Abstract
Fas-associated factor 1 (FAF1) is a scaffolding protein that plays multiple functions, and dysregulation of FAF1 is associated with many types of diseases such as cancers. FAF1 contains multiple ubiquitin-related domains (UBA, UBL1, UBL2, UAS, and UBX), each domain interacting with a specific partner. In particular, the interaction of UBL1 with heat shock protein 70 (Hsp70) is associated with tumor formation, although the molecular understanding remains unknown. In this study, the structural analysis revealed that His160 of FAF1 is important for its interaction with Hsp70. The association of Hsp70 with FAF1 is required for the interaction with IQGAP1. FAF1 negatively regulates RhoA activation by FAF1–Hsp70 complex formation, which then interacts with IQGAP1. These steps play a key role in maintaining the stability of cell-to-cell junction. We conclude that FAF1 plays a critical role in the structure and function of adherens junction during tissue homeostasis and morphogenesis by suppressing RhoA activation, which induces the activation of Rho-associated protein kinase, phosphorylation of myosin light chain, formation of actin stress fiber, and disruption of adherens junction. In addition, depletion of FAF1 increased collective invasion in a 3D spheroid cell culture. These results provide insight into how the FAF1–Hsp70 complex acts as a novel regulator of the adherens junction integrity. The complex can be a potential therapeutic target to inhibit tumorigenesis and metastasis.
Keywords: human Fas-associated factor 1 (FAF1), heat shock protein 70 (Hsp70), adherens junction, RhoA activation, IQGAP1, X-ray crystallography, FAF1–Hsp70 complex
Introduction
Fas-associated factor 1 (FAF1) was initially identified as a Fas-associated protein factor potentiating Fas-mediated apoptosis (Chu et al., 1995). FAF1 is a scaffolding protein that serves as a ubiquitin receptor and contains multiple ubiquitin-related domains, including the ubiquitin-associated (UBA) domain and three domains with ubiquitin-like folds, UBL1, UBL2, and ubiquitin-regulatory X (UBX) (Song et al., 2005). FAF1 is involved in diverse cell functions by interacting with various partners in each domain. The N-terminal UBA domain interacts with Lys48-linked polyubiquitinated proteins, and this interaction is required for FAF1-mediated apoptosis and stress response (Song et al., 2005, 2009). The C-terminal UBX domain interacts with valosin-containing protein (VCP, AAA ATPase p97) complexed with Npl4–Ufd1 heterodimer, and it plays a role in promoting ERAD substrate degradation in a VCP–Npl4–Ufd1-dependent manner (Song et al., 2005; Lee et al., 2013). The linker region between UBL2 and UAS interacts with interferon regulator factor 3 (IRF3), a key transcription factor of IFNβ signaling responsible for the host's innate immune response. FAF1 inhibits IRF3-mediated IFNβ production by reducing the interaction between IRF3 and IPO5/importin-β3 and by inhibiting nuclear translocation of IRF3 (Song et al., 2016). FAF1 plays a key role as a negative regulator of virus-induced IFNβ production. UBL1 domain is associated with heat shock protein 70 (Hsp70), and a previous report showed that FAF1 regulates the chaperone activity of Hsp70, but not the substrate of Hsp70 (Kim et al., 2005).
FAF1 is also a pro-apoptotic protein and acts as a tumor suppressor by reducing Hsp70, which is overexpressed in various cancer cells (Kim et al., 2005; Lee et al., 2012). FAF1 mRNA level was significantly downregulated in gastric cancer with distant metastasis (Bjørling-Poulsen et al., 2003). Decreased FAF1 expression levels in the advanced stage (IV) of both gastric and ovarian cancer have been reported (Bjørling-Poulsen et al., 2003; Kang et al., 2014). Recently, whole-exome sequencing analyses of hereditary colorectal cancer showed that FAF1 variants induce resistance to the apoptosis of colorectal cancer cells and increase the activity of β-catenin and NF-κB (Bonjoch et al., 2020). FAF1 inhibits tumor growth by regulating Hsp70 degradation, which has been identified as the molecular mechanism underlying low FAF1 expression in human cervical cancer tissues (Lee et al., 2012). FAF1 is known to promote β-TrCP-mediated degradation of cytosolic β-catenin and subsequently reduce breast cancer metastasis (Xie et al., 2017). These results implicate the correlation between FAF1 expression and carcinogenesis. However, further molecular studies are necessary to understand the specific role of FAF1 during cancer progression.
The integrity of the cellular barrier is important for maintaining the physiological environment of the underlying tissue. Adherens junctions regulate cell-to-cell adhesion, and their inhibition promotes cancer metastasis by facilitating cancer cell migration from the primary site to secondary sites (Farahani et al., 2014; Arnold et al., 2017; Cerutti and Ridley, 2017). Adherens junctions, composed of cadherins and cytosolic proteins, are connected to the actomyosin cytoskeleton. Therefore, the structure and functions of adherens junctions are tightly regulated by actomyosin dynamics. The Rho family of small GTPases including RhoA, Rac1, and Cdc42 are crucial regulators of actin dynamics, cell–substratum adhesion, and cell–cell adhesion. Rho activation leads to the assembly of contractile actin–myosin filaments (stress fibers) and associated focal adhesion complexes, while Rac1 activation induces the assembly of a meshwork of actin filaments at the cell periphery to produce lamellipodia and membrane ruffles, and Cdc42 activation induces actin-rich surface protrusions called filopodia (Hall, 1998). Rho GTPases function as molecular switches for a cycle between GTP-bound active state and GDP-bound inactive state. They are activated by guanine nucleotide exchange factors (GEFs), whereas GTPase-activating proteins (GAPs) inactivate Rho GTPases. Among these, RhoA directly enhances stress fiber formation by acting on its effector, Rho-associated protein kinase (ROCK). Activated ROCK phosphorylates myosin light chain (MLC) and increases the contractility of stress fibers (Cerutti and Ridley, 2017; Dyberg et al., 2017). RhoA has oncogenic properties and is upregulated in many cancers, including breast, lung, ovarian, and gastric cancers (Casteel et al., 2012; Jeong et al., 2016; Svensmark and Brakebusch, 2019). IQGAP1 is a scaffold protein involved in the assembly of adherens junctions. IQGAP1 interacts with signaling and structural molecules, thereby regulating many biological processes (Tanos et al., 2018). In epithelial cells, IQGAP1 localizes at the sites of cell–cell contact and colocalizes with p190RhoGAP and RhoA, inactivating RhoA and suppressing airway smooth muscle contraction (Bhattacharya et al., 2014). In the present study, we determined the crystal structure of FAF1 UBL1/Hsp70 complex and found the critical residues for this interaction. We identified additional interacting proteins of FAF1 such as nonmuscle myosin heavy chain IIA (NMIIA), IQGAP1, p190RhoGAP, and actin. We confirmed that the association of FAF1 UBL1 with Hsp70 is required for the interaction with IQGAP1 and p190RhoGAP. These interactions regulate FAF1-mediated RhoA activation to maintain the structure of adherens junction during morphogenesis. In addition, depletion of FAF1 increased collective invasion in transwell and 3D spheroid cell culture. These findings suggest that FAF1 negatively regulates RhoA activation by the interaction of the FAF1–Hsp70 complex with IQGAP1, thereby maintaining the stability of cell-to-cell junction. Therefore, FAF1 maintains the structure and function of adherens junction during tissue homeostasis and morphogenesis by regulating RhoA activation, stress fiber formation, and adherens junction assembly.
Results
Structural analysis of the FAF1–Hsp70 complex
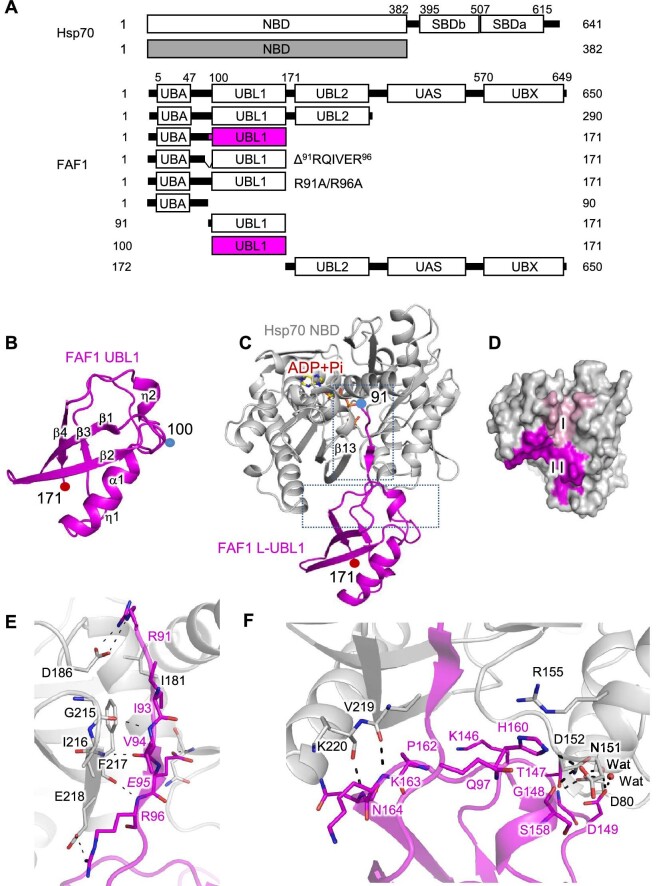
FAF1 plays a key role as a tumor suppressor by strongly interacting with Hsp70 (Kim et al., 2005; Lee et al., 2012). To understand the molecular mechanism, we examined the structural basis of the complex of FAF1 and Hsp70. Based on an earlier report (Kim et al., 2005), the nucleotide-binding domain of Hsp70 (Hsp70 NBD, aa 1–382) and various constructs of FAF1 were prepared (Figure 1A), and crystallization attempts were made. Crystals of FAF1 (aa 100–171) and FAF1 (aa 91–171) complexed with Hsp70 NBD were obtained. To distinguish the two FAF1 fragments, they are named FAF1 UBL1 and FAF1 L-UBL1, respectively. Diffraction data were collected to 1.2 Å and 2.2 Å resolution, respectively. The crystal structure of FAF1 UBL1 (aa 100–171) alone was determined using the multi-wavelength anomalous diffraction (MAD) technique, and the FAF1 UBL1–Hsp70 NBD complex structure was determined using FAF1 UBL1 alone and Hsp70 NBD structure (PDB code: 1S3X) (Sriram et al., 1997). Table 1 summarizes statistics on the data collection and refinement. FAF1 UBL1 adopts a tightly-packed globular structure comprising four β-strands, two η-helices, and one α-helix resembling the β-grasp fold of ubiquitin (Figure 1B), despite low sequence identity (18.6%). In the FAF1 L-UBL1–Hsp70 NBD complex structure, the UBL1 domain of FAF1 retained its globular fold, whereas the residues 91Arg–Arg96 of FAF1 adopted a β-structure (Figure 1C). FAF1 binds to Hsp70 in two parts. Figure 1D shows the two interfaces highlighted on the molecular surface of Hsp70 by coloring the residues of Hsp70 within 4.0 Å of FAF1. The detailed interactions are shown in Figure 1E and F,
Figure 1.
