Abstract
Genetic heterogeneity describes the occurrence of the same or similar phenotypes through different genetic mechanisms in different individuals. Robustly characterizing and accounting for genetic heterogeneity is crucial to pursuing the goals of precision medicine, for discovering novel disease biomarkers, and for identifying targets for treatments. Failure to account for genetic heterogeneity may lead to missed associations and incorrect inferences. Thus, it is critical to review the impact of genetic heterogeneity on the design and analysis of population level genetic studies, aspects that are often overlooked in the literature. In this review, we first contextualize our approach to genetic heterogeneity by proposing a high‐level categorization of heterogeneity into “feature,” “outcome,” and “associative” heterogeneity, drawing on perspectives from epidemiology and machine learning to illustrate distinctions between them. We highlight the unique nature of genetic heterogeneity as a heterogeneous pattern of association that warrants specific methodological considerations. We then focus on the challenges that preclude effective detection and characterization of genetic heterogeneity across a variety of epidemiological contexts. Finally, we discuss systems heterogeneity as an integrated approach to using genetic and other high‐dimensional multi‐omic data in complex disease research.
Keywords: complex disease, genetic heterogeneity, GWAS, precision medicine
1. INTRODUCTION
Further advancement in precision medicine necessitates robust characterizations of genetic heterogeneity in studies of complex disease. Heterogeneity is a ubiquitous theme in epidemiological research, offering compelling explanations for disease complexity, missing heritability, treatment resistance, and other phenomena. Heterogeneity is defined in a variety of ways, from “simple variation” to “a complex pattern of association.” Under the latter definition, genetic heterogeneity is commonly discussed but usually inadequately evaluated. Failing to properly account for genetic heterogeneity can result in missed associations, biased or incorrect inferences, and impedes the progress of personalized medicine. This review provides an overview of genetic heterogeneity and how it affects the design and analysis of genetic studies of complex disease.
Heterogeneity can appear both within and between explanatory and response variables, described in this review as “features” and “outcomes.” We focus on the unique nature of genetic heterogeneity as a heterogeneous pattern of association. To do so, we identify three “categories” that describe the various occurrences of heterogeneity in biomedical data. The proposed categorization draws on the definitions, challenges, and methods for analyzing heterogeneity within the epidemiological landscape. Most types of heterogeneity do not fall exclusively within one category; specific terms can fall under different categories based on the goals of a particular study or analysis. We do not attempt to comprehensively identify all types of heterogeneity, but rather identify commonly used terms to establish and describe the three categories before exploring genetic heterogeneity and its impact on genetic studies in more detail.
The three categories proposed in this review are feature heterogeneity, outcome heterogeneity, and associative heterogeneity. We briefly describe the first two categories, drawing on common examples from different epidemiological contexts. Next, we describe associative heterogeneity in detail and why genetic heterogeneity specifically falls in this third category. The “associative” nature of genetic heterogeneity makes it uniquely challenging to capture and characterize. We review in depth the many challenges that complicate the detection and characterization of genetic heterogeneity in genetic studies including power, noise, heterogeneity among common and rare variants, heritability, and epistasis. Because of these challenges, genetic heterogeneity often ends up as a potential explanation for less‐than‐ideal results in limitation sections. Furthermore, traditional epidemiology approaches usually emphasize homogeneity of the sample at the expense of sample size and generalizability. We propose investigators instead directly consider genetic heterogeneity in their analyses and we include a review of promising approaches to do so successfully. Lastly, we emphasize the importance of considering all three categories of heterogeneity together, that is, taking a “systems heterogeneity” approach to the genetic epidemiology of complex disease.
2. DEFINITIONS AND CATEGORIZATIONS OF HETEROGENEITY
In the most basic sense, the term “heterogeneity” commonly refers to types of variability in different forms of data, for example, in sample populations, cells, tissues, and phenotypes. This variability can be known and directly measured (observed) or unknown and not directly measured (unobserved). In the epidemiology literature, the term “heterogeneity” is used both in this generic sense but also in a context that goes beyond simple variation, for example, genetic heterogeneity (Ford et al., 1998). Genetic heterogeneity is uniquely defined in the context of an independent association of more than one locus or allele with the same or similar phenotypic outcome (McGinniss & Kaback, 2013). This definition differs from other usages of heterogeneity and warrants specific methodological approaches and study design considerations. Further, a clear understanding of how heterogeneity appears (i.e., observed, unobserved, or as part of an association) helps inform approaches to analysis.
The categories highlight the conceptual contrast between heterogeneity that is not part of an association and genetic heterogeneity as a pattern of association. We use the terms “feature” and “outcome” heterogeneity to describe variation that is independent of association. Features and outcomes are defined by the hypothesis of a given study; thus, some features may be investigated as outcomes in another study and vice versa. Associative heterogeneity specifically includes heterogeneous patterns of association, genetic heterogeneity being the primary example and the focus of this review. Two variables are associated if one variable provides information about another (Altman & Krzywinski, 2015). Statistical association can suggest true causal relationships, but can also result from confounding or other uncontrolled factors. Following robust observational studies, experimental validation is necessary to establish a causal role for the genetic mechanisms that give rise to genetic heterogeneity. Traditional causal models and inferential approaches should also be applied to better understand and describe genetic mechanisms underlying disease, including genetic heterogeneity (Madsen et al., 2011).
Thoughtful study design and strategic analysis choices can further facilitate these discoveries. Figure 1 offers conceptual illustrations of example observations from each of these three categories with their homogeneous counterpart. We further highlight examples of associative heterogeneity in Figure 2, showing the different alleles, loci, and environmental aspects associated with an outcome.
Figure 1.
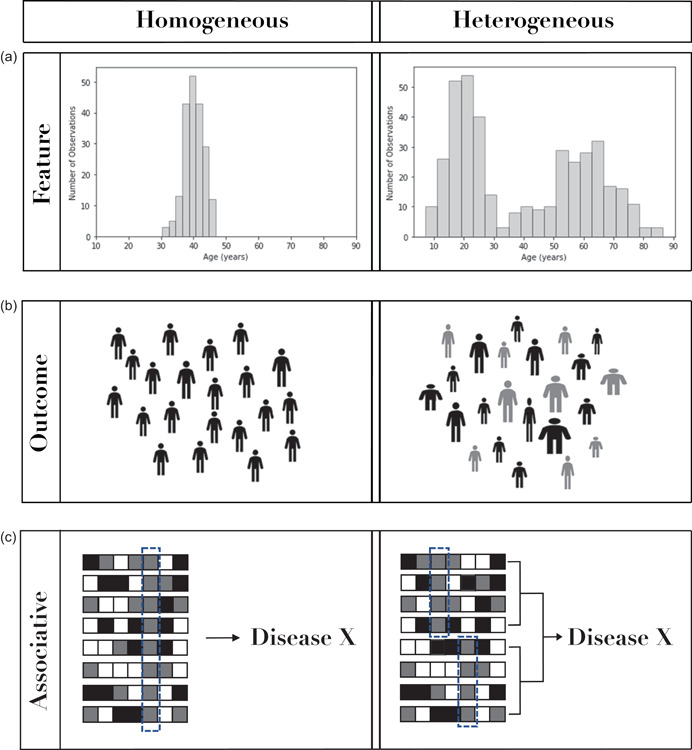
Conceptual illustrations contrasting homogeneity and heterogeneity using example observations within features, outcomes, or associations. Panel (a) depicts age as a feature with less variability on the left and with more variability on the right. Panel (b) depicts a phenotypic outcome, again with less variability on the left and more variability on the right. Panel (c) depicts subjects (rows) and features (columns) where features can have different values (shading). On the left, the feature highlighted by the dotted box is homogeneously associated with Disease X. On the right, associative heterogeneity is represented by two different features independently associated with Disease X within different groups of subjects.
Figure 2.
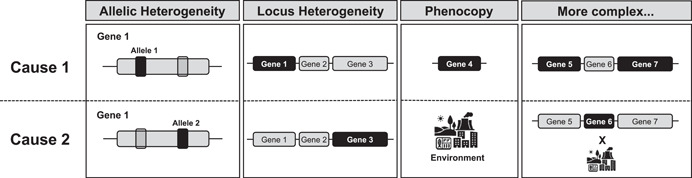
Examples of two independent causes of a single phenotype under four scenarios, allelic heterogeneity, locus heterogeneity, phenocopy, and more complex examples of heterogeneity. Causal factors are in black.
3. APPROACHES TO VARIATION IN FEATURES AND OUTCOMES
3.1. Feature heterogeneity
Variation is often synonymous with the concept of heterogeneity. Variability in explanatory variables (i.e., features) can be regarded as a mechanism of interest, noise, or more often, as a potential confounder. Heterogeneity of any kind necessarily relies on the existence of underlying variability. Feature heterogeneity, however, refers to feature value variation and its distribution among subjects or samples. Feature heterogeneity can include variation in risk factors such as age and family history; clinical variables such as blood pressure or tumor grades; or cellular‐level variables such as gene expression, epigenetics, and the cellular microenvironment. Feature heterogeneity is normal and expected across many scenarios, and has been widely explored (A. J. Holmes & Patrick, 2018; Lawson et al., 2020; Murphy et al., 2019). Still, in many epidemiological studies and especially in clinical trials, feature heterogeneity in certain covariates is strategically avoided and homogeneous samples are encouraged to minimize confounding and heterogeneous treatment effects (Kent et al., 2016, 2010). For example, in genome‐wide association studies (GWAS), failure to account for different allele frequencies of variants among different study populations can give rise to the illusion of genetic heterogeneity.
Methods applied to assess feature heterogeneity depend on the goals of a given analysis and whether the heterogeneity is being controlled for or analyzed directly. Sources of feature heterogeneity can be either observed, such as age, or unobserved, such as genetic ancestry. Most methods for feature heterogeneity can be divided along this margin. When feature heterogeneity is observable, samples or subjects are often stratified before implementing regression or other association approaches (M. Wang et al., 2016), for example by age or other risk factors. Computational methods to understand underlying variation are especially useful for identifying unobserved feature heterogeneity. Methods for analyzing gene expression must be able to differentiate potential contributors to heterogeneity; filtering out noise while preserving meaningful signal (e.g., from alternative splicing sites, etc.) (Wan & Larson, 2018). Principal component analysis (Patterson et al., 2006) or uniform manifold approximation and projection (Diaz‐Papkovich et al., 2019) can be used to capture genetic background variation or population substructure. Detailed discussions of these and other methods can be found elsewhere (Diaz‐Papkovich et al., 2019; Jaffe & Irizarry, 2014; Lu, 2019).
