several studies have tried to identify dysbiotic signals present in HIV, including a multicentre trial that described the presence of high levels of Tropheryma whipplei in the lungs of some individuals with HIV55; of note, however, this has also been observed in smokers and other cohorts, and thus may not be unique to HIV24. Further, the lung microbiome in people living with HIV, especially among those with advanced HIV, shows an increased burden of Prevotella and Veillonella species56. Even with the immune reconstitution that occurs after initiation of ART, there is persistent enrichment of the lower airway microbiota with these oral commensals56. This pattern of microbial dysbiosis is associated with subclinical inflammation, characterized by an increase in neutrophils and lymphocytes9,10. Further, patients with HIV who are treatment naive have been shown to have decreased biodiversity and greater inter-sample diversity compared to healthy controls, suggesting that this dysbiotic signature may be prominent in patients with HIV and may confer increased susceptibility to pneumonia57. In patients with HIV treated with ART, enrichment of the lower airways with oral taxa was associated with increased production of microbial SCFAs13. As shown in one longitudinal cohort study of South African patients with HIV treated with ART, SCFAs have direct inhibitory effects on T cell function via the suppression IFNγ and IL-17 production and are associated with an increased risk of tuberculosis. Given the well-known immunoregulatory properties of SCFAs and the evidence that these molecules could be produced through fermentation of aspirated oral anaerobes3, it is plausible that this may be one potential mechanism by which the lower airway microbiome impairs host immune surveillance to pathogens.
Cancer
Beyond associations with susceptibility to pathogens, several investigations have suggested a link between lower airway dysbiosis and lung cancer. Epidemiological studies and culture-dependent techniques have shown that chronic or recurrent bacterial infections are common in patients with lung cancer over the course of their disease, and post-obstructive pneumonia can negatively impact their response to therapy and overall survival58. Studies have reported a correlation between certain respiratory pathogens, such as Mycobacterium tuberculosis, with lung cancer, and a systematic review found a significantly increased lung cancer risk associated with pre-existing tuberculosis, independent of tobacco exposure59. This might be due to tuberculosis being associated with inflammatory mediators, such as TNF, and excessive extracellular matrix turnover. Mechanistically, the increase in pathogenic bacteria can lead to chronic inflammation through the persistent generation of inflammatory mediators, thereby affecting cellular apoptosis and increasing risk for DNA mutations60. Bacterial metabolites, such as reactive oxygen and nitrogen species, can cause direct DNA damage and disrupt a variety of signalling pathways, thereby generating a pro-carcinogenic environment60–62 (Fig. 3d). A number of bacterial metabolites, including SFCAs, have also been implicated in both the regulation of host metabolism in the lungs and in signalling pathways63. Thus, it is possible that alterations in bacteria-derived molecules can affect the tumour microenvironment and oncogenic signals. This delicate balance between tolerance and immune activation in the lung microenvironment can be disrupted by the overuse of antibiotics for the treatment of infections. For example, preclinical models using broad-spectrum antibiotics demonstrated the importance of commensal bacteria in supporting the host immune response against cancer, defined an important role for γδ TH17 cell responses in the mechanism and showed deleterious effects of antibiotic treatment on cancer progression64.
With the growing use of culture-independent methods, we can now appreciate microbiome differences between patients with and without lung cancer diagnoses. The microbiota of lung tumours is characterized by a lower alpha diversity compared to non-malignant lung tissue samples, an association that seems to be independent of tobacco use, environmental exposures and clinical staging65. Among taxonomic signatures, associations have been described between lung malignancy and increased relative abundance of oral commensals such as Streptococcus, Prevotella, Veillonella, Rothia, Megasphaera and Acidovorax17,66. Other less common respiratory pathogens, such as Staphylococcus and bacteria from the Firmicutes and Saccharibacteria (formerly known as TM7) phyla, have also been found in lower airway samples from patients with lung cancer67. Microbial differences have also been noted among various types of cancer. One study found that Brevundimonas, Acinetobacter and Propionibacterium were more enriched in lung adenocarcinoma whereas Enterobacter was more enriched in squamous cell carcinoma68. Another study demonstrated an association between the Acidovorax genus and squamous cell carcinoma among smokers, particularly in tumours harbouring TP53 mutations69; it is also likely that some of these microbial exposures contribute to a host immune response that may affect carcinogenesis. Indeed, human data show that enrichment with supraglottic-predominant taxa, such as Streptococcus, Prevotella and Veillonella, is associated with upregulation of transcriptomic pathways that are known to have a major pathogenic role in carcinogenesis such as ERK and PI3K17. Upregulation of the PI3K pathway occurs early in lung carcinogenesis and may affect proliferation, apoptosis and cell survival as well as the induction of PDL1 expression70,71. Thus, these data demonstrate how a dysregulated microbiota might increase the risk of lung cancer development. Furthermore, exposure of KRAS-mutated bronchial epithelial cells to either the supernatant of these taxa or their heat-killed products leads to similar upregulation of transcriptomic signatures to those seen in human data17. Overall, although the specific bacterial taxa identified varied somewhat from one study to another depending on the sample type and patient cohort, the consistent observation is that lung cancer is associated with a dysregulated local microbiome characterized by reduced alpha diversity and altered bacterial composition (Fig. 3d).
