Abstract
Background:
The epithelial Na+ channel (ENaC) is intrinsically linked to fluid volume homeostasis and blood pressure. Specific rare mutations in SCNN1A, SCNN1B, and SCNN1G, genes encoding the α, β, and γ subunits of ENaC, respectively, are associated with extreme blood pressure phenotypes. No associations between blood pressure and SCNN1D, which encodes the δ subunit of ENaC, have been reported. A small number of sequence variants in ENaC subunits have been reported to affect functional transport in vitro and/or blood pressure. The effects of the vast majority of rare and low frequency ENaC variants on blood pressure are not known.
Methods:
We explored the association of low frequency and rare variants in the genes encoding ENaC subunits, with systolic blood pressure (SBP), diastolic blood pressure (DBP), mean arterial pressure (MAP), and pulse pressure (PP). Using whole-genome sequencing data from fourteen studies participating in the TOPMed Whole-Genome Sequencing Program and sequence kernel association tests.
Results:
We found that variants in SCNN1A and SCNN1B were associated with DBP and MAP (p < 0.00625). Although SCNN1D is poorly expressed in human kidney tissue, SCNN1D variants were associated with SBP, DBP, MAP, and PP (p < 0.00625). ENaC variants in two of the four subunits (SCNN1B, and SCNN1D) were also associated with estimated glomerular filtration rate (p < 0.00625), but not with stroke.
Conclusions:
Our results suggest that variants in extrarenal ENaCs, in addition to ENaCs expressed in kidneys, influence blood pressure and kidney function.
Keywords: ENaC, blood pressure, estimated glomerular filtration rate, stroke, gene variants
Graphical Abstract
Introduction
The regulated absorption of filtered Na+ along the nephron helps govern extracellular fluid volume and blood pressure. The aldosterone-sensitive distal nephron (ASDN) has a critical role in Na+ absorption and is where key volume regulatory hormones and signaling pathways affect the absorption of filtered Na+. The epithelial Na+ channel (ENaC) is expressed in the distal aspects of the ASDN where it mediates the absorption of filtered Na+ across the luminal membrane of principal cells1,2. In addition to its role in Na+ absorption, ENaC function is required for K+ secretion in the ASDN1,3. ENaCs are also found at other tissues that affect total body Na+ and blood pressure. It functions as a salt sensor in lingual epithelia and influences salt intake, whereas expression in the distal colon has a role, albeit minor, in absorption of ingested Na+4,5. ENaC expression in antigen presenting immune cells has been proposed to facilitate release of cytokines in response to increased salt intake, which contribute to an increase in blood pressure6,7. ENaCs in vascular smooth muscle and endothelial cells may influence vascular tone7,8. Finally, ENaCs are expressed at specific sites in the central nervous system where they influence autonomic tone and blood pressure9.
ENaCs are formed from structurally related subunits, termed α, β, γ, and δ, which are encoded by the genes SCNN1A, SCNN1B, SCNN1G, and SCNN1D, respectively. These are Na+ selective channels that are primarily αβγ or δβγ heterotrimers, which exhibit differences in functional and regulatory properties2. ENaCs in human kidney are primarily a αβγ heterotrimer10, while ENaCs containing the δ subunit are expressed in other tissues11,12. Each ENaC subunit has two transmembrane domains. The second transmembrane domain from each subunit within a channel complex contributes to the channel pore, cation selectivity filter, and gate. The transmembrane domains are connected by a large, structurally complex extracellular domain that functions as a sensor, where specific extracellular factors, including monovalent cations and anions (Na+, Cl−, H+), peptides, proteases, and shear stress interact with the extracellular domain to regulate channel activity2. Short cytoplasmic amino (N)- and carboxyl (C)-termini also have key regulatory sites. For example, a Pro-Tyr (PY) motif in the cytoplasmic C-termini of ENaC subunits is a binding site for the ubiquitin ligase NEDD4–2 that facilitates channel ubiquitination at the cell surface and subsequent internalization13. The cytoplasmic N-termini of ENaC subunits have a His-Gly motif that affects channel gating14.
Several rare ENaC variants that have large effects on blood pressure with Mendelian inheritance have been identified in SCNN1A, SCNN1B, and SCNN1G. These disorders include (i) Liddle syndrome, an autosomal dominant disorder where specific gain-of-function mutations in SCNN1B, and SCNN1G are associated with hypertension and hypokalemia, and (ii) pseudohypoaldosteronism type I (PHA1), an autosomal recessive disorder where specific loss-of-function mutations in SCNN1A, SCNN1B, or SCNN1G are associated with hypotension and hyperkalemia15. Liddle syndrome mutations primarily result in disruption of the PY motif, significantly increasing channel residency time at the cell surface13. PHA1 mutations have been described that result in specific deletions or amino acid substitutions that cause a profound loss of function in vitro14,15.
Our group and others have identified a growing number of sites, including rare single nucleotide variants (SNVs) within the extracellular domains, where mutations affect channel gating activity2,12. This is congruent with the extracellular domains’ role in sensing extracellular factors and regulating channel gating in response to these factors. For most variants that alter ENaC function and are not associated with Liddle syndrome or PHA1, it is unclear whether they influence blood pressure, serum [K+], or the prevalence of blood pressure-associated disorders including stroke, myocardial infarction, and chronic kidney disease in specific populations.
Using whole-genome sequencing (WGS) and phenotype data available through the Trans-Omics in Precision Medicine (TOPMed) Whole-Genome Sequencing Project, we examined common functional human ENaC variants and arrays of low-frequency and rare ENaC variants for association with blood pressure levels and related traits and health outcomes.
Materials and Methods
WGS and harmonized blood pressure phenotype data were available for analysis from 28,058 participants in fourteen studies from TOPMed (selected from the > 142,000 individuals in forty-one studies in TOPMed) (Table 1). After excluding 300 individuals < 20 years old or > 90 years old and including 641 individuals from the Samoan Soifua Manuia Study that were not included in the TOPMed cohort, 28,399 individuals from the harmonized blood pressure phenotype dataset were included in our main (blood pressure) analyses. Subsets of this sample were included in our secondary analyses: 9,090 for total strokes (4,399 cases and 4,691 controls), and 14,557 for eGFR. The individuals included in our main analyses were from six self-reported ancestry groups (58.3% European, 29.9% African, 6.3% Asian, 4.1% Samoan, 0.2% Native American, and 1.2% other) (Table S2). To account for underlying population substructure, we used principal components of ancestry (PCAs) calculated by TOPMed. Based on the spaghetti plot (Figure S1), we concluded that PCAs 1–11 accounted for the vast majority of inter-ancestry variance in our population and should be included as covariates in our analyses. Relatedness in the analysis cohorts was controlled for using a genetic relatedness matrix (GRM) calculated by TOPMed.
