Abstract
Controlled primary cell wall remodeling allows plant growth under stressful conditions, but how these changes are conveyed to adjust cellulose synthesis is not understood. Here, we identify the TETRATRICOPEPTIDE THIOREDOXIN-LIKE (TTL) proteins as new members of the cellulose synthase complex (CSC) and describe their unique and hitherto unknown dynamic association with the CSC under cellulose-deficient conditions. We find that TTLs are essential for maintaining cellulose synthesis under high-salinity conditions, establishing a stress-resilient cortical microtubule array, and stabilizing CSCs at the plasma membrane. To fulfill these functions, TTLs interact with CELLULOSE SYNTHASE 1 (CESA1) and engage with cortical microtubules to promote their polymerization. We propose that TTLs function as bridges connecting stress perception with dynamic regulation of cellulose biosynthesis at the plasma membrane.
Peripheral membrane TTL proteins maintain the integrity of cellulose synthase complexes at the plasma membrane under stress.
INTRODUCTION
The cellulose content of land plants constitutes around 150 to 200 gigatons of carbon, roughly 1.5- to 2-fold more than the rest of life forms on Earth combined (1). This remarkable amount of cellulose, the main load-bearing component of plant cell walls, is synthesized by cellulose synthase complexes (CSCs). The CSC is a sophisticated molecular machinery that propels itself forward through the plasma membrane with its own catalytic activity while using cortical microtubules as steering devices (2). To date, four components of the cellulose synthase machinery have been identified: (i) the catalytic Cellulose Synthases (CESAs) (3); (ii) KORRIGAN, an integral membrane endoglucanase whose exact role is unknown (4); (iii) CSI1, which establishes a physical association between membrane-localized CSCs and cortical microtubules (5); and (iv) the CESA-COMPANION (CC) proteins, essential for the stability of CESAs and cortical microtubules upon salt stress and fungal interaction (6). Stress-sensing mechanisms enable rapid and controlled remodeling of the cell wall and mitigate perturbations on plant cellulose synthesis under environmental cues to alleviate growth reduction (6, 7). However, how fine-tuning of the highly dynamic CSC is achieved under stress remains largely unknown.
Tetratricopeptide Thioredoxin-Like (TTL) genes were identified through forward genetic screens for mutants with reduced root growth and isotropic cell expansion under elevated NaCl concentrations (8, 9). Various signaling pathways stimulate the recruitment of cytosolic TTL proteins to the plasma membrane where, acting as scaffolds, they amplify and stabilize the initial signals (10). In this study, we report that TTLs are peripheral membrane proteins that show a dual localization, i.e., cytosolic and at the plasma membrane as part of the CSC. TTLs are essential for CSC dynamics and cortical microtubule regulation to allow for cellulose biosynthesis during salinity stress. Conditions that caused altered cellulose content, such as high salinity or genetically induced CSC alteration, triggered the recruitment of TTLs from the cytosol to the CSC, allowing its stabilization. Our study identifies a new component of the CSC that aids in maintaining cellulose synthesis during stress conditions.
RESULTS
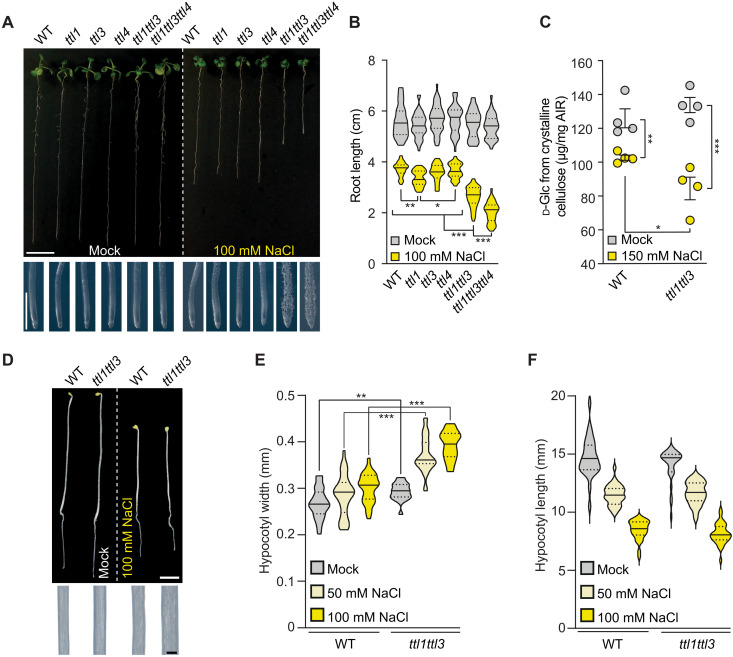
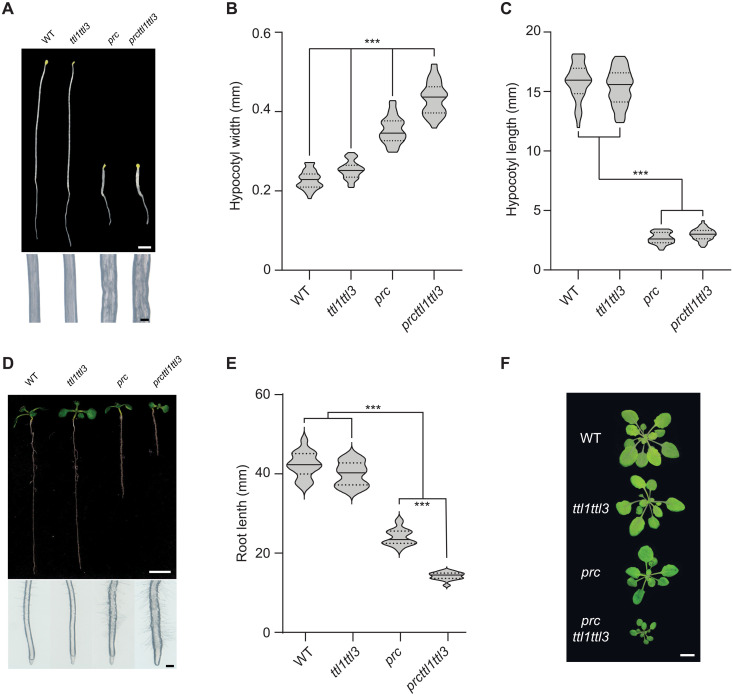
Reduced and isotropic growth are common phenotypes of cellulose-deficient mutants (6, 11, 12). Therefore, we explored potential cellulose synthesis defects in ttl mutants under salt stress. ttl1, ttl1ttl3, and ttl1ttl3ttl4 roots were significantly shorter than wild-type (WT; Col-0) roots under high–(200 mM) NaCl concentrations, going along with exacerbated isotropic cell expansion in ttl1ttl3 and ttl1ttl3ttl4 roots compared to WT or ttl1 (Fig. 1, A and B). Because the phenotypes of the ttl1ttl3 double mutant were only mildly enhanced in comparison to WT by adding the ttl4 knockout (Fig. 1A), we focused on ttl1ttl3 for further characterization of the TTLs. High NaCl concentrations led to less crystalline cellulose content in ttl1ttl3 as compared to WT roots (Fig. 1C) and isotropic growth defects in dark-grown hypocotyls from the former but not the latter (Fig. 1, D to F). This phenotype could be replicated with the primary wall cellulose synthesis inhibitor isoxaben (fig. S1, A to C) (6). Collectively, these results link TTL1 and TTL3 to cellulose synthesis under salt stress.
Fig. 1. TTL1 and TTL3 are required for salt stress tolerance and cellulose synthesis.
(A) WT (Col-0) and ttl mutants were germinated and grown for 3 days under control conditions and then transferred to control media or media supplemented with 100 mM NaCl and grown for eight additional days (top). Magnifications show distinctive root tip swelling in ttl double- and triple-mutant plants under salt stress, indicative of defective anisotropic growth (bottom). Scale bars, 1 cm (top) and 1 mm (close-up root tips). (B) Root length quantification of seedlings as in (A). Violin plots: Centerlines show the medians; dotted lines indicate the 25th and 75th percentiles; N ≥ 27 roots and three independent experiments. Welch’s analysis of variance (ANOVA) between 100 mM NaCl-treated groups: P ≤ 0.001; Dunnett’s T3 multiple comparison: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. (C) Cellulose represented as micrograms of d-glucose derived from cellulose per milligram of dried alcohol-insoluble residue (AIR) of roots grown as depicted in (A) but using 150 mM NaCl. Values are means ± SEM; N = 4 biological replicates; two technical replicates per biological replicate. Two-tailed, unpaired t test; *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. (D) Dark-grown WT and ttl1ttl3 seedlings germinated for 2 days under control conditions and transferred to either control plates or plates supplemented with 100 mM NaCl and grown for three additional days in the dark (top). Magnifications show distinctive hypocotyl width in ttl1ttl3 double-mutant plants under salt stress (bottom). Scale bars, 2 mm (top) and 200 μm (close-up hypocotyls). (E and F) Quantification of hypocotyl width and length of seedlings as depicted in (D). Violin plots: Centerlines show the medians; dotted lines indicate the 25th and 75th percentiles; N ≥ 18 seedlings from three independent experiments. Welch’s unpaired t test; **P ≤ 0.01 and ***P ≤ 0.001.
