Abstract
Burkholderia cepacia is an opportunistic pathogen that causes severe systemic infections in patients with chronic granulomatous disease (CGD) or with cystic fibrosis (CF), but its mechanisms of virulence are poorly understood. We developed a murine model of systemic infection in wild-type (WT) and gamma interferon knockout (GKO) BALB/c mice to facilitate dissection of components of pathogenicity and host defense. Both WT and GKO mice were susceptible to chronic splenic infection with B. cepacia, but not with Pseudomonas aeruginosa. B. cepacia strains from patients with CGD persisted longer than those from CF patients. C57BL/6 mice were the most susceptible murine strain; bacteria persisted in the spleen for 2 months. DBA/2, BALB/c, and A/J strains of mice were relatively resistant to infection. Certain strains of B. cepacia complex can persist in the murine spleen after systemic infection; this may provide clues to its virulence in compromised hosts, such as those with CGD and CF.
Burkholderia cepacia was originally isolated in 1952 as the cause of sour skin rot in onions (1), and since that time has become recognized as an important pathogen in certain compromised hosts. Whereas it is one of the recognized pathogens in patients with cystic fibrosis (CF) (6, 11) or with chronic granulomatous disease (CGD) (23), a unifying explanation for its predilection for these two dissimilar diseases remains elusive. A clearer understanding of the pathogenicity of this evolving pathogen would be greatly aided by a suitable animal model of infection.
B. cepacia is an extremely diverse class of bacteria, and in fact, is probably comprised of at least five discrete species (27). The term “genomovar” is used to describe the five potential novospecies within the B. cepacia complex (27). New species designations have recently been proposed for those genomovars which have characteristic phenotypes; genomovar II has been designated Burkholderia multivorans, and genomovar V is Burkholderia vietnamiensis. Most strains of B. cepacia from patients with CF are in genomovar III, and this genomovar contains all strains in the epidemic ET/12 cable pilus-possessing lineage and most of the strains which carry the B. cepacia epidemic strain marker (13a, 14).
Patients with CGD are at risk of infection with certain classes of bacteria and fungi which are able to evade the limbs of phagocytic defense which operate independent of an intact NADPH oxidase system required for production of oxidative radicals. These patients are susceptible to infection with B. cepacia, but not another common gram-negative opportunistic pathogen (Pseudomonas aeruginosa), because the former but not the latter can resist the nonoxidative bactericidal activity of polymorphonuclear leukocytes, upon which CGD neutrophils are dependent for killing (23). Furthermore, B. cepacia is responsible for particularly devastating infections in some patients with CF. Certain of these patients succumb to “cepacia syndrome,” which is characterized by necrotizing pneumonia and septicemia (11). This type of infection is notably different from P. aeruginosa infections in CF, which are characteristically slowly progressive and noninvasive. An explanation for the striking differences between the infections of these two organisms in CF remains elusive.
To date, one of the most effective prophylactic strategies for patients with CGD is human recombinant gamma interferon (IFN-γ) (10). IFN-γ is an important cytokine produced by natural killer cells and T helper cells. It exerts a number of regulatory effects during the course of infection and is an important positive regulator for the T helper cell type 1 (Th1) immune response and inhibits the Th2 response (12). Perhaps the principal role of IFN-γ in host defense against infection is its capacity to activate macrophages with subsequent production of reactive oxidative radicals, which are toxic to phagocytosed organisms (17). Nonetheless, phagocytic cells from patients with CGD, treated with IFN-γ, do not appear to have an enhanced ability to generate reactive oxygen radicals (10). The mechanism by which IFN-γ decreases the rate of serious infections in patients with CGD is currently not understood.
In order to begin to explore the virulence determinants of B. cepacia and the normal host defenses against this evolving human pathogen, we established a murine model of systemic infection. This model has facilitated the investigation of the relative virulence of different strains of B. cepacia and P. aeruginosa, as well as providing insight into the role of IFN-γ in host defense. These studies yielded data suggesting that B. cepacia (but not P. aeruginosa) is able to persist for up to 2 months in the murine spleen, that there are substantial differences in virulence among strains of B. cepacia, and that IFN-γ knockout (GKO) mice were not substantially compromised in their capacity to defend themselves against systemic challenge.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study and their characteristics are listed in Table 1. They were stored at −70°C in Mueller-Hinton broth with 8.0% dimethyl sulfoxide. All strains were cultured on blood agar plates (PML Microbiologicals, Richmond, British Columbia, Canada), and all colony counts were performed on tryptic soy agar (TSA) plates (Becton Dickinson, Cockeysville, Md.).
TABLE 1.