Table 1. Studies included in the blood pressure analyses.
The Genetics of Cardiometabolic Health in Amish and Women’s Health Initiative (WHI) studies are subdivided according to the abbreviated study name provided in the harmonized dataset.
| Study | n | Abbreviation |
|---|---|---|
| Genetics of Cardiometabolic Health in the Amish | 732 | Amish_FC13 |
| 269 | Amish_FC2 | |
| Atherosclerosis Risk in Communities Study VTE cohort | 3,039 | ARIC |
| Cleveland Family Study | 744 | CFS |
| Framingham Heart Study | 3,270 | FHS |
| Genetic Studies of Atherosclerosis Risk | 1,626 | GeneSTAR |
| Genetic Epidemiology Network of Arteriopathy | 1,138 | GENOA |
| Genetic Epidemiology Network of Salt Sensitivity | 1,616 | GenSalt |
| Hypertension Genetic Epidemiology Network | 1,635 | HyperGen |
| Jackson Heart Study | 3,124 | JHS |
| Samoan | 1,167 | Samoan |
| Women’s Health Initiative | 4,692 | WHI_ctr |
| 4,403 | WHI_stroke_case | |
| 944 | WHI_VTE_case | |
| Total | 28,399 |
We conducted single variant analyses for three common (minor allele frequency (MAF) ≥ 0.05) ENaC variants, two of which are functional variants16,17. We also performed sequence kernel association tests (SKAT) and burden tests for all low-frequency and rare variants (MAF < 0.05), as well as separate SKAT for rare (MAF < 0.01) and low-frequency (0.01 ≤ MAF < 0.05) SNVs within the genomic regions of SCNN1A, SCNN1B, SCNN1D, and SCNN1G. In each analysis, we have tested for association with systolic blood pressure (SBP), diastolic blood pressure (DBP), pulse pressure (PP), mean arterial pressure (MAP), estimated glomerular filtration rate (eGFR), and stroke.
Finally, we assessed SCNN1D expression in monocytes from eleven healthy patients. Monocytes isolated from heparinized blood were cultured in media containing either 150 mM NaCl or 190 mM NaCl for 72 hours. Total RNA isolated from the monocytes was sequenced, and expression analysis was performed to determine whether SCNN1D was expressed in those cells.
Additional methods details are presented in the Supplemental Materials18–27.
Results
Three common functional ENaC variants—αA334T, αT663A, and βG165R—are not associated with SBP, DBP, MAP, or PP
Two common ENaC α subunit variants, αA334T (rs11542844, MAF = 0.17) and αT663A (rs2228576, MAF = 0.27) alter channel function in heterologous expression systems16,17, although previous studies largely suggest that these variants do not affect blood pressure phenotypes28–30. There is also a common ENaC variant in the β subunit, βG165R (rs2303157, MAF = 0.24), although its effect on ENaC function has not been described. To account for the impact of these common variants in our analyses of low frequency variants, we tested whether any of these common variants were associated with blood pressure phenotypes. We conducted single-variant analyses for the association of each variant with SBP, DBP, MAP and PP using age, sex, BMI, study, and the first eleven PCAs as covariates and controlled for relatedness with a GRM. No association was found between any of the variants and SBP, DBP, MAP, or PP (Table 2). These results are largely consistent with previous findings for αA334T and αT663A16,17,28–30.
Table 2. Single-variant association tests of common ENaC variants with SBP, DBP, and PP.
Results of single-variant analyses of two common functional ENaC α subunit variants and one common ENaC β subunit variant for associations with SBP, DBP, and PP.
| Variant | rs ID | MAF | p value | |||
|---|---|---|---|---|---|---|
| SBP | DBP | MAP | PP | |||
| αA334T | rs11542844 | 0.18 | 0.9 | 0.9 | 0.9 | 0.7 |
| αT663A | rs2228576 | 0.75 | 0.1 | 0.6 | 0.9 | 0.06 |
| βG165R | rs2303157 | 0.24 | 0.3 | 0.9 | 0.6 | 0.07 |
Low frequency and rare ENaC variation in specific subunits is, in aggregate, associated with SBP, DBP, MAP or PP
We performed SKAT to determine the impact of aggregated low frequency (MAF < 0.05) variation within the genomic regions of SCNN1A, SCNN1B, SCNN1D, and SCNN1G on four blood pressure measures (SBP, DBP, PP, and MAP) using age, sex, BMI, and PCAs 1–11 as covariates, and controlled for relatedness using a GRM. The threshold for significance, based on the number of genes tested, was 0.00625 (see Methods). There were 3,972 SCNN1A, 12,334 SCNN1B, 2,405 SCNN1D, and 4,093 SCNN1G variants with a MAF < 0.05 within the promoter regions and gene boundaries of each gene that also had genotype missingness rates < 0.15. These variants present across the 14 studies were included in the analyses. There was a significant association of SCNN1A aggregate variants with DBP (p = 0.00004) and MAP (p = 0.0002), but not with SBP or PP (Table 4). SCNN1B variants were significantly associated with DBP (p = 0.002) and MAP (p = 0.003). SCNN1D variants were significantly associated with all of four blood pressure phenotypes: SBP (p = 0.0008), DBP (p = 0.002), PP (p = 0.003), and MAP (p = 0.0008). Finally, those at SCNN1G were not significantly associated with any of the four blood pressure phenotypes. As a negative control, DBP was permuted by reassignment at random in the population and no associations were observed between any of the subunit genes and the permuted phenotype values. We also performed SKAT with the combined variants from SCNN1A, SCNN1B, and SCNN1G (20,399 in total) and each of the four blood pressure measures. Because of the associations with SBP, DBP, PP, and MAP, SCNN1D variants were not included in the combined analyses of blood pressure phenotypes to avoid biasing the results. The combined analysis showed association with both DBP (p = 0.0002) and MAP (p = 0.0006, see Table 3).
Table 4. Sequence kernel association tests of low frequency and rare ENaC variants with eGFR.
