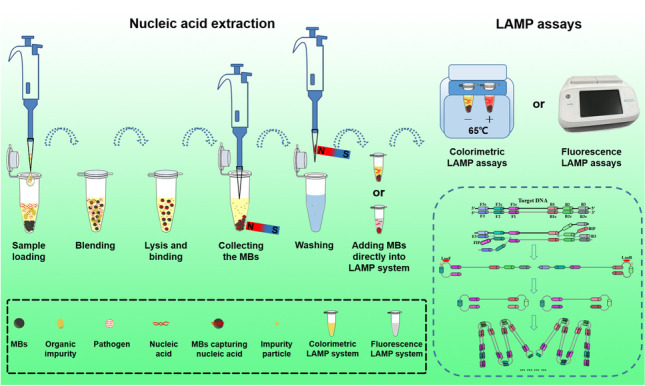
Abstract
Nucleic acid amplification tests (NAATs) have become an attractive approach for pathogen detection, and obtaining high-quality nucleic acid extracts from biological samples plays a critical role in ensuring accurate NAATs. In this work, we established an elution-free magnetic bead (MB)-based method by introducing polyethylene-polypropylene glycol (PEPPG) F68 in lysis buffer and using NaOH solution instead of alcohols as the washing buffer for rapid nucleic acid extraction from multiple types of biological samples, including nasopharyngeal swabs, serum, milk, and pork, which bypassed the nucleic acid elution step and allowed the nucleic acid/MB composite to be directly used as the template for amplification reactions. The entire extraction process was able to be completed in approximately 7 min. Even though the nucleic acid/MB composite could not be used for quantitative real-time PCR (qPCR) assays, this elution-free MB-based method significantly improved the sensitivity of the loop-mediated isothermal amplification (LAMP) assay. The sensitivity of the quantitative real-time LAMP (qLAMP) assays combined with this elution-free MB-based method showed an improvement of one to three orders of magnitude compared with qLAMP or qPCR assays combined with the traditional MB-based method. In addition to manual operation, like the traditional MB-based method, this universal, rapid, and facile nucleic acid extraction method also has potential for integration into automated robotic processing, making it particularly suitable for the establishment of an analysis platform for ultrafast and sensitive pathogen detection in various biological samples both in centralized laboratories and at remote sites.
Graphical abstract
Keywords: Elution-free, Magnetic beads, Nucleic acid extraction, Pathogen detection, Nucleic acid amplification tests, Polyethylene-polypropylene glycol F68
Introduction
Nucleic acids, antigens, and antibodies are major biomarkers used for pathogen detection [1]. Although antigen-based tests can be easily carried out with test strips even by untrained users, their poor sensitivity makes it difficult for them to meet the demands of containing pathogen transmission [2, 3]. Additionally, antibody-based tests are not suitable for early and timely screening of pathogens, since antibodies are generally produced 2 weeks after infection [4]. In contrast, molecular detection of nucleic acids exhibits excellent sensitivity and specificity, while being effective in the earliest phase of the infection [5]. Thus, nucleic acid amplification tests (NAATs) based on polymerase chain reaction (PCR) and loop-mediated isothermal amplification (LAMP) have increasingly become the detection modality of choice in clinical diagnostics and food safety inspection [6, 7]. In particular, NAATs are the gold standard for COVID-19 diagnosis at present [8, 9]. Nucleic acid extraction is the key starting point for NAATs including two steps of nucleic acid isolation from samples and nucleic acid purification, which plays an extremely critical role in ensuring highly sensitive NAATs [10–12], as high concentrations of organic components that inhibit nucleic acid amplification are normally present in complex biological samples [13, 14]. In this context, the need for universal and efficient nucleic acid extraction methods for accurate disease diagnosis and foodborne pathogen detection cannot be overstated.
Magnetic beads (MBs) with a functionalized surface that enables nucleic acid capture have been widely used for nucleic acid extraction from biological samples, based on which a variety of formats have been developed, such as manual extraction by a magnetic rack or microfluidic chips [15, 16], as well as automated robotic processing [17, 18]. Compared with the traditional nucleic acid extraction approaches, such as spin column-based methods, thermal lysis, and alkaline lysis, MB-based methods combine the advantages of simple processing, short time consumption, high product purity, and easy integration for high-throughput detection, making them the preferred nucleic acid extraction methods for pathogen detection [19, 20]. In the process of using the traditional MB-based method, once the nucleic acids are captured on MBs, they are isolated from samples, followed by washing with alcohols and eluting into nuclease-free water, and then application as templates for NAATs. However, the elution process will lead to dilution and the loss of target nucleic acids, which may cause a reduction in sensitivity and false-negative results. In contrast, the approach in which nucleic acids together with MBs are directly used for downstream NAATs concentrates nucleic acid molecules and produces rapid and highly sensitive tests, thereby avoiding the elution step and simplifying the testing protocols. However, the commonly used MBs demonstrate nonspecific adsorption of organic components, inhibiting nucleic acid amplification in the biological samples and introducing them into the reaction systems, especially samples rich in organic ingredients, such as serum and meat [21, 22]. Therefore, elution-free MB-based methods are rarely reported, and those that have been reported have only been utilized for nucleic acid extraction from samples with low content of organic ingredients, such as nasopharyngeal swabs [23].
Herein, we developed a universal elution-free MB-based nucleic acid extraction method for multiple types of biological samples, which allowed the MBs to capture nucleic acids directly, adding to the available reaction systems for downstream NAATs. Polyethylene-polypropylene glycol (PEPPG) F68 was employed to reduce the nonspecific adsorption of organic interference components by the MBs. Moreover, sodium hydroxide solution instead of alcohol was utilized as the washing buffer to avoid the inhibition of downstream amplification reactions by alcohols, as well as their unpleasant smell. The ingredients of lysis buffers and extraction procedure were also optimized to improve the extraction efficiency and reduce residual organic interference components. Furthermore, the performance of this proposed elution-free MB-based method for pathogen detection was examined in four types of simulated samples, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-positive nasopharyngeal swabs, Streptococcus aureus-infected serum, Escherichia coli O157:H7-contaminated milk, and Salmonella typhimurium-contaminated pork, in comparison with a commercial kit based on the traditional MB-based method. Our work will provide a universal, rapid, and facile nucleic acid extraction method applicable for both automated robotic processing and manual operation, which can potentially be deployed for ultrafast and sensitive pathogen detection in various biological samples via NAATs both in the professional laboratory and at remote sites lacking instruments.