Characteristics of the bacterial strains used in this study
| Strain designation | Genomovar | Presence of cable pilusa | Colonial morphology | Presence of B. cepacia epidemic strain markerab | Serum sensitivityc | Source |
|---|---|---|---|---|---|---|
| B. cepacia | ||||||
| JTC | II | − | − | R | CGD | |
| C2303 | III | − | + | R/S | CF | |
| C1257 | III | − | + | R/S | CF | |
| C5424 | III | + | + | R | CF | |
| C2389 | III | − | + | R | CF | |
| C5491 | III | − | + | R | CF | |
| FC441 | V | − | − | ND | CGD | |
| P. aeruginosa | ||||||
| M2 | − | NMd | − | R | Murine intestine (16) | |
| P1 | − | NM | − | S | CF | |
| C4020 | − | NM | − | R | CF | |
| C0489 | − | NM | − | R | CF | |
| C0262 | − | NM | − | S | CF | |
| C0295 | − | NM | − | R | CF | |
| C0487 | − | NM | − | S | CF |
+, present; −, absent.
See reference 14 for details.
R, resistant to serum after 3 h; S, sensitive to serum; R/S, resistant after 2 h of exposure, but sensitive after 3 h; ND, not determined.
NM, non-mucoid.
Serum sensitivity testing.
A fresh overnight culture of bacteria grown in Luria-Bertani broth at 37°C was centrifuged and resuspended in Hank’s balanced salt solution with 0.1% (wt/vol) gelatin (gHBSS [Gibco BRL, Grand Island, N.Y.]) and adjusted to a concentration of 8 × 108 bacteria per ml. Bacteria were cultured at 105 per ml in the presence of 10% (vol/vol) pooled human serum at 37°C in gHBSS. Samples were taken at zero, 2, and 3 h, diluted serially in gHBSS, and quantitated by plating on TSA and incubation at 37°C for up to 72 h. Serum sensitivity was assessed on the following scale: resistant if there was no loss in viability after 3 h; resistant/sensitive if there was no change in log CFU at 2 h, but 1 log10 drop in CFU after 3 h; and sensitive if there was 1 log10 drop in CFU observed after 2 h.
Mice.
These studies were approved by the University of British Columbia Animal Care Committee (A96-0373). Mating pairs of heterozygote BALB/c GKO mice were kindly provided by Genentech, Inc. (San Francisco, Calif.). The mice were bred for the homozygous recessive GKO and homozygous normal dominant wild-type (WT) progeny. The construction of these mouse strains is the same as the original description of GKO mice (5). C57BL/6, DBA/2, and BALB/c mice were purchased from Charles River, St. Constant, Canada, and A/J mice were purchased from Jackson Laboratories, Bar Harbor, Maine. All mice were housed and cared for in accordance with regulations of the University of British Columbia Animal Care Committee and The Canadian Council on Animal Care. Mice weighed between 20 and 25 g and were 6 to 8 weeks old at the commencement of each experiment.
PCR genotyping of mice.
Progeny from the original heterozygote mating pairs were screened for the GKO and WT genotypes by PCR amplification primed for the normal IFN-γ gene (5′-AGAAGTAAGTGGAAGGGCCCAGAAG-3′ and 5′-AGGGAAACTGGGAGAGGAGAAATAT-3′) and the disrupted gene (5′-TCAGCGCAGGGGCGCCCGGTTCTTT-3′ and 5′-ATCGACAAGACCGGCTTCCATCCGA-3′). Oligonucleotides were synthesized by the Nucleic Acids-Protein Service Unit at the University of British Columbia. Approximately 3.0 mm of mouse ear was snipped from each animal postmortem. These were incubated in 20 μg of proteinase K per ml (Boehringer Mannheim, GmbH, Mannheim, Germany) with 1.0% (wt/vol) sodium dodecyl sulfate and 50 mM Tris-Cl. Zirconial silica beads (0.5 ml [0.5-mm diameter]) (Biospec Products, Inc., Bartlesville, Okla.) were added, and the tubes were placed in a bead beater for 1 min (Fisher, Nepean, Canada). The beads were removed by microcentrifugation. Potassium acetate (3.0 M) was added at 0.1 volumes to the supernatant, followed by phenol-chloroform extraction and precipitation by ethanol. DNA was resuspended in Tris-EDTA buffer (10 mM Tris-HCl, 1.0 mM EDTA [pH 8.0]) and stored at 4°C. DNA was amplified in the presence of the specific primers (Perkin-Elmer DNA Thermocycler). The amplified DNA was separated by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide.
Infection of mice.
