Abstract
Major depressive disorder (MDD) is a devastating mental disorder that affects up to 17% of the population worldwide. Although brain-wide network-level abnormalities in MDD patients via resting-state functional magnetic resonance imaging (rsfMRI) exist, the mechanisms underlying these network changes are unknown, despite their immense potential for depression diagnosis and management. Here, we show that the astrocytic calcium-deficient mice, inositol 1,4,5-trisphosphate-type-2 receptor knockout mice (Itpr2−/− mice), display abnormal rsfMRI functional connectivity (rsFC) in depression-related networks, especially decreased rsFC in medial prefrontal cortex (mPFC)–related pathways. We further uncover rsFC decreases in MDD patients highly consistent with those of Itpr2−/− mice, especially in mPFC-related pathways. Optogenetic activation of mPFC astrocytes partially enhances rsFC in depression-related networks in both Itpr2−/− and wild-type mice. Optogenetic activation of the mPFC neurons or mPFC-striatum pathway rescues disrupted rsFC and depressive-like behaviors in Itpr2−/− mice. Our results identify the previously unknown role of astrocyte dysfunction in driving rsFC abnormalities in depression.
Astrocyte dysfunction drives functional MRI connectivity aberrations in depressed brains.
INTRODUCTION
The high lifetime prevalence (~17%) and the leading cause of disability and suicide make major depressive disorder (MDD) a devastating psychiatric disease (1). Efforts to identify pathophysiological mechanisms of depression implicate neuronal communication within specific brain circuits and networks (2, 3). Resting-state functional magnetic resonance imaging (rsfMRI) is a powerful noninvasive tool to investigate neuronal communication on a whole-brain scale by quantifying resting-state functional connectivity (rsFC), the correlation of infraslow (~0.1 Hz) spontaneous blood oxygen level–dependent (BOLD) MRI signal fluctuations, among spatially segregated but functionally related regions (4–8). Mounting evidence has established that depressed patients display abnormal rsFC in key functional hubs involved in emotional processing, such as the ventromedial prefrontal cortex (vmPFC), dorsolateral PFC (dlPFC), anterior cingulate (ACC), amygdala (AMY), thalamus (TH), striatum (Str), and hippocampus (HP) (2, 9, 10). Moreover, based on the distinct patterns of dysfunctional rsFC, reclassification of depression has improved the prediction of treatment response (11). Furthermore, rsFC-guided neuromodulation therapy is associated with remission for treatment-resistant depression (12). Despite the immense potential of rsfMRI for diagnosing depression and guiding depression treatment, the mechanisms underlying rsfMRI connectivity abnormalities in depression remain poorly understood.
Astrocytes, the most abundant glial cell type in the mammalian brain, are strongly implicated in depression (13–17). Evidence from postmortem analyses of depressed patients and animal studies found changes in astrocytic density, markers, and gliotransmitters in emotion-related brain regions (13, 15–18), including vmPFC, dlPFC, ACC, AMY, TH, Str, and HP. Moreover, astrocytes regulate synaptic transmission and plasticity (19), mediate myelinated axon excitability and conduction speed (20), synchronize the activity of their neighboring neurons (21), and modulate neuronal communication across brain regions (22). Furthermore, astrocytic intrinsic Ca2+ events are correlated with brain-wide infraslow spontaneous BOLD fluctuations (23). These studies suggest that astrocytes can present a key candidate for mechanisms underpinning rsfMRI connectivity. However, the role of astrocytes in rsfMRI connectivity remains understudied for both normal and depressed brains.
Astrocytic activation is manifested by elevated intracellular Ca2+ signals mainly mediated by the inositol 1,4,5-trisphosphate (IP3) pathway, and IP3 receptor type 2 (IP3R2) is the predominant functional IP3R isoform in astrocytes (24). IP3R2 knockout (Itpr2−/−) mice exhibit strongly attenuated Ca2+ signals in astrocytes, but not in neurons (13, 24–27). We previously established that Itpr2−/− mice displayed depressive-like behaviors in the forced swimming test (FST) and sucrose preference test (SPT) (13). Astrocytic Ca2+ signals can be effectively increased by astrocyte-specific optogenetic stimulation (24). Thus, we integrated whole-brain rsfMRI with cell type–specific optogenetic tools in Itpr2−/− mice, in conjunction with rsfMRI analysis in human patients with depression. We hypothesized that astrocyte dysfunction is a contributor to rsfMRI connectivity abnormalities in depression. To elucidate whether astrocyte dysfunction drives abnormal rsfMRI connectivity in depression, we examined the effects of astrocyte-specific functional loss and gain on brain-wide rsFC, as well as the association between abnormal rsFC and depressive-like behaviors in Itpr2−/− mice.
RESULTS
Astrocyte dysfunction disrupts brain-wide rsFC
Astrocyte calcium-deficient mice, the Itpr2−/− mice, displayed expected depressive-like behaviors in the tail-suspension test (TST) and FST (fig. S1, A and B), as we demonstrated previously (13). To investigate whether astrocyte dysfunction contributes to rsfMRI connectivity abnormalities in depression, we first examined how a decrease of astrocytic Ca2+ activity affects rsFC, by comparing the brain-wide rsFC between Itpr2−/− and wild-type (WT) mice. We performed independent component analysis (ICA) on all the rsfMRI data and identified 53 robust rsFC components (fig. S2A). On the basis of these ICA-detected rsFC components and atlas, 94 spatially separated and left-right symmetrical brain units (i.e., 47 brain regions) were defined as the regions of interest (ROIs) to calculate the pairwise correlation matrices (fig. S2B and table S1).
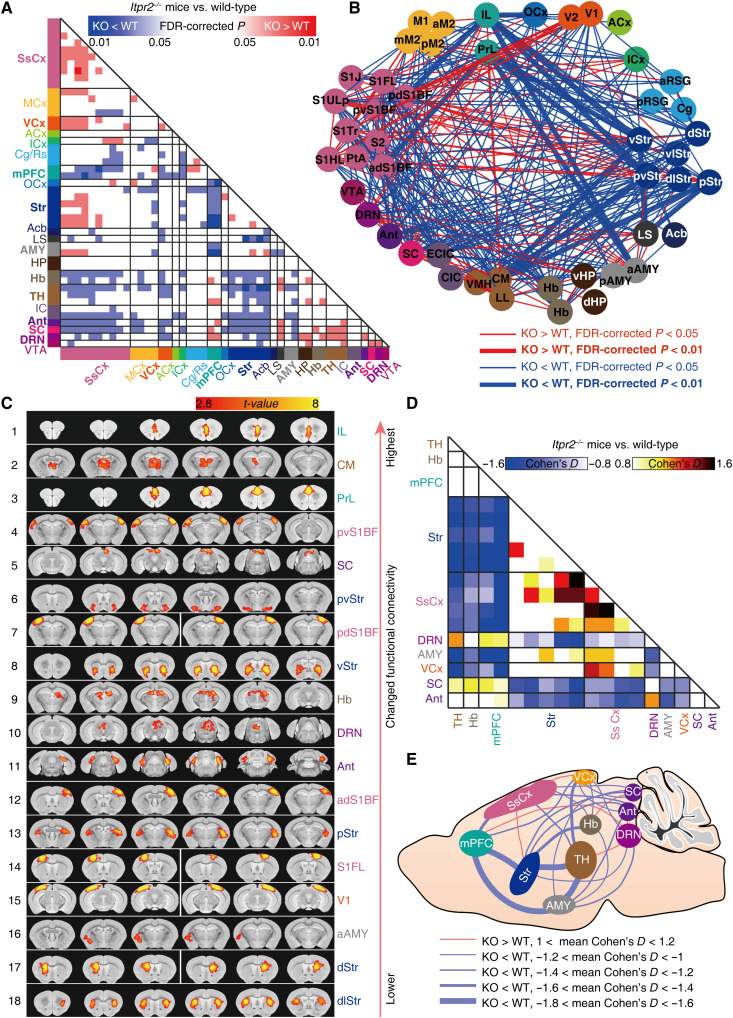
Intergroup comparison of the whole-brain pairwise correlation matrices revealed significant changes in the brain-wide rsFC of Itpr2−/− mice, compared with WT mice (Fig. 1, A and B). We ranked the ROIs in the order of the overall connectivity changes that link to each ROI, which were quantified as the “node modulation index” (NMI) from the mean effect size (Cohen’s D) of rsFC change (28, 29). Eighteen ROIs exhibited strong effect with NMI > 0.7 (fig. S3, E and F) (28, 29). These 18 ROIs corresponded to subregions within the medial PFC (mPFC), TH, Str, somatosensory cortex (SsCx), habenula (Hb), AMY, dorsal raphe nucleus (DRN), visual cortex (VCx), superior colliculus (SC), and anterior lobe cerebellum (Ant; Fig. 1C and fig. S3F), which were previously shown to be associated with depression (hereafter referred to as depression-related networks) (13, 14, 30, 31). The HP and ventral tegmental area (VTA), two other key hubs of depression (2), showed no obvious changes in their rsFC with other brain regions (Fig. 1, A and B, and fig. S3F).
Fig. 1. Brain-wide functional connectivity mapping of Itpr2−/− and WT mice by rsfMRI.
(A and B) Intergroup statistical comparison of rsFC of Itpr2−/− and WT mice (n = 11 and 13 mice for Itpr2−/− and WT mice, respectively), shown as matrix (A) and network graph (B). The nodes correspond to 47 brain regions detected from ICA (listed in table S1). Independent sample t test, P < 0.05, false discovery rate (FDR) correction for multiple comparisons; only the significantly changed pathways (FDR-corrected P < 0.05) are shown. (C) The 47 brain regions are ranked in the order of the overall rsFC changes, and the top 18 ROIs (mean effect size > 0.7) are shown in (C) to (E). The effect size matrix (D) and schematic diagram (E) showing the changed rsFC between top 18 ROIs. MCx, motor cortex; ACx, auditory cortex; ICx, insular cortex; Cg/Rs, cingulate/retrosplenial cortex; OCx, orbital cortex; Acb, accumbens nucleus; LS, lateral septal nucleus; Hb, habenular nucleus; IC, inferior colliculus; IL, infralimbic cortex; PrL, prelimbic cortex; CM, central medial thalamus nucleus. For the abbreviation details, see table S1.
To quantify the changed rsFC among the mostly affected brain regions in Itpr2−/− mice, we calculated the effect sizes (Itpr2−/− mice versus WT mice) within the depression-related networks (Fig. 1D). Effect size analysis showed that the rsFC was markedly decreased in mPFC-Str, mPFC-AMY, mPFC-SsCx, mPFC-VCx, TH-Str, TH-AMY, TH-SsCx, TH-VCx, Str-SC, Str-Ant, SsCx-SC, Ant-SsCx, AMY-SC, Ant-VCx, Hb-Str, Hb-AMY, Hb-SsCx, Hb-VCx, DRN-Str, DRN-AMY, and DRN-VCx pathways in Itpr2−/− mice relative to WT mice (Fig. 1, D and E). Whereas IP3R2 knockout increased the rsFC in DRN-Ant, mPFC-DRN, DRN-TH, Hb-SC, and SsCx-VCx pathways (Fig. 1, D and E). Because the TST and rsfMRI experiments were performed on the same animals, we further examined the correlation between rsFC and behavioral performances in the TST. Notably, six pathways exhibited negative correlation between rsFC and immobility time in TST, including the mPFC-Str [r = −0.5790, false discovery rate (FDR)–corrected P = 0.0171], mPFC-AMY (r = −0.5892, FDR-corrected P = 0.0171), mPFC-SsCx (r = −0.5912, FDR-corrected P = 0.0171), TH-Str (r = −0.5698, FDR-corrected P = 0.0171), TH-AMY (r = −0.5765, FDR-corrected P = 0.0171), and TH-SsCx (r = −0.5861, FDR-corrected P = 0.0171) pathways (fig. S1D). These results show that astrocyte dysfunction can lead to brain-wide rsfMRI connectivity abnormalities that predicted depressive-like behavior, particularly in depression-related networks.
