Abstract
This study presents a database of joint angles, moments, and forces of the lower extremity from distance running at a submaximal speed in recreational runners. Twenty recreational runners participated in two experimental sessions, specifically pre and post a 5k treadmill run, with a synchronous collection of markers trajectories and ground reaction forces for both limbs in walking and running trials. The raw data in C3D files could be used for musculoskeletal modelling. Extra datasets of joint angles, moments, and forces are presented ready-for-use in MAT files, which could be as reference for study of biomechanical alterations from distance running. Applying advanced data processing techniques (Machine Learning algorithms) to these datasets (C3D&MAT), such as Principal Component Analysis, could extract key features of variation, thus potentially being applied for correlation with accelerometric and gyroscope parameters from wearable sensors during field running. Dataset of multi-segmental foot could be another contribution for the investigation of foot complex biomechanics from distance running. The dataset from Asian males may also be used for population-based studies of running biomechanics.
Keywords: Running biomechanics, Lower extremity, Contact forces, Principal component analysis, Statistical parametric mapping
Running biomechanics; Lower extremity; Contact forces; Principal component analysis; statistical parametric mapping.
1. Introduction
Running has attracted extensive participation over the past decades for the benefits of physical fitness and health improvement. Due to the easy accessibility, running has become the most convenient physical activity around the globe, showing lower risks of all-cause mortality and increased life longevity [1, 2]. Specifically, being a most efficient form of exercise, running was proven with positive effect in prevention of chronic disease, such as over-weight (high BMI), cardiovascular disease, cancer mortality and several other healthy issues [2, 3, 4, 5]. Runners of elite or recreational levels increased substantially across all age cohorts, either during casual or official (marathon) running events [6, 7, 8].
However, high incidences of running-related injuries were documented and studied [9], aiming to investigate the potential mechanism for injury prevention [10]. Recent studies about distance running found redistributed plantar pressure in the foot [6], increased loadings to the ankle and knee joints [11], and distally shifted mechanical output (power and work) from the hip to ankle joint [12]. Further changes of the ankle cartilage [13, 14], running stability [15], foot strikes [16], and inter-limb symmetry [17, 18] were reported during distance running. Thus, factors of foot strikes [16, 19], strength training [20] and footwear [21] were analysed to help improve performance and prevent potential injuries.
Running often involves rapid and cyclical energy store-release in the muscles and tendons while walking gait is like a pendulum mechanics [22]. Development of joint kinematics and kinetics was assumed to be typically consistent or within a small variation range between steps and inter-limbs. However, sagittal angles became symmetrical while asymmetry was observed in the angular velocity between dominant and non-dominant limbs from an incremental speed induced running fatigue test [23]. Alterations of biomechanical loading (even the accumulation) may contribute to potential running-related injuries [11, 24, 25], thus affecting the running performance [26].
Approaches of musculoskeletal modelling, such as the Visual 3D [23, 27], Anybody [28, 29], and OpenSim [30, 31, 32] musculoskeletal simulation, are widely employed in the biomechanics community. Apart from traditionally reported variables of kinematics, kinetics, and muscle activities, the above-mentioned techniques could calculate extra in-vivo parameters, providing plausible and reliable methods to reveal alterations in the neuro-musculoskeletal system. In particular, the calculated muscle forces and joint forces during motions are difficult to measure in-vivo from experiments, which are believed to be key reasons leading to potential injuries [11, 33]. Our previous investigation of foot protonation and lower extremity biomechanical loadings reported that pronated foot postures in the right limb were observed following a 5k run, which was an early indicator of increased peak knee and ankle moments [11].
Apart from the discrete variables, biomechanical parameters typically vary over-time [34], thus requiring temporally understanding. Techniques of data processing and statistical analysis in biomechanics developed from traditionally employed comparisons of discrete values, such as maximum, minimum, or discrete time-point (i.e., foot strike angle), to multidimensional levels, such as 1-dimensional (1D) time-varying joint angles, moments and contact forces. The one-dimensional Statistical Parametric Mapping (SPM1d) is one of the emerging and widely employed approaches for statistical analysis [11, 35, 36, 37, 38]. This technique was originated and developed from the Statistical Parametric Mapping [39], to check significance between variables based on random field theory (RFT) [36, 37, 40]. Similarly, the Principal Component Analysis (PCA) [41] and Functional Data Analysis (FDA) [42] are recently employed to reveal knee angle variations in pathological gait [43], gait coordination and variability [44], strike patterns [45], running level classification [46], ground reaction forces [47], foot postures and function [48], jumping performance and force output [49, 50, 51, 52], bilateral limb asymmetry [53], and kinematic patterns from injury [54].
With the development of musculoskeletal modelling and statistical analysis techniques, scarce dataset of gait biomechanics from distance running were publicly available in the literature, though this physical activity gained great popularity in the recent years. Authors proposed the current dataset, including standardised walking and running data from Asian male recreational runners during a pre 5k and post 5k run at a submaximal speed [55]. The first part of raw C3D dataset could be used to explore the changes and symmetry from distance running as a pilot example for future studies to expand the database. Another probable application of this raw dataset is that the multi-segment foot model could be applied to investigate the intersegmental foot biomechanics from a distance run, which is scarcely reported in current literature. Our previous studies have reported the dynamic inter-segmental foot biomechanics from walking speeds [56], forward versus backward walking [57], and toes manipulation [58, 59]. A second part of the MAT dataset is the pre-processed (ready-for-use) joint angles, moments, and forces, which could be directly integrated into the PCA (we took PCA as an example for analysis in this study) or other machine learning algorithms as a baseline example for the accelerometer and gyroscope signals from IMU (inertial measurement units) correlation for field predictions.
2. Methods
2.1. Participants
Twenty male recreational runners of Chinese ethnicity (age: 25.8 ± 1.6 years, body mass: 67.8 ± 5.3 kg, body height: 1.73 ± 0.05 m, detailed information presented in the supplementary Table s1) were recruited to join in this project, investigating the biomechanical changes in the lower extremity during distance running, as per the inclusion criteria. All included runners are right leg dominant, which was determined as the preferred leg of ball-kicking [60], avoiding inter-limb discrepancy and asymmetry [23]. All participants had a history of overground or treadmill running activities (with 5–8km per session and 3–6 sessions per week), with details described in the supplementary Table s1. They were free from any lower extremity injuries or foot deformities in the past six months prior to the test. The study was approved by the ethics committee from the Research Institute in Ningbo University (RAGH20161208). Written consent was obtained from all participants, and they were informed of the objectives, procedures, and requirements of this running test.
2.2. Protocol and instruments
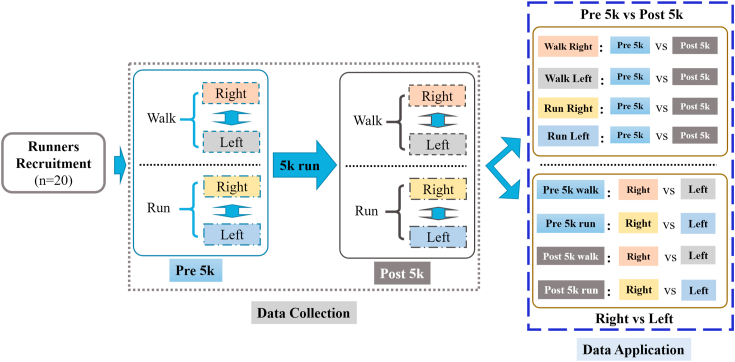
As the outlined in Figure 1, this project mainly included four parts. Firstly, the recreational runners of male Chinese ethnicity were recruited following the above-mentioned inclusion criteria. The pre 5k session (second part) was conducted to collect the right and left (separately) lower limbs during walking and running. Then (third part), all runners performed a 5k run on a treadmill at a submaximal speed [55]. The last (fourth) part repeated the second step with collection of the right and left limbs during walking and running. As shown in Figure 1 (Data Application), the collected dataset could be employed to investigate the biomechanical changes in the right and left limbs (respectively) and inter-limb symmetrical/asymmetrical responses from a 5k run.
Figure 1.
Outline of the experimental protocols (including Runners Recruitment, Data Collection during Pre 5k and Post 5k Sessions, and Data Application).
The pre and post 5k session were conducted on a 20m indoor pathway in a motion capture lab. The indoor running distance was similar with recent running studies [61, 62, 63]. Specifically, the lab was facilitated with an eight-camera Vicon Motion capture system (Vicon Metrics Ltd., Oxford, UK) and an embedded AMTI force platform (AMTI, Watertown, MA, United States) fixed in the middle of the pathway. They were synchronously employed to record the markers trajectories and ground reaction forces at a frequency of 200 Hz and 1000 Hz, respectively. The cameras and lab setting were calibrated before each experimental session with stable maker trajectory and less noise [64].