SKAT was performed on variants with a MAF < 0.05 in the genomic regions of SCNN1A, SCNN1B, SCNN1D, and SCNN1G with eGFR, separately, and then combined. SKAT was separately performed on rare (MAF < 0.01) and low frequency (0.01 ≤ MAF < 0.05) variants. Significant p values are in bold.
| Gene. Chromosome:Start–End | Variants with MAF < 0.05 | Variants with MAF < 0.01 | Variants with 0.01 ≤ MAF < 0.05 | |||
|---|---|---|---|---|---|---|
| n | p value | n | p value | n | p value | |
| SCNN1A. 12:6346824–6379751 | 2,739 | 0.01 | 2,626 | 0.008 | 113 | 0.07 |
| SCNN1B. 16:2329812–23381320 | 8,770 | 0.005 | 8,365 | 0.007 | 405 | 0.03 |
| SCNN1D. 1:1280417–1292050 | 1,758 | 0.002 | 1,648 | 0.2 | 110 | 0.001 |
| SCNN1G. 16:23184696–23216904 | 2,868 | 0.1 | 2,797 | 0.01 | 71 | 0.1 |
| Combined (SCNN1A, SCNN1B, SCNN1D, SCNN1G) | 16,135 | 0.0003 | 15,436 | 0.002 | 699 | 0.003 |
p values < 0.00625 are significant
Table 3. Sequence kernel association tests of low frequency and rare ENaC variants with blood pressure measures.
SKAT was performed on variants with a MAF < 0.05 in the genomic regions of SCNN1A, SCNN1B, SCNN1D, and SCNN1G with SBP, DBP, PP, and MAP, separately, and then combined (SCNN1A, SCNN1B and SCNN1G). SKAT was separately performed on rare (MAF < 0.01) and low frequency (0.01 ≤ MAF < 0.05) variants. Significant p values are in bold.
| Gene. Chromosome:Start–End | Variants with MAF < 0.05 | Variants with MAF < 0.01 | Variants with 0.01 ≤ MAF < 0.05 | ||||
|---|---|---|---|---|---|---|---|
| Measure | n | p value | n | p value | n | p value | |
|
SCNN1A. 12:6346824–6379751 |
SBP | 3,972 | 0.02 | 3,859 | 0.03 | 113 | 0.07 |
| DBP | 3,972 | 0.00004 | 3,859 | 0.0004 | 113 | 0.0007 | |
| PP | 3,972 | 0.7 | 3,859 | 0.3 | 113 | 0.9 | |
| MAP | 3,972 | 0.0002 | 3,859 | 0.002 | 113 | 0.002 | |
|
SCNN1B. 16:2329812–23381320 |
SBP | 12,334 | 0.02 | 11,925 | 0.04 | 409 | 0.04 |
| DBP | 12,334 | 0.002 | 11,925 | 0.003 | 409 | 0.01 | |
| PP | 12,334 | 0.3 | 11,925 | 0.3 | 409 | 0.04 | |
| MAP | 12,334 | 0.003 | 11,925 | 0.006 | 409 | 0.01 | |
|
SCNN1D. 1:1280417–1292050 |
SBP | 2,405 | 0.0008 | 2,293 | 0.2 | 112 | 0.0005 |
| DBP | 2,405 | 0.002 | 2,293 | 0.03 | 112 | 0.003 | |
| PP | 2,405 | 0.003 | 2,293 | 0.04 | 112 | 0.005 | |
| MAP | 2,405 | 0.0008 | 2,293 | 0.1 | 112 | 0.0007 | |
|
SCNN1G. 16:23184696–23216904 |
SBP | 4,093 | 0.5 | 4,022 | 0.3 | 71 | 0.6 |
| DBP | 4,093 | 0.2 | 4,022 | 0.1 | 71 | 0.4 | |
| PP | 4,093 | 0.06 | 4,022 | 0.1 | 71 | 0.09 | |
| MAP | 4,093 | 0.5 | 4,022 | 0.2 | 71 | 0.8 | |
| Combined (SCNN1A, SCNN1B, SCNN1G) |
SBP | 20,399 | 0.01 | 19,806 | 0.02 | 593 | 0.03 |
| DBP | 20,399 | 0.0002 | 19,806 | 0.0004 | 593 | 0.004 | |
| PP | 20,399 | 0.3 | 19,806 | 0.1 | 593 | 0.4 | |
| MAP | 20,399 | 0.0006 | 19,806 | 0.001 | 593 | 0.005 | |
| Combined (SCNN1A, SCNN1B, SCNN1D, SCNN1G) |
SBP | 22,099 | 0.01 | 705 | 0.006 | ||
| DBP | 22,099 | 0.0002 | 705 | 0.001 | |||
| PP | 22,099 | 0.08 | 705 | 0.14 | |||
| MAP | 22,099 | 0.0009 | 705 | 0.001 | |||
p values < 0.00625 are significant
We repeated the analyses with rare (MAF < 0.01) and low frequency (0.01 ≤ MAF < 0.05) variants separately to determine if one class of variants was driving the significance of our findings. In our analyses of rare variants: SCNN1A (3,859 variants) were significantly associated with DBP (p = 0.0004) and MAP (p = 0.002); SCNN1B (11,925 variants) were also associated with DBP (p = 0.003) and MAP (p = 0.006); the combined analysis of SCNN1A, SCNN1B, and SCNN1G (19,806 variants) was associated with DBP (p = 0.0004) and MAP (p = 0.001); and the combined analysis of SCNN1A, SCNN1B, SCNN1D, and SCNN1G (22,099 variants) was associated with DBP (p = 0.0002) and MAP (p = 0.0009). Neither SCNN1D (2,293 variants) nor SCNN1G (4,022 variants) were associated with any of the four phenotypes (See Table 3).
In our analyses of low frequency variants: SCNN1A (113 variants) were significantly associated with DBP (p = 0.0007) and MAP (p = 0.002); SCNN1D (112 variants) were associated with SBP (p = 0.0005), DBP (p = 0.003), PP (p = 0.005), and MAP (p = 0.0007); the combined analysis of SCNN1A, SCNN1B, and SCNN1G (593 variants) were associated with DBP (p = 0.004) and MAP (p = 0.005); and the combined analysis of SCNN1A, SCNN1B, SCNN1D, and SCNN1G (705 variants) were associated with SBP (p = 0.006), DBP (p = 0.001), and MAP (p = 0.001). Neither SCNN1B (409 variants) nor SCNN1G (71 variants) were associated with any of the four phenotypes (See Table 3).