Characteristically consistent rsFC changes in MDD patients and Itpr2−/− mice
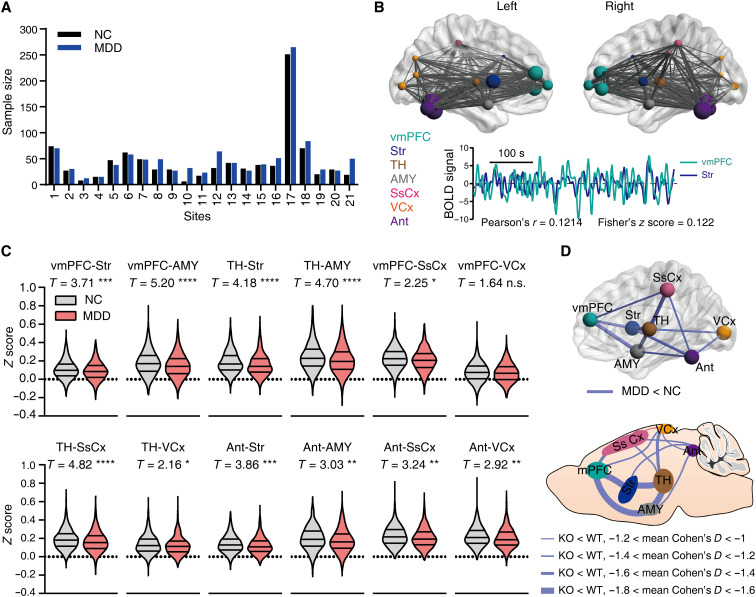
The Itpr2−/− mice showed changed rsFC mainly in depression-related networks, including mPFC, Str, AMY, TH, SsCx, VCx, Ant, SC, DRN, and Hb (Fig. 1, C to E). Guided by these results, we next assessed whether similar changes within depression-related networks also existed in MDD patients by analyzing the rsfMRI dataset from the REST-meta-MDD consortium (32). The human rsfMRI dataset provided us with time series extracted from human atlas–defined ROIs, which include most of the brain regions within the abovementioned depression-related networks except SC, DRN, and Hb. After preprocessing and data exclusion to ensure data quality (see Materials and Methods for details), a total of 1080 MDD patients (MDD) and 931 normal controls (NC), recruited from 21 study sites across China, were used for subsequent statistical analysis (Fig. 2A and table S2). The vmPFC rsFC was analyzed as human vmPFC is homologous to rodent mPFC (33, 34). We found that the rsFC of vmPFC-Str, vmPFC-AMY, TH-Str, TH-AMY, vmPFC-SsCx, TH-SsCx, TH-VCx, Ant-Str, Ant-AMY, Ant-SsCx, and Ant-VCx pathways was significantly decreased in the MDD compared with the NC group, consistent with the altered rsFC in Itpr2−/− mice (Fig. 2, C and D, and fig. S4). The rsFC of the mPFC-VCx pathway, which displayed a significant decrease in Itpr2−/− mice, was not changed in depressed patients (Fig. 2, C and D, and fig. S4). The rsFC of the Str-SsCx and AMY-SsCx pathways was significantly increased in Itpr2−/− mice but significantly decreased in MDD patients (fig. S4). The rsFC of the Str-AMY, vmPFC-TH, vmPFC-Ant, and TH-Ant pathways was significantly decreased in MDD patients but not changed in Itpr2−/− mice (fig. S4).
Fig. 2. Attenuated rsFC in MDD patients consistent with that in Itpr2−/− mice.
(A) Sample size for MDD patients (1080 MDD) and normal controls (931 NC). The horizontal axis represents 21 study sites from China. See also table S2. (B) Locations of ROIs and data analysis schematic. For each subject, rsFC was generated by calculating the Pearson’s correlation coefficient (r) of the BOLD signals and then transformed to Fisher’s z score, as in this representative subject. (C) Violin figures showing the distribution of rsFC for MDD and NC subjects. Linear mixed models. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; FDR for multiple comparison correction, n.s., no significance. (D) Similar rsFC changes in MDD patients and Itpr2−/− mice. See table S3 for MNI coordinates for ROIs in (B).
Although the head motion was regressed out at the individual level and then used as a covariate in linear mixed model (LMM) analysis, we additionally performed analyses after excluding subjects with large motion to validate our findings. We found that most of the rsFC decreases, except that of TH-VCx, in MDD patients remained significant regardless of using moderate [excluding the subjects with mean framewise displacement (FD) > 0.2 mm] or strict (excluding the subjects with mean FD > 0.1 mm) head motion control (fig. S5, E and F). In addition, we reran the analysis with a different parcellation scheme using the Craddock’s functional clustering atlas (35). We again found that the rsFC of most pathways, including vmPFC-Str, vmPFC-AMY, vmPFC-SsCx, Ant-AMY, Ant-Str, Ant-SsCx, and Ant-VCx, remained significantly decreased in the MDD group, albeit the rsFC changes in TH-Str, TH-AMY, TH-SsCx, TH-VCx, and vmPFC-VCx became nonsignificant (fig. S6B). Together, these results show that the altered rsFC in depressed patients and Itpr2−/− mice is highly consistent, and the changes in mPFC-related rsFC are the most robust and consistent when compared to those in other brain regions.
Optogenetic activation of mPFC astrocytes enhances rsFC in depression-related networks
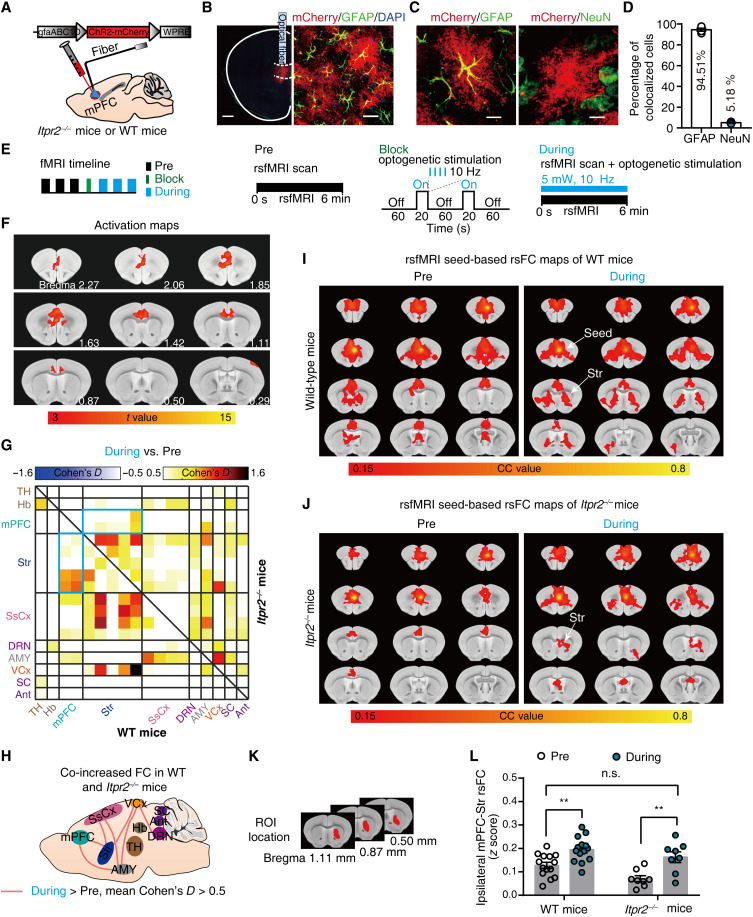
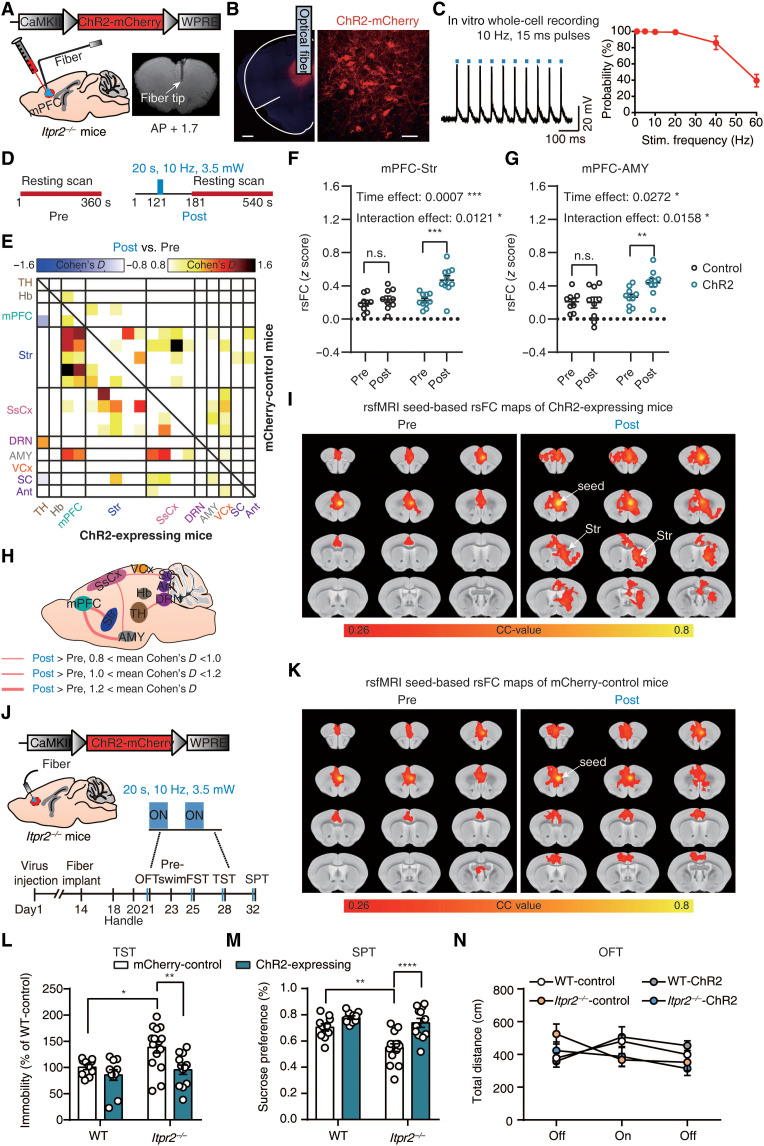
Cell type–specific optogenetic stimulation is an effective approach to selectively increase astrocytic Ca2+ signals (24). To further determine the role of astrocytes in rsFC in depression, we directly examined the effects of optogenetic stimulation of mPFC astrocytes on depression-related networks. We selectively stimulated astrocytes by injecting adeno-associated viruses (AAV) expressing the ChR2 under the glial fibrillary acidic protein (GFAP) promoter (gfaABC1D) into mPFC of WT or Itpr2−/− mice (Fig. 3, A and B). Confocal images showed that most of the mCherry-positive cells coexpressed GFAP (94.51%), an astrocytic marker (Fig. 3, C and D).
Fig. 3. Optogenetic stimulation of mPFC astrocytes increases rsFC within depression-related networks.
(A) Schematic showing the virus injection and optical fiber implantation. (B) Confocal images demonstrate ChR2 expression in mPFC astrocytes. Red, ChR2-mCherry; green, GFAP; blue, 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 500 μm (left) and 25 μm (right). (C) Representative histology of ChR2-mCherry colocalized with GFAP (left) or NeuN (right). Scale bars, 10 μm. (D) Quantitative analysis of the percentage of ChR2-positive cells that colocalized with GFAP (n = 290 mCherry-positive cells; 12 fields of view; four mice) and NeuN (n = 409 mCherry-positive cells; 17 fields of view; four mice). (E) Schematics of fMRI scanning timeline and corresponding paradigms. (F) Representative activation maps at 10-Hz stimulation. (G) Effect size matrix (Cohen’s D > 0.5) showing effect of optogenetic stimulation on ipsilateral rsFC within depression-related networks in WT (n = 14; bottom-left part) and Itpr2−/− mice (n = 8; top-right part). The blue matrix box indicates the effect size of mPFC-Str rsFC. (H) Effect size schematic diagram shows co-increased ipsilateral rsFC in WT and Itpr2−/− mice. (I and J) Resting-state correlation maps of ipsilateral mPFC of WT and Itpr2−/− mice before and during optogenetic stimulation. (K) Str ROI used to extract the CC value in the rsFC maps of mPFC (I and J). (L) Quantification of ipsilateral mPFC-Str rsFC in WT and Itpr2−/− mice. Two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparison test, data are expressed as means ± SEM, **P < 0.01.
We then performed rsfMRI experiments before and during 10-Hz blue light (473 nm) optogenetic activation of mPFC astrocytes (Fig. 3E). The fMRI activation maps displayed an increase of BOLD activity in the mPFC (Fig. 3F). Effect size analysis revealed that optogenetic stimulation of astrocytes increased the rsFC of mPFC-Str, mPFC-AMY, DRN-VCx, Str-SsCx, Str-VCx, AMY-SsCx, and AMY-VCx pathways in both Itpr2−/− and WT mice, albeit with moderate effect (effect size > 0.5; Fig. 3, G and H). Comparing the trend of rsFC changes due to Itpr2 knockout and optogenetic activation of mPFC astrocytes in Itpr2−/− mice, we found that 51% (78 of 153) of pathways showed rsFC changes in opposite directions (fig. S7A). In these 78 pathways, the rsFC differences between Itpr2−/− and WT mice were negatively correlated with the rsFC differences in Itpr2−/− mice before and during optogenetic activation of mPFC astrocytes (fig. S7B; r = −0.6546, P < 0.0001). Among them, 67 of 78 pathways showed rsFC decreases with Itpr2 knockout but increases with astrocyte activation (the second quadrant in fig. S7B), whereas the remaining pathways showed rsFC increases with Itpr2 knockout but decreases with astrocyte activation (the fourth quadrant in fig. S7B). These results suggested that optogenetic activation of mPFC astrocytes could alleviate rsFC alterations induced by Itpr2 knockout.
We next performed seed-based analysis (SBA) with seed defined in the ipsilateral mPFC. The correlation coefficient (CC) maps and quantitative analysis also showed that ipsilateral mPFC-Str rsFC was significantly increased during optogenetic activation of the mPFC astrocytes in both Itpr2−/− and WT mice (Fig. 3, I to L). These results demonstrate that increased Ca2+ signals in mPFC astrocytes by optogenetic stimulation can mitigate rsFC alterations in depression-related networks, especially partially enhance the decreased rsFC because of astrocyte dysfunction.