The lung microbiome may have a role in the progression of lung cancer or its risk for recurrence. Among patients with surgically resected early-stage non-small-cell lung cancer (NSCLC), recurrence was associated with a higher diversity and enrichment of Bacteroidaceae, Lachnospiraceae and Ruminococcaceae in the lung tissue microbiome72. In another study of patients who underwent surgical resection of NSCLC, bronchoalveolar lavage enrichment with Sphingomonas, Psychromonas and Serratia and a reduced abundance of Cloacibacterium, Geobacillus and Brevibacterium were observed in patients who had early lung cancer recurrence compared to those without recurrence in the subsequent 3 years73. Importantly, pre-surgery bronchoalveolar lavage microbiome was associated with the tumour expression of genes involved in cell proliferation, immunity and signal transduction. Distinct microbial differences can also be noted among patients with different stage of disease. For example, the genus Thermus was reported to be more abundant in tumour tissues from patients with advanced stage disease (TNM stages IIIB–IV), and Legionella abundance was higher in patients who developed metastases65. Further, in a recent study, a lower airway dysbiotic signature characterized by enrichment with oral commensals was more prevalent in individuals with TNM stage IIIB–IV NSCLC compared to those with the disease at earlier stages, and a similar microbial signal was associated with poor prognosis and increased risk for mortality74. Mechanistically, multi-omics integration of the microbiome and host transcriptomic data from the lower airways of patients with lung cancer showed that several of these oral commensals, including Veillonella and Streptococcus species, were associated with upregulation of the IL-17, PI3K, MAPK and ERK pathways in the airway transcriptome75. Preclinical mouse models of lung adenocarcinoma show that lower airway dysbiosis, induced by intratracheal aspiration of some of these oral commensals, can lead to TH17 inflammation, increased expression of immune-checkpoint inhibitor molecules (PD1) by T cells, accelerated tumour progression and mortality74. Using germ-free preclinical lung cancer models, others have shown that commensal taxa stimulate Myd88-dependent IL-1β and IL-23 production from myeloid cells, activate IL-17 production by γδ T cells and lead to a pro-inflammatory state that induces tumour proliferation76. Furthermore, preclinical evidence suggests that direct blocking of the IL-17 pathway through anti-IL-17 monoclonal antibodies can slow tumour growth in the setting of lower airway dysbiosis induced by aspiration of oral commensals74. These findings clearly link local microbiota–immune crosstalk to lung tumour development and thereby identify candidate cellular and molecular mediators that can be targeted for anticancer therapy.
The evidence described above shows some lack of consistency in the specific microbial taxa associated with pulmonary malignancy. Inconsistencies between studies may be due to socio-demographic differences inpatient cohorts or differences in research techniques employed, a phenomenon similarly noted in studies of gut microbiome signals associated with malignancies. For example, while gut microbiomic signatures have been found to be associated with inflammatory tone and susceptibility to immunotherapy in melanoma and NSCLC, the specific taxa identified have been inconsistent among different studies77–80. It is also possible that taxonomic signals are markers of functional microbial differences but evaluation of this hypothesis is prevented by a lack of studies focusing on microbial metabolism in the lung (for example, using approaches such as metagenome, metatranscriptome, metabolome or proteome). However, despite these different taxonomic signals, the host signals associated with lower airway dysbiosis seem to similarly point towards a dysregulated immune response with a pro-inflammatory TH17 state and increased expression of immune-checkpoint inhibition, which might impair effective immune surveillance of nascent malignant cells81. Importantly, tumour microbiome studies are often limited by a very low microbial biomass and are thus prone to sequencing noise and DNA contamination from kits and reagents, although various methods have been proposed on how to effectively deal with both of these issues82. Thus, although recent studies indicate that the lung microbiome may soon become a critical diagnostic and preventive biomarker for lung cancer stage, genotype and risk factor stratification83, further mechanistic and large-scale clinical studies are required to dissect its precise role in tumorigenesis.
