Abstract
We used isobaric mass tagging (iTRAQ) and lectin affinity capture mass spectrometry (MS)-based workflows for global analyses of parotid saliva (PS) and whole saliva (WS) samples obtained from patients diagnosed with primary Sjögren’s Syndrome (pSS) who were enrolled in the Sjögren’s International Collaborative Clinical Alliance (SICCA) as compared with two control groups. The iTRAQ analyses revealed up- and down-regulation of numerous proteins that could be involved in the disease process (e.g., histones) or attempts to mitigate the ensuing damage (e.g., bactericidal/permeability increasing fold containing family (BPIF) members). An immunoblot approach applied to independent sample sets confirmed the pSS associated up-regulation of β2-microglobulin (in PS) and down-regulation of carbonic anhydrase VI (in WS) and BPIFB2 (in PS). Beyond the proteome, we profiled the N-glycosites of pSS and control samples. They were enriched for glycopeptides using lectins Aleuria aurantia and wheat germ agglutinin, which recognize fucose and sialic acid/N-acetyl glucosamine, respectively. MS analyses showed that pSS is associated with increased N-glycosylation of numerous salivary glycoproteins in PS and WS. The observed alterations of the salivary proteome and N-glycome could be used as pSS biomarkers enabling easier and earlier detection of this syndrome while lending potential new insights into the disease process.
Keywords: Sjögren’s Syndrome, isobaric mass tagging, iTRAQ, lectin affinity capture, parotid saliva, whole saliva, N-glycosylation, glycosite
Graphical Abstract
INTRODUCTION
Sjögren’s syndrome (SS) is a chronic, multisystem autoimmune disease that predominantly affects salivary and lacrimal function with the potential to cause substantial morbidity.1 Estimates of disease prevalence range from 0.5 to 1.5% worldwide, and, like most autoimmune diseases, SS predominantly affects women.2,3 SS may occur either as a primary disease process (pSS) or in the context of a connective tissue disease (typically rheumatoid arthritis (RA) or systemic lupus erythematosus (SLE)), commonly referred to as secondary SS. Overall, pSS is characterized by progressive destruction of the exocrine glands, with subsequent mucosal and conjunctival dryness.4,5 However, extraglandular manifestations are frequently present, particularly autoantibody production. New classification criteria for pSS, based on the weighted sum of five objective tests, have recently been approved by both the American College of Rheumatology (ACR) and the European League Against Rheumatism (EULAR).6 The objective tests considered in the criteria have been used in previous criteria sets and include serology to detect anti-SSA (Ro), a labial salivary gland (LSG) biopsy to assess and quantify focal lymphocytic sialadenitis, ocular staining and a Schirmer test to quantify dryness of the eye, and a salivary function test.7,8 Classification criteria are used to determine entry into clinical trials and studies. However, diagnosis of pSS is still problematic because it requires involvement of different clinical specialties and often is made with clinicians relying on nonspecific signs and symptoms, the most prevalent of which is salivary hypofunction resulting in severe xerostomia (patient reported dry mouth) and conjunctival dryness. This reflects the fact that there are no specific biomarkers for diagnosing SS. Despite being commonly found in sera and saliva of SS patients, autoantibodies to SSA/Ro and SSB/La are not a unique characteristic of the disease.9,10 Likewise, antibodies in saliva against α-fodrin, which are present in the majority of SS cases, are also observed in patients diagnosed with other autoimmune diseases (e.g., SLE, RA, and multiple sclerosis11). Furthermore, anti-α-fodrin antibodies are neither sensitive nor specific serum biomarkers of pSS.12 For these reasons, a diagnostic test that uses whole saliva as the sample would be ideal because it could be ordered by any clinician among the three clinical specialties (rheumatology, oral medicine, and ophthalmology) involved in the diagnosis and management of SS patients. Furthermore, oral medicine clinicians with expertise in performing the LSG biopsy with a minimal incision and oral pathologists with the expertise in interpreting the biopsy slide in the presence of focal lymphocytic sialadenitis are not available in certain regions, which is another reason why salivary biomarkers would be very attractive in diagnosing pSS in clinical practice. Because salivary gland hypofunction is a primary manifestation of SS, it stands to reason that saliva can be considered a potential noninvasive source of biomarkers for this disease. Comprehensive proteomic analyses of the salivary proteome13 provide a starting point for investigating the consequences of disease processes that include salivary gland pathologies, of which SS is an interesting example.14 We think that the salivary gland pathologies that are the hallmark of this disease will be reflected in changes in salivary protein composition and that these alterations can be exploited to eventually develop a noninvasive test for this condition that significantly reduces the ambiguities associated with current diagnostic criteria.
Historically, quantitative salivary research at the proteome level has relied heavily on two-dimensional gel electrophoresis (2DE) often combined with mass spectrometry (MS)-based methods. Global untargeted proteomics studies have been performed on human saliva with the goal of discovering putative protein biomarkers of pSS (reviewed in refs 15–17). Typically, samples from 6 to 12 pSS patients were evaluated (sample number range 2–41). The studies differed in a number of aspects, including specimen types (whole saliva, glandular secretions, gland biopsies; pooled vs individual samples), methods of sample collection (stimulated vs nonstimulated), as well as workflows used for the proteomics analyses. Except for a study that evaluated the effects of pilocarpine treatment on salivary peptides/proteins in pSS patients that employed high-performance liquid chromatography (HPLC) separation,18 a MudPit analysis of parotid saliva,19 and analysis of intact salivary proteins20 and endogenous salivary peptides21 by matrix-assisted laser desorption time-of-flight mass spectrometry, all other studies employed 2DE for protein separation and relative quantification.22–29
As a body fluid, saliva is notable for the number of highly glycosylated proteins it contains and the complexity of its carbohydrate posttranslational modifications.30–32 Several studies have shown that the exocrinopathy of SS impacts the glycosylation of salivary or blood proteins. In this regard, the majority of investigators have relied on assays that employ either antibodies or lectins that react with specific elements of oligosaccharide structures. Staining tissue sections of labial salivary glands with a panel of lectins that recognize the basic building blocks of oligosaccharide structures—wheat germ agglutinin (WGA: [GlcNAcβ1-4]2–5, Neu5Acα2-3Manβ1-4GlcNAcβ1-4GlacNAc), Sambucus nigra agglutinin (SNA: Neu5Acα2-6Gal/GalNAc), and Concanavalin A (Con A: branched N-linked saccharides)—demonstrated an SS-associated decrease in sialylation.18 Interestingly, immunoblotting showed that the cell surface receptor for advanced glycation end products (RAGE) was overexpressed in labial salivary glands of SS patients relative to control samples,33 with the opposite pattern observed in serum.34 In addition, serum levels of the soluble (s) forms of highly glycosylated adhesion molecules, sICAM-1, sE-selectin, neopterin, and sL-selectin, were significantly elevated in SS patients as compared with a control group.35,36 Recently, it was reported that the concentration of MUC5B and MUC7 in whole saliva were similar between pSS patients and healthy individuals. Nevertheless, immunoblotting and glycan staining revealed a pSS-associated reduction in mucin glycosylation that was particularly prominent in association with MUC7.37
The Sjögren’s International Collaborative Clinical Alliance (SICCA) comprised a longitudinal multisite study that recruited and enrolled a large cohort of individuals who had already been diagnosed with SS or had signs and symptoms that may be suggestive of the disease.5,38 In addition to the standardized collection of relevant clinical, laboratory, and histological information, a set of biospecimens relevant to this study was also obtained from the participants including unstimulated whole saliva (WS) and parotid saliva (PS) samples and LSG biopsies. Here we present the results of an MS-based effort to discover and verify pSS-associated changes in salivary protein abundance, or glycosylation using the well-characterized SICCA PS and WS samples. We chose to analyze PS because SS primarily attacks serous (not mucous) salivary glands. Because the parotid was composed primarily of serous glands, we reasoned that there might be greater evidence of disease than in WS, which is a product of both mucous and serous glands. Specifically, we used an isobaric mass-tagging (iTRAQ) approach, at the peptide level, to discover salivary proteins that were differentially expressed between disease and control samples obtained from the SICCA biorepository as well as saliva obtained from healthy individuals who were not SICCA participants. Validation by immunoblotting was performed on a second set of SICCA saliva samples. In addition, we used lectins, carbohydrate-binding proteins that recognized specific carbohydrate structures, as affinity matrices for capturing the N-glycome, which was also profiled in the same sample sets. The results showed interesting differences at the level of protein abundance and glycosylation, which lend insights into the disease process and could be exploited as biomarkers of this difficult-to-diagnose condition.
MATERIAL AND METHODS
Saliva Samples
For the iTRAQ and lectin affinity capture experiments, PS and WS samples were obtained from the SICCA biospecimen repository maintained at the University of California, San Francisco (http://sicca-online.ucsf.edu). To reduce heterogeneity, we focused on samples obtained from a subset of individuals: premenopausal, Caucasian women. Saliva samples (PS and WS) from pSS patients (n = 15) were obtained from individuals who were SSA- or SSB-positive, had focal lymphocytic sialadenitis based on LSG biopsy (i.e., >1 foci/4 mm2), and had keratoconjunctivitis sicca established by ocular staining (i.e., ocular staining score ≥3).8 SICCA control (SC) samples (n = 15) were acquired from individuals who had no objective evidence of pSS at the time of collection based on the aforementioned (and other) objective tests. Although gathered at multiple sites, the SICCA saliva samples were collected using a vetted standard operating procedure (SOP, available at http://sicca-online.ucsf.edu) to ensure uniformity of sample quality and storage. A third set of PS and WS samples (n = 14) was obtained from an independent bank maintained in the laboratory of Dr. Susan Fisher. These salivas, designated healthy control (HC) samples, were collected from healthy premenopausal Caucasian women who were not part of the SICCA study. Immunoblot validation studies were performed on a second cohort of PS and WS samples obtained from the SICCA and independent banks (n = 5/group). All saliva samples were stored at −80 °C until they were analyzed.
Reagents
All standard reagents were obtained from Fisher Scientific unless otherwise specified.
Protein Concentration Determinations
The protein concentration of all saliva samples was determined by using the Micro Bicinchoninic Acid (μBCA) assay (Pierce). Each sample was assayed in triplicate at two different dilutions. For samples that showed a coefficient of variation (CV) > 20% the assay was repeated.
Trypsin Digestion and Isobaric Mass Tag (iTRAQ) Labeling
The PS and WS samples (15 pSS, 15 SC, and 14 HC) were individually digested with trypsin and labeled with iTRAQ reagents. An aliquot of each saliva sample corresponding to 25 μg total protein was diluted with 1 M triethylammonium bicarbonate (TEAB) to a final protein concentration of 0.625 μg/μL. Proteins were denatured with 0.1% sodium dodecyl sulfate (SDS), and cysteine residues were reduced using 5 mM aqueous tris(2-carboxyethyl)phosphine (TCEP) and alkylated with aqueous 200 mM iodoacetamide in the dark for 10 min. Sequencing-grade trypsin (porcine, Promega) was added at an enzyme/substrate ratio of 1:50, and samples were incubated overnight at 37 °C. Immediately following digestion, samples were labeled with iTRAQ-reagents corresponding to a randomly assigned unique reporter ion m/z value (e.g., m/z = 113, 114, 115, 116, 118, 119, or 121). The iTRAQ reagents were removed from the freezer and allowed to warm to ambient temperature. Then, 50 μL of 2-propanol was added to each reagent vial, followed by thorough mixing. Next, the entire iTRAQ reagent mixture was added to the digested samples, followed by a 2 h incubation at room temperature. The labeling reaction was quenched by adding 1 μL of 1 M Tris-HCl, pH 8. Next, the digested and labeled samples were randomly distributed among seven 8-plexes. Each 8-plex contained seven digested saliva samples and one reference pool aliquot (see below). Desalting and SDS removal from each 8-plex was performed by using Detergent Removal Spin Columns (Pierce) according to the protocol provided by manufacturer. The detergent and salt-free samples were eluted in 50 mM TEAB and stored at −80 °C.
