Highlights
-
•
We highlight the importance of generalizability in the validation of predictive models.
-
•
Pretreatment structural brain MRI was suggested as biomarker for drug treatment outcome in MDD.
-
•
The random forest model could predict treatment response above chance in the NeuroPharm dataset.
-
•
However, none of the models generalized to an independent cohort (EMBARC dataset).
Keywords: MDD, Structural MRI, Prediction, Treatment response, Remission, SSRI
Abstract
Brain morphology has been suggested to be predictive of drug treatment outcome in major depressive disorders (MDD). The current study aims at evaluating the performance of pretreatment structural brain magnetic resonance imaging (MRI) measures in predicting the outcome of a drug treatment of MDD in a large single-site cohort, and, importantly, to assess the generalizability of these findings in an independent cohort. The random forest, boosted trees, support vector machines and elastic net classifiers were evaluated in predicting treatment response and remission following an eight week drug treatment of MDD using structural brain measures derived with FastSurfer (FreeSurfer). Models were trained and tested within a nested cross-validation framework using the NeuroPharm dataset (n = 79, treatment: escitalopram); their generalizability was assessed using an independent clinical dataset, EMBARC (n = 64, treatment: sertraline). Prediction of antidepressant treatment response in the Neuropharm cohort was statistically significant for the random forest (p = 0.048), whereas none of the models could significantly predict remission. Furthermore, none of the models trained using the entire NeuroPharm dataset could significantly predict treatment outcome in the EMBARC dataset. Although our primary findings in the NeuroPharm cohort support some, but limited value in using pretreatment structural brain MRI to predict drug treatment outcome in MDD, the models did not generalize to an independent cohort suggesting limited clinical applicability. This study emphasizes the importance of assessing model generalizability for establishing clinical utility.
1. Introduction
Major Depressive Disorder (MDD) is one of the most prevalent and severe brain disorders in the world with 6.9 % of the European population estimated to suffer from the disease, making it the most burdensome disease in Europe (Wittchen et al., 2011). MDD is a highly heterogeneous disorder where the diagnosis is based on the presence of a set of symptoms leading to 227 unique ways to meet the criteria for the MDD based on the Diagnostic Statistical Manual (DSM-5) (Zimmerman et al., 2015). MDD can be treated in many different ways, including pharmaceuticals, psychotherapy, electroconvulsive therapy, and other somatic therapies, or combinations thereof (Gartlehner et al., 2017). Whereas selective serotonin/noradrenaline reuptake inhibitors (SSRI/SNRI) are first-line pharmaceutical treatment for MDD, only 40–60 % of patients respond clinically, and only 30–45 % achieve clinical remission (Carvalho et al., 2007, Khin et al., 2011, Thase et al., 2001). Moreover, up to a third of patients will not respond to a second-line medication either (Rush et al., 2006). Identifying biomarkers that enable the clinician to choose the optimal medication for the patient is seen as key for a precision medicine approach and accordingly, many attempts to define such a biomarker have been made (Fu et al., 2012). Such a biomarker would be valuable to guide the treatment of MDD and lower the rate of unsuccessful therapies, which would be of immense benefit to patients and reduce the associated economic burden.
Magnetic resonance imaging (MRI) is a prevalent and non-invasive method for measuring brain structure and function. Structural properties of the brain derived from MRI modalities, such as T1-weighted images, have the advantage of being relatively easy to acquire and, therefore, carry a strong potential as biomarkers for clinical application. The structure of the brain has reliably been shown to be altered in MDD patients compared to healthy controls. Although the reported effect sizes are small (Cohen’s d < 0.2) and it seems to be mainly associated with late-onset MDD (Eker and Gonul, 2010), many studies have reported reduced hippocampal volume in MDD (Eker and Gonul, 2010). Furthermore, several studies have identified associations between treatment outcome and regional cortical thickness and volume, as well as whole brain volume (Fonseka et al., 2018, Frodl et al., 2008, Järnum et al., 2011, Phillips et al., 2015). As reviewed in (Enneking et al., 2020), multiple studies have thus investigated if structural brain characteristics can be used as biomarkers of response to antidepressant drug treatment in MDD.
A handful of studies have attempted to use morphological features of the brain derived from pre-treatment MRI to predict the outcome (i.e., response or remission) to antidepressant treatment in MDD with varying study protocols (e.g., choice of antidepressant drug) and methodology (Bartlett et al., 2018, Costafreda et al., 2009, Gong et al., 2011, Grzenda et al., 2021, Liu et al., 2012, Nouretdinov et al., 2011). Earlier studies in small cohorts (n = 18 to 46) adopted similar approaches and classified treatment outcome using variants of voxel-based morphometry (features) combined with support vector machine classifiers (Costafreda et al., 2009, Gong et al., 2011, Liu et al., 2012, Nouretdinov et al., 2011). In two studies, sparse clusters showed high predictive accuracy for drug treatment response, but these clusters could not readily be associated with neurobiologically meaningful functions (Gong et al., 2011, Liu et al., 2012). Two additional reports by the same group applying a different methodology on the same data reported greater gray matter density in the right rostral anterior cingulate cortex, left posterior cingulate cortex, left middle frontal gyrus, and right occipital cortex as a predictive marker of treatment remission (Costafreda et al., 2009, Nouretdinov et al., 2011). The predictive performances of these studies were evaluated using leave-one-out cross-validation designs, and accuracy ranged from 70 % to 89 %. However, the use of leave-one-out cross-validation has since been strongly discouraged, particularly for small cohorts, due to an increased risk of performance misestimation (Flint et al., 2021, Varoquaux et al., 2017). In a more recent study including a large (n = 184) multi-center cohort (Establishing moderators and biosignatures of antidepressant response in clinical care; EMBARC), the prediction of remission 8 weeks after initiation of sertraline treatment (n = 87) or placebo (n = 97) was assessed using a random forest classifier applied to estimates of regional cortical thickness and subcortical volumes (Bartlett et al., 2018). Notably, the features were obtained from structural MRIs acquired both before and one week after the intervention was initiated. When the performance of this approach was evaluated using repeated fivefold cross-validation, an accuracy of 64 % was achieved. Post hoc analyses also revealed that thickening of the rostral anterior cingulate was associated with better responses to sertraline. In spite of these promising initial findings on the clinical usefulness of brain structural information for predicting the outcome of antidepressant treatment in MDD, these results remain to be replicated in an independent cohort.
Generalizability, the applicability of a model to an unseen dataset, is a core principle of machine learning and is essential to establishing the clinical usefulness of predictive models. Even when all proper procedures for establishing evidence for prediction are followed within a dataset (Poldrack et al., 2020), the real litmus test for a model is the out-of-sample prediction performance (Cohen et al., 2021). Recent large-scale efforts have been made toward determining the generalizability of models for the classification of MDD patients from healthy controls using structural MRI (Belov et al., Unpublished results). However, to our knowledge, the generalizability of models using pretreatment structural MRI to predict pharmacological treatment outcome in MDD has never been evaluated. Given the wide phenotypic heterogeneity of MDD, it is crucial to determine whether these models can be used to predict the treatment outcome of MDD patients from new sites.
