Abstract
Objective
Prolonged postictal generalized EEG suppression (PGES) is a potential biomarker for sudden unexpected death in epilepsy (SUDEP), which may be associated with dysfunctional autonomic responses and serotonin signaling. To better understand molecular mechanisms, PGES duration was correlated to 5HT1A and 5HT2A receptor protein expression and RNAseq from resected hippocampus and temporal cortex of temporal lobe epilepsy (TLE) patients with seizures recorded in preoperative evaluation.
Methods
Analyses included 36 cases (range: age 14–64 years, epilepsy onset 0–51, epilepsy duration 2–53, PGES 0–93 seconds), with 13 cases in all hippocampal analyses. 5HT1A and 5HT2A protein was evaluated by western blot and histologically in hippocampus (n=16) and temporal cortex (n=9). We correlated PGES duration to our previous RNAseq dataset for serotonin receptor expression and signaling pathways, as well as weighted gene correlation network analysis (WGCNA) to identify correlated gene clusters.
Results
In hippocampus, 5HT2A protein by western blot positively correlated with PGES duration (p=0.0024, R2=0.52) but 5HT1A did not (p=0.87, R2=0.0020). In temporal cortex, 5HT1A and 5HT2A had lower expression and did not correlate with PGES duration. Histologically, PGES duration did not correlate with 5HT1A or 5HT2A expression in hippocampal CA4, dentate gyrus, or temporal cortex. RNAseq identified two serotonin receptors with expression that correlated to PGES duration in an exploratory analysis: HTR3B negatively correlated (p=0.043, R2=0.26) and HTR4 positively correlated (p=0.049, R2=0.25). WGCNA identified 4 modules correlated to PGES duration, including positive correlation to synaptic transcripts (p=0.040, corr.=0.52), particularly potassium channels (KCNA4, KCNC4, KCNH1, KCNIP4, KCNJ3, KCNJ6, KCNK1). No modules were associated with serotonin receptor signaling.
Significance
Higher hippocampal 5HT2A receptor protein and potassium channel transcripts may reflect underlying mechanisms contributing to or resulting from prolonged PGES. Future studies with larger cohorts should assess functional analyses and additional brain regions to elucidate mechanisms underlying PGES and SUDEP risk.
Keywords: serotonin, PGES, SUDEP, hippocampus
Introduction
Postictal generalized EEG suppression (PGES) may occur after a generalized tonic-clonic seizure (GTCS), and prolonged PGES is associated with impaired arousal, respiration, and other autonomic functions.1–3 PGES has been suggested as a potential sudden unexpected death in epilepsy (SUDEP) biomarker,1–5 and occurs with autonomic dysfunction and respiratory arrest in SUDEP animal models.6,7
Potential mechanisms underlying PGES and SUDEP include dysfunctional serotonin signaling,8 as serotonin modulates respiration, arousal, and seizures.9 Serotonergic neurons and serotonergic receptors are present in various regions throughout the brain, and thus serotonin receptor modulators may have opposing effects on seizure threshold related to factors like ligand specificity, dose, and cell type.10,11 In temporal lobe epilepsy (TLE) patients, MRI and PET imaging revealed decreased 5HT1A receptor binding.12,13 In animal models, elevated serotonin reduced seizure frequency, seizures reduced serotonergic firing, low serotonin or serotonin receptor deletion (5HT1A, 5HT2C, 5HT4, 5HT7) promoted seizures, and 5HT1A overexpression resulted in sporadic autonomic dysfunction and death.14–21 The 5HT2 receptor family generally facilitates excitatory effects,17 thus 5HT2A antagonists may provide improvement of epilepsy.10,22 In high-risk SUDEP patients, hippocampal serotonin transporter (SERT) protein expression was increased.21 The brainstem contains serotonergic neurons that project to various brain regions,23,24 and we recently identified altered signaling pathways in the serotonergic dorsal raphe of the midbrain in SUDEP cases compared to controls.25 Further, an animal model linked PGES to dysfunctional serotonin signaling and dorsal raphe stimulation reduced PGES duration.26 With increasing PGES duration, interictal serum serotonin is decreased in epilepsy patients,27 although serotonin levels have not been studied in brain tissue.
We evaluated whether PGES duration recorded during presurgical evaluation correlated to serotonin receptor expression and RNAseq data in resected hippocampus and temporal cortex of TLE patients.
Materials and Methods
Human Brain Tissue.
Surgical brain tissue was obtained with approval by the New York University School of Medicine Institutional Review Board (IRB, #17–00398). Frozen brain tissue and formalin fixed paraffin embedded (FFPE) tissue for protein analyses were available from TLE patients undergoing surgical resection at the NYU Epilepsy Brain Bank, Amsterdam UMC, Royal Melbourne Hospital, University College of London, and Thomas Jefferson University. Informed consent was provided by each patient, and patients were enrolled in the brain tissue repositories from 2003 to 2019. Patients were considered for analyses who had a GTCS recorded by EEG prior to surgical resection. PGES was quantified at each center blinded to the molecular analysis results through expert visual review by an experienced clinical neurophysiologist (DF, BD, RT, MN, TOB). PGES was defined as suppression of the EEG activity ≤ 10μV across all head regions on scalp EEG following a seizure and termination was defined as the return of any cerebral activity < 10uV on any electrode.3,28 Review of video was used to determine if observed signals were related to obvious artifacts. When artifact precluded the determination of the offset PGES, PGES duration was defined as the duration of the interpretable portion of the post-ictal EEG. When multiple PGES readings were available from one patient, the longest PGES duration was evaluated in analyses. Patients were selected to provide coverage of the PGES spectrum, including no PGES (0 seconds), PGES < 50 seconds associated with low-risk SUDEP, and PGES ≥ 50 seconds associated with high-risk SUDEP.3 Case history is summarized in Table 1 and detailed in Supplemental Table 1, with 13 cases overlapping in hippocampus for protein, histology, and RNAseq analyses.
Table 1.
Case History Summary
| PGES | Cases (n) | Sex (M / F) |
PGES (seconds) | Age at Surgery (years) | Epilepsy Onset (years) | Epilepsy Duration (years) | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Western Blot | Hippocampus | < 50 sec | 9 | 2M / 7F | 13.3 ± 20.1 | 37.9 ± 14.1 | 18.3 ± 18.0 | 20.1 ± 11.0 |
| ≥ 50 sec | 7 | 3M / 4F | 58.0 ± 8.1 | 39.6 ± 11.7 | 14.9 ± 8.2 | 22.1 ± 16.5 | ||
| Temporal Cortex | < 50 sec | 6 | 1M / 5F | 21.2 ± 15.1 | 34.8 ± 14.5 | 20.8 ± 19.0 | 14.0 ± 9.2 | |
| ≥ 50 sec | 3 | 2M / 1F | 64.3 ± 13.6 | 30.0 ± 11.4 | 19.0 ± 20.3 | 11.0 ± 11.5 | ||
| Histology | ||||||||
| Hippocampus | < 50 sec | 9 | 2M / 7F | 13.3 ± 20.1 | 37.9 ± 14.1 | 18.3 ± 18.0 | 20.1 ± 11.0 | |
| ≥ 50 sec | 7 | 3M / 4F | 58.0 ± 8.1 | 39.6 ± 11.7 | 14.9 ± 8.2 | 22.1 ± 16.5 | ||
| Temporal Cortex | < 50 sec | 6 | 3M / 3F | 19.5 ± 13.3 | 28.5 ± 15.4 | 14.7 ± 14.3 | 13.8 ± 9.7 | |
| ≥ 50 sec | 3 | 2M / 1F | 64.3 ± 24.8 | 36.0 ± 16.1 | 17.3 ± 15.3 | 18.7 ± 3.5 | ||
| RNAseq | Hippocampus | < 50 sec | 8 | 2M / 6F | 14.8 ± 21.0 | 43.9 ± 14.0 | 24.4 ± 21.4 | 20.6 ± 12.2 |
| ≥ 50 sec | 8 | 4M / 4F | 57.1 ± 7.9 | 37.8 ± 12.0 | 15.3 ± 7.7 | 20.3 ± 16.2 | ||
The same hippocampal cases were used in protein and histology analyses. There are 13 cases that overlap in hippocampus for protein, histology, and RNAseq analyses. n = number of cases; M = male; F = female; mean ± standard deviation is indicated
Western blot.
