Abstract
Cyclin-dependent kinase-like 5 (CDKL5) deficiency disorder (CDD) is caused by heterozygous or hemizygous variants in CDKL5 and is characterized by refractory epilepsy, cognitive and motor impairments, and cerebral visual impairment. CDKL5 has multiple transcripts, of which the longest transcripts, NM_003159 and NM_001037343, have been used historically in clinical laboratory testing. However, the transcript NM_001323289 is the most highly expressed in brain and contains 170 nucleotides at the 3’ end of its last exon that are non-coding in other transcripts. Two truncating variants in this region have been reported in association with a CDD phenotype. To clarify the significance and range of phenotypes associated with late truncating variants in this region of the predominant transcript in the brain, we report detailed information on two individuals, updated clinical information on a third individual, and a summary of published and unpublished individuals reported in ClinVar. The two new individuals (one male and one female) each had a relatively mild clinical presentation including periods of pharmaco-responsive epilepsy, independent walking and limited purposeful communication skills. A previously reported male continued to have a severe phenotype. Overall, variants in this region demonstrate a range of clinical severity consistent with reports in CDD but with the potential for milder presentation.
Keywords: CDKL5, alternative transcript, phenotypic spectrum, whole genome sequencing, transcripts, epileptic encephalopathy
1. INTRODUCTION
Heterozygous or hemizygous pathogenic loss-of-function variants in CDKL5 are associated with Developmental and Epileptic Encephalopathy 2 (MIM# 300672), which includes early infantile epileptic encephalopathy (EIEE), hypotonia, developmental delay, intellectual disability, motor disabilities, and cerebral visual impairment (CVI) (Fehr et al., 2013; Olson et al., 2019; Tao et al., 2004; Weaving et al., 2004). As an X-linked disorder, CDKL5 Deficiency Disorder (CDD) is more common in females (Demarest et al., 2019; Fehr et al., 2013). However, an increasing number of males with CDKL5 deficiency have been reported with a high rate of mosaicism and variable phenotypes (Siri et al., 2021; Stosser et al., 2018). While genotype-phenotype correlations in CDD are not yet well-defined, late truncations and some specific missense variants have been associated with a milder developmental phenotype but not necessarily milder epilepsy (Fehr et al., 2015; Fehr, Wong, et al., 2016; MacKay et al., 2021).
The interpretation of CDKL5 variants is complicated by the multiple transcripts that result from alternative splicing of the gene (Hector et al., 2016). Historically, the longest transcripts (e.g., NM_003159 and NM_001037343) were used by several clinical genetic testing laboratories for variant reporting. However, two recent reports of de novo pathogenic variants in CDKL5 were not reported initially on clinical testing reports due to the lack of evidence for the clinical relevance of this transcript at the time of testing (Bodian, Schreiber, Vilboux, Khromykh, & Hauser, 2018; Schoch et al., 2020). Each variant was located in non-coding regions of the NM_003159 transcript but led to premature stop codons in the last exon of the NM_001323289 transcript, which is the most abundant isoform expressed in the brain (Hector et al., 2016; Williamson et al., 2012).
In this report, we characterize the variants and phenotype of individuals with truncating variants identified in the region of CDKL5 unique to the 3’ end of the predominant transcript in the brain. We add two newly reported individuals with such variants identified by research or clinical whole genome sequencing (WGS), provide updated clinical information for a previously published individual, and describe one additional published individual reported in ClinVar.
2. METHODS
Study Approval.
Patient 1 and his parents were enrolled in the Undiagnosed Diseases Network (UDN) at Baylor College of Medicine (BCM). This study was approved by the Institutional Review Board (IRB) at the National Institutes of Health and Baylor College of Medicine. Patients 2 and 3 were enrolled in a CDKL5 clinic-based observational research study approved by the Boston Children’s Hospital IRB, and patient 2 was enrolled in the Rare Genomes Project approved by the Massachusetts General Brigham IRB. Informed consent for each participant was obtained prior to all research procedures under these protocols.
WGS (Patient 1).
Trio clinical WGS was performed through the Illumina iHope Program on extracted DNA with sequencing-by-synthesis (SBS) next generation sequencing (NGS). The data were aligned and reported according to build 37.1 (Hg19) of the Human Reference Genome (http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/). The genome was sequenced to an average of ≥30 fold coverage. Over 99% of the genome was covered at 10-fold coverage or more and at least 97% of the genome was callable (passes all quality filters). Small insertion and deletion events (e.g., insertions up to 31 bases and deletions up to 27 bases) were detected and reported for this case. Variants were filtered and evaluated based on multiple factors including population allele frequency, variant consequence, evolutionary conservation, occurrence in a gene with a well-established gene-disease relationship, occurrence in a gene whose disease association overlaps with the patient’s reported phenotype, and inheritance mode, as appropriate. Variant nomenclature is based on standardized Human Genome Variation Society (HGVS) conventions. Copy number variants (CNVs) greater than 10 kb were also assessed and are reported with standardized International System for Human Cytogenetic Nomenclature (ISCN) nomenclature. Clinical interpretation was performed on small variants and CNVs of interest in accordance with the American College of Medical Genetics and Genomics guidelines. Further research reanalysis of the trio WGS data was performed within the UDN with Codified Genomics variant analysis software (www.codifiedgenomics.com). Data were filtered to identify a list of rare variants with a frequency of <0.01 in gnomAD (https://gnomad.broadinstitute.org/) and <0.01 in our local UDN exome and genome database, consisting of more than 750 samples. Initial analysis was focused on variants that were either de novo or biallelic in genes in which at least one variant was within the coding region or involved a canonical splice site.
