Abstract
Respiratory complex I is a ~1-MDa proton pump in mitochondria. Its structure has been revealed in great detail, but the structural basis of its assembly, in humans involving at least 15 assembly factors, is essentially unknown. We determined cryo–electron microscopy structures of assembly intermediates associated with assembly factor NDUFAF1 in a yeast model system. Subunits ND2 and NDUFC2 together with assembly factors NDUFAF1 and CIA84 form the nucleation point of the NDUFAF1-dependent assembly pathway. Unexpectedly, the cardiolipin remodeling enzyme tafazzin is an integral component of this core complex. In a later intermediate, all 12 subunits of the proximal proton pump module have assembled. NDUFAF1 locks the central ND3 subunit in an assembly-competent conformation, and major rearrangements of central subunits are required for complex I maturation.
Cryo-EM reveals the key function of an assembly factor and binding of the acyl transferase tafazzin to an early intermediate.
INTRODUCTION
Mitochondrial NADH:ubiquinone oxidoreductase (respiratory complex I) (NDUFAF1) is the largest membrane protein complex of the respiratory chain, consisting of 44 different subunits in mammals with an aggregate mass of ~1 MDa (1–3). The structure of respiratory complex I has been determined by x-ray crystallography and more recently by cryo–electron microscopy (cryo-EM) at increasingly high resolution (2, 4, 5). Complex I has four functional modules: the N module for NADH (reduced form of nicotinamide adenine dinucleotide) oxidation, the Q module for ubiquinone (Q) reduction, and the proximal and distal P modules, PP and PD, for proton pumping.
The assembly of mammalian complex I is a convoluted process. A loss of complex I assembly factors can result in severe human diseases (6–8). Five major building blocks with a total of more than 15 different assembly factors have been identified (9–14). So far, little is known about the structural basis of complex I assembly, apart from cryo-EM structures of assembly intermediates associated with the canonical factor NDUFAF2 (15) and the plant-specific factor GLDH (L-galactono-1,4-lactone dehydrogenase) (16). A low-resolution model for an ACAD9/ECSIT assembly factor dimer was recently reported (17).
We here report the structures of complex I assembly intermediates linked to the assembly factor NDUFAF1. NDUFAF1 was first described as CIA30 in the filamentous fungus Neurospora crassa along with a second assembly factor called CIA84 (18). While CIA84 is found only in fungi (19), the CIA30 ortholog in humans has become known as NDUFAF1 (20). Consistent with a central role of NDUFAF1 in complex I assembly, mutations in the NDUFAF1 gene result in mitochondrial disease (21, 22). In humans, NDUFAF1 is a component of the early MCIA (mitochondrial complex I assembly) intermediate (10), which is the starting point for the step-by-step assembly of the PP module (10, 12, 13). The structure of NDUFAF1 and its role in assembly are unknown.
Taking advantage of our well-characterized and genetically accessible Yarrowia lipolytica yeast model system (23), we purified assembly intermediates associated with NDUFAF1 and determined their structures by cryo-EM. We show that NDUFAF1 locks the ND3 subunit in a conformation that differs distinctly from the native structure, preventing disruption of the assembly by uncontrolled positioning of the N-terminal ND3 helix. Unexpectedly, we find that the transacylase tafazzin (24) is an integral component of the early PP module intermediate. Tafazzin is responsible for the remodeling of cardiolipin (CL), the signature lipid of mitochondria (25, 26). The remodeling is a stepwise process that typically results in CL with a higher proportion of unsaturated acyl chains. After formation of monolysocardiolipin (MLCL) by a specific lipase, tafazzin mediates transfer of a new acyl chain from a donor lipid. CL interacts tightly with membrane proteins including complex I (27) and is critical for the formation and stability of supercomplexes (28–30). It is thought to play a role in cristae formation (31) and to relieve packing stress in the crowded inner mitochondrial membrane (32). Tafazzin dysfunction causes Barth syndrome, an X-linked disease characterized by cardiac and skeletal myopathy, neutropenia, and abnormal growth (24, 30). Tafazzin is related to glycerolipid acyltransferases and shares a motif comprising a histidine and an aspartate residue (HisX4Asp) that is essential for the catalytic activity of this protein family (33–35). So far, only homology models based on acyltransferase structures have been reported (36), but no experimental structure of tafazzin is known.
RESULTS AND DISCUSSION
Impact of NDUFAF1 deletion on complex I assembly
We deleted the gene for assembly factor NDUFAF1 in Y. lipolytica by homologous recombination. The complex I content of mitochondrial membranes as assessed by NADH:hexaammineruthenium activity decreased to 14%, and ubiquinone reductase activity was below the detection limit, indicating that the ndufaf1Δ strain did not assemble functional complex I (table S1). After complementation with the NDUFAF1 gene on a plasmid, complex I expression was similar to that in the parental strain. This applied also to a variant coding for NDUFAF1 carrying a Twin-Strep-tag extension (37) for affinity purification.
To find out whether incomplete precursors of complex I assemble in the absence of NDUFAF1, we performed a complexome profiling analysis (10) of intact mitochondria of the deletion strain (fig. S1). The wild-type control contained assembly intermediates of the PP and PD modules, but their identification was compromised by low abundance and the known difficulties in detecting some of the hydrophobic components in the PP module (38). Nevertheless, it was clear that the ndufaf1Δ strain was able to assemble the PD module, whereas all other modules were virtually absent (fig. S1B). The absence of PP module intermediates agrees with studies on mammalian complex I assembly, demonstrating that NDUFAF1 is essential for formation of the MCIA complex (12, 39). The absence of N and Q module assembly intermediates in the Y. lipolytica deletion strain further indicates that their assembly is tightly controlled by the presence of the PP module, while the PD module is assembled independently. The underlying regulatory mechanism is unknown and may include translational control and/or proteolytic degradation of peripheral arm intermediates that cannot be processed further. This control appears to be less stringent in the filamentous fungus N. crassa, as assembly of the peripheral arm including the N and Q modules was still observed in a NDUFAF1 knockout strain (18).
Single-particle cryo-EM (see below) indicated that tafazzin was associated with an assembly intermediate of the PP module. Our structure-guided manual analysis was consistent with a migration behavior of tafazzin, both as a monomer and as a component of higher–molecular weight complexes in the mass range of complex I assembly intermediates. Comigration with assembly factor CIA84 was evident. By contrast, in the ndufaf1Δ mutant, only the tafazzin monomer was detected.
Purification and cryo-EM structure determination of assembly intermediates
We purified complex I assembly intermediates by affinity chromatography, making use of a Twin-Strep-tag extension (37) of assembly factor NDUFAF1. Cryo-EM analysis resulted in maps for two complex I assembly intermediates of clearly different shape and size (Fig. 1, figs. S2 and S3, and table S2). The map of the smaller intermediate had a resolution of 3.1 Å. In addition to the central complex I subunit ND2 and accessory subunit NDUFC2, it contained the assembly factor NDUFAF1 and the C-terminal half of assembly factor CIA84, which were both built with AlphaFold predictions (40) as templates. In the remaining map density, we identified a polypeptide stretch that we were able to assign to the acyltransferase tafazzin. The tafazzin structure was subsequently built manually in Coot.
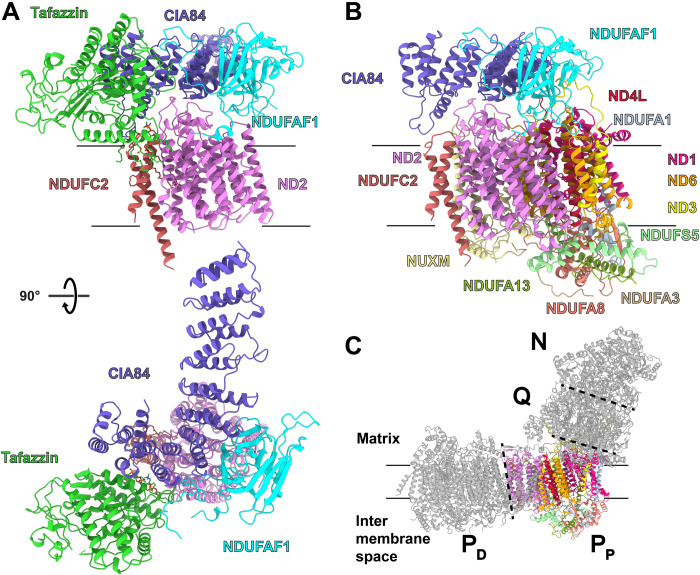
Fig. 1. Early and late PP module intermediates.
(A) Side view (top) and matrix view (below) of the early module PP intermediate. (B) Side view of the late PP module intermediate. (C) Functional modules of complex I; N module (NADH oxidation), Q module (ubiquinone reduction), and PP and PD modules (proximal and distal modules for proton pumping) [Protein Data Bank (PDB): 6rfr]. See also movies S1 and S2.
The map of the larger intermediate permitted straightforward assignment of density for the complete PP module of complex I. Further focused three-dimensional (3D) classifications (fig. S2) delivered maps for three different variants of the larger intermediate with resolutions ranging between 2.9 and 3.2 Å—one with both NDUFAF1 and CIA84, one with NDUFAF1 only, and one without assembly factors. Although the complexome profiling analysis (fig. S1) indicated comigration of tafazzin and CIA84, none of the variants showed binding of tafazzin.
