Abstract
Background and purpose
Deep learning contouring (DLC) has the potential to decrease contouring time and variability of organ contours. This work evaluates the effectiveness of DLC for prostate and head and neck across four radiotherapy centres using a commercial system.
Materials and methods
Computed tomography scans of 123 prostate and 310 head and neck patients were evaluated. Besides one head and neck model, generic DLC models were used. Contouring time using centres’ existing clinical methods and contour editing time after DLC were compared. Timing was evaluated using paired and non-paired studies. Commercial software or in-house scripts assessed dice similarity coefficient (DSC) and distance to agreement (DTA). One centre assessed head and neck inter-observer variability.
Results
The mean contouring time saved for prostate structures using DLC compared to the existing clinical method was 5.9 ± 3.5 min. The best agreement was shown for the femoral heads (median DSC 0.92 ± 0.03, median DTA 1.5 ± 0.3 mm) and the worst for the rectum (median DSC 0.68 ± 0.04, median DTA 4.6 ± 0.6 mm). The mean contouring time saved for head and neck structures using DLC was 16.2 ± 8.6 min. For one centre there was no DLC time-saving compared to an atlas-based method. DLC contours reduced inter-observer variability compared to manual contours for the brainstem, left parotid gland and left submandibular gland.
Conclusions
Generic prostate and head and neck DLC models can provide time-savings which can be assessed with paired or non-paired studies to integrate with clinical workload. Reducing inter-observer variability potential has been shown.
Keywords: Auto-contouring, Deep learning contouring, Multi-centre, Organs at risk
1. Introduction
It is important to have accurate organ at risk (OAR) contours for radiotherapy planning to ensure healthy tissue is spared. Techniques such as volumetric modulated radiotherapy or intensity modulated radiotherapy allow highly conformal dose distributions with steep dose gradients to be created so it is imperative that the contours are accurate. The accuracy of OAR contouring has also been shown to correlate with toxicity [1], [2]. Manual methods of contouring are very labour intensive and there is significant inter and intra-observer variation [3], [4], [5]. In addition, the contour quality and contouring time can depend on the experience of the user [6].
An alternative method is to use atlas-based auto-contouring which can reduce contouring time and improve consistency for sites such as head and neck, prostate, and lung [7], [8], [9]. However, atlas-based contouring is limited by the accuracy of the deformable image registration [10] and the limited range of patients within an atlas [11]. In addition, atlas-based auto-contouring has been shown to be inferior for small and thin OARs [12], as for small structures a minor error in deformable registration would make a bigger difference than for larger structures.
Recent advances in artificial intelligence have created further techniques in the form of deep learning. These use neural networks trained on large datasets of contoured images to improve contouring accuracy and once trained are quicker to use than atlas-based methods [13]. Although also limited by numbers of patients, deep learning is based on larger datasets than atlas-based methods, with larger datasets improving model accuracy without reducing the speed. Atlas-based models however, will reduce in speed when more datasets are added. Studies have shown that deep learning outperforms manual and atlas-based contouring [14], [15], [16], [17].
There is little evaluation of the same deep learning contouring (DLC) system at multiple centres within the literature. Kiljunen et al. [15] assessed DLC for prostate OARs on computed tomogrpahy (CT) scans at six centres but only five patients were analysed at each clinic. Oktay et al. [18] assessed DLC for 83 prostate and 26 head and neck CT scans from three centres but only ten scans were assessed for time-saving and individual images were assessed rather than clinical evaluation at each centre. Wong et al. [19] analysed DLC for 36 head and neck, 60 prostate and 21 central nervous system OARs at two centres. Studies from two further institutes [14], [20], [21] investigated DLC head and neck models on 217 and 58 patient CTs respectively but only over two sites.
This study aimed to evaluate a commercial DLC system for prostate and head and neck across four radiotherapy centres, including time-saving evaluation.
2. Materials and methods
2.1. Patient selection
The prostate study included 123 patient CTs from three radiotherapy centres. The head and neck study included 310 patient CTs from four centres. Each image came from a different patient and this was a retrospective study. Local approval was granted for this work and written informed consent was obtained from all patients.
Centres 2 (both sites), 3 (head and neck) and 4 (head and neck) selected consecutive patients. The time to outline OARs using the existing clinical method was recorded. A different set of clinical patients were outlined using DLC and the editing time recorded. Patients from centres 1 (both sites) and 3 (prostate) were consecutive, with times to outline OARs using the existing clinical method and to edit after using DLC recorded for the same patients. Centre 4 timings were collected using an SQL query of the ARIA database (Varian Medical Systems, Palo Alto, USA) for all patients that had a head and neck planning task competed within two weeks of an import/OAR outlining task being completed. All other centres recorded timings manually.
Where a paired method was used, every effort was made for the same observer to do both sets of contouring separated by a significant gap (4–6 weeks). For the unpaired methods, different patients were compared and were potentially contoured by different observers.
Each centre used its own local protocols for scanning and outlining. Full details are given in Table 1, Table 2.
Table 1.