Deletion of the Itpr2 attenuates the mPFC-to-Str neuronal communication
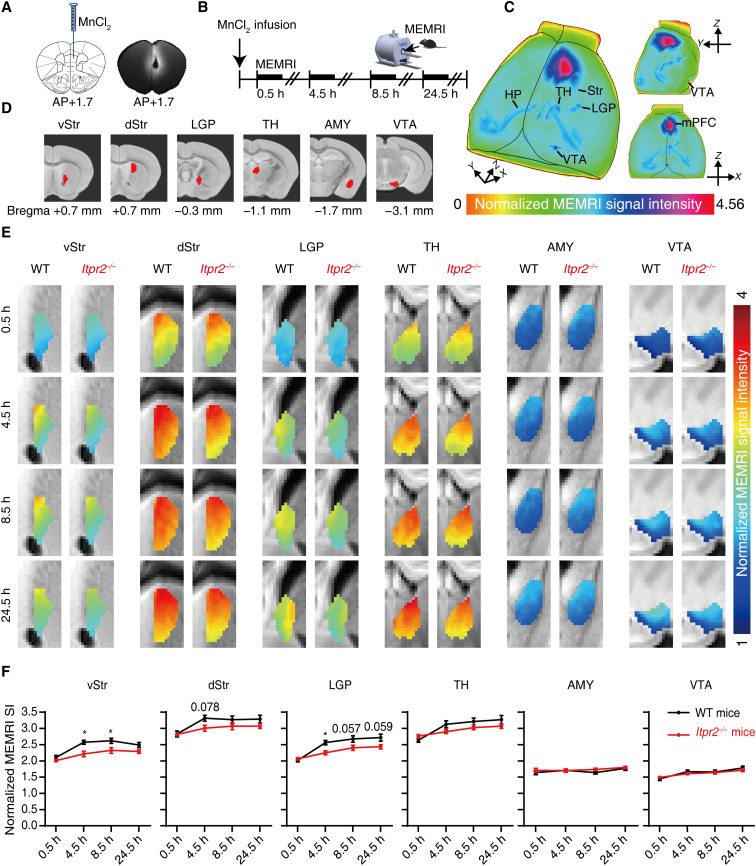
Functional connectivity is related to neural communication (36), which can be regulated by astrocytes (22). To understand how astrocytes modulate rsFC and whether it is by regulating neuronal communication (22), we used manganese-enhanced MRI (MEMRI) to determine whether astrocyte dysfunction affected neuronal communication and, if so, which of the mPFC-related pathways were affected the most. MEMRI is an effective method for evaluating neuronal communication across the whole brain (37). Mn2+, a calcium ion analog, can be absorbed by excitable neurons through voltage-gated calcium channels and then transported along the axons in an activity-dependent manner (37).
We first infused MnCl2 into the mPFC of Itpr2−/− and WT mice and tracked the accumulation of Mn2+ during the first 24.5 hours after injection (Fig. 4, A and B). We observed Mn2+-induced signal enhancement in the Str [ventral Str (vStr), dorsal Str (dStr), and lateral globus pallidus (LGP)], TH, AMY, and VTA (Fig. 4C). We then performed quantitative analysis of the signal intensities extracted from defined ROIs covering these brain regions (Fig. 4D). Compared with the WT mice, we found a reduced Mn2+ accumulation along the fiber tracts from mPFC to the vStr, dStr, and LGP 4.5 hours after Mn2+ administration, and a continuous reduction in both vStr and LGP at 8.5 hours in Itpr2−/− mice. By contrast, no apparent difference was observed in the Mn2+ accumulation at TH, AMY, and VTA (Fig. 4, E and F). These results demonstrate that astrocyte dysfunction decreases neuronal communication, and that the mPFC-Str pathway is the only significantly affected neural circuit among the mPFC-related pathways displaying altered rsFC in Itpr2−/− mice and depressed patients.
Fig. 4. Deletion of Itpr2 impairs the mPFC-to-Str neuronal communication.
(A) Schematic and a representative T1-weighted image illustrate the infusion site of MnCl2. (B) Experimental paradigm for dynamic manganese-enhanced MRI (MEMRI). (C) Representative three-dimensional maximum density projection images of a WT mouse showing the spread of Mn2+ enhancement 8.5 hours after infusion. (D) Definitions of ROIs. (E) Average color-coded MEMRI images. (F) Quantitative analysis of Mn2+ dynamic accumulation in the defined ROIs. Two-way repeated-measures ANOVA followed by Sidak’s multiple comparison test; *P < 0.5, n = 9 mice in each group. Data are expressed as means ± SEM.
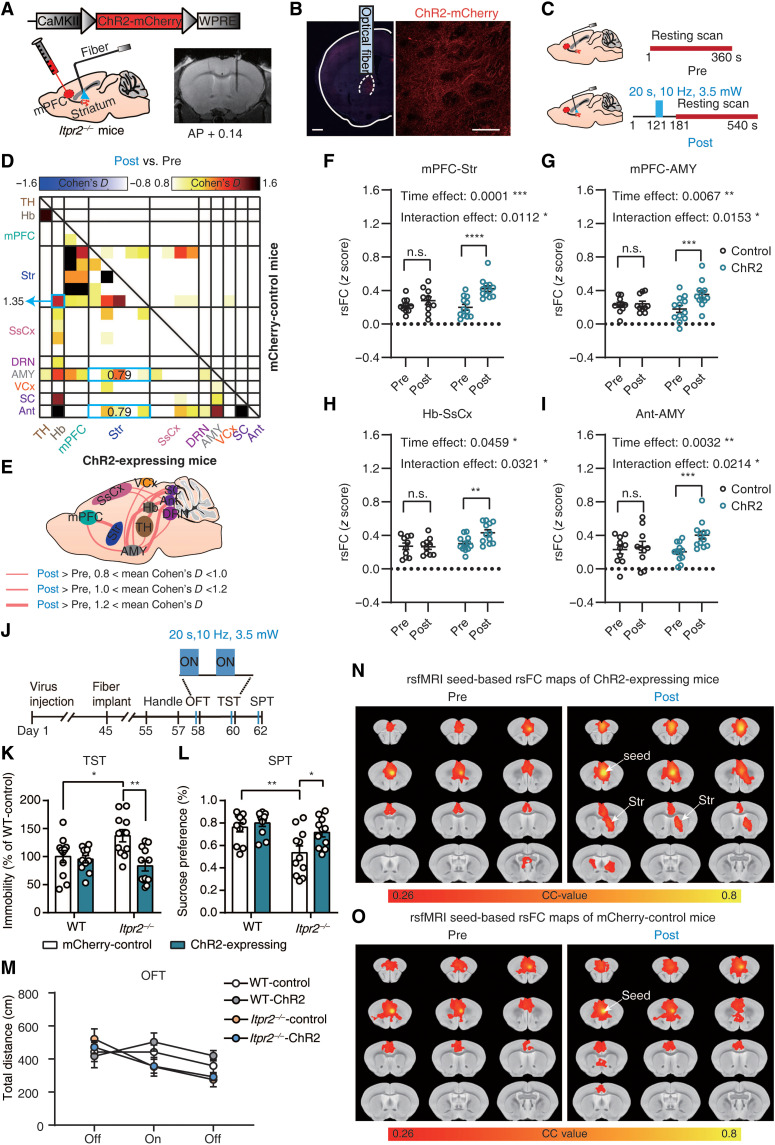
Optogenetic activation of mPFC neurons rescues rsFC and depressive-like behaviors
Using optogenetic stimulation to directly modulate neuronal communication of mPFC-related pathways, we sought to further determine the relationship between abnormal rsFC caused by astrocyte dysfunction and depressive-like behaviors in Itpr2−/− mice. We injected AAV-CamkII-hChR2-mCherry or AAV-CamkII-mCherry in the mPFC of Itpr2−/− mice and inserted an optical fiber above the infected cells (Fig. 5, A and B). In vitro whole-cell recording showed that 10-Hz optogenetic stimulation evoked robust action potential in the ChR2-expressing neurons from brain slices (Fig. 5C). We performed fMRI scans before and after 10-Hz optogenetic stimulation. Before each post-stimulation session, a 20-s optogenetic stimulation was delivered to activate ChR2-expressing neurons (Fig. 5D). Unilateral optogenetic stimulation of the mPFC evoked ipsilateral increases of BOLD activity mainly in the mPFC and Str, and also in other brain regions including the cingulate cortex, Hb, and VTA (fig. S8A). No activation was observed in mCherry-control mice (fig. S8B). The evoked BOLD signal in mPFC started to rise at ~3 s after stimulus onset and returned to baseline in ~40 s after the stimulus offset (fig. S8C).
Fig. 5. Optogenetic activation of mPFC neurons rescues partial rsFC and depressive-like behaviors in Itpr2−/− mice.
(A) Schematic showing the virus injection and optical fiber implantation. (B) Confocal images displaying ChR2 expression in neurons within mPFC. Red, ChR2-mCherry; blue, DAPI. Scale bars, 500 μm (left) and 50 μm (right). (C) In vitro slice recording (n = 4 neurons from three mice). (D) Optogenetic fMRI scanning paradigm. Pre, pre-stimulation; Post, post-stimulation. (E) Effect size matrix showing effect of optogenetic stimulation on ipsilateral rsFC within depression-related networks of ChR2-expressing (n = 10; bottom-left part) and control mice (n = 9; top-right part). (F and G) Quantitative analyses of ipsilateral mPFC-Str and mPFC-AMY rsFC before and after optogenetic stimulation. Two-way ANOVA with group (control versus ChR2) and time (Pre versus Post) as factors followed by Sidak’s multiple comparison test. (H) Effect size schematic diagram of ipsilateral rsFC of ChR2-expressing mice. (I and K) Average resting-state correlation maps of ipsilateral mPFC. (J) Schematic (top) and timeline (bottom) of behavioral tests. (L to N) Behavior performances of Itpr2−/− and WT mice in the TST, sucrose preference (SPT), and open-field test (OFT) after optogenetic stimulation (TST and SPT: n = 10, 10, 15, and 12, OFT: n = 10, 10, 13, and 12 for WT control, WT ChR2, Itpr2−/− control, and Itpr2−/− ChR2 mice, respectively). Two-way ANOVA followed by Sidak’s multiple comparison test. All data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Next, we analyzed the effect sizes (post-stimulation versus pre-stimulation) among the 18 ROIs identified in mouse rsfMRI results (Fig. 1, C to E). We found that rsFC in mPFC-Str, mPFC-AMY, AMY-SsCx, DRN-TH, and SsCx-SC pathways was increased in ChR2-expressing mice after optogenetic stimulation (Fig. 5, E and H). No obvious changes were found in other pathways, including TH-Str, TH-AMY, and Hb-Str (Fig. 5, E and H). Compared to the mCherry-control group, the ChR2-expressing group exhibited significantly larger increases of rsFC in mPFC-Str and mPFC-AMY pathways [Fig. 5, F and G; mPFC-Str: main effect of time: F1,17 = 16.99, P = 0.0007; time × group interaction: F1,17 = 7.877, P = 0.0121; mPFC-AMY: main effect of time: F1,17 = 5.841, P = 0.0272; time × group interaction: F1,17 = 7.183, P = 0.0158; two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparison test]. The rsFC in ChR2-expressing mice but not mCherry-control mice was significantly increased in AMY-SsCx, DRN-TH, and SsCx-SC pathways, albeit the differences in rsFC alterations were not significant between groups (fig. S8, D to F; AMY-SsCx: main effect of time: F1,17 = 11.25, P = 0.0038; time × group interaction: F1,17 = 1.583, P = 0.2254; DRN-TH: main effect of time: F1,17 = 10.81, P = 0.0043; time × group interaction: F1,17 = 3.999, P = 0.0618; SsCx-SC: main effect of time: F1,17 = 7.099, P = 0.0163; time × group interaction: F1,17 = 2.234, P = 0.1533; two-way ANOVA followed by Sidak’s multiple comparison test). Further quantitative analysis of rsFC in two most important pathways, namely, the mPFC-Str and mPFC-AMY pathways, showed that the increased rsFC was close to the level of WT mice (fig. S8, G and H). The SBA also showed optogenetic stimulation–induced increases of mPFC-Str rsFC in the ChR2-expressing mice, but not in the mCherry-control mice (Fig. 5, I and K). Furthermore, comparing the trend of rsFC changes due to Itpr2 knockout and optogenetic activation of mPFC neurons, we also found that 51% (78 of 153) of pathways showed rsFC changes in opposite directions (fig. S9A). In these 78 pathways, optogenetic activation of mPFC neurons caused the same direction of rsFC changes as optogenetic activation of mPFC astrocytes in 71.79% (56 of 78) of pathways (fig. S9A). Similarly, the rsFC differences between Itpr2−/− and WT mice were negatively correlated with the rsFC differences in Itpr2−/− mice before and after optogenetic activation of mPFC neurons (fig. S9B; r = −0.7507, P < 0.0001). Among them, 60 of 78 pathways showed rsFC decreases with Itpr2 knockout but increases with mPFC neuron activation (the second quadrant in fig. S9B), whereas the remaining pathways showed rsFC increases with Itpr2 knockout but decreases with mPFC neuron activation (the fourth quadrant in fig. S9B).