In agreement with what is known about complex I assembly in mammals (10, 12, 13), the intermediate containing ND2 and NDUFC2 is the seed complex of the NDUFAF1-dependent assembly pathway, while the intermediate with the complete PP module is a more advanced stage. We here use the terms early and late PP module intermediate to describe these complexes.
Structure of the early PP module intermediate
The transmembrane domain of the early PP module intermediate contains the central subunit ND2 with its 14 transmembrane helices (TMHs) and the accessory subunit NDUFC2 with two TMHs (Fig. 1A and movie S1). Assembly factors NDUFAF1 and CIA84 and the acyltransferase tafazzin are located on the matrix side of the complex.
The structures of ND2 and NDUFC2 in the intermediate are virtually identical to those in mature complex I (15, 41). Some density is missing for the 22-residue loop connecting TMH6 and seven of ND2 on the intermembrane space (IMS) side, and TMH6 is slightly displaced.
The central domain of the 27-kDa NDUFAF1 folds into a curved sandwich of two six-stranded β sheets. The N-terminal part of NDUFAF1 forms a short α helix on the membrane surface next to the C-terminal ends of TMH9 and TMH13 of ND2. NDUFAF1 further interacts with the loops connecting TMH5 and TMH6 as well as TMH11 and TMH12 in ND2.
CIA84 is a large protein with 852 residues and a mass of ~97 kDa. Its C-terminal end forms the core of the early PP module intermediate, binding to both ND2 and NDUFC2 as well as NDUFAF1 and tafazzin (Figs. 1A and 2). In the cryo-EM map, density for the N-terminal 430 residues is missing and is weak for the next ~220 residues, suggesting that this part of CIA84 is flexible. Its C-terminal half is folded into 10 tandem repeats of antiparallel α helices. The last two helices are connected by a long loop and are approximately orthogonal to each other. The complete sequence as analyzed with the program HHpred (42) indicated structural similarity with pentatricopeptide repeat (PPR) proteins, but a search using the Pfam (43) and PPR databases (44) did not return any hits for PPR sequence motifs in CIA84.
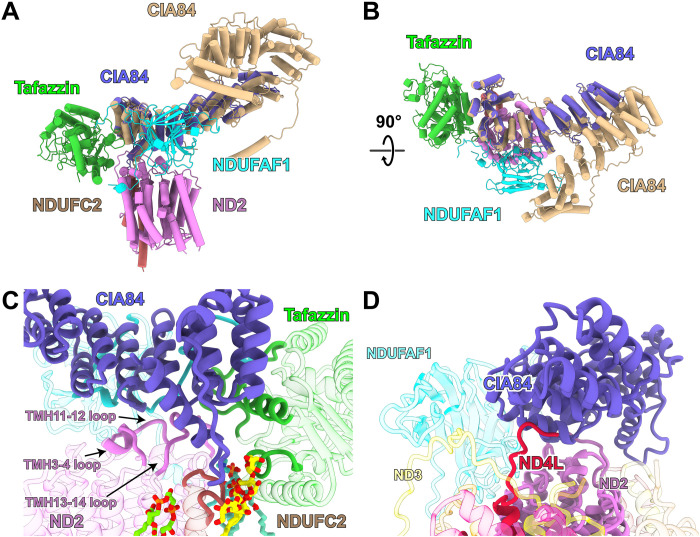
Fig. 2. Structure and interactions of CIA84.
(A and B) Overlay of the cryo-EM structure of the early PP module intermediate (compare Fig. 1) with the AlphaFold structure prediction for Y. lipolytica CIA84 (wheat) in two different orientations. (C) Interactions of CIA84 with ND2 and tafazzin. (D) In the late PP module intermediate of complex I, CIA84 interacts with the C terminus of subunit ND4L, which undergoes substantial rearrangements after release of CIA84 and upon the maturation complex I.
CIA84 interacts with ND2 loops connecting TMH3 and TMH4, TMH11 and TMH12, and TMH13 and TMH14 (Fig. 2C). These loops are folded as in mature complex I, apart from a small shift of the TMH12-13 loop that forms a β sheet with accessory subunit NDUFB4 in mature complex I. The long loop between the two C-terminal helices of CIA84 contacts the loop connecting the two TMHs of NDUFC2 and is involved in binding the head group of a lauryl maltose neopentyl glycol (LMNG) detergent molecule. CIA84 has an extensive interface with the β-sandwich domain and the C-terminal loop of NDUFAF1 and interacts closely with tafazzin.
The AlphaFold structure prediction (40) for complete CIA84 matches the structure of the C-terminal half well (Fig. 2, A and B) and suggests that the unresolved N-terminal part of the protein likewise has a tandem helix repeat fold. Clearly, CIA84 forms the core of the assembly intermediate and supports binding of NDUFAF1 and tafazzin. However, the function of the unresolved and most likely flexible N-terminal region remains to be established.
NDUFAF1 is the only assembly factor of the PP module that is conserved in fungi, plants, and animals, while there are clear differences in the other assembly factors of the PP module (45). In addition to NDUFAF1, the MCIA complex described for human complex I assembly (10, 12, 13) also contains ACAD9, ECSIT, and COA1. TMEM126B and TMEM186 are added in subsequent assembly steps. Assembly factor ECSIT (46) of mammalian complex I was predicted to include a PPR motif (47) and is thought to interact tightly with NDUFAF1 (17). The AlphaFold prediction for ECSIT (40) shows an N-terminal tandem helix repeat domain and thus suggests a possible structural similarity to CIA84. However, mammalian complex I has an additional large accessory subunit, NDUFA10, attached to the matrix side of ND2 (48). In human cell culture, it has been shown that NDUFA10 can coexist in an assembly intermediate with NDUFAF1 and ECSIT (13). Comparison of the structures shows that the binding site of NDUFA10 overlaps with that of CIA84 in the Y. lipolytica assembly intermediates (fig. S4), making it very unlikely that ECSIT binds there. In contrast, NDUFA10 and NDUFAF1 have complementary surfaces and no overlap (fig. S4), strongly suggesting that the binding site of NDUFAF1 on ND2 is conserved.
The early PP module intermediate contains the CL remodeling enzyme tafazzin
Tafazzin is evidently an integral part of the early PP module intermediate, sharing an extensive interface with CIA84, interacting tightly with the C-terminal loop of NDUFAF1, and binding to both ND2 and NDUFC2 (Fig. 1A). Note that tafazzin is bound to the matrix side of the inner mitochondrial membrane, in contrast to earlier reports proposing that the lipid target of tafazzin resides in the IMS-facing leaflet (49). The topology observed in our cryo-EM maps is consistent with the synthesis (50) and deacylation (51) of CL taking place on the matrix side of the inner mitochondrial membrane.
The tafazzin core is a β sheet, surrounded by α helices and connecting loops. The active site containing residues His76 and Asp81 (33, 34) is in a central position and in a cavity facing the membrane (Fig. 3). The histidine residue of the canonical HisX4Asp motif deprotonates the sn2 hydroxyl group of the lysophospholipid, aided by the aspartate to enable a nucleophilic attack on the ester bond of the acyl donor. Phospholipids at the active site of acyltransferases are coordinated by the positively charged side chains of arginines or lysines (52, 53). Tafazzin contains two fully conserved arginine residues, Arg101 and Arg130 (fig. S5), both located in the substrate cleft, ~14 Å apart where they are well placed for coordinating the two phosphates of (ML)CL (Fig. 3). Mutation of Arg94 in human tafazzin, corresponding to Arg101 in Y. lipolytica, causes Barth syndrome (54). Mutation of the strictly conserved Ser71 (78 in Y. lipolytica) close to the catalytically active histidine also results in Barth syndrome, most likely by interference with the active site. Further pathogenic mutations (55) involve conserved residues that cluster around the active site such as Phe148 and Val153 (fig. S6 and movie S3).
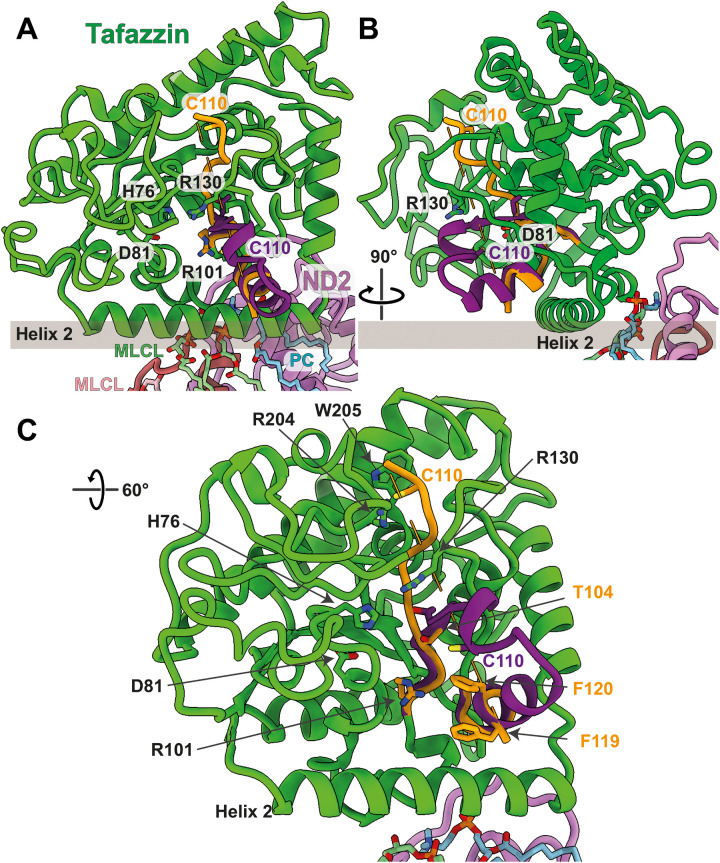
Fig. 3. The acyltransferase tafazzin in the early PP module intermediate.