Reference Pool Generation and Distribution Among the 8-Plexes
The reference pool was created by combining aliquots corresponding to 25 μg of protein from each HC saliva sample. Then, the protein concentration of the pool was determined by μBCA assay. The pooled sample was reduced, alkylated, digested with trypsin, and labeled with iTRAQ reagent (m/z = 117), as described above. An aliquot corresponding to 25 μg of protein from the digested pool was added to each 8-plex, thus ensuring an identical reference for each sample.
Offline High pH iTRAQ-Labeled Peptide Fractionation
To maximize the number of identified proteins, each 8-plex was subjected to extensive fractionation by using reversed-phase high-performance liquid chromatography (RP HPLC) performed at pH 10. An aliquot of each 8-plex tryptic digest was vacuum concentrated and resuspended in aqueous 0.1% NH4OH. Fractionation of the iTRAQ-labeled tryptic peptides was carried out by using a Paradigm MS4 HPLC System (Michrom BioResources) equipped with a binary pump. UV detection was performed at 230 nm. Peptides were separated on a Zorbax Extend-C18 column, 4.6 × 100 mm, 3.5 μm particle size (Agilent Technologies) by using mobile phase A (aqueous 0.1% NH4OH) and mobile phase B (0.1% NH4OH in acetonitrile/water, 80:20, v/v) paired in a stepwise linear gradient: 6% B for 10 min, 6–32% B over 30 min, 32–100% B over 6 min, 100% B for 2 min, followed by a 10 min of re-equilibration of the column at 6% B. The overall flow rate was 0.7 mL/min. A total of 18 1.4 mL fractions were collected. The fractions collected at the beginning and end of the gradient were combined, which resulted in 10 fractions per 8-plex, that is, a total of 70 samples for LC–MS analysis. The samples were vacuum-dried and reconstituted in aqueous 0.1% formic acid prior to storage at −80 °C.
Mass Spectrometry Analysis of iTRAQ-Labeled Peptides
Each high pH fraction was analyzed by RP HPLC MS/MS using a nanoLC Ultra system (Eksigent Technologies) interfaced with a 5600 TripleTOF mass spectrometer (SCIEX). Peptides were separated using an Acclaim PepMap100 C18 column (75 μm i.d. × 15 cm, 3 μm particle size, 100 Å pores) using mobile phase A (aqueous 0.1% formic acid) and mobile phase B (acetonitrile/water, 98:2, v/v, 0.1% formic acid) in conjunction with a linear gradient of 2–40% B over 60 min. The flow rate was 300 nL/min. The 5600 TripleTOF was operated in independent data acquisition (IDA) ion selection mode for MS/MS data collection. An initial survey scan was acquired (m/z 400–1600) for 250 ms, followed by MS/MS spectra (m/z 100–1600) for 150 ms in unit mode for the 20 most abundant precursor ions with an exclusion time of 15 s. Rolling collision energy was used while activating the with iTRAQ option.
iTRAQ Data Analyses
For protein identification and quantification, the data files generated by the Analyst TF 1.6 software, were submitted to database searching using ProteinPilot v4.5 (SCIEX). For each iTRAQ 8-plex, the MS data from the 10 fractions were analyzed together as one sample. The data were searched against the human subset of the SwissProt database (2014/01/22), which contained protein isoforms. Search parameters included sample type: iTRAQ 8-plex (peptide labeled), cysteine alkylation: iodoacetamide, digestion enzyme: trypsin, instrument: TripleTOF 5600, ID focus: Biological modification and Thorough search effort. The false discovery rate (FDR) was assessed by concomitant searching a decoy database of reversed protein sequences that was automatically generated by the ProteinPilot software. Reported proteins were identified with a local FDR of ≤5%. Protein Alignment Template v2.001 (SCIEX)39 was used to align the ProteinPilot generated protein summaries across the seven iTRAQ 8-plexes.
Immunoblotting
Saliva samples (20 μg) were electrophoretically separated on 10–20% Tris-glycine gels (1.5 mm thick, Novex by Life Technologies) and transferred to nitrocellulose membranes, followed by blocking for 1 h with 5% nonfat powdered milk in PBS-Tween before incubating overnight at 4 °C with the primary antibodies. Primary antibodies used were anti-BPIFA2 (Novus Biologicals), anti-μ2 M (Santa Cruz Biotechnology), and anti-CAH6 (Abcam). Then, the membranes were washed three times in PBS-Tween and incubated with a secondary antibody (Peroxidase AffiPure Donkey Anti-Rabbit IgG or Peroxidase AffiPure Donkey Anti-Mouse IgG from Jackson ImmunoResearch) at a 1:5000 dilution for 1 h. Protein bands were detected with ECL 2 Western Blotting Substrate (Thermo Scientific, Pierce) and ECL high-performance chemiluminescence film (Amersham, GE Healthcare).
Preparation of Saliva Samples for Lectin Affinity Enrichment
Aliquots of PS and WS from the three groups (pSS, SC, HC; n = 15/groups) were mixed to form six pools containing 500 μg protein each. Replicate pools were prepared for each saliva type (six pools total). The saliva samples were denatured with 6 M urea, reduced with 20 mM DTT (30 min at 37 °C), alkylated with 50 mM iodoacetamide (30 min at RT), and incubated overnight at 37 °C with sequencing grade trypsin (Promega, Madison, WI) added at a 1:50 (w/w) enzyme/substrate ratio. Following digestion, the samples were acidified with formic acid and desalted using HLB Oasis SPE cartridges (Waters, Milford, MA) that were prepared by washing with 80% acetonitrile in 1% formic acid prior to equilibration with 0.1% formic acid. Samples were eluted with 80% acetonitrile in 0.1% formic acid, neutralized by the addition of 0.5 M ammonium bicarbonate, and concentrated by vacuum centrifugation. Peptides were stored at −80 °C until use.
Lectin Column Preparation
Lectin columns were prepared according to our previously published method.40 In brief, WGA and Aleuria aurantia lectin (AAL) were purchased from Vector Laboratories (Burlingame, CA). Lectins (1.5–6 mg) were suspended at 10–20 mg/mL in PBS. One hundred mg POROS-AL beads (Applied Biosystems, Foster City, CA) was washed two times in 1 mL of PBS before the addition of the lectin solution. Sodium cyanoborohydride was added to a final concentration of 50 mM, and the sample was tumbled overnight at room temperature. The beads were washed once with 1 mL of 1 M Tris–HCl, pH 7.4. The remaining aldehydes were blocked by incubation for 30 min at room temperature in the same buffer containing 50 mM sodium cyanoborohydride. Unconjugated protein was removed by washing the beads (5 × 1 mL of 1 M sodium chloride). Then, the lectin-conjugated beads, ~300 μL, were packed into 4.6 × 50 mm PEEK HPLC columns and stored in PBS with 0.02% sodium azide at 4 °C. Prior to use, the lectin columns were assessed, according to our previously published protocol, for their capacity to bind N-linked glycopeptides using human lactoferrin and bovine fetuin as standards for AAL and WGA, respectively.40
Lectin Chromatography
A Paradigm MG4 HPLC system equipped with a CTC PAL robot configured as an autosampler and fraction collector (Michrom BioResources) was employed using mobile phase A: 25 mM Tris, pH 7.4, 10 mM calcium chloride, 10 mM magnesium chloride and mobile phase B: 0.5 M acetic acid. Lectin chromatography was performed on AAL and WGA columns prepared as described above. Each saliva tryptic digest was diluted into mobile phase A containing 50 mM sodium chloride. Then, the entire digest was applied to the lectin column and separated by using a three-step gradient: (1) collection of the unbound fraction, mobile phase A for 9.0 min at 50 μL/min; (2) elution, mobile phase B for 4.8 min at 500 μL/min; and (3) re-equilibration, mobile phase A for 6.0 min at 3000 μL/min. The bound fraction was desalted by using Oasis HLB cartridges as described above. Eluted samples were neutralized by the addition of 0.5 M ammonium bicarbonate and concentrated to <100 μL by vacuum centrifugation. N-Linked glycopeptides in the bound fractions were deglycosylated by treatment with PNGase F (glycerol-free, New England Biolabs, Ipswich, MA). A few microliters were used to estimate pH. If outside the desired range of 7.0 to 8.0, the pH was adjusted with 50 mM ammonium bicarbonate. Five microliters (2500 U) of glycosidase was added to each sample in a volume of 50–100 μL prior to incubation overnight at 37 °C. Following deglycosylation, samples were desalted and concentrated by using C18 Zip-Tips (Millipore, Billerica, MA).
Mass Spectrometry Analysis of PNGase F-Treated Samples
The PNGase-F-treated samples were analyzed by RP HPLC MS/MS using a nanoLC Ultra system (Eksigent Technologies) interfaced with a LTQ Orbitap Velos mass spectrometer (ThermoFisher Scientific). Peptides were separated by using an Acclaim PepMap100 C18 column (75 μm i.d. × 15 cm, 3 μm particle size, 100 Å pores) using mobile phase A (0.1% formic acid) and mobile phase B (acetonitrile/water, 98:2, v/v, 0.1% formic acid) in conjunction with a step gradient of 0–40% B (40 min), 40% B (10 min), 40–80% B (5 min), and 80% B (10 min). The flow rate was 500 nL/min. The LTQ Orbitrap Velos was operated in a data-dependent acquisition mode for MS and MS/MS data collection. An initial survey scan was acquired (m/z 350–1500) in the Orbitrap analyzer at mass resolution 30 000, followed by collision-induced dissociation of precursor ions in the ion trap to produce MS/MS spectra for the 20 most abundant precursor ions. The instrument was operated with an isolation width of 1.0, a normalized collision energy of 35, an Act Q of 0.25, and an MS/MS acquisition time of 30 ms.
N-Glycopeptide Assignment
Deglycosylated peptides were identified as previously described40 on the basis of several criteria, including the consensus motif NxS/T, x ≠ proline, in which Asn was converted to Asp (reported by the search engine as Asn deamidation), and the presence of at least one fragment ion encompassing the glycosite. To ensure inclusion of glycosites containing Lys or Arg in the X position (e.g., NKT), which were likely to have been cleaved by trypsin, the amino-acid residue following the carboxy-terminal cleavage site was also considered. Peptides containing the motif NGS or NGT were excluded due to the fact that asparagine residues in that sequence are prone to chemical deamidation during overnight trypsin digestion.41
RESULTS AND DISCUSSION
iTRAQ Analysis
We analyzed banked PS and WS samples from three groups of donors. The first group consisted of SICCA participants who were diagnosed with pSS according to classification criteria defined by the American College of Rheumatology.8 The second group was individuals who were evaluated for possible pSS at the SICCA clinic but had no objective signs of the disease at the time of enrollment. The third group was healthy individuals, with no signs of autoimmune disease, who were not part of the SICCA study.