3.2. Outcome heterogeneity
Outcome heterogeneity reflects the variation in outcomes or dependent variables. Types of heterogeneity approached from the perspective of “outcomes” include clinical, phenotype, disease, and trait heterogeneity. While epidemiological studies primarily focus on disease phenotypes, outcome heterogeneity is also inherent in healthy individuals and phenotypes. Understanding healthy variation is a key component of a diverse range of studies from healthy aging (S. Kim & Jazwinski, 2018) to immunogenetics (De Jager et al., 2015). In epidemiological and clinical research, clinical heterogeneity denotes variability in symptoms and clinical presentation of disease phenotypes, irrespective of underlying genetic architecture (Milaneschi et al., 2016). In the context of this review, phenotypes include diseases and/or disease characteristics measured as outcomes. Phenotype heterogeneity is often used interchangeably with clinical heterogeneity. Phenotype heterogeneity most often refers to the variation in symptoms among individuals with the same disease (Figure 1b) (Chiò et al., 2011). For many diseases, phenotype heterogeneity has been captured using descriptive studies, disease registries, and electronic health record (EHR) data (Shivade et al., 2014). With improvements in imaging, biomarkers, and other diagnostic tests, phenotypes have become increasingly well‐defined (Haffner et al., 2021; Ryan et al., 2018). Still, for complex or late‐onset diseases, certain characteristics remain unobserved and contribute to outcome heterogeneity. Trait or disease heterogeneity are special cases of outcome heterogeneity that describe traits, phenotypes, or diseases which are defined with insufficient specificity such that they are actually two or more separate traits (Thornton‐Wells et al., 2006; Wray & Maier, 2014). Trait and outcome heterogeneity more generally may suggest the existence of underlying genetic or other associative heterogeneity (Swinnen & Robberecht, 2014). Many investigators have suggested that certain diseases are more accurately defined by their phenotypic subtypes, including autism (Stevens et al., 2019) and Alzheimer's disease (Mitelpunkt et al., 2020). Analyses focused on subphenotypes are especially common in psychiatric epidemiology (Wendt et al., 2020).
Similar to feature heterogeneity, methods accounting for outcome heterogeneity share the methodological goal of understanding the contributors of variation, whether observed or unobserved. When readily observable, outcome heterogeneity can often be explicitly accounted for in study design, e.g., phenotype heterogeneity guiding decisions about disease subtype analyses. Begg and colleagues suggest guidelines for systematically addressing outcome heterogeneity in epidemiologic studies and offer two strategies: risk prediction (when subtypes are known) and hierarchical clustering (subtypes unknown) (Begg et al., 2013). Both supervised and unsupervised machine learning methods are employed for classification or subtyping of observed complex phenotypes (Athey & Imbens, 2015; Huang et al., 2018; Jacob et al., 2019; Kourou et al., 2015). Clustering algorithms, latent class analyses, factor analyses, and network approaches are other unsupervised approaches that attempt to identify unobserved outcome heterogeneity (Lubke & Muthén, 2005). When disease subtypes are unknown, hierarchical clustering minimizes within‐group variation and maximizes between‐group variation to establish an optimal set of subtypes (Van Rooden et al., 2010). Similarly, Thornton‐Wells and colleagues described three unsupervised clustering methods for analyzing trait heterogeneity in the presence of genetic heterogeneity and epistasis: Bayesian classification, hypergraph‐based clustering, and fuzzy k‐modes clustering (Thornton‐Wells et al., 2006).
4. ASSOCIATIVE HETEROGENEITY
Associative heterogeneity refers to any heterogeneous pattern of association between different features and an outcome. Associative heterogeneity is synonymous with etiologic heterogeneity, and differs from the other two categories by requiring heterogeneity in the relationship between features and outcomes. Examples include epigenetic heterogeneity in cancer (Guo et al., 2019), heterogeneous EHR data (i.e., clinical notes and laboratory values) and clinical outcomes (Pivovarov et al., 2015), and specific to this review, genetic heterogeneity (Urbanowicz et al., 2013). Locus and allelic heterogeneity are the two commonly described subtypes of genetic heterogeneity, although more complex scenarios also exist. We will use the definitions of genes, loci, and alleles reviewed in Elston et al. (2012).
Locus heterogeneity occurs when mutations at different loci result in the same disease (Figure 2). A common example of locus heterogeneity is breast cancer, where mutations in BRCA1 or BRCA2 can independently cause disease. Allelic heterogeneity arises when different alleles at the same locus cause the same or similar expression of a phenotype (Figure 2). While individual variants also have alleles, allelic heterogeneity specifically references different variants at the same locus, not alternative forms of the same variant. For example, cystic fibrosis exhibits widespread allelic heterogeneity, with over 100 possible causal mutations at the CFTR gene (Audrézet et al., 2004). Allelic heterogeneity also plays a role in more complex traits such as gene expression levels and schizophrenia (Hormozdiari et al., 2017). In some instances, locus or allelic heterogeneity can manifest phenotype heterogeneity—the variability in presentation of a disease reflecting the different underlying genetic etiologies (Wood et al., 2011). Cystic fibrosis again provides a useful example, exhibiting extensive clinical heterogeneity arising from allelic heterogeneity (Paranjapye et al., 2020). Traditional methods reviewed in Section 4.1 for investigating genetic heterogeneity are often underpowered or focus on removing heterogeneity from the subjects and/or data. More recently, machine learning approaches are increasingly applied to this problem and offer potential improvements in high‐dimensional data and multi‐omic data (D. Kim et al., 2015). We examine the specific challenges of genetic heterogeneity and the methodological hurdles that result in Section 5.
Phenocopy is another phenomenon that falls under the umbrella of associative heterogeneity. Phenocopies are affected individuals whose disease is not caused by the same (genetic) factors as other individuals with the disease (Lescai & Franceschi, 2010), but are rather the result of one or more environmental factors (Figure 2). The presence of phenocopies effectively increases associative heterogeneity by introducing additional associated variables spanning other genetic, epigenetic, and environmental factors, thereby decreasing the power to detect any one of them (Lescai & Franceschi, 2010).
4.1. Methods for associative heterogeneity
In the latter half of the twentieth century, linkage methods for family studies led to the discovery of now‐prominent examples of genetic heterogeneity, followed more recently by findings from GWAS and whole genome and whole exome sequencing. Despite many successes, GWAS findings are often constrained by small effect sizes and failures to replicate in other studies, especially among complex diseases (O'Connor, 2021). The still‐unmet promises of the “GWAS era” provide opportunities for improvements in methodologies to address gaps in understanding associative heterogeneity in genetic data and beyond.
Identifying and characterizing genetic heterogeneity remains imperative as the information gained is invaluable for therapeutic advancement (Lohr et al., 2014; J. Zhang et al., 2018) and improved predictive accuracy (Rahman et al., 2017). Ideal methods for addressing associative heterogeneity should seek to combine their capacity for improved prediction with an explanatory component that can provide clinically relevant interpretations. Heidema and colleagues (2006) offer an excellent framework to evaluate the strength of new models for analysis of associative heterogeneity, including the following components: the ability to handle high dimensionality, power to detect true effects, performance in the presence of complex genetic architectures, and open‐source availability of any software. Below we explore methods for associative heterogeneity in more detail, highlighting some of these successes and continuing challenges and limitations for population‐level genetic studies. We also suggest that methods development efforts prioritize clinical applicability and interpretability.
4.1.1. Epidemiologic approaches to associative heterogeneity
Before the GWAS era, family studies were considered the most informative strategy for mapping casual variants. Linkage analysis aims to identify genetic markers that cosegregate with the phenotype of interest, thereby mapping the location of a linked gene or genomics region (Cantor, 2019). A number of linkage methods directly address heterogeneity, for example, multi‐locus models that aid in the detection of linkage for non‐Mendelian phenotypes (Risch, 1990) and novel stratification approaches (Talebizadeh et al., 2013). Described in terms of population prevalence and the ratio of risk for relatives, Risch's additive model corresponds to genetic heterogeneity (as related to identity‐by‐descent sharing in affected sibling pairs), while the multiplicative model accounts for interaction between loci (Risch, 1990). Genetic heterogeneity has been identified using linkage analysis in a variety of diseases, including multiple sclerosis (Haines et al., 1998), hereditary lymphedema (Ferrell, 1998), and rheumatoid arthritis (Cordell, 2003). With the availability of next‐generation sequencing, there are increasing efforts to utilize the strengths of linkage methods to explore whole exome and whole genome sequencing and similar approaches for investigating complex traits (Xiao et al., 2019).
A variety of hypotheses about the genetic contribution to disease have dominated the literature, especially the ‘common disease‐common variant' hypothesis, which is typically interrogated using a GWAS approach. This hypothesis postulates that common diseases are caused by combinations of common alleles (minor allele frequency 5%) with individually small effect sizes (Risch & Merikangas, 1996). This is reflected by the average effect size of 1.33 for candidate SNPs among most published GWAS results (Hindorff et al., 2009). However in the presence of genetic heterogeneity, these small effect sizes are unsurprising (Kulminski et al., 2016). Increasing sample sizes in many epidemiological studies have helped to bolster the power of GWAS approaches, but these benefits often have been outweighed by the multiple testing burden and lack of replication, an inevitable consequence of testing hundreds of thousands to millions of variants. Detecting underlying genetic heterogeneity requires even greater statistical power, especially when individual effect sizes are small. We discuss the issue of power further in Section 5.
Many common diseases such as cardiovascular disease, psychiatric diseases, and cancer are highly complex, exhibiting genetic, phenotypic, and other types of heterogeneity. Complex diseases are caused by a combination of genetic and environmental factors (Stessman et al., 2014). GWAS has been an invaluable step in our understanding of complex diseases such as autoimmune disorders (Sawcer et al., 2011), metabolic traits (Polychronakos & Li, 2011), and psychiatric disorders (International Schizophrenia Consortium, 2009), among others, illuminating mechanisms and pathways for ongoing research. Furthermore, nearly 88% of GWAS hits (NHGRI catalog) are in noncoding regions (Edwards et al., 2013), suggesting that regulatory elements and noncoding RNAs play an important role in complex diseases and contribute to genetic heterogeneity (Boyle et al., 2017; Castelnuovo & Stutz, 2015). Despite many successes, GWAS and other large‐scale methods have delivered mixed results overall, including varying effect sizes (Han & Eskin, 2012) and variants with no known biological significance (Nishizaki & Boyle, 2017).