We here assess the ability of structural brain MRI measured prior to treatment to predict the response and remission to 8 weeks of treatment with the SSRI escitalopram among complying patients. To this end, we leveraged data from the NeuroPharm cohort (Köhler-Forsberg et al., 2020), the largest single-site study of drug treatment outcome in MDD. We hypothesized that in MDD, structural brain MRI scans acquired prior to treatment-onset would have predictive value in determining the response to drug treatment. Thereafter, we evaluate the generalizability of the models trained using the complete NeuroPharm cohort in an independent dataset (EMBARC).
2. Patients & methods
2.1. Participants
The current study is based on the NeuroPharm dataset, a large, single-site, non-randomized, single-treatment, naturalistic, open-label clinical trial evaluating neuroimaging, biochemical, EEG, and neuropsychological measures as biomarkers of antidepressant treatment outcome in depressed patients (https://www.clinicaltrials.gov, NCT02869035).
Here we summarize relevant study design elements, which are described elsewhere in greater detail (Köhler-Forsberg et al., 2020). One hundred untreated patients with MDD were initially included in the study. Patients were recruited from a central referral center within the Mental Health Services, Capital Region of Denmark or referred directly from one of five general practitioners. At inclusion, patients had to be between 18 and 65 years old and were required to meet the DSM-5 criteria for single or recurrent unipolar depression. Patients were required to be moderately to severely depressed, i.e., a score greater than 17 on the Hamilton Depression Rating Scale-17 item (HAMD-17) (Hamilton, 1960). Clinical diagnosis was confirmed by an experienced psychiatrist and confirmed with the “Mini-International Neuropsychiatric Interview” (Sheehan et al., 1998).
The study protocol was approved by the Ethics Committee (H-15017713), the Danish Data Protection Agency, and Danish Medicines Agency (protocol number: NeuroPharm-NP1, EudraCT-number 2016-001,626–34). The study complies with the Declaration of Helsinki II, and a Good Clinical Practice unit in the Capital Region of Denmark monitored the project. All participants signed written informed consent after an oral and written description of the study. Patients did not receive compensation for participation.
2.2. Treatment protocol
The full treatment protocol is detailed elsewhere (Köhler-Forsberg et al., 2020). Briefly, after inclusion, MDD individuals underwent, among other things, baseline assessments of depression- and anhedonic severity, neuropsychological testing, and a baseline MRI scan. Patients subsequently entered a treatment protocol with escitalopram to last up to twelve weeks. Escitalopram was the primary pharmacological treatment (flexible dosages: 5–20 mg/day), administered in alignment with current clinical practice. Patients not responding (<25 % decrease in HAMD-6) or with unacceptable side effects could switch to duloxetine from week 4 and onward (flexible dosages: 30–120 mg/day), consistent with clinical guidelines. Antidepressant medication was provided free of charge. Non-compliant patients, i.e., reporting to have taken <2/3 of their tablets or with serum concentrations of medicine below the detection limit at week 8, were excluded. Depression severity and side effects were assessed by a study physician or supervised research assistant during clinical follow-up sessions at 1, 2, 4, 8, and 12 weeks after treatment. Patients did not receive any other form of treatment than the described antidepressant regiment, including psychotherapy, for the duration of the trial.
2.3. Clinical assessment
Depression-severity at baseline depression severity and throughout the study was monitored by the Hamilton Depression Rating Scale 17 items (HAMD-17) and its subscale of 6 items (HAMD-6) (Timmerby et al., 2017). HAMD-6 was chosen a priori as the primary outcome for the evaluation of depression based on recent evidence of superior sensitivity to change in depression severity and clinimetric properties compared to HAMD-17 (Dunlop et al., 2019). Response to antidepressant treatment was defined as reduction from baseline of at least 50 % in HAMD-6 score at week 8, and remission was defined as HAMD-6 ≤ 4 at week 8.
2.4. MRI data acquisition
All MRI scans were acquired using a Siemens (Erlangen, Germany) MAGNETOM Prisma 3T scanner with a 64-channel head coil. High-resolution, structural T1-weighted magnetization-prepared rapid gradient-echo structural scans were acquired (repetition time = 1900 ms, echo time = 2.58 ms, inversion time = 900 ms, flip angle = 9°, number of slices = 224, slice thickness = 0.9 mm, matrix = 256 × 256, in-plane resolution 0.9 × 0.9 mm, no gap, and acquisition time = 4 min 26 s).
2.5. Processing of structural MRI
The structural MRI data was processed using FastSurfer (FreeSurfer) (Henschel et al., 2020) (https://github.com/Deep-MI/FastSurfer). An overall quality check of the processing was performed to ensure that no large error in the segmentation process was present, however, no manual editing was performed to provide a fully automated and better clinically applicable evaluation. The average cortical thickness of the 34 regions, per hemisphere, defined by the Desikan-Kiliany atlas (29) and the volume of eight subcortical regions (i.e., accumbens, amygdala, caudate, cerebellar gray matter, hippocampus, pallidum, putamen, and thalamus), as well the volume of the lateral ventricles and the mean cortical thickness of each hemisphere were extracted to be used for comparison between the different groups and as features in the classification models. Intracranial volume was quantified using SPM12 (v7219, https://www.fil.ion.ucl.ac.uk/spm) in Matlab (R2019a, MathWorks, Natick,MA, USA) by segmenting the gray matter, white matter, and cerebrospinal fluid and summing their combined volume (Malone et al., 2015).
2.6. Assessment of group differences
Group differences in demographic characteristics between responders and non-responders or remitters and non-remitters, within and between datasets, were assessed using Wilcoxon rank sum tests and Pearson’s chi-squared test where appropriate. Similarly, group associations with cortical thickness or subcortical volumes were evaluated independently for each region of interest using linear regression models, with age and sex as covariate for estimates of cortical thickness, and additionally with intracranial volume for subcortical volumes (Malone et al., 2015). P-values were adjusted for multiple comparisons using the false discovery rate (FDR) method (Benjamini and Hochberg, 1995) and q < 0.05 was considered statistically significant. All statistical tests and classification analyses were performed using R v4.1.0 (R Core Team, 2013).