Materials and methods
Materials and reagents
SARS-CoV-2 pseudovirus was purchased from Fubio Biological Technology Co., Ltd (Shanghai, China). Staphylococcus aureus (ATCC 25923), E. coli O157:H7 (ATCC 35150), S. typhimurium (ATCC 14028), Vibrio parahaemolyticus (ATCC 17802), Pseudomonas aeruginosa (ATCC 27853), nonpathogenic E. coli (ATCC 25922) strains, inactivated Mycoplasma pneumoniae, inactivated influenza A (H1N1) virus, and inactivated influenza B (Victoria) virus, as well as colorimetric/fluorescence LAMP detection master mix, were provided by Navid Biotechnology Co., Ltd. (Qingdao, China). Hydroxyl (Si-OH) MBs with diameters of 100, 200, 500, and 1000 nm were purchased from BioMag Biotech Co., Ltd. (Wuxi China). PEPPG F68 and sodium hydroxide were purchased from Macklin Biochemical Co., Ltd. (Shanghai China). Guanidine hydrochloride, guanidine isothiocyanate, sodium chloride, EDTA-Na2, Tris, SDS, Triton X-100, Tween-20, and dNTPs were purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Taq DNA polymerase, 10× FastTaq buffer, and HiScript II Reverse Transcriptase were purchased from Vazyme Biotechnology Co., Ltd. (Nanjing, China). 31000 EvaGreen dye (20× in water) was purchased from Biotium, Inc. (CA, USA). Pig serum was obtained from Solarbio Life Sciences Co., Ltd. (Beijing, China). The TIANamp DNA/RNA Kit was purchased from Tiangen Biotech Co., Ltd. (Beijing, China). The BCA Protein Assay Kit was purchased from Beyotime Biotechnology Co., Ltd. (Shanghai, China). All the other chemicals and reagents were of analytical grade.
Preparation of the biological samples
Simulated SARS-CoV-2-positive nasopharyngeal swabs were prepared by spiking 100 μL SARS-CoV-2 pseudovirus suspensions with the target gene at concentrations of 1.0×104, 1.0×103, 1.0×102, and 1.0×101 copies/mL on fresh swabs of the nasal cavity or throat of volunteers. Simulated S. aureus-infected serum was prepared by mixing 100 μL S. aureus cell suspensions at concentrations of 1.0×108, 1.0×107, 1.0×106, 1.0×105, 1.0×104, 1.0×103, 1.0×102, and 1.0×101 CFU/mL into 900 μL pig serum. Simulated E. coli O157:H7-contaminated milk was prepared by mixing 100 μL E. coli O157:H7 cell suspensions at concentrations of 1.0×108, 1.0×107, 1.0×106, 1.0×105, 1.0×104, 1.0×103, and 1.0×102 CFU/mL into 900 μL sterilized commercial milk. Simulated S. typhimurium contaminated pork was prepared by immersing 10 mg sterilized commercial pork in 1 mL S. typhimurium cell suspensions at concentrations of 1.0×108, 1.0×107, 1.0×106, 1.0×105, 1.0×104, 1.0×103, and 1.0×102 CFU/mL for 15 min, which was then ground into tissue homogenate for subsequent use.
Elution-free nucleic acid extraction based on MBs
The protocol of the elution-free MB-based method was determined after optimization. Briefly, 200 μL of samples and 25 μL MB (diameter: 1000 nm) suspension (50 μg/μL) were thoroughly mixed with the lysis buffer (50 mM Tris, 10 mM EDTA, 0.7 M NaCl, 4 M guanidine isothiocyanate, 3 M guanidine hydrochloride, 0.1% [w/v] SDS, 5% [v/v] Tween-20, 3% [v/v] Triton X-100, 30% [w/v] PEPPG F68) in a 1.5-mL centrifuge tube by vortexing. Then the tube was placed at room temperature for 5 min, followed by placement on a magnetic rack and waiting 1 min until the MBs were concentrated. After removal of the supernatant, the MBs were washed with 600 μL NaOH solution (3 mM) for 15 s and collected again with the help of a magnetic rack. The MBs were directly transferred into reaction systems for downstream NAATs. Moreover, for comparison, the prepared biological samples were also treated with a TIANamp DNA/RNA Kit based on the traditional MB-based method according to the manufacturer’s instruction.
Primer design
Primer sets specific to the SARS-CoV-2 ORF1ab polyprotein (orf1ab) gene, S. aureus strain G9 thermonuclease precursor (nuc) gene, E. coli O157:H7 strain Shiga-toxin 1 (stx1) gene, and S. typhimurium strain invasion protein (invA) gene were designed and optimized by NCBI primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast) and NUPACK software (http://www.nupack.org/), and synthesized by Sangon Biotech (Shanghai, China). The sequences of the primer sets and fragments used in this work are shown in Table 1.
Table 1.