The iTRAQ workflow yielded two types of data. First, we identified and quantified the salivary proteomes of WS (345 proteins) and PS (230 proteins) across all 8-plexes at an FDR of 5% (Table S-1). Eighty-four percent of PS proteins and 89% of proteins in WS were identified in all of the 8-plexes. Second, the quantification results allowed for comparisons among the donor groups (pSS vs HC, pSS vs SC, and SC vs HC) and saliva types (PS and WS). Table 1 lists the number of up- and down-regulated proteins (p < 0.05) for each comparison. For both saliva types, the greatest number of differentially expressed (DE) proteins was observed between the pSS vs HC samples (44 PS, 49 WS) with pSS vs SC (20 PS, 21 WS) and SC vs HC (27 PS, 17 WS) having substantially lower numbers. This observation is despite the fact that relatively equal numbers of DE proteins were identified between PS and WS, regardless of which two donor groups were compared. Figure 1 illustrates the data according to saliva types and the number of shared and unique identifications. When the pSS vs HC group was compared to the pSS vs SC group, both PS and WS showed approximately equal numbers of shared identifications (12 vs 13 for PS and WS, respectively) and similar or identical numbers of unique identifications (32 vs 36 and 8 vs 8 for PS and WS, respectively). When the pSS vs HC group was compared to the SC vs HC group, the shared identifications diverged (19 vs 10, for PS and WS, respectively). Only in the pSS vs HC comparison were more unique identifications made in WS vs PS. Finally, when the pSS vs SC group was compared with the SC vs HC group, few shard proteins were identified. The bulk of the identifications were unique. Table S-2 lists the differentially expressed proteins for all comparisons (p ≤ 0.05).
Table 1.
Numbers of Differentially Expressed (p < 0.05) Proteins Observed in Disease (pSS), SICCA Control (SC), and Healthy Control (HC) Salivas
| saliva type | pSS vs HC | pSS vs SC | SC vs HC | |
|---|---|---|---|---|
| parotid | total | 44 | 20 | 27 |
| ↑ | 23 | 11 | 14 | |
| ↓ | 21 | 9 | 13 | |
| whole | 49 | 21 | 17 | |
| ↑ | 21 | 10 | 3 | |
| ↓ | 28 | 11 | 14 |
Figure 1.
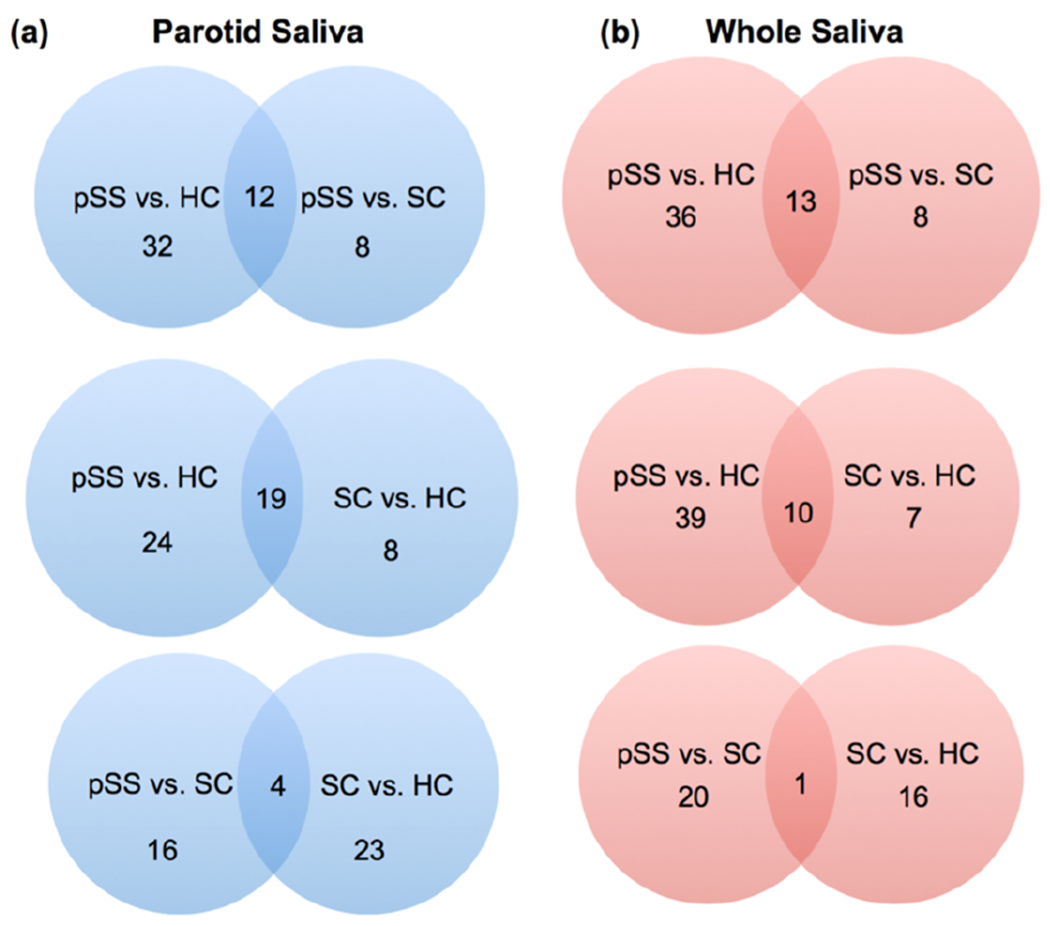
Many of the differentially expressed salivary proteins in saliva samples from pSS patients and control individuals were unique to the donor group. Shown is the number of shared and unique proteins among the groups: pSS vs HC, pSS vs SC, and SC vs HC. Data are shown for both saliva types PS (a) and WS (b) that were analyzed.
Table 2a contains the 13 most highly DE proteins from the pSS vs HC comparison for both saliva types. The identification, in WS, of several histones was unanticipated due to the fact that they are nuclear proteins rather than secretory products. We took this as evidence of the disease process, which involves B-cell-induced apoptosis of salivary (and lacrimal) glandular epithelium.42 Moreover, antihistone autoantibodies are detected, by protein array, in whole saliva from women diagnosed with pSS and at significantly higher levels as compared with saliva samples obtained from patients with systemic lupus erythematosus (SLE) and healthy individuals.43 The presence of histones in the salivary extracellular compartment of pSS patients, as indicated by their detection in WS, is likely a key component of the process by which the antihistone autoantibodies are generated.
Table 2.
iTRAQ-Based Comparison of Relative Abundances of Proteins Observed in PS and WS from pSS Patients versus (a) Healthy Individuals and (b) SICCA Controls
| UniProt accession number | protein name | saliva type | p value | fold change |
|---|---|---|---|---|
| (a) Healthy Individuals | ||||
| P62805 | histone H4 | WS | 1.1 × 10−2 | ↑ 9.6 |
| Q6FI13 | histone H2A type 2-A | WS | 1.8 × 10−2 | ↑ 8.4 |
| Q5QNW6 | histone H2B type 2-F | WS | 6.3 × 10−4 | ↑ 6.7 |
| Q71DI3 | histone H3.2 | WS | 4.3 × 10−2 | ↑ 4.1 |
| P61769 | beta-2-microglobulin | PS | 1.5 × 10−2 | ↑ 4.1 |
| P04083 | annexin A1 | WS | 1.1 × 10−2 | ↑ 3.7 |
| Q08380 | galectin-3-binding protein | PS | 1.0 × 10−4 | ↑ 2.8 |
| Q96DR5 | BPI fold-containing family A member | PS | 6.9 × 10−4 | ↓ 6.7 |
| P23280-2 | isoform 2 of carbonic anhydrase 6 | WS | 1.1 × 10−3 | ↓ 7.7 |
| P23280 | carbonic anhydrase 6 | PS | 7.6 × 10−4 | ↓ 8.3 |
| Q96DR5 | BPI fold-containing family A member 2 | WS | 7.7 × 10−4 | ↓ 9.1 |
| Q8N4F0 | BPI fold-containing family B member 2 | WS | 2.9 × 10−5 | ↓ 10 |
| Q8TDL5 | BPI fold-containing family B member 1 | WS | 1.2 × 10−5 | ↓ 14 |
| (b) SICCA Controls | ||||
| Q6FI13 | histone H2A type 2-A | WS | 8.0 × 10−3 | ↑ 10 |
| P01857 | Ig gamma-1 chain C region | PS | 1.6 × 10−4 | ↑ 9.5 |
| Q92817 | envoplakin | WS | 2.6 × 10−2 | ↑ 7.9 |
| P62805 | histone H4 | WS | 2.4 × 10−2 | ↑ 6.7 |
| Q15782 | chitinase-3-like protein 2 | PS | 2.7 × 10−2 | ↑ 5.7 |
| P07476 | involucrin | PS | 3.4 × 10−2 | ↑ 4.3 |
| P63104 | 14–3—3 protein zeta/delta | PS | 3.6 × 10−2 | ↑ 4.2 |
| P05164 | myeloperoxidase | WS | 3.5 × 10−2 | ↑ 3.4 |
| P06870 | kallikrein-1 | WS | 1.9 × 10−2 | ↓ 4.0 |
| PS | 1.5 × 10−2 | ↓ 4.1 | ||
| P01877 | Ig alpha-2 chain C region | WS | 5.3 × 10−3 | ↓ 4.3 |
| Q96DR5 | BPI fold-containing family A member 2 | WS | 3.4 × 10−3 | ↓ 7.1 |
| P23280 | carbonic anhydrase 6 | PS | 1.1 × 10−2 | ↓ 3.8 |
| P23280-2 | isoform 2 of carbonic anhydrase 6 | WS | 4.8 × 10−4 | ↓ 7.7 |
| P15515 | histatin-1 | PS | 1.1 × 10−2 | ↓ 3.8 |
| WS | 5.2 × 10−4 | ↓ 8.3 | ||
β-2-Microglobulin (β2m) has also been reported to be up-regulated in saliva from individuals diagnosed with pSS.28,44,45 This nonglycosylated protein, the light-chain component of the major histocompatibility class I heterodimer, is present on the surface of all nucleated cells. In this location, it plays a major role in immune responses. Salivary levels of β2m correlate with the extent of lymphocytic infiltration of the salivary glands in pSS patients.46 Therefore, our results corroborate previous observations, adding strength to our novel protein identifications.
Carbonic anhydrase VI (CAH6), a glycoprotein, is produced and secreted by all salivary glands. This zinc-containing metalloenzyme is responsible for regulating pH,47 a particularly important function in the oral cavity because tooth enamel erodes under acidic conditions. Accordingly, by binding to the enamel pellicle of salivary proteins that coat the tooth surface, CAH6 catalyzes the conversion of salivary bicarbonate and bacteria produced acids to carbon dioxide and water, thus preventing demineralization of the underlying surface hydroxyapatite.48 Significant down-regulation of CAH6 in the saliva of pSS patients relative to healthy individuals, which has been reported in multiple studies,22,28,49 was confirmed by our results. Thus reduced levels of CAH6 in the context of hyposalivation, a hallmark of pSS, creates a chronically acidic environment in the oral cavity of pSS patients, which likely contributes to the pathologies that are associated with this syndrome, including caries and other infections.
