Abstract
Background:
Brain cancer incidence and mortality rates are greater in males. Understanding the molecular mechanisms that underlie those sex differences could improve treatment strategies. Although sex differences in normal metabolism are well described, it is currently unknown whether they persist in cancerous tissue.
Methods:
Using PET imaging and mass spectrometry, we assessed sex differences in glioma metabolism in patient samples. Furthermore, we assessed the role of glutamine metabolism in male and female murine transformed astrocytes using isotope labeling, metabolic rescue experiments, and pharmacological and genetic perturbations to modulate pathway activity.
Findings:
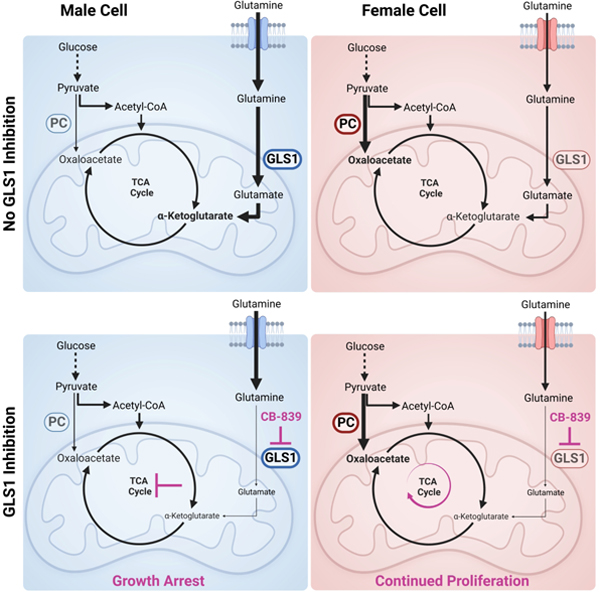
We found that male glioblastoma surgical specimens are enriched for amino acid metabolites, including glutamine. Fluoroglutamine PET imaging analyses showed that gliomas in male patients exhibit significantly higher glutamine uptake. These sex differences were well-modeled in murine transformed astrocytes, in which male cells imported and metabolized more glutamine and were more sensitive to glutaminase 1 (GLS1) inhibition. The sensitivity to GLS1 inhibition in males was driven by their dependence on glutamine-derived glutamate for α-ketoglutarate synthesis and TCA cycle replenishment. Females were resistant to GLS1 inhibition through greater pyruvate carboxylase (PC)-mediated TCA cycle replenishment, and knockdown of PC sensitized females to GLS1 inhibition.
Conclusion:
Our results show that clinically important sex differences exist in targetable elements of metabolism. Recognition of sex-biased metabolism may improve treatments for all patients through further laboratory and clinical research.
Graphical Abstract
eTOC blurb:
Sponagel et al. demonstrate that metabolite abundance differs in male and female brain tumors and that male gliomas have greater glutamine uptake. Furthermore, they show that targeting glutaminase 1 attenuates growth in male cells only, as growth is driven by their dependence on glutamine-derived glutamate for TCA cycle replenishment.
Introduction:
Cancerous growth requires extensive adaptations at the cellular, tissue, and systemic levels. Some adaptations create not only a growth advantage but also vulnerabilities that can be targeted. Targetable cancer cell adaptations include dependency on certain nutrients (e.g., glucose and glutamine), ATP production, reactive oxygen species (ROS) management, and macromolecule synthesis. These adaptations are collectively referred to as metabolic reprogramming1.
Metabolic reprogramming affects carbohydrates, amino acids, fatty acids, and mitochondrial function. Importantly, each of these elements of metabolism exhibit sex differences at the organismal, tissue, and cellular level from the moment of fertilization throughout adulthood and across many species2. Furthermore, sex-specific metabolic phenotypes have been linked to sex differences in diabetes and cardiovascular disease3,4 as well as cancer. In renal cell carcinoma, colon cancer, and low-grade glioma, certain metabolic phenotypes correlate with poor survival in one sex but not the other5–8, suggesting that metabolic reprogramming and its effects on survival can differ in male and female cancers. This may be especially important as sex differences in cancer incidence and mortality rate are evident throughout the world, across all ages, and in many different cancer types9–12. To date, the molecular mechanisms underlying sex-specific metabolic phenotypes in cancer and how these differences might affect metabolic treatment approaches in male and female patients remain unknown.
Here, we show that male and female glioblastoma (GBM) surgical specimens differ in their metabolic profile and that glutamine uptake is greater in male patient glioma and male murine transformed astrocytes. Furthermore, inhibition of glutaminase 1 (GLS1) is more effective against male transformed astrocytes and GLS1 inhibition affects the TCA cycle and glutathione in a sex-specific manner. Moreover, while male transformed astrocytes are more dependent on glutathione to maintain their redox balance, the underlying reason for their sensitivity to GLS1 inhibition is decreased TCA cycle influx from glutamine-derived glutamate. Lastly, we show that resistance to GLS1 inhibition in female cells results from pyruvate carboxylase (PC)-mediated carbon flux from glucose into the TCA cycle. Understanding how metabolism differs in male and female GBM will not only reveal novel elements of GBM biology but will also help stratify patients for metabolic treatment approaches and improve survival for both men and women with GBM and other cancers.
Results:
Sex differences in GBM metabolite abundance parallel those found in serum
Physiologic sex differences in metabolism are well described. A large-scale metabolomics analysis of over 500 metabolites in the blood serum of 1756 participants found that 1/3 of all metabolites showed significant sex differences13. Furthermore, amino acid and carbohydrate pathways were most significantly enriched in males. To determine whether such sex differences persist in cancerous tissue, we assessed metabolite abundance in a previously published dataset of 757 metabolites in 44 male and 32 female newly diagnosed GBM surgical specimens14. We removed metabolites that were constant across all samples, log transformed individual metabolite data, and derived a z-score for each metabolite (Fig. S1A). In a pathway-based approach, we averaged the z-scores across metabolites that belonged to the same pathway to represent metabolic pathway activity. First, we assessed the “super-pathways,” including lipids, carbohydrates, amino acids, xenobiotics, nucleotides, energy, peptides, and cofactors and vitamins. While none of these were significantly enriched in males versus females at the 5% false discovery rate (FDR) level (Table S1), we found that all super-pathways were male-biased (Fig. 1A). Similar to the published serum metabolite data, the amino acid and carbohydrate super-pathways were most strongly enriched in males (FDR adjusted p-value q=0.11). Each super-pathway was then subdivided into two or more “sub-pathways.” While no amino acid or carbohydrate sub-pathways were significantly enriched in males versus females at the 5% FDR level (Table S1), we found that similar to the super-pathways, most sub-pathways were enriched in male GBM (Fig. 1B). Notably, the lipid super-pathway was the only pathway that exhibited a female bias in multiple sub-pathways, again paralleling the findings in human serum. Although underpowered to detect significant sex differences in pathway enrichment, these data suggest that sex differences in metabolite abundance in GBM tissue and healthy serum are similar.
Figure 1. Sex differences in GBM metabolite abundance parallel those found in serum.
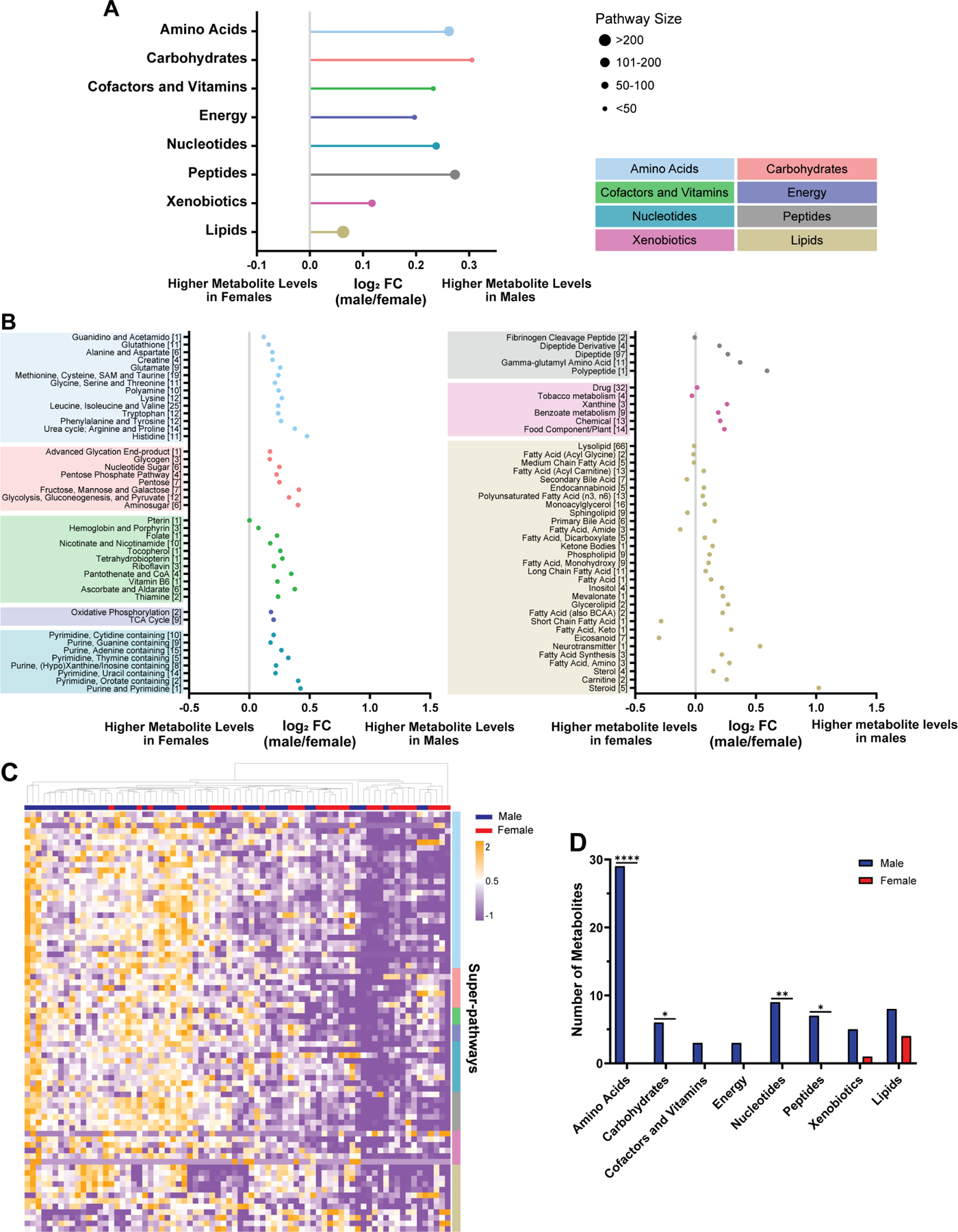
A, Log2 fold change (log2 FC, indicating male/female mean difference) of metabolic super-pathways in male (n=44) and female (n=32) GBM surgical specimens. Log2 FC>0 and log2 FC<0 correspond to super-pathway enrichment in male and female GBM, respectively. Pathways are organized based on Storey’s FDR adjusted q-values, with the smallest q-value at the top. The size of the dots represents the number of metabolites in each super-pathway. B, Log2 FC of metabolic sub-pathways in GBM surgical specimens. Log2 FC>0 and log2 FC<0 correspond to sub-pathway enrichment in male and female GBM, respectively. Sub-pathways are organized based on Storey’s FDR adjusted q-values, with the smallest q-value at the bottom of each super-pathway. The numbers in brackets refer to the number of metabolites in each sub-pathway. C, Hierarchical clustering of the top 75 metabolites (determined by greatest mean difference) of male and female GBM surgical specimens. Patient specimens and metabolites are represented in columns and rows, respectively. Rows were sorted based on super-pathway classification (indicated by colors on the right). D, Distribution of the top 75 metabolites with differential abundance between male and female GBM surgical specimens into super-pathways. *p<0.05, **p<0.01, ****p<0.0001; two-tailed Fisher’s exact test. See also Figure S1 and Table S1.
Next, we assessed sex differences in individual metabolite abundance. We selected 75 metabolites with the highest mean difference between male and female GBM surgical specimens (Table S1) and performed unsupervised hierarchical clustering (Fig. 1C). Most patient specimens with high metabolite abundance were males. To further evaluate this observation, we performed unbiased k-means clustering (n=2)8 on the same data (Fig. S1B). Cluster 1 included the low, and cluster 2 the high metabolite abundance groups. We found that females and males were associated with clusters 1 and 2, respectively (p=0.0018; Fisher’s exact test) (Fig. S1C), indicating that differences in metabolite abundance allow for sex-specific clustering of GBM. Lastly, we assigned each of the 75 metabolites to a super-pathway and assessed whether metabolites of certain super-pathways were enriched in males or females. Most metabolites belonged to the amino acid super-pathway and were significantly enriched in males (p<0.0001) (Fig. 1D). Similar to the sub-pathway analysis, most of the metabolites enriched in females belonged to the lipids super-pathway. Together, these data indicate that male and female GBM differ in their metabolic profile. Super-pathway and individual metabolite analyses identified amino acids as key metabolites enriched in males.
Males exhibit greater glutamine uptake in glioma and brain tissue
The clinical metabolite data identified amino acids as a super-pathway enriched in male GBM. Interestingly, the metabolite with the greatest mean difference (i.e., male-biased) was pyroglutamine (Table S1), a cyclic derivative of glutamine. Glutamine was also enriched in males, suggesting that glutamine metabolism might differ between male and female gliomas. This has clinical importance, as inhibition of glutamine metabolism is being evaluated in cancer clinical trials (NCT02071927, NCT04250545, NCT03528642, NCT03965845). To further investigate sex differences in glioma glutamine utilization, we analyzed published and unpublished [18F]fluoroglutamine ([18F]FGln) positron emission tomography (PET) data15–17 from a heterogeneous group of male and female glioma patients separately (Table S2). PET imaging showed marked [18F]FGln uptake that was associated with the known brain tumor lesion on magnetic resonance imaging (MRI) in both male and female patients (Fig. 2A). To quantify sex differences in [18F]FGln uptake, we measured maximum standardized uptake value normalized to lean body mass (SULmax), a method for normalizing tracer uptake in tissues to sex-specific differences in body composition18. Concordant with our metabolomic profiling, male gliomas exhibited significantly higher [18F]FGln uptake than female gliomas (Fig. 2B).
Figure 2. Males exhibit greater glutamine uptake in glioma and brain tissue.
