Abstract
Telomerase is a reverse transcriptase that adds telomeric repeats to chromosomal ends. In most normal human somatic cells, telomerase is repressed and telomeres progressively shorten, leading to limited proliferative life-span. Telomerase reactivation is associated with cellular immortalization and is a frequent event during tumorigenesis. The telomerase ribonucleoprotein complex consists of two essential components, a catalytic protein subunit [human telomerase reverse transcriptase (hTERT)] and a template RNA (hTR). hTR is constitutively expressed, while hTERT is almost universally absent in telomerase-negative cells. Although repression of telomerase is transcriptional in telomerase-negative cells, post-transcriptional and assembly processes are likely to play important roles in regulating telomerase activity in those that are telomerase-positive. The telomerase transcript can also be alternatively spliced into a variety of non-functional forms. To establish the quantitative relationships between telomerase activity and its various components, we determined the numbers of molecules of hTR and hTERT mRNA, and the levels of alternatively spliced hTERT mRNA variants in normal, in vitro immortalized and cancer cell lines. We report here that there is surprisingly little variation in the proportion of alternatively spliced forms of hTERT in different cell lines. The only variation observed occurred when a change in splicing to non-functional forms appeared in response to conditions that repress telomerase activity in IDH4 cells. We also found that most telomerase-positive cell lines only contain a few molecules of potentially functional hTERT mRNA, and there is a correlation between telomerase activity and the levels of both hTR and hTERT +α+β mRNA.
INTRODUCTION
Telomeres are specialized DNA–protein complexes located at the ends of the chromosomes in eukaryotic cells. The progressive shortening of telomeres after each cell division (1–3) is believed to be the basis for the limited life-span of normal human cells (4). Human telomerase is a ribonucleoprotein complex in which a catalytic reverse transcriptase protein subunit (hTERT) uses an integral RNA (hTR) as a template for the addition of telomeric repeat sequences to the ends of chromosomes (5–10). Telomerase is repressed in most normal human somatic cells, while the reactivation of telomerase to maintain telomere length is necessary for the unlimited growth potential of most cancer cells (11–15).
hTR is present in all cell types (5,16), while hTERT mRNA is not detected in telomerase-negative cells (8,9). Mixing in vitro transcribed hTR with in vitro translated hTERT in a rabbit reticulocyte lysate resulted in detectable telomerase activity (17,18), suggesting that hTERT might be the only missing or rate-limiting component for telomerase activity in normal human somatic cells. This was confirmed by showing that the ectopic expression of hTERT in cultured normal human cells was sufficient to produce telomerase activity, bypass cellular senescence and extend life-span in several cell types (19–22). Although hTR and hTERT are the minimal essential components of telomerase, the human telomerase RNP is a very large complex (23) containing many additional unidentified proteins (24). The level of hTERT expression has been reported to be an indicator of cancer progression (25–31). The amount of hTR is also increased when telomerase is reactivated, due to both increased transcription and increased stability (16).
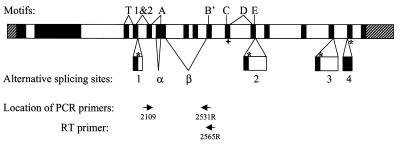
The hTERT transcript has been shown to contain at least six alternate splicing sites (four insertion sites and two deletion sites have been identified) (Fig. 1) (GenBank accession nos AF128893 and AF128894) (32). Due to the distance between the alternate splicing sites and low abundance of hTERT mRNA, only seven variants (six using a single alternate splicing site and one containing both deletion sites) have been shown to exist in human cDNA libraries or by RT–PCR (32,33). One deletion (β site, 182 bp) and one insertion (insertion 1, 38 bp) cause premature translation terminations upstream of the essential hTERT RT motifs (9,10,34). The other deletion site (α site) causes a 36 bp deletion within RT motif A without causing premature translation termination (32,33). Because the RT motifs are required for telomerase activity (9,10,34), these variants do not have catalytic activity. The other three insertions occur downstream of the RT motifs and produce truncations of the protein. Since some C-terminal modifications have been shown to interfere with the ability of telomerase to maintain telomeres (35,36), these insertion variants may not produce biologically functional proteins.
Figure 1.
Genomic organization of the hTERT gene. RT motifs (T, 1&2, A, B′, C, D and E) (9) and alternate splicing sites (insertions 1–4 and deletions α and β) are marked (32,33). Filled boxes are translated regions; shaded boxes are 5′- and 3′-UTR. In cases of alternative splicing, an asterisk marks the location of termination codon for the insertions. A cross marks the location of the termination codon for the variant containing deletion β. The location of the PCR primers used to determine splicing patterns is shown at the bottom.
Changes in splicing patterns have been shown to occur during development (37) and between normal and tumor tissue (38). However, there is essentially no information available about relative levels of alternate hTERT splicing variants in different cancer cells. In addition, it is not known if the expression levels of hTERT and hTR are directly related to each other or to telomerase enzyme activity. Furthermore, very little data is available on the actual numbers of hTR or hTERT mRNA molecules per cell in different cell lines. In the present study, hTR levels and hTERT transcript splicing patterns were quantitated and compared to telomerase activity. The results of these experiments indicate that there is a highly uniform pattern of splicing among all nine telomerase-positive cell lines examined, with the potentially functional +α+β form accounting for only 5% of the total message. A change in the splicing pattern accompanied the down regulation of hTERT in IDH4 but not HT1080 cells. The typical cultured cancer cell contains more than 10 000 molecules of hTR and less than one molecule of potentially active hTERT mRNA. Both the hTR and hTERT mRNA levels correlate with telomerase activity level. These results suggest that once hTERT transcription is activated, telomerase enzymatic activity is primarily regulated at the level of transcription of both its core components in most cell lines.
