Summary
Interphotoreceptor matrix proteoglycan 2 (IMPG2) mutations cause a severe form of early-onset retinitis pigmentosa (RP) with macular involvement. IMPG2 is expressed by photoreceptors and incorporated into the matrix that surrounds the inner and outer segments (OS) of rods and cones, but the mechanism of IMPG2-RP remains unclear. Loss of Impg2 function in mice produces a mild, late-onset photoreceptor phenotype without the characteristic OS loss that occurs in human patients. We generated retinal organoids (ROs) from patient-derived induced pluripotent stem (iPS) cells and gene-edited embryonic stem cells to model human IMPG2-RP in vitro. All ROs harboring IMPG2 mutations lacked an OS layer, in contrast to isogenic controls. Subsequent protein analyses revealed that this phenotype arises due to a loss of IMPG2 expression or its inability to undergo normal post-translational modifications. We hypothesized that loss of IMPG2 function destabilizes the interphotoreceptor matrix and renders the OS vulnerable to physical stressors, which is accentuated in the tissue culture environment. In support of this mechanism, transplantation of IMPG2 mutant ROs into the protected subretinal space of immunocompromised rodents restored OS production. Beyond providing a robust platform to study IMPG2-RP, this human RO model system may serve a broader role in honing strategies to treat advanced photoreceptor-based diseases.
Keywords: retinal organoid, disease modeling, stem cells, IMPG2, photoreceptors, retinitis pigmentosa
Highlights
-
•
Normal hPSC-ROs produce an interphotoreceptor matrix similar to non-human primates
-
•
Loss of IMPG2 function in ROs causes near absence of photoreceptor outer segments
-
•
Mutations in IMPG2 disrupt protein expression or post-translational modifications
-
•
Mutant ROs can form outer segments when placed in the protected subretinal space
Gamm and colleagues describe the first stem cell-derived retinal organoid model of an interphotoreceptor matrix-based blinding disorder, which displays a robust phenotype characteristic of late-stage retinitis pigmentosa.
Introduction
Retinitis pigmentosa (RP) and its related disorders comprise a diverse group of rare inherited retinal degenerative diseases caused by defects in genes involved in photoreceptor development, structure, health, and/or function (Hartong et al., 2006; Verbakel et al., 2018). Understanding the mechanisms of these genetic disorders is important, as it can reveal promising treatment strategies and shed light on basic molecular and cellular processes of vision. However, the heterogeneity of RP and the frequent lack of robust model systems that mimic human disease make it challenging to unravel gene-specific disease mechanisms. One such example is RP associated with mutations in genes involved in the structure and function of the interphotoreceptor matrix (IPM).
The IPM is a highly organized extracellular matrix that surrounds photoreceptor inner and outer segments (IS and OS) and adjoining apical processes of the retinal pigment epithelium within the subretinal space. Given its composition and strategic location, the IPM has many known and postulated roles in the outer retina, including retinoid transport, intercellular communication, neural retina adhesion, nutrient and oxygen transport, cytoskeletal organization, and structural support (Hageman and Johnson, 1991; Ishikawa et al., 2015). As such, it is not surprising that disruption of the IPM causes a variety of outer retinal disorders with profound visual consequences.
A prominent component of the IPM is interphotoreceptor matrix proteoglycan 2 (IMPG2), an abundant protein that is secreted by both rod and cone photoreceptors (Acharya et al., 2000; Chen et al., 2003; Foletta et al., 2001; Kuehn and Hageman, 1999). IMPG2 is heavily glycosylated with glycosaminoglycans (GAGs) as well as N- and O-linked oligosaccharides, which together contribute to approximately 60% of its mass (Acharya et al., 2000). These post-translational modifications (PTMs) are believed to be important for binding other IPM constituents, stabilizing the matrix, and supporting the growth and maintenance of OS (Chen et al., 2004). Mutations in IMPG2 cause multiple retinal pathologies, including a severe form of autosomal recessive RP that also affects the cone photoreceptor-rich macula early in the course of the disease, leading to irreversible vision loss in childhood (Bandah-Rozenfeld et al., 2010; Bocquet et al., 2013; Brandl et al., 2017; Huet et al., 2014; Khan and Al Teneiji, 2019; Meunier et al., 2014). Structurally, noninvasive imaging of affected patients by optical coherence tomography shows a reduction in retinal thickness and a loss of outer retinal bands corresponding to the photoreceptor OS layer (Brandl et al., 2017; Khan and Al Teneiji, 2019). Deletion of Impg2 in mice also shows progressive functional changes associated with RP along with histopathological findings reflective of rod and cone dysfunction, including OS disorganization (Salido and Ramamurthy, 2020; Xu et al., 2020). However, Impg2 knockout mice display a significantly milder phenotype than human patients with IMPG2-RP, even after long-term follow-up (Salido and Ramamurthy, 2020; Xu et al., 2020).
Human pluripotent stem cell (hPSC)-based “disease-in-a-dish” models are increasingly relied upon as tools to corroborate animal model findings, to fill the void when no animal models exist, or to provide clinical relevance when human-specific genetic backgrounds are needed (Singh et al., 2013; Sinha et al., 2020). This need is counterbalanced by inherent limitations in hPSC model systems, particularly those employing three-dimensional organoids whose phenotypic characteristics can vary considerably not only within the same culture population, but also within the same organoid (Capowski et al., 2019; Meyer et al., 2011; Nakano et al., 2012; O’Hara-Wright and Gonzalez-Cordero, 2020; Sinha et al., 2016). It is thus imperative to rigorously assess organoid phenotypes to assure that they are robust and reproducible across organoids, differentiations, and hPSC lines. In addition, the experimental throughput of organoid model systems is often low and unwieldy, which translates into higher implementation costs.
Human PSC-derived retinal organoids (ROs) proceed through a reproducible series of differentiation stages over the course of many months, ultimately achieving stage 3, which is demarcated by the light microscopic appearance of hair-like OS covering their surface (Capowski et al., 2019). The ability of these long, thin OS to grow and persist indefinitely in an unstable tissue culture environment implies the presence of a protective IPM. Indeed, hPSC-ROs have been shown to produce an IPM (Felemban et al., 2018), although expression of IMPG2 has not yet been demonstrated. Interestingly, IMPG2 was not detected in the matrix surrounding OS in mice; rather, by immunohistochemistry it appeared to be restricted to the inner segment region only (Salido and Ramamurthy, 2020). Nonetheless, loss of IMPG2 in mice did alter the composition of the distal IPM as indicated by the abnormal distribution of a related IPM protein, IMPG1 (Salido and Ramamurthy, 2020). Given these findings, we hypothesized that loss of IMPG2 function would destabilize the IPM of hPSC-ROs, leading to secondary disruption of OS and perhaps an observable phenotype in live cultures.
Using patient-derived and gene-edited hiPSC and hESC lines, we discovered that, in stark contrast to their isogenic wild-type controls, IMPG2 mutant or knockout ROs invariably lacked observable OS by light microscopy. Furthermore, western analysis of ROs harboring a missense mutation in IMPG2 revealed an absence of PTMs, offering mechanistic insight into the disease. All IMPG2-deficient hPSC-ROs, including those with heterozygous mutations, displayed a more severe and temporally advanced OS phenotype than expected for humans with IMPG2-RP at an equivalent stage of development. To test whether the RO culture environment contributed to these findings, we transplanted partially dissociated IMPG2 mutant and isogenic wild-type control ROs into the subretinal space of immunodeficient rats. When allowed to mature in this physically protected location, both mutant and control ROs were able to generate OS.