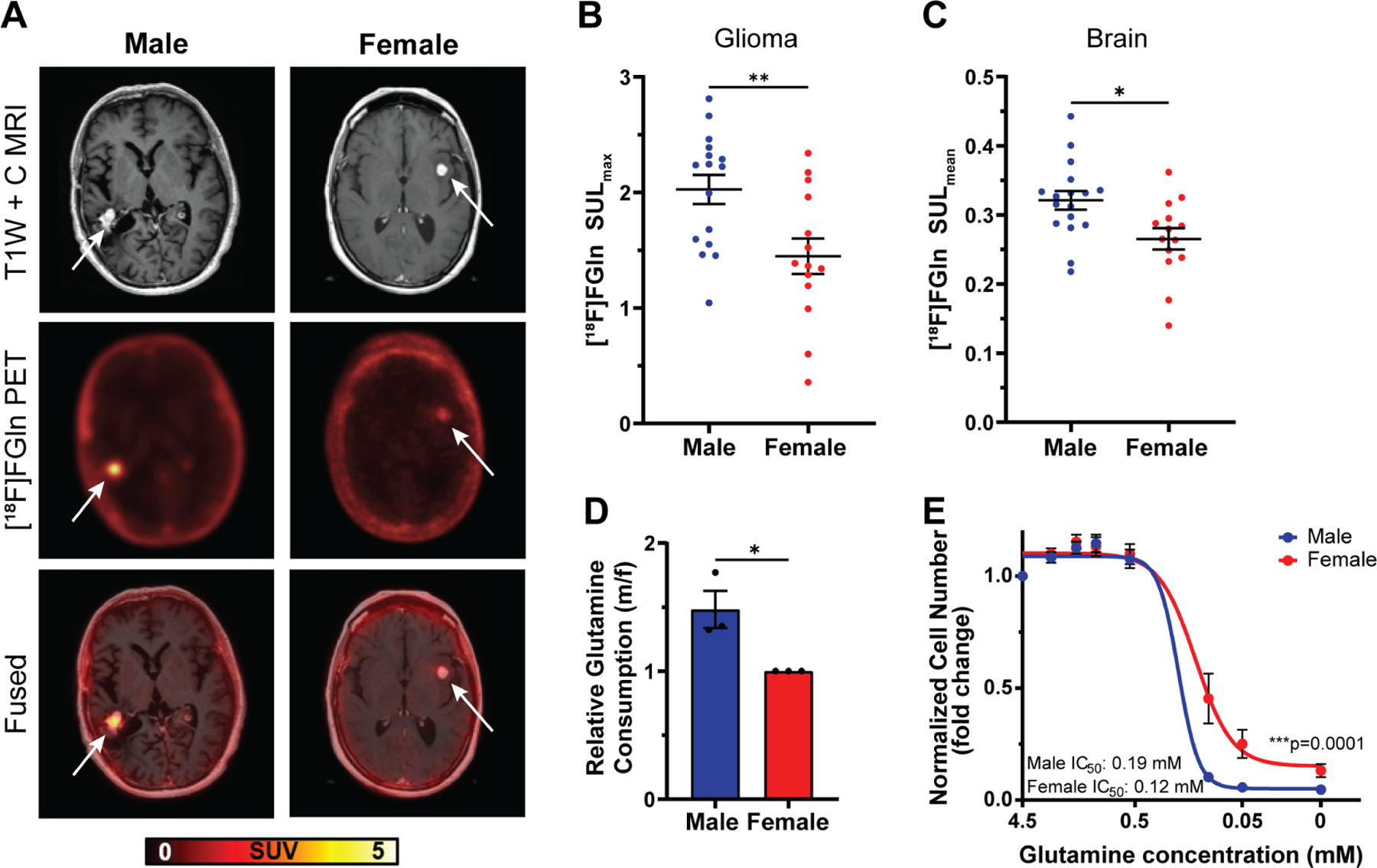
A, Representative T1W contrast MRI image, [18F]fluoroglutamine ([18F]FGln) PET image, and fused image of a male and female high-grade IDH wild-type glioma patient. Representative images were chosen based on tumor location, MRI contrast enhancement, and tumor size. Tumor location is indicated by a white arrow. B-C, Quantification of [18F]FGln uptake in human gliomas (B) and normal grey matter (C). D, Relative glutamine consumption in transformed astrocytes. E, Glutamine dose-response curves of transformed astrocytes. Data are mean +/− SEM (B-C) of n=15 male and n=13 female patients. *p<0.05, **p<0.01, t-test. Data are mean +/− SEM of n=3/sex (three independent experiments, one male and female cell line). *p<0.05; linear mixed effects model (D). Data are mean +/− SEM of n=9/sex (three independent experiments, three separate male and female cell lines). ***p=0.0001; two-way ANOVA (E). See also Figure S2 and Table S2.
Sex differences in glutamine uptake persisted even when the analysis was restricted to IDH wild-type (WT) glioma patients (Fig. S2A), indicating that the difference in glutamine uptake was not skewed by unequal numbers of IDH mutant or WT gliomas in the male or female patient groups. A similar trend was evident in the IDH mutant patients, but the sample size was too small for a statistically reliable conclusion (Fig S2B). When we restricted the analysis to either GBM (Fig. S2C) or all high-grade gliomas (HGG) (Fig. S2D), we continued to see a similar trend. While in GBM patients, the sample size was too small for a statistically reliable conclusion, HGG showed a strong trend (p=0.06) towards higher uptake in males, indicating that these sex differences were not entirely driven by unequal distribution of HGG and low-grade gliomas (LGG) in the male or female patient groups. Due to the small sample size, we could not analyze the LGG patient cohort by itself.
To determine whether sex differences in glutamine uptake were restricted to tumor tissue, we measured [18F]FGln uptake in the grey matter of the contralateral cortex of the same patients. The mean SUL (SULmean) value of [18F]Gln was significantly higher in male versus female brains (Fig. 2C). Together, these data suggest that sex differences in physiologic glutamine metabolism may be recapitulated in glioma.
To explore the mechanisms underlying sex differences in glutamine metabolism, we took advantage of a previously developed in vitro transformed mouse astrocyte model involving combined loss of function of Nf1 and p5319,20. We assessed glutamine consumption in male and female transformed astrocytes and found that male cells consumed ≈1.5-fold more glutamine than female cells (Fig. 2D). Increased glutamine consumption suggests that male cells are more dependent on glutamine for growth. To test this hypothesis, we cultured male and female cells under varying glutamine concentrations and measured cell number. Lowered glutamine levels led to greater reductions in cell number in males compared to females (Fig. 2E). These data indicate that sex differences in glutamine metabolism are present across species and that our transformed astrocyte model is appropriate to study the molecular mechanisms underlying sex differences in glutamine dependency in glioma.
Male transformed astrocytes exhibit higher glutamine utilization
To delineate specific glutamine requirements in male and female transformed astrocytes, we performed [13C515N2]glutamine ([13C515N2]Gln) tracing experiments. Upon cellular uptake, glutamine is rapidly metabolized to glutamate by the enzyme glutaminase (GLS). The γ-nitrogen that is released during this reaction is required for nucleotide synthesis (Fig. 3A). Furthermore, various other enzymes, including CAD protein, phosphoribosyl pyrophosphate amidotransferase (PPAT), guanosine monophosphate synthetase (GMPS), and phosphoribosylformylglycinamidine synthase (PFAS), can hydrolyze glutamine to incorporate the released γ-nitrogen into nucleotide synthesis (Fig. 3A). We assessed nitrogen label incorporation from [13C515N2]Gln into the purine guanine and the pyrimidine thymine using solid-state NMR and found that male cells incorporated approximately 20% more nitrogen from [13C515N2]Gln into nucleotides than female cells (Fig. 3B).
Figure 3. Male transformed astrocytes exhibit higher glutamine utilization.
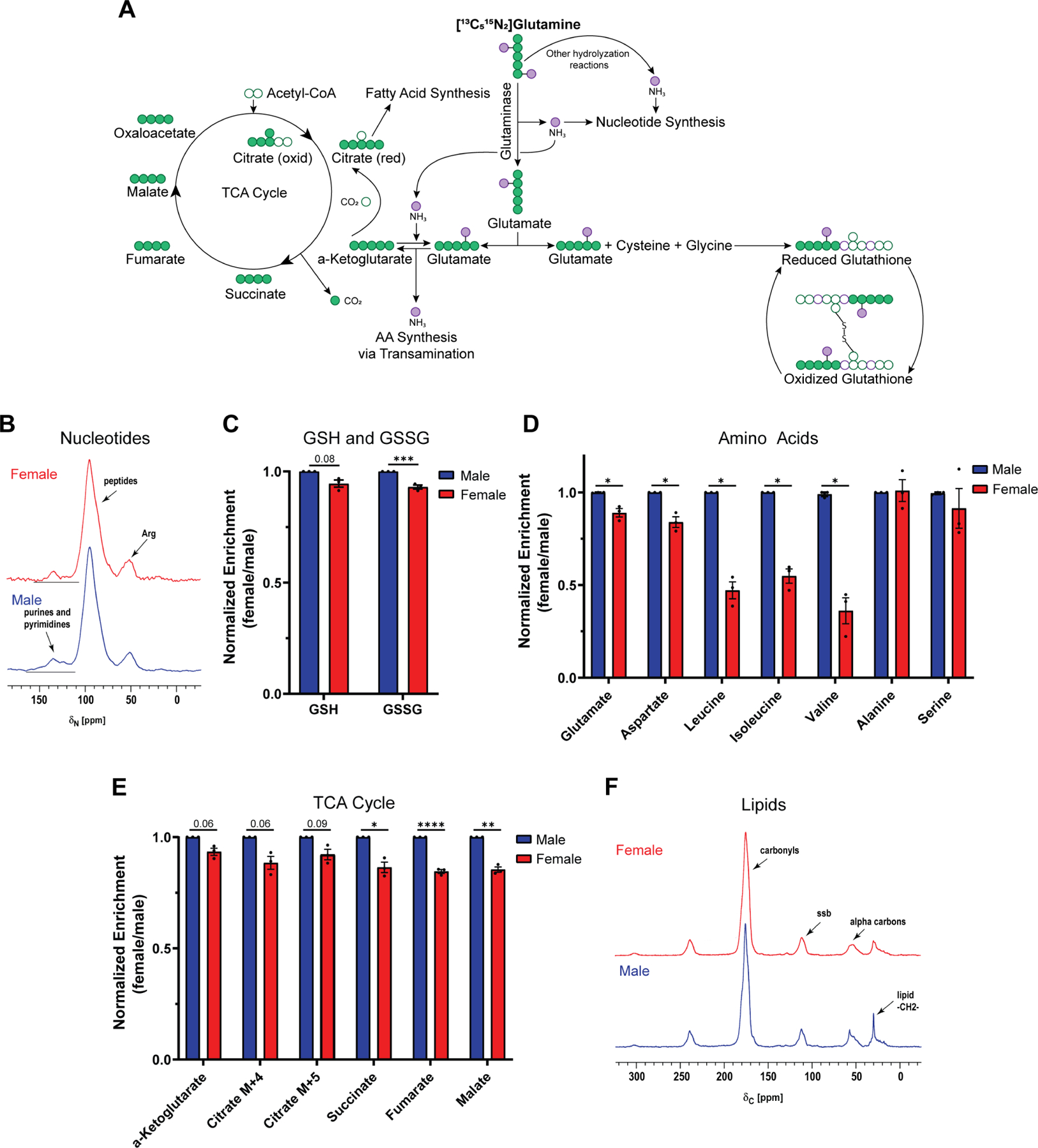
A, Schematic of major downstream metabolic pathways of glutamine. B-F, Label incorporation of [13C515N2]glutamine ([13C515N2]Gln) in transformed astrocytes. Label incorporation into nucleotides (B) and lipids (F) was measured via solid-state NMR. 50.3-MHz 15N NMR spectra (B) and 125.7-MHz cross-polarization magic-angle spinning 13C NMR spectra (F) are shown. Label incorporation into reduced (GSH) and oxidized (GSSG) glutathione (C) was measured via LC/MS. Prominent (fully labeled) isotopologues for GSH (m+5) and GSSG (m+10) are shown. Label incorporation into amino acids (D) and TCA cycle intermediates (E) was measured via GC/MS. Prominent isotopologues for glutamate (m+6), aspartate (m+5), and all other amino acids (m+1) are shown. Prominent labels (one turn of the TCA cycle) for α-KG (m+5), citrate (oxidative TCA cycle (m+4); reductive TCA cycle (m+5)), succinate (m+4), fumarate (m+4), and malate (m+4) are shown. Data are mean +/− SEM of n=3/sex (three independent experiments, one male and female cell line). ***p<0.001; linear mixed effects model (C). *q<0.05, **q<0.01, ****q<0.0001; linear mixed effects model, FDR adjusted p-values (D-E). See also Tables S3 and S4.
As a key component of glutamine metabolism, glutamate feeds into two major downstream pathways, glutathione and α-ketoglutarate (α-KG) synthesis. Together with glycine and cysteine, glutamate can form glutathione. Glutathione is the most abundant small molecular weight thiol in cells and acts as a free radical scavenger as well as a substrate for electrophile detoxification. Thus, glutathione plays a critical role in maintaining cellular redox balance and has been associated with chemotherapy resistance in cancer21. Glutathione exists primarily in its reduced form (GSH) in cells, but under oxidizing conditions it is converted to glutathione disulfide (GSSG). We assessed carbon label incorporation from [13C515N2]Gln into GSH and GSSG using LC/MS and found that male cells incorporated significantly more carbons from [13C515N2]Gln into the GSSG pool than female cells (Fig. 3C). While carbon incorporation into the GSH pool was not significantly greater in males, it did trend towards significance (p=0.08).
Glutamate can also be converted into α-KG via oxidative deamination. The liberated α-nitrogen from this reaction serves as the donor for amino acid synthesis via transamination. In addition, the γ-nitrogen liberated during glutamine hydrolyzation can be incorporated to convert α-KG to glutamate via reverse glutamate dehydrogenase (GDH)22 (Fig. 3A). Thus, both glutamine-derived ammonia groups can contribute to amino acid synthesis via transamination. We assessed total nitrogen label incorporation from [13C515N2]Gln into multiple amino acids using GC/MS. It is of note that assessing nitrogen incorporation from [13C515N2]Gln into amino acids does not allow us to distinguish between γ- and α-nitrogen. Aspartate and particularly the branched-chain amino acids (BCAAs) leucine, isoleucine, and valine showed higher nitrogen label incorporation from [13C515N2]Gln in male cells compared to female cells (Fig. 3D). This was not the case for serine and alanine, highlighting a potentially unique nitrogen requirement from glutamine for BCAA synthesis in male cells.
In addition to providing an amino group, the conversion from glutamate to α-KG replenishes the TCA cycle. We assessed incorporation of [13C515N2]Gln into TCA cycle metabolites using GC/MS and found that male cells incorporated more carbons from glutamine into the TCA cycle than female cells (Fig. 3E). Citrate is an important substrate in fatty acid synthesis. Glutamine can replenish the cellular citrate pool either via the oxidative direction (m+4), or the reductive direction (m+5) of the TCA cycle23. Both citrate isotopologues trended towards higher label incorporation in male cells, suggesting that males might utilize more glutamine for fatty acid synthesis. We assessed [13C515N2]Gln incorporation into lipids using solid-state NMR and found that in male cells carbon incorporation from glutamine is approximately 2-fold higher than in female cells (Fig. 3F). This extensive accounting of glutamine utilization in male and female transformed astrocytes shows that males are more dependent on glutamine for downstream cellular processes than their female counterparts.
GLS1 expression and dependency are greater in male transformed astrocytes and gliomas
Targeting glutamine metabolism continues to evolve as an attractive cancer treatment. Targeting GLS, which hydrolyzes glutamine to glutamate, was effective in pre-clinical cancer models24–28 and its clinical efficacy is currently being evaluated. The GLS family consists of two members, GLS1 and GLS2, which are expressed in the mitochondria and the cytosol, respectively. In brain tissue, GLS1 is expressed at higher levels than GLS229. Accordingly, the predominant enzyme in gliomas is GLS130. To determine if there was a sex difference in GLS expression, we examined mRNA expression levels in male and female GBM samples from the TCGA Firehose Legacy GBM patient dataset. We found that in both, male and female IDH WT GBM, GLS1 expression levels were significantly higher than GLS2 expression levels (Fig. S3A). Interestingly, GLS1, but not GLS2, was expressed at significantly higher levels in male versus female GBM (Fig. 4A and S3A). Next, we assessed GLS mRNA expression levels in male and female human glioma cell lines from the Broad Institute Cancer Cell Line Encyclopedia and found that GLS1 expression was significantly higher in male glioma cell lines (Fig. 4B). GLS2 expression was low and did not differ by sex (Fig. S3B). These data suggest that GLS1 may play an important role in the sex differences we observed in glutamine metabolism in glioma.
Figure 4. GLS1 expression and dependency are greater in male transformed astrocytes and gliomas.