MATERIALS AND METHODS
Cell culture
Table 1 describes the cell lines and strains included in this study. All cells were cultured at 37°C under 5% CO2 in a 4:1 mixture of Dulbecco’s MEM and medium 199 supplemented with 10% Defined Supplemented Bovine Calf Serum (Hyclone, Logan, UT) and 50 µg/ml of gentamicin (Sigma, St Louis, MO). Quiescent HT1080 cells were obtained by culturing HT1080 cells in the above medium without serum for 7 days (39). IDH4 cells [immortal telomerase-positive cells derived from IMR90 fibroblasts stably transfected with Simian Virus 40 (SV40) large-T antigen under the control of a dexamethasone-inducible promoter] (40,41), were cultured in serum-containing medium supplemented with 1 µg/ml dexamethasone to provide growing cells or without dexamethasone to produce quiescent cells.
Table 1. Description of cell lines used in this study.
| Cell line | Telomerase activity | Description |
|---|---|---|
| TRAP and hTERT +α+β mRNA were determined for all samples. The number of hTR molecules per cell and alternate splicing patterns of hTERT mRNA were also determined for cell types marked with an asterisk. | ||
| BJ* | – | Normal human foreskin fibroblast (obtained from James Smith, Baylor University Medical Center) |
| IMR90* | – | Normal human lung embryo fibroblasts (ATCC no. CCL-186) |
| BJ hTERT* | + |
BJ cells transfected with hTERT cDNA using a plasmid (19) |
| BJ puro hTERT | + |
BJ cells transfected with hTERT cDNA using retroviral vector pBabe puro (this study) |
| SW13* | – | SV40 large T-antigen immortalized ALT pathway cells derived from IMR90 (38,39) |
| SW39* | + | SV40 large T-antigen immortalized IMR90 cells, telomerase-postive (38,39) |
| IDH4* | + | SV40 T-antigen immortalized IMR90 cells (38,39) |
| HT1080* | + | Human fibrosarcoma cells (ATCC no. CCL-121) |
| HL60* | + | Human promyelocytic leukemia cells (ATCC no. CCL-240) |
| DU145* | + | Human prostate carcinoma cells (ATCC no. HTB-81) |
| RCC23* | + | Human renal cell carcinoma cells [Yoshida et al. (50)] |
| 293* | + | Human kidney cells, transformed with adenovirus 5 DNA (ATCC no. 45504) |
| HeLa* | + | Human cervical adenocarcinoma (ATCC no. CCL-2) |
| H1299* | + | Human non-small lung carcinoma cells (ATCC no. CRL-5803) |
| SK-BR-3 | + | Human mammary adenocarcinoma cells (ATCC no. HTB-30) |
| HCC95 | + | Human non-small lung carcinoma cells (obtained from John Minna, UT Southwestern) |
| BT-20 | + | Human mammary adenocarcinoma cells (ATCC no. HTB-19) |
| H358 | + | Human non-small lung carcinoma cells (ATCC no. CRL-5807) |
| MCF-7 | + | Human mammary adenocarcinoma cells (ATCC no. HTB-22) |
| MDA-MB- | + | Human mammary adenocarcinoma cells (ATCC no. HTB-26) |
| MDA-MB- | + | Human ductal carcinoma cells (ATCC no. HTB-129) |
| MDA-MB- | + | Human mammary adenocarcinoma cells (ATCC no. HTB-130) |
| MDA-MB- | + | Human mammary adenocarcinoma cells (ATCC no. HTB-132) |
Determination of the number of hTR molecules per cell and the number of hTERT mRNA molecules per cell by quantitative RT–PCR
Cells were trypsinized, counted using a Coulter ZM cell counter (Beckman Coulter, Fullerton, CA) and collected in PBS. Total RNA from these cells was prepared by a modification of the guanidium isothiocyanate/cesium chloride technique (42) using TriPure isolation reagent (Roche Molecular Biochemicals, Indianapolis, IN). The amount of total RNA per cell was calculated based on OD260 measurements.
The number of hTR molecules per cell for each cell type was determined using a competitor RNA as previously described (Table 1) (16). In a first step, the number of hTR molecules is estimated from the relative intensities of the amplified hTR and competitor bands. In the second step, the amount of competitor is adjusted to equal that of hTR, and the number of PCR cycles modified to ensure that the PCR amplification is in the linear range. The numbers of hTERT (+α+β) mRNA molecules were determined using 0.05–0.1 µg of total RNA from each sample by real-time RT–PCR using the LightCycler and LightCycler TeloTAGGG hTERT Kit from Roche Molecular Biochemicals according to the manufacturer’s instructions. The primers and fluorescent probes in this assay are designed so that only the potentially functional +α+β form of hTERT is measured.