Crystal structures of FAF1 UBL and FAF1 L-UBL1 complexed with Hsp70 NBD. (A) Domain structures of FAF1 and Hsp70 and the constructs prepared in this study. (B) Ribbon presentation of FAF1 UBL1 alone (aa 100–171). The secondary structures are indicated. (C) Ribbon representation of the complex of FAF1 L-UBL1 (aa 91–100) and Hsp70 NBD (aa 1–362) shown in magenta and silver, respectively, with the bound adenosine diphosphate (ADP) and phosphate (Pi) shown in the stick model. Subdomains of Hsp70 are indicated. (D) The molecular surface of Hsp70 within 4.0 Å from FAF1 is colored. Residues in contact with the linker region of FAF1 (aa 91–99) are colored in pink, while residues in contact with the UBL1 domain (aa 100–171) are in magenta. (E and F) Interactions between FAF1 L-UBL1 and Hsp70 NBD are shown in the stick model. Hydrogen bonds are indicated as dashed lines.
Table 1.
Statistics on the data collection and refinement.
| FAF1 UBL1 | FAF1 L-UBL1 complexed to | ||||
|---|---|---|---|---|---|
| Hsp70 NBD with ADP + Pi | |||||
| Native | SeMet MAD | Native | |||
| Peak | Edge | Remote | |||
| Data collection | |||||
| Beam line | PLS BL-5C | ||||
| Wavelength, Å | 1.0000 | 0.9794 | 0.9796 | 0.9717 | 1.0000 |
| Space group | P42212 | P3121 | |||
| Cell dimensions | |||||
| a, Å | 65.02 | 65.02 | 103.33 | ||
| b, Å | 65.02 | 65.02 | 103.33 | ||
| c, Å | 33.56 | 33.55 | 127.09 | ||
| α, ° | 90.00 | 90.00 | 90.00 | ||
| β, ° | 90.00 | 90.00 | 90.00 | ||
| γ, ° | 90.00 | 90.00 | 120.00 | ||
| a Resolution, Å | 50.0–1.2 | 50.0–1.6 | 50.0–2.2 | ||
| (1.28–1.20) | (1.66–1.60) | (2.28–2.20) | |||
| Total reflections | 659957 | 321179 | 321309 | 32847 | 1609110 |
| Unique reflections | 23153 | 18815 | 18830 | 18813 | 40388 |
| a Completeness, % | 99.7 (99.6) | 99.3 (99.0) | 99.2 (98.8) | 99.2 (98.8) | 99.9 (99.9) |
| a I/σ(I) | 49.6 (6.9) | 37.9 (6.8) | 37.3 (6.9) | 37.8 (7.1) | 24.4 (5.2) |
| b R merge, % | 5.0 (21.6) | 8.3 (20.7) | 6.5 (19.6) | 6.4 (19.7) | 9.8 (32.2) |
| Refinement | |||||
| Resolution, Å | 50.0–1.2 | 50.0–2.2 | |||
| c R cryst/Rfree, % | 22.2/24.3 | 17.4/20.6 | |||
| No. of protein atoms | 591 | 3638 | |||
| No. of water atoms | 169 | 454 | |||
| RMS deviation | |||||
| Bond length, Å | 0.005 | 0.007 | |||
| Bond angle, ° | 0.771 | 0.876 | |||
| Ramachandran analysis | |||||
| Favored region, % | 98.63 | 96.92 | |||
| Allowed region, % | 1.37 | 3.08 | |||
| PDB code | 7FGN | 7FGM | |||
a Values in parentheses are for the outer most resolution shell.
b R merge = ∑h∑i|I(h,i)−<I(h)>|/∑h∑iI(h,i), where I(h,i) is the intensity of the ith measurement of reflection h and <I(h)> is the mean value of I(h,i) for all i measurements.
c R free was calculated from randomly selected 5% set of reflections not included in the calculation of the R value.
respectively. The first interface involves the residues 91Arg–Arg96 of FAF1 adopting a β-structure and binding tightly to the cleft, making backbone–backbone interaction with the last β-strand of the β-sheet in the Hsp70 NBD (Figure 1E). The second interface involves 146Lys–Asp149, Ser158, His160, and 162Pro–Ser166 of FAF1 (Figure 1F). The residues involved in the intermolecular interactions were both polar and hydrophobic. There are three salt bridges between the two molecules (FAF1Arg91 with Hsp70Asp186, FAF1Arg96 with Hsp70Glu218, and FAF1His160 with Hsp70Asp152). FAF1His160 with Hsp70Asp152 interaction is of particular interest. The side chains of Hsp70Arg155 and FAF1Gln97 make a π–π interaction with imidazole of FAF1His160 (Figure 1F). In addition to these direct protein–protein interactions, three well-ordered water molecules bridged the two proteins. The interface area in the complex was ∼2400 Å2, consisting of ∼1000 Å2 for the linker and ∼1400 Å2 for the UBL1 domain of FAF1. The overall structures of FAF1 UBL1 and Hsp70 in the complex were the same as those in the free form, e.g. r.m.s.d. values were 0.65 Å for FAF1 UBL1 and 0.45 Å for Hsp70 NBD (Supplementary Figure S1A and B). However, the loop spanning 146Lys–Asp149 of FAF1, which is involved in the Hsp70 binding, showed a shift of 4 Å. These differences are in the same order of magnitude as those seen for the solution structure. The binding mode of the linker region of FAF1 in the complex seen here resembled that of the inter-domain linker in the crystal structure of ATP-bound DnaK, Escherichia coli Hsp70 (PDB entry: 4B9Q) (Kityk et al., 2012, 2018; Supplementary Figure S2A). In this state, the β-domain of the substrate-binding domain (SBD) of DnaK occupies the same space as the UBL1 domain of FAF1. However, the FAF1 UBL1 binding site of Hsp70 identified here was quite different from that seen in the complex structures of Hsp70 NBD with auxilin JD (Jiang et al., 2007), Hsp110/sse1 (Polier et al., 2008), Sil1 (Yan et al., 2011), Bag1 (Sondermann et al., 2001), Bag2 (Xu et al., 2008), or Bag5 (Arakawa et al., 2010) (Supplementary Figure S2B).
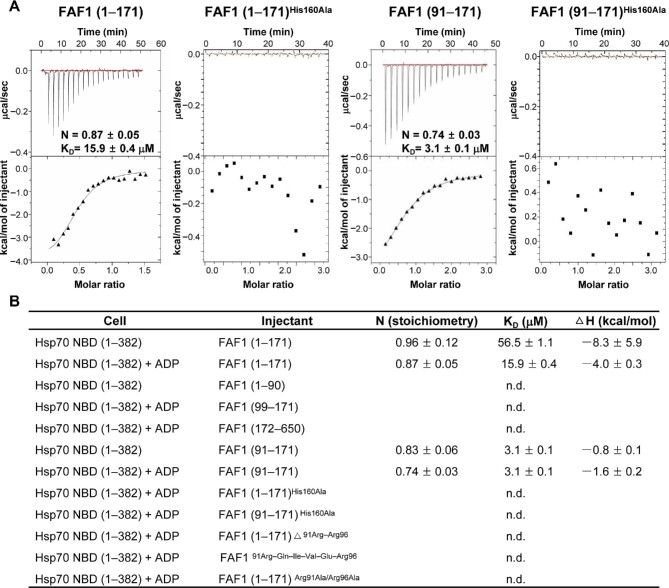
Binding affinity between FAF1 UBL1 and Hsp70 using ITC
Next, we examined the interactions between Hsp70 NBD and various fragments of FAF1 using isothermal titration calorimetry (ITC) (Figure 2A and B). First, both FAF1 (aa 1–171) and FAF1 (aa 91–171) bound to Hsp70 NBD in the presence of ADP at a 1:1 molar ratio, as in the crystal structure, with binding affinities (KD) of 15.9 ± 0.4 μM and 3.1 ± 0.1 μM, respectively (Figure 2A), while the corresponding values in the absence of ADP were 56.5 ± 1.1 μM and 3.1 ± 0.1 μM, respectively (Figure 2B). However, other constructs tried, including FAF1 (aa 1–90), FAF1 (aa 99–171), and FAF1 (aa 172–650), showed no detectable binding. Also, the peptide corresponding to 91Arg–Gln–Ile–Val–Glu–Arg96 (91RQIVER96) and FAF1 (aa 1–171) missing 91Arg–Arg96 exhibited no detectable binding. Therefore, these results suggest that both the linker region and the UBL1 (aa 100–171) domain of FAF1 are critical in binding to Hsp70. Comparison of FAF1 in the two structures, i.e. FAF1 alone and the complex, showed a large shift at the 146Lys–Asp149 region of FAF1, suggesting that this region is quite flexible (Supplementary Figure S1A and B). As such, we hypothesized that the FAF1His160–Hsp70Asp152 interaction is critical for the interaction between the two proteins, as suggested in the complex structure (Figure 1F). To test the idea, we constructed the FAF1His161Ala mutant in both FAF1 (aa 1–171) and FAF1 (aa 91–171). To our surprise, both mutants showed no binding to Hsp70 NBD (Figure 2A), thus confirming that the interaction is critical for the binding of the two proteins.