Methods to reduce the multiple testing burden in the context of genetic heterogeneity and other complex architectures have also been developed for GWAS data, including reducing the stringency using false discovery rates instead of the Bonferroni correction (Benjamini & Hochberg, 1995), genomic interval search (Llinares‐López et al., 2015), and use of haplotypes (Guinot et al., 2018). Additionally, incorporating “expert knowledge” into the analysis of GWAS data can be used to prioritize the most informative variants (Urbanowicz et al., 2012). Expert knowledge can refer to biological pathways, gene ontologies, phenotype networks, and informative feature selection methods (Harari et al., 2012; Ritchie, 2011). Still, key challenges remain that limit the ability of the GWAS methodology to characterize genetic heterogeneity, and is among the likely explanations for lack of replication in GWAS studies (Hodge et al., 2016; Sirugo et al., 2019).
The standard epidemiologic approach strives to find the best disease model in a homogeneous sample of the population. Rothman and colleagues assert the ‘necessary avoidance of representativeness' (i.e., collecting a homogeneous sample rather than a heterogeneous one) in scientific studies (Rothman et al., 2013). This approach is especially salient in establishing the efficacy of a new treatment in randomized controlled trials. Minimizing heterogeneity among study subjects is necessary to achieve an unconfounded analysis of the relationship between their exposure(s) and outcome(s) of interest. Randomization or other strategies to reduce bias can give the illusion of homogeneity, but despite these efforts, unobserved or unmeasured heterogeneity is often unavoidable. In the context of GWAS and other genetic studies, homogeneity allows for the most robust evidence for association, however it can also limit the generalizability of results (Martin et al., 2019). For example, ancestry‐related genetic differences are one explanation for the failure of risk alleles from European‐derived GWAS studies (homogeneous) to replicate in other populations (Kraft et al., 2009).
Genetic ancestry can influence the design and analysis of genetic studies in multiple ways. First, as mentioned in Section 3.1, it can result in feature heterogeneity—population substructure that is independent of disease and must be controlled for to prevent spurious associations. Second, differences in variant and disease frequencies between ethnic groups can help identify disease‐causing genes or variants. Mapping by admixture linkage disequilibrium was an early method for identifying disease‐associated variants, especially among recently admixed populations with large differences in allele frequencies (Smith & O'Brien, 2005). Joint approaches testing both genotype and ancestry association further improved statistical efficiency in studies of admixed populations (Szulc et al., 2017; Tang et al., 2010). These and other admixture approaches have the most power when differences in phenotype frequency are highest, such as multiple sclerosis which is most prevalent in individuals of northern European ancestry (Chi et al., 2019). Lastly, genetic ancestry can give rise to genetic heterogeneity as more specifically defined, that is, as a heterogeneous pattern of association. For example, a recent study of systemic lupus erythematosus identified different risk variants by ancestry group (Y.‐F. Wang et al., 2021). Genetic ancestry continues to be an important factor in discovering genetic associations and increasingly for illuminating health disparities (Shriner, 2017).
Despite precise phenotyping, stratification, rigorous subtype analyses, and large sample sizes, failing to fully account for heterogeneity can preclude the discovery of true associations. Instead, it may be useful to embrace heterogeneity and employ analysis strategies that can account for various levels and categories of heterogeneity. This may include implementing random‐effects models in meta‐analyses (Langan et al., 2019), investigating evidence for interactions (Park et al., 2018), utilizing admixed populations (Hou et al., 2021), and assessing etiologic (associative) heterogeneity in the context of environmental exposures (Peterson et al., 2018).
4.1.2. Associative heterogeneity in the prediction era
Two main paradigms dominate the methodological goals of population level genetic research—prediction and inference. In the era of big data and machine learning, the two are often combined. For example, GWAS represents an attempt at large‐scale inference, but has recently been employed to inform prediction tools such as polygenic risk scores (PRS) (Albiñana et al., 2021). Still, if heterogeneity is unaccounted for, the predictive capabilities of models will likely remain suboptimal (Ng et al., 2014). Efforts to improve predictive capabilities in the context of feature, outcome, and associative heterogeneity continue to expand, especially in cancer research and in multi‐omic datasets (Kourou et al., 2015).
Empirical evidence strongly suggests that polygenicity underlies the genetic component of many complex diseases (Visscher et al., 2012), leading to the development of metrics such as PRSs aimed at consolidating estimates of genetic risk using GWAS data. However, polygenic models can have poor generalizability across populations due to a combination of factors such as sample size (Dudbridge, 2013), differences in genetic variation between populations, genetic heterogeneity, and large environmental contributions (Torkamani et al., 2018). Some PRSs have been integrated into clinical practice, but have been met with criticism regarding the potential for European‐derived scores to exacerbate health disparities due to reduced predictive performance in minority populations (Martin et al., 2019). An expanded consideration of genetic heterogeneity in genomics‐derived models such as PRSs may produce improved and less biased predictions. Additional challenges to identifying genetic heterogeneity include lags in developing high‐dimensionality computational approaches compared to other areas of methods development. While strategies to reduce the multiple testing burden have offered improvements, machine learning methods may offer advantages over traditional hypothesis testing approaches (Rodgers, 2010). For example, D. Kim et al. (2015) applied an integrative framework to TCGA breast cancer survival data and showed improved predictive performance using grammatical evolutionary neural networks. This approach showed improvement in handling heterogeneity over methods such as survival multifactor dimensionality reduction and both detect gene–gene interactions (Motsinger‐Reif et al., 2008), however, these models face limitations in the presence of additional noise such as missingness or phenocopy. A novel multifactor dimensionality reduction approach sought to account for genetic heterogeneity using phenotype covariates (Mei et al., 2007), highlighting the potential utility of phenotypic heterogeneity as an indicator of underlying genetic heterogeneity. Epigenetic age estimators that rely on the "epigenetic clock” (Ashapkin et al., 2019) (i.e., underlying epigenetic heterogeneity) have also shown to be reliable predictors in both aging and cancer (Jones et al., 2015; Yu et al., 2020). Deep learning approaches such as convolutional neural networks have also been used to classify genetically heterogeneous cancer types using imaging data (Chang et al., 2018). Deep learning methods extract patterns in data using multiple “layers” or networks, where each layer learns progressively more abstract patterns (Truong et al., 2020).
One promising family of machine learning methods for the detection, modeling, and interpretable characterization of associative heterogeneity are learning classifier systems (LCS). LCSs are a type of rule‐based machine learning particularly suited to complex problem domains (Urbanowicz & Moore, 2009). LCSs have been applied to epidemiological surveillance (J. H. Holmes et al., 2000) and biomedical data mining (Bacardit et al., 2009; Urbanowicz et al., 2013). They conduct a form of piece‐wise modeling by evolving a set of human interpretable (IF:THEN) rules to cover the problem space (Urbanowicz & Browne, 2017). The algorithm ExSTraCS is one example of an LCS that has been developed to detect and characterize complex associations including both epistatic interactions and genetic heterogeneity (Urbanowicz & Moore, 2015).
5. OTHER CHALLENGES IN CAPTURING GENETIC HETEROGENEITY
Genetic heterogeneity plays an extensive role in biological processes and poses various analytical challenges that can limit advancements in complex disease epidemiology. Various errors and biases in study design and implementation can amplify feature and outcome heterogeneity, undermining or confounding subsequent analyses (Clayton et al., 2005). Challenges in approaches to genetic heterogeneity echo those found in most association analyses, including low statistical power (Manchia et al., 2013), a high multiple testing burden (Llinares‐López et al., 2015), rare variants (Betancur & Coleman, 2013), missing heritability (Ehret et al., 2012), and lack of replication (Yashin et al., 2015). Also, given that genetic heterogeneity is not the only factor complicating patterns of association involved in complex disease, it may be useful to jointly consider both genetic heterogeneity and epistasis.
5.1. Heritability and power
As previously suggested, genetic heterogeneity is believed to represent a component of the “missing heritability” problem cited as one of multiple phenomena contributing to the incomplete understanding of genetic risk of many diseases (Ehret et al., 2012; Van Der Sluis et al., 2010). Heritability is the proportion of variation in a phenotype that is due to genetic factors (Wray & Maier, 2014). Genetic heterogeneity can reduce power to detect associations by effectively reducing the overall population into smaller unobserved “subgroups” representing the different genetic etiologies. This reduces the ability of population‐level approaches to explain the full heritability of a disease. If phenotype heterogeneity is present in population studies, heritability estimates are reduced compared to estimates from family studies (Wray & Maier, 2014). Phenotype heterogeneity can also affect heritability estimates if it arises from underlying genetic heterogeneity. Further, when diseases are genetically correlated or exhibit shared heritability, power to detect associations increases for loci that contribute to both diseases but decreases for variants that are heterogeneous between them. The existence of multiple casual variants at the same locus, allelic heterogeneity, can also go undetected and contribute to reduced heritability estimates (Wood et al., 2011). Multiple studies have demonstrated that accounting for allelic heterogeneity explains additional variation for a variety of phenotypes including gene expression (Jansen et al., 2017), height (G. Zhang et al., 2012), BMI (Ehret et al., 2012), and lipid levels (Y. Wu et al., 2013).
Most approaches that tackle complex patterns of association in genetic studies are plagued by issues of insufficient power and the multiple testing burden that arises in high‐dimensional data analysis. The presence of heterogeneity (of any kind) can have a substantial effect on power. Manchia et al. (2013) demonstrate that phenotypic heterogeneity of 50% in a case‐control study (i.e., 50% of the cases are controls that have been misclassified or are cases with a different casual factor influencing their disease) increases the required sample size nearly threefold. Feature or outcome heterogeneity due to misclassification, measurement error, and selection biases (Sutton et al., 2000) also contribute to reductions in power (Cheng et al., 2010). Simply increasing sample size to improve power may have the opposite effect if additional heterogeneity is introduced into the study population (Kulminski et al., 2016). Conversely, despite reductions in sample size, subset analyses may boost statistical power if they accurately capture underlying genetic differences (Bhattacharjee et al., 2012). Many strategies have been proposed to improve power in the presence of genetic heterogeneity, in linkage analysis, (Bureau et al., 2008; Risch, 1990), via meta‐analyses (Bhattacharjee et al., 2012; Zintzaras & Ioannidis, 2005), and in the context of epistasis (Urbanowicz et al., 2013).