Conclusions and future directions
Studying the lung microbiome in varying disease states has identified microbial signatures associated with diagnosis, illness severity and prognosis. However, a lack of consistent findings across studies makes it difficult to clearly define lung dysbiosis. It is quite possible that even within a single disease process there are many forms of dysbiosis, a concept that has previously been referred to as the ‘Anna Karenina principle’ in reference to Tolstoy’s writings that “happy families are all alike; every unhappy family is unhappy in its own way.”84 Even less established is how we define lung microbial eubiosis. Part of the difficulty may reside in the nature of the lung microbiome, with its low biomass resulting in considerable background noise in sequencing data85,86, and the significant impact that the dynamic pendulum between immigration and elimination of microbes in the lung environment has on our assessments. Thus, contrasting with the Anna Karenina principle, all happy families (eubiosis) may not look alike, at least based on static snapshots of the lung microbiome. To add further complexity, even within the same individual, topographical differences in the lung microbiome exist9,14. Defining microbial resilience in such a dynamic environment is thus inherently difficult. As the use of culture-independent and multi-omics methods becomes more widespread, understanding these principals is essential for the successful development of novel diagnostic and therapeutic modalities targeting microbial–host interactions.
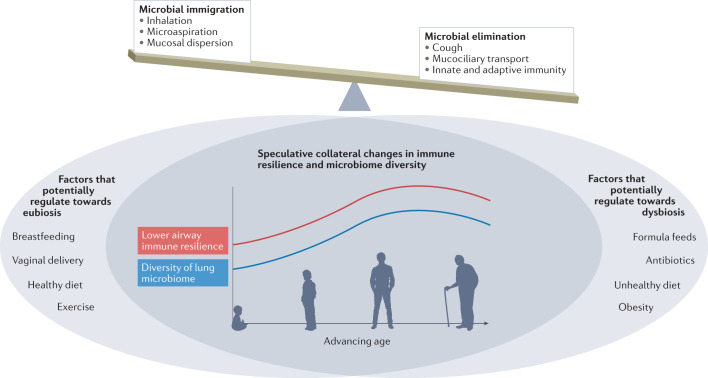
In Fig. 5, we illustrate an overarching speculative view of how the delicate balance between microbial immigration (driven by inhalation, microaspiration and mucosal dispersion) and microbial elimination (driven by cough, mucociliary transport and immune mechanisms) establishes a complex microbial–host interface. This interface is likely affected by many factors, including those related to both the host and the environment. The exact effects that each of these factors has on the microbiome are relatively unknown, although we can speculate about the existence of some beneficial and other detrimental factors that might change throughout the course of our lives (Fig. 5). We postulate that factors such as vaginal delivery87, exercise and a healthy diet contribute towards lower airway eubiosis, either directly or via the gut–lung axis. Conversely, obesity, an unhealthy diet and frequent antibiotic use could disrupt the lower airway microbiome in an unfavourable manner. The effect of breastfeeding versus formula feeds is also unknown, with only a single randomized controlled trial showing no difference in the incidence of gastrointestinal and respiratory infections in breastfed versus formula-fed infants, although this study did not specifically investigate the microbiome88. Finally, we surmise that lower airway immune resilience and the diversity of the lung microbiome likely change as we age, and the effect of external factors on the lung microbiome may therefore change across the lifespan. Well-designed controlled studies are needed to evaluate the potential effects of these environmental factors on the lung microbiome and host immune response. Careful consideration of age, sex and other important clinical conditions are key to a better understanding of how the lower airway microbiota influences lower airway immune tone. Thus, future investigations will need to move away from cross-sectional designs and correlative studies towards longitudinal sampling approaches that account for topographical characteristics and experimental conditions (Box 2). Further, although understanding the human lung microbiome requires human studies, preclinical models allow for better control of microbial and host conditions as well as the implementation of experimental approaches under controlled circumstances. These mechanistic approaches are needed to understand how the lung microbiome affects host susceptibility to inflammatory processes, infections and malignancy.
Fig. 5. Potential modifiers of lower airway microbiome course.
The dynamic nature of the lower airway microbiome is ever evolving. The overarching factors that control the balance of the microbiota are microbial immigration via inhalation and microaspiration and microbial elimination via cough, mucociliary transport and immune responses. We postulate that factors such as vaginal delivery87, exercise and a healthy diet may contribute towards eubiosis in the lower airway, whereas obesity, an unhealthy diet and frequent antibiotic use could disrupt the lower airway microbiome in an unfavourable manner. Finally, we surmise that lower airway immune resilience and the diversity of the lung microbiome (a symbol of health in its own right) follows a quasi-sinusoidal path, peaking in adulthood.
Next, we must consider the potential therapeutic impact that modulating the lung microbiome might have on respiratory diseases. Currently, many existing and widely used interventions beyond antibiotics, such as inhaled corticosteroids and cystic fibrosis transmembrane conductance regulator modulators, already alter the respiratory microbiota89–91. In fact, there are now several potential mechanisms that have been proposed to explain how corticosteroid therapy might impact epithelial barrier function and consequently alter airway ecology, including the increased expression of epithelial genes involved in tight junction promotion and the upregulation of IL-17 and TNF inflammatory pathways92. Beyond corticosteroids and cystic fibrosis transmembrane conductance regulator modulators, several other interventions have been trialled to modulate the lung microbiota with varied success. For example, oral probiotics have been studied in the context of both cystic fibrosis and ventilator-associated pneumonia, although these likely target shifts in the gut microbiome and their effects on the respiratory microbiome remain unknown93. Bacteriophage therapy has similarly been trialled, although its use is still in its infancy94. Other studies have employed both inhaled and systemic antibiotics to modulate the respiratory microbiota but results are mixed across various pulmonary diseases95.
