Abstract
Acute kidney injury (AKI) develops frequently in the course of patients with sepsis and strongly associates with in-hospital mortality. However, diagnosing AKI involves a considerable lag-time because it depends on assessing an increase in serum creatinine, and offers no insight in the underlying pathophysiology. Consequently, identifying a set of proteins reflecting the development of AKI may improve earlier recognition of AKI and the understanding of its pathophysiology. A targeted plasma proteomic approach was performed in early sepsis patients with and without subsequent AKI development in a matched pair design (n = 19 each). Principal component analysis identified 53 proteins associated with development of AKI, which were further analysed using Enrichr gene ontology and pathway analysis. Nine differentially expressed proteins from the targeted proteomics were increased among patients who subsequently developed AKI and correlated with principal components, namely CALCA, CALR, CA12, CLEC1A, PTK7, KIM-1, NPPC, NUCB2 and PGF. We demonstrated the biological insight in the development of AKI in early sepsis compared to non-AKI sepsis.
Subject terms: Sepsis, Acute kidney injury
Introduction
Sepsis is a life-threatening syndrome of organ dysfunction encompassing a dysregulated host response to infection. The incidence of sepsis varies across the world and has been estimated at approximately 200 cases per 100,000 inhabitants in high-income countries, with an estimated in-hospital mortality rate of 17–26%1–5. The incidence of sepsis has more than doubled in the last decade and is expected to keep rising due to increased lifespan, chronic disease, use of immunosuppressive therapy, chemotherapy, and invasive procedures (e.g. mechanical ventilation, renal replacement therapy)2,3. Sepsis is frequently accompanied with organ dysfunction involving kidneys, liver, lungs, cardiovascular system and central nervous system. The incidence of AKI is approximately 22–51% among patients with sepsis and is strongly associated with subsequent failure of other organs and increased mortality. Moreover, patients with AKI have a higher risk for developing chronic kidney disease and end-stage renal disease, as well as long-term mortality after sepsis4–12.
The pathophysiology of sepsis-AKI is complex and incompletely understood, which both hampers prompt recognition of AKI and the development of novel treatments to prevent or treat AKI. The KDIGO criteria are commonly used to diagnose and classify AKI13,14. Importantly, the KDIGO criteria depend on measurement of urine output, which is usually lacking at and during early hospitalization, and changes in serum creatinine, which induces considerable lag-time in diagnostics because of the time needed to accumulate above the threshold13. Moreover, a recent pre-existent serum creatinine to determine the change in serum creatinine, is often not available. Here, we used targeted plasma proteomics to analyse a set of proteins involved in the pathophysiology of early sepsis, that contribute to the development of AKI and offer insight into the pathophysiology of AKI development. To this end, we performed a targeted proteomic analysis of plasma obtained from patients with early sepsis admitted to the emergency department (ED), either with or without subsequent development of AKI according to the KDIGO criteria.
Methods
Study population
Data were collected at the emergency department of the University Medical Center Groningen (UMCG), a tertiary care hospital with approximately 34,000 emergency department visits per year. Adult non-trauma patients (> 18 years of age) diagnosed with sepsis using the sepsis-2 criteria (i.e. two or more SIRS criteria combined with the clinical suspicion of an infection) with sepsis recognition and blood withdrawal within 30 min were included prospectively from 16th of August 2012 to 31st of June 2013 (n = 94). Development of AKI within the first 24 h was defined according to the KDIGO criteria14 by an increase of at least 50% of serum creatinine as compared to baseline creatinine or an absolute rise of more than 26.4 µmol/l (n = 19). Identified cases were matched with patients from the same cohort without AKI based on age, sex, infection focus and sepsis-2 criteria. Finally, technical exclusion of 2 samples during plasma proteomics resulted in a non-AKI group of 19 patients and an AKI group of 17 patients. Written (deferred) informed consent was obtained from all participating patients. The study protocols were approved by the medical ethical committee of the University of Groningen (METc 2012/077).
Data collection
Collected data included patient characteristics, vital parameters and laboratory measurements at admission, and mortality at follow-up. Follow-up mortality was obtained from the Municipal Personal Records Database, containing full registration of all Dutch citizens. Patient characteristics consisted of age, gender, presence of diabetes mellitus, chronic kidney disease, liver disease, active cancer (radiotherapy or chemotherapy treatment received up to two years prior to the current hospitalization), and immunocompromised status. Vital parameters consisted of heart frequency, systolic and diastolic blood pressure, mean arterial pressure, body temperature (tympanic), oxygen saturation, and respiratory rate at admission. The recorded laboratory measurements consisted of haemoglobin, leukocyte count, thrombocyte count and plasma levels of lactate, creatinine, urea, and C-reactive protein (CRP). Other data included the source of infection, and length of hospital and intensive care unit (ICU) stay. Baseline serum creatinine and urea values (most recent measurement during the year preceding inclusion in the study) and those during hospitalization were collected from the electronic patient records. The estimated glomerular filtration rate (eGFR) was determined using the MDRD-4 formula15.
Plasma proteome analysis
Plasma protein expression was assessed using the aptamer-based multiplex proximity extension assay (PEA) from Olink Proteomics AB (Uppsala, Sweden), allowing for quantification of a substantial number of proteins in a small amount of plasma16. In this assay, proteins are recognised by pairs of oligonucleotide-labelled antibodies (“probes”). When the two probes are in close proximity, a new PCR target sequence is formed by a proximity-dependent DNA polymerisation reaction, resulting in relative numbers. The resulting sequence is subsequently detected and quantified using a standard real-time PCR. Proteins from the Organ Damage panel were measured, containing 92 different proteins. Data from 25 protein analytes were omitted from the analysis, as the measured levels were below the LOD in ≥ 25% of the patients. Next, data were subjected to Olink proteomics performed quality control per sample during which samples that deviate less than 0.3 NPX from the median pass the quality control. Two individuals had to be excluded, as these failed quality control (QC) analysis by the manufacturer. Final data analysis comprised 67 protein analytes from 36 patients (n = 17 AKI and n = 19 without AKI).