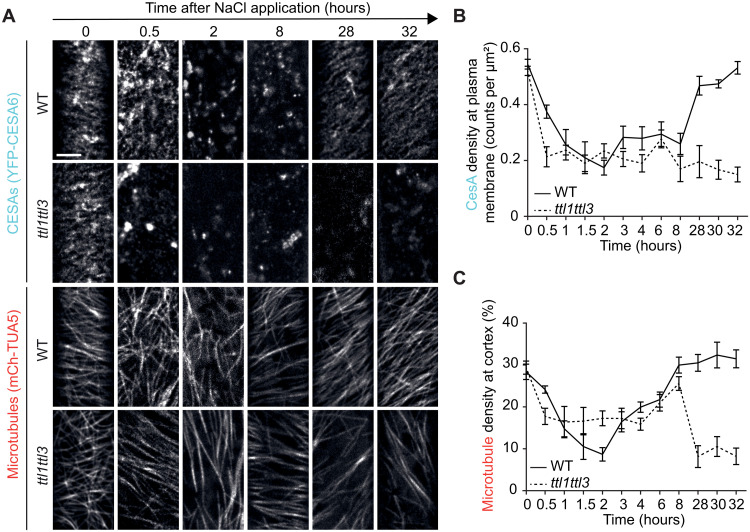
To explore whether TTL1/3 loss of activity might alter CESA removal and subsequent plasma membrane repopulation under salt stress, we generated a ttl1ttl3 YFP-CESA6 mCh-TUA5 line [encoding TUBULIN ALPHA-5; (13)]. CESAs and microtubules in etiolated hypocotyls were imaged between 30 min and 32 hours after NaCl treatment. As reported, the microtubule array in WT plants depolymerized 2 hours after treatment and reassembled within 8 hours thereof under NaCl treatment, in line with a decreased CESA6 density at the plasma membrane within 2 hours and subsequent recovery within 28 hours (Fig. 2, A to C) (1, 2, 8). In contrast, cortical microtubules showed incomplete depolymerization in ttl1ttl3 and, following a plateau phase after ~8 hours, failed to reassemble (Fig. 2, A to C). Concurrently, a rapid decrease in CESA density at the plasma membrane within 30 min of NaCl treatment was observed in ttl1ttl3, and CESAs’ recovery at the plasma membrane over the experiment’s time course never occurred (Fig. 2, A and B). Yellow fluorescent protein (YFP)–CESA fluorescence’s recovery after photobleaching was similar in WT and ttl1ttl3, ruling out a TTL3-dependent CESA6 delivery to the plasma membrane (fig. S2) and suggesting that TTLs stabilize cortical microtubules and the CSC at the plasma membrane during cell adjustment to salt stress.
Fig. 2. TTL3 regulates CESA activity and cortical microtubule polymerization under salt stress.
(A) Microtubule and CESA coverage in WT and ttl1ttl3 plants coexpressing mCh-TUA5 and YFP-CESA6 exposed to 200 mM NaCl. Images represent single frames taken at the indicated times of exposure. Time 0 shows cells just before NaCl treatment. Scale bar, 5 μm. (B) CESA density at the plasma membrane quantified from images as in (A). Time indicates the time after salt exposure. Two-way ANOVA analysis of CESA density; P ≤ 0.001 (genotype), P ≤ 0.001 (time), and P ≤ 0.001 (genotype × time). n ≥ 27 cells from at least three seedlings per time point and three independent experiments. Values are means ± SEM. (C) Microtubule coverage at the cell cortex quantified from images as in (A). Two-way ANOVA analysis of microtubule coverage is exactly as described for CESA density in (B).
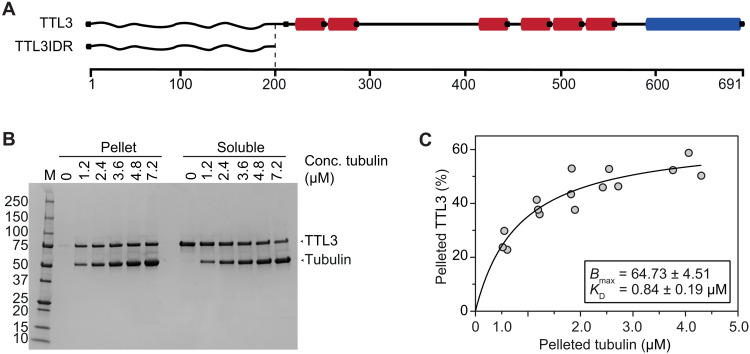
The disturbed cortical microtubule recovery under NaCl stress in ttl1ttl3 and high isoelectric point (>10) of its N-terminal intrinsically disordered region (IDR) hinted at possible direct interactions between TTL3 and the negatively charged tubulin surface, which was tested with microtubule spin-down assays (14) using Escherichia coli–expressed, full-length TTL3 (TTL3; Fig. 3A and fig. S3A). Saturation binding experiments (15) revealed a dissociation constant (KD) of 0.84 ± 0.19 μM (Fig. 3, B and C), a microtubule-binding affinity within the range reported for CELLULOSE SYNTHASE INTERACTIVE 1 and Companion of Cellulose Synthase 1, known to interact directly with microtubules and CESAs (6, 7, 16, 17). We could also detect a direct interaction of a 6x His–tagged version of the TTL3 IDR (TTL3IDR; fig. S3B) with microtubules, albeit at lower affinity (KD = 1.6 ± 0.4 μM; fig. S3, C and D), suggesting that the IDR plays a major role in the microtubule interaction of TTLs, but other parts of the TTLs might also be involved in this physical connection. To assess the possible role of TTL3 in microtubule polymerization, we performed microtubule turbidity assays. The TTL3IDR induced an atypical polymerization curve with a short exponential phase followed by a linear increase (fig. S3E), contrasting taxol- and microtubule-associated protein–rich fraction (MAPF)–based treatments, leading to an exponential growth phase followed by a stationary phase of equal microtubule polymerization and depolymerization (6). To exclude that protein denaturation caused the linear phase in TTL3IDR-treated samples, pelleted reactions were analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE). Intact tubulin bands at 55 kDa could be observed in the TTL3IDR-, MAPF-, and taxol-treated samples, while degradation products accumulated in both bovine serum albumin (BSA)– and buffer-treated samples (fig. S3F), indicating bona fide microtubule lattice stabilization by the TTL3IDR. The weaker microtubule binding strength of TTL3IDR in comparison to full-length TTL3 suggests that, in addition to the IDR, other parts of TTL3 play a role in microtubule binding.
Fig. 3. TTL3 is a microtubule-associated protein.
(A) Schematics of TTL3 and truncations thereof used here, with the predicted intrinsically disordered N terminus (wavy line; IDR), six tetratricopeptide repeats (red), and one C-terminal thioredoxin-like domain (blue). (B) Representative Coomassie gel used to estimate the dissociation constants for TTL3 and microtubules. Tubulin and TTL3 molecular sizes are indicated with arrowheads. (C) Binding affinity of TTL3 to microtubules determined by calculating their dissociation constant (KD) (0.84 ± 0.19 μM; best-fit values ± SE). TTL3 levels were kept constant while the amount of microtubules was increased. KD value was calculated by fitting a saturation binding curve onto the data, obtained from images as in (B); N = 3 independent experiments.
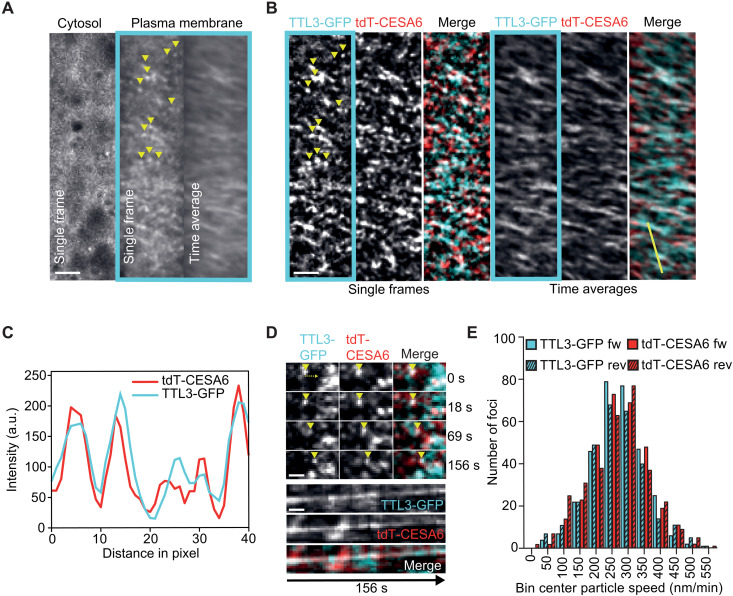
Next, we explored the potential colocalization between TTL1/3 and the CSC. The salt-sensitive phenotypes of ttl1ttl3ttl4 and ttl1ttl3 could be rescued in pTTL3:TTL3-GFP and pTTL1:TTL1-GFP complemented lines, respectively (fig. S4, A to D), indicating functional green fluorescent protein (GFP) fusions of both proteins. Spinning disk confocal microscopy in etiolated hypocotyls revealed a prominent cytoplasmic localization for TTL3-GFP and a weaker signal of motile foci at the plasma membrane (fig. S5A and movie S1). This dual localization of TTL3-GFP was confirmed by comparative immunoblot analyses between cytosolic and microsomal fractions (fig. S5B). TTL1-GFP displayed a similar dual localization but mainly in the root elongation zone (fig. S5C), in contrast to the broader expression of TTL3-GFP in roots and hypocotyls (3). The motile plasma membrane–localized TTL1/3-GFP foci were reminiscent of CESAs (13), prompting us to test whether TTLs indeed track together with them. As previously observed for pTTL3:TTL3-GFP and pTTL1:TTL1-GFP complemented lines, in a dual-labeled ttl1ttl3 line expressing TTL3-GFP and tdTomato (tdT)-CESA6 (14), the motile TTL3-GFP foci were substantially obscured by strong cytosolic signals (Fig. 4A). An in-house image modification workflow (see Materials and Methods) used to extract the foci from the cytosolic signal (Fig. 4B and movie S1) revealed that TTL3-GFP foci colocalized and comigrated with tdT-CESA6 at the plasma membrane (Pearson coefficient of 0.54 ± 0.11; 26 cells from 14 seedlings in three experiments; Fig. 4, B to D; fig. S5D; and movie S1). In addition, TTL3-GFP and tdT-CESA6 foci moved bidirectionally with similar speeds (271 ± 86 and 267 ± 93 nm/min, respectively; Fig. 4E). Some foci (both CESA and TTL3) did not obviously colocalize, which could stem from the image modification workflow to allow analysis of the foci in the first place or could indicate a broader role of TTL3 at the plasma membrane. Furthermore, we did not observe TTLs associated with Golgi-derived or other vesicular structures nor with stressed-induced small CesA compartments/microtubule-associated CesA compartments [smaCCs/MASCs; (13, 18)], indicating that the TTL-CESA interaction exclusively occurs at the plasma membrane.