We then investigated behavioral performances of Itpr2−/− and WT mice 1 min after optogenetic activation of mPFC neurons in parallel behavioral experiments (Fig. 5J). Compared to mCherry-control Itpr2−/− mice, the ChR2-expressing Itpr2−/− mice exhibited a significant decrease in immobility time in TST (P = 0.0095) and FST (P = 0.0197) and a significant increase in sucrose preference (P < 0.0001) after optogenetic activation of mPFC neurons such that their depressive-like behavioral performances were rescued to the levels similar to those of WT mice (Fig. 5, L and M, and fig. S8I). Optogenetic stimulation had no significant effect on the behavioral performances of WT mice in TST (P = 0.9597) and SPT (P = 0.4394; Fig. 5, L and M). No significant difference was observed in general locomotion in different groups (Fig. 5N; main effect of group: F3,123 = 0.9709, P = 0.4088). Together, these results show that optogenetic activation of mPFC neurons partially rescued rsFC, especially the mPFC-Str and mPFC-AMY rsFC, and rescued depressive-like behaviors in Itpr2−/− mice.
Optogenetic activation of mPFC-Str projection rescues rsFC and depressive-like behaviors
Combining the results of MEMRI and optogenetic activation of mPFC neurons (Figs. 4 and 5), we show that the mPFC-Str pathway is likely the most important neural circuit in mediating depressive-like behaviors in Itpr2−/− mice, as Mn2+ accumulation in mPFC-AMY pathway is unchanged (Fig. 4, E and F). We next selectively modulated mPFC-Str projection in Itpr2−/− mice using optogenetic stimulation. AAV-CamkII-hChR2-mCherry or AAV-CamkII-mCherry was unilaterally injected in the mPFC of Itpr2−/− mice, and the optical fiber was implanted above the ipsilateral Str to allow for delivery of blue light to the axons of mPFC (Fig. 6, A and C). Confocal images confirmed ChR2 expression on mPFC axon terminals in the Str (Fig. 6B). The fMRI activation maps displayed an increase of BOLD activity at the bilateral Str and ipsilateral mPFC during optogenetic stimulation in ChR2-expressing mice, but not in mCherry-control mice (fig. S10, A and B).
Fig. 6. Optogenetic activation of mPFC terminals in the Str partially rescues rsFC and depressive-like behaviors.
(A) Schematic showing the virus injection and optical fiber implantation. (B) Confocal images demonstrate ChR2 expression in mPFC projection to Str. Red, ChR2-mCherry; blue, DAPI. Scale bars, 500 μm (left) and 50 μm (right). (C) Schematic of optogenetic fMRI scan paradigm. Pre, pre-stimulation; Post, post-stimulation. (D) Effect size matrix showing effect of optogenetic stimulation on ipsilateral rsFC within depression-related networks of ChR2-expressing mice (n = 12; bottom-left part) and control mice (n = 10; top-right part). (E) Effect size schematic diagram of ipsilateral rsFC of ChR2-expressing mice. (F to I) Quantitative analyses of ipsilateral mPFC-Str, mPFC-AMY, Hb-SsCx, and Ant-AMY rsFC before and after optogenetic stimulation. Two-way ANOVA with Sidak’s multiple comparison test. (J) Experimental timeline for behavioral tests. (K to M) Behavioral performances of Itpr2−/− and WT mice in TST (n = 11, 11, 11, and 12 for WT control, WT ChR2, Itpr2−/− control, and Itpr2−/− ChR2 mice, respectively), SPT (n = 11 mice in each group), and OFT (n = 11, 11, 11, and 12 for WT control, WT ChR2, Itpr2−/− control, and Itpr2−/− ChR2 mice, respectively) after optogenetic activation of mPFC-Str projection. Two-way ANOVA followed by Sidak’s multiple comparison test. All data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (N and O) Average resting-state correlation maps of ipsilateral mPFC before and after optogenetic activation of mPFC-Str projection.
Next, we analyzed the effect sizes among the 18 ROIs, and data showed that rsFC in the mPFC-Str, mPFC-AMY, Hb-SsCx, Ant-AMY, AMY-SC, Hb-AMY, TH-AMY, and DRN-AMY pathways was increased in ChR2-expressing mice after optogenetic stimulation (Fig. 6, D and E). No obvious changes were found in other pathways, including TH-Str and Hb-Str (Fig. 6, D and E). The ChR2-expressing group displayed significantly larger increases in rsFC in the mPFC-Str, mPFC-AMY, Hb-SsCx, and Ant-AMY pathways relative to the mCherry-control group (Fig. 6, F to I; mPFC-Str: main effect of time: F1,20 = 23.29, P = 0.0001; time × group interaction: F1,20 = 7.809, P = 0.0112; mPFC-AMY: main effect of time: F1,20 = 9.161, P = 0.0067; time × group interaction: F1,20 = 7.025, P = 0.0153; Hb-SsCx: main effect of time: F1,20 = 4.529, P = 0.0459; time × group interaction: F1,20 = 5.309, P = 0.0321; Ant-AMY: main effect of time: F1,20 = 11.20, P = 0.0032; time × group interaction: F1,20 = 6.235, P = 0.0214; two-way ANOVA followed by Sidak’s multiple comparison test). The rsFC in ChR2-expressing mice, but not in mCherry-control mice, was significantly increased in the AMY-SC, Hb-AMY, TH-AMY, and DRN-AMY pathways, albeit the differences in rsFC alterations were not significant between groups (fig. S10, C to F; AMY-SC: main effect of time: F1,20 = 13.66, P = 0.0014; time × group interaction: F1,20 = 3.600, P = 0.0723; Hb-AMY: main effect of time: F1,20 = 14.24, P = 0.0012; time × group interaction: F1,20 = 3.319, P = 0.0835; TH-AMY: main effect of time: F1,20 = 9.733, P = 0.0054; time × group interaction: F1,20 = 2.964, P = 0.1006; DRN-AMY: main effect of time: F1,20 = 10.37, P = 0.0043; time × group interaction: F1,20 = 1.124, P = 0.3016; two-way ANOVA followed by Sidak’s multiple comparison test). Quantitative analysis of rsFC in the mPFC-Str and mPFC-AMY pathways showed that the increased rsFC was close to the level of WT mice (fig. S10, G and H). Compared with the results of optogenetic stimulation of mPFC neurons (Fig. 5, E and H), more rsFC was increased by the activation of mPFC-Str projection, and these increased rsFC was mainly concentrated in AMY-, Ant-, and Hb-related neural circuits (Fig. 6, D and E). These results suggest that changes in rsFC of AMY-, Ant-, and Hb-related neural circuits in Itpr2−/− mice are related to Str activity. We next performed SBA with seed defined in the ipsilateral mPFC. The CC maps also revealed increase in mPFC-Str rsFC after the selective activation of the mPFC-Str projection in ChR2-expressing mice compared with the mCherry-control group (Fig. 6, J and K). Notably, we found that 56% (86 of 153) of pathways showed rsFC changes in opposite directions when comparing the trend of rsFC changes due to Itpr2 knockout and optogenetic activation of mPFC-Str projection (fig. S11A). In these 86 pathways, optogenetic activation of mPFC-Str projection led to the same direction of rsFC changes as optogenetic activation of mPFC astrocytes in 79.09% (68 of 86) of pathways (fig. S11A). Similarly, the rsFC differences between Itpr2−/− and WT mice were negatively correlated with the rsFC differences in Itpr2−/− mice before and after optogenetic activation of mPFC-Str projection (fig. S11B; r = −0.6764, P < 0.0001). Among them, 72 of 86 pathways showed rsFC decreases with Itpr2 knockout but increases with activation of the mPFC-Str projection (the second quadrant in fig. S11B), whereas the remaining pathways showed rsFC increases with Itpr2 knockout but decreases with activation of the mPFC-Str projection (the fourth quadrant in fig. S11B).
We then investigated behavioral performances of Itpr2−/− and WT mice 1 min after selective optogenetic activation of the mPFC-Str pathway in parallel behavioral experiments (Fig. 6J). We found that relative to mCherry-control Itpr2−/− mice, the ChR2-expressing Itpr2−/− mice exhibited a significant decrease in immobility time in TST (P = 0.0012) and a significant increase in sucrose preference (P = 0.039) after optogenetic activation of mPFC-Str projection such that their behavioral performances were rescued to the levels comparable to those of WT mice (Fig. 6, K and L). No significant differences between the behavioral performances of two WT mice groups were observed in TST (P = 0.9996) and SPT (P = 0.9914; Fig. 6, K and L). Note that no significant difference was observed in general locomotion in different groups (Fig. 6M; main effect of group: F3,123 = 1.433, P = 0.2364). Together, these results suggest that the optogenetic activation of mPFC-Str projection rescued most of the abnormal rsFC, especially the mPFC-Str and mPFC-AMY rsFC, and produced an antidepressant effect in Itpr2−/− mice.
DISCUSSION
Here, we showed that Itpr2−/− mice exhibit a decreased rsFC signature in depression-related networks. MDD patients exhibited highly consistent rsFC changes with Itpr2−/− mice. We could partially rescue this signature by astrocytic activation. Moreover, enhancing mPFC-related rsFC using optogenetic stimulation, especially the mPFC-Str rsFC, was paralleled by the improvement of depressive-like behaviors in Itpr2−/− mice. These results indicate that astrocyte dysfunction drives aberrant rsfMRI connectivity in depression. Our work provides a previously unidentified and mechanistic insight into the plausible causal relationship between astrocytic mechanisms and rsfMRI-measured network aberration in depression.
Many studies have examined rsfMRI networks in both depressed patients and animal models, yielding highly quantitative and objective measurements that are of theranostic utility (2, 32, 38, 39). However, the relationship between astrocyte dysfunction and these rsfMRI measurements in depression remained unknown. Here, our results showed that aberrant rsfMRI connectivity in Itpr2−/− mice can be partially rescued by astrocyte-specific activation using optogenetic stimulation. We conclude that astrocyte dysfunction represents a contributing factor for developing rsfMRI connectivity abnormalities in depression. This conclusion aligns with work showing that astrocyte-specific genes exhibited strong spatial association to rsfMRI connectivity variability in depression (17).
In the present study, modulating rsfMRI connectivity in depression-related pathways (e.g., the mPFC-Str pathways) affect depressive-like behaviors in Itpr2−/− mice, which is supported by prior human neuroimaging studies (2, 39, 40). For example, the rsFC between vmPFC and ventral Str was reduced in depressed patients (40–43) and increased after effective treatment for depression (44). Note that although increases in mPFC-Str rsFC in MDD were also reported (11, 45), this discrepancy may emerge from small sample sizes (2) or different ROI selections (39). Our study identified a reduction of mPFC-Str rsFC in MDD patients with a large sample size (n = 1080 patients; n = 931 controls) and two different ROI parcellation schemes, corroborating our results in astrocyte dysfunction animal models. We revealed that enhancing mPFC-Str rsFC through optogenetic activation of mPFC neurons or the mPFC-Str pathway rescued depressive-like behaviors in Itpr2−/− mice but did not affect behavioral performances in WT mice. Another optogenetic study on normal rats showed reduced sucrose preference and social interaction after asynchronously enhancing mPFC excitability using step function opsins (SSFOs) (31). The contradictory findings here may be due to the different frequencies of the modulated neuronal activities in mPFC and its downstream regions (46), i.e., enhanced 70- to 80-Hz gamma activity after SSFO activation (31) versus directly activated 10-Hz neural activity in our study. Nevertheless, our observed antidepressant effect of 10-Hz mPFC stimulation is consistent with previous studies in both depressed patients and animal models (47–49). Together, our findings uncovered the astrocytic mechanism underlying mPFC-Str rsFC reduction in depression. We propose that astrocyte function may serve as a target for normalizing aberrant brain-wide functional connectivity networks to improve behavioral deficits in depression. Our integrative approach, combining human neuroimaging observations, gene knockout animal model, rsfMRI, optogenetic stimulation, and behavioral tests, allows us to uncover the mechanisms underlying human neuroimaging observations in depression and possibly other psychiatric diseases.