Sequences of nucleic acids used in this work
| Name | Sequence (5′–3′) | |
|---|---|---|
| SARS-CoV-2 orf1ab gene (aOP102293.1) | ||
| LAMP primer set [24] | F3 | CGGTGGACAAATTGTCAC |
| B3 | CTTCTCTGGATTTAACACACTT | |
| FIP | TCAGCACACAAAGCCAAAAATTTATTTTTCTGTGCAAAGGAAATTAAGGAG | |
| BIP | TATTGGTGGAGCTAAACTTAAAGCCTTTTCTGTACAATCCCTTTGAGTG | |
| LF | TTACAAGCTTAAAGAATGTCTGAACACT | |
| LB | TTGAATTTAGGTGAAACATTTGTCACG | |
| PCR primer set | F | TCACTAGGTTTCAAACTTTACTT |
| R | AGCTGTCCAACCTGAAG | |
| 77-nucleotide DNA fragment | TCACTAGGTTTCAAACTTTACTTGCTTTACATAGAAGTTATTTGACTCCTGGTGATTCTTCTTCAGGTTGGACAGCT | |
| S. aureus nuc gene (aDQ399678.1) | ||
| LAMP primer set [25] | F3 | ATGCAAAGAAAATTGAAGTCGA |
| B3 | GCGTTGTCTTCGCTCCAAAT | |
| FIP | CGTTTACCATTTTTCCATCAGCATAGTTTGACAAAGGTCAAAGAACT | |
| BIP | TCAAGGCTTGGCTAAAGTTGCTTATTTTCGCTTGTGCTTCACTT | |
| LF | TACGCTAAGCCACGTCCATA | |
| LB | CCTAACAATACACATGAACAAC | |
| PCR primer set | F | GTGCTGGCATATGTATGGC |
| R | CACTAAGCAACTAGTAGCG | |
| E. coli O157:H7 stx1 gene (aLC388498.1) | ||
| LAMP primer set [26] | F3 | TGTTGGAAGAATTTCTTTTGGA |
| B3 | GCTAATAGCCCTGCGTATC | |
| FIP | CGCGATGCATGATGATGACAATAGTGTTAATGCAATTCTGGGT | |
| BIP | GAGCTTCCTTCTATGTGCCCGCAGAGTGGATGAGTCCCA | |
| LF | TCGCACCGTAATTATGACT | |
| LB | AGATGGAAGAGTGCGTGGG | |
| PCR primer set | F | ATTCTGGGTAGCGTGGCATT |
| R | CTCTTCCATCTACCGGGCAC | |
| S. typhimurium invA gene (aM90846.1) | ||
| LAMP primer set [27] | F3 | CTGGACATTGTTGATTCAGGTA |
| B3 | ACATCACGGTAGCTCAGA | |
| FIP | GAAGGTGCCGAGAATAGCCAGCAAGTTCAACGCGCAATT | |
| BIP | TTCGTGAAACGCTGAAYGGAACTGACGACGGGTTAAATTAG | |
| LF | TCCGCCTGGTAACGAGTA | |
| LB | TCACCAGGAGATTACAACATGG | |
| PCR primer set | F | CAATGGCGGCGAATTACGAG |
| R | AGGAAGGTACTGCCAGAGGT | |
aGenBank accession number
LAMP reactions
The collected MBs containing the captured nucleic acids were directly used for performing colorimetric and fluorescence quantitative real-time LAMP (qLAMP). In brief, all the collected MBs (approximately 1250 μg) were transferred into a 50-μL amplification mixture containing 40.6 μL colorimetric/fluorescence LAMP detection master mix, 1 μL F3 and B3 (10 μM), 0.8 μL FIP and BIP (100 μM), 0.4 μL LF and LB (100 μM), and 0.6 μL HiScript II Reverse Transcriptase (200 U/μL, for RNA templates only). Additionally, 5 μL of nucleic acid products prepared by the commercial kit were added into the same amplification mixture for comparison. Fluorescence qLAMP was performed using a CFX Connect™ Real-Time PCR System (Bio-Rad, CA, USA) at 65°C for 45 min, and monitored at 1-min intervals. The threshold was set at 10 times the standard deviation of the fluorescence values of the baseline. The time threshold (Tt) value was defined as the time point when the fluorescence values exceeded the threshold. Colorimetric LAMP assays were performed at 65°C for 50 min in a heating block, and the results were directly read by the naked eye after placing the tubes on white cardboard.
PCR reactions
Besides LAMP assays, the nucleic acid products prepared using the commercial kit were also employed as templates for fluorescence quantitative real-time PCR (qPCR) to simulate the most commonly used NAAT process at present. Briefly, 2 μL of the nucleic acid products prepared by the commercial kit were added into a 20-μL reaction mixture containing 0.4 μL dNTPs (10 mM), 0.8 μL each for forward and backward primer (10 μM), 0.2 μL Taq DNA polymerase (5 U/μL), 3 μL 10× FastTaq buffer, 0.5 μL EvaGreen dye, and 0.15 μL HiScript II Reverse Transcriptase (200 U/μL, for RNA templates only). The reaction procedure included denaturation at 95°C for 3 min and 40 cycles consisting of 95°C for 10 s, and 58°C for 30 s for amplification. For RNA templates, a reverse transcription step of incubating the reaction system at 55°C for 5 min was added before the denaturation step in the reaction procedure. Fluorescence real-time PCR was performed by a CFX Connect™ Real-Time PCR System (Bio-Rad, CA, USA) and monitored after each thermal cycling. The threshold was set at 10 times the standard deviation of the fluorescence values of the baseline. The cycle threshold (Ct) value was defined as the number of cycles required for the fluorescence values to exceed the threshold.
Protein and nucleic acid absorption capability assays
The influence of PEPPG F68 on the protein and nucleic acid absorption capability of the MBs was investigated. In brief, S. aureus cell suspensions with a concentration of 3.0×108 CFU/mL were mixed with the MBs and lysis buffer with/without PEPPG F68, respectively. After incubation for 5 min, the residual protein content of the mixtures was measured by a BCA protein assay kit. Moreover, the protein content of the mixtures without the MBs was also measured to evaluate the influence of PEPPG F68 on BCA protein assays. For nucleic acid absorption capability assay, a 77-nucleotide nucleic acid fragment containing the partial sequence of SARS-CoV-2 orf1ab gene was designed, and then the nucleic acid fragment solutions at concentrations of 1.0×10−12, 1.0×10−11, 1.0×10−10, 1.0×10−9, and 1.0×10−8 M were mixed with the MBs and lysis buffer with/without PEPPG F68, respectively. The MBs were washed with the washing buffer and transferred into the PCR reaction system. The influence of PEPPG F68 on nucleic acid absorption was evaluated by comparing Ct values for the MBs incubated in the lysis buffer with/without PEPPG F68.
Results and discussion
Optimization of the MB-based elution-free method
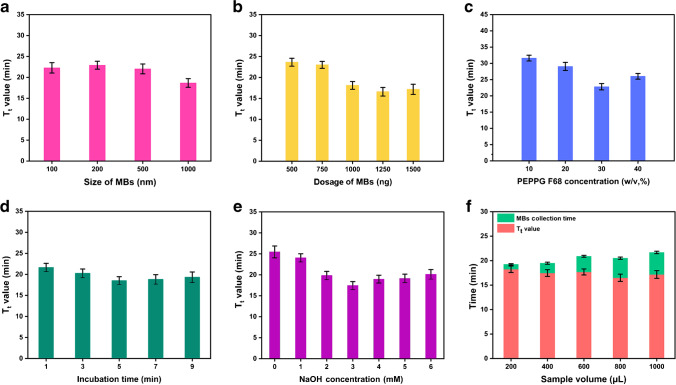
Before the protocol of the elution-free method was determined, it was optimized using fluorescence qLAMP with S. typhimurium (1.0×106 CFU/mL) as a template. First, the effects of particle size of the MBs on the nucleic acid extraction efficiency were explored. As shown in Fig. 1a, MBs with a diameter of 1000 nm exhibited the best performance with the lowest Tt value, while the performance of smaller beads showed no evidence of differences. Despite the larger MBs possessing relatively smaller specific surface area, a higher amount of Si-OH was coated on a single magnetic bead, which may be beneficial for the macromolecules like nucleic acid to be more stably adsorbed on the MBs. The lower performance of the 100-nm, 200-nm, and 500-nm MBs was likely due to the lower amount of Si-OH on their surface. Besides appropriate particle size, a sufficient dosage of the MBs would also improve the extraction efficiency. The Tt value decreased significantly with the increase in the dosage of MBs from 500 μg to 1250 μg, and failed to decrease further and increased slightly when the dosage of MBs was increased to 1500 μg (Fig. 1b). This was because the higher dosage of MBs introduced more organic interference components into the reaction system, leading to excessive absorption of LAMP components like primers and polymerase and resulting in reduced amplification efficiency. Thus, 1250 μg was determined as the optimal dosage of MBs.