Together, these results show that IMPG2-deficient hPSC-ROs provide a robust, physiologically relevant, and accelerated model of IMPG2-RP. As such, they should prove useful for testing strategies to treat this rare disorder. Moreover, given the unique and desirable characteristics of this disease model, we foresee its utility in a broader, gene-agnostic context to develop and screen a range of photoreceptor-targeted therapeutics and delivery systems.
Results
IPM protein localization in normal stage 3 hPSC-ROs is similar to that of non-human primate and mouse retina
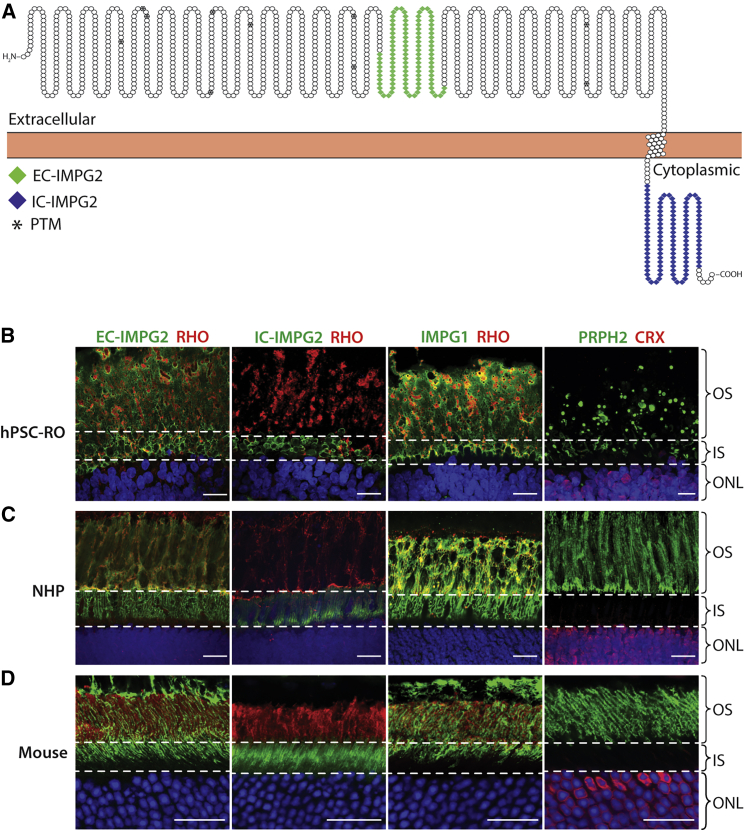
To model IMPG2 in hPSC-ROs, we first sought to compare the localization of key IPM and photoreceptor proteins in stage 3 hPSC-ROs relative to adult non-human primate (NHP) and mouse retina. Recent reports in mouse suggested that IMPG2 is exclusively found in photoreceptor inner segments (IS) (Salido and Ramamurthy, 2020). Since an understanding of IMPG2 localization is critical to studying its function, two primary antibodies were used to investigate its expression via immunohistochemistry (IHC) (Figures 1A, S1A, and S1B). The first antibody binds to an extracellular epitope of IMPG2 (EC-IMPG2), while the second recognizes an intracellular epitope (IC-IMPG2). In stage 3 hPSC-ROs, EC-IMPG2 immunofluorescence was found surrounding both photoreceptor IS and OS, while IC-IMPG2 immunofluorescence was localized only to photoreceptor IS (Figure 1B). IHC analysis of IMPG1 mirrored that of EC-IMPG2 (Figure 1B). To ensure that our findings were not an artifact of hPSC-RO culture, we repeated the IHC analyses using NHP and mouse retina sections, which revealed the same EC-IMPG2, IC-IMPG2, and IMPG1 expression patterns found in hPSC-ROs (Figures 1C and 1D). These results provided confidence that we could model normal and mutant IMPG2 function and dysfunction, respectively, using hPSC-RO cultures. Furthermore, they suggest that IMPG2 undergoes processing as it is transported from IS to OS, so the intracellular epitope is no longer detectable in OS, either due to protein cleavage or epitope masking.
Figure 1.
IMPG2 localizes to photoreceptor inner and outer segments in stage 3 hPSC-ROs, non-human primates, and mice
(A) Schematic depicting the location of the extracellular and intracellular epitopes used to generate the two IMPG2 antibodies employed in this study (EC-IMPG2 and IC-IMPG2, respectively). Sites of predicted post-translational modifications (PTM) are marked with an asterisk (∗). IMPG2 schematic was designed using Protter.
(B–D) IHC analysis showing the expression of EC-IMPG2, IC-IMPG2, and IMPG1 relative to the photoreceptor markers rhodopsin (RHO), peripherin 2 (PRPH2), and cone-rod homeobox (CRX) in regions corresponding to inner and outer segments (IS and OS; delineated by dotted lines). In stage 3 hPSC-ROs (B), NHP retina (C), and mouse retina (D), EC-IMPG2 and IMPG1 immunostaining is present in the IS and OS regions, whereas IC-IMPG2 immunostaining is restricted to the IS region. ONL = outer nuclear layer; scale bars, 20 μm.
hPSC-ROs harboring mutations in IMPG2 universally lack visible outer segments by light microscopy
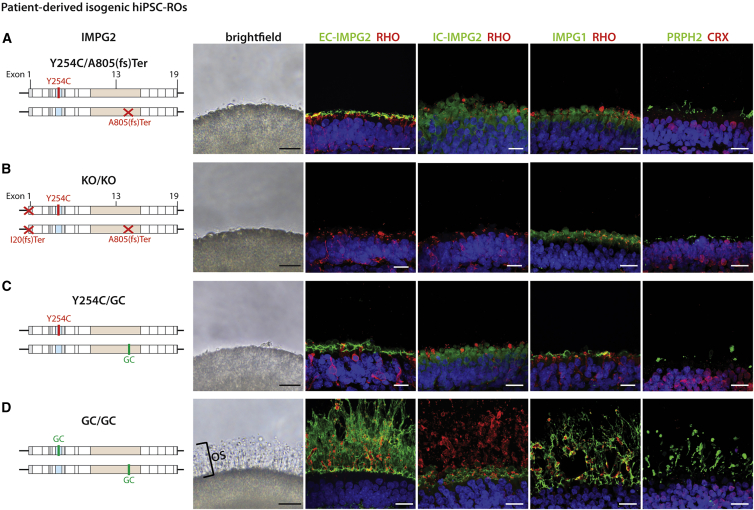
To model IMPG2-RP using hPSC-ROs, a total of seven different hPSC lines were employed: four isogenic hiPSC lines and three isogenic hESC lines. The founder hiPSC line was derived from a subject diagnosed with RP caused by compound heterozygous mutations in IMPG2 (IMPG2Y254C/A805(fs)ter). The remaining three hiPSC lines are isogenic to the founder, having undergone CRISPR-Cas9-mediated editing to gene correct (GC) one or both IMPG2 alleles (IMPG2Y254C/GC or IMPG2GC/GC) or to knock out (KO) both IMPG2 alleles (IMPG2KO/KO). All four hiPSC lines properly expressed the pluripotency markers OCT3/4, NANOG, and SOX2 by immunocytochemistry, possessed normal karyotypes, and had their expected IMPG2 genotype confirmed by Sanger sequencing (Figure S2).