Annexin A1 (ANXA1) plays an important role in the innate immune response as an effector of glucocorticoid-mediated processes.50 It also regulates inflammation by suppressing phospholipase A2 production.51 ANXA1 is a known salivary protein and was detected, along with other annexin isoforms, as a major component of microvesicles isolated from WS.52 With regard to autoimmune diseases, antiannexin A1 antibodies have been found in association with SLE, RA, and cutaneous lupus erythematosus.53 In addition, in vitro studies suggest that ANXA1 has antiviral properties, for example, inhibition of cytomegalovirus infection of human foreskin fibroblasts.54 Our observation that ANXA1 was up-regulated in pSS WS adds to the growing body of evidence that this protein plays an important role in the pathology of several autoimmune diseases. Given its role in repair of epithelial barriers,55 we speculate that salivary ANXA1 expression is up-regulated in an attempt to compensate for disease-related ulcerations that occur in the oral mucosa.
Galectin-3-binding protein (LG3BP), which promotes integrin-mediated cell adhesion, is a powerful stimulator of host immune defense.56,57 Its physical association with galectin-3, another immune modulator, is carbohydrate-mediated.58 Furthermore, LG3BP is up-regulated in microparticles isolated from platelet-poor plasma of SLE patients56 and synovial tissues from individuals diagnosed with RA.59 As with ANXA1, higher levels in the saliva of pSS patients may indicate a heightened response to oral infections, which are more common in these individuals.
Several members of bactericidal/permeability increasing fold containing family (BPIF) proteins were down-regulated in PS and WS of pSS patients as compared with HC saliva samples. The BPIF family is encoded by nine genes located on chromosome 20, and myriad members are detected in secretions recovered from the upper airways, nose, and mouth.60–62 They are key players in the systemic innate immune response to Gram-negative bacteria.63 BPIF A member 2 (BPIFA2), also known as parotid secretory protein, which is produced by serous cells of the parotid, submandibular, and sublingual glands,62 is unique to the oral cavity, where it has bactericidal effects on Pseudomonas aeruginosa, as shown in a colony-forming assay.64 As with CAH6, it stands to reason that hyposalivation in pSS patients results in decreased levels of salivary BPIFA2 such that bacteria can proliferate in the oral cavity, thus stimulating a host of secondary infections. In line with this theory, BPIFA2 was reduced in saliva samples of individuals diagnosed with periodontal disease.65
Table 2b lists the 13 most highly DE proteins from the pSS vs SC comparison for both saliva types. Again, CAH6 and BPIFA2 were significantly down-regulated in PS and WS, as was observed in the pSS vs HC comparison, suggesting that reduced levels of these two proteins severely impact the ecology of the oral cavity of pSS patients. In addition, histone H2A type 2-A and histone H4 were among the most highly up-regulated proteins detected in WS (also mirroring the pSS vs HC results), possibly reflecting the nature of the pSS disease process with respect to B-cell-induced apoptosis of salivary (and lacrimal) glandular epithelium.
Envolplakin and involucrin were up-regulated in the pSS vs SC saliva samples. These proteins along with periplakin comprise the protein scaffold of the cornified envelope that forms the outermost layers of the epidermis.66,67 The cornified envelope is composed of corneocytes (dead keratinocytes). Normal skin physiology entails controlled shedding of the cornified layers. The observation that both envolplakin and involucrin were significantly up-regulated in pSS WS and PS, respectively, might be indicative of an abnormal, accelerated breakdown of the cornified layers of the oral epidermis in the pSS patients compared with the SC individuals. In a possibly related phenomenon, the up-regulation of chitinase-3-like protein 2 could be related to abnormal fibrosis.68
Histatin-1 is a histidine-rich polypeptide secreted by the parotid salivary gland and is known to play a role in regulating hydroxyapatite crystal growth on the enamel pellicle of teeth.69,70 It is also a component of the microbial/fungal host defense system of the oral cavity in that it inhibits the growth of Candida albicans.69 In addition, it has recently been shown that histatin-1 promotes cell–substrate and cell–cell adhesion.71 Significant down-regulation of histatin-1 was observed in pSS vs SC WS and PS saliva samples and could be due to reduced secretion secondary to parotid gland damage. Perhaps the up-regulation of myeloperoxidase plays a compensatory role because it has known antimicrobial properties.72 In addition, myeloperoxidase activity was found to be elevated in plasma isolated from pSS patients compared with gender- and age-matched controls.73 Finally, 14–3–3 zeta/delta levels were shown to be altered in crevicular fluid in the setting of chronic periodontitis.74
Kallikrein-1 is a member of a family of serine proteases secreted by the parotid and submandibular salivary glands.75 Kallikriens cleave kininogen to release active kinins and have also been shown to play a role in the initiation and maintenance of the inflammatory response.76,77 Tissue kallikriens were up-regulated in salivary glands isolated from a mouse model of SS78 and in saliva isolated from whole SS patients.79,80 We observed kallikrein-1 to be down-regulated in pSS WS and PS compared with SC. The reason for the discrepancy between our results and the previously published findings was not obvious but could be due to species differences, the use of different diagnostic criteria, or the patients being at different stages in the disease process.
Validation of iTRAQ DE Proteins
Immunoblot analyses were performed on sets of PS and WS samples (n = 5) that were not used in the iTRAQ experiments. Figure 2 shows the results obtained for β2m, CAH6, and BPIFB2. Lanes 1–5 and 6–10 were pSS and HC samples, respectively (refer to Figure S-1 for images of the entire immunoblot). On average, significantly higher levels of β2m were detected in the five PS samples from pSS patients (Figure 2a) as compared with those from HCs, thus confirming the up-regulation that was observed in the iTRAQ experiment. pSS-associated down-regulation of CAH6 in PS (Figure 2b) and BPIFB2 in WS (Figure 2c) was also confirmed. CAH6 and BPIFB2 had higher estimated molecular weights than predicted by their amino sequences, most likely due to glycosylation.81,82
Figure 2.
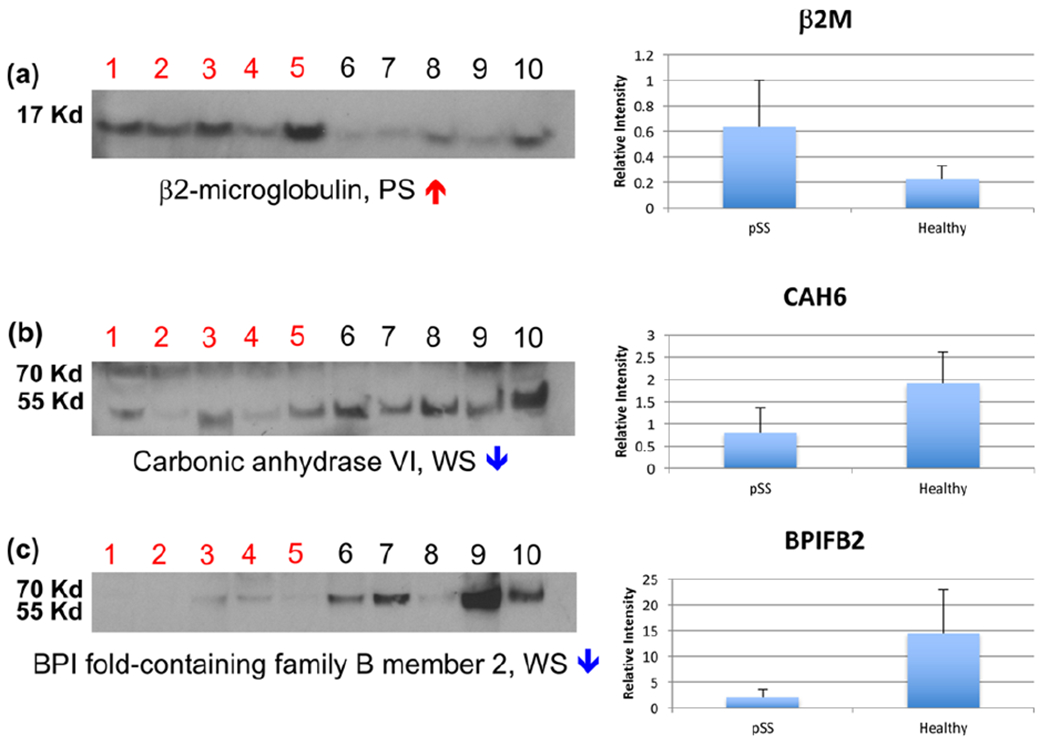
Validation of the proteins identified as differentially expressed in the iTRAQ experiments. Immunoblotting was used to verify the iTRAQ results in an independent cohort of PS and WS samples collected from pSS and HC donors (n = 5 per group). Overall, significantly higher amounts of β2-microglobulin were observed in pSS PS salivas as compared with HC samples (a), thus verifying the up-regulation detected in the iTRAQ experiment. Down-regulation of carbonic anhydrase VI in PS (b) and BPI fold-containing family B member 2 in WS was also confirmed. Lanes 1–5: pSS; lanes 6–10: HC. Refer to Figure S-1 for images of the entire immunoblot.
Pathway Analysis
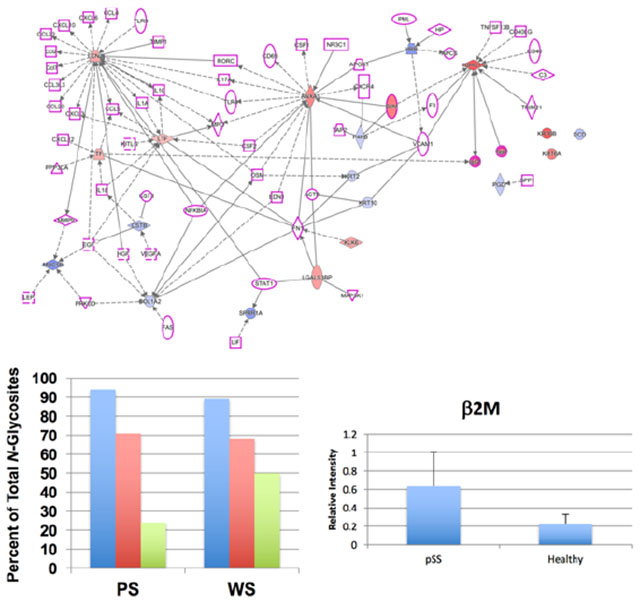
To gain additional insights into the possible roles of the observed DE proteins in pSS disease etiology, we employed Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com). The list of salivary DE proteins was uploaded to IPA, which linked them via their known physical and functional interactions. Interestingly, an initial analysis revealed a relationship with networks that were implicated in another autoimmune disease, psoriasis. It is not uncommon for pSS patients to also present with psoriasis, and reports suggest that the two conditions have similar disease etiologies.83–85 Accordingly, IPA was used to create network diagrams that highlighted DE salivary proteins that are involved in psoriasis and their direct connections to proteins that have been implicated in pSS. HC and SC data, which were used for comparison purposes, yielded nearly identical networks. Figure 3 maps the pSS vs HC data for WS and PS. The results suggested that many of the observed alterations in the pSS salivary protein repertoire were networked with pivotal immune and signaling pathways that are also implicated in the disease etiology of psoriasis.
Figure 3.