Bacteria were cultured from frozen stocks for 24 to 48 h on blood agar plates at 37°C. Several isolated colonies were picked and grown in 5.0 ml of LB broth for 16 h and harvested by centrifugation (10 min at 3,000 × g) and then resuspended in gHBSS. Bacteria were adjusted to approximately 8 × 108 CFU/ml with a Beckman Du 640 spectrophotometer. Bacteria were then diluted in gHBSS to 8 × 106 CFU/ml for B. cepacia and 7 × 105 CFU/ml for P. aeruginosa. Mice were challenged intraperitoneally with 0.5 ml of either B. cepacia or P. aeruginosa through a 25G needle. At preselected times after infection, groups of four mice were anesthetized with sodium pentobarbital and killed by cervical dislocation. During the necropsy, the gall bladder was excised, as were the spleen, liver, and lungs, by sterile technique. Three samples from liver, lung, and spleen were preserved for histology by freezing in liquid nitrogen by fixation in 10% buffered Formalin (Starplex Scientific) and in 3% buffered glutaraldehyde (Canemco, Inc.). The remaining tissues were then weighed, diluted 10-fold in gHBSS, and homogenized for up to 30 s in sterile 10-ml glass tubes with a sterilized Teflon pestel (Glas-col, Terre Haute, Ind.). During the necropsy, an examination for any lesion or abscess was performed. The homogenates and serial dilutions of the homogenates were plated on TSA, and viable bacterial counts were enumerated after 24, 48, and 72 h of incubation at 37°C.
Histology.
Formalin-fixed tissues were processed in a Fisher Histomatic 266 tissue processor and embedded in paraffin wax blocks. The tissues were cut into 2-μm sections, stained with Gram’s stain and hematoxylin and eosin stain, and inspected for pathological changes and the presence of bacteria.
Statistical methods.
Data were analyzed by using SPSS for Windows version 7.5 software. For each experiment, assumptions for normal distribution were violated, since data for CFU per gram of tissue were skewed. Therefore, the nonparametric tests by Wilcoxon and Kruskal-Wallis were used for comparing groups. Wilcoxon’s test was used for comparison of two groups, and the Kruskal-Wallis test was used for comparisons of more than two groups. For each experiment, the overall type 1 error was held at 0.05 by use of Bonferroni’s correction. Plots were generated with Sigma Plot version 3.0 software (Jandel Scientific).
RESULTS
Genotyping of mice.
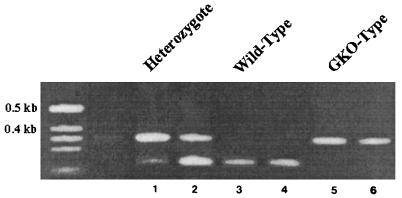
As shown in Fig. 1, the PCR amplification using primers specific for the intact IFN-γ gene gave a product 220 bp in size, and the primers specific for the neomycin cassette in the GKO mutation resulted in a product 375 bp in size. WT BALB/c mice gave a single 220-bp band, mice homozygous for the mutation in the IFN-γ gene gave a single 375-bp band, and mice with both bands were heterozygous for the mutation. The genotype for each mouse was confirmed by PCR after each experiment.
FIG. 1.
PCR products of the murine IFN-γ gene. Lanes: 1 and 2, heterozygotic mice; 3 and 4, WT mice; 5 and 6, GKO mice. See Materials and Methods for details. The unnumbered leftmost lane contains molecular size markers.
Intraperitoneal infection of WT and GKO mice with P. aeruginosa or B. cepacia.
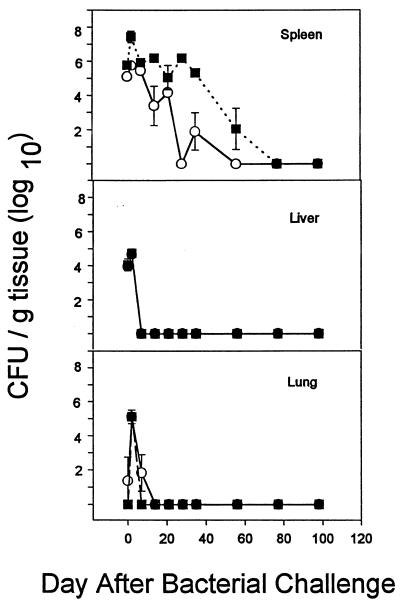
Figure 2 shows the results of experiments in which we determined the relative susceptibility of WT versus GKO mice to infection with bacterial species which were either susceptible (P. aeruginosa) or resistant (B. cepacia) to nonoxidative neutrophil-mediated killing. P. aeruginosa M2 is traditionally used for mouse pathogenicity studies (25), and B. cepacia JTC was from a fatal infection in a patient with CGD (23). Pilot experiments demonstrated that P. aeruginosa was cleared more rapidly from WT mice than was B. cepacia, so different time courses were evaluated for the two different pathogens.