(A and B) Tafazzin assumes two alternative conformations in the complex I assembly intermediate. Residues 101 to 110 and 119 to 124 of the cryo-EM structure are shown in orange; residues 111 to 118 are not resolved. The map indicates that the conformation suggested by the AlphaFold model (residues 101 to 124 are shown in purple) is present in a small proportion of tafazzin molecules. Residues mentioned in the text are indicated. MLCL and PC sandwiched between tafazzin and NDUFC2 are shown in pale green and cyan, respectively; MLCL interacting with NDUFC2 is pink. The membrane surface is shown as a gray bar. NDUFAF1 and CIA84 are removed for clarity. (C) Close-up of the active site.
Helix 2 of tafazzin is on the membrane surface (Fig. 3). Although it is very hydrophobic in most species (fig. S5) and has been previously modeled as a TMH (56), an AlphaFold model of human tafazzin shows the same orientation of helix 2 as observed in Y. lipolytica (Fig. 3 and fig. S6), which is indicative of its attachment to the membrane surface. This interaction orients the active site optimally for substrate transfer from the lipid phase of the membrane (Fig. 3). The region between the membrane anchor and NDUFC2 contains a number of nonprotein densities in the detergent micelle that we interpreted as a phosphatidylcholine (PC) and a MLCL molecule, i.e., the two substrates of the tafazzin transacylation reaction. An LMNG molecule is located next to MLCL at the interface of the NDUFC2 loop and CIA84, while another MLCL binds on the other side of NDUFC2 at its interface with ND2 (Fig. 4A). No MLCL molecules are found in the structure of the late PP module intermediate and in the mature complex I (15, 41). In the early PP module intermediate, the binding site at the NDUFC2/ND2 interface appears to be specific for MLCL, as there is no space for the fourth acyl chain (Fig. 4A). In the late PP module intermediate and in mature complex I, the binding site is disturbed by subunit NUXM and phosphatidylinositol is bound instead [Protein Data Bank (PDB): 7o71] (Fig. 4C) (15, 41). However, the MLCL binding site close to tafazzin would easily accommodate CL (Fig. 4A). CL may not bind in this site because MLCL is enriched in the membrane environment of the early PP module intermediate. Alternatively, the catalytic activity of tafazzin drives the conversion of CL to MLCL during the purification process.
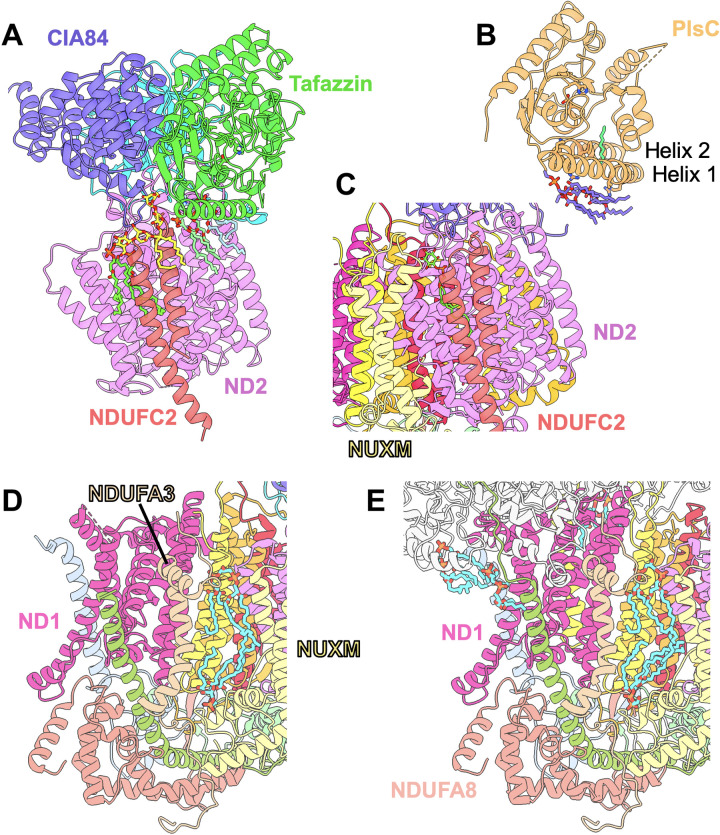
Fig. 4. Lipid binding during assembly and in mature complex I.
(A) In the early PP module intermediate, MLCL (light green) and PC (light blue) are bound at the tafazzin/ND2 interface, an LMNG (yellow) between NDUFC2 and CIA84, and a second MLCL (chartreuse) at the NDUFC2/ND2 interface. (B) In the crystal structure of the bacterial acyltransferase PlsC (PDB: 5kym), four PC molecules (purple) and an acyl chain (green) are bound to the N-terminal helices. The catalytic His and Asp residues are shown as sticks in (A) and (B). (C) The late PP module intermediate has lost the tafazzin-associated lipids and the LMNG; subunit NUXM is bound near NDUFC2, and LMNG is replaced by a smaller phosphatidylinositol molecule (chartreuse). (D) In the late PP module intermediate, two CL molecules (cyan) are bound between NUXM and NDUFA3 as in mature complex I (PDB: 7o71) (E).
A membrane attachment by a surface helix was also found in the related bacterial acyltransferase PlsC (52). In the PlsC crystal structure (PDB: 5kym), a number of lipids were bound to the membrane in a comparable position to the lipids bound to tafazzin (Fig. 4, A and B).
For the fold at the opening of the substrate binding site of tafazzin to the membrane, the map gives evidence for a mixture of different conformations. For residues 112 to 118, there is no density in the cryo-EM map, indicating that this stretch is flexible. In AlphaFold predictions for tafazzin from different species, including Y. lipolytica and Homo sapiens, this region folds into an α helix (residues 114 to 123) that follows a β strand (residues 103 to 105) and a short helix (residues 107 to 110). The map shows density for the end of the α helix at the membrane surface, but unexpectedly, the fold of residues 104 to 111 is different and forms an extended polypeptide structure including an ~45o angle with the predicted β strand (Fig. 3). Weak density extending from Thr104 suggests that the conformation predicted by AlphaFold is actually present in a small proportion of tafazzin molecules. The flexible region contains a conserved stretch of amino acids Asp/Asn-Ile/Val-Cys-Phe/Tyr, including the fully conserved cysteine, Cys110 (fig. S5). Cys110 moves by ~18 Å between both conformations (Fig. 3). In the cryo-EM structure, it is located near the substrate cleft, but the side chain is buried between the conserved Arg204 and Trp205. In the AlphaFold model, it is part of the short helix, near the membrane surface and exposed in the cleft, not far from Arg101, Arg130, and His76. The polypeptide structure in this vicinity, notably at the two phenylalanines, Phe119 and Phe120 (fig. S5), is highly conserved. Typically, acyltransferases rely on acyl chains covalently attached to the sulfur atom of phosphopantetheine groups of either an acyl carrier protein (ACP) or coenzyme A (CoA), but uniquely in this family, tafazzin is known to be independent of both CoA and ACP (57). It is tempting to speculate that the conserved Cys110 on a flexible loop takes the role of an acyl carrier and binds an acyl chain from PC to deliver it to the active site for transfer to MLCL.
It has long been known that CL is essential for complex I activity (58), and recent molecular dynamics simulations suggest a central role of this lipid in redox-coupled proton translocation (59). Biochemical experiments have shown that 10 CL molecules are tightly bound in mammalian complex I (60). We have modeled six CL-binding sites in the cryo-EM structure of complex I of Y. lipolytica (movie S4) (15), and five CL binding sites were identified in the structure of human complex I (61). Several of the resolved CL molecules are buried in the protein structure. Thus, it is plausible that CL in its mature configuration is necessary for complex I assembly. The interaction of tafazzin with an assembly intermediate might ensure that CL is present where and when it is needed. Consistent with this, in the structure of the late PP module intermediate, we find two CL molecules in the same binding site as in mature complex I (Fig. 4, D and E). Decreased complex I assembly in a tafazzin-deficient human cell line has been reported recently (62). However, a direct interaction of tafazzin with assembly intermediates of human complex I was not observed in a complexome profiling study (13), and our structure does not give evidence for conserved contact sites between tafazzin and proteins of the mammalian MCIA complex. In Drosophila, a correlation between the assembly of oxidative phosphorylation enzyme complexes and CL remodeling was demonstrated (32). However, this effect was not specific for respiratory complex I. In mitochondria of the yeast Saccharomyces cerevisiae, which does not have respiratory complex I, tafazzin has been found to form assemblies with other mitochondrial complexes, such as adenosine 5′-triphosphate synthase and the adenine nucleotide carrier (63). Together, there is mounting evidence that the assembly of mitochondrial protein complexes and the remodeling of CL by tafazzin are tightly linked. Our structure shows the structural basis of this functional relationship in an aerobic yeast. Future work will reveal the extent to which this can be generalized to other organisms.
