Abstract
Huntington’s disease (HD) is a fatal, hereditary neurodegenerative disorder that causes chorea, cognitive deficits, and psychiatric symptoms. It is characterized by accumulation of mutant Htt protein, which primarily impacts striatal medium-sized spiny neurons (MSNs), as well as cortical pyramidal neurons (CPNs), causing synapse loss and eventually cell death. Perturbed Ca2+ homeostasis is believed to play a major role in HD, as altered Ca2+ homeostasis often precedes striatal dysfunction and manifestation of HD symptoms. In addition, dysregulation of Ca2+ can cause morphological and functional changes in MSNs and CPNs. Therefore, Ca2+ imaging techniques have the potential of visualizing changes in Ca2+ dynamics and neuronal activity in HD animal models. This minireview focuses on studies using diverse Ca2+ imaging techniques, including two-photon microscopy, fiber photometry, and miniscopes, in combination of Ca2+ indicators to monitor activity of neurons in HD models as the disease progresses. We then discuss the future applications of Ca2+ imaging to visualize disease mechanisms and alterations associated with HD, as well as studies showing how, as a proof-of-concept, Ca2+imaging using miniscopes in freely-behaving animals can help elucidate the differential role of direct and indirect pathway MSNs in HD symptoms.
Keywords: Huntington’s disease, miniscope, fiber photometry, calcium imaging, two-photon microscopy (2-PM)
Introduction
Huntington’s Disease (HD) is a dominantly inherited neurodegenerative disorder that causes chorea, cognitive deficits, and psychiatric symptoms (Bates et al., 2015). HD is caused by an expansion of CAG triplet repeats (encoding glutamine) in the huntingtin gene (The Huntington’s Disease Collaborative Research Group, 1993; Walker, 2007). While the huntingtin protein (Htt) is typically expressed throughout the brain and body and is thought to play a role in gene expression and signal transduction, the mutant form (mHtt) preferentially targets striatal medium-sized spiny neurons (MSNs) and cortical pyramidal neurons (CPNs). At the cellular level, mHtt causes the HD phenotype through a toxic gain-of-function, leading to synapse and spine loss, cortical hyperexcitability, and striatal and cortical cell degeneration (Vonsattel and Difiglia, 1998; Tobin and Signer, 2000; Waldvogel et al., 2015).
Mouse models are widely used for investigation of HD pathogenesis. R6/2 and R6/1 are transgenic models that express a fragment of mHtt, leading to a rapidly progressing phenotype that develops symptoms as early as 4–5 weeks, and is considered a good model of juvenile HD (Li et al., 2005; Cepeda et al., 2010). Bacterial artificial chromosome (BAC) and yeast artificial chromosome (YAC) model mice, such as BACHD and YAC128, are transgenic models containing the full-length mutant Htt gene, often displaying neuropathology that is highly consistent with human HD (Slow et al., 2003; Gray et al., 2008). Knock-in mice (including CAG140 and Q175), carry the CAG expansion in the endogenous mouse Htt gene, and thus exhibit the progressive, symptomatic phenotype that replicates most faithfully human HD (Menalled, 2005).
Studies in animal models have suggested that perturbations in Ca2+ homeostasis play a role in HD, as altered Ca2+ dynamics often precede striatal dysfunction and manifestation of HD symptoms (Bezprozvanny, 2009; Wu et al., 2016; Raymond, 2017; Mackay et al., 2018). For example, mHtt can disrupt Ca2+ homeostasis through association with other proteins that regulate intracellular Ca2+ levels. mHtt also increases N-methyl-D-aspartate (NMDA) receptor-mediated excitotoxicity (Zeron et al., 2002; Bezprozvanny and Hayden, 2004), causing increased extracellular Ca2+ to enter the cell via extrasynaptic NMDARs (Cepeda et al., 2001). Pioneering Ca2+ imaging studies demonstrated that mHtt binds to the inositol 1,4,5-trisphosphate (InsP3) receptor 1, making it more sensitive to InsP3 upon its release when glutamate binds to its receptors. InsP3R1, when activated by InsP3, causes Ca2+ release from the endoplasmic reticulum (Bezprozvanny and Hayden, 2004; Wu et al., 2016; Czeredys, 2020), and low endoplasmic reticulum Ca2+ levels activate neuronal store-operated channels (SOCs), probably as a compensatory mechanism. SOCs are membrane-bound Ca2+-conducting channels that activate in response to depletion of Ca2+ stores (Putney, 1986). Because Ca2+ signaling is fundamentally disturbed early on in HD progression and potentially leads to development of disease symptoms, Ca2+ imaging techniques have the potential to visualize changes in neuronal activity associated with HD in animal models. They also provide a tool to test various hypotheses about HD pathogenesis, and visualize how intracellular Ca2+ dynamics differs in mouse models.