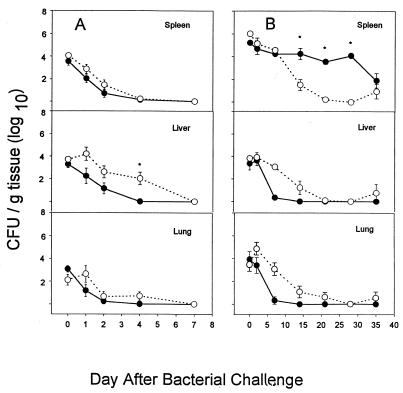
FIG. 2.
Intraperitoneal infection of BALB/c WT (●) and GKO (○) mice with P. aeruginosa M2 (A) or B. cepacia JTC (B). Mice were infected with ∼3 × 105 CFU of P. aeruginosa and then sacrificed at 0, 1, 2, 4, and 7 days or with ∼4 × 106 CFU of B. cepacia and sacrificed at 0, 2, 7, 14, 21, 28, and 35 days. Quantitative bacteriology of the spleen, liver, and lung was assessed at each time point. Data are the mean and standard error of the mean from 12 animals at each time point. An asterisk indicates a significant difference (P < 0.05) between WT and GKO mice at that time point.
P. aeruginosa, at a challenge dose of ∼3 × 105 CFU, administered intraperitoneally, was cleared from both the WT and GKO mice by 7 days with only modest differences between the two (Fig. 2A). At higher infectious challenges (5 × 105), there was substantial mortality (data not shown). At the sublethal dose chosen, there were no gross or microscopic pathological changes in any of the three organs.
The results with B. cepacia were strikingly different (Fig. 2B). A dose of 4 × 106 CFU was used for all experiments and resulted in bacteria being recovered from the spleen consistently after intraperitoneal challenge. The results were similar after intravenous challenge (data not shown), so the intraperitoneal route was used for simplicity and high reproducibility. Unlike P. aeruginosa, B. cepacia could be recovered from BALB/c mice for at least 35 days after systemic challenge. Whereas the bacteria were rapidly cleared from the lung and liver, they were recovered at approximately 104 CFU/g of spleen for the first 4 weeks of infection. Unexpectedly, the GKO mice cleared this strain of B. cepacia more rapidly from the spleen than did the WT mice, suggesting that IFN-γ was not critically important in defense against this opportunistic pathogen.
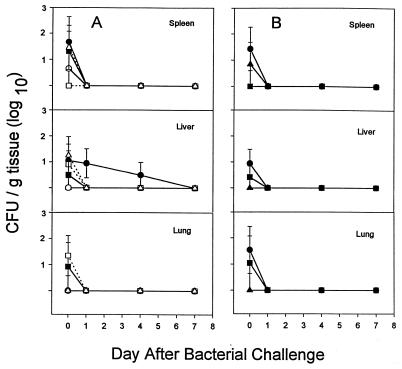
To determine if the observations of relative susceptibility to these two pathogens could be extended to other strains in these two species of bacteria, we evaluated six other strains of P. aeruginosa in WT BALB/c mice (Fig. 3A) and three in GKO mice (Fig. 3B). The sources and serum susceptibilities (in human serum) are given in Table 1. Serum-resistant versus serum-susceptible strains were tested to identify any possible correlation with virulence. The same infecting dose was used for the strains in these experiments as was used with strain M2, and in all cases, the bacteria were cleared from spleen, liver, and lung by day 7. There was no consistent difference between serum-resistant and serum-sensitive strains; however, the one strain which was still recoverable after 4 days (C4020) was serum resistant. As with strain M2, there was no substantial difference between the WT and GKO mice in their capacity to clear these strains of P. aeruginosa.
FIG. 3.
Intraperitoneal infection of wild-type BALB/c mice with ∼3 × 105 CFU of six different strains of P. aeruginosa (A) or GKO BALB/c mice with three different strains of P. aeruginosa (B). Quantitative bacteriology of the spleen, liver, and lung was assessed at 0, 1, 4, and 7 days. Data are the mean and standard error of the mean from four animals at each time point. ●, C4020; ■, P1; ▴, C0489; ○, C0262; □, C0295; ▵, C0487.