The distribution of hTERT mRNA alternate splicing patterns in some cell types was determined by RT–PCR (Table 1). A portion (1 µg) of each RNA sample was reverse transcribed in 20 µl with 0.2 pmol of the oligonucleotide TERT2565R (Table 2 and Fig. 1) using SuperScript First-strand Synthesis System for RT–PCR (catalog no. 11904-018) according to protocol using gene-specific primers recommended by the manufacturer (Gibco BRL, Gaithersburg, MD). Aliquots (2 µl) from each reverse transcription were PCR amplified with primers TERT2109 and TERT2531R (Table 2) (94°C for 20 s, 60°C for 20 s, 72°C for 40 s), using 2 U of Taq DNA polymerase (Gibco BRL). One of the PCR primers was 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Gibco BRL). In order to keep the PCR assay within the linear range, different samples were amplified for different numbers of cycles. PCR products were separated on a 5% non-denaturing polyacrylamide gel in 0.5× TBE buffer. The gel was dried and exposed to a PhosphorImager storage screen. Gel images were analyzed using ImageQuant version 5.0 (Molecular Dynamics, CA) to estimate the proportion of each hTERT alternate splicing variant mRNA molecules in each sample.
Table 2. Oligonucleotides used in this study.
| Name | Purpose | Sequence (5′→3′) |
|---|---|---|
| T7hTR+1 | Top primer to make template for in vitro T7 transcription of hTR competitor | CGTAATACGACTCACTATAGGGTTGCGGAGGGTGGGCCT |
| hTR+451rev | Bottom primer to make template for in vitro T7 transcription of hTR competitor | GCATGTGTGAGCCGAGTCCT |
| F3B | Forward primer for hTR | TCTAACCCTAACTGAGAAGGGCGTAG |
| hTR189R | Reverse primer for hTR | CCAGCAGCTGACATTTTTTG |
| TERT2565R | Gene-specific reverse transcription primer for hTERT | CGCAAACAGCTTGTTCTCCATGTC |
| TERT2109 | Forward primer for hTERT | GCCTGAGCTGTACTTTGTCAA |
| TERT2531R | Reverse primer for hTERT | AGGCTGCAGAGCAGCGTGGAGAGG |
The number of hTR molecules per cell and the number of hTERT mRNA molecules per cell for each cell type were calculated from two independent sets of samples based on the number of cell equivalents used in the respective quantitative RT–PCR for each sample.
Determination of telomerase activity
Whole-cell lysates (1000 cells/µl) of each cell type were prepared as described previously (12,43). Telomerase activity was determined using the TRAP-eze Telomerase Detection Kit (Intergen, Boston, MA) according to the protocol recommended by manufacturer, using 32P- or Cy-5-labeled TS primer (AATCCGTCGAGCAGTT). In brief, 32P- or Cy-5-labeled TS primer was mixed with cell lysate (1 µl) and extended by telomerase in cell lysate (30 min at room temperature). The extension product of TS primer was amplified by PCR in the presence of an internal standard. PCR products were then resolved on a 10% polyacrylamide gel, exposed to PhosphorImager screen and visualized by scanning on a Storm 860 system (Molecular Dynamics). Telomerase activity was calculated as the ratio of the intensity of telomerase ladder to the intensity of the 36 bp internal standard (ITAS) band using the ImageQuant version 5.0 software (Molecular Dynamics). Relative TRAP (telomere repeat amplification protocol) activities of two independent sets of samples were determined.
RESULTS
Telomerase activity and hTERT +α+β mRNA were determined for all cell types in Table 1. In addition, the number of hTR molecules per cell and the distribution of variant hTERT mRNA molecules per cell were quantitated for some of the cell types. These cell types included normal diploid telomerase-negative fibroblasts; telomerase-positive cells derived from normal telomerase-negative cells by introducing an exogenous hTERT cDNA; telomerase-negative cells immortalized by ectopic expression of SV40 large T-antigen (ALT pathway cells); and telomerase-positive cells that have activated their endogenous hTERT gene during the process of spontaneous immortalization driven by the expression of SV40 large T-antigen in culture or tumor formation in vivo.
Quantitation of telomerase activity
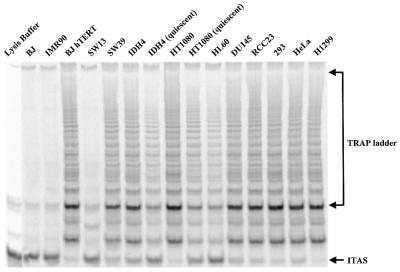
Telomerase activities of the cell strains and cell lines listed in Table 1 were determined by the TRAP assay (Fig. 2 and Table 3). For each lane, the ratio of the sum of the TRAP ladder versus the internal standard (ITAS) was calculated using ImageQuant version 5.0. The relative telomerase activity per cell for each cell line is presented as the percentage of the ratio of TRAP ladder/ITAS per cell versus the value of H1299, which was arbitrarily set as 100. The results are summarized in Table 3. BJ hTERT cells, which express an exogenous hTERT on a transfected plasmid, have 30% of the telomerase activity of H1299 cells; BJ puro hTERT which expresses an exogenous hTERT using retroviral construct has almost 100% of the telomerase activity of H1299 cells. The other cancer cell lines tested have between 5 and 159% of the telomerase activity of H1299, covering a wide range of telomerase activity; while quiescent HT1080 and quiescent IDH4 showed lowered telomerase activity (15–20% of the growing cells) as previously described (39).
Figure 2.
Telomerase activity levels of the cell types included in this study were determined by the TRAP assay as described in Materials and Methods.