Fig. 4. TTL3 shows a peripheral localization and comigrates with the CSC.
(A) TTL3-GFP in etiolated hypocotyl epidermal cells of ttl1ttl3 TTL3-GFP tdT-CESA6 seedlings. Single frames and time-average projections show that TTL3-GFP is not only cytosolic but also forms motile particles at the plasma membrane (framed in cyan, yellow arrowheads). Scale bar, 2.5 μm. (B) Plasma membrane focal plane of the cell as in (A). TTL3-GFP particles colocalize with tdT-CESA6 at the plasma membrane (single frames). Time-average projections of TTL3-GFP and tdT-CESA6 reveal fluorescent foci’s comigration. Scale bar, 2.5 μm. (C) Intensity plot of TTL3-GFP and tdT-C6 along the yellow line shown in (B). a.u., arbitrary units. (D) Time frames of individual TTL3-GFP and tdT-CESA6 plasma membrane foci from corresponding movies. Upon their visualization (yellow arrowhead at time 0 s), foci comigrate for at least 156 s. Scale bar, 1 μm. (E) Bidirectional motility and speed distribution of TTL3-GFP and tdT-CESA6 foci. Forward denotes movement from left to right; reverse denotes from right to left in relation to the major axes of analyzed cells.
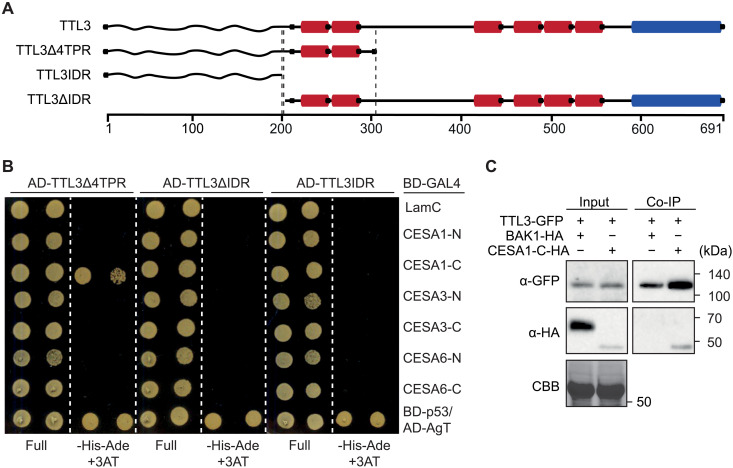
To evaluate a direct physical interaction between TTL3 and the primary CESAs, we used yeast two-hybrid assays (Y2H). As full-length TTL3 is unstable in yeast (10), we used three different TTL3 fragments fused to the activation domain of Gal4 (AD-Gal4): two N-terminal fragments encompassing the intrinsically disordered region (TTL3IDR), the IDR and the first two TPRs (TTL3∆4TPR), or a C-terminal region containing all six TPRs and the TRLX motif (TTL3∆IDR; Fig. 5A). As baits, we used the cytosolic N-terminal domains (CESA-N) or the cytosolic catalytic domains (CESA-C) of CESA1, CESA3, or CESA6 (CESA1-C, CESA3-C, and CESA6-C, respectively). TTL3∆4TPR interacted exclusively with CESA1-C, and neither TTL3IDR nor TTL3-∆IDR interacted with any CESA domain (Fig. 5B) despite of all constructs being expressed (fig. S6, A and B). To exclude auto/self-activation of BD-Gal4 or AD-Gal4 fusion proteins, BD-LamC and AD-AgT were included in the Y2H assays as negative controls (Fig. 5B and fig. S6C). Overall, this analysis indicates that the N terminus of TTL3, which includes the IDR and the first two TPRs, is required for the interaction with the N-terminal domain of CESA1. Whether the first two TPRs are required for direct interaction or for potential IDR interaction/folding requires further investigation. We then investigated the interaction of TTL3 and CESA1 in vivo by performing coimmunoprecipitation (Co-IP) assays. We used transient expression of TTL3-GFP and CESA1-C-HA in Nicotiana benthamiana using BAK1-HA as a negative control. All proteins were detected in the input (Fig. 5C), confirming their solubilization out of the membrane in the extraction buffer. After IP of TTL3-GFP using GFP-Trap beads, we detected a clear interaction of TTL3-GFP with CESA1-C-HA but not with BAK1-HA (Fig. 5C). Next, we used free (soluble) GFP as a negative control. TTL3-GFP but not GFP was coimmunoprecipitated with CESA1-C-HA, which confirmed the specificity of the TTL3-GFP interaction with CESA1-C-HA (fig. S6D). In agreement with the results obtained for TTL3∆4TPR, we found that TTL1∆4TPR interacted with CESA1-C but not with CESA1-N using Y2H (fig. S6E). We therefore conclude that TTL1 and TTL3 bind to CESA1-C via their N-terminal IDR and the first two TPRs.
Fig. 5. TTL3 interacts with CESA1.
(A) Schematics of TTL3 and truncations thereof used here, with the predicted intrinsically disordered N terminus (wavy line; IDR), six tetratricopeptide repeats (red; TPR), and one C-terminal thioredoxin-like domain (blue). (B) Y2H assays between TTL3 fragments fused to the AD-GAL4 and the cytosolic CESA-N and CESA-C BD domains fused to BD-GAL4. BD-LamC was included to discard the autoactivation of the AD-GAL4–fused proteins. The BD-p53/AD-AgT pair was used as a positive control. Growth on nonselective media (full) and interaction-selective media [lacking histidine and adenine (–His–Ade) and supplemented with 3-amino-1,2,4-triazole (+3AT2mM)] is shown. Photographs were taken after 3 days at 30°C. Three independent experiments yielded similar results. (C) TTL3-GFP was transiently coexpressed with CESA1-C-HA or BAK1-HA as a negative control in N. benthamiana. Total (Input) and Co-IP proteins were analyzed by immunoblotting using anti-GFP and anti-HA antibodies. Equal loading was confirmed by Coomassie brilliant blue (CBB) staining of input samples. Two independent experiments yielded similar results.
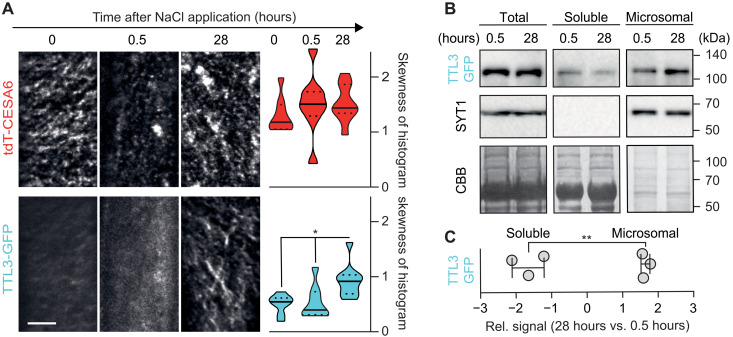
Given the hypersensitivity of ttl mutants to NaCl and direct interaction of TTLs with CESA1, we investigated the TTL3 localization and dynamics under control conditions or at 0.5 and 28 hours after NaCl treatment, time points at which CSC removal and subsequent plasma membrane repopulation are optimally observed (6). In etiolated hypocotyls of ttl1ttl3 TTL3-GFP tdT-CESA6 plants, TTL3-GFP was predominantly cytosolic under control conditions and at 0.5 hours after NaCl treatment, while it strongly relocalized to tdT-CESA6–rich foci at the plasma membrane at 28 hours after treatment, with a concomitant depletion of its cytosolic signal (Fig. 6A and see Materials and Methods for quantification). Furthermore, TTL3-GFP was significantly depleted from the soluble and enriched in the microsomal fraction at 28 hours after NaCl treatment, unlike the control microsomal protein SYT1 (Fig. 6, B and C) (19). Thus, TTL3 is a peripheral membrane protein that relocalizes to the CSC selectively upon stress.
Fig. 6. Salt stress induces TTL3 localization at the plasma membrane.
(A) Left: TTL3-GFP and tdT-CESA6 coverage at the plasma membrane in 3-day-old ttl1ttl3 hypocotyl epidermal cells prior (time 0) and after exposure to 200 mM NaCl for the indicated hours. Images represent single frames taken at the indicated times. Scale bar, 5 μm. Right: Quantification of histogram skewness of images as shown on the left. Centerlines in violin plots: medians; dotted lines: 25th and 75th percentiles. Welch’s ANOVA between CESA time points: P = 0.50; Welch’s ANOVA between TTL3-GFP time points: P ≤ 0.01. Dunnett’s T3 multiple comparisons: *P ≤ 0.05. n ≥ 18 cells from >8 seedlings per time point in three independent experiments. (B) Fractionation analysis (total, soluble, and microsomal fractions) of TTL3-GFP in ttl1ttl3ttl4 as in (G) at 0.5 or 28 hours of 200 mM NaCl treatment. TTL3-GFP and the microsomal control SYT1 were detected with anti-GFP and anti-SYT1 antibodies, respectively. Coomassie brilliant blue staining provides a loading control. (C) TTL3-GFP signal quantification from immunoblots as in (B). Band intensity data are expressed as the fold change of 28 versus 0.5 hours of 200 mM NaCl treatment in soluble and microsomal subcellular fractions. Values are individual replicates ± SEM; Welch’s unpaired t test; **P ≤ 0.01.