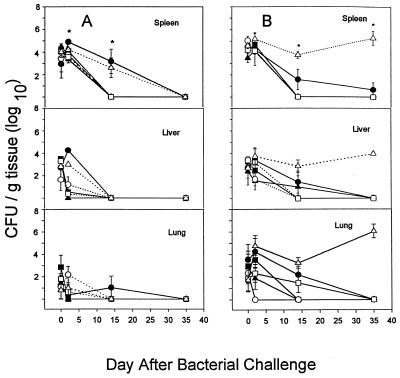
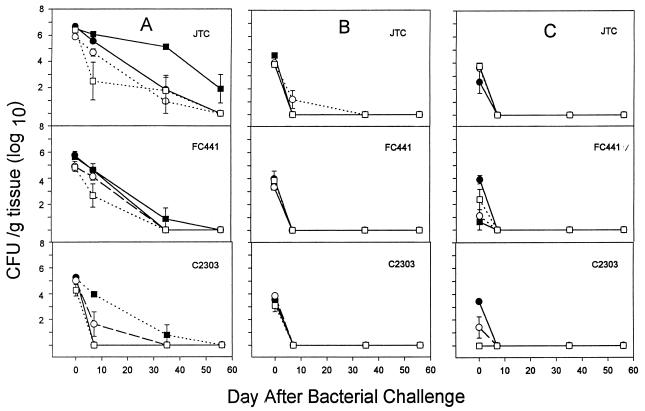
Six additional strains of B. cepacia were evaluated in WT (Fig. 4A) and GKO (Fig. 4B) BALB/c mice. Strains were from patients with CGD or CF and were from genomovar V or III (Table 1). All six strains were cleared from the WT animals by day 35, and at day 14, only two strains (C2303 and FC441) were present, at approximately 104 CFU/g of spleen. FC441, the only genomovar V strain tested, was cleared poorly by GKO mice. It was recovered at high number from spleen, liver, and lung at 35 days, and at necropsy, an intraperitoneal abscess and splenomegaly were found. This strain was recovered from a prolonged septicemic illness in a patient with X-linked recessive CGD (unpublished observation). The pattern of illness in this patient was atypical, because he was persistently bacteremic, in contrast to most patients with CGD, whose blood cultures are consistently negative in the face of severe invasive infection (unpublished data).
FIG. 4.
Intraperitoneal infection of wild-type BALB/c mice with ∼4 × 106 CFU of six different strains of B. cepacia (A) and GKO BALB/c mice with six different strains of B. cepacia (B). Quantitative bacteriology of the spleen, liver, and lung was assessed at 0, 2, 14, and 35 days. Data are the mean and standard error of the mean from four animals at each time point. An asterisk indicates a significant difference (P < 0.05) among strains at the indicated time point in the spleen only. ●, C2303; ■, C1257; ▴, C5424; ○, C2389; □, C5491; ▵, FC441.
Susceptibility of different murine strains to infection with B. cepacia.
Notable differences in susceptibility to intracellular and extracellular bacterial infection have been observed among inbred strains of mice. In order to begin to understand the host determinants of protection against B. cepacia, we evaluated two murine strains which have been shown to differ in their susceptibilities to infection with P. aeruginosa (16). Figure 5 shows the results of an experiment in which BALB/c and C57BL/6 mice were challenged with B. cepacia JTC. As observed in previous experiments, the bacteria were rapidly cleared from liver and lung, but could be cultured from the spleen for 35 days. C57BL/6 mice appeared to be more susceptible to infection; B. cepacia could be recovered from the spleen for 56 days (Fig. 5). These observations were extended to infection with a total of three strains of B. cepacia (from genomovars II, III, and V) and four strains of mice (Fig. 6). All strains of B. cepacia were cleared rapidly from liver and lung (Fig. 6B and 6C). In Fig. 6A (spleen), A/J and DBA/2 mice appeared to be more resistant to B. cepacia infection (similar to BALB/c mice). C57BL/6 mice were the most susceptible strain tested in this study. However, this observation appears to be unique to B. cepacia JTC (genomovar II), which persisted the longest in murine strain C57BL/6, with 105 CFU/g of spleen recovered at 35 days and approximately 102 CFU/g recovered after 56 days. Although the latter number is too small to be considered quantitatively reliable, it demonstrates the extraordinary capacity of this species of bacteria to persist in the spleen after systemic infection. Bacterial strains FC441 and C2303 showed no significant difference in resistance or susceptibility for any of the mouse strains tested.
FIG. 5.
Intraperitoneal infection of wild-type BALB/c (○) or C57BL/6 (■) mice with B. cepacia JTC. Quantitative bacteriology of the spleen, liver, and lung was assessed at 0, 2, 7, 14, 21, 28, 35, 56, 77, and 98 days. Data are the mean and standard error of the mean from four animals at each time point.
FIG. 6.
Intraperitoneal infection of wild-type A/J (●), C57BL/6 (■), BALB/c (○), or DBA/2 (□) mice with B. cepacia JTC, FC441, and C2303. Quantitative bacteriology of the spleen (A), liver (B), and lung (C) was assessed at 0, 7, 35, and 56 days. Data are the mean and standard error of the mean from four animals at each time point.