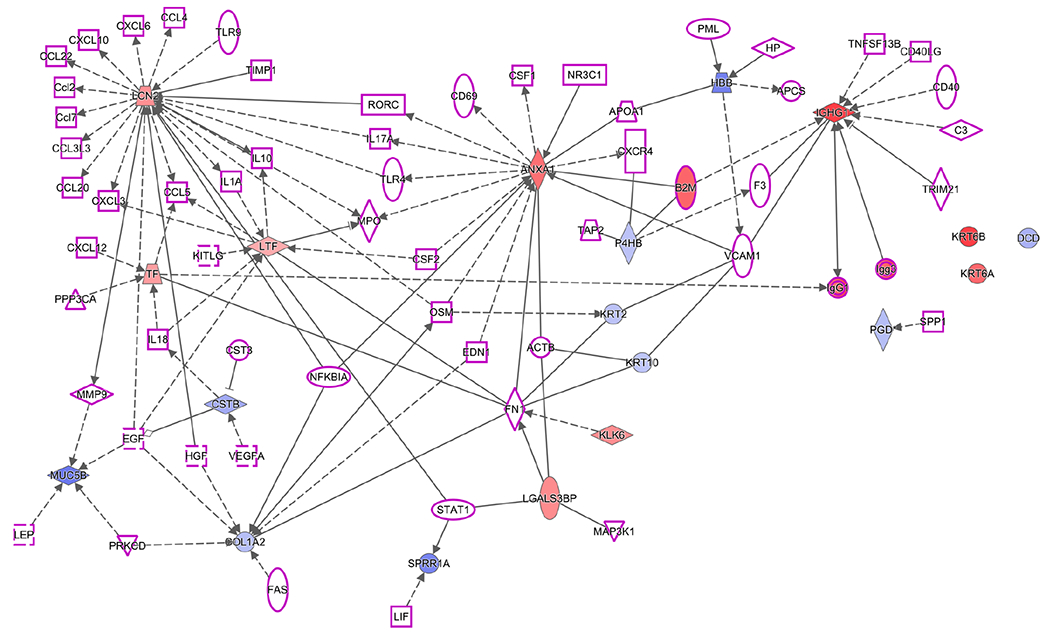
Ingenuity Pathway Analysis showed that the proteins, which were DE in Sjogren’s Syndrome pSS vs HC for PS and WS samples, were networked with proteins that are involved in this disease and in psoriasis, another autoimmune condition. (1) Salivary DE proteins (red, up-regulated; blue down-regulated) identified in the iTRAQ analyses that are involved with psoriasis; (2) proteins associated with Sjögren’s Syndrome (purple outline); (3) direct (physical ± functional; solid lines) or indirect (functional; dashed lines) interactions; and (4) shapes outlining the protein names denote the family to which they belong (refer to Table S-6 for shape definitions). The results suggested that many of the observed alterations were networked with pivotal immune and signaling pathway that also implicated in the disease etiology of psoriasis.
In addition, we performed an IPA that included top genes from pSS genome-wide association studies86 and the DE genes implicated in pSS and psoriasis, as described above and shown in Figure 3. The most significant pathways that emerged from this analysis were antigen signaling and dendritic cell maturation; dendritic cells process antigens and present them on the cell surface to T lymphocytes (data not shown). These pathways included β2 M (described above), IGHG1 (immunoglobulin heavy constant gamma 1), and several HLA genes. IGHG1 binds with Fc-gamma receptors on the surface of effector cells such as macrophages, monocytes, and natural killer cells, which then triggers antibody-dependent cell-mediated cytotoxicity.
Glycosylation
A lectin affinity capture workflow developed in our laboratory was utilized to study the impact of pSS on N-glycosylation of salivary proteins. WGA and AAL lectins were employed to selectively enrich for glycopeptides carrying carbohydrate motifs to which they specifically bind: [GlcNAcβ1-4]2–5, Neu5Acα2-3Manβ1-4GlcNAcβ1-4GlacNAc for WGA and fucose linked to α-(1,6)-GlcNAc and α-(1,3)-GlcNAc for AAL. These lectins were chosen based on a previous screen done by our laboratory, which demonstrated that WGA and AAL reacted with the largest number of salivary proteins spanning the greatest molecular weight range.87 We note that Sondej et al. observed poor reactivity of WGA with WS glycoproteins as compared with other lectins.88 However, WGA reacted strongly with both WS and PS samples in our previous study.87 This discrepancy could be due to differences in immunoblotting procedures used by both laboratories. Table 3 lists the entire set of glycoproteins that was detected according to the lectin that was used for separation and the saliva type. Of the 28 glycoproteins that were identified, 26 (93%) were also detected in the iTRAQ experiments. In addition, 89% of the 29 proteins were identified in WS, and this number decreased to 46% in PS. This is consistent with the fact that WS is a mixture of mucous and serous secretions; PS is primarily the product of serous glands. Furthermore, mucin-5B was only detected in WS because it is not secreted by the parotid gland.
Table 3.
Glycoproteins Detected in pSS, SC, and HC WS and PS Saliva Samples by Lectin Affinity Capture with AAL and WGA Lectinsa
| AAL |
WGA |
|||||
|---|---|---|---|---|---|---|
| accession number | protein name | gene name | PS | WS | PS | WS |
| P01833 | polymeric immunoglobulin receptor | PIGR_HUMAN | × | × | × | × |
| P01876 | Ig alpha-1 chain C region | IGHA1_HUMAN | × | × | × | × |
| P01877 | Ig alpha-2 chain C region | IGHA2_HUMAN | × | × | × | × |
| P02788 | lactotransferrin | TRFL_HUMAN | × | × | × | × |
| P04220* | Ig mu heavy chain disease protein | MUCB_HUMAN | × | |||
| P22079 | lactoperoxidase | PERL_HUMAN | × | × | × | |
| P25311 | zinc-alpha-2-glycoprotein | ZA2G_HUMAN | × | × | × | |
| Q08380 | galectin-3-binding protein | LG3BP_HUMAN | × | × | × | × |
| P04745 | alpha-amylase 1 | AMY1_HUMAN | × | |||
| P00738 | haptoglobin | HPT_HUMAN | × | × | ||
| P01033 | metalloproteinase inhibitor 1 | TIMP1_HUMAN | × | |||
| P01871 | Ig mu chain C region | IGHM_HUMAN | × | × | × | |
| P02787 | serotransferrin | TRFE_HUMAN | × | |||
| P02790 | hemopexin | HEMO_HUMAN | × | × | ||
| P05164 | myeloperoxidase | PERM_HUMAN | × | |||
| P10909 | clusterin | CLUS_HUMAN | × | × | ||
| P13646* | keratin, type I cytoskeletal 13 | K1C13_HUMAN | × | |||
| P20061 | transcobalamin-1 | TCO1_HUMAN | × | |||
| P32926 | desmoglein-3 | DSG3_HUMAN | × | |||
| P80188 | neutrophil gelatinase-associated lipocalin | NGAL_HUMAN | × | × | × | |
| Q02413 | desmoglein-1 | DSG1_HUMAN | × | × | ||
| Q02487 | desmocollin-2 | DSG2_HUMAN | × | |||
| Q9HC84 | mucin-5B | MUC5B_HUMAN | × | × | ||
| Q9UGM3 | deleted in malignant brain tumors 1 | DMBT1_HUMAN | × | × | ||
| Q9Y6R7 | IgGFc-binding protein | FCGBP_HUMAN | × | |||
| P06870 | kallikrein-1 | KLK1_HUMAN | × | |||
| Q8N4F0 | BPI fold-containing family B member 2 | BPIB2_HUMAN | × | |||
| P01009 | alpha-1-antitrypsin | A1AT_HUMAN | × | |||
Identification was made by MS. Accession numbers with an “*” indicate proteins that were not identified in the iTRAQ experiments.
In all, 52 N-glycosites were detected in this analysis. Figure 4 subsets this number by the lectin used for separation, saliva type, and disease or control status. In both the AAL and the WGA separations, the highest number of N-glycosites was identified in the pSS samples. The HC group had the lowest, with SCs generally at an intermediate level. This was true for both saliva types with the exception of the WGA-bound fraction from WS, in which the number of SC identifications was less than the HC group. This observation suggested that the salivary proteins of women diagnosed with pSS were N-glycosylated to a greater extent compared with those from the two control groups. To our knowledge, this is the first time this phenomenon has been reported in an unbiased global analysis.
Figure 4.
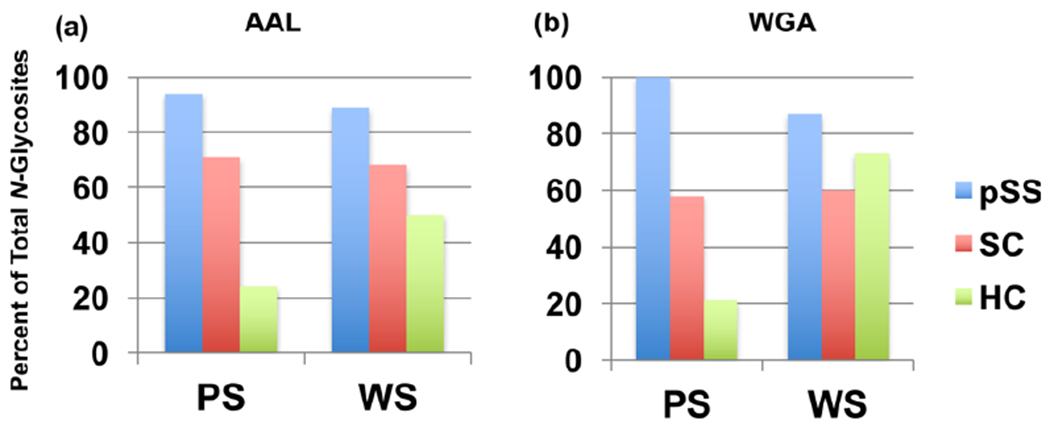
pSS is associated with increased N-glycosylation of salivary proteins. Shown is the percent of total N-glycosites observed in pSS, SC, and HC pooled PS and WS samples. Chromatography on AAL (a) or WGA (b) captured the highest number of N-glycosites from pSS (blue) samples as compared with the SC (red) and HC (green) salivas. Overall, the HC group had the lowest number with SCs at an intermediate level. An exception was the WGA-bound fraction from WS in which the number of N-glycosites observed in the HC group was greater than the SC group.
In addition, our experimental workflow identified individual N-glycosites, the asparagine residue to which a carbohydrate structure was attached in the context of a tryptic peptide that was sequenced to identify the modified protein. Examples of the data obtained are shown in Table 4, which lists the glycoproteins and N-glycosites, which were identified in the WGA bound fraction of PS from each donor group. Nineteen identifications were made in the pSS samples as compared with 11 and 4, in the SC and the HC groups, respectively. With the exception noted above, the same phenomenon was observed in all of the other samples (Tables S-3–S-5). Overall, the differences, which were greatest in PS, were less dramatic in WS.
Table 4.