Prior to the test, participants performed 10 min walking or running on treadmill (to confirm a submaximal speed range) and ground (to control similar speed with treadmill run with timing gate) as warm-up. During the pre 5k session, two trials of static standing with both legs shoulder-width apart were firstly captured, followed by three trials of walking and running tests with right and left foot (each) consecutively landing on the force platform. Upon the completion of the pre 5k test session, the reflective markers were removed from participants. Participants ran 5k run on the treadmill with their own running footwear at a submaximal speed (∼12 km/h, 80% of their personal best speed), which was to ‘mimic’ a casual run. A 5-min preparation was left for the post 5k test setup (putting reflective markers back). The post 5k session was performed following the same testing procedures as the pre 5k session.
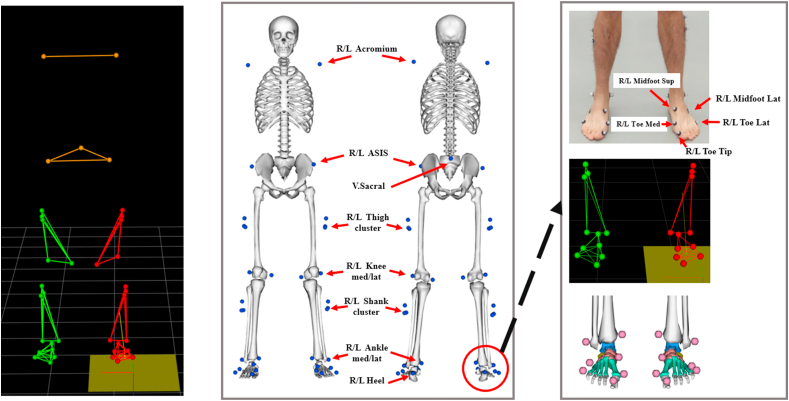
As illustrated in Figure 2, this dataset includes markers in both acromion, pelvis, bilateral thigh, bilateral knee, bilateral shank, and multi-segment foot. The details of collected data in raw C3D files are presented in the supplementary Table s2, including information of labels, dimension, unit, description (anatomical location) and capturing frequency.
Figure 2.
Illustration of marker-set (left & central) and multi-segmental foot model (right).
3. Data record and process
The collected data were then manually labelled in a Vicon Nexus software (version 1.8.5A, Vicon Metrics Ltd., Oxford, UK) following the maker-set model [11, 31, 65] as shown in Figure 2. In terms of the static and dynamic (walking and running) markers' trajectories, the gaps were visually checked and manually filled (using ‘pattern fill’ according to the shape of another trajectory without a gap to fill the selected gap) to avoid inconsistent trajectories [64]. As for the walking and running ground reaction forces, a threshold of 20N in the vertical direction was employed for the detection of foot strike and toe off to define the stance [38, 47, 66, 67]. A pipeline was set in the workstation of Nexus to export the raw C3D files. Consequently, a total of 28 successful trials per participant were obtained from the experimental tests, including trials from static, walking, and running of the left and right limbs during the pre 5k and post 5k sessions. The collected data could be applied for the pre 5k versus post 5k comparison and the right versus left limb (or dominant versus non-dominant) comparison.
3.1. Data store
One gait cycle (heel contact to next heel contact) was defined as per the vertical ground reaction force from the force platform during data processing of the left and right walking trials. While considering the high dynamics of running activity, one stance phase (heel contact to toe-off) for running trials was included during data processing of the left and right running trials. The threshold for the vertical ground reaction force was set at 20N [38, 47, 66, 67]. The collected experimental data were presented as raw in C3D files, which is public available in our figshare repository and SimTK repository.
3.2. Musculoskeletal model
This study employed an OpenSim full-body musculoskeletal model adapted from a recent study [31], and updated extra range of motion in the coronal and horizontal planes of the knee joint [11]. The current model added the muscle path into cylindrical wrapping surfaces adapting from the previously utilized ellipsoidal wrapping surface [68]. This model included 80 musculo-tendon parameters, derived from measured 21 cadaver specimens and MRI of 24 young healthy subjects, made it reasonable while scaling into other cohorts of young and healthy populations [31]. The model is available from the OpenSim full-body project repository (https://simtk.org/projects/full_body), including a combined head, torso, pelvis, and right and left lower extremity with femur, patella, tibia/fibula, and foot (talus, calcaneus, tarsus, metatarsal and toes). The hip joint was modelled as a ball-and-socket joint with three degree-of-freedoms (DOFs) in relative to the x-axis, y-axis, and z-axis. Apart from original DOF in the sagittal plane (flexion-extension, z-axis) of knee joint, extra two degree-of-freedoms in the abduction-adduction (x-axis) and internal-external rotation (y-axis) motions were updated, following previously validated studies [11, 69]. Ankle was modelled as a pin joint with dorsiflexion and plantarflexion in relative to z-axis. The subtalar and metatarsophalangeal joints were locked during the data processing.
The raw C3D files were exported as ‘trc’ (markers trajectories) and ‘mot’ (ground reaction force) using customized MATLAB scripts accompanying with the OpenSim software resource package (https://simtk-confluence.stanford.edu:8443/display/OpenSim/Scripting+with+Matlab). The data were filtered using a low pass (4th-order Butterworth) filter for ‘trc’ files at 6 Hz and ‘mot’ files at 30 Hz as per the capturing frequency [11].
The coordinates system from lab collected (Vicon) datasets (c3d files) were rotated to fit the OpenSim global and local coordinate system, thus positive x to the anterior, positive y to the superior, and positive z to the right. Following steps were performed in the OpenSim v4.0 as per previously published workflow [11, 30, 70], including the ‘Scale’, ‘Inverse Kinematics (IK)’, ‘Inverse Dynamics (ID)’, ‘Static Optimization (SO)’, and ‘Joint Reaction (JR)’ analysis. The surface EMG recorded muscle activities were compared against the simulated muscle activation from SO in OpenSim for model validation [11].
A customized Python function with a cubic spline was employed for the data interpolation (normalization) (https://spm1d.org/doc/Preliminaries/processing.html#interpolation), thus registering the data of left and right limbs into 101 data-point for walking gait cycle and 51 data-point for running stance. Averaged waveforms of joint angles, joint moments, and joint contact forces for each participant during pre-5k and post-5k sessions were plotted and stored as ‘fig’ files presented in the FIGURE folder for reference, respectively.
The pre-processed dataset was stored as ‘mat’ files in the MAT folder. In terms of the walking trials, nine matrices (pre 5k: 20∗101 vs post 5k: 20∗101; and right: 20∗101 vs left: 20∗101) (3 angles, 3 moments, and 3 contact forces) were created for the hip joint. Nine matrices (pre 5k: 20∗101 vs post 5k: 20∗101; and right: 20∗101 vs left: 20∗101) (3 angles, 3 moments and 3 contact forces) were created for the knee joint. Five matrices (pre 5k: 20∗101 vs post 5k: 20∗101) (1 angle, 1 moment and 3 contact forces) were created for the ankle joint. In terms of the running trials, nine matrices (pre 5k: 20∗51 vs post 5k: 20∗51) (3 angles, 3 moments, and 3 contact forces) were created for the hip joint. Nine matrices (pre 5k: 20∗51 vs post 5k: 20∗51) (3 angles, 3 moments and 3 contact forces) were created for the knee joint. Five matrices (pre 5k: 20∗51 vs post 5k: 20∗51) (1 angle, 1 moment and 3 contact forces) were created for the ankle joint.
In this dataset, a Principal Component Analysis (PCA) was taken as example for the advanced statistical analysis. The PCA would be conducted to reduce the high dimensionality data and project onto principal components (PCs) [47, 71], thus extracting the key features of variation.
| (1) |
As presented in Eq. (1), the original matrices (X = x1, x2, x3, … xn-1, xn) were orthogonally transformed into uncorrelated principal components (Z = z1, z2, z3, …, zm) (m < n), corresponding loading vectors (T2 = T1, T2, T3, …Tm) and residuals (Q), thus forming a relationship of Z = X∗T2.
This study mainly accounted for the main variations of the first key (3–4) PCs (z1, z2, z3, & z4) with explained variations of over 95%, and variations in the (3–4) PCs were plotted against the mean for visualization of the key features. The n was 101 data-point for walking trials and 51 data-point for running trials, respectively, during the PCA modelling. All the matrices of joint angles, joint moments, and joint contact forces firstly run statistical analysis, and then the PCA modelling in the MATLAB software (R2019a, The MathWorks Inc., MA, USA).
3.3. Usage notes
All data from this study are stored in c3d, mat and fig formats. The ‘c3d’ file can be read using the open-access software, MOKKA BTK (http://biomechanical-toolkit.github.io/mokka/index.html). The ‘mat’ and ‘fig’ files can be read using the commercial software MATLAB (The MathWorks, USA). The Musculoskeletal model in the current study is freely available from the SimTK forum (https://simtk.org/projects/opensim).
