Summary
Isolation of viable immune cells from tissues is critically important to characterize cellular and molecular processes during homeostasis and disease. Here, we provide an optimized protocol to achieve high yields of viable intestinal lamina propria mononuclear cells (LPMCs). We describe steps for intestinal tissue collection from humans and nonhuman primates, followed by mechanical disruption and enzymatic digestion. Furthermore, we detail characterization of the mononuclear phagocyte (MP) subtypes by flow cytometry analysis. The protocol is repeatable and scalable for downstream applications.
For complete details on the use and execution of this protocol, please refer to Cavarelli et al. (2022).
Subject areas: Cell isolation, Flow Cytometry/Mass Cytometry, Immunology
Graphical abstract
Highlights
-
•
Collection and identification of the portions of the lower intestinal tract
-
•
Detailed protocol for intestinal lamina propria mononuclear cells isolation
-
•
Detailed protocol for lamina propria mononuclear cells staining
-
•
Procedures to identify mononuclear phagocyte subsets by flow cytometry
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Isolation of viable immune cells from tissues is critically important to characterize cellular and molecular processes during homeostasis and disease. Here, we provide an optimized protocol to achieve high yields of viable intestinal lamina propria mononuclear cells (LPMCs). We describe steps for intestinal tissue collection from humans and nonhuman primates, followed by mechanical disruption and enzymatic digestion. Furthermore, we detail characterization of the mononuclear phagocyte (MP) subtypes by flow cytometry analysis. The protocol is repeatable and scalable for downstream applications.
Before you begin
The protocol reported here describes the exact procedure for the isolation and analysis of non-human primate (NHP) lamina propria mononuclear cells (LPMCs) from the sigmoid colon collected at animal necropsies. We have also used this exact protocol for the efficient isolation of LPMCs from human colonic and rectal tissue of variable size and weight including those obtained from resections and surgeries. In addition, we used this protocol to isolate cells for rectum, descending colon and small intestine (duodenum, ileum and jejunum) from cynomolgus macaques (CMs) (unpublished results). The protocol focuses on the analysis of mucosa at homeostasis, but we also used it to describe changes in LPMCs numbers and phenotype following SIV infection of CMs.
The protocol is highly reproducible and is scalable depending on the size of intestinal sample available. For optimal results, tissue samples should be stored immediately in cold Transport Solution (refer to materials and equipment section) and processed within 1–2 h of necropsies (NHP tissues) or of surgical retrieval (human tissues). If not immediately processed after removal from the patient/NHP, samples should be stored at 4°C prior to processing. We didn’t test samples stored longer than 2 h. We would not recommend to store samples for longer than 12–16 h.
Isolated LPMCs can be used for ex vivo cell culture or phenotypic and functional profiling by flow cytometry or mass cytometry. Additional applications, such as cell sorting and transcriptomic analyses are suitable as well. Here we focus on the identification of mononuclear phagocytes (MPs) by flow cytometry, and provide a comprehensive list of titrated antibody clones used to build an optimized panel to discriminate dendritic cells (DCs) from macrophages (Mφs) in both species. Finally, this protocol can be also used to characterize lymphocyte populations.
Institutional permissions
All work related to Cynomolgus macaques (Macaca fascicularis) was conducted in compliance with institutional guidelines and protocols approved by the local ethics committee “Comité d’Ethique en Expérimentation Animale du Commissariat à l’Energie Atomique et aux Energies Alternatives” (CEtEA #44). The study was authorized by the “Research, Innovation and Education Ministry”.
The study protocol for human mucosal tissues was approved by the institutional Ethics committee of IRCCS Ospedale San Raffaele (MUCHIV protocol Ethical Committee approval #35 06/02/2014 and extension #40 09/11/2015). Patients were followed at the Department of General and Gastrosurgery, IRCCS Ospedale San Raffaele, and signed the informed consent form.
Patients inform consent and ethical approval from your institution is required for the use of patient or NHP-derived samples.
Preparation of equipment and reagents
Timing: 0.5–1 h
-
1.
Switch on Class II or Class III Biological Safety Cabinet (BSC), to work with uninfected or infected tissues, respectively.
-
2.
Disinfect equipment (scissors and forceps) and BSC with 70% ethanol (or equivalent).
-
3.
Prepare a sterile container filled with an appropriate volume of transport solution (TS) (refer to materials and equipment section) and store it at 4°C for the collection and transport of the tissue.
-
4.
Prepare aliquots of enzymes (DNase I & Collagenase VIII) (refer to materials and equipment section).
-
5.
Prepare aliquots of LIVE/DEAD Fixable Blue/Near-IR Dead Cell Stain (refer to the staining section).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45-V500 (clone HI30) dil. 1/20 | BD Biosciences | 560779 |
| CD45-PerCp (clone D058-1283) dil. 1/40 | BD Biosciences | 558411 |
| CD3-PER-CP-Cy5.5 (clone UCHT1) dil. 1/100 | BD Biosciences | 560835 |
| CD3-V500 (clone SP34-2) dil. 1/33 | BD Biosciences | 560770 |
| CD8-BV650 (clone RPA-T8) dil. 1/50 | BD Biosciences | 563821 |
| CD14-Alexafluor700 (clone M5E2) dil. 1/100 | BD Biosciences | 557923 |
| CD16-PER-CP-Cy5.5 (clone 3G8) dil. 1/80 | BD Biosciences | 560717 |
| CD16-PE-CF594 (clone 3G8) dil. 1/50 | BD Biosciences | 562293 |
| CD19-PER-CP-Cy5.5 (clone SJ25C1) dil. 1/10 | BD Biosciences | 332780 |
| CD20-BV711 (clone 2H7) dil. 1/20 | BD Biosciences | 563126 |
| CD56-PER-CP-Cy5.5 (clone B159) dil. 1/50 | BD Biosciences | 560842 |
| HLA-DR APC (clone G46-6) dil. 1/40 | BD Biosciences | 559866 |
| HLA-DR APC-H7 (clone G46-6) dil. 1/20 | BD Biosciences | 561358 |
| CD11c-PB (clone 3.9) dil. 1/80 | BioLegend | 301625 |
| CD11c-APC (clone S-HCL-3) dil. 1/10 | BD Biosciences | 333144 |
| CD123-PE-Cy7 (clone 7G3) dil. 1/40 | BD Biosciences | 560826 |