Fig. 1.
Optimization of the protocol for elution-free MB-based method. Effect of (a) the MB size, (b) the MB dosage, (c) the PEPPG F68 concentration in lysis buffer, (d) the incubation time for MBs and samples in lysis buffer, (e) the NaOH concentration in washing buffer, and (f) the sample volume applied for nucleic acid extraction on the efficiency of LAMP assays
In this work, PEPPG F68 was applied in lysis buffer to reduce the absorption of protein ingredients of the samples, the major organic interference components that inhibit the amplification reaction, such as immunoglobulin, protease, mucin, and hemoglobin [28–30]. Therefore, the concentration of PEPPG F68 in the lysis buffer was also optimized. As shown in Fig. 1c, the Tt value of the nucleic acids isolated by the lysis buffer with 30% (w/v) PEPPG F68 was 22.55 min, whereas the values for nucleic acids isolated by the lysis buffer with 10% and 20% (w/v) PEPPG F68 were approximately 8 min greater, demonstrating that the application of PEPPG F68 indeed promoted NAAT efficiency. However, the lysis buffer with 40% (w/v) PEPPG F68 failed to further reduce the Tt value, likely because, in addition to proteins, this surfactant can also weaken the electrostatic interaction between nucleic acids and Si-OH on the surface of the MBs. Moreover, the application of PEPPG F68 would increase the viscosity of the lysis buffer, and thus excessive PEPPG F68 content would lead to inconvenient liquid handling and prolonged MB gathering time. Sufficient incubation time is of great importance for the complete lysis of cells and virions, and the release of genomic materials. The Tt value of the samples decreased from 21.65 min to 18.53 min with the increase in the incubation time from 1 min to 5 min. However, no further decrease in the Tt value was observed when the incubation time was extended to 7 min or 9 min (Fig. 1d). Accordingly, the 30% (w/v) PEPPG F68 was applied in the lysis buffer, and the optimal incubation time in lysis buffer was determined to be 5 min.
Besides the lysis buffer, the washing buffer was also optimized. The results illustrated that the Tt value decreased sharply with the increase in the NaOH concentration when the MBs were washed with the NaOH solution of no more than 3 mM, and increased continuously with the increase in the NaOH concentration from 3 mM to 6 mM since a high-pH environment is not beneficial for the capture of nucleic acid by the Si-OH on MBs (Fig. 1e). Finally, considering that the MBs carrying nucleic acids are directly used for downstream amplification, we explored the influence of the volume of initial samples on extraction efficiency. As expected, the Tt value was decreased slightly with increased sample input from 200 μL to 1000 μL, revealing that the nucleic acids captured by the MBs increased. However, the Tt value decreased only 1.74 min after the total input sample volume was increased fourfold, while the time consumption for MB collection by the magnetic rack was sharply increased from 1 min to 4.5 min due to the more dispersed MBs in larger-volume systems (Fig. 1f). Since both the total time and sample consumption were the lowest, the 200-μL sample was applied for each test. In all, the protocol was determined after the optimization, as shown in section 2.3. The total time consumption of the extraction process was only approximately 7 min, which was 20–25 min less than the traditional MB-based approaches, such as the commercial kit involved in this work.
Extraction efficiency evaluation of the elution-free MB-based method
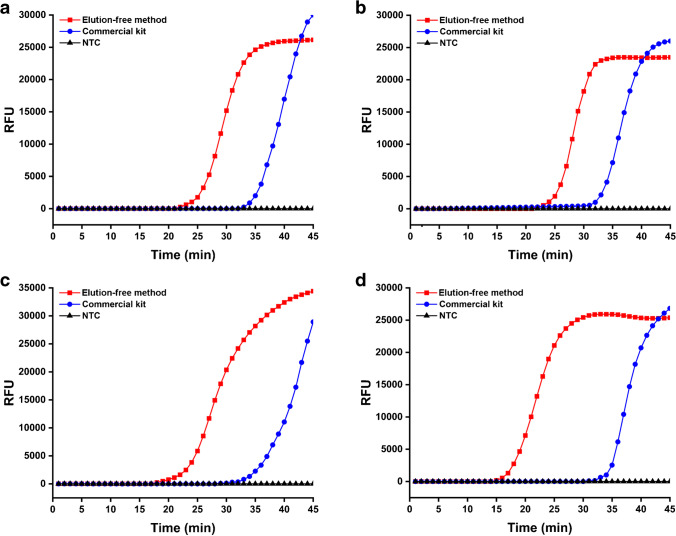
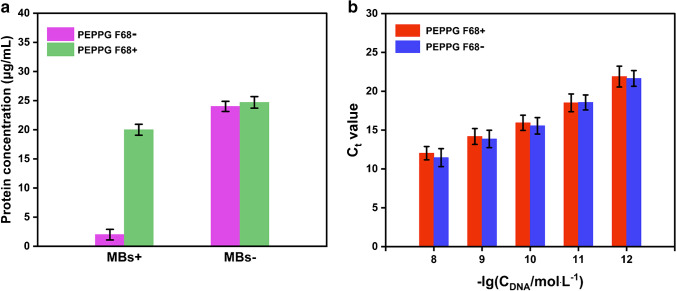
Nucleic acid extracts from the four kinds of biological samples prepared by both the elution-free MB-based method and the commercial kit were employed as the templates for fluorescence qLAMP assays. As shown in Fig. 2, fluorescence signal accumulation occurred in all assays, demonstrating that both approaches successfully isolated target nucleic acids from the four kinds of biological samples. Moreover, the Tt values for the nucleic acids prepared by the elution-free MB-based method were significantly smaller than those prepared by the commercial kit. Specifically, the average Tt values for the nucleic acids prepared by the elution-free MB-based method were 22.38 min for the simulated SARS-CoV-2-positive nasopharyngeal swabs (104 copies/mL), 22.86 min for the simulated S. aureus-infected serum (106 CFU/mL), 17.92 min for the simulated E. coli O157:H7-contaminated milk (106 CFU/mL), and 15.58 min for the simulated S. typhimurium-contaminated pork (106 CFU/mL), while those for the nucleic acids prepared by the commercial kit were 34.64 min, 35.11 min, 35.08 min, and 39.24 min, respectively (Fig. 2), indicating that the elution-free MB-based method could greatly improve the efficiency of LAMP assays. Notably, the elution-free MB-based extraction method showed better performance for the nucleic acid isolation from the samples with high content of organic interference components, as the Tt values were approximately 17 min and 24 min earlier for milk and pork samples, respectively, but only approximately 12 min earlier for swab and serum samples. For one thing, this method could more effectively enrich the nucleic acids and thereby increase the amount of target nucleic acids added to the reaction system. Moreover, the addition of PEPPG F68 in the lysis buffer effectively reduced the organic interference components introduced into the reaction systems, while having no effect on the enrichment of the target nucleic acids on the MBs. To validate our conjecture, the influence of PEPPG F68 on protein and nucleic acid adsorption capacity of MBs was evaluated. The results of BCA protein assays showed that the addition of PEPPG F68 strongly inhibited the absorption of protein on MBs, exhibiting the significantly higher protein content of the S. aureus cell suspensions of 3.0×108 CFU/mL treated by lysis buffer with PEPPG F68 than those treated by lysis buffer without PEPPG F68. Moreover, the protein content of the mixtures with and without PEPPG F68 showed no significant differences when no MBs were added to the mixtures, demonstrating that the presence of PEPPG F68 did not promote cell lysis or influence BCA protein assays (Fig. 3a). In contrast, the Ct values for MBs incubated in lysis buffer with or without PEPPG F68 showed no significant differences, demonstrating that this surfactant at this dose would not affect the nucleic acid absorption on MBs (Fig. 3b). With the help of this low-cost surfactant (approximately $0.13/g), this elution-free MB-based method involved no specially designed MBs and required only ordinary commercial Si-OH MBs (approximately $0.2/mg), ensuring that the cost of this method was about the same as that of traditional MB-based methods.