3.4. Code availability
The code for data interpolation and the Principal Component Analysis and Statistical Parametric Mapping analysis following the previously established studies [36, 37, 38, 47], which are available in the repositories (http://www.spm1d.org/; https://github.com/0todd0000/spm1dmatlab). The MOKKA BTK software is available in the repository (http://biomechanical-toolkit.github.io). The code for the OpenSim musculoskeletal model is available from the online repository (https://github.com/opensim-org/opensim-core).
4. Key results and findings
The dataset in raw data of ‘c3d’, pre-processed results and PCA modelling of the ‘mat’ and ‘fig’ files are available from the figshare repository and SimTK repository of our long-distance running project (https://auckland.figshare.com/projects/Dataset_of_Lower_Extremity_Joint_Angles_Moments_and_Forces_in_Distance_Running/136708, https://simtk.org/projects/longdistrun). Specific data organization and records are presented below.
4.1. Data records
The recorded markers trajectory and analog (ground reaction force) data were stored in C3D files (https://www.c3d.org), which are summarized in supplementary Table s2. Additional analog information was Fx1, Fx1, Fz1, Mx1, My1, and Mz1, which represented ground reaction force and moment at the x, y, and z coordinates of the foot in the current dataset. This dataset is available under the Creative Commons Attribution 4.0 International (CC BY 4.0) - Non-Commercial License.
4.1.1. Raw data
The C3D.zip folder contains the raw datasets collected from the motion capture experiments. The subfolders of ‘S01, S02, …, S20’ contain the data of twenty participants during the Pre 5k (Pre), and Post 5k (Post) sessions, respectively. The static trial is as ‘Sxx(number)_(Pre/Post)Static.c3d’, and the walking and running trials for right and left limbs are described as ‘Sxx(number)_(Pre/Post) (Walk/Run) (R/L)_xx(trials).c3d’.
4.1.2. Processed matrix data
The MAT.zip folder contains the processed (ready-for-use) dataset in well-organized matrices. The subfolders of ‘Pre5k_vs_Post5k’ and ‘Right_vs_Left’ contain the normalized data, which could be employed to investigate changes in Pre 5k versus Post 5k sessions and Right versus Left lower limbs difference during a submaximal 5k run.
‘PrePost_(walk/run) (/M/JR)_(R/L) (Ankle/Knee/Hip) (X/Y/Z).mat’ in the Pre5k_vs_Post5k folder of the MAT.zip represents the well-organized matrices comparison of walking (walk) or running (run) joint (Ankle/Knee/Hip) angles (), moment (M) or reaction force (JR) around the X, Y, and Z-axis during pre 5k (1–20 row) and post 5k (21–40 row) sessions.
‘(Pre/Post) (walk/run) (/M/JR)_LeftRight_(Ankle/Knee/Hip) (X/Y/Z).mat’ in the Right_vs_Left folder of the MAT.zip represents the well-organized matrices comparison of Right (1–20 row) and Left (21–40 row) joint (Ankle/Knee/Hip) angles (), moment (M) or reaction force (JR) during the Pre 5k or Post 5k walking (walk) or running (run) sessions.
4.1.3. Processed figure data
The FIGURE.zip folder contains the plotted figures of joint angles, moments, and forces from walking (Walk) and running (Run) trials. In terms of the walking (Walk) trials, the ‘Pre5k_vs_Post5k’ folder includes walking variables between the Pre 5k session and Post 5k session, as ‘(L/R)walk(/M/JR)_PrePost_(Ankle/Knee/Hip) (X/Y/Z)_raw.fig’.
The ‘Right_vs_Left’ folder includes variables between the Right limb and Left limb, as ‘(Pre/Post)Walk(/M/JR)_LeftRight_(Ankle/Knee/Hip) (X/Y/Z)_raw.fig’.
In terms of the running (Run) trials, the ‘Pre5k_vs_Post5k’ folder includes running variables between the Pre 5k and Post 5k sessions, as ‘(L/R)run(/M/JR)_PrePost_(Ankle/Knee/Hip) (X/Y/Z)_raw.fig’.
The ‘Right_vs_Left’ folder includes variables between the Right limb and Left limb, as ‘(Pre/Post)Run(/M/JR)_LeftRight_(Ankle/Knee/Hip) (X/Y/Z)_raw.fig’.
4.1.4. Processed PCA data
The PCA_mat.zip folder contains the matrix of PC coefficients, percentage explained, PC scores and reconstructed 4PCs. Specifically, the PCA modelling of pre 5k and post 5k in the right and left limbs are presented, as ‘PCA_PrePost_run(walk)_(/M/JR)_Left(right)_(Ankle/Knee/Hip) (X/Y/Z).mat’, during walking and running. The PCA modelling of the left and right limbs are presents, as ‘PCA_Pre(Post)run(walk)_(/M/JR)_LeftRight_(Ankle/Knee/Hip) (X/Y/Z).mat’.
The PCA_fig.zip folder contains the scores of the 4PCs and percentage explanation and line of accumulation in the first 10PCs. The specific first 4PCs were plot against the mean of joint angle, moment, or forces. Specifically, the pre 5k and post 5k in the right and left limbs are presented, as ‘PrePost_run(walk)_(/M/JR)_(R/L)_(Ankle/Knee/Hip) (X/Y/Z)_score_PCs.fig’ for the scores and explained percentage, and the ‘PrePost_run(walk)_(/M/JR)_(R/L)_(Ankle/Knee/Hip) (X/Y/Z)_4PCs.fig’ for the first 4PCs. The right and left limbs during pre 5k and post 5k are presented, as ‘Pre(Post)run(walk)_(/M/JR)_LeftRight_(Ankle/Knee/Hip) (X/Y/Z)_score_PCs.fig’ for the scores and explained percentage, and the ‘Pre(Post)run(walk)_(/M/JR)_LeftRight_(Ankle/Knee/Hip) (X/Y/Z)_ 4PCs.fig’ for the first 4PCs.
4.2. Results validation
The motion capture system and lab setting were calibrated before each experimental session to avoid noise and ensure high markers visibility. In terms of the marker gaps, a manual gap filling was performed using ‘pattern fill’ according to the shape of another trajectory without a gap to fill the selected gap, and force plate was checked (and zero-level) before each individual session to ensure an accurate measurement of ground reaction forces. The similar steps were followed as the recently published data descriptor articles [64, 72, 73, 74, 75, 76].
All the walking and running marker trajectories and ground reaction forces as stored in the raw C3D files were visually inspected and checked in the Motion Kinematics & Kinetics Analyzer (MOKKA) BTK software. The processed (ready-for-use) kinematics, kinetics, and forces results (presented in the FIGURE folder) were compared against recent studies of walking or running and confirmed with similar shape trends and magnitude from the authors.