WGA Lectin Affinity Capture and MS Identification of Glycoproteins and N-Glycosites from pSS, SC, and HC Parotid Saliva Samples
| pSS | SC | HC | ||||
|---|---|---|---|---|---|---|
| Accession Number | Protein | Glycosite | Sequence* | |||
| P01009 | Alpha-1-antitrypsin | A1AT_HUMAN@107 | ADTHDEILEGLNFNLTEIPEAQIHEGFQELLR | x | ||
| P01833 | Polymeric immunoglobulin receptor | PIGR_HUMAN@135 | GLSFDVSLEVSQGPGLLNDTK | x | x | |
| PIGR_HUMAN@186 | QIGLYPVLVIDSSGYVNPNYTGR | x | x | x | ||
| PIGR_HUMAN@469 | VPGNVTAVLGETLK | x | x | |||
| PIGR_HUMAN@83 | ANLTNFPENGTFVVIAQLSQDDSGR | x | x | x | ||
| P01871 | Ig mu chain C | IGHM_HUMAN@439 | STGKPTLYNVSLVMSDTAGTCY | x | ||
| P01876 | Ig alpha-1 chain C | IGHA1_HUMAN@144 | HRPALEDLLLGSEANLTCTLTGLR | x | x | x |
| IGHA1_HUMAN@340 | LAGKPTHVNVSVVMAEVDGTCY | x | x | x | ||
| P01877 | Ig alpha-2 chain C region | IGHA2_HUMAN@131 | HRPALEDLLLGSEANLTCTLTGLR | x | x | |
| IGHA2_HUMAN@327 | PTHVNVSVVMAEVDGTCY | x | x | |||
| IGHA2_HUMAN@47 | LVQGFFPQEPLSVTWSESGQNVTAR | x | x | |||
| P02788 | Lactotransferrin | TRFL_HUMAN@156 | PFLNWTGPPEPIEAAVAR | x | ||
| TRFL_HUMAN@497 | TAGWNIPMGLLFNQTGSCK | x | ||||
| P10909 | Clusterin | CLUS_HUMAN@354 | MLNTSSLLEQLNEQFNWVSR | x | ||
| CLUS_HUMAN@374 | LANLTQGEDQYYLR | x | ||||
| P22079 | Lactoperoxidase | PERL_HUMAN@106 | ASLTNVTDPSLDLTSLSLEVGCGAPAPVVR | x | x | |
| P25311 | Zinc-alpha-2-glycoprotein | ZA2G_HUMAN@109 | DIVEYYNDSNGSHVLQGR | x | x | |
| P80188 | Neutrophil gelatinase-associated lipocalin | NGAL_HUMAN@85 | SYNVTSVLFR | x | ||
| Q08380 | Galectin-3-binding protein | LG3BP_HUMAN@69 | ALGFENATQALGR | x |
Several published reports support our observations. First, a recent study analyzed IgG and IgA isolated from parotid gland salivary biopsies following somatic hypermutation in the context of pSS. The results showed that this process is accompanied by the acquisition of substantial numbers of new N-glycosylation sites that were not detected in comparable samples from control individuals.89 This observation led the authors to suggest that B cell hyperproliferation within the diseased glands of pSS patients may be the result of antigen-independent interactions such as those between these new N-glycosites and lectins within the microenvironment of the oral cavity.89 Second, lectin blotting with Sambucus nigra agglutinin (SNA) and an ELISA showed hyper-sialylation of serum IgA1 involving N-linked versus O-linked saccharides in patients diagnosed with pSS.90 Finally, using an immunoblot approach, the receptor for advanced glycation end products (RAGE) was shown to be expressed at 100-fold greater levels in labial salivary glands of pSS patients compared with control tissue.33 This overexpression could be a pathological response to increased N-glycosylation of salivary glycoproteins, which our data suggest is part of the pSS disease process.
CONCLUSIONS
We used untargeted (iTRAQ) and targeted (lectin affinity capture) MS-based workflows for global analyses of PS and WS samples from patients diagnosed with pSS as compared with two control groups—individuals with no evidence of disease who were examined in the SICCA clinic and donors with no history or symptoms of this condition. The iTRAQ analyses and subsequent validation experiments revealed the up- and down-regulation of numerous proteins that could be involved in the disease process (e.g., histone) or attempts to mitigate the ensuing damage (e.g., BPIF family members). An immunoblot approach applied to independent sample sets was used to confirm the pSS associated up-regulation of β2-microglobulin (in PS) and down-regulation of CAH6 (in WS) and BPIFB2 (in PS). The lectin affinity capture workflow we used to profile N-glycosylation sites in pSS vs control samples showed that this disease is associated with increased glycosylation of numerous salivary glycoproteins in PS and WS. The differentially expressed proteins or the changes in N-glycosylation could be biomarkers of pSS, enabling easier and earlier detection. The results of this study also lend potential new insights into the disease process, which suggest evidence of N-glycan up-regulation on glycoproteins present in the saliva of pSS patients.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health/National Institute of Dental and Craniofacial Research grant R01DE022031 and NIDCR HHSN268201300057C. We acknowledge Dr. Kimberley Taylor for her assistance with the pathway analysis involving Sjögren’s syndrome GWAS and psoriasis genes. We gratefully acknowledge the University of California, San Francisco Sandler-Moore Mass Spectrometry Core Facility, which received support from the Sandler Family Foundation and the Gordon and Betty Moore Foundation. In addition, we thank the staff of the Sjögren’s International Collaborative Clinical Alliance (SICCA, http://sicca-online.ucsf.edu).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.6b01051.
Figure S-1. Whole image immunoblots for validation of iTRAQ DE proteins. Table S-1. Parotid and whole salivary proteins identified and quantified in the iTRAQ experiment at an FDR of 5%. Table S-2. Comparison of differentially expressed proteins among donor groups (p < 0.05). Table S-3. WGA lectin affinity capture and MS identification of glycoproteins and N-glycosites from pSS, SC, and HC whole saliva samples. Table S-4. AAL lectin affinity capture and MS identification of glycoproteins and N-glycosites from pSS, SC, and HC parotid saliva samples. Table S-5. AAL lectin affinity capture and MS identification of glycoproteins and N-glycosites from pSS, SC, and HC whole saliva samples. Table S-6. Legend matching shapes to protein families for Figure 3a,b. (XLSX)
The authors declare no competing financial interest.
All mass spectrometry data and search results are available at the MassIVE data depository Web site. iTRAQ data set: http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=afdbf437cb0643f1ab8057b35c37c4c3 (password: pSSiTRAQ). Lectin affinity capture data set: http://massive.ucsd.edu/ProteoSAFe/status.jsp?task=dd274f44174b4d17b59cace036ea8c61 (password: pSSLAC).
REFERENCES
- (1).Fox PC Autoimmune diseases and Sjogren’s syndrome: an autoimmune exocrinopathy. Ann. N. Y. Acad. Sci 2007, 1098, 15–21. [DOI] [PubMed] [Google Scholar]
- (2).Thomas E; Hay EM; Hajeer A; Silman AJ Sjogren’s syndrome: a community-based study of prevalence and impact. Br J. Rheumatol 1998, 37 (10), 1069–76. [DOI] [PubMed] [Google Scholar]
- (3).Bolstad AI; Jonsson R Genetic aspects of Sjogren’s syndrome. Arthritis Res. 2002, 4 (6), 353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bloch KJ; Buchanan WW; Wohl MJ; Bunim JJ Sjoegren’s Syndrome. A Clinical, Pathological, and Serological Study of Sixty-Two Cases. Medicine (Philadelphia, PA, U. S.) 1965, 44, 187–231. [PubMed] [Google Scholar]
- (5).Daniels TE; Criswell LA; Shiboski C; Shiboski S; Lanfranchi H; Dong Y; Schiodt M; Umehara H; Sugai S; Challacombe S; Greenspan JS Sjogren’s International Collaborative Clinical Alliance Research, G., An early view of the international Sjogren’s syndrome registry. Arthritis Rheum. 2009, 61 (5), 711–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Shiboski CH; Shiboski SC; Seror R; Criswell LA; Labetoulle M; Lietman TM; Rasmussen A; Scofield H; Vitali C; Bowman SJ; Mariette X International Sjogren’s Syndrome Criteria Working, G., 2016 American College of Rheumatology/European League Against Rheumatism classification criteria for primary Sjogren’s syndrome: A consensus and data-driven methodology involving three international patient cohorts. Ann. Rheum. Dis 2017, 76 (1), 9–16. [DOI] [PubMed] [Google Scholar]
- (7).Vitali C; Bombardieri S; Jonsson R; Moutsopoulos HM; Alexander EL; Carsons SE; Daniels TE; Fox PC; Fox RI; Kassan SS;Pillemer SR;Talal N;Weisman MH European Study Group on Classification Criteria for Sjogren’s, S., Classification criteria for Sjogren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann. Rheum. Dis 2002, 61 (6), 554–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Shiboski SC; Shiboski CH; Criswell L; Baer A; Challacombe S; Lanfranchi H; Schiodt M; Umehara H; Vivino F; Zhao Y; Dong Y; Greenspan D; Heidenreich AM; Helin P; Kirkham B; Kitagawa K; Larkin G; Li M; Lietman T; Lindegaard J; McNamara N; Sack K; Shirlaw P; Sugai S; Vollenweider C; Whitcher J; Wu A; Zhang S; Zhang W; Greenspan J; Daniels T Sjogren’s International Collaborative Clinical Alliance Research, G., American College of Rheumatology classification criteria for Sjogren’s syndrome: a data-driven, expert consensus approach in the Sjogren’s International Collaborative Clinical Alliance cohort. Arthritis Care Res. 2012, 64 (4), 475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ben-Chetrit E; Fischel R; Rubinow A Anti-SSA/Ro and anti-SSB/La antibodies in serum and saliva of patients with Sjogren’s syndrome. Clin. Rheumatol 1993, 12 (4), 471–4. [DOI] [PubMed] [Google Scholar]
- (10).Baer AN; McAdams DeMarco M; Shiboski SC; Lam MY; Challacombe S; Daniels TE; Dong Y; Greenspan JS; Kirkham BW; Lanfranchi HE; Schiodt M; Srinivasan M; Umehara H; Vivino FB; Vollenweider CF; Zhao Y; Criswell LA; Shiboski CH Sjogren’s International Collaborative Clinical Alliance Research, G., The SSB-positive/SSA-negative antibody profile is not associated with key phenotypic features of Sjogren’s syndrome. Ann. Rheum. Dis 2015, 74 (8), 1557–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Witte T. Antifodrin antibodies in Sjogren’s syndrome: a review. Ann. N. Y. Acad. Sci 2005, 1051, 235–9. [DOI] [PubMed] [Google Scholar]