2.7. Prediction of treatment outcome
Prediction of treatment response or remission was evaluated using some of the most commonly used classifiers (caret, v6.0.92) (Kuhn, 2008): the random forest (randomForest, v4.7.1.1) (Breiman, 2001, Liaw and Wiener, 2002), boosted trees (xgboost, xgbTree, v1.6.0.1), support vector machines (SVM) (kernlab, svmRadialWeights, v0.9.31), and elastic net (glmnet, v4.1.4). Class weights were set to 1 - p, where p is the proportion of a given class among all samples. These weights were used to ensure equal representation of each class during training. Estimates of regional cortical thickness, subcortical volumes, mean cortical thickness of the whole hemisphere, and intracranial volumes, as well as age, sex, HAMD-6 at baseline (week 0), and recurrence status (i.e, first-episode or recurrent) were and used as input features. Numerical features were centered and scaled independently per training set and applied to the respective test set. Model performance was assessed using the area under the ROC Curve (AUC), balanced accuracy, sensitivity, and specificity evaluated within a stratified nested cross-validation with a repeated cross-validation with 5-fold and 25 repeats as outer loop and a 5-fold cross-validation as nested loop for hyperparameters optimization, as recommended in (Varoquaux et al., 2017). Hyperparameters optimization for all classifiers aside from the random forest was performed according to the default implementation in caret. For the random forest, the optimization was performed for both the number of variables randomly sampled as candidates at each split (mtry) and the number of trees to grow (ntree). The statistical significance of the AUCs were empirically derived using a null distribution estimated from 1,000 permutations for the elastic net and random forest and 100 permutations for the boosted trees and SVM. A smaller number of permutations were used for boosted trees and SVM due to the computational load required to train these models.
2.8. Generalizability of the classifiers on the EMBARC dataset
EMBARC is a large, multicenter, double-blind, randomized, placebo-controlled trial evaluating the treatment response to the SSRI sertraline in patients with MDD. The rationale and design of this study have been previously described (Trivedi et al., 2016). Individuals from the EMBARC dataset, meeting the same inclusion criteria as for NeuroPharm, were identified. Notably, only patients having received sertraline treatment,with HAMD-17 greater than 17 at week 0, and with pre-treatment structural MRI at week 0 were included. Pre-treatment structural MRIs were processed using the approach described above and the corresponding brain morphological features were extracted. Models trained on NeuroPharm data were evaluated in the task of predicting treatment response and remission in the EMBARC dataset using the same input features (i.e., pre-treatment MRI measures, age, sex, HAMD-6 at week 0, and recurrence status). The EMBARC data was normalized to match the data used for training using the parametric ComBat (sva, v4.32.0) procedure (Johnson et al., 2007). Model performance was estimated using bootstrap with 1,000 resamples and the statistical significance of the mean AUC was empirically derived using a null distribution estimated from 1,000 permutations.
2.9. Voxel-based morphometry
As previous studies have largely been performed using voxel-based morphometry (VBM) (Costafreda et al., 2009, Gong et al., 2011, Liu et al., 2012, Nouretdinov et al., 2011), we briefly evaluated this approach in the NeuroPharm dataset. The standard VBM protocol was applied using SPM12: 1) the structural MR images were segmented into gray matter, white matter and cerebrospinal fluid and imported into a rigidly aligned space, 2) gray and white matter segmentations were iteratively registered by non-linear warping to a group template generated from all images by the Diffeomorphic Anatomical Registration Through Exponentiated Lie algebra (DARTEL) toolbox (Ashburner, 2007), 3) the gray matter segmentations were normalized to MNI space, and scaled by the Jacobian determinants of the nonlinear deformation (modulation) to preserve the overall amount of each tissue class, and 4) the spatially normalized and modulated gray matter segmentations were smoothed with a 8 mm full width at half maximum (FWHM) Gaussian kernel. These final images were used as measures of volume in subsequent VBM analyses.
Voxel-wise group differences in VBM volumes were assessed using F-tests. P-values were corrected for family-wise error and considered significant at p < 0.05. For classification, significant voxels to be included as features were identified using an uncorrected voxel-level threshold of p < 0.005 (Costafreda et al., 2009). Significant voxels were identified independently for each training sets and used to extract VBM volumes in both the training and test sets. Classification using VBM volumes (in place of cortical thickness or subcortical volumes) was otherwise performed as described in Section 2.7.
2.10. Data and code availability
The NeuroPharm data can be made available upon request through an application to the Cimbi database (https://www.cimbi.dk). The EMBARC dataset can be accessed through the National Institute of Mental Health Data Archive (https://nda.nih.gov). The source code for this manuscript is freely available at https://github.com/vbeliveau/MDD_SSRI_structural_prediction.
3. Results
3.1. Demographics
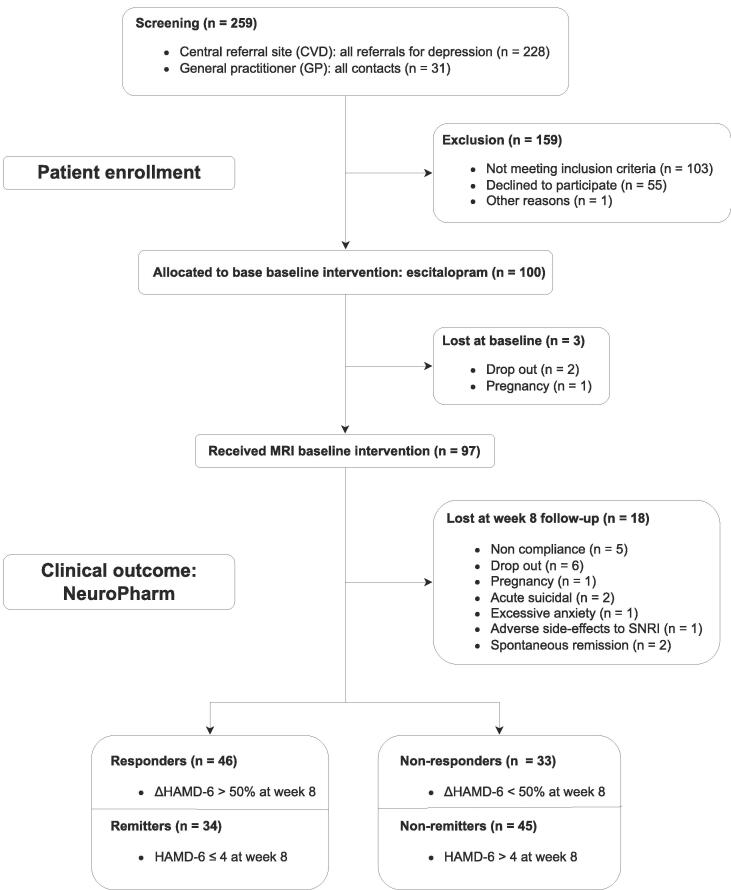
A flow chart of the participants in the study is shown in Fig. 1. Of the 100 patients enrolled in the NeuroPharm study, three did not complete a pre-treatment structural MRI scan. Eighteen of the 97 remaining patients were lost at follow-up (n = 18), resulting in 79 MDD patients having both structural MRI data and clinical assessments at baseline and week 8. Out of 336 participants, 64 patients were included from the EMBARC dataset.
Fig. 1.
Study flowchart for the NeuroPharm dataset presenting the assignment to the response and remission groups based on the clinical assessment of HAMD-6 at week 8.