Methods of assessing auto-contouring using deep learning contouring (DLC) for prostate structures.
| Centre 1 | Centre 2 | Centre 3 | |
|---|---|---|---|
| Number of patients for timing | 9 | 42 manual, 42 DLC | 10 |
| Number of patients for quantitative analysis | 9 | 10 | N/A |
| Same patients for timing and quantitative analysis? | Y | N | N/A |
| Timing Study design | Paired | Unpaired | Paired |
| Number of staff | 2 (both outlined 9 each) | 4 | 10 for manual, 1 for DLCExpert |
| Staff group | Radiotherapy Planners | Radiotherapy Planners | Radiotherapy Planners and clinicians. One clinician edited DLCExpert contours |
| Existing method | Manual | Manual | Manual |
| CT scanner/slice thickness | Philips Brilliance Big Bore (3 mm) | GE Discovery (2.5 mm) | GE Discovery (2.5 mm) |
| DLCExpert model name | Prostate_CT_NL006_MO | Prostate_CT_NL005_GN (bladder and rectum)Prostate_CT_NL010_NN (femoral heads) | Prostate_CT_NL006_MO |
| DLCExpert model description | Generic model based on data from a centre in the Netherlands. | Generic models based on data from a centre in the Netherlands. | Generic model based on data from a centre in the Netherlands. |
| Number of images for training model | 437 | 242 (bladder and rectum)337 (femoral heads) | 437 |
| OARs | Bladder, femoral heads, rectum | Bladder, femoral heads, rectum | Bladder, femoral heads, rectum |
| Targets | None | None | Prostate, seminal vesicles |
| Editing software | Pinnacle v16 (Philips, NL) | RayStation v7 (RaySearch, Sweden) | Eclipse v13.7 (Varian Medical Systems, Palo Alto, USA) |
| Outlining protocol basis | CHHiP | CHHiP, RTOG atlases | CHHiP |
| Timing method | Manual- time to draw or edit timed by the staff member | Manual- time to draw or edit timed by the staff member | Manual- time to draw or edit timed by the staff member Aria (Varian Medical Systems, Palo Alto, USA)-time to draw CTVs |
Table 2.
Methods of assessing auto-contouring using deep learning contouring (DLC) for head and neck organs at risk.
| Centre 1 | Centre 2 | Centre 3 | Centre 4 | |
|---|---|---|---|---|
| Number of patients for timing | 10 Manual, 10 DLC | 20 manual, 20 DLC | 9 manual, 7 DLC | 40 manual, 169 DLC |
| Number of patients for quantitative analysis | 10 | 10 | 10 | 15 |
| Same patients for timing and quantitative analysis? | Y | N | N | N |
| Timing Study design | Paired | Unpaired | Unpaired | Unpaired |
| Number of staff | 4 (all 4 outlined all 10 patients) | 3 | 1 | 6 |
| Staff group | Clinicians | Radiotherapy Planners | Clinicians | Radiotherapy Planners |
| CT scanner and slice thickness | Philips Brilliance Big Bore (3 mm) | GE Discovery (2.5 mm) | GE Discovery (2.5 mm) | Philips Brilliance Wide Bore, Siemens Confidence (3 mm) |
| Existing method | Manual | Atlas | Manual | Manual |
| DLCExpert model name | Combined: 2 local and 1 generic H&N_CT_NL004_GN | H&N_CT_NL004_GN | H&N CT NL004 GN | H&N_CT_NL004_GN |
| DLCExpert model description | Generic model based on data from a centre in the Netherlands used for mandible and SMGs. Models based on local data used for other contours. | Generic model based on data from a centre in the Netherlands. | Generic model based on data from a centre in the Netherlands. | Generic model based on data from a centre in the Netherlands. |
| Number of images for training model | 698 (generic),72 (local model A), 69 (local model B) | 698 | 698 | 698 |
| DLC OARs | Eyes, parotids, submandibular glands, brainstem, larynx, mandible, oral cavity, spinal cord | Brainstem mandible, parotids, spinal cord | Brainstem, parotids, spinal cord | Mandible, parotids, submandibular glands |
| OARs from other source | Atlas for orbits, optic nerves, lens | Atlas for orbits, optic nerves, lens | ||
| Editing software | Pinnacle v16 (Philips, NL) | RayStation v7 (RaySearch, Sweden) | Eclipse v13.7 (Varian Medical Systems, Palo Alto, USA) | Eclipse v15.6 MR5 (Varian Medical Systems, Palo Alto, USA) |
| Outlining protocol basis | DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines | DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines | DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines | DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines |
| Timing method | Manual - time to draw or edit OARs timed by the staff member | Manual- time to draw or edit OARs timed by the staff member | Manual-time to draw or edit OARs timed by the staff member | Aria (with greater than 3 h and < 10 min removed) |
2.2. Deep learning contouring software
A commercial DLC system (DLCExpertTM, Mirada Medical ltd, UK) was used for this study. Each centre was given the freedom to implement DLCExpert as was applicable to their workload and patients. DLCExpert comprises two stages of multi-layer central neural networks (CNNs) to classify OARs on a CT scan. The first stage coarsely segments the volume into OARs. This is implemented via a 14-layer multiclass CNN. The output of the first CNN is passed into multiple specialised 10-layer CNNs, which each perform binary classification for different organs. After correction of any discontinuities, the result is a prediction of the full resolution contours [14].
