Abstract
We show that DNA molecules amplified by PCR from DNA extracted from animal bones and teeth that vary in age between 25 000 and over 50 000 years carry C→T and G→A substitutions. These substitutions can reach high proportions among the molecules amplified and are due to the occurrence of modified deoxycytidine residues in the template DNA. If the template DNA is treated with uracil N-glycosylase, these substitutions are dramatically reduced. They are thus likely to result from deamination of deoxycytidine residues. In addition, ‘jumping PCR’, i.e. the occurrence of template switching during PCR, may contribute to these substitutions. When DNA sequences are amplified from ancient DNA extracts where few template molecules initiate the PCR, precautions such as DNA sequence determination of multiple clones derived from more than one independent amplification are necessary in order to reduce the risk of determination of incorrect DNA sequences. When such precautionary measures are taken, errors induced by damage to the DNA template are unlikely to be more frequent than ∼0.1% even under the unlikely scenario where each amplification starts from a single template molecule.
INTRODUCTION
When an organism dies, chemical damage starts to accumulate in its DNA (1). This makes the retrieval of DNA sequences by PCR progressively more difficult due to strand breaks as well as DNA modifications that may block strand elongation by DNA polymerases or cause incorrect nucleotides to be incorporated during PCR. For example, DNA extracted from ancient remains is generally of low average molecular weight (2) and both hydrolytic and oxidative damage has been shown to occur in macromolecules extracted from ancient tissues (3–5). In particular, the numbers of oxidised DNA bases in ancient DNA samples have been shown to correlate with amplification success (3) and often many misincorporated nucleotides are observed in DNA sequences retrieved by PCR from ancient DNA extracts (6).
One of the most common forms of hydrolytic DNA damage in living organisms is deamination. Deamination is particularly rapid for cytosine (7), which results in the conversion of cytosine to uracil in the DNA. When DNA containing such lesions is used as a template for PCR, C→T and G→A transitions in the DNA sequences recovered will result, since deoxyuridine residues in the template DNA strand cause DNA polymerases to incorporate deoxyadenosine residues at positions where a deoxyguanosine would normally have been incorporated. Here, we demonstrate that deoxyuridine residues are a frequent miscoding lesion in DNA extracted from a variety of ancient bone samples and investigate the extent to which this may result in the incorrect determination of nucleotide sequences from paleontological and archaeological remains.
MATERIALS AND METHODS
DNA extraction, PCR and sequencing
Eleven cave bear (Ursus spelaeus) bones and teeth excavated from nine caves (Hohle Fels, Germany; Gailenreuth, Germany; Grotte Merveilleuse, France; Geissenklösterle, Germany; Vindija, Croatia; Potocka, Slovenia; Conturines, Italy; Ramesch, Austria; Nixloch, Austria) were used in this study. From each sample, ∼50 mg bone/tooth powder were incubated in 1 ml of 0.5 M EDTA, pH 8.0, 5% N-laurylsarcosine, 2% cetyl trimethylammonium bromide, 0.3 mg/ml proteinase K and 5 mM N-phenacylthiazolium bromide (8) for 96 h at room temperature. DNA was extracted by binding to silica as described (9,10) and PCR was performed using AmpliTaq Gold (Perkin Elmer, USA) with a MgCl2 concentration of 2.5 mM, following the supplier’s instructions, except for the addition of bovine serum albumin to a final concentration of 0.25 mg/ml. Amplifications were done in a MJ Thermocycler with a 3 min activation step at 94°C, followed by 60 cycles of 93°C for 30 s, 41°C for 60 s and 72°C for 30 s. PCR products were isolated from 2.8% agarose gels and melted in 100 µl double distilled water, of which 5 µl were used for reamplification for 30 cycles under the PCR conditions described, except that the activation step was prolonged to 7 min and the annealing temperature was 45°C. Reamplification products were cloned using the Topo TA cloning kit (Invitrogen, The Netherlands). In cases where no primer dimers were observed in the first amplification, the product was cloned directly. Primer pairs used were: CBHVR1F1, 5′-CTA TTT AAA CTA TTC CCT GGT ACA TAC-3′, with CBHVR1R1, 5′-ATG GGG GCA CGC CAT TAA TGC-3′; CBHVR1F2, 5′-CAT CTC ATG TAC TGT ACC ATC ATA GT-3′, with CBHVR1R2, 5′-TAA ACT TTC GAA ATG TAG GTC CTC ATG-3′; CBHVR1F3, 5′-GCC CCA TGC ATA TAA GCA TG-3′, with CBHVR1R3, 5′-GGA GCG AGA AGA GGT ACA CGT-3′. These yielded products of 167, 166 and 176 bp (including primers), respectively. Mock extractions without sample and PCR blanks were performed in all experiments to monitor contamination.