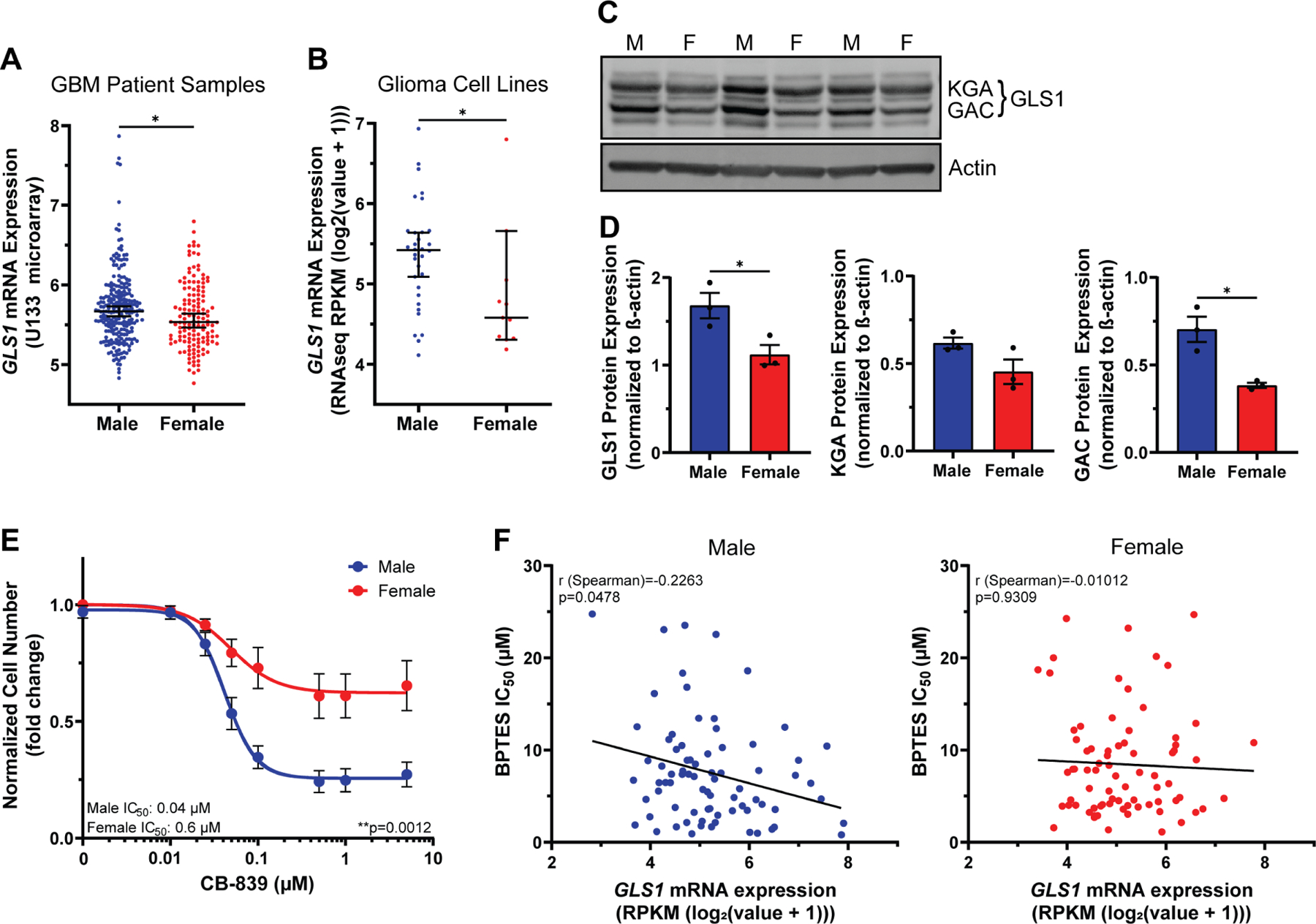
A-B, GLS1 mRNA expression levels in n=232 male and n=139 female IDH wild-type GBM patient samples (A) and n=32 male and n=11 female human glioma cell lines (B). C, Western blot showing the two GLS1 isoforms (KGA and GAC) in transformed astrocytes from three independent experiments. The lanes to the left and the right of the samples contained protein ladder and were digitally cropped from the image. M=male, F=female. D, Western blot quantification of GLS1 and its isoforms KGA and GAC. Expression values are normalized to β-actin. E, CB-839 dose-response curves of transformed astrocytes. F, Nonparametric Spearman correlation between GLS1 mRNA expression levels and BPTES IC50 values in n=77 male (left) and n=76 female (right) tumor cell lines. Data are median +/− 95% CI. *p<0.05; Mann-Whitney test (A-B). Data are mean +/− SEM of n=3/sex (three independent experiments using one male and female cell line). *p<0.05; t-test (D). Data are mean +/− SEM of n=9/sex (three independent experiments, three separate male and female cell lines). **p=0.0012; two-way ANOVA (E). See also Figure S3 and Table S5.
GLS1 protein expression was also significantly higher in male versus female transformed astrocytes (Fig. 4C–D). GLS1 exists in two isoforms, kidney glutaminase A (KGA) and glutaminase C (GAC). KGA protein levels were higher in male cells, but the difference was statistically insignificant. GAC protein levels were significantly higher in male cells (Fig. 4D). This is important, as GAC might play a more tumorigenic role than KGA31,32. To determine whether greater GLS1 expression was coupled to greater dependency on its function, we treated male and female transformed astrocytes with varying doses of the GLS1 inhibitor CB-839. Notably, male cells displayed a steep dose-dependent sensitivity to GLS1 inhibition, whereas female cells were almost entirely resistant (Fig. 4E). This was true despite similar reductions in glutamate abundance to less than 50% of vehicle in treated cells of both sexes (Fig. S3C). Although the amount of residual glutamate in female cells treated with CB-839 was higher in female cells, this difference was not significant (p=0.18, student’s t-test) (Fig. S3C). We confirmed sex differences in GLS1 dependency using BPTES, a less potent analog to CB-839, which also reduced male, but not female, cell number (Fig. S3D). To further validate sex-specific responses to GLS1 inhibition, we examined publicly available BPTES IC50 values in male and female tumor lines, spanning multiple cancer types33 (Table S5). Although female cell lines exhibited slightly higher IC50 values, these were not significantly different from the male lines (Fig. S3E). We then correlated BPTES IC50 values with GLS1 mRNA expression, as described by Yang et al.34, and found that only male tumor lines exhibited a significant, but modest negative correlation between GLS1 expression and BPTES IC50 values (Fig. 4F), indicating that high GLS1 expression is associated with greater sensitivity to GLS1 inhibition in males, but not females.
The clinical and laboratory data thus far indicate that male glioma express higher levels of GLS1 and are more sensitive to GLS1 inhibition, suggesting they are more dependent on glutamine-derived glutamate. In addition, GLS1 expression is associated with BPTES sensitivity in male, but not female tumor cell lines, suggesting that sex differences in GLS1 dependency may extend beyond gliomas.
Male transformed astrocytes are more dependent on glutathione for redox balance
To determine which metabolic pathways underlie the male-specific sensitivity to GLS1 inhibition, we assessed the effects of CB-839 on three metabolite groups linked with glutamine metabolism: TCA cycle, amino acids, and glutathione (Fig. 5A). While male and female cells both showed a reduction in TCA cycle metabolites upon GLS1 inhibition, the reduction was more profound in males. CB-839 treated cells exhibited heterogeneous responses in amino acid levels. Aspartate, glutamate, and proline, like TCA cycle metabolites, were more substantially reduced in male cells. In contrast, alanine, serine, asparagine, and threonine, were more substantially reduced in female cells. The remaining amino acids were increased in male and decreased in female cells. CB-839 treatment resulted in a significantly different (FDR<0.05) change in glutathione levels in male versus female cells. Males showed increased, and females showed decreased levels of glutathione, paralleling two of its amino acid components, glycine and cysteine. Thus, in male and female transformed astrocytes GLS1 inhibition has different metabolic effects.
Figure 5. Male transformed astrocytes are more dependent on glutathione for redox balance.
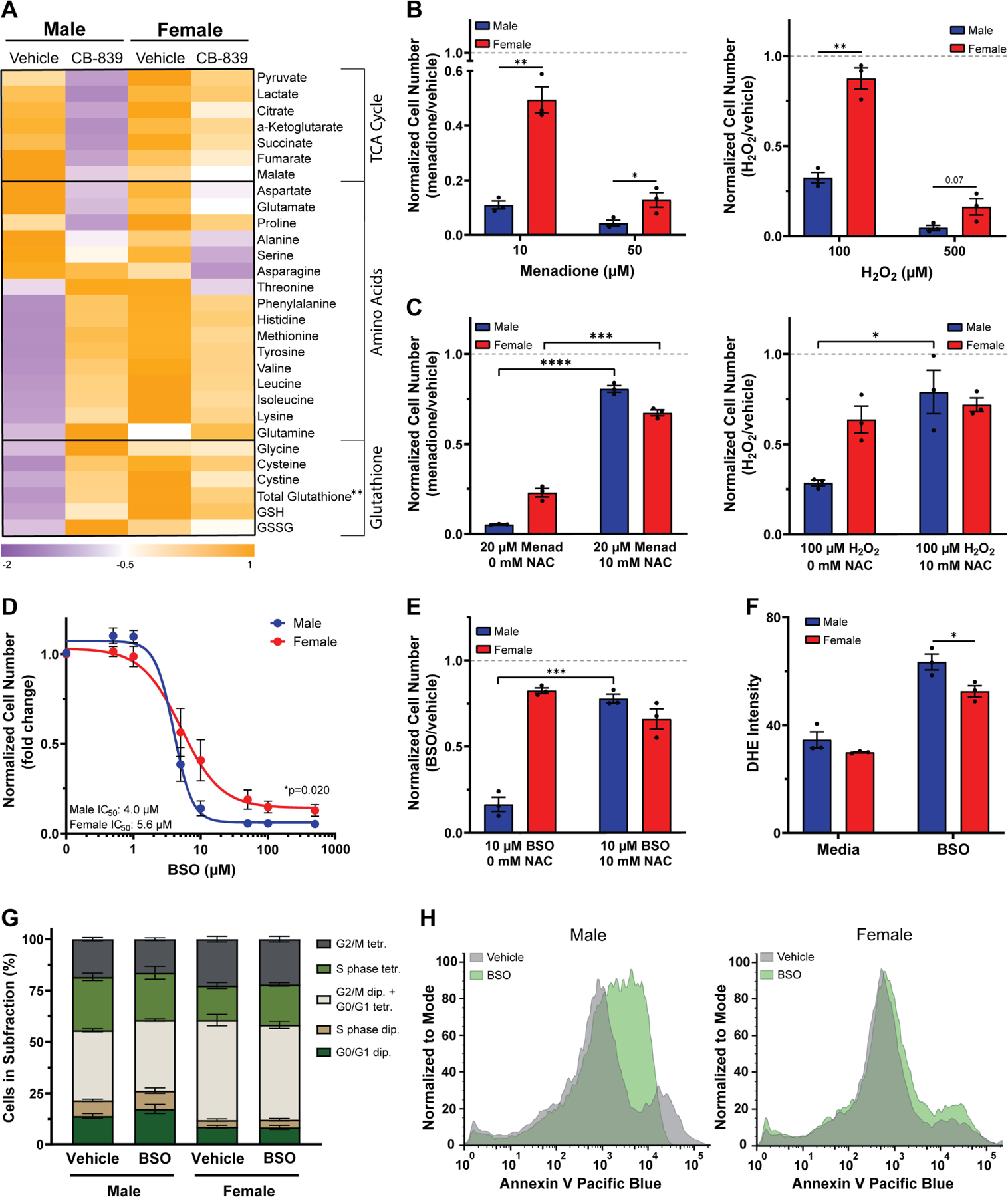
A, Heatmap of metabolite levels in transformed astrocytes treated with CB-839 or vehicle for 24 hrs. TCA cycle and amino acid metabolite abundance were measured via GC/MS; glutathione levels were measured using a spectrophotometric recycling assay. B, Cell number assay of transformed astrocytes treated with menadione or vehicle (left) or H2O2 or vehicle (right). C, Cell number assay of transformed astrocytes treated with menadione or vehicle and N-acetyl cysteine (NAC) (left) or H2O2 or vehicle and NAC (right). D, Buthionine sulfoximine (BSO) dose-response curves of transformed astrocytes. E, Cell number assay of transformed astrocytes treated with BSO or vehicle and NAC. F, Quantification of dihydroethidium (DHE) fluorescence intensity per cell in transformed astrocytes treated with BSO or vehicle. G, Quantification of propidium iodide histograms (Fig. S4D) of transformed astrocytes treated with BSO or vehicle. H, Representative Annexin V histograms of transformed astrocytes treated with BSO or vehicle. Histograms shown are representative examples chosen from three independent. Data are mean +/− SEM of n=3/sex (three independent experiments, one male and female cell line), or for (D) mean +/− SEM of n=9/sex (three independent experiments, three separate male and female cell lines). **q<0.01; t-test, FDR adjusted p-values (A, G). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; t-test (B-C, E-F). *p=0.020; two-way ANOVA (D). See also Figure S4.
The greater reduction in TCA cycle metabolites suggests that male cells may be more dependent on glutamine for TCA cycle replenishment than female cells. The increase in glutathione levels and its amino acid components, further suggests that glutathione might play a more important role in the male metabolic stress response induced by GLS1 inhibition. Notably, male cells exhibited lower levels of glutathione and its amino acid components at baseline, suggesting that male cells may be more sensitive to oxidative stress. Sex differences in glutathione and ROS regulation have been described35. Thus, we hypothesized that ROS regulation differs in male and female transformed astrocytes and that GLS1 inhibition has sex-specific effects on cellular redox balance.
To test our hypothesis, we first assessed whether male and female transformed astrocytes differ in their sensitivity to oxidative stress. We induced oxidative stress with menadione or hydrogen peroxide (H2O2) and found that both treatments resulted in a dose-dependent and significantly greater reduction in cell number in male versus female cells (Fig. 5B). Adding the thiol antioxidant N-acetylcysteine (NAC), significantly restored cell numbers in menadione treated male and female cells, and in H2O2 treated male cells (Fig. 5C), indicating that ROS accumulation and thiol oxidation are the major underlying reasons for the reduction in cell number upon menadione and H2O2 treatment. This suggests that male transformed astrocytes are more sensitive to oxidative stress than females.
To assess whether male cells are more dependent on glutathione to regulate redox balance, we depleted glutathione using buthionine sulfoximine (BSO), an inhibitor of glutamate-cysteine ligase. Male cells were significantly more sensitive to glutathione depletion than female cells (Fig. 5D). This was not due to differences in glutathione levels upon treatment, as BSO treatment almost completely depleted glutathione in both sexes (Fig. S4A). This indicates that although female transformed astrocytes have significantly higher total glutathione levels at baseline, their greater resistance to BSO is not due to a greater abundance of cellular glutathione. We utilized a second strategy to deplete glutathione by inhibiting the glutamate/cystine antiporter XCT36 with two XCT inhibitors, sulfasalazine (SAS) and erastin, and found that male cells were significantly more sensitive to both inhibitors (Fig. S4B–C).
To determine whether glutathione depletion affected cell number as a consequence of ROS accumulation and thiol oxidation, we treated male and female transformed astrocytes with vehicle or BSO with or without NAC. NAC significantly restored cell numbers in male and female BSO treated cells (Fig. 5E), and BSO induced significantly more dihydroethidium (DHE) oxidation – a measurement for superoxide accumulation – in males compared to females (Fig. 5F), suggesting increased levels of ROS. Thus, thiol oxidation may be the basis for cell number reduction in BSO treated cells. Lastly, we assessed whether BSO reduces cell number through cell cycle arrest or apoptosis. Neither male nor female cells underwent cell cycle arrest upon BSO treatment (Fig. 5G). However, in male cells, BSO increased Annexin V staining (Fig. 5H), indicating greater apoptosis. These data show that male transformed astrocytes are more dependent on glutathione to maintain their cellular redox balance, which might make them more dependent on GLS1 for the glutamate required in glutathione synthesis.