The brain regions with notably altered rsfMRI connectivity in MDD patients and Itpr2−/− mice can be associated with depression (2, 13, 14, 38, 42), such as mPFC, TH, Str, AMY, SsCx, VCx, SC, Hb, DRN, and Ant (Figs. 1 and 2). These regions are critically involved in positive affect and motivational processes (mPFC and Str), negative sensory and emotional experiences (AMY, SsCx, VCx, Hb, and DRN), and aversion processes (SC and TH), all of which may affect behaviors under depressive conditions (50). Our findings and previous studies showed that the mPFC/vmPFC is one of the most consistently impaired regions in depression (2, 39). In addition to the reduced mPFC-Str rsFC discussed above, decreased mPFC-AMY rsFC has also been consistently reported in MDD patients in previous studies (39, 51), supporting our findings. We found that the rsFC between vmPFC and TH was decreased in MDD patients as in a previous study (10), but not altered in Itpr2−/− mice. One possibility is that the rsFC of this circuit may be altered in only a subset of depression subgroups (10). Moreover, we found that the rsFC of TH-AMY and TH-Str pathways was decreased in both MDD patients and Itpr2−/− mice, in line with previous findings that depression is related to the hypoconnectivity of TH, AMY, and Str (10, 39). It is noteworthy that we found the altered rsFC in somatosensory and visual cortices in both MDD patients and Itpr2−/− mice, which may be linked to exteroceptive alterations in depression, such as increased pain tolerance for exteroceptive stimulation (52) and reduced visual contrast sensitivity (53). Our results of Itpr2−/− mice and previous studies suggested that the dysfunction of many small subcortical regions, such as Hb and DRN, plays an important role in depression (14, 54, 55). Because of the limited spatial resolution, it is difficult to examine human rsfMRI data to study the roles of these regions in depression, highlighting the importance of animal studies (2).
In this study, we used optogenetic stimulation to selectively manipulate the activity of the mPFC, rather than other brain regions in depression-related networks, because previous studies and our results (Figs. 1 and 2) suggested that the mPFC exhibits the most consistent and notable rsFC alterations due to astrocyte dysfunction in depression. On the one hand, previous postmortem analyses of MDD patients and animal studies have consistently found astrocytic abnormalities, such as morphological and numerical changes, in the PFC (13, 16, 18). Meanwhile, the PFC is one of the regions that exhibited highly consistent notable rsFC alterations in depression (2, 39) and also an effective target region for treating depression through neuromodulation therapy (12). On the other hand, our rsfMRI results of Itpr2−/− mice showed that the mPFC has the largest rsFC changes (IL rank 1 and PrL rank 3), which correlated to depressive-like behavior (Fig. 1C and fig. S1D). Combining the rsfMRI results of Itpr2−/− mice and MDD patients, we revealed that mPFC-related rsFC pathways are the most consistently altered pathways. Our results showed that optogenetic activation of mPFC activity can rescue the rsFC in mPFC-related pathways and alleviate depressive-like behaviors in Itpr2−/− mice. However, we do not preclude that optogenetic manipulation of other regions could generate rescuing effects due to the potential similar contribution of astrocytes in regulating network homeostasis in depression-related networks. The human PFC consists of multiple subdivisions and is much more complex than the rodent mPFC. So far, only the vmPFC subdivision of the human PFC has been shown to have the homologous structure in the rodent mPFC (33, 34). Guided by the well-controlled rodent mPFC rsFC findings, our study focused on analyzing rsFC in vmPFC-related pathways in MDD patients. However, we do not preclude the possibility that rsFC of other subdivisions of the human PFC (i.e., beyond vmPFC) may also be altered. Previous studies have shown that some other subdivisions of the human PFC (e.g., the dlPFC: Brodmann cytoarchitectonic areas 9, 46) also show astrocytic abnormalities and rsFC changes in MDD patients (2, 16). While those subdivisions do not have homologous structures within the mouse brain (33, 34), our study provides a valuable approach for the future optogenetic rsfMRI studies in other animal models that have these subdivisions, such as nonhuman primates.
Evidence from postmortem analyses of MDD patients showed that astrocytic abnormalities were mainly located in the PFC (vmPFC, dlPFC, and ACC), TH, AMY, and Str (15, 16, 18). Studies of astrocytic function in rodents support this notion, showing that astrocyte dysfunctions in mPFC (13, 55), AMY (56, 57), Hb (14, 54), Str (58), and DRN (55) drive depressive-like behaviors in animals. The high consistency between brain regions reported by our rsFC study and those with astrocytic abnormalities reported in postmortem analyses of depressed patients (15, 18) indicates that depressive symptomology and decreased rsfMRI connectivity in MDD patients are at least partially driven by astrocyte dysfunction. This evidence advances human neuroimaging observations toward identifying its underlying mechanisms. However, our finding likely shares a limitation common to all neuroimaging studies, in that well-characterized neuroimaging signature underlying depression could not be paired with postmortem analysis of the same patients (18). Longitudinal investigations on MDD patients integrating repeated neuroimaging observations with postmortem analysis of the same subjects in the end would be required in future studies.
We also showed that optogenetic manipulations of neuronal communication affect rsfMRI connectivity with accompanying changes in behaviors in Itpr2−/− mice (Figs. 5 and 6). These observations suggest that the deletion of IP3R2 in astrocytes leads to disruption of the functional network and its depressive-like behavioral output, probably via alterations in neural connectivity (59). Prior research supports this notion (60), showing that attenuated IP3-mediated astrocytic signaling reduces astrocytic coverage of synapses (61). IP3R2 knockout mice failed to modulate synaptic transmission and plasticity (26, 27, 62–64). Ultimately, these knockout mice failed to bridge the disconnected neuronal circuits within its territory (60). These observations support a view in which astrocyte dysfunction induced by IP3R2 knockout results in maladaptive astrocyte-neuron interactions, which may contribute to etiologically relevant abruption of rsfMRI connectivity and depression-related behaviors in Itpr2−/− mice. Previous study revealed that neuronal Ca2+ fluctuations correlate with infraslow BOLD fluctuations (65). However, we know of no evidence to directly examine the relationship between astrocytic activities and rsfMRI connectivity as in the present study. Note that our results do not exclude the possibility that the deletion of IP3R2 in astrocytes may lead to maladaptive metabolic activity (66) or altered vascular activity (67), both of which may affect rsfMRI BOLD fluctuations. Although the Itpr2−/− mice is a whole-brain IP3R2 knockout model, the altered rsFC in Itpr2−/− mice is mainly located in the brain regions associated with depression. It might involve regional heterogeneity of astrocyte functions, neuronal subtypes, and astrocyte-neuron interactions (68). Nevertheless, our results present the first evidence that astrocytic Ca2+ contributes to rsfMRI connectivity in both normal and depressed brains.
In summary, our results reveal the astrocytic mechanisms underlying functional MRI connectivity aberrations in depression, unifying two distinct and important concepts in the pathophysiology of depression, namely, microscopic astrocyte dysfunction and macroscopic functional network abruptions, using cell type–specific neuromodulation approaches alongside whole-brain imaging. These results will help advance a more specific and mechanistic interpretation of the rsfMRI-measured functional networks as a highly quantitative and reproducible imaging biomarker and theranostic tool for depression.
MATERIALS AND METHODS
Mice
All experiments were approved by the Southern Medical University Animal Ethics Committee and conformed to the Regulations for the Administration of Affairs Concerning Experimental Animals (China). The mice were bred in-house and grew in a temperature- and humidity-controlled room on a 12-hour light/dark cycle (lights on from 7:00 to 19:00) with ad libitum access to food and water. All mice were handled for 5 min/day for 3 days before behavioral tests. All behavioral tests were performed by observers blinded to group allocations between 1:00 p.m. and 5:00 p.m.
IP3R2 knockout mice were generated by crossing germline-heterozygous-null mutant Itpr2+/− mice as a gift from J. Chen (69). The mouse line was maintained on a C57BL/6J background. The offspring were genotyped by polymerase chain reaction (PCR) using mouse tail DNA and the allele-specific primers of the WT (5′-GCTGTGCCCAAAATCCTAGCACTG-3′; 3′-CATGCAGAGGTCGTGTCAGTCATT-5′) and mutant (5′-AGTGATACAGGGCAAGTTCATAC-3′; 3′-AATGGGCTGACCGCTTCCTCGT-5′).
Viruses and chemical reagent
The viruses AAV2/9-mCaMKIIα-hChR2(H134R)-mCherry-WPRE-pA (4.06 × 1012 particles ml−1), AAV2/9-mCaMKIIα-mCherry-WPRE-pA (1.60 × 1013 particles ml−1), and AAV2/5-gfaABC1D-hChR2(H134R)-mCherry-WPRE-pA (3.96 × 1012 particles ml−1) were purchased from Taitool Bioscience (China). Manganese chloride tetrahydrate (M109464) was purchased from Aladdin (China).
Stereotactic injection and optic fiber implantation
The mice (male, ~8 weeks) were anesthetized with 1.5% isoflurane and fixed in a stereotactic frame (RWD Life Science, China) before surgery. The viruses were stereotactically injected into the mPFC (AP: +1.70, ML: +0.35, DV: −2.70 mm relative to bregma; AP, ML, and DV denote the anteroposterior, mediolateral, and dorsoventral distance from the bregma, respectively). A volume of 300 nl of virus was injected into each location at a rate of 100 nl/min using a 5-μl microsyringe (Hamilton, USA) fitted with a 33-gauge needle and a microsyringe pump (Stoelting, USA). The needle was left in place for 5 min to allow diffusion of the viruses and then slowly withdrawn. Then, the scalp was concreted by tissue glue (No.03-396, DentKist, Korea).
For optogenetic fMRI scanning, custom-made right-angle banded fibers (diameter = 220 μm, numerical aperture = 0.5) were implanted above the infected cells of mPFC (AP: +1.70; ML: +0.35; DV: −2.60) or axon terminals of mPFC neurons in Str (AP: +0.16; ML: +1.70; DV: −3.50) 3 or 8 weeks after virus injection, respectively. The banded fiber was wrapped in a black opaque heat-shrinkable sleeve to prevent light leakage during optogenetic stimulation to elicit undesired visual stimulation. To minimize damage to brain tissue, the fiber tip was clipped to create an inclined plane to facilitate insertion. Before optogenetic experiments, the laser power of each fiber was measured by a power meter (PM100D and S142C, Thorlabs, USA).
Data acquisition of resting-state fMRI and optogenetic fMRI experiments
The mice were anesthetized with 3.5% isoflurane for induction. A bolus of dexmedetomidine (0.02 mg/kg) was intraperitoneally injected. One drop of 2.5% lidocaine was then applied to the chords to provide local anesthesia. The animals were endotracheally intubated and positioned on an MRI-compatible cradle. Isoflurane was then reduced to 2% and remained at this level until their heads were fixed by ear bars and their teeth were secured with a bite bar. A bolus of pancuronium bromide (0.2 mg/kg; TargetMol, USA), a neuromuscular blocking agent, was intraperitoneally injected to reduce the motion of mice. Note that the head motion of animals was smaller than previous study under light anesthesia (70): Session-averaged FD before and after regressing out the head motion parameters was 1.24 ± 0.02 and 0.07 ± 0.002 μm, respectively, equivalent to 0.5 and 0.03% of the voxel size (fig. S3A). No significant difference in mean FD was found between Itpr2−/− and WT mice before (P = 0.16) and after regression (P = 0.76; fig. S3, B and C). Ophthalmic ointment was applied to protect the eyes. Throughout the course of the experiment, the animals were mechanically ventilated using a small animal ventilator (TOPO, Kent Scientific, USA) at a rate of 80 breaths per minute, with a respiration cycle of 25% inhalation and 75% exhalation. Note that compared to free-breathing, ventilation did not lead to higher mean heart rates in mice, suggesting no increase in stress level due to our ventilation preparation (fig. S2D; P = 0.98). After the setup, isoflurane was reduced and maintained at 0.4%, and continuous subcutaneous infusion of dexmedetomidine (0.04 mg/kg per hour) was started to maintain the sedation level. Note that our study was designed to combine optogenetic fMRI under light sedation and behavioral assessment in awake condition, and our anesthetic regimen (0.04 mg kg−1 hour−1 dexmedetomidine + 0.4% isoflurane) has been shown to remain stable and robust evoked BOLD response and rsFC (71), approaching the awake state (72). Animal rectal temperature was maintained at 37 ± 0.1°C with an MR-compatible heater system (Heater system, Small Animal Instruments, USA). Continuous physiological monitoring (rectal temperature, breathing rate, heart rate, and oxygen saturation) was performed throughout the duration of the experiments using an MR-compatible system (Small Animal Instruments, USA). The above setup of mice took approximately 15 min. The following localization and anatomical MRI scans took approximately 15 min. During this period, the physiological states of mice gradually stabilized to normal ranges (rectal temperature: 37 ± 0.1°C, breathing: 80 breaths per minute, heart rate: 350 to 420 beats per minute, oxygen saturation: >95%) before the start of the functional MRI acquisition.
All functional MRI experiments were performed on a Bruker 7T MRI scanner (Bruker BioSpin, Germany) using a mouse head cryocoil (MRI CryoProbe, Bruker, Germany). The anatomical images were acquired using a spin echo (Turbo-RARE) sequence [field of view (FOV) = 16 × 16 mm2, matrix = 256 × 256, RARE factor = 8, repetition time (TR)/echo time (TE) = 2500/35 ms, slice thickness = 0.4 mm]. Local field homogeneity was optimized in the ROI using field maps scanned in advance. Functional data were then obtained using a single-shot gradient echo planar imaging (GE-EPI) sequence with FOV = 16 × 16 mm2, matrix = 64 × 64, flip angle = 54.7°, slice thickness = 0.4 mm, dummy scans = 10, TE = 15 ms, TR = 750 ms (rsfMRI) or 1000 ms (optogenetic fMRI).