Models used in this study were generic (excluding centre 1 head and neck model), meaning they had not been trained on data from any of the centres in this study. The generic prostate model was trained on CT scans contoured using outlining guidelines from a clinic in the Netherlands and the generic head and neck model was trained on CT scans contoured using guidelines from Brouwer et al. [22]. All centres were using Workflow Box (WBx) 2.0 (Mirada Medical ltd, UK) from September 2019 to June 2020 and WBx 2.2 (Mirada Medical ltd, UK) from June 2020 to November 2020. WBx is a DICOM based tool which enables images to be routed directly from the scanner where they are automatically outlined using DLCExpert and then routed to the planning system.
2.3. Contouring time
Each centre recorded the time to outline OARs using their existing clinical protocol. OARs were also contoured using DLC and the time taken to edit these was recorded. The time taken by DLC to contour was not included. This was automated when the image was sent from the CT scanner to the treatment planning system and hence was not contributing to contouring time. The methods for each centre are summarised in Table 1, Table 2.
Some centres implemented a paired study, where the same set of patients were timed using DLC and the centre’s existing clinical method. This data was assessed for statistical significance using a paired T-test. Other centres implemented an unpaired study, where a different set of patients were analsyed using DLC and the centre’s existing clinical method. This data was assessed for statistical significance using an unpaired T-test. Statistical analysis was performed in SPSS (V25, IBM).
2.4. Quantitative evaluation
Dice Similarity Coefficient (DSC) and distance to agreement (DTA) were calculated to compare the existing clinical method to contours created with DLC before editing. DTA was taken as the shortest distance from a point on one contour surface to the other contour surface. Mean DTA was used which was the mean of all the distances calculated. Each centre decided upon the calculation method of DSC and DTA. This included ADMIRE software (v3.21.2, Elekta AB, Sweden) and in-house scripts within planning systems.
2.5. Subjective scoring
Subjective scoring was not part of the original study but was recorded by some centres when performing their own clinical evaluations. It has been included here for completeness. For prostate DLC, centre 3 subjectively scored all patients and centre 2 subjectively scored the last ten sequential patients. For head and neck DLC, centre 1 subjectively scored all patients and centres 2 and 3 subjectively scored the last ten sequential patients. No scoring was recorded for centre 4.
Centres 1 and 2 scored 1–7 using Greenham et al. [23] based on agreement to an acceptable clinical contour where 1 = good agreement through to 7 = gross error.
Centre 3 scored 1–5 where 1 = clinically acceptable through to 5 = would be easier to start from scratch.
2.6. Inter-Observer variability
Centre 1 assessed inter-observer variability for head and neck outlining. Each clinician’s manual and DLC edited contours were compared with all of the remaining clinicians giving twelve comparisons in all. This was repeated for five patients, for brainstem, larynx, left parotid and left submandibular gland (SMG). Box plots of the mean DTA and DSC were compared for manual and DLC edited contours.
3. Results
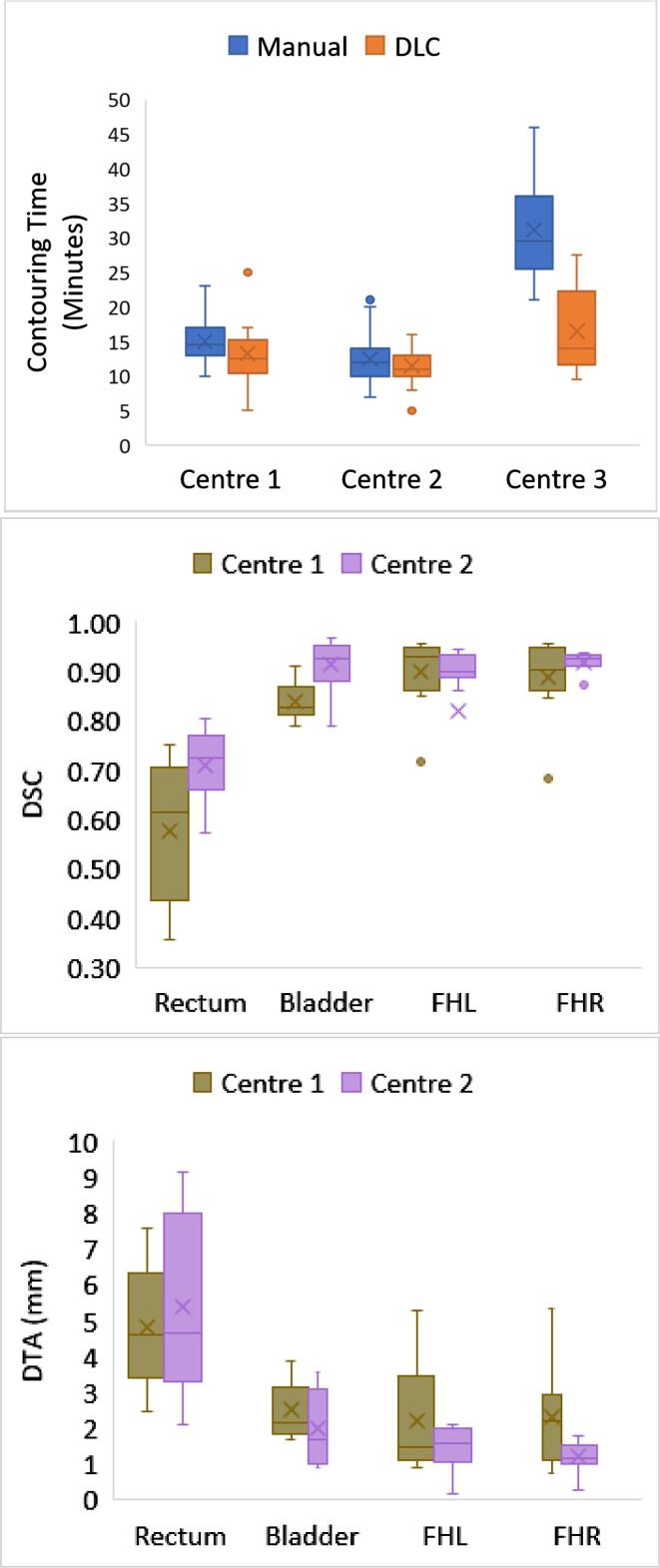
All three centres found a time-saving for prostate patient contouring using DLC compared to the existing clinical method. The total mean time saved per patient for contouring of all prostate structures assessed across the three centres was 5.9 ± 3.5 min (23 % time-saving on average). Fig. 1 shows the distribution of results for all centres. The paired T-tests and unpaired T-test indicated that the time differences between the manual and DLC methods were only significant at the 5 % level for centre 3 (supplementary table S1).
