Abstract
Chronic exposure to airborne carbon black ultrafine (nCB) particles generated from incomplete combustion of organic matter drives IL-17A–dependent emphysema. However, whether and how they alter the immune responses to lung cancer remains unknown. Here, we show that exposure to nCB particles increased PD-L1+ PD-L2+ CD206+ antigen-presenting cells (APCs), exhausted T cells, and Treg cells. Lung macrophages that harbored nCB particles showed selective mitochondrial structure damage and decreased oxidative respiration. Lung macrophages sustained the HIF1α axis that increased glycolysis and lactate production, culminating in an immunosuppressive microenvironment in multiple mouse models of non–small cell lung cancers. Adoptive transfer of lung APCs from nCB-exposed wild type to susceptible mice increased tumor incidence and caused early metastasis. Our findings show that nCB exposure metabolically rewires lung macrophages to promote immunosuppression and accelerates the development of lung cancer.
Air pollutant particle, ultrafine carbon black, impairs lung macrophage mitochondrial function and increases lung cancer risk.
INTRODUCTION
Long-term inhalation of ultrafine particles causes one in eight deaths worldwide (1). The harmful effects of inhaled particles are driven by the particles’ physical characteristics, including their size and polarity (2, 3). In addition to environmentally (e.g., wildfire and incinerating organic matter in burn pits) and industrially generated hydrophobic nano-sized (15 to 75 nm) carbon black (nCB) particles (4, 5), tobacco combustion generates nCB accumulation in the lung macrophages and induces double-stranded DNA breaks (6, 7). Mice exposed to nCB develop emphysema through mechanisms that depend on differentiation of T helper 17 (TH17) cells and inflammasome activation (6, 7). Although exposure to ultrafine particles is associated with increased mortality (8), whether and how ultrafine particles play a role in lung cancer development and metastasis remains unclear.
Chronic exposure to ultrafine particles leads to recruitment of inflammatory cells and increased abundance of proinflammatory cytokines, including interleukin-1β (IL-1β), IL-6, and tumor necrosis factor (TNF) in the lungs and circulation (6). High concentrations of IL-6, IL-1β, and IL-17A in the lungs of cigarette smokers and individuals exposed to nCB drive emphysema (6, 9). We have previously shown that nCB-driven inflammasome activation in the lung phagocytic cells is essential for emphysema progression (6); however, it remains unclear how nCB changes macrophage cellular functions. In addition, reactive oxygen species (ROS) promote inflammation in animal models of ultrafine particle–mediated lung disease (10) by altering macrophage and epithelial cell function (11). Although anti-inflammatory and antioxidant interventions have been used to treat lung injury (12), they fail to resolve inflammation because ultrafine particles persist in the lungs long after exposure ceases.
Mitochondria function as signaling and energy hubs that are sensitive to oxidative stress (13). Mitochondrial ROS (mtROS) can initiate inflammasome assembly and activate downstream caspases (14), damaging mitochondria and amplifying the inflammatory responses (15). Conversely, the anti-inflammatory cytokine, IL-10, induces autophagy to remove damaged mitochondria, restore oxidative phosphorylation (OXPHOS), and resolve inflammation (16).
Metabolites provide critical environmental cues that reprogram immune cell effector functions. For instance, lactate, a tumor-derived oncometabolite, acidifies the tumor microenvironment, providing a selective advantage for the development of regulatory T (Treg) cells (17) and thus assisting tumor-mediated immune escape (18). Mice bearing lactate dehydrogenase A (Ldha)–deficient tumors have fewer myeloid-derived suppressive cells and harbor a high number of cytotoxic natural killer (NK) cells compared with controls (19). Lactate also fuels the tricarboxylic acid cycle in non–small cell lung cancer (NSCLC) (20), and excessive lactate in the tumor microenvironment triggers tumor invasion (18) and hinders treatment efficacy (21).
We examined whether and how nCB present in the lungs of heavy smokers and a highly ubiquitous pollutant generated from combustion of organic matter promotes lung cancer. We found that, independent of tumor subtype, nCB increased expression of glycolytic pathways in lung antigen-presenting cells (APCs) and macrophages. We also identified mammalian target of rapamycin complex 1 (mTORC1)–hypoxia-inducible factor 1 α (HIF1α) as the key nCB-mediated mechanism that facilitates lactate expression in macrophages, which supports tolerogenic immune cells in the lungs, decreases cancer latency, and promotes metastasis. Together, our findings show how nCB accumulation in the lungs promotes NSCLC and poses a danger to public health.
RESULTS
nCB in the lungs accelerates NSCLC
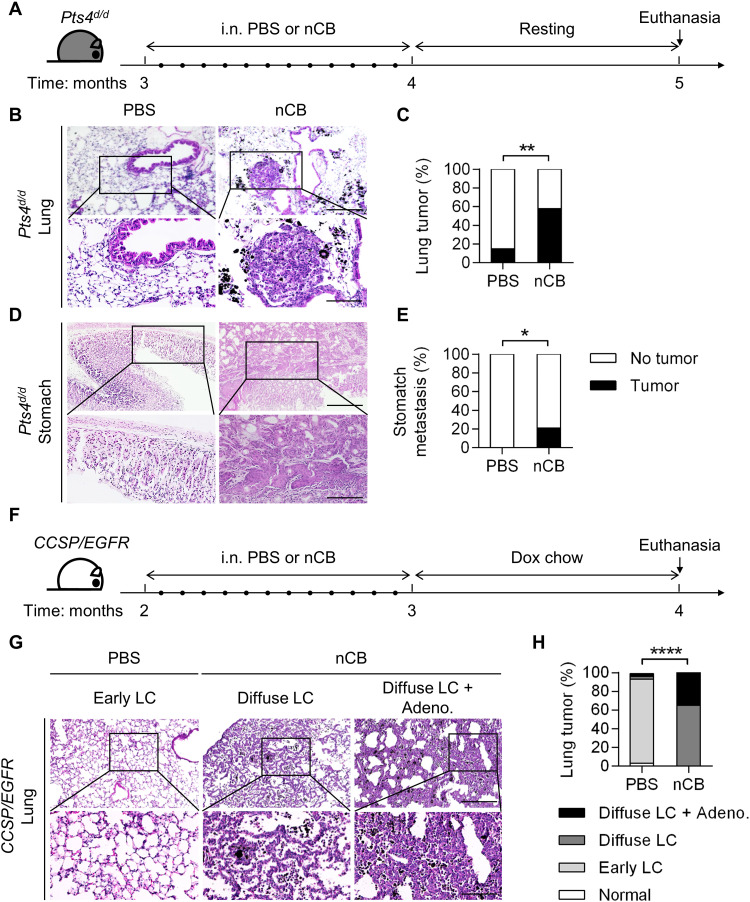
Mice with Cre recombinase under the club cell secretory protein (CCSP) promoter specifically delete floxed Pten and Smad4 in the airway (Pts4d/d) and spontaneously develop metastatic NSCLC with adenosquamous features (22). Approximately 20% of Pts4d/d mice develop localized tumors by 5 months of age, and 100% develop metastatic tumors by 1 year of age (22). Therefore, to determine whether nCB promotes lung cancer, we exposed 3-month-old mice intranasally to concentrations of nCB equal to those found in the lungs of heavy smokers (6). Control mice of the same age and genotype received intranasal phosphate-buffered saline (PBS). The mice were euthanized at 5 months of age, and their organs were harvested for histology (Fig. 1A). The nCB-exposed mice showed an increased incidence of lung tumors (58%, 11 of 19) when compared with control mice of the same genotype (15%, 3 of 20; Fig. 1, B and C). We found that 21% (4 of 19) of the nCB-exposed mice developed distant metastases, which were not expected in 5-month-old Pts4d/d mice (22) and were absent in the PBS-treated Pts4d/d mice (Fig. 1, D and E).
Fig. 1. nCB accelerates NSCLC development and metastasis in mice.
(A) Schematic diagram of nCB exposure in Pts4d/d mice. Three-month-old Pts4d/d mice (C57BL/6 background) were exposed to sterile PBS or nCB (20 mg/ml in sterile PBS) 12 times over a period of 1 month and euthanized after an additional month. i.n., intranasal (B and C) Representative images and (D and E) summary statistics of hematoxylin and eosin (H&E)–stained lung and stomach tissues from the indicated groups of mice at 5 months of age. Scale bars, (top) 300 μm and (bottom) 250 μm. PBS, n = 20; and nCB, n = 19. (F) Schematic diagram of nCB exposure in CCSP/EGFR mice. Two-month-old bitransgenic mice (FVB background) were exposed to sterile PBS or nCB (20 mg/ml in sterile PBS) 12 times over a period of 1 month, followed by a diet of doxycycline chow for an additional month to induce lung cancer. The mice were euthanized at 4 months of age. (G) Representative images of H&E staining showing lung cancer development. Early lepidic carcinoma (LC) was found in the PBS group. Diffuse LC alone or with adenocarcinoma (Adeno.) was found in the nCB group. Scale bars, (top) 300 μm and (bottom) 250 μm. (H) Summary statistics of the results shown in (G). PBS, n = 30; and nCB, n = 26. Significance was determined by chi-square and Fisher’s tests. *P < 0.05, **P < 0.01, and ****P < 0.0001.
We next examined whether nCB could promote tumors in a different model of lung cancer. We used mice expressing mutant human EGFRL858R under control of the CCSP promoter governed by a tet operator (TetO), which is regulated by the reverse tetracycline transactivator (rtTA) (23). This bitransgenic mouse model (CCSP/EGFR) develops nonmucinous lepidic carcinoma (LC), in which tumor cells arise from alveolar type II cells and extend into the alveolar walls. Two-month-old CCSP/EGFR mice were intranasally exposed to nCB or PBS for 1 month, followed by doxycycline administration for another month to induce EGFRL858R activation (Fig. 1F). We found that the nCB-exposed CCSP/EGFR mice had a higher incidence of diffuse LC and adenocarcinoma (35%, 9 of 26) than the PBS-exposed mice (3%, 1 of 30; Fig. 1, G and H). Our findings in both mouse models of lung cancer indicate that nCB accelerates NSCLC progression by a mechanism that is independent of the tumor genotype.
nCB increases lactate in lungs and macrophages
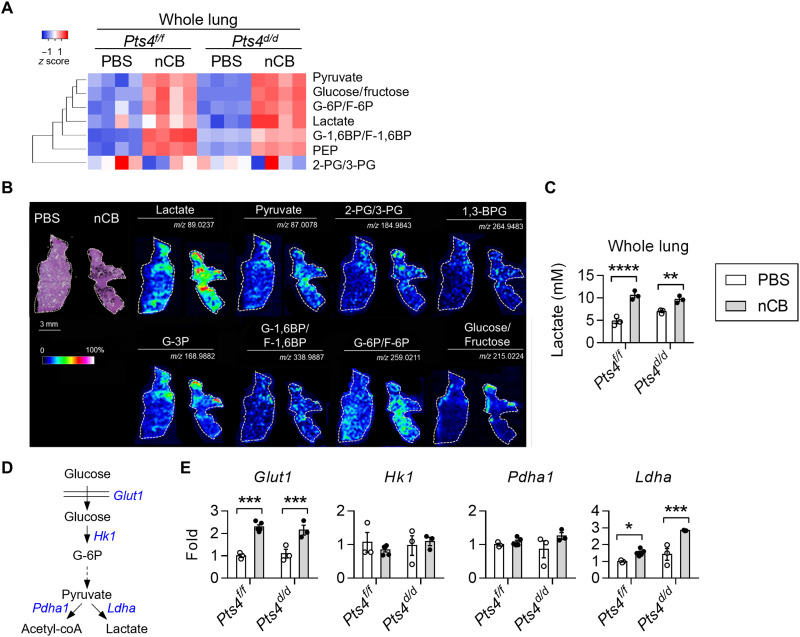
Because metabolic reprogramming is a hallmark of cancer, we next asked whether nCB exposure alters the lung metabolome. An unbiased whole liquid chromatography–mass spectrometry (LC-MS) analysis of control (floxed Pten and Smad4; Pts4f/f) and Pts4d/d mice treated with either nCB or PBS showed substantial increases in multiple metabolic systems, including glycolysis in the nCB-exposed lungs (Fig. 2A and fig. S1A). We next visualized the metabolite distribution in lung tissues by using desorption electrospray ionization (DESI) MS imaging. Lung sections were used to develop metabolite maps reconstructed from MS data and then stained with hematoxylin and eosin (H&E). We found increased abundance of glycolysis metabolites in the nCB-exposed lungs as compared to PBS-treated controls (Fig. 2B). Lactate exhibited the strongest signal that colocalized with nCB deposition. We validated these findings by direct measurement of lactate in the lung supernatants, which consistently showed increased lactate concentrations in nCB-exposed lungs (Fig. 2C). Similarly, nCB-exposed CCSP/EGFR mice showed increased lactate production (fig. S1B). Interrogation of key enzymes in the glycolysis pathway showed that gene expression of Glut1 and Ldha, but not Hk1 and Pdha1, was increased in both nCB-exposed Pts4f/f and Pts4d/d lungs compared with PBS-exposed lungs of the same genotypes (Fig. 2, D and E). Collectively, our findings suggest that increased lactate production occurs in the lungs in response to nCB exposure independently of tumors or genes associated with lung cancer.
Fig. 2. Increased lung lactate level in response to nCB exposure.