Male transformed astrocytes require glutamine to replenish their TCA cycle
To assess whether sex-specific effects of GLS1 inhibition on cell number are driven by a cellular redox imbalance, we measured DHE oxidation in CB-839 treated male and female transformed astrocytes. Similar to glutathione depletion, CB-839 resulted in a significantly greater DHE oxidation in male cells (Fig. 6A). However, in contrast to BSO, the CB-839 growth phenotype was not rescued by NAC (Fig. 6B). Thus, thiol oxidation is not the underlying reason for the male-specific sensitivity to CB-839. To gain greater insight, we assessed the effects of GLS1 inhibition on the cell cycle and apoptosis. Unlike their response to glutathione depletion, male transformed astrocytes underwent cell cycle arrest in response to GLS1 inhibition (Fig. 6C) without an increase in apoptosis (Fig. S5A). Thus, the mechanisms underlying changes in cell number are different upon glutathione depletion (apoptosis) and GLS1 inhibition (cell cycle arrest), indicating that while male cells are more dependent on glutathione, redox balance is not the basis for their greater sensitivity to GLS1 inhibition.
Figure 6. Male transformed astrocytes require glutamine to replenish their TCA cycle.
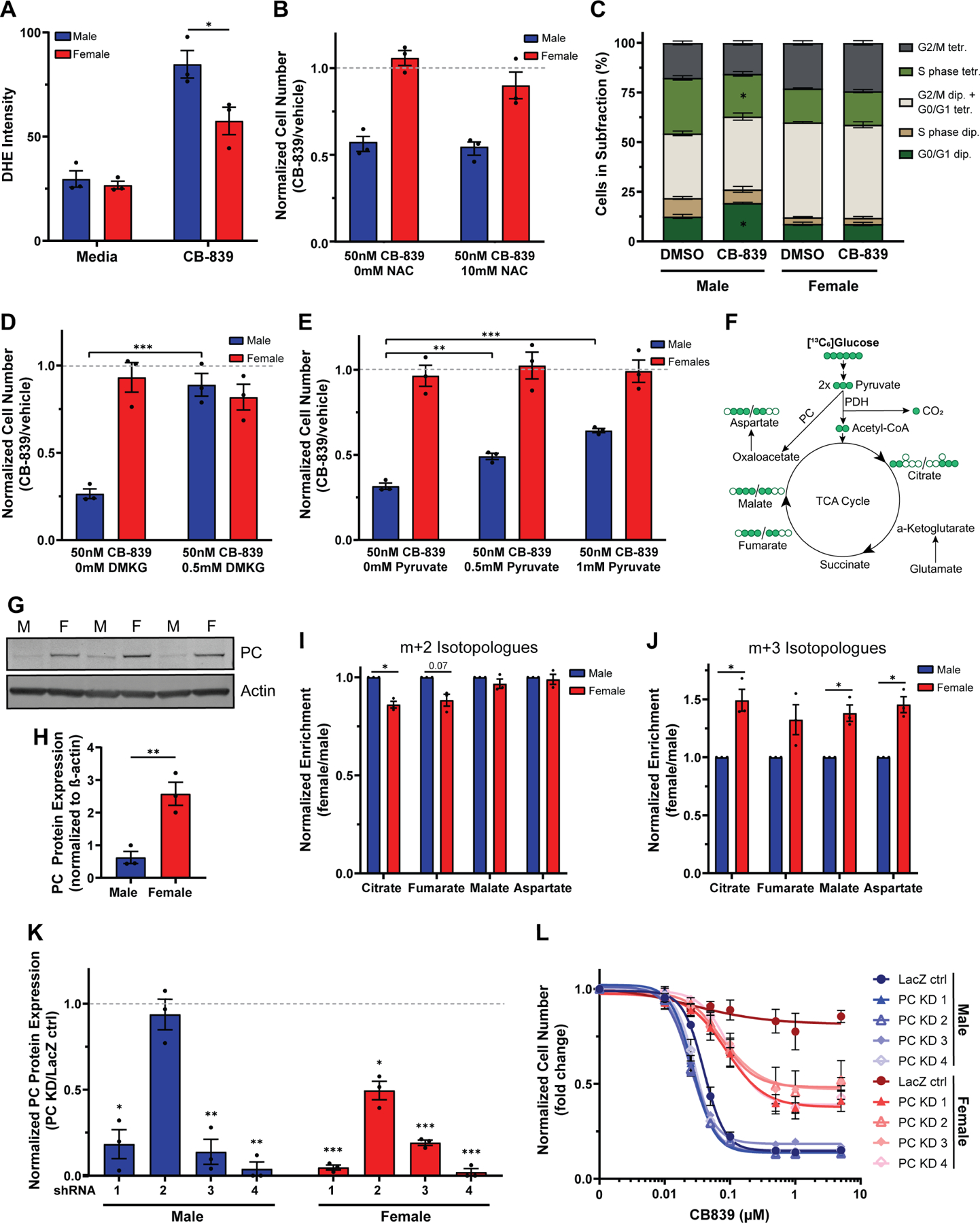
A, Quantification of dihydroethidium (DHE) fluorescence intensity per cell in transformed astrocytes treated with CB-839 or vehicle. B, Cell number assay of transformed astrocytes treated with CB-839 or vehicle and N-acetyl cysteine (NAC). C, Quantification of propidium iodide histograms (Fig. S5C) of transformed astrocytes treated with CB-839 or vehicle. D, Cell number assay of transformed astrocytes treated with CB-839 or vehicle and dimethyl-α-ketoglutarate (DMKG). E, Cell number assay of transformed astrocytes treated with CB-839 or vehicle and pyruvate. F, TCA cycle flux schematic. G, Western blot showing PC expression in transformed astrocytes under basal media conditions from three independent experiments. The lanes to the left and the right of the samples contained protein ladder and were digitally cropped from the image. M=male, F=female. H, Western blot quantification of PC. Expression values are normalized to β-actin. I-J, Label incorporation of [13C6]glucose in transformed astrocytes. Label incorporation into metabolites was measured via GC/MS. m+2 (I) and m+3 isotopologues (J) after one turn of the TCA cycle are shown. K, Western blot quantification of transformed astrocyte control cells (LacZ ctrl) and transformed astrocyte PC knockdown cells (PC KD). All expression values are normalized to β-actin. Protein levels in knockdown samples were normalized to their respective control. L, CB-839 dose-response curves of male and female transformed astrocyte control cells (LacZ ctrl) and transformed astrocyte PC knockdown cells (PC KD 1–4). IC50 values are not depicted since no IC50 value could be calculated for the female LacZ ctr. Data are mean +/− SEM of n=3/sex (three independent experiments, one male and female cell line). *p<0.05, **p<0.01, ***p<0.001; t-test (A, B, D, E, H, K). *q<0.05; t-test, FDR adjusted p-values (C). *q<0.05; linear mixed effects model, FDR adjusted p-values (I-J). Male LacZ ctrl compared to male PC KD cell lines: PC KD 1: p=0.09, PC KD 2: p=0.03, PC KD 3: p=0.71, PC KD 4: p=0.40. Female LacZ ctrl compared to female PC KD cell lines: PC KD 1–4: p<0.001; two-way ANOVA, Dunnetťs multiple comparisons test (L). See also Figure S5 and Tables S3, S6, and S7.
Glutamate is also required for α-KG synthesis, which is important in multiple cellular processes, including replenishing the TCA cycle. Since GLS1 inhibition with CB-839 resulted in a greater reduction in TCA cycle metabolites in male transformed astrocytes, we hypothesized that TCA cycle metabolite depletion triggers growth arrest in male cells. To test this hypothesis, we attempted to rescue CB-839 treated male and female transformed astrocytes with two metabolites commonly used for TCA cycle restoration, dimethyl-α-KG (DMKG) and pyruvate. DMKG fully rescued the male growth phenotype (Fig. 6D). Pyruvate had a weaker, but dose-dependent and significant rescue effect (Fig. 6E). DMKG is a TCA cycle metabolite and therefore a more direct rescue of TCA cycle function compared to pyruvate, which may explain the greater potency of DMKG. Thus, male transformed astrocytes are sensitive to GLS1 inhibition due to its inhibitory effects on TCA cycle activity.
TCA cycle metabolites can also be replenished by glucose-derived pyruvate, which can enter the TCA cycle via two enzymatic reactions. Most pyruvate entering the TCA cycle is converted to acetyl-CoA by pyruvate dehydrogenase (PDH). However, pyruvate carboxylase (PC) can also directly convert pyruvate to oxaloacetate (Fig. 6F). PC activity can support glutamine-independent cancerous growth as it provides a compensatory anaplerotic mechanism37. We assessed PC protein expression in transformed astrocytes and found that females have ≈4-fold higher expression of PC (Fig. 6G–H), which was unaffected by CB-839 treatment (Fig. S5C). Next, we assessed PC activity in male and female cells at baseline. We incubated cells in [13C6]glucose and assessed label enrichment. Upon its conversion from [13C6]glucose, [13C3]pyruvate can replenish the TCA cycle via PDH or PC. PDH transfers two, and PC three, labeled carbons into the TCA cycle, producing either m+2 or m+3 isotopologues on the cycle’s first turn (Fig. 6F)38. We measured m+2 and m+3 isotopologues of aspartate, malate, fumarate, and citrate. Label enrichment of the citrate m+2 isotopologue was significantly higher in males, suggesting a slightly higher utilization of PDH for glucose input into the TCA cycle in male cells (Fig. 6I). In contrast, females exhibited ≈1.5-fold greater label incorporation into all m+3 isotopologues (Fig. 6J), which was significant for citrate, malate, and aspartate, indicating they have greater PC capacity for feeding glucose into the TCA cycle. Thus, male cells are more reliant on glutamate for TCA cycle replenishment.
To confirm that PC mediates resistance to GLS1 inhibition in female transformed astrocytes, we knocked down PC protein expression in male and female transformed astrocytes using four different short hairpin RNAs (shRNAs) targeting murine Pc. Each shRNA significantly reduced PC protein expression in female transformed astrocytes, and three shRNAs significantly reduced PC protein expression in male transformed astrocytes (Fig. 6K, Fig. S5D). To assess whether PC knockdown (PC KD) sensitizes female transformed astrocytes to GLS1 inhibition, we treated male and female transformed astrocyte control and PC knockdown cells with varying doses of CB-839. While male PC KD cell lines showed a similar response to CB-839 treatment as the male control cell line, all female PC KD cell lines were significantly more sensitive to CB-839 than the female control cell lines (Fig. 6L). Despite similar PC protein expression levels in male and female PC KD cell lines, male cells continued to show greater sensitivity to CB-839 compared to female cells (Fig. 6L), indicating that PC is a mechanism of resistance to CB-839, but not the only mechanism that supports glutamine-independent growth in female transformed astrocytes.
Although PC mRNA expression was not significantly different in male and female GBM samples from the TCGA Firehose Legacy GBM patient dataset (Fig. S5E), we found that GLS1 and PC mRNA expression levels in the TCGA Pan-Cancer Atlas were negatively correlated in male, but not female pan-cancer samples (Fig. S5F). This finding suggests that male cancers with higher GLS1 expression and therefore greater sensitivity to CB-839 would have the least amount of PC-mediated resistance. Thus, we conclude that female cancer cells possess greater metabolic flexibility for TCA cycle replenishment, resulting in resistance to therapeutic targeting of glutamine metabolism.
Discussion:
Metabolic reprogramming supports the high energetic and biosynthetic demands of cancerous growth. Targeting cancer’s unique metabolic adaptations holds great promise as a treatment with a high therapeutic index. For this approach to work optimally for all patients, it must take sex differences in energy and biosynthetic pathways into account. Here, we report for the first time that cancer cells retain normal sex differences in glutamine metabolism. Sex differences in glutamine uptake, glutamine metabolism, and GLS1 expression were evident in our mouse transformed astrocyte cell lines, human glioma cell lines, and GBM patient samples, as well as in normal brain tissue, suggesting that sex differences in glutamine metabolism are conserved across species and between cancerous and non-cancerous tissue. Similar sex differences in glutamate levels and glutamine metabolism have been reported in normal brain tissues of rats39,40 as well as in the serum of healthy individuals13, indicating fundamental cellular and systemic sex differences in glutamine metabolism. The underlying reasons for these sex differences are currently unknown. However, since sex differences in glutamine metabolism exist under normal physiological conditions in the brain, it may be that these sex differences foster glutamine dependency in male gliomas. As circulating glutamine and glutamate levels are high in males, increased supply from systemic circulation may contribute to the development of glutamine-dependent gliomas in males specifically. However, a potential synergy between cell-intrinsic glutamine dependency in gliomas and increased supply from systemic circulation needs to be established.
We focused on glutamine metabolism because it is being targeted in early phase cancer clinical trials (NCT02071927, NCT04250545, NCT03528642, NCT03965845). We found that male cells were significantly more dependent on glutamine, which they shuttled into all its major downstream processes. GLS1 inhibition resulted in greater growth arrest in male cells, and sex-dependent effects on the TCA cycle and glutathione metabolism, which played a greater role in male redox balance. The underlying reason for sex differences in sensitivity to GLS1 inhibition was PC-mediated TCA cycle replenishment by glucose, which was more active in female cells.
In normal metabolism, sex differences are evident throughout life41–44. Here, we show for the first time that they are also a feature of cancer. There was substantial concordance between the male-biased metabolic pathways we identified in GBM and those described in male serum13. For example, amino acid and carbohydrate metabolites were increased in males, while females were enriched in certain lipids in both datasets. While these findings were highly significant in serum, they did not reach significance in the GBM dataset, which is most likely due to its small sample size. Overall, these findings suggest that fundamental sex differences in metabolism are present in GBM and may even extend beyond gliomas.
As glutamine metabolism differs in male and female cells, response to targeting glutamine metabolism might need to be adapted to sex-biased mechanisms of resistance. Male glioma patients may respond better to CB-839 treatment than female glioma patients and co-targeting PC activity might block resistance to GLS1 in female cells. Although PC knockdown significantly increased sensitivity to CB-839 in female transformed astrocytes, it did not increase sensitivity to levels comparable to male cells. Thus, it is likely that female transformed astrocytes employ additional mechanisms for glutamine-independent growth. These additional mechanisms remain to be determined. One potential mechanism may be that females have a glutamine-independent source of glutamate. Although glutamate levels were reduced by more than 50% in male and female transformed astrocytes in response to CB-839 (Fig. S3C), residual glutamate levels were slightly higher in females treated with CB-839. Whether this difference contributes to the CB-839 resistance remains to be determined.
We also showed that redox regulation differs in male and female transformed astrocytes. Sex differences in sensitivity to ROS, pro-oxidant levels, and ROS regulation are well described in healthy brain tissue2. Here, we provide evidence that these sex differences might persist in cancerous tissue and could result in sex-specific responses to treatments that depend on ROS accumulation. This study is not the first study providing evidence that novel anti-cancer therapeutics currently in clinical trials might have sex-specific effects. It was recently reported that clinically utilized bromodomain inhibitors could produce sex-specific responses45. Together, these studies emphasize the importance of including sex as a variable in basic research and clinical settings.
Consistent sex differences in metabolite abundance in cancerous and non-cancerous tissue suggest that sex differences in glutamine metabolism might be conserved across other cancers, a possibility that should be explored for optimized clinical use of GLS1 inhibition. Sex differences in other metabolic pathways may be similarly important. All branched-chain amino acids (BCAAs) were included in the top 75 enriched metabolite list and exhibited a male bias. Our glutamine tracing experiments showed that male transformed astrocytes incorporated ≈2-fold more label into BCAAs, indicating that targeting BCAA metabolism may be a unique vulnerability in male cells. Finally, our data suggest that other metabolic pathways, such as carbohydrates and fatty acids, exhibit sex differences in GBM. It remains to be determined how sex differences in these metabolic groups affect cancer biology and treatment response.
Limitations of the study
While the sex-specific analysis of human GBM metabolite abundance shows remarkable similarities with previously published studies, it is underpowered to detect significant sex differences. A greater number of patient samples would be necessary to validate those findings.