WGS (Patient 2).
Research trio WGS and data processing were performed through the Broad Center for Mendelian Genomics by the Genomics Platform at the Broad Institute of MIT and Harvard. PCR-free preparation of sample DNA (350 ng input at >2 ng/ul) was accomplished with Illumina HiSeq X Ten v2 chemistry, and libraries were sequenced to a mean target coverage of >30x. WGS data were processed through a pipeline based on Picard with base quality score recalibration and local realignment at known indels. The BWA aligner mapped reads to the human genome build 38. Single nucleotide variants (SNVs) and insertions/deletions (indels) are called jointly across all samples with the Genome Analysis Toolkit (GATK) HaplotypeCaller package version 4.0. Default filters were applied to SNV and indel calls by the GATK Variant Quality Score Recalibration (VQSR) approach. Structural variants were called with GATK-SV (Collins et al., 2020). Annotation was performed with Variant Effect Predictor (VEP). Lastly, the variant call set was uploaded to seqr (seqr.broadinstitute.org) for analysis where standard searches were applied filtering gnomAD and internal dataset allele frequencies. The identified variant in CDKL5 was confirmed by CLIA Sanger sequencing using standard methods.
WGS for patient 3 was published previously (Bodian et al., 2018).
Reported variants in CDKL5 were converted to the RefSeq transcript NM_001323289.2 for consistency with Alamut Visual Plus version v1.4 ∣ © 2021 SOPHiA GENETICS. This transcript contains one additional exon in the 5’-UTR compared to NM_001323289.1, which was an earlier version of the same transcript, used in a previous publication describing patient 3 (Bodian et al., 2018). Exon 1 is present in biologically relevant transcripts and when disrupted has been associated with a CDD phenotype (Bahi-Buisson et al., 2010; Hector et al., 2016; Liang et al., 2011; Nemos et al., 2009). Exon 18 in NM_001323289.2 is equivalent to exon 17 in NM_001323289.1.
3. PATIENT DATA
3.1. Patient 1
Presentation and history (birth to 5 months):
A 5-month-old boy was referred to the genetics clinic for evaluation of epilepsy, hypotonia, developmental delay, and poor visual response. Relevant perinatal history included intrauterine growth restriction during pregnancy and term birth via caesarean section due to non-reassuring fetal heart rate tracing. Concerns were noted at age 3 months due to limited eye contact and poor visual tracking. Shortly thereafter, the child was hospitalized with abnormal movements concerning for seizures and hypotonia. During this hospitalization, an MRI identified a subdural hematoma. History of trauma was ruled out. Electroencephalogram (EEG) during hospitalization showed focal slowing and recurrent bilateral occipital spike discharge consistent with epileptogenic foci. Levetiracetam was initiated, and the child remained seizure-free until the time of initial evaluation (age 5 months).
Evolution (5 to 33 months):
The patient’s development was delayed. He sat unassisted at 14 months and walked at 28 months. At 30 months, he had a raking grasp and used two words (“mom, dad”) and one sign (“more”). He was diagnosed with autism spectrum disorder and CVI by 27 months. When evaluated at 33 months, the child had remained seizure-free for over two years on levetiracetam. Weight and height were below the 10th percentile. He had diffuse hypotonia and reduced deep tendon reflexes. Repeat EEG and MRI were interpreted as normal.
Diagnostic testing:
Chromosomal microarray (CMA) detected a 0.254 Mb maternally inherited deletion involving NELL1 (maternally inherited). An epilepsy panel detected a heterozygous variant of uncertain significance (VUS) in CLN8. Plasma and cerebrospinal fluid (CSF) analyses were non-diagnostic. CSF neurotransmitter studies and metabolomics revealed low 5-hydroxyindoleacetic acid (5-HIAA) (161 nmol/L, normal range 179-711) and low homovanillic acid (HVA) (329 nmol/L, normal range 450-1132), but normal 3-O-methyldopa (3OMD) (153 nmol/L). From these results and the clinical picture, a diagnosis of aromatic L-amino acid decarboxylase (AADC) deficiency was considered. However, trio WGS did not identify causative variants in DDC, the gene related to AADC deficiency, and the clinical WGS was initially reported as non-diagnostic with no pathogenic or likely pathogenic variants and no variants of uncertain significance in other known disease genes consistent with the phenotype. Research reanalysis of trio WGS identified a hemizygous, de novo variant in CDKL5 (NM_001323289.2: c.2762_2763delCA (p.Thr921ArgfsTer7)) and confirmed the maternally inherited deletion involving NELL1 (Table 1). Given that this is a WGS and given limitations with the current tools available for evaluating noncoding variants, it is unclear whether one of the many noncoding variants of uncertain significance detected may be contributing to or modifying the phenotype.