Fig. 2.
Application of the elution-free MB-based method on pathogen detection from multiple types of simulated samples. Fluorescence curves of the qLAMP assays for the nucleic acid products prepared by the elution-free MB-based method and the commercial kit from simulated (a) SARS-CoV-2-positive nasopharyngeal swabs, (b) S. aureus-infected serum, (c) E. coli O157:H7-contaminated milk, and (d) S. typhimurium-contaminated pork. NTC represents no template control
Fig. 3.
Influence of PEPPG F68 in lysis buffer on the absorption capability of the Si-OH MBs to (a) protein and (b) nucleic acid content. PEPPG F68+ and PEPPG F68− represent lysis buffers with and without PEPPG F68, respectively. MBs+ and MBs− represent mixtures with and without MBs, respectively
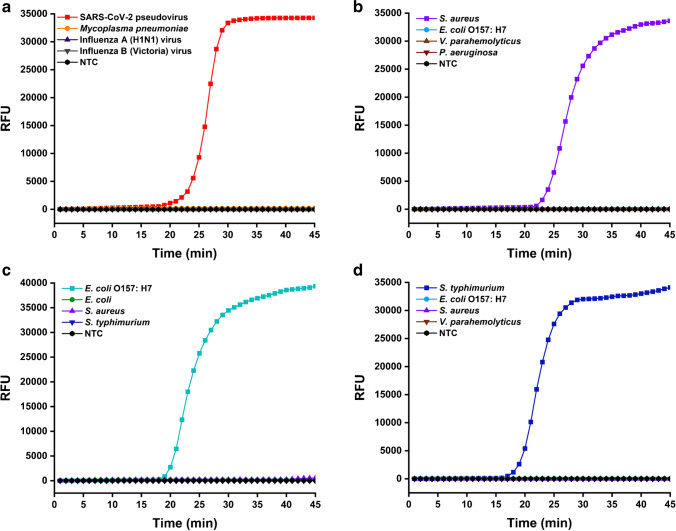
Besides introducing PEPPG F68 in lysis buffer, the alternation of washing buffer from alcohol to NaOH solution also contributed to the improvement in LAMP assay efficiency, as the cells or virions in the samples could be further lysed by NaOH during the washing process. Additionally, the interaction between nucleic acids and Si-OH of the MBs weakened in alkaline solution, allowing the nucleic acids to easily detach from MBs and participate in the LAMP reaction when they were transferred into the reaction system. Furthermore, since the LAMP reaction has better tolerance to 3 mM NaOH solution than alcohols, the MBs could be directly added into the reaction system without a drying step, which further simplified the protocol and reduced the probability of aerosol pollution and nonspecific amplification. As shown in Fig. 4, by combination with this elution-free MB-based method, the LAMP assays could specifically identify the target pathogen from the nucleic acid products of multiple types of biological samples spiked with the same amount of other pathogens that are frequently detected in corresponding biological samples, illustrating that the treatment of this method did not cause nonspecific amplification. In summary, by combination with the elution-free MB-based method, the entire pathogen detection process from sample to result could be completed within 30 min while exhibiting excellent specificity, which was approximately 40 min faster than the detection process involving traditional MB-based methods.
Fig. 4.