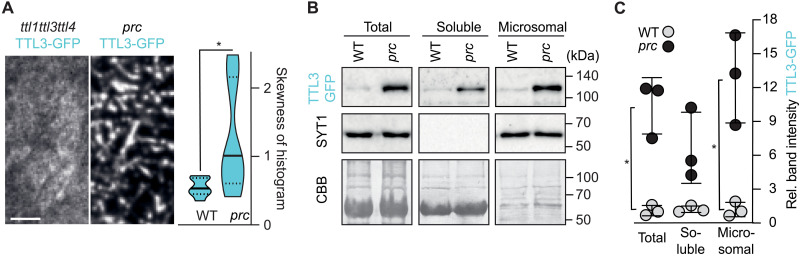
Next, we tested the role of TTLs in the cellulose-deficient mutant procuste 1 (prc1-1), a null mutant of CESA6 (20). When introduced into prc1-1, ttl1ttl3 aggravated the developmental defects, supporting a genetic interaction between CESA6 and TTL1/3 (Fig. 7, A to F, and fig. S7). Unexpectedly, TTL3-GFP overaccumulated and formed predominantly motile clusters at the plasma membrane in prc1-1 even under nonstressed conditions (Fig. 8A). Furthermore, TTL3-GFP levels were increased by ~6- and ~12-fold in the cytosolic and microsomal fractions, respectively, of prc1-1 compared to WT plants (Fig. 8, B and C). These results are reminiscent of those we found at 28 hours after NaCl treatment, highlighting a prominent role of TTL3 in cellulose synthesis, especially under stressed conditions.
Fig. 7. prc1-1 exacerbates the developmental defects of ttl1ttl3.
(A) Dark-grown WT (Col-0), ttl1ttl3, prc1-1, and ttl1ttl3prc1-1 seedlings grown under control conditions for 5 days (top). Magnifications show distinctive hypocotyl width in prc1-1 and prc1-1ttl1ttl3 triple-mutant plants compared to WT and ttl1ttl3, indicative of defective anisotropic growth (bottom). Scale bars, 2 mm (top) and 200 μm (close-up hypocotyls). (B and C) Hypocotyl width (B) and length (C) quantification of seedlings as depicted in (A). Violin plots: Centerlines show the medians; dotted lines indicate the 25th and 75th percentiles; Welch’s ANOVA on hypocotyl width: P ≤ 0.001; Welch’s ANOVA on hypocotyl length: P ≤ 0.001; Dunnett’s T3 multiple comparisons: ***P ≤ 0.001; N ≥ 35 seedlings. Two independent experiments yielded similar results. (D) Ten-day-old WT (Col-0) and ttl1ttl3, prc1-1, and ttl1ttl3prc1-1 seedlings grown on control media in the light (top). Magnifications show distinctive root tip swelling in prc1-1 and ttl1ttl3prc1-1 triple-mutant plants indicative of isotropic growth defects (bottom). Scale bars, 1 cm (top) and 250 μm (close-up root tips). (E) Quantification of root length of seedlings in (D). Violin plots: Centerlines show the medians; dotted lines indicate the 25th and 75th percentiles; Welch’s ANOVA on hypocotyl length: P ≤ 0.001; Dunnett’s T3 multiple comparisons: ***P ≤ 0.001; N ≥ 16 roots. Two independent experiments yielded similar results. (F) Morphological phenotypes of 5-week-old plants (rosettes) of WT (Col-0), ttl1ttl3, prc1-1, and ttl1ttl3prc1-1 grown in short days. Scale bar, 1 cm.
Fig. 8. The prc1-1 mutation increases the amount of TTL3 and its localization at the plasma membrane.
(A) Left: TTL3-GFP coverage at the plasma membrane in 3-day-old ttl1ttl3ttl4- or prc1-1–etiolated seedlings. Scale bar, 5 μm. Right: Histogram skewness of images as shown on the left. Violin plots: Centerlines show the medians; dotted lines indicate the 25th and 75th percentiles; n = 13 cells from at least eight seedlings from three independent experiments. Welch’s unpaired t test; *P ≤ 0.05. (B) Fractionation analysis (total, soluble, and microsomal fractions) of TTL3-GFP in WT and prc1-1. TTL3-GFP and the microsomal control SYT1 were detected with anti-GFP and anti-SYT1 antibodies, respectively. Coomassie brilliant blue staining provides a loading control. (C) TTL3-GFP signal quantification from immunoblots as in (B). Band intensity data are expressed as relative band intensity in total, soluble, and microsomal subcellular fractions. The TTL3-GFP immunoblot signal intensity was normalized to that of SYT1 for the input and microsomal fractions and to the Coomassie brilliant blue for soluble fractions. N = 3 independent experiments. Values are individual replicates ± SEM; Welch’s unpaired t test; *P ≤ 0.05.
DISCUSSION
Our work describes TTL proteins as new and unique members of the CSC, required for the stabilization of the cellulose synthesis machinery under stress. The TTL-CESA interaction at the plasma membrane significantly increased under conditions that cause reduced cellulose content, such as salt stress and structural alterations of the CSCs as observed in the prc1-1 mutant (20). This cellulose stress triggered the relocalization of cytosolic TTLs to the active CSCs, allowing for their stabilization and integrity (Fig. 9). The dynamic TTL recruitment from the cytosol to the plasma membrane–localized CSCs is unique since all previously described components of the cellulose synthase machinery are permanently associated with the complex (4).
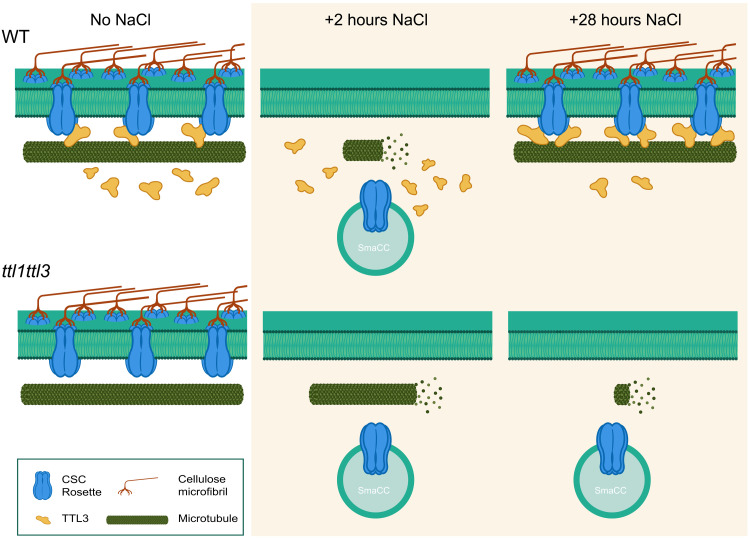
Fig. 9. Model for TTL function in cellulose biosynthesis under salt stress.
The CSCs produce cellulose at the plasma membrane tracking along cortical microtubules under control conditions in both WT (Top) and ttl mutants (Bottom). Two hours of exposure to elevated NaCl concentrations cause microtubule depolymerization and CSC displacement from the plasma membrane into SmaCCs (also called MASCs) in both genotypes. However, microtubule depolymerization is not as pronounced in ttl1ttl3 as compared to WT. After 28 hours of NaCl treatment, TTL proteins establish a salt-tolerant microtubule array and allow the repopulation of the CSC pool at the plasma membrane to restore cellulose synthesis in WT plants. However, in the absence of TTL proteins, microtubules disintegrate at later stages of treatment, while the CSCs do not repopulate the plasma membrane, leading to defective cellulose synthesis.
In addition to their role in cellulose synthesis, TTLs are scaffold components of the brassinosteroid (BR) signaling machinery at the plasma membrane (10). These two biological processes are interconnected via the GSK3-like kinase BR INSENSITIVE 2, which negatively regulates BR signaling and cellulose synthesis through phosphorylation of specific transcription factors and of the CESA1-N domain, respectively (21, 22). Our data cannot discard the possibility of TTL3 interacting with other plasma membrane–localizing proteins, which might be potential molecular linkers of the BR-cellulose synthesis processes. Whether the roles of TTLs in these two processes are mutually dependent deserves further investigation.