(A) Heatmap of glycolysis metabolites in the lungs measured by targeted liquid chromatography–mass spectrometry (LC-MS). Rows are clustered hierarchically. n = 4 per group. (B) Desorption electrospray ionization (DESI) MS images of lungs from PBS-exposed or nCB-exposed Pts4f/f mice at 5 months of age; left: H&E staining of lung sections. The intensity of lactate, pyruvate, 2-phosphoglyceric acid (2-PG) or 3-PG, 1,3-bisphosphoglycerate (1,3-BPG), glyceraldehyde-3-phosphate (G-3P), glucose-1,6-bisphosphate (G-1,6BP) or fructose-1,6-bisphosphate (F-1,6BP), glucose-6-phosphate (G-6P) or fructose-6-phosphate (F-6P), and glucose or fructose detection is displayed with measured mass/charge ratio (m/z) values. Scale bars, 3 mm for H&E stains and 5 mm for metabolite images. Phosphoenolpyruvate (PEP). (C) Representative lactate expression in whole-lung supernatants from three independent experiments. n = 3 per group. (D) A simplified glycolysis diagram. Glucose is transported into cells by glucose transporter 1 (Glut1) and then catalyzed by the rate-limiting enzyme hexokinase 1 (Hk1) to form G-6P. After the formation of pyruvate, the pyruvate is catalyzed by pyruvate dehydrogenase 1 (Pdha1) to form acetyl–coenzyme A (coA) or by lactate dehydrogenase A (Ldha) to form lactate. (E) Representative real-time quantitative PCR (qPCR) quantification of the whole-lung mRNA expression from three independent experiments. n = 3 to 4 per group. Two-way ANOVA with Holm’s correction was applied to determine P values. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
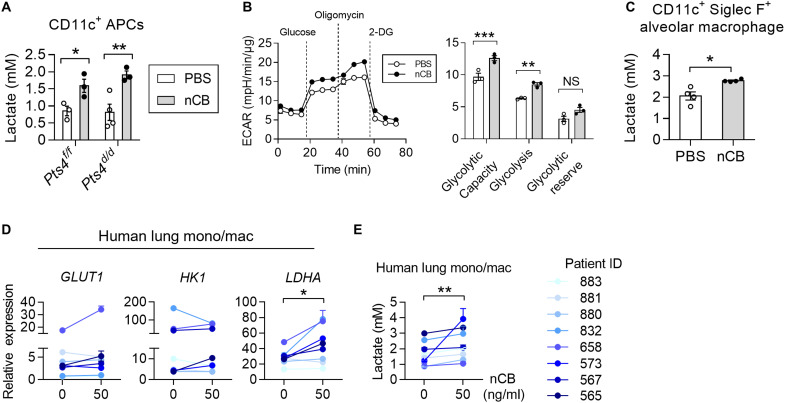
Tumor-derived lactate facilitates immune evasion by promoting M2 macrophage polarization that inhibits T cell effector functions, blocks monocyte trafficking, and hinders NK cell cytotoxicity (24, 25). We found that lactate expression increased in Pts4f/f control mice and tumor-bearing Pts4d/d mice after exposure to nCB, indicating additional sources of lactate production in the lungs in response to nCB. Because foreign particles are phagocytosed in the distal airway, we examined alveolar macrophage lactate production in response to nCB exposure. Because lung macrophages (i.e., alveolar and interstitial macrophages) express CD11c and >80% of the CD11c+ APCs are macrophages (26), we next isolated lung CD11c+ APCs from nCB-exposed Pts4f/f and Pts4d/d mice. These cells cultured in vitro showed higher lactate concentrations than cells isolated from PBS-exposed mice (Fig. 3A), suggesting that macrophages are a source of elevated lactate in nCB-exposed lungs.
Fig. 3. Increased lactate in mouse and human lung macrophages.
(A) Representative lactate production in lung CD11c+ APCs from three independent experiments. Cells were isolated from lungs of PBS-exposed or nCB-exposed Pts4f/f and Pts4d/d mice. n = 3 each group. (B) Representative Seahorse measurements from three independent experiments. CD11c+ Siglec F+ alveolar macrophages were treated with PBS or nCB (50 μg/ml) for 24 hours and used to measure ECAR with chemicals at various time points. ECAR values were normalized by total protein in the cells. n = 3 biological replicates per group. (C) Representative lactate expression in nCB-treated alveolar macrophages from three independent experiments. Lactate concentration in the supernatants was measured 24 hours after nCB treatment. n = 3 each group. (D) Quantitative measurements of glycolytic capacity, glycolysis, and glycolytic reserve are displayed on the right. Quantitative RNA transcripts of GLUT1, HK1, and LDHA in PBS-treated or nCB-treated human monocytes/macrophages (mono/macs). Human mono/macs were isolated from human lung tissues by negative selection with magnetic beads. Cells were treated with nCB (50 μg/ml) for 24 hours and collected to measure mRNA expression. Individual patient IDs are shown. (E) Lactate expression in whole-lung supernatants of human mono/macs. Paired t test and one-way or two-way ANOVA with Holm’s correction were applied to determine P values. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; NS, not significant.
Next, to determine whether glycolysis is increased in nCB-exposed CD11c+ Siglec F+ alveolar macrophages, we measured the extracellular acidification rate (ECAR) in real time by Seahorse analysis. We found increased glycolytic capacity and glycolysis but no alteration in glycolytic reserve in nCB-treated alveolar macrophages as compared to PBS-treated cells (Fig. 3B). Furthermore, alveolar macrophages consistently increased lactate secretion in response to nCB exposure (Fig. 3C). These results demonstrate that nCB up-regulates glycolysis in macrophages. We next examined the effect of nCB on human lung macrophages using magnetic beads to enrich monocytes/macrophages and exposed them to nCB. We found that nCB exposure resulted in significant increases in lactate production and LDHA expression and trends for elevated GLUT1 and HK1 expression in the human lung macrophages (Fig. 3, D and E).
Because dendritic cells (DCs) and lung epithelial cells also encounter and phagocytize ultrafine particles, we next determined their metabolic responses to nCB. Bone marrow–derived DCs (BMDCs) treated with nCB did not exhibit increased ECAR or lactate production compared with PBS-treated controls (fig S1, C and D). Similarly, BEAS-2B human lung bronchial epithelial cells and H1299 human lung cancer cells showed no significant change in glycolysis when exposed to PBS or nCB (fig. S1, E to G). Together, these data indicate that nCB induces lactate expression in macrophages but not in BMDCs or lung epithelial cells.
nCB promotes T cell exhaustion and FOXP3+ Treg cells
Lactate has been shown to direct T cell development toward tolerogenic FOXP3+ Treg cells and to inhibit effector T cell function (17, 27). We found increased infiltration of immune cells (fig. S2, A and B), and markers of CD4 T cell activation (CD25+ CD44+ and CD25+ CD69+) were increased in lung lymphocytes of Pts4f/f and Pts4d/d mice exposed to nCB (fig. S2, C and D), suggesting that T cell responses were independent of the presence of lung tumors. We next examined the relative abundance of programmed cell death protein 1 (PD-1), a primary inhibitory receptor and exhaustion marker (28), in the lungs of Pts4f/f and Pts4d/d mice exposed to nCB or PBS. Compared with the PBS-exposed lungs, the nCB-exposed lungs had increased infiltration by PD-1+ CD4 T cells (fig. S2, E and F). PD-1+ CD8+ T cells were up-regulated in the nCB-exposed lungs of Pts4f/f mice, but not in those of Pts4d/d mice, compared with the respective PBS-exposed controls (fig. S2, G and H). We also detected increased relative abundance of FOXP3+ Treg cells in response to nCB exposure (fig. S2, I and J). Exposure to nCB had no significant effect on the abundance of transforming growth factor–β (TGFβ), an anti-inflammatory cytokine required for Treg cell differentiation (29), in the lungs of Pts4f/f and Pts4d/d mice (fig. S2K). In contrast, IL-10, another anti-inflammatory cytokine, was elevated in nCB-exposed Pts4f/f and Pts4d/d lungs (fig. S2L). We examined the ratio of CD8+ T cells to Treg cells (CD8/Treg) as this cell ratio has been used as a prognostic indicator in solid tumors (30). The CD8/Treg ratio was decreased in nCB-exposed Pts4d/d lungs but not in nCB-exposed Pts4f/f lungs (fig. S2M). Similarly, expression of Gzmb, encoding the cytotoxic molecule granzyme B, was decreased in lung tissues from nCB-exposed Pts4d/d mice as compared to PBS-treated controls (fig. S2N).
In accordance with our previous findings (6), nCB selectively promoted TH17, but not TH1, responses in Pts4f/f mice and Pts4d/d mice (fig. S2, O and P). We previously showed that exposure to cigarette smoke or nCB induces IL-6, IL-1β, and TH17 responses in the lungs of mice and humans (6, 9, 31). In line with these results, we found that nCB-exposed Pts4f/f and Pts4d/d mice had elevated concentrations of TNF, IL-6, and IL-1β in bronchoalveolar lavage fluid (BAL; fig. S2, Q to S). Similarly, the numbers of PD-1+ CD4+ and CD8+ T cells were increased (fig. S2, T and U) in CCSP/EGFR lungs. Notably, TGFβ and IL-10 were increased, but Gmzb showed a downward trend in nCB-exposed CCSP/EGFR mice (fig. S2, V to X).
To further characterize CD4 T cell populations in our model, we performed cytometry by time of flight (CyTOF) using mice exposed to the same experimental conditions (fig. S3A). Our analysis showed increased relative abundance of CD4+ and CD8+ PD-1+ cytotoxic T lymphocyte–associated protein 4 (CTLA4+) T cells and CD25+ FOXP3+ Treg cells in Pts4f/f and Pts4d/d mice exposed to nCB (fig. S3, B to D). Although the relative abundance of CD4 T cells expressing RORγT+ or T-bet+, the canonical transcription factors for TH17 and TH1, respectively, was unaltered, nCB induced PD-1 and CTLA4 expression in RORγT+ cells and T-bet+ CD4+ cells (fig. S3, E to H). We found a trend for increased relative abundance of RORγT+ FOXP3+ CD4+ T cells (fig. S3I). These findings suggest that nCB induces PD-1+ CTLA4+ T cell and Treg cells in the lungs.
nCB activates the HIF1α pathway in the macrophages
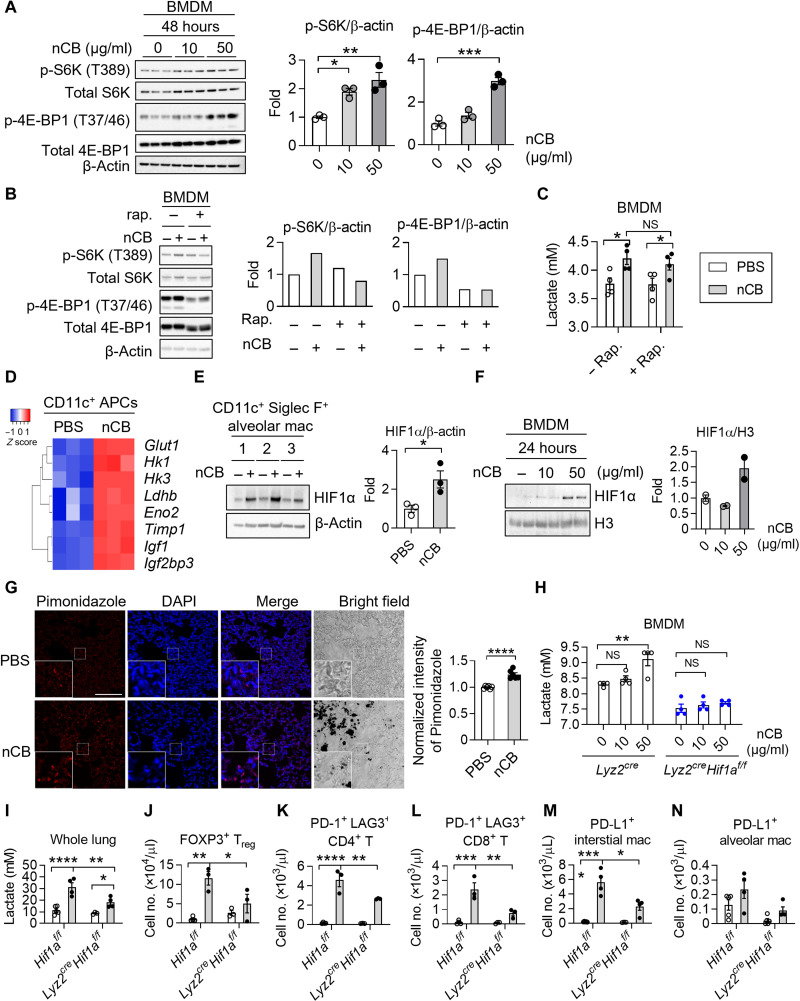
Prior studies showed that, in response to bacteria, the mTORC1 transiently up-regulates glycolysis in macrophages (16). To determine whether nCB changes mTORC1 activation in macrophages, we measured two downstream targets of mTORC1: p-S6K (T389) and p-4E-BP1 (T37/46). We found that nCB exposure resulted in dose-dependent and time-dependent increases in p-S6K and p-4E-BP1 levels in bone marrow–derived macrophages (BMDMs) that persisted after 48 hours (Fig. 4A and fig. S4A). As expected, nCB-treated BMDCs did not exhibit mTORC1 activation, indicating macrophage-specific responses (fig. S4B). We next used rapamycin to inhibit mTORC1 activation in BMDMs to determine whether mTORC1 activation is required for lactate production. The rapamycin treatment caused a decrease in mTORC1 activation, as determined by reduced p-4E-BP1 and p-S6K expression (Fig. 4B). Despite a reduced mTORC1 signaling, the rapamycin treatment failed to suppress lactate production in nCB-treated BMDMs (Fig. 4C). These findings suggest that although nCB activates mTORC1, other pathways control lactate production and glycolysis in nCB-exposed macrophages. To identify the upstream mechanism that controls glycolysis, we analyzed a microarray dataset (GSE72734) (7) showing gene expression in lung CD11c+ APCs from wild-type (WT) mice exposed to either PBS or nCB. Pathway analysis revealed that several high-expressing genes downstream of HIF1α were up-regulated in CD11c+ APCs isolated from nCB-exposed lungs (Fig. 4D). Under normal conditions, the HIF1α protein undergoes rapid turnover (32); however, its stability was increased in the nuclei of nCB-exposed CD11c+ Siglec F+ alveolar macrophages and BMDMs, concurrent with increased whole-lung hypoxia (Fig. 4, E to G). HIF1α was essential to controlling lactate expression because BMDMs isolated from myeloid cell–specific Hif1a-deficient (Lyz2creHif1af/f) mice treated with nCB failed to increase lactate production (Fig. 4H). Similarly, nCB-treated Hif1a-deficient alveolar macrophages showed decreased glycolysis (fig. S4C). Together, these findings suggest that HIF1α plays a critical role in increasing the rate of glycolysis in nCB-exposed macrophages.