Protein was isolated from frozen brain tissue (40 mg/sample) at 20% weight/volume in Tris-NaCl buffer (20 mM Tris base, 150 mM NaCl, 0.1% Triton-X 100, protease and phosphatase inhibitors at pH 7.5) using a hand held homogenizer equipped with a pestle on ice. Samples were incubated on ice for 15 minutes, centrifuged for 15 minutes at 14,000g, 4°C, and supernatant was isolated. Protein concentration was determined by BCA assay according to the manufacturer’s protocol (Pierce). Hippocampal (30 μg/lane) and temporal cortex (40 μg/lane) lysates were boiled in Bolt LDS Sample Buffer and DTT. For the hippocampus, one sample was included on both gels to allow for normalization across blots for all samples. Proteins were resolved on a 4–12% Bis-Tris gel (Invitrogen) and transferred onto nitrocellulose membranes. After blocking in 5% milk TBST, blots were probed for 5HT1A (1:500, Abcam ab227165), 5HT2A (1:500, Santa Cruz sc-166775), or actin (1:3000, Sigma A5441) in 5% milk TBST overnight at 4°C. Blots were incubated with corresponding HRP-conjugated secondary antibodies (1:3000, GE Healthcare) for 1 hour at room temperature. Bands were visualized after ECL (Pierce) on a BioRad ChemiDoc with the NYU Small Instrument Fleet. Blot images were analyzed in Fiji ImageJ for quantification with intensity normalized to actin.
Immunohistochemistry.
Subregional protein expression was assessed by immunohistochemistry as described.29 FFPE blocks were sectioned (8 μm) by the NYU Center for Biospecimen Research and Development (CBRD). Sections were deparaffinized and rehydrated through a series of xylenes and ethanol dilutions, followed by heat-induced antigen retrieval with 10 mM sodium citrate and 0.05% Triton-x 100 at pH 6. Sections were blocked with 10% normal donkey serum and incubated with 5HT1A (1:100, Abcam ab227165) or 5HT2A (1:100, Santa Cruz sc-166775) primary antibodies overnight at 4°C. Corresponding secondary antibodies were used (donkey anti-rabbit Alexa-Fluor 568, donkey anti-mouse Alexa-Fluor 488; Thermofisher) with DAPI counterstain and slides were coverslipped. Whole slide scanning was performed on each section at 20X magnification on a NanoZoomer HT2 (Hamamatsu) microscope with the NYU Experimental Pathology Research Laboratory (hippocampus) or the Leica Aperio Versa 8 microscope (temporal cortex). We analyzed one image in each hippocampal subregion and three images in the temporal cortex at 5X magnification in Fiji ImageJ by the same binary threshold for all images to determine the number of positive pixels in each image, reported as percentage of total image area.
RNAseq.
We analyzed our previous RNAseq dataset29,30 in the European Genome-phenome Archive (EGAS00001003922) in hippocampus of TLE patients for whom we could obtain an evaluation of PGES duration. Briefly30, RNA was isolated from surgical brain tissue by Qiazol Lysis Reagent with the miRNeasy Mini kit (Qiagen), RNA quality determined by Fragment Analyzer (Agilent Technologies), library preparation with NEBNext Ultra Directional RNA Library prep Kit for Illumina, ribosomal RNA depleted (New England Biolabs), and sequencing with the Illumina cBot and HiSeq 4000 paired-end with 151 nucleotide read length and depth of 50 million reads. DESeq2 in the R environment was used for library normalization. Case histories were previously detailed,29,30 are summarized in Table 1, and detailed in Supplemental Table 1.
Weighted Gene Correlation Network Analysis (WGCNA).
WGCNA was performed on our RNAseq dataset29,30 to determine whether PGES duration correlated to RNAseq in the R environment with the WGCNA package with defaults as described,31 except as stated. Soft threshold power beta was determined at R2 = 0.8 (power = 8), minModuleSize = 150, and deepSplit = 4. Gene ontology (GO) annotations for modules were determined following WGCNA with the anRichment package in the R environment with Entrez IDs against the human GOcollection (Supplemental Table 2). GO annotations were considered with a false discovery rate (FDR) < 5%.
Statistical analyses.
Statistical analyses used GraphPad Prism (version 9) and the R environment (http://www.r-project.org/). Western blot and histology correlation analyses were calculated by a Pearson correlation. A p value < 0.05 was considered significant.
Results
Case History
Clinical history is summarized in Table 1 and detailed in Supplemental Table 1 for each analysis, including cases with coverage of the PGES duration spectrum (range PGES 0 – 93 seconds): no PGES (0 seconds), PGES < 50 seconds associated with a lower risk of SUDEP, and PGES ≥ 50 seconds associated with higher risk of SUDEP. There were a total of 36 cases (range: age at surgery 14 – 64, epilepsy onset 0 – 51, epilepsy duration 2 – 53 years of age) evaluated across all analyses, and 13 of the same cases with hippocampal tissue were evaluated in protein, histology, and RNAseq analyses.
5HT1A and 5HT2A Protein Expression in Whole Brain Homogenate
Western blot of whole brain homogenate from resected hippocampus (containing combined dentate gyrus and CA1–4, n = 16, Figure 1A) and temporal cortex (n = 9, Figure 1B) revealed variable 5HT1A (55 kDa) expression among TLE patients. In the hippocampus, 5HT1A did not correlate with PGES duration (p = 0.87, R2 = 0.0020; Figure 1C). In temporal cortex, 5HT1A was expressed at a lower level than in hippocampus and did not correlate with PGES duration (p = 0.45, R2 = 0.085; Figure 1D). Hippocampal 5HT2A (55 kDa) positively correlated with PGES duration (p = 0.0024, R2 = 0.52; Figure 1E). Temporal cortex 5HT2A was expressed at lower levels than in hippocampus and there was no correlation with PGES duration (p = 0.20, R2 = 0.22; Figure 1F).
Figure 1. 5HT1A and 5HT2A protein expression in whole homogenate from the hippocampus and temporal cortex by PGES duration.