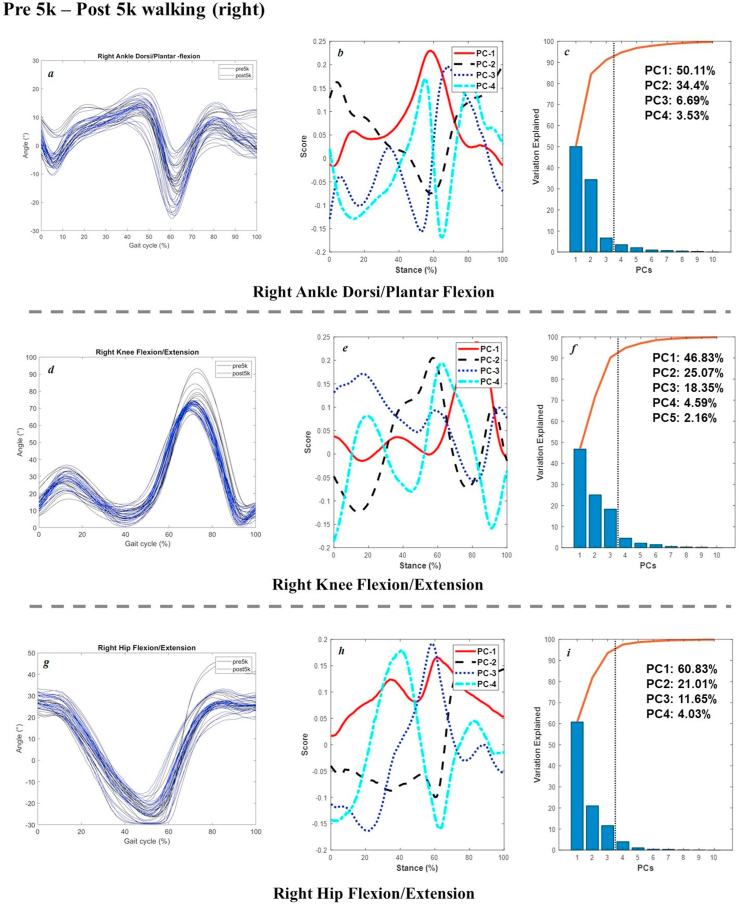
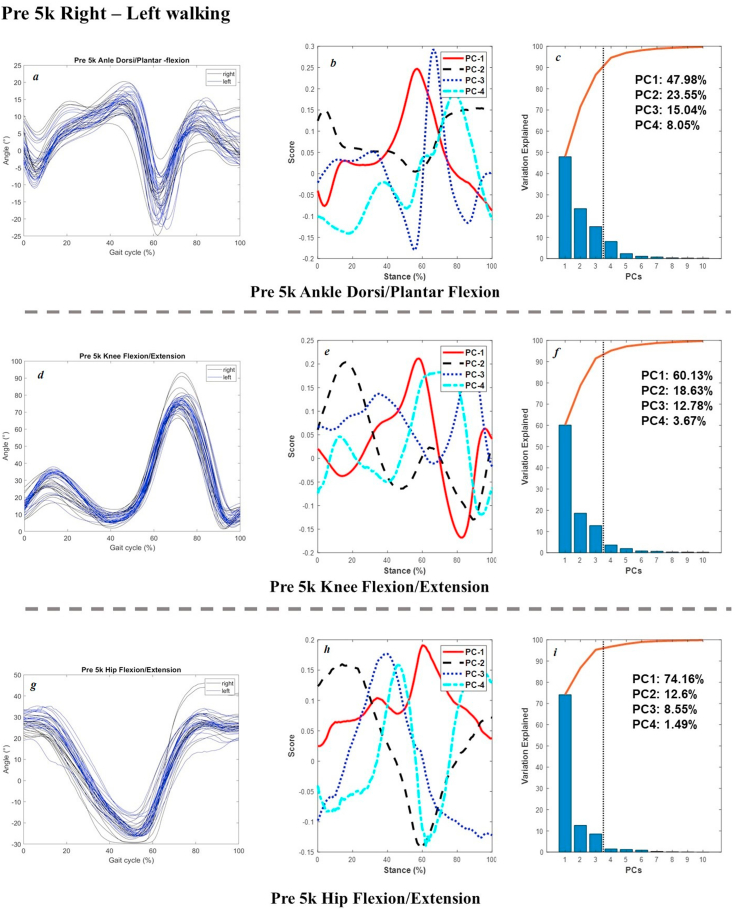
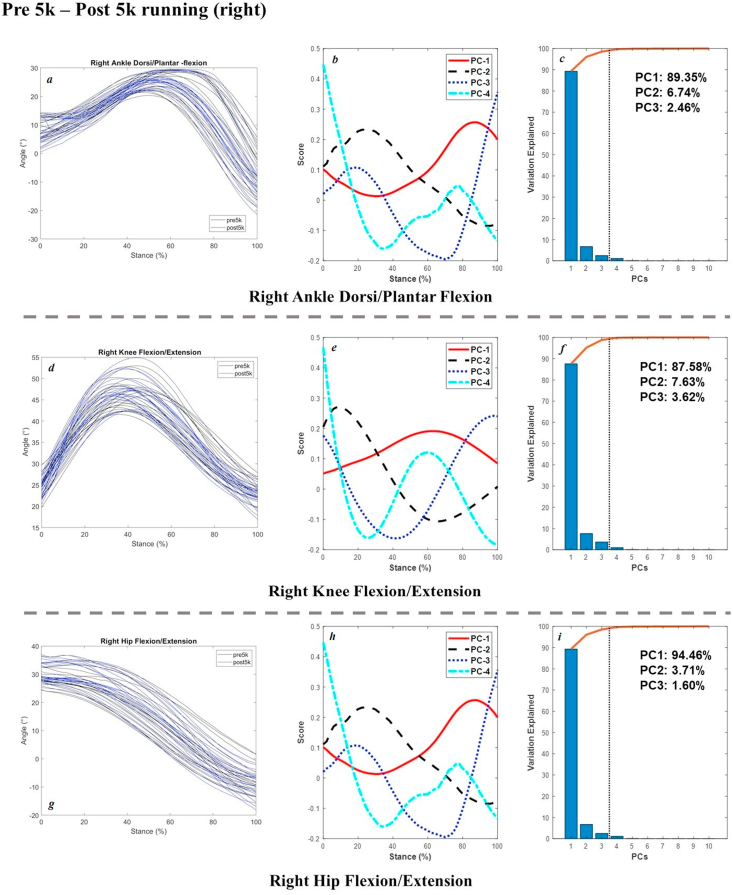
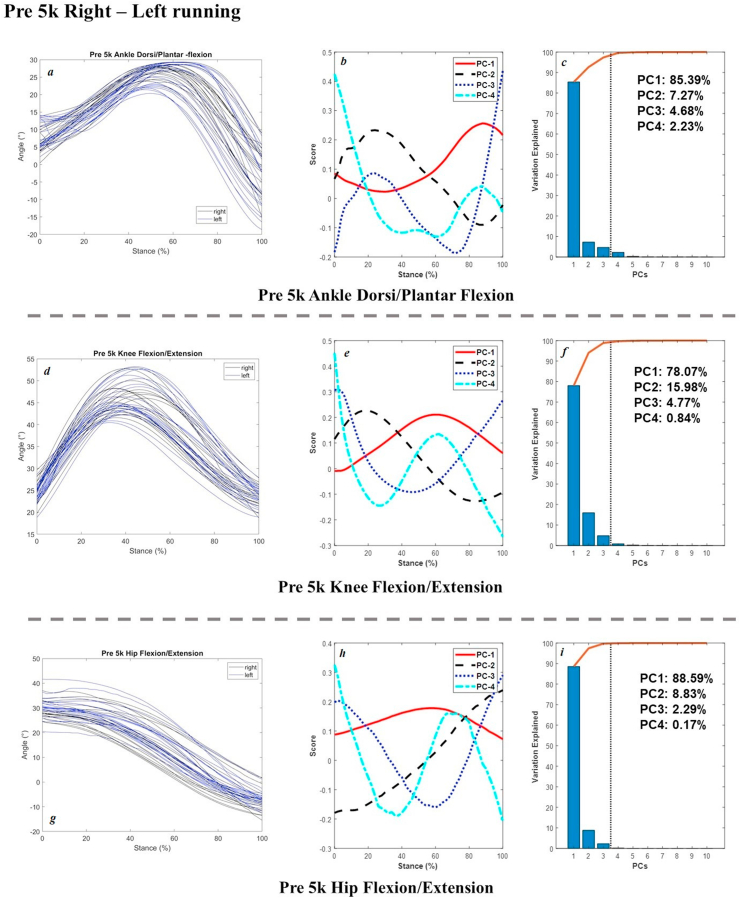
The joint angles in the sagittal plane, scores of 4 PCs, and percentage of explanation were plotted in Figure 3 (pre 5k and post 5k walking of the right limb), Figure 4 (left and right limb during pre 5k walking), Figure 5 (pre 5k and post 5k running of the right limb), and Figure 6 (left and right limb during pre 5k running) below as example for results validation. Extra dataset of joint kinetics and forces are available at our online project repository.
Figure 3.
Raw Joint angles in the sagittal plane (left: a, ankle; d, knee; g, hip) and scores of 4 PCs (central: b, e, h) and percentage explanation (right: c, f, i) during pre 5k and post 5k walking of the right limb.
Figure 4.
Raw Joint angles in the sagittal plane (left: a, ankle; d, knee; g, hip) and scores of 4 PCs (central: b, e, h) and percentage explanation (right: c, f, i) of left and right limb during pre 5k walking.
Figure 5.
Raw Joint angles in the sagittal plane (left: a, ankle; d, knee; g, hip) and scores of 4 PCs (central: b, e, h) and percentage explanation (right: c, f, i) during pre 5k and post 5k running of the right limb.
Figure 6.
Raw Joint angles in the sagittal plane (left: a, ankle; d, knee; g, hip) and scores of 4 PCs (central: b, e, h) and percentage explanation (right: c, f, i) of left and right limb during pre 5k running.
Specifically for the walking joint angles during pre 5k and post 5k sessions from the distance run, the first four PCs took up the main account of variances (94.73%) in the ankle dorsiflexion and plantarflexion, with PC1 of 50.11% showing variances from early stance (20%) to late stance (80%), PC2 of 34.4% during initial landing till mid stance, PC3 of 6.69% in the late stance, and PC4 of 3.53% in the late stance as well. The knee flexion showed main variances during push-off (PC1 of 46.83%) and mid stance (PC2 of 25.07%). The PC3 had variations during early stance while landing, with 18.35%. The hip flexion also presented main variances during late stance (PC1 of 60.83%, PC2 of 21.01%), and push-off (PC3 of 11.65%).