Here, we show that male transformed astrocytes are dependent on glutamine for their growth because they depend on α-KG for TCA cycle replenishment. However, α-KG plays additional metabolic and epigenetic roles. It is an essential co-factor for multiple demethylases which mediate histone and DNA demethylation. It is also an important component of transamination and amino acid synthesis. Whether the disruption of other cellular processes downstream of α-KG also contributes to the observed growth arrest in male cells following GLS1 inhibition requires further investigation. Furthermore, the [13C515N2]Gln tracing experiments in Fig. 3 revealed that male transformed astrocytes utilize considerably more glutamine into multiple other pathways, including lipid synthesis and nucleotide synthesis. Whether GLS1 inhibition disrupts other metabolic pathways in male or female cells, and whether this disruption contributes to the observed growth arrest in male cells remains to be determined.
All mechanistic in vitro studies were performed in the same model of transformed astrocytes. While we repeated several key experiments (glutamine deprivation, CB-839 treatment, BSO treatment) in three separately established male and female transformed astrocyte cell lines, the translational relevance of the work could be improved by using additional in vitro models. Despite this limitation, we believe that the validation of our in vitro data using human samples and cell line data throughout the paper is strong and suggests that sex differences in GLS1 dependency are not likely to be a model-specific phenotype. Lastly, in vivo studies including human xenograft models and spontaneous tumor models should be conducted to confirm and extend our findings to other types of glioma and other cancer models.
STAR★Methods:
Resource Availability
Lead Contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact Joseph E. Ippolito (ippolitoj@wustl.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and Code Availability
Original data for Fig. 1 in the paper is available in Prabhu et al.14. The tumor cell line dataset used in Fig. 4H in the paper is available in Daemen et al.33. Targeted GC/MS metabolite data and isotopologue data generated for this publication are included in the published article. Original western blot images, MRI images, PET images, and microscopy data reported in this paper will be shared by the lead contact upon request.
No original code was generated in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Subject Details
Cell culture
The male and female Nf1−/− DNp53 astrocytes (transformed astrocytes) were previously generated and published by our lab20. Briefly, primary cultures of male and female Nf1−/− astrocytes were isolated from the neocortices of individual postnatal day 1 Nf1fl/fl GFAP-Cre mice as described previously20,46 and cultured in Gibco Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12), supplemented with 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin (PS) at 37°C and 5% CO2. The sex of each mouse was determined from tail DNA by PCR for Jarid1c and Jarid1d 47,48. Cells from at least three male and three female mice across multiple litters were pooled. Male and female Nf1−/− astrocytes were then infected with retrovirus encoding a flag-tagged dominant-negative form of p53 (DNp53) and EGFP as previously described20. The DNp53 plasmid consists of amino acids 1–14 of the transactivation domain followed by amino acids 303–393, thus lacking the DNA binding domain. These male and female Nf1−/− DNp53 are referred to as male and female transformed astrocytes. Multiple astrocyte harvests and retroviral infections were performed, which led to multiple lines of male and female GBM astrocytes, which are referred to as “lots” of male and female transformed astrocytes. Experiments were performed with up to three different lots of transformed astrocytes. All transformed astrocytes were cultured in DMEM/F-12, supplemented with 10% FBS at 37°C and 5% CO2. All experiments were performed at passages 5 to 12.
Patient samples
As part of a prospective research protocol approved by the Institutional Review Board, patients with glioma were imaged with [18F]FGln PET/CT. All procedures were performed in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards (MSKCC IRB protocol number 12–168). All patients provided written informed consent [NCT01697930]. The following patient information was collected: sex, age (at the time of PET imaging), race, ethnicity, diagnosis, tumor grade, IDH status. Participants information on sex, age, race, and ethnicity were self-reported. Diagnosis was based on the 2016 WHO classification of CNS tumor49. Information on socioeconomic status was not collected.
Method Details
Human GBM metabolite analysis
The male and female human GBM metabolite dataset14 and the sex of each surgical specimen was kindly provided by Dr. Prakash Chinnaiyan. The dataset included 44 male and 32 female GBM surgical specimens and 757 metabolites. Upon initial normalization described previously14,50, metabolites showing no variation (same expression across all samples) were removed, leaving 718 metabolites for further analysis. Then, the data was log2 transformed and the z-score values were derived for each metabolite. The average z-score across metabolites belonging to each sub- or super-pathway was calculated to represent the metabolic pathway activity of the sub- or super-pathway. The R/Bioconductor software package limma version 3.48.351 was used for sex-differential analysis at the individual metabolite level, the sub-pathway level, and the super-pathway level separately.
Heatmap
Z-score values from each metabolite were represented in a heatmap using Morpheus (https://software.broadinstitute.org/morpheus). Hierarchical clustering was performed using Morpheus and samples were clustered based on Euclidean distance and average linkage. K-means clustering was performed using Morpheus and samples were clustered based on Euclidean distance.
Glutamine tracer PET imaging
[18F]fluoroglutamine (FGln) PET/CT imaging protocol
[18F]FGln was synthesized by Memorial Sloan Kettering Cancer Center’s Radiochemistry and Molecular Imaging Probe Core Facility as described15. Each [18F]FGln dose met drug product acceptance specifications, including radiochemical purity and identity, residual solvent content, endotoxin content, radionuclidic identity, pH, and appearance. The median administered activity was 241.98 MBq (6.54 mCi); IQR: 22.2 MBq (0.60 mCi). The tracer was given as a rapid intravenous bolus injection. Static PET scans were performed at 78 min ± 9 min after injection (15 to 30 min acquisition time) on a Discovery STE, 690, or 710 camera (GE Healthcare). A CT scan (120 kVp, 70 mA, and 3.8 mm slice thickness) was obtained for attenuation correction, anatomic localization, and coregistration purposes. PET emission data were acquired in 3-dimensional mode; corrected for attenuation, scatter, and random events; and iteratively reconstructed into a 256 × 256 × 47 matrix (voxel dimensions, 1.95 × 1.95 × 3.27 mm) for brain lesions using the ordered-subset expectation maximization algorithm provided by the manufacturer.
[18F]FGln PET image interpretation and data analysis
A board-certified nuclear medicine physician defined regions of interest (ROIs) for the lesions and normal brain on a GE Advantage workstation using PET VCAR software. The [18F]FGln PET/CT was windowed to visualize the focal [18F]FGln avidity associated with the known brain lesion on MRI then an ROI was placed to encompass the entire area of abnormal [18F]FGln avidity. For normal brain tracer uptake in the grey matter, an ROI was placed contralaterally to the brain lesion. Lesion volumes were measured by performing three-dimensional threshold-based volume of interest (VOI) analyses for the [18F]FGln uptake in all patients. Tracer uptake was quantified by SUVs and corrected for lean body mass (SUL) using the sex-specific Janmahasatian formulation18. For lesion VOI, SULmax was recorded. SULmax referred to the voxel with the maximum intensity of [18F]FGln uptake in the VOI. For patients with multiple lesions, we used the lesion with the highest SULmax. For brain VOI, SULmean was recorded. SULmean referred to the mean intensity of [18F]FGln uptake in the VOI. Representative axial images were chosen from a male and from a female high-grade glioma patient (IDH WT, MGMT unmethylated, recurrent tumor biopsy confirmed 12 days and within 1 month, respectively, after scan). Axial contrast-enhanced T1-weighted MRI (T1W + C MRI) axial images, PET and fused PET/MRI images are shown.
Drug preparation
CB-839 was purchased as a powder and diluted in DMSO to a stock concentration of 10 mM. BPTES was purchased as a powder and diluted in DMSO to a stock concentration of 100 mM. Menadione was purchased as a powder and diluted in DMSO to a stock concentration of 150 mM. Buthionine sulfoximine (BSO) was purchased as a powder and diluted in water to a stock concentration of 100 mM.
Cell number assays
Glutamine dose-response curve
Cell number was assessed using the colorimetric sulforhodamine B assay52. Briefly, cells were plated in triplicates in a 96-well plate at a density of 1,000 cells/well in DMEM/F-12 media supplemented with 10% FBS. For each experiment, a standard curve for male and female cells was plated at the same time. 6 hrs after plating, standard curves were fixed as described below. 24 hrs after plating, media was changed to DMEM/F-12 (USBiological) containing 1.2 g/L sodium bicarbonate, 10 mM HEPES, 10% dialyzed FBS, 10 mM glucose, and glutamine at various concentrations (4.5, 2.5, 1.5, 1, 0.5, 0.1, 0 mM). Cells were incubated for 4 days. On day 4, plates were fixed with ice cold 5% trichloroacetic acid at 4°C for 1 hr. Then, plates were washed 4 times with tap water and allowed to dry at room temperature overnight. Next, plates were stained with 0.057% (wt/vol) sulforhodamine B (SRB) in 1% (vol/vol) acetic acid at room temperature for 30 min. The plates were washed quickly 3 times with 1% (vol/vol) acetic acid and allowed to dry at room temperature overnight. SRB was dissolved in 10 mM unbuffered Tris-Base solution (pH 10.5) on a shaker at room temperature for at least 30 min. Absorbance was measured at 510 nm using a Synergy HT microplate reader (BioTek). Absorbance values were converted to interpolated cell numbers in GraphPad Prism, using the standard curve absorbance values for each experiment.
CB-839, buthionine sulfoximine (BSO), H2O2, and menadione treatment
Cell number was assessed as described above with minor modifications. Cells were plated in triplicates in a 96-well plate at a density of 2,000 cells/well in DMEM/F-12 media supplemented with 10% FBS. 24 hrs after plating, cells were treated with various doses of CB-839 (0.01, 0.025, 0.05, 0.1, 0.5, 1, 5 µM) or vehicle (DMSO), BSO (0.5, 1, 5, 10, 50, 100, 500 µM) or vehicle (water), H2O2 (100 µM and 500 µM) or vehicle (water), or menadione (10 µM and 50 µM) or vehicle (DMSO). Both male and female cells were around 50% confluent when treatment was started. Cells were incubated for 48 hrs.
Rescue assays
Cell number was assessed as described above with minor modifications. Cells were plated in triplicates in a 96-well plate at a density of 2,000 cells/well in DMEM/F-12 media supplemented with 10% FBS. 24 hrs after plating, cells were treated with either CB-839 (50 nM) or vehicle (DMSO), menadione (20 µM) or vehicle (DMSO), or H2O2 (100 µM) or vehicle (water) and either N-acetyl-cysteine (NAC) (10 mM), or dimethyl-α-ketoglutarate (DMKG) (0.5 mM), or pyruvate (0.5 mM and 1 mM). Both male and female cells were around 50% confluent when treatment was started. Cells were incubated for 48 hrs.
A 1 M NAC stock solution was prepared fresh before each experiment. 188 mg NaHCO3 and 326 mg NAC were dissolved in 2 mL molecular grade water. pH was adjusted to 7.0 and solution was filter sterilized with a 70 µm filter. An 800 mM pyruvate stock solution in media was prepared fresh and filter sterilized with a 70 µm filter before each experiment. DMKG was directly added to the media.
Glutamine consumption assay
Sample preparation
Cells were plated in a 6-well plate at a density of 50,000 cells/well in DMEM/F-12 media supplemented with 10% FBS. 6 technical replicates were plated. 24 hrs after plating, media was changed to DMEM/F-12 (USBiological) containing 1.2 g/L sodium bicarbonate, 10 mM HEPES, 10% dialyzed FBS, 10 mM glucose, and 4.5 mM glutamine. Cells were incubated for another 24 hrs. Then, media was collected from each well. Next, 20 µL of each media sample was added to 180 µL of a 2:2:1 methanol:acetonitrile:water (−20°C) mixture. Then, samples were vortexed for 1 min and placed at −20°C for 1 hr. Next, samples were centrifuged at 16,000 g at 4°C for 10 min and supernatant was transferred to LC glass vials. Samples were stored at −80°C until LC/MS analysis (see below).
Stable isotope tracing with LC/MS
Sample preparation
Cells were plated in 100 mm dishes at a density of 250,000 cells/well in DMEM/F-12 media supplemented with 10% FBS. 6 technical replicates were plated. 24 hrs after plating, media was changed to DMEM/F-12 (USBiological) containing 1.2 g/L sodium bicarbonate, 10 mM HEPES, 10% dialyzed FBS, 10 mM glucose, and 4.5 mM glutamine or [13C515N2]glutamine. Cells were incubated for another 24 hrs. Then metabolites were extracted at 75–85% confluence. For metabolite extraction, media was removed, cells were washed on dry ice 3 times with ice-cold 1X PBS, followed by 3 washing steps with water. Then, cells were scraped into methanol (−20°C) and transferred to Eppendorf tubes. Next, samples were dried at room temperature using a Labconco CentriVap Concentrator. Cells were lysed by adding a 2:2:1 methanol:acetonitrile:water (−20°C) mixture to the dried pellet. Then, cells were vortexed for 30 sec, placed into a liquid N2 bath for 1 min, and bath sonicated at 25°C for 10 min. This process was repeated 3 times. Then, samples were stored at −20°C overnight. The next day, samples were centrifuged at 12,000 g at 4°C for 10 min53. The supernatant was transferred to a new tube and dried at room temperature using a Labconco CentriVap Concentrator. Total protein content of pellets was measured by Pierce BCA protein assay kit, following the manufacturer’s microplate assay protocol.
Then, 1 µL of a 2:1 acetonitrile:water (−20°C) mixture was added to the dried pellet per 2.5 µg of protein. Next, the samples were bath sonicated at 25°C for 5 min and vortexed for 1 min. This process was repeated 2 times. Then, samples were stored at 4°C overnight. The next day, samples were centrifuged at 16,000 g at 4°C for 10 min and the supernatant was transferred to LC glass vials and stored at −80°C until LC/MS analysis.
LC/MS analysis
Ultra-high performance LC (UHPLC)/MS was performed with a Thermo Fisher Scientific Vanquish Horizon UHPLC system interfaced with a Thermo Fisher Scientific Orbitrap ID-X Tribrid Mass Spectrometer. Hydrophilic interaction liquid chromatography (HILIC) was conducted with a HILICON iHILIC-(P) Classic guard column (20 mm x 2.1 mm, 5 µm) connected to a HILICON iHILIC-(P) Classic HILIC column (100 mm x 2.1 mm, 5 µm). Mobile-phase solvents were composed of A=20 mM ammonium bicarbonate, 0.1% ammonium hydroxide and 2.5 µM medronic acid in water:acetonitrile (95:5) and B=2.5 µM medronic acid in acetonitrile:water (95:5). The column compartment was maintained at 45°C for all experiments. The following linear gradient was applied at a flow rate of 250 µL min-1: 0–1 min: 90% B, 1–12 min: 90–35% B, 12–12.5 min: 35–25% B, 12.5–14.5 min: 25% B. The column was re-equilibrated with 20 column volumes of 90% B. The injection volume was 3 µL for cell samples and 4 µL for media samples54.
Data were collected with the following settings: spray voltage, −3 kV; sheath gas, 35; auxiliary gas, 10; sweep gas, 1; ion transfer tube temperature, 250 °C; vaporizer temperature, 300 °C, mass range, 70–1000 Da, and two narrow-mass ranges, 270–400 Da and 550–700 Da; resolution, 240,000 (MS1), 30,000 (MS/MS); maximum injection time, 500 ms; isolation window, 1.5 Da; polarity, negative. LC/MS data were processed and analyzed with the open-source Skyline software55,56. Natural-abundance correction of 13C was performed with AccuCor57.