Given the important role of the IPM in OS maintenance, combined with the known structural and functional phenotype of patients with IMPG2-RP, we hypothesized that mutant IMPG2 hPSC-ROs would possess OS defects. Strikingly, all IMPG2Y254C/A805(fs)ter hiPSC-ROs failed to develop any observable OS by light microscopy, even after >200 days of differentiation (Figures 2A and S3A and Table 1). IHC analysis revealed the presence of EC-IMPG2 immunostaining in photoreceptor IS and on the immediate surface of IMPG2Y254C/A805(fs)Ter ROs (Figure 2A). The same pattern of expression was observed for IMPG1 (Figure 2A), consistent with a general lack of an IPM in the IMPG2 mutant ROs. Lastly, immunostaining for rhodopsin and PRPH2 confirmed the near absence of OS in IMPG2Y254C/A805(fs)Ter hiPSC-ROs (Figure 2A). Light microscopic and IHC findings in isogenic IMPG2KO/KO hiPSC-ROs were identical to those in IMPG2Y254C/A805(fs)ter hiPSC-ROs, with the exception that IMPG2 expression was completely undetectable in the IMPG2 knockout line as expected (Figures 2B and S3B and Table 1).
Figure 2.
hiPSC-ROs harboring patient-specific or gene-edited mutations in IMPG2 fail to extend outer segments or produce an IPM
(A) ROs from a founder hiPSC line containing compound heterozygous mutations in IMPG2 lack visible OS by bright field microscopy. Immunocytochemistry reveals a thin layer of EC-IMPG2, IC-IMPG2, and IMPG1 expression along the RO surface, along with severely stunted RHO and PRPH2 expression.
(B–D) Identical phenotypes are observed in ROs from isogenic IMPG2 knockout (KO/KO) (B) and heterozygous gene-corrected (Y254C/GC) (C) hiPSC lines, with the exception that IMPG2 immunostaining is absent in double knockout ROs. (D) Biallelic IMPG2 gene correction (GC/GC) of the founder hiPSC line fully rescues OS and IPM production in ROs as determined by bright field microscopy and immunocytochemistry. All ROs were differentiated >200 days. RHO = rhodopsin; PRPH2 = peripherin 2; CRX = cone-rod homeobox; scale bars, 20 μm.
Table 1.
Quantification of hPSC-RO differentiations in which photoreceptor OS layers were discernible by light microscopy (>200 days in culture)
| Cell line | Number of differentiations performed | Number of differentiations resulting in discernible OS layersa |
|---|---|---|
| IMPG2Y254C/A805(fs)Ter | 44 | 0 |
| IMPG2KO/KO | 44 | 0 |
| IMPG2Y254C/GC | 44 | 0 |
| IMPG2GC/GC | 38 | 38 |
| WA09-IMPG2+/+ | 92 | 92 |
| WA09-IMPG2Y254C/Y254C | 49 | 0 |
| WA09-IMPG2KO/KO | 49 | 0 |
A discernible OS layer was uniformly present or absent in all ROs from each differentiation (≥10 ROs per differentiation).
Since IMPG2-RP is a recessive disorder with no phenotype in young heterozygous carriers, we predicted that the OS defect present in the founder IMPG2 mutant ROs would be alleviated by correction of a single mutant allele. Accordingly, we differentiated ROs from the single gene-corrected IMPG2Y254C/GC hiPSC line for >200 days, expecting to see restoration of the OS layer. However, IMPG2Y254C/GC ROs also lacked visible OS by light microscopy and displayed immunostaining patterns for IMPG2, IMPG1, rhodopsin, and PRPH2 indistinguishable from IMPG2Y254C/A805(fs)ter hiPSC-ROs (Figures 2C and S3C and Table 1). To verify that the lack of OS was specifically due to IMPG2 dysfunction and not to an unknown deficiency of the founder line, we also differentiated the biallelic gene-corrected IMPG2GC/GC line for >200 days. All IMPG2GC/GC ROs produced a thick OS layer that was readily identifiable by light microscopy (Figures 2D and S3D and Table 1). IHC analysis of IMPG2GC/GC ROs revealed normalized IMPG2 and IMPG1 expression patterns indicative of IPM restoration and also confirmed the presence of OS based on rhodopsin and PRPH2 immunostaining (Figure 2D).
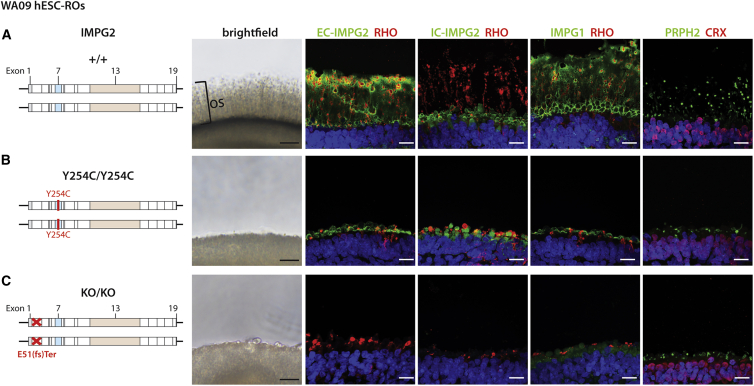
To further ensure that the OS phenotype observed in the mutant IMPG2 hiPSC-ROs was not a cell line artifact, we engineered homozygous Y254C IMPG2 mutations into the widely used WA09 hESC line (WA09-IMPG2Y254C/Y254C; Figure 3B). In addition, we separately knocked out both IMPG2 alleles in the same line (WA09-IMPG2KO/KO; Figure 3C). As with the hiPSC lines, the wild-type WA09 line and both isogenic CRISPR-Cas9 gene-edited WA09 lines expressed the pluripotency markers OCT3/4, NANOG, and SOX2, possessed normal karyotypes, and had their expected IMPG2 genotype confirmed by Sanger sequencing (Figure S4).
Figure 3.
Introduction of IMPG2 mutations into wild-type WA09 hESCs abolishes outer segment extension and IPM production in ROs
(A) ROs differentiated from wild-type (+/+) WA09-IMPG2 hESCs produce a thick OS layer visible by bright field microscopy. Immunofluorescence confirms the presence of elongated RHO+/PRPH2+ OS and a corresponding IPM with proper localization of EC-IMPG2, IC-IMPG2, and IMPG1.
(B and C) Biallelic introduction of pathogenic Y254C mutations (Y254C/Y254C) (B) or knockout mutations (KO/KO) (C) into IMPG2 by CRISPR-Cas9 gene editing eliminates OS as observed by bright field microscopy. A thin layer of EC-IMPG2, IC-IMPG2, and IMPG1 expression is present by immunocytochemistry along the surface of the Y254C/Y254C mutant ROs, along with severely stunted RHO and PRPH2 expression. The KO/KO ROs have an identical phenotype with the exception that no IMPG2 expression is observed. All ROs were differentiated >200 days. RHO = rhodopsin; PRPH2 = peripherin 2; CRX = cone-rod homeobox; scale bars, 20 μm.
As shown previously, all wild-type WA09 hESC-ROs universally achieved stage 3 of differentiation (Capowski et al., 2019), hallmarked by a thick OS layer visible by light microscopy (Figures 3A and S3E and Table 1). Immunostaining of wild-type WA09 hESC-ROs showed proper IMPG2 and IMPG1 expression in the IPM and rhodopsin and PRPH2 expression in OS (Figure 3A). In contrast, all ROs generated from the WA09-IMPG2Y254C/Y254C and WA09-IMPG2KO/KO isogenic lines failed to demonstrate OS by light microscopy after >200 days of differentiation (Figures 3B, 3C, S3F, and S3G and Table 1). Consistent with the light microscopic findings, WA09-IMPG2Y254C/Y254C and WA09-IMPG2KO/KO ROs displayed similar IMPG2, IMPG1, rhodopsin, and PRPH2 immunostaining patterns as their corresponding IMPG2 mutant and knockout hiPSC-ROs, respectively (Figures 3B and 3C). Of note, IMPG2 mutations did not affect general photoreceptor survival over the time course of our studies, since all hPSC-ROs demonstrated a uniform, compacted outer nuclear layer with no significant difference in thickness between normal and mutant lines (Figure S5).