Fig. 1.
Box and whisker plot of contouring times, dice similarity coefficient (DSC) and distance to agreement (DTA) for prostate structures from 3 centres using manual and deep learning contouring (DLC). The boxes indicate the interquartile range (IQR), the line indicates the median and the cross indicates the mean. The whiskers indicate the highest and lowest values within 1.5 times the IQR and data outside this range indicated by circles. Axes for DTA and DSC have been limited to exclude one centre 2 patient left femoral head result as the DLC contour was outside of the external volume.
At centres 1 and 2, the DTA and DSC comparing manual contours to DLC contours showed the best agreement for the femoral heads (median DSC 0.92 ± 0.03, median DTA 1.5 ± 0.3 mm) and the worst agreement for the rectum (median DSC 0.68 ± 0.04, median DTA 4.6 ± 0.6 mm) (Fig. 1 middle and bottom panels and supplementary tables S2 and S3). This reflected the subjective scores (supplementary table S4).
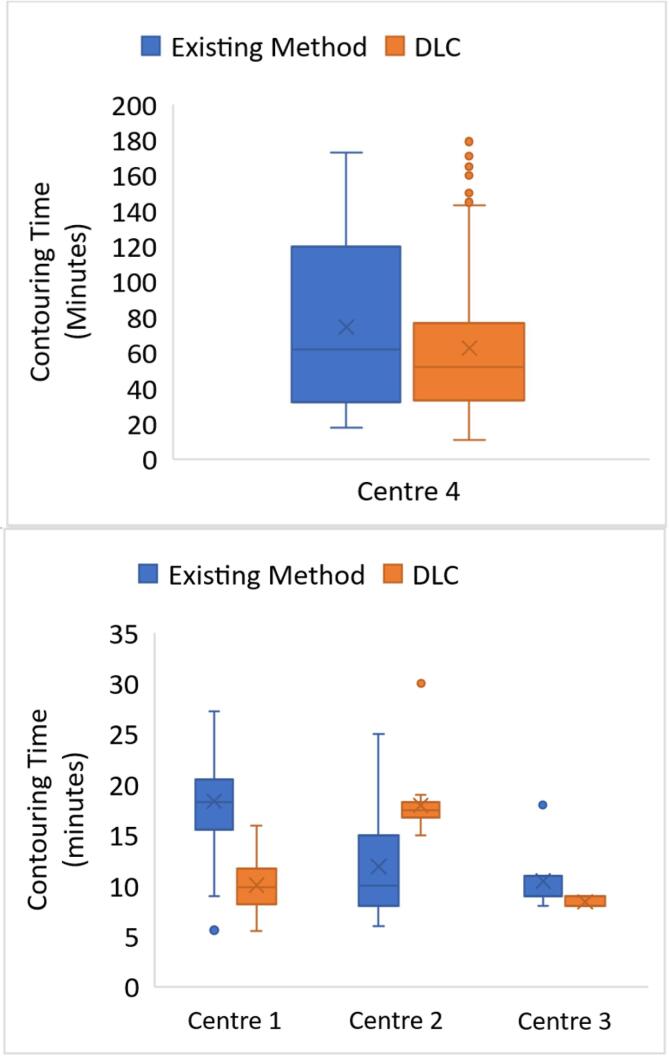
The total mean time saved for head and neck OAR contouring using DLC compared to the existing clinical method across the four centres was 16.2 ± 8.6 min (7 % time-saving on average) (Fig. 2). Where the existing clinical method was manual, (excluding centre 2) the average time-saving was 22.5 ± 8.4 min (27 %). The p-values indicated that the difference between the existing and DLC contouring methods were significant at the 5 % level for centres 1 and 2 (supplementary table S5). The distribution of all results showed a time saving for centre 4 (12.2 ± 8.2 min) and a time increase for centre 2 of 6.1 ± 1.3 min and smaller interquartile ranges for all centres using DLC (Fig. 2).