For optogenetic experiments, an Arduino programming board was used to synchronize the scanner trigger and the optogenetic stimulation laser. Blue light was delivered using a 473-nm laser transmitter via an optical patch cable (5 to 10 m). The optogenetic stimulation (10 Hz; light power: 3.5 mW at the fiber tip; 15% duty cycle) was delivered with a duration of 20 s.
rsfMRI and optogenetic fMRI data preprocessing
For each animal, the anatomical and fMRI images were skull-stripped using an in-house automatic brain-extracted algorithm, and the effect was checked in ITK-SNAP (www.itksnap.org/). Preprocessing was performed using SPM12 (Wellcome Department of Imaging Neuroscience, University College, London, UK). Functional images were slice timing–corrected, realigned with six rigid body parameters, registered, and smoothed (0.2-mm isotropic Gaussian kernel). During the registration, the functional images were first registered to the anatomical images of each mouse and then to a mouse brain template (www.imaging.org.au/AMBMC/Model) using transformation matrix obtained by registering the anatomical images and the template. Therefore, the fMRI images were resliced into a common group space with the same resolution and physical voxel size (final resolution: 156 × 100 × 47 voxels at 0.1 × 0.1 × 0.5 mm3). A total of 248 sessions were scanned. Among them, 15 sessions (6.05%) were excluded because of motion artifacts (>0.5 voxel shifts detected by realignment), 4 sessions (1.61%) were excluded because of sudden changes in respiration pattern, and 4 sessions (1.61%) were excluded because of sudden changes in oxygen saturation level.
rsfMRI data analysis
The 12 head motion parameters (roll, pitch, yaw, translation in three dimensions, and the first derivative of each of those six parameters) (73) were regressed out from the time series of each voxel. This was followed by a temporal band-pass filtering (0.001 to 0.1 Hz). The preprocessed rsfMRI images were decomposed using spatial ICA with GIFT toolbox (Group ICA/IVA of fMRI Toolbox, v3.0a, www.nitrc.org/projects/gift/). The estimated number of components for all rsfMRI data was found to be 67 using the minimum description length criterion as implemented in the GIFT. After discarding 14 components related to cerebrospinal fluid, motion-evoked, or vascular-evoked pseudo-activations, the remaining 53 ICA maps were then visually inspected and labeled on the basis of the spatial patterns in reference to known anatomical and functional locations (74), resulting in 94 labels of spatially separated and left-right symmetrical brain units corresponding to 47 brain regions (fig. S2 and table S1).
Pearson’s CCs between each pair of brain units were calculated to generate pairwise correlation matrices for each mouse. CCs for left and right ROIs were averaged to form 47 × 47 correlation matrices. Correlation matrices were transformed using Fisher’s z transformation for ease of comparison. Group comparisons of correlation matrices were performed using two-tailed independent sample t tests with FDR multiple comparisons correction. Changed rsFC was visualized in BrainNet Viewer (www.nitrc.org/projects/bnv/). The effect size was calculated as the standardized difference between the group means (Cohen’s D). The overall connectivity change in each brain region was quantified by NMI, which was calculated as the average effect size.
For SBA, an atlas-defined 4 × 4 region in the right mPFC was chosen as seed. The individual CC map was generated by calculating the Pearson’s CCs between the time series of the seed and the BOLD signal of every other voxel using MATLAB 2017b (MathWorks, USA). The CC maps of all repeated sessions of each mouse were averaged. Each group averaged CC maps were generated by averaging CC maps across all mice in the group.
Optogenetic fMRI data analysis
For each session after optogenetic stimulation, the six head motion parameters and their first derivative were regressed out from the time course of each voxel. Then, a standard general linear model was applied to calculate the response maps, and the Student’s t test was performed to identify activated voxels using a threshold of P < 0.01 (FDR-corrected) in SPM12. For ChR2-expressing mice, only those sessions with obvious BOLD activations at the fiber tip were used in the subsequent SBA. The t maps of all mice in each group were averaged to detect the BOLD activations at the group level. For the fMRI data with optogenetic stimulation of mPFC astrocytes, the second-level one-sample t test analysis was performed to generate the group activation maps with FDR-corrected P < 0.05 and cluster size > 30 voxels.
SBA was performed to generate CC maps of right mPFC in ChR2-expressing and mCherry-control mice. The CC maps of sessions after stimulation were calculated using only the data acquired 40 s after the stimulus offset, when the evoked BOLD signals have already returned to baseline. The rsFC within depression-related networks was generated using the ROIs in ipsilateral cortex from ICA maps in fig. S2. As fiber implantation causes slight artifacts, the ROIs of the mPFC were replaced by 6 × 6 voxels at the tip of the fiber in experiments with optogenetically activated mPFC neurons or astrocytes. The CC values were then transformed using Fisher’s z transformation for group comparison.
RsfMRI data analysis of MDD patients
A resting-state fMRI dataset consisting of 1276 patients with MDD and 1104 normal controls was obtained from the REST-meta-MDD consortium (32). Dataset was preprocessed using DPARSF software with a standardized pipeline at local sites (32). Briefly, after discarding the initial 10 volumes, all fMRI images were slice timing–corrected, realigned with six rigid body parameters, and temporal-filtered (0.01 to 0.1 Hz). Furthermore, the Friston-24 head motion parameters, white matter and cerebrospinal fluid signals, and linear trend were regressed out from the images to minimize head motion confounds, physiological noises, and BOLD signal drifts. Last, the images were registered to the Montreal Neurological Institute (MNI) space.
This dataset consisted of 1276 patients with MDD and 1104 normal controls from 25 study sites in China. All data were deidentified and anonymized, and related studies were approved by local Institutional Review Boards. All subjects signed the informed consent at each local institution. The criteria used to control data quality before group statistical analyses included the following (please also see table S2): (i) The data of site 3, site 18, and site 24 were excluded because of low image quality after preprocessing (by visual inspection), resulting in 1196 MDD and 1016 NC; (ii) 97 MDD and 75 NC were excluded because of their age less than 18 or more than 65, resulting in 1099 MDD and 941 NC; (iii) 10 MDD and 6 NC were excluded because their PFC or parietal cortex was not imaged during scanning (by visual inspection), resulting in 1089 MDD and 935 NC; (iv) 8 MDD and 4 NC were excluded because their images were not registered to the MNI space (by visual inspection), resulting in 1081 MDD and 931 NC; and (v) 1 MDD was excluded because of the images showing structural abnormalities in the occipital lobe. Last, a total of 1080 MDD and 931 NC recruited from 21 study sites across China met the above criteria. To control the effect of head motion on our findings, we reran the analysis after excluding subjects with mean FD more than 0.2 or 0.1 mm: i.e., excluding the subjects with mean FD > 0.2 mm, resulting in 1059 MDD and 917 NC; excluding the subjects with mean FD > 0.1 mm, resulting in 879 MDD and 733 NC.
The dataset provided us with time series extracted from human atlas–defined ROIs instead of the raw images, and these ROIs did not include DRN, SC, and Hb regions. To assess whether the rsFC alterations within depression-related networks found in Itpr2−/− mice (i.e., the top 18 ROIs, corresponding to subregions within mPFC, Str, AMY, TH, SsCx, VCx, Ant, SC, DRN, and Hb; Fig. 1, C to E) also existed in MDD patients, the mean time courses of these brain regions (some regions consisted of multiple human atlas–defined ROIs) were used to calculate the rsFC of the corresponding pathways except the DRN-, SC-, and Hb-related pathways. The mean time course of mPFC, Str, TH, SsCx, VCx, and Ant was generated using the Dosenbach functional atlas (75). The mean time series of AMY was generated using the Automated Anatomical Labeling atlas (76). See table S3 for the coordinates in MNI space of these human atlas–defined ROIs. The rsFC was quantified by the average Pearson’s CCs between the time series of two brain regions and then transformed to z score.
We used LMM to assess the association of depression (here, diagnosis) with rsFC. Demographic information (i.e., age, sex, education, and head motion) was added into LMM as covariates to control their confounding effects. The head motion here referred to mean FD derived from Jenkinson’s relative root mean square algorithm (32). The LMM analysis was performed using the MATLAB function “fitlme” (www.mathworks.com/help/stats/fitlme.html), with the formula as follows (32):
y ~ 1 + Diagnosis + Age + Sex + Education + Motion + (1|Site) + (Diagnosis|Site), which yields t and P values for the fixed effect of Diagnosis. It is noteworthy that the LMM contained random effects specific to site and fixed effects independent of site, which helps with controlling the potential systematic site-related confounding factors (32). FDR correction was applied for multiple comparisons. The effect size was calculated as the Cohen’s D.
In validation analysis, the mean time series of each brain region was generated using the Craddock’s functional clustering atlas (35). See table S4 for the coordinates in MNI space of these ROIs.
MnCl2 administration, MEMRI data acquisition, and analysis
For MnCl2 administration, 5 nl of MnCl2 solution (600 mM) was injected into mPFC of WT and Itpr2−/− mice, and the scalp was healed with tissue glue. All mice were scanned on a 7T Bruker scanner, with a 72-mm-diameter volume coil for radiofrequency pulse transmission and a 10-mm loop coil for signal detection. It took approximately 30 min to prepare for the scan, so the dynamic MEMRI data of each mouse were acquired at 0.5, 4.5, 8.5, and 24.5 hours from MnCl2 infusion. For consistent coil positioning among multiple scans, we marked the precise position of the coil on the scalp at the first scan of each mouse, which was shaved during the MnCl2 administration. During imaging, the mouse head was secured in a plastic cradle with a bite bar and ear bars. Mice breathed freely with a nosecone. Anesthesia was maintained at 1% isoflurane, and body temperature was maintained at 36.5 ± 0.5°C via a warm-water circuit integrated into the plastic cradle. We used a T1-weighted three-dimensional fast low angle shot (FLASH) sequence with flip angle = 35°, 9 averages, TR/TE = 35/4 ms, FOV = 16 mm × 16 mm × 10 mm, matrix size = 160 × 160 × 50, yielding 100 μm × 100 μm × 200 μm voxels within 47 min.
The MEMRI data from four mice were discarded because of abnormal brain morphology (WT: n = 1) and deviated injection position from mPFC (WT: n = 1; Itpr2−/−: n = 2). The MR images were skull-stripped manually within ITK-SNAP and registered to a mouse brain template. At the individual level, image intensities in each scan were normalized using those in surrounding muscles, which should not be affected by Mn2+ transport. We used the maximum density projection in ImageJ (http://rsb.info.nih.gov/ij/) to trace the spread of Mn2+ at 8.5 hours after infusion. Six ROI masks (vStr, dStr, LGP, TH, AMY, and VTA), each of which spans three contiguous slices, were drawn on the basis of the spread of Mn2+ in the template space using ITK-SNAP. The ROI masks were then applied to the normalized images of each scan to extract the mean MEMRI signal intensities.
Immunofluorescence
Animals that had undergone behavioral analysis were deeply anesthetized and perfused transcardially with saline followed by 4% paraformaldehyde (PFA). Brains were removed, postfixed overnight at 4% PFA at 4°C, and transferred to 30% sucrose in phosphate-buffered saline (PBS; pH 7.4). Three days later, brains were cut into 40-μm-thick sections using freezing microtome (Leica CM1950, Germany). Sections were washed with 0.1 M PBS three times and then incubated in blocking buffer containing 5% normal goat serum in 0.5% Triton X-100/PBS for 1.5 hours at room temperature. After blocking, sections were incubated with primary antibodies in PBS overnight at 4°C. The primary antibodies used were GFAP (an astrocytic marker, 1:300; Cell Signaling Technology, 3670S) and NeuN (a neuronal marker, 1:500; Millipore, MABN140). After three washes with PBS, sections were incubated with either Alexa Fluor 488 goat anti-mouse (1:500; Invitrogen, R37120) or Alexa Fluor 488 donkey anti-rabbit (1:500; Invitrogen, A21206) secondary antibodies at room temperature for 1 hour. Sections were washed for another three times. 4′,6-Diamidino-2-phenylindole (DAPI) was used for fluorescent nuclear counterstain. The coverslips were mounted onto glass slides and visualized using a Nikon C2 confocal microscope (Japan).
Behavioral assays
For behavioral tests, mice were implanted unilaterally with optical fibers (200 μm diameter, numerical aperture = 0.37; Inper, China) over the infected cells of mPFC or vStr. The optical fiber was connected to a laser source (Inper B1465, China) using an optical fiber sleeve. During the optogenetic stimulation, 20-s blue light (473 nm, 10 Hz, 15% duty cycle) was delivered as 3.5 mW at the fiber tip. Behavioral tests were performed after habituation. The fiber positions of all mice were checked with confocal images, ensuring that data used for further analysis were from mice with the correct fiber positioning.
Tail-suspension test
After optogenetic stimulation, the mice were transferred to home cages for a minute of break. Then, the mice were taped 1 cm from the tip of their tails and then suspended on a horizontal bar, with their heads about 15 cm above the surface during suspension. The immobility within a 5-min period was recorded using Mouse Tail Suspension (MED-TSS-MS, USA).