Histological evaluation of murine infection with B. cepacia.
No consistent histological abnormalities were seen in liver, lung, or spleen from mice infected with any of the bacterial strains. Specimens from 27 animals, with high or low bacterial counts, were examined. Liver, spleen, and lung were available from 26 of these animals, and liver and lung only were available from 1 animal.
DISCUSSION
B. cepacia is an important evolving pathogen in patients with CF or with CGD, but its mechanisms of pathogenesis are very poorly understood. The purpose of these studies was to develop an animal model of infection which would permit a systematic investigation of potential virulence determinants and host defense mechanisms. We chose to pursue our investigations with a systemic challenge model because B. cepacia infections in these two dissimilar disease states are characteristically invasive, as opposed to the mucosal respiratory tract disease characteristic of P. aeruginosa infections in patients with CF. Indeed, one of the strains of B. cepacia (FC441) was recovered from a 15-year-old boy with X-linked CGD at British Columbia’s Children’s Hospital, who suffered a prolonged bout of septicemia with multiple organ infection from which he recovered after a long course of aggressive antimicrobial therapy (unpublished data). B. cepacia is also a particularly virulent pathogen in patients with CF, with a characteristic syndrome of necrotizing pneumonia, septicemia, and death (11). Furthermore, patients with CF who are colonized with B. cepacia and undergo lung transplantation are at very high risk of dying postoperatively from B. cepacia septicemia (22).
IFN-γ therapy is very effective at preventing serious infections (including those with B. cepacia) in patients with CGD, but its mechanism of action is poorly understood. One of the goals of this study was to try to determine a role for IFN-γ in defense against this bacterium. We chose to utilize a GKO model, hypothesizing that these mice would be compromised in their capacity to clear B. cepacia. We were surprised to observe that the GKO mice were no worse at clearing B. cepacia than were their WT isogenic littermates. The mouse model was designed by using B. cepacia JTC. We found that the GKO mice were able to clear this strain of B. cepacia more efficiently than the WT BALB/c mice. Whereas a clear role for IFN-γ in defense against infection with intracellular pathogens has been demonstrated by other investigators (12), the same may not be the case for B. cepacia. IFN-γ is an important immune modulator early in infection. It regulates the proliferation and function of activated T lymphocytes. It acts as a promoter of Th1 response and down-regulates the Th2 cellular immune response (29). Thus, a role for Th1 cells in host defense against B. cepacia or P. aeruginosa infection cannot be inferred from these data. In the GKO mouse, a more profound activation of the Th2 response may be occurring in the absence of inhibition by IFN-γ, suggesting that resistance in the GKO mice to B. cepacia JTC may correlate with a Th2-dominated immune response. A formal evaluation of Th1 versus Th2 cells in host defense against infection with either P. aeruginosa or B. cepacia was beyond the scope of these studies.
It has been suggested by other investigators that B. cepacia may be an intracellular pathogen. This conclusion is drawn from several observations, including its propensity to cause chronic infections and its capacity to enter and replicate within respiratory epithelial cells (6). Our data support the possibility of intracellular pathogenicity. We observed that the bacteria could persist for up to 2 months in the murine spleen and that in so doing, an insubstantial inflammatory response was induced. This murine model provides an ideal opportunity to investigate the nature of the persistent infection. We are currently attempting to determine the cell(s) in which it persists by infecting mice with green fluorescent-tagged bacteria and then evaluating splenic tissues en bloc with fluorescent microscopy or after single-cellular dispersion followed by fluorescence-activated cell sorting.
This model may also permit the identification of microbial virulence determinants. We evaluated a range of bacterial strains in order to attempt to draw conclusions about two fairly broad and diverse bacterial species. The strains of P. aeruginosa were recovered from mouse intestine (25) or from the respiratory tract of patients with CF. Recent observations have suggested that there is a difference in the capacity of different strains of P. aeruginosa to invade epithelial cells (2, 8), and these differences may be due, in part, to products exported by a type III secretion apparatus (28). Our data do not support a difference among these strains in their capacity to cause serious murine infection; at the dose studied, ∼3 × 105 CFU, all animals (both WT and GKO) cleared the bacteria completely by 7 days. However, our model differs from those reported by the other investigative groups (8, 28); we were interested in infection rather than invasion as an endpoint.