Structure of the late PP module intermediate: TMHs of central subunits are rearranged during assembly
While the three variants of the late PP module intermediate differ in the binding of assembly factors, the complete set of subunits of the PP module is present throughout; i.e., central subunits ND1, ND3, ND4L, and ND6 plus six accessory subunits and lipids were added to ND2 and NDUFC2 of the early PP module intermediate (Fig. 1B and movie S2). Our structure indicates that, in Y. lipolytica, the hydrophobic ND1 subunit is directly attached to the nascent PP module, whereas in mammals, ND1 is first linked to the Q module, and the resulting PP-a module is attached to the PP-b module lacking ND1 (13). In this respect, the assembly of the PP module in Y. lipolytica differs from that in mammals and resembles the pathway described in plants more closely (14).
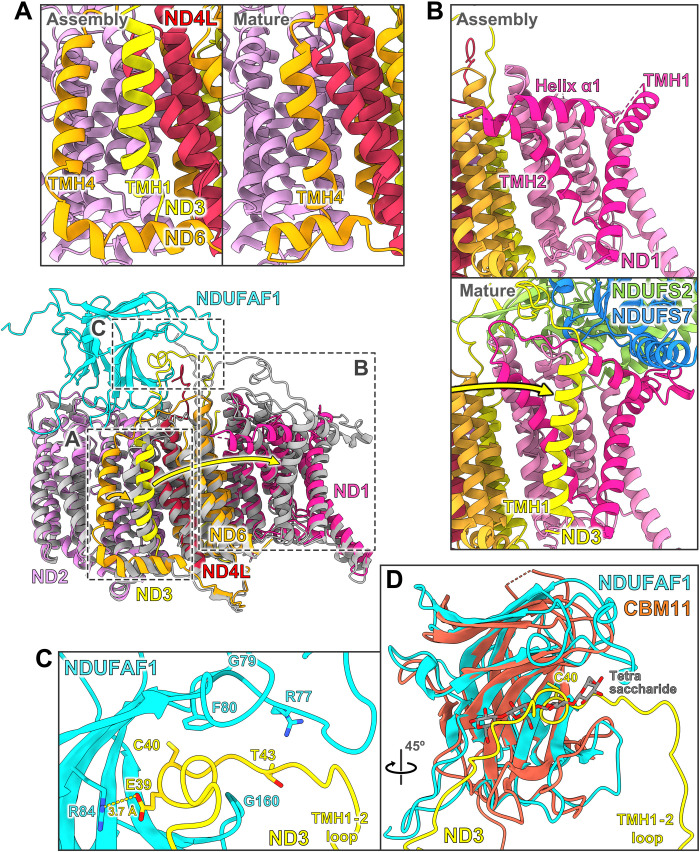
In the two variants that include assembly factors, their interactions with ND2 and NDUFC2 are unchanged as compared with the early PP module intermediate. Whereas the TMH6- loop on the IMS side of ND2 was disordered in the early PP module intermediate, it interacts with accessory subunit NDUFS5 and is now folded as in the mature complex. In contrast to ND2, other central subunits differ considerably when compared to their structures in mature complex I (movie S5). This is particularly notable for subunit ND3, which consists of three TMHs with a special topology (Fig. 5). In mature complex I, TMH1 and TMH2 are more than 35 Å apart and located on different sides of the membrane arm. TMH1ND3 binds to the ND1 subunit, and the TMH1-2 loop runs along the interface between P and Q modules (Figs. 5B and 6B). In the late PP module intermediate, TMH2 and TMH3 are in their final positions, but the TMH1-2 loop runs in the opposite direction (Fig. 5) and is locked in a binding cleft of NDUFAF1 (see below). TMH1ND3 is shifted by ~40 Å toward the distal end of the membrane arm and is located near subunit ND4L (Fig. 6A and movie S5). Notably, this position overlaps with that of TMH4ND6 in mature complex I. As a result, TMH4ND6 is displaced by 10 to 12 Å in the assembly intermediate, with a major rearrangement of its TMH3-4 loop on the matrix side and the loop following TMH4 on the IMS side (Fig. 5). The C-terminal end of ND4L is bound by CIA84 in a position distinctly different from that in the mature complex (Fig. 2D). Switching ND4L to its mature conformation requires rearrangement of the TMH3-4 loop of ND6. The absence of TMH1ND3 from its final position near ND1 in the late PP module intermediate relates to a considerable distortion of ND1. The amphipathic surface helix α1 of ND1 is shifted to a position where it would clash with TMH1ND3, and the loops on the matrix side are disordered (Fig. 6B and movie S5). These loops form the interface with the Q module in mature complex I and include the TMH5-6 loop that is thought to play a key role in coupling redox chemistry to proton translocation (41). Folding of this ND1 region might occur through binding of TMH1ND3 and/or of the Q module, during later steps of complex I assembly.
Fig. 5. Topology of subunit ND3 and helix rearrangements upon assembly.
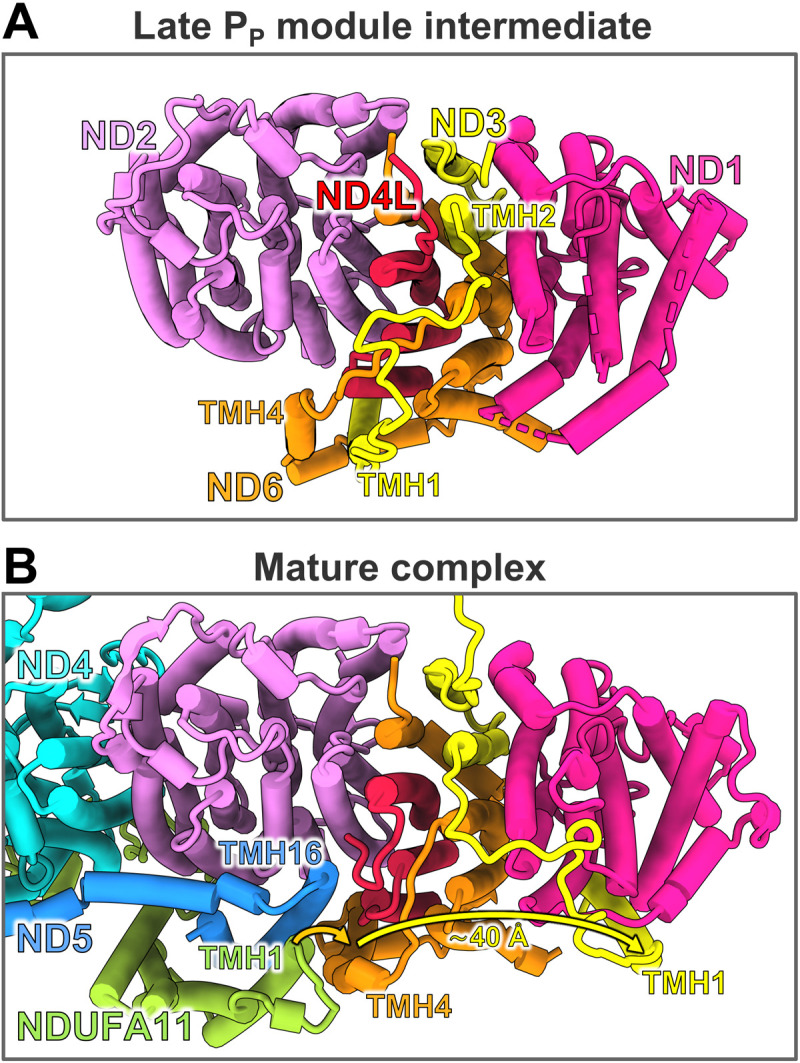
Late PP module intermediate (A) and membrane arm section of mature complex I (B) viewed from the matrix side. Assembly factors, accessory subunits except NDUFA11, and matrix arm are removed for clarity. TMH1 and TMH2 of ND3 are connected by a long loop on the matrix side. In both the late PP module intermediate and mature complex I, the two helices are more than 35 Å apart. During maturation of complex I, TMH1ND3 moves from a site close to ND4L over a distance of ~40 Å to its final position next to subunit ND1. The position of TMH1ND3 in the late PP module intermediate is assumed by TMH4ND6 in the mature complex. This rearrangement occurs in concert with binding of the PD module as TMH16ND5 and TMH1NDUFA11 get in close proximity to the position of TMH4ND6 in the assembly intermediate.
Fig. 6. The TMH1-2 loop of ND3 binds to assembly factor NDUFAF1 in the late PP module intermediate.