Forced swimming test
The FST was performed in a 45 cm × 19 cm clear glass cylinder filled with water to a height of 23 cm (23° to 25°C). The mice underwent 4 min of preswimming 48 hours before the test. The immobility within a 4-min period was recorded by a video tracking system (EthoVision XT 11.5, Noldus, The Netherlands). In the optogenetic test, the mice were transferred to home cages for a minute of break after optogenetic stimulation, and then the immobility time within a 4-min period was recorded.
Sucrose preference test
Mice were habituated to 60-ml tubes with stoppers (a two-bottle choice) filled with drinking water for 2 days. After habituation, the mice underwent optogenetic stimulation and were then given access to one bottle of water and one bottle of 1% sucrose solution for 24 hours. Bottles containing water and sucrose were weighed at 12:00 and 18:00 on the first day and weighed at 12:00 on the second day. After each fluid weight measurement, bottle positions were switched to ensure that the mice did not develop a position preference. The sucrose preference for each mouse was calculated as follows: sucrose preference (%) = 100 × (total consumption of sucrose/total consumption of both water and sucrose). In the behavioral test of optogenetic activation of mPFC-Str projection, data of one ChR2-expressing Itpr2−/− mouse were excluded because this mouse has a place preference.
Open-field test
The open-field apparatus consisted of a rectangular chamber (40 cm × 40 cm × 30 cm), which was made of gray polyvinyl chloride. The mice were gently placed in the center and permitted to explore freely for 5 min. In the optogenetic experiments, the free exploration time was 9 min, with 3 min each for stimulation off, stimulation on, and stimulation off conditions. The digitized images of the path taken by each mouse were stored, and the locomotion activity was analyzed using EthoVision 7.0 software.
Ex vivo electrophysiology
Mice were anesthetized with isoflurane and decapitated, and brains were rapidly removed. The brains were quickly placed into ice-cold modified artificial cerebrospinal fluid (ACSF) containing 250 mM sucrose, 26 mM NaHCO3, 10 mM glucose, 10 mM MgSO4, 2 mM KCl, 1.3 mM NaH2PO4, and 0.2 mM CaCl2. Slices containing the mPFC (300 μm) were prepared in ice-cold modified ACSF using a VT-1200S vibratome (Leica, Germany), transferred to the storage chamber containing regular ACSF containing 126 mM NaCl, 26 mM NaHCO3, 10 mM glucose, 3 mM KCl, 2 mM CaCl2, 1.25 mM NaH2PO4, and 1 mM MgSO4, and allowed to recover at 34°C for 30 min and then at room temperature (25 ± 1°C) for 1 hour before recording. During the slice preparation, all solutions were saturated with 95% O2/5% CO2 (v/v).
The brain slices were placed in the recording chamber, with continuous perfusion of ACSF at a flow rate of 2 ml/min. Whole-cell patch-clamp recording from mCherry+ cells was visualized under a Nikon Eclipse FN1 microscope (Nikon) equipped with a 40× water-immersion lens and illuminated with a mercury lamp. For optogenetic experiments, ChR2-expressing soma was activated with 15-ms pulses of 473-nm blue light generated by a Smart Light Source (Inper, China) under the control of the digital output of the amplifier. Pipette resistance ranged from 3 to 5 megohms. The intracellular solution contained 140 mM K-gluconate, 9 mM Hepes, 4.4 mM phosphocreatine disodium, 4 mM adenosine triphosphate–Mg, 4.5 mM MgCl2, 0.3 mM guanosine triphosphate, and 5 mM EGTA, at 290 to 300 mOsm (pH 7.2 to 7.3), adjusted with KOH. Data were acquired with an EPC 10 amplifier (HEKA Elektronik, Germany), filtered at 2.9 kHz with a Bessel filter, digitized at 10 kHz, and analyzed using pClamp10.2 software (Molecular Devices, USA).
Statistical analysis
All statistical analyses were carried out using SPSS 22.0. Data were confirmed to be normally distributed using the Shapiro-Wilk test except where noted. Normally distributed data were tested by one-way ANOVA, two-way ANOVA, and two-way repeated-measures ANOVA, followed by Sidak’s multiple comparisons. We used the independent sample t test to compare the rsFC between Itpr2−/− and WT mice. We used the LMM to compare the rsFC between MDD patients and normal controls, with demographic information (i.e., age, sex, education, and head motion) added into LMM as covariates to control their confounding effects. Statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We would like to thank the members of the REST-meta-MDD Consortium for providing the human fMRI data.
Funding: This work was supported by Ministry of Science and Technology China Brain Initiative Grant 2022ZD0204702 (X.C.), National Natural Science Foundation of China 81871349 (Y.F.), Guangzhou Key Research Program on Brain Science 202206060001 (X.C.), Guangzhou Science and Technology Project 201904020039 (X.C.), Key-Area Research and Development Program of Guangdong Province 2018B030334001 and 2018B030333001 (X.C.), National Program for Support of Top-notch Young Professionals (X.C.), Hong Kong Research Grant Council C7048-16G (E.X.W.), Hong Kong Research Grant Council GRF17112120 and GRF17127121 (E.X.W.), Guangdong Key Technologies Project 2018B030336001 (E.X.W.), and National Natural Science Foundation of China U21A6005 (Y.F.).
Author contributions: Conceptualization: X.C., Y.F., E.X.W., J.L., J.-W.M., and X.W. Animal fMRI experiments: J.L. and Z.A. Animal and human fMRI data analysis: J.L. and X.W. Surgery, behavioral experiments, and analysis: J.-W.M. Animal fMRI setup: S.Z. In vitro whole-cell recordings: C.-Y.Z. MEMRI scan and analysis: J.L., Y.X., and P.Y. PCR and immunofluorescence experiments: J.R., L.-Y.C., and R.M. Suggestions: T.-M.G., W.C., Q.F., X.W., and A.T.L.L. Visualization: J.L., J.-W.M., and X.W. Supervision: X.C., Y.F., and E.X.W. Writing—original draft: J.L., J.-W.M., and X.W. Writing—review and editing: X.C., Y.F., E.X.W., J.L., J.-W.M., and X.W.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All data needed to evaluate the conclusions in the paper are available on dataverse (https://doi.org/10.7910/DVN/YBKBZF).
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S3 and S4
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.Kessler R. C., Berglund P., Demler O., Jin R., Koretz D., Merikangas K. R., Rush A. J., Walters E. E., Wang P. S.; National Comorbidity Survey Replication , The epidemiology of major depressive disorder: Results from the National Comorbidity Survey Replication (NCS-R). JAMA 289, 3095–3105 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Spellman T., Liston C., Toward circuit mechanisms of pathophysiology in depression. Am. J. Psychiatry 177, 381–390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castren E., Is mood chemistry? Nat. Rev. Neurosci. 6, 241–246 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Fox M. D., Raichle M. E., Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 8, 700–711 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Biswal B., Yetkin F. Z., Haughton V. M., Hyde J. S., Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Leong A. T., Chan R. W., Gao P. P., Chan Y. S., Tsia K. K., Yung W. H., Wu E. X., Long-range projections coordinate distributed brain-wide neural activity with a specific spatiotemporal profile. Proc. Natl. Acad. Sci. U.S.A. 113, E8306–E8315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leong A. T. L., Gu Y., Chan Y. S., Zheng H., Dong C. M., Chan R. W., Wang X., Liu Y., Tan L. H., Wu E. X., Optogenetic fMRI interrogation of brain-wide central vestibular pathways. Proc. Natl. Acad. Sci. U.S.A. 116, 10122–10129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan R. W., Leong A. T. L., Ho L. C., Gao P. P., Wong E. C., Dong C. M., Wang X., He J., Chan Y. S., Lim L. W., Wu E. X., Low-frequency hippocampal-cortical activity drives brain-wide resting-state functional MRI connectivity. Proc. Natl. Acad. Sci. U.S.A. 114, E6972–E6981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen K. R., Westlund M. K., Klimes-Dougan B., Mueller B. A., Houri A., Eberly L. E., Lim K. O., Abnormal amygdala resting-state functional connectivity in adolescent depression. JAMA Psychiatry 71, 1138–1147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lui S., Wu Q., Qiu L., Yang X., Kuang W., Chan R. C. K., Huang X., Kemp G. J., Mechelli A., Gong Q., Resting-state functional connectivity in treatment-resistant depression. Am. J. Psychiatry 168, 642–648 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Drysdale A. T., Grosenick L., Downar J., Dunlop K., Mansouri F., Meng Y., Fetcho R. N., Zebley B., Oathes D. J., Etkin A., Schatzberg A. F., Sudheimer K., Keller J., Mayberg H. S., Gunning F. M., Alexopoulos G. S., Fox M. D., Pascual-Leone A., Voss H. U., Casey B. J., Dubin M. J., Liston C., Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat. Med. 23, 28–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole E. J., Stimpson K. H., Bentzley B. S., Gulser M., Cherian K., Tischler C., Nejad R., Pankow H., Choi E., Aaron H., Espil F. M., Pannu J., Xiao X., Duvio D., Solvason H. B., Hawkins J., Guerra A., Jo B., Raj K. S., Phillips A. L., Barmak F., Bishop J. H., Coetzee J. P., DeBattista C., Keller J., Schatzberg A. F., Sudheimer K. D., Williams N. R., Stanford accelerated intelligent neuromodulation therapy for treatment-resistant depression. Am. J. Psychiatry 177, 716–726 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Cao X., Li L. P., Wang Q., Wu Q., Hu H. H., Zhang M., Fang Y. Y., Zhang J., Li S. J., Xiong W. C., Yan H. C., Gao Y. B., Liu J. H., Li X. W., Sun L. R., Zeng Y. N., Zhu X. H., Gao T. M., Astrocyte-derived ATP modulates depressive-like behaviors. Nat. Med. 19, 773–777 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Cui Y., Yang Y., Ni Z., Dong Y., Cai G., Foncelle A., Ma S., Sang K., Tang S., Li Y., Shen Y., Berry H., Wu S., Hu H., Astroglial Kir4.1 in the lateral habenula drives neuronal bursts in depression. Nature 554, 323–327 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Torres-Platas S. G., Nagy C., Wakid M., Turecki G., Mechawar N., Glial fibrillary acidic protein is differentially expressed across cortical and subcortical regions in healthy brains and downregulated in the thalamus and caudate nucleus of depressed suicides. Mol. Psychiatry 21, 509–515 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Zhou X., Xiao Q., Xie L., Yang F., Wang L., Tu J., Astrocyte, a promising target for mood disorder interventions. Front. Mol. Neurosci. 12, 136 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson K. M., Collins M. A., Kong R., Fang K., Li J., He T., Chekroud A. M., Yeo B. T. T., Holmes A. J., Convergent molecular, cellular, and cortical neuroimaging signatures of major depressive disorder. Proc. Natl. Acad. Sci. U.S.A. 117, 25138–25149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Czeh B., Nagy S. A., Clinical findings documenting cellular and molecular abnormalities of glia in depressive disorders. Front. Mol. Neurosci. 11, 56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen N. J., Lyons D. A., Glia as architects of central nervous system formation and function. Science 362, 181–185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lezmy J., Arancibia-Cárcamo I. L., Quintela-López T., Sherman D. L., Brophy P. J., Attwell D., Astrocyte Ca2+-evoked ATP release regulates myelinated axon excitability and conduction speed. Science 374, eabh2858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fellin T., Pascual O., Gobbo S., Pozzan T., Haydon P. G., Carmignoto G., Neuronal synchrony mediated by astrocytic glutamate through activation of extrasynaptic NMDA receptors. Neuron 43, 729–743 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Kol A., Adamsky A., Groysman M., Kreisel T., London M., Goshen I., Astrocytes contribute to remote memory formation by modulating hippocampal-cortical communication during learning. Nat. Neurosci. 23, 1229–1239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M., He Y., Sejnowski T. J., Yu X., Brain-state dependent astrocytic Ca2+ signals are coupled to both positive and negative BOLD-fMRI signals. Proc. Natl. Acad. Sci. U.S.A. 115, E1647–E1656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X., Nagai J., Khakh B. S., Improved tools to study astrocytes. Nat. Rev. Neurosci. 21, 121–138 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Wang Q., Kong Y., Wu D. Y., Liu J. H., Jie W., You Q. L., Huang L., Hu J., Chu H. D., Gao F., Hu N. Y., Luo Z. C., Li X. W., Li S. J., Wu Z. F., Li Y. L., Yang J. M., Gao T. M., Impaired calcium signaling in astrocytes modulates autism spectrum disorder-like behaviors in mice. Nat. Commun. 12, 3321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petravicz J., Fiacco T. A., McCarthy K. D., Loss of IP3 receptor-dependent Ca2+ increases in hippocampal astrocytes does not affect baseline CA1 pyramidal neuron synaptic activity. J. Neurosci. 28, 4967–4973 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agulhon C., Fiacco T. A., McCarthy K. D., Hippocampal short- and long-term plasticity are not modulated by astrocyte Ca2+ signaling. Science 327, 1250–1254 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Zerbi V., Floriou-Servou A., Markicevic M., Vermeiren Y., Sturman O., Privitera M., von Ziegler L., Ferrari K. D., Weber B., de Deyn P. P., Wenderoth N., Bohacek J., Rapid reconfiguration of the functional connectome after chemogenetic locus coeruleus activation. Neuron 103, 702–718.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 29.J. Cohen, Statistical Power Analysis for the Behavioral Sciences (Routledge, 1988). [Google Scholar]