5.2. Rare variant heterogeneity
Common variants are not alone in their contribution to genetic heterogeneity in complex common diseases; rare variants must also be considered. Rare variants have a minor allele frequency of less than 1%–5% and are thus excluded from most GWAS (Gorlov et al., 2008). Hence, rare variants are a likely source of some of the unexplained variability (heritability) of some diseases (Lee et al., 2014), although the debate over their relative contribution is ongoing (Gibson, 2012). Both rare and common variants can lead to the same disease (McClellan & King, 2010). Schizophrenia is a common example of a genetically heterogeneous disease likely caused by a “spectrum of risk variants” including rare copy number variants at multiple different loci, rare alleles at the same locus, or common variants with small or modest effects (Sebat et al., 2009; Sullivan et al., 2018). Multiple studies have suggested that the interplay between rare and common variants also contributes to certain diseases, including cancer and epilepsy (Dibbens et al., 2007; Hahn et al., 2016).
Different rare variants associated with a given disease outcome could be viewed as the most extreme form of genetic heterogeneity. As a worst‐case scenario, it's possible that some disease phenotypes could result from unique rare genetic variants specific to small groups of individuals. Studies are increasingly capturing rare variants via whole genome or whole exome sequencing, and some apply gene‐level or other “binning” approaches to improve power (Moore et al., 2016; Povysil et al., 2019). Other gene‐based methods include burden tests first introduced by B. Li and Leal (2008) and further improved by others (Sun et al., 2013), kernel‐based tests (Dutta et al., 2019; M. C. Wu et al., 2011), and functional regression approaches (Fan et al., 2016; Svishcheva et al., 2019). While associative heterogeneity nearly always places increased demand on power (Cirulli & Goldstein, 2010), gene‐level rare variant analyses may benefit from the presence of allelic heterogeneity (Povysil et al., 2019). The cumulative effect of multiple risk variants in the same gene (i.e., allelic heterogeneity) across different subjects can aid in detecting the causal gene (Povysil et al., 2019).
5.3. Epistasis
Epistasis, alongside genetic heterogeneity, is a key contributor to the genetic landscape of complex diseases (Monir & Zhu, 2017). Epistasis is the interaction between genes at different loci (i.e., not alleles). Epistasis is also difficult to capture and model and is another likely source of some "missing heritability” of various phenotypes (Ritchie, 2015; Zhu & Fang, 2018). While the definition of genetic heterogeneity includes an assumption of independence between loci, epistasis could exist alongside heterogeneity as an additional complex pattern of association. Traditionally, associative heterogeneity and epistasis represent mutually exclusive patterns of association (between the same set of loci) (Cordell, 2002). Applying Rothman's sufficient component cause framework, Madsen et al. (2011) demonstrated that in fact, additive and multiplicative penetrance models can correspond to both genetic heterogeneity and epistasis, unless highly specific assumptions are made. Thus, while strict genetic heterogeneity and epistasis cannot occur between the same pair of loci, both mechanisms (between a different set of loci) could give rise to the same phenotype.
Others have also sought to jointly identify associative heterogeneity and epistasis. For example, Fenger et al. utilized latent class analysis and structural equation modeling to uncover heterogeneous subpopulations, and suggested that inclusion of epistasis increased the likelihood of detecting a true association (between genetic variants and complex traits) in a homogeneous population (Fenger et al., 2008). Li and colleagues combined clustering with deep learning to analyze datasets with both genetic heterogeneity and epistasis (X. Li et al., 2018). Turner and Bush (Turner & Bush, 2011) demonstrated the conceptual overlap between epistasis and genetic heterogeneity in an analysis of regulatory SNPs. They suggest that multiple different epistatic combinations that influence a disease also fall under the umbrella of heterogeneity.
6. SYSTEMS HETEROGENEITY
Given the pervasiveness of all categories of heterogeneity in complex diseases, the concept of systems heterogeneity has been proposed to offer a more integrated view (D. C. Wang & Wang, 2017). This approach is especially salient as genetic data is increasingly supplemented by transcriptomic, proteomic, metabolomic, and other data. Notably, in addition to heterogeneity within data types, a systems approach takes into account interactions between them. Simultaneously considering the interacting factors from the single‐cell to between‐patient level has potential to greatly improve identification of biomarkers and treatment targets. These types of integrated approaches are becoming widespread as data mining and other bioinformatics tools improve (Gomez‐Cabrero et al., 2014; Liu et al., 2019). For example, D. Kim et al. (2015) introduced an integrative framework to combine heterogeneous data sources (e.g., multi‐omics) to identify interactions in survival data. Additional work by Kim and colleagues demonstrated that integration of multi‐omics data can improve outcome prediction (D. Kim et al., 2014).
Many of the recent advancements in analyzing heterogeneity have been made in cancer research by integrating multiple data types (Bareche et al., 2018). Song et al. (2016) reviewed methods for the clinical detection of underlying genetic heterogeneity in breast cancer using molecular imaging, next‐generation sequencing, and expression profiling. Natrajan et al. (2016) modeled tumor microenvironment using a “tumor ecosystem diversity index” derived from histology image analysis. Knowledge of spatial and temporal intratumor heterogeneity has drastically improved using single‐cell approaches including detailed characterization of branched evolution, signaling networks, gene expression and other mechanisms (Gupta & Somer, 2017; Patel et al., 2014). For example, scRNA‐seq data has helped resolve cell type composition from bulk RNAseq data (Chu et al., 2022; X. Wang et al., 2019). Capturing multi‐omic heterogeneity is also imperative for understanding phenotypic plasticity and uncovering mechanisms of treatment resistance (Sheng et al., 2018).
Others have developed integration methods targeted to cancer research and beyond, with various methods and packages available for application to “big data” (Dong & Srivastava, 2013; Karczewski & Snyder, 2018; Rohart et al., 2017). Network analysis (Sudhakar et al., 2020), multi‐view clustering (Rappoport & Shamir, 2018; Shi et al., 2019), factor analyses (Argelaguet et al., 2018) and Bayesian approaches (Ray et al., 2014) are only a handful of methods aimed at analyzing heterogeneous multi‐omic data. While these approaches integrate heterogeneous data types, few have a specific focus on elucidating underlying associative heterogeneity. A full consideration of feature, outcome, and associative approaches to heterogeneity as part of study design and during analysis of multi‐omic data types would be useful for pursuing a systems approach in population level studies.
7. SUMMARY
“Intuitively, the concept of heterogeneity is clear, but as we scrutinize it, our initial impression fractures into complexity” (Kolasa & Rollo, 1991). Using an epidemiological framework, this complexity can be condensed to heterogeneity in features, outcomes, and in the relationships between them. As a heterogeneous pattern of association between features and an outcome, genetic heterogeneity falls into the latter category, termed “associative heterogeneity” in this review. Detecting and characterizing genetic heterogeneity is key for advancing our knowledge of complex diseases, but key challenges and limitations remain. Well‐known issues of power, heritability, and variant frequency limit the ability of methods to detect genetic heterogeneity. Understanding genetic heterogeneity as a heterogeneous pattern of association is essential for choosing the most effective approaches and methods. Other complex patterns of association such as epistasis and gene‐environment interactions can also exist alongside genetic heterogeneity and should also be considered. Additionally, ever‐growing sample sizes and an increased emphasis on representativeness and generalizability conflicts with standard assumptions of homogeneity and demands further attention. Expert consideration of these and other ways in which genetic heterogeneity impacts the design and analysis of genetic studies can help us confront these and other ongoing methodological challenges.
Detecting genetic heterogeneity at the population level warrants thoughtful study design and the development of computational approaches that can tackle complex scenarios in genetic data and beyond, especially as multi‐omic analyses and integrated datasets become increasingly common. Causal inference frameworks for genetic heterogeneity (Madsen et al., 2011) and other “genetically informed” inference methods (Pingault et al., 2018) can be used to support these efforts. Prediction modeling for disease risks, diagnoses, and treatment decisions can likely also benefit from a consideration of genetic heterogeneity and its challenges during model development, allowing for enhancing accuracy and generalizability. Cancer and other complex diseases that are known to be highly heterogeneous are a valuable resource for interrogating underlying genetic heterogeneity and for developing and testing new methods. New approaches to analyzing genetic heterogeneity should capitalize and build on the strengths of traditional epidemiological frameworks to improve risk estimates, capture missing heritability, increase prediction accuracy, provide mechanistic insights, and identify biomarkers and targets for treatment. We are confident that these avenues for research on genetic heterogeneity will aid in advancing the goals of personalized medicine.
ACKNOWLEDGMENTS
We would like to thank Van Truong, Trang Le, and Stephen Bagley for their helpful comments and feedback. This study was supported by National Institutes of Health grants LM010098, LM012601, F31‐LM013583‐01, and NHGRI training grant HG000046.