| CD64-PE-CF594 (clone) dil. 1/20 | BD Biosciences | 565389 |
| CD64-V450 (clone 10.1) dil. 1/20 | BD Biosciences | 561202 |
| CD103-PE (clone Bly7) dil. 1/20 | eBioscience | 12-1038 |
| CX3CR1-FITC (clone 2A9-1) dil. 1/20 | BioLegend | 341605 |
| Biological samples | ||
| Human colon samples | IRCCS Ospedale San Raffaele, Milan, Italy | https://www.hsr.it |
| Macaque colon samples | ICEA, MVA-HB/IDMIT, Fonteney-aux-Roses, France | https://www.idmitcenter.fr |
| Chemicals, peptides, and recombinant proteins | ||
| Deoxyribonuclease (DNase) I from bovine pancreas | Sigma-Aldrich | D4263 |
| Collagenase VIII from Clostridium histolyticum – 500 mg | Sigma-Aldrich | C2139 |
| EDTA 0.5 M, ph 8 | Sigma-Aldrich | 03690 |
| DL-Dithiothreitol solution (DTT) | Sigma-Aldrich | 646563 |
| Gentamycin 50 mg/mL | Gibco- Thermo Fisher Scientific | 15750-037 |
| HEPES-buffer solution 1 M | Gibco- Thermo Fisher Scientific | 15630-056 |
| Penicillin-Streptomycin-Neomycin (PSN) | Gibco- Thermo Fisher Scientific | 15640-055 |
| 1× HBSS with calcium chloride and magnesium chloride [HBSS+Ca/+Mg] | Gibco- Thermo Fisher Scientific | 24010-091 |
| 1× HBSS without calcium chloride and magnesium chloride [HBSS-Ca/-Mg]] | Gibco- Thermo Fisher Scientific | 14170-088 |
| 1× Roswell Park Memorial Institute 1640 medium (RPMI-1640) + GlutaMAXTM | Gibco- Thermo Fisher Scientific | 61870-010 |
| 1× Phosphate-buffered saline (PBS) | Gibco- Thermo Fisher Scientific | 14190-094 |
| Dimethyl sulfoxide (DMSO) - 100 mL | Sigma-Aldrich | D2650 |
| Ethanol | Various | N/A |
| Heat-inactivated fetal calf serum (Hi-FCS) | Lonza | DE14-801F |
| Macaque serum | In-house produced | N/A |
| BD FACS™ Lysing solution – 100 mL | BD Biosciences | 349202 |
| Critical commercial assays | ||
| LIVE/DEADTM Fixable Blue Dead Cell Stain kit | Thermo Fisher Scientific | L23105 |
| LIVE/DEADTM Fixable Near-IR for 633 or 635 excitation Dead Cell Stain kit | Thermo Fisher Scientific | L10119 |
| Software and algorithms | ||
| BD FACSDiva | BD Biosciences | https://www.bdbiosciences.com |
| FlowJo | BD Biosciences | https://www.flowjo.com |
| Deposited data | ||
| FCS raw data | FlowRepository | http://flowrepository.org/id/FR-FCM-Z5Q9 |
| Other | ||
| Scissors | Coveto | 804568 |
| Forceps | Coveto | 719328 |
| 50 mL FALCON tubes | Fisher Scientific | 352070 |
| Falcon® 100 mm TC-treated Cell Culture Dish (100 mm × 20 mm Petri dish) | Corning Life Sciences | 353003 |
| Falcon® 150 mm × 15 mm Not TC-treated Bacteriological Petri Dish (150 mm × 15 mm Petri dish) | Corning Life Sciences | 351058 |
| 0.45-μm filter unit, 200 mL | Thermo Fisher Scientific | 126-0045 |
| 70-μm cell strainer | Corning Life Sciences | 352350 |
| CorningTM 10 mL serological pipettes | Fisher Scientific | 356551 |
| CorningTM 25 mL serological pipettes | Fisher Scientific | 356525 |
| Cryovials 1.8 mL | Thermo Fisher Scientific | 375418PK |
| FACS tubes 5 mL | Corning Life Sciences | 352063 |
| 96-well plate U-bottom | Corning Life Sciences | 353077 |
| Corning® CoolCell™ LX Cell Freezing Container | Sigma-Aldrich | CLS432001 |
| VorTemp 1550 shaking incubator | Labnet International | S2050A |
| LSRFortessa flow cytometer | BD Biosciences | N/A |
| Beckman Coulters NaviosEx flow cytometer | Beckman Coulter | N/A |
Materials and equipment
Transport solution
The transport solution (TS) consist -of RPMI 1640 + GlutaMAXTM supplemented with 1% Penicillin-Streptomycin-Neomycin (PSN) and 50 μg/mL gentamycin. TS should be kept at 4°C and can be stored for up to 1 week.
| Reagent | Final concentration | Amount |
|---|---|---|
| PSN | 1% (v/v) | 5 mL |
| Gentamycin (50 mg/mL) | 50 μg/mL (0.001%) | 500 μL |
| RPMI 1640 | N/A | 494.5 mL |
| Total | N/A | 500 mL |
Pre-digestion solution
Dilute the 500 mM EDTA solution and the 1 M DTT solution in 25 mL of HBSS-Ca/-Mg to achieve a final concentration of 5 mM and 1 mM respectively. Pre-warm HBSS-Ca/-Mg at 37°C before preparing the pre-digestion solution. We recommend to prepare a fresh solution at the beginning of each experiment.
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA (500 mM) | 5 mM (0.01%) | 250 μL |
| DTT (1 M) | 1 mM (0.001%) | 25 μL |
| 1× HBSS-Ca/-Mg | N/A | 24.73 mL |
| Total | N/A | 25 mL |
Note: We suggest to prepare 25 mL of pre-digestion solution for each portion of about 12 cm2 colonic tissue (refers to step 5 of this protocol). Prepare as much tubes of pre-digestion solution as needed, according to the size of the tissue. The HBSS used should not contain calcium and magnesium, which would interfere with EDTA activity.
Enzyme stocks
Reconstitute 500 mg of Collagenase VIII in 50 mL of HBSS+Ca/+Mg to obtain a 10 mg/mL stock solution.
Reconstitute 1 vial of lyophilized DNase I with 10 mL of sterile 0.15 M NaCl to obtain a 2,000 U/mL solution.
Note: To prepare 200 mL of a 0.15 M NaCl solution, resuspend 1.75 g of NaCl in 200 mL of distilled water. Sterilize by filtering through a 0.45-μm filter and store at 18°C–25°C.
CRITICAL: We recommend to prepare aliquots of DNase I & Collagenase VIII and to store them at −20°C for up to 1 year to reduce freeze-thaw cycles. Date aliquots prior to freezing.
Enzyme digestion solution
Dilute the 10 mg/mL Collagenase VIII and 2,000 U/mL DNase I stocks in 25 mL of HBSS+Ca/+Mg to achieve a final concentration of 0.25 mg/mL and 5 U/mL respectively. Pre-warm HBSS+Ca/+Mg at 37°C before preparing the pre-digestion solution. We recommend to prepare a fresh enzyme digestion solution at the beginning of each experiment.
CRITICAL: We recommend to prepare 630 μL aliquots for Collagenase VIII and 65 μL aliquots for DNase I, to avoid repeated freeze/thaw cycles. The volumes reported here are optimized for preparation of 25 mL digestion solution.
CRITICAL: Enzyme concentration described here refers to a specific lot of Collagenase VIII. We recommend to titrate the concentration of enzyme needed for optimal tissue digestion each time a new lot is used.
| Reagent | Final concentration | Amount |
|---|---|---|
| Collagenase VIII (10 mg/mL) | 0.25 mg/mL (0.025%) | 625 μL |
| Dnase I (2,000 U/mL) | 5 U/mL (0.0025%) | 62.5 μL |
| 1× HBSS+Ca/+Mg | N/A | 24.31 mL |
| Total | N/A | 25 mL |
Note: Enzymatic activity of the Collagenase VIII and DNase I is supported by the presence of calcium and magnesium in the HBSS medium.