Table 1.
Summary of clinical phenotypes for probands with CDKL5 variants within the distal 3’ end of exon 18 unique to the brain- expressed transcript, NM_001323289.2
| Patient 1, this publication | Patient 2, this publication | Patient 3, Bodian et al., 2018 with updates (2-9 years) | Schoch et al., 2020 | |
|---|---|---|---|---|
| Age at publication | 2 years 6 months old | 2 years 11 months old | 2 years old (Bodian et al., 2018), 9 years old (this publication) | 3 years 4 months old |
| Sex | Male | Female | Male | Male |
| Variant | c.2762_2763delCA (p.Thr921ArgfsTer7) | c.2842C>T (p.Arg948Ter) | c.2828_2829delGA (p.Arg943AsnfsTer11) | c.2842C>T (p.Arg948Ter) |
| ACMG Classification | Likely Pathogenic | Pathogenic | Likely Pathogenic | Pathogenic |
| ACMG/ClinGen Rett/Angelman Expert panel Criteria (McKnight et al., 2021) | PVS1_moderate, PS2_strong, PM2_supporting | PVS1_very strong, PS2_strong, PM2_supporting | PVS1_moderate, PS2_strong, PM2_supporting | PVS1_very strong, PS2_strong, PM2_supporting |
| Phenotype: | ||||
| Seizure onset | 3 months old | 2 months old | 2 weeks old | Not reported |
| Seizure type | Focal onset seizures (3 months); seizure free and off medication (current) | Focal onset tonic-clonic; infantile spasms; atonic seizures; seizure free period of 2 years, controlled on monotherapy | Bilateral tonic (2 weeks); focal non-motor (behavioral arrest, 6 months); infantile spasms (7 months); generalized tonic mixed with spasms; myoclonic | Infantile spasms and atonic seizures |
| Developmental delay | Yes, sat at 14 months, walked at 28 months; raking grasp; says mom/dad and signs “more” | Yes, walked at 25 months; unilateral pincer grasp; limited receptive/expressive language but single words and gestures | Yes, lack of head control, no hand use, few gestures and rare word approximations | Yes, development at 9- to 12-month-old level |
| Developmental regression | No regression or loss of skills | No | No | Yes, regression with onset of seizures |
| Hypotonia | Yes | Normal to borderline low tone | Profound hypotonia | Not reported |
| Visual Impairment | Cerebral visual impairment | Cerebral visual impairment (mild) | Cerebral visual impairment (severe, no visual tracking) | Not reported |
| Brain MRI findings | 4 mm left subdural fluid collection (3 months); normal (2 years 6 months) | Normal (2 months, 2 years 9 months) | Normal (2 months); mild diffuse volume loss (25 months) | Not reported |
| Other medical history | Mild elevation in transaminases, autism spectrum disorder | Autism spectrum disorder | Jejunostomy-tube dependence, chronic lung disease, Nissen fundoplication | Not reported |
3.2. Patient 2
Presentation and history (birth to 3 months):
A 3-month-old girl was referred to the epilepsy clinic for a second opinion regarding seizures. Relevant perinatal history included term vaginal birth, induced due to maternal hypertension. Concern for seizures began at age 8 weeks. Seizure semiology at that time was described as focal tonic (bilateral arm stiffening, right head and eye deviation evolving to bilateral tonic-clonic. EEG at time of seizure-onset was abnormal due to left frontal sharp waves and a single right central onset seizure with spread to the left central region, consisting of bilateral tonic-clonic activity.
Evolution (3 months to 3 years):
Phenobarbital was initiated with remission of the clonic seizure component. After 1 month of treatment, seizures remitted entirely for 3 months. At 7 months, the child developed infantile spasms with hypsarrhythmia that was refractory to adrenocorticotropic hormone (ACTH) but with subsequent electroclinical remission on vigabatrin. Vigabatrin was discontinued after 3 weeks due to concerns for oculogyric crisis. The oculogyric crisis resolved, infantile spasms did not recur, and repeat EEG was normal. She had a 2-year period of seizure freedom and then developed atonic seizures (head drops). An EEG at this time showed generalized and multifocal spikes, continuous generalized slowing, a discontinuous pattern in sleep, and absence of normal sleep architecture. Atonic seizures were treated effectively with valproic acid. As of the last follow-up at 3 years, she remained seizure-free on treatment with valproic acid and levetiracetam.