The demographics and clinical variables are presented in Table 1 for the NeuroPharm data and in Table S1 for the EMBARC dataset. No significant differences in demographic characteristics between the treatment outcome groups were found (all p-values > 0.05), aside from the expected difference in HAMD-6 at week 8 (p < 0.001). Based on the relative change in HAMD-6 at week 8 compared to baseline, a total of 46 (58.2 %) patients were labeled as responders and 33 (41.8 %) non-responders, and 34 (43.0 %) patients were labeled as remitters and 45 (57.0 %) as non-remitters. As expected, responder and remitter groups overlapped: all remitters were also classified as responders, 12 (15.2 %) responders were classified as non-remitters, and the remaining non-responders were labeled non-remitters. The number of participants in each group is presented as a contingency table (Table 2) for the NeuroPharm dataset and in Table S2 for the EMBARC dataset.
Table 1.
Demographics and clinical variables for remission and response in the NeuroPharm dataset. HAMD-6: Hamilton Depression Rating Scale-6 item.
| Response | Remission | |||||
|---|---|---|---|---|---|---|
| Characteristic | Non-responder, N = 331 | Responder, N = 461 | p-value2 | Non-remitter, N = 451 |
Remitter, N = 341 |
p-value2 |
| Age | 24 (22, 27) | 25 (23, 29) | 0.2 | 24 (22, 27) | 26 (23, 29) | 0.14 |
| Sex | 0.4 | 0.13 | ||||
| Female | 26 (79 %) | 32 (70 %) | 36 (80 %) | 22 (65 %) | ||
| Male | 7 (21 %) | 14 (30 %) | 9 (20 %) | 12 (35 %) | ||
| HAMD-6 - Baseline | 12 (11, 13) | 12 (12, 13) | 0.9 | 13 (12, 13) | 12 (11.25, 13) | 0.2 |
| HAMD-6 - Week 8 | 9 (7, 11) | 3 (2, 5) | <0.001 | 8 (6, 10) | 2 (1, 3) | <0.001 |
| Recurrence status | 0.5 | 0.6 | ||||
| First-episode | 12 (36 %) | 20 (43 %) | 17 (38 %) | 15 (44 %) | ||
| Recurrent | 21 (64 %) | 26 (57 %) | 28 (62 %) | 19 (56 %) | ||
Median (IQR); n (%)
Wilcoxon rank sum exact test; Pearson's Chi-squared test; Wilcoxon rank sum test
Table 2.
Contingency table presenting the number of participants from the NeuroPharm dataset in each of the treatment outcome groups.
| Non-remitter | Remitter | Total | |
|---|---|---|---|
| Non-responder | 33 | 0 | 33 |
| Responder | 12 | 34 | 46 |
| Total | 45 | 34 | 79 |
Significant differences in age (W = 1143, p < 0.0001) and recurrence status (χ2 = 12.409, df = 1, p < 0.0001) were observed between the NeuroPharm and EMBARC datasets, where as sex was not different (χ2 = 0.395, df = 1, p = 0.530).
3.2. Group differences
No significant group difference in regional cortical thickness or volume pretreatment was found between responders and non-responders nor between remitters and non-remitters in the NeuroPharm dataset. Detailed results are presented in Tables S1-2. Similarly, no significant group difference was identified in the pretreatment EMBARC data, aside from the volume of the right putamen between responders and non-responders (t = 3.75, q = 0.035, FDR-corrected), see Tables S3-4 for details.
3.3. Prediction of treatment outcome within the NeuroPharm dataset
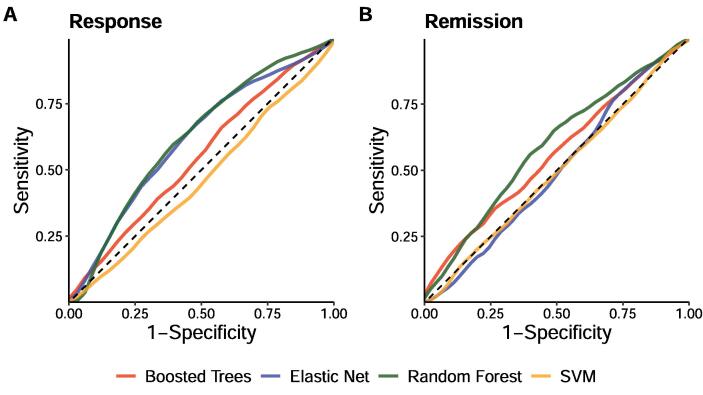
Detailed information on the performance of the models in predicting treatment response and remission in the NeuroPharm dataset is reported in Table 3. For treatment response, only the random forest classifiers achieved mean AUCs significantly greater than the null (p = 0.048). For the prediction of treatment remission, none of the models obtained mean AUCs significantly greater than the null. Fig. 2 presents the mean receiver operating characteristic (ROC) curves of the models.
Table 3.
Performance metrics of the different classifiers for predicting treatment response or remission in the NeuroPharm dataset. AUC: Area under the ROC Curve. Values are given as mean (SD), aside from the p-values.
| Response | Remission | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Classifier | AUC | Balanced Accuracy | Sensitivity | Specificity | AUC p-value | AUC | Balanced Accuracy | Sensitivity | Specificity |
AUC p-value |
| Elastic Net | 0.61 (0.04) | 59.4 (2.5) | 60.0 (7.0) | 58.8 (6.5) | 0.067 | 0.50 (0.05) | 48.4 (4.7) | 54.5 (7.2) | 42.2 (8.0) | 0.440 |
| Random Forest | 0.62 (0.03) | 58.0 (3.2) | 37.1 (5.4) | 79.0 (2.7) | 0.048 | 0.59 (0.03) | 55.2 (3.0) | 75.3 (4.6) | 35.1 (4.4) | 0.115 |
| SVM | 0.46 (0.04) | 48.0 (3.5) | 41.0 (13.3) | 55.0 (16.8) | 1.000 | 0.50 (0.03) | 49.5 (4.8) | 84.0 (9.9) | 15.1 (9.6) | 0.470 |
| Boosted Trees | 0.55 (0.04) | 53.8 (3.7) | 40.4 (4.1) | 67.2 (5.5) | 0.220 | 0.56 (0.06) | 54.2 (6.0) | 66.0 (6.6) | 42.4 (9.1) | 0.160 |
Fig. 2.
Mean receiver operating characteristic (ROC) curves of the different classifiers for the classification of (A) response and (B) remission in the NeuroPharm dataset.
3.4. Assessment of generalizability
To evaluate the generalizability of the identified predictive model derived based on the NeuroPharm data, we tested the models for predicting response or remission in the EMBARC dataset. To this end, classification models were trained to predict treatment response and remission using all available data (i.e., n = 79) from the NeuroPharm dataset. Applied to the EMBARC dataset, neither the elastic net nor the random forest achieved AUCs significantly greater than the null for treatment response (p = 0.57, and p = 0.53, respectively) and remission (p = 0.69, and p = 0.83, respectively). Detailed performance metrics are reported in Table S7. Due to their poor performance in the NeuroPharm dataset, we did not evaluate the boosted trees and SVM models.
As a post hoc analysis, we evaluated the performance of classification models trained on only the EMBARC dataset using the cross-validation framework previously described. These models were not able to predict treatment response and remission better than chance (all mean AUCs < 0.5). Fig. S1 presents the corresponding ROC curves and the associated performance metrics are reported in Table S8.