Table 3. Quantitation results of telomerase activity, hTR and hTERT mRNAa.
| Cell type | hTR (×10–3) | hTERT +α+β mRNA per cell | Total hTERT mRNA | Relative TRAP per cellb |
|---|---|---|---|---|
| ahTR levels were quantitated by competitive RT–PCR while hTERT +α+β mRNA levels were determined by LightCycler RT–PCR. The total number of hTERT mRNA molecules was calculated using the measured fractions of hTERT +α+β from Table 4 where available. | ||||
| ND, not determined; NA, not available, since the percentage of hTERT +α+β mRNA in this sample was too low to be quantitated. | ||||
| BJ | 13.5 ± 0.5 | 0.00 | 0.00 | 0 |
| IMR90 | 11 ± 2 | 0.00 | 0.00 | 0 |
| BJ hTERT | 23 ± 8 | 220.00 | 220.00 | 30 |
| BJ puro hTERT | ND | 2000.00 | 2000.00 | 96 |
| SW13 | 11 ± 1 | 0.00 | 0.00 | 0 |
| SW39 | 18 ± 4 | 0.41 | 6.70 | 14 |
| IDH4 | 38 ± 10 | 0.12 | 1.54 | 10 |
| Quiescent IDH4 | 70 ± 36 | 0.02 | NA | 2 |
| HT1080 | 34 ± 15 | 0.66 | 13.00 | 48 |
| Quiescent HT1080 | 23 ± 3 | 0.29 | 5.40 | 7 |
| HL60 | 13 ± 2 | 0.44 | 15.00 | 25 |
| DU145 | 17 ± 5 | 4.45 | 78.20 | 16 |
| RCC23 | 22 ± 9 | 0.69 | 32.00 | 23 |
| 293 | 69 ± 19 | 1.34 | 17.50 | 69 |
| HeLa | 54 ± 15 | 0.24 | 5.60 | 16 |
| SK-BR-3 | ND | 0.57 | ND | 64 |
| BT-20 | ND | 0.26 | ND | 27 |
| H358 | ND | 0.12 | ND | 13 |
| MDA-MB-231 | ND | 1.03 | ND | 26 |
| MDA-MB-436 | ND | 0.84 | ND | 125 |
| H1299 | 112 ± 43 | 1.34 | 23.00 | 100 |
| MCF-7 | ND | 1.68 | ND | 159 |
| MDA-MB-435S | ND | 0.73 | ND | 24 |
| MDA-MB-468 | ND | 0.41 | ND | 5 |
| HCC95 | ND | 2.80 | ND | 78 |
bTelomerase activity levels are in reference to H1299 level, which is taken as 100.
Quantitation of the telomerase RNA component
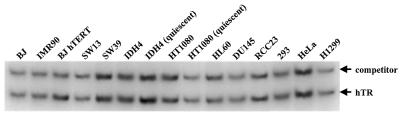
The telomerase RNA component in some cell types was quantitated by competitive RT–PCR using a competitor RNA with a 20 bp insert between the two RT–PCR primers (Fig. 3). The number of cells equivalent to 1 µg of input total RNA for each cell type was calculated from the number of cells and amount of total RNA recovered in each RNA preparation. The number of hTR molecules per cell was determined by the ratio of PCR products corresponding to competitor RNA and hTR, the amount of competitor RNA added to each RT reaction and the number of cells equivalent to 1 µg of total RNA input for each cell type. The results are summarized in Table 3. The telomerase-negative cells (BJ, IMR90 and SW13) have a range of 11 000–13 500 molecules of hTR per cell. hTR levels increase (up to 112 000 molecules per cell) in cancer and immortalized cells due to both increased transcription and increased stability as reported previously (16).
Figure 3.
Determination of hTR levels by competitive RT–PCR. Results of the quantitation step are shown, in which the amount of competitor has been adjusted to be equivalent to the endogenous transcript. The PCR product corresponding to the competitor RNA is 20 bp longer than that of hTR. The equivalent intensity of the two bands in each sample demonstrates that an accurate quantitation of the level of hTR has been obtained. Results are summarized in Table 3.
Quantitation of the hTERT mRNA
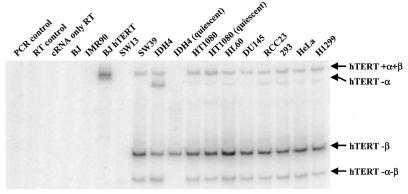
The distribution patterns of the alternate splicing variants containing one or both of the deletions (α and β) for some cell types were analyzed by RT–PCR (Fig. 4 and Table 4). The mRNA of the +α+β form of hTERT of all cell types listed in Table 1 was quantitated using LightCycler and LightCycler TeloTAGGG hTERT Kit. The results are summarized in Table 3, along with the estimate of total hTERT mRNA levels calculated from the distribution of alternate splicing variants shown in Table 4.
Figure 4.
Determination of the mRNA levels of the +α+β and three alternate splicing variants of hTERT. The products corresponding to hTERT –α, hTERT –β, hTERT –α–β are 36, 182 and 218 bp shorter than that of hTERT +α+β, respectively. Distribution among the alternate splicing variants is summarized in Table 4.