In terms of the left and right walking joint angles during pre 5k session for interlimb symmetry of asymmetry, the main variances of ankle dorsiflexion and plantarflexion were observed during mid stance (PC1 of 47.98%), and early landing and late push-off (PC2 of 23.55%), PC3 of 15.04%. The knee flexion had main variations from mid-stance to push-off in PC1 of 60.13%, followed by initial landing angles in PC2 of 18.63%, and final push-off in PC3 of 12.78%. The hip flexion showed variances across the stance in PC1 of 74.16%, and mid-stance in PC2 of 12.6% and PC3 of 8.55%.
As for the running joint angles during pre 5k and post 5k sessions from distance run, the main variances of ankle dorsiflexion and plantarflexion were observed in push-off (PC1 of 89.35%, PC2 of 6.74%). The knee flexion was mainly reported in the mid-stance as well in PC1 of 87.58%, and early landing to late push-off in PC2 of 7.63%. The variances of hip flexion were mainly observed in late push-off as well in PC1 of 94.46%. For the running joint angles comparison between the left and right limb during pre 5k run for interlimb symmetry of asymmetry, the main variations were observed in the midstance (PC1 of 78.07%) and followed by the landing and push-off in PC2 of 15.98% and PC3 of 4.77%.
5. Discussion
A public dataset on the walking and running biomechanics of recreational runners during a pre 5k and post 5k run was presented in the current study, which explicitly included raw data in c3d format, and pre-processed data (joint angles, moments and contact forces readily for reuse or reference) in mat format using a popular Open-source musculoskeletal modelling software (OpenSim). Extra details on the metadata were provided concerning runners' demographics (mass, height, running experience and limb dominance) and information of raw C3D files, which gave open options for the biomechanical researchers and sport scientists to reuse this dataset. In terms of the pre-processed ready-for-use data, authors ran a preliminary PCA modelling on the dataset to extra the key features of variations in the joint angles, moments, and contact forces from a 5k run. This is an example of using advanced statistics to generate key variations of the biomechanical parameters from distance running, apart from statistical significance of being different and larger or smaller. The current dataset of male Asian runners in Chinese ethnicity may provide comparison reference for the study of population-based long-distance running biomechanics. Another important contribution is to analyse the multi-segmental foot biomechanical changes from distance running, which is scarcely reported and documented in the literature. The current dataset also investigates biomechanical response from distance running with self-preferred shoes, which may be used as baseline (or comparison) to investigate the biomechanical alterations with improved footwear technology [77].
While validating the raw dataset from the current study, the detailed information concerning the experimental protocol and instruments employed for data collection showed consistency with recent literature of running studies [11, 24, 46, 78]. The gap in the marker trajectory and ground reaction forces were visually checked in the Vicon Nexus and Mokka BTK software [79] in C3D files. In terms of the pre-processed ready-for-use dataset in well-organized matrix, the joint angles in the sagittal plane of lower extremity joints (hip, knee and ankle) were plotted and compared against literatures. This was due to the high waveform similarity was observed and documented from recent studies of distance runners [80]. While appropriate similarity was recorded in the other two planes, the sagittal angles were believed to be consistent considering the variations from distance running [81, 82] or inter-limb asymmetry [23, 83]. In terms of the potential similarity or difference between different MSK software, the current study presented ready-for-use data from OpenSim platform following well-established pipeline [30, 31, 32], and comparison between different MSK software was conducted for the calculation of angles, moments, and muscle activations and forces [28, 29]. The current dataset also includes extra the marker trajectory (trc) and ground reaction force (mot) files, allowing users to employ different MSK approaches.
Advanced statistics have been employed recently for the classification of runner cohorts (distance and gender) while analysing the kinematics [46], ground reaction forces [47] and running performance [84]. Following an example of principal component analysis, a PCA of ground reaction forces between gender at incremental running speeds was conducted to extract key features of variances for understanding the impact characteristics [47]. As illustrated in the figures of this study, the joint angles, PCA scores and percentage of explanations showed the regions of key variations from the score values, and main contributions. Extra information of PCA results on the joint moments and contact forces during pre 5k and post 5k and right and left limbs were included in the online repository. Users may refer to this results in raw to understand biomechanical responses from a single submaximal distance running [85], and investigate potential biomechanical loading redistribution across joints and between limbs.
Apart from the PCA statistical modelling, users of biomechanics and sports science community may run hypothetical statistics on the ready-for-use dataset. An example was provided for reference using the popular open-source one-dimensional statistical parametric mapping (SPM1d) package. Prior to statistical analysis, the normality (‘spm1d.stats.normality.ttest_paired’) distribution could be checked using the built-in function in the open-source SPM1d package (www.spm1d.org), which was previously reported the applications in analysis of the time-varying 1D biomechanical data [36, 37]. As per the results of the normality check, Statistical Parametric Mapping (SPM, ‘spm1d.stats.ttest_paired’) or Statistical non-Parametric Mapping (SnPM, ‘spm1d.stats.nonparam.ttest_paired’) analysis would be taken for the statistical comparison in the current study, following a previous example [38]. The paired sample t-test in SPM was previously used to check the difference between Pre 5k and Post 5k running and examine the inter-limb (Right against Left) symmetry or asymmetry [11, 86, 87, 88].
There are few limitations for the processed datasets and experimental procedures which should be noted. Firstly, the open-source OpenSim software was employed for all the data processing, which was due to that this software is free and popular among all the biomechanics community [32]. These datasets may need further validation via comparisons against other musculoskeletal platforms, such as Anybody [23, 27] and Visual 3D [28, 29]. Secondly, this study presented the dataset of the male recreational runners' cohort all in Chinese ethnicity. While considering the population-based running biomechanical performance and related injuries [89, 90, 91, 92, 93], this database of 20 participants (relatively small sample size) may be a foundation and provide a pilot example for future studies to expand the samples of runners from different ethnicities. Thirdly, the 5k run was conducted on a treadmill at a submaximal (80% PB) speed, while the data collection was overground running without strictly controlling running speeds [47] and surfaces [61]. As reported in the literature, different running surface may alter the biomechanical responses [94, 95, 96]. However, the current dataset mainly focuses on the response from a distance run with particular interests of the biomechanical changes in nature. Lastly, this study collected datasets of barefoot walking and running in a lab setting during a pre 5k and post 5k run with self-preferred shoes without standardizing the running footwear. This would be a first public available database to investigate inter foot segmental biomechanical changes from distance running.
6. Conclusion
In summary, this study presented a validated database in raw (C3D files) and pre-processed ready-for-use (mat files) matrix, explicitly including the joint angles, moments, and forces of the lower extremity from distance running at a submaximal speed in recreational runners. The dataset is from Asian population of Chinese ethnicity, which may be used for the analysis of population-based running biomechanics. The raw database included multi-segmental foot marker-set, which may add up the information of inter-segmental foot biomechanical changes from distance running. The extracted key variances and weighted scores from PCA modelling of joint angles, moments and contact forces may be reference as training database for advanced statistics and wearable technology prediction in the real-scenarios analysis of distance running.
Declarations
Author contribution statement
Qichang Mei: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Justin Fernandez: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Liangliang Xiang; Zixiang Gao; Peimin Yu: Performed the experim-ents.
Julien S Baker: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Yaodong Gu: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This study was sponsored by National Natural Science Foundation of China (No. 12202216), NSFC (Natural Science Foundation of China) - RSE (The Royal Society of Edinburgh) Joint Project (No. 81911530253) and K. C. Wong Magna Fund in Ningbo University. QM of this paper was supported by the New Zealand-China. Doctoral Research Scholarship issued from the Ministry of Foreign Affairs and Trade (New Zealand). LX, ZG and PM are currently supported by the China Scholarship Council (CSC).
Data availability statement
Data associated with this study has been deposited at https://auckland.figshare.com/projects/Dataset_of_Lower_Extremity_Joint_Angles_Moments_and_Forces_in_Distance_Running/136708.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
This study was sponsored by National Natural Science Foundation of China (No. 12202216), NSFC (Natural Science Foundation of China) - RSE (The Royal Society of Edinburgh) Joint Project (No. 81911530253) and K. C. Wong Magna Fund in Ningbo University. QM of this paper was supported by the New Zealand-China Doctoral Research Scholarship issued from the Ministry of Foreign Affairs and Trade (New Zealand). LX, ZG and PM are currently supported by the China Scholarship Council (CSC).
Contributor Information
Qichang Mei, Email: meiqichang@outlook.com, qmei907@aucklanduni.ac.nz.