Figure 2.
ITC analysis of FAF1 and Hsp70 NBD. (A) ITC results of FAF1 binding to Hsp70 NBD. FAF1 (aa 1–171), FAF1 (aa 91–171) [His160Ala], FAF1 (aa 91–171), and FA1 (aa 91–171) [His160Ala] were titrated to Hsp70 (aa 1–362) with ADP. The top panel shows the thermal effects associated with the injection, while the bottom panel shows the binding isotherm corresponding to the data in the top panel and the best-fitted curve with a one-site binding model. (B) Summary of ITC measurement.
FAF1 interacts with protein complexes in the cell adherens junction
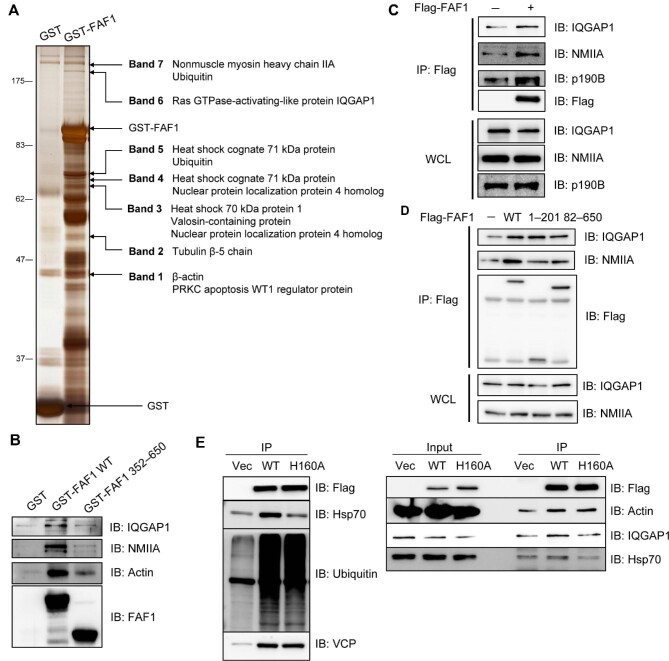
To investigate the function of the FAF1–Hsp70 complex in cells, we identified the FAF1-interacting proteins by a pull-down assay. Proteins directly binding to GST-FAF1 were separated and subjected to in-gel digestion with trypsin followed by peptide sequencing employing nanoUPLC-ESI-q-TOF tandem mass spectrometry (MS) (equipped at Drug Development Research Core Center) (Figure 3A). In addition to the already reported interacting proteins, e.g. Hsp70, VCP, Npl4, and ubiquitin, we found NMIIA, IQGAP1, tubulin, β-actin, and PRKC apoptosis WT1 regulator protein (PAWR) as FAF1-interacting proteins (Supplementary Table S1). NMIIA, a constituent of actin filament, is important for the dynamics of adherens junction, as it regulates cell morphogenesis, tissue homeostasis, and healing by regulating stress fiber formation with actin filaments (Heuzé et al., 2019). IQGAP1, a scaffolding protein involved in the assembly of adherens junctions, co-localizes with p190-RhoGAP and RhoA (Bhattacharya et al., 2014; Xu et al., 2018). To identify the FAF1-binding site of these proteins, we performed a GST-pull-down using the full-length FAF1 (aa 1–650) and FAF1 (aa 352–650) lacking UBA, UBL1, and UBL2. We found that the binding to actin, NMIIA, and IQGAP1 was abolished in FAF1 (aa 352–650), unlike full-length FAF1 (Figure 3B), suggesting that the N-terminal region of FAF1, i.e. FAF1 (aa 1–351), may be involved in regulating adherens junction. To validate the newly identified interacting proteins, we examined cellular interactions by immunoprecipitation. Lysates from HeLa and HEK293T cells overexpressing Flag-FAF1 were immunoprecipitated with anti-Flag antibodies. Immune complexes were detected by western blotting analysis using anti-IQGAP1, anti-NMIIA, and anti-actin antibodies (Figure 3C; Supplementary Figure S3A). FAF1 further interacted with p190B-RhoGAP, known as IQGAP1-interacting protein (Supplementary Figure S3B). However, FAF1 interacted with p190B-RhoGAP and actin only when the cells were lysed in a mild hypotonic solution containing octyl β-D-glucopyranoside but not NP-40 (Supplementary Figure S3B), suggesting that p190B-RhoGAP and actin weakly interact with FAF1.
Figure 3.
FAF1 forms a complex with IQGAP1 and actomyosin. (A and B) GST and GST-FAF1 (A) or GST, GST-FAF1 WT, and GST-FAF1 (aa 362–650) (B) were incubated with glutathione-agarose beads for 3 h and washed six times with washing buffer. GST-fused protein-bound beads were further incubated with HeLa cell lysates for 3 h, and then the beads were washed six times. Protein complexes were separated by SDS–PAGE and detected by silver staining (A) or analysed by western blotting (B). (C) HeLa cells were transfected with Flag or Flag-FAF1. After 24 h, cell lysates were immunoprecipitated with anti-Flag antibody, and the immune complex was analysed by western blotting. (D) HeLa cells were transfected with Flag, Flag-FAF1 WT, Flag-FAF1 (aa 1–201), or Flag-FAF1 (aa 82–650). After 24 h, cell lysates were immunoprecipitated with anti-Flag antibody, and the immune complex was analysed by western blotting. (E) HeLa cells were transfected with FAF1 WT or FAF1H160A mutant and then immunoprecipitated with anti-Flag antibodies. Immunoprecipitated proteins were immunoblotted with the indicated antibodies.
To further identify which specific region of FAF1 is involved in recruiting IQGAP1 and NMIIA, we performed an immunoprecipitation assay after the transfection with Flag-FAF1 full-length (aa 1–650), FAF1 (aa 1–201) containing UBA and UBL1 domain, and FAF1 (aa 82–650) deleting UBA domain. All three, the full-length and the two truncated mutants, commonly containing the UBL1 domain, interacted with IQGAP1 and NMIIA, indicating that the UBL1 domain is necessary to bind to these proteins (Figure 3D). Since UBL1 interacts with Hsp70, we investigated whether the association of FAF1 with Hsp70 is a prerequisite for various interactions of FAF1 with IQGAP1, NMIIA, and actin by employing Flag-FAF1His160Ala mutant, which is unable to bind to Hsp70. First, we examined whether His160 of FAF1 is important for the interaction with Hsp70. As shown in Figure 3E, FAF1 wild-type (WT) interacted with Hsp70, poly-ubiquitin, and VCP as reported previously (Song et al., 2005), whereas FAF1His160Ala mutant interacted with poly-ubiquitin and VCP but not with Hsp70. Intriguingly, this mutant could not interact with IQGAP1 as well as Hsp70.
Silencing FAF1 induces significant morphological changes through RhoA activation
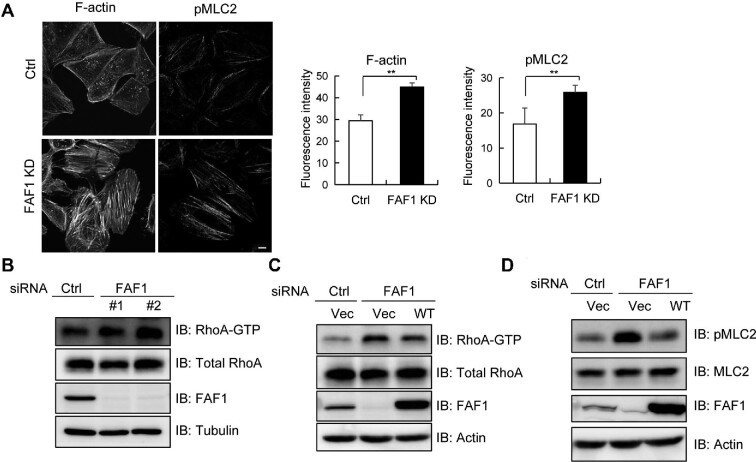
To investigate whether FAF1 interacting with protein complexes in the cell adherens junction plays a role in morphogenesis, we examined morphological changes in HeLa cells silencing FAF1. We found that the silencing of FAF1 increased the cell size and transformed the cells to a more spherical shape. Since polymerized cytoskeletons are responsible for cell morphology, we investigated whether FAF1 alters the formation of actin stress fibers, which are contractile actomyosin bundles containing actin filaments and active NMII. We examined the stress fibers in HeLa cells after silencing FAF1 by confocal microscopy along with immunostaining with anti-actin and anti-pMLC2. HeLa cells with silenced FAF1 (FAF1 KD) showed significantly increased centrally located F-actin stress fibers and pMLC2 that conformationally altered NMII into bipolar filaments (Figure 4A). These filaments associate with actin filaments in bundles (Vicente-Manzanares et al., 2009), suggesting that FAF1 regulates stress fiber formation. In addition, HeLa cells were stained with antibodies for paxillin and NMIIA, which are expressed in focal adhesions and a constituent of actin filament, respectively. Consistent with Figure 4A, paxillin and NMIIA levels were also increased in HeLa cells after FAF1 silencing (Supplementary Figure S4).