The patient’s development was delayed. She achieved head control at 1 month, rolled at 3 months, sat at 14 months, crawled at 19 months, walked at 25 months and began to climb stairs with assistance at 3 years. By age 3 years, she had a unilateral right pincer grasp, used four words, and understood simple commands. She used head and mouth movements for non-verbal communication. She was diagnosed with autism spectrum disorder and CVI by age 2 years.
Diagnostic testing:
Urine amino acid analysis showed elevations of homocysteine (6.5 umol/g, normal range 0-2.5) and argininosuccinate (139 umol/g, normal range 0-74) in the setting of a negative newborn screen that included argininosuccinate lyase deficiency. An epilepsy gene panel (sequencing and deletion-duplication), which included CDKL5, and CMA were non-diagnostic. Research trio WGS revealed a heterozygous de novo CDKL5 variant resulting in a premature stop codon in the alternative NM_001323289.2 transcript, c.2842C>T, p.Arg948Ter, classified as pathogenic and confirmed clinically with Sanger sequencing. No pathogenic or likely pathogenic variants in ASL or concerning variants of uncertain significance in this gene were detected in the WGS.
3.3. Patient 3
Presentation and history (birth to 2 years, previously published (Bodian et al., 2018)):
Baby boy, born at term by uncomplicated delivery, presented at 2 weeks with multiple daily bilateral tonic seizures, followed by a one-month period of seizure freedom at age 4 months on levetiracetam, topiramate and clonazepam. At age 6 months, he developed focal seizures (behavioral arrest, “gurgling” sound, variable eye deviation), and at 7 months he developed infantile spasms. The EEG background was initially normal but then deteriorated, showing disorganization, high-amplitude delta activity, and multifocal epileptiform discharges. Infantile spasms did not respond to standard first line treatment (ACTH and prednisolone) or ketogenic diet. MRI was initially interpreted as normal; repeat imaging at age 25 months demonstrated mild diffuse volume loss.
At age 2 years, the child’s development was severely delayed and at the level of a 1-2 month-old. He exhibited profound hypotonia, intellectual disability, and CVI. Notable medical surgical history included gastrostomy tube dependence, chronic lung disease, and Nissen fundoplication.
Evolution (2 to 8 years):
At last follow-up at 8 years, drug-resistant epilepsy continued with two main seizure types: clusters of epileptic spasms and brief tonic seizures and clusters of myoclonic jerks (independently or following clusters of spasms and tonic seizures). Seizure frequency was five per day on average, duration was up to 5-10 minutes, and the child had been hospitalized recently for status epilepticus. He continued on treatment with levetiracetam, valproic acid, clobazam and ketogenic diet. Additional failed anti-seizure medications over time included oxcarbazepine, cannabidiol, and rufinamide. EEGs at age 2.6 and 3.2 years showed hypsarrhythmia, and an EEG at 6.8 years confirmed seizure type as clusters of epileptic spasms with intermixed brief tonic seizures.
At approximately 2-3 years of age, he developed some head control and ability to sit with support, but these skills were not sustained. At last follow-up, fine motor skills were limited, lacking grasping or purposeful hand movements. He was able to communicate with a few gestures, facial expressions, and rare word approximations. He regarded faces inconsistently but lacked visual tracking. He had alternating periods of sleeplessness and periods of prolonged sleep for up to 24 hrs. He was fed through a jejunostomy tube from age 3 years, had hepatomegaly of unclear etiology, and had recurrent aspiration pneumonias and chronic obstructive pulmonary disease.
Diagnostic testing:
Metabolic and additional genetic testing were negative as previously reported (Bodian et al., 2018). Trio WGS identified a de novo 2 base-pair deletion (c.2828_2829delGA (p.Arg943AsnfsTer11)) in exon 18 of the CDKL5 NM_001323289.2 transcript.
3.4. Variants in ClinVar
The variant (c.2842C>T (p.Arg948Ter)) identified in patient 2 is classified as pathogenic in Clinvar and was previously reported in a male with refractory epilepsy and developmental regression (Table 1) (Schoch et al., 2020). Four additional unpublished variants producing premature truncations in the 3’ region unique to this predominant transcript in the brain are reported in ClinVar with limited clinical information (Supplementary Table 1). Three were classified as pathogenic, all in the transcript NM_001323289.2. The first was c.2716C>T (p.Gln906Ter), de novo, in a patient reported with early onset epileptic encephalopathy 2. Two variants, c.2785C>T (p.Gln929Ter) and c.2821del (p.Tyr941IlefsTer12), were each reported with unknown inheritance and reported phenotype of early infantile epileptic encephalopathy 2 and Angelman syndrome-like. One of these variants (c.2821del (p.Tyr941IlefsTer12)) was also submitted to Clinvar as a variant of uncertain significance with limited clinical information by another submitter. The other variant was reported as likely benign, c.2733G>A (p.Trp911Ter), with unreported sex, inheritance and phenotype.