Fig. 4. nCB activates the HIF1α pathway to increase lactate in macrophages.
(A) Representative Western blots showing expression of the mTORC1 downstream targets p-S6K (T389) and p-4E-BP1 (T37/46). n = 3 biological replicates per group. (B and C) Effects of rapamycin on nCB-treated BMDMs. Cells were exposed to nCB (50 μg/ml) for 24 hours, followed by 100 nM rapamycin (rap.) for another 24 hours. n = 4 biological replicates per group. (D) Heatmap of selected HIF1α-regulated genes in CD11c+ lung APCs from PBS-exposed or nCB-exposed WT mice. n = 3 per group. (E and F) Representative Western blots showing HIF1α in nCB-treated macrophages. n = 2 or 3 biological replicates per group. (G) Detection of hypoxia by pimonidazole in PBS- or nCB-exposed lungs. Scale bars, 100 μm. n = 5 biological replicates per group. (H) Lactate production in supernatants of nCB-treated BMDMs. Supernatants were collected 24 hours after nCB treatment. n = 4 per group. Representative (I) lactate production in the lungs and quantification of total live (J) FOXP3+ Treg cells; (K) PD-1+ LAG3+ CD4 T cells, (L) PD-1+ LAG3+ CD8 T cells, (M) PD-L1+ interstitial macrophages, and (N) PD-L1+ alveolar macrophages. n = 3 to 5 per group. All experiments have been repeated for two to three times and shown as representatives. Significance was determined by Student’s t test, one-way ANOVA, and two-way ANOVA with Holm’s correction. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To determine whether Hif1α in macrophages is critical for the increased lactate production in macrophages in vivo, we exposed Lyz2creHif1af/f mice to nCB. Compared with nCB-treated control mice (Hif1af/f), the nCB-treated Lyz2creHif1af/f mice had significantly less lactate in their lungs (Fig. 4I). These findings suggest that HIF1α plays a critical role in lactate production in macrophages in vivo. We next examined whether macrophage-derived lactate is responsible for recruitment of immunosuppressive cells in the lungs. As expected, nCB exposure resulted in increased lung infiltration by PD-1+ CD4 and CD8 T cells in Hif1af/f mice treated with nCB, but not in Lyz2creHif1af/f mice (Fig. 4, J to L). nCB-treated Hif1af/f mice showed an increase in expression of PD-L1 by interstitial macrophages, which was attenuated in the nCB-treated Lyz2creHif1af/f mice (Fig. 4, M and N).
We next examined whether the physical property of nCB is critical for the observed increased activation of mTORC1 and HIF1α in macrophages. We stimulated BMDMs with Polyethylene glycol (PEG)ylated nCB (PEG-nCB), a hydrophilic form of nCB that does not cause inflammation (6). BMDMs phagocytosed nCB or PEG-nCB (fig. S4D), but the PEG-nCB did not enhance mTORC1 activation or HIF1α stabilization (fig. S4, E and F). We next exposed WT mice to nCB and PEG-nCB (fig. S4G). We found that PEG-nCB neither increased whole-lung lactate abundance nor induced T cells exhaustion and macrophage recruitment (fig. S4, H to J). These results suggest that the hydrophobic property of ultrafine nCB is critical in activating mTORC1 and promoting HIF1α activity.
nCB aggregates disrupt mitochondrial homeostasis and induce autophagy
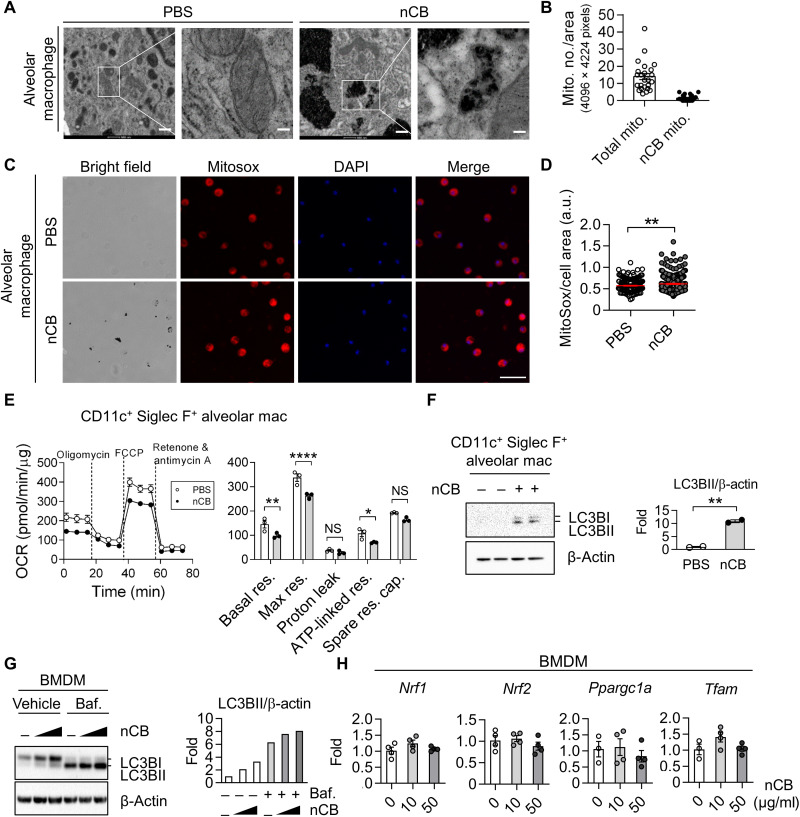
Mitochondria are critical energy and signaling hubs that control immune cell function (33). High-resolution transmission electron microscopy (TEM) revealed nCB aggregates in the cytoplasm of alveolar macrophages (Fig. 5A and fig. S4K). TEM also detected nCB particles within the mitochondria of these cells, suggesting that nCB can penetrate the lipid bilayer of the mitochondrial membrane. Quantification of macrophages mitochondria showed that ~10% of mitochondria in the nCB-exposed alveolar macrophages contained nCB particles (Fig. 5B). Furthermore, we found evidence that nCB causes cellular stress due to increased mtROS levels in alveolar macrophages (Fig. 5, C and D). As expected, OXPHOS was reduced in the nCB-treated macrophages, as evidenced by reduced rates of basal respiration, maximum respiration, and adenosine triphosphate production (Fig. 5E).
Fig. 5. nCB impairs mitochondrial homeostasis and induces autophagy in macrophages.
(A) Representative micrographs showing nCB aggregates in the cytoplasm and mitochondria. Scale bars, 500 and 100 nm (enlarged area). Images are representative of at least 10 microscopic fields per sample. (B) Quantification of mitochondria with nCB aggregates per image area, n = 24. (C and D) Representative images and quantification of mtROS in nCB-treated BMDMs. BMDMs were treated with nCB at the indicated concentrations for 24 hours. Scale bar, 50 μm. Images are representative of 20 pictures from four mice. a.u., arbitrary units. (E) Representative measurements of OCR in alveolar macrophages. Quantitative measurements of basal respiration (basal res.), maximal respiration (max res.), proton leakage, adenosine triphosphate (ATP) production, and spare respiratory capacity (spare res. cap.) are shown. n = 3 independent biological replicates per condition. (F and G) Representative Western blots of autophagy and quantification in nCB-treated macrophages. BMDMs were treated with nCB for 24 hours and then with 10 nM bafilomycin (baf.) 2 hours before harvesting. (H) Gene expression of mitochondrial biogenesis in BMDMs after treatment with nCB for 24 hours. Nuclear respiratory factor 1 (Nrf1) and 2 (Nrf2), PPARG coactivator 1 α (Ppargc1a), and transcription factor A mitochondrial (Tfam) were measured by real-time qPCR. n = 4 biological replicates per condition. All experiments have been repeated for three times and shown as representatives. Significance was determined by two-way ANOVA with Holm’s correction. *P < 0.05, **P < 0.01, and ****P < 0.0001.
Because we found that intracellular nCB penetrates the mitochondria, we next examined whether it induces autophagy. Total amounts of LC3B protein and its cleavage product LC3BII increased in a dose-dependent manner in nCB-treated alveolar macrophages and BMDMs (Fig. 5, F and G), indicating enhanced phagosome synthesis or reduced phagosome turnover in response to nCB exposure. Bafilomycin A1 treatment to block autophagosome and lysosome fusion resulted in LC3BII accumulation in the nCB-treated macrophages (Fig. 5G, right), suggesting an increase in phagosome synthesis. Furthermore, mitochondrial biogenesis was unaltered by the nCB treatment because the regulatory genes, Nrf1, Nrf2, Ppargc1a, and Tfam, were not affected (Fig. 5H). Together, these data suggest that activation of autophagy and phagosome synthesis occur directly in response to mitochondrial damage in nCB-treated macrophages.
nCB-exposed macrophages accelerate NSCLC progression
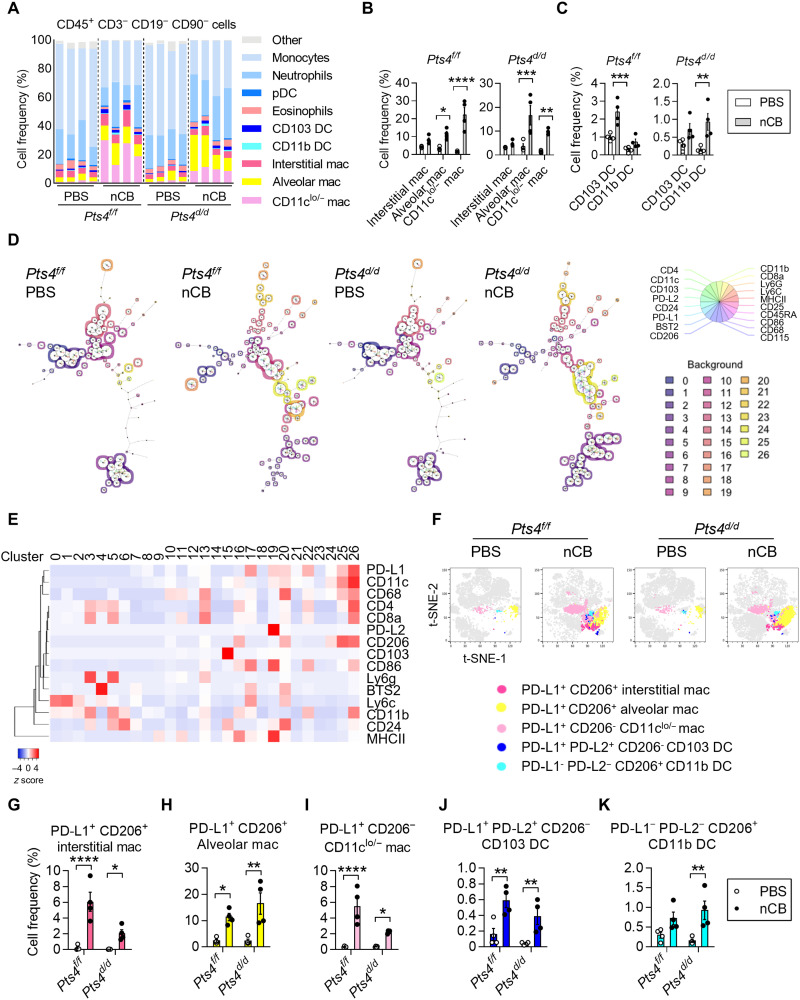
To determine how nCB changes myeloid cell populations in the lungs, we analyzed CD45+ CD3− CD19− CD90− cells using 22 distinct surface markers. FlowSOM (34), an unsupervised clustering method, identified 27 distinct cell clusters (fig. S5, A to C). Notably, a high relative abundance of macrophages in the lungs of nCB-exposed Pts4f/f and Pts4d/d mice did not conform to the conventional interstitial or alveolar macrophage phenotypes, with some macrophages showing low or negative CD11c expression (Fig. 6, A and B). nCB exposure altered DCs, resulting in increased relative abundance of CD103+ DCs in Pts4f/f mice and CD11b+ DCs in Pts4d/d mice (Fig. 6, A and C). There were also several myeloid clusters specific to the nCB-exposed lungs in comparison with the PBS-exposed lungs (Fig. 6D). These nCB-associated myeloid clusters showed increased expression of CD206, PD-L1, and PD-L2 markers (Fig. 6, E to K) consistent with tumor-associated immunosuppressive macrophages and DCs (35). nCB-exposed CCSP/EGFR tumor mice also showed increased relative abundance of PD-L1+ monocytes/macrophages (fig. S5, D and E). Together, our results suggest that, independent of tumors, nCB promotes PD-L1+, PD-L2+, and CD206+ myeloid cells in the lungs.
Fig. 6. nCB increases relative abundance of CD206+ PD-L1+ macrophages and DC in the lungs.