In summary, disruption of IMPG2 in either hiPSCs or hESCs reproducibly resulted in an unequivocal light microscopic phenotype consisting of the near absence of an IPM and markedly stunted photoreceptor OS. Conversely, homozygous correction of IMPG2 mutations fully restored a normal stage 3 phenotype with a thick IPM and elongated OS in all ROs examined.
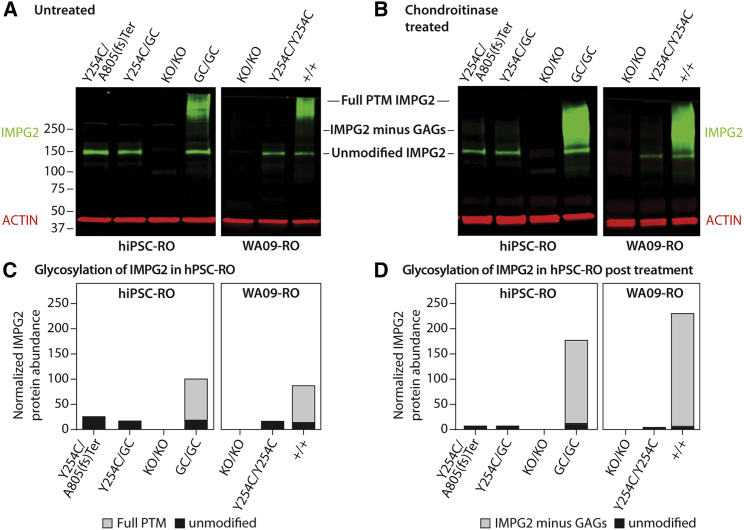
The Y254C IMPG2 mutation prevents post-translational modifications of the expressed protein
IMPG2 is a chondroitin sulfate proteoglycan with a core molecular weight of 138 kDa that undergoes extensive PTM via attachment of glycosaminoglycans (GAGs), N-linked glycosylation, and O-linked glycosylation. These PTMs more than double the mass of the core protein (Acharya et al., 2000) and are presumed to be critical for its role in organizing and maintaining the IPM. To investigate the mechanism of IPM disruption and OS loss in hPSC-ROs expressing the Y254C mutation (i.e., hiPSC-IMPG2Y254C/A805(fs)ter, hiPSC-IMPG2Y254C/GC, and WA09-IMPG2Y254C/Y254C), we evaluated the PTM status of the mutant protein by western blot. As controls, we also examined the PTM status of normal IMPG2 from fully gene-corrected or wild-type isogenic ROs. It was previously shown using a custom polyclonal antibody that IMPG2 is largely undetectable on western blots unless it is first treated with chondroitinase to remove GAG attachments (Acharya et al., 2000). Interestingly, using the EC-IMPG2 antibody, a band corresponding to unmodified IMPG2 (138 kDa) was detectable in RO lysates at >200 days of differentiation even without chondroitinase treatment (Figure 4A). In addition, we detected a broad, very high molecular weight protein band in untreated lysates from mature hiPSC-IMPG2GC/GC and WA09-IMPG2+/+ ROs, consistent IMPG2 with PTMs (Figure 4A). Incubation with chondroitinase resulted in the appearance of a more prominent, intermediate-sized band in the hiPSC-IMPG2GC/GC and WA09-IMPG2+/+ RO lysates, corresponding to the predicted molecular weight of IMPG2 without GAG attachments (230 kDa; Figure 4B). These results demonstrate that IMPG2 undergoes extensive PTM in wild-type and fully gene-corrected hPSC-ROs.
Figure 4.
The Y254C missense mutation produces full length IMPG2 protein that lacks post-translational modifications
(A and B) Immunoblot analysis of untreated (A) or chondroitinase-treated (B) protein samples from hPSC-ROs differentiated >200 days reveals a band corresponding to unmodified IMPG2 in all samples except the knockout lines. Only lysates from wild-type or fully gene-corrected ROs contain detectable levels of post-translationally modified (PTM) IMPG2 protein before or after chondroitinase treatment.
(C and D) Quantification of the levels of PTM (gray bars) versus unmodified (black bars) IMPG2 protein (normalized to ACTIN) in the immunoblots of untreated (C) or chondroitinase-treated (D) hPSC-RO lysates. GAG = glycosaminoglycans.
In contrast to the control hPSC lines, western analysis of ROs from the hiPSC-IMPG2Y254C/A805(fs)ter, hiPSC-IMPG2Y254C/GC, and WA09-IMPG2Y254C/Y254C lines with and without chondroitinase treatment exclusively showed the presence of unmodified IMPG2 (Figures 4A and 4B). To ensure that the western blot bands were specific to IMPG2, we also analyzed ROs from the hiPSC IMPG2KO/KO line, which yielded no bands as expected (Figures 4A and 4B). Thus, the Y254C mutation alone is capable of prohibiting proper PTM of IMPG2. Of note, the lack of a detectable post-translationally modified IMPG2 band in western blots from heterozygous gene-corrected hiPSCY254C/GC ROs suggests that the vast majority of fully PTM IMPG2 is localized to the IPM, which fails to form in these ROs.
Mutant IMPG2 hPSC-ROs extend and maintain OS when differentiated in the physically stable environment of the subretinal space
The OS phenotype observed in IMPG2 mutant and knockout hPSC-ROs is considerably more severe and earlier in onset than that of human patients, who are born with fully formed and functional photoreceptor OS that are subsequently lost over the course of years. Furthermore, the presence of one normal IMPG2 allele did not correct the mutant RO phenotype (Figures 2C and S3C), in contrast to the human condition in which IMPG2 haploinsufficiency has either no significant consequence or causes a late-onset vitelliform macular dystrophy (Brandl et al., 2017). The advanced phenotype observed in mutant ROs could be due to an inability of IMPG2-deficient ROs (1) to produce elongated OS under any circumstances or (2) to stabilize and maintain elongated OS due to factors exacerbated by the tissue culture environment. To distinguish between these possibilities, stage 1 (day 45) mutant IMPG2 or isogenic control hPSC-ROs were partially dissociated into aggregates and transplanted into the subretinal space of immunocompromised Foxn1-S334ter-3 rats (Figure 5A) (Martinez-Navarrete et al., 2011). This animal model was chosen because it supports long-term xenograft survival and demonstrates a near-complete lack of photoreceptors, which permits clear delineation of transplanted donor photoreceptor populations.