4. DISCUSSION
CDD more commonly affects females in whom X-inactivation results in a mixture of affected and unaffected cells; however, an increasing number of affected hemizygous males with germline or mosaic variants in CDKL5 have been described (Olson et al., 2019; Siri et al., 2021; Stosser et al., 2018). A wide variety of pathogenic variants in CDKL5 have been associated with the disorder, including missense variants in the N-terminal catalytic domain and frameshift or truncating variants located throughout the gene (Hector et al., 2017; Olson et al., 2019). These variant types in relationship to functional domains in the protein are visualized in figures in the literature, including well-established late truncations (Hector et al., 2017; Olson et al., 2019). This report expands the spectrum of CDKL5 variants to include late truncations unique to the predominantly expressed transcript in the brain (NM_001323289), which are associated with a range of severity in clinical presentation from mild to severe in both males and females. We describe one male and one female with a developmental and epilepsy phenotype at the mild end of the spectrum for CDD and also a previously published male with a persistent severe phenotype (Bodian et al., 2018).
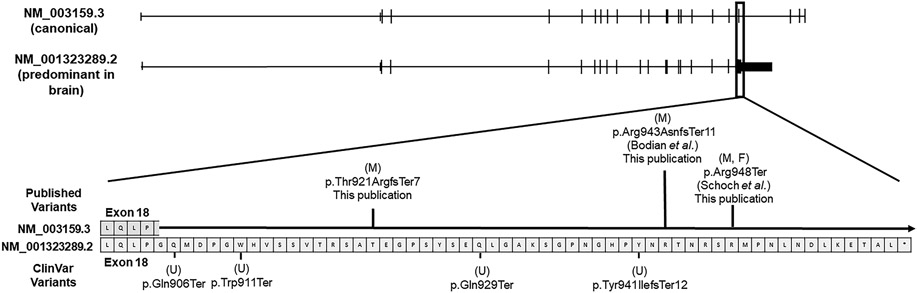
CDKL5 has several known transcripts and alternatively spliced exons (Hector et al., 2016). The CDKL5 transcripts used previously in clinical testing laboratories (NM_003159 or NM_001037343) contain 21 or 22 exons and differ only in the 5’ non-coding exons. However, a truncated isoform of CDKL5 (NM_001323289) is the primary isoform expressed in the brain and is considered to be more biologically relevant and also has higher expression in other tissues relative to other transcripts (Williamson et al., 2012) (Supplementary Figure 1). These three CDKL5 transcripts are similar in their 5’ coding sequences; however, the NM_001323289 isoform with highest expression in brain does not contain the last three coding exons (exons 19-21 in NM_003159 or exons 20-22 in NM_001037343). Pathogenic variants in these three exons have not been reported previously (Hector et al., 2017; Olson et al., 2019). Moreover, the NM_001323289.2 transcript has an additional 170 nucleotides at the 3’ end of exon 18 compared to exon 18 of NM_003159 or exon 19 of NM_001037343 and is most similar to the transcript with highest abundance in the central nervous system, hCDKL5_1, described by Hector et al. (Hector et al., 2016; Williamson et al., 2012). The additional 170 nucleotides included in exon 18 of NM_00132389.2 are contained within an intron of these other two transcripts (Figure 1), and as a result, variants within this region were not reported previously by clinical testing laboratories (Bodian et al., 2018; Schoch et al., 2020). Of note, no frameshift or stop-gained variants in this 170 bp region appear in gnomAD supporting the hypothesis that loss of function variants in this region are deleterious (Karczewski et al., 2020). Little is known about the biological significance of these additional 170 amino acids except that these residues reside in a disordered region that is enriched in polar amino acids.
Figure 1.
Cartoon representation of two of the relevant CDKL5 transcripts with magnification of the 170 bp region at the 3’ end of exon 18 in CDKL5 (NM_001323289.2). The canonical transcript, which was historically used by many reference laboratories (NM_003159.3), is shown in comparison to the predominant transcript in the brain (NM_001323289.2). The location of the Clinvar variants are provided below the transcript and the location of published variants are above the transcript. Boxes represent individual amino acids. Letters in parentheses indicate the sex of the patient if known. (F = female, M = Male, U = Unknown).