The focus of this minireview is not to describe in detail diverse Ca2+ imaging techniques as excellent, comprehensive reviews are already available (Werner et al., 2019; Broussard and Petreanu, 2021; Wang et al., 2021; Grienberger et al., 2022; Kim and Schnitzer, 2022; Lake and Higley, 2022). Instead, we discuss studies using some of these techniques to monitor the activity of neurons, astrocytes, and mitochondria in HD models (Table 1). Compared to imaging studies in other disease conditions, use of Ca2+ imaging to examine HD progression has taken longer to implement. This is partly due to the overwhelming reliance on electrophysiological recording techniques and also to the slow development of genetic mouse models of HD, associated to the fact that some models display a very severe phenotype, e.g., R6/2, and others show only very mild, protracted symptoms (BACHD, Q175). In the last section, we discuss future applications of Ca2+ imaging to visualize HD disease mechanisms, as well as some of our own studies showing how, as a proof-of-concept, Ca2+ imaging using miniscopes in freely-behaving animals can help elucidate the role of CPNs and direct and indirect pathway MSNs in HD symptoms. We apologize for any omissions of the already vast literature on Ca2+ imaging in HD.
TABLE 1.
Ca2+ imaging methods applied to the study of Huntington’s disease.
| Imaging technique | Type of preparation | Spatial resolution | Equipment cost | Tissue damage | HD papers published |
| Confocal or 2-PLSM | Cell cultures or slices | Individual cells, organelles | Tissue no longer intact in in vitro systems of dissociated cells or brain slices | Fernandes et al., 2007; Lim et al., 2008; Wang et al., 2013; Jiang et al., 2016; Fernandez-Garcia et al., 2020; Oikonomou et al., 2021 | |
| Head-fixed (freely moving limbs) | Individual neurons (depth of ∼1 mm) | Inflammation; potential damage to dura and cortical surface during craniotomy and cranial window implantation | Burgold et al., 2019; Donzis et al., 2020 | ||
| Fiber photometry | Freely-moving | Population signal (depth ∼2 mm) | $ | Inflammation; tissue displacement by optic fiber implantation (200 μm diameter fiber) | Koch et al., 2022 |
| Miniscope | Freely-moving | Individual neurons (depth ∼150 μm from GRIN lens) | $– | Inflammation; tissue displacement due to GRIN lens implantation (0.5–1.0 mm diameter lens) | No papers yet Proof-of-concept |
Fluorescence confocal and 2-photon laser scanning microscopy
Past investigations into circuits and pathways involved in HD development have been limited by difficulty recording from large neuron populations and constraints in temporal and spatial resolution. While a leading tool for investigating neuronal dynamics has been electrophysiological recordings of neuron electrical activity in the form of action potential firing (both extracellularly and intracellularly) due to high temporal resolution, these methods can be limited in spatial resolution, specificity of neuron types, or are typically able to record from only a few neurons. This presents challenges in investigating altered circuits in HD due to the relatively isolated neuronal recordings.
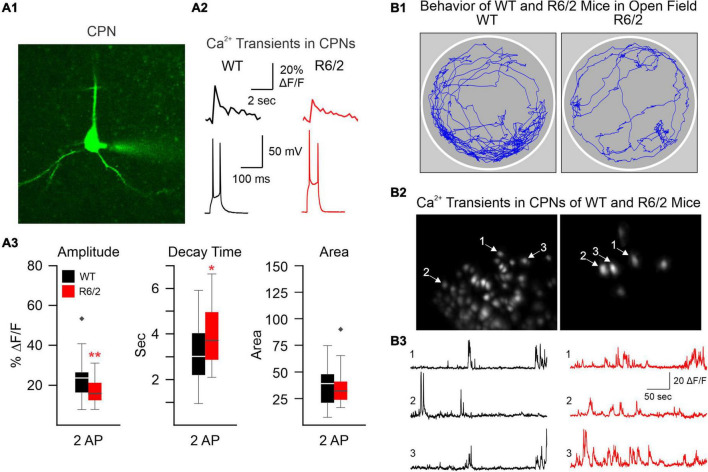
The development of confocal and two-photon laser scanning microscopy (2-PLSM) has opened the door to neuron recordings in deeper tissue, using laser excitation methods that are able to reduce background fluorescence noise and discern individual neurons (Svoboda and Yasuda, 2006). Pioneering groups developed 2-PLSM and recognized its potential to solve problems with photobleaching, depth, and spatial resolution present in other imaging methods, such as confocal and light microscopy (Denk et al., 1996; Svoboda et al., 1997). When utilized for Ca2+ imaging, fluorescent genetically encoded Ca2+ indicators (GECIs) bind to intracellular Ca2+ and emit fluorescent signals upon excitation, which can be acquired through the microscope. Since Ca2+ influx is associated with action potentials, it provides an indirect indication of neuronal firing activity. Cell type-specific expression of the Ca2+ indicator can be used to limit fluorescence to neurons of interest. In brain slices, 2-PLSM allows for recording from 50 to 100 cells simultaneously and extracting spike estimates from Ca2+ signals (Deneux et al., 2016), opening the possibility to compare neuronal Ca2+ activity in HD and wildtype (WT) mice. Fernandez-Garcia et al., for example, used fluorescence imaging of striatal and cortical cell cultures from R6/1 and WT mice to examine Ca2+ dynamics. Notably, spontaneously active neurons and coordinated ensemble activity were reduced in striatal but not in cortical cultures, suggesting reduced ability to integrate excitatory inputs (Fernandez-Garcia et al., 2020). However, altered Ca2+ influx may occur in the cerebral cortex of HD model mice. We used 2-PLSM to examine, in brain slices, Ca2+ influx associated with action potentials evoked by depolarizing current pulses in CPNs of R6/2 mice. We found reduced amplitude of somatic Ca2+ transients in R6/2 mice compared with controls (Figure 1A). This reduction appeared compensated by increased decay times, which could be deleterious due to reduced Ca2+ buffering capacity in HD neurons (Oikonomou et al., 2021).