Woodward, A. A. , Urbanowicz, R. J. , Naj, A. C. , & Moore, J. H. (2022). Genetic heterogeneity: Challenges, impacts, and methods through an associative lens. Genetic Epidemiology, 46, 555–571. 10.1002/gepi.22497
REFERENCES
- Albiñana, C. , Grove, J. , McGrath, J. J. , Agerbo, E. , Wray, N. R. , Bulik, C. M. , Nordentoft, M. , Hougaard, D. M. , Werge, T. , Børglum, A. D. , Mortensen, P. B. , Privé, F. , & Vilhjálmsson, B. J. (2021). Leveraging both individual‐level genetic data and GWAS summary statistics increases polygenic prediction. The American Journal of Human Genetics, 108(6), 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman, N. , & Krzywinski, M. (2015). Points of significance: Association, correlation and causation. Nature Methods, 12(10), 899–900. [DOI] [PubMed] [Google Scholar]
- Argelaguet, R. , Velten, B. , Arnol, D. , Dietrich, S. , Zenz, T. , Marioni, J. C. , Buettner, F. , Huber, W. , & Stegle, O. (2018). Multi‐omics factor analysis—A framework for unsupervised integration of multi‐omics data sets. Molecular Systems Biology, 14(6), e8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin, V. V. , Kutueva, L. I. , & Vanyushin, B. F. (2019). Epigenetic clock: Just a convenient marker or an active driver of aging? Reviews on Biomarker Studies in Aging and Anti‐Aging Research, 1178, 175–206. [DOI] [PubMed] [Google Scholar]
- Athey, S. , & Imbens, G. W. (2015). Machine learning methods for estimating heterogeneous causal effects. Stat, 1050(5), 1–26. [Google Scholar]
- Audrézet, M.‐P. , Chen, J.‐M. , Raguénes, O. , Chuzhanova, N. , Giteau, K. , Maréchal, C. L. , Quéré, I. , Cooper, D. N. , & Férec, C. (2004). Genomic rearrangements in the cftr gene: Extensive allelic heterogeneity and diverse mutational mechanisms. Human Mutation, 23(4), 343–357. [DOI] [PubMed] [Google Scholar]
- Bacardit, J. , Burke, E. K. , & Krasnogor, N. (2009). Improving the scalability of rule‐based evolutionary learning. Memetic Computing, 1(1), 55–67. [Google Scholar]
- Bareche, Y. , Venet, D. , Ignatiadis, M. , Aftimos, P. , Piccart, M. , Rothe, F. , & Sotiriou, C. (2018). Unravelling triple‐negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Annals of Oncology, 29(4), 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg, C. B. , Zabor, E. C. , Bernstein, J. L. , Bernstein, L. , Press, M. F. , & Seshan, V. E. (2013). A conceptual and methodological framework for investigating etiologic heterogeneity. Statistics in Medicine, 32(29), 5039–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y. , & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1), 289–300. [Google Scholar]
- Betancur, C. , & Coleman, M. (2013). Etiological heterogeneity in autism spectrum disorders: Role of rare variants. In Buxbaum & Hof P. R. (Eds.), The neuroscience of autism spectrum disorder (pp. 113–144). Academic Press. [Google Scholar]
- Bhattacharjee, S. , Rajaraman, P. , Jacobs, K. , Wheeler, W. , Melin, B. , Hartge, P. , Yeager, M. , Chung, C. , Chanock, S. , & Chatterjee, N. (2012). A subset‐based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. The American Journal of Human Genetics, 90(5), 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, E. A. , Li, Y. I. , & Pritchard, J. K. (2017). An expanded view of complex traits: From polygenic to omnigenic. Cell, 169(7), 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau, A. , Labbe, A. , Croteau, J. , & Mérette, C. (2008). Using disease symptoms to improve detection of linkage under genetic heterogeneity. Genetic Epidemiology, 32(5), 476–486. [DOI] [PubMed] [Google Scholar]
- Cantor, R. M. (2019). Analysis of genetic linkage. In Pyeritz R. E., Korf B. R., & Grody W. W. (Eds.), Emery and Rimoin's Principles and Practice of Medical Genetics and Genomics (pp. 227–236). Elsevier. [Google Scholar]
- Castelnuovo, M. , & Stutz, F. (2015). Role of chromatin, environmental changes and single cell heterogeneity in non‐coding transcription and gene regulation. Current Opinion in Cell Biology, 34, 16–22. [DOI] [PubMed] [Google Scholar]
- Chang, P. , Grinband, J. , Weinberg, B. , Bardis, M. , Khy, M. , Cadena, G. , Su, M.‐Y. , Cha, S. , Filippi, C. , Bota, D. , Baldi, P. , Poisson, L. M. , Jain, R. , & Chow, D. (2018). Deep‐learning convolutional neural networks accurately classify genetic mutations in gliomas. American Journal of Neuroradiology, 39(7), 1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, D. , Branscum, A. J. , & Stamey, J. D. (2010). Accounting for response misclassification and covariate measurement error improves power and reduces bias in epidemiologic studies. Annals of Epidemiology, 20(7), 562–567. [DOI] [PubMed] [Google Scholar]
- Chi, C. , Shao, X. , Rhead, B. , Gonzales, E. , Smith, J. B. , Xiang, A. H. , Graves, J. , Waldman, A. , Lotze, T. , Schreiner, T. , Weinstock‐Guttman, B. , Aaen, G. , Tillema, J. M. , Ness, J. , Candee, M. , Krupp, L. , Gorman, M. , Benson, L. , Chitnis, T. , … Barcellos, L. F. (2019). Admixture mapping reveals evidence of differential multiple sclerosis risk by genetic ancestry. PLoS Genetics, 15(1), e1007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiò, A. , Calvo, A. , Moglia, C. , Mazzini, L. , Mora, G. , & PARALS Study Group . (2011). Phenotypic heterogeneity of amyotrophic lateral sclerosis: A population based study. Journal of Neurology, Neurosurgery & Psychiatry, 82(7), 740–746. [DOI] [PubMed] [Google Scholar]
- Chu, T. , Wang, Z. , Pe'er, D. , & Danko, C. G. (2022). Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single‐cell RNA sequencing in oncology. Nature Cancer, 3(4), 505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli, E. T. , & Goldstein, D. B. (2010). Uncovering the roles of rare variants in common disease through whole‐genome sequencing. Nature Reviews Genetics, 11(6), 415–425. [DOI] [PubMed] [Google Scholar]
- Clayton, D. G. , Walker, N. M. , Smyth, D. J. , Pask, R. , Cooper, J. D. , Maier, L. M. , Smink, L. J. , Lam, A. C. , Ovington, N. R. , Stevens, H. E. , Nutland, S. , Howson, J. M. , Faham, M. , Moorhead, M. , Jones, H. B. , Falkowski, M. , Hardenbol, P. , Willis, T. D. , & Todd, J. A. (2005). Population structure, differential bias and genomic control in a large‐scale, case‐control association study. Nature Genetics, 37(11), 1243–1246. [DOI] [PubMed] [Google Scholar]
- Cordell, H. J. (2002). Epistasis: What it means, what it doesn't mean, and statistical methods to detect it in humans. Human Molecular Genetics, 11(20), 2463–2468. [DOI] [PubMed] [Google Scholar]
- Cordell, H. J. (2003). Affected‐sib‐pair data can be used to distinguish two‐locus heterogeneity from two‐locus epistasis. The American Journal of Human Genetics, 73(6), 1468–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager, P. L. , Hacohen, N. , Mathis, D. , Regev, A. , Stranger, B. E. , & Benoist, C. (2015). Immvar project: Insights and design considerations for future studies of “healthy” immune variation. Seminars in Immunology, 27, 51–57. [DOI] [PubMed] [Google Scholar]
- Diaz‐Papkovich, A. , Anderson‐Trocmé, L. , & Gravel, S. (2019). Umap reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS genetics, 15(11), e1008432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens, L. M. , Heron, S. , & Mulley, J. (2007). A polygenic heterogeneity model for common epilepsies with complex genetics. Genes, Brain and Behavior, 6(7), 593–597. [DOI] [PubMed] [Google Scholar]
- Dong, X. L. , & Srivastava, D. (2013). Big data integration. 2013 IEEE 29th International Conference on Data Engineering (ICDE) (pp. 1245–1248). IEEE. [Google Scholar]
- Dudbridge, F. (2013). Power and predictive accuracy of polygenic risk scores. PLoS Genetics, 9(3), e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, D. , Scott, L. , Boehnke, M. , & Lee, S. (2019). Multi‐skat: General framework to test for rare‐variant association with multiple phenotypes. Genetic epidemiology, 43(1), 4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, S. , Beesley, J. , French, J. , & Dunning, A. (2013). Beyond GWAS: Illuminating the dark road from association to function. The American Journal of Human Genetics, 93(5), 779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret, G. , Lamparter, D. , Hoggart, C. , Whittaker, J. , Beckmann, J. , & Kutalik, Z. (2012). A multi‐SNP locus association methods reveals a substantial fraction of the missing heritability. The American Journal of Human Genetics, 91(5), 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston, R. C. , Satagopan, J. M. , & Sun, S. (2012). Genetic terminology. In Elston R. C. (Ed.), Statistical Human Genetics (pp. 1–9). Springer. [Google Scholar]
- Fan, R. , Wang, Y. , Yan, Q. , Ding, Y. , Weeks, D. E. , Lu, Z. , Ren, H. , Cook, R. J. , Xiong, M. , Swaroop, A. , Chew, E. Y. , & Chen, W. (2016). Gene‐based association analysis for censored traits via fixed effect functional regressions. Genetic Epidemiology, 40(2), 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger, M. , Linneberg, A. , Werge, T. , & Jørgensen, T. (2008). Analysis of heterogeneity and epistasis in physiological mixed populations by combined structural equation modelling and latent class analysis. BMC Genetics, 9(1), 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell, R. (1998). Hereditary lymphedema: Evidence for linkage and genetic heterogeneity. Human Molecular Genetics, 7(13), 2073–2078. [DOI] [PubMed] [Google Scholar]
- Ford, D. , Easton, D. F. , Stratton, M. , Narod, S. , Goldgar, D. , Devilee, P. , Bishop, D. T. , Weber, B. , Lenoir, G. , Chang‐Claude, J. , Sobol, H. , Teare, M. D. , Struewing, J. , Arason, A. , Scherneck, S. , Peto, J. , Rebbeck, T. R. , Tonin, P. , Neuhausen, S. , … Zelada‐Hedman, M. (1998). Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. The American Journal of Human Genetics, 62(3), 676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, G. (2012). Rare and common variants: Twenty arguments. Nature Reviews Genetics, 13(2), 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Cabrero, D. , Abugessaisa, I. , Maier, D. , Teschendorff, A. , Merkenschlager, M. , Gisel, A. , Ballestar, E. , Bongcam‐Rudloff, E. , Conesa, A. , & Tegnér, J. (2014). Data integration in the era of omics: current and future challenges. BMC Systems Biology, 8(Suppl. 2), I1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlov, I. P. , Gorlova, O. Y. , Sunyaev, S. R. , Spitz, M. R. , & Amos, C. I. (2008). Shifting paradigm of association studies: Value of rare single‐nucleotide polymorphisms. The American Journal of Human Genetics, 82(1), 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinot, F. , Szafranski, M. , Ambroise, C. , & Samson, F. (2018). Learning the optimal scale for GWAS through hierarchical SNP aggregation. BMC Bioinformatics, 19(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Peng, Y. , Gao, A. , Du, C. , & Herman, J. G. (2019). Epigenetic heterogeneity in cancer. Biomarker Research, 7(1), 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R. G. , & Somer, R. A. (2017). Intratumor heterogeneity: Novel approaches for resolving genomic architecture and clonal evolution. Molecular Cancer Research, 15(9), 1127–1137. [DOI] [PubMed] [Google Scholar]