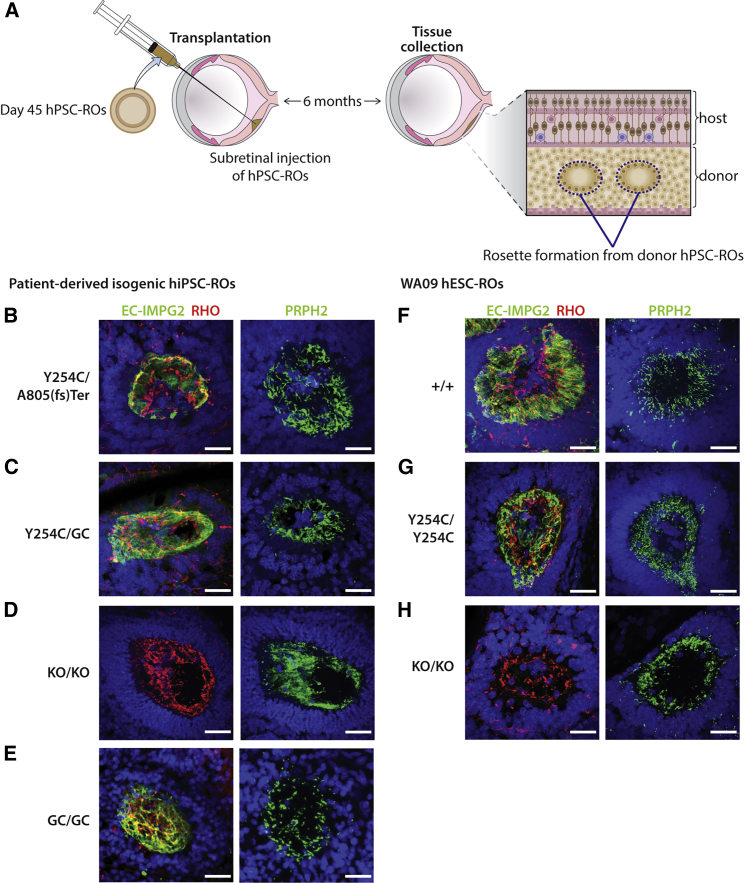
Figure 5.
Transplantation of mutant IMPG2 hPSC-ROs into the subretinal space of immunocompromised rats restores their capacity to extend and maintain OS
(A) Schematic summarizing the experimental approach used to transplant day 45 hPSC-ROs into the subretinal space of Foxn1-S334ter immunocompromised rats, a time point when this model is nearly devoid of host photoreceptors.
(B–H) Immunocytochemical analysis of human donor photoreceptor rosettes 6 months after RO transplantation reveals extensive RHO+/PHPR2+ OS in all mutant and control lines: IMPG2Y254C/A805(fs)ter (B), IMPG2Y254C/GC (C), IMPG2KO/KO (D), IMPG2GC/GC (E), WA09-IMPG2+/+ (F), WA09-IMPG2Y254C/Y254C (G), and WA09-IMPG2KO/KO (H). Scale bars, 25 μm.
The transplanted RO aggregates were allowed to mature in vivo in the rat subretinal space for 6 months, whereupon they formed photoreceptor rosettes with central lumina into which OS could extend (Figures 5A and S6) (Iraha et al., 2018; McLelland et al., 2018; Tu et al., 2019). We hypothesized that, if mechano-physical forces present in the tissue culture environment are responsible for the advanced disease state in IMPG2-deficient ROs, maturing them in the protected subretinal space should mitigate the phenotype. Indeed, photoreceptor rosettes generated from transplanted IMPG2Y254C/A805(fr)ter or IMPG2Y254C/GC hiPSC-ROs possessed inwardly oriented, elongated rhodopsin+/PRPH2+ OS with a surrounding IMPG2+ IPM, similar to what was present in rosettes from isogenic control hiPSC-ROs (Figures 5B, 5C, and 5E). Interestingly, OS were also found in photoreceptor rosettes from transplanted hiPSC-IMPG2KO/KO ROs, indicating that the presence of IMPG2 is not necessary for the initial production of OS in the subretinal space (Figure 5D), consistent with observations in early-stage IMPG2-RP patients. These findings were reproduced using transplanted WA09-IMPG2 ROs (Figures 5F–5H).
Discussion
A growing number of studies have shown that hPSC-derived cells, tissues, and organoids can model cellular and molecular features of human diseases. Advantages of hPSC-based model systems include their human genotype, theoretically unlimited supply, and amenability to experimental manipulation and long-term investigation in an accessible tissue culture environment. Human PSC systems are particularly valuable when animal models are either not available or fail to recapitulate key aspects of human disease. However, simply having a human genotype does not ensure that a cell type or organoid derived from hPSCs will manifest a disease-relevant phenotype. Challenges in this regard are numerous and result from limitations unique to hPSCs or inherent to all cell culture models, such as (1) extent of structural and functional maturation achievable, (2) variability between and within cultures, (3) reliance on cell or tissue autonomous effects, and (4) time needed to manifest natural disease processes.
Advancements in production, differentiation, and staging methods have lessened many of the challenges for using hPSC-derived ROs (Capowski et al., 2019). For example, a recent study showed that cone photoreceptors in stage 3 ROs from multiple hPSC lines exhibited complex, intrinsic light responses and membrane electrophysiological properties akin to adult rhesus macaque foveal cones (Saha et al., 2022). Another report showed the potential for ROs to reproduce the neuronal circuitry of a full thickness retina (Cowan et al., 2020). Thus, ROs are capable of modeling normal human retinal physiology when allowed to mature in long-term culture. However, even early-onset retinal disorders can take years to cause structural or functional deficits, and although ROs can be maintained for many months in culture, it is a costly and inefficient process that still falls well short of the time needed to observe most human phenotypes in vivo. Efforts to accelerate or circumvent natural disease processes in vitro have been described, but they rely largely on mechanistically nonspecific interventions such as exposure to mitochondrial stressors or toxic chemicals or introduction of genetic mutations unrelated to the disease of interest (Fathi et al., 2022; Mertens et al., 2018).
Other important considerations in hPSC disease modeling are the ease, rigor, and reproducibility with which a phenotype can be assessed and monitored. ROs are complex, dynamic structures whose morphology and cellular constituency vary spatially and temporally. Most methods to assess effects of inherited retinal disease genes require RO sacrifice (fixation or dissociation) followed by multi-step imaging analyses or other assays that employ specific tools, reagents, or expertise. In addition, such readouts are often subject to sampling error and/or provide only partial or indirect measures of disease activity.
In an attempt to address these issues and establish a robust, user-friendly, and mechanistically germane RO disease model, we chose to focus on an inherited disorder affecting the IPM. A major factor influencing this choice was the observation that fragile photoreceptor OS survive indefinitely in culture on the exposed surface of ROs, which suggests the presence of a functional, protective IPM. Indeed, the existence of an IPM was previously reported in late-stage ROs, although IMPG2 was not observed in that study (Felemban et al., 2018). Therefore, it stood to reason that disruption of the IPM could lead to an early-onset OS phenotype discernible in live ROs by light microscopy. Another impetus for this study rested in the general importance of OS layer thickness measurements as sensitive indicators of inherited retinal disease severity and treatment response in vivo (Cideciyan et al., 2021; Huang et al., 2021; Sousa et al., 2019).
We first confirmed that normal stage 3 hPSC-ROs elaborate a thick IPM that surrounds OS and showed for the first time that it contains IMPG2. Subsequent IHC analyses in ROs, mouse retina, and NHP retinal tissue revealed that antibodies directed against intracellular or extracellular epitopes of IMPG2 exhibited distinct but overlapping expression patterns. More specifically, intracellular IMPG2 immunostaining localized solely to IS, while extracellular IMPG2 immunostaining localized to regions surrounding both IS and OS. This finding suggests that IMPG2 is altered as it passes through the IS, either through protein cleavage or epitope masking. Our results also explain why a prior mouse study that solely used an antibody targeting the intracellular terminus of IMPG2 reported that it was expressed only in IS (Salido and Ramamurthy, 2020).