Enzyme-linked immuno-absorbent analysis (ELISA)
To perform a technical validation, we measured plasma levels of kidney injury molecule-1 (KIM-1), a biomarker for human renal proximal tubule injury, by ELISA (Quantikine; DSKM100). Results were read at an absorbance of 450 nm with wavelength correction at 570 nm using a Bio-Tek Synergy H4 microplate reader.
Differential expression analysis
Differential expression analysis of identified proteins between AKI and non-AKI groups were performed using the R package limma, applying a linear model with age and sex as covariates. Limma uses an empirical Bayes method to moderate the standard errors of the estimated log fold changes17.
Gene ontology and pathway enrichment analysis
Molecular function and pathway analysis were performed by Enrichr enrichment analysis. Enrichr for gene ontology (GO) molecular function and the NCI-Nature 2016 pathways analytic tools18,19. Enrichr is a comprehensive resource for composed gene sets and a search engine that accumulates biological knowledge for further biological discoveries. Proteins from PC1 and PC2 analyses with a fold change > 1 between sepsis and sepsis-AKI were used to identify GO and pathways (n = 48), and were used to perform GO and pathways analyses. P-values were calculated using Fisher exact test and corrected for multiple testing by Benjamini–Hochberg adjustment and pathways with a corrected p-value < 0.05 were considered significant. The combined score of the pathway analysis (Table 5) is calculated using the Fisher exact test and the Z-score to order the pathways.
Table 5.
Pathway analysis by Enrichr/NCI Nature 2016.
| Pathway name | p-value | Adjusted p-value | Odds ratio | Combined score |
|---|---|---|---|---|
| Signalling events mediated by VEGFR1 and VEGFR2 | < 0.001 | < 0.001 | 36.71 | 526.02 |
| Angiopoietin receptor Tie2-mediated signalling | < 0.001 | < 0.001 | 41.13 | 500.00 |
| PDGF receptor signalling network | 0.014 | 0.049 | 84.88 | 360.44 |
| Class I PI3K signalling events | < 0.001 | 0.003 | 29.49 | 250.03 |
| Alpha9 beta1 integrin signalling events | 0.002 | 0.015 | 37.67 | 241.75 |
| Glypican 1 network | 0.002 | 0.015 | 34.66 | 217.05 |
| Thromboxane A2 receptor signalling | < 0.001 | 0.004 | 25.03 | 200.74 |
| Ephrin B reverse signalling | 0.002 | 0.015 | 30.94 | 187.27 |
| Regulation of p38-alpha and p38-beta | 0.002 | 0.015 | 30.94 | 187.27 |
| CXCR4-mediated signalling events | < 0.001 | 0.002 | 18.80 | 173.82 |
| Alpha-synuclein signalling | 0.003 | 0.016 | 28.87 | 171.07 |
| EPHA forward signalling | 0.003 | 0.016 | 27.07 | 157.11 |
| Signalling events mediated by Hepatocyte Growth Factor Receptor (c-Met) | < 0.001 | 0.009 | 17.91 | 126.87 |
| amb2 Integrin signalling | 0.004 | 0.019 | 22.78 | 124.96 |
| Signalling events mediated by PTP1B | 0.007 | 0.030 | 17.31 | 86.05 |
| Signalling events mediated by focal adhesion kinase | 0.009 | 0.033 | 15.45 | 73.54 |
| Fc-epsilon receptor I signalling in mast cells | 0.009 | 0.033 | 15.45 | 73.54 |
| PDGFR-beta signalling pathway | 0.003 | 0.018 | 10.57 | 59.53 |
| CDC42 signalling events | 0.012 | 0.044 | 12.71 | 55.94 |
Proteins that correlated with PC1 and/or PC2 and which were positive differentially expressed proteins among patients with sepsis-AKI as compared to patients without AKI. The proteins were used to identify pathways affected by sepsis-AKI, only pathways with an adjusted p-value < 0.05 are shown.
Statistical analysis
Statistical analyses were performed using SPSS 23 (IBM, USA). Normality was tested using a Shapiro–Wilk test. Since most data was non-normally distributed, categorical variables were compared using a using a z-test, while a Mann–Whitney U or Kruskal–Wallis test was used for continuous variables. PC were extracted from the principal component analysis (PCA) based on an Eigenvalue cut-off > 1, no rotation method was applied. Factors that correlated with the PCs, using a two-tailed Spearman test, were selected for further analysis. The correlation between proteomics and KIM-1 measurements were calculated using a two-tailed Pearson test. To identify factors associated with the course of renal function, PCs that were significantly different between patients with AKI and without AKI were entered in a stepwise linear regression analysis along with age, gender, sepsis severity as well as serum creatinine at baseline and upon admission (probability-of-F-to-enter < /= 0.05, probability-of-F-to-remove > /= 0.1). The Benjamini–Hochberg false discovery rate correction method was used where applicable to correct for multiple comparison testing. A p-value < 0.05 was considered significantly different.
Statement of methods used
All methods were carried out in accordance with relevant guidelines and regulations.
Statement of approval
The protocol was reviewed by the ethical review board of the UMCG and approved (METc 2012/077). Written informed consent was obtained from all participating patients.
Results
Characterisation of the early sepsis groups
From August 2012 to June 2013, we included in total 94 patients with sepsis at the ED. Of those patients, 19 (20%) developed AKI according to the KDIGO criteria in the first 24 h after admission. These 19 patients were matched with 19 sepsis patients without AKI from this cohort. Patients who later on developed AKI had a higher systolic blood pressure upon presentation to the ED, while mean arterial and diastolic blood pressure were not different from patients without subsequent development of AKI (Table 1). Other baseline and disease characteristics were not different between both groups. Upon admission, patients that subsequently developed AKI had increased serum creatinine and urea and decreased eGFR. Importantly, pre-existent renal function and the renal function at hospital discharge was not different between both groups (Table 2). Hence, at hospital admission, patients with subsequent sepsis-AKI only displayed decreased renal function and higher systolic blood pressure as compared to patients without AKI development.