CRITICAL: Enzyme digestion solution should be prepared a few minutes before the digestion step. We suggest 25 mL of enzyme digestion solution for each intestinal tissue of about 12 cm2 (refers to steps 10 and 18 of this protocol). Prepare as much tubes of enzyme digestion solution as needed, according to the size of the tissue and the number of tissues being processed. Pre-warm the digestion solution at 37°C prior to use to allow maximal enzymatic activity.
Note: The volume of both pre-digestion and enzyme digestion solution reported here refers to tissues of about 12 cm2. The same volume of the solutions can be used for tissues of 6–12 cm2, whereas for tissues smaller than 6 cm2 we suggest to the scale-down the volume. For instance, when working with biopsies from colonoscopy/endoscopy, the use of 12 mL of solutions is recommended.
Wash buffer
The wash buffer solution consists -of RPMI 1640 + GlutaMAXTM supplemented with 5% FCS, 1% PSN, and HEPES 1 M. Prepare 500 mL of wash buffer. Pre-warm the RPMI medium at 37°C before using it. Wash buffer should be stored at 4°C for up to 2 weeks.
| Reagent | Final concentration | Amount |
|---|---|---|
| FCS | 5% (v/v) | 50 mL |
| PSN | 1% (v/v) | 5 mL |
| HEPES (1 M) | 10 mM (0.01%) | 5 mL |
| RPMI 1640 | N/A | 440 mL |
| Total | N/A | 500 mL |
Cell freezing solution
Prepare 100 mL aliquots of 10% DMSO solution in FCS. Aliquots can be stored at 4°C for up to 2 months or at 20°C for up to 1 year.
| Reagent | Final concentration | Amount |
|---|---|---|
| FCS | 90% (v/v) | 90 mL |
| DMSO | 10% (v/v) | 10 mL |
| Total | N/A | 100 mL |
Note: FCS used to prepare both Wash buffer and Cell freezing solution should be heat inactivated 30 min at 56°C before use.
Alternatives: Cell culture media and some reagents described here are available from other manufactures and are compatible with this protocol, as listed in Table 1.
Table 1.
Alternative resources table
| Reagent or resource | Alternative | Source | Identifier |
|---|---|---|---|
| EDTA 0.5 M, ph 8 | EDTA 0.5 M, ph 8 | Merck Millipore | 324504 |
| EDTA 0.5 M, ph 8 | Thermo Fisher Scientific | 15575020 | |
| HEPES-buffer solution 1 M | HEPES buffer 1 M | Lonza | BEBP17-737E |
| 1× HBSS with calcium chloride and magnesium chloride [HBSS+Ca/+Mg] | HBSS Modified, with calcium, with magnesium, without phenol red | Sigma-Aldrich | 55037C |
| 1× HBSS without calcium chloride and magnesium chloride [HBSS-Ca/-Mg]] | HBSS Modified, with phenol red, without calcium, without magnesium | Sigma-Aldrich | 55021C |
| 1× Roswell Park Memorial Institute 1640 medium (RPMI-1640) + GlutaMAXTM | RPMI-1640 | Sigma-Aldrich | R8754 |
| RPMI-1640 | Thermo Fisher Scientific | 11875093 |
BD LSRFortessa & Beckman Coulters NaviosEx flow cytometer
The antibody panel described here was conceived to be run on a 5 laser BD LSRFortessa flow cytometer to identify NHP MPs and on a 3 laser Beckman Coulters NaviosEx flow cytometer to identify human MPs. The respective configurations are detailed in Tables 2 and 3.
Table 2.
LSRFortessa configuration
| Laser | Filter | Fluorochromes example |
|---|---|---|
| 350 | 450/50 | BlueVid |
| 740/35 | BUV737 | |
| 405 | 450/50 | vioBlue, Pacific Blue, V450, BV421, eFluor450 |
| 525/50 | AmCyan, V500, BV521, BV510 | |
| 610/20 | BV605 | |
| 660/20 | BV650 | |
| 710/50 | BV711 | |
| 780/60 | BV786, BV785 | |
| 488 | 530/30 | FITC, GFP, AF488, BB515 |
| 695/40 | PercP, PerCP5.5, 7-AA, BB700 | |
| 561 | 585/15 | PE |
| 610/20 | PE-TR, Pedazzle, PECF594, ECD | |
| 670/30 | PE-Cy5 | |
| 710/50 | PE-Cy5.5 | |
| 780/60 | PE-Cy7, PEvio770 | |
| 633 | 670/14 | APC, AF647 |
| 730/45 | AF700 | |
| 780/60 | APC-Cy7, APC-H7, APCvio770 |
Table 3.
NaviosEx configuration
| Laser | Filter | Fluorochromes example |
|---|---|---|
| 405 | 450/50 | Pacific Blue, V450, BV421, BFP, VioBlue |
| 550/40 | Krome Orange, Pacific Orange, AmCyan, V500, | |
| BV 510, VioGreen | ||
| 488 | 525/40 | FITC, GFP, Alexa Fluor 488, BB515, VioBright 515 |
| 575/25 | PE, PI | |
| 620/20 | PE Texas Red, ECD, PE-Vio 615 | |
| 695/30 | PerCP, PerCP Cy5.5, PE Cy5, 7AAD, PerCP-Vio700 | |
| 755 LP | PE Cy7, PE-Vio770 | |
| 633 | 660/20 | APC, AF647 |
| 725/20 | APC Cy5.5, APC R700, AF700 | |
| 755 LP | APC Cy7, APC H7, APC Alexa 750, APC-Vio770 |
The antibody panel detailed in the staining section of this protocol (see Tables 4 and 5) can be used to identify MP populations (e.g., monocytes, DCs and Mφ) with optimally titrated antibodies. The macaque panel also allows to identify lymphocyte population (e.g., CD3+ and CD8+ T lymphocytes and CD20+ B lymphocytes). In case different leukocyte populations need to be probed, alternative antibodies and fluorochrome combinations can be used, however, those will require titration and optimization.
Note: Antibody panels must be optimized for each flow cytometer. The type of lasers, filter combinations and voltage setting each cytometer is equipped with, may affect the signal.
Table 4.
BD LSRFortessa macaque mononuclear phagocyte phenotypic panel
| Marker | Fluorochrome | Concentration (μL/100 μL)a | Purpose |
|---|---|---|---|
| Viability | Blue for UV excitation | 10 | Live cells |
| CD45 | PerCp | 2,5 | Leukocytes |
| CD3 | V500 | 3 | T lymphocytes |
| CD8 | BV650 | 2 | T lymphocytes/NK cells |
| CD14 | Alexa fluor -700 | 1 | Monocytes/macrophages |
| CD16 | PE-CF594 | 2 | Monocytes/macrophages |
| CD20 | BV711 | 5 | B lymphocytes |
| HLA-DR | APC-H7 | 5 | Myeloid cells |
| CD11c | APC | 10 | Myeloid cells |
| CD123 | PE-Cy7 | 2,5 | pDCs |
| CD64 | V450 | 5 | Macrophages |
| CD103 | PE | 5 | mDCs |
| CX3CR1 | FITC | 5 | mDCs & macrophages |
Add FACS buffer up to 100 μL.
Table 5.