Clones were sequenced in Alf Express automated sequencers directly from colony PCRs (11) using a Thermo Sequenase Kit (Amersham Pharmacia Biotech, Germany) or in an ABI 3700 capillary sequencer following plasmid preparation. For each PCR product at least three clones were sequenced.
Single primer extensions
To assess the substitution pattern on only one strand of the ancient DNA template, single primer extensions were performed prior to some amplifications. In order to increase the chance that the reactions would start from several molecules, extracts from all 11 samples were pooled and concentrated 20-fold by ethanol precipitation following standard protocols (12). The number of starting molecules for one reaction was ∼200, as determined by real time PCR using a 99 bp amplification of the 12S rDNA (13). As the sequences of all 11 samples have been determined previously (Hofreiter et al., unpublished data), substitutions due to DNA damage could be readily identified. Single primer extension PCR was done as described above, except that only one of the primers was added at the beginning and the Taq Gold enzyme was fully activated for 10 min at 94°C. Twenty-five cycles of extension were done, followed by addition of the second primer and 45 PCR cycles. Products were analysed on an agarose gel, reamplified and cloned as described above. As a control, the same experiment was performed using a 99 bp piece of the cave bear 12S rDNA sequence cloned in a plasmid. The plasmid concentration was measured by UV absorbance and diluted to ∼200 starting molecules to match the concentration of starting molecules in the ancient DNA extract. Amplification was as above, except that the annealing temperature was 59°C. Primers used were CB12S1f, 5′-ACC CCA CTA TGC TTA GCC TTA AA-3′, and CB12S1r, 5′-ACC GCC AAG TCC TTT GAG TT-3′.
UNG treatment
One aliquot of the cave bear DNA pool was treated with Escherichia coli uracil N-glycosylase (UNG) before amplification while another aliquot was incubated with UNG that had been heat-inactivated for 10 min at 95°C. Among the clones sequenced, the number of positions where substitutions occurred were counted.
Determination of the deamination rate
For each position in the cave bear DNA sequence carrying a C or G, we calculated the misincorporation rate λI such that each consistent substitution, i.e. positions where all clones from one amplification differed from all clones from two or more additional amplifications, was counted as a full event and given the value 1. For non-consistent substitutions, i.e. substitutions which were observed in one or several but not all clones from an amplification, we divided the number of clones carrying a substitution by the total number of clones sequenced from that PCR product. For example, if two of eight clones from a PCR contained a T instead of a C at a certain position, the value 0.25 was assigned to that position. Finally, to calculate λI we summed all values for a given position and divided by the number of PCRs done for that position. At variable positions, i.e. positions at which some cave bear samples carry a C (or a G) and others carry a T (or an A), only the number of PCRs which could possibly yield a C/G→T/A substitution were counted. Finally, a second parameter, λ′, was calculated by summing all values λi and dividing by the total number of positions analysed (115), yielding the average substitution rate over all positions.
Deamination hotspots
To determine if deamination hotspots occur in the DNA sequences studied, consistent substitutions affecting the 99 positions that carry a G or a C in all cave bears studied were analysed from the 79 amplifications performed. These positions were scored a total of 3243 times in independent amplifications. Assuming that each substitution and each PCR are independent, an average substitution rate λm of 0.0139 was calculated and used as the null hypothesis H0 of equal substitution probability across sequence positions. For the alternative hypothesis H1, a specific substitution probability λi for each position was estimated by dividing the number of substitutions by the number of amplifications covering that position. Finally, the difference in the log-likelihood of arriving at the observed data given the parameters from H0 and H1, assuming a binomial distribution of substitutions, was compared to an empirical distribution of log-likelihood differences from 1000 simulated amplifications with homogeneous λm.
RESULTS
Pleistocene bone samples, PCR and cloning
Eight bones and three teeth from cave bears (U.spelaeus) found in nine caves in central Europe were dated by accelerator mass spectroscopy and found to range in age from >49 500 to ∼26 500 years before present. DNA was extracted from each bone and three fragments of the mitochondrial control region, varying in length between 166 and 176 bp (including primers), were amplified by PCR. The amplifications were repeated multiple times for each extract. The rates with which amplification products that could be visualised by ethidium staining in agarose gels were obtained from the different extracts varied from 100 (7 of 7 amplifications) to 24% (11 of 45 amplifications). Each product selected for further study was cloned in a plasmid vector and the inserts of between 3 and 15 clones were sequenced. In total, 484 clones from 79 PCR amplification products were analysed.