Table 1.
Baseline and disease characteristics.
| No AKI (n = 19) (median ± IQR) or n (%) | AKI (n = 17) (median ± IQR) or n (%) | |
|---|---|---|
| Age (years) | 65 (59–75) | 60 (52–76) |
| Sex (male) | 12 (63%) | 13 (76%) |
| History of chronic kidney disease | 6 (32%) | 7 (41%) |
| History of cardiovascular disease | 13 (68%) | 10 (59%) |
| History of diabetes mellitus | 9 (47%) | 7 (41%) |
| Infection focus: airways/pneumonia | 6 (32%) | 5 (29%) |
| Infection focus: urogenital | 9 (47%) | 7 (41%) |
| Infection focus: abdominal | 2 (11%) | 3 (18%) |
| Infection focus: unknown | 2 (11%) | 2 (12%) |
| Systolic blood pressure (mmHg) | 132 (125–150) | 105 (90–130)* |
| Diastolic blood pressure (mmHg) | 70 (65–80) | 66 (52–80) |
| Mean arterial pressure (mmHg) | 93 (88–107) | 78 (67–97) |
| Heart frequency (bpm) | 110 (95–120) | 110 (100–125) |
| Respiratory frequency (per minute) | 22 (18–28) | 20 (18–29) |
| Oxygen saturation (%) | 94 (91–98) | 95 (90–97) |
| Glasgow coma score | 15 (15–15) | 15 (15–15) |
| Body temperature (°C) | 38.5 (38.2–38.8) | 38.6 (38.0–39.4) |
| Haemoglobin (mmol/L) | 7.1 (6. 1–8.1) | 7.2 (6.1–8.6) |
| Leukocytes (106/mL) | 12.2 (10–15.3) | 8.5 (5–15.6) |
| Bandemia (> 5%) | 2 (11%) | 1 (6%) |
| Thrombocytes (109/mL) | 205 (174–317) | 189 (146–217) |
| CRP (mg/L) | 63 (22–197) | 146 (80–335) |
| Lactate (mmol/L) | 1.5 (1.1–1.7) | 1.7 (1.2–2.1) |
| Multiple organ failure | 3 (16%) | 3 (18%) |
| Liver failure | 0 (0%) | 0 (0%) |
| Respiratory failure | 6 (32%) | 3 (18%) |
| ICU admittance | 1 (5%) | 3 (18%) |
| Length-of-stay in hospital (days) | 6 (2–8) | 8 (5–14) |
| Deceased in hospital | 1 (5%) | 1 (6%) |
| Oncology | 2 (11%) | 6 (35%) |
| Nefrotoxic drugs (i.e. cytostatics) | 2 (11%) | 5 (29%) |
IQR interquartile range, CRP c-reactive protein, ICU intensive care unit, */**means p < 0.05/0.01.
Table 2.
Course of renal function.
| No AKI (n = 19) (median ± IQR) or n (%) | AKI (n = 17) (median ± IQR) or n (%) | |
|---|---|---|
| Baseline creatinine (μmol/L) | 77 (62–102) | 96 (84–132) |
| Baseline urea (mmol/L) | 6 (5.3–8.4) | 7.5 (6–10.0) |
| Baseline eGFR (ml/min/1.73m2) | 90.7 (53.6–103.9) | 68.4 (46.1–80.3) |
| CKD class: 1 (> 90) | 10 (53%) | 3 (18%) |
| CKD class: 2 (60–90) | 4 (21%) | 7 (41%) |
| CKD class: 3a (45–60) | 1 (5%) | 1 (6%) |
| CKD class: 3b (30–45) | 0 (0%) | 0 (0%) |
| CKD class: 4 (15–30) | 3 (16%) | 3 (18%) |
| CKD class: 5 (< 15) | 1 (5%) | 3 (18%) |
| Creatinine upon admission (μmol/L) | 81 (65–105) | 160 (104–222)** |
| eGFR upon admission (ml/min/1.73 m2) | 80.9 (49.5–95.3) | 40.3 (29.3–53.6)** |
| Urea/creatinine ratio at admission | 90.2 (72.7–108.9) | 76.4 (53.8–102.6) |
| Urea upon admission (mmol/L) | 7.4 (5.5–10.3) | 11.7 (8.9–16.5)* |
| Maximum creatinine < 48 h (μmol/L) | 87 (66–105) | 176 (118–222)** |
| Creatinine at discharge (μmol/L) | 65 (57–90) | 103 (77–134) |
| eGFR at discharge (ml/min/1.73m2) | 88.3 (59.1–127.5) | 69.6 (46.1–98.4) |
| Urea at discharge (mmol/L) | 5.3 (3.7–8.6) | 7.6 (6.8–9.4) |
| AKI at discharge | 0 (0%) | 1 (6%) |
| Creatinine after discharge (μmol/L) | 65 (57–90) | 103 (77–134) |
| eGFR after discharge (ml/min/1.73 m2) | 88.3 (59.1–127.5) | 69.6 (46.1–98.4) |
Although no changes in baseline renal function existed between patients with and without AKI, the serum creatinine level, eGFR upon admission, as well as the maximum serum creatinine reached during the first 48 h after hospitalization are different between groups. AKI acute kidney injury, CKD chronic kidney disease, eGFR estimated glomerular filtration rate, IQR interquartile range, */**means p < 0.05/0.01.