- Haffner, M. C. , Zwart, W. , Roudier, M. P. , True, L. D. , Nelson, W. G. , Epstein, J. I. , De Marzo, A. M. , Nelson, P. S. , & Yegnasubramanian, S. (2021). Genomic and phenotypic heterogeneity in prostate cancer. Nature Reviews Urology, 18(2), 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines, J. L. , Terwedow, H. A. , Burgess, K. , Pericak‐Vance, M. A. , Rimmler, J. B. , Martin, E. R. , Oksenberg, J. R. , Lincoln, R. , Zhang, D. Y. , Banatao, D. R. , Gatto, N. , Goodkin, D. E. , & Hauser, S. L. (1998). Linkage of the MHC to familial multiple sclerosis suggests genetic heterogeneity. Human Molecular Genetics, 7(8), 1229–1234. [DOI] [PubMed] [Google Scholar]
- Hahn, M. , De Voer, R. , Hoogerbrugge, N. , Ligtenberg, M. , Kuiper, R. , & van Kessel, A. G. (2016). The genetic heterogeneity of colorectal cancer predisposition‐guidelines for gene discovery. Cellular Oncology, 39(6), 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, B. , & Eskin, E. (2012). Interpreting meta‐analyses of genome‐wide association studies. PLoS Genetics, 8(3), e1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari, O. , Wang, J.‐C. , Bucholz, K. , Edenberg, H. J. , Heath, A. , Martin, N. G. , Pergadia, M. L. , Montgomery, G. , Schrage, A. , Bierut, L. J. , Madden, P. F. , & Goate, A. M. (2012). Pathway analysis of smoking quantity in multiple GWAS identifies cholinergic and sensory pathways. PLoS One, 7(12), e50913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidema, A. G. , Boer, J. M. , Nagelkerke, N. , Mariman, E. C. , van der A, D. L. , & Feskens, E. J. (2006). The challenge for genetic epidemiologists: How to analyze large numbers of SNPs in relation to complex diseases. BMC Genetics, 7(1), 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff, L. A. , Sethupathy, P. , Junkins, H. A. , Ramos, E. M. , Mehta, J. P. , Collins, F. S. , & Manolio, T. A. (2009). Potential etiologic and functional implications of genome‐wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences, 106(23), 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge, S. E. , Hager, V. R. , & Greenberg, D. A. (2016). Using linkage analysis to detect gene‐gene interactions. 2. Improved reliability and extension to more‐complex models. PLOS One, 11(1), e0146240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, A. J. , & Patrick, L. M. (2018). The myth of optimality in clinical neuroscience. Trends in Cognitive Sciences, 22(3), 241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, J. H. , Durbin, D. R. , & Winston, F. K. (2000). The learning classifier system: an evolutionary computation approach to knowledge discovery in epidemiologic surveillance. Artificial Intelligence in Medicine, 19(1), 53–74. [DOI] [PubMed] [Google Scholar]
- Hormozdiari, F. , Zhu, A. , Kichaev, G. , Ju, C. J.‐T. , Segré, A. V. , Joo, J. W. J. , Won, H. , Sankararaman, S. , Pasaniuc, B. , Shifman, S. , & Eskin, E. (2017). Widespread allelic heterogeneity in complex traits. The American Journal of Human Genetics, 100(5), 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, K. , Bhattacharya, A. , Mester, R. , Burch, K. S. , & Pasaniuc, B. (2021). On powerful GWAS in admixed populations. Nature genetics, 53(12), 1631–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Cai, N. , Pacheco, P. P. , Narrandes, S. , Wang, Y. , & Xu, W. (2018). Applications of support vector machine (svm) learning in cancer genomics. Cancer Genomics‐Proteomics, 15(1), 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium . (2009). Common polygenic variation contributes to risk of schizophrenia that overlaps with bipolar disorder. Nature, 460(7256), 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob, S. , Wolff, J. J. , Steinbach, M. S. , Doyle, C. B. , Kumar, V. , & Elison, J. T. (2019). Neurodevelopmental heterogeneity and computational approaches for understanding autism. Translational Psychiatry, 9(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, A. E. , & Irizarry, R. A. (2014). Accounting for cellular heterogeneity is critical in epigenome‐wide association studies. Genome Biology, 15(2), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. , Hottenga, J.‐J. , Nivard, M. G. , Abdellaoui, A. , Laport, B. , De Geus, E. J. , Wright, F. A. , Penninx, B. W. , & Boomsma, D. I. (2017). Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Human Molecular Genetics, 26(8), 1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. J. , Goodman, S. J. , & Kobor, M. S. (2015). DNA methylation and healthy human aging. Aging Cell, 14(6), 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski, K. J. , & Snyder, M. P. (2018). Integrative omics for health and disease. Nature Reviews Genetics, 19(5), 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, D. M. , Nelson, J. , Dahabreh, I. J. , Rothwell, P. M. , Altman, D. G. , & Hayward, R. A. (2016). Risk and treatment effect heterogeneity: Re‐analysis of individual participant data from 32 large clinical trials. International Journal of Epidemiology, 45(6), 2075–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, D. M. , Rothwell, P. M. , Ioannidis, J. P. , Altman, D. G. , & Hayward, R. A. (2010). Assessing and reporting heterogeneity in treatment effects in clinical trials: A proposal. Trials, 11(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Li, R. , Dudek, S. M. , & Ritchie, M. D. (2015). Predicting censored survival data based on the interactions between meta‐dimensional omics data in breast cancer. Journal of Biomedical Informatics, 56, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Shin, H. , Sohn, K.‐A. , Verma, A. , Ritchie, M. D. , & Kim, J. H. (2014). Incorporating inter‐relationships between different levels of genomic data into cancer clinical outcome prediction. Methods, 67(3), 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , & Jazwinski, S. M. (2018). The gut microbiota and healthy aging: A mini‐review. Gerontology, 64(6), 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasa, J. , & Rollo, C. D. (1991). Introduction: The heterogeneity of heterogeneity: A glossary. Ecological Heterogeneity, 86, 1–23. [Google Scholar]
- Kourou, K. , Exarchos, T. P. , Exarchos, K. P. , Karamouzis, M. V. , & Fotiadis, D. I. (2015). Machine learning applications in cancer prognosis and prediction. Computational and Structural Biotechnology Journal, 13, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, P. , Zeggini, E. , & Ioannidis, J. P. A. (2009). Replication in genome‐wide association studies. Statistical Science, 24(4), 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulminski, A. M. , Loika, Y. , Culminskaya, I. , Arbeev, K. G. , Ukraintseva, S. V. , Stallard, E. , & Yashin, A. I. (2016). Explicating heterogeneity of complex traits has strong potential for improving GWAS efficiency. Scientific Reports, 6(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan, D. , Higgins, J. P. , Jackson, D. , Bowden, J. , Veroniki, A. A. , Kontopantelis, E. , Viechtbauer, W. , & Simmonds, M. (2019). A comparison of heterogeneity variance estimators in simulated random‐effects meta‐analyses. Research Synthesis Methods, 10(1), 83–98. [DOI] [PubMed] [Google Scholar]
- Lawson, A. R. J. , Abascal, F. , Coorens, T. H. H. , Hooks, Y. , O'Neill, L. , Latimer, C. , Raine, K. , Sanders, M. A. , Warren, A. Y. , Mahbubani, K. T. A. , Bareham, B. , Butler, T. M. , Harvey, L. M. R. , Cagan, A. , Menzies, A. , Moore, L. , Colquhoun, A. J. , Turner, W. , Thomas, B. , … Martincorena, I. (2020). Extensive heterogeneity in somatic mutation and selection in the human bladder. Science, 370(6512), 75–82. [DOI] [PubMed] [Google Scholar]
- Lee, S. , Abecasis, G. R. , Boehnke, M. , & Lin, X. (2014). Rare‐variant association analysis: study designs and statistical tests. The American Journal of Human Genetics, 95(1), 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescai, F. , & Franceschi, C. (2010). The impact of phenocopy on the genetic analysis of complex traits. PLoS One, 5(7), e11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , & Leal, S. M. (2008). Methods for detecting associations with rare variants for common diseases: Application to analysis of sequence data. The American Journal of Human Genetics, 83(3), 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Liu, L. , Zhou, J. , & Wang, C. (2018). Heterogeneity analysis and diagnosis of complex diseases based on deep learning method. Scientific Reports, 8(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Mi, Y. , Mueller, T. , Kreibich, S. , Williams, E. G. , Van Drogen, A. , Borel, C. , Frank, M. , Germain, P. L. , Bludau, I. , Mehnert, M. , Seifert, M. , Emmenlauer, M. , Sorg, I. , Bezrukov, F. , Bena, F. S. , Zhou, H. , Dehio, C. , Testa, G. , … Aebersold, R. (2019). Multi‐omic measurements of heterogeneity in HeLa cells across laboratories. Nature Biotechnology, 37(3), 314–322. [DOI] [PubMed] [Google Scholar]
- Llinares‐López, F. , Grimm, D. G. , Bodenham, D. A. , Gieraths, U. , Sugiyama, M. , Rowan, B. , & Borgwardt, K. (2015). Genome‐wide detection of intervals of genetic heterogeneity associated with complex traits. Bioinformatics, 31(12), i240–i249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr, J. G. , Stojanov, P. , Carter, S. L. , Cruz‐Gordillo, P. , Lawrence, M. S. , Auclair, D. , Sougnez, C. , Knoechel, B. , Gould, J. , Saksena, G. , Cibulskis, K. , McKenna, A. , Chapman, M. A. , Straussman, R. , Levy, J. , Perkins, L. M. , Keats, J. J. , Schumacher, S. E. , Rosenberg, M. , … Golub, T. R. (2014). Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell, 25(1), 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M. (2019). Embedded feature selection accounting for unknown data heterogeneity. Expert Systems with Applications, 119, 350–361. [Google Scholar]
- Lubke, G. H. , & Muthén, B. (2005). Investigating population heterogeneity with factor mixture models. Psychological Methods, 10(1), 21–39. [DOI] [PubMed] [Google Scholar]