Specificity validation of the qLAMP assays combined with the elution-free MB-based method for pathogen detection in multiple types of biological samples. (a) SARS-CoV-2 pseudovirus detection in nasopharyngeal swabs spiked with SARS-CoV-2 pseudovirus, M. pneumoniae, influenza A (H1N1) virus, and influenza B (Victoria) virus. (b) S. aureus detection in serum spiked with S. aureus, E. coli O157:H7, V. parahaemolyticus, and P. aeruginosa. (c) E. coli O157:H7 detection in milk contaminated with E. coli O157:H7, nonpathogenic E. coli, S. aureus, and S. typhimurium. (d) S. typhimurium detection in pork contaminated with S. typhimurium, E. coli O157:H7, S. aureus, and V. parahaemolyticus. NTC represents no template control
Sensitivity of the LAMP assays combined with the elution-free MB-based method
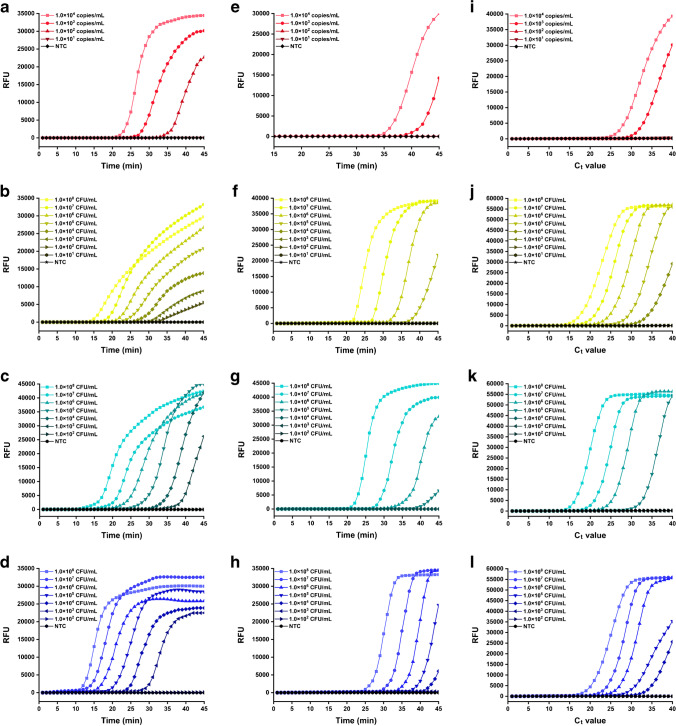
Apart from excellent efficiency and specificity, high sensitivity is another important property of NAATs to ensure successful diagnoses. Thus, the sensitivity of the LAMP assays combined with the elution-free MB-based method was explored next. Biological samples described in section 2.2 were treated with the elution-free MB-based method and the commercial kit for nucleic acid extraction, and the products were employed as templates for the following fluorescence qLAMP and colorimetric LAMP assays. As shown in Fig. 5a and e, the qLAMP assays with the nucleic acid products prepared by the elution-free MB-based method as templates showed higher sensitivity than those with the products prepared by the commercial kit. The products of the simulated SARS-CoV-2-positive nasopharyngeal swabs with 1.0×104, 1.0×103, and 1.0×102 copies/mL of target nucleic acid were detected as positive by the fluorescence qLAMP assays combined with the elution-free MB-based method, while only the samples with 1.0×104 and 1.0×103 copies/mL target nucleic acid were detected as positive by the fluorescence qLAMP assays combined with the commercial kit. The results demonstrated that the elution-free MB-based method could improve the sensitivity of fluorescence qLAMP assays to this RNA target by one order of magnitude. Moreover, fluorescence qLAMP assays combined with the elution-free MB-based method exhibited much better performances for DNA targets detection, as the limit of detection (LOD) for the S. aureus nuc gene, E. coli O157:H7 stx1 gene, and S. typhimurium invA gene in different simulated biological samples was 1.0×102 CFU/mL, 1.0×103 CFU/mL, and 1.0×103 CFU/mL (Fig. 5b–d), respectively, while the LOD of the fluorescence qLAMP assays combined with the commercial kit to the same targets was 1.0×105 CFU/mL, 1.0×106 CFU/mL, and 1.0×105 CFU/mL, respectively (Fig. 5f–h), suggesting the elution-free MB-based extraction method could improve the sensitivity to these DNA targets by two to three orders of magnitude as compared with the traditional MB-based method. Besides lower LOD, the Tt values of the fluorescence qLAMP assays combined with the elution-free MB-based method were significantly smaller than those combined with the commercial kit for the same targets, which was consistent with the results of extraction efficiency evaluation in section 3.2.
Fig. 5.
Sensitivity validation of (a–d) the fluorescence qLAMP assays combined with the elution-free MB-based method, (e–h) the fluorescence qLAMP assays combined with the commercial kit, and (i–l) the fluorescence qPCR assays combined with the commercial kit for pathogen detection in multiple types of biological samples. (a, e, and i) SARS-CoV-2 pseudovirus detection in nasopharyngeal swabs. (b, f, and j) S. aureus detection in serum. (c, g, and k) E. coli O157:H7 detection in milk. (d, h, and l) S. typhimurium detection in pork. NTC represents no template control
Unfortunately, the products prepared by the elution-free MB-based method could not be used as templates for qPCR assays, as almost no fluorescence signal accumulation was observed in the reactions, which might be attributed to the inhibition of the reaction by the Fe3+ released from the MBs during the high-temperature thermal cycling process, since this heavy metal ion can strongly inhibit qPCR at a concentration of 3 mg/L [31]. Therefore, only the nucleic acid products prepared by the commercial kit based on the traditional MB-based method were employed as templates for qPCR assays for the comparison, which is the most commonly used NAATs protocol at present [32]. As shown in Fig. 5i–l, by combination with the elution-free MB-based method, the sensitivity of the qLAMP assays to the target bacteria in the biological samples was even higher than the qPCR assays, whose LOD to SARS-CoV-2, S. aureus, E. coli O157:H7, and S. typhimurium was 1.0×103 copies/mL, 1.0×104 CFU/mL, 1.0×105 CFU/mL, and 1.0×104 CFU/mL, respectively. The results further indicated that the elution-free MB-based method was more effective for nucleic acid extraction from samples with high content of organic interference components, such as serum, milk, and pork, involved in this work than the traditional ones.
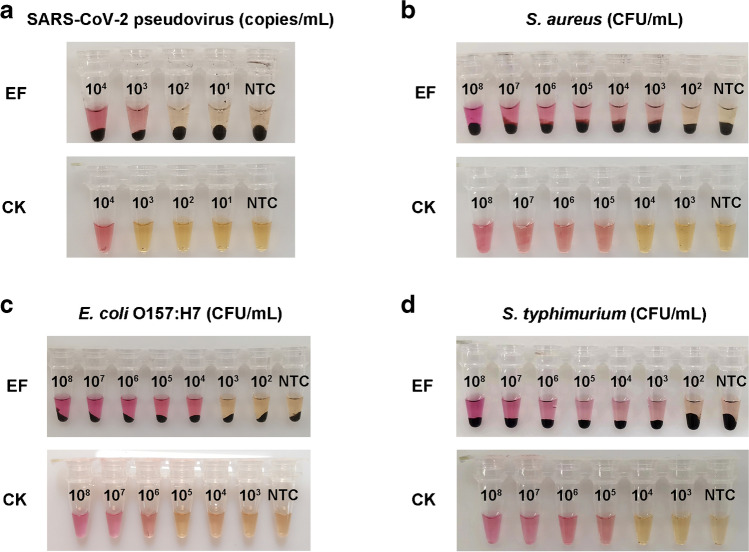
In addition to fluorescence qLAMP assays, the elution-free MB-based method improved the sensitivity of the colorimetric LAMP assays by one order of magnitude for the RNA targets and two to three orders of magnitude for DNA targets (Fig. 6). While the sensitivity of the colorimetric LAMP assays was still lower than the fluorescence qLAMP assays and qPCR in general, neither the colorimetric LAMP nor the elution-free MB-based method requires sophisticated bulky instruments or complicated protocols, and their combination provides a fast and facile approach suitable for on-site pathogen detection from sample to result in remote areas lacking instruments or professional staff, which is of great importance for the timely control of pathogen transmission. In all, the more concentrated nucleic acids and lower levels of organic interference components in the products prepared by the elution-free MB-based methods as compared with the traditional methods not only resulted in higher detection efficiency but also improved the sensitivity of the LAMP assays.