Table 4. Distribution of alternate splicing variants of hTERT mRNA.
| Cell type | hTERT +α+β per cell (%) | hTERT –α+β per cell (%) | hTERT +α–β per cell (%) | HTERT –α–β per cell (%) |
|---|---|---|---|---|
| All values are average percentages of each alternatively spliced hTERT mRNA determined in two independent samples for each cell type ± S.D. | ||||
| BJ | 0.0 | 0.0 | 0.0 | 0.0 |
| IMR90 | 0.0 | 0.0 | 0.0 | 0.0 |
| BJ hTERT | 100.0 ± 0.0 | 0.0 | 0.0 | 0.0 |
| SW13 | 0.0 | 0.0 | 0.0 | 0.0 |
| SW39 | 6.1 ± 1.0 | 0.0 | 85 ± 2.0 | 9.0 ± 1.2 |
| IDH4 | 7.8 ± 3.3 | 10.0 ± 14.0 | 68 ± 23.0 | 15.0 ± 6.0 |
| Quiescent IDH4 | 0.0 | 0.0 | 100 ± 0.0 | 0.0 |
| HT1080 | 5.1 ± 3.6 | 0.5 ± 0.6 | 88 ± 5.0 | 6.9 ± 0.7 |
| Quiescent HT1080 | 5.4 ± 5.9 | 0.3 ± 0.4 | 89 ± 6.0 | 5.5 ± 0.7 |
| HL60 | 3.0 ± 0.1 | 0.2 ± 0.2 | 88 ± 1.0 | 9.3 ± 1.8 |
| DU145 | 5.7 ± 2.6 | 0.3 ± 0.4 | 85 ± 5.0 | 9.5 ± 2.5 |
| RCC23 | 2.2 ± 3.0 | 0.6 ± 0.9 | 90 ± 3.0 | 7.2 ± 1.3 |
| 293 | 7.7 ± 0.7 | 0.6 ± 0.9 | 80 ± 4.0 | 11.0 ± 4.0 |
| HeLa | 4.3 ± 2.1 | 0.4 ± 0.6 | 80 ± 5.0 | 15.0 ± 8.0 |
| H1299 | 5.9 ± 4.7 | 0.6 ± 0.9 | 83 ± 0.2 | 10.3 ± 5.4 |
Telomerase-negative cell types have no detectable hTERT mRNA, confirming that hTERT expression in these cells is repressed. For all cells expressing endogenous hTERT, ∼5% of the total hTERT mRNA is in the potentially functional +α+β form, therefore the cells that have higher hTERT (+α+β) mRNA also have higher total hTERT mRNA levels (Tables 3 and 4). Among the telomerase-positive cell types, most cells have only 1–30 copies of total hTERT mRNA per cell (Table 3).
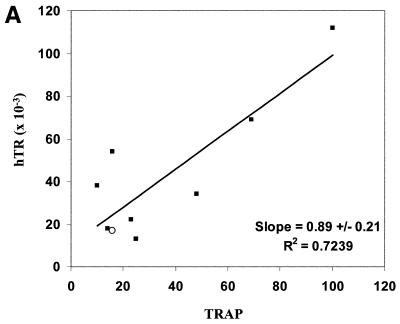
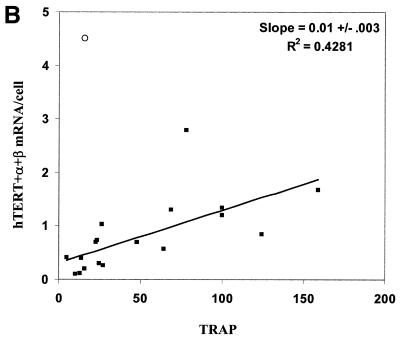
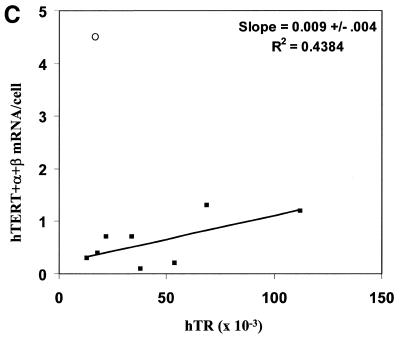
Protein products of the hTERT alternate splicing variants hTERT –α+β, hTERT +α–β and hTERT –α–β do not reconstitute telomerase activity (44,45). We compared potentially active hTERT (+α+β) mRNA to the average values of two independent determinations of hTR levels and TRAP activities in different combinations to assess the relationship between each pair of variants (Fig. 5, only data from non-quiescent cells expressing endogenous telomerase are shown). There is a weak correlation between TRAP activity and the abundance of either hTR (Fig. 5A) or hTERT (+α+β) mRNA (Fig. 5B). The value for DU145 cells lies well out of the range of the others when hTERT (+α+β) mRNA is compared to telomerase activity (open circle data points in Fig. 5). When this cell type is excluded from the analysis (see Discussion), there is also a weak correlation between hTR and hTERT levels (Fig. 5C).
Figure 5.
(A and C) Data for all cell types marked with an asterisk in Table 1 for which number of hTR molecules per cell were determined. (B) Data for all telomerase-positive cell types in Table 1. In each panel, the open circle denotes DU145 data. The standard error of the slope was calculated by the formula: S.E. = {S.D. of Y × [(1 – r2)/(n – 2)]/S.D.of X}, and was determined after excluding the data from DU145.
DISCUSSION
In the present study, the telomerase activity levels, hTR levels and hTERT mRNA levels in a number of normal and immortal and cancer cell lines were quantitated. As previously reported (16), the hTR level increases in the immortal and cancer cell lines. The present report is the first report quantitating hTERT mRNA molecules per cell in immortal human cells.
The cell types used in the current study that express endogenous telomerase show a similar distribution pattern among the alternate splicing variant forms. About 5% of total hTERT mRNA is in hTERT +α+β form, 80–90% in hTERT +α–β form, 5–15% in hTERT –α–β form and <1% in hTERT –α+β. The similar distribution of these splicing variants indicates that, at least in culture, the ∼20-fold variation in telomerase activity that is described in different tumor cells cannot be explained by alternative splicing patterns at the α and β sites. However, it is possible that alternate splicing elsewhere in the hTERT mRNA molecules that are +α+β could contribute to the variations in telomerase activity. Furthermore, these results indicate a common splicing pattern among diverse tumor types ranging from fibrosarcomas to leukemias and adenocarcinomas, indicating that a common splicing mechanism among different cell types determines the splicing of hTERT mRNA.