We now report a total of 8 individuals with protein truncating variants in this 3’ region of the predominant transcript in the brain, at least 7 of whom have a phenotype consistent with CDD (the phenotype for the eighth individual is not reported). By updated ACMG classification, these are classified as pathogenic or likely pathogenic (Table 1). For each of the published patients including the individuals in this report, the CDKL5 variants were missed on initial genetic evaluation due to the intronic location of the variants within the canonical NM_003159 transcript used by clinical genetic testing laboratories (Bodian et al., 2018; Schoch et al., 2020). Thus, it is critical that this region be considered in diagnostic testing. To date, all reported pathogenic variants in this region are truncating, which is consistent with prior literature suggesting that pathogenic missense variants are limited to the N-terminal catalytic domain except for a single variant that disrupts splicing leading to exon skipping (Hector et al., 2017). As these variants affect the last exon of the isoform, they would be predicted to escape nonsense-mediated decay and produce a truncated protein.
The phenotypic spectrum of individuals with variants in the 3’ region of the brain-specific transcript is consistent with that reported for CDD more broadly. However, each of our two newly reported individuals (patients 1 and 2) has a relatively mild phenotype with pharmaco-responsive epilepsy and developmental skills at the higher end of the reported spectrum (MacKay et al., 2021; Olson et al., 2019; Siri et al., 2021). Both are independently mobile and have purposeful communication skills, which are achievements reached by only a minority of individuals with CDD (Demarest et al., 2019; Fehr, Downs, et al., 2016; Fehr et al., 2015; Olson et al., 2021). Tone is normal or mildly low in both patients. In contrast, patient 3 continues to have drug-resistant epilepsy, severe developmental impairment, and severe hypotonia. Notably, three of the four variants within the 170 bp region at the most 3’ end of exon 18 in CDKL5 that have been submitted to Clinvar are associated with early infantile epileptic encephalopathy (Supplementary Table 1). CVI is a feature reported in all three patients presented here and is a common feature in CDD with a range of severity (Brock et al., 2021; Olson et al., 2021).
Limited literature supports a milder developmental phenotype in association with late truncations CDKL5 in females. However, a recent report found that individuals (females and males) with late truncating variants had higher CDD clinical severity scores (more severe phenotype) despite better functional abilities, and seizure frequency was higher in one study (Fehr et al., 2015; Fehr, Wong, et al., 2016; MacKay et al., 2021). A recent review by Siri et al. summarized the clinical findings of 50 males with variants in CDKL5 (Siri et al., 2021). Although none of these patients had reported variants in the extra 170 nucleotide region of the CDKL5 NM_001323289.2 isoform, two of the patients were described with a milder developmental phenotype, four had seizures responsive to anti-epileptic medication, and many patients were reported to have autism spectrum disorder. Additional studies have not shown differences in developmental scores of males overall compared to females, but power was limited by small samples size, and there was a high degree of variability in males (Brock et al., 2021; MacKay et al., 2021; Olson et al., 2021; Olson et al., 2019). It has been reported that post-zygotic CDKL5 variants occur in 9% of patients with CDD overall but in 32% of males (Olson et al., 2019; Siri et al., 2021). Mosaicism may account, at least partially, for the phenotypic variability in males, although no evidence of somatic mosaicism was noted in the trio WGS sequencing of patient 1. While the relatively mild phenotype of patients 1 (male) and 2 (female) may be attributed to the late truncation, a mild phenotype is not universal for variants in this region. In patient 2, the milder phenotype relative to the previously published patient with the same genotype may be explained, at least partly, by her sex given the X-linked inheritance pattern associated with this disorder. Conversely, the variant observed in patient 2 was previously published in a male with severe phenotype, and the severe phenotype in patient 3 has persisted as the patient has grown older (Bodian et al., 2018; Schoch et al., 2020). Additionally both patients with a mild phenotype are at an age where a honeymoon period with relative seizure control has been reported (median 20-24 months of age, median duration 4-6 months) and thus additional time is needed to determine epilepsy outcomes (Bahi-Buisson et al., 2008; Fehr, Wong, et al., 2016; Kobayashi et al., 2021; Olson et al., 2019).
In conclusion, truncations in the 3’ region of the predominant CDKL5 transcript in the brain (NM_00132389) are relevant clinically and associated with a phenotype consistent with CDD with a range of severities. It is critical that clinical diagnostic laboratories assess for variants in this transcript.
Supplementary Material
Supplementary Figure 1. CDKL5 transcript-specific expression patterns. This screen capture from GTEX (https://gtexportal.org) shows the expression of various CDKL5 isoforms across different tissues. The ENST00000623535 transcript is equivalent to NM_001323289 and is the transcript with highest expression in the brain and in other tissues.