- Madsen, A. M. , Ottman, R. , & Hodge, S. E. (2011). Causal models for investigating complex genetic disease ii. what causal models can tell us about penetrance for additive, heterogeneity, and multiplicative two‐locus models. Human Heredity, 72(1), 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchia, M. , Cullis, J. , Turecki, G. , Rouleau, G. A. , Uher, R. , & Alda, M. (2013). The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One, 8(10), e76295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, A. R. , Kanai, M. , Kamatani, Y. , Okada, Y. , Neale, B. M. , & Daly, M. J. (2019). Clinical use of current polygenic risk scores may exacerbate health disparities. Nature Genetics, 51(4), 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan, J. , & King, M.‐C. (2010). Genetic heterogeneity in human disease. Cell, 141(2), 210–217. [DOI] [PubMed] [Google Scholar]
- McGinniss, M. J. , & Kaback, M. M. (2013). Heterozygote testing and carrier screening. In Pyeritz R. E., Korf B. R., & Grody W. W. (Eds.), Emery and Rimoin's principles and practice of medical genetics (pp. 1–10). Elsevier. [Google Scholar]
- Mei, H. , Cuccaro, M. , & Martin, E. (2007). Multifactor dimensionality reduction‐phenomics: A novel method to capture genetic heterogeneity with use of phenotypic variables. The American Journal of Human Genetics, 81(6), 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milaneschi, Y. , Lamers, F. , Peyrot, W. J. , Abdellaoui, A. , Willemsen, G. , Hottenga, J. J. , Jansen, R. , Mbarek, H. , Dehghan, A. , Lu, C. , CHARGE Inflammation Working Group, Boomsma, D. I. , & Penninx, B. W. J. H. (2016). Polygenic dissection of major depression clinical heterogeneity. Molecular Psychiatry, 21(4), 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelpunkt, A. , Galili, T. , Kozlovski, T. , Bregman, N. , Shachar, N. , Markus‐Kalish, M. , & Benjamini, Y. (2020). Novel Alzheimer's disease subtypes identified using a data and knowledge driven strategy. Scientific Reports, 10(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monir, M. M. , & Zhu, J. (2017). Comparing GWAS results of complex traits using full genetic model and additive models for revealing genetic architecture. Scientific Reports, 7(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, C. C. B. , Basile, A. O. , Wallace, J. R. , Frase, A. T. , & Ritchie, M. D. (2016). A biologically informed method for detecting rare variant associations. BioData Mining, 9(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motsinger‐Reif, A. A. , Fanelli, T. J. , Davis, A. C. , & Ritchie, M. D. (2008). Power of grammatical evolution neural networks to detect gene‐gene interactions in the presence of error. BMC Research Notes, 1(1), 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, N. , Ward, H. A. , Jenab, M. , Rothwell, J. A. , Boutron‐Ruault, M.‐C. , Carbonnel, F. , Kvaskoff, M. , Kaaks, R. , Kühn, T. , Boeing, H. , Aleksandrova, K. , Weiderpass, E. , Skeie, G. , Borch, K. B. , Tjønneland, A. , Kyrø, C. , Overvad, K. , Dahm, C. C. , Jakszyn, P. , … Gunter, M. J. (2019). Heterogeneity of colorectal cancer risk factors by anatomical subsite in 10 European countries: A multinational cohort study. Clinical Gastroenterology and Hepatology, 17(7), 1323–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan, R. , Sailem, H. , Mardakheh, F. K. , AriasGarcia, M. , Tape, C. J. , Dowsett, M. , Bakal, C. , & Yuan, Y. (2016). Microenvironmental heterogeneity parallels breast cancer progression: A histology‐genomic integration analysis. PLOS Medicine, 13(2), e1001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C. K. , Weigelt, B. , A'Hern, R. , Bidard, F.‐C. , Lemetre, C. , Swanton, C. , Shen, R. , & Reis‐Filho, J. S. (2014). Predictive performance of microarray gene signatures: impact of tumor heterogeneity and multiple mechanisms of drug resistance. Cancer Research, 74(11), 2946–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizaki, S. S. , & Boyle, A. P. (2017). Mining the unknown: Assigning function to noncoding single nucleotide polymorphisms. Trends in Genetics, 33(1), 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, L. J. (2021). The distribution of common‐variant effect sizes. Nature Genetics, 53(8), 1243–1249. [DOI] [PubMed] [Google Scholar]
- Paranjapye, A. , Ruffin, M. , Harris, A. , & Corvol, H. (2020). Genetic variation in CFTR and modifier loci may modulate cystic fibrosis disease severity. Journal of Cystic Fibrosis, 19, S10–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C. , Kim, J. , Kim, J. , & Park, S. (2018). Machine learning‐based identification of genetic interactions from heterogeneous gene expression profiles. PloS One, 13(7), e0201056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A. P. , Tirosh, I. , Trombetta, J. J. , Shalek, A. K. , Gillespie, S. M. , Wakimoto, H. , Cahill, D. P. , Nahed, B. V. , Curry, W. T. , Martuza, R. L. , Louis, D. N. , Rozenblatt‐Rosen, O. , Suva, M. L. , Regev, A. , & Bernstein, B. E. (2014). Single‐cell RNA‐seq highlights intratumoral heterogeneity in primary glioblastoma. Science, 344(6190), 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, N. , Price, A. L. , & Reich, D. (2006). Population structure and eigenanalysis. PLoS Genetics, 2(12), e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, R. E. , Cai, N. , Dahl, A. W. , Bigdeli, T. B. , Edwards, A. C. , Webb, B. T. , Bacanu, S.‐A. , Zaitlen, N. , Flint, J. , & Kendler, K. S. (2018). Molecular genetic analysis subdivided by adversity exposure suggests etiologic heterogeneity in major depression. American Journal of Psychiatry, 175(6), 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault, J.‐B. , O'reilly, P. F. , Schoeler, T. , Ploubidis, G. B. , Rijsdijk, F. , & Dudbridge, F. (2018). Using genetic data to strengthen causal inference in observational research. Nature Reviews Genetics, 19(9), 566–580. [DOI] [PubMed] [Google Scholar]
- Pivovarov, R. , Perotte, A. J. , Grave, E. , Angiolillo, J. , Wiggins, C. H. , & Elhadad, N. (2015). Learning probabilistic phenotypes from heterogeneous EHR data. Journal of Biomedical Informatics, 58, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polychronakos, C. , & Li, Q. (2011). Understanding type 1 diabetes through genetics: advances and prospects. Nature Reviews Genetics, 12(11), 781–792. [DOI] [PubMed] [Google Scholar]
- Povysil, G. , Petrovski, S. , Hostyk, J. , Aggarwal, V. , Allen, A. S. , & Goldstein, D. B. (2019). Rare‐variant collapsing analyses for complex traits: Guidelines and applications. Nature Reviews Genetics, 20(12), 747–759. [DOI] [PubMed] [Google Scholar]
- Rahman, R. , Matlock, K. , Ghosh, S. , & Pal, R. (2017). Heterogeneity aware random forest for drug sensitivity prediction. Scientific Reports, 7(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappoport, N. , & Shamir, R. (2018). Multi‐omic and multi‐view clustering algorithms: review and cancer benchmark. Nucleic Acids Research, 46(20), 10546–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, P. , Zheng, L. , Lucas, J. , & Carin, L. (2014). Bayesian joint analysis of heterogeneous genomics data. Bioinformatics, 30(10), 1370–1376. [DOI] [PubMed] [Google Scholar]
- Risch, N. (1990). Linkage strategies for genetically complex traits. I. Multilocus models. American Journal of Human Genetics, 46(2), 222. [PMC free article] [PubMed] [Google Scholar]
- Risch, N. , & Merikangas, K. (1996). The future of genetic studies of complex human diseases. Science, 273(5281), 1516–1517. [DOI] [PubMed] [Google Scholar]
- Ritchie, M. D. (2011). Using biological knowledge uncover the mystery in the search for epistasis in genome‐wide association studies: Biology used to search for epistasis. Annals of Human Genetics, 75(1), 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M. D. (2015). Finding the epistasis needles in the genome‐wide haystack. In Moore J. H. & Williams S. M. (Eds.), Epistasis (pp. 19–33). Springer. [DOI] [PubMed] [Google Scholar]
- Rodgers, J. L. (2010). The epistemology of mathematical and statistical modeling: A quiet methodological revolution. American Psychologist, 65(1), 1. [DOI] [PubMed] [Google Scholar]
- Rohart, F. , Gautier, B. , Singh, A. , & LêCao, K.‐A. (2017). mixOmics: An R package for omics feature selection and multiple data integration. PLOS Computational Biology, 13(11), e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman, K. J. , Gallacher, J. E. , & Hatch, E. E. (2013). Why representativeness should be avoided. International Journal of Epidemiology, 42(4), 1012–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, J. , Fransquet, P. , Wrigglesworth, J. , & Lacaze, P. (2018). Phenotypic heterogeneity in dementia: A challenge for epidemiology and biomarker studies. Frontiers in Public Health, 6, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer, S. , Hellenthal, G. , Pirinen, M. , Spencer, C. C. , Patsopoulos, N. A. , Moutsianas, L. , Dilthey, A. , Su, Z. , Freeman, C. , Hunt, S. E. , Edkins, S. , Gray, E. , Booth, D. , Potter, S. C. , Goris, A. , Band, G. , Oturai, A. B. , Strange, A. , Saarela, J. , … Compston, A. (2011). Genetic risk and a primary role for cell‐mediated immune mechanisms in multiple sclerosis. Nature, 476(7359), 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat, J. , Levy, D. L. , & McCarthy, S. E. (2009). Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends in Genetics, 25(12), 528–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, S. , Bernardo, M. M. , Dzinic, S. H. , Chen, K. , Heath, E. I. , & Sakr, W. A. (2018). Tackling tumor heterogeneity and phenotypic plasticity in cancer precision medicine: Our experience and a literature review. Cancer and Metastasis Reviews, 37(4), 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Q. , Hu, B. , Zeng, T. , & Zhang, C. (2019). Multi‐view subspace clustering analysis for aggregating multiple heterogeneous omics data. Frontiers in Genetics, 10, 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivade, C. , Raghavan, P. , Fosler‐Lussier, E. , Embi, P. J. , Elhadad, N. , Johnson, S. B. , & Lai, A. M. (2014). A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association, 21(2), 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner, D. (2017). Overview of admixture mapping. Current Protocols in Human Genetics, 94(1), 1–23. [DOI] [PubMed] [Google Scholar]
- Sirugo, G. , Williams, S. M. , & Tishkoff, S. A. (2019). The missing diversity in human genetic studies. Cell, 177(1), 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. W. , & O'Brien, S. J. (2005). Mapping by admixture linkage disequilibrium: Advances, limitations and guidelines. Nature Reviews Genetics, 6(8), 623–632. [DOI] [PubMed] [Google Scholar]
- Song, J.‐L. , Chen, C. , Yuan, J.‐P. , & Sun, S.‐R. (2016). Progress in the clinical detection of heterogeneity in breast cancer. Cancer Medicine, 5(12), 3475–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stessman, H. A. , Bernier, R. , & Eichler, E. E. (2014). A genotype‐first approach to defining the subtypes of a complex disease. Cell, 156(5), 872–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, E. , Dixon, D. R. , Novack, M. N. , Granpeesheh, D. , Smith, T. , & Linstead, E. (2019). Identification and analysis of behavioral phenotypes in autism spectrum disorder via unsupervised machine learning. International Journal of Medical Informatics, 129, 29–36. [DOI] [PubMed] [Google Scholar]