Telomerase activity decreases in quiescent cells (39). Overall hTERT mRNA levels decrease in differentiating HL60 myeloleukemia cells that are becoming postmitotic, suggesting transcriptional control (30). The present results show that similar decreases can occur by at least two additional mechanisms. In HT1080 cells, total transcription only decreased slightly and no changes in splicing patterns were found, suggesting that post-translational changes, increased protein degradation or alteration of some other telomerase component is reducing telomerase activity levels. In contrast, the change in IDH4 cells can be explained by a shift in splicing patterns (Tables 3 and 4). The total hTERT mRNA level in quiescent IDH4 cells decreased only slightly (2-fold, data not shown), while the hTERT mRNA alternate splicing pattern changed dramatically, with quiescent IDH4 cells containing mostly the hTERT +α–β form. The significantly decreased amount of +α+β hTERT mRNA is sufficient to explain the decrease in telomerase activity (Tables 3 and 4).
Competitive RT–PCR data indicate that most cancer cells contain 15 000–60 000 molecules of hTR and 1–30 molecules of hTERT mRNA. Although the number of hTR molecules is consistent with the ability to detect hTR by in situ hybridization (46,47), the number of hTERT messages is sufficiently low that it would be difficult to detect without a PCR amplification step.
The telomerase ribonucleoprotein is a complex molecule, with multiple factors involved in the proper folding and assembly of both RNA and protein components. In order to understand the efficiency and dynamics of this process, it is important to understand the quantitative relationships among the different participants. We have attempted to determine the amount of hTERT protein by western blotting. Although we can detect an overexpressed hTERT driven by a retroviral vector (in which there are ∼2000 molecules of +α+β hTERT message per cell by real-time RT–PCR; Table 3), we have been unable to see the endogenous protein in H1299 cancer cells (in which there are only ∼1.3 molecules of +α+β hTERT message per cell by competitive RT–PCR). Western blotting with dilutions of highly purified hTERT (kindly provided by the Geron Corporation) indicates that we should have been able to detect ∼600 molecules of hTERT protein per cell. Given the half-life of the mRNA (∼2–3 h) (48) and the half-life of the active telomerase complex (∼24 h) (39), the amount of hTERT mRNA that we detect is a reasonable value for producing even this upper limit for hTERT protein per cell. The great excess of hTR over protein, coupled with the correlation of hTR levels with TRAP activity (Fig. 5A), suggests that most of the hTR is sequestered by other more abundant hTR-binding proteins such as hnRNP C1/C2 and La, misfolded, or otherwise incompetent for assembly into active telomerase.
We have previously demonstrated that the rate of hTR transcription is increased in cells that have activated their endogenous hTERT expression (16), suggesting that there is some link between hTERT transcription and hTR transcription. The present results examine steady state levels rather than transcription, and show that there is a weak correlation between the levels of the two factors. Given that the half-life of hTR in cancer cells is ∼3 weeks (16) and the half-life of the hTERT mRNA is only ∼2 h, the correlation between steady state levels and transcriptional levels may be indirect.
There are weak correlations between hTR levels and TRAP activity (Fig. 5A), and between +α+β hTERT message levels and telomerase activity (Fig. 5B), indicating that, although neither factor is completely limiting, higher levels of either may facilitate increased rates of assembly of active complexes. The scatter in the data is certainly partially explained by the fundamental inaccuracies of comparing two PCR-based analyses. However, given the multiple factors involved in the assembly of active telomerase complexes, it probably also reflects the relative contributions of different limiting factors in different cell types. For example, the amount of +α+β hTERT message in DU145 cells is clearly much greater than expected based on the expression in other tumor cell lines (Fig. 5B), suggesting that something other than hTERT mRNA is limiting TRAP activity. This is consistent with observations that retroviral vectors overexpressing hTERT produce a much smaller increase in TRAP activity in DU145 cells compared to other cell lines (49).
In summary, the distribution among the alternate splicing forms in telomerase positive cell types are strikingly similar, suggesting that the distribution among hTERT alternate splicing forms is determined by common mechanisms in tumor cells that do not change significantly with tissue origin. In some but not all cultured cells, conditions that produce a decreased telomerase activity induce a change in hTERT splicing patterns. Using a diverse set of immortal cell types, we observed a correlation between the level of telomerase activity and the levels of both hTR and the potentially active hTERT +α+β mRNA. We conclude that telomerase repression in normal and ALT pathway cells is transcriptional, that there are very few molecules of hTERT mRNA per cell, and that the telomerase activity level in telomerase-positive cells is influenced by both hTR and hTERT +α+β mRNA levels in the majority of human immortal and cancer cells.
Acknowledgments
ACKNOWLEDGEMENTS
We acknowledge technical excellent assistance by Dennis M. White and Bryan C. Frank. BJ neonatal foreskin fibroblasts were kindly provided by James Smith, Baylor University Medical Center, Houston, TX. The hTR and hTERT cDNAs were provided by Geron Corporation (Menlo Park, CA). This work was supported by NIA grant AG07992, and the Ellison Medical Foundation (to J.W.S.).