TTL orthologs can be identified in all embryophytes down to the liverwort Marchantia polymorpha, the most basal lineage of extant plants (fig. S7), while all other intrinsic components of the CSC emerged earlier in evolution (23). Thus, TTL genes are likely a key evolutionary acquisition that might allow plants to withstand stresses that are inherent to land colonization and enable a fine-tuned stabilization of the cellulose synthesis machinery. We thus propose that TTLs could be the end point of a cellulose sensing pathway used by the plants to maintain cell wall integrity during stress adaptation since dynamic changes in cellulose properties are essential for plants to withstand stresses.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana Columbia (Col-0) ecotype was used in this study. The transferred DNA insertions for TTL genes used in this study have been described previously: ttl1-2 mutant allele, referred to as ttl1 (AT1G53300) Salk_063943 in this study (5); ttl3 (AT2G42580) Sail_193_B05; ttl4 (AT3G58620) Salk_026396; and ttl1ttl3 double and ttl1ttl3ttl4 triple mutant (9). prc1-1 single mutant was obtained from Nottingham Arabidopsis Stock Centre (NASC) and was described previously (24). The ttl1ttl3ttl4 TTL3-GFP (ttl1ttl3ttl4 pTTL3:TTL3g-GFP 2.4) stable transgenic Arabidopsis line used in this study was described previously (10).The ttl1ttl3 TTL1-GFP (ttl1ttl3 pTTL1:TTL1g-GFP) stable transgenic line was generated as described in the “Plasmid constructs” and “Generation of transgenic plants” sections. ttl1ttl3 YFP-CESA6 mCherry-TUA5 was obtained by crossing ttl1ttl3 with a YFP-CESA6 mCherry-TUA5 dual-labeled line (2, 13), ttl1ttl3 TdT-CESA6 TTL3-GFP was obtained by crossing ttl1ttl3ttl4 TTL3-GFP with TdT-CESA6 (25), prc1-1 TTL3-GFP was obtained by crossing ttl1ttl3ttl4 TTL3-GFP with prc1-1, and prc1-1ttl1ttl3 was obtained by crossing ttl1ttl3 with prc1-1. Seeds were surface sterilized using the chlorine gas method (26) and cold treated for 4 days at 4°C for stratification. For root elongation assays, seeds were sowed onto half-strength Murashige and Skoog (MS) agar-solidified medium [1% (w/v) for vertical growth] containing 1.5% sucrose and subsequently grown under cool-white light (approximately 120 μmol photon m−2 s−1) at 22°C, with a long-day photoperiod (16-hour-light/8-hour-dark cycle). For etiolated seedling analyses, seeds were sowed onto half-strength MS agar-solidified medium [1% (w/v) for vertical growth] containing 1% sucrose, stratified for 4 days, exposed to light (approximately 120 μmol photon m−2 s−1) for 3 hours, and subsequently grown under dark conditions at 22°C for 4 days. When required, seedlings were transferred to soil after 7 days of in vitro growth and watered every 2 days. In soil, plants were grown in a mixture of organic substrate and vermiculite (4:1, v/v) at 22°C, with a long-day photoperiod (approximately 120 μmol photon m−2 s−1).
Isoxaben and NaCl treatment
For in vitro assays, seeds were sowed in half-strength MS agar-solidified medium supplemented with 1% sucrose and 1 nM isoxaben (Santa Cruz Biotechnology), and 5-day-old etiolated seedlings were photographed and analyzed for hypocotyl length and width. For salt stress, seedlings were transferred from half-strength MS agar-solidified medium (1.5% sucrose) plates to half-strength MS agar-solidified medium (1.5% sucrose) for control plates and plates supplemented with 50 and 100 mM NaCl. Seedlings were transferred to NaCl plates after 2 days (etiolated seedlings) or 3 days (light-grown seedlings) and photographed 3 or 7 to 8 days later, respectively.
Plasmid constructs
A genomic fragment spanning the 1.4-kb TTL1 promoter region upstream of the start codon and the TTL1 genomic region without stop codon was polymerase chain reaction (PCR) amplified using the primers detailed in table S1 and cloned into the pENTR/D-TOPO vector (Invitrogen). The CESA1-C coding sequence for expression in N. benthamiana was PCR amplified using specific primers using pGBKT7-CesA1-C domain as the template and the attB1 adapter universal primer (Invitrogen) and introduced into the pDONR/Zeo vector (vector) using the BP Clonase II Gateway Cloning Kit (Invitrogen). The truncated version of TTL3IDR was generated using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) to remove base pairs 610 to 2071 of the TTL3 coding sequence (CDS) using pENTR/D-TOPO-TTL3 full-length CDS as the template. All the resulting pENTR clones were verified by diagnostic PCR, restriction analysis, and sequencing. These pENTR clones in combination with the appropriate destination vectors (pDEST) were used to create the final Gateway expression constructs by Gateway LR Clonase II reaction (Invitrogen).
The pGWB4, pGWB5, and pGWB14 vectors, from the pGWB vector series, were provided by T. Nakagawa (Department of Molecular and Functional Genomics, Shimane University) and were used as pDEST for either transient expression in N. benthamiana or for generating stable lines in A. thaliana. Last, the Y2H pGADT7(GW) and pGBKT7(GW) destination vectors were provided by S. Prat (Centro Nacional de Biotecnología-Consejo Superior de Investigaciones Científicas).
The 35S::TTL3-GFP, 35S::BAK1-HA, and 35S::GFP as well as the TTL3, TTL3∆4TPR, and TTL3∆IDR Y2H constructs were described previously (10) and used in this study. The CESA constructs used for the Y2H assay were generated by amplifying the N-terminal and internal catalytic loop of CESA1 (CESA1-N: 1 to 270 amino acids; CESA1-C: 320 to 856 amino acids), CESA3 (CESA3-N: 1 to 260 amino acids; CESA3-C: 304 to 842 amino acids), and CESA6 (CESA6-N: 1 to 277 amino acids; CESA6-C: 321 to 868 amino acids), identified on the basis of transmembrane topology predictions, from A. thaliana Col-0 complementary DNA with primers listed in table S1. The fragments and the matchmaker Gold Y2H vector pGBKT7 were digested (Nde I/Sfi I), ligated, and transformed into Saccharomyces cerevisiae strain AH109 as described in the “Yeast two-hybrid assay” section.
Plasmids for heterologous protein expression of full-length TTL3 and 6xHis-TTL3IDR were constructed as follows. The vector petM11SUMO3GFP was cut with Bam HI, and the full-length TTL3 CDS, amplified with specific overhangs (table S1), was inserted using the Gibson assembly method (27). The TTL3IDR (amino acids 1 to 200) was amplified with specific overhangs (table S1) for the pPROEX HTb vector (Invitrogen, USA) and fused with the linearized vector (Eco RI + Xba I) using the Gibson assembly technique.
Generation of transgenic plants
Expression clones were transformed into Agrobacterium tumefaciens strain GVG3101::pMP90. The pGWB4 harboring the full-length line pTTL1::TTL1g-GFP construct was transformed into ttl1ttl3 Arabidopsis plants by floral dipping (28) to generate stable transgenic plants. Seeds obtained were sowed in half-strength MS agar-solidified medium supplemented with 1.5% sucrose (w/v) and with hygromycin B (50 μg/ml; Duchefa). T4 homozygous transgenic plants were used in this study.
Transient expression in N. benthamiana
Transient expression in N. benthamiana was performed following the protocol described in (29). A. tumefaciens (GV3101::pMP90) carrying the different constructs was used together with the p19 strain. Cultures were grown overnight in LB medium containing rifampicin (50 mg/ml), gentamycin (25 mg/ml), and antibiotic specific for the positive selection of the plasmid constructs. Individual colonies were used for inoculation on LB for a given construct. After overnight incubation at 28°C, Agrobacterium cells were collected by centrifugation (15 min at 3000g in 50-ml falcon tubes) at room temperature, and pellets were resuspended in agroinfiltration solution [10 mM MES (pH 5.6) and 10 mM MgCl2 and supplemented with 100 μM acetosyringone]. Subsequently, cultures were incubated for 2 to 4 hours at room temperature under dark conditions. Bacterial suspension was adjusted at an optical density at 600 nm (OD600) of 0.8 for the constructs and 0.2 for the p19 strain to reach a final OD600 of approximately 1 for agroinfiltration. For double-infiltration experiments, Agrobacterium strains were infiltrated at OD600 of 0.4 for each construct and at OD600 of 0.2 for the p19 strain. Cultures were infiltrated in 4-week-old N. benthamiana leaves using 1-ml syringes (BD Plastipak) at the abaxial side of the leaf. After infiltration, all plants were kept in the greenhouse and analyzed 2 days later.
Protein extraction and Co-IP in N. benthamiana
Protein extraction and Co-IP in N. benthamiana were performed as described previously (30) with some modifications. Briefly, 4-week-old N. benthamiana plants were used for transient expression assays as described in the “Transient expression in N. benthamiana” section. N. benthamiana leaves were mainly ground to fine powder in liquid nitrogen. Approximately 0.5 g of ground leaves per sample was used, and total proteins were then extracted with extraction buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 10% glycerol, 10 mM EDTA (pH 8), 1 mM NaF, 1 mM Na2MoO4·2H2O, 10 mM 1,4-dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 1% (v/v) P9599 protease inhibitor cocktail (Sigma-Aldrich)]; 0.5% (v/v) NP-40 (CAS: 9036-19-5, USB Amersham Life Science) was added at 2 ml/g of powder using an end-over-end rocker for 30 min at 4°C. Samples were centrifuged for 20 min at 4°C and 9000 rpm (9056g). Supernatants (approximately 4 mg/ml of protein) were filtered by gravity through Poly-Prep chromatography columns (731-1550, Bio-Rad), and 100 μl was used as input. The remaining supernatants were incubated for 2 hours at 4°C with 15 μl of GFP-Trap coupled to agarose beads (Chromotek) in an end-over-end rocker. During incubation of protein samples with GFP-Trap beads, the final concentration of detergent (NP-40) was adjusted to 0.2% (v/v) to avoid nonspecific binding to the matrix as recommended by the manufacturer. For subsequent incubation, beads were collected and washed four times with the wash buffer (similar to extraction buffer but without detergent). Last, beads were resuspended in 75 ml of 2× concentrated Laemmli sample buffer and heated at 70°C for 40 min to dissociate immunocomplexes from the beads. Total (input), immunoprecipitated (IP), and Co-IP proteins were separated in a 10% SDS-PAGE gel and analyzed as described in the “Immunoblot analysis” section.