Classes of nucleotide substitutions
When the nucleotide substitutions observed among the clones were scrutinised, it was found that for 23 of the amplification products all clones that had been sequenced carried substitutions that distinguished them from all the clones of two or more other independent amplifications from the same DNA extracts. Such substitutions that occurred consistently among clones within one amplification but were not reproducible in other amplifications were observed at a total of 48 nucleotide positions (Fig. 1). They are likely to be the result of nucleotide misincorporations that occurred during the first cycle of the PCR in cases where the amplification started from a single DNA molecule. In such cases, all molecules cloned from the PCR product will carry any nucleotide misincorporation that occurred in the first cycle of the PCR when a single DNA molecule extracted from the bone served as a template (10,14). Thus, the consistent substitutions represent errors induced during replication of the ancient DNA molecules.
Figure 1.
DNA sequences from five different amplifications from one cave bear sample. Dots indicate identity to the consensus sequence, given at the top of the alignment. The first number in front of a sequence gives the number of the amplification, the second the number of the sequenced clone. Four of the five amplifications carry consistent substitutions (indicated by an asterisk) relative to the consensus sequence at one to five positions. Clone 4-6 represents a jumping PCR artifact.
Among the clones from the amplification products that carried consistent substitutions, 23 additional substitutions were observed in single clones. Assuming that these amplifications started from single DNA molecules, these singleton substitutions are due to Thermus aquaticus (Taq) DNA polymerase misincorporations in later cycles of the PCR. During most of these cycles, the vast majority of template molecules are the result of replications in previous PCR cycles. Thus, the nucleotide misincorporations that occur as singletons in clones that carry consistent changes are not likely to be due to modifications present in the original DNA molecules extracted from the bones. Instead, they represent errors that occur during replication of newly synthesised molecules.
For the 56 PCR amplifications where no consistent substitutions were observed, a total of 126 substitutions were found in a single or a few clones from a total of 351 clones analysed. Since it is not known if these amplifications started from single or multiple template molecules, these misincorporations can be explained either by DNA damage present in the ancient DNA template or by subsequent PCR errors during amplification.
Thus, in order to understand the origin of nucleotide misincorporations seen among cloned PCR products, we distinguish between three classes of substitutions (Table 1): first, 48 consistent substitutions, inferred to be derived from the first or early cycles of PCR when ancient template molecules are replicated; secondly, 23 singleton substitutions in clones carrying consistent substitutions, inferred to be due to misincorporations in later cycles of the PCR when template molecules predominantly derived from previous cycles of the PCR serve as a template; thirdly, 126 singleton substitutions in clones lacking consistent substitutions which could be due to damage in the original template as well as misincorporations in subsequent PCR cycles.
Table 1. Total number of consistent and singleton substitutions found in 484 clones from 79 PCR products.
| Total number | G/C→T/A | Per cent | |
|---|---|---|---|
| The number and percentage of G/C→T/A substitutions compared to the total number of changes are shown in columns three and four. | |||
| Consistent substitutions | 48 | 48 | 100 |
| Singletons in clones with consistent substitutions | 23 | 3 | 13 |
| Singletons in clones without consistent substitutions | 126 | 82 | 65 |
Patterns of nucleotide substitutions
Among the 48 consistent substitutions observed, 13 were G→A substitutions and 35 were C→T substitutions. Thus, the substitutions inferred to have occurred in the first cycle of the PCR show an extreme bias where all substitutions are G/C→A/T changes. In contrast, among the singleton substitutions found in the clones that carry consistent substitutions only 3 of 23 substitutions are G/C→A/T changes. Finally, of the 126 singleton substitutions found in the clones that carry no consistent substitutions, 82 (65%) were G/C→A/T changes. Thus, while the singleton substitutions inferred to have occurred during later stages of the amplification process are in the range that would be expected if all possible base substitutions were equally likely, the pattern of singleton substitutions which could be due either to PCR errors or to base modifications in the template DNA is drastically different from what one would expect under this assumption, in that G/C→A/T substitutions predominate (t-test, P < 0.0005). The same pattern is also obvious in the case of the consistent changes, which are all likely to be the result of miscorporations in the first cycle of the PCR when the ancient DNA molecules serve as templates. Here, all substitutions seen are G/C→A/T changes.
It is noteworthy that the total number of singleton substitutions found in PCRs with no consistent changes (126 singletons in 351 clones) is significantly higher than that for PCRs with consistent changes (23 singletons in 123 clones) (t-test, P < 0.0005), although the latter are likely to have been initiated from a single template molecule and thus to have gone through more replications per molecule. However, if one removes G/C→A/T substitutions from the two data sets, the numbers are no longer different (44 singletons in 351 clones versus 20 singletons in 123 clones, P > 0.15). This supports the idea that a large proportion of the G/C→A/T substitutions among the singleton substitutions are due to misincorporations that result from modifications present in the ancient DNA molecules.