Beckman Coulters NaviosEx human mononuclear phagocyte phenotypic panel
| Marker | Fluorochome | Concentration (μL/100 μL)a | Purpose |
|---|---|---|---|
| Viability | APC-Cy7 | 10 | Live cells |
| CD45 | V500 | 5 | Leukocytes |
| CD3 | PerCp-Cy5.5 | 1 | T lymphocytes |
| CD14 | Alexa fluor -700 | 10 | Monocytes/macrophages |
| CD16 | PerCp-Cy5.5 | 1,25 | Monocytes/macrophages |
| CD19 | PerCp-Cy5.5 | 10 | B lymphocytes |
| CD56 | PerCp-Cy5.5 | 2 | NK cells |
| HLA-DR | APC | 5 | Myeloid cells |
| CD11c | PB | 1,25 | Myeloid cells |
| CD123 | PE-Cy7 | 1,25 | pDCs |
| CD64 | PE-CF594 | 5 | Macrophages |
| CD103 | PE | 5 | mDCs |
| CX3CR1 | FITC | 5 | mDCs & macrophages |
Add FACS buffer up to 100 μL.
Step-by-step method details
Tissue collection and transport
Timing: 30–40 min
Intestinal tissues collected at animal necropsies or during patient surgery should be processed within 2 h of collection, as to avoid major cellular damages (troubleshooting 1). If not processed immediately after collection, we recommend storage of the tissue at 4°C in TS.
Colonic tissues should be transported in a sterile container filled with 150 mL of cold TS. This volume can be scaled-up or down according to the size of the intestinal tissue. For instance, if the whole NHP descending colon is collected, transport in 250 mL of TS is recommended. Human tissues of up to 5 cm length, should be transported in 50 mL of TS.
CRITICAL: Keep the TS in ice during the “waiting” time of surgical retrieval.
Colonic tissue enzymatic digestion to obtain a single cell suspension
Timing: 3.5–4 h
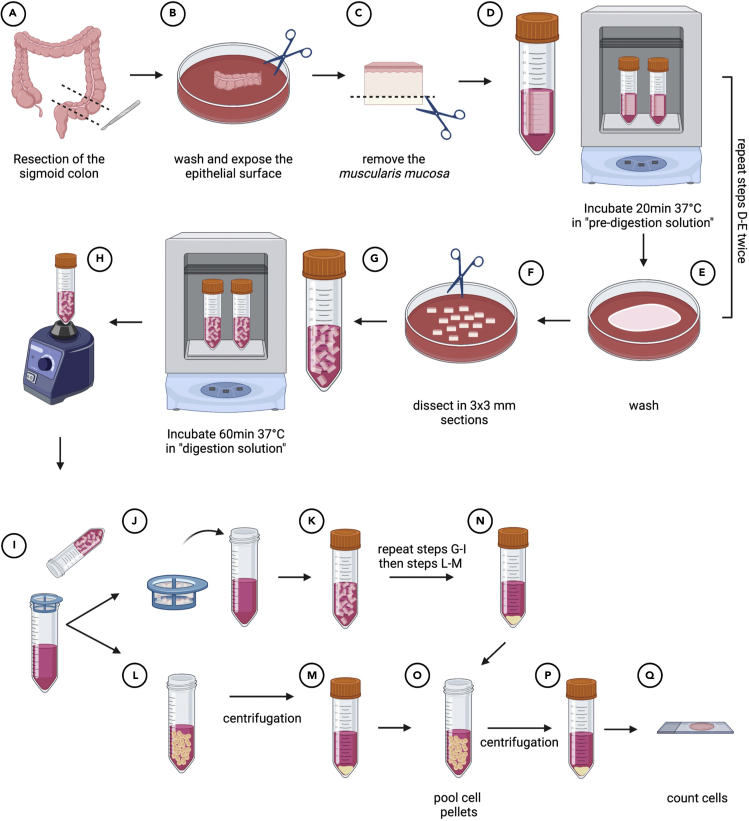
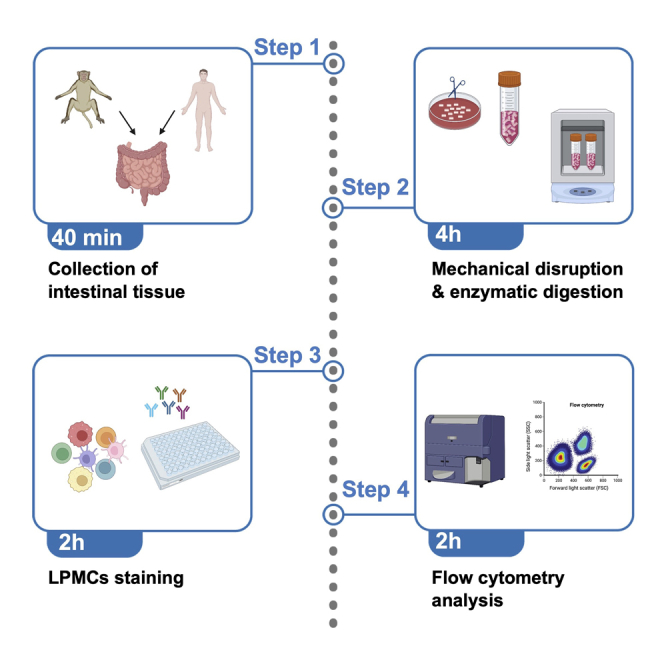
This section provides information for optimal dissection and digestion of the sigmoid colon to liberate LPMCs. An overview of the whole protocol is shown in Figure 1.
CRITICAL: For optimal results, ensure that the tissue is kept in solution at each step of this protocol.
-
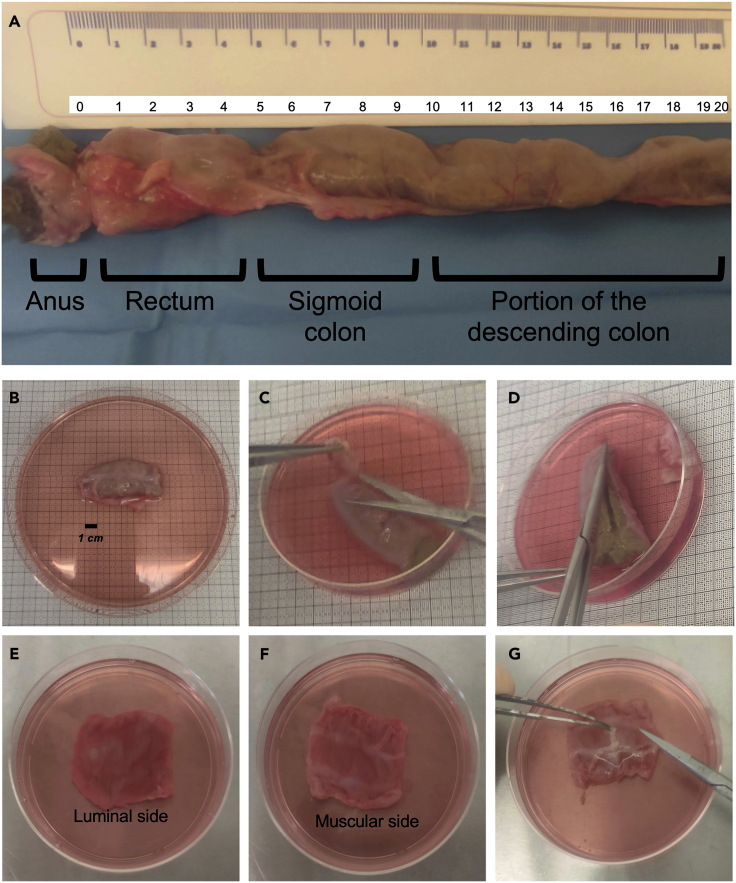
1.