Figure 4.
FAF1 silencing promotes actin stress fiber formation and FAF1 negatively regulates RhoA activation. (A) HeLa cells transfected with control siRNA or FAF1 siRNA#2 for 72 h were fixed and stained for F-actin (phalloidin) or double phosphorylated MLC2 (Thr18/Ser19) to investigate stress fiber formation. The bar graph indicates the fluorescence intensities for F-actin or double phosphorylated MLC2 in the left panel. The experiments were conducted in quadruplet. Scale bar, 10 μm. (B) HeLa cells were transfected with two types of FAF1 siRNAs, #1 and #2. After 72 h, cells were lysed, and the lysates were incubated with GST-RBD of Rhotekin to measure the level of GTP-bound RhoA. Cell extracts were used to analyse the amount of total RhoA by western blotting. The relative amount of RhoA-GTP in control and FAF1 KD cells was determined by western blotting followed by densitometry analysis. (C) HeLa cells with silenced FAF1 (by transfecting with control siRNA or FAF1 siRNA#2 for 48 h) were transfected with Flag or Flag-FAF1 as indicated for adding back FAF1. At 24 h post-transfection, cells were lysed, and the lysates were incubated with GST-RBD of Rhotekin to measure the level of GTP-bound RhoA. Cell extracts were analysed using the indicated antibody. The relative amount of RhoA-GTP was determined by western blotting followed by densitometry analysis. (D) HeLa cells were transfected with control siRNA or FAF1 siRNA#2 for 48 h and transfected with Flag or Flag-FAF1 as indicated. At 24 h post-transfection, cells were lysed with gel sample buffer and analysed using the indicated antibodies.
FAF1 KD cells clearly showed an increase in actin stress fiber formation, but not lamellipodia or filopodia, which is a well-known phenotype of Rho activation (Hall, 1998; Maekawa et al., 1999). To investigate how FAF1 inhibits stress fiber formation, we examined RhoA activation because active RhoA activates ROCK and then phosphorylates MLC, which leads to the assembly of contractile actomyosin bundles with unbranched polymerized actin (Hall, 1998). We quantified GTP-bound active RhoA through a pull-down assay using the GST-RBD of Rhotekin that interacts with active RhoA and then performed western blotting analysis using a specific antibody. We used two different siRNAs to silence endogenous FAF1 expression. As shown in Figure 4B, silencing of endogenous FAF1 increased GTP-bound active RhoA but not Rac1 (Supplementary Figure S5A–C). Then, to confirm the restoration of RhoA activation in FAF1-silenced cells by adding FAF1, we measured the RhoA activation in HeLa cells with endogenously silenced FAF1 followed by transfections with control or Flag-FAF1. Active GTP-bound RhoA was increased in FAF1 KD cells, and the increase in active RhoA level was reduced by adding Flag-FAF1 compared to control (Figure 4C). The effects were verified by quantifying pMLC2 as a target protein of active RhoA via ROCK activation. As shown in Figure 4D, the pMLC2 level significantly increased in cells silencing FAF1, and this increase was abolished by adding Flag-FAF1 via transfection. These results confirmed that FAF1 suppresses RhoA activation and its downstream signaling, ROCK, and phosphorylation of MLC2.
FAF1 is vital for stabilizing adherens junction
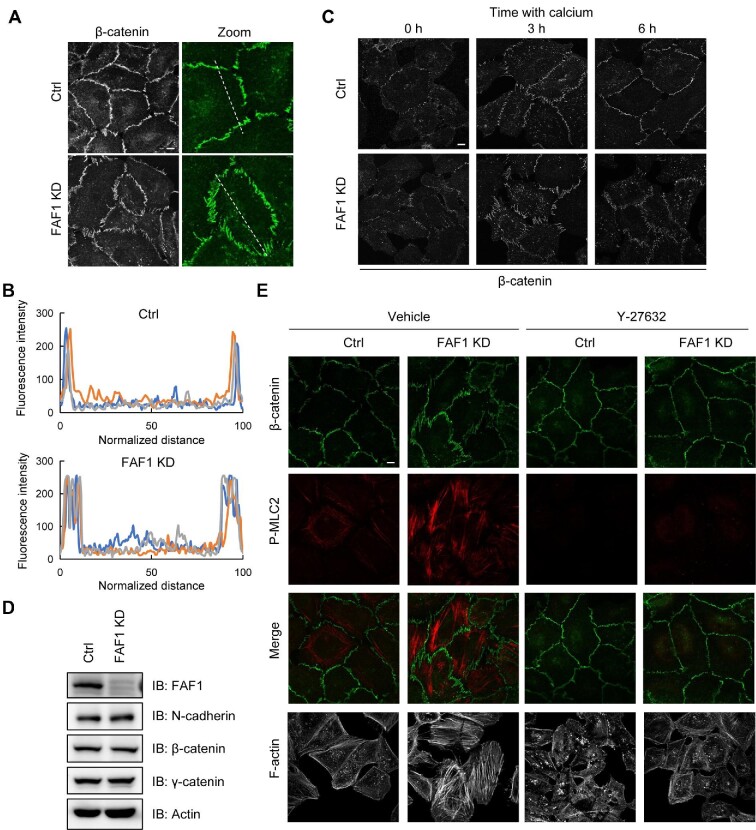
Rho GTPases are regulators of the dynamic organization and contractility of junctional actomyosin. Since silencing of FAF1 activated RhoA (Figure 4), we examined whether FAF1 affects the integrity of the cell adherens junction. The adherens junction integrity in control and FAF1-depleted HeLa cells was visualized by staining the cells with β-catenin, since β-catenin is a cytosolic adapter protein between extracellular cadherin and intracellular β-catenin/F-actin. Image analysis showed that β-catenin in control cells with a stable adherens junction had a thin linear appearance, whereas β-catenin in the FAF1-depleted cells showed a stretched and diffused appearance (Figure 5A). When the fluorescence intensities of β-catenin (dash lines) were plotted, the peak of adherens junction was sharp in control cells but broad in FAF1 KD cells (Figure 5B). These results demonstrated that silencing of FAF1 impairs the integrity of adherens junction.
Figure 5.
FAF1 alters the integrity of adherens junction. (A) HeLa cells were transfected with control siRNA or FAF1 siRNA #2 and replated on coverslips for 72 h. Cells were fixed and stained with β-catenin to investigate the integrity of adherens junction. Scale bar, 10 μm. (B) The fluorescence intensities of β-catenin are represented along the dashed line in A. Normalization was performed with respect to both end points of the dashed line in three different experiments. (C) HeLa cells were transfected with control siRNA or FAF1 siRNA for 48 h and replated on coverslips in a low-calcium medium. After 30 h, cells were returned to the normal-calcium medium to initiate junction assembly. The cells were then fixed and stained with β-catenin at the indicated time points. Scale bar, 10 μm. (D) HeLa cells were transfected with control siRNA or FAF1 siRNA for 72 h and lysed with gel sample buffer, and the lysates were analysed by western blotting. (E) HeLa cells were transfected with control siRNA or FAF1 siRNA and replated on coverslips for 72 h. Cells were treated with vehicle or Y-27632 (10 μM) for 6 h and then stained with F-actin (phalloidin) and β-catenin. Scale bar, 10 μm.
Next, to confirm the effects of FAF1 on the formation of cell–cell junctions, we conducted a calcium switch assay. A low calcium level disrupts adherens junction by removing calcium from the binding sites on E-cadherin extracellular domains, causing a conformational change and interrupting cell–cell adhesion. Confluent HeLa cells with endogenous silencing of FAF1 were cultured in a low Ca2+ medium (∼2 μM Ca2+) for 30 h, and the assembly of cell–cell junctions was initiated by switching the medium to a normal Ca2+ level (∼2 mM Ca2+). The degree of junction formation and maturation was evaluated by staining for β-catenin with an anti-β-catenin antibody. When calcium was depleted from the medium for 30 h, β-catenin staining in both control and FAF1-depleted cells was dispersed as shown in disrupted adherens junctions (0 h). β-catenin in control cells appeared stretched at 3 h after re-incubation with the medium containing a normal concentration of calcium (2 mM), but it had a thin linear appearance after 6 h incubation. However, in FAF1-depleted cells, β-catenin remained discontinuous and stretched even after 6 h incubation in the medium containing a normal calcium concentration (Figure 5C). To investigate whether these changes were due to the protein levels in adherens junction, we examined the expression levels of actin, N-cadherin, β-catenin, and γ-catenin in control and FAF1-depleted cells. Silencing of FAF1 did not affect the protein expression levels in adherens junction (Figure 5D). These findings confirmed that FAF1 actively regulates the stability and dynamics of adherens junctions.