(A) Mass cytometry analysis of myeloid populations among CD45+ CD3− CD19− CD90− cells from PBS-exposed or nCB-exposed lungs of Pts4f/f and Pts4d/d mice. n = 4 biological replicates per group. (B) Summary data from interstitial, alveolar, and other types of macrophages in the indicated groups. (C) Summary data from CD103 DCs and CD11b DCs in the indicated groups. (D) Representative FlowSOM minimal decision trees of clusters in PBS-exposed or nCB-exposed lungs of Pts4f/f and Pts4d/d mice. Each cluster is colored with a distinct background. Color codes are shown on the bottom right with a total of 27 clusters. For each cluster, the expression of markers is indicated in the pie charts. The size of the pie corresponds to the average size of the population in the sample. (E) Heatmap of markers in each cluster identified by FlowSOM. Unsupervised clustering was applied to the columns. (F) Representative t-SNE plots showing emerging CD206+ PD-L1+ macrophages (three types) and DCs (two types) in nCB-exposed lungs. Summary data of (G) CD206+ PD-L1+ interstitial macrophages; (H) CD206+ PD-L1+ alveolar macrophages; (I) PD-L1+ macrophages; (J) PD-L1+ PD-L2+ CD103 DCs; and (K) CD206+ CD11b DCs in PBS-exposed or nCB-exposed lungs of Pts4f/f and Pts4d/d mice. Significance was determined by two-way ANOVA with Holm’s correction. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
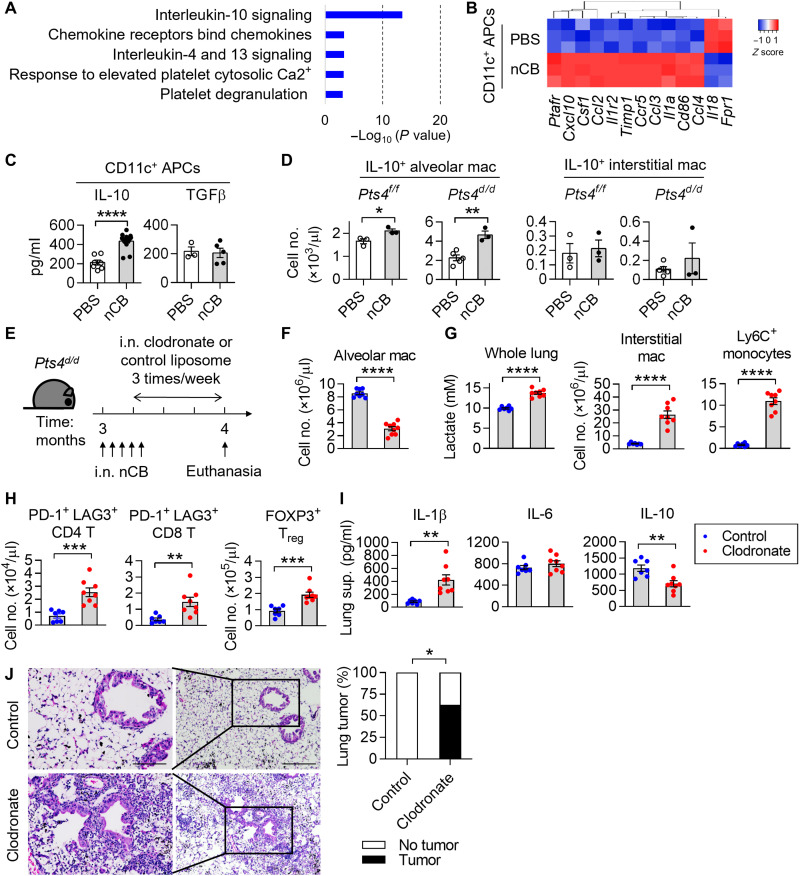
To determine the mechanism of nCB-induced altered gene expression in lung macrophages, we next performed a pathway enrichment analysis using a published dataset (GSE72734) (7). The results revealed increased expression of IL-10 signaling pathway in the CD11c+ APCs from nCB-exposed mice (Fig. 7, A and B). We validated these findings in CD11c+ APCs, which showed that nCB exposure increased IL-10 expression but did not affect TGFβ expression (Fig. 7C). Similarly, we found that nCB exposure resulted in increased numbers of IL-10+ SiglecF+ alveolar macrophages, but not IL-10+ CD11b+ interstitial macrophages, in the lungs of nCB-exposed Pts4f/f and Pts4d/d mice (Fig. 7D and fig. S5F).
Fig. 7. Loss of alveolar macrophages exaggerates inflammation and tumors in response to nCB.
(A) Top five pathways altered in CD11c+ APCs from nCB-exposed lungs. (B) Heatmap of the IL-10–regulated genes shown in (A). n = 3 per group. (C) IL-10 and TGFβ in lung CD11c+ APCs. n = 3 to 15 per group. (D) Representative IL-10 expression in lung macrophages. n = 3 per condition. (E) Schematic diagram of alveolar macrophage depletion in Pts4d/d mice. Three-month-old Pts4d/d mice received five doses of nCB intranasally in the first 1.5 week. Clodronate or control liposomes were intranasally delivered to the mice three times per week for 3 weeks after the third dose of nCB. Mice were euthanized at 4 months of age. (F) Total live alveolar macrophages. (G) Whole-lung lactate levels and total cell numbers of interstitial macrophages and Ly6C+ monocytes. (H) Total cell numbers of live PD-1+ LAG3+ CD4 or CD8 T cells and FOXP3+ Treg cells. (I) IL-1β, IL-6, and IL-10 expression in lungs. n = 7 or 8 per group. (J) Representative H&E stains and summary statistics of tumor foci found in liposome-treated nCB lungs. All experiments have been repeated for two to three times and shown as representatives. Significance was determined by Student’s t test and Fisher’s tests. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To determine whether IL-10 derived from myeloid cells dampens lactate expression in response to nCB exposure, we next measured lactate abundance in nCB-exposed Lyz2creIl10f/f mice. Myeloid-specific loss of Il10 showed a slight increase in whole-lung lactate level at baseline (fig. S5G). Notably, the concentrations of IL-β and IL-6 were higher in the lungs of nCB-exposed Lyz2creIl10f/f mice compared with nCB-exposed Il10f/f mice (fig. S5H). These results suggest that IL-10 secreted by macrophages can dampen lactate and inflammation in the lungs. In addition, PBS-exposed IL-10–deficient Pts4d/d mice exhibited higher tumor incident (50%, 5 of 10) than PBS-exposed Pts4d/d mice (15%, 3 of 20). nCB exposure and Il-10 depletion in Pts4d/d mice accelerated NSCLC as ~90% (10 of 11) of the mice developed tumors at 5 months of age (fig. S5I). These results indicate that, in response to nCB, lung macrophages express IL-10 to inhibit inflammation in this model.
To evaluate the role of CD11c+ Siglec F+ alveolar macrophages in NSCLC, we used the clodronate liposome depletion strategy (Fig. 7E). We found a significant decrease of CD11c+ Siglec F+ alveolar macrophages that validated our clodronate liposome treatment (Fig. 7F and fig. S5J). We found increased concentration of lactate, interstitial macrophages, and Ly6c+ monocytes in clodronate-treated nCB-exposed Pts4d/d mice (Fig. 7G). Clodronate-treated lungs also showed increased PD-1+ LAG3+ CD4 and CD8 T cells and FOXP3+ Treg cells (Fig. 7H). These changes were accompanied by increased IL-1β and decreased IL-10, but IL-6 abundance was unchanged (Fig. 7I). Lung tumor incidents in the clodronate group (62.5%, 5 of 8) were higher than in the control (0%, 0 of 7; (Fig. 7J). Together, these results demonstrated that alveolar macrophages in nCB-exposed Pts4d/d lungs are important to suppressing inflammatory response and cancer progression.
nCB-exposed APCs accelerate NSCLC progression
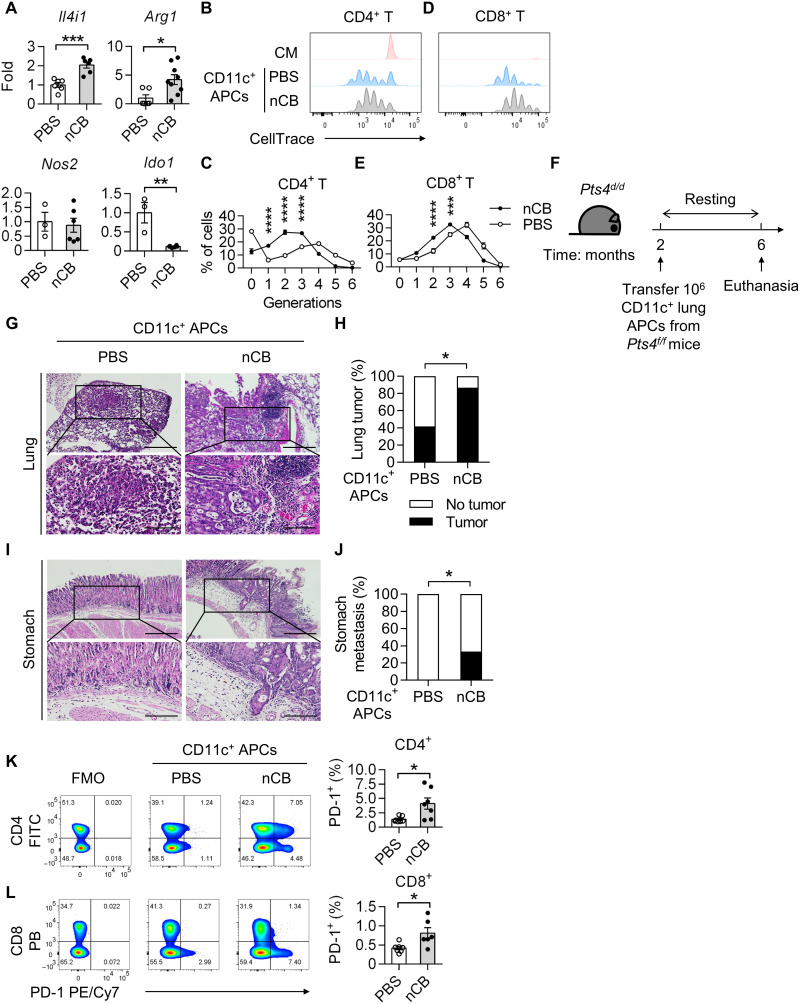
Because we observed an increase in the abundance of immunosuppressive T cells in response to nCB exposure, we next determined the expression of the immunosuppressive enzymes Il4i1, Arg1, Nos2, and Ido1, in CD11c+ APCs isolated from nCB-exposed lungs. We found that nCB exposure resulted in increases in Il4i1 and Arg1 expression, a decrease in Ido1 expression, and no change in Nos2 expression (Fig. 8A). Consistent with these results, we found that CD11c+ APCs from nCB-exposed lungs dampened CD4 T cell proliferation (Fig. 8, B and C). Similarly, splenic CD8 T cells exhibited decreased proliferation when cocultured with CD11c+ APCs from nCB-exposed lungs (Fig. 8, D and E). These results demonstrate that CD11c+ APCs from nCB-exposed lungs are immunosuppressive and impede T cell proliferation.
Fig. 8. Lung CD11c+ nCB APCs are immunosuppressive and promote NSCLC.
(A) Real-time qPCR detecting IL-4–induced 1 (Il4i1), arginase 1 (Arg1), nitric oxide synthase 1 (Nos1), and indoleamine 2,3-dioxygenase 1 (Ido1). n = 3 to 9 per group. Histograms statistics of T lymphocyte proliferation assessed by flow cytometry after coculture of splenic (B and C) CD4+or (D and E) CD8+ T cells with CD11c+ lung APCs. Expansion of the lymphocytes was determined by CellTrace. n = 4 per condition. (F) Schematic diagram of adoptive CD11c+ lung APCs transfer in Pts4d/d mice. Two-month-old Pts4d/d mice received 106 magnetic bead–isolated CD11c+ lung APCs from either PBS-exposed or nCB-exposed Pts4f/f mice. The Pts4d/d mice were euthanized at 6 months of age. Representative H&E stains and summary statistics of (G and H) lungs and (I and J) stomachs from the indicated groups. Scale bars, (top) 300 μm and (bottom) 250 μm. PBS CD11c+ APCs, n = 12; nCB CD11c+ APCs, n = 15. Representative flow cytometry analysis of live (K) CD4+ or (L) CD8+ PD-1+ lymphocytes. Numbers in the boxes indicate percentages of cells positive for the indicated markers. Summary statistics are shown on the right. n = 7 per group. All experiments have been repeated for three times and shown as representatives. Significance was determined by Student’s t test, Fisher’s test, and two-way ANOVA with Holm’s correction. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To further determine the role of CD11c+ APCs in nCB-induced NSCLC progression, we adoptively transferred lung CD11c+ APCs from PBS-exposed or nCB-exposed Pts4f/f mice into 2-month-old Pts4d/d mice (Fig. 8F). At 6 months of age, ~87% (13 of 15) of mice that received nCB-exposed CD11c+ APCs had developed lung cancer, when compared with ~42% (5 of 12) of mice that received PBS-exposed CD11c+ cells (Fig. 8, G and H). Notably, ~33% (5 of 15) of the mice that received nCB-exposed CD11c+ APCs developed distant metastasis, whereas there was no distant metastasis in the PBS group (Fig. 8, I and J). Consistent with these results, the relative abundance of PD-1+ CD4 and CD8 T cells in the mice that received the nCB-exposed CD11c+ APCs was increased compared with those in the control group (Fig. 8, K and L). Together, these results demonstrated that lung CD11c+ macrophages from nCB-exposed mice are immunosuppressive and directly facilitate lung cancer progression.
DISCUSSION
Exposure to ultrafine particles generated from incomplete combustion of organic matter (e.g., cigarette smoke, burn pit, forest fire, and other environmental pollutants) causes lung inflammation and emphysema (6, 36). We show that nCB fosters early and more invasive lung cancer in mice, as summarized in Fig. 9. We used two distinct NSCLC models: one in which the airway-specific loss of two tumor suppressors, Pten and Smad4, renders mice prone to adenosquamous carcinoma development and another in which overexpression of mutant EGFR leads to lepidic adenocarcinomas. In both models, exposure to nCB, a prototypic ultrafine particle, accelerated NSCLC progression. These results suggest that, in addition to the multiple well-known carcinogens in tobacco smoke, nCB exerts an additive effect to further promote lung cancer development and metastasis.
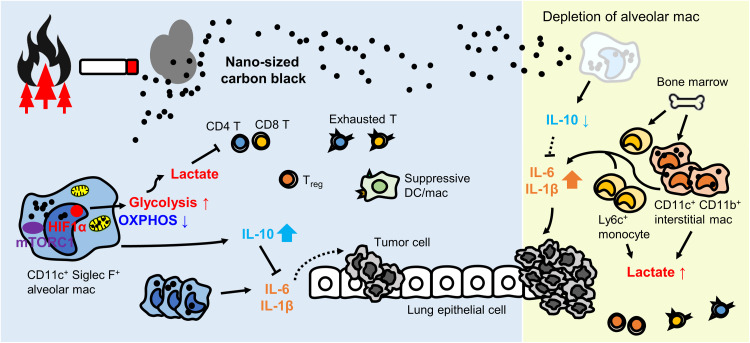
Fig. 9. Summary model: nCB reprograms CD11c+ lung macrophages to accelerate cancer.