Spinning disk live-cell imaging and data processing
Three-day-old plant hypocotyls or 5-day-old roots were covered with a 1% agarose cushion and mounted onto the imaging system as described previously (31). Fluorescence-tagged proteins were imaged with a CSU-W1 T1 Yokogawa spinning disk head fitted to a Nikon Eclipse Ti2-E inverted microscope with a CFI Apo TIRF × 100 numerical aperture (NA), 1.49 oil immersion objective, and two iXon Life 888 EM-CCD cameras (Andor, GB). The system was assembled by 3i (www.intelligent-imaging.com). GFP was imaged using a 488-nm solid-state diode laser (150 mW) and a 525/30- to 25-nm emission filter, and red fluorescent protein (RFP) was detected with a 561-nm solid-state diode laser (150 mW) and a 617/73- to 25-nm emission filter. Alternatively, a CSU-W1 Yokogawa spinning disk head was fitted to a Nikon Eclipse Ti-E inverted microscope with a CFI PlanApo × 100 NA, 1.40 oil immersion objective, and two iXon Ultra EM-CCD cameras (Andor, GB). For this visitron system (www.visitron.de), GFP was imaged using a 488-nm solid-state diode laser (200 mW) and a 525/50-nm emission filter; RFP was detected with a 561-nm solid-state diode laser (200 mW) and a 630/75-nm emission filter. Time-lapse images were processed and analyzed with Fiji (32). Drifts were corrected by using the plugin StackReg or MultiStackReg in cases where two channels were imaged (33). When the drift of samples could not be corrected in this way, they were excluded from the analysis. Backgrounds were subtracted by the “subtract background” tool (rolling ball radius, 30 to 50 pixels). To quantify CESA velocities, three frames were averaged by “WalkingAverage,” and kymograph analysis was performed with the kymograph tool of FIESTA (34).
Microtubule quantification, particle detection, and particle clustering at the plasma membrane
Particle density at the plasma membrane of cells was determined as described before (6, 7). Microtubule density measurements were done as described in detail before (35). Colocalization analysis was performed using the JACoP plugin in Fiji. Particle clustering at the plasma membrane (e.g., by recruitment of cytosolic TTL3-GFP to the plasma membrane–localized CESA particles) was assessed by measuring the skewness of the image histogram with Fiji (36). Skewness is a measure for the asymmetry of an image histogram. Since the rather evenly distributed cytosolic signal of TTL3-GFP resulted in a low skewness of the histogram, the relocalization of TTL3-GFP to the CSC upon stress results in an enhanced skewness due to signal-cluster formation. A predefined cell area was cut out from all analyzed cells. The area was designed to fit into all analyzed cells, thereby excluding all non-cell signals (e.g., the area outside of the analyzed cell and the outer cell lines). All images were taken with constant settings, and raw images were used for this analysis to not affect the histogram.
Yeast two-hybrid assay
The Gal4-based Y2H system (Clontech Laboratories) was used for testing the interaction of TTL3∆4TPR, TTL3∆IDR, and TTL3IDR with the N-terminal domains (CESA-N) and the cytosolic catalytic domains (CESA-C) of CESA1, CESA3, and CESA6. The bait and prey constructs are detailed in the “Plasmid constructs” section. Bait and prey plasmids were transformed into S. cerevisiae strain AH109 with the lithium acetate/single-stranded carrier DNA/polyethylene glycol method, and transformants were grown on plasmid-selective media [synthetic defined (SD)/–Trp–Leu]. Plates were incubated at 28°C for 4 days, and independent colonies for each bait-prey combination were resuspended in 200 μl of sterile water. Ten-fold serial dilutions were made, and 5 μl of each dilution was spotted onto three alternative interaction-selective medium [SD/–Trp–Leu–His+3-AT (for 3-amino-1,2,4-triazole, 2 mM), SD/–Trp–Leu-Ade, and SD/–Trp–Leu–Ade+3-AT]. Plates were incubated at 28°C and photographed 3 or 7 days later.
Yeast two-hybrid protein extraction
For immunoblot analysis, one or two independent yeast cotransformants (a and b) for each bait-prey plasmid combination were grown in 50 ml of SD/–Leu–Trp to an OD600 of 0.7 to 1. Cultures were centrifuged at 4000 rpm for 3 min. The resulting pellet was washed once with cold water and resuspended in 200 ml of radioimmunoprecipitation assay (RIPA) buffer [2 mM sodium phosphate buffer (pH 7), 0.2% Triton X-100, 0.02% (w/v) SDS, 0.2 mM EDTA (pH 8), and 10 mM NaCl] containing protease inhibitor (1 tablet/10 ml; cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail, Roche). Glass beads (500 ml, 425 to 600 mm; Sigma-Aldrich) were added, and the sample was vortexed in FastPrep FP120 (BIO 101) at a power setting of 5.5 for two 15-s intervals separated by 1-min interval on ice. Next, 400 ml of RIPA buffer with protease inhibitors was added, and the sample was vigorously vortexed. The supernatant was recovered, and the protein concentration was determined using Bradford assays. Total protein (50 μg) was resolved on 10% polyacrylamide/SDS gels and analyzed by immunoblotting to detect the Gal4AD-HA fusion proteins with an anti-hemagglutinin (HA) antibody (1:5000; Sigma-Aldrich, H3663) as described in the “Immunoblot analysis” section.
Preparation of total, soluble, and microsomal fractions for immunoblot
Three-day-old etiolated ttl1ttl3ttl4 TTL3-GFP and prc1-1 TTL3-GFP seedlings grown in half-strength solidified MS agar with 1% (w/v) sucrose were used for subcellular fractionation analysis. For the subcellular fractionation analysis under salt stress, ttl1ttl3ttl4 TTL3-GFP–etiolated seedlings were grown for 3 days in half-strength MS agar-solidified medium with 1% (w/v) sucrose and then transferred to half-strength MS liquid medium with 1% (w/v) sucrose (control) or supplemented with 200 mM NaCl. Samples were collected 0.5 and 28 hours after the salt stress imposition. For the subcellular fractionation analysis without salt stress of TTL3-GFP in WT and prc1-1, etiolated seedlings were grown for 3 days in half-strength MS agar-solidified medium with 1% (w/v) sucrose.
The total, microsomal, and soluble fractions were prepared using a modified method that allows the use of a limited amount of tissue sample (37) with some modifications. Briefly, 100 mg of 3-day-old etiolated seedlings was grounded in 1.5-ml microcentrifuge tubes using polypropylene pestles with 1.5× EB buffer on ice. The concentrated 1.5× EB was used to account for the tissue water content of etiolated seedlings. The 1.0× EB consisted of 100 mM tris-HCl (pH 7.5), 25% (w/w) (0.81 M) sucrose, 5% (v/v) glycerol, 10 mM EDTA (pH 8.0), 5 mM EGTA (pH 8.0), 5 mM KCl, and 1 mM DTT. To inhibit proteolysis and phosphatase activity, the 1.5× EB was supplemented with 0.2% (w/v) BSA, 50 mM sodium fluoride (NaF), 2 mM sodium molybdate (Na2MoO4), 1 mM Pefabloc-SC (AEBSF), and 1% (v/v) protease inhibitor cocktail (ref: P9599, Sigma-Aldrich). The homogenate was transferred to the prepared Polyvinylpolypyrrolidone (PVPP) pellets that were previously equilibrated in 200 mM tris-HCl (pH 7.5), 40% (w/w) (1.37 M) sucrose, 20 mM EDTA (pH 8.0), 20 mM EGTA (pH 8.0), 10 mM KCl, and 0.4% BSA; mixed (vortex for 30 s); and left for 5 min for the PVPP to adsorb phenolic compounds. Samples were cleared by centrifugation (600g, 3 min, 4°C). The supernatants were saved, and pellets were reextracted twice using 1.1× EB and 1.0× EB, respectively. Combined supernatants were recleared (600g, 3 min at 4°C), aliquots for total protein were saved, and combined supernatants were centrifuged (21,000g for 1 hour and 40 min at 4°C) to yield total membrane pellets. Supernatants were saved for the soluble protein fraction analysis. The membrane pellets were washed with wash buffer that consists of 20 mM tris-HCl (pH 7.5), 5 mM EDTA, 5 mM EGTA, 1 mM PMSF, and 1% (v/v) protease inhibitor cocktail and centrifuged (21,000g, 45 min). Then, pellets were resuspended in storage buffer that consists of 20 mM tris-HCl (pH 7.5), 5 mM EDTA, 5 mM EGTA, 20% glycerol, 1 mM DTT, 50 mM NaF, 2 mM Na2MoO4, 1 mM AEBSF, and 1% (v/v) protease inhibitor cocktail. For immunoblot analysis, total and soluble fractions were 1× concentrated compared with microsomal protein extract. Samples were diluted in 2× Laemmli buffer [125 mM tris-HCl (pH 6.8), 4% (w/v) SDS, 20% (v/v) glycerol, 2% (v/v) β-mercaptoethanol, and 0.01% (w/v) bromophenol blue], denatured by heating samples for 45 min at 70°C, separated in a 10% SDS-PAGE gel, and analyzed as described in the “Immunoblot analysis” section.