3.5. Voxel-based morphometry
No significant group differences in VBM volumes were identified between responders and non-responders nor between remitters and non-remitters. The predictive performance of the classifiers using VBM volumes were limited at best, with mean AUCs ranging from 0.42 to 0.58. We note that p-values for the AUCs were not here estimated given their closeness to 0.5 and the large computations this would entail. ROC curves are presented in Fig. S2 and the associated performance metrics are included in Table S9.
4. Discussion
Previous studies have claimed some, but limited success in predicting the outcome of drug treatments in MDD using structural MRI, but so far, no study has demonstrated the generalizability of such models in an independent cohort. Generalizability is a cornerstone to any form of clinical application and is a prerequisite to claims of usefulness beyond research interest. This point is critical for assessing the results presented in our study.
In this work, we evaluated the potential of pre-treatment structural brain MRI for the prediction of antidepressant treatment outcome in patients with MDD following an eight-week treatment program starting with escitalopram. In the NeuroPharm dataset, no significant group difference in pretreatment regional cortical thickness or subcortical volume was observed between responders and non-responders, nor between remitters and non-remitters. Our classification models were first trained and tested in-sample (NeuroPharm dataset only), and their generalizability was subsequently assessed using a second independent clinical test dataset (EMBARC). At first glance, the predictive models evaluated in-sample using cross-validation suggested that distinct profiles of brain structure could be captured by our models enabling us to predict treatment response above chance. In contrast to earlier, smaller studies, the classification performances of our models were noticeably reduced both for response and remission (Bartlett et al., 2018, Costafreda et al., 2009, Gong et al., 2011, Liu et al., 2012, Nouretdinov et al., 2011), but were still comparable to that of (Bartlett et al., 2018) (Bartlett et al., 2018). However, our models did not generalize to a new dataset which clearly undermines their clinical value. Subsequent evaluations using VBM, an approach adopted by most earlier studies (Costafreda et al., 2009, Gong et al., 2011, Liu et al., 2012, Nouretdinov et al., 2011), also revealed no group differences in VBM volumes between the treatment outcomes and the models derived from these data achieved only limited predictive performances. As such, these results further perpetuate the current and unfortunate situation where strong predictive performance of small studies using machine learning cannot be replicated by studies using larger datasets (Flint et al., 2021). Indeed, it is worth emphasizing that the training (NeuroPharm) and test (EMBARC) dataset used here represent two of the largest studies with MRI investigating treatment outcome following an SSRI intervention. Even though the primary evaluation of the models was performed within a cross-validation framework, which is generally accepted as unbiased (Varoquaux et al., 2017), estimating the models’ performance using a single dataset (i.e., NeuroPharm dataset only) would have been misleading concerning their generalizability and clinical usefulness.
It is unclear whether the range of predictive performances observed across previous studies and the lack of generalizability from our models on the EMBARC dataset stem from different choices of drugs, intensity of clinical follow-up, study design, methodology, or phenotypic heterogeneity (i.e., MDD subtypes) which may not have been well captured in smaller cohorts. In fact, evidence of disease heterogeneity is readily apparent in both the NeuroPharm and EMBARC cohorts with 27 % and 21 % of the responders, respectively, showing response to the treatment, but not achieving remission, while the remaining responders also achieved remission. Interestingly, our treatment response model achieved numerically better performances compared to that of remission. Although many studies have proposed different definitions for subtypes of MDD (Musil et al., 2017, van Loo et al., 2012), there is currently no overwhelming evidence justifying their application in clinical diagnosis and, as of yet, their usage in conjunction with brain structural measures for the prediction of antidepressant treatment outcome remains to be investigated. Without more information it becomes difficult to identify the specific factors driving these discrepancies across studies and future research should concentrate on establishing model generalizability across different cohorts. One possible approach is to establish models that are invariant and robust to dataset shift (Quiñonero-Candela et al., 2008). However, obtaining the data necessary to train such models can only be accomplished through data sharing efforts by the research community. To this end, we make our models and code publicly available and access to the data can be requested (see the Data and code availability section).
When looking at model interpretability, there is little agreement across previous studies concerning which brain regions provide structural information predictive of treatment outcome following antidepressant intervention in MDD. Across the NeuroPharm and EMBARC datasets, only the volume of the right putamen in the EMBARC dataset was significantly different between responders and non-responders. The orbitofrontal cortex and the hippocampus are two brain regions thought to be primarily implicated in the drug treatment of MDD (Bartlett et al., 2018). Early pathological studies have demonstrated a decrease in cortical thickness, neuronal size, and neuronal and glial densities in the rostral orbitofrontal cortex of depressed individuals (Rajkowska et al., 1999). In a mixed-treatment study, greater hippocampal volume at baseline was found in remitters compared to non-remitters (MacQueen et al., 2008), a finding which we could not reproduce. Longitudinal studies have also revealed that orbitofrontal cortex thickness and hippocampal volume are increased in remitters and decreased in non-remitters over the treatment period (Phillips et al., 2015) and that greater hippocampal volume is associated with better clinical outcome (Frodl et al., 2008). Although similar inferences could likely be drawn if one was to look at the interpretability of our models trained on the NeuroPharm dataset, e.g., using Shapley values, interpreting the importance of features within models which do not generalize is likely to be misleading. Ultimately, our results regarding regional differences are in line with recent evidence suggesting that structural alterations in MDD may be relatively small and heterogeneous (Schmaal et al., 2017, Schmaal et al., 2016) and indicate that more research is needed to disentangle the complex interplay between the physiological mechanisms leading to changes in brain morphology in MDD.
In this study, we have purposefully avoided including additional features which were available from the NeuroPharm and EMBARC cohorts, such as clinical characteristics, functional MRI, positron emission tomography or quantitative electroencephalogram (qEEG) parameters, to be able to draw conclusions centered on the predictive capabilities of structural MRI and basic demographics. Other studies utilizing the NeuroPharm cohort have investigated prediction of treatment response by reward processing using functional MRI (Brandt et al., 2021) and alpha asymmetry from qEEG (Ip et al., 2021), but with limited success. Future work should concentrate on combining the information of multiple domains to improve the prediction of outcome following antidepressant treatment in MDD.
A few limitations for this work have to be acknowledged. Firstly, some individuals (n = 10) included in the NeuroPharm dataset switched their SSRI treatment to the SNRI duloxetine during the course of the treatment. Although this deviates from a perspective focused solely on the treatment of MDD with a unique drug, it does reflect the naturalistic clinical course of a drug intervention in the treatment of MDD and is therefore closer to clinical application outside of the research setting. Secondly, some individuals from the NeuroPharm dataset were excluded as they did not follow the complete treatment protocol up to week 8 due to reasons which could potentially be linked to the treatment, i.e., acute suicidal, excessive anxiety, adverse side-effect to SNRI, or spontaneous remission. These individuals should not be excluded in the context of a randomized controlled trial, nonetheless, they were not included here so that the models trained using NeuroPharm data could be applied to the EMBARC dataset where this information is not available, and to make our results relatable to previous studies where this exclusion is also performed. Finally, it is worth noting that there are important intrinsic differences between the NeuroPharm and the EMBARC datasets, with some of the most striking being a difference in mean age of 14 years between the two datasets and the usage of a different SSRI drugs (i.e., escitalopram and sertraline) which may not have identical treatment effects.