Unlike the gut microbiome, in which probiotics can be used to restore normal gastrointestinal flora, no such respiratory probiotic currently exists. However, in a preclinical model, episodic instillation of oral commensals into the lower respiratory tract modified host susceptibility to respiratory pathogens4. While these types of investigations may provide the biological plausibility that microbiome modifications in the lung could provide health benefits, mechanistic studies that include both preclinical work and individuals with respiratory diseases are needed. These type of investigations in turn might allow us to identify microbial signatures associated with less inflammation or decreased pathogen susceptibility. Until we understand what a beneficial ecological target might look like — a so-called ‘ideal’ respiratory microbiome — we are limited in our ability to implement safe and meaningful clinical trials.
In conclusion, there is a need to embrace the dynamic nature of microbes visiting the lower airways, particularly as it relates to host immune tone and the pathogenesis of multiple different disease processes. Novel clinical applications need to account for the evolutionary nature of the lung microbiome where eubiosis and dysbiosis truly exist on a spectrum.
Box 2 Lung transplantation as an opportunity to examine longitudinal changes in the microbiome.
Despite the dramatic increase in studies utilizing culture-independent methods to profile the lung microbiome, most research to date provides only a static view of the lung microbiome, usually at a given stage of disease. Prior studies have shown differences after lung transplantation in both bacterial load and composition in comparison to healthy controls and non-lung transplant recipients with chronic lung disease109–112. However, even in lung transplantation, where patients routinely undergo surveillance bronchoscopies to monitor for signs of infection and rejection, most studies have only explored whether the composition and function of the lung microbiome at a pre-specified time point is associated with subsequent disease development and progression. For example, a recent prospective study demonstrated increased risk for chronic lung allograft dysfunction (CLAD) and death among asymptomatic lung transplant recipients that develop increased lung bacterial burden113. In addition, prior observational studies have suggested that Actinobacteria dominance (Propionibacterium and Corynebacterium species) early after lung transplantation is associated with lower rates of chronic and acute rejection and less inflammation114, whereas infection with Chlamydia pneumoniae is associated with higher rates of acute and chronic rejection115. Furthermore, one study found that the presence of certain pathogens from the Firmicutes and Proteobacteria phyla was associated with increased inflammatory gene expression (such as TNF and COX2) whereas enrichment with Bacteroidetes was associated with a tissue remodelling profile (such as MMP12, TIMP1 and PDGFD)116. Collectively, these data suggest that microbial dysbiosis likely has implications on allograft function and survival after lung transplantation. How the dynamic nature of the microbial–host interface changes over time remains unclear.
Longitudinal evaluation of changes occurring in the lung microbiome may provide further insight into the evolving microbial–host interaction and how it affects disease progression. A number of studies have demonstrated that dynamic changes to the lung microbiome occur after transplantation, in which a reduction of alpha diversity correlates with a decline in lung function, increase in infection rates and allograft failure109,110,112,117. Different taxa tend to dominate the lung microbiome in the first year after transplantation, with Bacteroidetes being the most abundant phylum, followed by Firmicutes, Proteobacteria and Actinobacteria116,117. Over time, there is often enrichment with Proteobacteria, particularly Pseudomonas, which has been linked to development of CLAD in multiple studies110,118–120. One study in particular revealed two species within the genus Pseudomonas — P. aeruginosa and P. fluorescens — and observed lower biodiversity, more bacterial DNA, greater neutrophilia and increased clinical symptoms among patients with a higher relative abundance of these taxa112. Interestingly, another study found that recolonization of the allograft with Pseudomonas in individuals with cystic fibrosis was not associated with CLAD109. Thus, further longitudinal investigations are needed to better understand the factors that determine risks for complications among patients undergoing lung transplantation.
Given the frequency with which lung transplant recipients undergo surveillance bronchoscopies, there is an untapped opportunity to longitudinally examine the evolutionary nature of the lung microbiome after transplantation and study its relationship to lower airway immune tone and clinical outcomes. Further, there are several unique factors occurring after lung transplantation that make it a highly useful disease model to study microbial–host interactions. First, sample acquisition can be made prior to development of disease, including both bacterial, viral, and fungal infections and inflammatory complications (that is, acute and chronic rejection). Second, the samples are often rich in microbes due to their impaired mechanical clearance, such due to a defective afferent cough reflex, anastomotic mechanical barriers and mucociliary dysfunction. Third, frequent microaspiration results from a high incidence of oropharyngeal dysphagia, gastroesophageal reflux disease and gastroparesis after lung transplantation. Finally, significant heterogeneity in prescribed immunosuppressive regimens exists, thus altering the host response to varying degrees.