Substitutions on one DNA strand
Fourteen of 23 clones that carry consistent substitutions have two or more such substitutions in the same amplification product. Interestingly, all such substitutions seen in any single amplification product are either C→T or G→A substitutions, respectively (see for example Fig. 1), i.e. in no case was a consistent C→T substitution found together with a consistent G→A change in the same PCR product. Thus, all substitutions on one and the same DNA strand were of the same type. This lends support to the idea that the consistent substitutions are the result of misincorporations that occurred during the first cycle of amplifications where a single template DNA strand initiated the amplification. It furthermore suggests that a single type of molecular modification in the ancient template molecules is the cause of the substitutions seen. However, since it is not possible to know which strand served as a template when the misincorporations occurred, it cannot be determined if a T was inserted instead of a C or an A instead of a G in the first cycle.
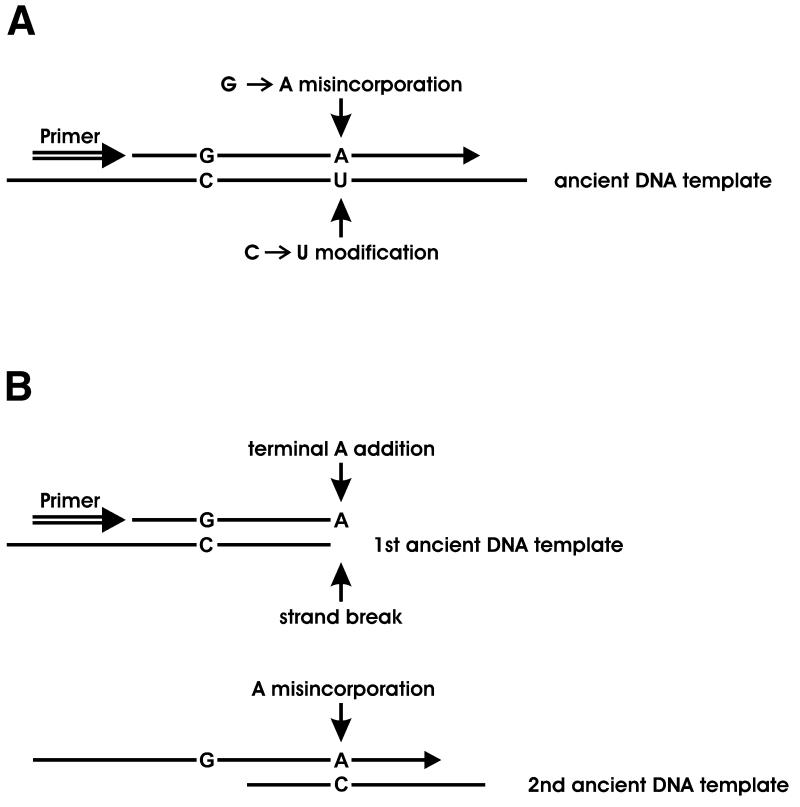
In order to resolve the latter question, an amplification was performed where only one primer was present during the first 25 cycles, after which the second primer was added and an additional 45 cycles performed. Since during the first 25 cycles only one of the template strands served as a template, misincorporations due to modifications in that strand should predominate among clones analysed from the final amplification product. The nucleotides incorporated as a result of modifications in the ancient DNA molecules will consequently be seen on the strand generated from the primer used during the first 25 cycles of the PCR (Fig. 2A). When the left-hand primer was used in such an experiment, 36 out of a total of 52 substitutions (69%) seen were G→A changes on that strand. When the right-hand primer was used, 39 substitutions out of a total of 45 (87%) were G→A changes on the primer strand. Thus, when the original template molecules extracted from the Pleistocene bones are copied by the Taq polymerase, a high frequency of deoxyadenosine residues are incorporated opposite positions where the unmodified template carries deoxycytidine residues. When modern DNA was used as the template, one T→C substitution was observed among 84 clones sequenced.
Figure 2.
Proposed mechanisms explaining the incorporation of deoxyadenosine residues opposite to deoxycytidine residues in the template molecule. (A) Cytosine deamination. The U in the ancient DNA template represents a deoxyuridine residue, a 5-hydroxydeoxyuridine residue or any other modified deoxycytidine residue read as T by Taq polymerase. (B) Jumping PCR. In the first step, the primer is extended on a template that does not include the second primer site and a terminal deoxyadenosine residue is added at the end of the extension product. In the second step, this product is extended on a second template. If the terminal A primes where the template carries a C, the result is incorporation of a deoxyadenosine residue opposite a deoxycytidine residue.
Uracil N-glycosylase treatment
A modification that could explain the incorporation of deoxyadenosine instead of deoxyguanosine during amplification is deamination of deoxycytidine residues in the template DNA. Such deamination results in modified residues which are read as deoxythymidine residues by the Taq polymerase and therefore result in G→A misincorporations.