REFERENCES
- 1.Olovnikov A.M. (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol., 41, 181–190. [DOI] [PubMed] [Google Scholar]
- 2.Watson J.D. (1972) Origin of concatemeric T7 DNA. Nature New Biol., 239, 197–201. [DOI] [PubMed] [Google Scholar]
- 3.Harley C.B., Fletcher,A.B. and Greider,C.W. (1990) Telomeres shorten during aging. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 4.Wright W.E. and Shay,J.W. (1992) The two-stage mechanism controlling cellular senescence and immortalization. Exp. Gerontol., 27, 383–389. [DOI] [PubMed] [Google Scholar]
- 5.Feng J., Funk,W.D., Wang,S.S., Weinrich,S.L., Avilion,A.A., Chiu,C.P., Adams,R.R., Chang,E., Allsopp,R.C., Yu,J. et al. (1995) The RNA component of human telomerase. Science, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- 6.Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- 7.Greider C.W. and Blackburn,E.H. (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature, 337, 331–337. [DOI] [PubMed] [Google Scholar]
- 8.Meyerson M., Counter,C.M., Eaton,E.N., Ellisen,L.W., Steiner,P., Caddle,S.D., Ziaugra,L., Beijersbergen,R.L., Davidoff,M.J., Liu,Q., Bacchetti,S., Haber,D.A. and Weinberg,R.A. (1997) hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell, 90, 785–795. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- 10.Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 11.Shay J.W. and Bacchetti,S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer, 5, 787–791. [DOI] [PubMed] [Google Scholar]
- 12.Kim N.W., Piatyszek,M.A., Prowse,K.R., Harley,C.B., West,M.D., Ho,P.L., Coviello,G.M., Wright,W.E., Weinrich,S.L. and Shay,J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 13.Hahn W.C., Counter,C.M., Lundberg,A.S., Beijersbergen,R.L., Brooks,M.W. and Weinberg,R.A. (1999) Creation of human tumour cells with defined genetic elements. Nature, 400, 464–468. [DOI] [PubMed] [Google Scholar]
- 14.Hahn W.C., Stewart,S.A., Brooks,M.W., York,S.G., Eaton,E., Kurachi,A., Beijersbergen,R.L., Knoll,J.H., Meyerson,M. and Weinberg,R.A. (1999) Inhibition of telomerase limits the growth of human cancer cells. Nature Med., 5, 1164–1170. [DOI] [PubMed] [Google Scholar]
- 15.Herbert B., Pitts,A.E., Baker,S.I., Hamilton,S.E., Wright,W.E., Shay,J.W. and Corey,D.R. (1999) Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl Acad. Sci. USA, 96, 14276–14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi X., Tesmer,V.M., Savre-Train,I., Shay,J.W. and Wright,W.E. (1999) Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol., 19, 3989–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beattie T.L., Zhou,W., Robinson,M.O. and Harrington,L. (1998) Reconstitution of human telomerase activity in vitro. Curr. Biol., 8, 177–180. [DOI] [PubMed] [Google Scholar]
- 18.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W., Trager,J.B., Taylor,R.D., Carlos,R., Andrews,W.H., Wright,W.E., Shay,J.W., Harley,C.B. and Morin,G.B. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 19.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 20.Vaziri H. and Benchimol,S. (1998) Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol., 8, 279–282. [DOI] [PubMed] [Google Scholar]
- 21.Wyllie F.S., Jones,C.J., Skinner,J.W., Haughton,M.F., Wallis,C., Wynford-Thomas,D., Faragher,R.G. and Kipling,D. (2000) Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nature Genet., 24, 16–17. [DOI] [PubMed] [Google Scholar]
- 22.Ouellette M.M., McDaniel,L.D., Wright,W.E., Shay,J.W. and Schultz,R.A. (2000) The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum. Mol. Genet., 9, 403–411. [DOI] [PubMed] [Google Scholar]
- 23.Schnapp G., Rodi,H.P., Rettig,W.J., Schnapp,A. and Damm,K. (1998) One-step affinity purification protocol for human telomerase. Nucleic Acids Res., 26, 3311–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt S.E., Aisner,D.L., Baur,J., Tesmer,V.M., Dy,M., Ouellette,M., Trager,J.B., Morin,G.B., Toft,D.O., Shay,J.W., Wright,W.E. and White,M.A. (1999) Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev., 13, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dome J.S., Chung,S., Bergemann,T., Umbricht,C.B., Saji,M., Carey,L.A., Grundy,P.E., Perlman,E.J., Breslow,N.E. and Sukumar,S. (1999) High telomerase reverse transcriptase (hTERT) messenger RNA level correlates with tumor recurrence in patients with favorable histology Wilms’ tumor. Cancer Res., 59, 4301–4307. [PubMed] [Google Scholar]
- 26.Takahashi S., Kitamoto,M., Takaishi,H., Aikata,H., Kawakami,Y., Nakanishi,T., Shimamoto,F., Tahara,E., Tahara,H., Ide,T. and Kajiyama,G. (2000) Expression of telomerase component genes in hepatocellular carcinomas. Eur. J. Cancer, 36, 496–502. [DOI] [PubMed] [Google Scholar]
- 27.Choi S., Kang,H., Im,E., Kim,Y.J., Bae,Y.T., Choi,Y.H., Lee,K.H., Chung,H., Chang,H. and Kim,N.D. (2000) Inhibition of cell growth and telomerase activity of breast cancer cells in vitro by retinoic acids. Int. J. Oncol., 17, 971–976. [DOI] [PubMed] [Google Scholar]