Acknowledgements
This work was supported by grant R37 CA244775 (LNS, NCI/NIH).
Author contributions
The authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Microbiology thanks Christopher Brightling, Michael Surette and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
All authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Steenhuijsen Piters WAA, Binkowska J, Bogaert D. Early life microbiota and respiratory tract infections. Cell Host Microbe. 2020;28:223–232. doi: 10.1016/j.chom.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Sulaiman I, et al. Functional lower airways genomic profiling of the microbiome to capture active microbial metabolism. Eur. Respir. J. 2021 doi: 10.1183/13993003.03434-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu BG, et al. Episodic aspiration with oral commensals induces a MyD88-dependent, pulmonary T-helper cell type 17 response that mitigates susceptibility to Streptococcus pneumoniae. Am. J. Respir. Crit. Care Med. 2021;203:1099–1111. doi: 10.1164/rccm.202005-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pattaroni C, et al. Early-life formation of the microbial and immunological environment of the human airways. Cell Host Microbe. 2018;24:857–865.e4. doi: 10.1016/j.chom.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Thorsen J, et al. Infant airway microbiota and topical immune perturbations in the origins of childhood asthma. Nat. Commun. 2019;10:5001. doi: 10.1038/s41467-019-12989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickson RP, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann. Am. Thorac. Soc. 2015;12:821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charlson ES, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RP, et al. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio. 2017 doi: 10.1128/mBio.02287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal LN, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YJ, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J. Allergy Clin. Immunol. 2015;136:874–884. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson RP, et al. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am. J. Respir. Crit. Care Med. 2018 doi: 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal LN, et al. Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host Microbe. 2017;21:530–537.e4. doi: 10.1016/j.chom.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erb-Downward JR, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev. Respir. Med. 2013;7:245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Nuñez M, et al. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014;52:4217–4223. doi: 10.1128/JCM.01967-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsay JJ, et al. Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 2018;198:1188–1198. doi: 10.1164/rccm.201710-2118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gustafson AM, et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Transl. Med. 2010;2:26ra25. doi: 10.1126/scitranslmed.3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006;173:991–998. doi: 10.1164/rccm.200509-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soler N, et al. Airway inflammation and bronchial microbial patterns in patients with stable chronic obstructive pulmonary disease. Eur. Respir. J. 1999;14:1015–1022. doi: 10.1183/09031936.99.14510159. [DOI] [PubMed] [Google Scholar]
- 21.Bresser P, Out TA, van Alphen L, Jansen HM, Lutter R. Airway inflammation in nonobstructive and obstructive chronic bronchitis with chronic haemophilus influenzae airway infection. Comparison with noninfected patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2000;162:947–952. doi: 10.1164/ajrccm.162.3.9908103. [DOI] [PubMed] [Google Scholar]
- 22.Parameswaran GI, Wrona CT, Murphy TF, Sethi S. Moraxella catarrhalis acquisition, airway inflammation and protease-antiprotease balance in chronic obstructive pulmonary disease. BMC Infect. Dis. 2009;9:178. doi: 10.1186/1471-2334-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rangelov K, Sethi S. Role of infections. Clin. Chest Med. 2014;35:87–100. doi: 10.1016/j.ccm.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris A, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segal LN, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opron K, et al. Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort. NPJ Biofilms Microbiomes. 2021;7:14. doi: 10.1038/s41522-021-00185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris A, et al. Longitudinal analysis of the lung microbiota of cynomolgous macaques during long-term SHIV infection. Microbiome. 2016;4:38. doi: 10.1186/s40168-016-0183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sze MA, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012;185:1073–1080. doi: 10.1164/rccm.201111-2075OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One. 2012;7:e47305. doi: 10.1371/journal.pone.0047305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dicker AJ, et al. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2020 doi: 10.1016/j.jaci.2020.02.040. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, et al. Inflammatory endotype-associated airway microbiome in chronic obstructive pulmonary disease clinical stability and exacerbations: a multicohort longitudinal analysis. Am. J. Respir. Crit. Care Med. 2021;203:1488–1502. doi: 10.1164/rccm.202009-3448OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tufvesson E, Bjermer L, Ekberg M. Patients with chronic obstructive pulmonary disease and chronically colonized with Haemophilus influenzae during stable disease phase have increased airway inflammation. Int. J. Chron. Obstruct. Pulmon. Dis. 2015;10:881–889. doi: 10.2147/COPD.S78748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sze MA, et al. The host response to the lung microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2015 doi: 10.1164/rccm.201502-0223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabello H, et al. Bacterial colonization of distal airways in healthy subjects and chronic lung disease: a bronchoscopic study. Eur. Respir. J. 1997;10:1137–1144. doi: 10.1183/09031936.97.10051137. [DOI] [PubMed] [Google Scholar]