In order to test if deoxyuridine residues may be responsible for the misincorporations seen, an aliquot of the cave bear DNA was treated with UNG from E.coli. This enzyme removes uracil from DNA (1) and the resulting abasic site is subsequently hydrolysed by β-elimination resulting in a strand break. Thus, UNG treatment of the template DNA is expected to reduce the number of G/C→A/T substitutions if these are due to deamination of cytosine.
When the template was treated with UNG, no G/C→A/T substitutions and only two other substitutions were seen among 87 clones sequenced. In contrast, in a control experiment, where heat-inactivated UNG was used under otherwise identical conditions, 19 G/C→A/T substitutions and no other types of substitutions were seen among 79 clones sequenced. Thus, UNG eliminates the bases responsible for the G→A misincorporations seen during replication of the ancient DNA strands.
Positional distribution of misincorporations
In order to test if misincorporations occur more frequently at certain positions in the DNA sequences analysed than at others, the average misincorporation rate per position carrying a C or a G (1.39%, see Materials and Methods) was compared to the observed misincorporation rate for each position. When tested using a parametric bootstrap procedure, the null hypothesis of a homogeneous substitution rate was not rejected for the data. Thus, there is no evidence for ‘hotspots’ for misincorporation in these sequences. However, it should be noted that the power to detect such hotspots may be low.
DISCUSSION
Several lines of evidence indicate that cytosine deamination is common in the DNA extracted from these late Pleistocene bones. First, all 48 consistent substitutions as well as a large proportion of singleton substitutions observed in clones without consistent substitutions can be explained by cytosine deamination. In contrast, very few of the singleton substitutions representing PCR errors during later cycles of amplification can be explained by this mechanism. Secondly, when single primer extensions were performed, deoxyadenosine residues were incorporated opposite positions where an undamaged template DNA strand would carry deoxycytidine residues. Thirdly, treatment with UNG eliminated the G/C→A/T substitutions, showing that a modified base recognised by that enzyme is responsible for the high substitution rate in the amplifications of the ancient DNA molecules.
The frequency of deaminated deoxycytidine residues is inferred to be high among the ancient DNA molecules since 2.2-fold more singleton substitutions per position are observed in the clones without consistent substitutions as compared to the clones with consistent substitutions (Table 1). Furthermore, deamination of cytosine predominated over other forms of damage since if one removed all G/C→A/T substitutions from the two sets of clones, no statistically significant difference in the relative numbers of substitutions was observed. Finally, after UNG treatment the amplification products did not contain significantly more substitutions than when contemporary undamaged DNA was used as template.
Cytosine deamination seems to be common in DNA extracted from many ancient specimens. First, the cave bear bones and teeth analysed here come from nine different caves and differ in age by ∼25 000 years and in all cases G/C→A/T substitutions are seen. Secondly, PCR products cloned from Neolithic human remains have been shown to display a similar pattern of substitutions and the G/C→A/T substitutions in those cases can similarly be removed by UNG treatment (15). Thirdly, others have recently shown that the substitution spectrum among clones derived from ancient DNA extracts is compatible with cytosine deamination (6). Fourthly, among 20 consistent substitutions seen among clones from 47 amplifications of three Neanderthal remains, 18 are G/C→A/T substitutions (10,16; D.Serre, unpublished observation). The two consistent substitutions that were not G/C→A/T transitions were C/G→A/T transversions. A likely explanation for the latter misincoporations is that the template strands carried 8-hydroxydeoxyguanosines, opposite which deoxyadenosines will be incorporated (1). Thus, the second most frequent type of misincorporation accounts for <3% (2/68) of all consistent changes observed to date. In conclusion, cytosine deamination is the predominant miscoding modification in many if not most ancient DNA samples.
It should be noted, however, that other mechanisms in addition to cytosine deamination may cause G/C→A/T substitutions in amplifications of ancient DNA. One such mechanism results from the tendency for Taq polymerase to add deoxyadenosine residues when it reaches the ends of templates (17). This has been shown to cause substitutions when degraded templates are used in PCR and ‘jumping’ or ‘template switching’ occurs (18) and it can be expected to occur particularly frequently opposite deoxycytidine residues (19; Fig. 2B). Obviously, these two mechanisms may both occur in the same sample. However, on balance, although jumping may be responsible for some of the G/C→A/T changes found and the prevalence of the two mechanisms may vary from sample to sample, the fact that G/C→AT changes can be removed by UNG treatment suggests that cytosine deamination predominates in generating substitutions in amplifications from ancient DNA.
It should also be noted that the substrate specificity of E.coli UNG includes not only DNA molecules containing uracil but also 5-hydoxyuracil (20). Thus, it is not clear whether direct hydrolytic deamination of deoxycytidine residues resulting in deoxyuridine residues or deamination in combination with oxidation resulting in 5-hydroxydeoxyuridine residues is responsible for the substitutions seen. Since the latter modified base has been observed in ancient DNA extracts (3), it is likely that it is at least partly involved in the generation of the G/C→A/T substitutions.