Stable isotope tracing with GC/MS
Sample preparation
Cells were plated in a 6-well plate at a density of 50,000 cells/well in DMEM/F-12 media supplemented with 10% FBS. 6 technical replicates were plated. For glutamine labeling experiments, media was changed to DMEM/F-12 (USBiological) containing 1.2 g/L sodium bicarbonate, 10 mM HEPES, 10% dialyzed FBS, 10 mM glucose, and 4.5 mM glutamine or [13C515N2]glutamine 24 hrs after plating. For glucose labeling experiments, media was changed to DMEM/F-12 (USBiological) containing 1.2 g/L sodium bicarbonate, 10 mM HEPES, 10% dialyzed FBS, 4.5 mM glutamine, and 10 mM glucose or [13C6]glucose 24 hrs after plating. Cells were incubated for another 24 hrs. Then metabolites were extracted at 75–85% confluence. For metabolite extraction, media was removed, and cells were washed on dry ice 2 times with ice-cold 1X PBS. Then, cells were scraped into 200 µL of an 80:20 methanol:water (−20°C) mixture. Two wells were combined per one technical replicate. Next, samples were centrifuged at 12,000 g at 4°C for 20 min. The supernatant was transferred to a flat bottom GC insert vial and dried at room temperature using a Labconco CentriVap Concentrator. Then, the dried pellets were derivatized with 14 µL of 20 mg/mL methoxyamine HCl in pyridine and incubated on at 38°C for 90 min. Immediately following the 90 min incubation, 56 µL of MTBSTFA (with 1% t-BDMCS were added and samples were incubated at 38°C for 30 min58.
GC/MS analysis
Derivatized samples were run on an Agilent 7890A GC coupled to an Agilent 5975C MS and data was acquired and analyzed in MSD ChemStation E.02.02.1431 (Agilent). The GC temperature program was set to: initial temperature: 60°C, fold for 0.65 min; temperature rate 1: 11.5°C/min to 201°C, hold for 0.65 min; temperature rate 2: 3.5°C/minute to 242°C, hold for 0 min; temperature rate 3: 16°C/min to 300°C, hold for 7 minutes. Data on all isotopologues was collected in SIM mode (see Table S3 for m/z monitored). Isotopologue peak areas in each sample were normalized in FluxFix 59 to isotopologues of unlabeled samples to adjust for the natural abundance of each isotopologue.
Solid-state NMR
Sample preparation
Experimental design and data analysis and interpretation were consistent with previous publications60,61. Cells were plated in 150 mm dishes at a density of 300,000 cells/dish in DMEM/F-12 media supplemented with 10% FBS. 24 hours after plating, media was changed to DMEM/F-12 (USBiological) containing 1.2 g/L sodium bicarbonate, 10 mM HEPES, 10% dialyzed FBS, 10 mM glucose, and 4.5 mM [13C515N2]glutamine. Cells were incubated for 48 hrs. Then, cells were trypsinized and centrifuged at 200 g at 4°C for 5 min. Media was aspirated and the cell pellet was washed with ice cold 1X PBS. Washing was repeated two times and the cell pellet was stored at −80°C. Before NMR analysis the cell pellet was dried in a lyophilizer at −20°C. The total weight of the male and female dried cell pellet submitted for solid-state NMR analysis was 20 mg each and was accumulated over three sample preparations.
Solid-state NMR spectrometer and pulse sequences
Experiments were performed at 12 Tesla with a transmission-line probe having a 12-mm long, 6-mm inner-diameter analytical coil, and a Chemagnetics/Varian ceramic spinning module. Samples were spun using a thin-wall Chemagnetics/Varian (Fort Collins, CO/Palo Alto, CA) 5-mm outer diameter-zirconia rotor at 8000 Hz, with the speed under active control and maintained to within ±2 Hz. A Tecmag Libra pulse programmer (Houston, TX) controlled the spectrometer. Two-kW American Microwave Technology (AMT) power amplifiers were used to produce radio-frequency pulses for 13C (125.7 MHz) and 15N (50.3 MHz). The 1H (500 MHz) radio-frequency pulses were generated by a 2-kW Creative Electronics tube amplifier driven by a 50-W AMT amplifier. The π-pulse lengths were 8 μs for both 13C and 1H, and 9 µs for 15N. Proton-carbon-matched cross-polarization transfers were made in 2 ms at 56 kHz. Proton dipolar decoupling was 100 kHz during data acquisition. The 13C spectra were obtained with 80-μs interrupted decoupling which enhances signals from non-protonated carbonyl carbons (175 ppm), and from protonated carbons whose dipolar coupling to protons is reduced by molecular motion (30 ppm). The 13C chemical-shift scale was referenced to tetramethylsilane. The 15N chemical-shift scale was referenced to solid ammonium sulfate. (To switch to liquid ammonia as a secondary reference, add 20 ppm.) All of the 13C and 15N NMR signals were due to labeling. Natural abundance signals were negligible.
mRNA expression data
RNA-Sequencing and mRNA microarray data were downloaded from cBioPortal for cancer genomics (http://www.cbioportal.org/)62,63.
For glioma cell lines and tumor cell lines, mRNA expression data from the Cancer Cell Line Encyclopedia 64,65 were downloaded and graphed as mRNA expression (RNA Seq RPKM (log2(value + 1))) data. Only cell lines that had a sex assigned in the Cancer Cell Line Encyclopedia were downloaded. Then, the sex of the cell lines was confirmed using the Cellosaurus data resource (https://web.expasy.org/cellosaurus/). Cell lines with a discrepancy between the sex assigned by the Cancer Cell Line Encyclopedia and by the Cellosaurus data resource were excluded. For tumor cell lines, only cell lines with a measurable IC5033 were included in the analysis.
For GBM patient samples, mRNA expression data from the TCGA Firehose Legacy GBM patient dataset66,67 were downloaded and graphed as mRNA expression (microarray) data. Only samples with a known sex and IDH wild-type status were included in the analysis. IDH status of each sample was determined using the IDH mutational status from the TCGA Firehose Legacy GBM patient dataset and the TCGA Merged Cohort of LGG and GBM dataset68.
For pan-cancer patient samples, mRNA expression data from the TCGA Pan-Cancer Atlas69 were downloaded and graphed as mRNA expression (RNA Seq RSEM (log2(value + 1))) data. Only samples with a known sex status were included in the analysis.
Western blotting
To extract protein, cells were harvested at 95% confluence. First, plates were washed once with ice cold 1X PBS, then cells were lysed with RIPA buffer, containing cOmplete Protease Inhibitor Cocktail, PhosStop Phosphatase Inhibitor Cocktail, and PMSF. Cell lysates were left on ice for 30 min, and vortexed every 5 min. Then, cells were stored at −80°C. Before use, samples were thawed and centrifuged at 12,000 g at 4°C for 20 min. Then, supernatant was transferred to a new Eppendorf tube. Protein concentration was measured with the DC Protein Assay, following the manufacturer’s microplate assay protocol, and using an Infinite 200 PRO microplate reader (Tecan) to measure absorbance. 25 µg protein/sample was combined with RIPA buffer, NuPAGE LDS Sample Buffer, and NuPAGE Sample Reducing Agent, and samples were incubated at 95°C for 10 min. Then, samples were run on a NuPAGE 10% Bis-Tris Gel in MOPS SDS Running Buffer. Next, proteins were transferred to an Odyssey nitrocellulose membrane. Membranes were blocked in Odyssey Blocking Buffer diluted 1:1 with PBS at room temperature for 1 hour. Membranes were incubated in primary antibody diluted in blocking buffer (1:2000 anti-GLS1; 1:1000 anti-pyruvate carboxylase) at 4°C overnight. The following day, membranes were washed with PBS-T and incubated in anti-β-actin antibody diluted in blocking buffer (1:60,000) for 1 hr at room temperature. Then, membranes were washed with PBS-T and incubated in secondary antibody diluted in blocking buffer for 45 min at room temperature (1:50,000 IRDye 680RD Donkey anti-Mouse, 1:5,000 IRDye 800CW Donkey anti-Rabbit). Membranes were washed with PBS-T again and proteins were visualized using a ChemiDoc MP Imaging System (Bio-Rad). Band intensity was quantified using Image Lab Software (Bio-Rad).
Targeted GC/MS
Sample preparation
Cells were plated in a 6-well plate at a density of 50,000 cells/well in DMEM/F-12 media supplemented with 10% FBS. 6 technical replicates were plated. 24 hrs after plating, cells were treated with CB-839 (50 nM) or vehicle (DMSO). After 24 hrs, media was removed and cells were washed on dry ice twice with ice-cold 1X PBS and scraped into 200 µL of a cold 80:20 methanol:water mixture together with 10 µL of a cocktail of labeled amino acids and TCA cycle internal standards. All internal standards concentrations were 2.5 mM, except for cystine which was 1.25 mM. Two wells were combined per one technical replicate. Next, cell suspension was centrifuged at 12,000 g for 20 min at 4°C. The supernatant was transferred to a flat bottom GC insert vial and dried at room temperature using a Labconco CentriVap Concentrator. The cell debris pellet was stored at −80°C for protein quantification. Then, the dried pellets were derivatized following the method described in58. Briefly, the dried pellets were derivatized with 20 µL of 20 mg/mL methoxyamine HCl in pyridine and incubated on at 38°C for 90 min. Immediately following the 90 min incubation, 80 µL of MTBSTFA (with 1% t-BDMCS were added and samples were incubated at 38°C for 30 min.
To assess protein concentration in each sample, cell debris pellets were resuspended in 200 µL of RIPA lysis buffer cocktail, containing protease inhibitor cocktail, 2 mM PMSF, and 1 mM sodium orthovanadate. Cell lysates were left on ice for 30–60 min, and periodically vortexed vigorously. Protein concentration was measured with the Pierce BCA assay, following the manufacturer’s microplate assay protocol, and using a Synergy HT microplate reader (BioTek) to measure absorbance.
GC/MS analysis
Derivatized samples were run on an Agilent 7890A GC coupled to an Agilent 5975C MS and data was acquired and analyzed in MSD ChemStation E.02.02.1431 (Agilent). The GC temperature program was set to: initial temperature: 80°C, hold for 2 min; temperature rate 1: 10°C/min to 300°C, hold for 6 min. Data was collected in SIM mode (see Table S6 for ions). Metabolite peak areas in each sample were quantified relative to nmole internal standard amounts and normalized to µg protein.
Glutathione assay
Oxidized, reduced, and total glutathione was assessed using a spectrophotometric recycling assay. Briefly, cells were plated in 100 mm dishes at a density of 250,000 cells/dish in DMEM/F-12 media supplemented with 10% FBS. 3 technical replicates were plated. 24 hrs after plating, cells were treated with vehicle, CB-839 (50 nM), or BSO (10 µM). Cells were incubated for 24 hrs. Then, cells were scraped into 5% 5-sulfosalicylic acid and frozen. Both male and female cells were 70–80% confluent at time of cell harvest. Total glutathione was quantified using the 5,5′-dithiobis-2-nitrobenzoic acid recycling assay as described previously by Anderson70. Reduced and oxidized glutathione were distinguished using 2-vinylpyridine (2VP) mixed 1:1 (vol:vol) with ethanol as previously described by Griffith71. 20 µL of 2VP/ethanol was added for each 100 µL sample. All sample data were normalized to protein content, using the method of Lowry72.
DHE stain
Superoxide accumulation was assessed using the dihydroethidium (DHE) stain. Briefly, cells were plated in duplicates in a 96-well plate at a density of 2000 cells/well in DMEM/F-12 media supplemented with 10% FBS. 24 hrs after plating, cells were treated with vehicle, CB-839 (50 nM), or BSO (10 µM). Cells were treated for 24 hrs. Then, media was carefully removed from wells and cells were incubated in fresh culture media containing 5 µM DHE (10 mM stock solution in DMSO) at 37°C for 30 min (protected from light). Then, media was carefully removed, and cells were fixed with ice cold 4% paraformaldehyde at 4°C for 15 min. Next, plates were washed 3 times with 1X PBS and nuclei were counterstained with Hoechst 33342 (1 µg/mL in PBS). Plates were stored at 4°C protected from light until analysis. Images were acquired using an Operetta CLS High-Content Analysis System (PerkinElmer) and the Harmony High-Content Imaging and Analysis Software. DHE staining intensity in the nucleus was quantified by an individual blinded to treatment conditions by ImageJ analysis of mean fluorescent intensity. For each experiment, staining across 3 fields of view per well was averaged across 2 wells per treatment condition. Each field of view contained at least 20 cells.
Flow Cytometry
Cells were harvested and plated in 150 mm dishes at a density of 300,000 cells/dish in DMEM/F-12 media supplemented with 10% FBS. 24 hrs after plating, cells were treated with vehicle, CB-839 (50 nM), or BSO (10 μM). 48 hrs after treatment, cells were harvested with Accutase, counted, and aliquoted to be stained with either propidium iodide (cell cycle analysis) or Annexin V, Pacific Blue Conjugate (apoptotic cell detection). For propidium iodide staining, 1.0 x 106 cells per condition were fixed for at least 15 min in 70% EtOH, washed with 1X PBS/1% FBS, strained through a 70 μm filter, and resuspended in 200 μL of propidium iodide solution containing 15 μL 500X propidium iodide, 9.9 mL 1X PBS, 100 μL 10% Triton-X-100, and 25 μL RNAse A. Cells were then incubated in the dark at 37°C for 20 min and then kept on ice (protected from light) until analysis. For Annexin V staining, cells were first resuspended at a concentration of 1.0 x 106 cells in 1 mL of 1X Annexin Binding Buffer. 100 μL of each suspension was then transferred to a 5 mL flow cytometry tube, to which 5μL of Annexin V, Pacific Blue Conjugate were added. Cells were incubated for 15 min at room temperature in the dark, resuspended with an additional 400μL of 1X Annexin Binding Buffer, and kept on ice (protected from light) until analysis. All samples were run on the same ZE5 Cell Analyzer (Bio-Rad) with unstained/untreated and single positive controls. Apoptosis and cell cycle analyses were each performed three times on independent sample preparations.
FlowJo software was utilized to analyze all flow cytometry data. For cell cycle analysis, lines were drawn to demarcate which regions of each propidium iodide histogram corresponded to the following phases of the aneuploid cell cycle: G0/G1 for the diploid population, S phase for the diploid population, G2/M for the diploid population + G0/G1 for the tetraploid population (these peaks are indistinguishable), S phase for the tetraploid population, and G2/M for the tetraploid population (for representative cell cycle histograms see Fig. S4D and S5C). The percentage of the entire cell population in each phase was calculated according to the area of each demarcated region. For apoptosis analysis, Annexin V, Pacific Blue Conjugate histograms for each condition were compared by overlaying them with one another. Positive shift in staining intensity correlates with an increased number of cells stained with the Annexin V, Pacific Blue Conjugate.
Knockdown of pyruvate carboxylase (PC) in transformed astrocytes
PC knockdown (PC-KD) cell lines were generated from male and female transformed astrocytes by transducing them with lentiviral short hairpin RNAs (shRNAs) against murine Pc. Four different shRNAs against Pc were used. shRNAs were purchased from Sigma (St. Louis, USA) and the target sequences are as follows: shRNA 1: CCCTTCAGCTATTTGTCCTTT, shRNA 2: CCGAGGTGTAAAGACCAACAT, shRNA 3: GCGTCTGGAGTATAAGCCTAT, shRNA 4: GCACTACTTCATCGAGGTCAA. Transformed astrocyte control cell lines were made by viral transduction with a lentiviral shRNA against LacZ. Its target sequence is GTCGGCTTACGGCGGTGATTT and was provided by the McDonnell Genome Institute at Washington University in St. Louis. All shRNAs were provided in the pLKO.1-puromycin lentivirus plasmid vector.
Lentiviral production
Lentivirus was produced by transfection of HEK 293T cells with the FuGENE 6 Transfection Reagent according to the manufacturers protocol. Briefly, HEK 293T cells were plated in a 6-well plate at a density of 300,00 cells/well in DMEM/F-12 media supplemented with 10% FBS. 24 hrs after plating, the media was changed to OptiMEM media, containing FuGENE 6 Transfection Reagent (FuGENE:DNA ratio of 2.4:1), 2.5 µg lentivirus of interest and 2.5 µg and 250 ng of the lentiviral packaging vectors Delta8.9 and VSV-G, respectively. Cells were incubated for 5 hrs. Then, media was changed to DMEM/F-12 media supplemented with 10% FBS and 0.6 mM sodium butyrate. 48 hrs post-transfection, lentiviral media was collected, filtered with a 0.45μm syringe filter, and aliquoted and stored at −80°C.