FIGURE 1.
(A) We used two-photon microscopy to examine Ca2+ influx induced by action potentials (APs) in layer II cortical pyramidal neurons (CPNs) from R6/2 model mice. (A1) A CPN was patched and filled with a Ca2+ indicator (OGB-1). (A2) Action potentials (APs) evoked by 50 ms depolarizing current steps from the resting membrane potential. Accompanying Ca2+ transients are shown above the APs. (A3) The amplitude of somatic Ca2+ transients was reduced in R6/2 mice compared to controls. This reduction was compensated by increased decay times, which could lead to reduced Ca2+ buffering capacity [modified from Oikonomou et al. (2021)]. (B) A miniscope and a video camera above a cylindrical arena were used to visualize Ca2+ transient activity and behavior of WT and R6/2 mice. (B1) Movement tracking during 5 min behavior video recording. Notice the sparsity of movement in the symptomatic R6/2 mouse. (B2) Images of fluorescent CPNs in layer V of the same WT and symptomatic R6/2 mice. (B3) Ca2+ transients from three selected CPNs shown in B2. *p < 0.05, **p < 0.01, and black diamonds are outliers for the box and whisker plots.
Reduced Ca2+ buffering capacity may be associated with mitochondrial dysfunction. Indeed, mitochondrial Ca2+ levels also are dysregulated in HD. Using Ca2+ probes that do not permeate the mitochondrial membrane as indicators of extramitochondrial Ca2+, it is possible to indirectly monitor influxes of Ca2+ into the mitochondria, as well as total Ca2+ retained in the mitochondria upon opening of permeability transition pore (PTP) and Ca2+ release into the cytosol (Ferreira et al., 2018). Additionally, small molecule fluorescent Ca2+ indicators (e.g., Fura-2, Fura-FF) and GECIs (e.g., GCaMP, Cameleon, Pericam) can be made to specifically target organelles such as the mitochondria, directly providing insights into mitochondrial Ca2+ transport (De Nadai et al., 2021). Many of these experiments are done on neural cell cultures, though mitochondrial Ca2+ imaging can also be used on isolated brain mitochondria, with potential to be used in vivo with multiphoton microscopy (Calvo-Rodriguez et al., 2021). Using diverse methods to visualize mitochondrial Ca2+ dynamics, it was found that increased mitochondrial Ca2+ uptake promoted the oxidative stress and accumulation of mitochondrial DNA mutations in striatal neurons associated with HD (Wang et al., 2013). Indeed, the authors observed more mitochondrial Ca2+ loading in YAC128 compared to WT cells, and suggested reducing mitochondrial Ca2+ uptake as a potential therapeutic target for HD. In a similar study, it was reported that mitochondria in YAC transgenic mice are more sensitive to Ca2+-induced activation of PTP, leading to enhanced NMDA-induced apoptosis (Fernandes et al., 2007). Enhanced sensitivity of PTP in HD knock-in mouse models also has been observed (Lim et al., 2008). The contribution of reactive astrocytes to cell death in HD, and the changes in Ca2+ activity in HD model mice, also indicate the potential utilization of Ca2+ imaging on astrocytes as a potential investigative topic. Notably, even before overt astrogliosis, striatal astrocytes from symptomatic R6/2 mice display reduced spontaneous Ca2+ signals compared to astrocytes from WT animals (Jiang et al., 2016).
When recording in cell cultures or brain slices it is difficult to ascertain more complete neural circuits as they function in living mice. Fortunately, 2-PLSM imaging can also be utilized on head-restrained, behaving mice. This is done by performing a craniotomy to expose the area of interest, placing a glass cover over the cranial window, and recording through a 2-PLSM lens placed above the window (Donzis et al., 2018), which can record depths up to 1 mm. During recording, the head must be secured in place to avoid motion artifacts, and mouse behavior is constrained to movement on a ball treadmill. In live mice, two-photon microscopy allows for longitudinal imaging, potentially being able to visualize the differing Ca2+ dynamics during mouse behavior in various neuronal populations, e.g., CPNs and interneurons, as the disease progresses. Burgold et al. used 2-PLSM to monitor the activity of CPNs in layers II/III of the primary motor cortex in awake, behaving R6/2 mice and WT littermates. CPNs in HD mice showed increased activity within 1 week before the onset of motor deficits, suggesting reduced cortical inhibition (Burgold et al., 2019).
In our lab, Donzis et al. utilized 2-PLSM to examine cortical network dynamics in R6/2 and Q175 mice and evaluate changes in cortical microcircuits. They observed Ca2+ transients during motion and non-motion epochs and found significant alterations in behavior and cortical neuron activity in R6/2 and Q175 mouse models compared to WTs (Donzis et al., 2020). Some alterations were similar in the two HD models, such as a reduction in Ca2+ transient amplitude, indicative of decreased bursting activity and consistent with electrophysiological findings (Walker et al., 2008). Other alterations included changes in Ca2+ transient frequency during motion and non-motion epochs, with decreased frequencies in both epoch types in R6/2 mice and increased frequencies in motion epochs in Q175 mice. This indicates that neuronal activity from these populations is more significantly altered during motion in HD models. Both models also displayed decreased CPN interneuronal correlations, which suggests disrupted communication as the disease progresses (Donzis et al., 2020).