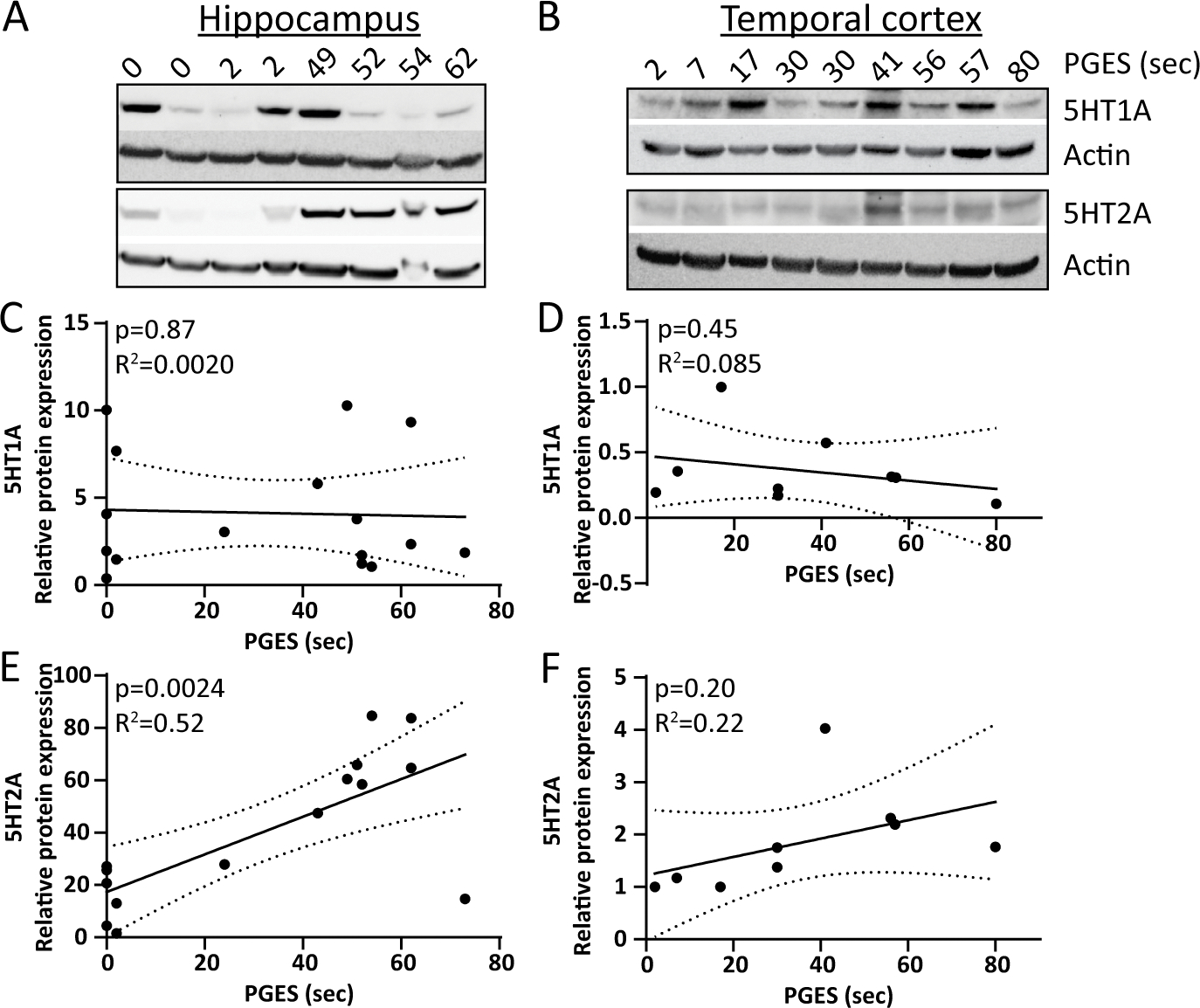
A) Representative western blot in hippocampus of 5HT1A (55 kDa), 5HT2A (55 kDa), and actin. B) Representative western blot in temporal cortex of 5HT1A (55 kDa), 5HT2A (55 kDa), and actin. C) Quantification of 5HT1A relative to actin in hippocampus (n = 16) indicates no correlation to PGES duration. D) Quantification of 5HT1A relative to actin in temporal cortex (n = 9) indicates no correlation to PGES duration. E) Quantification of 5HT2A relative to actin in hippocampus (n = 16) indicates a positive correlation to PGES duration. F) Quantification of 5HT2A relative to actin in temporal cortex (n = 9) indicates no correlation to PGES duration.
5HT1A and 5HT2A Protein Expression Histologically
5HT1A and 5HT2A expression was evaluated histologically in hippocampus (n = 16, dentate gyrus and CA4 subsector) and temporal cortex (n = 9) by PGES duration. In dentate gyrus, CA4, and temporal cortex (Figure 2A–F), 5HT1A expression was not correlated with PGES duration (Figure 2G–I). In dentate gyrus, CA4, and temporal cortex (Figure 3A–F), there was no correlation of 5HT2A expression and PGES duration (Figure 3G–I).
Figure 2. 5HT1A protein expression in subregions of the hippocampus and temporal cortex by PGES duration.
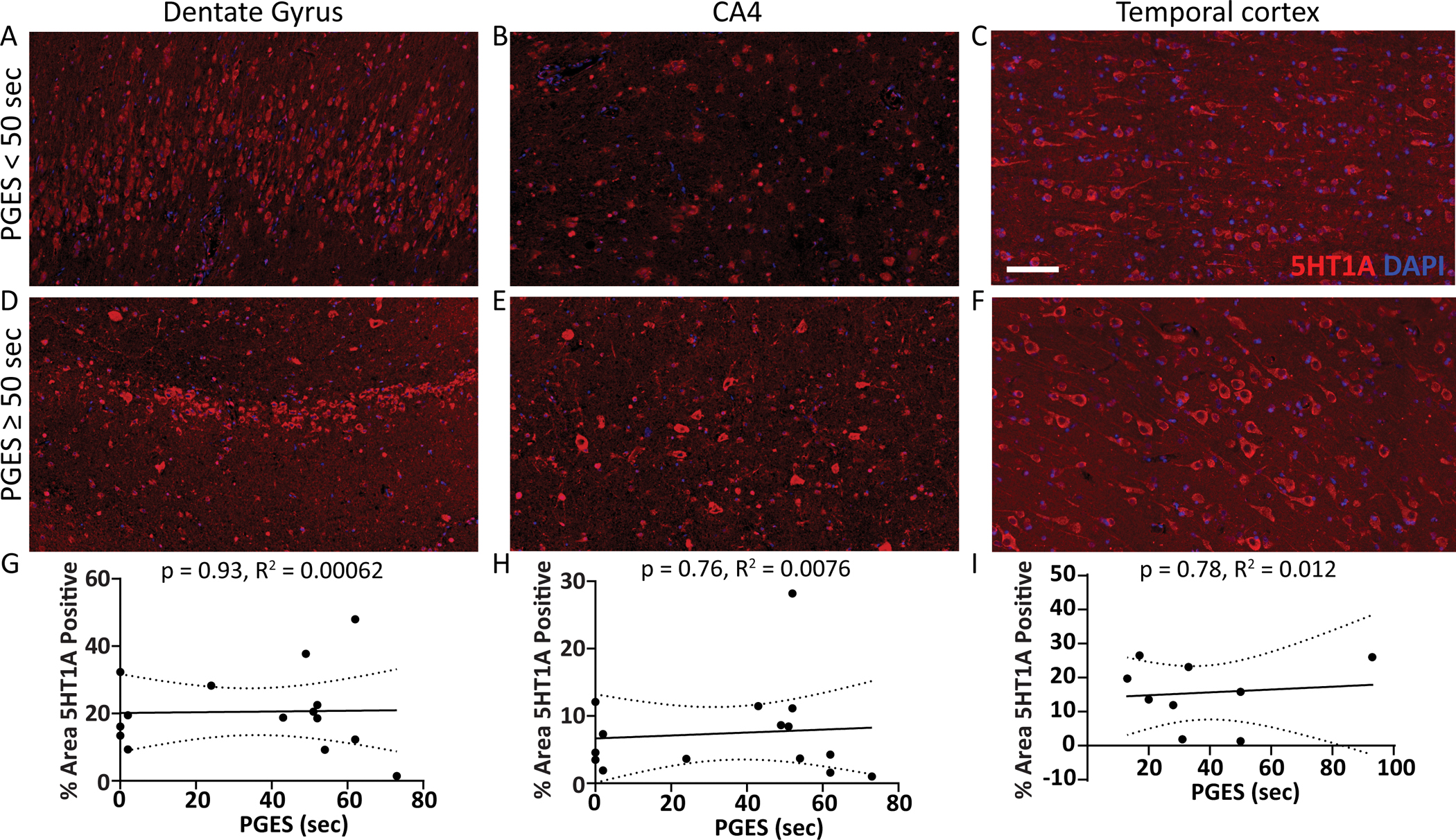
Representative images show 5HT1A expression (red) in surgical brain tissue from epilepsy cases in the hippocampal dentate gyrus, CA4 subsector, and temporal cortex A-C) with PGES < 50 seconds and D-F) PGES ≥ 50 seconds. G-I) Semiquantification of 5HT1A expression in subregions across the PGES duration spectrum indicates no correlation in the hippocampus (n = 16) or temporal cortex (n =9). Scale bar represents 100 μm for all histology panels.