Environmental airborne nCB particles (e.g., wildfire and cigarette smoke) are phagocytosed by lung CD11c+ Siglec F+ alveolar macrophages. nCB selectively damages mitochondria in macrophages and decreases OXPHOS. To compensate for the ATP loss, alveolar macrophages up-regulate HIF1α signaling to fuel glycolysis. These metabolic changes lead to the accumulation of the glycolysis by-product lactate, in the microenvironment. Increased lactate in the lungs inhibits effector T cell activation, promotes Treg cells, and increases suppressive DC/macrophages and exhausted T cells, which can promote tumors. The insoluble nature of nCB phagocytosed and released by local and recruited macrophages exacerbates inflammation, while alveolar macrophages secrete IL-10 to decrease inflammation. Depletion of alveolar macrophages reduces IL-10 level and triggers recruitment of inflammatory Ly6C+ monocyte and interstitial macrophage from the bone marrow. In the absence of alveolar macrophages, increased lactate and proinflammatory cytokines further strengthen immunosuppression and tumor growth.
Several immunomodulating metabolic products of glucose, particularly lactate, accumulate in the lungs in response to nCB exposure. Lactate has been shown to polarize immune cells to favor tumor progression (24, 37). Consistently, we show here that nCB promotes recruitment of lymphoid and myeloid immunosuppressive cells in the lungs irrespective of tumors because the response was similar in Pts4f/f and Pts4d/d mice. Specifically, lactate production in nCB-exposed macrophages caused increased expression of immune checkpoint inhibitors in T cells, macrophages, and DCs in the lungs. HIF1α has been shown to differentiate myeloid-derived suppressor cells from tumor-associated macrophages (38). nCB-exposed lungs with HIF1α-deficient myeloid cells show reduced whole-lung lactate level, T cells expressing exhaustion markers, and Treg cells, suggesting that HIF1α controls the suppressive function of myeloid cells in nCB-exposed lungs, which in part, is controlled by IL-10. In support of our findings, lactate has been shown to accumulate at sites of chronic inflammation and induce TH17 cells differentiation that perpetuates the inflammation (39, 40). Our prior work has shown that exposure to cigarette smoke or nCB increases TH17 cell numbers that are critical in emphysema development in murine models and are associated with human chronic obstructive pulmonary disease (6, 9, 31, 41). In the present study, we observed TH17 cell recruitment to the lung tumors in nCB-exposed mice. One intriguing question remains regarding how TH17 cell recruitment can modify the tumor microenvironment in response to nCB or smoke. Cancer cells use glycolysis to generate lactate in an aerobic environment, which is known as the Warburg effect. While tumors can be a primary source of lactate (42), we found that macrophages harboring ultrafine particles also play a critical role in up-regulating lactate production independently of the tumors. These results suggest that ultrafine particles can modify the lung environmental metabolic landscape, accelerating lung cancer development and metastasis.
Alveolar macrophages are one of the first responders to clear foreign material in the lungs. We found increased relative abundance of macrophages in nCB-exposed lungs regardless of the presence or absence of tumors. Whereas bacteria-infected macrophages show a transient elevation of mTORC1 signaling and glycolysis (16), macrophages harboring hydrophobic nCB displayed sustained activation of mTORC1. Furthermore, we showed that nCB penetrates the mitochondrial membrane and interferes with mitochondrial function. Disruption of the electron transport chain induces ROS, which can inhibit Prolyl hydroxylases and Cullin-3 and thus stabilize HIF1α to promote glycolysis (43). Chronic mTORC1 activation in macrophages promotes glycolysis, increases phagocytic function, and reduces apoptosis (44, 45). Our results demonstrate how chronic mTORC1 signaling drives glycolysis and increases lactate production in macrophages. Alveolar macrophages engulf insoluble nCB and eventually lyse and release necrotic cellular debris to activate the Apoptosis-associated speck-like protein containing a CARD (ASC)-dependent inflammasome (6). We found that alveolar macrophages are critical in controlling lung inflammation and NSCLC progression in response to ultrafine particles. nCB increases infiltration of immunosuppressive lung macrophages decorated with CD206 and PD-L1 that inhibit T cell activation and promotes NSCLC, as shown in our coculture and adoptive transfer experiments. Depletion of alveolar macrophages fuels inflammation, showing increased Ly6C+ monocytes and interstitial macrophages in nCB-exposed lungs. Simultaneously, we observed increased T cell exhaustion and Treg cells, indicating activation of a compensatory response. Future direction includes investigating the effect of nCB on CD8 T and NK cell cytotoxicity. Inflammation fuels tumor progression (46); consistently, we showed that alveolar macrophages removal unleashed tumor growth.
Apoptotic cell–derived arginine is required for continual macrophages efferocytosis (47). Consistent with that, we observed an increase in Arg1 expression in CD11c+ APCs from nCB-exposed lungs, suggesting increased phagocytosis to remove nCB from the lungs. Although glycolysis is often linked to M1-type macrophages (48), macrophages can react to different stimuli in their environment (49), and under inflammatory conditions, M1 and M2 markers coexist (50), as shown in our CyTOF analysis. Notably, in response to nCB, we found increased relative abundance of lung macrophages expressing low/no CD11c that are neither alveolar nor interstitial macrophages. A caveat of our studies includes an inability to harvest distinct macrophage populations because of low viability using flow sorting techniques. Therefore, the exact roles of different subset of newly recruited macrophages in lung cancer await further studies. Furthermore, an innate immune memory response, termed “trained immunity,” has been described in bacteria-infected macrophages (51). Specifically, Bacille Calmette-Guérin–trained macrophages require mTORC1 and glycolysis for histone remodeling and functional changes (51). Future studies are needed to examine whether nCB affects trained immunity in macrophages.
We showed that nCB reaches the alveoli and that nCB aggregates are predominantly found in the mitochondria of alveolar macrophages but not in other distal lung cells. The reason for these findings is unclear but may be related to the function of resident alveolar macrophages that efficiently phagocytose particles shielding the airway epithelia. Previous studies showed that nanoparticles target and damage mitochondria (52) and also indirectly induce oxidative stress in macrophages (53). The damaged organelles are subsequently recycled by autophagy, which is a typical response to intracellular nanoparticles (54). Accordingly, we found an increase in autophagosomes in nCB-treated macrophages; however, although mTORC1 is a known negative regulator of autophagy (55), we also found augmented mTORC1 expression in nCB-treated macrophages. Consistent with our findings, increased autophagy has been described in macrophages with constitutive mTORC1 activation to suppress inflammation in response to bacterial infection (56). Our data suggest that lung macrophages maintain cellular metabolism and energy homeostasis to oppose nCB-induced inflammatory signals. Together, our findings support the notion that sustained mTORC1 activation is critical for immunosuppressive cytokine secretion in response to nCB.
We showed that nCB-mediated activation of the HIF1α axis supports glycolysis and lactate production in lung macrophages, forming a constellation of metabolic alterations that promote NSCLC progression and invasiveness. Although in vitro nCB treatment failed to increase lactate production in BMDCs and lung epithelial cells, lung DCs and epithelial cells may respond differentially to ultrafine particles in vivo. One intriguing question is whether nCB-induced local effects in lungs can spread systemically to distal organs. Some evidence suggests that inhaled nCB can migrate to secondary organs and is genotoxic to the liver (57). Black carbon (polycyclic aromatic hydrocarbons containing CB) can translocate from mothers’ lungs to the placenta (58), supporting the hypothesis that ambient ultrafine particles can be transported from lungs to distal organs. Future studies are needed to determine whether nCB in the lungs can induce systemic immune suppression or DNA changes that can reprogram the microenvironment and promotes tumors outside the lungs.
In response to nCB, HIF1α is stabilized in lung macrophages, and massive infiltration of interstitial macrophages, compared to other immune cells, strongly supports a role for HIF1α in this cell population in NSCLC progression. However, future studies using HIF1α with alternative immune cell–specific knockout could further expand on the current findings.
In conclusion, our findings demonstrate that accumulation of nCB, the main component of anthracotic pigments found in lung macrophages, promotes NSCLC. Notably, nCB impairs mitochondrial homeostasis and rewires metabolism in lung macrophages, causing decreased OXPHOS and increased glycolysis. Furthermore, macrophages harboring nCB secrete lactate and prompt recruitment of tolerogenic cells that create an immunosuppressive microenvironment in the lungs. In response to nCB exposure, sustained HIF1α signaling in lung macrophages supports metabolic changes and induces an immunosuppressive environment that accelerates NSCLC.
MATERIALS AND METHODS
Animals
All animal work was performed in accordance with the protocol approved by the Institutional Animal Use and Care Committees at Baylor College of Medicine. Mice were bred and maintained under pathogen-free conditions. Two lung adenocarcinoma strains, CCSPicrePtenf/fSmad4f/f (Pts4d/d) and tetO-EGFRmutCCSPrtTA (CCSP/EGFR), have been described (22, 23). Littermates were randomly assigned to experimental groups. Il-10 KO, Lyz2cre, and Hif1af/f mice were from the Jackson Laboratory. Il-10f/f mice were obtained from G. Diehl. For in vivo nCB challenge experiments, 3-month-old Pts4d/d and 2-month-old CCSP/EGFR mice were used. For other in vivo experiments, 2-month-old mice were used. For bone marrow cell isolation, 6- to 8-week-old mice were used for generating BMDMs or BMDCs.
Cell cultures
BMDMs and BMDCs were isolated by suspending syringe-flushed marrow from murine femurs and tibias as described (59). Briefly, both ends of femurs and tibias were cut; cells were flushed from the marrow with a 26.5-gauge needle. Marrow cells were then subject to lysis of red blood cells (RBCs) by using homemade ammonium chloride–potassium bicarbonate (ACK) buffer. For BMDMs, marrow cells were resuspended in complete RPMI 1640 medium with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1% penicillin-streptomycin, 1 mM sodium pyruvate, Non-essential amino acid (NEAA), and 0.05 mM 2-mercaptoethanol supplemented with mouse recombinant protein, granulocyte-macrophage colony-stimulating factor (GM-CSF, 20 ng/ml; R&D Systems) or M-CSF (10 ng/ml; R&D Systems). Because GM-CSF can promote differentiation of both DCs and macrophages, we remove DCs and their progenitors by rigorously washing with PBS and replacing with fresh supplemented medium every 2 to 3 days. For BMDCs, the medium was supplemented with mouse GM-CSF (20 ng/ml) and IL-4 (10 ng/ml; R&D Systems). Media were replaced every 2 days. On day 7, BMDMs or BMDCs were harvested and seeded in a plate. Isolation of alveolar macrophages from BAL is as described (60). Briefly, lavages were pooled from BAL washes (PBS with 2 mM EDTA). Alveolar macrophages were purified by adherence for 2 hours by RPMI 1640 complete medium. The nonadherent cells were washed off with warm PBS. The macrophage viability decreased when nCB concentration is higher than 50 μg/ml, so we used concentrations lower than 50 μg/ml for the experiments. One day after seeding, BMDMs or BMDCs were treated with the indicated amount of nCB, PEG-nCB, or PBS for 24 or 48 hours. For some experiments, 10 nM bafilomycin A (Sigma-Aldrich) or 100 nM rapamycin (Sigma-Aldrich) was given to the cells 2 or 24 hours before harvest. Human lung epithelial cell lines, BEAS-2B and H1299, were purchased from American Type Culture Collection (ATCC). BEAS-2B cells were cultured in a precoated vessel as suggested on the ATCC website. Briefly, the coating solution containing fibronectin (0.01 mg/ml; Gibco), bovine collagen type I (0.03 mg/ml; Gibco), and bovine serum albumin (BSA; 0.01 mg/ml; GenDEPOT) in BEGM basal medium (Lonza) was added into a plate overnight in a 5% CO2 incubator at 37°C. The coating solution was removed before seeding BEAS-2B cells with BEGM bronchial epithelial cell growth medium (Lonza) in the plate. For NSCLC cell lines, H1299 cells were cultured in RPMI 1640 medium (Gibco) that is supplemented with 10% FBS, 2 mM l-glutamine, and 1% penicillin-streptomycin. All cells were cultured in a 5% CO2 incubator at 37°C. The indicated amount of nCB or PBS was added 24 hours after seeding. Supernatants and cells were harvested 24 or 48 hours after the treatment.
Nanoparticle nCB murine models
Characterization and preparation of 15-nm nCB and PEG-nCB have been described (6). Pts4f/f, Pts4d/d, and Pts4d/dIl-10 KO mice age at 3 months were challenged with nCB (0.5 mg/50 μl) or sterile PBS for 1 month, three times per week. Mice were anesthetized with isoflurane and then challenged with 50 μl of nCB or PBS. Mice at 5 months of age were euthanized. WT C57BL/6J, Lyz2creIl-10f/f, Lyz2creHif1af/f, and CCSP/EGFR mice were exposed to nCB or PBS at 2 months of age. After the last challenge of PBS or nCB, CCSP/EGFR mice were given ad libitum access to doxycycline-containing food pellets (625 parts per million; Envigo) for 1 month to induce lung cancer and then euthanized at 4 months of age. For PEG-nCB in vivo experiments, eight doses of PBS, nCB, or PEG-nCB were given to the C57BL/6 WT mice. We used equal weight of nCB and PEG-nCB.
Alveolar macrophage depletion nCB model
Pts4d/d mice age at 3 months were exposed to five doses of nCB (0.5 mg/50 μl) in 1.5 weeks. Mice were given 50 μl of clodronate liposome or control liposome (Liposoma) intranasally for 3 weeks (three times/week) after the third dose of nCB. Mice were euthanized for analysis 2 days after the final clodronate treatment.
CD11c+ lung APC adoptive transfer model
CD11c+ lung APCs from Pts4f/f mice were isolated using anti-CD11c–conjugated magnetic beads (Miltenyi Biotec) as instructed in the manufacturer’s protocol. Pts4d/d mice received 106 isolated cells intraperitoneally at 2 months of age. Recipients at 6 months were euthanized for analyses.