Although 2-PLSM can provide much greater context in a moving mouse than in slices, the fixation of the headbar during experiments and the limitations of movement in such experiments may alter mouse behavior and brain activity, and is thus not representative of fully free movement. For example, symptomatic R6/2 mice tend to move less in natural conditions compared to WTs. In contrast, when head-fixed and placed on a ball, they tend to move more. This can be caused by R6/2 mice being less able to balance on the ball and thus compensating by adjusting position to maintain balance (Donzis et al., 2020). Additionally, certain mouse models, such as the R6/2, are more susceptible to stress, and their altered behavior when moving on the ball could reflect the stressful environment. Advancements such as the use of an air-lifted track (Kislin et al., 2014; Nashaat et al., 2016) or linear self-paced treadmills (Muldoon et al., 2015) could potentially cause less stress and balance challenges to the head-fixed mouse than the spherical treadmill.
Many new implementations of 2-PLSM are currently in development, such as creation of a two-photon microendoscope, in which an endoscopic (GRIN or fiber-based) lens is surgically implanted into the mouse brain expanding imaging to deep tissues inaccessible by traditional two-photon benchtop microscopes, and allowing use in freely-moving animals (Jung and Schnitzer, 2003; Kucikas et al., 2021). Challenges of endoscopic systems lie in minimizing tissue damage when implanting into the brain, which is comparatively higher than recording through a cranial window as GRIN lenses cause damage to tissue around the implant site (Meng et al., 2019; Zhang et al., 2019; Kucikas et al., 2021).
Fiber photometry
Fiber photometry is another way to measure bulk neuronal activity, using a fiber optic cannula probe implanted into the brain carrying pulses to excite Ca2+ indicators, whose fluorescence is then carried back through the fibers (Li et al., 2019). It provides a way to record neuronal activity in freely-moving animals due to the flexibility of the fiber optic cable. The small size of the optic fibers (∼200 μm) also reduces damage to surrounding brain regions (a disadvantage in other Ca2+ imaging modalities), and allows for recordings from multiple brain regions simultaneously, as well as in deep tissue (Girven and Sparta, 2017; Wang et al., 2021). These recordings can then be correlated with animal behavior (Martianova et al., 2019). The stability of fiber photometry (when implanted in the brain) also allows for longitudinal recordings, correlating changes in Ca2+ dynamics as HD progresses. The popularity of fiber photometry has been promoted by groups such as the Deisseroth’s Lab, who have used this technique to record and reveal real-time, behavior-related circuits (Gunaydin et al., 2014; Kim et al., 2016).
Fiber photometry has been used to record Ca2+ fluorescence from the striatum of YAC128 model mice while undergoing a series of motor learning and coordination tasks, specifically the rotarod test (Koch et al., 2022). The authors investigated signaling changes by studying Ca2+ dynamics at different stages of the disease and found that even pre-manifest YAC128 mice display altered striatal activity associated with rotarod performance compared to WT. These alterations include weaker inverse correlation between latency to fall and striatal activity and different patterns of brain activity accompanying paw slip events, suggesting decreased synaptic plasticity associated with motor learning.
However, a major drawback of fiber photometry is its inability to record from individual cells. Instead, fiber photometry can record from populations of neurons (defined genetically to express GECIs), which has much lower spatial resolution and does not allow for visualization of specific neuronal interactions. It has also been observed that fiber photometry is more correlated with non-somatic Ca2+, as opposed to somatic Ca2+, and thus it is not a one-to-one representation of spiking activity (Legaria et al., 2022). Despite these limitations, fiber photometry has the potential to be used in conjunction with other techniques, such as in vivo electrophysiology (Patel et al., 2020), allowing for Ca2+ transients from specific subpopulations of neurons to be combined with measurements of electrophysiological activity and optogenetics. This would allow for in vivo recordings of Ca2+ dynamics in specific circuits, correlated with how firing patterns change with different behaviors or disease progression. Its simplicity, versatility, and low entry barrier are major draws to fiber photometry. Therefore, although fiber photometry has been used sparingly to investigate changes associated with HD, the many recent developments make it a promising tool.
Current areas of development for improving fiber photometry include, aside from simultaneous use with other imaging modalities, improving multi-fiber photometry, which allows for increased flexibility of probe placement and higher density channel placement (Sych et al., 2019), improving visualization of dynamics in various circuits (such as striatal direct/indirect pathways for HD), as well as optic cannulas with thinner tips to allow for more flexible placement. As techniques to separate signals continue to improve, simultaneous visualization of two neuronal populations using Ca2+ indicators of different colors (Meng et al., 2018) could potentially detect changes in activity of striatal direct and indirect pathway neurons associated with HD symptoms.
Miniscopes for use in Huntington’s disease mouse models: Proof-of-concept
Miniscopes represent another method that builds upon confocal microscopy and utilizes miniature head-mounted fluorescence microscopes to visualize Ca2+ transients in the brain via GECIs. Similar to other Ca2+ imaging methods, neuron cell types selected by genetic expression of GECI-GFP protein can be visualized using the miniscope. Miniscopes use GRIN lenses in the brain, allowing for visualization of individual fluorescent cells expressing GECI-GFP (Zhang et al., 2019). Pioneered by the Schnitzer group, the lightweight, stable miniscope allows for neuronal activity to be recorded with minimal motion artifacts from a freely-moving mouse with unrestricted movement, time-correlated with behaviors, providing an advantage over traditional two-photon microscopy methods (Ziv and Ghosh, 2015). Additionally, experiments comparing miniscopes with stationary 2-PLSM show that there is little variability in signal amplitude and signal-to-noise ratio between the two methods (Glas et al., 2019), and miniscopes are a highly reliable method for neuronal activity recordings.