- 30.Hare B. D., Duman R. S., Prefrontal cortex circuits in depression and anxiety: Contribution of discrete neuronal populations and target regions. Mol. Psychiatry 25, 2742–2758 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferenczi E. A., Zalocusky K. A., Liston C., Grosenick L., Warden M. R., Amatya D., Katovich K., Mehta H., Patenaude B., Ramakrishnan C., Kalanithi P., Etkin A., Knutson B., Glover G. H., Deisseroth K., Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science 351, aac9698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan C. G., Chen X., Li L., Castellanos F. X., Bai T. J., Bo Q. J., Cao J., Chen G. M., Chen N. X., Chen W., Cheng C., Cheng Y. Q., Cui X. L., Duan J., Fang Y. R., Gong Q. Y., Guo W. B., Hou Z. H., Hu L., Kuang L., Li F., Li K. M., Li T., Liu Y. S., Liu Z. N., Long Y. C., Luo Q. H., Meng H. Q., Peng D. H., Qiu H. T., Qiu J., Shen Y. D., Shi Y. S., Wang C. Y., Wang F., Wang K., Wang L., Wang X., Wang Y., Wu X. P., Wu X. R., Xie C. M., Xie G. R., Xie H. Y., Xie P., Xu X. F., Yang H., Yang J., Yao J. S., Yao S. Q., Yin Y. Y., Yuan Y. G., Zhang A. X., Zhang H., Zhang K. R., Zhang L., Zhang Z. J., Zhou R. B., Zhou Y. T., Zhu J. J., Zou C. J., Si T. M., Zuo X. N., Zhao J. P., Zang Y. F., Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc. Natl. Acad. Sci. U.S.A. 116, 9078–9083 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bicks L. K., Koike H., Akbarian S., Morishita H., Prefrontal cortex and social cognition in mouse and man. Front. Psychol. 6, 1805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts A. C., Clarke H. F., Why we need nonhuman primates to study the role of ventromedial prefrontal cortex in the regulation of threat- and reward-elicited responses. Proc. Natl. Acad. Sci. U.S.A. 116, 26297–26304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craddock R. C., James G. A., Holtzheimer P. E. III, Hu X. P., Mayberg H. S., A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 33, 1914–1928 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park H. J., Friston K., Structural and functional brain networks: From connections to cognition. Science 342, 1238411 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Yu X., Wadghiri Y. Z., Sanes D. H., Turnbull D. H., In vivo auditory brain mapping in mice with Mn-enhanced MRI. Nat. Neurosci. 8, 961–968 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ray D., Bezmaternykh D., Mel’nikov M., Friston K. J., Das M., Altered effective connectivity in sensorimotor cortices is a signature of severity and clinical course in depression. Proc. Natl. Acad. Sci. U.S.A. 118, e2105730118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaiser R. H., Andrews-Hanna J. R., Wager T. D., Pizzagalli D. A., Large-scale network dysfunction in major depressive disorder: A meta-analysis of resting-state functional connectivity. JAMA Psychiatry 72, 603–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felger J. C., Li Z., Haroon E., Woolwine B. J., Jung M. Y., Hu X., Miller A. H., Inflammation is associated with decreased functional connectivity within corticostriatal reward circuitry in depression. Mol. Psychiatry 21, 1358–1365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furman D. J., Hamilton J. P., Gotlib I. H., Frontostriatal functional connectivity in major depressive disorder. Biol. Mood Anxiety Disord. 1, 11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulders P. C., van Eijndhoven P. F., Schene A. H., Beckmann C. F., Tendolkar I., Resting-state functional connectivity in major depressive disorder: A review. Neurosci. Biobehav. Rev. 56, 330–344 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Rupprechter S., Romaniuk L., Series P., Hirose Y., Hawkins E., Sandu A. L., Waiter G. D., McNeil C. J., Shen X., Harris M. A., Campbell A., Porteous D., Macfarlane J. A., Lawrie S. M., Murray A. D., Delgado M. R., McIntosh A. M., Whalley H. C., Steele J. D., Blunted medial prefrontal cortico-limbic reward-related effective connectivity and depression. Brain 143, 1946–1956 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z., Wang X., Liu J., Chen J., Liu X., Nie G., Jorgenson K., Sohn K. C., Huang R., Liu M., Liu B., Kong J., Acupuncture treatment modulates the corticostriatal reward circuitry in major depressive disorder. J. Psychiatr. Res. 84, 18–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabbay V., Ely B. A., Li Q., Bangaru S. D., Panzer A. M., Alonso C. M., Castellanos F. X., Milham M. P., Striatum-based circuitry of adolescent depression and anhedonia. J. Am. Acad. Child Adolesc. Psychiatry 52, 628–641.e13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang C., Hu Y., Talishinsky A. D., Potter C. T., Calva C. B., Ramsey L. A., Kesner A. J., Don R. F., Junn S., Tan A., Pierce A. F., Nicolas C., Arima Y., Lee S. C., Su C., Coudriet J. M., Mejia-Aponte C. A., Wang D. V., Lu H., Yang Y., Ikemoto S., Medial prefrontal cortex and anteromedial thalamus interaction regulates goal-directed behavior and dopaminergic neuron activity. Nat. Commun. 13, 1386 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchikami M., Thomas A., Liu R., Wohleb E. S., Land B. B., DiLeone R. J., Aghajanian G. K., Duman R. S., Optogenetic stimulation of infralimbic PFC reproduces ketamine’s rapid and sustained antidepressant actions. Proc. Natl. Acad. Sci. U.S.A. 112, 8106–8111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C. T., Cheng C. M., Chen M. H., Juan C. H., Tu P. C., Bai Y. M., Jeng J. S., Lin W. C., Tsai S. J., Su T. P., Antidepressant efficacy of prolonged intermittent theta burst stimulation monotherapy for recurrent depression and comparison of methods for coil positioning: A randomized, double-blind, sham-controlled study. Biol. Psychiatry 87, 443–450 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Hare B. D., Shinohara R., Liu R. J., Pothula S., DiLeone R. J., Duman R. S., Optogenetic stimulation of medial prefrontal cortex Drd1 neurons produces rapid and long-lasting antidepressant effects. Nat. Commun. 10, 223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mechling A. E., Arefin T., Lee H. L., Bienert T., Reisert M., Ben Hamida S., Darcq E., Ehrlich A., Gaveriaux-Ruff C., Parent M. J., Rosa-Neto P., Hennig J., von Elverfeldt D., Kieffer B. L., Harsan L. A., Deletion of the mu opioid receptor gene in mice reshapes the reward-aversion connectome. Proc. Natl. Acad. Sci. U.S.A. 113, 11603–11608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Y., Kong L., Wu F., Womer F., Jiang W., Cao Y., Ren L., Wang J., Fan G., Blumberg H. P., Xu K., Wang F., Decreased functional connectivity between the amygdala and the left ventral prefrontal cortex in treatment-naive patients with major depressive disorder: A resting-state functional magnetic resonance imaging study. Psychol. Med. 43, 1921–1927 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Thompson T., Correll C. U., Gallop K., Vancampfort D., Stubbs B., Is pain perception altered in people with depression? A systematic review and meta-analysis of experimental pain research. J. Pain 17, 1257–1272 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Bubl E., Kern E., Ebert D., Bach M., van Elst L. T., Seeing gray when feeling blue? Depression can be measured in the eye of the diseased. Biol. Psychiatry 68, 205–208 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Hu H., Cui Y., Yang Y., Circuits and functions of the lateral habenula in health and in disease. Nat. Rev. Neurosci. 21, 277–295 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Fullana M. N., Ruiz-Bronchal E., Ferrés-Coy A., Juárez-Escoto E., Artigas F., Bortolozzi A., Regionally selective knockdown of astroglial glutamate transporters in infralimbic cortex induces a depressive phenotype in mice. Glia 67, 1122–1137 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Roberts A. J., Khom S., Bajo M., Vlkolinsky R., Polis I., Cates-Gatto C., Roberto M., Gruol D. L., Increased IL-6 expression in astrocytes is associated with emotionality, alterations in central amygdala GABAergic transmission, and excitability during alcohol withdrawal. Brain Behav. Immun. 82, 188–202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.John C. S., Sypek E. I., Carlezon W. A., Cohen B. M., Öngür D., Bechtholt A. J., Blockade of the GLT-1 transporter in the central nucleus of the Amygdala induces both anxiety and depressive-like symptoms. Neuropsychopharmacology 40, 1700–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skupio U., Tertil M., Bilecki W., Barut J., Korostynski M., Golda S., Kudla L., Wiktorowska L., Sowa J. E., Siwiec M., Bobula B., Pels K., Tokarski K., Hess G., Ruszczycki B., Wilczynski G., Przewlocki R., Astrocytes determine conditioned response to morphine via glucocorticoid receptor-dependent regulation of lactate release. Neuropsychopharmacology 45, 404–415 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duman R. S., Sanacora G., Krystal J. H., Altered connectivity in depression: GABA and glutamate neurotransmitter deficits and reversal by novel treatments. Neuron 102, 75–90 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volterra A., Liaudet N., Savtchouk I., Astrocyte Ca2+ signalling: An unexpected complexity. Nat. Rev. Neurosci. 15, 327–335 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Tanaka M., Shih P. Y., Gomi H., Yoshida T., Nakai J., Ando R., Furuichi T., Mikoshiba K., Semyanov A., Itohara S., Astrocytic Ca2+ signals are required for the functional integrity of tripartite synapses. Mol. Brain 6, 6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin-Fernandez M., Jamison S., Robin L. M., Zhao Z., Martin E. D., Aguilar J., Benneyworth M. A., Marsicano G., Araque A., Synapse-specific astrocyte gating of amygdala-related behavior. Nat. Neurosci. 20, 1540–1548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen N., Sugihara H., Sharma J., Perea G., Petravicz J., le C., Sur M., Nucleus basalis-enabled stimulus-specific plasticity in the visual cortex is mediated by astrocytes. Proc. Natl. Acad. Sci. U.S.A. 109, E2832–E2841 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J.-H., Zhang M., Wang Q., Wu D.-Y., Jie W., Hu N.-Y., Lan J.-Z., Zeng K., Li S.-J., Li X.-W., Yang J.-M., Gao T.-M., Distinct roles of astroglia and neurons in synaptic plasticity and memory. Mol. Psychiatry 27, 873–885 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Gu X., Chen W., Volkow N. D., Koretsky A. P., du C., Pan Y., Synchronized astrocytic Ca2+ responses in neurovascular coupling during somatosensory stimulation and for the resting state. Cell Rep. 23, 3878–3890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonvento G., Bolanos J. P., Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 33, 1546–1564 (2021). [DOI] [PubMed] [Google Scholar]
- 67.Attwell D., Buchan A. M., Charpak S., Lauritzen M., MacVicar B. A., Newman E. A., Glial and neuronal control of brain blood flow. Nature 468, 232–243 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ben Haim L., Rowitch D. H., Functional diversity of astrocytes in neural circuit regulation. Nat. Rev. Neurosci. 18, 31–41 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Li X., Zima A. V., Sheikh F., Blatter L. A., Chen J., Endothelin-1-induced arrhythmogenic Ca2+ signaling is abolished in atrial myocytes of inositol-1,4,5-trisphosphate(IP3)-receptor type 2-deficient mice. Circ. Res. 96, 1274–1281 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Grandjean J., Corcoba A., Kahn M. C., Upton A. L., Deneris E. S., Seifritz E., Helmchen F., Mansuy I. M., Mann E. O., Rudin M., Saab B. J., A brain-wide functional map of the serotonergic responses to acute stress and fluoxetine. Nat. Commun. 10, 350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sumiyoshi A., Keeley R. J., Lu H., Physiological considerations of functional magnetic resonance imaging in animal models. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 4, 522–532 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Grandjean J., Schroeter A., Batata I., Rudin M., Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. Neuroimage 102 (Pt. 2), 838–847 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Satterthwaite T. D., Elliott M. A., Gerraty R. T., Ruparel K., Loughead J., Calkins M. E., Eickhoff S. B., Hakonarson H., Gur R. C., Gur R. E., Wolf D. H., An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64, 240–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.G. Paxinos, K. B. J. Franklin, The Mouse Brain in Stereotaxic Coordinates (Academic Press, Cambridge, MA, USA, ed. 4, 2001). [Google Scholar]
- 75.Dosenbach N. U. F., Nardos B., Cohen A. L., Fair D. A., Power J. D., Church J. A., Nelson S. M., Wig G. S., Vogel A. C., Lessov-Schlaggar C. N., Barnes K. A., Dubis J. W., Feczko E., Coalson R. S., Pruett J. R. Jr., Barch D. M., Petersen S. E., Schlaggar B. L., Prediction of individual brain maturity using fMRI. Science 329, 1358–1361 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M., Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15, 273–289 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S3 and S4
Tables S1 and S2