Yaodong Gu, Email: guyaodong@hotmail.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Lee D., Brellenthin A.G., Thompson P.D., Sui X., Lee I.-M., Lavie C.J. Running as a key lifestyle medicine for longevity. Prog. Cardiovasc. Dis. 2017;60:45–55. doi: 10.1016/j.pcad.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Oja P., Kelly P., Pedisic Z., Titze S., Bauman A., Foster C., Hamer M., Hillsdon M., Stamatakis E. Associations of specific types of sports and exercise with all-cause and cardiovascular-disease mortality: a cohort study of 80 306 British adults. Br. J. Sports Med. 2017;51:812–817. doi: 10.1136/bjsports-2016-096822. [DOI] [PubMed] [Google Scholar]
- 3.Lear S.A., Hu W., Rangarajan S., Gasevic D., Leong D., Iqbal R., Casanova A., Swaminathan S., Anjana R.M., Kumar R., Rosengren A., Wei L., Yang W., Chuangshi W., Huaxing L., Nair S., Diaz R., Swidon H., Gupta R., Mohammadifard N., Lopez-Jaramillo P., Oguz A., Zatonska K., Seron P., Avezum A., Poirier P., Teo K., Yusuf S. The effect of physical activity on mortality and cardiovascular disease in 130,000 people from 17 high-income, middle-income, and low-income countries: the PURE study. Lancet. 2017;390:2643–2654. doi: 10.1016/S0140-6736(17)31634-3. [DOI] [PubMed] [Google Scholar]
- 4.Pedisic Z., Shrestha N., Grgic J., Kovalchik S., Stamatakis E., Liangruenrom N., Titze S., Biddle S., Bauman A.E., Virgile A., Oja P. Infographic. Is running associated with a lower risk of all-cause, cardiovascular and cancer mortality, and is more better? A systematic review and meta-analysis. Br. J. Sports Med. 2020;54:817–818. doi: 10.1136/bjsports-2019-101793. [DOI] [PubMed] [Google Scholar]
- 5.Moreira J.B.N., Wohlwend M., Wisløff U. Exercise and cardiac health: physiological and molecular insights. Nat Metab. 2020;2:829–839. doi: 10.1038/s42255-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 6.Mei Q., Gu Y., Sun D., Fernandez J. How foot morphology changes influence shoe comfort and plantar pressure before and after long distance running? Acta Bioeng. Biomech. 2018;20:179–186. [PubMed] [Google Scholar]
- 7.Besson T., Macchi R., Rossi J., Morio C.Y.M., Kunimasa Y., Nicol C., Vercruyssen F., Millet G.Y. 2022. Sex Differences in Endurance Running, Sports Medicine. [DOI] [PubMed] [Google Scholar]
- 8.Snyder K.L., Hoogkamer W., Triska C., Taboga P., Arellano C.J., Kram R. Effects of course design (curves and elevation undulations) on marathon running performance: a comparison of Breaking 2 in Monza and the INEOS 1:59 Challenge in Vienna. J. Sports Sci. 2021;39:754–759. doi: 10.1080/02640414.2020.1843820. [DOI] [PubMed] [Google Scholar]
- 9.Hulme A., Nielsen R.O., Timpka T., Verhagen E., Finch C. Risk and protective factors for middle- and long-distance running-related injury. Sports Med. 2017;47:869–886. doi: 10.1007/s40279-016-0636-4. [DOI] [PubMed] [Google Scholar]
- 10.Taunton J.E., Ryan M.B., Clement D.B., McKenzie D.C., Lloyd-Smith D.R., Zumbo B.D. A retrospective case-control analysis of 2002 running injuries. Br. J. Sports Med. 2002;36:95–101. doi: 10.1136/bjsm.36.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mei Q., Gu Y., Xiang L., Baker J.S., Fernandez J. Foot pronation contributes to altered lower extremity loading after long distance running. Front. Physiol. 2019;10:573. doi: 10.3389/fphys.2019.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanno M., Willwacher S., Epro G., Brüggemann G.-P. Positive work contribution shifts from distal to proximal joints during a prolonged run. Med. Sci. Sports Exerc. 2018;50:2507–2517. doi: 10.1249/MSS.0000000000001707. [DOI] [PubMed] [Google Scholar]
- 13.Kim H.K., Fernandez J., Logan C., Tarr G.P., Doyle A., Mirjalili S.A. T2 relaxation time measurements in tibiotalar cartilage after barefoot running and its relationship to ankle biomechanics. J. Biomech. 2019;90:103–112. doi: 10.1016/j.jbiomech.2019.04.046. [DOI] [PubMed] [Google Scholar]
- 14.Kim H.K., Mirjalili A., Doyle A., Fernandez J. Tibiotalar cartilage stress corresponds to T2 mapping: application to barefoot running in novice and marathon-experienced runners. Comput. Methods Biomech. Biomed. Eng. 2019;22:1153–1161. doi: 10.1080/10255842.2019.1645133. [DOI] [PubMed] [Google Scholar]
- 15.Hoenig T., Hamacher D., Braumann K.M., Zech A., Hollander K. Analysis of running stability during 5000 m running. Eur. J. Sport Sci. 2019;19:413–421. doi: 10.1080/17461391.2018.1519040. [DOI] [PubMed] [Google Scholar]
- 16.Larson P., Higgins E., Kaminski J., Decker T., Preble J., Lyons D., McIntyre K., Normile A. Foot strike patterns of recreational and sub-elite runners in a long-distance road race. J. Sports Sci. 2011;29:1665–1673. doi: 10.1080/02640414.2011.610347. [DOI] [PubMed] [Google Scholar]
- 17.Ueberschär O., Fleckenstein D., Warschun F., Kränzler S., Walter N., Hoppe M.W. Measuring biomechanical loads and asymmetries in junior elite long-distance runners through triaxial inertial sensors. Sports Orthop. Traumatol. 2019;35:296–308. [Google Scholar]
- 18.Hanley B., Tucker C.B. Gait variability and symmetry remain consistent during high-intensity 10,000 m treadmill running. J. Biomech. 2018;79:129–134. doi: 10.1016/j.jbiomech.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa H., Yamauchi T., Kraemer W.J. Foot strike patterns of runners at the 15-km point during an elite-level half marathon. J. Strength Condit Res. 2007;21:888–893. doi: 10.1519/R-22096.1. [DOI] [PubMed] [Google Scholar]
- 20.Blagrove R.C., Howatson G., Hayes P.R. Effects of strength training on the physiological determinants of middle- and long-distance running performance: a systematic review. Sports Med. 2018;48:1117–1149. doi: 10.1007/s40279-017-0835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller J.T., Bellenger C.R., Thewlis D., Tsiros M.D., Buckley J.D. The effect of footwear on running performance and running economy in distance runners. Sports Med. 2015;45:411–422. doi: 10.1007/s40279-014-0283-6. [DOI] [PubMed] [Google Scholar]
- 22.Troy K.L., Tetreault K., Goodworth A.D., Ji S., Popovic M.B. Biomechanics and biomechatronics in sports, exercise, and entertainment. Biomechatronics, Elsevier. 2019:451–494. [Google Scholar]
- 23.Gao Z., Mei Q., Fekete G., Baker J.S., Gu Y. The effect of prolonged running on the symmetry of biomechanical variables of the lower limb joints. Symmetry (Basel). 2020;12:720. [Google Scholar]
- 24.Mei Q., Xiang L., Sun D., Li J., Fernandez J., Gu Y. Foot pronation after prolonged running increased the medical contact force in the knee joint: a study based on OpenSim modelling and machine learning prediction. China Sport Sci. 2019;39:51–59. [Google Scholar]
- 25.Kim H.K., Mei Q., Gu Y., Mirjalili A., Fernandez J. Reduced joint reaction and muscle forces with barefoot running. Comput. Methods Biomech. Biomed. Eng. 2021;24:1263–1273. doi: 10.1080/10255842.2021.1880572. [DOI] [PubMed] [Google Scholar]
- 26.Folland J.P., Allen S.J., Black M.I., Handsaker J.C., Forrester S.E. Running technique is an important component of running economy and performance. Med. Sci. Sports Exerc. 2017;49:1412–1423. doi: 10.1249/MSS.0000000000001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soares T.S.A., de Oliveira C.F., Pizzuto F., Manuel Garganta R., Vila-Boas J.P., Paiva M.C. da A. Acute kinematics changes in marathon runners using different footwear. J. Sports Sci. 2018;36:766–770. doi: 10.1080/02640414.2017.1340657. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y., Jung Y., Choi W., Lee K., Koo S. Similarities and differences between musculoskeletal simulations of OpenSim and AnyBody modeling system. J. Mech. Sci. Technol. 2018;32:6037–6044. [Google Scholar]
- 29.Trinler U., Schwameder H., Baker R., Alexander N. Muscle force estimation in clinical gait analysis using AnyBody and OpenSim. J. Biomech. 2019;86:55–63. doi: 10.1016/j.jbiomech.2019.01.045. [DOI] [PubMed] [Google Scholar]
- 30.Delp S.L., Anderson F.C., Arnold A.S., Loan P., Habib A., John C.T., Guendelman E., Thelen D.G. OpenSim: open-source software to create and analyze dynamic simulations of movement. IEEE Trans. Biomed. Eng. 2007;54:1940–1950. doi: 10.1109/TBME.2007.901024. [DOI] [PubMed] [Google Scholar]