We repeated these analyses using another gene-based test, the burden test, and found no association between SCNN1A, SCNN1B, SCNN1D, or SCNN1G and SBP, DBP, MAP, or PP, suggesting that our analyses include variants that are increasing blood pressure as well as variants that are decreasing blood pressure (Table S3). We also performed SKAT analyses on SCNN1A (MAF < 0.05), SCNN1B (MAF < 0.05), and SCNN1D (0.01 ≤ MAF 0.05) upstream, downstream, missense, synonymous, and intronic variants, separately, with DBP to determine if any class of variant was contributing to our results. We found no association between DBP and SCNN1A, SCNN1B, or SCNN1D upstream, downstream, missense, synonymous, and intronic variants (Table S4).
Low frequency and rare ENaC variation in specific subunits is, in aggregate, associated with eGFR
High blood pressure is associated with chronic kidney disease (CKD) and stroke31,32. ENaC variants associated with altered blood pressure may also be associated with health outcomes such as stroke, eGFR and CKD. We hypothesized that ENaC variants are associated with eGFR and that SBP and DBP mediate these associations. Stepwise regression indicated that DBP was contributing to the variance of the null model for eGFR and therefore, we included DBP as a covariate in our eGFR analyses. We performed SKAT analyses using data from 14,557 individuals from seven TOPMed studies (Tables S5 and S6) to test for associations between eGFR (from which CKD is diagnosed) and variants with MAF < 0.05 in SCNN1A, SCNN1B, SCNN1D, and SCNN1G. In addition to DBP, models were adjusted for age, sex, BMI, and PCs 1–11 as covariates. We identified associations between eGFR and SCNN1B (n = 8,770) and SCNN1D (n = 1,758) variants (p = 0.005, p = 0.002, respectively), but not the SCNN1A (n = 2,739) and SCNN1G (n = 2,868) variants included in the analyses (p = 0.01, and p = 0.1, respectively) (Table 4). When combining ENaC variants (MAF < 0.05) from all four subunits (16,135 variants), we observed a significant association between the variants and eGFR (p=0.0003). As a negative control, eGFR phenotypes were permuted to randomize eGFR across samples and the analyses were repeated. There were no associations with permuted eGFR.
We repeated these analyses separately with rare and low frequency variants. Only low frequency variants in SCNN1D (110 variants) were significantly associated with eGFR (p = 0.001). There was no association between eGFR and rare variants in individual subunits, SCNN1A (2,626 variants), SCNN1B (8,365 variants), SCNN1D (1,648 variants), and SCNN1G (2,797 variants) (p = 0.008, p = 0.007, p = 0.2, and p = 0.01, respectively, see Table 4). However, when combining ENaC variants of all four subunits (15,436 variants) we observed significant associations between rare variants and low frequency variants and eGFR (p = 0.002 and 0.003, respectively).
These analyses were repeated using burden tests. Of the conditions tested, only SCNN1D low frequency variants (0.01 ≤ MAF < 0.05) were significantly associated with eGFR, suggesting strong directionality of effect for these variants (Table S7).
Low-frequency and rare variation in SCNN1A, SCNN1B, SCNN1D, and SCNN1G is not associated with stroke
We also performed SKAT analyses using data from a subset of individuals from the blood pressure analyses consisting of 9,090 individuals (4,399 cases and 4,691 controls) from the Women’s Health initiative (Tables S8 and S9) to test for associations between stroke and low frequency and rare variation (MAF < 0.05) in SCNN1A, SCNN1B, SCNN1D, and SCNN1G using age, sex, BMI, and PCAs 1–11 as covariates. After removing variants monomorphic in this subset, 2,330 SCNN1A variants 6,965 SCNN1B variants, 1,621 SCNN1D, and 2,208 SCNN1G variants remained in the analyses. There was no significant association of combined variants with MAF < 0.05 in SCNN1A, SCNN1B, SCNN1D, or SCNN1G with overall stroke (p = 0.08, p = 0.5, p = 0.5, and p = 0.2, respectively) (Table 5). Additionally, we performed SKAT with 13,124 variants from SCNN1A, SCNN1B, SCNN1D, and SCNN1G combined. The combined ENaC variation SKAT was not associated with stroke (p = 0.3). We repeated these analyses with rare and low frequency variants separately. There were no associations between SCNN1A (2,216 and 114 variants, respectively), SCNN1B (6,556 and 409 variants, respectively), SCNN1D (1,497 and 124 variants, respectively), SCNN1G (2,137 and 71 variants, respectively), or all four subunits combined (12,406 and 718 variants, respectively) (See Table 5). We repeated these analyses using burden tests, and found no association between SCNN1A, SCNN1B, SCNN1D, or SCNN1G and stroke (Table S10).
Table 5. Sequence kernel association tests of low frequency and rare ENaC variants with stroke.
SKAT was performed on variants with a MAF < 0.05 in the genomic regions of SCNN1A, SCNN1B, SCNN1D, and SCNN1G with stroke, separately, and then combined. SKAT was separately performed on rare (MAF < 0.01) and low frequency (0.01 ≤ MAF < 0.05) variants.