- (12).Sordet C; Gottenberg JE; Goetz J; Bengoufa D; Humbel RL; Mariette X; Sibilia J Anti-{alpha}-fodrin autoantibodies are not useful diagnostic markers of primary Sjogren’s syndrome. Ann. Rheum. Dis 2005, 64 (8), 1244–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Denny P; Hagen FK; Hardt M; Liao L; Yan W; Arellanno M; Bassilian S; Bedi GS; Boontheung P; Cociorva D; Delahunty CM; Denny T; Dunsmore J; Faull KF; Gilligan J; Gonzalez-Begne M; Halgand F; Hall SC; Han X; Henson B; Hewel J; Hu S; Jeffrey S; Jiang J; Loo JA; Ogorzalek Loo RR; Malamud D; Melvin JE; Miroshnychenko O; Navazesh M; Niles R; Park SK; Prakobphol A; Ramachandran P; Richert M; Robinson S; Sondej M; Souda P; Sullivan MA; Takashima J; Than S; Wang J; Whitelegge JP; Witkowska HE; Wolinsky L; Xie Y; Xu T; Yu W; Ytterberg J; Wong DT; Yates JR 3rd; Fisher SJ The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res 2008, 7 (5), 1994–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lee JM; Garon E; Wong DT Salivary diagnostics. Orthod Craniofac Res. 2009, 12 (3), 206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ferraccioli G; De Santis M; Peluso G; Inzitari R; Fanali C; Bosello SL; Iavarone F; Castagnola M Proteomic approaches to Sjogren’s syndrome: a clue to interpret the pathophysiology and organ involvement of the disease. Autoimmun. Rev 2010, 9 (9), 622–6. [DOI] [PubMed] [Google Scholar]
- (16).Baldini C; Giusti L; Bazzichi L; Lucacchini A; Bombardieri S Proteomic analysis of the saliva: a clue for understanding primary from secondary Sjogren’s syndrome? Autoimmun. Rev 2008, 7 (3), 185–91. [DOI] [PubMed] [Google Scholar]
- (17).Chen W; Cao H; Lin J; Olsen N; Zheng SG Biomarkers for Primary Sjogren’s Syndrome. Genomics, Proteomics Bioinf. 2015, 13 (4), 219–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Peluso G; De Santis M; Inzitari R; Fanali C; Cabras T; Messana I; Castagnola M; Ferraccioli GF Proteomic study of salivary peptides and proteins in patients with Sjogren’s syndrome before and after pilocarpine treatment. Arthritis Rheum. 2007, 56 (7), 2216–22. [DOI] [PubMed] [Google Scholar]
- (19).Ambatipudi KS; Swatkoski S; Moresco JJ; Tu PG; Coca A; Anolik JH; Gucek M; Sanz I; Yates JR 3rd; Melvin JE Quantitative proteomics of parotid saliva in primary Sjogren’s syndrome. Proteomics 2012, 12 (19–20), 3113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zoukhri D; Rawe I; Singh M; Brown A; Kublin CL; Dawson K; Haddon WF; White EL; Hanley KM; Tuse D; Malyj W; Papas A Discovery of putative salivary biomarkers for Sjogren’s syndrome using high resolution mass spectrometry and bioinformatics. J. Oral Sci 2012, 54 (1), 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wei P; Kuo WP; Chen F; Hua H Diagnostic model of saliva peptide finger print analysis of primary Sjogren’s syndrome patients by using weak cation exchange magnetic beads. Biosci. Rep 2013, 33 (4), 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ryu OH; Atkinson JC; Hoehn GT; Illei GG; Hart TC Identification of parotid salivary biomarkers in Sjogren’s syndrome by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry and two-dimensional difference gel electrophoresis. Rheumatology (Oxford, U. K.) 2006, 45 (9), 1077–86. [DOI] [PubMed] [Google Scholar]
- (23).Giusti L; Baldini C; Bazzichi L; Ciregia F; Tonazzini I; Mascia G; Giannaccini G; Bombardieri S; Lucacchini A Proteome analysis of whole saliva: a new tool for rheumatic diseases–the example of Sjogren’s syndrome. Proteomics 2007, 7 (10), 1634–43. [DOI] [PubMed] [Google Scholar]
- (24).Fleissig Y; Deutsch O; Reichenberg E; Redlich M; Zaks B; Palmon A; Aframian DJ Different proteomic protein patterns in saliva of Sjogren’s syndrome patients. Oral Dis 2009, 15 (1), 61–8. [DOI] [PubMed] [Google Scholar]
- (25).Hjelmervik TO; Jonsson R; Bolstad AI The minor salivary gland proteome in Sjogren’s syndrome. Oral Dis 2009, 15 (5), 342–53. [DOI] [PubMed] [Google Scholar]
- (26).Hu S; Zhou M; Jiang J; Wang J; Elashoff D; Gorr S; Michie SA; Spijkervet FK; Bootsma H; Kallenberg CG; Vissink A; Horvath S; Wong DT Systems biology analysis of Sjogren’s syndrome and mucosa-associated lymphoid tissue lymphoma in parotid glands. Arthritis Rheum. 2009, 60 (1), 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Deutsch O; Krief G; Konttinen YT; Zaks B; Wong DT; Aframian DJ; Palmon A Identification of Sjogren’s syndrome oral fluid biomarker candidates following high-abundance protein depletion. Rheumatology (Oxford, U. K.) 2015, 54 (5), 884–90. [DOI] [PubMed] [Google Scholar]
- (28).Baldini C; Giusti L; Ciregia F; Da Valle Y; Giacomelli C; Donadio E; Sernissi F; Bazzichi L; Giannaccini G; Bombardieri S; Lucacchini A Proteomic analysis of saliva: a unique tool to distinguish primary Sjogren’s syndrome from secondary Sjogren’s syndrome and other sicca syndromes. Arthritis Res. Ther 2011, 13 (6), R194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Baldini C; Giusti L; Ciregia F; Da Valle Y; Giacomelli C; Donadio E; Ferro F; Galimberti S; Donati V; Bazzichi L; Bombardieri S; Lucacchini A Correspondence between salivary proteomic pattern and clinical course in primary Sjogren syndrome and non-Hodgkin’s lymphoma: a case report. J. Transl. Med 2011, 9, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Loomis RE; Prakobphol A; Levine MJ; Reddy MS; Jones PC Biochemical and biophysical comparison of two mucins from human submandibular-sublingual saliva. Arch. Biochem. Biophys 1987, 258 (2), 452–64. [DOI] [PubMed] [Google Scholar]
- (31).Prakobphol A; Levine MJ; Tabak LA; Reddy MS Purification of a low-molecular-weight, mucin-type glycoprotein from human submandibular-sublingual saliva. Carbohydr. Res 1982, 108 (1), 111–22. [DOI] [PubMed] [Google Scholar]
- (32).Prakobphol A; Thomsson KA; Hansson GC; Rosen SD; Singer MS; Phillips NJ; Medzihradszky KF; Burlingame AL; Leffler H; Fisher SJ Human low-molecular-weight salivary mucin expresses the sialyl lewisx determinant and has L-selectin ligand activity. Biochemistry 1998, 37 (14), 4916–27. [DOI] [PubMed] [Google Scholar]
- (33).Katz J; Stavropoulos F; Bhattacharyya I; Stewart C; Perez FM; Caudle RM Receptor of advanced glycation end product (RAGE) expression in the minor salivary glands of patients with Sjogren’s syndrome: a preliminary study. Scand. J. Rheumatol 2004, 33 (3), 174–8. [DOI] [PubMed] [Google Scholar]
- (34).Stewart C; Cha S; Caudle RM; Berg K; Katz J Decreased levels of soluble receptor for advanced glycation end products in patients with primary Sjogren’s syndrome. Rheumatol. Int 2008, 28 (8), 771–6. [DOI] [PubMed] [Google Scholar]
- (35).Andrys C; Krejsek J; Slezak R; Drahosova M; Kopecky O Serum soluble adhesion molecules (sICAM-1, sVCAM-1, sE-selectin) and neopterin in patients with Sjogren’s syndrome. Acta Medica (Hradec Kralove) 1999, 42 (3), 97–101. [PubMed] [Google Scholar]
- (36).Garcia-Carrasco M; Pizcueta P; Cervera R; Ramos-Casals M; Siso A; de La Red G; Ingelmo M; Font J; Engel P Circulating concentrations of soluble L-selectin (CD62L) in patients with primary Sjogren’s syndrome. Ann. Rheum. Dis 2000, 59 (4), 297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chaudhury NM; Proctor GB; Karlsson NG; Carpenter GH; Flowers SA Reduced Mucin-7 (Muc7) Sialylation and Altered Saliva Rheology in Sjogren’s Syndrome Associated Oral Dryness. Mol. Cell. Proteomics 2016, 15 (3), 1048–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Malladi AS; Sack KE; Shiboski SC; Shiboski CH; Baer AN; Banushree R; Dong Y; Helin P; Kirkham BW; Li M; Sugai S; Umehara H; Vivino FB; Vollenweider CF; Zhang W; Zhao Y; Greenspan JS; Daniels TE; Criswell LA Primary Sjogren’s syndrome as a systemic disease: a study of participants enrolled in an international Sjogren’s syndrome registry. Arthritis Care Res. 2012, 64 (6), 911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Tambor V; Hunter CL; Seymour SL; Kacerovsky M; Stulik J; Lenco J CysTRAQ - A combination of iTRAQ and enrichment of cysteinyl peptides for uncovering and quantifying hidden proteomes. J. Proteomics 2012, 75 (3), 857–67. [DOI] [PubMed] [Google Scholar]
- (40).Drake PM; Schilling B; Niles RK; Braten M; Johansen E; Liu H; Lerch M; Sorensen DJ; Li B; Allen S; Hall SC; Witkowska HE; Regnier FE; Gibson BW; Fisher SJ A lectin affinity workflow targeting glycosite-specific, cancer-related carbohydrate structures in trypsin-digested human plasma. Anal. Biochem 2011, 408 (1), 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Krokhin OV; Antonovici M; Ens W; Wilkins JA; Standing KG Deamidation of -Asn-Gly- sequences during sample preparation for proteomics: Consequences for MALDI and HPLC-MALDI analysis. Anal. Chem 2006, 78 (18), 6645–50. [DOI] [PubMed] [Google Scholar]
- (42).Varin MM; Guerrier T; Devauchelle-Pensec V; Jamin C; Youinou P; Pers JO In Sjogren’s syndrome, B lymphocytes induce epithelial cells of salivary glands into apoptosis through protein kinase C delta activation. Autoimmun. Rev 2012, 11 (4), 252–8. [DOI] [PubMed] [Google Scholar]
- (43).Hu S; Vissink A; Arellano M; Roozendaal C; Zhou H; Kallenberg CG; Wong DT Identification of autoantibody biomarkers for primary Sjogren’s syndrome using protein microarrays. Proteomics 2011, 11 (8), 1499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Castro J; Jimenez-Alonso J; Sabio JM; Rivera-Civico F; Martin-Armada M; Rodriguez MA; Jaimez L; Castillo MJ; Sanchez-Roman J Grupo Lupus Virgen de las N, Salivary and serum beta2-microglobulin and gamma-glutamyl-transferase in patients with primary Sjogren syndrome and Sjogren syndrome secondary to systemic lupus erythematosus. Clin. Chim. Acta 2003, 334 (1–2), 225–31. [DOI] [PubMed] [Google Scholar]
- (45).Asashima H; Inokuma S; Nakachi S; Matsuo Y; Rokutanda R; Hagiwara K; Kobayashi S Extremely high salivary beta(2)-microglobulin and Na(+) levels in a Sjogren syndrome patient. Int. J. Rheum Dis 2012, 15 (2), e31–3. [DOI] [PubMed] [Google Scholar]
- (46).Michalski JP; Daniels TE; Talal N; Grey HM Beta2 microglobulin and lymphocytic infiltration in Sjogren’s syndrome. N. Engl. J. Med 1975, 293 (24), 1228–31. [DOI] [PubMed] [Google Scholar]
- (47).Bournia VK; Vlachoyiannopoulos PG Subgroups of Sjogren syndrome patients according to serological profiles. J. Autoimmun 2012, 39 (1–2), 15–26. [DOI] [PubMed] [Google Scholar]