Figure 3. 5HT2A protein expression in subregions of the hippocampus and temporal cortex by PGES duration.
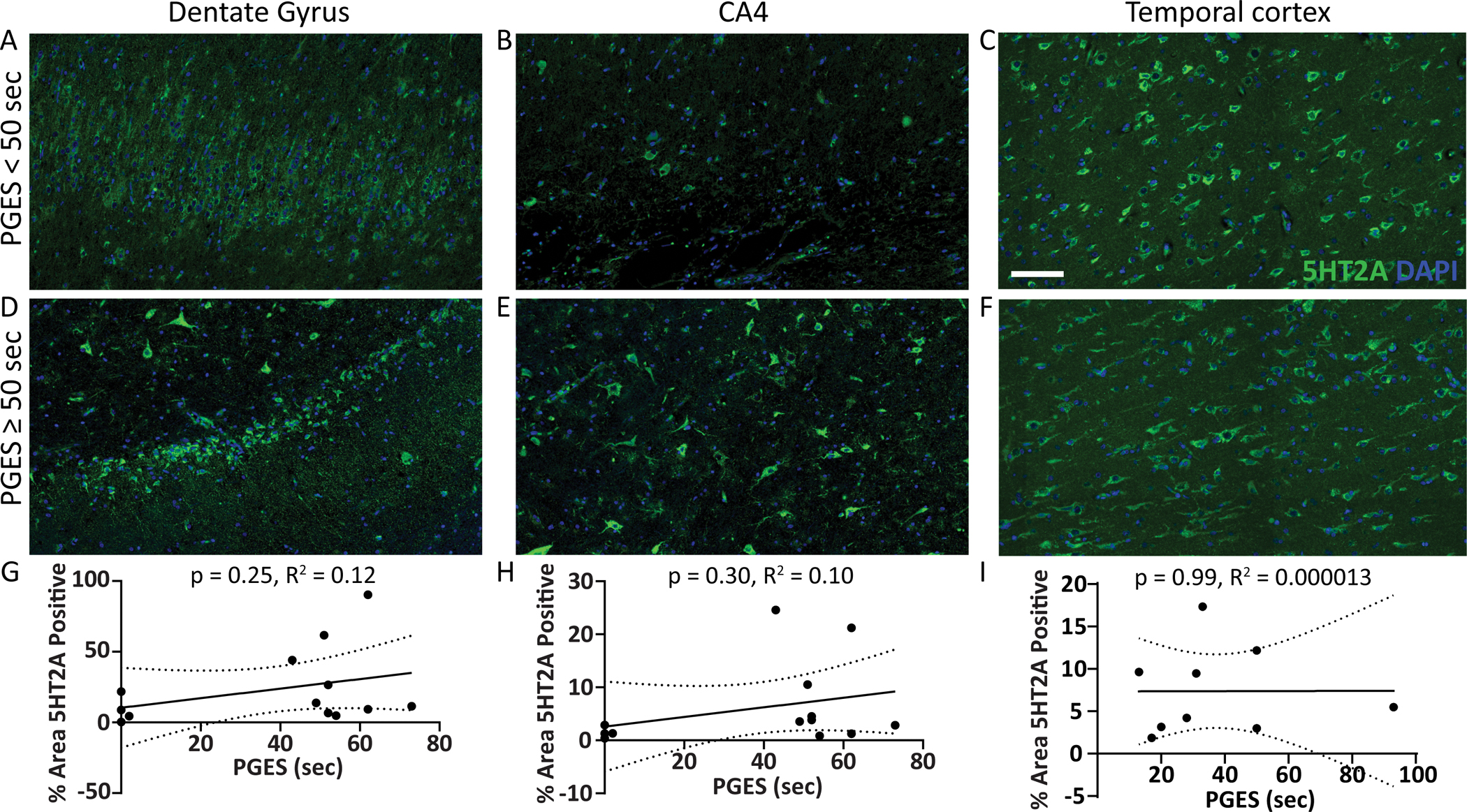
Representative images show 5HT2A expression (green) in surgical brain tissue from epilepsy cases in the hippocampal dentate gyrus, CA4 subsector, and temporal cortex A-C) with PGES < 50 seconds and D-F) PGES ≥ 50 seconds. G-I) Semiquantification of 5HT2A expression in subregions across the PGES duration spectrum indicates no correlation in the hippocampus (n = 16) or temporal cortex (n =9). Scale bar represents 100 μm for all histology panels.
RNAseq in Hippocampus
To determine whether 5HT1A and 5HT2A (encoded by HTR1A and HTR2A) as well as other serotonin receptors correlated to PGES duration, we analyzed our previous RNAseq dataset29,30 (n = 16), overlapping with 13 patients with hippocampal protein data (Table 1, Supplemental Figure 1A). There were 14 serotonin receptors detected in the hippocampus (Figure 4A), with an exploratory analysis identifying two receptors significantly correlated to PGES duration: HTR3B (also known as 5HT3B) with a negative correlation (p = 0.043, R2 = −0.26) and HTR4 (also known as 5HT4) with a positive correlation (p = 0.049, R2 = 0.25; Figure 4B–C).
Figure 4. Serotonin receptor transcript expression from RNAseq in the hippocampus by PGES duration.