| Gene. Chromosome:Start–End | Variants with MAF < 0.05 | Variants with MAF < 0.01 | Variants with 0.01 ≤ MAF < 0.05 | |||
|---|---|---|---|---|---|---|
| n | p value | n | p value | n | p value | |
| SCNN1A. 12:6346824–6379751 | 2,330 | 0.08 | 2,216 | 0.05 | 114 | 0.2 |
| SCNN1B. 16:2329812–23381320 | 6,965 | 0.5 | 6,556 | 0.7 | 409 | 0.4 |
| SCNN1D. 1:1280417–1292050 | 1,621 | 0.5 | 1,497 | 0.6 | 124 | 0.4 |
| SCNN1G. 16:23184696–23216904 | 2,208 | 0.2 | 2,137 | 0.1 | 71 | 0.3 |
| Combined (SCNN1A, SCNN1B, SCNN1D, SCNN1G) | 13,124 | 0.3 | 12,406 | 0.3 | 718 | 0.3 |
SCNN1D is expressed in human monocytes
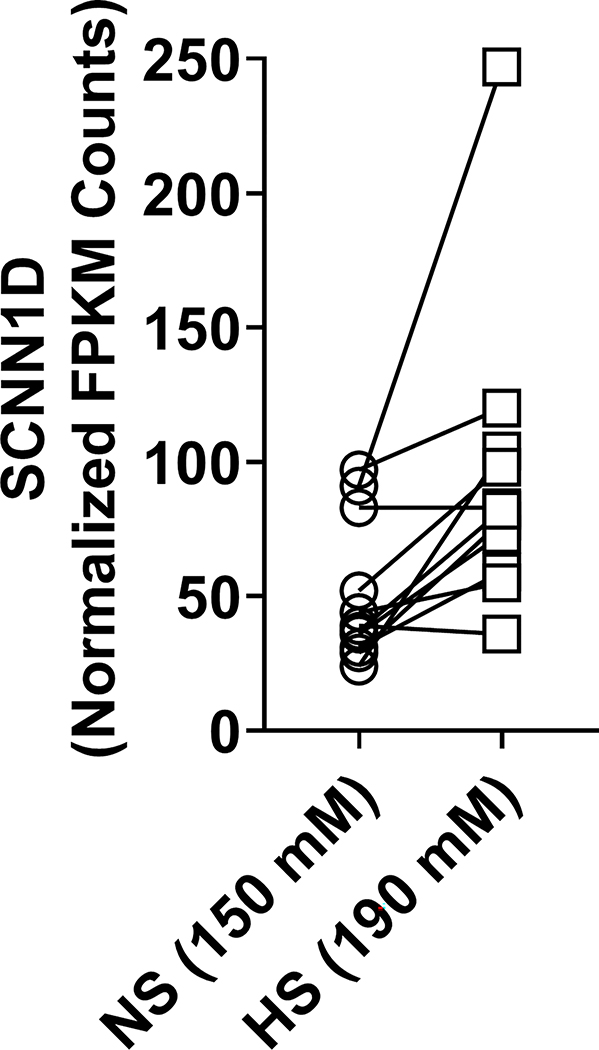
The ENaC δ subunit is poorly expressed in transporting epithelia that regulate total body Na+ (kidney and colon)10,33, in contrast to the α, β, and γ subunits. Our results showing association of SCNN1D low-frequency and rare variants with blood pressure phenotypes, and not SCNN1G, suggest that ENaC function in Na+ sensing cells influence blood pressure variation in the general population. SCNN1D is expressed in the central nervous system, where rare variants could influence blood pressure parameters. Antigen-presenting cells have recently been reported to have an ENaC-dependent response to increased Na+ intake, leading to an increase in blood pressure6. We previously showed that SCNN1A and SCNN1G, but not SCNN1B, are expressed in monocyte-derived dendritic cells in mice6. To determine whether SCNN1D may have a role in Na+ sensing in human monocytes and monocyte-derived cells, we performed RNA sequencing on human monocytes isolated from 11 volunteers and exposed to either normal or elevated sodium. We detected SCNN1D expression in cells from each of the volunteers (Figure 1). We did not detect an effect of elevated sodium exposure on SCNN1D expression in these cells. Nonetheless, expression of SCNN1D suggests the δ subunit of ENaC may be involved in Na+ sensing in these cells.
Figure 1. Expression of ENaC subunits in human monocytes.
Human monocytes were exposed to a normal (150 mM) or high (190 mM) NaCl solution for 72 h, and RNA was isolated and sequenced. Normalized fragments per kilobase of transcript per million mapped reads (FPKM) are shown. Similar levels of expression of δ ENaC were seen between the two exposures (p = 0.06, unpaired Student’s t test).
Discussion
The consequences of variants that lead to extreme increases or decreases in ENaC activity, which manifest as Liddle syndrome and PHA1, respectively, are well known monogenic disorders. There are few studies examining the effects of other ENaC variants on complex traits such as blood pressure variation or hypertension-related health outcomes. We found that, in aggregate, low frequency and rare variants in SCNN1A and SCNN1B are associated with DBP and MAP, with both rare and low frequency variants contributing to the associations of SCNN1A with DBP and MAP and rare variants driving the associations of SCNN1B with MAP and DBP. Variants in SCNN1D are associated with SBP, DBP, PP, and MAP, and are driven by low-frequency variants. Variants in SCNN1B and SCNN1D are associated with eGFR, which, for SCNN1D, is, again, driven by low frequency variants. While there has been speculation about the impact of common and rare functional ENaC variants on blood pressure7,34, we found no evidence that these common functional variants (αA334T, αT663A) are associated with SBP, DBP, PP, or MAP, consistent with the evidence provided by ClinVar35. These results are largely in agreement with previous findings16,17,28–30. We did not find an association of low frequency and rare variants in SCNN1A, SCNN1B, SCNN1D, and SCNN1G, assessed either separately or together, with stroke, despite the associations of hypertension with stroke31,32. Potential reasons for the lack of associations are small effect of these combined variants on blood pressure variation or heterogeneity of effects across ancestries that may have reduce power to detect effects, since these outcomes are likely mediated through increases in blood pressure. In addition, the SKAT test combines variants that increase and decrease blood pressure and therefore does not provide estimates for associations. As analyses of a large database may result in spurious results, and it will be important to confirm our key findings using other large databases.
ENaCs in human kidney are primarily αβγ channels10. We expected to find associations of low frequency and rare variants in SCNN1A, SCNN1B, and SCNN1G with blood pressure traits based on the known roles of ENaC in regulating the reabsorption of filtered Na+ in the ASDN and whole-body Na+ content, which are major determinants of extracellular fluid volume and blood pressure1,2. ENaC-dependent Na+ absorption also has an important role in regulating serum [K+], as it is tightly coupled to renal K+ secretion mediated by the K+ channels Kir1.1 and BK1,3. A lower serum [K+] in the setting of increased in ENaC activity, coupled with increased K+ secretion, is predicted to enhance activity of the Na+–Cl− cotransporter (NCC) in the DCT36. It was surprising that variants in SCNN1G were not associated with blood pressure traits, given what is known regarding the roles of specific sites in the γ subunit in regulating channel activity. We and others have identified sites in the γ subunit where specific mutations affect ENaC activity in heterologous expression systems2,37,38. Cleavage of the γ subunit at defined sites, with the release of an imbedded inhibitory tract, has a large effect on channel activity. Modification of specific γ subunit cytoplasmic residues by palmitoylation, and the interaction of specific acidic phospholipids with γ subunit residues also affect channel activity2.