Immunoblot analysis
Proteins separated by SDS-PAGE were blotted using Trans-blot Turbo Transfer System (Bio-Rad) or wet electroblotting systems (Bio-Rad) onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P, Millipore) following the instructions by the manufacturer. PVDF membranes, containing electroblotted proteins, were then incubated with the appropriate primary antibody followed by the appropriate secondary peroxidase-conjugated antibody. The following primary antibodies were used for detection of epitope-tagged proteins: mouse monoclonal anti-GFP clone B-2 (1:1000; catalog no. sc-9996, Santa Cruz Biotechnology), mouse monoclonal anti-HA clone HA-7 (1:3000; catalog no. H3663, Sigma-Aldrich), and rabbit polyclonal anti-SYT1 antibody (1:5000). The secondary antibodies used in this study were anti-mouse immunoglobulin G (IgG) whole-molecule peroxidase (1:80,000; catalog no. A9044, Sigma-Aldrich) and anti-rabbit IgG whole-molecule peroxidase (1:80,000; catalog no. A0545, Sigma-Aldrich). Proteins and epitope-tagged proteins on immunoblots were detected using the Clarity ECL Western Blotting Substrate or SuperSignal West Femto Maximum Sensitivity Substrate according to the manufacturer’s instructions, and images of different time exposures were acquired using the Chemidoc XRS1 System (Bio-Rad). Only images with no saturated pixels were used for protein quantification. Immunoblotted PVDF membranes were stained with Coomassie brilliant blue R-250 to confirm equal loading of the different samples in each experiment.
Heterologous protein expression
6xHis-TTL3IDR and 6xHis-SUMO3-TTL3 were expressed in ArcticExpress (DE3) E. coli cells (Agilent). A starter culture was grown overnight at 28°C and used to inoculate the main cultures at a ratio of 1:10. Cultures were grown at 37°C until an OD600 of ~0.6 was reached. Cultures were moved to 12°C, and protein expression was induced when cultures were cooled down by the addition of isopropyl β-d-1-thiogalactopyranoside at a final concentration of 1 mM. Cells were collected after overnight incubation by centrifugation at 5000g and washed in 150 mM NaCl. Pellets were resuspended in lysis buffer [50 mM tris-HCl, 200 mM NaCl, and 20 mM imidazole (pH 7.4) for 6xHis-TTL3IDR and 50 mM Hepes, 200 mM NaCl, and 20 mM imidazole (pH 7.2) for 6xHis-SUMO3-TTL3]. From here on, all following tris-based buffers were used for 6xHis-TTL3IDR and Hepes-based buffers for 6xHis-SUMO3-TTL3. Cell lysates were prepared by passing the solution through a Microfluidics M-110P homogenizer (Microfluidics Corp., USA) at 151.7 MPa for four times. Cellular debris was spun down at 20,000g, and supernatant was collected and filtered through a 0.2-μm syringe filter. Recombinant proteins were purified using Ni Sepharose High Performance HisTrap HP columns (GE Healthcare Life Sciences, USA) and an ÄKTA pure 25L (GE Healthcare Life Sciences, USA) equipped with a sample pump. Buffers for protein purification were prepared as follows: buffer A: 50 mM tris-HCl (pH 7.4) and 200 mM NaCl or 50 mM Hepes (pH 7.2) and 200 mM NaCl; buffer B: 50 mM tris-HCl (pH 7.4), 200 mM NaCl, and 500 mM imidazole or 50 mM Hepes (pH 7.2), 200 mM NaCl, and 500 mM imidazole. Sample application onto the column was performed at 1 ml/min with 96% A and 4% B. The column was washed stepwise at 1 ml/min as follows: 20 column volumes (CVs) of 96% A and 4% B and 20 CV of 90% A and 10% B. Final protein sample was eluted at 1 ml/min for 20 CV with a linear gradient starting at 90% A and 10% B until 0% A and 100% B were reached. Elution was performed in upflow mode. Samples were collected in 2-ml fractions. Fractions enriched with protein were combined and gel filtered to remove imidazole using PD-10 Desalting Columns (GE Healthcare, USA) according to the gravity protocol in the manual. The column was equilibrated with 50 mM tris-HCl (pH 7.4) and 200 mM NaCl or 50 mM Hepes (pH 7.2) and 200 mM NaCl. Proteins were concentrated at 12°C and 3000g using Macrosep Advance Centrifugal Devices with a 3 K or 30 K cutoff (Pall Corporation, USA). 6xHis-TTL3IDR was snap-frozen and stored at −80°C until further use. 6xHis-SUMO3-TTL3 was cut with SUMO protease [bdSENP1 (38)] in a ratio of 500:1 at 12°C for 4 hours under gentle agitation. The cleavage products were separated by size exclusion chromatography using a HiLoad 16/600 Superdex 200-pg (Cytiva, U.S.A.) column. The column was equilibrated in storage buffer [50 mM Hepes (pH 7.2), 200 mM NaCl, and 10% glycerol], and cleavage products were separated at a flow rate of 1 ml/min for 1.5 CV. Fraction collection was initiated after 0.2 CV. The final protein was concentrated as described above and snap-frozen until further use. Isoelectric point (pI) of proteins was calculated using the Expasy ProtParam tool (https://web.expasy.org/protparam/).
Microtubule affinity and turbidity assays
Microtubule spin-down and turbidity assays were performed as described in detail before (1, 10). Briefly, to determine the dissociation constant, a constant amount (7.5 μg) of 6xHis-TTL3IDR or TTL3 was incubated with increasing concentrations of polymerized microtubules ranging from 0 to 7.2 μM (calculated on the basis of the molecular weight of a tubulin dimer). Please note that all microtubule dilutions were derived from the same 18 μM taxol-stabilized microtubule stock solution. Thus, microtubule length or composition does not change with varying concentrations. Tween 20 (0.05%) was added to all reactions. The samples were incubated for 30 min at room temperature and subsequently spun at 25,000g for 30 min at room temperature to pellet microtubules and bound proteins. Supernatant and pellet fractions were subjugated to SDS-PAGE, and protein levels in both supernatant and pellet fractions were analyzed using the Gel-function of Fiji. Final dissociation constant (KD) was estimated by fitting a saturation binding curve onto the data points with GraphPad Prism v9.0 (GraphPad Software Inc., USA).
Phylogenetic analysis
M. polymorpha and Physcomitrella patens were used as representatives from embryophytes (earliest land plants). A. thaliana TTL3 protein sequence was used as input for a PSI-BLAST analysis against M. polymorpha, P. patens, and all the National Center for Biotechnology Information organisms tagged as chlorophyta (taxid:3041) and charophytes (taxid:3146). In the case of M. polymorpha and P. patens, results with the smallest E value with IDR, TPR, and TRXL (a region with homology to class h thioredoxins but lacking essential Cys residues required for thioredoxin activity) domains present in the same sequence were used as representatives. For chlorophyta and charophytes, no results were obtained with IDR, TPR, and TRXL domains. Thus, in the case of chlorophyta, the four results with the smallest E value and with TPR and TRX (a region with homology to class h thioredoxins and conserving the essential Cys residues required for thioredoxin activity) domains were used as representatives; meanwhile, for charophytes, the only results came from Chara braunii. Domains were retrieved from InterPro using the outputs from MOBIdb_Lite (IDR), SMART (TPR), CDD (TRX), and InterPro (C. braunii domains). Domains were illustrated using IBS 1.0.3 (38) and Adobe Illustrator CC 2017 21.0.0. Alignment was carried out using the web version of MAFFT v.7 (https://mafft.cbrc.jp/alignment/server) (39, 40) with standard parameters. Phylogenetic analysis was performed with MEGA-X (4142) [version 10.1.7, neighbor-joining method, bootstrap method, 500 replications (numbers from 0 to 100), substitution model: p-distance (numbers from 0 to 1), uniform rate among sites, gaps treatment: pairwise deletion].
Statistical analysis and experimental design
For statistical analyses, Welch’s unpaired t test, two-tailed unpaired t test, one-way analysis of variance (ANOVA), Welch’s ANOVA, or two-way ANOVA was performed using GraphPad Prism 9.3. If appropriate, Dunnett’s T3 multiple comparisons test or Tukey’s multiple comparisons test was performed afterward. A P value of <0.05 was considered as statistically significant. Statistical methods used to calculate P values are described in the figure legends. Data analysis (especially for images) was either done automatically, or file names were removed before the analysis. For the measurement of plant size, investigators were not blinded. In this case, data were always collected according to the genotype of plants. Sample size was determined for each experiment based on similar data reported in scientific literature.
Acknowledgments
We are grateful to K. Scklodowski for generating the CESA constructs used for the Y2H assays and to O. Voinnet for critical reading of the manuscript. Live-cell imaging was performed with equipment maintained by the Scientific Center for Optical and Electron Microscopy (ScopeM, ETH-Zürich) and by the Center for Advanced Bioimaging (CAB) Denmark.
Funding: This work was funded by the Spanish Ministry for Science and Innovation (MCIN/AEI/ 10.13039/501100011033 1-PID2020-114419RB-I00, PGC2018-098789-B-I00, and PID2019-107657RB-C22 to M.A.B., N.R.-L., and A.G.C., respectively), the Andalusian Research Plan cofinanced by the European Union (PAIDI 2020-PY20_00084 and UMA20-FEDERJA-023) to M.A.B. and Junta de Andalucía UMA-FEDER project (UMA18-FEDERJA-154) to N.R.-L., and the Swiss National foundation to C.S.-R. (SNF 31003A_163065/1 to AM). C.K. was supported by a Peter und Traudl Engelhorn-Stiftung fellowship, an ETH Career Seed Grant (SEED-05 19-2) of the ETH Foundation, an Emerging Investigator grant (NNF20OC0060564) of the Novo Nordisk Foundation, and an Experiment grant (R346-2020-1546) of the Lundbeck foundation. Á.G.-M. was supported by BES-2015-071256 and EMBO Short-Term 7632 Fellowships. F.P. was supported by and FPU19/02219 fellowship. V.A.-S. was supported by an Emerging Investigator research project (UMA20-FEDERJA-007) and cofinanced by the “Programa Operativo FEDER 2014-2020” and by the “Consejería de Economía y Conocimiento de la Junta de Andalucía”. Y.J. was funded by the Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 101001097).