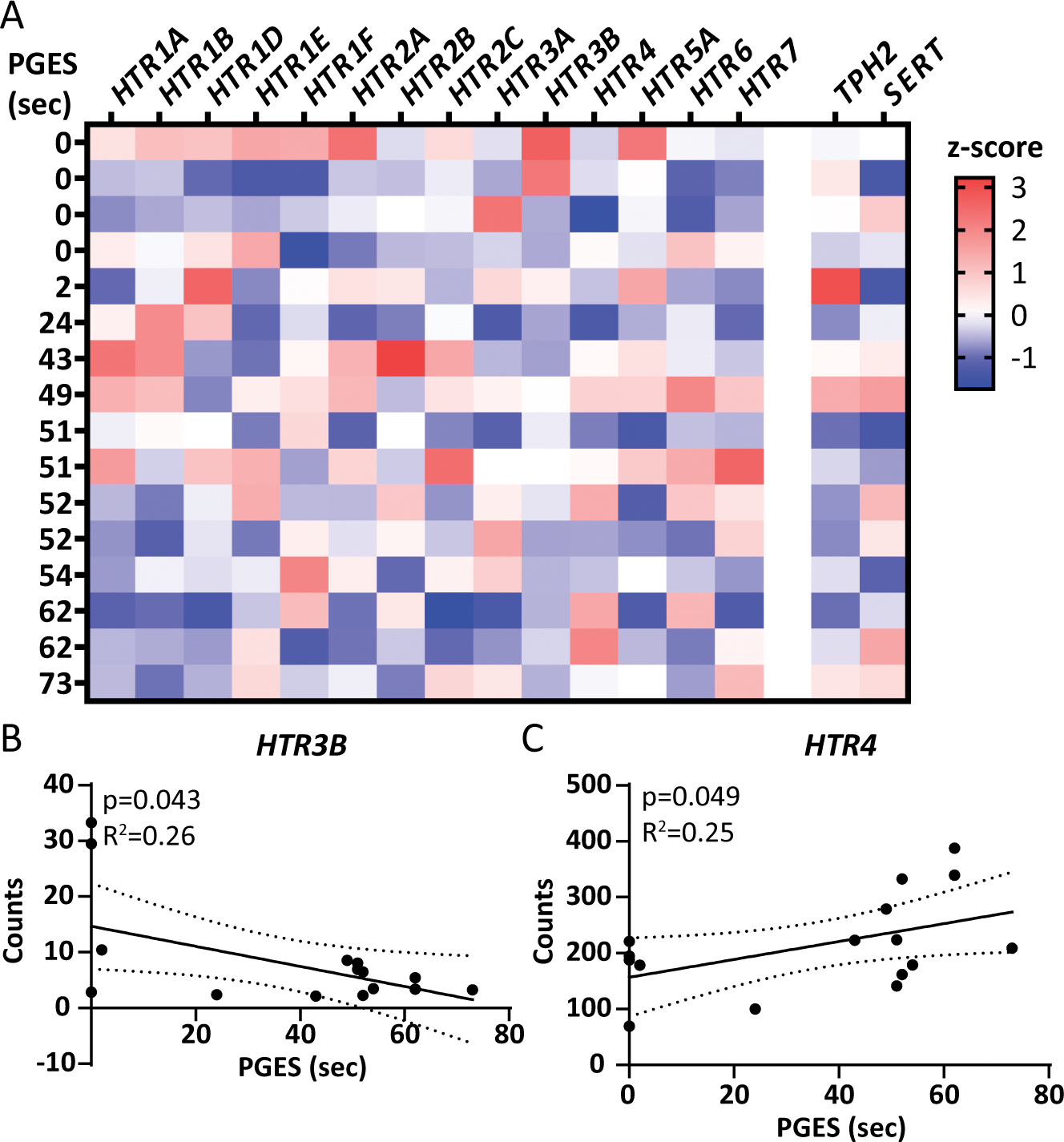
A) Expression of each of the serotonin receptors detected in the hippocampus (n = 16) and 2 related serotonergic transcripts, TPH2 the rate limiting enzyme in serotonin synthesis and SERT the serotonin transporter. PGES duration is indicated on the left, with increasing value from top to bottom. Z-score is indicated on the heatmap, with higher values represented in red and lower values in blue. B) Of the 14 serotonin receptors detected by RNAseq, 2 correlated to PGES duration. HTR3B had a negative correlation with PGES duration (p = 0.043, R2 = 0.26). C) HTR4 had a positive correlation with PGES duration (p = 0.049, R2 = 0.25).
To assess enriched serotonin signaling pathway transcripts associated with PGES duration, WGCNA was performed on our hippocampal dataset29,30 and revealed no enrichment in this brain region (Supplemental Tables 2–3).
WGCNA identified additional transcripts that correlated with PGES duration (Supplemental Figure 1). There were 2,597 of 42,753 transcripts that correlated with PGES (p < 0.05), distributed among all 27 modules (Supplemental Table 2). The top two transcripts negatively correlated: PPP1R17 (protein primarily expressed in cerebellum, p = 1.16 × 10−5, R2 = 0.76; M-white module; Supplemental Figure 1B) and DHRS7 (p = 1.48 × 10−5, R2 = 0.75; M-ivory module; Supplemental Figure 1C). Among all 27 modules, 4 significantly correlated to PGES (Supplemental Figure 1D). PGES duration positively correlated with synapse transcripts (p = 0.040, corr. = 0.52; included 7 potassium channels: KCNA4, KCNC4, KCNH1, KCNIP4, KCNJ3, KCNJ6, KCNK1; Supplemental Figure 2; Supplemental Table 3) and stimulus detection (taste receptors, p = 0.044, corr. = 0.51). PGES negatively correlated with cellular localization (p = 0.0084, corr. = −0.63) and developmental process transcripts (p = 0.020, corr. = −0.57).
Five modules significantly correlated to epilepsy onset, duration, age, and sex, regardless of PGES duration, and with no overlap of PGES correlated modules. Epilepsy onset positively correlated with metabolic processes related to ribosomal transcripts (p = 0.0075, corr. = 0.64), mitochondrial transcripts (p = 0.0016, corr. = 0.72), and localization related to neuronal projection (p = 0.022, corr. = 0.57). Epilepsy duration positively correlated with stimulus detection related to olfactory receptors (p = 0.010, corr. = 0.62). Age positively correlated with metabolic processes related to mitochondrial transcripts (p = 0.0026, corr. = 0.55). Sex (male gender) positively correlated to a module (p = 1.09 × 10−7, corr. = 0.94) that did not have a significant GO annotation. There were insufficient cases with temporal cortex samples to perform WGCNA.
Discussion
PGES duration positively correlated with 5HT2A receptor protein expression in hippocampal homogenate from TLE patients. 5HT1A receptor protein expression did not correlate to PGES duration in hippocampus or temporal lobe. On hippocampal RNAseq, PGES duration negatively correlated with HTR3B and positively correlated with HTR4, but was not correlated with the serotonin signaling pathway. By WGCNA, PGES duration positively correlated with synaptic transcripts (including potassium channels) and stimulus detection, and negatively correlated with cellular localization and developmental process transcripts.