Fig. 2.
Box and whisker plot of contouring times for head and neck structures from 4 centres using manual and deep learning contouring (DLC). The boxes indicate the interquartile range (IQR), the line indicates the median and the cross indicates the mean. The whiskers indicate the highest and lowest values within 1.5 times the IQR and data outside this range indicated by circles.
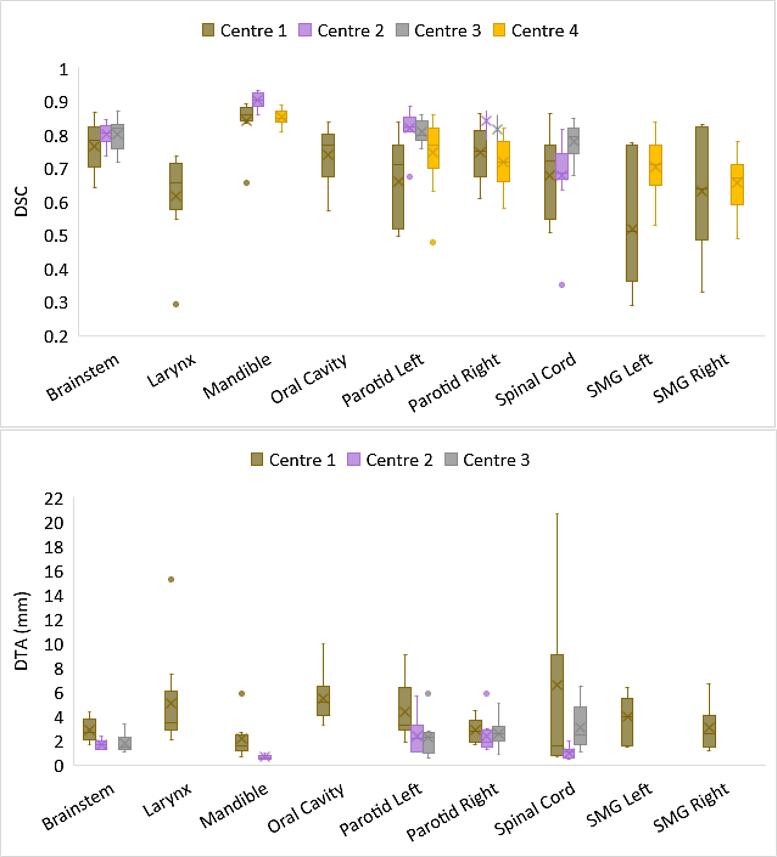
The mandible, brainstem and right parotid scored highly for DSC and DTA across all centres where analysed (Fig. 3 and supplementary tables S6 and S7). DSC and DTA values were calculated prior to editing the DLC contour.
Fig. 3.
Box and whisker plot of dice similarity coefficient (DSC) and distance to agreement (DTA) for head and neck organs at risk from 4 centres using an existing clinical method and deep learning contouring (DLC). The boxes indicate the interquartile range (IQR), the line indicates the median and the cross indicates the mean. The whiskers indicate the highest and lowest values within 1.5 times the IQR and data outside this range indicated by circles. Axis has been limited to exclude one patient spinal cord result for centre 1.
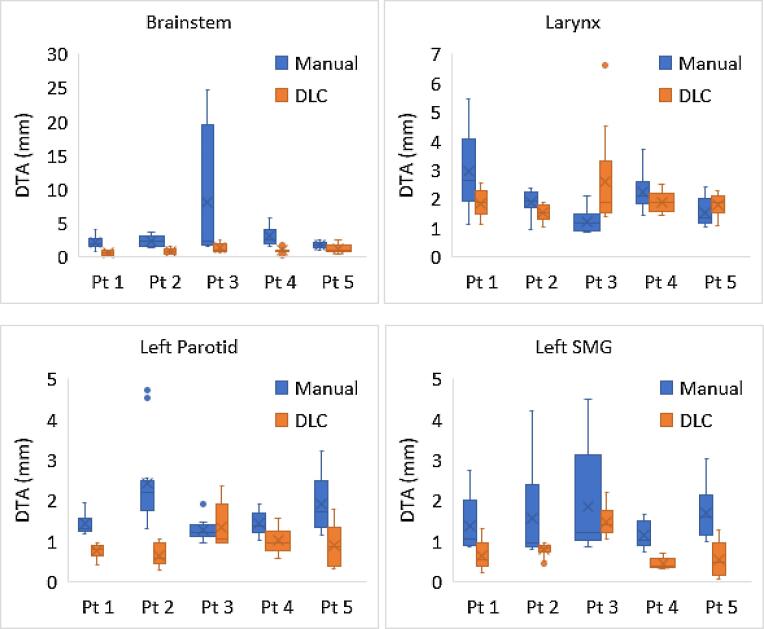
The inter-observer variability assessment showed that DLC generated contours reduced inter-observer variability compared to manual contours for the brainstem, left parotid gland and left SMG, with lower median DTA and higher DSC for the DLC contours (Fig. 4 and supplementary tables S9 and S10). There were also smaller interquartile ranges for the majority of the contours using DLC (Fig. 4). The difference between the two methods was statistically significant at the 5 % level using a Wilcoxon signed rank test (supplementary tables S9 and S10). No statistical difference in observer variability between DLC and manual contours was observed for the larynx DTA.