Remove the tissue from the TS and identify the different portions of the intestine. Figure 2A helps to identify portions and corresponding size of the distal NHP intestine. We suggest to place the intestine in a 150 mm × 15 mm Petri dish containing 30 mL of TS.
Note: This step might not be required when processing human colonic tissue. In our experience, portions of sigmoid/descending colon of about 3–5 cm in length were usually provided.
-
2.
Dissect the sigmoid colon from the rest of the large intestine (Figure 2B) and, using sterile forceps, place it inside a 100 mm × 20 mm Petri dish containing 10 mL of TS.
-
3.Using scissors, remove the external fat (Figure 2C) and carefully open the intestinal tube (Figure 2D) to expose the luminal side (Figure 2E).
-
a.Remove the fecal material by gently washing the tissue with TS.
-
b.Transfer the tissue to a new 150 mm × 15 mm Petri dish filled with 10 mL of TS.
-
c.Repeat the washing steps 2–3 times, until the fecal material is completely removed.
-
a.
-
4.
Using scissors and forceps, carefully remove the muscular layer, without cutting the mucosae (Figure 2G).
Note: The epithelial luminal side of the tissue (Figure 2E) can be easily distinguished from the muscular side (Figure 2F) because of the pinkish tinge of the mucosae. The muscular layer appears white (Figures 2F and 2G).
Figure 1.
Step by step illustration of the procedures required to obtain a single-cell suspension of LPMCs
Figure 2.
Representative example of a portion of the lower NHP intestinal tract and of the sigmoid colon resection used for processing
(A) Identification of the anus, rectum, sigmoid colon and the distal portion of the descending colon.
(B) Sigmoid colon resection.
(C–G) Initial steps of the protocol: removal of the external fat (C); opening of the intestinal tube (D); view of the mucosa from the luminal side (E); view from the muscularis side (F); removal of the muscular layer (G).
The removal of the underlying muscular layer will allow a better enzymatic access to the mucosal tissue.
-
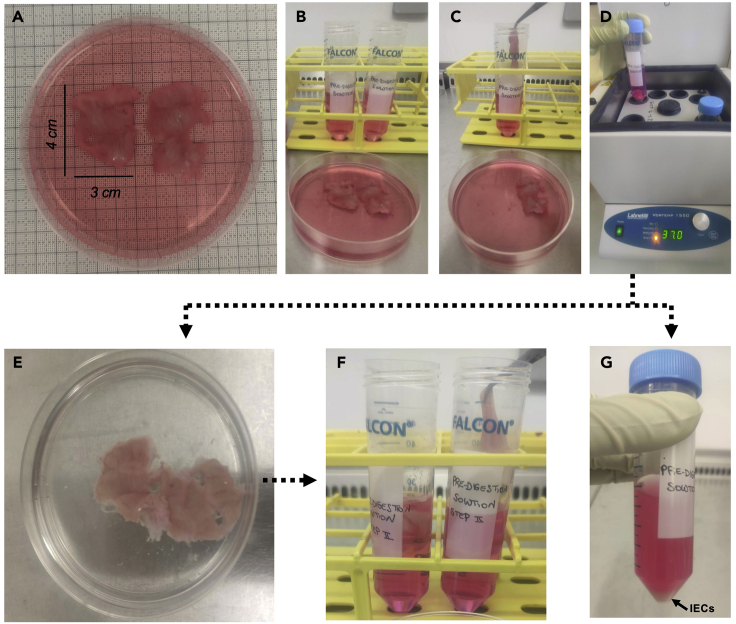
5.
Cut the tissue in two sections no greater than 3 × 4 cm (12 cm2, Figure 3A) and transfer each of them in a 50 mL Falcon tube pre-filled with 25 mL of pre-digestion solution (Figures 3B and 3C).
Note: Treatment with DTT and EDTA during the “pre-digestion” step allows to remove any remaining mucus and to liberate intestinal epithelial cells (IECs).
-
6.
Incubate at 37°C for 20 min inside a shaking incubator (Figure 3D).
Note: We used a VorTemp 1550 shaking incubator set at 800 RPM. If not available, a similar instrument can be used.
-
7.Using forceps, collect the tissue, place it inside a Petri dish.
-
a.Wash with 10 mL of 1× PBS (Figure 3E).
-
b.Transfer it inside a 50 mL Falcon tube pre-filled with 25 mL of pre-digestion solution (Figure 3F) (refers to materials and equipment section).
-
c.Incubate at 37°C for 20 min inside a shaking incubator.
-
a.
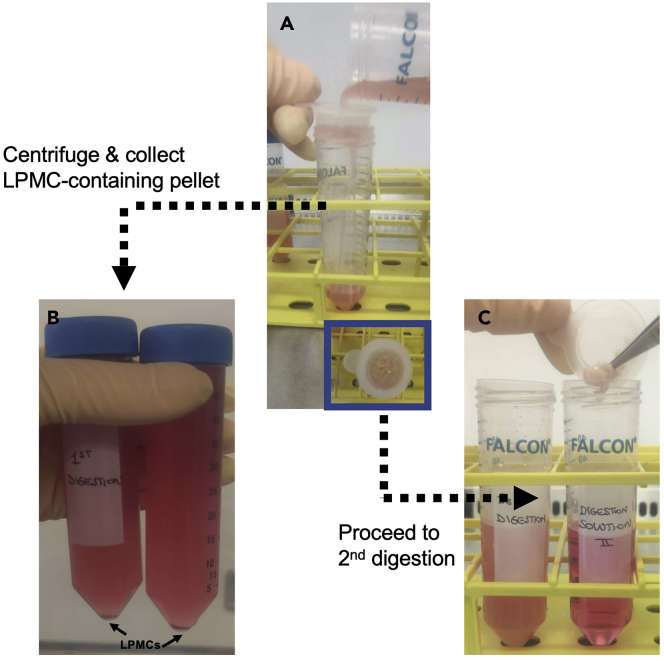
CRITICAL: The pre-digestion step here is repeated twice to completely remove mucus and liberate IECs. Those are visible as a pellet deposit in Figure 3G.
Figure 3.
A representative example ofthe pre-digestion phase of the protocol to liberate intestinal epithelial cells (IECs) at steps 5–7
(A–D) The tissue is cut in two portions of 12 cm2 (A, B), transferred to a Falcon tube pre-filled with digestion solution (C, first enzymatic digestion step), and incubated at 37°C for 20 min (D). (E, F) After the enzymatic digestion, wash the tissue (E) and transfer it to a Falcon tube pre-filled with digestion solution (F, second enzymatic digestion step). (G) IECs are visible as a pellet deposit.
Following the pre-digestion step, the IECs remain in the pre-digestion solution and can be either discarded, or collected for further phenotypic and functional assays. However, in the latter case, the protocol may require further adjustment because it was not optimized to preserve IECs viability.
-
8.Using forceps, collect the tissue, place it inside a Petri dish.
-
a.Wash with 10 mL of 1× PBS.
-
b.Wash with 10 mL of HBSS+Ca/+Mg.
-
a.