- 35.Monso E, et al. Risk factors for lower airway bacterial colonization in chronic bronchitis. Eur. Respir. J. 1999;13:338–342. doi: 10.1034/j.1399-3003.1999.13b20.x. [DOI] [PubMed] [Google Scholar]
- 36.Teo SM, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teo SM, et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe. 2018;24:341–352.e5. doi: 10.1016/j.chom.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosch A, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. a prospective cohort study. Am. J. Respir. Crit. Care Med. 2017;196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat. Commun. 2019;10:5714. doi: 10.1038/s41467-019-13698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang YJ, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011;127:372–381. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudey SN, Ledford DK, Cardet JC. Mechanisms of non-type 2 asthma. Curr. Opin. Immunol. 2020;66:123–128. doi: 10.1016/j.coi.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Dwyer DN, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1127–1138. doi: 10.1164/rccm.201809-1650OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molyneaux PL, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molyneaux PL, et al. Host-microbial interactions in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2017 doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, et al. Microbes are associated with host innate immune response in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2017;196:208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang D, et al. Dysregulated lung commensal bacteria drive interleukin-17B production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity. 2019;50:692–706.e7. doi: 10.1016/j.immuni.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Dickson RP, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sulaiman I, et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat. Microbiol. 2021;6:1245–1258. doi: 10.1038/s41564-021-00961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitsios GD, et al. Respiratory tract dysbiosis is associated with worse outcomes in mechanically ventilated patients. Am. J. Respir. Crit. Care Med. 2020;202:1666–1677. doi: 10.1164/rccm.201912-2441OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl Acad. Sci. Usa. 2012;109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carmody LA, et al. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann. Am. Thorac. Soc. 2013;10:179–187. doi: 10.1513/AnnalsATS.201211-107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oriano M, et al. Sputum neutrophil elastase associates with microbiota and Pseudomonas aeruginosa in bronchiectasis. Eur. Respir. J. 2020 doi: 10.1183/13993003.00769-2020. [DOI] [PubMed] [Google Scholar]
- 53.Sulaiman I, et al. Evaluation of the airway microbiome in nontuberculous mycobacteria disease. Eur. Respir. J. 2018 doi: 10.1183/13993003.00810-2018. [DOI] [PubMed] [Google Scholar]
- 54.Alexandrova Y, Costiniuk CT, Jenabian MA. Pulmonary immune dysregulation and viral persistence during HIV infection. Front. Immunol. 2021;12:808722. doi: 10.3389/fimmu.2021.808722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lozupone C, et al. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am. J. Respir. Crit. Care Med. 2013;187:1110–1117. doi: 10.1164/rccm.201211-2145OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Twigg HL, III, et al. Effect of advanced HIV infection on the respiratory microbiome. Am. J. Respir. Crit. Care Med. 2016;194:226–235. doi: 10.1164/rccm.201509-1875OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Selwyn PA, et al. Increased risk of bacterial pneumonia in HIV-infected intravenous drug users without AIDS. AIDS. 1988;2:267–272. doi: 10.1097/00002030-198808000-00005. [DOI] [PubMed] [Google Scholar]
- 58.Qiao D, et al. A retrospective study of risk and prognostic factors in relation to lower respiratory tract infection in elderly lung cancer patients. Am. J. Cancer Res. 2015;5:423–432. [PMC free article] [PubMed] [Google Scholar]
- 59.Liang HY, et al. Facts and fiction of the relationship between preexisting tuberculosis and lung cancer risk: a systematic review. Int. J. Cancer. 2009;125:2936–2944. doi: 10.1002/ijc.24636. [DOI] [PubMed] [Google Scholar]
- 60.Ho LJ, et al. Increased risk of secondary lung cancer in patients with tuberculosis: a nationwide, population-based cohort study. PLoS One. 2021;16:e0250531. doi: 10.1371/journal.pone.0250531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gomes M, Teixeira AL, Coelho A, Araujo A, Medeiros R. The role of inflammation in lung cancer. Adv. Exp. Med. Biol. 2014;816:1–23. doi: 10.1007/978-3-0348-0837-8_1. [DOI] [PubMed] [Google Scholar]