Fig. 4.
Distance to agreement (DTA) for manual and deep learning contouring (DLC) edited head and neck organs at risk. Results are from centre 1 for different permutations of 4 observers compared to each other for 5 patients (pt).
4. Discussion
This work assessed the use of DLC across four centres within the NHS where the contouring method would ideally be standardised. Prostate contouring showed a time-saving using DLC for all centres although this was only statistically significant for centre 3. This may be due to a larger time-saving for the prostate and seminal vesicles which were only analysed by centre 3. Kiljunen et al. [15] found a larger time-saving of twelve minutes (46 %) with a range of 1.5 % to 70.9 %. Their study included the prostate, seminal-vesicles and penile bulb in addition to the OARs analysed in this study which may increase the time-saving. Zabel et al. [24] found a time-saving of 8.5 min (44 %) using DLC for outlining bladder and rectum on 15 prostate CT scans. However, the manual times included contouring by a radiation therapist and editing by a radiation oncologist. The times in the current study from centres 1 and 2 include manual contouring by one person, hence manual contouring for these sites will be faster.
Time-savings using DLC for head and neck were seen for centres using manual contouring as their existing clinical method. Oktay et al. [18] found a time-saving of 93 % using DLC compared to manual contouring for head and neck CT scans. A maximum time-saving of 45 % was found in this current study. However, Oktay et al. timed an average manual contouring of 87 min for experts which was longer than the average time of any centres in this current study. Also, time-saving will likely increase with the number of structures contoured, with sites who outline more OARs gaining more time. Oktay et al. outlined nine structures using DLC whereas three out of the four centres in the current study outlined less than this. Hence it would be expected that the time-saving in the current study would be less.
The manual timings in this study were significantly lower than in other studies, meaning such a high percentage time-saving was not achievable. For example, Kiljunen et al. [15] reported prostate OAR manual contouring times of approximately 20 min for most centres with two centres taking over 40 min. In contrast, the longest manual prostate contouring time in the current study was 15 min, possibly because less structures were outlined. This current work has also shown that implementation significantly affects the times quoted. Centre 4 reported longer head and neck manual contouring times compared to the other centres and this was due to timing using ARIA. Care should be taken when interpreting and comparing results from studies analysing auto-contouring as there may be differences such as contour numbers, current outlining method, timing methods, staff expertise and knowledge that a measurement was being performed. Although larger time-savings imply the initial DLC quality was better, with variation across centres a conclusion cannot be drawn regarding this for the current study.
Time-saving using DLC for head and neck was statistically significant for centre 1 that had a combined model using structures selected from three models, two trained on their own data and one generic model. This suggests that DLC models developed with local data may provide a larger time-saving benefit. In addition, centre 1 tested more structures than the other centres which may contribute to the larger time-saving. Centre 2 was the only centre to observe a time increase using DLC, possibly because the centre uses a well-established atlas-based method and timing was only recorded for a limited number of structures. Staff also had significant prior experience adjusting atlas contours and did not have experience of DLC.
The DSCs obtained show the generic model performed well for the bladder and femoral heads (Fig. 1 and supplementary table S11). The DSC for rectum and bladder were slightly lower than other studies where generic models were used but the femoral heads were better than or comparable to other studies [15], [18], [24]. The largest variation between centres was for the rectum, potentially due to the small sample size or differing local contouring protocols. This again shows that care should be taken when interpreting results as the same DLC model can produce different results at different centres.
The majority of DSCs obtained for head and neck OARs were comparable to other studies. The DSC for the mandible in literature was between 0.89 and 0.94 and the left parotid between 0.77 and 0.84 [18], [25]. The DSCs for spinal cord were lower than other studies (e.g. 0.806 and 0.87 [18], [25]). However, the model by Ibragimov et al. [25] was tested on data from the same centre so was not a generic model.
Each centre chose their own method of contouring timing, resulting in paired and non-paired studies with different patient numbers. A sample size calculation performed showed 51 patients required in each group for an unpaired study and 26 patients if it was paired [26]. However, an unpaired study requires no additional clinical work through repetition of measurements. The calculation suggests a larger sample size is required in the current study but this was not possible due to the clinical workload.
The results from centre 1 suggest that inter-observer variability using DLC was improved, agreeing with other studies [15]. Kiljunen et al. found improved inter-observer variability for prostate OARs, with more benefit for large, challenging structures such as lymph nodes. Even in centres with existing efficient contouring methods, DLC could provide the benefit of improved consistency. Further assessment is needed across a larger multi-centre dataset which has currently not been carried out within the literature. However, this evaluation is very resource intensive. The inter-observer DSCs for manual contouring (supplementary table S9) were comparable to the DLC vs manual contouring DSCs for head and neck OARs (supplementary table S6). This suggests that a clinician is likely to agree as much with DLC as they do with another clinician.
Different software was used to calculate the DSC and DTA at each centre so the values cannot be directly compared. This again highlights the need for caution when comparing data in literature as the calculation of these is not standardised. However, they do give an indication of which contours were closer to clinical ones. It has been shown that a DSC larger than 0.65 should give a time-saving [27] as the time to edit the auto-contours would probably be quicker than manual delineation. Although no individual contour times were calculated in the current study, the majority of contours had a DSC greater than 0.65. However, it has recently been shown for lungs [28] that surface DSC and added path length provide a better indicator of time-saving using auto-segmentation.