Accuracy of DNA sequence retrieval
It is noteworthy that cytosine deamination causes G/C→A/T substitutions to occur in the final amplification product with a remarkable frequency from some specimens. In almost one-third of all amplifications analysed here consistent G/C→A/T substitutions would have caused an incorrect DNA sequence to be determined if only one single PCR product had been sequenced either directly or from multiple clones. The fact that the large predominance of G/C→A/T substitutions has not previously been seen in studies of cloned amplification products from ancient remains (6) is likely to be due to the fact that the introduction of modified Taq polymerases that allow ‘hot start PCR’ to be performed (21) has made amplifications that start from few molecules easier to achieve. Under such circumstances, chemical modifications in the original template are more likely to influence the results since when a few or a single DNA molecule initiates PCR, any damage present in the molecules can potentially cause misincorporation at a particular position to occur in all molecules amplified. In fact, in all cases where consistent changes were observed, real-time PCR quantitation of the extract indicated that the amplifications had started from less than 100 template molecules (data not shown).
The easiest way to avoid misincorporations caused by cytosine deamination would seem to be to treat the ancient DNA with UNG prior to amplification. However, since UNG creates abasic sites which rapidly results in strand breaks upon heating during PCR, this may result in the removal of the last endogenous DNA molecules in cases where few molecules (which may all carry deoxyuridine residues) survive in an extract. On the other hand, in extracts that contain many starting molecules, consistent changes are very unlikely to occur and UNG treatment is unnecessary. Thus, when extracts of ancient specimens that contain few template DNA molecules are used for PCR, we suggest that DNA sequences are determined from at least two independent amplification products. It is especially useful if this is done by cloning of the amplification products and sequencing of the inserts of multiple clones, since this allows DNA sequence heterogeneity in the amplification product to be determined in an unambiguous way (22). If a consistent difference between the two amplifications is observed, at least one more amplification should be performed to determine which of the two sequences is reproducible (22–24), as outlined in Figure 3.
Figure 3.
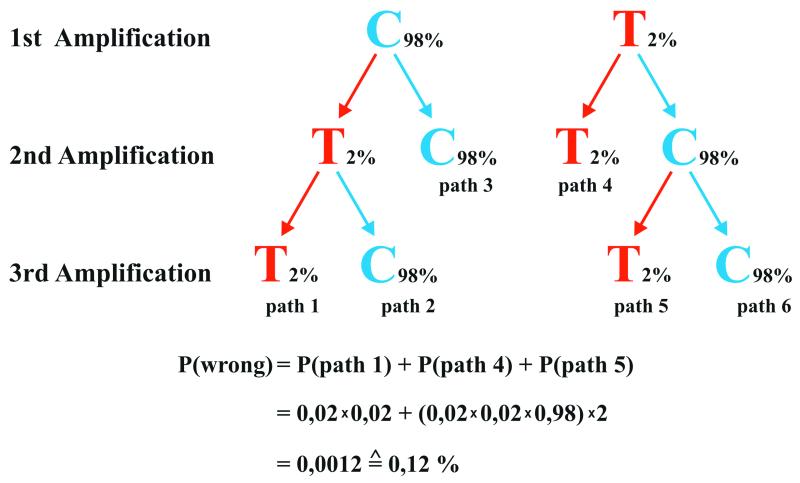
Schematic illustration of the strategy where two or three amplifications are used to determine an ancient DNA sequence. The correct base (C) is blue and the incorrect base (T) red. An error rate of 2% is assumed and the probability of determining an incorrect base is the sum of the three paths that arrive at the incorrect base after two or three amplifications (see text for details).
In order to evaluate how often incorrect DNA sequences may be determined when this strategy is used, we make the conservative assumption that each amplification starts from a single molecule. If the strategy of using two or three independent amplifications is employed, the observed average misincorporation rate for positions carrying G and C nucleotides of ∼2% results in a likelihood of determining a C/G position incorrectly as a T/A of ∼0.12%. Assuming a G/C content of 50%, the average error rate due to G/C→A/T substitutions in determination of ancient DNA sequences is then ∼0.06% per position. Thus, the risk of determining an incorrect base at any particular position is small even when amplifications start from single molecules. However, when longer sequences are determined, the risk that some positions are incorrectly determined obviously increases. For example, the 11 cave bear sequences determined here each contain 115 G and C residues. Assuming that each amplification started from a single molecule, this results in the expectation that 1.5 positions could be incorrectly determined among the 11 DNA sequences. Similarly, among the Neanderthal DNA sequences determined to date by the strategy described here (10,16,25,26), around 800 positions carry G or C, which results in an expectation of ∼0.96 incorrectly determined sequence positions. In conclusion, even under the worst case scenario where each PCR starts from one single DNA strand, the overall error rate is ∼0.06% and thus not more than approximately one order of magnitude higher than the 0.01% regarded as good practice for DNA sequencing of contemporary DNA (27), and not high enough to affect most biological conclusions drawn from their analysis.