- 31.Rajagopal A., Dembia C.L., DeMers M.S., Delp D.D., Hicks J.L., Delp S.L. Full-body musculoskeletal model for muscle-driven simulation of human gait. IEEE Trans. Biomed. Eng. 2016;63:2068–2079. doi: 10.1109/TBME.2016.2586891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seth A., Hicks J.L., Uchida T.K., Habib A., Dembia C.L., Dunne J.J., Ong C.F., DeMers M.S., Rajagopal A., Millard M., Hamner S.R., Arnold E.M., Yong J.R., Lakshmikanth S.K., Sherman M.A., Ku J.P., Delp S.L. OpenSim: simulating musculoskeletal dynamics and neuromuscular control to study human and animal movement. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelburne K.B., Torry M.R., Pandy M.G. Muscle, ligament, and joint-contact forces at the knee during walking. Med. Sci. Sports Exerc. 2005;37:1948–1956. doi: 10.1249/01.mss.0000180404.86078.ff. [DOI] [PubMed] [Google Scholar]
- 34.Bowser B.J., Fellin R., Milner C.E., Pohl M.B., Davis I.S. Reducing impact loading in runners: a one-year follow-up. Med. Sci. Sports Exerc. 2018;50:2500. doi: 10.1249/MSS.0000000000001710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanrenterghem J., Venables E., Pataky T., Robinson M.a. The effect of running speed on knee mechanical loading in females during side cutting. J. Biomech. 2012;45:2444–2449. doi: 10.1016/j.jbiomech.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 36.Pataky T.C., Vanrenterghem J., Robinson M.A. Zero- vs. one-dimensional, parametric vs. non-parametric, and confidence interval vs. hypothesis testing procedures in one-dimensional biomechanical trajectory analysis. J. Biomech. 2015;48:1277–1285. doi: 10.1016/j.jbiomech.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 37.Pataky T.C., Vanrenterghem J., Robinson M.A. The probability of false positives in zero-dimensional analyses of one-dimensional kinematic, force and EMG trajectories. J. Biomech. 2016;49:1468–1476. doi: 10.1016/j.jbiomech.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 38.Mei Q., Xiang L., Li J., Fernandez J., Gu Y. Analysis of running ground reaction forces using the one-dimensional statistical parametric mapping (SPM1d) J. Med. Biomechanics. 2021;36:684–691. [Google Scholar]
- 39.Friston K., Ashburner J., Kiebel S., Nichols T., Penny W. Elsevier/Academic Press; Amsterdam: 2007. Statistical Parametric Mapping: the Analysis of Functional Brain Images. [Google Scholar]
- 40.Pataky T.C. Generalized n-dimensional biomechanical field analysis using statistical parametric mapping. J. Biomech. 2010;43:1976–1982. doi: 10.1016/j.jbiomech.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Lever J., Krzywinski M., Altman N. Points of significance: principal component analysis. Nat. Methods. 2017;14:641–642. [Google Scholar]
- 42.Ramsay J.O., Silverman B.W. second ed. Springer New York; New York, NY: 2005. Functional Data Analysis. [Google Scholar]
- 43.Deluzio K.J., Wyss U.P., Zee B., Costigan P.A., Sorbie C. Principal component models of knee kinematics and kinetics: normal vs. pathological gait patterns. Hum. Mov. Sci. 1997;16:201–217. [Google Scholar]
- 44.Daffertshofer A., Lamoth C.J.C., Meijer O.G., Beek P.J. PCA in studying coordination and variability: a tutorial. Clin. BioMech. 2004;19:415–428. doi: 10.1016/j.clinbiomech.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Osis S.T., Hettinga B.A., Leitch J., Ferber R. Predicting timing of foot strike during running, independent of striking technique, using principal component analysis of joint angles. J. Biomech. 2014;47:2786–2789. doi: 10.1016/j.jbiomech.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Clermont C.A., Phinyomark A., Osis S.T., Ferber R. Classification of higher- and lower-mileage runners based on running kinematics. J Sport Health Sci. 2019;8:249–257. doi: 10.1016/j.jshs.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu L., Mei Q., Xiang L., Liu W., Mohamad N.I., István B., Fernandez J., Gu Y. Principal component analysis of the running ground reaction forces with different speeds. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.629809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behling A.V., Nigg B.M. Relationships between the foot posture Index and static as well as dynamic rear foot and arch variables. J. Biomech. 2020;98 doi: 10.1016/j.jbiomech.2019.109448. [DOI] [PubMed] [Google Scholar]
- 49.Harrison A.J., Ryan W., Hayes K. Functional data analysis of joint coordination in the development of vertical jump performance. Sports BioMech. 2007;6:199–214. doi: 10.1080/14763140701323042. [DOI] [PubMed] [Google Scholar]
- 50.Warmenhoven J., Harrison A., Robinson M.A., Vanrenterghem J., Bargary N., Smith R., Cobley S., Draper C., Donnelly C., Pataky T. A force profile analysis comparison between functional data analysis, statistical parametric mapping and statistical non-parametric mapping in on-water single sculling. J. Sci. Med. Sport. 2018;21:1100–1105. doi: 10.1016/j.jsams.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Warmenhoven J., Bargary N., Liebl D., Harrison A., Robinson M., Gunning E., Hooker G. PCA of waveforms and functional PCA: a primer for biomechanics. J. Biomech. 2021;116 doi: 10.1016/j.jbiomech.2020.110106. [DOI] [PubMed] [Google Scholar]
- 52.Cushion E.J., Warmenhoven J., North J.S., Cleather D.J. Principal component analysis reveals the proximal to distal pattern in vertical jumping is governed by two functional degrees of freedom. Front. Bioeng. Biotechnol. 2019;7:193. doi: 10.3389/fbioe.2019.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.da Silva Soares J., Carpes F.P., de Fátima Geraldo G., Bertú Medeiros F., Roberto Kunzler M., Sosa Machado Á., Augusto Paolucci L., Gustavo Pereira de Andrade A. Functional data analysis reveals asymmetrical crank torque during cycling performed at different exercise intensities. J. Biomech. 2021;122 doi: 10.1016/j.jbiomech.2021.110478. [DOI] [PubMed] [Google Scholar]
- 54.Donoghue O.A., Harrison A.J., Coffey N., Hayes K. Functional data analysis of running kinematics in Chronic Achilles tendon injury. Med. Sci. Sports Exerc. 2008;40:1323–1335. doi: 10.1249/MSS.0b013e31816c4807. [DOI] [PubMed] [Google Scholar]
- 55.Enoksen E., Rønning Tjelta A., Tjelta L.I., Bosch A. Distribution of training volume and intensity of elite male and female track and marathon runners. Int. J. Sports Sci. Coach. 2011;6:273–293. [Google Scholar]
- 56.Sun D., Fekete G., Mei Q., Gu Y. The effect of walking speed on the foot inter-segment kinematics, ground reaction forces and lower limb joint moments. PeerJ. 2018;6:e5517. doi: 10.7717/peerj.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun D., Fekete G., Baker J.S., Gu Y. Foot motion character during forward and backward walking with shoes and barefoot. J. Mot. Behav. 2020;52:214–225. doi: 10.1080/00222895.2019.1605972. [DOI] [PubMed] [Google Scholar]
- 58.Xiang L., Mei Q., Fernandez J., Gu Y. A biomechanical assessment of the acute hallux abduction manipulation intervention. Gait Posture. 2020;76:210–217. doi: 10.1016/j.gaitpost.2019.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Xiang L., Mei Q., Xu D., Fernandez J. Multi-segmental motion in foot during counter-movement jump with toe manipulation. Appl. Sci. 2020;10:1893. [Google Scholar]
- 60.Chapman J.P., Chapman L.J., Allen J.J. The measurement of foot preference. Neuropsychologia. 1987;25:579–584. doi: 10.1016/0028-3932(87)90082-0. [DOI] [PubMed] [Google Scholar]