The model was designed by using strain JTC, which was isolated from a CGD patient and belongs to genomovar II. Strain JTC showed persistence in the spleen for up to 56 days in the WT C57BL/6 mice (Fig. 6A) and for 20 days in the GKO mice (Fig. 2B). This suggested that IFN-γ does not play an essential role in host defense against this strain. Strain C2303 was the only one of five CF isolates from genomovar III (including cable pilus-positive strain C5424) that persisted past 14 days, and no significant difference was seen between the WT and GKO BALB/c mice in clearance of any CF strains. The second CGD isolate tested in this model, FC441, is a member of genomovar V. This strain was eliminated from the spleen of WT BALB/c mice, but persisted in the spleen, liver, and lung of GKO mice (Fig. 4B), suggesting that IFN-γ may play a role in the immune response against this strain of B. cepacia. These data suggest that strains of B. cepacia from different genomovars may have substantially different pathogenic potentials in mice and that this range of pathogenicities may correlate with human disease severity. Evaluation of many more strains will be required before any firm conclusions about virulence can be drawn. Potential virulence determinants, as opposed to colonizing factors, have not yet been identified, but their characterization may be greatly facilitated by use of this animal model.
A number of bacterial determinants have been identified which may play a role in virulence of B. cepacia. These include lipopolysaccharide (9), pili (13, 19, 26), protease (15), hemolysin, phospholipase C, siderophores, and exopolysaccharide (6, 18). Although each of these has been shown to facilitate a potential step in pathogenesis in vitro, none has been shown to enhance disease per se. One characteristic of B. cepacia which appears to enhance its virulence is resistance to nonoxidative phagocytic killing (23) and to cationic peptides (24). This is likely a result of an unusual lipopolysaccharide which is relatively impermeable due to a low number of phosphate residues (4). We have recently engineered a mutant of B. cepacia which is susceptible to cationic peptides and which appears to have a disruption of one of the genes involved in LPS biosynthesis (24). The animal model described in this paper should be well suited to evaluate the relative virulence of strains of B. cepacia with mutations leading to alterations in production of these different potential virulence determinants. We are also attempting to identify novel virulence determinants by using transconjugation and signature-tagged mutagenesis (7).
Clues to defense against microbial challenge have been gained from the study of different inbred strains of mice (12). For instance, specific strains are susceptible or resistant to infection with Mycobacterium bovis bacillus Calmette-Guerin and are designated bcgr or bcgs (21). The bcg gene has been identified in the mouse (20), and a human homologue has recently been cloned and codes for a protein named the “human natural resistance-associated macrophage protein” (3). A similar approach may be possible in determining the gene(s) involved in resistance or susceptibility to infection with different B. cepacia strains by comparing susceptible (C57BL/6) to resistant (DBA/2, BALB/c, and A/J) mice. Such studies are planned and may shed new light on strategies used by this pathogen to persist and/or cause devastating infection in certain compromised hosts.
In summary, we have developed a new animal model of invasive infection with B. cepacia. This model should aid in the dissection of microbial determinants of infectivity as well has determinants of host defense. Such studies may aid in the design of novel anti-infective strategies for this highly adapted opportunistic pathogen.
ACKNOWLEDGMENTS
This work was supported with funds from Genentech, Inc., the British Columbia Lung Association, and the Canadian Cystic Fibrosis Foundation. Barbara Steen was supported by a studentship from the Canadian Cystic Fibrosis Foundation.
We thank Laurie Ainsworth for statistical analysis, Fergall Magee for histological evaluation, Karen Chu and Jacqueline Chung for technical assistance, and Eshwar Mahenthiralingam and Richard Stokes for advice.
REFERENCES
- 1.Burkholder W H. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1952;40:115–117. [Google Scholar]
- 2.Burns J L, Jonas M, Chi E Y, Clark D A, Berger A, Griffith A. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect Immun. 1996;64:4054–4059. doi: 10.1128/iai.64.10.4054-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cellier M, Govoni G, Vidal S, Kwan T, Groulx N, Liu J, Sanchez F, Skamene E, Schurr E, Gros P. Human natural resistance-associated macrophage protein: cDNA cloning, chromosomal mapping, genomic organization, and tissue-specific expression. J Exp Med. 1994;180:1741–1752. doi: 10.1084/jem.180.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox A D, Wilkinson S G. Ionizing groups in lipopolysaccharides of Pseudomonas cepacia in relation to antibiotic resistance. Mol Microbiol. 1991;5:641–646. doi: 10.1111/j.1365-2958.1991.tb00735.x. [DOI] [PubMed] [Google Scholar]