Middle: Overlay of the late PP module intermediate (colors as in Fig. 1; accessory subunits and CIA84 are omitted for clarity) with the PP module of mature complex I (gray; PDB: 7o71). The considerable displacements of TMH1ND3 and TMH4ND6 are indicated by yellow and orange arrows. (A) In the assembly intermediate, TMH1ND3 binds close to subunit ND4L. In mature complex I, this helix position is occupied by TMH4ND6. (B) Rearrangements of ND1. During assembly, TMH1ND3 moves by more than 40 Å to its position near ND1 in the fully assembled complex. TMH1 and TMH2 and surface helix α1 of ND1 undergo major rearrangements. NDUFS2 and NDUFS7 are central subunits of the Q module. (C) In the assembly intermediate, the TMH1-2 loop of subunit ND3 binds tightly to the assembly factor NDUFAF1. Cys40 of ND3 is buried in the binding cleft. (D) NDUFAF1 closely resembles CBM11. An overlay of both structures (CBM11, brown; PDB: 6r3m) reveals that the binding cleft of CBM11 for carbohydrates matches the binding cleft of NDUFAF1 for the TMH1-2 loop of ND3 (compare fig. S9).
The question arises as to what could trigger the helix rearrangement. A comparison with mature complex I shows that TMH16 of central subunit ND5 and TMH1 of accessory subunit NDUFA11 in the PD module would collide with TMH4ND6 in the position assumed in the late PP intermediate (Fig. 5). Joining of the PD and PP modules thus requires the ND6 helix to move to its final position occupied by TMH1ND3. We therefore propose that addition of the PD module and rearrangements in the PP module are concerted processes. In mammalian complex I, assembly of an intermediate comprising PP and Q modules was observed while NDUFAF1 was still bound (13). This would allow a safe transfer of the TMH1-2 loop of ND3 to its final position at the interface between the two modules. However, assuming that NDUFAF1 is similarly positioned on the PP module in mammals, it would clash with the long N-terminal extension of NDUFS2, which could therefore only adopt its final structure on the membrane arm after release of the assembly factor (fig. S7). Moreover, our structure suggests a major clash with the position of accessory subunits NDUFA6 and ACPM1 on the Q module of mature complex I (fig. S7), which would explain why these subunits are added at a later stage after NDUFAF1 detaches (13).
NDUFAF1 locks the long matrix loop of ND3 in a binding cleft
In mature complex I, the long TMH1-2 loop of ND3 lines the interface of PP and Q modules (Fig. 6B). Modifications of a conserved cysteine residue in this loop (Cys40ND3 in Y. lipolytica) (64) have a strong impact on complex I activity (65), and locking loop movements by an engineered disulfide bond disengages the proton pumps (66). In the late PP module intermediate, the TMH1-2 loop of ND3 is embedded in a binding cleft of NDUFAF1 (Fig. 6C). The architecture of the binding cleft at the concave side of the β-sandwich structure closely resembles that for carbohydrate chains in carbohydrate-binding domain family 11 (CBM11) proteins (Fig. 6D and fig. S9) (67). The observed structural conservation is consistent with a previous proposal (45). None of the four critical tyrosine residues involved in carbohydrate binding (67) are conserved in NDUFAF1, but one of them (Tyr53) is replaced by Phe80FAF1, and an aromatic residue is conserved at this position in NDUFAF1 (figs. S9 and S10). Only Gly79FAF1 and Gly160FAF1 are strictly conserved in the binding cleft of NDUFAF1, and both residues are located in loops that form its narrowest part. In the first loop of residues 77 to 80, Phe80FAF1 is close to the conserved Cys40ND3 and binds to a short hydrophobic segment of the TMH1-2 loop of ND3. The strictly conserved Glu39ND3 forms an ion pair with Arg84FAF1 (Fig. 6C). The arginine residue is not conserved in mammals (fig. S10); however, in human and mouse NDUFAF1, an arginine residue is conserved two positions downstream, which may also engage in a salt bridge with the ND3 loop.
Our structure of the late PP module intermediate provides a clear picture of the function of NDUFAF1. Because neither the ND1 subunit nor the Q module is present immediately after addition of the ND3 subunit, the long matrix loop and the preceding TMH1 of this subunit cannot be directly incorporated into the nascent structure. Thus, NDUFAF1 might support the initial attachment of ND3 and stabilizes a conformation of ND3 that is clearly different from mature complex I. Fixing the long TMH1-2 loop of ND3 prevents the adjacent TMH1 from moving freely during assembly, as this might otherwise interfere with the addition of new subunits or trap the loop/helix in an incorrect position without the possibility for later correction. The alkylation of a conserved cysteine residue in the TMH1-2 loop of ND3 in catalytically inactive complex I causes irreversible loss of Q reductase activity (65). Thus, another benefit of securing the loop in the binding cleft of an assembly factor might be the protection of Cys40ND3 from unwanted modifications during assembly. The conversion of a binding cleft for carbohydrate chains into one for a peptide loop follows a well-known evolutionary phenomenon whereby structural scaffolds can assume unexpected new functions.
Note that, while the loss of CIA84 has no impact on ND3 or ND6 in the late PP module intermediate, density for TMH1ND3 and TMH4ND6 is weak in the variant lacking all assembly factors (fig. S8). Because a strep-tag on NDUFAF1 was used for purification, detachment of this assembly factor must have occurred after affinity chromatography and may thus be an artefact of sample preparation. Nevertheless, this observation supports our conclusion that NDUFAF1 is necessary and sufficient for the positioning of TMH1ND3 and consequently also of TMH4ND6 in the late PP intermediate.
Dysfunction of NDUFAF1 interferes with complex I assembly, and patients carrying mutations in the NDUFAF1 gene suffer from hypertrophic cardiomyopathy (22) and cardioencephalomyopathy (21). An overlay of the AlphaFold model of human NDUFAF1 with our structure indicates that the pathogenic mutations cluster on β strands 6 and 9 of the outer leaflet of the β-sandwich structure and in an adjacent loop (fig. S6, B and D). None of these positions are in direct contact with the binding site for the ND3 loop. We can therefore assume that these disease-related mutations interfere with the structure of the assembly factor.
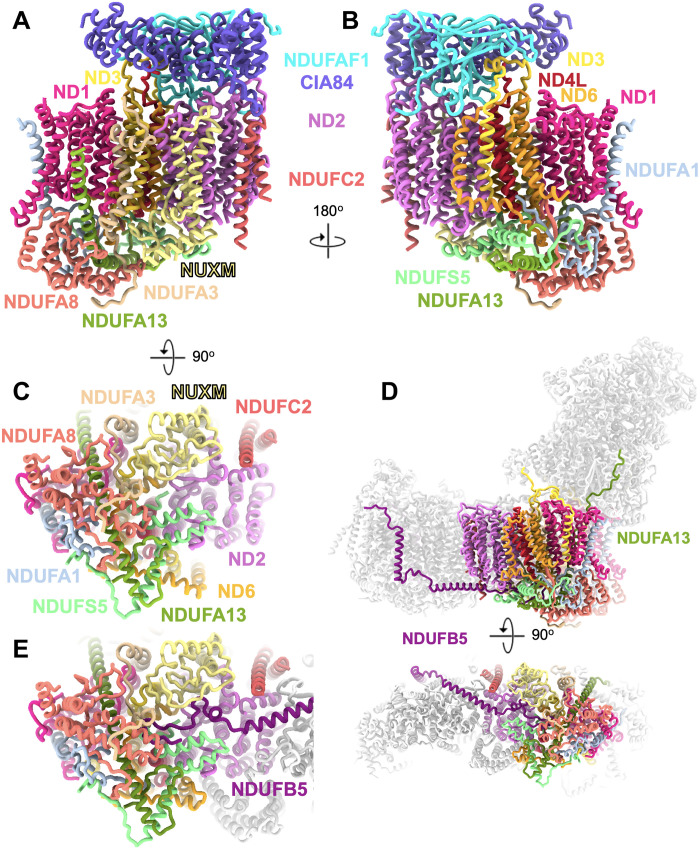
Accessory subunits in the late PP module intermediate
The PP module contains seven accessory subunits: apart from NDUFC2 that is already present in the early PP module intermediate NDUFA1, NDUFA2, NDUFA8, NDUFA13, NDUFS5, and NUXM (the latter is missing in metazoans). Five of them are involved in the formation of a prominent structure on the IMS side of the protein complex. This IMS domain contains a binding cavity for the C-terminal end of the NDUFB5 subunit, which, in mature complex I, forms a staple along the IMS side surface from ND4L to the PD domain, with its N-terminal TMH at the interface of ND4 and ND5 (Fig. 7). This suggests that the recruitment of NDUFB5 to its binding cavity plays a role in joining the PP and PD modules. The only accessory subunit not fully folded in the late PP intermediate is NDUFA13, which consists of a long, bent helix with its C terminus in the IMS followed by a transmembrane stretch. The N-terminal 30 residues are disordered in the assembly intermediate; in the mature complex, they form an extended loop that runs along the surface of the central subunit NDUFS2 of the Q module to 65 Å above the membrane surface (Fig. 7). Thus, NDUFA13 may play a role in joining the P and Q modules.