-
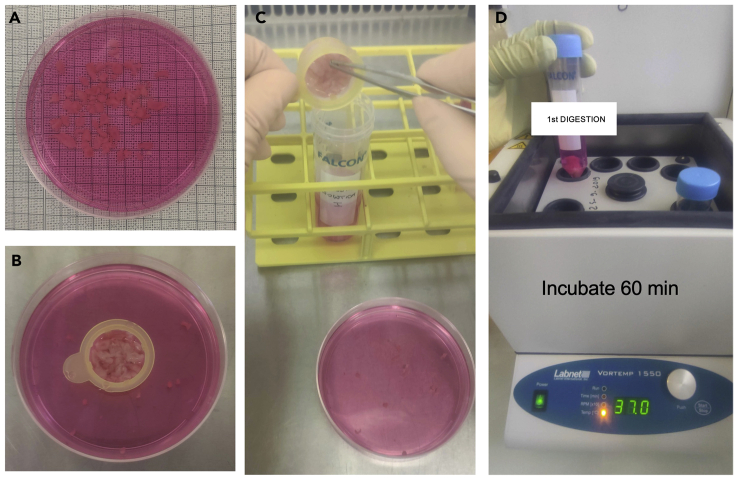
9.
Dissect, using scissors and forceps, each 12 cm2 tissue portions into small pieces no greater than 0.5 × 0.5 cm (Figure 4A).
Note: We suggest to directly cut the tissue inside a cell strainer to facilitate collection and subsequent transfer of the small pieces to a 50 mL tube (Figures 4B and 4C).
-
10.
Transfer to a 50 mL Falcon tube filled with 25 mL of pre-warmed enzyme digestion solution (Figure 4C) (refers to materials and equipment section).
-
11.
Incubate at 37°C for 60 min inside a shaking incubator (Figure 4D).
Note: Prepare the same number of enzyme digestion solution tubes as for the pre-digestion step. Here, we prepared 2 tubes of enzyme digestion solution, one for each 12 cm2 tissue portion.
-
12.At the end of the incubation:
-
a.Place a 70-μm cell strainer on top of a new 50 mL Falcon tube.
-
b.Take the tube containing the digested tissue, vortex it and filter the solution through the cell strainer as shown in Figure 5A.
-
a.
Note: The filtered solution contains the LPMCs whereas the undigested tissue remains inside the cell strainer. Do not discard the undigested tissue (see step 18).
-
13.
Add 25 mL of wash buffer to the 25 mL of LPMCs-containing solution and centrifuge at 500 g for 10 min, 18°C–25°C.
-
14.At the end of the centrifugation:
-
a.Discard the supernatant using a 25 mL pipette.
-
b.Resuspend the LPMCs-containing pellet (Figure 5B) in 1 mL of wash buffer using 1 mL pipette.
-
c.Transfer it to a new 50 mL Falcon tube and add 49 mL of wash buffer.
-
a.
-
15.
Centrifuge the tube at 500 g for 10 min, 18°C–25°C.
-
16.
Using a 25 mL serological pipette, aspirate and discard the supernatant without disturbing the LPMCs pellet.
-
17.
Resuspend the LPMCs-containing pellet in 10 mL of wash buffer.
-
18.
Any undigested residual tissue in the cell strainer at step 12 should be transferred in a new 50 mL Falcon tube containing 25 mL of digestion solution (Figure 5C) and incubated for 30 min at 37°C inside a shaking incubator.
-
19.
Repeat steps 12–17.
Pause point: The LPMCs obtained after the first enzymatic digestion at step 17 of the protocol should be keep on ice or at 4°C while completing the second enzymatic digestion of the tissue (steps 18 and 19).
CRITICAL: Enzymatic digestion allows to liberate cells from the tissue, however the incubation time is a critical factor to be considered. Indeed, surface expression of certain markers can be impacted by digestions longer than 1 h. On the other side, shorter digestions may be not enough to completely liberate cells from the tissue. We have optimized this protocol to perform two sequential digestions (60 min followed by 30 min) rather than one extended digestion to reduce the time cells are exposed to the enzyme, and to replenish the enzyme for optimal activity. Note that the first digestion results in significantly higher yield of LPMCs than the second one.
-
20.Add the LPMCs obtained after the second digestion to those obtained after the first digestion.
-
a.Add an appropriate volume of RPMI-1640 medium supplemented with 10% FCS and 1% PSN.
-
b.Count the cells.
-
a.
Note: We recommend to use an automated cell counter to determine cell count and viability. If not available, trypan blue staining and counting in a hemocytometer is suitable as well.
-
21.LPMCs can be used immediately for culture or can be cryopreserved for long-term storage.
-
a.For cryopreservation, resuspend the LPMC pellet in freezing media (refer to materials and equipment section).
-
b.Quickly aliquot 1 mL of the cell suspension per cryovial.
-
c.Place cryovials into a freezing container and transfer to a - 80°C freezer.
-
d.Transfer cryovials to liquid nitrogen after 24 h.
-
a.
Note: For optimal results, we recommend to freeze a minimum concentration of 5 × 106 cells per 500 mL of freezing media and a maximum of 30 × 106 cells per 1 mL of freezing media. Freezing containers, such as CoolCellTM or Mr.FrostyTM, provides the critical 1°C/min cooling rate required for successful cryopreservation of cells.
Figure 4.
A representative example of the dissectedintestine section in small pieces (step 9) and of the digestion phase (step 10) to liberate LPMCs
The tissue is dissected into small pieces (A), that are collected inside a cell strainer (B) and transferred to a Falcon tube filled with enzyme digestion solution (D).
Figure 5.
Filtration step after the first enzymatic digestion
A representative example of the filtration step 12 through a 70-μm cell strainer viewed from different angles (A) to collect LPMC (panel B shows the cell pellet after centrifugation) and transfer of undigested tissue to a Falcon tube filled with enzyme digestion solution (C) in step 18.
Staining of LPMCs for flow cytometry identification of mononuclear phagocyte subsets
Timing: 1.5 h
This section describes how to phenotypically characterize LPMCs by flow cytometry. Staining is performed in a 96 well U-bottom tissue culture treated plate, but canonical 5 mL FACS tubes can be used as well.
-
22.
Pre-cool centrifuge to 4°C.
-
23.
Prepare a viability dye solution in 1× PBS at a suitable concentration – we recommend a dilution factor ranging between 1/1,000 and 1/3,000 for optimal detection of live/dead cells.
CRITICAL: To obtain a final 1/3,000 dilution, we recommend to prepare an intermediate 1 in 300 dilution by adding 1 μL of the Fixable Dead Cell Stain in 300 μL of 1× PBS. Use 10 μL of this solution to prepare 100 μL of the antibody master mix as described in Tables 4 and 5.
Note: We recommend to use a marker of cell viability to detect dead cells by flow cytometry analysis. This protocol has been designed for use with the LIVE/DEADTM Fixable Blue Dead Cell Stain kit when working with NHP cells and the LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit for 633 or 635 nm excitation, when working with human cells.
The dye should be reconstituted with 50 mL of DMSO as per manufacturer’s instructions (https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0006891_LIVE_DEAD_Fixable_Dead_Cell_Stain_Kits_QR.pdf) and aliquots should be frozen at 20°C protected from light.
Alternative flow cytometry viability stains can be used with fixable and non-fixable cell samples, but fluorochrome choice and concentration should be adapted by the users.