Treatment with a ROCK inhibitor abolishes the impairment of adherens junction due to the silencing of FAF1
Impairment of adherens junction in the FAF1-silenced cells occurs via RhoA activation, which activates ROCK, upregulates pMLC, and triggers stress fiber formation. To confirm this, we examined β-catenin and pMLC levels in control and FAF1-depleted HeLa cells by treating the cells with the ROCK inhibitor Y-27632. As shown in Figure 5E, silencing of FAF1 impaired adherens junction detected by the elevated β-catenin and pMLC levels, whereas Y-27632 treatment inhibited the disassembly of adherens junction by suppressing pMLC induction. The ROCK inhibitor abolished the impairment of adherens junction induced by FAF1 knockdown. These results suggested that FAF1 depletion impairs adherens junction by activating RhoA and then increases stress fiber formation by activating ROCK.
The FAF1–Hsp70 complex is required for suppressing RhoA activation or suppressing stress fiber formation
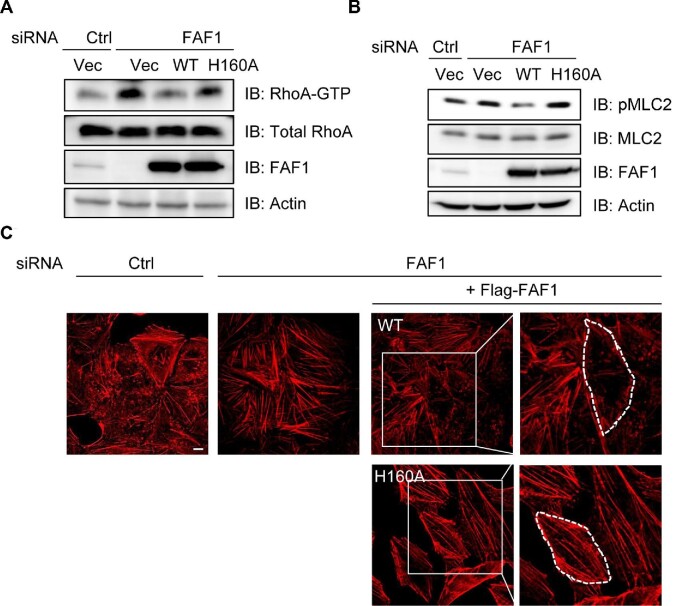
Elevated Hsp70 expression is highly correlated with increased cancer cell proliferation as well as metastasis and thus is suggested as a therapeutic target for cancer (Hall, 1998; Juhasz et al., 2013; Yi et al., 2017). The regulation of cell adherence by Hsp70 is also proposed as one of the mechanisms contributing to metastasis (Juhasz et al., 2013; Boudesco et al., 2018). FAF1 interacts with IQGAP1, NMIIA, and actin through the UBL1 domain, which interacts with Hsp70. These findings encouraged us to investigate whether the binding of Hsp70 to FAF1 is required for its interaction with IQGAP1, NMIIA, and actin and for maintaining the integrity of adherens junction. As shown in Figure 3E, FAF1 WT interacted with Hsp70, poly-ubiquitin, and VCP as reported previously (Song et al., 2005), whereas FAF1His160Ala mutant interacted with poly-ubiquitin and VCP but not with Hsp70. Intriguingly, this mutant could not interact with IQGAP1 as well as Hsp70. We examined RhoA activation by adding back FAF1 WT and FAF1His160Ala mutant in FAF1-depleted cells, as recent studies demonstrated that IQGAP1 inhibits RhoA activation. As shown earlier in Figure 4, silencing of FAF1 induced RhoA activation, which was abolished by overexpressing FAF1 WT but not FAF1His160Ala mutant (Figure 6A). The quantification of pMLC, a downstream target of RhoA activation, further confirmed these effects (Figure 6B). FAF1 activates RhoA by interacting with IQGAP1 via the association of Hsp70 with the UBL1 domain of FAF1. Stress fiber formation in FAF1-silenced cells was diminished by adding back FAF1 WT but not FAF1His160Ala mutant (Figure 6C).
Figure 6.
FAF1–Hsp70 interaction is crucial for FAF1 regulation of RhoA activation. HeLa cells with FAF1 knockdown were transfected with FAF1 WT and FAF1H160A mutant. Then, RhoA activation (A) and MLC2 phosphorylation (B) levels were measured, and actin stress fiber formation was detected by confocal microscopy (C). Scale bar, 10 μm.
Silencing of FAF1 increases collective invasion in transwell and 3D spheroid cell culture
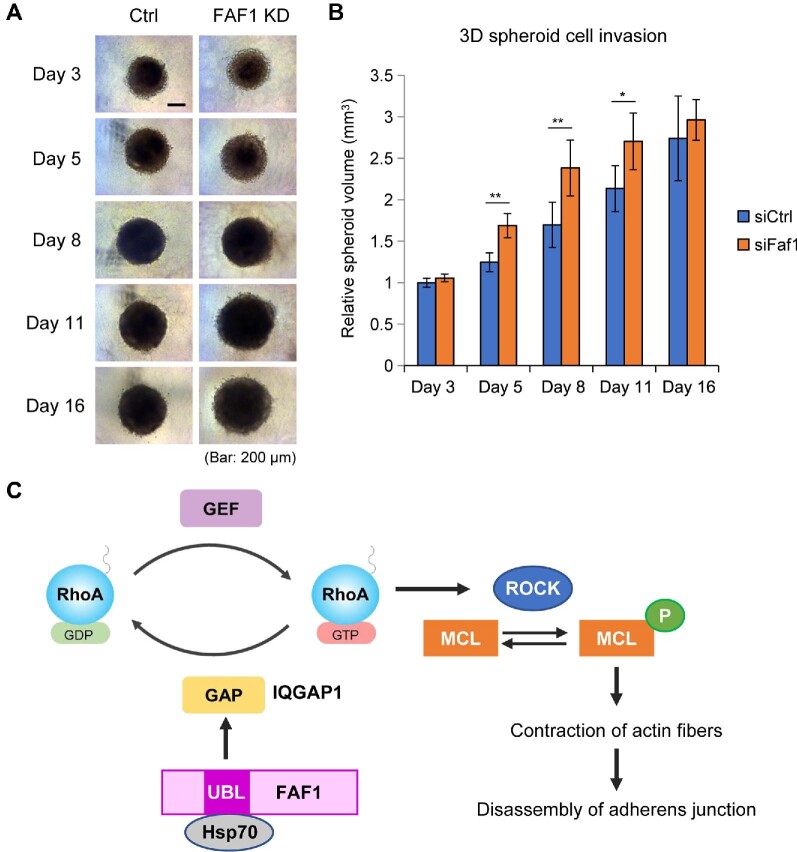
To investigate whether morphological changes induced by FAF1 depletion affect cancer metastasis, we utilized in vitro transwell invasion and a 3D spheroid model of MDA-MB-231 breast cancer cells. FAF1 depletion leads to increased transwell invasion (Supplementary Figure S6). A 3D spheroid model reflects the in vivo microenvironment more precisely than an in vitro monolayer cell culture model and is, hence, called an ‘in vivo-like model’. The 3D arrangement of cells allows us to study the cell–cell interactions and other in vivo processes. As shown in Figure 7A and B, FAF1 silencing in this model promoted the invasion of cancer cells. The results verified our hypothesis that the expansion of cell size and weakening of adherens junction affect the invasive potential of cancer cells.
Figure 7.
FAF1 inhibits cancer cell invasion in MDA-MB-231 cells by regulating the integrity of adherens junction. (A and B) MDA-MB-231 cells were transfected with control siRNA or FAF1 siRNA for 48 h and then spheroid formation was initiated in 2.5% Matrigel. Then, size kinetics were measured by calculating volume changes based on the radius of each spheroid. Representative images (A) and relative volume of the 3D spheroid cell invasion (B) are shown. Error bars represent standard deviation (n = 3). Scale bar, 200 μm. (C) Schematic diagram of the effect of FAF1 on stabilizing adherens junction by suppressing RhoA activation.
Discussion
In this study, we determined the crystal structure of a complex of FAF1 UBL1 and Hsp70 NBD and found that this complex plays a key role in stabilizing adherens junction by suppressing RhoA activation through its interaction with IQGAP1 via the UBL1 domain. Silencing of FAF1 in HeLa cells induces stress fiber formation by promoting RhoA activation, ROCK activation, and MLC phosphorylation as downstream signaling components (Figure 7C).