Serotonin and serotonin receptors are relevant to SUDEP because they modulate seizure activity, arousal and respiration,9,10,14 and preclinical models link PGES to serotonergic signaling.26 Recent clinical studies indicate fenfluramine, active at multiple serotonin receptors, SERT, and potentially at other receptors, modulates seizure activity in Dravet syndrome patients, which is also seen in animal studies along with decreased seizure associated respiratory arrest.9 In animal models, serotonin receptors have opposing effects on seizure threshold, partly reflecting ligand specificity and dose, downstream pathways (e.g., 5HT2A couples with G-proteins and GPCRs), seizure etiology, and in cell type specific expression by brain region; e.g., excitatory/inhibitory neurons, astrocytes, oligodendrocytes.10,32 The 5HT2 receptors facilitate excitation,17 and 5HT2A antagonists reduce seizures in animal models.10,22 When activated, 5HT2A can inhibit calcium and sodium conductance, release hippocampal arachidonic acid, and influence neuronal morphology and plasticity.11 Activation of presynaptic autoreceptor and postsynaptic 5HT1A receptors can increase potassium and decrease calcium conductance, decreasing neurotransmitter release.11 Imaging studies identified decreased 5HT1A binding in TLE compared to controls.13,33 Among TLE patients, we did not observe 5HT1A changes in this brain region by PGES duration. Our prior proteomics studies comparing SUDEP to non-SUDEP epilepsy cases did not detect serotonin receptors in the frontal cortex, hippocampus, or brainstem nuclei, and thus differences could not be evaluated.25,29 Future studies should explore mechanistic implications of the association between increased hippocampal 5HT2A and PGES duration, particularly whether 5HT2A expression is increased in other hippocampal subregions like CA1, whether 5HT2A antagonism reduces PGES duration and whether seizure burden or prolonged PGES upregulate 5HT2A receptor protein expression as a compensatory mechanism or epiphenomenon.
At the RNA level, several transcripts were associated with PGES duration. HTR3B decreased with PGES duration. The 5HT3 receptor is a ligand-gated heteromeric ion channel, and activation results in fast depolarization.34 5HT3 receptor agonism increases seizure duration and antagonism decreases severity of convulsions and afterdischarge duration.14 HTR4 increased with PGES duration. When activated, 5HT4 inhibits potassium channel conductance that results in longer hyperexcitability, and is implicated in cell survival and spine growth.11 In epilepsy animal models, 5HT4 brainstem protein expression was decreased,35 5HT4 deletion promoted seizures and increased mortality,18 and 5HT4 agonism decreased seizure-induced respiratory arrest and tonic seizures.36 DHRS7 (dehydrogenase/reductase 7) was a lead transcript correlating with PGES duration. Related proteins metabolize prostaglandins, lipids, and steroids37,38 and interact with cannabinoid receptor 2 (CB2).39 It is unclear the effect that these transcript expression levels may have on PGES and SUDEP risk, whether they are altered as a result of prolonged PGES or contribute to PGES duration, and thus should be investigated further.
Synaptic transcripts, including potassium channels, were positively correlated with PGES duration (4 voltage-gated, 2 inward rectifiers, 1 two pore domain). Voltage-gated channels facilitate repolarization and modify duration and delay of action potentials, inward rectifiers maintain resting membrane potential, and two pore domain channels contribute to leak current important for resting membrane potential.40,41 Potassium channels are not implicated in PGES, but potassium conductance, expression, and gain and loss-of-function mutations can cause epilepsy and may increase SUDEP risk.40,42,43 A comparison of the same TLE cases in this study to controls in another of our studies did not identify an alteration to these 7 potassium channels in hippocampus by RNAseq.30 Our SUDEP proteomics studies25,29 did not detect the same potassium channels in the current study. Five different potassium channels were detected but were similar in frontal cortex and hippocampus,29 and three potassium channels in brainstem nuclei were detected and not altered between SUDEP and non-SUDEP epilepsy.25 Follow up studies should investigate whether the altered potassium channel RNA corresponds to functional changes, whether excitatory or inhibitory neurons are impacted differently, and evaluate additional brain regions like the brainstem. Increased voltage-gated potassium channels may be a compensatory response to hyperexcitability or shift voltage activation to more negative potentials contributing to hyperexcitability, similar to gain-of-function KCNH143 mutations. Alternatively, they may reflect an overcompensation coupled with dysregulation that suppresses brain activity in PGES.
There were several other correlations of WGCNA identified modules to clinical history. PGES duration negatively correlated with cellular localization and developmental process transcripts, and positively correlated with stimulus detection related to taste receptors. Neuronal migration defects occur in epilepsy44 and taste receptors45 are expressed in extra-oral regions and altered in some disease states. Four other modules with GO annotations were associated with clinical history. Localization related to neuronal projection and metabolic processes positively correlated with epilepsy onset, indicating these transcripts were more elevated at a later age of epilepsy onset than in patients with an earlier onset. Epilepsy onset age is positively related to age at surgery, in which a positive correlation was similarly seen for mitochondrial transcripts. Further, epilepsy duration positively correlated with stimulus detection related to olfactory receptors, indicating these transcripts were more elevated in patients with prolonged epilepsy duration than those with a shorter duration. Hippocampal olfactory receptors have non-sensory functions, ligands include endogenous molecules, and expression is abnormally regulated in neurodegeneration.46 The meaning of these clinical history associations requires further investigation at the protein and functional levels.
Our study had several limitations. PGES duration can vary in the same patient during different seizures and the number of convulsive seizures captured on video-EEGs was limited.1,5,47 5HT2A and 5HT1A protein levels may vary between western blot and histology detection techniques due to subregional differences and differences in detection method based on solubility and other factors as we and others have reported (time to tissue processing/freezing).48,49 RNAseq did not correlate HTR1A or HTR2A to PGES. Protein changes may not correspond to transcript changes,31 although overall pathway analysis may be more comparable. Hippocampal sclerosis was present in most cases for protein analyses (n=13/16, predominantly ILAE type 1), however neuronal loss was not associated with decreased serotonin receptors. Patients were not evaluated for pathogenic gene variants. SUDEP cases were not evaluated in the current study, but future studies will be of interest in SUDEP cases with PGES information available to further investigate the association of serotonin receptor and potassium channel expression related to SUDEP risk.
In summary, higher 5HT2A receptor protein by western blot and potassium channel transcript expression in the hippocampus of TLE patients was associated with prolonged PGES. Future studies should investigate serotonin receptor (5HT1A, 5HT2A, 5HT3B, 5HT4) and potassium channel protein expression in patients with PGES and in SUDEP cases, particularly in implicated brain regions like the hippocampal subregions and brainstem, as well as the mechanistic implications of these expression changes.
Supplementary Material
Key Points:
The 5HT2A receptor protein positively correlated with PGES duration in the hippocampus of TLE patients by western blot.
The transcripts HTR3B negatively correlated and HTR4 positively correlated with PGES duration in hippocampus.
Potassium channel transcripts positively correlated with PGES duration in hippocampus.
Acknowledgements:
This research was supported by funding from the National Institutes of Health (NIH) National Institute of Neurological Disorders and Stroke (NINDS) UO1 NS090415 05 Center for SUDEP Research: The Neuropathology of SUDEP, Finding A Cure for Epilepsy and Seizures (FACES), NIH National Institute of Aging (NIA) P30AG066512, European Union’s Seventh Framework Program (FP7/2007–2013) under grant agreement 602102 (EPITARGET), and the Top Sector Life Sciences & Health via a PPP Allowance made available to the Dutch Epilepsy Foundation to stimulate public-private partnerships, and European Union Horizon 2020 WIDESPREAD-05–2020-Twinning, EpiEpiNet under grant agreement 952455.