-
24.
Prepare FACS buffer by supplementing 1× PBS with 2%–5% FCS and 2,5 mM EDTA.
-
25.
Prepare an antibody master mix to stain cells of interest.
Note: An example panel of fluorochrome-labeled antibodies to identify MPs among NHP and human LPMCs is shown in Tables 4 and 5, respectively (the antibody concentrations are titrated for staining in a volume of 100 μL of FACS buffer).
CRITICAL: Prepare an appropriate volume of master mix according to the number of samples being stained. Always add an extra 1–2 samples to your master mix to account for pipetting error, so a 10× master mix will be good for 8–9 samples.
Note: The LIVE/DEADTM Fixable Dead Cell Stain can either be used prior to staining cells with the master mix or added directly to the master mix, as shown in Tables 4 and 5. We observed a similar separation of live and dead cells by using either protocols.
-
26.
Adjust cell concentration to 1–4 × 106 LPMCs per 200 μL of FACS buffer and transfer them to a 96 well U-bottom tissue culture treated plate.
CRITICAL: We recommend to prepare -an unstained control and fluorescence minus one (FMO) controls. Do not add any antibodies to the unstained control. Ideally the FMO cell concentration must be the same as that of the sample concentration, however if unavoidable, lesser cell concentration can be used for the FMOs.
-
27.
Centrifuge the plate at 500 g at 4°C for 5 min and aspirate the supernatant by using a multi-channel pipette, without disturbing the cell pellet. Alternatively, supernatant can be removed by flipping the plate.
-
28.Resuspend the cell pellet with 100 μL of “saturation solution” per well.
-
a.Incubate for 20 min at 4°C.
-
b.Centrifuge at 500 g at 4°C for 5 min.
-
a.
Note: Prepare “saturation solution” by supplementing 1× PBS with 5% macaque serum (if NHP cells are used) or with 5% FCS (if human cells are used).
-
29.
Aspirate the supernatant and resuspend cells in 100 μL of the antibody master mix prepared in step 25.
-
30.
Incubate cells for 15 min at 4°C. Cover the plate with aluminum foil to protect from light.
-
31.
Add 150 μL 1× PBS to each well and centrifuge at 500 g at 4°C for 5 min.
-
32.
Remove the supernatant and repeat step 31.
Optional: If a high contamination by red blood cells (RBC) is observed, a lysis step can be performed by incubating cells during 5 min with 200 μL of 1× BD FACS lysing solution TM (troubleshooting 4). After incubation wash twice with 200 μL of 1× PBS.
-
33.
Aspirate the supernatant and resuspend the cell pellet in 200 μL BD CytofixTM buffer per well. To ensure a single cell suspension, pipette up and down several times.
-
34.
Incubate for 20 min at 4°C protected from light.
Note: A solution of 1%–4% PFA can be also used for fixation of samples for flow cytometry.
CRITICAL: For biosafety reasons, fixation of human tissues is mandatory.
-
35.
Add 100 μL 1× PBS to each well and centrifuge at 500 g at 4°C for 5 min.
-
36.
Aspirate the supernatant and resuspend the cell pellet in 200 μL 1× PBS.
-
37.
Transfer the cells into a 5 mL FACS tube for acquisition at a flow cytometer.
-
38.
Add 100–200 μL of 1× PBS according to the number of cells being stained.
Pause point: Samples may be kept at 4°C protected from light and be run the following day if samples have been fixed.
Note: Samples can be left in the 96-well plate if the flow cytometer is equipped with an HTS attachment for automated sample collection.
-
39.
Collect data with a- BD LSRFortessa or a Beckman Coulters NaviosEx flow cytometer and analyze by using FlowJo software.
Expected outcomes
Expected LPMCs yield
The procedures described here allows to obtain a high yield of viable and pure LPMCs from tissues of varying size, including larger tissue samples typically obtained at animal’s necropsies or smaller tissues obtained from human colorectal surgeries. The protocol can be scaled according to the size of the intestine available and can be adapted to digest very small tissue biopsies.
Using this protocol, we obtained a mean yield of 96 × 106 LPMCs by processing tissues with a size of 2–5 cm of length (from n= 30 NHP), with a mean viability of 89% (as determined by trypan blue viability assay and counting with an automated cell counter) and 95% (as determined by flow cytometric assessment).
Accurate identification of intestinal MPs by flow cytometry
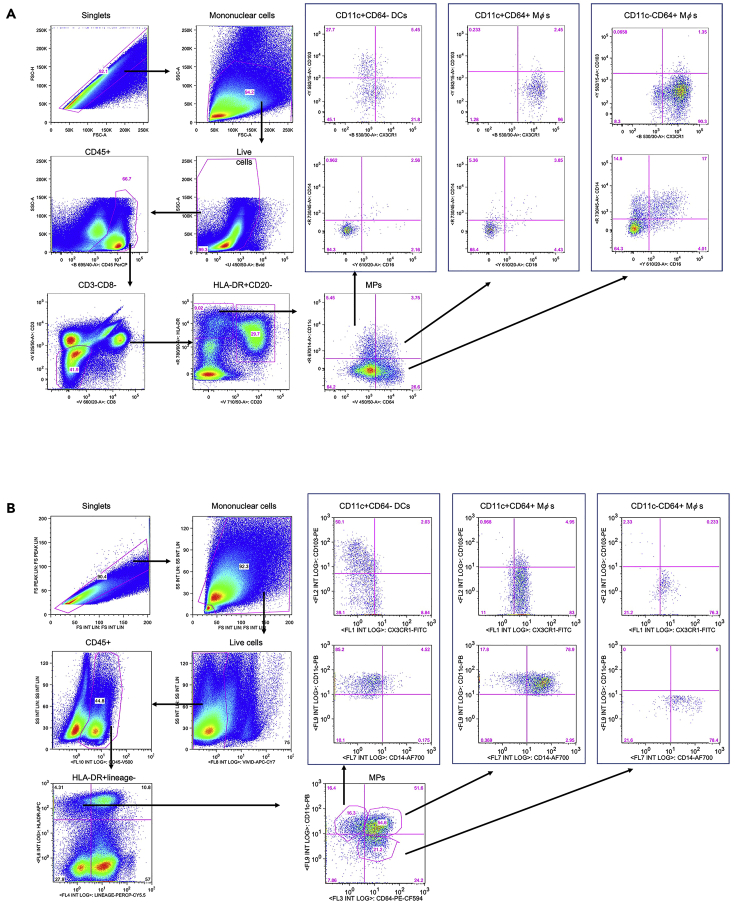
The identification and characterization of intestinal MP populations is challenging because DCs and Mφs share several common markers, including HLA-DR, CD11c and CX3CR1. Here we describe a flow cytometry panel optimized to identify collagenase-liberated MPs in intestinal tissues from human beings and NHPs, as well as the expected MPs composition and frequency (Figure 6). By using this protocol, five main MP populations can be identified in both species: three populations of CD64- DCs (tolerogenic CD103+, non-tolerogenic CD103-, and CX3CR1+ DC) and two of CD64+ Mφs (immature CD11c+ and mature CD11c-, both expressing different levels of CX3CR1.) In addition, expression of CD14 and CD16 by each population can be assessed.
Figure 6.