The crystal structure of the complex revealed that both the 10 or so residues preceding the UBL1 domain as well as residues 146–149 and 158–166 of FAF1 are important in the binding to Hsp70 NBD. Furthermore, ITC analysis and structural data suggested that His160 of FAF1 is critical for the binding to Hsp70. Residues lining the Hsp70 surface for the β-structured linker binding are highly conserved. Surprisingly, the interdomain linker of Hsp70, forming a β-structure, binds to the same binding cleft as the FAF1 linker, resulting in the β-domain of SBD positioned near the UBL1 of FAF in the ATP-bound DnaK (Supplementary Figure S2A). Therefore, once FAF1 binds to Hsp70, the interdomain linker of Hsp70 cannot bind to the cleft, i.e. the two will be competing for the same binding surface. The interdomain linker modulates various structure–function features of Hsp70, such as its global conformation, the affinity for peptide substrates, and the interaction with co-chaperones (Kampinga and Craig, 2010; Clerico et al., 2019; Mayer and Gierasch, 2019; Rosenzweig et al., 2019). In other words, this interdomain linker facilitates interdomain communication between the NBD and SBD of Hsp70. Unlike the highly conserved interdomain linker of Hsp70, the sequence identity on the linker region of FAF1 is low, and there is no sequence homology to the inter-domain linker of Hsp70. However, this is not critical, since backbone-to-backbone interaction occurs at ‘interface I’ between Hsp70 and FAF1 (Figure 1E). However, Gln97 and His160 of FAF1 are highly conserved amongst the species. Although FAF1 and Hsp70 in the crystal structure are not in their full forms, FAF1 binding certainly can inhibit Hsp70 function. With the recent improvement in omics technologies, a vast number of post-translational modifications (PTMs) have been uncovered on the Hsp70 family of proteins (Nitika et al., 2020 and the references therein). Although the respective roles are not yet fully understood, the PTMs that lie close to the interaction site of Hsp70 and FAF1 may alter the interaction. The interaction between the two proteins is exothermic with a KD of 3–16 μM in the presence of nucleotides. Also, the interface area of ∼2400 Å2 suggests that the interaction between the two is not very strong but enough to function as a regulator.
Mechanical stress induces heat shock response, increasing the expression of heat shock proteins, Hsp70, Hsp90, and Hsp27. Inhibitors targeting heat shock proteins have been developed as anticancer drugs, since upregulated heat shock proteins are responsible for cancer progression and metastasis. For example, geldanamycin, an Hsp90 inhibitor, increases stress fiber formation by regulating RhoA-dependent cytoskeleton remodeling, thus inhibiting invasion and metastasis (Amiri et al., 2007). Hsp70 increases metastatic growth through the upregulation of cytoskeleton-dependent signaling as well as chaperone and anti-apoptotic functions (Juhasz et al., 2013). Hsp70 regulates many cell adhesion molecules, among which upregulation of RhoA expression by extracellular Hsp70 to increase cell migration and invasion in hepatocarcinoma has been reported (Yi et al., 2017). In addition, the expression of FAF1 and Hsp70 is associated with cancer development. Most previous studies have been conducted on either FAF1 or Hsp70 individually. However, Kang et al. (2014) reported a correlation between FAF1 and Hsp70 expression in ovarian cancer. They showed that FAF1 expression decreases in advanced stages of ovarian cancer, while the expression of Hsp70 increases; therefore, FAF1 expression inversely correlates with Hsp70 expression. This report supported our findings that FAF1 structurally inhibits the Hsp70 cycle. We further investigated whether inhibition of Hsp70 activity is necessary for the regulation of RhoA by FAF1 via interaction with IQGAP1.
Dysregulation of cell adhesion plays a critical role in malignant transformation and metastasis. Despite the functional significance of RhoA activation, intact adherens junctions, and reorganization of the actin cytoskeleton machinery, details regarding how the associated proteins participate in the process are not yet known. RhoA has been widely studied to understand its role in the invasion and metastasis of malignant tumors and is considered a promising therapeutic target. Yoon et al. (2016) showed that RhoA activity is higher in gastric adenocarcinoma, and RhoA inhibition can reverse chemotherapy resistance. Both the expression level and activity of RhoA are important for the progression of cancers, e.g. breast cancer (Zheng et al., 2020). Using an active RhoA pull-down assay, we showed that FAF1 depletion promotes RhoA activity without altering the expression of RhoA. Additionally, FAF1 depletion increased breast cancer cell invasion in the transwell assay and 3D spheroid model. FAF1 is known to regulate the proteasomal degradation of cancer-related proteins recruiting the VCP and E3 ligase (Menges et al., 2009). For instance, FAF1 destabilizes TβRII on the cell surface by recruiting the VCP/E3 ligase complex, thereby limiting pro-oncogenic responses to TGF-β (Xie et al., 2017). TβRII regulates RhoA expression and activity (Ozdamar et al., 2005; Huang and Chen, 2012), implying that FAF1 regulate cancer cell proliferation and invasion through direct or indirect regulation of RhoA.
RhoA-mediated regulation of invasion and metastasis occurs through the stimulation of contractility by activating downstream molecules, ROCK and MLC, which leads to the formation of stress fibers and focal adhesions. Treatment with a ROCK inhibitor abolished the impairment of adherens junction induced by silencing of FAF1 (Figure 5E), suggesting that FAF1 regulates the integrity of adherens junction via the RhoA–ROCK pathway. RhoA activity is primarily regulated by GEFs, GAPs, and guanine nucleotide dissociation inhibitors. IQGAP1 is another essential factor regulating the RhoA activity. It is a scaffold protein that provides a platform for small GTPase proteins like RhoA, RhoC, and Rac1 and their regulators, such as p190RhoGAP, Cdc42, and Arf6 (Hedman et al., 2015). Therefore, the function of IQGAP1 as a scaffold may facilitate the rapid regulation of RhoA activity. In addition, IQGAP1 localizes to sites of cell–cell contact and colocalizes with p190RhoGAP and RhoA, inactivating RhoA and suppressing airway smooth muscle contraction (Bhattacharya et al., 2014). IQGAP1 is associated with diverse oncogenic functions like cell proliferation, extravasation, and metastasis (Hebert et al., 2020). IQGAP1 interacts with > 100 proteins and exhibits oncogenic functions either alone or in combination with binding proteins. Here, we found that FAF1 interacts with IQGAP1 and p190RhoGAP. The interaction of FAF1 with p190RhoGAP is significantly weak compared to that with IQGAP1. Therefore, IQGAP1 could mediate the interaction of p190RhoGAP with FAF1. The FAF1–Hsp70 complex was found to bind with IQGAP1, which means that this complex regulates RhoA activity by recruiting other modulators such as p190RhoGAP. Complex formation of FAF1 with Hsp70 may provide an interaction surface needed for IQGAP1. However, it is also possible that IQGAP1 interacts with only the FAF1 UBL1 domain in the Hsp70-bound conformation. The crystal structure of the complex showed that only a handful of residues (146–149 and 158–166) of FAF1 UBL1 were engaged in its interaction with Hsp70. Therefore, a large surface of UBL1 is available for other partner proteins (Figure 1C). The J domain binds onto the surface above the interdomain linker and contacts the β-domain of the SBD (Kityk et al., 2018). The UBL1 surface is somewhat negatively charged (Supplementary Figure S1C).
Here, we showed another novel function of FAF1 of using Hsp70 as an adapter protein and interacting with IQGAP1 to regulate the assembly and disassembly of adherens junction through RhoA signaling. However, further studies are required to investigate which is more important for the regulation of cell invasion by the FAF1–Hsp70 complex.
Materials and methods
Expression and purification of FAF1 and Hsp70
Several constructs, including full-length protein of human FAF1, were cloned into pET-28a with a His-tag added at the N-terminus (Figure 1A) and expressed in E. coli Rosetta-gami 2 (DE3) cells or Rosetta2 (DE3) cells (Novagen). After induction using 0.5 mM isopropyl β-D-1-thiogalactopyranoside, the cells were allowed to grow for 12 h at 18°C and then harvested. Proteins were purified employing the Ni-NTA affinity, Mono Q anion exchange chromatography followed by the Superdex-200S 26/60 or Superdex-75S 26/60 gel filtration and concentrated to 20–25 mg/ml. The tag was removed after the affinity purification. All mutants of FAF1 were generated and purified following the same procedure as for the WT. To obtain selenomethionine (SeMet)-substituted protein, FAF1 UBL1 was overexpressed using the same procedure described above except with E. coli B834 (DE3) cells and M9 cell culture medium as described previously (Hendrickson et al., 1990). The hexapeptide corresponding to 91RQIVER96 of FAF1 was synthesized. The NBD of human Hsp70 (aa 1–382) was cloned into the pET-28a vector (Novagen) with the TEV cleavage site and overexpressed in E. coli Rosetta2 (DE3) cells (Novagen). Purification was carried out employing a Ni-NTA column followed by removal of the tag and filtration through a Superdex-200S 26/60 gel filtration column (GE Healthcare). The final protein in 50 mM HEPES, pH 7.5, 150 mM NaCl, 2 mM MgCl2, and 1 mM DTT was concentrated to 20 mg/ml.
Crystallization, data collection, and structure determination of FAF1 UBL1 and FAF1 UBL1–Hsp70 NBD
Diffraction quality crystals of both the native and SeMet-substituted FAF1 UBL1 were obtained by mixing equal volumes of protein in 50 mM HEPES, pH 7.5, 150 mM NaCl at 20 mg/ml and reservoir solution containing 50 mM Bis–Tris, pH 6.5, 50 mM (NH4)2SO4, and 30% (v/v) pentaerythritol ethoxylate. Crystals were transferred into a cryoprotectant containing a reservoir solution with 20% ethylene glycol before data collection. The crystals of FAF1 L-UBL1 complexed with Hsp70 NBD with ADP and Pi were obtained using 200 mM imidazole malate, pH 8.5, and 12% PEG 10000 and stabilized in 25% ethylene glycol before data collection. Diffraction data were collected using synchrotron radiation (beamline 4A of Pohang Light Source, Korea) and were processed, integrated, and scaled using the HKL2000 program suite (Otwinowski and Minor, 1997). The structure of FAF1 UBL1 was determined by MAD using the programs SOLVE (Terwilliger and Berendzen, 1996) and RESOLVE (Terwilliger, 2000), while the structure of FAF1 L-UBL1–Hsp70 NBD complex was determined using molecular replacement with CNS (Brünger et al., 1998). Crystal structures of FAF1 UBL1 from this study and Hsp70 NBD with ADP (Sriram et al., 1997) were determined. Model building was done using COOT (Emsley and Cowtan, 2004) and refined with CNS (Brünger et al., 1998) and REFMAC5 (Murshudov et al., 1997). The ideality and geometry of the model were checked using PROCHECK (Laskowski et al., 1993). Table 1 summarizes statistics on the data collection and refinement. PyMOL (www.pymol.org/) was used to prepare figures.