Footnotes
Disclosure of Conflicts of Interest
The authors declare no conflicts of interest.
Daniel Friedman receives salary support for consulting and clinical trial related activities performed on behalf of The Epilepsy Study Consortium, a non-profit organization. Dr. Friedman receives no personal income for these activities. NYU receives a fixed amount from the Epilepsy Study Consortium towards Dr. Friedman’s salary. Within the past two years, The Epilepsy Study Consortium received payments for research services performed by Dr. Friedman from: Alterity, Baergic, Biogen, BioXcell, Cerevel, Cerebral, Jannsen, Lundbeck, Neurocrine, SK Life Science, and Xenon. He has also served as a paid consultant for Neurelis Pharmaceuticals and Receptor Life Sciences. He has received travel support from the Epilepsy Foundation. He has received research support from NINDS, CDC, Epitel, and Neuropace unrelated to this study. He holds equity interests in Neuroview Technology. He received royalty income from Oxford University Press. Shobi Sivathamboo is supported by a Bridging Postdoctoral Fellowship from Monash University (BPF20–3253672466) and the Victorian Medical Research Acceleration Fund. She reports salary support from Kaoskey and Optalert for clinical trial related activities; she receives no personal income for these activities. Terence J. O’Brien is supported by a Program Grant (APP1091593) and Investigator Grant (APP1176426) from the National Health and Medical Research Council of Australia and the Victorian Medical Research Acceleration Fund. He reports grants and consulting fees paid to his institution from Eisai, UCB Pharma, Praxis, Biogen, ES Theraputics and Zynerba.
Ethical Publication Statement
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Data Availability
The data that support the findings of this study are available in the public repositories listed in the methods, supplemental files, and from the authors upon reasonable request.
References
- 1.Kang JY, Rabiei AH, Myint L, Nei M. Equivocal significance of post-ictal generalized EEG suppression as a marker of SUDEP risk. Seizure. May 2017;48:28–32. doi: 10.1016/j.seizure.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 2.Ryvlin P, Nashef L, Lhatoo SD, et al. Incidence and mechanisms of cardiorespiratory arrests in epilepsy monitoring units (MORTEMUS): a retrospective study. Lancet Neurol. Oct 2013;12(10):966–77. doi: 10.1016/S1474-4422(13)70214-X [DOI] [PubMed] [Google Scholar]
- 3.Lhatoo SD, Faulkner HJ, Dembny K, Trippick K, Johnson C, Bird JM. An electroclinical case-control study of sudden unexpected death in epilepsy. Ann Neurol. Dec 2010;68(6):787–96. doi: 10.1002/ana.22101 [DOI] [PubMed] [Google Scholar]
- 4.Surges R, Strzelczyk A, Scott CA, Walker MC, Sander JW. Postictal generalized electroencephalographic suppression is associated with generalized seizures. Epilepsy Behav. Jul 2011;21(3):271–4. doi: 10.1016/j.yebeh.2011.04.008 [DOI] [PubMed] [Google Scholar]
- 5.Lamberts RJ, Gaitatzis A, Sander JW, Elger CE, Surges R, Thijs RD. Postictal generalized EEG suppression: an inconsistent finding in people with multiple seizures. Neurology. Oct 2013;81(14):1252–6. doi: 10.1212/WNL.0b013e3182a6cbeb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Zhao H, Zeng C, et al. Optogenetic activation of 5-HT neurons in the dorsal raphe suppresses seizure-induced respiratory arrest and produces anticonvulsant effect in the DBA/1 mouse SUDEP model. Neurobiol Dis. Feb 2018;110:47–58. doi: 10.1016/j.nbd.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patodia S, Somani A, Thom M. Review: Neuropathology findings in autonomic brain regions in SUDEP and future research directions. Auton Neurosci. 11 2021;235:102862. doi: 10.1016/j.autneu.2021.102862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrucci AN, Joyal KG, Purnell BS, Buchanan GF. Serotonin and sudden unexpected death in epilepsy. Exp Neurol. 03 2020;325:113145. doi: 10.1016/j.expneurol.2019.113145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilliam FG, Hecimovic H, Gentry MS. Serotonergic therapy in epilepsy. Curr Opin Neurol. 04 01 2021;34(2):206–212. doi: 10.1097/WCO.0000000000000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guiard BP, Di Giovanni G. Central serotonin-2A (5-HT2A) receptor dysfunction in depression and epilepsy: the missing link? Front Pharmacol. 2015;6:46. doi: 10.3389/fphar.2015.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masson JaEMBaHMaDM. Serotonergic signaling: multiple effectors and pleiotropic effects. Wiley Interdisciplinary Reviews: Membrane Transport and Signaling. 2012;1(6):685–713. doi: 10.1002/wmts.50 [DOI] [Google Scholar]
- 12.Theodore WH, Martinez AR, Khan OI, et al. PET of serotonin 1A receptors and cerebral glucose metabolism for temporal lobectomy. J Nucl Med. Sep 2012;53(9):1375–82. doi: 10.2967/jnumed.112.103093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovacchini G, Toczek MT, Bonwetsch R, et al. 5-HT 1A receptors are reduced in temporal lobe epilepsy after partial-volume correction. J Nucl Med. Jul 2005;46(7):1128–35. [PMC free article] [PubMed] [Google Scholar]
- 14.Bagdy G, Kecskemeti V, Riba P, Jakus R. Serotonin and epilepsy. J Neurochem. Feb 2007;100(4):857–73. doi: 10.1111/j.1471-4159.2006.04277.x [DOI] [PubMed] [Google Scholar]
- 15.Faingold CL. Neuronal networks in the genetically epilepsy-prone rat. Adv Neurol. 1999;79:311–21. [PubMed] [Google Scholar]
- 16.Audero E, Coppi E, Mlinar B, et al. Sporadic autonomic dysregulation and death associated with excessive serotonin autoinhibition. Science. Jul 04 2008;321(5885):130–3. doi: 10.1126/science.1157871 [DOI] [PubMed] [Google Scholar]
- 17.Crunelli V, Venzi M, De Deurwaerdère P, Di Giovanni G. Role of Serotonin2A (5-HT2A) Receptors in Epilepsy. 2018:375–394. [Google Scholar]
- 18.Compan V, Zhou M, Grailhe R, et al. Attenuated response to stress and novelty and hypersensitivity to seizures in 5-HT4 receptor knock-out mice. J Neurosci. Jan 14 2004;24(2):412–9. doi: 10.1523/JNEUROSCI.2806-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witkin JM, Baez M, Yu J, Barton ME, Shannon HE. Constitutive deletion of the serotonin-7 (5-HT(7)) receptor decreases electrical and chemical seizure thresholds. Epilepsy Res. Jun 2007;75(1):39–45. doi: 10.1016/j.eplepsyres.2007.03.017 [DOI] [PubMed] [Google Scholar]