Fig. 6.
Sensitivity validation of the colorimetric LAMP assays combined with the elution-free MB-based method and the commercial kit for pathogen detection in multiple types of biological samples. (a) SARS-CoV-2 pseudovirus detection in nasopharyngeal swabs. (b) S. aureus detection in serum. (c) E. coli O157:H7 detection in milk. (d) S. typhimurium detection in pork. EF and CK represent the elution-free MB-based method and commercial kit, respectively. NTC represents no template control
Conclusion
Herein, an elution-free MB-based nucleic acid extraction method was established by introducing PEPPG F68 in lysis buffer and using NaOH solution instead of alcohols as washing buffer, which was utilized for pathogen detection in multiple types of biological samples via NAATs. Following nucleic acid isolation and purification, the nucleic acid/Si-OH MB composite can be directly added to the reaction systems as templates for LAMP assays. Compared with the traditional MB-based method, this proposed elution-free approach not only simplified the extraction protocol and reduced the time consumption, but also enhanced the efficiency and sensitivity of LAMP assays. The LOD of both fluorescence qLAMP and colorimetric LAMP assays combined with this elution-free MB-based method showed an improvement of one to three orders of magnitude compared with those combined with the traditional MB-based method, and the fluorescence qLAMP assays even exhibited higher sensitivity than the most widely used qPCR gold-standard assay for pathogen detection in samples with high content of organic interference components, despite the inability to use the products of this extraction method for qPCR assays. Moreover, the time consumption of the entire detection process from sample to result was less than 30 min, which was much shorter than the most commonly used approaches at present. In addition, this elution-free MB-based method was robust and reliable for pathogen detection in a variety of biological samples, in contrast with those that were only applicable for samples with low content of organic interference components, such as nasopharyngeal swabs, thus contributing to the progress of analytical chemistry applied for nucleic acid analysis in biological samples. Finally, the execution of nucleic acid extraction via this method did not require sophisticated bulky equipment or specialized instruments, making it easy to integrate into automated robotic processing, as well as for manual extraction by microfluidic chips and a magnetic rack, supporting its potential for pathogen detection from various biological samples via NAATs in both the centralized laboratory and remote sites.
Acknowledgment
We highly appreciate the financial support of the Major Scientific and Technological Innovation Projects of Shandong Province, the Science and Technology Benefiting the People Demonstration Project of Qingdao, and the Key Project of Shandong Province Natural Science Foundation, as well as the volunteers helping us prepare the simulated SARS-CoV-2-positive nasopharyngeal swabs.
Authors' contributions
Qianqian Jiang, Jinling Guo, and Ailin Wang performed the experiments; Qianqian Jiang and Yang Li analyzed the data; Yang Li, Cuiping Ma, Chao Shi, and Lin Huang designed the study; Yang Li and Qianqian Jiang wrote the manuscript; and all authors contributed to the writing of the paper, had primary responsibility for the final content, and read and approved the final manuscript.
Funding
This work is supported by the Major Scientific and Technological Innovation Projects of Shandong Province (2022SFXGFY01), the Science and Technology Benefiting the People Demonstration Project of Qingdao (22-2-7-smjk-2-nsh), and the Key Project of Shandong Province Natural Science Foundation (ZR2020KH030).
Declarations
Ethics approval
The authorized Human Health and Ethics Committee of the Affiliated Hospital of Qingdao University approved this study, and all the volunteers helping us prepare the simulated SARS-CoV-2-positive nasopharyngeal swabs provided informed consent. In addition, all methods were carried out in accordance with the relevant guidelines and regulations.
Conflicts of interest
There are no conflicts of interest to declare.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qianqian Jiang and Yang Li contributed equally to this work.
References
- 1.Milavec M, Cleveland MH, Bae YK, Wielgosz RI, Vonsky M, Huggett JF. Metrological framework to support accurate, reliable, and reproducible nucleic acid measurements. Anal Bioanal Chem. 2022;414(2):791–806. doi: 10.1007/s00216-021-03712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mockel M, Corman VM, Stegemann MS, Hofmann J, Stein A, Jones TC, et al. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers. 2021;26(3):213–220. doi: 10.1080/1354750X.2021.1876769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, et al. Diagnostics for SARS-CoV-2 infections. Nat Mater. 2021;20(5):593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li K, Huang B, Wu M, Zhong A, Li L, Cai Y, et al. Dynamic changes in anti-SARS-CoV-2 antibodies during SARS-CoV-2 infection and recovery from COVID-19. Nat Commun. 2020;11(1):6044. doi: 10.1038/s41467-020-19943-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams NM, Bordelon H, Wang K-KA, Albert LE, Wright DW, Haselton FR. Comparison of Three Magnetic Bead Surface Functionalities for RNA Extraction and Detection. ACS Appl Mater Interfaces. 2015;7(11):6062–6069. doi: 10.1021/am506374t. [DOI] [PubMed] [Google Scholar]
- 6.Wei C, Zhong J, Hu T, Zhao X. Simultaneous detection of Escherichia coli O157:H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech. 2018;8(1):76. doi: 10.1007/s13205-018-1086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein S, Muller TG, Khalid D, Sonntag-Buck V, Heuser AM, Glass B, et al. SARS-CoV-2 RNA Extraction Using Magnetic Beads for Rapid Large-Scale Testing by RT-qPCR and RT-LAMP. Viruses. 2020;12(8). 10.3390/v12080863. [DOI] [PMC free article] [PubMed]
- 8.Niu C, Wang X, Gao Y, Qiao X, Xie J, Zhang Y, et al. Accurate quantification of SARS-CoV-2 RNA by isotope dilution mass spectrometry and providing a correction of reverse transcription efficiency in droplet digital PCR. Anal Bioanal Chem. 2022. 10.1007/s00216-022-04238-6. [DOI] [PMC free article] [PubMed]