ITC analysis
Binding was tested using the ITC200 instrument (MicroCal) at 25°C, and the data were analysed using ORIGIN 7.0. All samples were placed in 50 mM HEPES, pH 7.5, 150 mM NaCl, and 1 mM MgCl2, with or without ADP. They were centrifuged and degassed before the measurements at 25°C. All injections were added at an interval of 150 sec to the sample solution in the cell with gentle stirring using a computer-controlled micro-syringe. After making corrections for dilution by subtracting the values for buffer alone, we analysed the data with a one-site binding model using ORIGIN 7.0.
Plasmids and reagents
The following antibodies were used in this study: mouse monoclonal Flag antibody (Sigma), rabbit anti-FAF1 (AbFrontier), rabbit anti-NMIIA antibody (BioLegend), mouse anti-RhoA (Abcam), mouse anti-p190B RhoGAP, mouse anti-β-catenin, mouse anti-γ-catenin, mouse-anti-N-cadherin (BD Bioscience), rabbit anti-MLC2, anti-PP-MLC2 (Cell Signaling Technology), rabbit anti-IQGAP1, mouse anti-actin, and anti-tubulin (Santa Cruz Biotechnology). Y-27632 (Y0503) was purchased from Sigma. GST-FAF1 WT, GST-FAF1 UAS-UBX, pFlag-CMV-2-FAF1 WT, pFlag-CMV-2-FAF1 (82–650), and pFlag-CMV-2-FAF1 (1–120) were prepared as previously described (Murshudov et al., 1997; Song et al., 2005). pFlag-CMV-2-FAF1 H160A was generated by cloning employing mutagenesis. All plasmid constructs were verified by DNA sequencing.
Cell culture and transfection
HeLa cells were purchased from ATCC and cultured in Eagle's Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin G and 100 μg/ml streptomycin at 37°C in a 5% CO2-containing humidified incubator. HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented in the same manner mentioned above. For the transient overexpression of specific proteins, cells were transfected using LT-1 and analysed 24 h post-transfection. For gene silencing, FAF1 siRNAs were obtained from Dharmacon (ON-TARGETplus SMARTpool siRNA) and Bioneer. FAF1 siRNA #1 was obtained from Dharmacon (L-009106-00-0005), and FAF1 siRNA #2 (Cat No. 1049605) and control siRNA were from Bioneer. Cells were transfected with siRNAs using DharmaFECT1 following the manufacturer's protocol at a final concentration of 50 nM.
GST pull-down assay and identification of binding proteins
GST-FAF1 and GST-FAF1[UAS-UBX] were expressed in E. coli BL21 (DE3) cells and bound to glutathione-agarose beads for 3 h at 4°C with gentle rotation. The beads were then washed six times with phosphate-buffered saline containing 0.1% Triton X-100. HeLa cells were lysed with hypotonic buffer containing protease inhibitors (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1 mM PMSF, 5 μg/ml aprotinin, 10 μg/ml leupeptin, and 10 μg/ml pepstatin A) and incubated for 30 min on ice, followed by centrifugation at 12000 rpm for 15 min at 4°C. GST-FAF1 bound to beads were incubated with HeLa cell extracts for 3 h at 4°C, and then the beads were washed thrice with lysis buffer and additionally twice with lysis buffer without any detergent. The beads were resuspended in gel sample buffer, and binding proteins were separated by SDS–PAGE and visualized using a silver staining kit or western blotting analysis. Gel bands of binding proteins were de-stained and digested with trypsin, and the resulting peptides were extracted as previously described (Seo et al., 2008). Peptides were separated using trap column cartridge, injected into C18 reversed-phase analytical column (nanoACQUITY BEH300 particle size: 1.7 μm, ID: 75 μm and length: 250 mm) with integrated electrospray ionization PicoTipTM using nanoAcquityTM UPLC/ESI/q-TOF MS/MS (SYNAPTTM G2Si; Waters Co.) (equipped at Ewha Drug Development Research Core Center).
Immunoprecipitation
Cells were lysed with hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 60 mM octyl-β-D-glucopyranoside, pH 7.9) containing protease inhibitors (1 mM PMSF, 5 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 5 mM Na3VO4, 5 mM NaF, 0.1 mM EDAT, and 0.1 mM EGTA) and an HDAC inhibitor (10 mM sodium butyrate) for 30 min on ice. The cell lysates were centrifuged at 12000 rpm for 15 min at 4°C. The supernatant was incubated with anti-Flag antibody for 2 h at 4°C, and the lysate–antibody complexes were incubated with protein-G sepharose 4 Fast Flow beads for another 1 h at 4°C. The precipitated beads were washed six times with lysis buffer to remove non-specific binding and additionally twice with lysis buffer without any detergent. The immune complex was eluted with gel sample buffer, separated by SDS–PAGE, and analysed by western blotting.
Confocal microscopy
Cells were grown on the SecureSlip™ coverslip (Sigma) and fixed with 4% paraformaldehyde in Hanks' balanced salt solution (HBSS) for 10 min. After washing with HBSS, cells were permeabilized with 0.1% Triton X-100 in HBSS for 10 min. After HBSS washing, cells were incubated with 3% bovine serum albumin in HBSS for 1 h to block non-specific protein adsorption and then incubated with primary antibodies for 2 h at 37°C. After washing thrice with HBSS, the cells were stained for 1 h at 37°C with Alexa Fluor-conjugated secondary antibodies. After washing thrice with HBSS, the mounting medium for fluorescence with DAPI was used for staining the nuclei. After being mounted, the cells were observed under a Zeiss LSM510 Meta laser scanning microscope. Images were photographed and processed using the LSM510 software (Carl Zeiss).
RhoA activity assay
HeLa cells were cultured in a 100-mm dish, and FAF1 was depleted by transfecting the cells with control or FAF1 siRNA for 72 h. Active RhoA pull-down assays were performed according to the manufacturer's instructions (Thermo Scientific). Briefly, HeLa cell lysates were obtained by incubation with lysis buffer followed by centrifugation at 8000 rpm. Cell lysates (750 μg) were incubated with GST-RBD of Rhotekin for 1 h at 4°C with rocking. Precipitates were washed three times with lysis buffer, solubilized with gel sample buffer, separated by SDS–PAGE, and detected by using anti-RhoA antibody.
3D spheroid tumor invasion assay
The spheroid cell culture of MDA-MB-231 cells was prepared in an EMEM-conditioned medium (CM) containing 2.5% Matrigel. The cells were transfected with 100 nM control siRNA or FAF1 siRNA and cultured for 48 h. A total of 7500 transfected cells in 100 μl EMEM-CM were suspended in 100 μl 5% Matrigel in a CellCarrier-96 spheroid ULA plate (Perkin Elmer, ref. 6055330). Then, the cells were centrifuged at 13000× g for 10 min and incubated for 2–3 days for spheroid initiation. From the day of spheroid initiation, the radius of each spheroid was measured at indicated dates by observing under an Axiovert 200 microscope (Carl Zeiss). The volume of each spheroid was calculated by measuring the size using AxioVision Rel.4.7.
Accession numbers
The atomic coordinates and structure factors of FAF1 UBL1 and FAF1 L-UBL1 complexed with Hsp70 NBD with ADP and Pi have been deposited in the Protein Data Bank (http://www.rcsb.org/) under the accession codes of 7FGN and 7FGM, respectively.
Statistical analysis
Data were analyzed using Student's t-test for comparisons between two groups to determine the statistical significance (P-value). P < 0.05 was considered to indicate a significant difference.
Supplementary Material
Acknowledgements
The authors appreciate the technical support of MS analysis performed by Mr W. Kang (Ewha Womans University).
Contributor Information
Soonhwa Song, Graduate School of Pharmaceutical Sciences, College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea.
Joon Kyu Park, Biomedical Research Institute, Korea Institute of Science and Technology (KIST), Seoul 02792, Republic of Korea.
Sang Chul Shin, Biomedical Research Institute, Korea Institute of Science and Technology (KIST), Seoul 02792, Republic of Korea.
Jae-Jin Lee, Graduate School of Pharmaceutical Sciences, College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea.
Seung Kon Hong, Biomedical Research Institute, Korea Institute of Science and Technology (KIST), Seoul 02792, Republic of Korea.
In-Kang Song, Graduate School of Pharmaceutical Sciences, College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea.
Bokyung Kim, Graduate School of Pharmaceutical Sciences, College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea.
Eun Joo Song, Graduate School of Pharmaceutical Sciences, College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea.
Kong-Joo Lee, Graduate School of Pharmaceutical Sciences, College of Pharmacy, Ewha Womans University, Seoul 03760, Republic of Korea.
Eunice EunKyeong Kim, Biomedical Research Institute, Korea Institute of Science and Technology (KIST), Seoul 02792, Republic of Korea.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (2017R1A2B3007224 and 2020R1A4A4079494 to E.E.K.; 2020R1F1A1055369 to K.-J.L.; 2019R1A2C2004052 to E.J.S.). S.S. and I.-K.S. were supported by Brain Korea 21 Plus (BK21 Plus) Project.