- (48).Kivela J; Parkkila S; Parkkila AK; Leinonen J; Rajaniemi H Salivary carbonic anhydrase isoenzyme VI. J. Physiol 1999, 520 (2), 315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Hu S; Wang J; Meijer J; Ieong S; Xie Y; Yu T; Zhou H; Henry S; Vissink A; Pijpe J; Kallenberg C; Elashoff D; Loo JA; Wong DT Salivary proteomic and genomic biomarkers for primary Sjogren’s syndrome. Arthritis Rheum. 2007, 56 (11), 3588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Hannon R; Croxtall JD; Getting SJ; Roviezzo F; Yona S; Paul-Clark MJ; Gavins FN; Perretti M; Morris JF; Buckingham JC; Flower RJ Aberrant inflammation and resistance to glucocorticoids in annexin 1−/− mouse. FASEB J. 2002, 17 (2), 253–5. [DOI] [PubMed] [Google Scholar]
- (51).Arcone R; Arpaia G; Ruoppolo M; Malorni A; Pucci P; Marino G; Ialenti A; Di Rosa M; Ciliberto G Structural characterization of a biologically active human lipocortin 1 expressed in Escherichia coli. Eur. J. Biochem 1993, 211 (1–2), 347–55. [DOI] [PubMed] [Google Scholar]
- (52).Xiao H; Wong DT Proteomic analysis of microvesicles in human saliva by gel electrophoresis with liquid chromatography-mass spectrometry. Anal. Chim. Acta 2012, 723, 61–7. [DOI] [PubMed] [Google Scholar]
- (53).Iaccarino L; Ghirardello A; Canova M; Zen M; Bettio S; Nalotto L; Punzi L; Doria A Anti-annexins autoantibodies: their role as biomarkers of autoimmune diseases. Autoimmun. Rev 2011, 10 (9), 553–8. [DOI] [PubMed] [Google Scholar]
- (54).Derry MC; Sutherland MR; Restall CM; Waisman DM; Pryzdial EL Annexin 2-mediated enhancement of cytomegalovirus infection opposes inhibition by annexin 1 or annexin 5. J. Gen. Virol 2007, 88 (1), 19–27. [DOI] [PubMed] [Google Scholar]
- (55).Leoni G; Nusrat A Annexin A1: shifting the balance towards resolution and repair. Biol. Chem 2016, DOI: 10.1515/hsz-2016-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Sasaki T; Brakebusch C; Engel J; Timpl R Mac-2 binding protein is a cell-adhesive protein of the extracellular matrix which self-assembles into ring-like structures and binds beta1 integrins, collagens and fibronectin. EMBO J. 1998, 17 (6), 1606–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Ostergaard O; Nielsen CT; Iversen LV; Tanassi JT; Knudsen S; Jacobsen S; Heegaard NH Unique protein signature of circulating microparticles in systemic lupus erythematosus. Arthritis Rheum. 2013, 65 (10), 2680–90. [DOI] [PubMed] [Google Scholar]
- (58).Lin TW; Chang HT; Chen CH; Chen CH; Lin SW; Hsu TL; Wong CH Galectin-3 Binding Protein and Galectin-1 Interaction in Breast Cancer Cell Aggregation and Metastasis. J. Am. Chem. Soc 2015, 137 (30), 9685–93. [DOI] [PubMed] [Google Scholar]
- (59).Ohshima S; Kuchen S; Seemayer CA; Kyburz D; Hirt A; Klinzing S; Michel BA; Gay RE; Liu FT; Gay S; Neidhart M Galectin 3 and its binding protein in rheumatoid arthritis. Arthritis Rheum. 2003, 48 (10), 2788–95. [DOI] [PubMed] [Google Scholar]
- (60).Bingle CD; Bingle L; Craven CJ Distant cousins: genomic and sequence diversity within the BPI fold-containing (BPIF)/PLUNC protein family. Biochem. Soc. Trans 2011, 39 (4), 961–5. [DOI] [PubMed] [Google Scholar]
- (61).Bingle CD; Seal RL; Craven CJ Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a subfamily of the BPI fold-containing superfamily. Biochem. Soc. Trans 2011, 39 (4), 977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Bingle L; Bingle CD Distribution of human PLUNC/BPI fold-containing (BPIF) proteins. Biochem. Soc. Trans 2011, 39 (4), 1023–7. [DOI] [PubMed] [Google Scholar]
- (63).Prokopovic V; Popovic M; Andjelkovic U; Marsavelski A; Raskovic B; Gavrovic-Jankulovic M; Polovic N Isolation, biochemical characterization and anti-bacterial activity of BPIFA2 protein. Arch. Oral Biol 2014, 59 (3), 302–9. [DOI] [PubMed] [Google Scholar]
- (64).Geetha C; Venkatesh SG; Dunn BH; Gorr SU Expression and anti-bacterial activity of human parotid secretory protein (PSP). Biochem. Soc. Trans 2003, 31 (4), 815–8. [DOI] [PubMed] [Google Scholar]
- (65).Wu Y; Shu R; Luo LJ; Ge LH; Xie YF Initial comparison of proteomic profiles of whole unstimulated saliva obtained from generalized aggressive periodontitis patients and healthy control subjects. J. Periodontal Res 2009, 44 (5), 636–44. [DOI] [PubMed] [Google Scholar]
- (66).Sevilla LM; Nachat R; Groot KR; Klement JF; Uitto J; Djian P; Maatta A; Watt FM Mice deficient in involucrin, envoplakin, and periplakin have a defective epidermal barrier. J. Cell Biol 2007, 179 (7), 1599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Bouameur JE; Favre B; Borradori L Plakins, a versatile family of cytolinkers: roles in skin integrity and in human diseases. J. Invest. Dermatol 2014, 134 (4), 885–94. [DOI] [PubMed] [Google Scholar]
- (68).Recklies AD; White C; Ling H The Chitinase 3-like protein human cartilage glycoprotein 39 (HC-gp39) stimulates proliferation of human connective-tissue cells and activates both extracellular signal-regulated kinase- and protein kinase B-mediated signalling pathways. Biochem. J 2002, 365 (1), 119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Oppenheim FG; Yang YC; Diamond RD; Hyslop D; Offner GD; Troxler RF The primary structure and functional characterization of the neutral histidine-rich polypeptide from human parotid secretion. J. Biol. Chem 1986, 261 (3), 1177–82. [PubMed] [Google Scholar]
- (70).Amado F; Lobo MJ; Domingues P; Duarte JA; Vitorino R Salivary peptidomics. Expert Rev. Proteomics 2010, 7 (5), 709–21. [DOI] [PubMed] [Google Scholar]
- (71).van Dijk IA; Nazmi K; Bolscher JG; Veerman EC; Stap J Histatin-1, a histidine-rich peptide in human saliva, promotes cell-substrate and cell-cell adhesion. FASEB J. 2015, 29 (8), 3124–32. [DOI] [PubMed] [Google Scholar]
- (72).Klebanoff SJ Myeloperoxidase: friend and foe. J. Leukocyte Biol 2005, 77 (5), 598–625. [DOI] [PubMed] [Google Scholar]
- (73).Cay HF; Sezer I; Dogan S; Felek R; Aslan M Polymorphism in the TNF-alpha gene promoter at position −1031 is associated with increased circulating levels of TNF-alpha, myeloperoxidase and nitrotyrosine in primary Sjogren’s syndrome. Free Radical Biol. Med 2011, 51 (6), S104. [PubMed] [Google Scholar]
- (74).Huynh AH; Veith PD; McGregor NR; Adams GG; Chen D; Reynolds EC; Ngo LH; Darby IB Gingival crevicular fluid proteomes in health, gingivitis and chronic periodontitis. J. Periodontal Res 2015, 50 (5), 637–49. [DOI] [PubMed] [Google Scholar]
- (75).Jenzano JW; Daniel PA; Kent RT; Leal JL; Koth DL Evaluation of kallikrein in human parotid and submandibular saliva. Arch. Oral Biol 1986, 31 (9), 627–8. [DOI] [PubMed] [Google Scholar]
- (76).Proud D; Kaplan AP Kinin formation: mechanisms and role in inflammatory disorders. Annu. Rev. Immunol 1988, 6, 49–83. [DOI] [PubMed] [Google Scholar]
- (77).Bhoola K; Ramsaroop R; Plendl J; Cassim B; Dlamini Z; Naicker S Kallikrein and kinin receptor expression in inflammation and cancer. Biol. Chem 2001, 382 (1), 77–89. [DOI] [PubMed] [Google Scholar]
- (78).Takada K; Takiguchi M; Konno A; Inaba M Autoimmunity against a tissue kallikrein in IQI/Jic Mice: a model for Sjogren’s syndrome. J. Biol. Chem 2005, 280 (5), 3982–8. [DOI] [PubMed] [Google Scholar]
- (79).Friberg B; Jonsson R; Linde A Salivary kallikrein in Sjogren’s syndrome. Clin. Exp. Rheumatol 1988, 6 (2), 135–8. [PubMed] [Google Scholar]
- (80).Hernandez CC; Donadi EA; Reis ML Kininogen-kallikrein-kinin system in plasma and saliva of patients with Sjogren’s syndrome. J. Rheumatol 1998, 25 (12), 2381–4. [PubMed] [Google Scholar]
- (81).Ramachandran P; Boontheung P; Xie Y; Sondej M; Wong DT; Loo JA Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res 2006, 5 (6), 1493–503. [DOI] [PubMed] [Google Scholar]
- (82).Bingle L; Barnes FA; Lunn H; Musa M; Webster S; Douglas CW; Cross SS; High AS; Bingle CD Characterisation and expression of SPLUNC2, the human orthologue of rodent parotid secretory protein. Histochem. Cell Biol 2009, 132 (3), 339–49. [DOI] [PubMed] [Google Scholar]
- (83).Wu JJ; Nguyen TU; Poon KY; Herrinton LJ The association of psoriasis with autoimmune diseases. J. Am. Acad. Dermatol 2012, 67 (5), 924–30. [DOI] [PubMed] [Google Scholar]
- (84).Kurz C; Wunderlich S; Spieler D; Schwaiger BJ; Andres C; Traidl-Hoffmann C; Ilg R Acute transverse myelitis and psoriasiform dermatitis associated with Sjoegren’s syndrome: a case report. BMC Res. Notes 2014, 7, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Akiyama M; Ueno T; Kanzaki A; Kuwana M; Nagao M; Saeki H Association of psoriasis with Hashimoto’s thyroiditis, Sjogren’s syndrome and dermatomyositis. J. Dermatol 2016, 43 (6), 711–2. [DOI] [PubMed] [Google Scholar]
- (86).Taylor KE; Wong Q; Levine DM; McHugh C; Laurie C; Doheny K; Lam MY; Baer AN; Challacombe S; Lanfranchi H; Schiødt M; Srinivasan M; Umehara H; Vivino FB; Zhao Y; Shiboski S; Daniels TE; Greenspan JS; Shiboski CH; Criswell LA Genome-Wide Association Analysis Reveals Genetic Heterogeneity of Sjogren’s Syndrome According to Ancestry. Arthritis Rheumatol. 2017, DOI: 10.1002/art.40040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Albertolle ME; Hassis ME; Ng CJ; Cuison S; Williams K; Prakobphol A; Dykstra AB; Hall SC; Niles RK; Ewa Witkowska H; Fisher SJ Mass spectrometry-based analyses showing the effects of secretor and blood group status on salivary N-glycosylation. Clin. Proteomics 2015, 12, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Sondej M; Denny PA; Xie Y; Ramachandran P; Si Y; Takashima J; Shi W; Wong DT; Loo JA; Denny PC Glycoprofiling of the Human Salivary Proteome. Clin. Proteomics 2009, 5 (1), 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Hamza N; Hershberg U; Kallenberg CG; Vissink A; Spijkervet FK; Bootsma H; Kroese FG; Bos NA Ig gene analysis reveals altered selective pressures on Ig-producing cells in parotid glands of primary Sjogren’s syndrome patients. J. Immunol 2015, 194 (2), 514–21. [DOI] [PubMed] [Google Scholar]
- (90).Basset C; Durand V; Jamin C; Clement J; Pennec Y; Youinou P; Dueymes M; Roitt IM Increased N-linked glycosylation leading to oversialylation of monomeric immunoglobulin A1 from patients with Sjogren’s syndrome. Scand. J. Immunol 2000, 51 (3), 300–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