Currently, our lab is using the UCLA miniscope (v3) to record Ca2+ transients in neurons from layers V to VI in the cortex to image CPNs. Similar to the reductions in amplitude observed in our previous publication using 2-PLSM (Donzis et al., 2020), we found reduced amplitude of Ca2+ transients in symptomatic R6/2 mice (Figure 1B). However, in contrast to our previous study in head-fixed animals, we also found that R6/2 mice moved much less than WT mice, demonstrating that Ca2+ imaging using miniscopes better reflects the HD phenotype of symptomatic R6/2 mice, namely hypokinesia. In addition, using Cre-expressing mice in different neuronal populations we have been able to visualize specific types of interneurons (in PV- and SOM-Cre mice) as well as direct and indirect pathway MSNs (in D1- and A2A-Cre mice).
Miniscopes represent an improvement on challenges faced by other forms of Ca2+ imaging regarding low spatial resolution and limitations on behavior and movement. The remaining limitations with miniscopes, but also present with other imaging techniques, include general obstacles with Ca2+ imaging regarding the duration of the transient signal. Ca2+ imaging is in general slower to respond than voltage-based imaging due to delay in binding of Ca2+ indicators. Also, due to slow release of Ca2+ indicators, cell fluorescence can remain after the firing of action potentials. The extended decay time (in the order of seconds) of the signal limits visualization/deconvolution on the duration of Ca2+ responses, and only reliably represents the beginning of the signal. For some proposed mechanisms for disease pathology of HD (Raymond, 2017) that suggest that changes in NMDAR response duration are responsible for apoptosis, Ca2+ imaging can be less effective. Additionally, most miniscopes utilize single-photon Ca2+ imaging, which has higher background fluorescence and tissue scattering of light, which somewhat limits its spatial resolution compared to 2-PLSM. Recently, lightweight (2–3 gr) miniature 2-photon microscopes are being developed (Zong et al., 2017, 2021, 2022), applying the deep penetration and higher resolution of two-photon microscopy to a freely-moving animal.
Though still in development, these systems enable monitoring with two-photon imaging over weeks, with minimal restrictions on behavior. However, similar to other methods using implanted lenses, the GRIN lenses used in miniscope imaging can cause some brain tissue displacement or damage.
Future research with miniscopes is focused on imaging more cell types in a circuit, potentially using multiple GECIs with wavelengths that can be distinguished, thus visualizing Ca2+ activity from multiple neuron populations (Werner et al., 2019). Additionally, projects enabling simultaneous extracellular electrophysiology recordings (Aharoni and Hoogland, 2019) expand the current methods to investigate neural circuits. The development of pipelines such as MIN1PIPE and CaImAn with built-in motion correction and denoising features greatly improved accessibility to miniscope imaging (Lu et al., 2018; Giovannucci et al., 2019). Behavioral classification to aim for closed-loop miniscope systems are also in development to streamline data processing and allow easier extraction of animal’s behavior during recording (Aharoni and Hoogland, 2019). Current improvement of wireless miniscopes (Barbera et al., 2019) also continues to minimize behavioral interference during recording, as well as the damage produced by the GRIN lens.
Conclusion
This minireview has presented several different Ca2+ imaging modalities and their applications to investigate the development and disease progression of HD in mouse models. These methods have been used to compare alterations in Ca2+ dynamics, which are likely key in HD manifestation. Current advances in Ca2+ imaging will make it possible to use Ca2+ imaging in conjunction with other methods to corroborate and formulate hypotheses about specific neuronal networks, dynamic alterations, and circuits involved with HD. For example, use of miniscopes in different Cre mouse lines may be able to elucidate the potentially differential role of striatal direct and indirect pathway neurons during the onset of motor symptoms, as well as the role of different types of interneurons in striatum and cerebral cortex.
Author contributions
All authors wrote the first draft of the manuscript, worked on revision, read, and approved the submitted version of the manuscript.
Acknowledgments
We acknowledge Elissa J. Donzis, Ana María Estrada-Sánchez, and Samiksha Chopra for their participation in the initial stages of the miniscope project. We also acknowledge Baljit Khakh and Xinzhu Yu for their guidance in the 2-PLSM experiments.