Human lung monocytes/macrophage isolation and nCB treatment
Human lung tissues were obtained from patients undergoing lobectomy for the treatment of early-stage lung cancer (I and II; n = 8). All studies were approved by the institutional review board at Baylor College of Medicine. Informed consent was obtained from each patient. Human lung single cells were prepared from surgical resection and lungs in patients with lung cancer as described (61). Briefly, small pieces of uninvolved tumor lung tissues (10 cm away from the tumor) were digested in collagenase type D (2 mg/ml; Worthington) solution with Hanks’ balanced salt solution (HBSS) at 37°C incubator for 30 min. Cells were then passed through a 40-μm cell strainer followed by ACK lysis to remove RBCs. A pan monocyte isolation kit (Miltenyi Biotec) was used for isolating lung monocytes/macrophages. After counting the cells, Fc block and biotin-labeled antibodies were added as recommended in the manufacturer’s protocol. Without wash, avidin beads were added and then performed negative selection by autoMACS (Miltenyi Biotec). Cells were then counted and seeded in a 96-well plate (2 × 105 cells per well) with a complete RPMI medium. nCB (50 μg/ml) or PBS was added into the culture 24 hours after seeding. Supernatants and cells were collected 24 hours later for the following analyses.
Mouse whole-lung cell isolation for flow cytometry
To prepare single-cell suspension from mouse lungs, tissues were cut into 2-mm pieces and then digested by collagenase type D (2 mg/ml; Worthington) and deoxyribonuclease (DNase) I (0.04 mg/ml; Roche) for 1 hour in a 37°C incubator. Digests were filtered through 40-μm cell strainers and centrifuged at 400g for 5 min. RBCs were lysed by using ACK buffer. Single-cell suspension was filtered and washed and then resuspended in PBS. Cells were blocked with Fc blocker (BioLegend) on ice for 10 min, followed by staining with live/dead indicator and surface markers at 4°C for 30 min. Transcription factors were detected by intracellular staining; following surface staining, FOXP3/transcription factor staining buffer set (Invitrogen) was used as indicated in the manufacturer’s protocol. For IL-10 detection, cells were cultured in brefeldin A (10 μg/ml; Sigma-Aldrich) for 4 hours. After staining dead cells and cell surface markers, a Cytofix/Cytoperm kit (BD Biosciences) was used to fix and permeabilize the cells for staining intracellular IL-10 (BioLegend). For IL-17A and interferon-γ (IFN-γ) staining, cells were stimulated in the complete RPMI 1640 medium with phorbol 12-myristate 13-acetate (10 ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich) overnight and supplemented with brefeldin A (10 μg/ml; Sigma-Aldrich) for 4 hours. Cells were stained for surface markers and viability dye, fixed with 1% formaldehyde, permeabilized with 0.5% saponin, and stained with IL-17A antibodies (BD Biosciences) and IFN-γ (BD Biosciences). For CTLA4 staining, a Cytofix/Cytoperm kit (BD Biosciences) was used as described by the manufacturer. Stained cells were analyzed by BD LSR II flow cytometer (BD Biosciences) and FlowJo software. Gating methods for TH17, TH1, and alveolar and interstitial macrophages are as described (6, 62). Fluorophore-conjugated flow antibodies used in this study are as follows: Pacific Blue anti-mouse CD3 antibody (558214), phycoerythrin (PE)–Cy5 anti-mouse CD4 antibody (553050), and Percp-Cy5.5 rat anti-mouse Siglec-F (565526) from BD Biosciences; Percp-Cy5.5 anti-CD3 (100218), PE-Cy5 anti-CD4 (116003), fluorescein isothiocyanate (FITC) anti-mouse CD4 (100406), FITC anti-mouse CD45 (103108), Pacific Blue anti-mouse CD8a (100725), Alexa Fluor 700 anti-mouse CD4 (100430), Allophycocyanin anti-mouse CD274 (B7-H1, PD-L1) (124311), Allophycocyanin anti-mouse CD152 (CTLA4) (106309), PE/Cy7 anti-mouse CD279 (PD-1) (109110), Brilliant Violet 605 anti-mouse Ly6C (128036), Percp-Cy5.5 anti-mouse Ly-6G (127616), PE/Cy7 anti-mouse MER proto-oncogene tyrosine kinase (151522), PE anti-mouse CD64 (FcγRI) (161004), Pacific Blue anti-mouse I-A/I-E (107620), PE anti-mouse MER (151505), PE/Cy7 anti-mouse CD64 (139314), and Allophycocyanin/Cy7 anti-mouse/human CD11b (101225) from BioLegend; FOXP3 monoclonal antibody (mAb), Allophycocyanin (17-5773-82); eFluor 450 anti-mouse CD11c (48-0114-82); CD11b mAb, PE (12-0112-82); CD44 mAb, eFluor 450 (48-0441-82); CD69 mAb, Allophycocyanin (17-0699-42); and CD11c mAb, Allophycocyanin (17–0114-82) from Thermo Fisher Scientific.
Mass cytometry staining, data acquisition, and analysis
Single-cell suspensions were prepared using a GentleMACs dissociator (Milltenyi). Lungs were sliced into 2- to 3-mm pieces with scissors and processed using 6 ml of digestion buffer containing DNase I (0.1 mg/ml; Sigma-Aldrich) and collagenase IV (0.5 mg/ml; Thermo Fisher Scientific). Organs were ground on the dissociator and incubated for a total of 30 min at 37°C. The homogenates were chilled on ice, and enzymes were deactivated using 10 mM EDTA. The suspensions were filtered through a 40-μm cell strainer followed by RBC lysis for 5 min on ice. The homogenates were then Fc-blocked using mouse BD Fc Block (4 μg/ml; BD Biosciences) on ice for 15 min. The resulting homogenates were first stained with cell surface antigens using various antibodies conjugated to different metal isotopes suspended in Maxpar cell staining buffer (Fluidigm) for 1 hour at 4°C. The cells were then stained for viability using Cell-ID Cisplatin-198Pt (Fluidigm) as directed by the manufacturer. The metal-conjugated antibodies were either purchased directly from Fluidigm or custom-conjugated using either Maxpar X8 or Maxpar MCP9 Antibody Labeling Kits (Fluidigm) as directed by the manufacturer. Cells were washed using Maxpar cell staining buffer (Fluidigm) and were then fixed by freshly made 2.0% paraformaldehyde (Alfa Aesar) at room temperature for 10 min. Cells were then washed using Maxpar cell staining buffer and were permeabilized using methanol for 30 min on ice. Intracellular antigens were stained using metal-conjugated antibodies suspended in Maxpar cell staining buffer for 1 hour at 4°C. Cellular DNA was then stained with Intercalator-Ir in Maxpar Fix and Perm buffer (Fluidigm) overnight at 4°C. Before the acquisition, samples were washed with Maxpar cell staining buffer followed by Maxpar Cell Acquisition Solution (Fluidigm) and were resuspended in containing 10% (v/v) EQ Four Element Calibration Beads (Fluidigm). Data were acquired on a Helios mass cytometer (Fluidigm). Data were normalized against the EQ Four Element Calibration Beads and randomized using uniform negative distribution to FL Studio Score files (FSC) using CyTOF software version 7.0. FlowJo v10 was used for the subsequent analyses. First, bead data were removed and then gated the single-cell events based on event length and DNA content (191 + 193 Iridium signal). Next, dead cells were excluded by gating out the 198Pt signal. For automated analysis, data visualization and analysis were performed on pregated data, live CD45+ CD3− CD90− CD19− for analyzing myeloid cells. Clustering analysis was performed by Phenograph and FlowSOM plugin in the R environment (34) after merging 16-sample datasets (9000 cells per sample) by DownSample plugin. Initial 100 nodes were used for FlowSOM, and the parameters were shown in the figures. A total of 27 subsets were then defined and visualized by FlowSOM minimal decision trees or t-distributed stochastic neighbor embedding (t-SNE) plots. Annotation of populations was determined by the heatmap of median expression values and the clustering results. After acquiring the clustering and visualization parameters from the concatenated dataset, each sample was then analyzed with the same settings. Percentage of populations in each sample and tSNE plots were identified individually. Antibodies used for CyTOF analysis were as follows: anti-mouse CD90 (G7) 144Nd (105202), anti-mouse CD19 (6D5) 110Cd (115502), anti-mouse CD4 (H129.19) 114Cd (100506), anti-mouse CD11c (N418) 116Cd (117302), anti-mouse CD103 (2e7) 163Dy (121402), anti-mouse PD-L2 (Ty25) 166Er (107222), anti-mouse CD24 (M1/69) 170Er (101802), anti-mouse FOXP3 (150D) 168Er (320002), anti-mouse PD-L1 (10F.9G2) 151Eu (124302), anti-mouse BST2 (927) 165Ho (127030), anti-mouse CTLA4 (UC10-4B9) 139La (106202), anti-mouse CD206 (C068C2) 150Nd (141702), anti-mouse CD11b (M1/70) 113Cd (101248), anti-mouse CD8a (53-6.7) 112Cd (100746), anti-mouse Ly6G (IA8) 111Cd (127602), anti-mouse Ly6C (HK1.4) 106Cd (128002), anti-mouse CD25 (3C7) 141Pr (101902), anti-mouse CD45R (RA3-6B2) 176Yb (103202), anti-mouse CD86 (GL1) 172Yb (105002), anti-mouse CD68 (FA-11) 171Yb (137002), anti-mouse CD115 (AF5) 147Sm (135538), anti-mouse T-bet (4B10) 167Er (644825), anti-mouse CD38 (90) 169Tm (102702), and anti-mouse PD-1 (RMP1-30) 156Gd (109113) from BioLegend; anti-mouse CD3e (145-2C11) 152Sm (3152004), anti-mouse CD45 (30-F11) 89Y (3089005B), and anti-mouse I-A/I-E (Y3P) 209Bi (3209006B) from Fluidigm; and anti-mouse RORyt (Q31-378) 153Eu (624084) from BD Biosciences.
Lung CD11c+ macrophage isolation and splenic T cell cocultured
Lung CD11c+ macrophages from PBS or nCB-exposed WT mice and splenic CD4+ and CD8+ T cells from 6- to 8-week-old WT mice were enriched by magnetic beads (Miltenyi Biotec). Briefly, whole-lung cells were labeled with anti-CD11c–conjugated magnetic beads (Miltenyi Biotec) and separated using autoMACS cell separator, as indicated in the manufacturer’s protocol. For mouse splenic T cell isolation, spleens were passed through a 40-μm cell strainer and resuspended in the RPMI 1640 complete medium. RBC-free cells were labeled with anti-CD4 or CD8-conjugated magnetic beads for isolation. Splenic CD4+ and CD8+ T cells were stained with the CellTrace Violet dye (5 μM/106 cells/ml PBS as diluent; Invitrogen) at room temperature in dark. After 20 min, five times the original staining volume of RPMI 1640 complete medium was added to the cells and incubated for 5 min to quench the reaction, and then, cells were pelleted by centrifugation. Lung CD11c+ macrophages were cocultured with splenic CD4+ or CD8+ T cells (1:10 ratio) in the presence of anti-mouse CD3 (1 μg/ml; BD Biosciences) for 3 days. Cell proliferation of splenic CD4+ and CD8+ T cells was analyzed by BD LSR II flow cytometer (BD Biosciences) and FlowJo software.
Cell lysis and immunoblotting
Cells were rinsed with cold PBS twice and then lysed in radioimmunoprecipitation assay buffer (Santa Cruz Biotechnology) containing protease and phosphatase inhibiter cocktail (Thermo Fisher Scientific). Protein concentration was measured by DC protein assay (Bio-Rad), and proteins were denatured in a sample buffer before SDS–polyacrylamide gel electrophoresis (PAGE). For nuclear protein extraction, the Cell Fractionation Kit (Cell Signaling Technology) was used for detecting HIF1α and H3. Protein lysates (20 mg) were subjected to SDS-PAGE and transferred to nitrocellulose membrane using the iBlot Dry Blotting System (Invitrogen), probed with indicated antibodies: phospho-4E-BP1 (Thr37/46) (236B4) rabbit mAb (1:1000; Cell Signaling Technology, 2855), phospho-p70 S6 kinase (Thr389) antibody (1:1000; Cell Signaling Technology, 9205), HIF-1α (D2U3T) rabbit mAb (1:1000; Cell Signaling Technology, 14179), histone H3 (D1H2) XP rabbit mAb (1:1000; Cell Signaling Technology, 4499), and LC3B (D11) XP rabbit mAb (1:1000; Cell Signaling Technology, 3868). After detecting the phosphoproteins, the membranes were stripped in the stripping buffer (Thermo Fisher Scientific) and reblotted with other primary antibodies: 4E-BP1 (53H11) rabbit mAb (1:1000; Cell Signaling Technology, 9644), p70 S6 kinase (49D7) rabbit mAb (1:1000; Cell Signaling Technology, 2708), and β-actin (13E5) rabbit mAb (horseradish peroxidase conjugate) (1:1000; Cell Signaling Technology, 5125). Immunoblot signals were detected by a ChemiDoc XRS imager (Bio-Rad) with chemiluminescence substrate (Thermo Fisher Scientific) and quantified by ImageJ.
Whole-lung cell, BAL, and CD11c+ APC cytokine measurement
RBC-free single-cell suspension or CD11c+ magnetic bead isolated APCs from PBS or nCB-exposed lungs were suspended in complete RPMI 1640 medium and cultured overnight at 37°C. BAL fluid was collected as described (6). Cultured supernatants or BAL samples were collected for measuring cytokine expression by enzyme-linked immunosorbent assay kits.
In silico analysis
To analyze the pathway changes in CD11c+ lung macrophage, we used our published microarray dataset (GSE72734). Genes that were significantly changed (P ≤ 0.05, fold change ≥ 1.5) in PBS or nCB groups were selected and input into Reactome, an open-source database for pathway analysis. The top five pathways were plotted with P values. To identify HIF1α or IL-10–regulated genes, literature and Reactome database were used. HIF1α- or IL-10–regulated genes that were significantly changed (P ≤ 0.05, fold change ≥ 1.5) were plotted in the heatmaps using Heatmapper or ggplot2 in RStudio.
Lactate detection
Lactate concentrations were determined using the Lactate Assay Kit (BioVision) and followed the preparation procedures according to the manufacturer’s instruction. Briefly, culture supernatants were collected on ice and filtered through a 10-kDa molecular weight spin filter (Amicon) to deplete lactate dehydrogenase in the FBS-containing medium. Diluted samples were added to a 96-well plate with a reaction mix. Lactate concentrations were determined by comparison to standard curves.