Vandewinckele et al. [29] have produced recommendations for implementation and quality assurance of artificial intelligence applications in radiotherapy. In addition to the overlap and distance metrics used here, they recommend to compare the overall volume and the dosimetric impact of the delineation uncertainty. The impact of a DLC contour that is minimally different to a clinical contour may not have a clinically significant impact on the evaluated dose. van Rooij et al. [30] showed that differences in DLC contours for head and neck patients did not produce clinically significant differences in radiotherapy plans.
In conclusion, clinical implementation of non-centre specific DLC models for prostate and head and neck OARs can provide time-savings. Time-savings become more significant when a larger number of OARs are contoured. A generic model can be implemented and tested in different ways, using paired or unpaired studies to fit in with clinical workload. Unpaired studies take less time as they prevent contour replication. DSC and DTA showed good agreement with clinical contours for the majority of structures. The potential for reducing inter-observer variability has been indicated but further work is needed to confirm this.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to all staff members who performed outlining and data collection for the study including Liz Adams and Dan Love. We would like to thank Mirada for their technical support. Robert Chuter is supported by a Cancer Research UK Centres Network Accelerator Award Grant (A21993) to the ART-NET consortium.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2022.11.003.
Contributor Information
Zoe Walker, Email: zoe.walker@uhcw.nhs.uk.
Gary Bartley, Email: gary.bartley@uhcw.nhs.uk.
Christina Hague, Email: christina.hague@nhs.net.
Daniel Kelly, Email: daniel.kelly5@nhs.net.
Clara Navarro, Email: clara.navarro@nhs.net.
Jane Rogers, Email: Jane.Rogers2@uhcw.nhs.uk.
Christopher South, Email: csouth@nhs.net.
Simon Temple, Email: simon.temple@nhs.net.
Philip Whitehurst, Email: philip.whitehurst1@nhs.net.
Robert Chuter, Email: robert.chuter@nhs.net.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Walker G.V., Awan M., Tao R., Koay E.J., Boehling S., Grant J.D., et al. Prospective randomized double-blind study of atlas-based organ-at-risk auto segmentation-assisted radiation planning in head and neck cancer. Radiother Oncol. 2014;112:321–325. doi: 10.1016/j.radonc.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukesh M., Benson R., Jena R., Hoole A., Roques T., Scrase C., et al. Interobserver variation in clinical target volume and organs at risk segmentation in post-parotidectomy radiotherapy: can segmentation protocols help? Br J Radiol. 2012;85:e530–e536. doi: 10.1259/bjr/66693547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steenbakkers R.J.H.M., Duppen J.C., Fitton I., Deurloo K.E.I., Zijp L., Uitterhoeve A.L.J., et al. Observer variation in target volume delineation of lung cancer related to radiation oncologist-computer interaction: a ’Big Brother’ evaluation. Radiother Oncol. 2005;77:182–190. doi: 10.1016/j.radonc.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj A.K., Kehwar T.S., Chakarvarti S.K., Sastri G.J., Oinam A.S., Pradeep G., et al. Variations in inter-observer contouring and its impact on dosimetric and radiobiological parameters for intensity-modulated radiotherapy planning in treatment of localised prostate cancer. J Radiother Pract. 2008;2:77–88. doi: 10.1017/S1460396908006316. [DOI] [Google Scholar]
- 5.Brouwer C.L., Steenbakkers R.J.H.M., van den Heuvel E., Duppen J.C., Navran A., Bijl H.P., et al. 3D Variation in delineation of head and neck organs at risk. Radiat Oncol. 2012;7:32. doi: 10.1186/1748-717X-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schick K., Sisson T., Frantzis J., Khoo E., Middleton M. An assessment of OAR deli- neation by the radiation therapist. Radiography. 2011;17:183–187. doi: 10.1016/j.radi.2011.01.003. [DOI] [Google Scholar]
- 7.La Macchia M., Fe F., Amichetti M., Cianchetti M., Gianolini S., Paola V., et al. Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat Oncol. 2012;7:160. doi: 10.1186/1748-717X-7-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmat I., Georg P., Georg D., Birkfellner W., Goldner G., Stock M. Assessment of accuracy and efficiency of atlas-based auto- segmentation for prostate radiotherapy in a variety of clinical conditions. Strahlenther Onkol. 2012;188:807–815. doi: 10.1007/s00066-012-0117-0. [DOI] [PubMed] [Google Scholar]
- 9.Kim J., Han J., Ailawadi S., Baker J., Hsia A., Xu Z., et al. SU-F-J-113: multi-atlas based automatic organ segmentation for lung radiotherapy planning. Med Phys. 2016;43:3433. doi: 10.1118/1.4956021. [DOI] [Google Scholar]
- 10.Zhong H., Kim J., Chetty I.J. Analysis of deformable image registration accuracy using computational modeling. Med Phys. 2010;37:970–979. doi: 10.1118/1.3302141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larrue A., Gujral D., Nutting C., Gooding M. The impact of the number of atlases on the performance of automatic multi-atlas contouring. Phys Med. 2015;31:e30. [Google Scholar]