Funding
This work was partially supported by the Cell, Circuits and Systems Analysis Core (USPHS grant P50HD103557).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aharoni D., Hoogland T. M. (2019). Circuit investigations with open-source miniaturized microscopes: Past, present and future. Front. Cell. Neurosci. 13:141. 10.3389/fncel.2019.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera G., Liang B., Zhang L., Li Y., Lin D. T. (2019). A wireless miniScope for deep brain imaging in freely moving mice. J. Neurosci. Methods 323 56–60. 10.1016/j.jneumeth.2019.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates G. P., Dorsey R., Gusella J. F., Hayden M. R., Kay C., Leavitt B. R., et al. (2015). Huntington disease. Nat. Rev. Dis. Primers 1:15005. [DOI] [PubMed] [Google Scholar]
- Bezprozvanny I. (2009). Calcium signaling and neurodegenerative diseases. Trends Mol. Med. 15 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I., Hayden M. R. (2004). Deranged neuronal calcium signaling and Huntington disease. Biochem. Biophys. Res. Commun. 322 1310–1317. [DOI] [PubMed] [Google Scholar]
- Broussard G. J., Petreanu L. (2021). Eavesdropping wires: Recording activity in axons using genetically encoded calcium indicators. J. Neurosci. Methods 360:109251. 10.1016/j.jneumeth.2021.109251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold J., Schulz-Trieglaff E. K., Voelkl K., Gutierrez-Angel S., Bader J. M., Hosp F., et al. (2019). Cortical circuit alterations precede motor impairments in Huntington’s disease mice. Sci. Rep. 9:6634. 10.1038/s41598-019-43024-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo-Rodriguez M., Kharitonova E. K., Bacskai B. J. (2021). In vivo brain imaging of mitochondrial Ca(2+) in neurodegenerative diseases with multiphoton microscopy. Biochim. Biophys. Acta Mol. Cell Res. 1868:118998. 10.1016/j.bbamcr.2021.118998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda C., Ariano M. A., Calvert C. R., Flores-Hernandez J., Chandler S. H., Leavitt B. R., et al. (2001). Nmda receptor function in mouse models of Huntington disease. J. Neurosci. Res. 66 525–539. [DOI] [PubMed] [Google Scholar]
- Cepeda C., Cummings D. M., Andre V. M., Holley S. M., Levine M. S. (2010). Genetic mouse models of Huntington’s disease: Focus on electrophysiological mechanisms. ASN Neuro 2:e00033. 10.1042/AN20090058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeredys M. (2020). Dysregulation of neuronal calcium signaling via store-operated channels in Huntington’s disease. Front. Cell Dev. Biol. 8:611735. 10.3389/fcell.2020.611735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nadai A., Vajente N., Pendin D., Mattarei A. (2021). Mt-fura-2, a ratiometric mitochondria-targeted Ca(2+) sensor. Determination of spectroscopic properties and Ca(2+) imaging assays. Methods Mol. Biol. 2275 187–215. 10.1007/978-1-0716-1262-0_12 [DOI] [PubMed] [Google Scholar]
- Deneux T., Kaszas A., Szalay G., Katona G., Lakner T., Grinvald A., et al. (2016). Accurate spike estimation from noisy calcium signals for ultrafast three-dimensional imaging of large neuronal populations in vivo. Nat. Commun. 7:12190. 10.1038/ncomms12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W., Yuste R., Svoboda K., Tank D. W. (1996). Imaging calcium dynamics in dendritic spines. Curr. Opin. Neurobiol. 6 372–378. [DOI] [PubMed] [Google Scholar]
- Donzis E. J., Estrada-Sanchez A. M., Indersmitten T., Oikonomou K., Tran C. H., Wang C., et al. (2020). Cortical network dynamics is altered in mouse models of Huntington’s disease. Cereb. Cortex 30 2372–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzis E. J., Holley S. M., Cepeda C., Levine M. S. (2018). Neurophysiological assessment of Huntington’s disease model mice. Methods Mol. Biol. 1780 163–177. 10.1007/978-1-4939-7825-0_9 [DOI] [PubMed] [Google Scholar]
- Fernandes H. B., Baimbridge K. G., Church J., Hayden M. R., Raymond L. A. (2007). Mitochondrial sensitivity and altered calcium handling underlie enhanced Nmda-induced apoptosis in Yac128 model of Huntington’s disease. J. Neurosci. 27 13614–13623. 10.1523/JNEUROSCI.3455-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia S., Orlandi J. G., Garcia-Diaz Barriga G. A., Rodriguez M. J., Masana M., Soriano J., et al. (2020). Deficits in coordinated neuronal activity and network topology are striatal hallmarks in Huntington’s disease. BMC Biol. 18:58. 10.1186/s12915-020-00794-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira I. L., Carmo C., Naia L. S. I. M., Cristina Rego A. (2018). Assessing mitochondrial function in in vitro and ex vivo models of Huntington’s disease. Methods Mol. Biol. 1780 415–442. 10.1007/978-1-4939-7825-0_19 [DOI] [PubMed] [Google Scholar]
- Giovannucci A., Friedrich J., Gunn P., Kalfon J., Brown B. L., Koay S. A., et al. (2019). CaImAn an open source tool for scalable calcium imaging data analysis. Elife 8:e38173. 10.7554/eLife.38173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girven K. S., Sparta D. R. (2017). Probing deep brain circuitry: New advances in in vivo calcium measurement strategies. ACS Chem. Neurosci. 8 243–251. 10.1021/acschemneuro.6b00307 [DOI] [PubMed] [Google Scholar]