Author contributions: S.S. and K.-J.L. designed the study. S.S., J.-J.L., I.-K.S., B.K., E.J.S., and K.-J.L. conducted the biochemical/cell biology studies and analysed the results. J.K.P., S.C.S., S.K.H., and E.E.K. carried out structural and biophysical studies and analyzed the results. E.J.S., K.-J.L., and E.E.K. wrote the manuscript.
Conflict of interest: none declared.
References
- Amiri A., Noei F., Feroz T.et al. (2007). Geldanamycin anisimycins activate Rho and stimulate Rho- and ROCK-dependent actin stress fiber formation. Mol. Cancer Res. 5, 933–942. [DOI] [PubMed] [Google Scholar]
- Arakawa A., Handa N., Ohsawa N.et al. (2010). The C-terminal BAG domain of BAG5 induces conformational changes of the Hsp70 nucleotide-binding domain for ADP–ATP exchange. Structure 18, 309–319. [DOI] [PubMed] [Google Scholar]
- Arnold T.R., Stephenson R.E., Miller A.L. (2017). Rho GTPases and actomyosin: partners in regulating epithelial cell–cell junction structure and function. Exp. Cell Res. 358, 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M., Sundaram A., Kudo M.et al. (2014). IQGAP1-dependent scaffold suppresses RhoA and inhibits airway smooth muscle contraction. J. Clin. Invest. 124, 4895–4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørling-Poulsen M., Seitz G., Guerra B.et al. (2003). The pro-apoptotic FAS-associated factor 1 is specifically reduced in human gastric carcinomas. Int. J. Oncol. 23, 1015–1023. [PubMed] [Google Scholar]
- Bonjoch L., Franch-Expósito S., Garre P.et al. (2020). Germline mutations in FAF1 are associated with hereditary colorectal cancer. Gastroenterology 159, 227–240.e7. [DOI] [PubMed] [Google Scholar]
- Boudesco C., Cause S., Jego G.et al. (2018). Hsp70: a cancer target inside and outside the cell. Methods Mol. Biol. 1709, 371–396. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Clore G.M.et al. (1998). Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Casteel D.E., Turner S., Schwappacher R.et al. (2012). Rho isoform-specific interaction with IQGAP1 promotes breast cancer cell proliferation and migration. J. Biol. Chem. 287, 38367–38378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti C., Ridley A.J. (2017). Endothelial cell–cell adhesion and signaling. Exp. Cell Res. 358, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu K., Niu X., Williams L.T. (1995). A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc. Natl Acad. Sci. USA92, 11894–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerico E.M., Meng W., Pozhidaeva A.et al. (2019). Hsp70 molecular chaperones: multifunctional allosteric holding and unfolding machines. Biochem. J. 476, 1653–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyberg C., Fransson S., Andonova T.et al. (2017). Rho-associated kinase is a therapeutic target in neuroblastoma. Proc. Natl Acad. Sci. USA114, E6603–E6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Farahani E., Patra H.K., Jangamreddy J.R.et al. (2014). Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis 35, 747–759. [DOI] [PubMed] [Google Scholar]
- Hall A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509–514. [DOI] [PubMed] [Google Scholar]
- Hebert J.D., Tian C., Lamar J.M.et al. (2020). The scaffold protein IQGAP1 is crucial for extravasation and metastasis. Sci. Rep. 10, 2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman A.C., Smith J.M., Sacks D.B. (2015). The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 16, 427–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W.A., Horton J.R., LeMaster D.M. (1990). Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuzé M.L., Sankara Narayana G.H.N., D'Alessandro J.et al. (2019). Myosin II isoforms play distinct roles in adherens junction biogenesis. eLife 8, e46599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Chen Y.G. (2012). Regulation of TGF-β receptor activity. Cell Biosci. 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D., Park S., Kim H.et al. (2016). RhoA is associated with invasion and poor prognosis in colorectal cancer. Int. J. Oncol. 48, 714–722. [DOI] [PubMed] [Google Scholar]
- Jiang J., Maes E.G., Taylor A.B.et al. (2007). Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz K., Lipp A.M., Nimmervoll B.et al. (2013). The complex function of hsp70 in metastatic cancer. Cancers 6, 42–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., Craig E.A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.J., Moon H.S., Chung H.W. (2014). The expression of FAS-associated factor 1 and heat shock protein 70 in ovarian cancer. Obstet. Gynecol. Sci. 57, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Song E.J., Lee Y.S.et al. (2005). Human Fas-associated factor 1 interacts with heat shock protein 70 and negatively regulates chaperone activity. J. Biol. Chem. 280, 8125–8133. [DOI] [PubMed] [Google Scholar]
- Kityk R., Kopp J., Mayer M.P. (2018). Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69, 227–237.e4. [DOI] [PubMed] [Google Scholar]
- Kityk R., Kopp J., Sinning I.et al. (2012). Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874. [DOI] [PubMed] [Google Scholar]
- Laskowski R., Macarthur M.W., Moss D.S.et al. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Lee J.J., Kim Y.M., Jeong J.et al. (2012). Ubiquitin-associated (UBA) domain in human Fas associated factor 1 inhibits tumor formation by promoting Hsp70 degradation. PLoS One 7, e40361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.J., Park J.K., Jeong J.et al. (2013). Complex of Fas-associated factor 1 (FAF1) with valosin-containing protein (VCP)–Npl4–Ufd1 and polyubiquitinated proteins promotes endoplasmic reticulum-associated degradation (ERAD). J. Biol. Chem. 288, 6998–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Ishizaki T., Boku S.et al. (1999). Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285, 895–898. [DOI] [PubMed] [Google Scholar]
- Mayer M.P., Gierasch L.M. (2019). Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 294, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges C.W., Altomare D.A., Testa J.R. (2009). FAS-associated factor 1 (FAF1): diverse functions and implications for oncogenesis. Cell Cycle 8, 2528–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Nitika Porter, C.M., Truman A.W.et al. (2020). Post-translational modifications of Hsp70 family proteins: expanding the chaperone code. J. Biol. Chem. 295, 10689–10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Ozdamar B., Bose R., Barrios-Rodiles M.et al. (2005). Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 307, 1603–1609. [DOI] [PubMed] [Google Scholar]
- Polier S., Dragovic Z., Hartl F.U.et al. (2008). Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R., Nillegoda N.B., Mayer M.P.et al. (2019). The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20, 665–680. [DOI] [PubMed] [Google Scholar]
- Seo J., Jeong J., Kim Y.M.et al. (2008). Strategy for comprehensive identification of post-translational modifications in cellular proteins, including low abundant modifications: application to glyceraldehyde-3-phosphate dehydrogenase. J. Proteome Res. 7, 587–602. [DOI] [PubMed] [Google Scholar]
- Sondermann H., Scheufler C., Schneider C.et al. (2001). Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557. [DOI] [PubMed] [Google Scholar]
- Song E.J., Yim S.H., Kim E.et al. (2005). Human Fas-associated factor 1, interacting with ubiquitinated proteins and valosin-containing protein, is involved in the ubiquitin–proteasome pathway. Mol. Cell. Biol. 25, 2511–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Park J.K., Lee J.J.et al. (2009). Structure and interaction of ubiquitin-associated domain of human Fas-associated factor 1. Protein Sci. 18, 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Lee J.J., Kim H.J.et al. (2016). Fas-associated factor 1 negatively regulates the antiviral immune response by inhibiting translocation of interferon regulatory factor 3 to the nucleus. Mol. Cell. Biol. 36, 1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram M., Osipiuk J., Freeman B.et al. (1997). Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 5, 403–414. [DOI] [PubMed] [Google Scholar]
- Svensmark J.H., Brakebusch C. (2019). Rho GTPases in cancer: friend or foe? Oncogene 38, 7447–7456. [DOI] [PubMed] [Google Scholar]
- Tanos B.E., Yeaman C., Rodriguez-Boulan E. (2018). An emerging role for IQGAP1 in tight junction control. Small GTPases 9, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. (2000). Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Berendzen J. (1996). Correlated phasing of multiple isomorphous replacement data. Acta Crystallogr. D Biol. Crystallogr. 52, 749–757. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S.et al. (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Jin K., Shao L.et al. (2017). FAF1 phosphorylation by AKT accumulates TGF-β type II receptor and drives breast cancer metastasis. Nat. Commun. 8, 15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.-D., Diao M.-Q., Niu G.-J.et al. (2018). A small GTPase, RhoA, inhibits bacterial infection through integrin mediated phagocytosis in invertebrates. Front. Immunol. 9, 1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Page R.C., Gomes M.M.et al. (2008). Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat. Struct. Mol. Biol. 15, 1309–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Li J., Sha B. (2011). Structural analysis of the Sil1–Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor. Biochem. J. 438, 447–455. [DOI] [PubMed] [Google Scholar]
- Yi Z., Li Y., Liu D.et al. (2017). Extracellular HSP70/HSP70-PCs regulate hepatocarcinoma cell migration and invasion via RhoA. Oncol. Lett. 13, 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon C., Cho S.J., Aksoy B.A.et al. (2016). Chemotherapy resistance in diffuse-type gastric adenocarcinoma is mediated by RhoA activation in cancer stem-like cells. Clin. Cancer Res. 22, 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zheng C.W., Zeng R.J., Xu L.Y.et al. (2020). Rho GTPases: promising candidates for overcoming chemotherapeutic resistance. Cancer Lett. 475, 65–78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.