5. Conclusion
The usefulness of pre-treatment structural brain MRI in predicting the outcome of antidepressant treatment in MDD is not convincing. Our work utilizing the NeuroPharm cohort indicates that treatment response can be predicted above chance in-sample, but these results did not generalize to an independent test dataset. Future work should concentrate on improving the generalizability of the models across cohorts and combining features from structural brain MRI with information from other domains.
Funding and disclosure
Funding for this study was provided by the Innovation Fund Denmark (grant 4108-00004B, NeuroPharm), and the Lundbeck Foundation (grant R279-2018-1145, BrainDrugs); the Innovation Fund Denmark and the Lundbeck Foundation had no further role in the study design, collection, analysis and interpretation of data, the writing of the report and the decision to submit the paper for publication. GMK has received honoraria as a speaker for Sage Biogen and as a consultant for Sanos. All remaining authors declare that they have no potential conflicts of interest.
CRediT authorship contribution statement
Vincent Beliveau: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing – original draft, Visualization. Ella Hedeboe: Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing – original draft, Visualization. Patrick M. Fisher: Conceptualization, Writing – original draft. Vibeke H. Dam: Conceptualization, Writing – original draft. Martin B. Jørgensen: Conceptualization, Writing – original draft. Vibe G. Frokjaer: Conceptualization, Writing – original draft. Gitte M. Knudsen: Conceptualization, Writing – original draft, Funding acquisition. Melanie Ganz: Conceptualization, Writing – original draft, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2022.103224.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Bartlett E.A., DeLorenzo C., Sharma P., Yang J., Zhang M., Petkova E., Weissman M., McGrath P.J., Fava M., Ogden R.T., Kurian B.T., Malchow A., Cooper C.M., Trombello J.M., McInnis M., Adams P., Oquendo M.A., Pizzagalli D.A., Trivedi M., Parsey R.V. Pretreatment and early-treatment cortical thickness is associated with SSRI treatment response in major depressive disorder. Neuropsychopharmacology. 2018;43:2221–2230. doi: 10.1038/s41386-018-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov, V., Erwin-Grabner, T., Gonul, A.S., Amod, A.R., Ojha, A., Dols, A., Scharntee, A., Uyar-Demir, A., Harrison, B.J., Besteher, B., Klimes-Dougan, B., Zarate, C., Davey, C.G., Ching, C.R.K., Connolly, C.G., Stein, D.J., Dima, D., Linden, D.E.J., Mehler, D.M.A., Pozzi, E., Melloni, E., Benedetti, F., MacMaster, F.P., Grabe, J., Völzke, H., Gotlib, I.H., Soares, J.C., Evans, J.W., Sim, K., Wittfeld, K., Cullen, K., Reneman, L., Oudega, M.L., Portella, M.J., Sacchet, M.D., Li, M., Aghajani, M., Wu, M.-J., Jahanshad, N., Saemann, P., Bülow, R., Poletti, S., Whittle, S., Thomopoulos, S.I., van, J.A., Basgoze, Z., Veltman, D.J., Schmaal, L., Thompson, P.M., Unpublished results. Multi-site benchmark classification of major depressive disorder using machine learning on cortical and subcortical measures 56. https://doi.org/10.48550/arXiv.2206.08122.
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57(1):289–300. [Google Scholar]
- Brandt I.M., Köhler-Forsberg K., Ganz M., Ozenne B., Jorgensen M.B., Poulsen A., Knudsen G.M., Frokjaer V.G., Fisher P.M. Reward processing in major depressive disorder and prediction of treatment response – Neuropharm study. Eur. Neuropsychopharmacol. 2021;44:23–33. doi: 10.1016/j.euroneuro.2020.12.010. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Carvalho A.F., Cavalcante J.L., Castelo M.S., Lima M.C.O. Augmentation strategies for treatment-resistant depression: a literature review. J. Clin. Pharm. Ther. 2007;32:415–428. doi: 10.1111/j.1365-2710.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- Cohen, J.P., Cao, T., Viviano, J.D., Huang, C.-W., Fralick, M., Ghassemi, M., Mamdani, M., Greiner, R., Bengio, Y., 2021. Problems in the deployment of machine-learned models in health care. CMAJ 193, E1391–E1394. https://doi.org/10.1503/cmaj.202066. [DOI] [PMC free article] [PubMed]
- Costafreda S.G., Chu C., Ashburner J., Fu C.H.Y., Domschke K. Prognostic and diagnostic potential of the structural neuroanatomy of depression. PLoS ONE. 2009;4(7):e6353. doi: 10.1371/journal.pone.0006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop B.W., Parikh S.V., Rothschild A.J., Thase M.E., DeBattista C., Conway C.R., Forester B.P., Mondimore F.M., Shelton R.C., Macaluso M., Logan J., Traxler P., Li J., Johnson H., Greden J.F. Comparing sensitivity to change using the 6-item versus the 17-item Hamilton depression rating scale in the GUIDED randomized controlled trial. BMC Psychiatry. 2019;19:420. doi: 10.1186/s12888-019-2410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eker C., Gonul A.S. Volumetric MRI studies of the hippocampus in major depressive disorder: Meanings of inconsistency and directions for future research. World J. Biol. Psychiatry. 2010;11:19–35. doi: 10.3109/15622970902737998. [DOI] [PubMed] [Google Scholar]
- Enneking V., Leehr E.J., Dannlowski U., Redlich R. Brain structural effects of treatments for depression and biomarkers of response: a systematic review of neuroimaging studies. Psychol. Med. 2020;50:187–209. doi: 10.1017/S0033291719003660. [DOI] [PubMed] [Google Scholar]
- Flint C., Cearns M., Opel N., Redlich R., Mehler D.M.A., Emden D., Winter N.R., Leenings R., Eickhoff S.B., Kircher T., Krug A., Nenadic I., Arolt V., Clark S., Baune B.T., Jiang X., Dannlowski U., Hahn T. Systematic misestimation of machine learning performance in neuroimaging studies of depression. Neuropsychopharmacology. 2021;46(8):1510–1517. doi: 10.1038/s41386-021-01020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseka T.M., MacQueen G.M., Kennedy S.H. Neuroimaging biomarkers as predictors of treatment outcome in Major Depressive Disorder. J. Affect. Disord. 2018;233:21–35. doi: 10.1016/j.jad.2017.10.049. [DOI] [PubMed] [Google Scholar]
- Frodl T., Jäger M., Smajstrlova I., Born C., Bottlender R., Palladino T., Reiser M., Möller H.-J., Meisenzahl E.M. Effect of hippocampal and amygdala volumes on clinical outcomes in major depression: a 3-year prospective magnetic resonance imaging study. J. Psychiatry Neurosci. JPN. 2008;33:423–430. [PMC free article] [PubMed] [Google Scholar]