Fig. 7. Accessory subunits of the late PP module intermediate.
The intermediate consisting of the assembly factors NDUFAF1 and CIA84; the core subunits ND1, ND2, ND3, ND4L, and ND6; and the accessory subunits NDUFA1, NDUFA3, NDUFA8, NDUFA13, NDUFC2, NDUFS5, and NUXM is shown in side views (A and B) and from the IMS (C). (D) In mature complex I, NDUFB5 and NDUFA13 interlock the PP module with the PD module and the Q module, respectively. (E) In mature complex I, the PD module subunit NDUFB5 binds in a preformed pocket to the IMS side of the PP module.
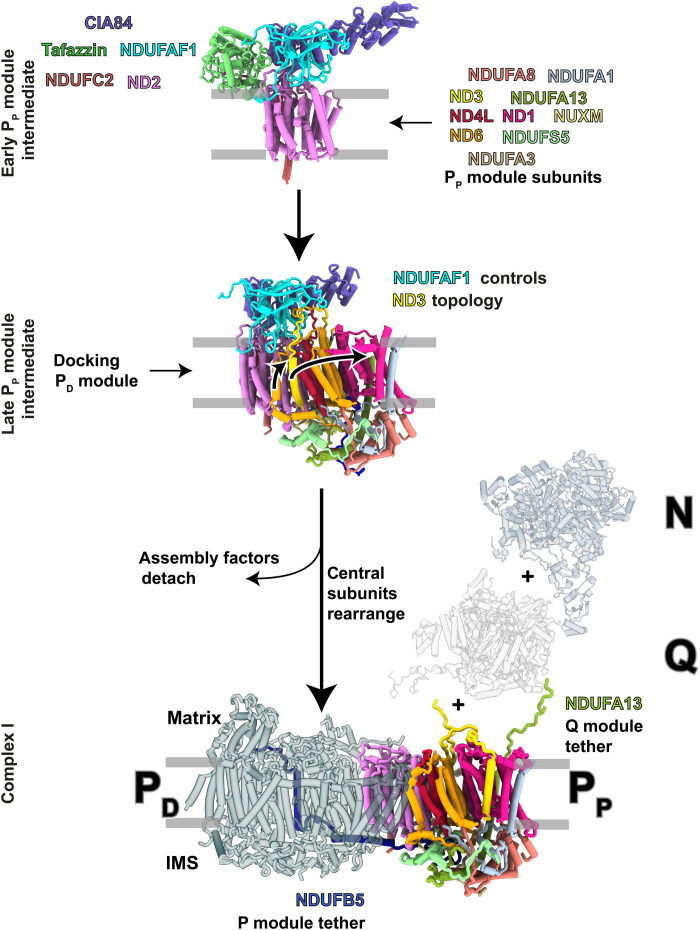
In summary, our work provides the first detailed insights into the intricate assembly of respiratory complex I (Fig. 8), a protein complex of central importance for cellular energy metabolism. The assembly factor NDUFAF1 locks the central ND3 subunit in an assembly-competent conformation that prevents uncontrolled positioning of the N-terminal ND3 helix during assembly of the proximal proton pump module. The pronounced rearrangement of ND3 into its final conformation is associated with structural changes in central subunits ND6, ND4L, and ND1 and presumably occurs in concert with the merging of the two building blocks (PP and PD) that constitute the membrane arm of complex I. Unexpectedly, our cryo-EM structures reveal a link between the assembly of complex I and the remodeling of CL, a key lipid of the inner mitochondrial membrane. Our structure shows how the transacylase tafazzin interacts with the inner mitochondrial membrane, allows conclusions to be drawn about the mechanism of transacylation, and will be the basis for a molecular understanding of pathogenic mutations causing Barth syndrome.
Fig. 8. NDUFAF1-dependent complex I assembly.
Assembly factors NDUFAF1 (cyan) and CIA84 (purple) form the core of the early PP module intermediate. The central subunit ND3 (yellow) consists of three TMHs with an unusually long loop connecting TMH1 and TMH2. NDUFAF1 locks this loop in a binding cleft and forces the adjacent TMH1ND3 into a position that is occupied by TMH4 of ND6 (orange) in mature complex I. Docking of the PD module to the late PP module intermediate is concerted with the transitions of ND6, ND3, and ND1 to their conformations in the fully assembled complex. NDUFB5 and NDUFA13 interlock the PP module with the PD and Q modules. Assembly of the N module is mediated by the assembly factor NDUFAF2 (15).
MATERIALS AND METHODS
Mass spectrometry
Detailed description of material and methods together with raw data from mass spectrometry and complexome data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database partner repository (68) with the dataset identifier (ID) PXD033522.
Gene deletion
The ndufaf1Δ strain was generated by homolog recombination as described (69). YALI0_D14564g with 1-kb flanks on both sides was amplified, and the coding region was exchanged for the ura3 gene by Gibson assembly (70). This construct was used to delete the gene from the haploid GB20 (mus51Δ, NUGM-Htg2, NDH2i, lys11-, leu2-, ura3- and MatB) strain (23). Substitution by the ura3 marker was confirmed by polymerase chain reaction, and absence of the protein was confirmed by complexome profiling (fig. S1).
Purification of NDUFAF1 complexes
For purification, we complemented the ndufaf1Δ strain in trans with the ndufaf1 gene carrying an extension coding for a C-terminal Twin-Strep-tag (37) appended to a AEAAAKEAAAKA linker (71). NDUFAF1 was purified using a Strep-Tactin XT column (IBA Lifesciences, Germany) essentially following instructions provided by the manufacturer. Mitochondrial membranes (1 to 2 g of membrane protein) were solubilized using 1 g of dodecyl maltoside per 1 g of mitochondrial protein for 15 min on ice, followed by ultracentrifugation. The clear supernatant was applied to the Strep-Tactin XT column and subsequent wash and elution steps were carried out using 0.025% LMNG in all buffers. Combined elution fractions were concentrated and applied to a Superose 6 Increase 10/300 column (Cytiva, Germany). A broad peak containing assemblies of high molecular mass was collected and concentrated, while the well-separated NDUFAF1 monomer peak was discarded. The buffer used for size exclusion contained 20 mM tris-HCl, 100 mM NaCl, 1 mM EDTA, and 0.002% LMNG at a pH of 7.5.
Sequence analysis and alignments
Sequences were aligned using the Clustal Omega algorithm (72) and are displayed using the Jalview software (73).
Sample vitrification and cryo-EM data acquisition
Three microliters of purified sample was applied onto freshly glow-discharged (15 mA for 45 s in a PELCO easiGlow system) C-flat 1.2/1.3 300-mesh copper grids (Science Services GmbH) at a final concentration of 1.2 mg/ml. Samples were blotted for 4.5 s with a blot force of 20 (595 Whatman paper) at 4°C in 100% humidity and plunge-frozen in liquid ethane by using a FEI Vitrobot Mark IV. Cryo-EM data were collected automatically using EPU software (Thermo Fisher Scientific) on a Titan Krios G3i microscope operating at 300 kV and equipped with a K3 detector operating in electron counting mode. Movies were acquired at a nominal magnification of ×105,000, resulting in a pixel size of 0.837 Å. Each movie was recorded for 3 s and subdivided into 60 frames. Electron flux rate was set to 14 e− per pixel per second at the detector, resulting in an accumulated exposure of 60 e−/Å2 at the specimen. An energy filter slit width of 30 eV was used, and a 70-μm C2 and 100-μm objective aperture were inserted during acquisition. A total of 6229 movies at a defocus range of −1 to −3 μm were collected.
Image processing
Image processing was performed in RELION4 (74), unless otherwise specified. Dose-fractionated movies were motion-corrected with RELION’s implementation of the MotionCor2 algorithm (75), and contrast transfer function (CTF) parameters were estimated using CTFFIND4.1 (76). Micrographs with an estimated CTF resolution worse than 7 Å were removed, and 2,048,808 particles were picked from 6218 micrographs by using crYOLO 1.7.3 (77). After 2D classification in cryoSPARC v3.3.1 (78), 235,444 particles were subjected to an ab initio reconstruction, which revealed the presence of two major particle populations, representing two distinct complexes of different size and shape. Ab initio reconstructions of these complexes were further refined by heterogeneous refinement, and each reconstruction was used as a reference for two separate 3D classifications in RELION, after removal of junk particles by 2D classification. A second round of 3D classification was performed, using the two ab initio reconstructions as references simultaneously. After CTF refinement and Bayesian polishing, particles of the smaller complex (early PP module assembly intermediate) were refined and reconstructed in cryoSPARC by nonuniform refinement, resulting in a map resolution of 3.2 Å. Particles of the larger complex (late PP module intermediate) was heterogeneous with respect to bound assembly factors on the matrix side. Two further 3D classifications without image alignment and with masks around the binding site separated the particles into three PP module assembly intermediates. One did not have any bound assembly factors, a second intermediate bound NDUFAF1, and a third intermediate bound both NDUFAF1 and CIA84. After nonuniform refinement in cryoSPARC, the map resolution improved to 3.0, 3.3, and 3.2 Å, respectively. Resolutions were estimated according to the Fourier shell correlation 0.143 cutoff criterion of two independently refined half maps (79). As a final step, three cycles of density modification with phenix.resolve_cryo_em (80) were carried out with all four maps, by using in each case the unsharpened map, two unfiltered half-maps, and an estimate of the protein mass as inputs. A detailed overview of the processing workflow is shown in fig. S2.