- 62.Dong Q, Chen ES, Zhao C, Jin C. Host-microbiome interaction in lung cancer. Front. Immunol. 2021;12:679829. doi: 10.3389/fimmu.2021.679829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vieira RS, et al. Butyrate attenuates lung inflammation by negatively modulating Th9 cells. Front. Immunol. 2019;10:67. doi: 10.3389/fimmu.2019.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng M, et al. Microbiota modulate tumoral immune surveillance in lung through a γδT17 immune cell-dependent mechanism. Cancer Res. 2014;74:4030–4041. doi: 10.1158/0008-5472.CAN-13-2462. [DOI] [PubMed] [Google Scholar]
- 65.Yu G, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. doi: 10.1186/s13059-016-1021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee SH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. doi: 10.1016/j.lungcan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 67.Liu HX, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer. 2018;142:769–778. doi: 10.1002/ijc.31098. [DOI] [PubMed] [Google Scholar]
- 68.Gomes S, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 2019;9:12838. doi: 10.1038/s41598-019-49195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greathouse KL, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee SK, et al. IFN-gamma regulates the expression of B7-H1 in dermal fibroblast cells. J. Dermatol. Sci. 2005;40:95–103. doi: 10.1016/j.jdermsci.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 71.Chen J, et al. Interferon-gamma-induced PD-L1 surface expression on human oral squamous carcinoma via PKD2 signal pathway. Immunobiology. 2012;217:385–393. doi: 10.1016/j.imbio.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 72.Peters BA, et al. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol. Biomark. Prev. 2019;28:731–740. doi: 10.1158/1055-9965.EPI-18-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patnaik SK, et al. Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2021;161:419–429.e6. doi: 10.1016/j.jtcvs.2020.01.104. [DOI] [PubMed] [Google Scholar]
- 74.Tsay JJ, et al. Lower airway dysbiosis affects lung cancer progression. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-20-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsay JJ, et al. Lower airway dysbiosis affects lung cancer progression. Cancer Discov. 2021;11:293–307. doi: 10.1158/2159-8290.CD-20-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin C, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176:998–1013.e16. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 78.Matson V, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gopalakrishnan V, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chaput N, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 81.Akbay EA, et al. Interleukin-17A promotes lung tumor progression through neutrophil attraction to tumor sites and mediating resistance to PD-1 blockade. J. Thorac. Oncol. 2017;12:1268–1279. doi: 10.1016/j.jtho.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh S, Natalini JG, Segal LN. Lung microbial-host interface through the lens of multi-omics. Mucosal Immunol. 2022 doi: 10.1038/s41385-022-00541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poore GD, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579:567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2017;2:17121. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 85.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Erb-Downward JR, et al. Critical relevance of stochastic effects on low-bacterial-biomass 16S rRNA gene analysis. mBio. 2020 doi: 10.1128/mBio.00258-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pattaroni C, et al. Early-life formation of the microbial and immunological environment of the human airways. Cell Host Microbe. 2018;24:857–865.e4. doi: 10.1016/j.chom.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 88.Kramer MS, et al. Promotion of breastfeeding intervention trial (PROBIT): a randomized trial in the Republic of Belarus. JAMA. 2001;285:413–420. doi: 10.1001/jama.285.4.413. [DOI] [PubMed] [Google Scholar]
- 89.Contoli M, et al. Long-term effects of inhaled corticosteroids on sputum bacterial and viral loads in COPD. Eur. Respir. J. 2017 doi: 10.1183/13993003.00451-2017. [DOI] [PubMed] [Google Scholar]
- 90.Durack J, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 2017;140:63–75. doi: 10.1016/j.jaci.2016.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hisert KB, et al. Restoring cystic fibrosis transmembrane conductance regulator function reduces airway bacteria and inflammation in people with cystic fibrosis and chronic lung infections. Am. J. Respir. Crit. Care Med. 2017;195:1617–1628. doi: 10.1164/rccm.201609-1954OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramsheh MY, et al. Lung microbiome composition and bronchial epithelial gene expression in patients with COPD versus healthy individuals: a bacterial 16S rRNA gene sequencing and host transcriptomic analysis. Lancet Microbe. 2021;2:e300–e310. doi: 10.1016/S2666-5247(21)00035-5. [DOI] [PubMed] [Google Scholar]
- 93.Johnstone J, et al. Effect of probiotics on incident ventilator-associated pneumonia in critically ill patients: a randomized clinical trial. JAMA. 2021;326:1024–1033. doi: 10.1001/jama.2021.13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Law N, et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection. 2019;47:665–668. doi: 10.1007/s15010-019-01319-0. [DOI] [PubMed] [Google Scholar]