- Glas A., Hubener M., Bonhoeffer T., Goltstein P. M. (2019). Benchmarking miniaturized microscopy against two-photon calcium imaging using single-cell orientation tuning in mouse visual cortex. PLoS One 14:e0214954. 10.1371/journal.pone.0214954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M., Shirasaki D. I., Cepeda C., Andre V. M., Wilburn B., Lu X. H., et al. (2008). Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J. Neurosci. 28 6182–6195. 10.1523/JNEUROSCI.0857-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C., Giovannucci A., Zeiger W., Portera-Cailliau C. (2022). Two-photon calcium imaging of neuronal activity. Nat. Rev. Methods Primers 2:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin L. A., Grosenick L., Finkelstein J. C., Kauvar I. V., Fenno L. E., Adhikari A., et al. (2014). Natural neural projection dynamics underlying social behavior. Cell 157 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R., Diaz-Castro B., Looger L. L., Khakh B. S. (2016). Dysfunctional calcium and glutamate signaling in striatal astrocytes from Huntington’s disease model mice. J. Neurosci. 36 3453–3470. 10.1523/JNEUROSCI.3693-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. C., Schnitzer M. J. (2003). Multiphoton endoscopy. Opt. Lett. 28 902–904. [DOI] [PubMed] [Google Scholar]
- Kim C. K., Yang S. J., Pichamoorthy N., Young N. P., Kauvar I., Jennings J. H., et al. (2016). Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat. Methods 13 325–328. 10.1038/nmeth.3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Schnitzer M. J. (2022). Fluorescence imaging of large-scale neural ensemble dynamics. Cell 185 9–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislin M., Mugantseva E., Molotkov D., Kulesskaya N., Khirug S., Kirilkin I., et al. (2014). Flat-floored air-lifted platform: A new method for combining behavior with microscopy or electrophysiology on awake freely moving rodents. J. Vis. Exp. 88:e51869. 10.3791/51869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E. T., Sepers M. D., Cheng J., Raymond L. A. (2022). Early changes in striatal activity and motor kinematics in a Huntington’s disease mouse model. Mov. Disord. 37 2021–2032. 10.1002/mds.29168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucikas V., Werner M. P., Schmitz-Rode T., Louradour F., Van Zandvoort M. (2021). Two-photon endoscopy: State of the art and perspectives. Mol. Imaging Biol. 10.1007/s11307-021-01665-2 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake E. M. R., Higley M. J. (2022). Building bridges: Simultaneous multimodal neuroimaging approaches for exploring the organization of brain networks. Neurophotonics 9:032202. 10.1117/1.NPh.9.3.032202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaria A. A., Matikainen-Ankney B. A., Yang B., Ahanonu B., Licholai J. A., Parker J. G., et al. (2022). Fiber photometry in striatum reflects primarily nonsomatic changes in calcium. Nat. Neurosci. 25 1124–1128. 10.1038/s41593-022-01152-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Y., Popovic N., Brundin P. (2005). The use of the R6 transgenic mouse models of Huntington’s disease in attempts to develop novel therapeutic strategies. NeuroRx 2 447–464. 10.1602/neurorx.2.3.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu Z., Guo Q., Luo M. (2019). Long-term fiber photometry for neuroscience studies. Neurosci. Bull. 35 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D., Fedrizzi L., Tartari M., Zuccato C., Cattaneo E., Brini M., et al. (2008). Calcium homeostasis and mitochondrial dysfunction in striatal neurons of Huntington disease. J. Biol. Chem. 283 5780–5789. [DOI] [PubMed] [Google Scholar]
- Lu J., Li C., Singh-Alvarado J., Zhou Z. C., Frohlich F., Mooney R., et al. (2018). Min1pipe: A miniscope 1-photon-based calcium imaging signal extraction pipeline. Cell Rep. 23 3673–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay J. P., Nassrallah W. B., Raymond L. A. (2018). Cause or compensation?-Altered neuronal Ca(2+) handling in Huntington’s disease. CNS Neurosci. Ther. 24 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianova E., Aronson S., Proulx C. D. (2019). Multi-fiber photometry to record neural activity in freely-moving animals. J. Vis. Exp. 10.3791/60278 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Menalled L. B. (2005). Knock-in mouse models of Huntington’s disease. NeuroRx 2 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C., Zhou J., Papaneri A., Peddada T., Xu K., Cui G. (2018). Spectrally resolved fiber photometry for multi-component analysis of brain circuits. Neuron 98 707–717.e4. 10.1016/j.neuron.2018.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G., Liang Y., Sarsfield S., Jiang W. C., Lu R., Dudman J. T., et al. (2019). High-throughput synapse-resolving two-photon fluorescence microendoscopy for deep-brain volumetric imaging in vivo. Elife 8:e40805. 10.7554/eLife.40805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon S. F., Villette V., Tressard T., Malvache A., Reichinnek S., Bartolomei F., et al. (2015). Gabaergic inhibition shapes interictal dynamics in awake epileptic mice. Brain 138 2875–2890. 10.1093/brain/awv227 [DOI] [PubMed] [Google Scholar]