ENaC function in non-renal tissues may drive the blood pressure associations we observed. Among these are sites in humans that express channels with a δ subunit. We found by RNA sequencing that human monocytes express ENaC δ subunits (Figure 2), which could contribute to the release of cytokines that affect blood pressure6. The δ subunit is expressed in human umbilical vein endothelial cells39. Previous studies have suggested that ENaC function in endothelial cells influence cellular stiffness and NO release7,8, raising the possibility that SCNN1D variants could influence blood pressure. Recently, Paudel, et al. identified SCNN1A, SCNN1B, SCNN1D, and SCNN1G mRNA and protein expression in human internal mammary artery and aorta and suggested that SCNN1D may be associated with hypertension40. ENaC δ subunits are also expressed in human taste buds and may have a role in mediating salt taste4 and influence Na+ intake. This could influence blood pressure by a number of mechanisms, including changes in extracellular and intravascular volume, release of cytokines from circulating dendritic cells6 and monocytes, and through alterations in the gut microbiota and the subsequent induction of TH17 cells41. Our observed association of low frequency and rare variants in SCNN1D with blood pressures suggest that the δ subunit has an unrecognized role in blood pressure regulation in human. This is consistent with a wider scope of SCNN1D expression as previously documented12, based on the recently released encyclopedias of DNA elements (ENCODE 3) data11. Expression of SCNN1D in B cells, T cells and monocytes suggests a potential role of the δ subunit in immunity and immunity-mediated diseases.
Aside from the well-defined Liddle syndrome variants, it is notable that few studies, to date, have found an association between low-frequency or rare ENaC variants and blood pressure measures. Five ENaC variants associated with increases or decreases in blood pressure salt-sensitivity were observed in the GenSalt study42. In addition, seven functional ENaC missense variants in the GenSalt study were identified by Ray et al.37, although these were not associated with differences in salt-sensitivity. Other variants have been associated with hypertension in specific populations, including variants in SCNN1B and SCNN1G43–45. Additionally, an ENaC gain-of-function variant in SCNN1A was associated with a modest Liddle syndrome-like phenotype and a blunting of the inhibitory effect of extracellular Na+ 2,46,47.
We noted that variants in SCNN1B and SCNN1D are associated with eGFR. While DBP is contributing to the variance of eGFR in our population, we controlled for DBP in our eGFR analyses and the associations of SCNN1B with eGFR are independent of DBP. The association with SCNN1B could reflect the effects of variants on ENaC function in the ASDN. Changes in extracellular fluid volume, blood pressure or volume regulatory hormones may influence glomerular filtration rate (GFR). Also, connecting tubule/collecting duct tubuloglomerular feedback, a phenomenon where ENaC dependent Na+ transport in the ASDN influences renal afferent arterial tone48 may be altered by ENaC variants. As mentioned above, associations with SCNN1D suggest that extrarenal ENaC influences GFR. Vascular ENaC influences renal vascular tone and blood flow, factors that will affect GFR49. ENaC-dependent release of cytokines from circulating dendritic cells6 and monocytes may also affect glomerular function50. Low-frequency variation (0.01 ≤ MAF < 0.05) in SCNN1D was also associated with eGFR via burden test. This may be caused by many of the variants exerting an effect in the same direction on eGFR, or one or several low-frequency variants with strong and similar directional effects on eGFR. These possibilities are not mutually exclusive and will require future study to elucidate the impact of SCNN1D variants on eGFR.
Perspectives
Low frequency and rare variants in SCNN1A, SCNN1B, and SCNN1D are, in aggregate, associated with key blood pressure parameters and eGFR in TOPMed data. The association of SCNN1D variants with BP and eGFR highlight the importance of ENaCs outside of the nephron in regulating these physiologic parameters. These observations raise the possibility that humans with hypertension and specific non–Liddle ENaC variants, particularly variants with a gain-of function phenotype, may benefit from a trial of ENaC inhibitors, such as amiloride, to lower blood pressure.
Supplementary Material
Novelty and Relevance.
What is New?
We found associations between low-frequency and rare variation in SCNN1A, SCNN1B, and SCNN1Dand blood pressure, and associations between SCNN1B and SCNN1D variation and eGFR.
What is Relevant?
Variation in SCNN1D, encoding the ENaC δ subunit that is poorly expressed in human kidney, is associated with blood pressure and eGFR.
Clinical/Pathophysiological Implications?
Humans with hypertension and specific non-Liddle ENaC variants, particularly variants with a gain-of function phenotype, may benefit from a trial of ENaC inhibitors, such as amiloride, to lower blood pressure.
Sources of Funding:
This work was supported by grants from the NIH/NIDDK (T32DK063922, T32DK061296, R01DK098204, P30DK079307, R01DK125439), from the NIH/NHLBI (K01HL130497, R01HL147818, R03HL155041, R01HL144941, R01HL046380, R01HL113338, R35HL135816, R01HL055673, R01HL093093, U01HL072507, R01HL087263, R01HL090682, R01HL120393, R01HL133040, U01HL072518, R01HL087698, R01HL112064, R01HL049762, R01HL058625, R01HL071025), from the NIH/NHGRI (R01HG011052), from the NIH/NINR (R01NR002241), and NIH/National Center for Research Resources (M01RR000052).
Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). See Supplemental Materials for additional information regarding TOPMed support, and Tables S11a and S11b for study-specific omics support information.
Non standard abbreviations:
- ASDN
aldosterone-sensitive distal nephron
- DBP
diastolic blood pressure
- ENaC
epithelial Na+ channel
- eGFR
estimated glomerular filtration rate
- GRM
genetic relatedness matrix
- MAP
mean arterial pressure
- MAF
minor allele frequency
- PCA
principal components of ancestry
- PHA1
pseudohypoaldosteronism type I
- PP
pulse pressure
- SKAT
sequence kernel association tests
- SNVs
single nucleotide variants
- SBP
systolic blood pressure
- TOPMed
Trans-Omics in Precision Medicine
- WGS
whole-genome sequencing
Footnotes
Disclosures: The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
See Table S1 for TOPMed banner authors.