Model building
The acyltransferase tafazzin was identified in the early PP module intermediate by modeling a polyalanine chain into the map density and then placing matching amino acids into residue-specific map features. This delivered short sequence fragments (e.g., YPFWR), which were used as query sequences for protein BLAST (81) searches in the protein database of Y. lipolytica, which indicated a hit for the acyltransferase tafazzin (UniProt entry: Q6CBZ7). The tafazzin sequence matched the map density perfectly, and 94% of the amino acids were modeled.
Models were built in Coot (82), using the cryo-EM structure of Y. lipolytica complex I (PDB ID: 7o71) as a template for the complex I subunits and AlphaFold predictions for CIA84 and NDUFAF1, while tafazzin was built manually. The structures were rebuilt in Coot and refined using phenix.real_space_refine (83). Refinement and validation statistics are summarized in table S2. Figures were made with UCSF ChimeraX (84).
Acknowledgments
We thank the Central Electron Microscopy Facility of the Max Planck Institute of Biophysics for cryo-EM infrastructure and technical support as well as J. Castillo-Hernández, Ö. Yildiz, the Central IT team, and the Max Planck Computing and Data Facility for support in cryo-EM data processing and calculation of AlphaFold models. We thank K. Siegmund for excellent technical assistance.
Funding: This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft) (ZI 552/4-2 to V.Z., WI 3728/1-1 to I.W., and CRC1507/P14 to V.Z. and J.V.) and by the German Federal Ministry of Education and Research (BMBF) grant to the German Network for Mitochondrial Disorders mitoNET- 01GM1906D (to I.W.). EM data were collected at the Central Electron Microscopy Facility of the MPI of Biophysics, funded by the Max Planck Society.
Author contributions: J.S. carried out the biochemical work, prepared the cryo-EM grids, acquired and processed the cryo-EM data, built and analyzed the atomic models, drew figures, and contributed to writing the manuscript. E.L. prepared the cryo-EM grids, acquired and processed the cryo-EM data, built and analyzed the atomic models, drew figures, and contributed to writing the manuscript. I.W. carried out the complexome profiling analysis, processed the mass spectrometry data, and contributed to writing the manuscript. W.K. provided the cryo-EM infrastructure, supervised the cryo-EM work, and contributed to writing the manuscript. J.V. built and analyzed the atomic models, interpreted the structures, drew figures, and contributed to writing the manuscript. V.Z. initiated the project, interpreted the mechanistic implications of the structures, drew figures, and wrote the first draft of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Cryo-EM maps and model coordinates have been deposited in the wwPDB with accession codes EMD-14765 and 7zkq for the early PP module assembly intermediate, EMD-14764 and 7zkp for the late intermediate with both assembly factors, EMD-14770 for the late intermediate with NDUFAF1, and EMD-14766 for the late intermediate without assembly factors. Complexome profiling data have been deposited to the Proteomics Identification Database (PRIDE) archive with the dataset identifier PXD033522. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 and S2
Other Supplementary Material for this : manuscript includes the following:
Movies S1 to S5
REFERENCES AND NOTES
- 1.Galemou Yoga E., Angerer H., Parey K., Zickermann V., Respiratory complex I - Mechanistic insights and advances in structure determination. Biochim. Biophys. Acta 1861, 148153 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Vercellino I., Sazanov L. A.,The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell Biol. 23,141–161 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Padavannil A., Ayala-Hernandez M. G., Castellanos-Silva E. A., Letts J. A.,The mysterious multitude: Structural perspective on the accessory subunits of respiratory complex I. Front. Mol. Biosci. 8,798353 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parey K., Wirth C., Vonck J., Zickermann V., Respiratory complex I - Structure, mechanism and evolution. Curr. Opin. Struct. Biol. 63, 1–9 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Agip A.-N. A., Blaza J. N., Fedor J. G., Hirst J., Mammalian respiratory complex I through the lens of cryo-EM. Annu. Rev. Biophys. 48, 165–184 (2019). [DOI] [PubMed] [Google Scholar]
- 6.McKenzie M., Ryan M. T.,Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life 62,497–502 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Nouws J., Nijtmans L. G. J., Smeitink J. A., Vogel R. O.,Assembly factors as a new class of disease genes for mitochondrial complex I deficiency: Cause, pathology and treatment options. Brain 135,12–22 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Hock D. H., Robinson D. R. L., Stroud D. A.,Blackout in the powerhouse: Clinical phenotypes associated with defects in the assembly of OXPHOS complexes and the mitoribosome. Biochem. J. 477,4085–4132 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sánchez-Caballero L., Guerrero-Castillo S., Nijtmans L.,Unraveling the complexity of mitochondrial complex I assembly: A dynamic process. Biochim. Biophys. Acta 1857,980–990 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Heide H., Bleier L., Steger M., Ackermann J., Dröse S., Schwamb B., Zörnig M., Reichert A. S., Koch I., Wittig I., Brandt U.,Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 16,538–549 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Formosa L. E., Dibley M. G., Stroud D. A., Ryan M. T.,Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Semin. Cell Dev. Biol. 76,154–162 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Formosa L. E., Muellner-Wong L., Reljic B., Sharpe A. J., Jackson T. D., Beilharz T. H., Stojanovski D., Lazarou M., Stroud D. A., Ryan M. T.,Dissecting the roles of mitochondrial complex i intermediate assembly complex factors in the biogenesis of complex I. Cell Rep. 31,107541 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Guerrero-Castillo S., Baertling F., Kownatzki D., Wessels H. J., Arnold S., Brandt U., Nijtmans L.,The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 25,128–139 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Ligas J., Pineau E., Bock R., Huynen M. A., Meyer E. H.,The assembly pathway of complex I in Arabidopsis thaliana. Plant J. 97,447–459 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Parey K., Haapanen O., Sharma V., Köfeler H., Züllig T., Prinz S., Siegmund K., High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease. Sci. Adv. 5, eaax9484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soufari H., Parrot C., Kuhn L., Waltz F., Hashem Y.,Specific features and assembly of the plant mitochondrial complex I revealed by cryo-EM. Nat. Commun. 11,5195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giachin G., Jessop M., Bouverot R., Acajjaoui S., Saidi M., Chretien A., Bacia-Verloop M., Signor L., Mas P. J., Favier A., Borel Meneroud E., Hons M., Hart D. J., Kandiah E., Boeri Erba E., Buisson A., Leonard G., Gutsche I., Soler-Lopez M.,Assembly of the mitochondrial complex I assembly complex suggests a regulatory role for deflavination. Angew. Chem. Int. Ed. Engl. 60,4689–4697 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Küffner R., Rohr A., Schmiede A., Krull C., Schulte U.,Involvement of two novel chaperones in the assembly of mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Mol. Biol. 283,409–417 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Schulte U.,Biogenesis of respiratory complex I. J. Bioenerg. Biomembr. 33,205–212 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Vogel R. O., Janssen R. J., Ugalde C., Grovenstein M., Huijbens R. J., Visch H. J., van den Heuvel L. P., Willems P. H., Zeviani M., Smeitink J. A., Nijtmans L. G.,Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 272,5317–5326 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Dunning C. J., McKenzie M., Sugiana C., Lazarou M., Silke J., Connelly A., Fletcher J. M., Kirby D. M., Thorburn D. R., Ryan M. T.,Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26,3227–3237 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fassone E., Taanman J. W., Hargreaves I. P., Sebire N. J., Cleary M. A., Burch M., Rahman S.,Mutations in the mitochondrial complex I assembly factor NDUFAF1 cause fatal infantile hypertrophic cardiomyopathy. J. Med. Genet. 48,691–697 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Kerscher S., Drose S., Zwicker K., Zickermann V., Brandt U.,Yarrowia lipolytica, a yeast genetic system to study mitochondrial complex I. Biochim. Biophys. Acta 1555,83–91 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Bione S., D’Adamo P., Maestrini E., Gedeon A. K., Bolhuis P. A., Toniolo D.,A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 12,385–389 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Ji J., Greenberg M. L.,Cardiolipin function in the yeast S. cerevisiae and the lessons learned for Barth syndrome. J. Inherit. Metab. Dis. 45,60–71 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlame M., Xu Y.,The function of tafazzin, a mitochondrial phospholipid-lysophospholipid acyltransferase. J. Mol. Biol. 432,5043–5051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musatov A., Sedlak E.,Role of cardiolipin in stability of integral membrane proteins. Biochimie 142,102–111 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Zhang M., Mileykovskaya E., Dowhan W.,Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277,43553–43556 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H., Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Chatzispyrou I. A., Guerrero-Castillo S., Held N. M., Ruiter J. P. N., Denis S. W., I. J. L, R. J. Wanders, M. van Weeghel, S. Ferdinandusse, F. M. Vaz, U. Brandt, R. H. Houtkooper,Barth syndrome cells display widespread remodeling of mitochondrial complexes without affecting metabolic flux distribution. Biochim. Biophys. Acta 1864,3650–3658 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Beltran-Heredia E., Tsai F. C., Salinas-Almaguer S., Cao F. J., Bassereau P., Monroy F.,Membrane curvature induces cardiolipin sorting. Commun. Biol. 2,225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y., Anjaneyulu M., Donelian A., Yu W., Greenberg M. L., Ren M., Owusu-Ansah E., Schlame M.,Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proc. Natl. Acad. Sci. U.S.A. 116,11235–11240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heath R. J., Rock C. O.,A conserved histidine is essential for glycerolipid acyltransferase catalysis. J. Bacteriol. 