Targeted plasma proteomic revealed protein profiles involved in the development of AKI
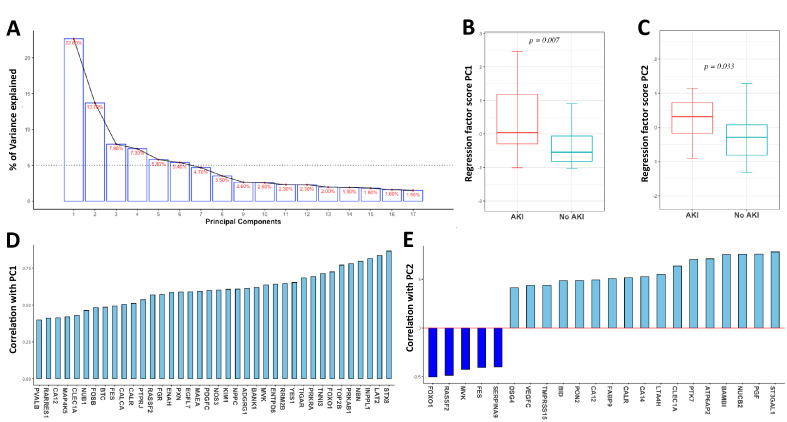
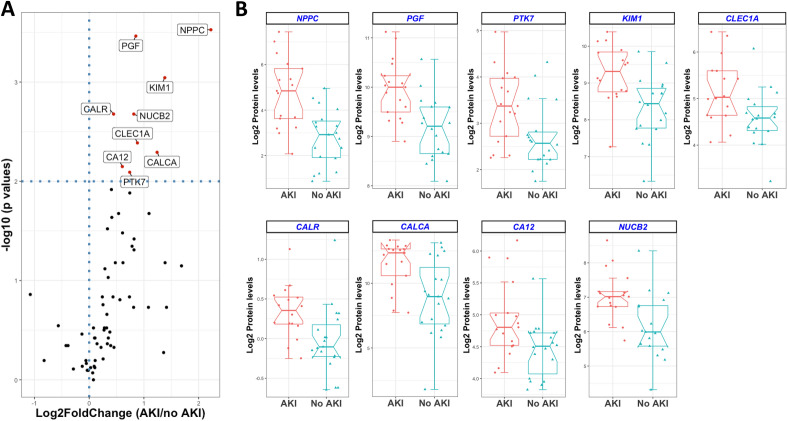
In order to identify a set of proteins involved in the development of AKI in patients with sepsis, a targeted plasma proteomic analysis was performed. The organ damage panel measured 92 proteins; after deploying a missingness threshold of ≥ 25%, the panel identified 67 proteins. PCA of the proteins revealed 17 components, of which the first 6 had an Eigenvalue above the cut-off of 1. These 6 individual components explained from 25 to 5% of the variation of the development of AKI during sepsis (Fig. 1A). Of these 6 PCs, PC1 and PC2 demonstrated a positive regression coefficient between sepsis and sepsis-AKI (Fig. 1B,C). In total, 38 proteins correlated significantly with PC1 (Fig. 1D), while 22 proteins correlated with PC2 (Table 3; Fig. 1E). Of all identified proteins (n = 67), nine were increased among patients with sepsis-AKI as compared to sepsis (DEP fold change > 2, p-value < 0.05) and at the same time correlated with PC1, PC2 or both (Fig. 2A), namely: calcitonin gene related peptide 1 (CALCA), calreticulin (CALR), carbonic anhydrase 12 (CA12), c-type lectin domain family 1 member (CLEC1A), inactive tyrosine protein kinase 7 (PTK7), kidney injury molecule-1 (KIM-1), natriuretic peptide precursor C (NPPC), nucleobindin-2 (NUCB2) and prostaglandin F2 alpha receptor (PGF). Boxplots of these nine proteins in patients with sepsis-AKI and sepsis are depicted in (Fig. 2B). An additional validation of the proteomic analysis was confirmed by measuring KIM-1 (R = 0.517, p < 0.05). Thus, targeted proteomics revealed nine DEPs to play a role in the development of AKI during sepsis.
Figure 1.
Summary of principal component analysis results (PCA). Result of the PCA, revealing approximately 23% of variance explained by PC1 (A). Analysis of all components that explained at least 5% variance (i.e. PC1–6) reveals a significant difference between patients with and without AKI for PC1 (B) and PC2 (C), but not for the other components. Extraction of the proteins associated with PC1 reveals 38 proteins (D), while 22 proteins were associated with PC2 (E). AKI Acute kidney injury, PC principal component, whiskers in box plot represent min–max.
Table 3.
Relative plasma levels of proteins extracted by principal component analysis.
| Symbol | Uniprot ID | Full name | Correlation PCA | FC AKI vs. no AKI | p-value | Adj. p-value |
|---|---|---|---|---|---|---|
| ADGRG1 | Q9Y653 | Adhesion G-protein coupled receptor G1 | PC1 | 1.48 | 0.156 | 0.285 |
| ATP6AP2 | O75787 | Renin receptor | PC2 | 1.06 | 1.000 | 0.982 |
| BAMBI | Q13145 | BMP and activin membrane-bound inhibitor homolog | PC2 | 1.45 | 0.021 | 0.093 |
| BANK1 | Q8NDB2 | B-cell scaffold protein with ankyrin repeats | PC1 | 1.26 | 0.433 | 0.547 |
| BID | P55957 | BH3-interacting domain death agonist | PC2 | 0.82 | 0.778 | 0.809 |
| BTC | P35070 | Probetacellulin | PC1 | 1.18 | 0.146 | 0.298 |
| CA12 | O43570 | Carbonic anhydrase 12 | PC1, PC2 | 1.52 | 0.007 | 0.042 |
| CA14 | Q9ULX7 | Carbonic anhydrase 14 | PC2 | 1.76 | 0.049 | 0.137 |
| CALCA | P06881 | Calcitonin gene-related peptide 1 | PC1 | 2.36 | 0.005 | 0.034 |
| CALR | P27797 | Calreticulin | PC1, PC2 | 1.36 | 0.002 | 0.022 |
| CLEC1A | Q8NC01 | C-type lectin domain family 1 member A | PC1, PC2 | 1.84 | 0.004 | 0.031 |
| DSG4 | Q86SJ6 | Desmoglein-4 | PC2 | 1.37 | 0.471 | 0.567 |
| EGFL7 | Q9UHF1 | Epidermal growth factor-like protein 7 | PC1 | 1.26 | 0.030 | 0.106 |
| ENAH | Q8N8S7 | Protein enabled homolog | PC1 | 1.25 | 0.219 | 0.352 |
| ENTPD6 | O75354 | Ectonucleoside triphosphate diphosphohydrolase 6 | PC1 | 1.25 | 0.300 | 0.454 |
| FABP9 | Q0Z7S8 | Fatty acid-binding protein 9 | PC2 | 1.27 | 0.093 | 0.197 |