References
- 1.Pearce D, Soundararajan R, Trimpert C, Kashlan OB, Deen PM, Kohan DE. Collecting duct principal cell transport processes and their regulation. Clin J Am Soc Nephrol. 2015;10:135–146. doi: 10.2215/CJN.05760513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleyman TR, Eaton DC. Regulating ENaC’s gate. Am J Physiol Cell Physiol. 2020;318:C150–C162. doi: 10.1152/ajpcell.00418.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrisoza-Gaytan R, Ray EC, Flores D, Marciszyn AL, Wu P, Liu L, Subramanya AR, Wang W, Sheng S, Nkashama LJ, et al. Intercalated cell BKalpha subunit is required for flow-induced K+ secretion. JCI Insight. 2020;5. doi: 10.1172/jci.insight.130553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu JJ, Elkaddi N, Garcia-Blanco A, Spielman AI, Bachmanov AA, Chung HY, Ozdener MH. Arginyl dipeptides increase the frequency of NaCl-elicited responses via epithelial sodium channel alpha and delta subunits in cultured human fungiform taste papillae cells. Sci Rep. 2017;7:7483. doi: 10.1038/s41598-017-07756-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malsure S, Wang Q, Charles RP, Sergi C, Perrier R, Christensen BM, Maillard M, Rossier BC, Hummler E. Colon-specific deletion of epithelial sodium channel causes sodium loss and aldosterone resistance. J Am Soc Nephrol. 2014;25:1453–1464. doi: 10.1681/ASN.2013090936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbaro NR, Foss JD, Kryshtal DO, Tsyba N, Kumaresan S, Xiao L, Mernaugh RL, Itani HA, Loperena R, Chen W, et al. Dendritic Cell Amiloride-Sensitive Channels Mediate Sodium-Induced Inflammation and Hypertension. Cell Rep. 2017;21:1009–1020. doi: 10.1016/j.celrep.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mutchler SM, Kleyman TR. New insights regarding epithelial Na+ channel regulation and its role in the kidney, immune system and vasculature. Curr Opin Nephrol Hypertens. 2019;28:113–119. doi: 10.1097/MNH.0000000000000479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warnock DG, Kusche-Vihrog K, Tarjus A, Sheng S, Oberleithner H, Kleyman TR, Jaisser F. Blood pressure and amiloride-sensitive sodium channels in vascular and renal cells. Nat Rev Nephrol. 2014;10:146–157. doi: 10.1038/nrneph.2013.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Wang HW, Ahmad M, Keshtkar-Jahromi M, Blaustein MP, Hamlyn JM, Leenen FHH. Central and peripheral slow-pressor mechanisms contributing to Angiotensin II-salt hypertension in rats. Cardiovasc Res. 2018;114:233–246. doi: 10.1093/cvr/cvx214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menon R, Otto EA, Hoover P, Eddy S, Mariani L, Godfrey B, Berthier CC, Eichinger F, Subramanian L, Harder J, et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight. 2020;5. doi: 10.1172/jci.insight.133267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium EP, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giraldez T, Rojas P, Jou J, Flores C, Alvarez de la Rosa D. The epithelial sodium channel delta-subunit: new notes for an old song. Am J Physiol Renal Physiol. 2012;303:F328–338. doi: 10.1152/ajprenal.00116.2012 [DOI] [PubMed] [Google Scholar]
- 13.Kamynina E, Staub O. Concerted action of ENaC, Nedd4–2, and Sgk1 in transepithelial Na(+) transport. Am J Physiol Renal Physiol. 2002;283:F377–387. doi: 10.1152/ajprenal.00143.2002 [DOI] [PubMed] [Google Scholar]
- 14.Grunder S, Firsov D, Chang SS, Jaeger NF, Gautschi I, Schild L, Lifton RP, Rossier BC. A mutation causing pseudohypoaldosteronism type 1 identifies a conserved glycine that is involved in the gating of the epithelial sodium channel. EMBO J. 1997;16:899–907. doi: 10.1093/emboj/16.5.899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanukoglu I, Hanukoglu A. Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases. Gene. 2016;579:95–132. doi: 10.1016/j.gene.2015.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samaha FF, Rubenstein RC, Yan W, Ramkumar M, Levy DI, Ahn YJ, Sheng S, Kleyman TR. Functional polymorphism in the carboxyl terminus of the alpha-subunit of the human epithelial sodium channel. J Biol Chem. 2004;279:23900–23907. doi: 10.1074/jbc.M401941200 [DOI] [PubMed] [Google Scholar]
- 17.Tong Q, Menon AG, Stockand JD. Functional polymorphisms in the alpha-subunit of the human epithelial Na+ channel increase activity. Am J Physiol Renal Physiol. 2006;290:F821–827. doi: 10.1152/ajprenal.00312.2005 [DOI] [PubMed] [Google Scholar]
- 18.Sofer T, Zheng X, Gogarten SM, Laurie CA, Grinde K, Shaffer JR, Shungin D, O’Connell JR, Durazo-Arvizo RA, Raffield L, et al. A fully adjusted two-stage procedure for rank-normalization in genetic association studies. Genet Epidemiol. 2019;43:263–275. doi: 10.1002/gepi.22188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, Kusek JW, Eggers P, Van Lente F, Greene T, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng X, Gogarten SM, Lawrence M, Stilp A, Conomos MP, Weir BS, Laurie C, Levine D. SeqArray-a storage-efficient high-performance data format for WGS variant calls. Bioinformatics. 2017;33:2251–2257. doi: 10.1093/bioinformatics/btx145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gogarten SM, Sofer T, Chen H, Yu C, Brody JA, Thornton TA, Rice KM, Conomos MP. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35:5346–5348. doi: 10.1093/bioinformatics/btz567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The NHLBI Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program. BRAVO variant browser: University of Michigan and NHLBI. 2018. [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S ML, Wu M. SKAT: SNP-Set (Sequence) Kernel Association Test; 2017.