Lentiviral transduction
24 hrs prior to viral transduction, transformed astrocytes were plated in a 6-well plate at a density of 100,00 cells/well in DMEM/F-12 media supplemented with 10% FBS. After 24 hrs, media was changed to DMEM/F-12 media supplemented with 10% FBS, containing lentiviral media (viral media:culture media ratio of 1:3) and 5 µg/mL polybrene and cells were incubated for 48 hrs. Then, successfully transduced cells were selected with puromycin (2.5 µg/mL) for 6 days. PC knockdown was confirmed via western blot analysis after the newly established cell lines were passaged three times.
Quantification and Statistical Analysis
Data representation and statistical analysis
Data in Figure 1 were analyzed using R version 4.1.173, the R/Bioconductor software package limma version 3.48.351, and GraphPad Prism software version 9.0. Data were graphed using GraphPad Prism software version 9.0. For statistical analysis of sex-specific enrichment of super-pathways, sub-pathways, and individual metabolites data were analyzed using LIMMA where the moderated two sample t-test was used to compare metabolite expression between sex, and p-values were adjusted with the false discovery rate (FDR) approach using the Storey’s q-value method. For statistical analysis of nonrandom associations between male and female sample enrichment or metabolite enrichment in males or female a two-tailed Fisher’s exact test was applied.
Data in Fig. 2–6 were graphed and analyzed using GraphPad Prism software version 9.0. Data are expressed as mean ± SEM or median ± 95% confidence interval (specified in figure legends). All data shown as normalized data were normalized independently within each experiment. Each technical replicate was divided by the average of the control treatment (glutamine deprivation and drug treatments), the male value (glutamine isotope tracing experiments), or the female value (glucose isotope tracing experiments and glutamine uptake assay). For statistical analysis of two groups for which one was the normalized group, raw data was analyzed by a linear mixed effects model for which experiments were considered repeated measurements. For all other analyses between two groups, test for normality distribution was first performed, using the D'Agostino-Pearson omnibus K2 test. If samples had a normal distribution an unpaired, two-tailed, parametric t-test was performed. If samples did not have a normal distribution an unpaired, two-tailed, non-parametric Mann-Whitney test was performed. Metabolite data and cell cycle analysis data were compared between two groups using two-tailed, parametric t-tests and raw p-values were adjusted using the FDR approach using the two-stage step-up method of Benjamini, Krieger, and Yekutielii. Dose-response curve concentrations were log10 transformed, curves were fit to normalized response versus log(inhibitor) concentration using the 4-parameters dose response logistic model, and absolute IC50 values were calculated when appropriate. Ordinary two-way ANOVAS were performed to compare the dose response curve data between sex. Raw p-values were adjusted using Dunnetťs multiple comparisons test when more than one cell line was compared to a control cell line. For correlation analyses, nonparametric Spearman correlation coefficient was calculated, and linear regression was fit to model the linear relationship between two markers. In all cases an FDR adjusted p-value or raw p-value<0.05 was considered statistically significant.
All experiments using male and female transformed astrocytes were carried out at least three times using male and female transformed astrocytes from one cell line (one lot). All dose-response curves in Fig. 1–5 were carried out three times using three separately established cell lines – each consisting of a corresponding male and female line generated at the same time (three lots). Dose-response curves in Fig. 6 were carried out three times using one control cell line and four separately established PC knock down cell lines (same lot of cells, four different shRNAs).
Supplementary Material
Z-scores, mean difference and adjusted p-values of individual metabolites, sub- and super pathways, related to Figure 1
Isotopologue enrichment of [13C515N2]glutamine in male and female transformed astrocytes, related to Figure 3
Sex and tissue annotation of human tumor cell lines, related to Figure 4
Isotopologue enrichment of [13C6]glucose in male and female transformed astrocytes, related to Figure 6
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GLS1 (KGA/GAC) polyclonal antibody | Proteintech | Cat# 12855-1-AP; RRID:AB_2110381 |
| anti-pyruvate carboxylase polyclonal antibody | Proteintech | Cat# 16588-1-AP; RRID:AB_1851513 |
| anti-β-actin monoclonal antibody | Sigma | Cat# A1978; RRID:AB_476692 |
| IRDye 680RD donkey anti-mouse | LI-COR | Cat# 926-68072; RRID:AB_10953628 |
| IRDye 800CW donkey anti-rabbit | LI-COR | Cat# 926-32213; RRID:AB_621848 |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| [18F]fluoroglutamine | Memorial Sloan Kettering Cancer Center’s Radiochemistry and Molecular Imaging Probe Core Facility | N/A |
| CB-839 | Selleckchem | Cat# S7655 |
| BPTES | Selleckchem | Cat# S7753 |
| Menadione | Selleckchem | Cat# S1949 |
| Buthionine sulfoximine | Sigma | Cat# 5.08228 |
| DMEM/F-12 | USBiological | Cat# D9807-06 |
| Dialyzed FBS | Gibco | Cat# 26400044 |
| D-glucose | Corning | Cat# 25-037-Cl |
| L-glutamine | Gibco | Cat# 25030081 |
| Sulforhodamine B | Sigma | Cat# S1402 |
| H2O2 | Sigma | Cat# H1009 |
| N-acetyl-cysteine | Sigma | Cat# A9165 |
| Dimethyl-α-ketoglutarate | Sigma | Cat# 349631 |
| Sodium pyruvate | Sigma | Cat# P5280 |
| [13C6]glucose | Cambridge Isotope Laboratories | Cat# CLM-1396-PK |
| [13C515N2]glutamine | Cambridge Isotope Laboratories | Cat# CNLM-1275-H-PK |
| Methanol (MS grade) | Thermo Fisher Scientific | Cat# A456 |
| Water (MS grade) | Sigma | Cat# 900682 |
| Acetonitrile (MS grade) | Sigma | Cat# 900667 |
| Methoxyamine hydrochloride | Sigma | Cat# 226904 |
| Pyridine anhydrous 99.8% | Sigma | Cat# 270970 |
| MTBSTFA (with 1% t-BDMCS) | Sigma | Cat# M-108 |
| cOmplete protease inhibitor cocktail | Roche | Cat# 11697498001 |
| PhosStop phosphatase inhibitor cocktail | Roche | Cat# 4906845001 |
| NuPAGE LDS sample buffer | Thermo Fisher | Cat# NP0007 |
| NuPAGE sample reducing agent | Thermo Fisher | Cat# NP0004 |
| MOPS SDS running buffer | Thermo Fisher | Cat# NP0001 |
| Odyssey nitrocellulose membrane | LI-COR | Cat# 926-31092 |
| Intercept blocking buffer PBS | LI-COR | Cat# 927-70003 |
| Hygromycin B | Goldbio | Cat# H-270-1 |
| D-luciferin firefly | Biosynth Carbosynth | Cat# L-8220 |
| hEGF | Sigma | Cat# E9644 |
| Matrigel basement membrane matrix | Corning | Cat# 354234 |
| [13C6]citric acid | Cambridge Isotope Laboratories | Cat# CLM-9021-PK |
| [13C4]α-ketoglutaric acid sodium salt | Cambridge Isotope Laboratories | Cat# CLM-4442 |
| [13C4]succinic acid | Cambridge Isotope Laboratories | Cat# CLM-1571 |
| [13C4]fumaric acid | Cambridge Isotope Laboratories | Cat# CLM-1529 |
| [13C4]L-malic acid | Cambridge Isotope Laboratories | Cat# CLM-8065 |
| Stable isotope labeled canonical amino acid mix | Cambridge Isotope Laboratories | Cat# MSK-CAA-1 |
| RIPA lysis buffer cocktail | Santa Cruz | Cat# sc-24948 |
| 5-sulfosalicylic acid | Sigma | Cat# S7408 |
| Dihydroethidium stain | Thermo Fisher | Cat# D11347 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat# 62249 |
| Propidium iodide | Sigma | Cat# P4170 |
| Annexin V, pacific blue conjugate | Thermo Fisher Scientific | Cat# A35122 |
| Triton-X-100 | Sigma | Cat# 9002-93-1 |
| RNAse A, DNase and protease-free | Thermo Fisher Scientific | Cat# EN0531 |
| Annexin binding buffer | Thermo Fisher Scientific | Cat# V13246 |
| OptiMEM | Gibco | Cat# 31985070 |
| FuGENE 6 Transfection Reagent | Promega | Cat# E2691 |
| Polybrene | Santa Cruz | Cat# SC-1342200 |
| Sodium butyrate | Sigma | Cat# B5887 |
| Puromycin | Sigma | Cat# P8833 |
| Critical commercial assays | ||
| DC protein assay kit | Bio-Rad | Cat# 5000111 |
| Pierce BCA assay kit | Thermo Fisher Scientific | Cat# 23225 and Cat# 23227 |
| Deposited data | ||
| Experimental models: Cell lines | ||
| Nf1−/−; DNp53 astrocytes | Rubin Laboratory | N/A |
| Nf1−/−; DNp53; PC KD astrocytes | This manuscript | N/A |
| HEK 293T cells | Kroll Laboratory | N/A |
| Experimental models: Organisms/strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| pLKO.1-puromycin-LacZ shRNA | McDonnell Genome Institute at Washington University in St. Louis | NA |
| pLKO.1-puromycin-PC_1 shRNA | Sigma | Cat# TRCN0000112425 |
| pLKO.1-puromycin-PC_2 shRNA | Sigma | Cat# TRCN0000112426 |
| pLKO.1-puromycin-PC_3 shRNA | Sigma | Cat# TRCN0000112427 |
| pLKO.1-puromycin-PC_4 shRNA | Sigma | Cat# TRCN0000112428 |
| Software and algorithms | ||
| R v4.1.1 | R Core Team, 2021 | https://www.r-project.org/ |
| Limma v3.48.3 | Bioconductor |
https://bioconductor.org/packages/release/bioc/html/limma.html
DOI:10.18129/B9.bioc.limma |
| PET VCAR software | GE Healthcare | https://www.gehealthcare.com/products/advanced-visualization/all-applications/pet-vcar |
| GraphPad Prism v9 | GraphPad | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| Skyline software | MacLean et al., 2009 | https://skyline.ms/project/home/software/Skyline/begin.view |
| ChemStation E.02.02.1431 | Agilent | https://www.agilent.com/en/product/software-informatics/analytical-software-suite |
| FluxFix: Isotopologue Analysis Tool v0.1 | Trefely et al., 2016 | http://fluxfix.science/http://fluxfix.science/ |
| Image Lab Software v6.1 | Bio-Rad |
https://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z; RRID:SCR_014210 |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus |
| Harmony High-Content Imaging and Analysis Software v4.5 | PerkinElmer | https://www.perkinelmer.com/product/harmony-4-9-office-license-hh17000010 |
| ImageJ v1.53a | Schneider et al., 2012 | https://imagej.nih.gov/ij/index.html; RRID:SCR_003070 |
| FlowJo v10.7.2 | FlowJo | https://www.flowjo.com/solutions/flowjo/downloads; RRID:SCR_008520 |
| Other | ||
Highlights:
Metabolite abundance differs between male and female gliomas
Glutaminase 1 inhibition attenuates growth in male, but not female, glioma cells
Male glioma cells depend upon glutaminase 1 for TCA cycle replenishment
Pyruvate carboxylase is required for glutamine-independent growth in female cells
Context and Significance:
Glioblastoma is the most common malignant brain tumor in adults. The prognosis of glioblastoma is extremely poor and new treatment strategies are needed. In males, glioblastoma is more common and associated with poorer outcomes; as such, understanding the mechanisms that underlie this sex disparity could improve treatment strategies for male and female patients. Cancer cells require metabolic reprogramming to fuel their metabolic demands and targeting cancer metabolism is an effective treatment strategy in multiple cancers; although sex differences in normal metabolism are well described, it is largely unknown whether they persist in cancerous tissue. Here, Sponagel at al. show that clinically important sex differences exist in glioblastoma metabolism and that targeting metabolic elements may have different effects in male and female patients.
Acknowledgments:
This work was supported by the Cancer Biology Pathway Molecular Oncology Training Grant NIH T32CA113275 (JSp), the NIH grants R01 CA174737-06 (JBR), K99/R00 CA218869 (JEI), R21 CA242221 (JEI), R35 ES028365 (GJP), P01 CA217797 (DRS), Joshua’s Great Things (JBR), the Alvin J. Siteman Cancer Center Investment Program / The Foundation for Barnes-Jewish Hospital (JBR, JEI) and the Barnard Research Fund (JBR, JEI). SK was supported in part by the NIH/NCI Paul Calabresi Career Development Award for Clinical Oncology K12 CA184746. The Radiation and Free Radical Research Core is supported by NIH/NCI Center Core Grant P30 CA086862. MSK core facilities are supported by NIH/NCI Cancer Center Support Grant P30 CA008748. The authors gratefully acknowledge the TREC Training Workshop R25CA203650 (PI: Melinda Irwin).
The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital in St. Louis for the use of the Siteman Flow Cytometry Core. The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant P30 CA091842. In particular, we thank William Eades for his help with flow cytometry. We thank the Biomedical Mass Spectrometry Core at Washington University School of Medicine for providing GC/MS equipment. The core is supported by the NIH grant R24 GM136766. We thank Michael McCormick and the Radiation and Free Radical Research Core at the University of Iowa for their help with assessing cellular GSH and GSSG levels.
Funding:
This work was supported by NIH grants, Joshua’s Great Things, the Siteman Investment Program, and the Barnard Research Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest:
GJP is a scientific advisor for Cambridge Isotope Laboratories. The Patti laboratory has a collaboration agreement with Agilent Technologies and Thermo Fisher Scientific.