| FES | P07332 | Tyrosine-protein kinase Fes/Fps | PC1, PC2 | 1.07 | 0.754 | 0.816 |
| FGR | P09769 | Tyrosine-protein kinase Fgr | PC1 | 1.08 | 0.639 | 0.736 |
| FOSB | P53539 | Protein fosB | PC1 | 1.22 | 0.076 | 0.168 |
| FOXO1 | Q12778 | Forkhead box protein O1 | PC1, PC2 | 1.08 | 0.379 | 0.515 |
| INPPL1 | O15357 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | PC1 | 1.21 | 0.300 | 0.442 |
| KIM1 | Q96D42 | Kidney Injury Molecule 1 | PC1 | 2.60 | < 0.001 | 0.008 |
| LAT2 | Q9GZY6 | Linker for activation of T-cells family member 2 | PC1 | 1.53 | 0.066 | 0.175 |
| LTA4H | P09960 | Leukotriene A-4 hydrolase | PC2 | 0.99 | 0.802 | 0.818 |
| MAEA | Q7L5Y9 | E3 ubiquitin-protein transferase MAEA | PC1 | 2.66 | 0.186 | 0.329 |
| MAP4K5 | Q9Y4K4 | Mitogen-activated protein kinase kinase kinase kinase 5 | PC1 | 0.68 | 0.285 | 0.444 |
| MVK | Q03426 | Mevalonate kinase | PC1, PC2 | 1.76 | 0.186 | 0.318 |
| NBN | O60934 | Nibrin | PC1 | 1.27 | 0.379 | 0.502 |
| NOS3 | P29474 | Nitric oxide synthase, endothelial | PC1 | 1.77 | 0.038 | 0.119 |
| NPPC | P23582 | C-type natriuretic peptide | PC1 | 4.67 | < 0.001 | 0.010 |
| NUB1 | Q9Y5A7 | NEDD8 ultimate buster 1 | PC1 | 1.16 | 0.471 | 0.555 |
| NUCB2 | P80303 | Nucleobindin-2 | PC2 | 1.76 | 0.002 | 0.019 |
| PDGFC | Q9NRA1 | Platelet-derived growth factor C | PC1 | 1.30 | 0.330 | 0.460 |
| PGF | P43088 | Prostaglandin F2-alpha receptor | PC2 | 1.81 | < 0.001 | 0.015 |
| PON2 | Q15165 | Serum paraoxonase/arylesterase 2 | PC2 | 0.98 | 0.802 | 0.802 |
| PRKAB1 | Q9Y478 | 5′-AMP-activated protein kinase subunit beta-1 | PC1 | 1.33 | 0.012 | 0.058 |
| PRKRA | O75569 | IFN-inducible dsRNA-dependent protein kinase activator A | PC1 | 1.02 | 0.754 | 0.799 |
| PTK7 | Q13308 | Inactive tyrosine-protein kinase 7 | PC2 | 1.67 | 0.008 | 0.043 |
| PTPRJ | Q12913 | Receptor-type tyrosine-protein phosphatase β | PC1 | 1.06 | 0.300 | 0.430 |
| PVALB | P20472 | Parvalbumin alpha | PC1 | 2.12 | 0.186 | 0.308 |
| PXN | P49023 | Paxillin | PC1 | 1.07 | 0.379 | 0.490 |
| RARRES1 | P49788 | Retinoic acid receptor responder protein 1 | PC1 | 1.33 | 0.023 | 0.087 |
| RASSF2 | P50749 | Ras association domain-containing protein 2 | PC1, PC2 | 0.96 | 0.684 | 0.771 |
| RRM2B | Q7LG56 | Ribonucleoside-diphosphate reductase subunit M2 B | PC1 | 2.13 | 0.021 | 0.086 |
| SERPINA9 | Q86WD7 | Serpin A9 | PC2 | 1.10 | 0.433 | 0.534 |
| ST3GAL1 | Q11201 | CMP-N-acetylneuraminate-β-galactosamide-α-2,3-sialyltransferase 1 | PC2 | 1.06 | 0.731 | 0.807 |
| STX8 | Q9UNK0 | Syntaxin-8 | PC1 | 1.53 | 0.033 | 0.110 |
| TIGAR | Q9NQ88 | Fructose-2,6-bisphosphatase | PC1 | 1.72 | 0.045 | 0.133 |
| TMPRSS15 | P98073 | Enteropeptidase | PC2 | 1.67 | 0.146 | 0.287 |
| TNNI3 | P19429 | Troponin I, cardiac muscle | PC1 | 3.22 | 0.071 | 0.164 |
| TOP2B | Q02880 | DNA topoisomerase 2-β | PC1 | 1.35 | 0.146 | 0.277 |
| VEGFC | P49767 | Vascular endothelial growth factor C | PC2 | 2.61 | 0.066 | 0.167 |
| YES1 | P07947 | Tyrosine-protein kinase Yes | PC1 | 1.39 | 0.066 | 0.159 |
Principal component analysis revealed 38 proteins to be correlated with PC1 and 22 proteins to be associated with PC2, of which nine proteins are different between AKI as compared to non-AKI controls (bold). AKI acute kidney injury, FC fold change.
Figure 2.
Plasma protein levels in sepsis patients with acute kidney injury as compared to patients without acute kidney injury. (A) Volcano plot demonstrates the log2 of the relative amount of plasma proteins and the − 10log p-value. Proteins with p-value < 0.01 are labelled in the volcano plot. (B) Boxplots demonstrating the nine upregulated proteins in patients with sepsis-AKI and sepsis. The boxplots represent the interquartile range of the protein levels and the whiskers represent the min/max range. AKI acute kidney injury.