- 5.Dalton D K, Pitts-Meek S, Keshav S, Figari I S, Bradley A, Stewart T A. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259:1739–1745. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 6.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hensel M, Shea J E, Gleeson C, Jones M D, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 8.Hirakata Y, Izumikawa K, Yamaguchi T, Igimi S, Furuya N, Maesaki S, Tomono K, Yamada Y, Kohno S, Yamaguchi K, Kamihira S. Adherence to and penetration of human intestinal Caco-2 epithelial cell monolayers by Pseudomonas aeruginosa. Infect Immun. 1998;66:1748–1751. doi: 10.1128/iai.66.4.1748-1751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes J E, Stewart J, Barclay G R, Govan J R W. Priming of neutrophil respiratory burst activity by lipopolysaccharide from Burkholderia cepacia. Infect Immun. 1997;65:4281–4287. doi: 10.1128/iai.65.10.4281-4287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The International Chronic Granulomatous Disease Cooperative Study Group. A controlled trial of interferon gamma to prevent infection in chronic granulomatous disease. N Engl J Med. 1991;324:509–516. doi: 10.1056/NEJM199102213240801. [DOI] [PubMed] [Google Scholar]
- 11.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann S H E. Immunity to intracellular microbial pathogens. Immunol Today. 1995;16:338–342. doi: 10.1016/0167-5699(95)80151-0. [DOI] [PubMed] [Google Scholar]
- 13.Krivan H C, Ginsburg V, Roberts D D. Pseudomonas aeruginosa and Pseudomonas cepacia isolated from cystic fibrosis patients bind specifically to gangliotetraosylceramide (Asialo GM1) and gangliotriaosylceramide (Asialo GM2) Arch Biochem Biophys. 1988;260:493–496. doi: 10.1016/0003-9861(88)90473-0. [DOI] [PubMed] [Google Scholar]
- 13a.Mahenthiralingam, E., et al. Unpublished observations.
- 14.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKevitt A I, Bajaksouzian S, Klinger J D, Woods D E. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect Immun. 1989;57:771–778. doi: 10.1128/iai.57.3.771-778.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morissette C, Skamene E, Gervais F. Endobronchial inflammation following Pseudomonas aeruginosa infection in resistant and susceptible strains of mice. Infect Immun. 1995;63:1718–1724. doi: 10.1128/iai.63.5.1718-1724.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray H W. Interferon-gamma, the activated macrophage, and host defense against microbial challenge. Ann Intern Med. 1988;108:595–608. doi: 10.7326/0003-4819-108-4-595. [DOI] [PubMed] [Google Scholar]
- 18.Nelson J W, Butler S L, Krieg D, Govan J R W. Virulence factors of Burkholderia cepacia. FEMS Immunol Med Microbiol. 1994;8:89–98. doi: 10.1111/j.1574-695X.1994.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 19.Sajjan S U, Forstner J F. Identification of the mucin-binding adhesin of Pseudomonas cepacia isolated from patients with cystic fibrosis. Infect Immun. 1992;60:1434–1440. doi: 10.1128/iai.60.4.1434-1440.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schurr E, Skamene E, Forget A, Gros P. Linkage analysis of the Bcg gene on mouse chromosome 1: identification of a tightly linked marker. J Immunol. 1989;142:4507–4513. [PubMed] [Google Scholar]
- 21.Skamene E, Gros P, Forget A, Kongshavn P A L, St. Charles C, Taylor B A. Genetic regulation of resistance to intracellular pathogens. Nature. 1982;297:506–509. doi: 10.1038/297506a0. [DOI] [PubMed] [Google Scholar]
- 22.Snell G I, de Hoyos A, Krajden M, Winton T, Maurer J R. Pseudomonas cepacia in lung transplant recipients with cystic fibrosis. Chest. 1993;103:466–471. doi: 10.1378/chest.103.2.466. [DOI] [PubMed] [Google Scholar]
- 23.Speert D P, Bond M, Woodman R C, Curnutte J T. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J Infect Dis. 1994;170:1524–1531. doi: 10.1093/infdis/170.6.1524. [DOI] [PubMed] [Google Scholar]
- 24.Steen B A, Hancock R E W, Speert D P. Characterization of a Burkholderia cepacia mutant with increased susceptibility to cationic peptides. Pediatr Pulmonol. 1998;17(Suppl.):380A. [Google Scholar]
- 25.Stieritz D D, Holder I A. Experimental studies of the pathogenesis of infections due to Pseudomonas aeruginosa: description of a burned mouse model. J Infect Dis. 1975;131:688–691. doi: 10.1093/infdis/131.6.688. [DOI] [PubMed] [Google Scholar]
- 26.Sun L, Jiang R-Z, Steinbach S, Holmes A, Campanelli C, Forstner J, Sajjan U, Tan Y, Riley M, Goldstein R. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat Med. 1995;1:661–666. doi: 10.1038/nm0795-661. [DOI] [PubMed] [Google Scholar]
- 27.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 28.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 29.Young H A, Hardy K J. Role of interferon-γ in immune cell regulation. J Leukoc Biol. 1995;58:373–381. [PubMed] [Google Scholar]