ACKNOWLEDGMENTS
The authors thank the patients and their families for their willingness to participate in this research study. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number U01HG007709. L.C.B. and H.T.C. each hold a Career Award for Medical Scientists from the Burroughs Wellcome Fund. H.T.C. is also supported by the Child Neurology Society and Foundation, Wallace Endowment Award, McNair Medical Institute, and NIH DP5OD26428. H.E.O, L.S. and I.H. are supported by the National Institute of Neurologic Disorders and Stroke (K23 NS107646-04, PI Olson) and the International Foundation for CDKL5 Research. A.O.D.L., E.H., and G.V., along with sequencing and analysis of patient 2 was supported by the National Human Genome Research Institute (NHGRI), the National Eye Institute, the National Heart, Lung and Blood Institute grant UM1 HG008900, NHGRI grants U01 HG011755 and R01 HG009141, and the Chan Zuckerberg Initiative to the Rare Genomes Project. Research reported in this publication was also supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number P50HD103555 for use of the Clinical Translational Core facilities. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Clinical WGS for patient 1 and his family members was provided free of cost by the iHope Program, a philanthropic program supported by Illumina Inc.
Footnotes
DISCLOSURE STATEMENT
Dr. Olson received consulting fees from Takeda Pharmaceuticals and Zogenix regarding clinical trial design, Ovid Therapeutics regarding clinical trial results, Marinus Pharmaceuticals regarding CDKL5 Deficiency Disorder, and has done consulting for the FOXG1 Research Foundation. Dr. O’Donnell-Luria receives consulting fees from Tome Biosciences and is a paid member of the Scientific Advisory Boards for Congenica and SPARK for Autism.
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics. Erin Thorpe, Akanchha Kesari, and Denise Perry are employees and stockholders of Illumina, Inc.
DATA AVAILABILITY STATEMENT
Data for patient 1 was submitted to Phenome Central (P0009410) and Clinvar (SCV001245579.1). Data for patient 2 is available via DUOS (https://duos.broadinstitute.org/) and AnVIL (https://anvilproject.org/). No new data analysis was performed for patient 3.
REFERENCES
- Bahi-Buisson N, Girard B, Gautier A, Nectoux J, Fichou Y, Saillour Y, … Bienvenu T (2010). Epileptic encephalopathy in a girl with an interstitial deletion of Xp22 comprising promoter and exon 1 of the CDKL5 gene. Am J Med Genet B Neuropsychiatr Genet, 153B(1), 202–207. doi: 10.1002/ajmg.b.30974 [DOI] [PubMed] [Google Scholar]
- Bahi-Buisson N, Kaminska A, Boddaert N, Rio M, Afenjar A, Gerard M, … Bienvenu T (2008). The three stages of epilepsy in patients with CDKL5 mutations. Epilepsia, 49(6), 1027–1037. doi: 10.1111/j.1528-1167.2007.01520.x [DOI] [PubMed] [Google Scholar]
- Bodian DL, Schreiber JM, Vilboux T, Khromykh A, & Hauser NS (2018). Mutation in an alternative transcript of CDKL5 in a boy with early-onset seizures. Cold Spring Harb Mol Case Stud, 4(3). doi: 10.1101/mcs.a002360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock D, Fidell A, Thomas J, Juarez-Colunga E, Benke TA, & Demarest S (2021). Cerebral Visual Impairment in CDKL5 Deficiency Disorder Correlates With Developmental Achievement. J Child Neurol, 36(11), 974–980. doi: 10.1177/08830738211019284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RL, Brand H, Karczewski KJ, Zhao X, Alfoldi J, Francioli LC, … Talkowski ME (2020). A structural variation reference for medical and population genetics. Nature, 581(7809), 444–451. doi: 10.1038/s41586-020-2287-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarest ST, Olson HE, Moss A, Pestana-Knight E, Zhang X, Parikh S, … Benke TA (2019). CDKL5 deficiency disorder: Relationship between genotype, epilepsy, cortical visual impairment, and development. Epilepsia, 60(8), 1733–1742. doi: 10.1111/epi.16285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr S, Downs J, Ho G, de Klerk N, Forbes D, Christodoulou J, … Leonard H (2016). Functional abilities in children and adults with the CDKL5 disorder. Am J Med Genet A, 170(11), 2860–2869. doi: 10.1002/ajmg.a.37851 [DOI] [PubMed] [Google Scholar]
- Fehr S, Leonard H, Ho G, Williams S, de Klerk N, Forbes D, … Downs J (2015). There is variability in the attainment of developmental milestones in the CDKL5 disorder. J Neurodev Disord, 7(1), 2. doi: 10.1186/1866-1955-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr S, Wilson M, Downs J, Williams S, Murgia A, Sartori S, … Christodoulou J (2013). The CDKL5 disorder is an independent clinical entity associated with early-onset encephalopathy. Eur J Hum Genet, 21(3), 266–273. doi: 10.1038/ejhg.2012.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr S, Wong K, Chin R, Williams S, de Klerk N, Forbes D, … Leonard H (2016). Seizure variables and their relationship to genotype and functional abilities in the CDKL5 disorder. Neurology, 87(21), 2206–2213. doi: 10.1212/WNL.0000000000003352 [DOI] [PubMed] [Google Scholar]