Biological processes in the development of AKI during early sepsis
To analyse important pathways that were associated with the future development of AKI in sepsis, PCA proteins with a fold change > 1 were used to perform GO and pathway analyses using Enrichr. GO analysis showed an increase in the following molecular functions in patients with sepsis-AKI compared to matched pairs: immunoglobulin receptor binding (GO:0034987) (p = 0.041), carbonate dehydratase activity (GO:0004089) (p = 0.009) and platelet-derived growth factor receptor binding (GO:0005161) (p = 0.009) (Table 4). Further, Enrichr pathway analysis revealed nineteen pathways to be associated with sepsis-AKI (Table 5). Ranking the pathways using the combined scores demonstrated seven pathways above 200, revealing activation of endothelium (i.e. VEGF signalling, Angiopoietin receptor Tie2 and PDGF signalling). Together, Enrichr GO showed three molecular functions and nineteen biological pathways to be associated with sepsis-AKI, demonstrating an important role for endothelium in early prediction of progression towards sepsis-AKI.
Table 4.
GO analysis molecular function by Enrichr.
| Gene ontology | p-value | Adjusted p-value | Odds Ratio | Combined score |
|---|---|---|---|---|
| Immunoglobulin receptor binding (GO:0034987) | < 0.001 | 0.004 | 216.83 | 2034.63 |
| Carbonate dehydratase activity (GO:0004089) | < 0.001 | 0.008 | 96.34 | 779.62 |
| Platelet-derived growth factor receptor binding (GO:0005161) | < 0.001 | 0.009 | 78.82 | 610.51 |
Proteins that correlated with PC1 or PC2 and were increased among patients with AKI as compared to patients without AKI were used to identify pathways affected by sepsis-AKI.
Discussion
Here, we used a targeted proteomic analysis of plasma derived from patients with sepsis admitted to the ED to explore the role of a set of proteins in the pathophysiology of sepsis-AKI. PCA revealed 53 proteins to be correlated with the development of sepsis-AKI, of which 48 proteins increased in sepsis-AKI and only 5 proteins decreased. Of these correlated proteins, nine also contributed to discrimination in PCA and were all significantly increased in sepsis-AKI (CLEC1A, CALR, KIM1, CA12, CALCA, NUCB2, PGF, PTK7 and NPPC). From the 48 proteins increased in sepsis-AKI, GO analysis demonstrated activation of immunoglobulin receptor binding, carbonate dehydratase activity and platelet-derived growth factor receptor binding in patients with sepsis-AKI. Enrichr pathway analyses assigned vascular endothelium activation and extracellular matrix remodelling during sepsis-AKI. The proteomic analysis of early sepsis patients at the ED demonstrated insights into pathways involved in the development of early AKI.
In the development of sepsis-AKI, PCA based GO analysis demonstrated three molecular functions of the proteins, associated with sepsis-AKI, namely immunoglobulin receptor binding, carbonate dehydratase activity and platelet-derived growth factor receptor binding. First, based on the molecular function (protein activity), the increased immunoglobulin receptors indicate altered immune cell activity by increased capacity of immune cells for immune regulatory functions. During sepsis-AKI, immune cells (i.e. neutrophils, macrophages and dendritic cells) infiltrate through the endothelial barrier to the kidney aggravating kidney injury by releasing ROS and proteases20–22. Second, given that carbonate dehydratase activity is mainly located inside the kidney and elevated during hypoxia, its increased circulating plasma levels likely indicate kidney injury because of increased circulating enzyme23,24. Third, the platelet-derived growth factor receptor binding exerts its stimulatory effects on cell growth, proliferation and fibrosis formation and it is released during kidney injury by endothelium and kidney cells25,26. Thus, GO indicates kidney injury by increased release of kidney carbonate dehydratase activity and increased vascular endothelial and kidney remodelling pathways via platelet-derived growth factor receptor binding during sepsis-AKI.
Pathway analyses support the role for endothelium in the development of AKI during sepsis27,28. The first three involved signalling events, namely VEGFR1 and VEGFR2, angiopoietin receptor Tie2-mediated signalling and PDGF receptor signalling network are mainly involved in vascular endothelial regulation. The pathophysiology of endothelium and kidney during sepsis-AKI is provoked by immune cells recruitment, leading to elevated ROS and cytokines release, abating endothelial function and aggravating sepsis-AKI21,22,27,29. Together, these pathways hint at a crucial role of endothelial signalling systems in subsequent development kidney dysfunction27,28,30. Further, the subsequent four signalling pathways, class I PI3K signalling events, alpha9 beta1 integrin signalling events, glypican 1 network and thromboxane A2 receptor signalling, reflect the response of endothelia and kidney cells due to tissue injury. Class I PI3K signalling is upregulated and has ambivalent effects in mediating kidney injury by supporting protective pathways in preclinical AKI models31,32,33, where Alpha9 beta1 integrin signalling events is used by endothelial and kidney cells to migrate and adhere to the extracellular matrix, thereby mediating remodelling and fibrosis formation, indicating the loss of functional endothelial and kidney tissue34,35. Circulating glypican 1 network originates from a dysfunctional glycocalyx, an endothelial barrier, associated with organ failure during infections. Sepsis-induced damage of the endothelial glycocalyx breaks down vascular barrier and contributes to microcirculatory changes in sepsis-AKI36,37. Further, an activated thromboxane A2 pathway causes platelet adhesion, capillary thrombosis and mediates vasoconstriction, decreasing kidney blood flow and function38–40. These findings are in line with other studies reporting the role of endothelial dysfunction in the development of sepsis-AKI41,42,43. Together, plasma proteomic profile of sepsis patients strongly suggest that endothelium dysfunction precedes development of acute kidney injury, suggesting that an endothelium protective strategy early in sepsis may protect from the development of sepsis-AKI.
We revealed underlying pathways involved in the development of AKI during sepsis compared to non-AKI sepsis. Current literature on sepsis-AKI described the microvascular network via leakage and signalling as critical contributor to the progression of kidney injury. The role of endothelial loss altering kidney function is thought to be due to hypoxia and endothelial barrier loss promoting extravasation of bacterial toxins and signalling molecules, in turn affecting underlying organs27,44,45,46. In this paper we emphasize the importance of the relation between endothelium and kidney function.