Lung hypoxia analysis
For probing lung hypoxia, pimonidazole (20 mg/kg in PBS; Hypoxyprobe) were administrated via intraperitoneally into mice 1 hour before euthanasia. Followed by tissue processing and paraffin embedding, 5-μm-thick sections were cut. Slides were then dewaxed in xylene, rehydrated, and subjected to antigen retrieval by citrate buffer [10 mM trisodium citrate with 0.05% Tween 20 (pH 6.0)] at 95°C for 10 min. PBS-rinsed slides were permeabilized with 1% BSA with 0.1% Triton X-100 solution (PBS as diluent) for 15 min and blocked in 10% goat serum (Equitech-Bio) with PBS for 30 min at room temperature. Slides were hybridized with primary antibody, anti-Mab1 (1:50), overnight at 4°C in dark, followed by PBS wash for three times and then counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 5 μg/ml; Invitrogen). After washing, slides were mounted on the Antifade mounting solution (Vectashield), cover-slipped, and imaged on a Leica TCS SP8X STED3x super-resolution microscope. Hypoxic signals were quantified by ImageJ.
TEM and mitochondrial analysis
TEM was performed at the Texas Children’s Hospital. Briefly, 1-mm-thick lung tissue samples were fixed in Trump’s buffer glutaraldehyde (4% formaldehyde and 1% glutaraldehyde in phosphate buffer) overnight at 4°C. Samples were postfixed in buffered 1% osmium tetroxide, dehydrated in ethanol, and embedded with the Poly/Bed 812 Embedding Kit (PolyScieneces). Sixty-nanometer sections were prepared using a Leica UC 6 ultramicrotome and stained with 2% aqueous uranyl acetate and Reynold’s lead citrate. Sections were examined with an FEI Tecnai Spirit transmission electron microscope equipped with an FEI Eagle digital acquisition system at an acceleration of 60 kV. Image analysis was performed by ImageJ.
Lung histopathology analysis
Lungs were fixed with instillation of 10% neutral-buffered formalin via a tracheal cannula at 25-cm H2O pressure after perfusion with PBS. Lungs were fixed at 4°C overnight, followed by paraffin embedding. Sections (5 μm thick) were stained with H&E and imaged by the EVOS M5000 Imaging System (Invitrogen).
DESI MS imaging
DESI imaging MS data were acquired in a quadrupole time-of-flight mass spectrometer (SYNAPT G2-XS, Waters Corporation) with a traveling wave ion mobility cell. MS data were acquired at 0.5 s per spectrum scan rate in the resolution mode of the mass spectrometer from mass/charge ratio of 50 to 1200. The mass spectrometer sample cone was set at 40 V, and StepWave radio frequency was lowered to 150 to enhance the transmission of the lower–molecular weight metabolites. DESI solvent (98% methanol and 2% water with 0.01% formic acid, v/v) was pumped through the sprayer at 2 μl/min using a binary high-performance LC pump (ACQUITY UPLC M-Class, Waters Corporation). A DESI sprayer emitter, based on precision-machined emitters, was kept between 800 and 900 V. Imaging MS experimental setup and data analysis were done using High Definition Imaging v1.5 software (Waters Corporation). Any potential mass drift during the MS acquisition was corrected by using leucine-enkephalin (200 pg/μl) dissolved in the spray solvent as a lock mass to maintain mass accuracy. The MS ion intensities were also normalized relative to the leucine-enkephalin internal standard peak. Lung tissues were flash-frozen in liquid nitrogen without embedding. The 10-μm tissue sections were stored in a −80°C freezer. Before DESI imaging, the sections were dried under a vacuum desiccator for 10 to 15 min at room temperature. The negative ion mode and the positive ion mode DESI imaging experiments were run sequentially on the same tissue section. Metabolites were putatively identified on the basis of the mass accuracy within ±10 mDa for ions that were localized only on the tissues. All negative ion mode ions were putatively assigned as M-H ions except for glucose/fructose, which was identified as M + Cl. The same section slides were then stained with H&E.
Targeted MS analysis
The metabolic effect of nCB in the lungs was assessed in collaboration with the Baylor College of Medicine Metabolomics Core. Fresh lung samples were flash-frozen and kept at −80°C for metabolite analysis. The metabolome extraction and MS details were as described (63).
Measurement of mtROS
mtROS were determined by staining cells with MitoSox Red mitochondrial superoxide indicator (Invitrogen). Cells were washed twice with HBSS and then stained with 5 μM MitoSox for 15 min at 37°C. Before fixing the cells with 4% formaldehyde, cells were washed three times with prewarmed HBSS to remove the extra dye. The slides were then mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen) and examined by Zeiss Imager (M2 microscope with Zeiss AxioCam MRm 1.4MP CCD Monochrome Microscope Camera). To quantify the MitoSox signal in each cell, QuPath and ImageJ were used.
OCR/ECAR analysis
The oxygen consumption rate (OCR) and ECAR were determined by the XFp extracellular flux analyzer (Agilent). For BEAS-2B cells, cells were treated with nCB (50 μg/ml) in a six-well plate. After 24 hours, cells were detached by 0.25% trypsin. Cell viability was checked by acridine orange/propidium iodide stain (Nexcelom). Cells with viability > 80% were used for this experiment. BEAS-2B cells (6 × 104) were seeded in the Cell-Tak (Corning) precoated XFp plate. For CD11c+ Siglec F+ alveolar macrophages, BAL cells were directly seeded in the plate (7 × 104 cells per well). After 2 hours of incubation, suspension cells were removed from the supernatant, leaving alveolar macrophages attached at the bottom. Alveolar macrophages were then treated with nCB (50 μg/ml) for 24 hours. For BMDCs, 7 × 104 cells were seeded in the precoated plate followed by nCB or PBS treatment. Before analyzing the metabolic changes, the media were replaced with Agilent Seahorse XF Base Medium (without Phenol Red) supplemented with l-glutamine (2 mM) alone or addition with l-glucose (10 mM), and the cells were incubated at 37°C in a non-CO2 incubator to equilibrate the CO2 level in the atmosphere. OCR and ECAR were then detected by XFp analyzer under baseline conditions and the treatment of metabolic drugs, glucose (10 mM), 2-deoxyglucose (50 mM), oligomycin (2 μM for the mito stress test and 1 μM for glycolysis stress test), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (0.5 μM), and rotenone/antimycin A (0.5/0.5 μM). Results were analyzed by Seahorse Wave Desktop Software.
RNA extraction and real-time quantitative PCR
Cells were lysed with TRIzol reagent (Invitrogen). RNA was then extracted by phenol/chloroform. Reverse transcription was performed according to the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). Real-time polymerase chain reaction (PCR) reaction and analysis were conducted in a 7500 Real-Time PCR system (Applied Biosystems) using TaqMan probes [Il4i1 (Mm00516786), Nos2 (Mm00440502), Arg1 (Mm00492586), Ido1 (Mm00492586), Glut1 (Mm00441480), Pdha1 (Mm00468675), Ldha (Mm01612132), Hk1 (Mm00439344), Nrf1 (Mm00447996), Nrf2 (Mm00477784), Tfam (Mm00447485), Ppargc1a (Mm01208835), GLUT1 (Hs00892681), HK1 (Hs00175976), and LDHA (Hs01378790); Invitrogen], Gzmb (Mm00442837_m1), and TaqMan Universal PCR Master Mix (Applied Biosystems).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 software. Statistical comparison between groups was performed using the unpaired Student’s t test, one-way or two-way analysis of variance (ANOVA) with Holm’s correction, Wilcoxon signed-rank test, and Fisher’s test when appropriate. A P value less than 0.05 was considered statistically significant; NS indicates not significant. Statistical significance values were set as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are presented as means ± SEM. P value and n can be found in the main and supplementary figure legends.
Acknowledgments
We thank J. Sederstrom in the Cytometry and Cell Sorting Core (CCSC) at Baylor College of Medicine for expert assistance. We thank G. Diehl, L. Burrage, and B. Stroup for providing mouse breeders, and A. Rodriguez and C.-H. Lin for assisting in silico analysis. The DESI mass spectrometry imaging analysis was contributed by B. Shestha and A. Midey in the Water’s Corporation. Electronic microscopy analysis was conducted by J. Barrish in the Pathology Department at the Texas Children’s Hospital.
Funding: This work was supported by the National Institutes of Health grants R01 ES029442-01 and R01 AI135803-01 (F.K.); T32 AI053831, R01 HL117181, HL140398, R01 AI135803, and R41 AI124997 (D.B.C.); R01 DK115454 (A.C.); T32 GM088129 (D.A.); T32 GM007330 (P.M.B.); and T32 DK060445-17 (L.M.); National Institutes of Health merit award R37 CA248478 (B.M.B.); National Institutes of Health Institutional Research and Career Development Award K12 GM 88010-12 to M.C.M.; the U.S. Veterans Administration (VA) merit grant CX000104 (F.K.); the U.S. VA Office of Research and Development grant I01BX004828 (D.B.C.); the Cancer Prevention and Research Institute of Texas (CPRIT) grant RP200443 (H.-S.L.); the American Thoracic Society unrestricted grant and American Heart Association Career Development award 19CDA34660279 (X.Y.); American Lung Association Catalyst award CA-622265 (X.Y.); the Center for Clinical and Translational Sciences, McGovern Medical School Pilot award 1UL1TR003167–01 (X.Y.); Parker B. Francis Fellowship (X.Y.); the Southwest Center for Occupational and Environmental Health Pilot Projects Research Training award CDC/NIOSH T42 OH008421 (C.-Y.C.); Baylor College of Medicine Comprehensive Cancer Training Program, CPRIT RP160283 (C.-Y.C.); and Baylor Research Advocates for Student Scientists (BRASS) scholarship (D.A.). CyTOF and flow cytometry experiments were conducted in the CCSC, funded by the CPRIT Core Facility Support Award (CPRIT-RP180672) and the NIH (P30 CA125123 and S10 RR024574). Targeted metabolomics was supported by the Metabolomics Core at Baylor College of Medicine funded by the NIH (P30 CA125123) and Dan L. Duncan Cancer Center.
Author contributions: Conceptualization: C.-Y.C., R.Y., and F.K. Methodology: C.-Y.C., R.Y., P.P., E.A.M., D.C.M., J.M.K., N.P., B.M.B., A.C., L.M., X.Y., and J.M.T. Investigation: C.-Y.C., R.Y., Y.-T.C., D.A., A.B., P.M.B., L.B.-D., P.E.P., P.P., H.-Y.T., Z.Z., L.M., L.S., and Y.W. Visualization: C.-Y.C. and F.K. Supervision: F.K. Analysis: C.-Y.C., R.Y., Y.-T.C., and H.-S.L. Writing—original draft: C.-Y.C. Writing—review and editing: C.-Y.C., R.Y., P.P., M.C.M., D.B.C., and F.K.