- 12.Teguh D.N., Levendag P.C., Voet P.W.J., Al-Mamgani A., Han X., Wolf T.K., et al. Clinical validation of atlas-based auto-segmentation of multiple target volumes and normal tissue (swallowing/mastication) structures in the head and neck. Int J Radiat Oncol Biol Phys. 2011;81:950–957. doi: 10.1016/j.ijrobp.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Meyer P., Noblet V., Mazzara C., Lallement A. Survey on deep learning for radiotherapy. Comput Biol Med. 2018;98:126–146. doi: 10.1016/j.compbiomed.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 14.van Dijk L.V., Van den Bosch L., Aljabar P., Peressutti D., Both S., Steenbakkers R.J., et al. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring. Radiother Oncol. 2020;142:115–123. doi: 10.1016/j.radonc.2019.09.022. [DOI] [PubMed] [Google Scholar]
- 15.Kiljunen T., Akram S., Niemelä J., Löyttyniemi E., Seppälä J., Heikkilä J., et al. A Deep Learning-Based Automated CT Segmentation of Prostate Cancer Anatomy for Radiation Therapy Planning-A Retrospective Multicenter Study. Diagnostics. 2020;10:959. doi: 10.3390/diagnostics10110959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolov S., Blackwell S., Mendes R., De Fauw J., Meyer C., Hughes C., et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. J Med Internet Res. 2021;23:e26151. doi: 10.2196/26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lustberg T., van Soest J., Gooding M., Peressutti D., Aljabar P., van der Stoep J., et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126:312–317. doi: 10.1016/j.radonc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Oktay O., Nanavati J., Schwaighofer A., Carter D., Bristow M., Tanno R., et al. Evaluation of Deep Learning to Augment Image-Guided Radiotherapy for Head and Neck and Prostate Cancers. JAMA Netw open. 2020;3:e2027426. doi: 10.1001/jamanetworkopen.2020.27426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong J., Huang V., Wells D.M., Giambattista J.A., Giambattista J., Kolbeck C., et al. Implementation of Deep Learning-Based Auto-Segmentation for Radiotherapy Planning Structures: A Multi-Center Workflow Study. Int J Radiat Oncol Biol Phys. 2020;108:S101. doi: 10.1016/j.ijrobp.2020.07.2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouwer C.L., Boukerroui D., Oliveira J., Looney P., Steenbakkers R.J., Langendijk J.A., et al. Assessment of manual adjustment performed in clinical practice following deep learning contouring for head and neck organs at risk in radiotherapy. Phys Imaging Radiat Oncol. 2020;16:54–60. doi: 10.1016/j.phro.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunenberg E.J., Steinseifer I.K., van den Bosch S., Kaanders J.H., Brouwer C.L., Gooding M.J., et al. External validation of deep learning-based contouring of head and neck organs at risk. Phys Imaging Radiat Oncol. 2020;15:8–15. doi: 10.1016/j.phro.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brouwer C.L., Steenbakkers R.J.H.M., Bourhis J., Budach W., Grau C., Grégoire V., et al. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines. Radiother Oncol. 2015;117:83–90. doi: 10.1016/j.radonc.2015.07.041. [DOI] [PubMed] [Google Scholar]
- 23.Greenham S., Dean J., Fu C.K.K., Goman J., Mulligan J., Deanna T., et al. Evaluation of atlas-based auto-segmentation software in prostate cancer patients. J Med Radiat Sci. 2014;61:151–158. doi: 10.1002/jmrs.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabel W.J., Conway J.L., Gladwish A., Skliarenko J., Didiodato G., Goorts-Matthews L., et al. Clinical evaluation of deep learning and atlas-based auto-contouring of bladder and rectum for prostate radiation therapy. Pract Radiat Oncol. 2020;11:e80–e89. doi: 10.1016/j.prro.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Ibragimov B., Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44:547–557. doi: 10.1002/mp.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sim J, Wright C, Research in healthcare, concepts designs and methods. Nelson Thomas:2020.
- 27.Langmack K.A., Perry C., Sinstead C., Mills J., Saunders D. The utility of atlas-based segmentation in the male pelvis is dependent on the interobserver agreement of the structures segmented. Br J Radiol. 2014;87:20140299. doi: 10.1259/bjr.20140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaassen F., Hazelaar C., Vaniqui A., Gooding M., van der Heyden B., Canters R., et al. Evaluation of measures for assessing time-saving of automatic organ-at- risk segmentation in radiotherapy. Phys Imaging Radiat Oncol. 2019;13:1–6. doi: 10.1016/j.phro.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandewinckele V., Claessens M., Dinkla A., Brouwer C., Crijns W., Verellen D., et al. Overview of artificial intelligence-based applications in radiotherapy: recommendations for implementation and quality assurance. Radiother Oncol. 2020;153:55–56. doi: 10.1016/j.radonc.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 30.van Rooij W., Dahele M., Brandao H.R., Delaney A.R., Slotman B.J., Verbakel W.F. Deep learning-based delineation of head and neck organs at risk: geometric and dosimetric evaluation. Int J Radiat Oncol Biol Phys. 2019;104:677–684. doi: 10.1016/j.ijrobp.2019.02.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.