Gating strategy showing the analysis of viable NHP and human LPMCs and identification of mononuclear phagocytes
Single cells are identified by plotting FSC-A against FSC-H. Mononuclear cells are identified by size by plotting FSC-A against SSC-A, and live cells are defined as LIVE/DEADTM Fixable Blue (A) or Near-IR (B) Dead Cell stain negative. (A) Macaque samples: among CD45+ leukocytes an initial gate on CD3-CD8- cells allowed to exclude T lymphocytes and NK cells, followed by a gate on HLA-DR+CD20- cells to exclude B lymphocytes. MPs were identified based on the expression of CD11c and CD64 and included CD11c+CD64- DCs, CD11c+CD64+ Mφs, and CD11c- CD64+ Mφs. Expression of CD103, CX3CR1, CD14 and CD16 by each of the three MP subsets is shown. (B) Human cells: among CD45+ leukocytes a gate on HLA-DR+/lineage – cells (lineage defined as CD3-CD56-CD19-CD16-) allowed to exclude T and B lymphocytes, NK cells and CD16+ cells. MPs were identified based on the expression of CD11c and CD64 and included CD11c+CD64- DCs, CD11c+CD64+ Mφs, and CD11c- CD64+ Mφs. Expression of CD103, CX3CR1, and CD14 by each of the three MP subsets is shown.
Further information on expected frequencies and phenotypic marker expression, such as CD103, CX3CR1 and CD14 by DCs and Mφs subsets, can be found on a previously published research article that have used this protocol (Cavarelli et al., 2022). By using the same digestion protocol and an antibody panel specifically designed for lymphocytes identification (not detailed here), T cell subsets composition of the macaque colon was previously described (Cavarelli et al., 2021).
Limitations
Enzymatic digestion of tissue is widely used and helps to increase cell yield, although cleavage of some cell surface markers is an associated risk. Such limitation can be overcome by stringently following the incubation time and the enzyme concentration described in this protocol. In a set of preliminary experiments, we compared collagenase IV versus collagenase VIII to digest NHP tissues (data not shown), obtaining better outcomes in terms of cell yield and viability by collagenase VIII digestion. In addition, collagenase activity might change from lot to lot. For optimal results, suitability of each lot of the enzyme must be determined empirically.
Troubleshooting
Problem 1
Delays in obtaining tissues after necropsy/surgery (step 1).
Potential solution
The intestinal tissue is extremely fragile and cells necrosis and/or apoptosis due to delays in necropsy/surgery timings might be an issue. This protocol has been successfully used to isolate LPMCs from samples processed immediately after collection and in a range of ‘waiting’ time between 30 min and 2 h. If a delay cannot be avoided, we recommend to keep the tissue in cold transport solution to optimize the results in terms of viable cell yield.
Problem 2
Incomplete removal of epithelial cells and/or mucus (steps 5 and 6).
Potential solution
Epithelial cells are more vulnerable to enzymatic digestion, with consequent cell death and poor quality of the entire sample. Also, complete removal of mucus, which is particularly abundant in the small intestine, is critical to preserve cell viability. Make sure to strictly respect incubation time and reagents concentration during the pre-digestion step and to abundantly wash the tissue.
Problem 3
Difficulty filtering cells through a cell strainer (step 12).
Potential solution
When a high number of cells is liberated from the tissues (more than 200–300 × 106 cells) it can be greatly difficult to filter them through a 70-μm cell strainer. We suggest to use a 1 mL plastic tip to facilitate cell passage through the filter and to regularly change the cell strainer if it becomes obstructed.
Problem 4
Low number of viable LPMCs (step 20).
Potential solution
Enzymatic digestion can have an impact on LPMCs yield and viability. We recommend to strictly respect the timing and concentration of collagenase indicated in this protocol. It’s important to prepare all reagents and to bring them at the appropriate temperature prior starting manipulating the tissue, however, collagenase should be thawed and added to the “digestion solution” immediately prior usage to guarantee maximal enzymatic efficiency. A low cell yield might be due to insufficient digestion time, or the use of HBSS without calcium and magnesium. If multiple tissues are processed at the same time, we suggest ensuring the incubation time for each of them to be respected. In addition, to preserve cell viability, we recommend to keep the tissue in solution at each step of this protocol.
Problem 5
Red blood cells contamination (steps 31 and 32).
Potential solution
If a high number of red blood cells is present in your LPMC pellet that might affect downstream experiments, we suggest to perform a lysis with an appropriate lysis solution (refer to key resources table).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mariangela Cavarelli (mariangela.cavarelli@cea.fr).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank present and past members of the Animal Science and Welfare and FlowCyTech facilities of the IDMIT infrastructure for their excellent expertise and useful discussions. We thank Dr. Saverio Di Palo and Dr. Ugo Elmore (IRCCS Ospedale San Raffaele) for providing human tissues. We also thank present and past members of G. Scarlatti’s laboratory at IRCCS Ospedale San Raffaele for their help with human tissue experiments. At IRCCS Ospedale San Raffaele, flow cytometry acquisitions of human cells were performed at FRACTAL facility.
This work was funded by the French Agence Nationale de Recherches sur le Sida et les Hépatites Virales (ANRS, decision n°14415/15516, recipients M.C. and G.S.). M.C. was a beneficiary of a Marie Curie Individual fellowship (grant agreement n° 658277 for the project DCmucoHIV).
This work was also supported by the “Programme Investissements d’Avenir” (PIA) managed by the ANR under reference ANR-11-INBS-0008, funding the Infectious Disease Models and Innovative Therapies (IDMIT, Fontenay-aux-Roses, France) infrastructure, and ANR-10-EQPX-02-01, funding the FlowCyTech facility (IDMIT, Fontenay-aux-Roses, France). The funders had no role in the design of the study, data collection or interpretation, or the decision to submit the work for publication.
Graphical abstract and Figure 1 were created with biorender.com.
Author contributions
K.B. helped with the experiments. B.D. and S.L. optimized tissues collection from NHPs. G.S. and R.L.G. provided financial support and intellectual input. M.C. provided financial support, designed the protocol, performed most of the experiments, analyzed the data, prepared the figures, and wrote the manuscript. All authors approved the submitted version.
Declaration of interests
The authors declare no competing interests.
Data and code availability
FCS raw data are available from the FlowRepository website (http://flowrepository.org/id/FR-FCM-Z5Q9). The authors declare that all data supporting the findings of this study are available within the article.
References
- Cavarelli M., Foglieni C., Hantour N., Schorn T., Ferrazzano A., Dispinseri S., Desjardins D., Elmore U., Dereuddre-Bosquet N., Scarlatti G., Le Grand R. Identification of CX3CR1 + mononuclear phagocyte subsets involved in HIV-1 and SIV colorectal transmission. iScience. 2022;25:104346. doi: 10.1016/J.ISCI.2022.104346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli M., Hua S., Hantour N., Tricot S., Tchitchek N., Gommet C., Hocini H., Chapon C., Dereuddre-Bosquet N., Le Grand R. Leukocytospermia induces intraepithelial recruitment of dendritic cells and increases SIV replication in colorectal tissue explants. Commun. Biol. 2021;4:861. doi: 10.1038/S42003-021-02383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
FCS raw data are available from the FlowRepository website (http://flowrepository.org/id/FR-FCM-Z5Q9). The authors declare that all data supporting the findings of this study are available within the article.