A potential limitation of the current study is that the organ damage panel is limited to quantification of 92 proteins. Further, the study design consists of a small cohort of 38 patients in a single tertiary hospital in the Netherlands, which allowed us to identify potential pathways involved in sepsis-AKI, but not to validate individual biomarkers. Proteins involved in the pathophysiology of AKI can after validation be selected for routine lab analyses, such as CRP. Additionally, we performed a pathway analysis on a targeted proteomic approach which may bias towards specific pathways (over)represented in the targeted proteomics. For future perspectives a larger cohort can be used to differentiate between transient and persistent AKI47. The values determined using the oLink multiarray are relative and, therefore, cannot be directly compared to the absolute values of different biomarkers or values obtained from other panels. Further, although our hospital serves a diverse patient population from a substantial geographical area of both urban and rural nature, this study was performed in an academic tertiary care teaching hospital, which may limit generalizability to other settings.
Conclusion
Principal component analyses of the targeted proteomics revealed 53 proteins to be associated with the development of AKI among patients with early sepsis. Gene ontology (GO) of the proteins from PC1 and PC2 analyses with a fold change > 1, indicated increased immune antibody binding capacity, release of kidney carbonate dehydratase activity and platelet-derived growth factor receptor binding. Pathway analyses reported endothelial activation during kidney dysfunction in the pathophysiology of sepsis-AKI. Further, unravelling the pathophysiology of sepsis-AKI could be of major relevance to identify targets that enable early recognition or may serve to develop novel strategies to prevent the development AKI and optimize the outcome after sepsis.
Author contributions
B.S.S. performed and designed experiments, analysed data, produced graphs, wrote the manuscript. C.K.B. analysed data, produced graphs, wrote the manuscript. E.C.S. performed and designed experiments and analysed data, produced graphs, wrote the manuscript. V.M.Q. designed study protocol, assisted with the data collection. J.C.M. designed study protocol, supervised the work. R.H.H. designed study protocol, supervised the work, revised the final version of the manuscript. V.K. supervised the work, designed experiments, assisted in writing the manuscript and revised the final version of the manuscript. H.R.B. supervised the work, designed experiments, analysed data, assisted in writing the manuscript and revised the manuscript.
Funding
The current work was financially supported by a Mandema stipend from the UMCG and a Kolff Grant (16OKG06) from the Dutch Kidney Foundation awarded to HRB, and by two MD/PhD program grants from the UMCG awarded to BSS and ECvdS.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fleischmann C, et al. Assessment of global incidence and mortality of hospital-treated sepsis: Current estimates and limitations. Am. J. Respir. Crit. Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 2.Kumar G, et al. Nationwide trends of severe sepsis in the 21st century (2000–2007) Chest. 2011;140:1223–1231. doi: 10.1378/chest.11-0352. [DOI] [PubMed] [Google Scholar]
- 3.Mayr FB, Yende S, Angus DC. Epidemiology of severe sepsis. Virulence. 2014;5:4–11. doi: 10.4161/viru.27372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudd KE, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoller J, et al. Epidemiology of severe sepsis: 2008–2012. J. Crit. Care. 2016;31:58–62. doi: 10.1016/j.jcrc.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 6.Pfister D, et al. Cerebral perfusion in sepsis-associated delirium. Crit. Care. 2008;12:R63. doi: 10.1186/cc6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent JL, et al. Sepsis in European intensive care units: Results of the SOAP study. Crit. Care Med. 2006;34:344–353. doi: 10.1097/01.ccm.0000194725.48928.3a. [DOI] [PubMed] [Google Scholar]
- 8.Poukkanen M, et al. Acute kidney injury in patients with severe sepsis in Finnish Intensive Care Units. Acta Anaesthesiol. Scand. 2013;57:863–872. doi: 10.1111/aas.12133. [DOI] [PubMed] [Google Scholar]
- 9.Gordon AC, et al. Effect of early vasopressin vs norepinephrine on kidney failure in patients with septic shock: The VANISH randomized clinical trial. JAMA. 2016;316:509–518. doi: 10.1001/jama.2016.10485. [DOI] [PubMed] [Google Scholar]
- 10.Flannery AH, et al. Sepsis-associated acute kidney disease and long-term kidney outcomes. Kidney Med. 2021;3:507–514. doi: 10.1016/j.xkme.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coca SG, Singanamala S, Parikh CR. Chronic kidney disease after acute kidney injury: A systematic review and meta-analysis. Kidney Int. 2012;81:442–448. doi: 10.1038/ki.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neyra JA, et al. Impact of acute kidney injury and CKD on adverse outcomes in critically ill septic patients. Kidney Int. Rep. 2018;3:1344–1353. doi: 10.1016/j.ekir.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poston JT, Koyner JL. Sepsis associated acute kidney injury. BMJ. 2019;364:k4891. doi: 10.1136/bmj.k4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron. Clin. Pract. 2012;120:c179–184. doi: 10.1159/000339789. [DOI] [PubMed] [Google Scholar]
- 15.Luo XQ, et al. Expressing the Modification of Diet in Renal Disease Study equation for estimating glomerular filtration rate with standardized serum creatinine values. Clin Chem. 2007;53:766–772. doi: 10.1373/clinchem.2006.077180. [DOI] [PubMed] [Google Scholar]