However, if cytosine deamination occurred at elevated rates at certain positions in ancient DNA molecules, a majority of ancient DNA molecules could carry a deaminated deoxycytidine residue at such positions. In such a situation, even repeated independent amplifications would cause a T or A residue instead of a correct C or G residue to be determined from ancient organisms. Although a likelihood ratio test gives no support for the occurrence of hotspots of deamination in the sequences, one case where misincorporations would have led to the determination of an incorrect sequence position under the strategy depicted in Figure 3 was observed in one of the samples analysed here (Fig. 1). In this case, the first amplification carried two consistent C→T changes, the second carried no consistent changes, while the third amplification carried two consistent C→T changes, one of which was also seen in the first amplification and would thus be regarded as representing the bona fide sequence. However, when two further amplifications were performed, the base seen at this site was in both cases a C. Consequently, this was deemed to be the correct base. Although this is a rare example that falls within the realm of what is expected under the assumption of equal rates of deamination per position, it nevertheless shows that even carefully designed experiments may yield misleading results when the damage in the original template is frequent. It also underscores the fact that it is essential that several amplifications are performed, especially if a conclusion relies on the finding of a particular nucleotide at a particular position in a DNA sequence.
In order to further evaluate whether inaccurate DNA sequence determination has occurred at any appreciable frequency in the ancient DNA sequences determined to date using the approach in Figure 3, we investigated whether any apparent acceleration of the rate of evolution of ancient DNA sequences compared to related extant organisms can be observed. No such acceleration is seen in cave bears (28), ground sloths (29) or Neanderthals, i.e. in any of the groups of late Pleistocene organisms for which DNA sequences from numerous individuals have been determined. We furthermore investigated whether a difference in substitution spectrum among extinct organisms from that found in related extant organisms can be observed. When the consensus DNA sequences of almost 9000 human mitochondrial DNA sequences are compared to the orthologous consensus DNA sequences from three Neanderthals and more than 400 chimps, 43.5% of substitutions (10 of 23) seen between contemporary humans and Neanderthals are G/C→A/T changes, while the same proportion (43.1%, 22 of 51 substitutions) is seen between human and chimp. The same proportions of G/C→A/T changes are also seen for similar comparisons involving cave bears and extant bears, and ground sloths and extant sloths (data not shown). This indicates that few if any of the substitutions that are fixed between the mitochondrial DNA sequences of Neanderthals and contemporary humans, between cave bears and brown bears or between ground sloths and extant tree sloths, respectively, are due to misincorporations induced by cytosine deamination.
Finally, it should be noted that the occurrence of artifacts in DNA sequence determination discussed here can be avoided if ancient DNA extracts where quantitation of the original template indicates that several hundred template DNA molecules initiate the PCR are used. Under such circumstances, consistent errors are not likely to occur (14). However, if a specimen is so interesting that extracts with fewer copies are studied, the examination of multiple amplifications (24) is imperative.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Michael Bölker for suggesting the single primer extension experiment, Birgit Nickel, Carsten Schwarz and Michaela Winkler for sequencing, Mark Stoneking for help with the statistical analyses and Melanie Kuch, Ivan Nasidze, Hendrik Poinar, Mark Stoneking and Linda Vigilant for helpful discussions. This work was funded by the Max Planck Society and the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- 2.Pääbo S. (1989) Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplification. Proc. Natl Acad. Sci. USA, 86, 1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Höss M., Jaruga,P., Zastawny,T.H., Dizdaroglu,M. and Pääbo,S. (1996) DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res., 24, 1304–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poinar H.N., Höss,M., Bada,J.L. and Pääbo,S. (1996) Amino acid racemization and the preservation of ancient DNA. Science, 272, 864–866. [DOI] [PubMed] [Google Scholar]