Author contributions: M.A.B., C.S.-R., C.K., Á.G.-M., and V.A.-S. designed the research. C.K. conducted imaging and image analysis, heterologous protein expression, and microtubule-based assays. Á.G.-M. and V.A.-S. generated the plant lines and the physiological and the biochemical data. A.M. conducted cellulose measurements. A.G.C. performed Y2H studies. F.P. performed the physiological and the phylogenetic analysis. L.A. and Y.J. contributed to lines generation. N.R.-L. contributed to biochemical analyses. Data analyses were led by C.K., Á.G.-M., V.A.-S., C.S.-R., and M.A.B. C.K., V.A.-S., C.S.-R., and M.A.B. wrote the manuscript with major contribution from Y.J. All authors read and provided feedback on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Table S1
Other Supplementary Material for this : manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Y. M. Bar-On, R. Phillips, R. Milo,The biomass distribution on Earth. Proc. Natl. Acad. Sci. 115,6506–6511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. R. Paredez, C. R. Somerville, D. W. Ehrhardt,Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312,1491–1495 (2006). [DOI] [PubMed] [Google Scholar]
- 3.J. R. Pear, Y. Kawagoe, W. E. Schreckengost, D. P. Delmer, D. M. Stalker,Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. 93,12637–12642 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.T. Vain, E. F. Crowell, H. Timpano, E. Biot, T. Desprez, N. Mansoori, L. M. Trindade, S. Pagant, S. Robert, H. Höfte, M. Gonneau, S. Vernhettes,The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 165,1521–1532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Y. Gu, N. Kaplinsky, M. Bringmann, A. Cobb, A. Carroll, A. Sampathkumar, T. I. Baskin, S. Persson, C. R. Somerville,Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. 107,12866–12871 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A. Endler, C. Kesten, R. Schneider, Y. Zhang, A. Ivakov, A. Froehlich, N. Funke, S. Persson,A mechanism for sustained cellulose synthesis during salt stress. Cell 162,1353–1364 (2015). [DOI] [PubMed] [Google Scholar]
- 7.C. Kesten, A. Wallmann, R. Schneider, H. E. McFarlane, A. Diehl, G. A. Khan, B.-J. van Rossum, E. R. Lampugnani, W. G. Szymanski, N. Cremer, P. Schmieder, K. L. Ford, F. Seiter, J. L. Heazlewood, C. Sánchez-Rodríguez, H. Oschkinat, S. Persson,The companion of cellulose synthase 1 confers salt tolerance through a tau-like mechanism in plants. Nat. Comm. 10,857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. Rosado, A. L. Schapire, R. A. Bressan, A. L. Harfouche, P. M. Hasegawa, V. Valpuesta, M. A. Botella,The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol. 142,1113–1126 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.N. Lakhssassi, V. G. Doblas, A. Rosado, A. E. del Valle, D. Posé, A. J. Jiménez, A. G. Castillo, V. Valpuesta, O. Borsani, M. A. Botella,The Arabidopsis tetratricopeptide thioredoxin-like gene family is required for osmotic stress tolerance and male sporogenesis. Plant Physiol. 158,1252–1266 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.V. Amorim-Silva, A. Garcia-Moreno, A. G. Castillo, N. Lakhssassi, A. E. D. Valle, J. Pérez-Sancho, Y. Li, D. Posé, J. Pérez-Rodriguez, J. Lin, V. Valpuesta, O. Borsani, C. Zipfel, A. P. Macho, M. A. Botella,TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in Arabidopsis. Plant Cell 31,1807–1828 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.S.-L. Xu, A. Rahman, T. I. Baskin, J. J. Kieber,Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20,3065–3079 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. K. Polko, J. J. Kieber,The regulation of cellulose biosynthesis in plants. Plant Cell 31,282–296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.R. Gutierrez, J. J. Lindeboom, A. R. Paredez, A. M. C. Emons, D. W. Ehrhardt,Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11,797–806 (2009). [DOI] [PubMed] [Google Scholar]
- 14.B. L. Goode, S. C. Feinstein,Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J. Cell Biol. 124,769–782 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.C. Kesten, R. Schneider, S. Persson,In vitro microtubule binding assay and dissociation constant estimation. Bio-protocol 6,e1759 (2016). [Google Scholar]
- 16.M. Bringmann, E. Li, A. Sampathkumar, T. Kocabek, M.-T. Hauser, S. Persson,POM-POM2/CELLULOSE SYNTHASE INTERACTING1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24,163–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. Li, L. Lei, C. R. Somerville, Y. Gu,Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc. Natl. Acad. Sci. 109,185–190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.E. F. Crowell, V. Bischoff, T. Desprez, A. Rolland, Y.-D. Stierhof, K. Schumacher, M. Gonneau, H. Höfte, S. Vernhettes,Pausing of golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21,1141–1154 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.N. Ruiz-Lopez, J. Pérez-Sancho, A. E. del Valle, R. P. Haslam, S. Vanneste, R. Catalá, C. Perea-Resa, D. V. Damme, S. García-Hernández, A. Albert, J. Vallarino, J. Lin, J. Friml, A. P. Macho, J. Salinas, A. Rosado, J. A. Napier, V. Amorim-Silva, M. A. Botella,Synaptotagmins at the endoplasmic reticulum–plasma membrane contact sites maintain diacylglycerol homeostasis during abiotic stress. Plant Cell 33,2431–2453 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.T. Desprez, M. Juraniec, E. F. Crowell, H. Jouy, Z. Pochylova, F. Parcy, H. Höfte, M. Gonneau, S. Vernhettes,Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 104,15572–15577 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.L. Xie, C. Yang, X. Wang,Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 62,4495–4506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Sánchez-Rodríguez, K. Ketelaar, R. Schneider, J. A. Villalobos, C. R. Somerville, S. Persson, I. S. Wallace,BRASSINOSTEROID INSENSITIVE2 negatively regulates cellulose synthesis in Arabidopsis by phosphorylating cellulose synthase 1. Proc. Natl. Acad. Sci. 114,3533–3538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.E. R. Lampugnani, E. Flores-Sandoval, Q. W. Tan, M. Mutwil, J. L. Bowman, S. Persson,Cellulose synthesis - central components and their evolutionary relationships. Trends Plant Sci. 24,402–412 (2019). [DOI] [PubMed] [Google Scholar]
- 24.T. Desnos, V. Orbović, C. Bellini, J. Kronenberger, M. Caboche, J. Traas, H. Höfte,Procuste1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively in dark- and light-grown Arabidopsis seedlings. Development 122,683–693 (1996). [DOI] [PubMed] [Google Scholar]
- 25.A. Sampathkumar, R. Gutierrez, H. E. McFarlane, M. Bringmann, J. Lindeboom, A.-M. Emons, L. Samuels, T. Ketelaar, D. W. Ehrhardt, S. Persson,Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 162,675–688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.B. E. Lindsey III, L. Rivero, C. S. Calhoun, E. Grotewold, J. Brkljacic,Standardized method for high-throughput sterilization of Arabidopsis seeds. J. Vis. Exp. e56587, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D. G. Gibson, L. Young, R.-Y. Chuang, J. C. Venter, C. A. Hutchison, H. O. Smith,Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6,343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 28.S. J. Clough, A. F. Bent,Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16,735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 29.M. Fenech, V. Amorim-Silva, A. E. del Valle, D. Arnaud, N. Ruiz-Lopez, A. G. Castillo, N. Smirnoff, M. A. Botella,The role of GDP-l-galactose phosphorylase in the control of ascorbate biosynthesis. Plant Physiol. 185,1574–1594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Y. Kadota, A. P. Macho, C. Zipfel,Immunoprecipitation of Plasma Membrane Receptor-Like Kinases for identification of phosphorylation sites and associated proteins. J. Signal Transduct. 1363,133–144 (2016). [DOI] [PubMed] [Google Scholar]
- 31.J. Verbančič, J. J. Huang, H. E. McFarlane,Analysis of cellulose synthase activity in Arabidopsis using spinning disk microscopy. Star Protoc. 2,100863 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona,Fiji: An open-source platform for biological-image analysis. Nat. Methods 9,676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.P. Thévenaz, U. E. Ruttimann, M. Unser,A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7,27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 34.F. Ruhnow, D. Zwicker, S. Diez,Tracking single particles and elongated filaments with nanometer precision. Biophys. J. 100,2820–2828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. Kesten, F. M. Gámez-Arjona, A. Menna, S. Scholl, S. Dora, A. I. Huerta, H.-Y. Huang, N. Tintor, T. Kinoshita, M. Rep, M. Krebs, K. Schumacher, C. Sánchez-Rodríguez,Pathogen-induced pH changes regulate the growth-defense balance in plants. EMBO J. 38,e101822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. Bolte, F. P. Cordelières,A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224,213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 37.L. Abas, C. Luschnig,Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal. Biochem. 401,217–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. Frey, D. Görlich,A new set of highly efficient, tag-cleaving proteases for purifying recombinant proteins. J. Chromatogr. A 1337,95–105 (2014). [DOI] [PubMed] [Google Scholar]
- 39.W. Liu, Y. Xie, J. Ma, X. Luo, P. Nie, Z. Zuo, U. Lahrmann, Q. Zhao, Y. Zheng, Y. Zhao, Y. Xue, J. Ren,IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31,3359–3361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.S. Kuraku, C. M. Zmasek, O. Nishimura, K. Katoh,aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41,W22–W28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.K. Katoh, J. Rozewicki, K. D. Yamada,MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20,1160–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. Kumar, G. Stecher, M. Li, C. Knyaz, K. Tamura,MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35,1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1
Movie S1