- 16.Assarsson E, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen EY, et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuleshov MV, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kara S, et al. Impact of plasma membrane domains on IgG Fc receptor function. Front. Immunol. 2020;11:1320. doi: 10.3389/fimmu.2020.01320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinsey GR, Okusa MD. Role of leukocytes in the pathogenesis of acute kidney injury. Crit. Care. 2012;16:214. doi: 10.1186/cc11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SA, Noel S, Sadasivam M, Hamad ARA, Rabb H. Role of immune cells in acute kidney injury and repair. Nephron. 2017;137:282–286. doi: 10.1159/000477181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waheed A, Sly WS. Carbonic anhydrase XII functions in health and disease. Gene. 2017;623:33–40. doi: 10.1016/j.gene.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supuran CT. Experimental carbonic anhydrase inhibitors for the treatment of hypoxic tumors. J. Exp. Pharmacol. 2020;12:603–617. doi: 10.2147/JEP.S265620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostendorf T, Boor P, van Roeyen CR, Floege J. Platelet-derived growth factors (PDGFs) in glomerular and tubulointerstitial fibrosis. Kidney Int. Suppl. 2014;2011(4):65–69. doi: 10.1038/kisup.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boor P, Ostendorf T, Floege J. PDGF and the progression of renal disease. Nephrol. Dial. Transplant. 2014;29(Suppl 1):i45–i54. doi: 10.1093/ndt/gft273. [DOI] [PubMed] [Google Scholar]
- 27.Molema G, Zijlstra JG, van Meurs M, Kamps J. Renal microvascular endothelial cell responses in sepsis-induced acute kidney injury. Nat. Rev. Nephrol. 2022;18:95–112. doi: 10.1038/s41581-021-00489-1. [DOI] [PubMed] [Google Scholar]
- 28.Ince C, et al. The endothelium in sepsis. Shock. 2016;45:259–270. doi: 10.1097/SHK.0000000000000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato W, et al. Selective stimulation of VEGFR2 accelerates progressive renal disease. Am. J. Pathol. 2011;179:155–166. doi: 10.1016/j.ajpath.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saharinen P, Eklund L, Alitalo K. Therapeutic targeting of the angiopoietin-TIE pathway. Nat. Rev. Drug. Discov. 2017;16:635–661. doi: 10.1038/nrd.2016.278. [DOI] [PubMed] [Google Scholar]
- 31.Meng F, et al. PI3K/AKT activation attenuates acute kidney injury following liver transplantation by inducing FoxO3a nuclear export and deacetylation. Life Sci. 2021;272:119119. doi: 10.1016/j.lfs.2021.119119. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, et al. Dexmedetomidine protects against lipopolysaccharide-induced acute kidney injury by enhancing autophagy through inhibition of the PI3K/AKT/mTOR pathway. Front. Pharmacol. 2020;11:128. doi: 10.3389/fphar.2020.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang G, et al. Protective effect of tempol on acute kidney injury through PI3K/Akt/Nrf2 signaling pathway. Kidney Blood Press. Res. 2016;41:129–138. doi: 10.1159/000443414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pozzi A, Zent R. Integrins in kidney disease. J. Am. Soc. Nephrol. 2013;24:1034–1039. doi: 10.1681/ASN.2013010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnittert J, Bansal R, Storm G, Prakash J. Integrins in wound healing, fibrosis and tumor stroma: High potential targets for therapeutics and drug delivery. Adv. Drug. Deliv. Rev. 2018;129:37–53. doi: 10.1016/j.addr.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Mahmoud M, et al. The glycocalyx core protein Glypican 1 protects vessel wall endothelial cells from stiffness-mediated dysfunction and disease. Cardiovasc. Res. 2021;117:1592–1605. doi: 10.1093/cvr/cvaa201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher J, Linder A, Bentzer P. Elevated plasma glypicans are associated with organ failure in patients with infection. Intensive Care Med. Exp. 2019;7:2. doi: 10.1186/s40635-018-0216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulkarni AP, Bhosale SJ. Epidemiology and pathogenesis of acute kidney injury in the critically ill patients. Indian J. Crit. Care Med. 2020;24:S84–S89. doi: 10.5005/jp-journals-10071-23394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boffa JJ, Just A, Coffman TM, Arendshorst WJ. Thromboxane receptor mediates renal vasoconstriction and contributes to acute renal failure in endotoxemic mice. J. Am. Soc. Nephrol. 2004;15:2358–2365. doi: 10.1097/01.ASN.0000136300.72480.86. [DOI] [PubMed] [Google Scholar]
- 40.Mederle K, Meurer M, Castrop H, Hocherl K. Inhibition of COX-1 attenuates the formation of thromboxane A2 and ameliorates the acute decrease in glomerular filtration rate in endotoxemic mice. Am. J. Physiol. Renal Physiol. 2015;309:F332–340. doi: 10.1152/ajprenal.00567.2014. [DOI] [PubMed] [Google Scholar]
- 41.Ebihara I, et al. Angiopoietin balance in septic shock patients with acute kidney injury: Effects of direct hemoperfusion with polymyxin B-immobilized fiber. Ther. Apher. Dial. 2016;20:368–375. doi: 10.1111/1744-9987.12468. [DOI] [PubMed] [Google Scholar]
- 42.Katayama S, et al. Markers of acute kidney injury in patients with sepsis: The role of soluble thrombomodulin. Crit. Care. 2017;21:229. doi: 10.1186/s13054-017-1815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson-Cohen C, et al. Association of markers of endothelial dysregulation Ang1 and Ang2 with acute kidney injury in critically ill patients. Crit. Care. 2016;20:207. doi: 10.1186/s13054-016-1385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basile DP. The endothelial cell in ischemic acute kidney injury: Implications for acute and chronic function. Kidney Int. 2007;72:151–156. doi: 10.1038/sj.ki.5002312. [DOI] [PubMed] [Google Scholar]
- 45.Gomez H, et al. A unified theory of sepsis-induced acute kidney injury: Inflammation, microcirculatory dysfunction, bioenergetics, and the tubular cell adaptation to injury. Shock. 2014;41:3–11. doi: 10.1097/SHK.0000000000000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldenberg NM, Steinberg BE, Slutsky AS, Lee WL. Broken barriers: A new take on sepsis pathogenesis. Sci. Transl. Med. 2011;3:8825. doi: 10.1126/scitranslmed.3002011. [DOI] [PubMed] [Google Scholar]
- 47.Luo XQ, et al. Machine learning for early discrimination between transient and persistent acute kidney injury in critically ill patients with sepsis. Sci. Rep. 2021;11:20269. doi: 10.1038/s41598-021-99840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.