Competing interests: B.M.B. has served as an advisor to AstraZeneca. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.“7 Million premature deaths annually linked to air pollution,” World Health Organization, 25 March 2014. [PubMed]
- 2.Di Q., Wang Y., Zanobetti A., Wang Y., Koutrakis P., Choirat C., Dominici F., Schwartz J. D., Air pollution and mortality in the medicare population. N. Engl. J. Med. 376, 2513–2522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen A. J., Brauer M., Burnett R., Anderson H. R., Frostad J., Estep K., Balakrishnan K., Brunekreef B., Dandona L., Dandona R., Feigin V., Freedman G., Hubbell B., Jobling A., Kan H., Knibbs L., Liu Y., Martin R., Morawska L., Pope C. A. III, Shin H., Straif K., Shaddick G., Thomas M., van Dingenen R., van Donkelaar A., Vos T., Murray C. J. L., Forouzanfar M. H., Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: An analysis of data from the Global Burden of Diseases Study 2015. Lancet 389, 1907–1918 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long C. M., Nascarella M. A., Valberg P. A., Carbon black vs. black carbon and other airborne materials containing elemental carbon: Physical and chemical distinctions. Environ. Pollut. 181, 271–286 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Blasch K. W., Kolivosky J. E., Heller J. M., Environmental air sampling near burn pit and incinerator operations at bagram airfield, Afghanistan. J. Occup. Environ. Med. 58, S38–S43 (2016). [DOI] [PubMed] [Google Scholar]
- 6.You R., Lu W., Shan M., Berlin J. M., Samuel E. L. G., Marcano D. C., Sun Z., Sikkema W. K. A., Yuan X., Song L., Hendrix A. Y., Tour J. M., Corry D. B., Kheradmand F., Nanoparticulate carbon black in cigarette smoke induces DNA cleavage and Th17-mediated emphysema. eLife 4, e09623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu W., You R., Yuan X., Yang T., Samuel E. L. G., Marcano D. C., Sikkema W. K. A., Tour J. M., Rodriguez A., Kheradmand F., Corry D. B., The microRNA miR-22 inhibits the histone deacetylase HDAC4 to promote TH17 cell-dependent emphysema. Nat. Immunol. 16, 1185–1194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pun V. C., Kazemiparkouhi F., Manjourides J., Suh H. H., Long-term PM2.5 exposure and respiratory, cancer, and cardiovascular mortality in older US adults. Am. J. Epidemiol. 186, 961–969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan M., Yuan X., Song L.-Z., Roberts L., Zarinkamar N., Seryshev A., Zhang Y., Hilsenbeck S., Chang S.-H., Dong C., Corry D. B., Kheradmand F., Cigarette smoke induction of osteopontin (SPP1) mediates TH17 inflammation in human and experimental emphysema. Sci. Transl. Med. 4, 117ra119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N., Xia T., Nel A. E., The role of oxidative stress in ambient particulate matter-induced lung diseases and its implications in the toxicity of engineered nanoparticles. Free Radic. Biol. Med. 44, 1689–1699 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saldiva P. H., Clarke R. W., Coull B. A., Stearns R. C., Lawrence J., Murthy G. G. K., Diaz E., Koutrakis P., Suh H., Tsuda A., Godleski J. J., Lung inflammation induced by concentrated ambient air particles is related to particle composition. Am. J. Respir. Crit. Care Med. 165, 1610–1617 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Romieu I., Castro-Giner F., Kunzli N., Sunyer J., Air pollution, oxidative stress and dietary supplementation: A review. Eur. Respir. J. 31, 179–197 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Mills E. L., Kelly B., O’Neill L. A. J., Mitochondria are the powerhouses of immunity. Nat. Immunol. 18, 488–498 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Sorbara M. T., Girardin S. E., Mitochondrial ROS fuel the inflammasome. Cell Res. 21, 558–560 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dan Dunn J., Alvarez L. A., Zhang X., Soldati T., Reactive oxygen species and mitochondria: A nexus of cellular homeostasis. Redox Biol. 6, 472–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ip W. K. E., Hoshi N., Shouval D. S., Snapper S., Medzhitov R., Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson M. J., Vignali P. D. A., Mullett S. J., Overacre-Delgoffe A. E., Peralta R. M., Grebinoski S., Menk A. V., Rittenhouse N. L., DePeaux K., Whetstone R. D., Vignali D. A. A., Hand T. W., Poholek A. C., Morrison B. M., Rothstein J. D., Wendell S. G., Delgoffe G. M., Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 591, 645–651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschhaeuser F., Sattler U. G., Mueller-Klieser W., Lactate: A metabolic key player in cancer. Cancer Res. 71, 6921–6925 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Husain Z., Huang Y., Seth P., Sukhatme V. P., Tumor-derived lactate modifies antitumor immune response: Effect on myeloid-derived suppressor cells and NK cells. J. Immunol. 191, 1486–1495 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Faubert B., Li K. Y., Cai L., Hensley C. T., Kim J., Zacharias L. G., Yang C., Do Q. N., Doucette S., Burguete D., Li H., Huet G., Yuan Q., Wigal T., Butt Y., Ni M., Torrealba J., Oliver D., Lenkinski R. E., Malloy C. R., Wachsmann J. W., Young J. D., Kernstine K., DeBerardinis R. J., Lactate metabolism in human lung tumors. Cell 171, 358–371.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apicella M., Giannoni E., Fiore S., Ferrari K. J., Fernández-Pérez D., Isella C., Granchi C., Minutolo F., Sottile A., Comoglio P. M., Medico E., Pietrantonio F., Volante M., Pasini D., Chiarugi P., Giordano S., Corso S., Increased lactate secretion by cancer cells sustains non-cell-autonomous adaptive resistance to MET and EGFR targeted therapies. Cell Metab. 28, 848–865.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Liu J., Cho S. N., Akkanti B., Jin N., Mao J., Long W., Chen T., Zhang Y., Tang X., Wistub I. I., Creighton C. J., Kheradmand F., DeMayo F. J., ErbB2 pathway activation upon smad4 loss promotes lung tumor growth and metastasis. Cell Rep. 10, 1599–1613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D. H., Lee H. S., Yoon D., Berry G., Wheeler T. M., Sugarbaker D. J., Kheradmand F., Engleman E., Burt B. M., Progression of EGFR-mutant lung adenocarcinoma is driven by alveolar macrophages. Clin. Cancer Res. 23, 778–788 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Colegio O. R., Chu N. Q., Szabo A. L., Chu T., Rhebergen A. M., Jairam V., Cyrus N., Brokowski C. E., Eisenbarth S. C., Phillips G. M., Cline G. W., Phillips A. J., Medzhitov R., Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer K., Hoffmann P., Voelkl S., Meidenbauer N., Ammer J., Edinger M., Gottfried E., Schwarz S., Rothe G., Hoves S., Renner K., Timischl B., Mackensen A., Kunz-Schughart L., Andreesen R., Krause S. W., Kreutz M., Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109, 3812–3819 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Becher B., Schlitzer A., Chen J., Mair F., Sumatoh H. R., Teng K. W. W., Low D., Ruedl C., Riccardi-Castagnoli P., Poidinger M., Greter M., Ginhoux F., Newell E. W., High-dimensional analysis of the murine myeloid cell system. Nat. Immunol. 15, 1181–1189 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Mendler A. N., Hu B., Prinz P. U., Kreutz M., Gottfried E., Noessner E., Tumor lactic acidosis suppresses CTL function by inhibition of p38 and JNK/c-Jun activation. Int. J. Cancer 131, 633–640 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Jin H. T., Ahmed R., Okazaki T., Role of PD-1 in regulating T-cell immunity. Curr. Top. Microbiol. Immunol. 350, 17–37 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Li M. O., Wan Y. Y., Sanjabi S., Robertson A. K., Flavell R. A., Transforming growth factor-β regulation of immune responses. Annu. Rev. Immunol. 24, 99–146 (2006). [DOI] [PubMed] [Google Scholar]
- 30.deLeeuw R. J., Kost S. E., Kakal J. A., Nelson B. H., The prognostic value of FoxP3+ tumor-infiltrating lymphocytes in cancer: A critical review of the literature. Clin. Cancer Res. 18, 3022–3029 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Shan M., Cheng H.-F., Song L.-Z., Roberts L., Green L., Hacken-Bitar J., Huh J., Bakaeen F., Coxson H. O., Storness-Bliss C., Ramchandani M., Lee S.-H., Corry D. B., Kheradmand F., Lung myeloid dendritic cells coordinately induce TH1 and TH17 responses in human emphysema. Sci. Transl. Med. 1, 4ra10 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Marxsen J. H., Stengel P., Doege K., Heikkinen P., Jokilehto T., Wagner T., Jelkmann W., Jaakkola P., Metzen E., Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem. J. 381, 761–767 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Neill L. A., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Gassen S., Callebaut B., Van Helden M. J., Lambrecht B. N., Demeester P., Dhaene T., Saeys Y., FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Conway E. M., Pikor L. A., Kung S. H. Y., Hamilton M. J., Lam S., Lam W. L., Bennewith K. L., Macrophages, inflammation, and lung cancer. Am. J. Respir. Crit. Care Med. 193, 116–130 (2016). [DOI] [PubMed] [Google Scholar]
- 36.McLean J., Anderson D., Capra G., Riley C. A., The potential effects of burn pit exposure on the respiratory tract: A systematic review. Mil. Med. 186, 672–681 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Angelin A., Gil-de-Gómez L., Dahiya S., Jiao J., Guo L., Levine M. H., Wang Z., Quinn W. J. III, Kopinski P. K., Wang L., Akimova T., Liu Y., Bhatti T. R., Han R., Laskin B. L., Baur J. A., Blair I. A., Wallace D. C., Hancock W. W., Beier U. H., Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 25, 1282–1293.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corzo C. A., Condamine T., Lu L., Cotter M. J., Youn J. I., Cheng P., Cho H. I., Celis E., Quiceno D. G., Padhya T., McCaffrey T. V., McCaffrey J. C., Gabrilovich D. I., HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J. Exp. Med. 207, 2439–2453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas R., Smith J., Rocher-Ros V., Nadkarni S., Montero-Melendez T., D’Acquisto F., Bland E. J., Bombardieri M., Pitzalis C., Perretti M., Marelli-Berg F. M., Mauro C., Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 13, e1002202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pucino V., Certo M., Bulusu V., Cucchi D., Goldmann K., Pontarini E., Haas R., Smith J., Headland S. E., Blighe K., Ruscica M., Humby F., Lewis M. J., Kamphorst J. J., Bombardieri M., Pitzalis C., Mauro C., Lactate buildup at the site of chronic inflammation promotes disease by inducing CD4+ T cell metabolic rewiring. Cell Metab. 30, 1055–1074.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan X., Chang C.-Y., You R., Shan M., Gu B. H., Madison M. C., Diehl G., Perusich S., Song L.-Z., Cornwell L., Rossen R. D., Wetsel R., Kimal R., Coarfa C., Eltzschig H. K., Corry D. B., Kheradmand F., Cigarette smoke-induced reduction of C1q promotes emphysema. JCI Insight 5, e124317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de la Cruz-Lopez K. G., Castro-Munoz L. J., Reyes-Hernandez D. O., Garcia-Carranca A., Manzo-Merino J., Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front. Oncol. 9, 1143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perillo B., di Donato M., Pezone A., di Zazzo E., Giovannelli P., Galasso G., Castoria G., Migliaccio A., ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 52, 192–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linke M., Pham H. T. T., Katholnig K., Schnöller T., Miller A., Demel F., Schütz B., Rosner M., Kovacic B., Sukhbaatar N., Niederreiter B., Blüml S., Kuess P., Sexl V., Müller M., Mikula M., Weckwerth W., Haschemi A., Susani M., Hengstschläger M., Gambello M. J., Weichhart T., Chronic signaling via the metabolic checkpoint kinase mTORC1 induces macrophage granuloma formation and marks sarcoidosis progression. Nat. Immunol. 18, 293–302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang C., Yu J., Luo Y., Chen S., Wang W., Zhao C., Sun Z., Wu W., Guo W., Han Z., Hu X., Liao F., Feng X., Tsc1 is a critical regulator of macrophage survival and function. Cell. Physiol. Biochem. 36, 1406–1418 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Yurdagul A. Jr., Subramanian M., Wang X., Crown S. B., Ilkayeva O. R., Darville L., Kolluru G. K., Rymond C. C., Gerlach B. D., Zheng Z., Kuriakose G., Kevil C. G., Koomen J. M., Cleveland J. L., Muoio D. M., Tabas I., Macrophage metabolism of apoptotic cell-derived arginine promotes continual efferocytosis and resolution of injury. Cell Metab 31, 518–533.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galvan-Pena S., O’Neill L. A., Metabolic reprograming in macrophage polarization. Front. Immunol. 5, 420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bleriot C., Chakarov S., Ginhoux F., Determinants of resident tissue macrophage identity and function. Immunity 52, 957–970 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Martinez F. O., Gordon S., The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arts R. J. W., Carvalho A., la Rocca C., Palma C., Rodrigues F., Silvestre R., Kleinnijenhuis J., Lachmandas E., Gonçalves L. G., Belinha A., Cunha C., Oosting M., Joosten L. A. B., Matarese G., van Crevel R., Netea M. G., Immunometabolic pathways in BCG-induced trained immunity. Cell Rep. 17, 2562–2571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallud A., Klöditz K., Ytterberg J., Östberg N., Katayama S., Skoog T., Gogvadze V., Chen Y. Z., Xue D., Moya S., Ruiz J., Astruc D., Zubarev R., Kere J., Fadeel B., Cationic gold nanoparticles elicit mitochondrial dysfunction: A multi-omics study. Sci. Rep. 9, 4366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Q., Wang N., Zhu M., Lu J., Zhong H., Xue X., Guo S., Li M., Wei X., Tao Y., Yin H., TiO2 nanoparticles cause mitochondrial dysfunction, activate inflammatory responses, and attenuate phagocytosis in macrophages: A proteomic and metabolomic insight. Redox Biol. 15, 266–276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohammadinejad R., Moosavi M. A., Tavakol S., Vardar D. Ö., Hosseini A., Rahmati M., Dini L., Hussain S., Mandegary A., Klionsky D. J., Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 15, 4–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dossou A. S., Basu A., The emerging roles of mTORC1 in macromanaging autophagy. Cancers 11, 1422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan H., Zhong X. P., Lee S., Sustained activation of mTORC1 in macrophages increases AMPKα-dependent autophagy to maintain cellular homeostasis. BMC Biochem. 17, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bourdon J. A., Saber A. T., Jacobsen N. R., Jensen K. A., Madsen A. M., Lamson J. S., Wallin H., Møller P., Loft S., Yauk C. L., Vogel U. B., Carbon black nanoparticle instillation induces sustained inflammation and genotoxicity in mouse lung and liver. Part. Fibre Toxicol. 9, 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bové H., Bongaerts E., Slenders E., Bijnens E. M., Saenen N. D., Gyselaers W., Van Eyken P., Plusquin M., Roeffaers M. B. J., Ameloot M., Nawrot T. S., Ambient black carbon particles reach the fetal side of human placenta. Nat. Commun. 10, 3866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landers C. T., Tung H. Y., Knight J. M., Madison M. C., Wu Y., Zeng Z., Porter P. C., Rodriguez A., Flick M. J., Kheradmand F., Corry D. B., Selective cleavage of fibrinogen by diverse proteinases initiates innate allergic and antifungal immunity through CD11b. J. Biol. Chem. 294, 8834–8847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu B., Wu Y., Huang S., Zhang R., Son Y. M., Li C., Cheon I. S., Gao X., Wang M., Chen Y., Zhou X., Nguyen Q., Phan A. T., Behl S., Taketo M. M., Mack M., Shapiro V. S., Zeng H., Ebihara H., Mullon J. J., Edell E. S., Reisenauer J. S., Demirel N., Kern R. M., Chakraborty R., Cui W., Kaplan M. H., Zhou X., Goldrath A. W., Sun J., Uncoupling of macrophage inflammation from self-renewal modulates host recovery from respiratory viral infection. Immunity 54, 1200–1218.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.You R., DeMayo F. J., Liu J., Cho S. N., Burt B. M., Creighton C. J., Casal R. F., Lazarus D. R., Lu W., Tung H. Y., Yuan X., Hill-McAlester A., Kim M., Perusich S., Cornwell L., Rosen D., Song L. Z., Paust S., Diehl G., Corry D., Kheradmand F., IL17A regulates tumor latency and metastasis in lung adeno and squamous SQ.2b and AD.1. Cancer Immunol. Res. 6, 645–657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svedberg F. R., Brown S. L., Krauss M. Z., Campbell L., Sharpe C., Clausen M., Howell G. J., Clark H., Madsen J., Evans C. M., Sutherland T. E., Ivens A. C., Thornton D. J., Grencis R. K., Hussell T., Cunoosamy D. M., Cook P. C., MacDonald A. S., The lung environment controls alveolar macrophage metabolism and responsiveness in type 2 inflammation. Nat. Immunol. 20, 571–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Putluri N., Shojaie A., Vasu V. T., Nalluri S., Vareed S. K., Putluri V., Vivekanandan-Giri A., Byun J., Pennathur S., Sana T. R., Fischer S. M., Palapattu G. S., Creighton C. J., Michailidis G., Sreekumar A., Metabolomic profiling reveals a role for androgen in activating amino acid metabolism and methylation in prostate cancer cells. PLOS ONE 6, e21417 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5