- Sudhakar, P. , Verstockt, B. , Cremer, J. , Verstockt, S. , Sabino, J. , Ferrante, M. , & Vermeire, S. (2020). Understanding the molecular drivers of disease heterogeneity in Crohn's disease using multi‐omic data integration and network analysis. Inflammatory Bowel Diseases, 27(6), 870–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, P. F. , Agrawal, A. , Bulik, C. M. , Andreassen, O. A. , Børglum, A. D. , Breen, G. , Cichon, S. , Edenberg, H. J. , Faraone, S. V. , Gelernter, J. , Mathews, C. A. , Nievergelt, C. M. , Smoller, J. W. , & O'Donovan, M. C. (2018). Psychiatric genomics: An update and an agenda. American Journal of Psychiatry, 175(1), 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , Zheng, Y. , & Hsu, L. (2013). A unified mixed‐effects model for rare‐variant association in sequencing studies. Genetic Epidemiology, 37(4), 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, A. J. , Abrams, K. R. , Jones, D. R. , Jones, D. R. , Sheldon, T. A. , & Song, F. (2000). Methods for meta‐analysis in medical research (Vol. 348). Wiley. [Google Scholar]
- Svishcheva, G. R. , Belonogova, N. M. , Zorkoltseva, I. V. , Kirichenko, A. V. , & Axenovich, T. I. (2019). Gene‐based association tests using GWAS summary statistics. Bioinformatics, 35(19), 3701–3708. [DOI] [PubMed] [Google Scholar]
- Swinnen, B. , & Robberecht, W. (2014). The phenotypic variability of amyotrophic lateral sclerosis. Nature Reviews Neurology, 10(11), 661–670. [DOI] [PubMed] [Google Scholar]
- Szulc, P. , Bogdan, M. , Frommlet, F. , & Tang, H. (2017). Joint genotype‐and ancestry‐based genome‐wide association studies in admixed populations. Genetic Epidemiology, 41(6), 555–566. [DOI] [PubMed] [Google Scholar]
- Talebizadeh, Z. , Arking, D. E. , & Hu, V. W. (2013). A novel stratification method in linkage studies to address inter‐and intra‐family heterogeneity in autism. PLoS One, 8(6), e67569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H. , Siegmund, D. O. , Johnson, N. A. , Romieu, I. , & London, S. J. (2010). Joint testing of genotype and ancestry association in admixed families. Genetic Epidemiology, 34(8), 783–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton‐Wells, T. A. , Moore, J. H. , & Haines, J. L. (2006). Dissecting trait heterogeneity: A comparison of three clustering methods applied to genotypic data. BMC Bioinformatics, 7(1), 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani, A. , Wineinger, N. E. , & Topol, E. J. (2018). The personal and clinical utility of polygenic risk scores. Nature Reviews Genetics, 19(9), 581. [DOI] [PubMed] [Google Scholar]
- Truong, A. H. , Sharmanska, V. , Limback‐Stanic, C. , & Grech‐Sollars, M. (2020). Optimization of deep learning methods for visualization of tumor heterogeneity and brain tumor grading through digital pathology. Neuro‐Oncology Advances, 2(1), vdaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S. D. , & Bush, W. S. (2011). Multivariate analysis of regulatory snps: empowering personal genomics by considering cis‐epistasis and heterogeneity. In Altman R. B., Dunker A. K., L. Hunter , T. A. Murray , & Klein T. E. (Eds.), Biocomputing 2011 (pp. 276–287). World Scientific. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz, R. J. , Andrew, A. S. , Karagas, M. R. , & Moore, J. H. (2013). Role of genetic heterogeneity and epistasis in bladder cancer susceptibility and outcome: A learning classifier system approach. Journal of the American Medical Informatics Association, 20(4), 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz, R. J. , & Browne, W. N. (2017). Introduction to learning classifier systems. SpringerBriefs in Intelligent Systems. Springer Berlin Heidelberg. [Google Scholar]
- Urbanowicz, R. J. , Granizo‐Mackenzie, D. , & Moore, J. H. (2012). Using expert knowledge to guide covering and mutation in a Michigan style learning classifier system to detect epistasis and heterogeneity. In Hutchison D., Kanade T., Kittler J., Kleinberg J. M., Mattern F., Mitchell J. C., Naor M., Nierstrasz O., PanduRangan C., Steffen B., Sudan M., Terzopoulos D., Tygar D., Vardi M. Y., Weikum G., Coello C. A. C., Cutello V., Deb K., Forrest S., Nicosia G., & Pavone M. (Eds.), Parallel problem solving from nature—PPSN XII. Series Title: Lecture Notes in Computer Science (Vol. 7491, pp. 266–275). Springer Berlin Heidelberg. [Google Scholar]
- Urbanowicz, R. J. , & Moore, J. H. (2009). Learning classifier systems: A complete introduction, review, and roadmap. Journal of Artificial Evolution and Applications, 2009, 1–25. [Google Scholar]
- Urbanowicz, R. J. , & Moore, J. H. (2015). Exstracs 2.0: Description and evaluation of a scalable learning classifier system. Evolutionary Intelligence, 8(2), 89–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Sluis, S. , Verhage, M. , Posthuma, D. , & Dolan, C. V. (2010). Phenotypic complexity, measurement bias, and poor phenotypic resolution contribute to the missing heritability problem in genetic association studies. PLoS One, 5(11), e13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooden, S. M. , Heiser, W. J. , Kok, J. N. , Verbaan, D. , Van Hilten, J. J. , & Marinus, J. (2010). The identification of Parkinson's disease subtypes using cluster analysis: A systematic review. Movement disorders, 25(8), 969–978. [DOI] [PubMed] [Google Scholar]
- Visscher, P. , Brown, M. , McCarthy, M. , & Yang, J. (2012). Five years of GWAS discovery. The American Journal of Human Genetics, 90(1), 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , & Larson, D. R. (2018). Splicing heterogeneity: Separating signal from noise. Genome Biology, 19(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D. C. , & Wang, X. (2017). Systems heterogeneity: An integrative way to understand cancer heterogeneity. Seminars in Cell & Developmental Biology, 64, 1–4. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Spiegelman, D. , Kuchiba, A. , Lochhead, P. , Kim, S. , Chan, A. T. , Poole, E. M. , Tamimi, R. , Tworoger, S. S. , Giovannucci, E. , Rosner, B. , & Ogino, S. (2016). Statistical methods for studying disease subtype heterogeneity. Statistics in Medicine, 35(5), 782–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Park, J. , Susztak, K. , Zhang, N. R. , & Li, M. (2019). Bulk tissue cell type deconvolution with multi‐subject single‐cell expression reference. Nature communications, 10(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. F. , Zhang, Y. , Lin, Z. , Zhang, H. , Wang, T. Y. , Cao, Y. , Morris, D. L. , Sheng, Y. , Yin, X. , Zhong, S. L. , Gu, X. , Lei, Y. , He, J. , Wu, Q. , Shen, J. J. , Yang, J. , Lam, T. H. , Lin, J. H. , Mai, Z. M. , … Yang, W. (2021). Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nature Communications, 12(1), 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt, F. R. , Pathak, G. A. , Tylee, D. S. , Goswami, A. , & Polimanti, R. (2020). Heterogeneity and polygenicity in psychiatric disorders: A genome‐wide perspective. Chronic Stress, 4, 2470547020924844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, A. R. , Hernandez, D. G. , Nalls, M. A. , Yaghootkar, H. , Gibbs, J. R. , Harries, L. W. , Chong, S. , Moore, M. , Weedon, M. N. , Guralnik, J. M. , Bandinelli, S. , Murray, A. , Ferrucci, L. , Singleton, A. B. , Melzer, D. , & Frayling, T. M. (2011). Allelic heterogeneity and more detailed analyses of known loci explain additional phenotypic variation and reveal complex patterns of association. Human Molecular Genetics, 20(20), 4082–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, N. R. , & Maier, R. (2014). Genetic basis of complex genetic disease: The contribution of disease heterogeneity to missing heritability. Current Epidemiology Reports, 1(4), 220–227. [Google Scholar]
- Wu, M. C. , Lee, S. , Cai, T. , Li, Y. , Boehnke, M. , & Lin, X. (2011). Rare‐variant association testing for sequencing data with the sequence kernel association test. The American Journal of Human Genetics, 89(1), 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Waite, L. L. , Jackson, A. U. , Sheu, W. H.‐H. , Buyske, S. , Absher, D. , Arnett, D. K. , Boerwinkle, E. , Bonnycastle, L. L. , Carty, C. L. , Cheng, I. , Cochran, B. , Croteau‐Chonka, D. C. , Dumitrescu, L. , Eaton, C. B. , Franceschini, N. , Guo, X. , Henderson, B. E. , Hindorff, L. A. , … Mohlke, K. L. (2013). Trans‐ethnic fine‐mapping of lipid loci identifies population specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genetics, 9(3), e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L. , Wei, F. , Liang, F. , Li, Q. , Deng, H. , Tan, S. , Chen, S. , Xiong, F. , Guo, C. , Liao, Q. , Li, X. , Zhang, W. , Wu, M. , Zhou, Y. , Xiang, B. , Zhou, M. , Li, X. , Xiong, W. , Zeng, Z. , & Li, G. (2019). Tsc22d2 identified as a candidate susceptibility gene of multi‐cancer pedigree using genome‐wide linkage analysis and whole‐exome sequencing. Carcinogenesis, 40(7), 819–827. [DOI] [PubMed] [Google Scholar]
- Yashin, A. I. , Wu, D. , Arbeeva, L. S. , Arbeev, K. G. , Kulminski, A. M. , Akushevich, I. , Kovtun, M. , Culminskaya, I. , Stallard, E. , Li, M. , & Ukraintseva, S. V. (2015). Genetics of aging, health, and survival: Dynamic regulation of human longevity related traits. Frontiers in Genetics, 6, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. , Hazelton, W. D. , Luebeck, G. E. , & Grady, W. M. (2020). Epigenetic aging: More than just a clock when it comes to cancer. Cancer Research, 80(3), 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Karns, R. , Sun, G. , Indugula, S. R. , Cheng, H. , Havas‐Augustin, D. , Novokmet, N. , Durakovic, Z. , Missoni, S. , Chakraborty, R. , Rudan, P. , & Deka, R. (2012). Finding missing heritability in less significant loci and allelic heterogeneity: Genetic variation in human height. PLoS One, 7(12), e51211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Späth, S. S. , Marjani, S. L. , Zhang, W. , & Pan, X. (2018). Characterization of cancer genomic heterogeneity by next‐generation sequencing advances precision medicine in cancer treatment. Precision clinical medicine, 1(1), 29–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , & Fang, G. (2018). Matrixepistasis: Ultrafast, exhaustive epistasis scan for quantitative traits with covariate adjustment. Bioinformatics, 34(14), 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintzaras, E. , & Ioannidis, J. P. A. (2005). Heterogeneity testing in meta‐analysis of genome searches. Genetic Epidemiology, 28(2), 123–137. [DOI] [PubMed] [Google Scholar]