- 5.Poinar H.N. and Stankiewicz,B.A. (1999) Protein preservation and DNA retrieval from ancient tissues. Proc. Natl Acad. Sci. USA, 96, 8426–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen A.J., Willerslev,E., Wiuf,C., Mourier,T. and Arctander,P. (2001) Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol., 18, 262–265. [DOI] [PubMed] [Google Scholar]
- 7.Lindahl T. (1996) The Croonian Lecture, 1996: Endogenous damage to DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci., 351, 1529–1538. [DOI] [PubMed] [Google Scholar]
- 8.Poinar H.N., Hofreiter,M., Spaulding,W.G., Martin,P.S., Stankiewicz,B.A., Bland,H., Evershed,R.P., Possnert,G. and Pääbo,S. (1998) Molecular coproscopy: dung and diet of the extinct ground sloth Nothotheriops shastensis.Science, 281, 402–406. [DOI] [PubMed] [Google Scholar]
- 9.Höss M. and Pääbo,S. (1993) DNA extraction from Pleistocene bones by a silica-based purification method. Nucleic Acids Res., 21, 3913–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krings M., Stone,A., Schmitz,R.W., Krainitzki,H., Stoneking,M. and Pääbo,S. (1997) Neandertal DNA sequences and the origin of modern humans. Cell, 90, 19–30. [DOI] [PubMed] [Google Scholar]
- 11.Kilger C., Krings,M., Poinar,H. and Pääbo,S. (1997) “Colony sequencing”: direct sequencing of plasmid DNA from bacterial colonies. Biotechniques, 22, 412–418. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J. and Russel,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Morin P.A., Chambers,K.E., Boesch,C. and Vigilant,L. (2001) Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Mol. Ecol., 10, 1835–1844. [DOI] [PubMed] [Google Scholar]
- 14.Handt O., Krings,M., Ward,R.H. and Pääbo,S. (1996) The retrieval of ancient human DNA sequences. Am. J. Hum. Genet., 59, 368–376. [PMC free article] [PubMed] [Google Scholar]
- 15.Krings M. (1998) Neandertaler DNA-Sequenzen und der Ursprung des modernen Menschen. PhD thesis, Ludwig-Maximilians University, Munich, Germany.
- 16.Krings M., Geisert,H., Schmitz,R.W., Krainitzki,H. and Pääbo,S. (1999) DNA sequence of the mitochondrial hypervariable region II from the Neandertal type specimen. Proc. Natl Acad. Sci. USA, 96, 5581–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark J.M. (1988) Novel non-templated addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res., 16, 9677–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pääbo S., Irwin,D.M. and Wilson,A.C. (1990) DNA damage promotes jumping between templates during enzymatic amplification. J. Biol. Chem., 265, 4718–4721. [PubMed] [Google Scholar]
- 19.Kwok S., Kellog,D.E., McKinney,N., Spasic,D., Goda,L., Levenson,C. and Sninsky,J.J. (1990) Effects of primer–template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res., 18, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatahet Z., Kow,Y.W., Purmal,A.A., Cunningham,R.P. and Wallace,S.S. (1994) New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA-glycosylase. J. Biol. Chem., 269, 18814–18820. [PubMed] [Google Scholar]
- 21.Moretti T., Koons,B. and Budowle,B. (1998) Enhancement of PCR amplification yield and specificity using AmpliTaq Gold™ DNA polymerase. Biotechniques, 25, 716–722. [PubMed] [Google Scholar]
- 22.Hofreiter M., Serre,D., Poinar,H.N., Kuch,M. and Pääbo,S. (2001) Ancient DNA. Nature Rev. Genet., 2, 353–359. [DOI] [PubMed] [Google Scholar]
- 23.Handt O., Höss,M., Krings,M. and Pääbo,S. (1994) Ancient DNA: methodological challenges. Experientia, 50, 524–529. [DOI] [PubMed] [Google Scholar]
- 24.Höss M., Handt,O. and Pääbo,S. (1994) Recreating the past by PCR. In Mullis,K., Ferre,F. and Gibbs,R. (eds), The Polymerase Chain Reaction. Birkhäuser, Boston, MA, pp. 257–264.
- 25.Ovchinnikov I.V., Gotherstrom,A, Romanova,G.P., Kharitonov,V.M., Liden,K. and Goodwin,W. (2000) Molecular analysis of Neanderthal DNA from the northern Caucasus. Nature, 404, 490–493. [DOI] [PubMed] [Google Scholar]
- 26.Krings M., Capelli,C., Tschentscher,F., Geisert,H., Meyer,S., von Haeseler,A., Grossschmidt,K., Possnert,G., Paunovic,M. and Pääbo,S. (2000) A view of Neandertal genetic diversity. Nature Genet., 26, 144–146. [DOI] [PubMed] [Google Scholar]
- 27.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 28.Loreille O., Orlando,L., Patou-Mathis,M., Philippe,M., Taberlet,P. and Hänni,C. (2001) Ancient DNA analysis reveals divergence of the cave bear, Ursus spelaeus, and brown bear, Ursus arctos, lineages. Curr. Biol., 11, 200–203. [DOI] [PubMed] [Google Scholar]
- 29.Greenwood A.D., Castresana,J., Feldmaier-Fuchs,G. and Pääbo,S. (2001) A molecular phylogeny of two extinct sloths. Mol. Phylogenet. Evol., 18, 94–103. [DOI] [PubMed] [Google Scholar]