- Nashaat M. A., Oraby H., Sachdev R. N., Winter Y., Larkum M. E. (2016). Air-Track: A real-world floating environment for active sensing in head-fixed mice. J. Neurophysiol. 116 1542–1553. 10.1152/jn.00088.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou K. D., Donzis E. J., Bui M. T., Cepeda C. T., Levine M. S. (2021). Calcium dysregulation and compensation in cortical pyramidal neurons of the R6/2 mouse model of Huntington’s disease. J. Neurophysiol. 126 1159–1171. 10.1152/jn.00181.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. A., Mcalinden N., Mathieson K., Sakata S. (2020). Simultaneous electrophysiology and fiber photometry in freely behaving mice. Front. Neurosci. 14:148. 10.3389/fnins.2020.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney J. W., Jr. (1986). A model for receptor-regulated calcium entry. Cell Calcium 7 1–12. 10.1016/0143-4160(86)90026-6 [DOI] [PubMed] [Google Scholar]
- Raymond L. A. (2017). Striatal synaptic dysfunction and altered calcium regulation in Huntington disease. Biochem. Biophys. Res. Commun. 483 1051–1062. [DOI] [PubMed] [Google Scholar]
- Slow E. J., Van Raamsdonk J., Rogers D., Coleman S. H., Graham R. K., Deng Y., et al. (2003). Selective striatal neuronal loss in a Yac128 mouse model of Huntington disease. Hum. Mol. Genet. 12 1555–1567. 10.1093/hmg/ddg169 [DOI] [PubMed] [Google Scholar]
- Svoboda K., Yasuda R. (2006). Principles of two-photon excitation microscopy and its applications to neuroscience. Neuron 50 823–839. [DOI] [PubMed] [Google Scholar]
- Svoboda K., Denk W., Kleinfeld D., Tank D. W. (1997). In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature 385 161–165. [DOI] [PubMed] [Google Scholar]
- Sych Y., Chernysheva M., Sumanovski L. T., Helmchen F. (2019). High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nat. Methods 16 553–560. 10.1038/s41592-019-0400-4 [DOI] [PubMed] [Google Scholar]
- The Huntington’s Disease Collaborative Research Group (1993). A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72 971–983. [DOI] [PubMed] [Google Scholar]
- Tobin A. J., Signer E. R. (2000). Huntington’s disease: The challenge for cell biologists. Trends Cell Biol. 10 531–536. 10.1016/s0962-8924(00)01853-5 [DOI] [PubMed] [Google Scholar]
- Vonsattel J. P., Difiglia M. (1998). Huntington disease. J. Neuropathol. Exp. Neurol. 57 369–384. [DOI] [PubMed] [Google Scholar]
- Waldvogel H. J., Kim E. H., Tippett L. J., Vonsattel J. P., Faull R. L. (2015). The neuropathology of Huntington’s disease. Curr. Top. Behav. Neurosci. 22 33–80. [DOI] [PubMed] [Google Scholar]
- Walker A. G., Miller B. R., Fritsch J. N., Barton S. J., Rebec G. V. (2008). Altered information processing in the prefrontal cortex of Huntington’s disease mouse models. J. Neurosci. 28 8973–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker F. O. (2007). Huntington’s disease. Lancet 369 218–228. [DOI] [PubMed] [Google Scholar]
- Wang J. Q., Chen Q., Wang X., Wang Q. C., Wang Y., Cheng H. P., et al. (2013). Dysregulation of mitochondrial calcium signaling and superoxide flashes cause mitochondrial genomic DNA damage in Huntington disease. J. Biol. Chem. 288 3070–3084. 10.1074/jbc.M112.407726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Demarco E. M., Witzel L. S., Keighron J. D. (2021). A selected review of recent advances in the study of neuronal circuits using fiber photometry. Pharmacol. Biochem. Behav. 201:173113. 10.1016/j.pbb.2021.173113 [DOI] [PubMed] [Google Scholar]
- Werner C. T., Williams C. J., Fermelia M. R., Lin D. T., Li Y. (2019). Circuit mechanisms of neurodegenerative diseases: A new frontier with miniature fluorescence microscopy. Front. Neurosci. 13:1174. 10.3389/fnins.2019.01174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Ryskamp D. A., Liang X., Egorova P., Zakharova O., Hung G., et al. (2016). Enhanced store-operated calcium entry leads to striatal synaptic loss in a Huntington’s disease mouse model. J. Neurosci. 36 125–141. 10.1523/JNEUROSCI.1038-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeron M. M., Hansson O., Chen N., Wellington C. L., Leavitt B. R., Brundin P., et al. (2002). Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron 33 849–860. 10.1016/s0896-6273(02)00615-3 [DOI] [PubMed] [Google Scholar]
- Zhang L., Liang B., Barbera G., Hawes S., Zhang Y., Stump K., et al. (2019). Miniscope grin lens system for calcium imaging of neuronal activity from deep brain structures in behaving animals. Curr. Protoc. Neurosci. 86:e56. 10.1002/cpns.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y., Ghosh K. K. (2015). Miniature microscopes for large-scale imaging of neuronal activity in freely behaving rodents. Curr. Opin. Neurobiol. 32 141–147. [DOI] [PubMed] [Google Scholar]
- Zong W., Obenhaus H. A., Skytoen E. R., Eneqvist H., De Jong N. L., Vale R., et al. (2022). Large-scale two-photon calcium imaging in freely moving mice. Cell 185 1240–1256.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W., Wu R., Chen S., Wu J., Wang H., Zhao Z., et al. (2021). Miniature two-photon microscopy for enlarged field-of-view, multi-plane and long-term brain imaging. Nat. Methods 18 46–49. [DOI] [PubMed] [Google Scholar]
- Zong W., Wu R., Li M., Hu Y., Li Y., Li J., et al. (2017). Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat. Methods 14 713–719. 10.1038/nmeth.4305 [DOI] [PubMed] [Google Scholar]