- 9.Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearlman SI, Leelawong M, Richardson KA, Adams NM, Russ PK, Pask ME, et al. Low-Resource Nucleic Acid Extraction Method Enabled by High-Gradient Magnetic Separation. ACS Appl Mater Interfaces. 2020;12(11):12457–12467. doi: 10.1021/acsami.9b21564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hapsianto BN, Kojima N, Kurita R, Yamagata H, Fujita H, Fujii T, et al. Direct Capture and Amplification of Small Fragmented DNAs Using Nitrogen-Mustard-Coated Microbeads. Anal Chem. 2022;94(21):7594–7600. doi: 10.1021/acs.analchem.2c00531. [DOI] [PubMed] [Google Scholar]
- 12.Katevatis C, Fan A, Klapperich CM. Low concentration DNA extraction and recovery using a silica solid phase. PLoS One. 2017;12(5):e0176848. doi: 10.1371/journal.pone.0176848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Point of Care Diagnostics: Status and Future. Anal Chem. 2012;84(2):487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- 14.Nacham O, Clark KD, Anderson JL. Extraction and Purification of DNA from Complex Biological Sample Matrices Using Solid-Phase Microextraction Coupled with Real-Time PCR. Anal Chem. 2016;88(15):7813–7820. doi: 10.1021/acs.analchem.6b01861. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Y, Zhang D, Qi P. Combination of a flow cytometric bead system with 16S rRNA-targeted oligonucleotide probes for bacteria detection. Anal Bioanal Chem. 2019;411(10):2161–2168. doi: 10.1007/s00216-019-01651-2. [DOI] [PubMed] [Google Scholar]
- 16.Ritzi-Lehnert M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert Rev Mol Diagn. 2012;12(2):189–206. doi: 10.1586/erm.11.98. [DOI] [PubMed] [Google Scholar]
- 17.Knepp Julia H, Geahr Melissa A, Forman Michael S, Valsamakis A. Comparison of Automated and Manual Nucleic Acid Extraction Methods for Detection of Enterovirus RNA. J Clin Microbiol. 2003;41(8):3532–3536. doi: 10.1128/JCM.41.8.3532-3536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi A, Matsuda K, Uehara M, Honda T, Saito Y. A novel automated device for rapid nucleic acid extraction utilizing a zigzag motion of magnetic silica beads. Anal Chim Acta. 2016;906:1–6. doi: 10.1016/j.aca.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Kang J, Li Y, Zhao Y, Wang Y, Ma C, Shi C. Nucleic acid extraction without electrical equipment via magnetic nanoparticles in Pasteur pipettes for pathogen detection. Anal Biochem. 2021;635:114445. doi: 10.1016/j.ab.2021.114445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathy S, Chalana AK, Talukdar A, Rajesh PV, Saha A, Pramanik G, et al. Limited-resource preparable chitosan magnetic particles for extracting amplification-ready nucleic acid from complex biofluids. Analyst. 2022;147(1):165–177. doi: 10.1039/d1an01150b. [DOI] [PubMed] [Google Scholar]
- 21.Hao Q, Xu Q, Niu S, Ding C, Luo X. Anti-Fouling Magnetic Beads Combined with Signal Amplification Strategies for Ultra-Sensitive and Selective Electrochemiluminescence Detection of MicroRNAs in Complex Biological Media. Anal Chem. 2021;93(30):10679–10687. doi: 10.1021/acs.analchem.1c02186. [DOI] [PubMed] [Google Scholar]
- 22.Leuko S, Goh F, Ibanez-Peral R, Burns BP, Walter MR, Neilan BA. Lysis efficiency of standard DNA extraction methods for Halococcus spp. in an organic rich environment. Extremophiles. 2008;12(2):301–308. doi: 10.1007/s00792-007-0124-8. [DOI] [PubMed] [Google Scholar]
- 23.He Y, Xie T, Tong Y. Rapid and highly sensitive one-tube colorimetric RT-LAMP assay for visual detection of SARS-CoV-2 RNA. Biosens Bioelectron. 2021;187:113330. doi: 10.1016/j.bios.2021.113330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suarez GD, Suarez DA, Kiu Tang YY, Zhang J-X, Li J, Nagl S, et al. Uncovering mechanisms of RT-LAMP colorimetric SARS-CoV-2 detection to improve assay reliability. Anal Methods-Uk. 2022;14(4):378–382. doi: 10.1039/d1ay01395e. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Li H, Wang Y, Zhang L, Xu J, Ye C. Loop-Mediated Isothermal Amplification Label-Based Gold Nanoparticles Lateral Flow Biosensor for Detection of Enterococcus faecalis and Staphylococcus aureus. Front Microbiol. 2017;8:192. doi: 10.3389/fmicb.2017.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara-Kudo Y, Nemoto J, Ohtsuka K, Segawa Y, Takatori K, Kojima T, et al. Sensitive and rapid detection of Vero toxin-producing Escherichia coli using loop-mediated isothermal amplification. J Med Microbiol. 2007;56(Pt 3):398–406. doi: 10.1099/jmm.0.46819-0. [DOI] [PubMed] [Google Scholar]
- 27.Kreitlow A, Becker A, Schotte U, Malorny B, Plötz M, Abdulmawjood A. Establishment and validation of a loop-mediated isothermal amplification (LAMP) assay targeting the ttrRSBCA locus for rapid detection of Salmonella spp. in food. Food Control. 2021;126:107973. doi: 10.1016/j.foodcont.2021.107973. [DOI] [Google Scholar]
- 28.Sidstedt M, Hedman J, Romsos EL, Waitara L, Wadsö L, Steffen CR, et al. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal Bioanal Chem. 2018;410(10):2569–2583. doi: 10.1007/s00216-018-0931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubo I, Kajiya M, Aramaki N, Furutani S. Detection of Salmonella Enterica in Egg Yolk by PCR on a Microfluidic Disc Device Using Immunomagnetic Beads. Sensors. 2020;20(4). 10.3390/s20041060. [DOI] [PMC free article] [PubMed]
- 30.Ozalp VC, Bayramoglu G, Kavruk M, Keskin BB, Oktem HA, Arica MY. Pathogen detection by core–shell type aptamer-magnetic preconcentration coupled to real-time PCR. Anal Biochem. 2014;447:119–125. doi: 10.1016/j.ab.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 31.Chen N-T, Chang C-W. Quantification of Legionella pneumophila by real-time quantitative PCR from samples with humic acid and ferric ion. Sci Total Environ. 2012;414:608–613. doi: 10.1016/j.scitotenv.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Scarabotto A, Balestro S, Gagliardi S, Trotti R. Comparison of Two RNA Extraction Methods for the Molecular Detection of SARS-CoV-2 from Nasopharyngeal Swab Samples. Diagnostics. 2022;12(7). 10.3390/diagnostics12071561. [DOI] [PMC free article] [PubMed]