After validating IMPG2 expression in normal ROs, we examined the phenotypic consequence of its loss of function in a series of patient-derived or gene-edited hiPSC and hESC lines. All mutant IMPG2-RP ROs demonstrated a near absence of OS and IPM by live light microscopy and/or IHC. Conversely, all homozygous isogenic control ROs possessed an IPM and were densely covered with elongated OS. While the phenotypic impact of IMPG2 knockout and frameshift mutations could be reasoned by the lack of protein expression, an explanation for the equally severe phenotype in ROs with the Y254C IMPG2 mutation, which is fully expressed, was not immediately apparent. However, further investigation demonstrated an absence of PTMs in the Y254C mutant protein, which are presumably necessary for stabilizing the IPM (Chen et al., 2004). The fact that a single point mutation eliminated most or all PTMs implies a pervasive conformational disturbance, which in turn provides a plausible mechanism for the loss of IMPG2 function in this and other missense mutants.
Additional insight into the mechanism underlying the advanced mutant IMPG2 RO disease phenotype was garnered after observing that correction of a single allele failed to re-establish an OS layer. Since IMPG2 haploinsufficiency either fails to produce a clinically meaningful phenotype or produces a late-onset vitelliform macular dystrophy in human carriers, our results suggest that the mechanically stressful RO culture conditions not only accelerate the onset of an IMPG2-RP phenotype, but they also reduce the threshold for manifestation. Confirmation of this theory came following transplantation of mutant IMPG2 RO aggregates into the mechanically stable subretinal space, which permitted growth of OS. While it is also possible that host-secreted factors helped stimulate OS elongation of transplanted photoreceptors, such influences are not required given the robust presence of OS on wild-type ROs in vitro. Overall, these results reconcile the RO disease phenotype with clinical observations and imaging analyses in patients with IMPG2-RP, who are born with a functional OS layer that is lost over the course of years. Similarly, the Impg2 knockout mouse models are born with a normal appearing OS layer that undergoes relatively mild structural changes into adulthood (Salido and Ramamurthy, 2020; Xu et al., 2020).
An interesting feature of retinal diseases caused by IMPG2 mutations is their predilection to affect the macula early in their course (Brandl et al., 2017; Huet et al., 2014), possibly due to a selective detrimental effect on cone versus rod OS stability. The stage 3 ROs employed in the present study, which exhibit a 4:1 rod:cone ratio (Capowski et al., 2019), showed uniform (i.e., nonselective) OS disruption. However, more insight into potential regional effects of IMPG2 mutations within the retina could be obtained in the future using cone-enriched ROs.
From a practical application standpoint, the availability of both null and haploinsufficient IMPG2 mutant human RO models provides a simple means to broadly assess therapeutic potency. More specifically, a treatment that generates a visible OS layer in homozygous null ROs indicates restoration of greater than 50% of normal IMPG2 levels. In contrast, formation of an OS layer in heterozygous—but not homozygous—mutant ROs would indicate that the treatment is capable of augmenting IMPG2 expression by less than 50% of normal levels. Following this initial live assessment, a more precise determination of IMPG2 protein levels and PTM status in treated versus untreated ROs could be carried out using quantitative protein analysis.
In conclusion, our results show that hPSC-ROs provide a rigorous and mechanistically relevant model of IMPG2 dysfunction capable of mimicking later stages of human disease that mutant mice do not achieve. Furthermore, the RO phenotype we observe represents an ideal treatment window for initial clinical trials; namely, after vision loss occurs due to OS disruption but before the onset of irreversible photoreceptor cell death. As such, we anticipate their use as a powerful tool to advance gene therapies for IMPG2-RP. Moreover, since this RO phenotype is likely to occur following introduction of any IMPG2 loss of function mutation, these models could serve as a generic testing platform for early-stage testing of gene or RNA editing strategies regardless of the ultimate disease target.
Experimental procedures
Human pluripotent stem cell lines
All work with hPSC lines was carried out in accordance with institutional, national, and international guidelines and approved by the Institutional Review Board and Stem Cell Research Oversight Committee at the University of Wisconsin-Madison. For details on hPSC reprograming, gene editing, and genotyping please see supplemental information.
hPSC culture and retinal organoid differentiation
All hPSC lines were cultured and differentiated for >200 days using a previously published protocol (Capowski et al., 2019). Additional details regarding the retinal organoid differentiation protocol are provided in supplemental information.
hPSC-RO immunocytochemistry
ROs were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) at room temperature (RT) with gentle agitation for 1 h, washed with 1X PBS, and incubated in 15% sucrose in PBS for 40 min, followed by equilibration in 30% sucrose for 40 min. ROs were cryopreserved immediately post-equilibration to preserve photoreceptor outer segments and then cryosectioned at a thickness of 15 μm. Cryosections were blocked in blocking solution (10% normal donkey serum, 5% BSA, 1% fish gelatin, and 0.5% Triton X-100 in 1X PBS) for 1 h at RT and incubated at 4°C overnight with primary antibodies prepared in blocking solution (see Table S3 for a list of the primary antibodies). After the incubation, cryosections were washed 3X in 1X PBS, then incubated for 30 min in the dark at RT with appropriate fluorophore-conjugated secondary antibodies (Alexa Fluor 488, Alexa Fluor 555, Thermo Fisher Scientific; Cat# A-21206, A-31570) prepared in blocking solution. Thereafter, the immunostained cryosections were washed 3X in PBS, mounted in Prolong Gold Antifade with DAPI (Thermo Fisher Scientific, Cat# P36931), and imaged using a Nikon A1 laser scanning confocal microscope with NIS Elements AR 5.0 software. For information regarding mouse and rat immunocytochemistry please see supplemental information.
Western blot analysis
hPSC-ROs were lysed in RIPA buffer containing Halt protease inhibitor (Thermo Fisher Scientific; Cat# P8340), followed by quantification of total protein using a DC Protein assay kit II (Bio-Rad, Cat# 5000112). For samples undergoing chondroitinase treatment, 20 μg of protein was resuspended in 0.1 M Tris acetate buffer (pH 7.3) containing 3 mU of chondroitinase ABC (Sigma-Aldrich; Cat# C3667-5UN) for 1 h at 37°C. 20 μg of each treated or untreated protein sample was then separated on a 4%–20% mini-Protean TGX gel (Bio-Rad; Cat# 4561094) and transferred onto a PVDF membrane (Millipore; Immobilon-FL). After transfer, membranes were dried at RT for 15 min, reactivated in methanol for 1 min, and then blocked for 1 h at RT with Intercept Blocking Buffer (LI-COR Biosciences; Cat# 927-70001). Membranes were then incubated overnight at 4°C with anti-EC-IMPG2 (1:500) and anti-ACTIN (1:3,000) primary antibodies diluted in blocking buffer with 0.1% Tween 20. Thereafter, blots were washed 4X in PBS +0.1% Tween 20 and incubated for 1 h at RT in appropriate secondary antibodies (LI-COR Biosciences Cat# 926-68022, 926-32213) diluted 1:10,000 in blocking buffer with 0.1% Tween 20 and 0.01% SDS. The blots were washed again 4X in 1X PBS +0.1% Tween 20, and protein bands were visualized using an Odyssey Infrared Imager (LI-COR Biosciences).
Animal care and subretinal transplantation of hPSC-RO aggregates
Athymic SD-Foxn1 Tg (S334ter)3LavRrrc (Foxn1-S334ter-3) rats (Seiler et al., 2014) were used for all RO transplantation experiments. Foxn1-S334ter-3 rats undergo rapid and near-complete loss of rod PRs by P30, with subsequent cone PR dysfunction and death (Hombrebueno et al., 2010; LaVail et al., 2018; Martinez-Navarrete et al., 2011). All transplantations were performed between 3 and 6 months of age, when only a single discontinuous layer of degenerating host cone PRs remains. Animal procedures were approved by the University of Wisconsin-Madison Institutional Animal Care and Use Committee and were conducted in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research, the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the laws and regulations of the United States Department of Agriculture. For further details regarding rat subretinal transplantation please see supplemental information.