- 28.Ferlicot S., Paradis,V., Dargere,D., Monges,G. and Bedossa,P. (1999) Detection of telomerase in hepatocellular carcinomas using a PCR ELISA assay: comparison with hTR expression. J. Clin. Pathol., 52, 725–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito H., Kyo,S., Kanaya,T., Takakura,M., Inoue,M. and Namiki,M. (1998) Expression of human telomerase subunits and correlation with telomerase activity in urothelial cancer. Clin. Cancer Res., 4, 1603–1608. [PubMed] [Google Scholar]
- 30.Koyanagi Y., Kobayashi,D., Yajima,T., Asanuma,K., Kimura,T., Sato,T., Kida,T., Yagihashi,A., Kameshima,H. and Watanabe,N. (2000) Telomerase activity is down regulated via decreases in hTERT mRNA but not TEP1 mRNA or hTERC during the differentiation of leukemic cells. Anticancer Res., 20, 773–778. [PubMed] [Google Scholar]
- 31.Takakura M., Kyo,S., Kanaya,T., Tanaka,M. and Inoue,M. (1998) Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res., 58, 1558–1561. [PubMed] [Google Scholar]
- 32.Wick M., Zubov,D. and Hagen,G. (1999) Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene, 232, 97–106. [DOI] [PubMed] [Google Scholar]
- 33.Kilian A., Bowtell,D.D., Abud,H.E., Hime,G.R., Venter,D.J., Keese,P.K., Duncan,E.L., Reddel,R.R. and Jefferson,R.A. (1997) Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet., 6, 2011–2019. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama J., Tahara,H., Tahara,E., Saito,M., Ito,K., Nakamura,H., Nakanishi,T., Ide,T. and Ishikawa,F. (1998) Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nature Genet., 18, 65–68. [DOI] [PubMed] [Google Scholar]
- 35.Counter C.M., Hahn,W.C., Wei,W., Caddle,S.D., Beijersbergen,R.L., Lansdorp,P.M., Sedivy,J.M. and Weinberg,R.A. (1998) Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc. Natl Acad. Sci. USA, 95, 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ouellette M.M., Aisner,D.L., Savre-Train,I., Wright,W.E. and Shay,J.W. (1999) Telomerase activity does not always imply telomere maintenance. Biochem. Biophys. Res. Commun., 254, 795–803. [DOI] [PubMed] [Google Scholar]
- 37.Ulaner G.A., Hu,J.F., Vu,T.H., Giudice,L.C. and Hoffman,A.R. (1998) Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res., 58, 4168–4172. [PubMed] [Google Scholar]
- 38.Ulaner G.A., Hu,J.F., Vu,T.H., Oruganti,H., Giudice,L.C. and Hoffman,A.R. (2000) Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium and myometrium. Int. J. Cancer, 85, 330–335. [PubMed] [Google Scholar]
- 39.Holt S.E., Aisner,D.L., Shay,J.W. and Wright,W.E. (1997) Lack of cell cycle regulation of telomerase activity in human cells. Proc. Natl Acad. Sci. USA, 94, 10687–10692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright W.E., Pereira-Smith,O.M. and Shay,J.W. (1989) Reversible cellular senescence: Implication for a two-stage model for the immortalization of normal human diploid fibroblasts. Mol. Cell. Biol., 9, 3088–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shay J.W., West,M.D. and Wright,W.E. (1992) Re-expression of senescent markers in deinduced reversibly immortalized cells. Exp. Gerontol., 27, 477–492. [DOI] [PubMed] [Google Scholar]
- 42.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 43.Wright W.E., Shay,J.W. and Piatyszek,M.A. (1995) Modifications of a telomeric repeat amplification protocol (TRAP) result in increased reliability, linearity and sensitivity. Nucleic Acids Res., 23, 3794–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi X., White,D.M., Aisner,D.L., Baur,J.A., Wright,W.E. and Shay,J.W. (2000) An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia, 2, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colgin L.M., Wilkinson,C., Englezou,A., Kilian,A., Robinson,M.O. and Reddel,R.R. (2000) The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia, 2, 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morales C.P., Burdick,J.S., Saboorian,M.H., Wright,W.E. and Shay,J.W. (1998) In situ hybridization for telomerase RNA in routine cytologic brushings for the diagnosis of pancreaticobiliary malignancies. Gastrointest. Endosc., 48, 402–405. [DOI] [PubMed] [Google Scholar]
- 47.Morales C.P., Lee,E.L. and Shay,J.W. (1998) In situ hybridization for the detection of telomerase RNA in the progression from Barrett’s esophagus to esophageal adenocarcinoma. Cancer, 83, 652–659. [PubMed] [Google Scholar]
- 48.Xu D., Gruber,A., Bjorkholm,M., Peterson,C. and Pisa,P. (1999) Suppression of telomerase reverse transcriptase (hTERT) expression in differentiated HL-60 cells: regulatory mechanisms. Br. J. Cancer, 80, 1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McChesney P.A., Aisner,D.L., Frank,B.C., Wright,W.E. and Shay,J.W. (2000) Telomere dynamics in cells with introduced telomerase: a rapid assay for telomerase activity on telomeres. Mol. Cell. Biol. Res. Commun., 3, 312–318. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida M.A., Shimizu,M., Ikeuchi,T., Tonomura,A., Yokota,J. and Oshimura,M. (1994) In vitro growth suppression and morphological change in a human renal cell carcinoma cell line by the introduction of normal chromosome 3 via microcell fusion. Mol. Carcinog., 9, 114––121.. [DOI] [PubMed] [Google Scholar]