- 20.Massey CA, Thompson SJ, Ostrom RW, et al. X-linked serotonin 2C receptor is associated with a non-canonical pathway for sudden unexpected death in epilepsy. Brain Commun. 2021;3(3):fcab149. doi: 10.1093/braincomms/fcab149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patodia S, Somani A, Liu J, et al. Serotonin transporter in the temporal lobe, hippocampus and amygdala in SUDEP. Brain Pathol. Apr 27 2022:e13074. doi: 10.1111/bpa.13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Oekelen D, Megens A, Meert T, Luyten WH, Leysen JE. Functional study of rat 5-HT2A receptors using antisense oligonucleotides. J Neurochem. Jun 2003;85(5):1087–100. doi: 10.1046/j.1471-4159.2003.01738.x [DOI] [PubMed] [Google Scholar]
- 23.Hornung J-P. Chapter 11 - Raphe Nuclei. In: Mai JK, Paxinos G, eds. The Human Nervous System (Third Edition). Academic Press; 2012:401–424. [Google Scholar]
- 24.Luchetti A, Bota A, Weitemier A, et al. Two Functionally Distinct Serotonergic Projections into Hippocampus. J Neurosci. 06 17 2020;40(25):4936–4944. doi: 10.1523/JNEUROSCI.2724-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leitner DF, Kanshin E, Askenazi M, et al. Raphe and ventrolateral medulla proteomics in epilepsy and sudden unexpected death in epilepsy. Brain Commun. 2022;4(4):fcac186. doi: 10.1093/braincomms/fcac186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrucci AN, Joyal KG, Chou JW, Li R, Vencer KM, Buchanan GF. Post-ictal Generalized EEG Suppression is reduced by Enhancing Dorsal Raphe Serotonergic Neurotransmission. Neuroscience. 01 15 2021;453:206–221. doi: 10.1016/j.neuroscience.2020.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murugesan A, Rani MRS, Hampson J, et al. Serum serotonin levels in patients with epileptic seizures. Epilepsia. 06 2018;59(6):e91–e97. doi: 10.1111/epi.14198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theeranaew W, McDonald J, Zonjy B, et al. Automated Detection of Postictal Generalized EEG Suppression. IEEE Trans Biomed Eng. 02 2018;65(2):371–377. doi: 10.1109/TBME.2017.2771468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitner DF, Mills JD, Pires G, et al. Proteomics and Transcriptomics of the Hippocampus and Cortex in SUDEP and High-Risk SUDEP Patients. Neurology. Apr 2021;doi: 10.1212/WNL.0000000000011999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills JD, van Vliet EA, Chen BJ, et al. Coding and non-coding transcriptome of mesial temporal lobe epilepsy: Critical role of small non-coding RNAs. Neurobiol Dis. Feb 2020;134:104612. doi: 10.1016/j.nbd.2019.104612 [DOI] [PubMed] [Google Scholar]
- 31.Seyfried NT, Dammer EB, Swarup V, et al. A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst. 01 2017;4(1):60–72.e4. doi: 10.1016/j.cels.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu T, Pandey SC. Cellular localization of serotonin(2A) (5HT(2A)) receptors in the rat brain. Brain Res Bull. Apr 2000;51(6):499–505. doi: 10.1016/s0361-9230(99)00278-6 [DOI] [PubMed] [Google Scholar]
- 33.Martinez A, Finegersh A, Cannon DM, et al. The 5-HT1A receptor and 5-HT transporter in temporal lobe epilepsy. Neurology. Apr 16 2013;80(16):1465–71. doi: 10.1212/WNL.0b013e31828cf809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes NM, Hales TG, Lummis SC, Peters JA. The 5-HT3 receptor--the relationship between structure and function. Neuropharmacology. Jan 2009;56(1):273–84. doi: 10.1016/j.neuropharm.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uteshev VV, Tupal S, Mhaskar Y, Faingold CL. Abnormal serotonin receptor expression in DBA/2 mice associated with susceptibility to sudden death due to respiratory arrest. Epilepsy Res. Feb 2010;88(2–3):183–8. doi: 10.1016/j.eplepsyres.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 36.Tupal S, Faingold CL. Serotonin 5-HT. Epilepsy Res. Nov 2021;177:106777. doi: 10.1016/j.eplepsyres.2021.106777 [DOI] [PubMed] [Google Scholar]
- 37.Štambergová H, Zemanová L, Lundová T, et al. Human DHRS7, promising enzyme in metabolism of steroids and retinoids? J Steroid Biochem Mol Biol. Jan 2016;155(Pt A):112–9. doi: 10.1016/j.jsbmb.2015.09.041 [DOI] [PubMed] [Google Scholar]
- 38.Araya S, Kratschmar DV, Tsachaki M, Stücheli S, Beck KR, Odermatt A. DHRS7 (SDR34C1) - A new player in the regulation of androgen receptor function by inactivation of 5α-dihydrotestosterone? J Steroid Biochem Mol Biol. 07 2017;171:288–295. doi: 10.1016/j.jsbmb.2017.04.013 [DOI] [PubMed] [Google Scholar]
- 39.Sharaf A, Mensching L, Keller C, et al. Systematic Affinity Purification Coupled to Mass Spectrometry Identified p62 as Part of the Cannabinoid Receptor CB2 Interactome. Front Mol Neurosci. 2019;12:224. doi: 10.3389/fnmol.2019.00224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köhling R, Wolfart J. Potassium Channels in Epilepsy. Cold Spring Harb Perspect Med. 05 02 2016;6(5)doi: 10.1101/cshperspect.a022871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kshatri AS, Gonzalez-Hernandez A, Giraldez T. Physiological Roles and Therapeutic Potential of Ca. Front Mol Neurosci. 2018;11:258. doi: 10.3389/fnmol.2018.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Lin ZJ, Liu L, et al. Epilepsy-associated genes. Seizure. Jan 2017;44:11–20. doi: 10.1016/j.seizure.2016.11.030 [DOI] [PubMed] [Google Scholar]
- 43.Niday Z, Tzingounis AV. Potassium Channel Gain of Function in Epilepsy: An Unresolved Paradox. Neuroscientist. 08 2018;24(4):368–380. doi: 10.1177/1073858418763752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stouffer MA, Golden JA, Francis F. Neuronal migration disorders: Focus on the cytoskeleton and epilepsy. Neurobiol Dis. 08 2016;92(Pt A):18–45. doi: 10.1016/j.nbd.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuzim K, Korolczuk A. An update on extra-oral bitter taste receptors. J Transl Med. 10 21 2021;19(1):440. doi: 10.1186/s12967-021-03067-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrer I, Garcia-Esparcia P, Carmona M, et al. Olfactory Receptors in Non-Chemosensory Organs: The Nervous System in Health and Disease. Front Aging Neurosci. 2016;8:163. doi: 10.3389/fnagi.2016.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajakulendran S, Nashef L. Postictal generalized EEG suppression and SUDEP: a review. J Clin Neurophysiol. Feb 2015;32(1):14–20. doi: 10.1097/WNP.0000000000000147 [DOI] [PubMed] [Google Scholar]
- 48.Cloughesy TF, Yoshimoto K, Nghiemphu P, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. Jan 22 2008;5(1):e8. doi: 10.1371/journal.pmed.0050008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leitner DF, Kanshin E, Askenazi M, et al. Pilot study evaluating everolimus molecular mechanisms in tuberous sclerosis complex and focal cortical dysplasia. PLoS One. 2022;17(5):e0268597. doi: 10.1371/journal.pone.0268597 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the public repositories listed in the methods, supplemental files, and from the authors upon reasonable request.