- 61.Hébert-Losier K., Mourot L., Holmberg H.C. Elite and amateur orienteers’ running biomechanics on three surfaces at three speeds. Med. Sci. Sports Exerc. 2015;47:381–389. doi: 10.1249/MSS.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 62.Schache A.G., Wrigley T.v., Baker R., Pandy M.G. Biomechanical response to hamstring muscle strain injury. Gait Posture. 2009;29:332–338. doi: 10.1016/j.gaitpost.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 63.Benson L.C., Clermont C.A., Ferber R. New considerations for collecting biomechanical data using wearable sensors: the effect of different running environments. Front. Bioeng. Biotechnol. 2020;8:86. doi: 10.3389/fbioe.2020.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ozkaya G., Jung H.R., Jeong I.S., Choi M.R., Shin M.Y., Lin X., Heo W.S., Kim M.S., Kim E., Lee K.K. Three-dimensional motion capture data during repetitive overarm throwing practice. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamner S.R., Delp S.L. Muscle contributions to fore-aft and vertical body mass center accelerations over a range of running speeds. J. Biomech. 2013;46:780–787. doi: 10.1016/j.jbiomech.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breine B., Malcolm P., Galle S., Fiers P., Frederick E.C., De Clercq D. Running speed-induced changes in foot contact pattern influence impact loading rate. Eur. J. Sport Sci. 2019;19:774–783. doi: 10.1080/17461391.2018.1541256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Drongelen S., Wesseling M., Holder J., Meurer A., Stief F. Knee load distribution in hip osteoarthritis patients after total hip replacement. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.578030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arnold E.M., Ward S.R., Lieber R.L., Delp S.L. A model of the lower limb for analysis of human movement. Ann. Biomed. Eng. 2010;38:269–279. doi: 10.1007/s10439-009-9852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meireles S., De Groote F., Van Rossom S., Verschueren S., Jonkers I. Differences in knee adduction moment between healthy subjects and patients with osteoarthritis depend on the knee axis definition. Gait Posture. 2017;53:104–109. doi: 10.1016/j.gaitpost.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 70.Meireles S., De Groote F., Reeves N.D., Verschueren S., Maganaris C., Luyten F., Jonkers I. Knee contact forces are not altered in early knee osteoarthritis. Gait Posture. 2016;45:115–120. doi: 10.1016/j.gaitpost.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 71.Lever J., Krzywinski M., Altman N. Points of significance: principal component analysis. Nat. Methods. 2017;14:641–642. [Google Scholar]
- 72.Schreiber C., Moissenet F. A multimodal dataset of human gait at different walking speeds established on injury-free adult participants. Sci. Data. 2019;6:111. doi: 10.1038/s41597-019-0124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreira L., Figueiredo J., Fonseca P., Vilas-Boas J.P., Santos C.P. Lower limb kinematic, kinetic, and EMG data from young healthy humans during walking at controlled speeds. Sci. Data. 2021;8:103. doi: 10.1038/s41597-021-00881-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hood S., Ishmael M.K., Gunnell A., Foreman K.B., Lenzi T. A kinematic and kinetic dataset of 18 above-knee amputees walking at various speeds. Sci. Data. 2020;7:1–8. doi: 10.1038/s41597-020-0494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lencioni T., Carpinella I., Rabuffetti M., Marzegan A., Ferrarin M. Human kinematic, kinetic and EMG data during different walking and stair ascending and descending tasks. Sci. Data. 2019;6:309. doi: 10.1038/s41597-019-0323-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Horst F., Slijepcevic D., Simak M., Schöllhorn W.I. Gutenberg Gait Database, a ground reaction force database of level overground walking in healthy individuals. Sci. Data. 2021;8:232. doi: 10.1038/s41597-021-01014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muniz-Pardos B., Sutehall S., Angeloudis K., Guppy F.M., Bosch A., Pitsiladis Y. Recent improvements in marathon run times are likely technological, not physiological. Sports Med. 2021;51:371–378. doi: 10.1007/s40279-020-01420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen T.L.W., Lam W.K., Wong D.W.C., Zhang M. A half marathon shifts the mediolateral force distribution at the tibiofemoral joint. Eur. J. Sport Sci. 2021:1–23. doi: 10.1080/17461391.2021.1938690. [DOI] [PubMed] [Google Scholar]
- 79.Barre A., Armand S. Biomechanical ToolKit: open-source framework to visualize and process biomechanical data. Comput. Methods Progr. Biomed. 2014;114:80–87. doi: 10.1016/j.cmpb.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 80.Garcia M.C., Taylor-Haas J.A., Ford K.R., Long J.T. Assessment of waveform similarity in youth long-distance runners. Gait Posture. 2020;77:105–111. doi: 10.1016/j.gaitpost.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 81.Chen T.L.W., Wong D.W.C., Wang Y., Tan Q., Lam W.K., Zhang M. Changes in segment coordination variability and the impacts of the lower limb across running mileages in half marathons: implications for running injuries. J. Sport Health Sci. 2022;11:67–74. doi: 10.1016/j.jshs.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willwacher S., Sanno M., Brüggemann G.P. Fatigue matters: an intense 10 km run alters frontal and transverse plane joint kinematics in competitive and recreational adult runners. Gait Posture. 2020;76:277–283. doi: 10.1016/j.gaitpost.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 83.Melo C.C., Carpes F.P., Vieira T.M., Mendes T.T., de Paula L.V., Chagas M.H., Peixoto G.H.C., de Andrade A.G.P. Correlation between running asymmetry, mechanical efficiency, and performance during a 10 km run. J. Biomech. 2020;109 doi: 10.1016/j.jbiomech.2020.109913. [DOI] [PubMed] [Google Scholar]
- 84.Liu Q., Mo S., Cheung V.C.K., Cheung B.M.F., Wang S., Chan P.P.K., Malhotra A., Cheung R.T.H., Chan R.H.M. Classification of runners’ performance levels with concurrent prediction of biomechanical parameters using data from inertial measurement units. J. Biomech. 2020;112 doi: 10.1016/j.jbiomech.2020.110072. [DOI] [PubMed] [Google Scholar]
- 85.Paquette M.R., Camelio K., Gruber A.H., Powell D.W. Influence of prolonged running and training on tibial acceleration and movement quality in novice runners. J. Athl. Train. 2020;55:1292–1299. doi: 10.4085/1062-6050-0491.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hughes-Oliver C.N., Harrison K.A., Williams D.S.B., Queen R.M. Statistical parametric mapping as a measure of differences between limbs: applications to clinical populations. J. Appl. Biomech. 2019;35:377–387. doi: 10.1123/jab.2018-0392. [DOI] [PubMed] [Google Scholar]
- 87.Serrien B., Goossens M., Baeyens J.P. Statistical parametric mapping of biomechanical one-dimensional data with Bayesian inference. Int Biomech. 2019;6:9–18. doi: 10.1080/23335432.2019.1597643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furlong L.A.M., Egginton N.L. Kinetic asymmetry during running at preferred and nonpreferred speeds. Med. Sci. Sports Exerc. 2018;50:1241–1248. doi: 10.1249/MSS.0000000000001560. [DOI] [PubMed] [Google Scholar]
- 89.Santos-Concejero J., Tam N., Coetzee D.R., Oliván J., Noakes T.D., Tucker R. Are gait characteristics and ground reaction forces related to energy cost of running in elite Kenyan runners? J. Sports Sci. 2016;414:1–8. doi: 10.1080/02640414.2016.1175655. [DOI] [PubMed] [Google Scholar]
- 90.Vitti A., Nikolaidis P.T., Villiger E., Onywera V., Knechtle B. The “New York City Marathon”: participation and performance trends of 1.2M runners during half-century. Res. Sports Med. 2020;28:121–137. doi: 10.1080/15438627.2019.1586705. [DOI] [PubMed] [Google Scholar]
- 91.Kong P.W., De Heer H. Anthropometric, gait and strength characteristics of Kenyan distance runners. J. Sports Sci. Med. 2008;7:499–504. [PMC free article] [PubMed] [Google Scholar]
- 92.Tam N., Santos-Concejero J., Tucker R., Lamberts R.P., Micklesfield L.K. Bone health in elite Kenyan runners. J. Sports Sci. 2018;36:456–461. doi: 10.1080/02640414.2017.1313998. [DOI] [PubMed] [Google Scholar]
- 93.Mei Q., Gu Y., Xiang L., Yu P., Gao Z., Shim V., Fernandez J. Foot shape and plantar pressure relationships in shod and barefoot populations. Biomech. Model. Mechanobiol. 2020;19:1211–1224. doi: 10.1007/s10237-019-01255-w. [DOI] [PubMed] [Google Scholar]
- 94.Kluitenberg B., Bredeweg S.W., Zijlstras S., Zijlstra W., Buist I. Comparison of vertical ground reaction forces during overground and treadmill running. A validation study. BMC Muscoskel. Disord. 2012;13:235. doi: 10.1186/1471-2474-13-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cronin N.J., Finni T. Treadmill versus overground and barefoot versus shod comparisons of triceps surae fascicle behaviour in human walking and running. Gait Posture. 2013;38:528–533. doi: 10.1016/j.gaitpost.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 96.Mileti I., Serra A., Wolf N., Munoz-Martel V., Ekizos A., Palermo E., Arampatzis A., Santuz A. Muscle activation patterns are more constrained and regular in treadmill than in overground human locomotion. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.581619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at https://auckland.figshare.com/projects/Dataset_of_Lower_Extremity_Joint_Angles_Moments_and_Forces_in_Distance_Running/136708.