- Fu C.H.Y., Steiner H., Costafreda S.G. Predictive neural biomarkers of clinical response in depression: A meta-analysis of functional and structural neuroimaging studies of pharmacological and psychological therapies. Neurobiol. Dis. 2012;52:75–83. doi: 10.1016/j.nbd.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Gartlehner, G., Wagner, G., Matyas, N., Titscher, V., Greimel, J., Lux, L., Gaynes, B.N., Viswanathan, M., Patel, S., Lohr, K.N., 2017. Pharmacological and non-pharmacological treatments for major depressive disorder: review of systematic reviews. BMJ Open 7, e014912. https://doi.org/10.1136/bmjopen-2016-014912. [DOI] [PMC free article] [PubMed]
- Gong Q., Wu Q., Scarpazza C., Lui S., Jia Z., Marquand A., Huang X., McGuire P., Mechelli A. Prognostic prediction of therapeutic response in depression using high-field MR imaging. NeuroImage. 2011;55:1497–1503. doi: 10.1016/j.neuroimage.2010.11.079. [DOI] [PubMed] [Google Scholar]
- Grzenda A., Speier W., Siddarth P., Pant A., Krause-Sorio B., Narr K., Lavretsky H. Machine learning prediction of treatment outcome in late-life depression. Front. Psychiatry. 2021;12:1783. doi: 10.3389/fpsyt.2021.738494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, M., 1960. A Rating Scale for Depression. J. Neurol. Neurosurg. Psychiatry 23, 56. http://dx.doi.org.ep.fjernadgang.kb.dk/10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed]
- Henschel L., Conjeti S., Estrada S., Diers K., Fischl B., Reuter M. FastSurfer - A fast and accurate deep learning based neuroimaging pipeline. NeuroImage. 2020;219 doi: 10.1016/j.neuroimage.2020.117012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip C.T., Olbrich S., Ganz M., Ozenne B., Köhler-Forsberg K., Dam V., Beniczky S., Jørgensen M., Frokjaer V., Søgaard B., Christensen S., Knudsen., G., Pretreatment qEEG biomarkers for predicting pharmacological treatment outcome in Major Depressive Disorder: Independent validation from the NeuroPharm study. Eur. Neuropsychopharmacol. 2021;49:101–112. doi: 10.1016/j.euroneuro.2021.03.024. [DOI] [PubMed] [Google Scholar]
- Järnum H., Eskildsen S.F., Steffensen E.G., Lundbye-Christensen S., Simonsen C.W., Thomsen I.S., Fründ E.-T., Théberge J., Larsson E.-M. Longitudinal MRI study of cortical thickness, perfusion, and metabolite levels in major depressive disorder: Longitudinal study of advanced MRI in depression. Acta Psychiatr. Scand. 2011;124:435–446. doi: 10.1111/j.1600-0447.2011.01766.x. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Khin N.A., Chen Y.-F., Yang Y., Yang P., Laughren T.P. Exploratory analyses of efficacy data from major depressive disorder trials submitted to the US food and drug administration in support of new drug applications. J. Clin. Psychiatry. 2011;72(04):464–472. doi: 10.4088/JCP.10m06191. [DOI] [PubMed] [Google Scholar]
- Köhler-Forsberg, K., Jorgensen, A., Dam, V.H., Stenbæk, D.S., Fisher, P.M., Ip, C.T., Ganz, M., Poulsen, H.E., Giraldi, A., Ozenne, B., Jørgensen, M.B., Knudsen, G.M., Frokjaer, V.G., 2020. Predicting Treatment Outcome in Major Depressive Disorder Using Serotonin 4 Receptor PET Brain Imaging, Functional MRI, Cognitive-, EEG-Based, and Peripheral Biomarkers: A NeuroPharm Open Label Clinical Trial Protocol. Front. Psychiatry 11, 641–641. https://doi.org/10.3389/fpsyt.2020.00641. [DOI] [PMC free article] [PubMed]
- Kuhn M. Building predictive models in R using the caret package. J. Stat. Softw. 2008;28:1–26. doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- Liaw A., Wiener M. Classification and regression by randomForest. R. News. 2002;2:18–22. [Google Scholar]
- Liu, F., Guo, W., Yu, D., Gao, Q., Gao, K., Xue, Z., Du, H., Zhang, J., Tan, C., Liu, Z., Zhao, J., Chen, H., 2012. Classification of Different Therapeutic Responses of Major Depressive Disorder with Multivariate Pattern Analysis Method Based on Structural MR Scans. PLoS ONE 7, e40968. https://doi.org/10.1371/journal.pone.0040968. [DOI] [PMC free article] [PubMed]
- MacQueen G.M., Yucel K., Taylor V.H., Macdonald K., Joffe R. Posterior hippocampal volumes are associated with remission rates in patients with major depressive disorder. Biol. Psychiatry Neurodegen. Dement. Depress. 2008;64:880–883. doi: 10.1016/j.biopsych.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Malone I.B., Leung K.K., Clegg S., Barnes J., Whitwell J.L., Ashburner J., Fox N.C., Ridgway G.R. Accurate automatic estimation of total intracranial volume: A nuisance variable with less nuisance. NeuroImage. 2015;104:366–372. doi: 10.1016/j.neuroimage.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musil, R., Seemüller, F., Meyer, S., Spellmann, I., Adli, M., Bauer, M., Kronmüller, K., Brieger, P., Laux, G., Bender, W., Heuser, I., Fisher, R., Gaebel, W., Schennach, R., Möller, H., Riedel, M., 2017. Subtypes of depression and their overlap in a naturalistic inpatient sample of major depressive disorder. Int. J. Methods Psychiatr. Res. 27, e1569. https://doi.org/10.1002/mpr.1569. [DOI] [PMC free article] [PubMed]
- Nouretdinov I., Costafreda S.G., Gammerman A., Chervonenkis A., Vovk V., Vladimir Vapnik F.u., C.h.y., Machine learning classification with confidence: Application of transductive conformal predictors to MRI-based diagnostic and prognostic markers in depression. NeuroImage. 2011;56:809–813. doi: 10.1016/j.neuroimage.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Phillips J.L., Batten L.A., Tremblay P., Aldosary F., Blier P. A prospective, longitudinal study of the effect of remission on cortical thickness and hippocampal volume in patients with treatment-resistant depression. Int. J. Neuropsychopharmacol. 2015;18(8):pyv037–pyv. doi: 10.1093/ijnp/pyv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack R.A., Huckins G., Varoquaux G. Establishment of best practices for evidence for prediction: a review. JAMA Psychiatry. 2020;77:534–540. doi: 10.1001/jamapsychiatry.2019.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñonero-Candela J., Sugiyama M., Schwaighofer A., Lawrence N.D. Mit Press; 2008. Dataset shift in machine learning. [Google Scholar]
- R Core Team, 2013. R: A Language and Environment for Statistical Computing.