Inclusion and diversity statement
We worked to ensure gender balance and ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. We worked to ensure sex balance in the selection of non-human subjects. We worked to ensure diversity in experimental samples through the selection of the cell lines. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
References:
- 1.Pavlova NN, and Thompson CB (2016). The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23, 27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin JB, Lagas JS, Broestl L, Sponagel J, Rockwell N, Rhee G, Rosen SF, Chen S, Klein RS, Imoukhuede P, et al. (2020). Sex differences in cancer mechanisms. Biol. Sex Differ 11, 17. doi: 10.1186/s13293-020-00291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tramunt B, Smati S, Grandgeorge N, Lenfant F, Arnal J-F, Montagner A, and Gourdy P (2020).Sex differences in metabolic regulation and diabetes susceptibility. Diabetologia 63, 453–461. doi: 10.1007/s00125-019-05040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy E, Amanakis G, Fillmore N, Parks RJ, and Sun J (2017). Sex differences in metabolic cardiomyopathy. Cardiovasc. Res 113, 370–377. doi: 10.1093/cvr/cvx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen GK, Mellnick VM, Yim AK-Y, Salter A, and Ippolito JE (2018). Synergy of Sex Differences in Visceral Fat Measured with CT and Tumor Metabolism Helps Predict Overall Survival in Patients with Renal Cell Carcinoma. Radiology 287, 884–892. doi: 10.1148/radiol.2018171504. [DOI] [PubMed] [Google Scholar]
- 6.Lopes-Ramos CM, Chen C-Y, Kuijjer ML, Paulson JN, Sonawane AR, Fagny M, Platig J, Glass K, Quackenbush J, and DeMeo DL (2020). Sex Differences in Gene Expression and Regulatory Networks across 29 Human Tissues. Cell Rep 31, 107795. doi: 10.1016/j.celrep.2020.107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Y, Rattray NJW, Zhang Q, Mironova V, Santos-Neto A, Hsu K-S, Rattray Z, Cross JR, Zhang Y, Paty PB, et al. (2020). Sex Differences in Colon Cancer Metabolism Reveal A Novel Subphenotype. Sci. Rep 10, 4905. doi: 10.1038/s41598-020-61851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ippolito JE, Yim AKY, Luo J, Chinnaiyan P, and Rubin JB (2017). Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI insight 2 doi: 10.1172/jci.insight.92142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgren G, Liang L, Adami H-O, and Chang ET (2012). Enigmatic sex disparities in cancer incidence. Eur. J. Epidemiol 27, 187–196. doi: 10.1007/s10654-011-9647-5. [DOI] [PubMed] [Google Scholar]
- 10.Zheng D, Trynda J, Williams C, Vold JA, Nguyen JH, Harnois DM, Bagaria SP, McLaughlin SA, and Li Z (2019). Sexual dimorphism in the incidence of human cancers. BMC Cancer 19, 684. doi: 10.1186/s12885-019-5902-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siegel RL, Miller KD, Fuchs HE, and Jemal A (2021). Cancer Statistics, 2021. CA. Cancer J. Clin 71, 7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 12.Dong M, Cioffi G, Wang J, Waite KA, Ostrom QT, Kruchko C, Lathia JD, Rubin JB, Berens ME, Connor J, et al. (2020). Sex Differences in Cancer Incidence and Survival: A Pan-Cancer Analysis. Cancer Epidemiol. Biomarkers Prev 29, 1389–1397. doi: 10.1158/1055-9965.EPI-20-0036. [DOI] [PubMed] [Google Scholar]
- 13.Krumsiek J, Mittelstrass K, Do KT, Stückler F, Ried J, Adamski J, Peters A, Illig T, Kronenberg F, Friedrich N, et al. (2015). Gender-specific pathway differences in the human serum metabolome. Metabolomics 11, 1815–1833. doi: 10.1007/s11306-015-0829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prabhu AH, Kant S, Kesarwani P, Ahmed K, Forsyth P, Nakano I, and Chinnaiyan P (2019). Integrative cross-platform analyses identify enhanced heterotrophy as a metabolic hallmark in glioblastoma. Neuro. Oncol 21, 337–347. doi: 10.1093/neuonc/noy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunphy MPS, Harding JJ, Venneti S, Zhang H, Burnazi EM, Bromberg J, Omuro AM, Hsieh JJ, Mellinghoff IK, Staton K, et al. (2018). In Vivo PET Assay of Tumor Glutamine Flux and Metabolism: In-Human Trial of 18 F-(2 S ,4 R )-4-Fluoroglutamine. Radiology 287, 667–675. doi: 10.1148/radiol.2017162610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grkovski M, Goel R, Krebs S, Staton KD, Harding JJ, Mellinghoff IK, Humm JL, and Dunphy MPS (2020). Pharmacokinetic Assessment of 18 F-(2 S, 4 R )-4-Fluoroglutamine in Patients with Cancer. J. Nucl. Med 61, 357–366. doi: 10.2967/jnumed.119.229740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venneti S, Dunphy MP, Zhang H, Pitter KL, Zanzonico P, Campos C, Carlin SD, La Rocca G, Lyashchenko S, Ploessl K, et al. (2015). Glutamine-based PET imaging facilitates enhanced metabolic evaluation of gliomas in vivo. Sci. Transl. Med 7, 274ra17–274ra17. doi: 10.1126/scitranslmed.aaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahari AK, Chien D, Azadi JR, and Wahl RL (2014). Optimum Lean Body Formulation for Correction of Standardized Uptake Value in PET Imaging. J. Nucl. Med 55, 1481–1484. doi: 10.2967/jnumed.113.136986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kfoury N, Sun T, Yu K, Rockwell N, Tinkum KL, Qi Z, Warrington NM, McDonald P, Roy A, Weir SJ, et al. (2018). Cooperative p16 and p21 action protects female astrocytes from transformation. Acta Neuropathol. Commun 6, 12. doi: 10.1186/s40478-018-0513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun T, Warrington NM, Luo J, Brooks MD, Dahiya S, Snyder SC, Sengupta R, and Rubin JB (2014). Sexually dimorphic RB inactivation underlies mesenchymal glioblastoma prevalence in males. J. Clin. Invest 124, 4123–4133. doi: 10.1172/JCI71048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bansal A, and Simon MC (2018). Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol 217, 2291–2298. doi: 10.1083/jcb.201804161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, and Haigis MC (2017). Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science (80-. ) 358, 941–946. doi: 10.1126/science.aam9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fendt S-M, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Heiden MG Vander, and Stephanopoulos G (2013). Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nat. Commun 4, 2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boysen G, Jamshidi-Parsian A, Davis MA, Siegel ER, Simecka CM, Kore RA, Dings RPM, and Griffin RJ (2019). Glutaminase inhibitor CB-839 increases radiation sensitivity of lung tumor cells and human lung tumor xenografts in mice. Int. J. Radiat. Biol 95, 436–442. doi: 10.1080/09553002.2018.1558299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wicker CA, Hunt BG, Krishnan S, Aziz K, Parajuli S, Palackdharry S, Elaban WR, Wise-Draper TM, Mills GB, Waltz SE, et al. (2021). Glutaminase inhibition with telaglenastat (CB-839) improves treatment response in combination with ionizing radiation in head and neck squamous cell carcinoma models. Cancer Lett 502, 180–188. doi: 10.1016/j.canlet.2020.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oizel K, Yang C, Renoult O, Gautier F, Do QN, Joalland N, Gao X, Ko B, Vallette F, Ge W-P, et al. (2020). Glutamine uptake and utilization of human mesenchymal glioblastoma in orthotopic mouse model. Cancer Metab 8, 9. doi: 10.1186/s40170-020-00215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin H, Wang S, Zaal EA, Wang C, Wu H, Bosma A, Jochems F, Isima N, Jin G, Lieftink C, et al. (2020). A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. Elife 9. doi: 10.7554/eLife.56749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Momcilovic M, Bailey ST, Lee JT, Fishbein MC, Braas D, Go J, Graeber TG, Parlati F, Demo S, Li R, et al. (2018). The GSK3 Signaling Axis Regulates Adaptive Glutamine Metabolism in Lung Squamous Cell Carcinoma. Cancer Cell 33, 905–921.e5. doi: 10.1016/j.ccell.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardona C, Sánchez-Mejías E, Dávila JC, Martín-Rufián M, Campos-Sandoval JA, Vitorica J, Alonso FJ, Matés JM, Segura JA, Norenberg MD, et al. (2015). Expression of Gls and Gls2 glutaminase isoforms in astrocytes. Glia 63, 365–382. doi: 10.1002/glia.22758. [DOI] [PubMed] [Google Scholar]
- 30.Szeliga M, Sidoryk M, Matyja E, Kowalczyk P, and Albrecht J (2005). Lack of expression of the liver-type glutaminase (LGA) mRNA in human malignant gliomas. Neurosci. Lett 374, 171–173. doi: 10.1016/j.neulet.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 31.van den Heuvel APJ, Jing J, Wooster RF, and Bachman KE (2012). Analysis of glutamine dependency in non-small cell lung cancer. Cancer Biol. Ther 13, 1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, Yin Y, Li Y, Chen X, Chang Y, Zhang H, Liu J, Beasley J, McCaw P, Zhang H, et al. (2021). A glutaminase isoform switch drives therapeutic resistance and disease progression of prostate cancer. Proc. Natl. Acad. Sci 118, e2012748118. doi: 10.1073/pnas.2012748118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daemen A, Liu B, Song K, Kwong M, Gao M, Hong R, Nannini M, Peterson D, Liederer BM, de la Cruz C, et al. (2018). Pan-Cancer Metabolic Signature Predicts Co-Dependency on Glutaminase and De Novo Glutathione Synthesis Linked to a High-Mesenchymal Cell State. Cell Metab 28, 383–399.e9. doi: 10.1016/j.cmet.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Yang W, Warrington NM, Taylor SJ, Whitmire P, Carrasco E, Singleton KW, Wu N, Lathia JD, Berens ME, Kim AH, et al. (2019). Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci. Transl. Med 11, 1–15. doi: 10.1126/scitranslmed.aao5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Ahn YJ, and Asmis R (2020). Sexual dimorphism in glutathione metabolism and glutathione-dependent responses. Redox Biol 31, 101410.doi: 10.1016/j.redox.2019.101410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobler L, Zhang H, Suri P, Park C, and Timmerman LA (2018). xCT inhibition sensitizes tumors to γ-radiation via glutathione reduction. Oncotarget 9, 32280–32297. doi: 10.18632/oncotarget.25794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng T, Sudderth J, Yang C, Mullen AR, Jin ES, Mates JM, and DeBerardinis RJ (2011). Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci 108, 8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen P-H, Cai L, Huffman K, Yang C, Kim J, Faubert B, Boroughs L, Ko B, Sudderth J, McMillan EA, et al. (2019). Metabolic Diversity in Human Non-Small Cell Lung Cancer Cells. Mol. Cell 76, 838–851.e5. doi: 10.1016/j.molcel.2019.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Suwailem E, Abdi S, and El-Ansary A (2018). Sex differences in the glutamate signaling pathway in juvenile rats. J. Neurosci. Res 96, 459–466. doi: 10.1002/jnr.24144. [DOI] [PubMed] [Google Scholar]
- 40.Davis AM, Ward SC, Selmanoff M, Herbison AE, and McCarthy MM (1999). Developmental sex differences in amino acid neurotransmitter levels in hypothalamic and limbic areas of rat brain. Neuroscience 90, 1471–1482. doi: 10.1016/S0306-4522(98)00511–9. [DOI] [PubMed] [Google Scholar]
- 41.Tiffin GJ, Rieger D, Betteridge KJ, Yadav BR, and King WA (1991). Glucose and glutamine metabolism in pre-attachment cattle embryos in relation to sex and stage of development. J. Reprod. Fertil 93, 125–32. [DOI] [PubMed] [Google Scholar]
- 42.Ray PF, Conaghan J, Winston RM, and Handyside AH (1995). Increased number of cells and metabolic activity in male human preimplantation embryos following in vitro fertilization. J. Reprod. Fertil 104, 165–71. [DOI] [PubMed] [Google Scholar]
- 43.Hedrington MS, and Davis SN (2015). Sexual Dimorphism in Glucose and Lipid Metabolism during Fasting, Hypoglycemia, and Exercise. Front. Endocrinol. (Lausanne) 6. doi: 10.3389/fendo.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamont LS, McCullough AJ, and Kalhan SC (2003). Gender differences in the regulation of amino acid metabolism. J. Appl. Physiol 95, 1259–1265. doi: 10.1152/japplphysiol.01028.2002. [DOI] [PubMed] [Google Scholar]
- 45.Kfoury N, Qi Z, Prager BC, Wilkinson MN, Broestl L, Berrett KC, Moudgil A, Sankararaman S, Chen X, Gertz J, et al. (2021). Brd4-bound enhancers drive cell-intrinsic sex differences in glioblastoma. Proc. Natl. Acad. Sci 118, e2017148118. doi: 10.1073/pnas.2017148118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dasgupta B (2005). Neurofibromin Regulates Neural Stem Cell Proliferation, Survival, and Astroglial Differentiation In Vitro and In Vivo. J. Neurosci 25, 5584–5594. doi: 10.1523/JNEUROSCI.4693-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J, Deng X, and Disteche CM (2008). Sex-Specific Expression of the X-Linked Histone Demethylase Gene Jarid1c in Brain. PLoS One 3, e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clapcote SJ, and Roder JC (2005). Simplex PCR assay for sex determination in mice. Biotechniques 38, 702–706. doi: 10.2144/05385BM05. [DOI] [PubMed] [Google Scholar]
- 49.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, and Ellison DW (2016). The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131, 803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 50.Chinnaiyan P, Kensicki E, Bloom G, Prabhu A, Sarcar B, Kahali S, Eschrich S, Qu X, Forsyth P, and Gillies R (2012). The Metabolomic Signature of Malignant Glioma Reflects Accelerated Anabolic Metabolism. Cancer Res 72, 5878–5888. doi: 10.1158/0008-5472.CAN-12-1572-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47– e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vichai V, and Kirtikara K (2006). Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc 1, 1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 53.Naser FJ, Jackstadt MM, Fowle-Grider R, Spalding JL, Cho K, Stancliffe E, Doonan SR, Kramer ET, Yao L, Krasnick B, et al. (2021). Isotope tracing in adult zebrafish reveals alanine cycling between melanoma and liver. Cell Metab 33, 1493–1504.e5. doi: 10.1016/j.cmet.2021.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stancliffe E, Schwaiger-Haber M, Sindelar M, and Patti GJ (2021). DecoID improves identification rates in metabolomics through database-assisted MS/MS deconvolution. Nat. Methods 18, 779–787. doi: 10.1038/s41592-021-01195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams KJ, Pratt B, Bose N, Dubois LG, St. John-Williams, L., Perrott, K.M., Ky, K., Kapahi, P., Sharma, V., MacCoss, M.J., et al. (2020). Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J. Proteome Res 19, 1447–1458. doi: 10.1021/acs.jproteome.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su X, Lu W, and Rabinowitz JD (2017). Metabolite Spectral Accuracy on Orbitraps. Anal. Chem 89, 5940–5948. doi: 10.1021/acs.analchem.7b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiehn O (2016). Metabolomics by Gas Chromatography–Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr. Protoc. Mol. Biol 114. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trefely S, Ashwell P, and Snyder NW (2016). FluxFix: automatic isotopologue normalization for metabolic tracer analysis. BMC Bioinformatics 17, 485. doi: 10.1186/s12859-016-1360-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao C-H, Fowle-Grider R, Mahieu NG, Liu G-Y, Chen Y-J, Wang R, Singh M, Potter GS, Gross RW, Schaefer J, et al. (2016). Exogenous Fatty Acids Are the Preferred Source of Membrane Lipids in Proliferating Fibroblasts. Cell Chem. Biol 23, 483–493. doi: 10.1016/j.chembiol.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y-J, Mahieu NG, Huang X, Singh M, Crawford PA, Johnson SL, Gross RW, Schaefer J, and Patti GJ (2016). Lactate metabolism is associated with mammalian mitochondria. Nat. Chem. Biol 12, 937–943. doi: 10.1038/nchembio.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov 2, 401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal 6, pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, Barretina J, Gelfand ET, Bielski CM, Li H, et al. (2019). Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, et al. (2020). Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402.e16. doi: 10.1016/j.cell.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. (2013). The Somatic Genomic Landscape of Glioblastoma. Cell 155, 462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cancer Genome Atlas Research Network (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. (2016). Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 164, 550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. (2018). Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 173, 291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson M (1985). Handbook of methods for oxygen radical research (CRC Press; ). [Google Scholar]
- 71.Griffith OW (1980). Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal. Biochem 106, 207–212. doi: 10.1016/0003-2697(80)90139-6. [DOI] [PubMed] [Google Scholar]
- 72.Lowry O, Rosebrough N, Farr AL, and Randall R (1951). PROTEIN MEASUREMENT WITH THE FOLIN PHENOL REAGENT. J. Biol. Chem 193, 265–275. doi: 10.1016/S0021-9258(19)52451-6. [DOI] [PubMed] [Google Scholar]
- 73.R Core Team (2021). R: A language and Environment for Statistical Computing
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Z-scores, mean difference and adjusted p-values of individual metabolites, sub- and super pathways, related to Figure 1
Isotopologue enrichment of [13C515N2]glutamine in male and female transformed astrocytes, related to Figure 3
Sex and tissue annotation of human tumor cell lines, related to Figure 4
Isotopologue enrichment of [13C6]glucose in male and female transformed astrocytes, related to Figure 6
Data Availability Statement
Original data for Fig. 1 in the paper is available in Prabhu et al.14. The tumor cell line dataset used in Fig. 4H in the paper is available in Daemen et al.33. Targeted GC/MS metabolite data and isotopologue data generated for this publication are included in the published article. Original western blot images, MRI images, PET images, and microscopy data reported in this paper will be shared by the lead contact upon request.
No original code was generated in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