180,1425–1430 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlame M.,Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 49,1607–1620 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewin T. M., Wang P., Coleman R. A.,Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38,5764–5771 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Hijikata A., Yura K., Ohara O., Go M.,Structural and functional analyses of Barth syndrome-causing mutations and alternative splicing in the tafazzin acyltransferase domain. Meta Gene 4,92–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt T. G., Batz L., Bonet L., Carl U., Holzapfel G., Kiem K., Matulewicz K., Niermeier D., Schuchardt I., Stanar K.,Development of the Twin-Strep-tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr. Purif. 92,54–61 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Kmita K., Wirth C., Warnau J., Guerrero-Castillo S., Hunte C., Hummer G., Kaila V. R., Zwicker K., Brandt U., Zickermann V.,Accessory NUMM (NDUFS6) subunit harbors a Zn-binding site and is essential for biogenesis of mitochondrial complex I. Proc. Natl. Acad. Sci. U.S.A. 112,5685–5690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stroud D. A., Surgenor E. E., Formosa L. E., Reljic B., Frazier A. E., Dibley M. G., Osellame L. D., Stait T., Beilharz T. H., Thorburn D. R., Salim A., Ryan M. T.,Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature 538,123–126 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D.,Highly accurate protein structure prediction with AlphaFold. Nature 596,583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parey K., Lasham J., Mills D. J., Djurabekova A., Haapanen O., Yoga E. G., Xie H., Kuhlbrandt W., Sharma V., Vonck J., Zickermann V.,High-resolution structure and dynamics of mitochondrial complex I–Insights into the proton pumping mechanism. Sci. Adv. 7,eabj3221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann L., Stephens A., Nam S. Z., Rau D., Kubler J., Lozajic M., Gabler F., Soding J., Lupas A. N., Alva V.,A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430,2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G. A., Sonnhammer E. L. L., Tosatto S. C. E., Paladin L., Raj S., Richardson L. J., Finn R. D., Bateman A.,Pfam: The protein families database in 2021. Nucleic Acids Res. 49,D412–D419 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutmann B., Royan S., Schallenberg-Rudinger M., Lenz H., Castleden I. R., McDowell R., Vacher M. A., Tonti-Filippini J., Bond C. S., Knoop V., Small I. D.,The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol. Plant 13,215–230 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Elurbe D. M., Huynen M. A.,The origin of the supernumerary subunits and assembly factors of complex I: A treasure trove of pathway evolution. Biochim. Biophys. Acta 1857,971–979 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Vogel R. O., Janssen R. J., van den Brand M. A., Dieteren C. E., Verkaart S., Koopman W. J., Willems P. H., Pluk W., van den Heuvel L. P., Smeitink J. A., Nijtmans L. G.,Cytosolic signaling protein Ecsit also localizes to mitochondria where it interacts with chaperone NDUFAF1 and functions in complex I assembly. Genes Dev. 21,615–624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giachin G., Bouverot R., Acajjaoui S., Pantalone S., Soler-Lopez M.,Dynamics of human mitochondrial complex I assembly: Implications for neurodegenerative diseases. Front. Mol. Biosci. 3,43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Agip A.-N. A., Blaza J. N., Bridges H. R., Viscomi C., Rawson S., Muench S. P., Hirst J., Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states. Nat. Struct. Mol. Biol. 25, 547–556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claypool S. M., McCaffery J. M., Koehler C. M.,Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 174,379–390 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlame M., Haldar D.,Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J. Biol. Chem. 268,74–79 (1993). [PubMed] [Google Scholar]
- 51.Baile M. G., Whited K., Claypool S. M.,Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Mol. Biol. Cell 24,2008–2020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson R. M., Yao J., Gajewski S., Kumar G., Martin E. W., Rock C. O., White S. W.,A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer. Nat. Struct. Mol. Biol. 24,666–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamada T., Feese M. D., Ferri S. R., Kato Y., Yajima R., Toguri T., Kuroki R.,Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacia oleracea. Acta Crystallogr. D 60,13–21 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Sakamoto O., Kitoh T., Ohura T., Ohya N., Iinuma K.,Novel missense mutation (R94S) in the TAZ (G4.5) gene in a Japanese patient with Barth syndrome. J. Hum. Genet. 47,229–231 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Whited K., Baile M. G., Currier P., Claypool S. M.,Seven functional classes of Barth syndrome mutation. Hum. Mol. Genet. 22,483–492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karkucinska-Wieckowska A., Trubicka J., Werner B., Kokoszynska K., Pajdowska M., Pronicki M., Czarnowska E., Lebiedzinska M., Sykut-Cegielska J., Ziolkowska L., Jaron W., Dobrzanska A., Ciara E., Wieckowski M. R., Pronicka E.,Left ventricular noncompaction (LVNC) and low mitochondrial membrane potential are specific for Barth syndrome. J. Inherit. Metab. Dis. 36,929–937 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Y., Malhotra A., Ren M., Schlame M.,The enzymatic function of tafazzin. J. Biol. Chem. 281,39217–39224 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Fry M., Green D. E.,Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J. Biol. Chem. 256,1874–1880 (1981). [PubMed] [Google Scholar]
- 59.Jussupow A., Di Luca A., Kaila V. R. I.,How cardiolipin modulates the dynamics of respiratory complex I. Sci. Adv. 5,eaav1850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharpley M. S., Shannon R. J., Draghi F., Hirst J.,Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry 45,241–248 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Guo R., Zong S., Wu M., Gu J., Yang M.,Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell 170,1247–1257.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Anzmann A. F., Sniezek O. L., Pado A., Busa V., Vaz F. M., Kreimer S. D., DeVine L. R., Cole R. N., Le A., Kirsch B. J., Claypool S. M., Vernon H. J.,Diverse mitochondrial abnormalities in a new cellular model of TAFFAZZIN deficiency are remediated by cardiolipin-interacting small molecules. J. Biol. Chem. 297,101005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Claypool S. M., Boontheung P., McCaffery J. M., Loo J. A., Koehler C. M.,The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol. Biol. Cell 19,5143–5155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galkin A., Meyer B., Wittig I., Karas M., Schagger H., Vinogradov A., Brandt U.,Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J. Biol. Chem. 283,20907–20913 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burger N., James A. M., Mulvey J. F., Hoogewijs K., Ding S., Fearnley I. M., Loureiro-Lopez M., Norman A. A. I., Arndt S., Mottahedin A., Sauchanka O., Hartley R. C., Krieg T., Murphy M. P.,ND3 Cys39 in complex I is exposed during mitochondrial respiration. Cell. Chem. Biol. 29,636–649.e14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cabrera-Orefice A., Yoga E. G., Wirth C., Siegmund K., Zwicker K., Guerrero-Castillo S., Zickermann V., Hunte C., Brandt U.,Locking loop movement in the ubiquinone pocket of complex I disengages the proton pumps. Nat. Commun. 9,4500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ribeiro D. O., Viegas A., Pires V. M. R., Medeiros-Silva J., Bule P., Chai W., Marcelo F., Fontes C., Cabrita E. J., Palma A. S., Carvalho A. L., Molecular basis for the preferential recognition of β1,3-1,4-glucans by the family 11 carbohydrate-binding module from Clostridium thermocellum. FEBS J. 287,2723–2743 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., Vizcaino J. A.,The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47,D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Angerer H., Radermacher M., Mankowska M., Steger M., Zwicker K., Heide H., Wittig I., Brandt U., Zickermann V.,The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc. Natl. Acad. Sci. U.S.A. 111,5207–5212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. 3rd, Smith H. O.,Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6,343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Chen X., Zaro J. L., Shen W. C.,Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 65,1357–1369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A. R. N., Potter S. C., Finn R. D., Lopez R.,The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47,W636–W641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J.,Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics 25,1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kimanius D., Dong L., Sharov G., Nakane T., Scheres S. H. W.,New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478,4169–4185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A.,MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14,331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rohou A., Grigorieff N.,CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192,216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S.,SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2,218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Rosenthal P. B., Henderson R.,Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333,721–745 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Terwilliger T. C., Ludtke S. J., Read R. J., Adams P. D., Afonine P. V.,Improvement of cryo-EM maps by density modification. Nat. Methods 17,923–927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L.,BLAST+: Architecture and applications. BMC Bioinformatics 10,421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Emsley P., Lohkamp B., Scott W. G., Cowtan K.,Features and development of Coot. Acta Crystallogr. D 66,486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D.,Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74,531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E.,UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27,14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 and S2
Movies S1 to S5