- Rajkowska G., Miguel-Hidalgo J.J., Wei J., Dilley G., Pittman S.D., Meltzer H.Y., Overholser J.C., Roth B.L., Stockmeier C.A. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression ∗. Biol. Psychiatry. 1999;45:1085–1098. doi: 10.1016/S0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- Rush A.J., Trivedi M.H., Wisniewski S.R., Nierenberg A.A., Stewart J.W., Warden D., Niederehe G., Thase M.E., Lavori P.W., Lebowitz B.D., McGrath P.J., Rosenbaum J.F., Sackeim H.A., Kupfer D.J., Luther J., Fava M. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: A STAR*D Report. Am. J. Psychiatry. 2006;163(11):1905–1917. doi: 10.1176/ajp.2006.163.11.1905. [DOI] [PubMed] [Google Scholar]
- Schmaal L., Veltman D.J., van Erp T.G.M., Sämann P.G., Frodl T., Jahanshad N., Loehrer E., Tiemeier H., Hofman A., Niessen W.J., Vernooij M.W., Ikram M.A., Wittfeld K., Grabe H.J., Block A., Hegenscheid K., Völzke H., Hoehn D., Czisch M., Lagopoulos J., Hatton S.N., Hickie I.B., Goya-Maldonado R., Krämer B., Gruber O., Couvy-Duchesne B., Rentería M.E., Strike L.T., Mills N.T., de Zubicaray G.I., McMahon K.L., Medland S.E., Martin N.G., Gillespie N.A., Wright M.J., Hall G.B., MacQueen G.M., Frey E.M., Carballedo A., van Velzen L.S., van Tol M.J., van der Wee N.J., Veer I.M., Walter H., Schnell K., Schramm E., Normann C., Schoepf D., Konrad C., Zurowski B., Nickson T., McIntosh A.M., Papmeyer M., Whalley H.C., Sussmann J.E., Godlewska B.R., Cowen P.J., Fischer F.H., Rose M., Penninx B.W.J.H., Thompson P.M., Hibar D.P. Subcortical brain alterations in major depressive disorder: Findings from the ENIGMA Major Depressive Disorder working group. Mol. Psychiatry. 2016;21(6):806–812. doi: 10.1038/mp.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal L., Hibar D.P., Sämann P.G., Hall G.B., Baune B.T., Jahanshad N., Cheung J.W., van Erp T.G.M., Bos D., Ikram M.A., Vernooij M.W., Niessen W.J., Tiemeier H., Hofman A., Wittfeld K., Grabe H.J., Janowitz D., Bülow R., Selonke M., Völzke H., Grotegerd D., Dannlowski U., Arolt V., Opel N., Heindel W., Kugel H., Hoehn D., Czisch M., Couvy-Duchesne B., Rentería M.E., Strike L.T., Wright M.J., Mills N.T., de Zubicaray G.I., McMahon K.L., Medland S.E., Martin N.G., Gillespie N.A., Goya-Maldonado R., Gruber O., Krämer B., Hatton S.N., Lagopoulos J., Hickie I.B., Frodl T., Carballedo A., Frey E.M., van Velzen L.S., Penninx B.W.J.H., van Tol M.-J., van der Wee N.J., Davey C.G., Harrison B.J., Mwangi B., Cao B., Soares J.C., Veer I.M., Walter H., Schoepf D., Zurowski B., Konrad C., Schramm E., Normann C., Schnell K., Sacchet M.D., Gotlib I.H., MacQueen G.M., Godlewska B.R., Nickson T., McIntosh A.M., Papmeyer M., Whalley H.C., Hall J., Sussmann J.E., Li M., Walter M., Aftanas L., Brack I., Bokhan N.A., Thompson P.M., Veltman D.J. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol. Psychiatry. 2017;22(6):900–909. doi: 10.1038/mp.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan, D.V., Lecrubier, Y., Sheehan, K.H., Amorim, P., Janavs, J., Weiller, E., Hergueta, T., Baker, R., Dunbar, G.C., 1998. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J. Clin. Psychiatry 59 Suppl 20, 22-33;quiz 34-57. [PubMed]
- Thase M.E., Entsuah A.R., Rudolph R.L. Remission rates during treatment with venlafaxine or selective serotonin reuptake inhibitors. Br. J. Psychiatry. 2001;178:234–241. doi: 10.1192/bjp.178.3.234. [DOI] [PubMed] [Google Scholar]
- Timmerby N., Andersen J.H., Søndergaard S., Østergaard S.D., Bech P. A systematic review of the clinimetric properties of the 6-item version of the hamilton depression rating scale (HAM-D6) Psychother. Psychosom. 2017;86:141–149. doi: 10.1159/000457131. [DOI] [PubMed] [Google Scholar]
- Trivedi M.H., McGrath P.J., Fava M., Parsey R.V., Kurian B.T., Phillips M.L., Oquendo M.A., Bruder G., Pizzagalli D., Toups M., Cooper C., Adams P., Weyandt S., Morris D.W., Grannemann B.D., Ogden R.T., Buckner R., McInnis M., Kraemer H.C., Petkova E., Carmody T.J., Weissman M.M. Establishing moderators and biosignatures of antidepressant response in clinical care (EMBARC): Rationale and design. J. Psychiatr. Res. 2016;78:11–23. doi: 10.1016/j.jpsychires.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loo H.M., de Jonge P., Romeijn J.-W., Kessler R.C., Schoevers R.A. Data-driven subtypes of major depressive disorder: a systematic review. BMC Med. 2012;10:156. doi: 10.1186/1741-7015-10-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G., Raamana P.R., Engemann D., Hoyos-Idrobo A., Schwartz Y., Thirion B. Assessing and tuning brain decoders: cross-validation, caveats, and guidelines. NeuroImage. 2017;145:166–179. doi: 10.1016/j.neuroimage.2016.10.038. [DOI] [PubMed] [Google Scholar]
- Wittchen H.U., Jacobi F., Rehm J., Gustavsson A., Svensson M., Jönsson B., Olesen J., Allgulander C., Alonso J., Faravelli C., Fratiglioni L., Jennum P., Lieb R., Maercker A., van Os J., Preisig M., Salvador-Carulla L., Simon R., Steinhausen H.-C. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur. Neuropsychopharmacol. 2011;21(9):655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- Zimmerman M., Ellison W., Young D., Chelminski I., Dalrymple K. How many different ways do patients meet the diagnostic criteria for major depressive disorder? Compr. Psychiatry. 2015;56:29–34. doi: 10.1016/j.comppsych.2014.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NeuroPharm data can be made available upon request through an application to the Cimbi database (https://www.cimbi.dk). The EMBARC dataset can be accessed through the National Institute of Mental Health Data Archive (https://nda.nih.gov). The source code for this manuscript is freely available at https://github.com/vbeliveau/MDD_SSRI_structural_prediction.
Data will be made available on request.