Author contributions
S.J.M.: experimental design, methodology, project supervision, data collection and formal analysis, writing, and data curation. S.B. and C.B.: production and maintenance of retinal organoids and data collection. A.L.L. and L.D.J.: subretinal transplantation and tissue collection. C.S.: genome engineering of WA09 lines. B.N.W.: data collection and data curation. D.S.: experimental design, and writing. B.D.P. and D.M.G.: conceptualization, funding acquisition, methodology, project administration, project supervision, writing, and data curation.
Acknowledgments
This work was supported by the Foundation Fighting Blindness (TA-RM-0618-0742; B.D.P. and D.M.G.), a T32 NIH grant for Postdoctoral Research in Neurodevelopmental Disorders Training (T32Act HD040127; B.N.W.), the McPherson Eye Research Institute Sandra Lemke Trout Chair in Eye Research (D.M.G.), the Retina Research Foundation Emmett A. Humble Distinguished Directorship (D.M.G.), NIH grant F30EY031230 (A.L.L.), and an unrestricted grant from Research to Prevent Blindness, Inc., to the UW-Madison Department of Ophthalmology and Visual Sciences. The authors also thank the University of Wisconsin Department of Pathology and Laboratory Medicine for use of its facilities and services, and the Waisman Center Imaging and hPSC/Gene Editing Cores (supported by NICHD U54 HD090255). We thank members of the Gamm lab for comments on the manuscript, and H. Adam Steinberg (Art for Science) for help in figure preparation.
Conflicts of interest
D.M.G. is an inventor on patents related to generation of 3D retinal organoids (US PTO no. 9328328). S.J.M. and D.M.G. are co-inventors on a patent related to the use of mutant IMPG2 hPSC-ROs as in vitro model systems filed by the Wisconsin Alumni Research Foundation, Madison, WI. D.M.G. has an ownership interest in and receives grant funding from Opsis Therapeutics, LLC, which has licensed the technology to generate 3D retinal organoids. L.D.J. is a consultant for Opsis Therapeutics, LLC. The terms of this arrangement have been reviewed and approved by the University of Wisconsin-Madison in accordance with its conflict-of-interest policies.
Published: October 6, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.09.004.
Supplemental information
References
- Acharya S., Foletta V.C., Lee J.W., Rayborn M.E., Rodriguez I.R., Young W.S., Hollyfield J.G. SPACRCAN, a novel human interphotoreceptor matrix hyaluronan-binding proteoglycan synthesized by photoreceptors and pinealocytes. J. Biol. Chem. 2000;275:6945–6955. doi: 10.1074/jbc.275.10.6945. [DOI] [PubMed] [Google Scholar]
- Bandah-Rozenfeld D., Collin R.W.J., Banin E., Ingeborgh van den Born L., Coene K.L.M., Siemiatkowska A.M., Zelinger L., Khan M.I., Lefeber D.J., Erdinest I., et al. Mutations in IMPG2, encoding interphotoreceptor matrix proteoglycan 2, cause autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2010;87:199–208. doi: 10.1016/j.ajhg.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet B., Marzouka N.A.D., Hebrard M., Manes G., Sénéchal A., Meunier I., Hamel C.P. Homozygosity mapping in autosomal recessive retinitis pigmentosa families detects novel mutations. Mol. Vis. 2013;19:2487–2500. [PMC free article] [PubMed] [Google Scholar]
- Brandl C., Schulz H.L., Charbel Issa P., Birtel J., Bergholz R., Lange C., Dahlke C., Zobor D., Weber B.H.F., Stöhr H. Mutations in the genes for interphotoreceptor matrix proteoglycans, IMPG1 and IMPG2, in patients with vitelliform macular lesions. Genes. 2017;8:170. doi: 10.3390/genes8070170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski E.E., Samimi K., Mayerl S.J., Phillips M.J., Pinilla I., Howden S.E., Saha J., Jansen A.D., Edwards K.L., Jager L.D., et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development. 2019;146:dev171686. doi: 10.1242/dev.171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Lee J.W., Nishiyama K., Shadrach K.G., Rayborn M.E., Hollyfield J.G. SPACRCAN in the interphotoreceptor matrix of the mouse retina: molecular, developmental and promoter analysis. Exp. Eye Res. 2003;76:1–14. doi: 10.1016/S0014-4835(02)00273-7. [DOI] [PubMed] [Google Scholar]
- Chen Q., Cai S., Shadrach K.G., Prestwich G.D., Hollyfield J.G. Spacrcan binding to hyaluronan and other glycosaminoglycans: molecular and biochemical studies. J. Biol. Chem. 2004;279:23142–23150. doi: 10.1074/jbc.M401584200. [DOI] [PubMed] [Google Scholar]
- Cideciyan A.V., Krishnan A.K., Roman A.J., Sumaroka A., Swider M., Jacobson S.G. Measures of function and structure to determine phenotypic features, natural history, and treatment outcomes in inherited retinal diseases. Annu. Rev. Vis. Sci. 2021;7:747–772. doi: 10.1146/annurev-vision-032321-091738. [DOI] [PubMed] [Google Scholar]
- Cowan C.S., Renner M., De Gennaro M., Gross-Scherf B., Goldblum D., Hou Y., Munz M., Rodrigues T.M., Krol J., Szikra T., et al. Cell types of the human retina and its organoids at single-cell resolution. Cell. 2020;182:1623–1640.e34. doi: 10.1016/j.cell.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi A., Mathivanan S., Kong L., Petersen A.J., Harder C.R.K., Block J., Miller J.M., Bhattacharyya A., Wang D., Zhang S.-C. Chemically induced senescence in human stem cell-derived neurons promotes phenotypic presentation of neurodegeneration. Aging Cell. 2022;21:e13541. doi: 10.1111/acel.13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felemban M., Dorgau B., Hunt N.C., Hallam D., Zerti D., Bauer R., Ding Y., Collin J., Steel D., Krasnogor N., et al. Extracellular matrix component expression in human pluripotent stem cell-derived retinal organoids recapitulates retinogenesis in vivo and reveals an important role for IMPG1 and CD44 in the development of photoreceptors and interphotoreceptor matrix. Acta Biomater. 2018;74:207–221. doi: 10.1016/j.actbio.2018.05.023. [DOI] [PubMed] [Google Scholar]
- Foletta V.C., Nishiyama K., Rayborn M.E., Shadrach K.G., Young W.S., Hollyfield J.G. SPACRCAN in the developing retina and pineal gland of the rat: spatial and temporal pattern of gene expression and protein synthesis. J. Comp. Neurol. 2001;435:354–363. doi: 10.1002/cne.1035. [DOI] [PubMed] [Google Scholar]
- Hageman G.S., Johnson L.V. Chapter 9 Structure, composition and function of the retinal interphotoreceptor matrix. Prog. Retin. Res. 1991;10:207–249. doi: 10.1016/0278-4327(91)90014-S. [DOI] [Google Scholar]