- Hector RD, Dando O, Landsberger N, Kilstrup-Nielsen C, Kind PC, Bailey ME, & Cobb SR (2016). Characterisation of CDKL5 Transcript Isoforms in Human and Mouse. PLoS One, 11(6), e0157758. doi: 10.1371/journal.pone.0157758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RD, Kalscheuer VM, Hennig F, Leonard H, Downs J, Clarke A, … Cobb SR (2017). CDKL5 variants: Improving our understanding of a rare neurologic disorder. Neurol Genet, 3(6), e200. doi: 10.1212/NXG.0000000000000200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, … MacArthur DG (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581(7809), 434–443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Tohyama J, Takahashi Y, Goto T, Haginoya K, Inoue T, … Matsumoto N (2021). Clinical manifestations and epilepsy treatment in Japanese patients with pathogenic CDKL5 variants. Brain Dev, 43(4), 505–514. doi: 10.1016/j.braindev.2020.12.006 [DOI] [PubMed] [Google Scholar]
- Liang JS, Shimojima K, Takayama R, Natsume J, Shichiji M, Hirasawa K, … Yamamoto T (2011). CDKL5 alterations lead to early epileptic encephalopathy in both genders. Epilepsia, 52(10), 1835–1842. doi: 10.1111/j.1528-1167.2011.03174.x [DOI] [PubMed] [Google Scholar]
- MacKay CI, Wong K, Demarest ST, Benke TA, Downs J, & Leonard H (2021). Exploring genotype-phenotype relationships in the CDKL5 deficiency disorder using an international dataset. Clin Genet, 99(1), 157–165. doi: 10.1111/cge.13862 [DOI] [PubMed] [Google Scholar]
- Nemos C, Lambert L, Giuliano F, Doray B, Roubertie A, Goldenberg A, … Philippe C (2009). Mutational spectrum of CDKL5 in early-onset encephalopathies: a study of a large collection of French patients and review of the literature. Clin Genet, 76(4), 357–371. doi: 10.1111/j.1399-0004.2009.01194.x [DOI] [PubMed] [Google Scholar]
- Olson HE, Costantini JG, Swanson LC, Kaufmann WE, Benke TA, Fulton AB, … Heidary G (2021). Cerebral visual impairment in CDKL5 deficiency disorder: vision as an outcome measure. Dev Med Child Neurol. doi: 10.1111/dmcn.14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson HE, Demarest ST, Pestana-Knight EM, Swanson LC, Iqbal S, Lal D, … Benke TA (2019). Cyclin-Dependent Kinase-Like 5 Deficiency Disorder: Clinical Review. Pediatr Neurol, 97, 18–25. doi: 10.1016/j.pediatrneurol.2019.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch K, Tan QK, Stong N, Deak KL, McConkie-Rosell A, McDonald MT, … Shashi V (2020). Alternative transcripts in variant interpretation: the potential for missed diagnoses and misdiagnoses. Genet Med, 22(7), 1269–1275. doi: 10.1038/s41436-020-0781-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siri B, Varesio C, Freri E, Darra F, Gana S, Mei D, … Alfei E (2021). CDKL5 deficiency disorder in males: Five new variants and review of the literature. Eur J Paediatr Neurol, 33, 9–20. doi: 10.1016/j.ejpn.2021.04.007 [DOI] [PubMed] [Google Scholar]
- Stosser MB, Lindy AS, Butler E, Retterer K, Piccirillo-Stosser CM, Richard G, & McKnight DA (2018). High frequency of mosaic pathogenic variants in genes causing epilepsy-related neurodevelopmental disorders. Genet Med, 20(4), 403–410. doi: 10.1038/gim.2017.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Van Esch H, Hagedorn-Greiwe M, Hoffmann K, Moser B, Raynaud M, … Kalscheuer VM (2004). Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am J Hum Genet, 75(6), 1149–1154. doi: 10.1086/426460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaving LS, Christodoulou J, Williamson SL, Friend KL, McKenzie OL, Archer H, … Gecz J (2004). Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet, 75(6), 1079–1093. doi: 10.1086/426462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson SL, Giudici L, Kilstrup-Nielsen C, Gold W, Pelka GJ, Tam PP, … Christodoulou J (2012). A novel transcript of cyclin-dependent kinase-like 5 (CDKL5) has an alternative C-terminus and is the predominant transcript in brain. Hum Genet, 131(2), 187–200. doi: 10.1007/s00439-011-1058-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. CDKL5 transcript-specific expression patterns. This screen capture from GTEX (https://gtexportal.org) shows the expression of various CDKL5 isoforms across different tissues. The ENST00000623535 transcript is equivalent to NM_001323289 and is the transcript with highest expression in the brain and in other tissues.
Data Availability Statement
Data for patient 1 was submitted to Phenome Central (P0009410) and Clinvar (SCV001245579.1). Data for patient 2 is available via DUOS (https://duos.broadinstitute.org/) and AnVIL (https://anvilproject.org/). No new data analysis was performed for patient 3.