- 95.Chotirmall SH, et al. Therapeutic targeting of the respiratory microbiome. Am. J. Respir. Crit. Care Med. 2022 doi: 10.1164/rccm.202112-2704PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang L, et al. Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study. Respir. Res. 2019;20:265. doi: 10.1186/s12931-019-1218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu N, et al. Rapid identification of pathogens associated with ventilator-associated pneumonia by Nanopore sequencing. Respir. Res. 2021;22:310. doi: 10.1186/s12931-021-01909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Endimiani A, et al. Are we ready for novel detection methods to treat respiratory pathogens in hospital-acquired pneumonia? Clin. Infect. Dis. 2011;52:S373–S383. doi: 10.1093/cid/cir054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee SH, et al. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J. Microbiol. Immunol. Infect. 2019;52:920–928. doi: 10.1016/j.jmii.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zacharioudakis IM, et al. Evaluation of a multiplex PCR panel for the microbiological diagnosis of pneumonia in hospitalized patients: experience from an Academic Medical Center. Int. J. Infect. Dis. 2021;104:354–360. doi: 10.1016/j.ijid.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 101.Langelier C, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl Acad. Sci. USA. 2018;115:E12353–E12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nelson MT, et al. Maintenance tobramycin primarily affects untargeted bacteria in the CF sputum microbiome. Thorax. 2020;75:780–790. doi: 10.1136/thoraxjnl-2019-214187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Segal LN, et al. Randomised, double-blind, placebo-controlled trial with azithromycin selects for anti-inflammatory microbial metabolites in the emphysematous lung. Thorax. 2017;72:13–22. doi: 10.1136/thoraxjnl-2016-208599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kingah PL, Muma G, Soubani A. Azithromycin improves lung function in patients with post-lung transplant bronchiolitis obliterans syndrome: a meta-analysis. Clin. Transpl. 2014;28:906–910. doi: 10.1111/ctr.12401. [DOI] [PubMed] [Google Scholar]
- 105.Drekonja D, et al. Fecal microbiota transplantation for clostridium difficile infection: a systematic review. Ann. Intern. Med. 2015;162:630–638. doi: 10.7326/M14-2693. [DOI] [PubMed] [Google Scholar]
- 106.Johnsen PH, et al. Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: a double-blind, randomised, placebo-controlled, parallel-group, single-centre trial. Lancet Gastroenterol. Hepatol. 2018;3:17–24. doi: 10.1016/S2468-1253(17)30338-2. [DOI] [PubMed] [Google Scholar]
- 107.Shi Y, et al. Fecal microbiota transplantation for ulcerative colitis: a systematic review and meta-analysis. PLoS One. 2016;11:e0157259. doi: 10.1371/journal.pone.0157259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trivedi R, Barve K. Gut microbiome a promising target for management of respiratory diseases. Biochem. J. 2020;477:2679–2696. doi: 10.1042/BCJ20200426. [DOI] [PubMed] [Google Scholar]
- 109.Willner DL, et al. Reestablishment of recipient-associated microbiota in the lung allograft is linked to reduced risk of bronchiolitis obliterans syndrome. Am. J. Respir. Crit. Care Med. 2013;187:640–647. doi: 10.1164/rccm.201209-1680OC. [DOI] [PubMed] [Google Scholar]
- 110.Borewicz K, et al. Longitudinal analysis of the lung microbiome in lung transplantation. FEMS Microbiol. Lett. 2013;339:57–65. doi: 10.1111/1574-6968.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Charlson ES, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am. J. Respir. Crit. Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dickson RP, et al. Changes in the lung microbiome following lung transplantation include the emergence of two distinct Pseudomonas species with distinct clinical associations. PLoS One. 2014;9:e97214. doi: 10.1371/journal.pone.0097214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Combs MP, et al. Lung microbiota predict chronic rejection in healthy lung transplant recipients: a prospective cohort study. Lancet Respir. Med. 2021;9:601–612. doi: 10.1016/S2213-2600(20)30405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schott C, et al. Bronchiolitis obliterans syndrome susceptibility and the pulmonary microbiome. J. Heart Lung Transpl. 2018;37:1131–1140. doi: 10.1016/j.healun.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Glanville AR, et al. Chlamydia pneumoniae infection after lung transplantation. J. Heart Lung Transpl. 2005;24:131–136. doi: 10.1016/j.healun.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 116.Bernasconi E, et al. Airway microbiota determines innate cell inflammatory or tissue remodeling profiles in lung transplantation. Am. J. Respir. Crit. Care Med. 2016;194:1252–1263. doi: 10.1164/rccm.201512-2424OC. [DOI] [PubMed] [Google Scholar]
- 117.Mouraux S, et al. Airway microbiota signals anabolic and catabolic remodeling in the transplanted lung. J. Allergy Clin. Immunol. 2018;141:718–729.e7. doi: 10.1016/j.jaci.2017.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Botha P, et al. Pseudomonas aeruginosa colonization of the allograft after lung transplantation and the risk of bronchiolitis obliterans syndrome. Transplantation. 2008;85:771–774. doi: 10.1097/TP.0b013e31816651de. [DOI] [PubMed] [Google Scholar]
- 119.Gregson AL, et al. Interaction between Pseudomonas and CXC chemokines increases risk of bronchiolitis obliterans syndrome and death in lung transplantation. Am. J. Respir. Crit. Care Med. 2013;187:518–526. doi: 10.1164/rccm.201207-1228OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vos R, et al. Pseudomonal airway colonisation: risk factor for bronchiolitis obliterans syndrome after lung transplantation? Eur. Respir. J. 2008;31:1037–1045. doi: 10.1183/09031936.00128607. [DOI] [PubMed] [Google Scholar]