- Hartong D.T., Berson E.L., Dryja T.P. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- Hombrebueno J.R., Tsai M.M., Kim H.-L., De Juan J., Grzywacz N.M., Lee E.-J. Morphological changes of short-wavelength cones in the developing S334ter-3 transgenic rat. Brain Res. 2010;1321:60–66. doi: 10.1016/j.brainres.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-W., Yang J.-J., Yang C.-H., Yang C.-M., Hu F.-R., Ho T.-C., Chen T.-C. The structure-function correlation analysed by OCT and full field ERG in typical and pericentral subtypes of retinitis pigmentosa. Sci. Rep. 2021;11:16883. doi: 10.1038/s41598-021-96570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet R.A.C.V., Collin R.W.J., Siemiatkowska A.M., Klaver C.C.W., Hoyng C.B., Simonelli F., Khan M.I., Qamar R., Banin E., Cremers F.P.M., et al. IMPG2-Associated retinitis pigmentosa displays relatively early macular involvement. Invest. Ophthalmol. Vis. Sci. 2014;55:3939–3953. doi: 10.1167/iovs.14-14129. [DOI] [PubMed] [Google Scholar]
- Iraha S., Tu H.-Y., Yamasaki S., Kagawa T., Goto M., Takahashi R., Watanabe T., Sugita S., Yonemura S., Sunagawa G.A., et al. Establishment of immunodeficient retinal degeneration model mice and functional maturation of human ESC-derived retinal sheets after transplantation. Stem Cell Rep. 2018;10:1059–1074. doi: 10.1016/j.stemcr.2018.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Sawada Y., Yoshitomi T. Structure and function of the interphotoreceptor matrix surrounding retinal photoreceptor cells. Exp. Eye Res. 2015;133:3–18. doi: 10.1016/j.exer.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Khan A.O., Al Teneiji A.M. Homozygous and heterozygous retinal phenotypes in families harbouring IMPG2 mutations. Ophthalmic Genet. 2019;40:247–251. doi: 10.1080/13816810.2019.1627467. [DOI] [PubMed] [Google Scholar]
- Kuehn M.H., Hageman G.S. Molecular characterization and genomic mapping of human IPM 200, a second member of a novel family of proteoglycans. Mol. Cell Biol. Res. Commun. 1999;2:103–110. doi: 10.1006/mcbr.1999.0161. [DOI] [PubMed] [Google Scholar]
- LaVail M.M., Nishikawa S., Steinberg R.H., Naash M.I., Duncan J.L., Trautmann N., Matthes M.T., Yasumura D., Lau-Villacorta C., Chen J., et al. Phenotypic characterization of P23H and S334ter Rhodopsin transgenic rat models of inherited retinal degeneration. Exp. Eye Res. 2018;167:56–90. doi: 10.1016/j.exer.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Navarrete G., Seiler M.J., Aramant R.B., Fernandez-Sanchez L., Pinilla I., Cuenca N. Retinal degeneration in two lines of transgenic S334ter rats. Exp. Eye Res. 2011;92:227–237. doi: 10.1016/j.exer.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland B.T., Lin B., Mathur A., Aramant R.B., Thomas B.B., Nistor G., Keirstead H.S., Seiler M.J. Transplanted hESC-derived retina organoid sheets differentiate, integrate, and improve visual function in retinal degenerate rats. Invest. Ophthalmol. Vis. Sci. 2018;59:2586–2603. doi: 10.1167/iovs.17-23646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J., Reid D., Lau S., Kim Y., Gage F.H. Aging in a dish: iPSC-derived and directly induced neurons for studying brain aging and age-related neurodegenerative diseases. Annu. Rev. Genet. 2018;52:271–293. doi: 10.1146/annurev-genet-120417-031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier I., Manes G., Bocquet B., Marquette V., Baudoin C., Puech B., Defoort-Dhellemmes S., Audo I., Verdet R., Arndt C., et al. Frequency and clinical pattern of vitelliform macular dystrophy caused by mutations of interphotoreceptor matrix IMPG1 and IMPG2 genes. Ophthalmology. 2014;121:2406–2414. doi: 10.1016/j.ophtha.2014.06.028. [DOI] [PubMed] [Google Scholar]
- Meyer J.S., Howden S.E., Wallace K.A., Verhoeven A.D., Wright L.S., Capowski E.E., Pinilla I., Martin J.M., Tian S., Stewart R., et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cell. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Ando S., Takata N., Kawada M., Muguruma K., Sekiguchi K., Saito K., Yonemura S., Eiraku M., Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- O’Hara-Wright M., Gonzalez-Cordero A. Retinal organoids: a window into human retinal development. Development. 2020;147:dev189746. doi: 10.1242/dev.189746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Capowski E., Fernandez Zepeda M.A., Nelson E.C., Gamm D.M., Sinha R. Cone photoreceptors in human stem cell-derived retinal organoids demonstrate intrinsic light responses that mimic those of primate fovea. Cell Stem Cell. 2022;29:487–489. doi: 10.1016/j.stem.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salido E.M., Ramamurthy V. Proteoglycan IMPG2 shapes the interphotoreceptor matrix and modulates vision. J. Neurosci. 2020;40:4059–4072. doi: 10.1523/JNEUROSCI.2994-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M.J., Aramant R.B., Jones M.K., Ferguson D.L., Bryda E.C., Keirstead H.S. A new immunodeficient pigmented retinal degenerate rat strain to study transplantation of human cells without immunosuppression. Graefes Arch. Clin. Exp. Ophthalmol. 2014;252:1079–1092. doi: 10.1007/s00417-014-2638-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Shen W., Kuai D., Martin J.M., Guo X., Smith M.A., Perez E.T., Phillips M.J., Simonett J.M., Wallace K.A., et al. iPS cell modeling of Best disease: insights into the pathophysiology of an inherited macular degeneration. Hum. Mol. Genet. 2013;22:593–607. doi: 10.1093/hmg/dds469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D., Phillips J., Joseph Phillips M., Gamm D.M. Mimicking retinal development and disease with human pluripotent stem cells. Invest. Ophthalmol. Vis. Sci. 2016;57:ORSFf1–ORSFf9. doi: 10.1167/iovs.15-18160. [DOI] [PubMed] [Google Scholar]
- Sinha D., Steyer B., Shahi P.K., Mueller K.P., Valiauga R., Edwards K.L., Bacig C., Steltzer S.S., Srinivasan S., Abdeen A., et al. Human iPSC modeling reveals mutation-specific responses to gene therapy in a genotypically diverse dominant maculopathy. Am. J. Hum. Genet. 2020;107:278–292. doi: 10.1016/j.ajhg.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa K., Fernandes T., Gentil R., Mendonça L., Falcão M. Outer retinal layers as predictors of visual acuity in retinitis pigmentosa: a cross-sectional study. Graefes Arch. Clin. Exp. Ophthalmol. 2019;257:265–271. doi: 10.1007/s00417-018-4185-4. [DOI] [PubMed] [Google Scholar]
- Tu H.-Y., Watanabe T., Shirai H., Yamasaki S., Kinoshita M., Matsushita K., Hashiguchi T., Onoe H., Matsuyama T., Kuwahara A., et al. Medium- to long-term survival and functional examination of human iPSC-derived retinas in rat and primate models of retinal degeneration. EBioMedicine. 2019;39:562–574. doi: 10.1016/j.ebiom.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbakel S.K., van Huet R.A.C., Boon C.J.F., den Hollander A.I., Collin R.W.J., Klaver C.C.W., Hoyng C.B., Roepman R., Klevering B.J. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 2018;66:157–186. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Xu H., Qu C., Gan L., Sun K., Tan J., Liu X., Jiang Z., Tian W., Liu W., Zhang S., et al. Deletion of the Impg2 gene causes the degeneration of rod and cone cells in mice. Hum. Mol. Genet. 2020;29:1624–1634. doi: 10.1093/hmg/ddaa062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.