- 26.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, Zhao S, Ye F, Sheng Q, Shyr Y. MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. Biomed Res Int. 2014;2014:248090. doi: 10.1155/2014/248090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambrosius WT, Bloem LJ, Zhou L, Rebhun JF, Snyder PM, Wagner MA, Guo C, Pratt JH. Genetic variants in the epithelial sodium channel in relation to aldosterone and potassium excretion and risk for hypertension. Hypertension. 1999;34:631–637. doi: 10.1161/01.hyp.34.4.631 [DOI] [PubMed] [Google Scholar]
- 29.Wang XF, Lu XM, Lin RY, Wang SZ, Zhang LP, Qian J, Lu DR, Wen H, Jin L. Lack of association of functional variants in alpha-ENaC gene and essential hypertension in two ethnic groups in China. Kidney Blood Press Res. 2008;31:268–273. doi: 10.1159/000151286 [DOI] [PubMed] [Google Scholar]
- 30.Yang W, Zhu Z, Wang J, Ye W, Ding Y. Evaluation of the relationship between T663A polymorphism in the alpha-epithelial sodium channel gene and essential hypertension. Saudi Med J. 2015;36:1039–1045. doi: 10.15537/smj.2015.9.11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosario RF, Wesson DE. Primary hypertension and nephropathy. Curr Opin Nephrol Hypertens. 2006;15:130–134. doi: 10.1097/01.mnh.0000214771.88737.ee [DOI] [PubMed] [Google Scholar]
- 32.Johnson RJ, Herrera-Acosta J, Schreiner GF, Rodriguez-Iturbe B. Subtle acquired renal injury as a mechanism of salt-sensitive hypertension. N Engl J Med. 2002;346:913–923. doi: 10.1056/NEJMra011078 [DOI] [PubMed] [Google Scholar]
- 33.Waldmann R, Champigny G, Bassilana F, Voilley N, Lazdunski M. Molecular cloning and functional expression of a novel amiloride-sensitive Na+ channel. J Biol Chem. 1995;270:27411–27414. doi: 10.1074/jbc.270.46.27411 [DOI] [PubMed] [Google Scholar]
- 34.Pavlov TS, Staruschenko A. Involvement of ENaC in the development of salt-sensitive hypertension. Am J Physiol Renal Physiol. 2017;313:F135–F140. doi: 10.1152/ajprenal.00427.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoorn EJ, Gritter M, Cuevas CA, Fenton RA. Regulation of the Renal NaCl Cotransporter and Its Role in Potassium Homeostasis. Physiol Rev. 2020;100:321–356. doi: 10.1152/physrev.00044.2018 [DOI] [PubMed] [Google Scholar]
- 37.Ray EC, Chen J, Kelly TN, He J, Hamm LL, Gu D, Shimmin LC, Hixson JE, Rao DC, Sheng S, et al. Human epithelial Na+ channel missense variants identified in the GenSalt study alter channel activity. Am J Physiol Renal Physiol. 2016;311:F908–F914. doi: 10.1152/ajprenal.00426.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winarski KL, Sheng N, Chen J, Kleyman TR, Sheng S. Extracellular allosteric regulatory subdomain within the gamma subunit of the epithelial Na+ channel. J Biol Chem. 2010;285:26088–26096. doi: 10.1074/jbc.M110.149963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Downs CA, Johnson NM, Coca C, Helms MN. Angiotensin II regulates delta-ENaC in human umbilical vein endothelial cells. Microvasc Res. 2018;116:26–33. doi: 10.1016/j.mvr.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 40.Paudel P, van Hout I, Bunton RW, Parry DJ, Coffey S, McDonald FJ, Fronius M. Epithelial Sodium Channel delta Subunit Is Expressed in Human Arteries and Has Potential Association With Hypertension. Hypertension. 2022:101161HYPERTENSIONAHA12218924. doi: 10.1161/HYPERTENSIONAHA.122.18924 [DOI] [PubMed] [Google Scholar]
- 41.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mahler A, Balogh A, Marko L, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551:585–589. doi: 10.1038/nature24628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu X, Gu D, He J, Rao DC, Hixson JE, Chen J, Li J, Huang J, Wu X, Rice TK, et al. Resequencing Epithelial Sodium Channel Genes Identifies Rare Variants Associated With Blood Pressure Salt-Sensitivity: The GenSalt Study. Am J Hypertens. 2018;31:205–211. doi: 10.1093/ajh/hpx169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones ES, Owen EP, Rayner BL. The association of the R563Q genotype of the ENaC with phenotypic variation in Southern Africa. Am J Hypertens. 2012;25:1286–1291. doi: 10.1038/ajh.2012.125 [DOI] [PubMed] [Google Scholar]
- 44.Nkeh B, Samani NJ, Badenhorst D, Libhaber E, Sareli P, Norton GR, Woodiwiss AJ. T594M variant of the epithelial sodium channel beta-subunit gene and hypertension in individuals of African ancestry in South Africa. Am J Hypertens. 2003;16:847–852. doi: 10.1016/s0895-7061(03)01016-1 [DOI] [PubMed] [Google Scholar]
- 45.Hannila-Handelberg T, Kontula K, Tikkanen I, Tikkanen T, Fyhrquist F, Helin K, Fodstad H, Piippo K, Miettinen HE, Virtamo J, et al. Common variants of the beta and gamma subunits of the epithelial sodium channel and their relation to plasma renin and aldosterone levels in essential hypertension. BMC Med Genet. 2005;6:4. doi: 10.1186/1471-2350-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salih M, Gautschi I, van Bemmelen MX, Di Benedetto M, Brooks AS, Lugtenberg D, Schild L, Hoorn EJ. A Missense Mutation in the Extracellular Domain of alphaENaC Causes Liddle Syndrome. J Am Soc Nephrol. 2017;28:3291–3299. doi: 10.1681/ASN.2016111163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Chen J, Shi S, Sheng S, Kleyman TR. Analyses of epithelial Na(+) channel variants reveal that an extracellular beta-ball domain critically regulates ENaC gating. J Biol Chem. 2019;294:16765–16775. doi: 10.1074/jbc.RA119.010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, D’Ambrosio MA, Garvin JL, Ren Y, Carretero OA. Connecting tubule glomerular feedback in hypertension. Hypertension. 2013;62:738–745. doi: 10.1161/HYPERTENSIONAHA.113.01846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drummond HA, Stec DE. betaENaC acts as a mechanosensor in renal vascular smooth muscle cells that contributes to renal myogenic blood flow regulation, protection from renal injury and hypertension. J Nephrol Res. 2015;1:1–9. doi: 10.17554/j.issn.2410-0579.2015.01.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heymann F, Meyer-Schwesinger C, Hamilton-Williams EE, Hammerich L, Panzer U, Kaden S, Quaggin SE, Floege J, Grone HJ, Kurts C. Kidney dendritic cell activation is required for progression of renal disease in a mouse model of glomerular injury. J Clin Invest. 2009;119:1286–1297. doi: 10.1172/JCI38399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.